Abstract
Checkpoint blockade therapies have improved cancer treatment, but such immunotherapy regimens fail in a large subset of patients. Conventional type 1 dendritic cells (DC1s) control the response to checkpoint blockade in preclinical models and are associated with better overall survival in patients with cancer, reflecting the specialized ability of these cells to prime the responses of CD8+ T cells1–3. Paradoxically, however, DC1s can be found in tumours that resist checkpoint blockade, suggesting that the functions of these cells may be altered in some lesions. Here, using single-cell RNA sequencing in human and mouse non-small-cell lung cancers, we identify a cluster of dendritic cells (DCs) that we name ‘mature DCs enriched in immunoregulatory molecules’ (mregDCs), owing to their coexpression of immunoregulatory genes (Cd274, Pdcd1lg2 and Cd200) and maturation genes (Cd40, Ccr7 and Il12b). We find that the mregDC program is expressed by canonical DC1s and DC2s upon uptake of tumour antigens. We further find that upregulation of the programmed death ligand 1 protein–a key checkpoint molecule–in mregDCs is induced by the receptor tyrosine kinase AXL, while upregulation of interleukin (IL)-12 depends strictly on interferon-? and is controlled negatively by IL-4 signalling. Blocking IL-4 enhances IL-12 production by tumour-antigen-bearing mregDC1s, expands the pool of tumour-infiltrating effector T cells and reduces tumour burden. We have therefore uncovered a regulatory module associated with tumour-antigen uptake that reduces DC1 functionality in human and mouse cancers.
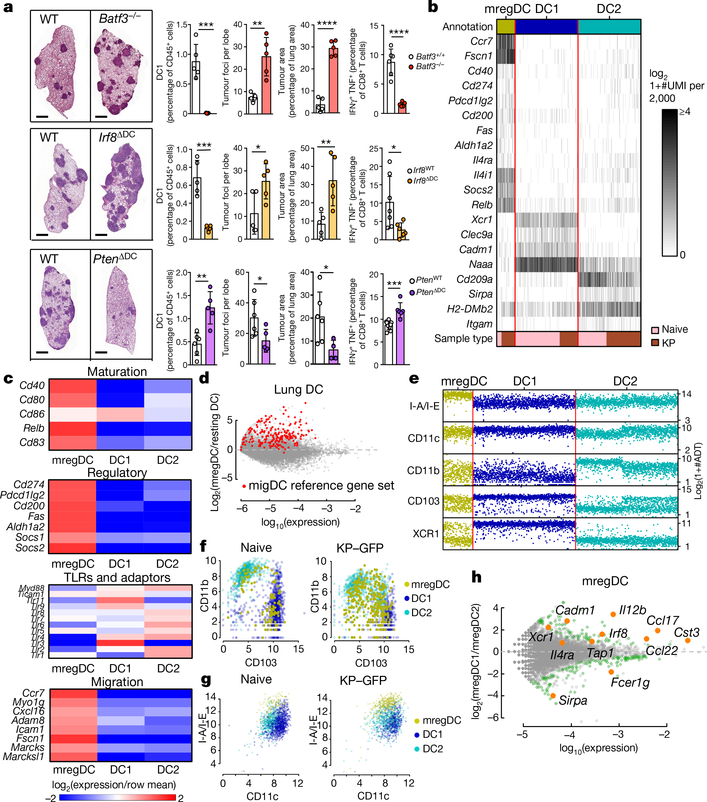
It has previously been found that numbers of DC1 are reduced in non-small-cell lung cancer (NSCLC) lesions compared with adjacent lung tissues3,4, prompting us to examine whether DC1 deficiency affects the growth of murine lung adenocarcinoma lesions that express the oncogene KrasG12D and lack the tumour suppressor Tp53 (also known as Trp53) (‘KP’ lesions). We used DC1-deficient Batf3−/− mice, as well as a lung DC1-deficient model (Irf8δDC mice), in which Irf8–a transcription factor required for the development of DC1s5–is deleted in CD207+ cells (which include lung DC1s and Langerhans cells), leading to the specific loss of DC1s in lungs (Fig. 1a). Batf3−/− and Irf8δDC mice showed a higher tumour burden and reduced numbers of CD8+ T cells producing tumour necrosis factor-α (TNF) and interferon (IFN)γ compared with control littermates (Fig. 1a). We also generated a mouse model with expanded lung DC1 numbers, as previously described6, by deleting Pten from CD207+ cells. These PtenδDC mice had a threefold expansion of lung s DC1s and a lower tumour burden, associated with higher numbers of TNF+ IFNγ+ CD8+ T cells (Fig. 1a).
Fig. 1 |. Identification of a dendritic-cell cluster enriched in immunoregulatory and maturation molecules.
a, Lung tumours were quantified in Batf3+/+ or Batf3−/−, Irf8WT or Irf8δDC, and PtenWT or PtenδDC mice (DC, dendritic cell; WT, wild type). At the left are images of the right-hand lungs; scale bar, 1 mm. At the right are quantifications (by flow cytometry) of DC1s as a proportion of CD45+ (immune) cells; tumour foci and area; and IFNγ+ TNF+ CD8+ T cells. Right lungs were digested for flow cytometry. The results shown are from one experiment, representative of three independent experiments (with five mice per experiment). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (Student’s t-test). Data are shown as means ± standard deviation (s.d.). b–h, CD45+ lin− MHCII+ CD11c+ cells from lungs of naive or KP-tumour-bearing mice were sorted for scRNA-seq and CITE-seq. b, c, Heat maps show unique molecular identifier (UMI) counts of selected genes, with key indicating sample type of origin (b), or relative cluster averages (c). d, Differential expression between mregDC and average tissue-resident dendritic cell (pooled DC1 and DC2) signatures, with genes belonging to a reference migratory dendritic-cell signature indicated in red. e–g, Protein expression levels detected by CITE-seq, grouped by transcriptome-defined cluster. h, Differential gene expression between mregDC1s versus mregDC2s in naive lungs. Genes indicated in green are significant with Benjamini–Hochberg-adjusted P-values of less than 0.15.
Although these results suggest that a paucity of DC1s contributes to reduced antitumour immunity, we hypothesized that additional molecular programs may also reduce DC1 functionality in vivo. Using single-cell RNA sequencing (scRNA-seq), we profiled lineage− (lin−) MHCII+ CD11c+ cells from naive and tumour-bearing lungs. Unsupervised clustering analysis revealed three clusters expressing canonical DC markers such as Flt3 and Cd11c (Fig. 1b, Extended Data Fig. 1a and Supplementary Table 1). DC1 genes included Xcr1, Clec9a and Cadm1, while DC2 genes included Itgam, Cd209a and Sirpa (Fig. 1b). The third DC cluster expressed maturation markers such as Cd80, Cd86, Cd40, Relb and Cd83, along with immunoregulatory genes including Cd274, Pdcd1lg2, Cd200, Fas, Socs1, Socs2 and Aldh1a2 (Fig. 1c). This cluster also upregulated transcripts associated with cytoskeletal rearrangement and cell migration, and markedly downregulated the expression of Toll-like-receptor signalling genes (Fig. 1c). This pattern of maturation markers along with regulatory molecules led us to annotate this transcriptionally defined cluster as ‘mature DCs enriched in immunoregulatory molecules’ (mregDCs).
