Abstract
Aim:
Resistance to methicillin and Macrolide–Lincosamide and Streptogramins B and their association with erm genes in Staphylococcus aureus are unknown in Nepal.
Materials & methods:
Nonduplicate nasal swabs from 160 school children were collected from April to September 2018 and processed using standard microbiological procedures.
Results:
Out of 160 samples, 64 (40%) were S. aureus in which 17 (26.6%) were methicillin-resistance Staphylococcus aureus (MRSA). D-test identified 15 (23.4%) as inducible clindamycin-resistant, which were more prevalent in MRSA (76.4%) than methicillin-sensitive S. aureus (MSSA; 4.2%). 18.7% of isolates harbored the ermC gene followed by ermA (15.6%) and ermB (3.1%), and were more in MRSA than MSSA.
Conclusion:
To prevent treatment failure by inducible resistance, D-test must be performed on erythromycin-resistant and/or clindamycin-sensitive isolates.
Keywords: : antibiotic resistance, D-test, erm, erythromycin, inducible clindamycin resistance, methicillin-resistant Staphylococcus aureus, Nepal
Lay abstract
With the increased prevalence of methicillin-resistant S. aureus in hospital- and community-acquired infections, there has been an upsurge in resistance toward Macrolide–Lincosamide–Streptogramin type B antibiotics. This has rendered therapy difficult, thereby increasing morbidity, length of hospital-stay and cost of treatment. Therefore, the correct identification and reporting of S. aureus isolates and their susceptibility patterns, more specifically, toward methicillin, clindamycin and erythromycin is very crucial.
Staphylococcus aureus, especially methicillin-resistant S. aureus (MRSA) was frequently isolated and identified to be the cause of nosocomial and community-acquired infections [1]. Previously, S. aureus was sensitive to glycopeptides such as vancomycin and teicoplanin but during recent years these organisms have developed resistance to these antibiotics, which has encouraged physicians to prescribe another family of antibiotics as an alternative; macrolide–lincosamide–streptogramin type B (MLSB) [2,3].
Both MRSA and methicillin-sensitive S. aureus (MSSA) can be treated by clindamycin, an MLSB antibiotic which is of low cost, and has fewer side effects and high bioavailability in both oral and parenteral forms [4]. Although they are chemically different, the MLSB group exhibits similar inhibitory effects in bacterial protein synthesis [3,5]. There are several mechanisms to MLSB resistance, which include macrolide efflux pump, target site modification and enzymatic antibiotic inactivation [3,6]. Among them target site modification is predominant, which is mediated by the erm gene [3,5]. Four genes, namely ermA, ermB, ermC and ermF, are frequently associated with resistance to MLSB [7–10]. These genes produce methylase, an enzyme that modifies the ribosomal target site preventing binding of the antibiotic and leading to constitutive and inducible resistance [8,11]. Inducible resistance is developed when a strong inducer of the methylase enzyme, erythromycin, is present. Such an isolate is susceptible to clindamycin but resistant to erythromycin. D-zone effect or erythromycin induction of clindamycin resistance using the disk-diffusion method provides proof of this statement [12]. Therefore, it is very crucial to identify actual MLSB resistance for prescribing appropriate therapy in infected patients [13]. If the patient is prescribed with clindamycin without the proper identification of MLSB resistance, it can lead to treatment failure [14].
We found multiple studies reporting variable rates of inducible clindamycin resistance in different places [2,15–22]. Moreover, prevalence rates of different erm genes are still understudied in Nepal. Therefore, the aim of this study is to give an exact picture of what the prevalence of these genes are in the Nepal community. This will provide a proper direction for future researchers as well as prevent health professional from prescribing antibiotics relating to this condition.
Materials & methods
Materials
All the microbiological media and antibiotic discs were purchased from HiMedia Pvt. Ltd., Co., Mumbai, India. Other chemicals were purchased as primer (integrated DNA technology), Master mix (Takara Bio Inc., Nojihigashi, Japan), DNA Ladder (GeneDirex, Inc., MD, USA), Good view nucleic acid stain- HGV II (SBS Genetech Co., Ltd, Beijing, China), agarose (GeneDireX, Inc.).
Sample collection
After the approval from Nepal Health Research Council, National Ethical Guidelines for Health Research in Nepal (reg. no. 195/2O18), a prospective cross-sectional study was conducted over 6 months of period to isolate S. aureus from nasal samples collected from students of two different schools of Kathmandu, namely Kirtipur Secondary and Mangal Secondary School. The informed consent was taken from guardian of students before sample collection. Only those participants (school children) who were not taking any medications were included in the study. Nasal swabs were collected by inserting a sterile moistened cotton swabs (HiMedia) into each nostril and transferred to the laboratory keeping in transport media. The laboratory tests were conducted from April–September 2018 in the Microbiology lab of Center for Health and Disease Studies, Nepal. A total of nonduplicate 160 samples were analyzed in the study.
Bacterial isolation & identification
The specimens collected were inoculated in mannitol salt agar (MSA), blood agar (BA) and incubated at 37°C aerobically for 24 h. Beta hemolytic colonies on blood agar and typical mannitol fermenting colonies in MSA were observed. Pin-point-sized colonies on blood agar with diameter of 2–3 mm were indicative of S. aureus. Gram stain, catalase, oxidase, O–F and coagulase (free and bound) test were performed for further identification using a standard microbiological techniques [23].
