Abstract
Although COVID-19 largely causes respiratory complications, it can also lead to various extrapulmonary manifestations resulting in higher mortality and these comorbidities are posing a challenge to the health care system. Reports indicate that 30–60% of patients with COVID-19 suffer from neurological symptoms. To understand the molecular basis of the neurologic comorbidity in COVID-19 patients, we have investigated the genetic association between COVID-19 and various brain disorders through a systems biology-based network approach and observed a remarkable resemblance. Our results showed 123 brain-related disorders associated with COVID-19 and form a high-density disease-disease network. The brain-disease-gene network revealed five highly clustered modules demonstrating a greater complexity of COVID-19 infection. Moreover, we have identified 35 hub proteins of the network which were largely involved in the protein catabolic process, cell cycle, RNA metabolic process, and nuclear transport. Perturbing these hub proteins by drug repurposing will improve the clinical conditions in comorbidity. In the near future, we assumed that in COVID-19 patients, many other neurological manifestations will likely surface. Thus, understanding the infection mechanisms of SARS-CoV-2 and associated comorbidity is a high priority to contain its short- and long-term effects on human health. Our network-based analysis strengthens the understanding of the molecular basis of the neurological manifestations observed in COVID-19 and also suggests drug for repurposing.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12035-020-02266-w.
Keywords: COVID-19, Neurologic comorbidity, Network biology, Disease-gene interaction, Hub genes, Drug repurposing
Introduction
As of the end of October 2020, the novel Coronavirus Disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has crossed over 50 million cases globally with more than 1 million deaths. While SARS-CoV-2 is known to cause substantial complications in respiratory and pulmonary systems, causing pneumonia and acute respiratory distress syndrome (ARDS), several COVID-19 cases present various extrapulmonary manifestations of COVID-19. These include manifestations of cardiovascular, renal, hematologic, gastrointestinal, hepatobiliary, endocrinological, ophthalmological dermatological, and neurological systems [1–4]. To date, increasing evidences are indicating the neurological manifestations of the central and peripheral nervous system in COVID-19 [5]. These neurological complications include headaches, dizziness, nausea, loss of consciousness, seizures, anorexia, anosmia, ageusia, encephalopathy, and meningo-encephalitis [6–11]. In addition, ischemic stroke [12], acute necrotizing encephalopathy (ANE) [8], and acute inflammatory demyelinating polyneuropathy (Guillain-Barré syndrome, GBS) [13] have also been reported to be associated with COVID-19. Moreover, COVID-19 patient autopsy studies have also identified viral RNA transcripts and viral proteins in brain tissues [14, 15].
The potential mechanisms underpinning the various neurological syndromes include direct or indirect viral neuronal injury [16], a secondary hyperinflammation syndrome related to cytokine storm [17], post-infectious immune-mediated disorders, or the effects of a severe systemic disorder with the neurological consequences of sepsis, hyperpyrexia, hypoxia, vasculopathy, and/or coagulopathy.
Recently, two studies utilized the 3D human brain organoids to study the neuroinvasive potentials of SARS-CoV-2. Ramani et al. [18] have revealed that SARS-CoV-2 readily targets cortical neuronal cells and induces tauopathies and neuronal cell death. The other study by Song et al. [19] models the SARS-CoV-2 infection of neuronal cells in hiPSC-derived brain 3D organoids. These comorbidities thus lead to higher risk of disease development and higher mortality associated with COVID-19. It is largely assumed that other neurological manifestations will likely surface in the near future. Thus, understanding the infection mechanisms of SARS-CoV-2 and associated comorbidity is a high priority to contain its short- and long-term effects on human health.
SARS-CoV-2 causes the disease by hijacking the host cell machinery and perturbs the highly organized cellular networks. Moreover, the highly coordinated interactions between molecules observed in a healthy cell are also gets altered in a disease condition [20]. This suggests that SARS-CoV-2 interaction with host human cells will be different in healthy and diseased cells, and thus could lead to different impacts of COVID-19 infection. Therefore, pre-existing clinical conditions can facilitate the appearance of another disease if they share the same or functionally related genes [21]. As SARS-CoV-2 has been shown to be associated with several neurological manifestations, we here predicted the risk of COVID-19 infection in patients with various neurological disorders.