We found identical clusters of DC1s, DC2s and mregDCs in lung metastases from B16 tumours (Extended Data Fig. 1c) and in public scRNA-seq datasets of CD45+ cells in MC38 tumours and in MCA-induced sarcoma (Extended Data Fig. 1d). Notably, the mregDC signature was consistent with a previously described signature in migratory DCs across different lymph nodes in naive mice7 (Fig. 1d), and accordingly was enriched in migratory DCs in tumour-draining lymph nodes (DLNs) (Extended Data Fig. 1e, f). These findings suggest that expression of the mregDC module may serve as a homeostatic mechanism to regulate adaptive responses against peripheral antigens8,9. Because mregDCs lacked DC1- and DC2-specific markers detectable by scRNA-seq, we performed ‘cellular indexing of transcriptomes and epitopes by sequencing’ (CITE-seq) analysis of lin- MHCII+ CD11c+ DCs, providing information about levels of marker proteins. The use of CITE-seq revealed that subsets of both DC1 (XCR1+ CD103+) and DC2 (XCR1− CD103− CD11b+) expressed the mregDC signature, suggesting that both DC1 and DC2 can differentiate into mregDCs (Fig. 1e, f). In addition, mregDCs expressed the highest levels of MHC class II protein among DCs (Fig. 1e, g). CITE-seq also revealed that CD103+ CD11b− mregDCs (mregDC1s) expressed higher Il12b, Ccl17, Irf8 and Cadm1 levels, whereas CD103− CD11b+ mregDCs (mregDC2s) expressed higher Sirpa and Fcer1g levels, among other genes (Fig. 1h). As unbiased clustering of transcripts did not identify distinct mregDC1 and mregDC2 clusters, we used a biased approach to detect cells expressing DC1 or DC2 marker genes within the mregDC cluster. Stratifying mregDCs by DC1 and DC2 gene scores and comparing these scores with the expression of CITE-seq markers showed that mregDCs that stained positively for CD103 versus CD11b were weakly stratified, whereas DC1s and DC2s were separated into two distinct populations–further demonstrating how the transcriptional programs of these two lineages largely converge upon differentiation into mregDCs (Extended Data Fig. 1g).
Because the mregDC signature was enriched in DLNs (Extended Data Fig. 1f), we asked whether extravasation into lymphatics controlled the induction of regulatory molecules in DCs. We found that the mregDC module was unaffected in Ccr7−/− mice compared with wild-type mice (Fig. 2a, b and Extended Data Fig. 2a, b), suggesting that CCR7-dependent extravasation through lymphatics10 was not required to trigger the mregDC program.
Fig. 2 |. The mregDC1 program is associated with uptake of tumour antigens.
a, b, CD45+ lin− MHCII+ CD11c+ cells were sorted from the lungs of Ccr7−/− mice for scRNA-seq. Expression profiles (a) and frequencies of mregDCs as a proportion of total dendritic cells (b) in WT and Ccr7−/− mice are shown. c, Flow cytometry of lungs and DLNs from WT mice bearing KP–GFP tumours. Results shown are from one experiment, representative of three independent experiments (n = 5). d, CD45+ lin− MHCIIhi CD11c+ CD24hi CD11b− CD103+ cells from WT mouse lungs were sorted using fluorescence-activated cell sorting and stained for EEA1 (an endosomal marker). e, Lung GFP+ and GFP− DC1 populations were sorted from mice bearing KP–GFP tumours and analysed by RNA-seq. Genes that are upregulated in mregDCs relative to DC1s (with a log2-transformed fold change (log2FC) of more than 2; Benjamini–Hochberg-adjusted P-value of less than 0.01) are shown in gold. P-values of signature association are less than 2.2 × 10−16 (Fisher’s exact test). f, Flow cytometry of DC1s from mouse lungs bearing KP–GFP tumours. g, Ultraviolet-irradiated KP–GFP cells were added to a bone-marrow-derived DC1 culture for 2 h before analysis of DC1s by flow cytometry. h, GFP+ and GFP− DC1s were sorted from lungs bearing KP–GFP tumours and cocultured with sorted naive CD62L+ CD44− CD4+ T cells. i, GFP+ and GFP− DC1s were sorted from lungs bearing KP–GFP tumours in B6D2 mice and cocultured with naive CD8+ JEDI T cells isolated from JEDI mouse spleens. JEDI T cells were analysed on day 5. The results shown are from one experiment, representative of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (Student’s t-test). Data are shown as means ± s.d (c, f–i).
The mregDC subset was more abundant in tumour lesions than in naive lungs (Extended Data Fig. 2c), leading us to hypothesize that mregDC induction may correlate with the load of apoptotic cells. To measure whether the DC1 regulatory program was associated with tumour-antigen uptake, we injected mice with KP cells expressing green fluorescent protein (KP–GFP cells). We found that while both DC1 and DC2 subsets acquired GFP in the tumour tissue, GFP remained detectable only in migratory DC1s in the DLN (Fig. 2c and Extended Data Fig. 2d). This is consistent with previous findings showing that DC1s have reduced proteolytic activity compared with DC2s2,7,11, perhap contributing to their enhanced cross-presentation potential12. Accordingly, GFP colocalized with cytosolic EEA1+ compartments in lung DC1s, indicating that tumour antigens were internalized and maintained intact in early endosomes by DC1s (Fig. 2d). RNA-seq of GFP+ DC1s and GFP–DC1s from KP–GFP-tumour-bearing lungs revealed an enrichment of mregDC genes in the GFP+ compartment (Fig. 2e). Using flow cytometry, we confirmed that GFP+ DC1s and DC2s upregulated many protein products of the mregDC transcriptional cluster, including programmed death ligand 1 (PD-L1), CD40 and IL-12 (Fig. 2f and Extended Data Fig. 2e, f). Similarly, markers of the mregDC gene module were upregulated in blue fluorescent protein (BFP)-expressing DC1s that populated B16–BFP/OVA lung metastases (Extended Data Fig. 2g). Using bone-marrow-derived DC1s, we tested whether in vitro uptake of ultraviolet-irradiated apoptotic KP–GFP cells was associated with upregulation of the mregDC program (Extended Data Fig. 2h). We found that DC1s upregulated the expression of PD-L1, CD40 and IL-12 upon capture of apoptotic KP–GFP tumour cells in vitro (Fig. 2g), establishing that uptake of tumour-cell-associated antigen is associated with induction of the mregDC program in DC1.
Tumour-antigen-charged GFP+ DC1s were more potent at driving the differentiation of naive T cells into regulatory T cells than were GFP–DC1s (Fig. 2h); however, mregDCs also expressed many immunostimulatory molecules. Thus, we cocultured GFP+ mregDC1s from KP–GFP tumour lesions with GFP-specific T-cell antigen receptor (TCR) ‘JEDI’ T cells. We found that GFP+ mregDC1s drove the activation of CD8+ JEDI T cells in vitro (Fig. 2i), underlining the capacity of DC1s to induce antigen-specific responses of CD8+ T cells and the dual regulatory and immunogenic program of mregDCs.