Antibiotic susceptibility testing
Antimicrobial susceptibility was studied by the Kirby–Bauer disk diffusion method on a Mueller–Hinton agar plate (12-cm diameter), following Clinical Laboratory Standard Institute (CLSI) guidelines [1,19]. The tested antimicrobial agents were: penicillin (10 U), cefoxitin (30 μg), gentamicin (10 μg), erythromycin (15 μg), clindamycin (2 μg) and ciprofloxacin (5 μg). Cefoxitin (30 μg) was used for the detection of methicillin resistance. Erythromycin (15 μg) and clindamycin (2 μg) discs placed 15 mm apart were used for detection of inducible clindamycin resistance as per recommended CLSI guidelines [23,24].
Detection of methicillin resistance
With cefoxitin disks, isolates with zone of inhibition ≥22 mm in diameter were considered methicillin resistance and those with ≤21 mm were considered as methicillin susceptible.
Detection of inducible clindamycin resistance
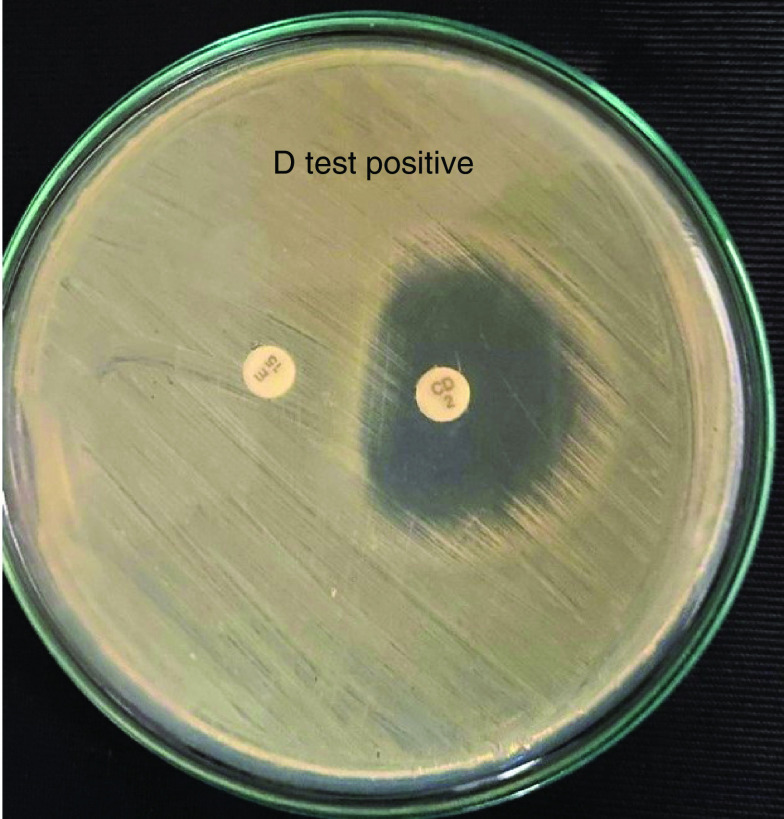
Formation of a flattening shape of the clindamycin inhibition zone ≥21 mm (D shape) around the erythromycin disk has shown in Figure 1, which indicates that erythromycin has induced clindamycin resistance.
Figure 1. . Inducible clindamycin resistant (D-test) positive showing MRSA in MHA media.
CD: Clindamycin; E: Erythromycin; MHA: Muller hinton agar; MRSA: Methicillin-resistant Staphylococcus aureus.
ATCC 25923 strains of S. aureus were used to perform quality control. A separate in-house strain that showed inducible clindamycin resistance was also used for quality control.
DNA extraction
Genomic DNA was extracted from S. aureus isolates from QIAamp DNA Mini Kit, Cat No./ID: 51304 (Qiagen). One to two colonies of isolates were taken with nichrome loop, suspended in nutrient broth and incubated for 24 h. The turbidity was checked and DNA was extracted according to the manufacturer’s protocol of QIAamp. The extracted DNA was kept in -20°C until used for PCR.
Amplification of ermA, ermB & ermC genes
DNA amplification was performed using specific primers for detection of erm genes. Primer sequences were designed using Primer-BLAST of NCBI using gene ID 13913675 for ermA and 24247827 for ermC. Primer sequence for ermB was used from research paper [8]. Sequences for primers used for PCR were as follows: ermA/F: 5′-AAGCGGTAAACCCCTCTGA-3, ermA/R: 5′-TTCGCAAATCCCTTCTCAAC-3 with amplicon size of 190 bp, ermB/F: 5′-CATTTAACGACGAAACTGGC-3′, ermB/R: 5′-GGAACATCTGTGGTATGGCG-3′ with amplicon size of 142 bp, ermC/F: 5′-AATCGTCAATTCCTGCATGT-3′, ermC/R: 5′-TAATCGTGGAATACGGGTTTG-3′ with amplicon size of 299 bp. Each reaction was carried out in final volume of 25 μl with Mastermix (12 μl), forward primer (1 μl), reverse primer (1 μl), DNA template (4 μl) and 7 μl nuclease-free water. PCR amplifications were adjusted according to conditions described in previous studies with some modification [12]. Amplification conditions were as follows: initial denaturation (94°C/4 min for ermA, 95°C/2 min for ermB, 95°C/2 min for ermC), denaturation (94°C/30 s for ermA, 95°C/30 s for ermB, 95°C/30 s for ermC), various annealing temperatures (55°C/30 s for ermA, 50.2°C/30 s for ermB, 52.4°C/30 s for ermC) and 72°C/30 s and final extension at 72°/5 min and hold at 4°C for infinity. PCR products were analyzed by separating on 1.8% agarose gel electrophoresis, stained with nucleic acid stain solution and finally visualized in gel documentation system [12]. A reaction containing all materials and nuclease-free water except template DNA was used as negative control. A native isolate harboring ermA, ermB and ermC gene was used as a positive control for erm genes. 100 bp ladder from GeneDireX, Inc. was used to identify the size of amplified products.