Over the last 20 years, numerous human-viral interactomes have been constructed to understand the mechanisms of viral entry, infection, and disease progression [22–26]. Investigating such interactomes has led to the discovery of shared and distinct molecular pathways associated with viral pathogenicity. In the present work, we have utilized a network-based system biological framework (Fig. 1) to investigate the molecular interplay between COVID-19 and various human neurological disorders. Here, we have constructed a brain-specific protein-protein interaction network of the 332 genes of human’s targeted by SARS-CoV-2 reported by Gordon et al. [27], with their neighboring genes and named the network as COVID-19 target network (CTN). The genes in the network were then further used to identify the neurological disease associated with them. Based on the shared genes, we have integrated the CTN with brain diseases and generated a disease-gene network of the brain (BDGN). This human brain disease-gene network consists of a total of 123 various brain disorders including COVID-19 interacting with 653 genes of CTN. Out of these 123 diseases, 28 diseases were directly linked with the COVID-19, indicating the comorbidity and complexity of COVID-19. Next, we have identified the functional modules and hub genes of the CTN network that can be considered as the hotspot for comorbidity and could be the target for drug repurposing. We therefore emphasized that targeting these functional modules will inhibit the viral replication and growth and thus will improve the medical conditions in comorbidity associated with COVID-19.
Fig. 1.
A strategic workflow adopted in this study with self-explained legends
Material and Methods
Brain-Specific Protein-Protein Interaction (PPI) Network for COVID-19 Target Genes
Interactome data of the human brain was retrieved from the TissueNet v.2 database with 165,240 interactions [28]. Quantitative tissue association for human PPI is provided by TissueNet. TissueNet gathers RNA-Seq and protein-based assay profiles from the genotype-tissue expression project (GTEX) and human protein atlas (HPA) for preparing extensive interaction networks. Experimentally validated protein interaction information extracted from four major databases (DIP, BioGrid, MINT, and IntAct) was also included for preparing PPI networks. A list of 332 genes of humans known to interact with COVID-19 was obtained from Gordon et.al [27] and was used for creating a subnetwork having interaction between the COVID-19 target genes and their neighboring genes. The subnetwork was called the COVID-19 target network (CTN). Out of 332 genes, 4 genes (GDF15, INHBE, SBNO1, and CEP43) did not show any interactions in the brain.
Assembling Brain-Specific Disease-Gene and Disease-Disease Interaction Network
After obtaining the list of COVID-19 target genes and their neighboring genes from CTN, Gene ORGANizer [29] tool was used to identify the brain-related disorders linked with these genes. Gene ORGANizer database allows us to analyze the relationship between the query genes and the organs affected by them. The database provides organ-specific disease-gene relation from highly curated DisGeNET and human phenotype ontology (HPO) tools. Disease-gene interactions having valid HPO identifiers were considered for further study. A total of 2002 disease-gene interactions related to brain disorder was obtained, which included 127 various brain disorders interacting with 653 genes. Forty-three out of 653 genes were the direct target of the COVID-19. Finally, a disease-gene interaction network was prepared which consists of 780 nodes (i.e., diseases/genes) and 2046 interactions. Disease-Disease interaction network was then prepared using the disease-gene interaction network in which two diseases are considered to be associated only if they share a common gene known to cause those diseases. A network of 123 diseases connected with each other was prepared, in which 28 diseases were directly connected to the COVID-19. The calculation of the topological coefficient of the disease-gene interaction network also indicated the tendency of nodes in the network to have shared neighbors [30]. The values of topological coefficient also suggest that CTN may have functional modules within the network, as the genes in the modules are more connected in-between rather than the genes present outside the module [31].