The ability of mregDC1s to activate CD8+ T cells despite the induction of many regulatory molecules prompted us to examine whether modulation of the regulatory program could further enhance the immunogenic function of DC1s. Of note, we observed that while mregDCs in naive and tumour-bearing lungs shared many genes, Cd274 and Pdcd1lg2 expression was increased while Il12b expression was reduced in tumour-associated mregDCs, suggesting the presence of a tumour-driven program that modulated the functionality of DCs (Extended Data Fig. 2i).
To identify drivers of the mregDC program, we probed the contribution of pathways known to regulate PD-L1 and IL-12 induction. The absence of type I and type II IFN signalling did not restrain PD-L1 upregulation upon tumour-antigen capture in vivo (Fig. 3a–c). Similarly, PD-L1 upregulation still occurred in the absence of inflammasome or TRIF/ MyD88 signalling (Extended Data Fig. 3a–c). By contrast, we found that IFN? was the main driver of IL-12 in DC1s, as absence of Ifng or Ifngr1 abolished IL-12 production by DC1s at baseline or upon tumour-antigen uptake in vivo (Fig. 3b, c), consistent with recent results13. However, in contrast with previous findings13, the absence of lymphocytes in Rag1–/– mice did not prevent DC1 induction of IL-12, nor did it prevent the upregulation of PD-L1 and CD40 upon capture of tumour antigens (Extended Data Fig. 3d). TLR signalling was also not required for IL-12 production in GFP+ DC1s (Extended Data Fig. 3c).
Fig. 3 |. IL-4 blockade enhances DC1 functionality and antitumour immunity.
a–d, Lungs of tumour-bearing Ifnar1−/− (a), Ifng−/− (b) Ifngr1−/− (c) and WT (d) mice were analysed by flow cytometry. The results shown are from one experiment, representative of three independent experiments (n = 3–5 per experiment). e, Ultraviolet-irradiated apoptotic KP–GFP cells were added to DC1s derived from Axl−/− or WT bone marrow for 2 h before analysis by flow cytometry. f, Flow cytometry of tumour-bearing lungs from mixed bone-marrow chimeric mice (transplanted with a 1/1 ratio of WT (CD45.1) and Axl−/− (CD45.2) bone marrow). g, The AXL inhibitor R428 was added to bone-marrow-derived WT DC1s before adding apoptotic KP–GFP cells. Results shown are from one experiment, representative of two independent experiments (n = 4; e–g). h, Differential expression of TH2 response genes identified by scRNA-seq, showing relative cluster average. i, Flow-cytometry analysis of lungs bearing KP–GFP tumours. j, o, p, Mice bearing KP–GFP tumours were injected with anti-IL-4 or control immunoglobulin G (IgG). Lungs were analysed by flow cytometry (j, p) and lung tumours were quantified (o). Scale bar, 1 mm (o). The results shown are from one experiment, representative of three independent experiments (n = 3–6 per experiment). k, Recombinant IL-4 (rIL4) was added to bone-marrow-derived WT DC1s before adding apoptotic KP–GFP cells. The results shown are from one experiment, representative of three independent experiments (n = 8). l, GFP+ DC1s were sorted from KP–GFP-tumour-bearing lungs of B6D2 mice treated with anti-IL-4 or control IgG and cocultured with naive CD8+ JEDI T cells. JEDI T cells were analysed on day 2. m, GFP+ DC1s were sorted from lungs bearing KP–GFP tumours from mice treated either with anti-IL-4 or control IgG. Dendritic cells were pulsed with ovalbumin peptide 323–339 and cocultured with OT-II cells. n, Mice were injected with KP–GFP and treated with anti-PD-L1. Scale bar, 1 mm. The results shown are from one experiment, representative of two independent experiments (n = 5). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (one-way analysis of variance (ANOVA) and Tukey’s test (a–c, f, l, m) or Student’s t-test (d, e, g, i–k, n–p). Data are shown as means ± s.d (a–g, i–p).
Phagocytic cell-surface receptors are known to contribute to immunomodulation in myeloid cells, prompting us to assess their effect on DC1s undergoing antigen uptake. Using scRNA-seq, we found that Axl was one of the few phagocytic cell-surface receptors that remained expressed in mregDC1s (Fig. 3d and Extended Data Fig. 3e). AXL activation can be induced by GAS6 and PROS1 proteins when they are bound to phosphatidylserine on the surface of apoptotic cells14,15. We found that Axl deficiency reduced PD-L1 upregulation upon tumour-antigen capture in bone-marrow-derived DC1s (Fig. 3e). To directly assess the cell-intrinsic ability of AXL to drive PD-L1 expression in DC1s, we reconstituted lethally irradiated mice with a 1/1 ratio of Axl+/+ and Axl−/− bone-marrow cells. We found that antigen-uptake-driven PD-L1 upregulation was reduced in lung Axl−/− DC1s compared with Axl+/+ DC1s in the same mice (Fig. 3f). These results were confirmed in vitro using the AXL kinase inhibitor R428, which reduced PD-L1 upregulation in bone-marrow-derived DC1s upon capture of tumour antigens (Fig. 3g). These results are consistent with prior studies showing that AXL modulates PD-L1 expression in tumour cells16,17 and is enriched in tumour lesions that resist immunotherapy18. However, AXL inhibition did not e modulate IL-12 production by DC1s (Fig. 3e, g), prompting us to search for additional regulators of IL-12 production.
Analysis of cytokine programs expressed by mregDCs revealed upregulation of T-helper-2 (TH2) response genes, including Il4i1 (showing a fold-change above resting dendritic-cell clusters (FC) of 27), Ccl22 (FC = 29), Tnfrsf4 (FC = 227) and Il4ra (FC = 2.6) (Fig. 3h and Extended Data Fig. 3f). Accordingly, IL-4Rα protein levels were increased in tumour-antigen-charged DC1s (Fig. 3i), prompting us to assess the consequences of IL-4 signalling on DC1 functionality. Use of an IL-4-blocking antibody in tumour-bearing mice doubled the number of IL-12-producing DC1s in the lungs compared with mice treated with isotype control antibody, without affecting PD-L1 levels (Fig. 3j). IL-12 levels were increased even more strongly in mregDC1s in DLNs upon IL-4 blockade (Extended Data Fig. 3g). Recombinant IL-4 acted directly on bone-marrow-derived DC1s to reduce IL-12 production upon capture of apoptotic KP–GFP cells, revealing a direct capacity for IL-4 to modulate DC1 function (Fig. 3k). Altogether, these results suggest that blocking an IL-4-induced program rescues IFNγ-induced IL-12 production by DC1s, without modulating PDL-1 expression. This contrasts with the effects of treatment with a CD40 agonist, which upregulated PD-L1 expression on DC1s in vivo (Extended Data Fig. 3h); and with the effects of the Toll-like receptor-3 (TLR-3) agonist poly(I:C), which upregulated PD-L1 and CD40 levels without increasing IL-12 production by DC1s in vivo (Extended Data Fig. 3i).