Statistical analyses
Microsoft Excel 2016 was used to keep the record of the data. The data were analyzed using SPSS version 25 for Windows. Pearson Chi-square test was used to find the association between the two variables. p-value of less than 0.05 was considered statistically significant.
Results
Isolation of S. aureus & antibiotic susceptibility pattern
In this study, out of 160 samples processed, 64 samples showed positive culture growth for S. aureus in which 17 (26.5%) were MRSA. The antimicrobial sensitivity tests among MRSA showed that all isolates were resistance to penicillin (ten units) and highly sensitive (88.2%) to gentamicin. The results of antibiotic susceptibility testing for other antibiotics are shown in Table 1.
Table 1. . Result of antibiogram test for methicillin-resistance Staphylococcus aureus isolates.
| Antibiotics used | Sensitive (%) | Intermediate (%) | Resistant (%) |
|---|---|---|---|
| Gentamycin (10 μg) | 16 (88.2) | – | 1 (5.8) |
| Erythromycin (15 μg) | 4 (23.5) | 3 (17.6) | 10 (58.8) |
| Clindamycin (2 μg) | 12 (70.5) | 2 (11.7) | 3 (17.6) |
| Penicillin (ten units) | 0 | 0 | 17 (100) |
| Ciprofloxacin (5 μg) | 6 (35.2) | 1 (5.8) | 10 (58.8) |
Inducible clindamycin resistance
Inducible clindamycin resistance was seen among 15 (23.4%) of the total isolates. The distribution of iMLSB among MRSA, MSSA and total isolates showed the higher rate of inducible clindamycin resistance in MRSA compared with MSSA (p ≤ 0.05) as shown in Table 2.
Table 2. . Distribution of inducible macrolide–lincosamide–streptogramin type B among methicillin-resistance Staphylococcus aureus, methicillin-sensitive S. aureus and total isolates.
| Property | MRSA (n = 17) | MSSA (n = 47) | Total isolates (n = 64) |
|---|---|---|---|
| Inducible clindamycin resistance (iMLSB) | 13 (76.4%) | 2 (4.2%) | 15 (23.4%) |
iMLSB: Inducible macrolide–lincosamide–streptogramin type B; MRSA: Methicillin-resistance Staphylococcus aureus; MSSA: Methicillin-resistance S. aureus.
Association of erm genes in MRSA & MSSA
As shown in Table 3 and Figures 2–4, the electrophoresis results of PCR amplicon showed that 15.6% were ermA positive, 3.1% were ermB positive and 18.7% were ermC positive in which three isolates had both ermA and ermC genes. In this study, the MRSA isolates harbored the erm genes in which ermB and ermC genes were only present in MRSA, whereas few MSSA also had ermA genes.
Table 3. . Result of erm genes in isolates according to sensitivity to methicillin.
| Genotype | PCR result | Sensitivity to methicillin | |
|---|---|---|---|
| MRSA (%) | MSSA (%) | ||
| ermA | Positive | 58.8 | 4.2 |
| Negative | 41.2 | 95.8 | |
| ermB [30] | Positive | 11.7 | 0 |
| Negative | 88.3 | 0 | |
| ermC | Positive | 70.5 | 0 |
| Negative | 29.5 | 0 | |
MRSA: Methicillin-resistance Staphylococcus aureus; MSSA: Methicillin-resistance S. aureus.
Figure 2. . Electrophoresis result of ermA gene.