Identification of Functional Protein Modules of CTN
Organization of biological function is believed to be in a modular and hierarchal manner [32]. It is believed that cellular function modular organization is reflected as modular structure in molecular network [33]. Protein modules are groups of highly connected proteins that tend to be co-localized and co-expressed. Proteins in protein-modules form a single multimolecular machine by interacting with each other at the same time and place [34, 35]. The MCODE plugin of the Cytoscape tool was used for identifying protein modules in CTN with degree cut-off value equals to 2, node score cut-off value equals to 0.2 and k-core value equals to 2. MCODE algorithm operates on three stages, i.e., vertex weighing, complex prediction, and optionally post-processing step, for identifying the locally highly connected region in the interaction network [36]. A total of 32 modules were obtained, from which the five largest modules in terms of their sizes have been selected for further studies. These selected five modules have the maximum number of genes and also have multiple COVID-19 target genes in it. These five functional modules thus cover a major part of the COVID-19 target network and most likely will play an important role in the progression of the disease. The functional modules identified in CTN are large, so it is important to identify which genes in each module best explains its behavior. A widely used approach is to identify highly connected genes (also known as hub genes) in the modules [37].
Identification of Hub Genes of CTN
Hub genes in the network are the genes that are highly connected with other genes in the network on a direct basis. Any change in the expression or activity of the hub gene has the potential to influence the working of the network. Hubs are frequently more relevant to the functionality of the network than other genes. We calculated the topological properties like the degree of connectivity (K), betweenness centrality value, and closeness centrality for all the 5 functional modules, similar to our previous study [26]. All these network topology parameters were calculated using the network analyzer plugin of the Cytoscape tool [38]. Briefly, degree (k) signifies the number of interactions made by nodes in a network and is expressed as:
where Ku is the node-set containing all the neighbors of node u, and w(u,v) is the edge weight connecting node u with node v.
Betweenness centrality (Cb) represents the degree to which nodes stand between each other based on the shortest paths. A node with higher betweenness centrality represents more control over the network. It is expressed as:
where p(k,u,f) is the number of interactions from k to f that passes through u, and p(k,f) denotes the total number of shortest interactions between node k and f.
Closeness centrality (Cc) is a measure of how fast information is traveled from one node to other nodes in the network. Closeness centrality value ranges from 0 to 1, and isolated genes have closeness centrality value equal to zero.
where z is the node for which the closeness value is calculated and L(z,m) is the length of the shortest path between two nodes z and m. It has been seen that genes having a high degree of connectivity also have high closeness centrality score.
Eccentricity (eG) is defined as shortest distance between one node from all the other nodes present in the network. The shortest the distance between the nodes, the faster the travel of information among them. If a node is an isolated node, its eccentricity value will be zero.
where v is the central node in a connected network G for which we are calculating the eccentricity and u are the nodes presented in the network.
Topological coefficients (Tf) indicate the tendency of the nodes in the network to have shared neighbors. Nodes having no or one neighbor are assigned a topological coefficient of zero. The topological coefficient of a node, n with kf neighbors is computed as:
where j(f,p) is the number of shared neighbors between f and p, plus 1 if there is an edge between f and p.
The top 35 genes having a high degree of connectivity and betweenness values were considered as hub genes of the CTN. Apart from topological parameters, we have also calculated the Dyadicity, which measures the connectedness of the nodes belonging to the same groups, and the Jaccard similarity coefficient to check the extent of molecular overlapping among COVID-19 and other neurological disorders.
Identification of Key Genes and Key Regulators of the Protein Modules
After identifying the hub genes of the CTN network, we have identified one key gene in each of the functional modules. A key gene is among the hub genes of the modules which also plays role in connecting the functional modules with each other and provides the idea about how these modules shares information with each other. Furthermore, for identifying the key regulator or driver genes which control the regulation of the modules, tracing of hub genes of each module up to motif level was performed. Key regulators are the genes that are highly connected in the network not only when seen from the top of the network but its connection reaches the lower levels in the network making the gene an important and influential part of the network. Apart from identifying the important genes in the modules, it is equally important to identify the processes and the pathways in which these modules play role, to better understand how the alterations in the function of these modules affects the host condition.