GFP+ mregDC1s isolated from mice treated with IL-4-blocking antibody were more potent at activating JEDI CD8+ T cells compared with GFP+ mregDCs isolated from mice treated with isotype antibody (Fig. 3l and Extended Data Fig. 3j). Similarly, CD4+ T cells expressing an ovalbumin-specific TCR (OT-II cells), activated with mregDCs from mice treated with IL-4-blocking antibody and pulsed with ovalbumin peptide, produced increased cytokine levels compared with OT-II cells activated with ovalbumin-peptide-pulsed mregDCs from control mice (Fig. 3m). Notably, IL-4-blocking antibodies reduced the growth of KP–GFP lesions that resisted PD-L1 blockade (Fig. 3n, o) and increased the numbers of IFNγ+ TNF+ CD8+ T cells in lung tumours (Fig. 3p) and in DLNs (Extended Data Fig. 3k). Together these results suggest that blockade of IL-4 during mregDC generation in vivo enhances mregDC immunogenicity and T-cell effector function.
It is likely that the increased antitumour response induced by IL-4-blocking antibodies is not only the result of enhanced DC1 function. Nonetheless our results–showing that: (1) mregDC1s express IL-4Rα and undergo upregulation of IL-4-inducible genes upon uptake of tumour antigens; (2) IL-4 acts directly on DC1s to modulate their IL-12 production; (3) mregDCs generated in the presence of IL-4-blocking antibodies result in enhanced T-cell activation; and (4) IL-4-blocking antibodies enhance IL-12 production by DC1s and expand the population of IFNγ+ CD8+ T effector cells in vivo–suggest an important role for DC1s in the antitumour responses mediated by IL-4 blockade.
To assess whether mregDCs were present in human tissues, we analysed immune cells from tumour and non-involved lung tissues of 35 patients with NSCLC by scRNA-seq. Unsupervised clustering identified a DC1 cluster expressing CLEC9A, XCR1 and IRF8 and a DC2 cluster expressing CD1C and FCER1A (Fig. 4a). Similar to our results in mice, we also identified a human mregDC cluster that expressed the maturation markers CCR7, CD40, RELB and CD83 and the regulatory molecules CD274, CD200, FAS and ALDH1A2, as well as low levels of TLR signalling genes and increased levels of migratory genes (Fig. 4a, b). Human mregDCs also expressed high levels of the TH2 response genes IL4R, IL4I1, CCL17, CCL22 and BCL2L1 (Fig. 4c). Direct stratification of mregDCs using DC1 and DC2 gene scores identified mregDC1 and mregDC2 subsets (Fig. 4d). Notably, this analysis confirmed that, as in mice, IL12B expression was specific to mregDC1s in humans (Fig. 4d).
Fig. 4 |. Human NSCLC lesions are populated by mregDCs.
a–c, scRNA-seq of CD45+ cells of matched non-involved lung (nLung) and tumour from resection specimens of 35 NSCLCs. After clustering, dendritic-cell clusters were selected for further analysis. Heat maps show downsampled UMI counts after evenly sampling dendritic-cell types (a), or relative cluster averages (b, c). d, Stratification of dendritic-cell transcriptomes using scores for human dendritic-cell subtypes. Single cells are coloured by cluster annotation (left) or expression of IL12B (right). Genes used to construct the scores are defined in Supplementary Table 2. e, Protein expression levels detected by CITE-seq. f, Homology analysis of gene expression across mouse and human dendritic-cell clusters.
CITE-seq analysis of seven NSCLC lesions and non-involved lung tissues confirmed that DC1s and DC2s contributed to the mregDC cluster (Fig. 4e). Among all DC clusters, mregDCs expressed the highest levels of HLA-DR, PD-L1, PD-L2, CD86 and CD40 proteins (Fig. 4e and Extended Data Fig. 4a). Human DC1s expressed high CD141 and XCR1 protein levels, whereas human DC2s expressed high CD1c levels (Fig. 4e and Extended Data Fig. 4a). We also identified mregDCs in a public scRNA-seq dataset of human NSCLC lesions (Extended Data Fig. 4b). To align the gene signatures expressed across DC subsets in mice and humans, we coclustered genes and cell types based on the transcriptional DC profile for each cell type in each species, using genes that were conserved and variable across DCs in the mouse and human datasets. Our analysis revealed that the mregDC program is conserved across the two species (Fig. 4f).
Together, our findings reveal a targetable immunoregulatory program–expressed by DCs across different tissues, tumour types and species–that restrains DC immunostumulatory function and controls the threshold of T-cell activation. This immunoregulatory program is associated with the capture of cell-associated antigens during normal or excessive cell death and is enriched in antigen-charged DC that migrate to the DLN to shape tissue and tumor-specific immunity. We show that this immunoregulatory program is partially driven by AXL and IL-4 signalling and that IL-4-blocking antibodies rescue DC1 functionality in tumour lesions and enhance cytolytic antitumour immunity. These results extend prior work showing that the IL-4/IL-13 pathway can promote tumour growth19,20, and emphasize the need to test combination therapies tha block both PD-L1 and IL-4 signalling in different cancer types.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Mice
C57BL/6 and B6D2F1/J mice were purchased from Charles River Laboratories at the age of seven weeks and housed in our facility for at least one week before being used in experiments. B6.129S(C)-Batf3tm1Kmm/J, B6(Cg)-Irf8tm1.1Hm/J, B6.129S4-Ptentm1Hwu/J, B6.129P2(C)-Ccr7tm1Rfer/J, B6.129S7-Ifngtm1Ts/J, B6.129S7-Ifngr1tm1Agt/J, B6(Cg)-Ifnar1tm1.2Ees/J, B6.129S7-Il1r1tm1lmx/J, B6.129P2(SJL)-Myd88tm1.1Befr/J, B6.129S7-Rag1tm1Mom/J and C57BL/6J-Ticam1Lps2/J mice were purchased from Jackson Laboratories and bred in our facility or used for experiments after at least one week of housing in our facility. Cd207-Cre mice were provided by B. Clausen21. JEDI mice were provided by B. Brown. Axl−/− and Axl−/− Mertk−/− bone marrow were provided by C. Rothlin and S. Ghosh. Asc−/− mice were provided by Millenium.
Floxed mice were crossed to Cd207-Cre mice in our facility. Mice were maintained at specified pathogen-free (SPF) health status in individually ventilated cages at 21–22 °C and 39–50% humidity. Male mice at the age of 8–12 weeks were used for experiments. All animal procedures were approved by the Institutional Animal Care and Use Committees (IACUCs) of the respective institutions.
KP and B16 mouse models
Eight-week-old mice were injected intravenously with 5 × 105 KP cells22, KP–GFP cells or B16-BFP/OVA cells. All cell lines tested negative for mycoplasma and were authenticated by phenotyping their potential for generating tumours in mice. Lungs and lymph nodes were analysed on day 28 (KP or KP–GFP) or on day 22 (B16-BFP/OVA), except when otherwise indicated. When indicated, mice were injected intraperitoneally (i.p.) with 25 μg anti-IL-4 antibody (BioXcell, clone 11B11) on days 21, 23 and 26; with 100 μg CD40 agonistic antibody (BioXcell clone FGK4.5/ FGK45) on day 27; with 200 μg poly(I:C) high-molecular-weight RNA (InvivoGen) on day 27; or with 200 μg anti-PD-L1 antibody (BioXcell, clone 10F.9G2) on days 15, 18, 21, 24 and 27.
To quantify tumours, we stained slides of paraffin-embedded left lung lobes with haematoxylin/eosin; we scanned slides using an Olympus digital scanner and analysed them using Panoramic Viewer software.