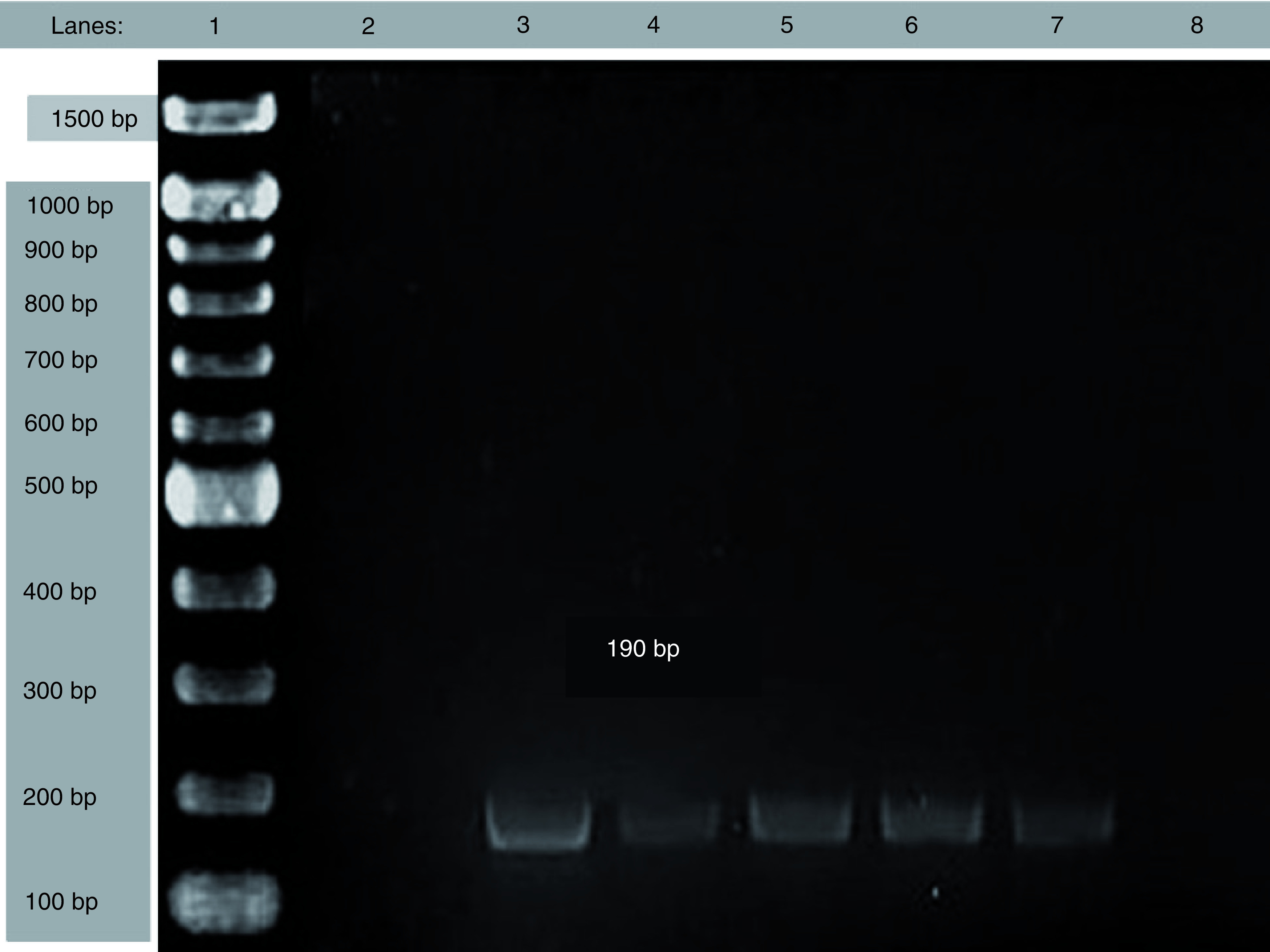
Lane 1:DNA marker, Lane 2: negative control; Lane 3: positive control; Lane 4-8: samples and 190 bp for ermA.
Figure 4. . Frequency of erm genes in S. aureus isolates.