Gene Ontology and Pathway Enrichment Analysis
For enrichment analysis of protein-modules, Database for Annotation Visualization and Integrated Discovery (David v.6.7) tool was used [39]. Gene Ontology (GO) enrichment analysis includes annotation at biological, cellular, and at molecular levels. DAVID uses the GO and Kyoto encyclopaedia of genes and genomes (KEGG) database for the enrichment analysis of the genes. Pathways and functions having P value less than 0.05 were considered significantly enriched.
Network-Based Drug Repurposing
For identifying the drug targets for hub genes, key genes, and key regulators, databases such as DrugBank, Clue.io [40], ChemblInteraction [41], and DGIdb [42] were screened out. A final drug-gene interaction network was prepared using the STITCH database [43]. STITCH is a database known to predict the physical and functional interaction between the query genes and drugs. The interactions in the STITCH database are derived from five sources namely, automated text mining, high throughput lab experiment data, co-expression interaction data, interaction prediction by genomic context, and by previous knowledge from other databases. For each interaction, STITCH calculated a combined score. A combined score is calculated by combining the corrected probability of observing an interaction randomly and probabilities of interaction from different evidence channels. The drug-gene interactions having a combined score value > 0.7 were considered high confidence interactions.
Apart from chemical molecules as drugs, we also identified the microRNAs (miRNAs) as a potential drug candidate, the miRNAs interacting with the 35 hub genes were retrieved from the miRTarBase database [44]. mirTarBase database provides miRNA interaction with several hosts organism including humans. This database uses several validation methods such as reporter assay, western blot, qPCR, microarray, NGS, and pSILAC to validate the interaction between host genes and miRNAs. For 35 hub genes, 1997 miRNA-gene interactions were retrieved. Interactions having at least one strong evidence of validation methods were used for further analysis.
Results
Protein-Protein Interaction Network Between COVID-19 and Human Host in the Brain
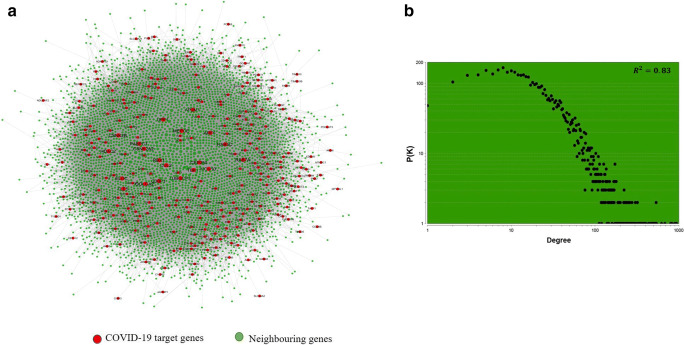
For constructing COVID-19 and its human interaction network, we retrieved the protein-protein interaction network of the brain from TissuevNet2.0 [28] database. A network consisting of 12,968 number of nodes and 165,241 number of edges was constructed using the brain’s PPI data (Supplementary Fig. 1). A list of 332 human target genes of COVID-19 was retrieved from Gordan et.al [27]. A subnetwork of these 332 genes with their neighboring genes was constructed from the brain’s PPI network and was named as the COVID-19 target network (CTN) (Fig. 2a, Supplementary Table 1). Out of the 332 COVID-19 target genes, 327 genes were part of the subnetwork. The subnetwork has 5061 number of nodes and 95,802 number of edges. More than 50% of interactions from the main network become the part of a subnetwork which clearly depicts how deeply the COVID-19 target genes are connected in the brain. The degree distribution of CTN indicates that the network has a scale-free property (Fig. 2b). A scale-free network is one that follows a power-law distribution and is independent of the size of the network (i.e., the number of nodes in the network), which means the basic structural foundation of the network remains the same even when the network grows [45, 46]. Next, we also calculated the dyadicity (D) between the COVID-19 target genes in CTN and obtain a dyadicity value as 24.2. Dyadicity refers to the connectedness of the nodes belonging to the same group in a network. In a dyadic network, nodes belonging to the same group are more connected with each other than in a random network. A network is called dyadic when D > 1 [47, 48]. A value of 24.2 in our analysis indicates that COVID-19 target genes are very well connected and making a complex in the network, which can hijack the host cellular machinery. The complexes in a network are likely to contribute to the progression of disease and comorbidity at the molecular level [49]. Proteins present in a community have a higher probability to play role in comorbidity as compared to the other proteins which are not part of the complex. Therefore, to understand the high risks of comorbidities associated with covid19, we have prepared and briefly analyzed the disease-gene and disease-disease interaction network of CTN.