Bone-marrow transplant
We injected 106 bone-marrow donor cells intravenously into lethally irradiated (2 × 6.5 Gy) recipient mice. Mice were maintained on sulfamethoxazole/trimethoprim (STI Pharma) for 3 weeks. KP–GFP cells were injected 8 weeks after bone-marrow transplant and mice were analysed 11 weeks after bone-marrow transplant.
Flow cytometry and fluorescence-activated cell sorting
Single-cell suspensions were obtained from lung and lymph nodes by digestion with collagenase IV (0.25 mg ml−1; Sigma) at 37 °C for 30 min (lung) or 25 min (lymph nodes), followed by passing through a 70-μm cell strainer and lysis of red blood cells (RBCs; using RBC lysis buffer, BioLegend) for 2 min at room temperature. For flow cytometry or FACS, cells were stained in FACS buffer (phosphate-buffered saline (PBS) supplemented with 2% bovine serum albumin (BSA) and 5 mM EDTA) with monoclonal antibodies specific to CD45 (clone 30-F11, BioLegend), Siglec F (clone E50–2440, BD Pharmingen), CD11c (clone N418, Invitrogen), CD24 (clone M1/69, Invitrogen), CD103 (clone 2E7, BioLegend), XCR1 (clone ZET, BioLegend), I-A/I-E (clone M5/114.15.2, eBioscience), CD11b (clone M1/70, eBioscience), CD40 (clone 1C10, ebioscience), PD-L1 (clone MIH5, Invitrogen), IL-12p40 (clone C17.8, eBioscience), IL-4Rα (clone I015F8, BioLegend), AXL (clone MAXL8DS, Invitrogen), FAS (clone SA367H8, BioLegend), CD47 (clone miap301, Invitrogen), CD107a (clone 1D4B, Biolegend), CD200 (clone OX90, eBioscience), CD70 (clone FR70, eBioscience), CD127 (clone A7R34, Biolegend), CD3 (clone 145–2C11, eBioscience), CD8 (clone 53–6.7), CD4 (clone GK1.5 ebioscience), Ki67 (clone 16A8, BioLegend), TNF (clone MP6-XT22, eBioscience), IFNγ (clone XMG1.2, eBioscience), CD25 (clone PC61.5, eBioscience) or FOXP3 (clone FJK-16 s, Invitrogen). For intracellular staining, cells were fixed with either BD Fix/Perm (for intracellular cytokine stains) or Invitrogen Fix/Perm (for nuclear stains) according to kit instructions. For T-cell cytokine stains, cells were incubated with 10 μg ml−1 brefeldin A, 0.2 μg ml−1 ionomycin and 0.5 μg ml−1 phorbol myristate acetate (PMA; all from Sigma) for 3 h at 37 °C followed by staining and fixation. For FACS, cells were prepared and stained as described and sorted on a BD FACSAria flow cytometer.
Human subjects
Samples of tumour and non-involved lungs were obtained from surgical specimens of patients undergoing resection at the Mount Sinai Medical Center in accordance with a protocol reviewed and approved by the Institutional Review Board (IRB) at the Icahn School of Medicine at Mount Sinai (IRB Human Subjects Electronic Research Applications 10–00472 and 10–00135) and in collaboration with the Biorepository and Department of Pathology. After rinsing in PBS, tissues were minced and incubated for 40 min at 37 °C in collagenase IV (0.25 mg ml−1), collagenase D (200 U ml−1) and DNase I (0.1 mg ml−1; all from Sigma). Cell suspensions were then aspirated through a 18G needle ten times and strained through 70-μm mesh before RBC lysis. Suspensions were enriched for CD45+ cells by bead-positive selection (Miltenyi) before processing for scRNA-seq or CITE-seq. A detailed unsupervised analysis of the human scRNA-seq and CITE-seq dataset will be published elsewhere (A.M.L. et al., manuscript in preparation).
ScRNA-seq
For each scRNA-seq or CITE-seq sample, we sorted 8,500 DCs as above and encapsulated them using the 10x Chromium 3’ v2 chemistry kit according to the manufacturer’s instructions. For scRNA-seq, libraries were prepared according to the manufacturer’s instructions. QC of cDNA and final libraries was performed by CyberGreen qPCR library quantification assay (KAPA). Samples were sequenced on an Illumina Nextseq 550 using the 75-cycle kit to a depth of 100 million reads per library.
CITE-seq
We carried out mouse CITE-seq experiments similarly to scRNA-seq of FACS-purified samples, with the following exceptions. Before sorting, cells were stained with a mix of fluorescent antibodies and antibodies that had been conjugated to oligonucleotide barcodes using Thunder-Link PLUS Oligo Conjugation kits (Expedeon) according to the manufacturer’s instructions. Sorted cells were encapsulated using the 10x Chromium platform, and libraries were prepared as previously described23, with minor modifications. In brief, amplification of complementary DNA was performed in the presence of 2 pM of an antibody-oligo-specific primer to increase the yield of antibody-derived tags (ADTs). The amplified cDNA was then separated by SPRI size selection into cDNA fractions containing messenger-RNA-derived cDNAs (larger than 300 base pairs) and ADT-derived cDNAs (smaller than 180 base pairs), which were further purified by additional rounds of SPRI selection. Independent sequencing libraries were generated from the mRNA and ADT cDNA fractions, which were quantified, pooled and sequenced together on an Illumina Nextseq to a depth of 80 million reads per gene expression library and 20 million reads per ADT library.
For human CITE-seq experiments, cells were prepared as above. Samples were split and barcoded using ‘Hashing’ antibodies24, staining β2-microglobulin and CD298, before pooling and staining with CITE-seq antibodies, allowing for distinct biological samples to be batched together to minimize technical batch effects and for improved detection of doublets. Human CITE-seq experiments used either panels of in-house antibodies conjugated as above, or antibodies purchased from the Biolegend TOTALseq catalogue.
scRNA and CITE-seq analysis
For mouse data, after library demultiplexing, gene-expression libraries were aligned to the mm10 reference transcriptome and count matrices were generated using the default Cell Ranger 2.1 workflow, using the ‘raw’ matrix output. CITE-seq library reads were directly queried for antibody and cell barcodes in the appropriate read positions, including antibody sequences within a Hamming distance of 1 from the reference sequence. For human data, gene-expression libraries were aligned to the GRCh38 reference transcriptome and CITE-seq features were detected using the ‘feature barcoding’ workflow in Cell Ranger 3.1. Where applicable, doublets were removed based on costaining of distinct sample-barcoding (‘Hashing’) antibodies (maximum staining antibody counts/second-most staining antibody counts = less than 5).
Cell clustering for human data will be described elsewhere (A.M.L. et al., manuscript in preparation). For analysis of mouse experiments, clustering proceeded similarly for both scRNA-seq and CITE-seq experiments, relying on gene-expression signatures for clustering and withholding CITE-seq protein signatures, when available, for validation and downstream analyses. After filtering for cells passing quality thresholds (mitochondrial gene content less than 25%; more than 800 gene-expression UMIs detected) and excluding plasmacytoid DCs (Supplementary Tables 1, 2), we implemented a protocol similar to that previously described for clustering single-cell transcriptional signatures25,26, with minor modifications. The clustering was based o modelling the probability of observing gene i in cell j as:
in which mapj is the assignment of cell j to cell type; αi,mapj is the probability that a molecule drawn from cell type mapj is of gene i; and Zj is a normalization factor equal to the total number of UMIs in cell j. Given this model and assuming a hard association of cells with types, the log-likelihood (LL) of the entire dataset is:
in which Uij is the number of UMIs of gene i observed in cell j.