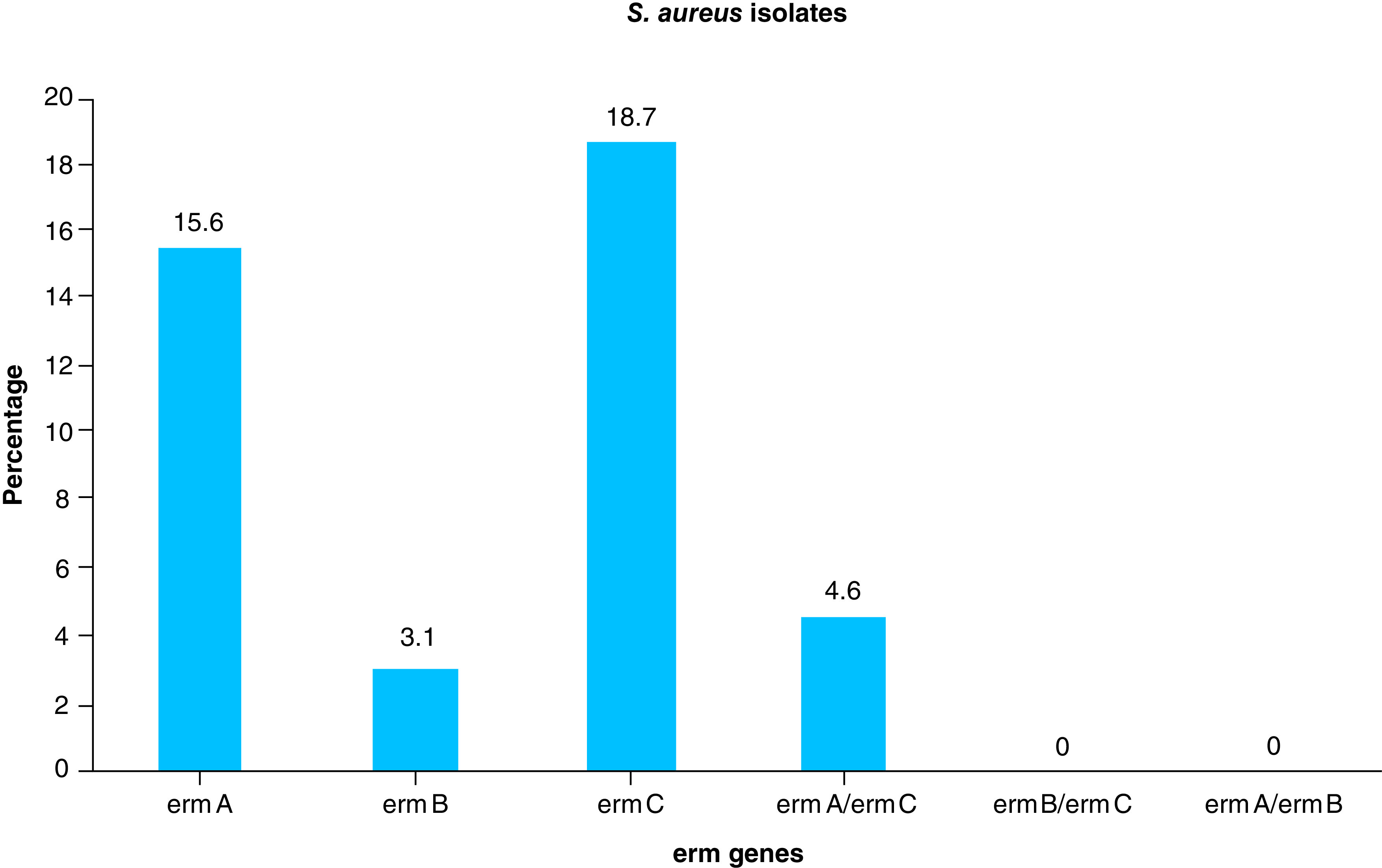
Figure 3. . Electrophoresis result of ermC gene.
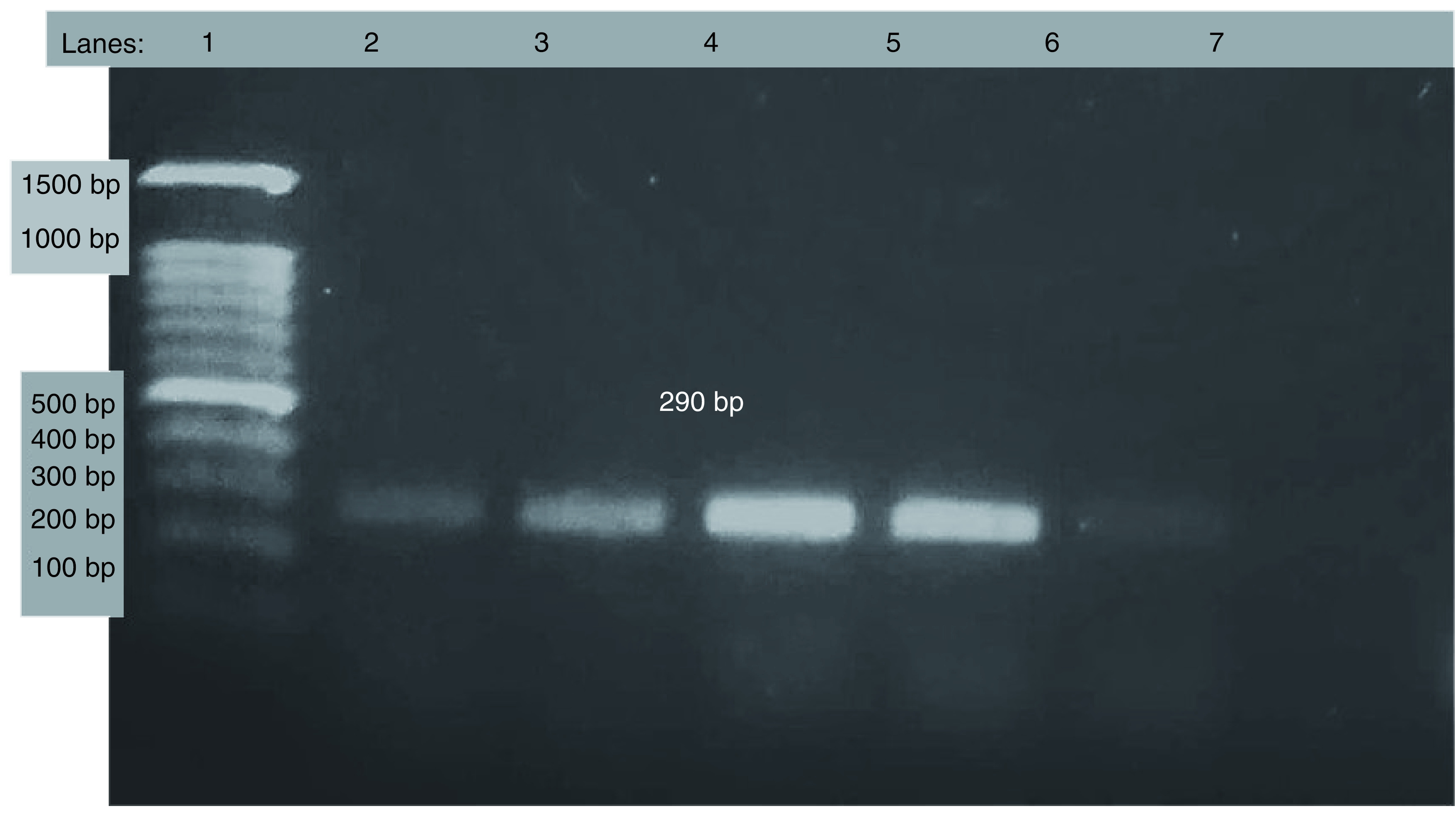
Lane 1: DNA marker, Lane 2: positive control; Lane 3-7:samples and 299 bp for ermC.