Fig. 2.
a COVID-19 target genes (in red) interaction network in the brain with neighboring genes (in green). b Scatter plot showing the distribution of degree (k) in the COVID-19 target network (CTN)
Disease-Gene and Disease-Disease Interaction Network in the Brain
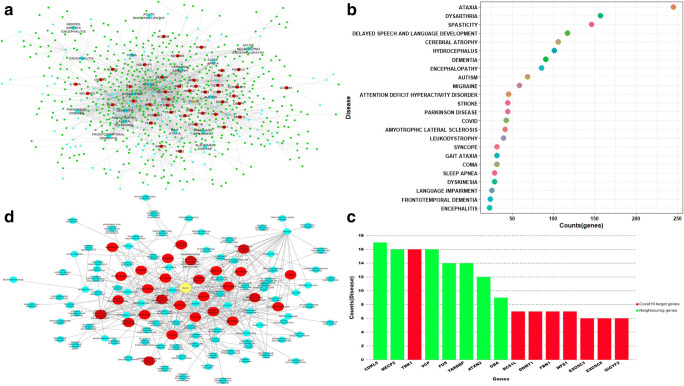
A disease-gene interaction network and disease-disease interaction network from CTN were prepared to understand the links of COVID-19 with neurological comorbidities. A disease-gene interaction map of CTN was constructed from the disease-gene interaction data of the brain obtained from the Gene ORGANizer database. A total of 313 brain diseases, 715 genes, and 2965 disease-gene pairs were considered for network construction. The disease-gene association map in CTN will then be constructed by connecting the associated node (genes) and brain disorder. The resulting network revealed 653 genes linked to a total of 127 disorders, which also includes COVID-19 (Fig. 3a, Supplementary Table 2). Figure 3a represents the disease-gene network map comprising of 780 nodes and 2002 disease-gene interactions and is termed as the brain disease-gene network (BDGN).
Fig. 3.
a Disease gene interaction network: the figure represents the interaction of CTN network genes with their related brain disorder. COVID-19 target genes are represented in red, neighboring genes of COVID-19 target genes are represented in green, diseases are represented by blue color, and COVID-19 disease is represented by yellow color. b Dot plot of highly connected diseases and the number of genes associated with disease in the brain’s gene-disease interaction network. c Bar plot of genes highly connected to multiple diseases in the brain’s gene-disease interaction network. d Brain’s disease-disease interaction network. Red color nodes represent the disease directly connected to COVID-19
The disease-gene interaction network showed that several disorders were connected with more than one gene in the network such as ataxia (k = 245), dementia (k = 91), autism (k = 69), and COVID-19 (k = 43) (Fig. 3b). Similarly, the disease-gene interaction network also showed that many of the disorder share the common genotype. For example, SARS-CoV-2 targets, TBK1 (k = 16), BCS1L (k = 7), DNMT1 (k = 7), FBN1 (k = 7), WFS1 (k = 7) and neighborhood nodes, CDKL5 (k = 17), MECP2 (k = 16), VCP (k = 16), TARDBP (k = 14), and ATXN2 (k = 12) are linked to multiple disorders (Fig. 3c, Supplementary Table 3). Also, the calculation of the topological coefficient of the BDGN network indicated that several nodes in the network have shared neighbors (Supplementary Fig. 2). Thus, the resulting BDGN reveals the molecular connection of COVID-19 with various brain disorders and also the close association of COVID-19 targets with the genes causing the brain disorders (Supplementary Fig. 3).