The updated algorithm outline was as follows: 1. Randomly sample without replacement 1,000 cells from each batch. Let the resulting genes-by-cells matrix be U. 2. Initialize the model. Repeat A to F 1,000 times: A. Randomly select a value Nds_umis from the (P1,P2 ) percentiles of the empirical distribution of the number of UMIs per cell of U. B. Down-sample U to Nds_umis UMIs per cell. The downsampled matrix is denoted as U’. C. Select highly variable genes (see below). D. Cluster the cells in U’ on the basis of the genes selected in step C using k-means++ (https://tanaylab.github.io/tglkmeans/index.html), with k seeds, following log2 (X + Kreg_ds) transformation, in which Kreg_ds is a regularization factor. E. Estimate α given k-means++ assignments map for the cells in U’ by setting αi,m equal to the proportion of UMI mapped to gene i in cells belonging to k-means cluster m to the total number of UMI observed in cells belonging to k-means cluster m. F. Calculate the maximum-likelihood assignments over the multinomial mixture models represented by the columns of α, and set map equal to these assignments. G. Compute the log-likelihood of U given the present initialized type assignments map. H. Select model parameters that correspond to the randomized seed that maximized the log-likelihood of U. 3. Estimate α given ‘map’ for the cells in U, as in step E. 4. Given the values of α, calculate the assignment for each cell in U and update the assignments of cells to clusters ‘map’. 5. Return to step 3 and repeat until the likelihood converges, or until a specified maximum number of iterations is reached. 6. Estimate ‘map’ given α for U.
For the joint clustering of the mouse samples, we included barcodes with more than 800 UMIs and used Kreg_ds = 0.1; (P1,P2 ) = (10th, 40th) percentiles; Kreg = 5 × 10−6; k = 5. To improve the initiation of the model, cell-cycle genes were excluded from the k-means clustering (step D).
To determine highly variable genes, as in other publications25–27, we selected genes with variability that was inconsistent with multinomial sampling. We calculated a loess curve for the log(variance/mean) versus log(mean) distribution and binned the log(variance/mean) values by intervals of 0.2 of log10 (mean). We selected genes with more than 50 UMIs in U’ from the 8th percentile of each bin and also required that their log2 (variance/mean) is 0.1 or higher above the loess curve.
Differential expression analysis
We tested for differential expression between two sets of cells by estimating the gene expression per set (similarly to the estimation of the model multinomial parameters), and calculated the observed log fold change between the two sets for each gene. We then randomly shuffled the cells of the two sets for at least 104 permutations while maintaining the sizes of the sets and calculating the log fold change between the permuted sets for each permutation. The empirical P-value was then defined as based on the rank of the absolute value of observed the log fold-change of each gene within its empirical fold-change distribution. Empirical P-values were adjusted for multiple-hypothesis testing with the Benjamini–Hochberg procedure using the R command p.adjust with the option ‘method = BH’.
Analysis of public datasets
M38 data13 were downloaded from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) using accession codes GSM3090155 and GSM3090156; T3 sarcoma data28 were downloaded from GEO using accession code GSE119352. After filtering expression matrices of low UMI and high mitochondrial gene events, we carried out a preliminary analysis to determine the broad diversity of cell types present across both datasets. We then used this analysis to construct gene lists for in silico sorting of red blood cells, mast cells, T cells, B cells and neutrophils. Furthermore, we used gene lists based on the results of our clustering analysis in KP and tumour-naive mice to construct gene lists for macrophages (any genes expressed a log2FC of more than 1 among macrophages in the sample compared with any dendritic-cell cluster) and plasmacytoid DCs (using the same gene list for in silico sorting before clustering). Gene scores were defined as the fraction of RNA in a cell belonging to genes in a gene list. All gene lists and gene-score thresholding parameters are defined in Supplementary Table 2.
For human public datasets, NSCLC scRNA-seq data from eight patients29 were downloaded from ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) using accession numbers E-MTAB-6149 and E-MTAB-6653. Cells were directly classified by a maximum-likelihood-like approach to the clusters generated for human samples (A.M.L. et al., manuscript in preparation).
Generation of dendritic-cell subtype scores
Gene lists for the stratification of dendritic-cell subtypes were generated as follows. We defined DC1 and DC2 genes as those with an absolute log2FC of more than 1 between the average expression of the DC1 and DC2 clusters. We defined mregDC genes as those for which the log2FC difference between mregDCs and both DC1s and DC2s was more than 1. For human mregDC genes, this threshold was increased to 1.5 to enhance the specificity of the gene list, thereby enhancing the separation of mregDCs with the resulting gene score. As with in silico sorting gene lists, we defined dendritic-cell gene scores as the fraction of RNA in a cell belonging to genes in the gene list.
Cross-species homology analysis
Variable genes for cells mapping to dendritic-cell clusters were identified independently for each species as above, including genes in the top 20th percentile of each bin to expand the number of genes for comparison. Dendritic-cell cluster averages were normalized within each species by dividing the average cluster expression plus a regularization constant (10−4) by the average of cluster averages plus the regularization constant. After selecting genes with conserved gene symbols, normalized dendritic-cell expression matrices were merged and log-normalized before hierarchical clustering of the cluster averages by Pearson correlation distance (1 – correlation)/2 or k-means clustering of the genes.
Ultra-low input RNA-seq
We sorted 104 DCs as described above into 700 μl trizol. We isolated RNA using RNeasy Micro Kits (Qiagen), and synthesized 0.5–1 ng of RNA into cDNA using the Smart-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara Bio). Sequencing libraries were prepared using the Low Input Library Prep Kit (Takara Bio). Libraries were sequenced on an Illumina NextSeq 550 system. Fastqs were aligned to the mm10 reference genome; reads were dereplicated for polymerase chain reaction (PCR) duplicates; and gene counts were generated using STAR v2.5 using ‘–quantMode GeneCounts’. Differential expression analyses were performed with the limma R package.
Immunofluorescence:confocal microscopy
DCs were sorted as above. We centrifuged 105 GFP+ DCs onto Alcian-blue-treated coverslips and fixed them in 1% paraformaldehyde. Cells were permeablized in 0.2% saponin/RPMI medium and stained overnight with anti-EEA1 antibody (ThermoFisher catalogue number PA1–063A). Coverslips were washed and stained with anti-rabbit Alexa Fluor 594. Coverslips were washed and stained with 4’,6-diamidino-2-phenylindole (DAPI; 1 ng ml−1) for 5 min. Coverslips were mounted using Prolong Gold Anti-Fade and imaged on a Zeiss 780 Confocal Microscope.