Discussion
S. aureus infection is one of the major causes of infection, mainly in low- and middle-income countries and the rate of emergence of antibiotic resistance is quite alarming [25].
We found a significant number of S. aureus from school children, which also show resistance to methicillin, erythromycin and clindamycin. It is very important to correctly identify and report S. aureus isolates, especially in clinical and diagnostic settings, including whether the isolates are truly susceptible to clindamycin when they are resistant to erythromycin. A simple D-test can be performed in the laboratory, so that inducible clindamycin-resistant isolates can be excluded for clindamycin therapy [26]. Prevalence of S. aureus in a community is multifactorial in nature, depending on the geographical location, socioeconomic status, patient age, species of bacteria, inconsistent use of erythromycin and source of the strain (community or nosocomial) [7,27].
The results of our study have shown that the prevalence of inducible clindamycin resistance was 23.4% among all the isolates. As shown in Table 4, several studies conducted in different parts of the world are inconsistent with the prevalence of iMLSB and have previously reported higher variability in prevalence, which ranged from 3.3 to 43% [4,11,13,15,16,21,25,28–33]. Also, the prevalence of iMLSB resistance in MRSA was 76.4%, which is much higher than the prevalence rate previously reported by different studies, which were 12.3–35.9% [4,10,25,29,30,34,35]. This indicates an alarming increase in antibiotic resistance. Although iMLSB resistance in MRSA was higher, it was within the range (4–68%) when compared with data of previously reported studies among MSSA [4,10,25,34,35].
Table 4. . Comparison of frequencies of ermA, ermB and ermC genes in different studies.
| Study (year) | Inducible resistance isolates | iMLSB (%) | Ref. | ||
|---|---|---|---|---|---|
| ermA (%) | ermB (%) | ermC (%) | |||
| Lina et al. (1999) | 63.2 | 0.7 | 25 | 30.2 | [24] |
| Hamilton-Miller et al. (2000) | – | – | – | 43 | [31] |
| Fiebelkorn et al. (2003) | – | – | – | 25.2 | [25] |
| Vivian et al. (2010) | 29.6 | 17.1 | 0.66 | 3.3 | [23] |
| Moosavian et al. (2014) | 41.1 | Nonprevalent | 17.7 | 32.3 | [12] |
| Aydeniz et al. (2015) | 81.9 | 0.9 | 10.8 | 27.47 | [26] |
| Havaei et al. (2016) | 11.11 | 22.22 | 44.44 | 4.18 | [22] |
| Fasihi et al. (2016) | 11 | 3.5 | 20.5 | 12.5 | [11] |
| Adhikari et al. (2017) | – | – | – | 11.48 | [17] |
| Khashei et al. (2018) | – | – | – | 8.6 | [9] |
| Our study | 66.67 | 13.33 | 73.38 | 23.4 | [30] |
Results from our PCR study showed that the prevalence of ermA, ermB and ermC genes were: 15.62, 3.12 and 18.75%, respectively, among inducible clindamycin-resistant isolates. Prevalence of ermA gene varied among different studies conducted at different geographic location, which ranged from 11 to 81.9% [11,15,16,28,29,31]. Our study was within this range obtained from those previously conducted. Our study also showed the prevalence of ermC gene to be within the range when compared with the rates previously reported by several studies [11,15,16,28,31], which ranged from 0.66 to 44.44%. Due to lack of the pertinent literature in different erm genes in Nepal, our study aimed to determine the prevalence of ermA, ermB and ermC genes from S. aureus isolates and inducible clindamycin-resistant MRSA isolates. The prevalence of ermA, ermB [36] and ermC genes among iMLSB isolates were 66.67, 13.33 and 73.38%, respectively. The comparison of the prevalence rates of ermA, ermB and ermC genes in different studies are given in Table 2.
Limitations
Regarding the limitations of our study, it was conducted in two different schools of Kirtipur, Kathmandu, Nepal and so while there may be a chance of us extrapolating our findings and applying it across Kathmandu valley, this may not be representative of the whole country. However, Kathmandu is the most populated city in Nepal. Hence, our findings could well represent the whole country. Due to lack of resources and funding, we were not able to perform genotyping or Staphylococcal cassette chromosome mec gene (SSCmec) typing of S. aureus. Furthermore, MIC of antibiotics used was not determined.
Conclusion & recommendations
To conclude, this study identified the significant presence of MRSA in nasal swabs from school children, which clearly emphasizes the importance of sanitation. Furthermore, there is an increase in inducible clindamycin resistance that directly impacts the treatment of the cases. Therefore, this highlights the importance of performing D-test to identify these isolates in detail, which ought to be followed by laboratory in routine identification of S. aureus.
Future perspective
As there is presence of significant numbers of S. aureus in Nepalese school children as well as resistance to several antibiotics, more research should be done in this area to identify the pathogens, their virulence factors and antibiotic-resistance patterns in order to mitigate the misuse of antibiotics. Furthermore, characterization of gene sequences and typing of strain will give more insight into the genes involved in resistance development.