To further demonstrate the association between COVID-19 and brain disorders, a disease-disease association network was then constructed, where two diseases were considered to be related if they share one common gene. The resulting disease-disease interaction network contains a total of 123 diseases (nodes) and 436 edges, representing a higher clustering between diseases (Fig. 3d and Supplementary Table 4). Out of 123 disease nodes in the network, 28 nodes (in red) were directly linked with COVID-19 (yellow node) (Fig. 3d). Jackard similarity coefficient was also calculated to check the extent of molecular overlapping between COVID-19 and other brain-related disorders. Several diseases like ataxia, dysarthria, spasticity, cerebral atrophy, autism, dementia, and stroke were closely related to the COVID-19 and are also connected with multiple genes in the brain (Supplementary Fig. 4A and 4B). Thus, because of these molecular overlapping, patients having these diseases are more prone to COVID-19 and vice-versa. The higher molecular similarities between brain diseases create a highly complex high-density comorbidity cluster which contributes to higher mortality in COVID-19 patients.
To further strengthen the above statement of the high-density comorbidity, we calculated the eccentricity value of the network. The eccentricity of a node in a biological network can be interpreted as the easiness of the node to be functionally reached by all other nodes in the network [50]. A node with a high eccentricity value compared to the average eccentricity value of the network will be more easily influenced by the activity of the other nodes and conversely could also easily influence the other nodes in the network by its activity. We observed that more than 80% of the nodes in the disease-disease network shows the eccentricity value greater than or equal to the network’s average eccentricity value (i.e., 4), which represents that the functionality of the nodes in the network is highly linked to each other making the network a highly complex cluster (Supplementary Table 5).