In vitro bone-marrow-derived dendritic-cell cultures
Bone-marrow cells from mice were isolated by flushing femurs, tibias and humeri with PBS, supplemented with 0.5% BSA, 2 nM EDTA, and 1% penicillin/streptomycin (P/S). Bone-marrow cells were strained through a 70-μm filter and centrifuged before resuspension in 1× RBC lysis buffer (BioLegend) for 5 min on ice. Lineage-negative progenitor cells were isolated using a lineage cell depletion kit (Miltenyi Biotec) and plated in DMEM medium with 10% fetal calf serum (FCS), 1% L-glutamine, 1% sodium pyruvate, 1% MEM non-essential amino acids, 1% P/S, 55 μM 2-mercaptoethanol and 200 ng ml−1 recombinant human Flt-3 ligand (R&D Systems). After three days of differentiation, cells were plated onto a monolayer of OP9-DL1 stromal cells and cocultured for an additional four days. DCs were analysed on day 7. OP9-DL1 cells were cultured in MEM-α medium with 20% FCS and 1% P/S. Prior to coculture with bone-marrow cells, OP9-DL1 cells were treated with 10 μg ml−1 mitomycin C (Sigma Aldrich) for 2 h and washed three times with PBS. DCs were stimulated on day 7 with apoptotic KP–GFP cells (ultraviolet-irradiated 24 h before stimulation) for 2 h. Where indicated, DCs were treated with 10 μg ml−1 recombinant IL-4 (Shenandoah) 24 h and 30 min before stimulation with apoptotic KP–GFP cells. Where indicated, DCs were treated with 1 μM of the AXL inhibitor R428 (ref. 30; Selleckchem S2841) 24 h and 30 min before stimulation with apoptotic KP–GFP cells.
JEDI T-cell assay
DCs from B6D2 mice were sorted as above. CD8+ T cells were isolated from the spleen of a JEDI mouse31 using a CD8+ enrichment kit (Invitrogen) and labelled with cell trace violet (Invitrogen). We plated 105 T cells in Click’s medium supplemented with 10% FCS, 1% P/S, 1% L-glutamine, 1% sodium pyruvate, 1% MEM non-essential amino acids, 2 mM HEPES and β-mercaptoethanol. We added 3 × 104 5 × 104 DCs, and analysed T cells on days 2 or 5 as indicated.
CD4+ T cell assay
DCs were sorted as above. Naive CD4+ T cells (CD3+ CD4+ CD44− CD62L+ cells) were sorted from the spleen of a naive mouse. We plated 105 T cells e in Click’s medium supplemented with 10% FCS, 1% P/S, 1% L-glutamine, 1% sodium pyruvate, 1% MEM non-essential amino acids, 2 mM HEPES and β-mercaptoethanol, and added 104 DCs. We added 5 ng ml−1 huIL-2 and 1 μg ml−1 anti-CD3 antibody (both from bioXcell) on days 2 and 4. T cells were analysed on day 5.
OT-II assay
We sorted DCs as above, plated 104 cells and pulsed them with 30 ng ml−1 ovalbumin peptide 323–339 (Sigma) for 30 min, followed by three PBS washes. OT-II cells were isolated from the spleen of an OT-II mouse using a CD4+ enrichment kit (Invitrogen). We added 105 T cells to DCs in Click’s medium supplemented with 10% FCS, 1% P/S, 1% L-glutamine, 1% sodium pyruvate, 1% MEM non-essential amino acids, 2 mM HEPES and β-mercaptoethanol. T cells were analysed on day 2.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
All mice sequencing data are publicly available (GEO accession code GSE131957). All human sequencing data is available on NCBI with Bio-Project ID PRJNA609924.
Code availability
Scripts to reproduce clustering and differential expression analyses, as well as for direct reproduction of figures related to computational results, are available at https://github.com/effiken/Maier_et_al_nature_2020.
Extended Data
Extended Data Fig. 1 |. mregDCs are a distinct dendritic-cell cluster present in numerous tumour models.
a, Digested lungs of naive or KP-tumour-bearing mice at day 28 post tumour-cell injection were stained with antibodies conjugated either to fluorophores for FACS or to oligonucleotides for CITE-seq analysis. CD45+ Siglec F− Ly6G− MHCII+ CD11c+ cells were sorted and loaded onto a 10x Chromium chip for scRNA-seq and CITE-seq analysis. Dendritic-cell clusters were identified according to marker-gene expression after clustering of transcriptomes. Heat maps show UMI counts of lineage genes across all clusters after downsampling to 2,000 UMIs per cell. b, Left, gene–gene correlation of highly variable genes, with relevant gene modules outlined and annotated; right, scRNA expression divided by cluster. Genes on left and right panels are aligned. c, CD45+ Siglec F− Ly6G− MHCII+ CD11c+ cells from lungs of naive or B16-BFP/OVA-tumour-bearing mice at day 22 were sorted and loaded onto a 10x Chromium platform for scRNA-seq. DCs were mapped to the clusters generated for the experiment shown inFig. 1 by maximum-likelihood classification. Heat maps show UMI counts of lineage genes across all clusters after downsampling to 2,000 UMIs per cell. d, Mouse-tumour public scRNA data for immune cells from an M38 model13 and a T3 sarcoma model28 were accessed from GEO. Top, broad cell types were sorted in silico using gene lists, resulting in pure DC populations. pDC, plasmacytoid DC. Bottom left, DC1s, DC2s and mregDCs were identified using scores generated from gene lists that defined these populations. Bottom middle, annotations in the heat map were derived from k-means clustering (k = 3) of coordinates in the dendritic-cell-score scatter plot. Bottom right, DCs of each annotation are quantified. Gene lists defining cell types for in silico sorting and stratification of dendritic-cell subtypes are in Supplementary Table 2. e, Lung DC1s and migratory DC1s (migDC1) from DLNs were sorted and analysed by RNA-seq. Genes highlighted in red identify a reference set of genes from migratory DCs7. f, Lung DC1s and migratory DC1s from DLNs in both naive and KP-tumour-bearing mice were sorted and analysed by RNA-seq. The plot compares migDC1 gene expression with lung DC1 expression by log2FC in naive (x-axis) and KP-tumour-bearing (y-axis) mice. Genes upregulated in mregDCs relative to DC1s (log2FC greater than 2; Benjamini–Hochberg-adjusted P-value less than 0.01), as assayed by scRNA-seq, are shown in gold. g, Stratification of dendritic-cell transcriptomes using dendritic-cell subtype scores in naive and KP-tumour-bearing lungs. Scores for each subtype were generated from gene lists that were differentially expressed among clusters. Single cells are coloured by cluster identification (left) or CITE-seq surface marker expression (colour-bar units are log10(1 + ADT counts)). Gene scores are the same as in d (lower left).