Summary points.
Methicillin-resistance Staphylococcus aureus and inducible clindamycin resistance are emerging as a public health threat, which could lead to increased morbidity and mortality if proper diagnosis and treatment are not done.
There is a significant association of ermA, ermB, ermC genes and inducible clindamycin resistance, which can be used for rapid diagnosis and treatment.
Inducible clindamycin resistance was found to be more among methicillin-resistance S. aureus.
Every diagnostic and microbiology laboratories should perform D-test to identify inducible clindamycin resistance if they encounter S. aureus isolates sensitive to clindamycin but resistant to erythromycin.
Acknowledgments
The authors sincere thank to Narayan Sharma Bashyal of Central Department of Microbiology and Samikshya Kafle of Center for Health and Disease Studies-Nepal (CHDS) for laboratory support. They are thankful to CHDS and Central Department of Microbiology for providing them the lab facilities.
Footnotes
Author contributions
R Timsina, B Timalsina and A Singh conceived and designed the study. B Timalsina designed the primers and PCR assay. R Timsina and U Shrestha processed the samples and performed microbiological tests. R Timsina, U Shrestha and B Timalsina performed molecular biology tests. B Timalsina and A Singh supervised the work and manuscript. All the authors made substantial contributions to editing and writing the manuscript. All the authors read and approved the final manuscript.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
A written informed consent from parents/guardians of all participants (all aged below sixteen years) was taken prior to the study. The ethical permission was sought from the Ethical Board of Nepal Health Research Council (NHRC), Ram Shah Path, Kathmandu, Nepal (reg no. 195/2018).
Availability of data & materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Open access
This work is licensed under the Creative Commons Attribution 4.0 License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Lima De Castro Nunes E, Dos Santos KRN, Mondino PJJ, De Freire Bastos MDC, Giambiagi-Demarval M. Detection of ileS-2 gene encoding mupirocin resistance in methicillin-resistant Staphylococcus aureus by multiplex PCR. Diagn. Microbiol. Infect. Dis. 34(2), 77–81 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Joshi PR, Acharya M, Kakshapati T, Leungtongkam U, Thummeepak R, Sitthisak S. Co-existence of bla OXA-23 and bla NDM-1 genes of Acinetobacter baumannii isolated from Nepal: antimicrobial resistance and clinical significance. Antimicrob. Resist. Infect. Control. 6(1), 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Provides information on resistance to Macrolide–Lincosamide–Streptogramin type B (MLSB).
- 3.Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34(4), 482–492 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Baral R, Khanal B. Inducible clindamycin resistance in Staphylococcus aureus strains isolated from clinical samples. Int. J. Biomed. Res. 8(2), 81–84 (2017). [Google Scholar]
- 5.Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43(12), 2823–2830 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasten MJ. Clindamycin, metronidazole, and chloramphenicol. : Mayo Clinic Proceedings. Elsevier Ltd, Mayo Clinic Rochester, MN, USA, 825–833 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Aktas Z, Aridogan A, Kayacan CB, Aydin D. Resistance to macrolide, lincosamide and streptogramin antibiotics in Staphylococci isolated in Istanbul, Turkey. J. Microbiol. 45(4), 286–290 (2007). [PubMed] [Google Scholar]; • Explains the association of ermA, ermB, ermC and ermF genes for resistance to MLSB.
- 8.Jensen LB, Frimodt-Møller N, Aarestrup FM. Presence of erm gene classes in Gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 170(1), 151–158 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Saderi H, Emadi B, Owlia P. Phenotypic and genotypic study of macrolide, lincosamide and streptogramin B (MLSB) resistance in clinical isolates of Staphylococcus aureus in Tehran, Iran. Med. Sci. Monit. 17(2), BR48–BR53 (2011). www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3524716&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin TP, Suh B, Axelrod P, Truant AL, Fekete T. Potential clindamycin resistance in clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus: report of a clinical failure. Antimicrob. Agents Chemother. 49(3), 1222–1224 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghanbari F, Ghajavand H, Havaei R. et al. Distribution of erm genes among Staphylococcus aureus isolates with inducible resistance to clindamycin in Isfahan, Iran. Adv. Biomed. Res. 5(1), 62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saffar H, Rajabiani A, Abdollahi A, Habibi S, Baseri Z. Frequency of inducible clindamycin resistance among gram-positive cocci in a tertiary hospital, Tehran, Iran. Iran. J. Microbiol. 8(4), 243–248 (2016). [PMC free article] [PubMed] [Google Scholar]; •• Provides mechanism of resistance caused by erm genes and D-zone effect.