Extended Data Fig. 2 |. The mregDC program is enriched in both canonical dendritic-cell subsets upon tumour-antigen uptake.
a, b, CD45+ Siglec F− Ly6G− MHCII+ CD11c+ cells were sorted from lungs of Ccr7−/− mice and loaded onto the 10x Chromium followed by scRNA-seq. Transcriptomes were mapped to the clusters generated for the wild-type experiment shown in Fig. 1 by maximum-likelihood classification. a, The heat map shows UMI counts of selected genes in dendritic-cell clusters after downsampling to 2,000 UMIs per cell, comparing cells from Ccr7−/− mice to cells from WT mice. b, Comparison of differential expression analyses between mregDCs and resting DCs in WT mice (x-axis) and Ccr7−/− mice ( y-axis) (b). c, Frequencies of mregDCs as a percentage of total DCs, as measured by scRNA-seq in naive and KP–GFP-tumour-bearing mice. d, Gating strategy for subsets of conventional lung DCs. e, Flow cytometry of GFP+ versus GFP− DC2s (CD11b+ CD103−) from KP–GFP-tumour- bearing mice. f, Flow cytometry of GFP+ versus GFP- DC1s or DC2s from KP–GFP-tumour-bearing mice. g, Flow cytometry of BFP+ versus BFP− DC1s from B16-BFP/OVA tumour-bearing mice. The experiment shown is representative of two independent experiments; *P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001 (Student’s t-test); data are means ± s.d. (e–g). h, KP–GFP cells were exposed to ultraviolet radiation for 30 min, rested for 24 h, and stained with annexin V and propidium iodide in order to confirm induction of apoptosis before experiments involving coculture of DCs. i, Differential expression between mregDCs identified by transcriptome from KP-tumour-bearing and naive mice. Genes in green are significantly differentially expressed (Benjamini–Hochberg-adjusted P-value of less than 0.15); selected immune genes are shown in orange.
Extended Data Fig. 3 |. The mregDC program is independent of MyD88/TRIF, inflammasome signalling and lymphocytes.
a–d, Flow-cytometry analysis of DC1s isolated from KP–GFP-tumour-bearing lungs in Asc−/− (a), Il1r−/− (b), Myd88−/−Trif−/− (c) and Rag1−/− (d) mice. e, Heat map showing average TAM receptor RNA expression in mregDC, DC1 and DC2 scRNA-seq clusters. f, Differential expression of TH2 response genes across dendritic-cell clusters identified by scRNA-seq, showing a log2FC between average mregDC expression and resting dendritic-cell expression. Genes in green are differentially expressed (Benjamini–Hochberg-adjusted P-value less than 0.15); TH2 response genes are in orange. g, k, Mice were injected with KP–GFP tumour cells, treated with anti-IL-4 (αIL-4) or control IgG on days 21, 24 and 26, and analysed on day 28. GFP+ DC1s carrying tumour antigens in lung and DLNs (g) and T cells in DLNs (k) were analysed by flow cytometry. h, KP–GFP-tumour-bearing mice were injected with an agonistic CD40 antibody (CD40a) on days 25 and 27; lungs were analysed on day 28. i, KP–GFP-tumour-bearing mice were injected with polyI:C on day 27, and lungs were analysed on day 28.j, GFP+ conventional DC1s were purified from KP–GFP-tumour-bearing lungs from B6D2 mice treated either with anti-IL-4 or control IgG and cocultured with naive CD8+ JEDI T cells isolated from JEDI mouse spleens. JEDI T cells were analysed on day 2. One experiment, representative of two independent experiments, is shown (a–d, g–k). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (Student’s t-test (a–d, g, k) or one-way ANOVA and Tukey’s test (h, i)). Data are shown as means ± s.d. (a–d, g–i, k).
Extended Data Fig. 4 |. Protein expression profile of mregDCs in human NSCLC lesions.
a, Average CITE-seq surface protein staining intensity of dendritic-cell clusters in non-involved lung (nLung) and tumour lesions isolated from human NSCLC resections (n = 7). b, scRNA-seq data from a published dataset29 of matched nLung and tumour from resection specimens of eight patients with NSCLC were mapped to the clusters generated for the NSCLC data in Fig. 4 by maximum-likelihood classification. Heat maps show downsampled UMI counts in dendritic-cell clusters after downsampling cells to 2,000 UMIs per cell and evenly sampling cells from dendritic-cell types.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health (NIH) grants R01 CA154947, R01 s (to M.M.), 1R01CA212376 (to S.G. and C.V.R), F30CA243210 (to S.T.C.) and 5T32CA078207 (to A.M.L.). We thank C. Berin for helpful discussions; D. Farber and P. Dogra for critical comments on the manuscript; and the Mount Sinai flow cytometry core, Human Immune Monitoring Center and Mount Sinai Biorepository for support. Research support was provided by Regeneron and Takeda.
Footnotes
Competing interests Research support for these studies was provided by Regeneron and Takeda. The authors declare no other competing financial interests.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-020-2134-y.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-020-2134-y.
Peer review information Nature thanks Cornels Melief and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
References
- 1.Sánchez-Paulete AR et al. Intratumoral immunotherapy with XCL1 and sFlt3L encoded in recombinant Semliki Forest Virus-derived fosters dendritic cell-mediated T-cell cross-priming. Cancer Res. 78, 6643–6654 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Salmon H et al. Expansion and activation of CD103+ dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition. Immunity 44, 924–938 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broz ML et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell 26, 638–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavin Y et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 169, 750–765 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginhoux F et al. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206, 3115–3130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sathaliyawala T et al. Mammalian target of rapamycin controls dendritic cell development downstream of Flt3 ligand signaling. Immunity 33, 597–606 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller JC et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13, 888–899 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Idoyaga J et al. Specialized role of migratory dendritic cells in peripheral tolerance induction. J. Clin. Invest. 123, 844–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinman RM, Hawiger D & Nussenzweig MC Tolerogenic dendritic cells. Annu. Rev. Immunol. 21, 685–711 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Ohl L et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity 21, 279–288 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Helft J et al. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J. Clin. Invest. 122, 4037–4047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delamarre L, Pack M, Chang H, Mellman I & Trombetta ES Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307, 1630–1634 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Garris CS et al. Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-γ and IL-12. Immunity 49, 1148–1161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stitt TN et al. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell 80, 661–670 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Rothlin CV, Carrera-Silva EA, Bosurgi L & Ghosh S TAM receptor signaling in immune homeostasis. Annu. Rev. Immunol. 33, 355–391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skinner HD et al. Integrative analysis identifies a novel AXL-PI3 kinase-PD-L1 signaling axis associated with radiation resistance in head and neck cancer. Clin. Cancer Res. 23, 2713–2722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukita Y et al. Axl kinase drives immune checkpoint and chemokine signalling pathways in lung adenocarcinomas. Mol. Cancer 18, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugo W et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coussens LM, Zitvogel L & Palucka AK Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science 339, 286–291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mantovani A & Allavena P The interaction of anticancer therapies with tumor-associated macrophages. J. Exp. Med. 212, 435–445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zahner SP et al. Conditional deletion of TGF-βR1 using Langerin-Cre mice results in Langerhans cell deficiency and reduced contact hypersensitivity. J. Immunol. 187, 5069–5076 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Xue W et al. Response and resistance to NF-κB inhibitors in mouse models of lung adenocarcinoma. Cancer Discov. 1, 236–247 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoeckius M et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaitin DA et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul F et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 163, 1663–1677 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Baran Y et al. MetaCell: analysis of single cell RNA-seq data using K-nn graph partitions. Genome Biol. 20, 206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gubin MM et al. High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell 175, 1443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambrechts D et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Holland SJ et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res. 70, 1544–1554 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Agudo J et al. GFP-specific CD8 T cells enable targeted cell depletion and visualization of T-cell interactions. Nat. Biotechnol. 33, 1287–1292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mice sequencing data are publicly available (GEO accession code GSE131957). All human sequencing data is available on NCBI with Bio-Project ID PRJNA609924.