- 13.Khashei R, Malekzadegan Y, Sedigh Ebrahim-Saraie H, Razavi Z. Phenotypic and genotypic characterization of macrolide, lincosamide and streptogramin B resistance among clinical isolates of staphylococci in southwest of Iran. BMC Res. Notes 11(1), 1–6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drinkovic D. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J. Antimicrob. Chemother. 48(2), 315–316 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Fasihi Y, Saffari F, Kandehkar Ghahraman MR, Kalantar-Neyestanaki D. Molecular detection of macrolide and lincosamide-resistance genes in clinical methicillin-resistant Staphylococcus aureus isolates from Kerman, Iran. Arch. Pediatr. Infect. Dis. 5(1), e37761 (2017). [Google Scholar]
- 16.Moosavian M, Shoja S, Rostami S, Torabipour M, Farshadzadeh Z. Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus due to erm genes, Iran. Iran. J. Microbiol. 6(6), 421–427 (2014). [PMC free article] [PubMed] [Google Scholar]
- 17.Gade ND, Qazi MS. Inducible clindamycin resistance among Staphylococcus aureus isolates. Indian J. Basic Appl. Med. Res. 2(8), 961–967 (2013). [Google Scholar]
- 18.Navidinia M. Detection of inducible clindamycin resistance (MLSBi) among methicillin- resistant Staphylococcus aureus (MRSA) isolated from health care providers. J. Paramed. Sci. 6(1), 91–96 (2015). [Google Scholar]
- 19.Govindan S, Mohammed CA, Bairy I. Inducible clindamycin resistance among the Staphylococcus aureus colonizing the anterior nares of school children of Udupi Taluk. Nepal J. Epidemiol. 4(1), 337–340 (2014). [Google Scholar]
- 20.Baiu SH, Al-Abdli NE. Inducible clindamycin resistance in methicillin resistant Staphylococcus aureus. Am. J. Infect. Dis. Microbiol. 4(1), 25–27 (2016). [Google Scholar]
- 21.Adhikari RP, Shrestha S, Barakoti A, Amatya R. Inducible clindamycin and methicillin resistant Staphylococcus aureus in a tertiary care hospital, Kathmandu, Nepal. BMC Infect. Dis. 17, 483 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Provided data on inducible clidamycin-resistance Staphylococcus aureus in clinical samples from Nepal.
- 22.Goudarzi M, Navidinia M, Beiranvand E, Goudarzi H. Phenotypic and molecular characterization of methicillin-resistant staphylococcus aureus clones carrying the panton-valentine leukocidin genes disseminating in iranian hospitals. Microb. Drug Resist. 24(10), 1543–1551 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Cheesbrough M. District Laboratory Practice in Tropical Countries (Part 2) (2nd Edition). Cambridge University Press (CUP); Cambridge, England: [Google Scholar]
- 24.CLSI. Performance Standards for Antimicrobial Susceptibility Testing (30th Edition). Clinical Laboratory Standard Institute, PA, USA [Google Scholar]
- 25.Gurung RR, Maharjan P, Chhetri GG. Antibiotic resistance pattern of Staphylococcus aureus with reference to MRSA isolates from pediatric patients. Futur. Sci. OA. 6(4), 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farzana K, Rashid Z, Akhtar N, Sattar A, Khan JA, Nasir B. Nasal carriage of staphylococci in health care workers: antimicrobial susceptibility profile. Pak. J. Pharm. Sci. 21(3), 290–294 (2008). [PubMed] [Google Scholar]; •• Explains a simple D-test that can be done in laboratory to idenfity inducible clindamycin resistance.
- 27.Merino-Díaz L, De La Casa AC, Torres-Sánchez MJ, Aznar-Martín J. Detección de resistencia inducible a clindamicina en aislados cutáneos de Staphylococcus spp. por métodos fenotípicos y genotípicos. Enferm. Infecc. Microbiol. Clin. 25(2), 77–81 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Aydeniz Ozansoy F, Cevahir N, Kaleli İ. Investigation of macrolide, lincosamide and streptogramin B resistance in Staphylococcus aureus strains isolated from clinical samples by phenotypical and genotypical methods. Mikrobiyol. Bul. 49(1), 1–14 (2015). [PubMed] [Google Scholar]
- 29.Abdalla AE, Kabashi AB, Elobaid ME. et al. Methicillin and inducible clindamycin-resistant Staphylococcus aureus isolated from postoperative wound samples. J. Pure Appl. Microbiol. 13(3), 1605–1609 (2019). [Google Scholar]
- 30.Junaid K, Mustafa AUL, Arshad S, Farraj DAAL, Younas S, Ejaz H. Burn wound infections: a serious threat of multidrug-resistant Staphylococcus aureus. Pak. J. Med. Health Sci. 13(3), 804–807 (2019). [Google Scholar]
- 31.De Lima V, Coutinho S, Paiva RM. et al. Distribution of erm genes and low prevalence of inducible resistance to clindamycin among staphylococci isolates. Braz. J. Infect. Dis. 14(6), 564–568 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Lina G, Quaglia A, Reverdy ME, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43(5), 1062–1066 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiebelkorn KR, Crawford SA, McElmeel ML, Jorgensen JH. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J. Clin. Microbiol. 41(10), 4740–4744 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chelae S, Laohaprertthisarn V, Phengmak M, Kongmuang U, Kalnauwakul S. Detection of inducible clindamycin resistance in staphylococci by disk diffusion induction test. J. Med. Assoc. Thai. 92(7), 947–951 (2009). [PubMed] [Google Scholar]
- 35.Seifi N, Kahani N, Askari E, Mahdipour S, Nasab Naderi M. Inducible clindamycin resistance in Staphylococcus aureus isolates recovered from Mashhad, Iran. Iran. J. Microbiol. 4(2), 82–86 (2012). [PMC free article] [PubMed] [Google Scholar]
- 36.Timsina R, Timalsina B, Singh A. Screening of erm gene of inducible clindamycin resistant Staphylococcus aureus. J. Inst. Sci. Technol. 24(2), 39–43 (2019). [Google Scholar]