Recent studies evidently showed the extra pulmonary associations in COVID-19 that contribute to higher mortality in COVID-19 patients. Thus, it is of utmost necessity to develop effective drugs to target the patient-specific risks of comorbidity during COVID-19 infection. However, presence of several overlapping molecular connection makes it difficult to identify and prioritize the targets for the treatment of the COVID-19 infection. We therefore focused next to target the host functional protein modules linked with diverse brain diseases.
A Broad Range of Disorders Are Linked to Functional Protein-Modules of Host Interaction Network
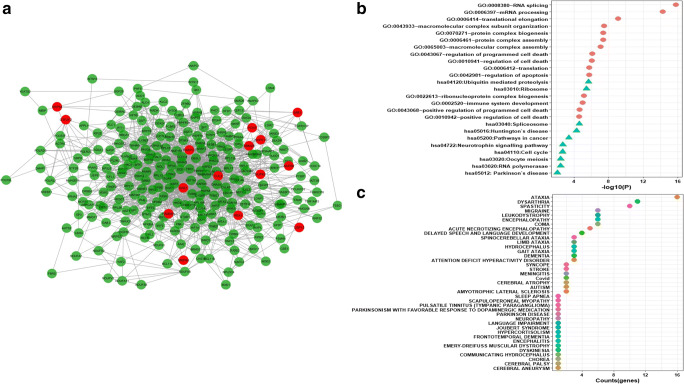
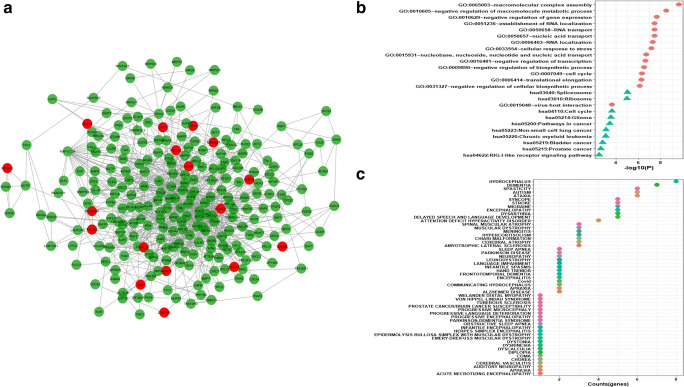
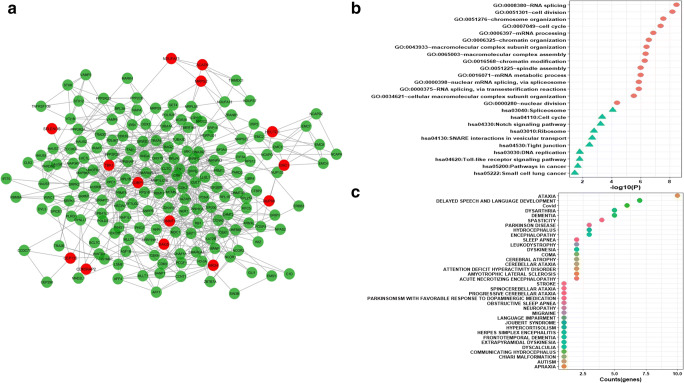
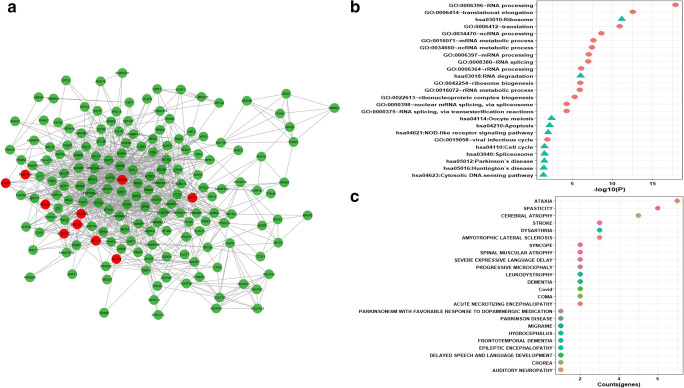
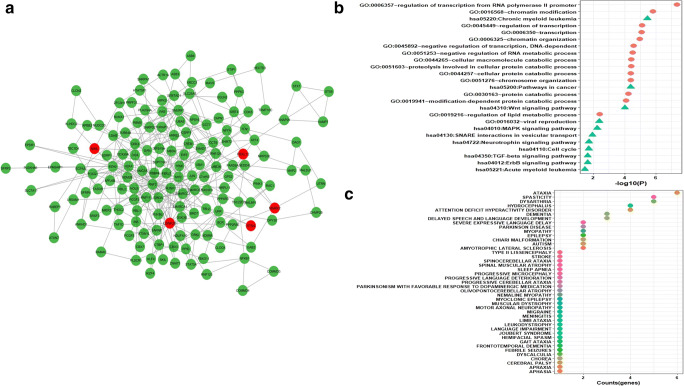
It is generally accepted that biological networks are not randomly connected and follow a structural pattern which gives rise to the modular structure and hierarchical organization. Modularity suggests nodes that are highly connected in a community are most likely to have the same biological functions and play a role in similar pathways [51]. Many complex networks exhibit modular structures, where in-between modules interactions are less dense as compared to interaction within modules. These modules reflect the organization of the functional unit in a network with relative independence [52]. Generally, diseases sharing similar genes are more predisposed to form disease modules and comorbidity. Similarly, genes related to similar diseases are likely to highly interact with each other [53]. For identifying protein modules, we used the MCODE module of the Cytoscape tool. A total of 32 protein modules in CTN were identified by MCODE. The top 5 modules having a larger number of nodes and edges were selected for further studies (Supplementary Table 6). Module-1 (Fig. 4a) had 257 nodes and 1542 edges including 15 COVID-19 target genes (in red). Module-2 (Fig. 5a) had 253 nodes and 921 edges including 17 COVID-19 target genes. Module-3 (Fig. 6a) had 183 nodes and 619 edges including 14 COVID-19 target genes. Module-4 (Fig. 7a) had 175 nodes and 899 edges including 10 COVID-19 target genes and module-5 (Fig. 8a) had 155 nodes and 340 edges including 5 COVID-19 target genes. The presence of a high number of COVID-19 target genes in these protein modules indicated that during the infection, these modules might be hijacked and strongly altered as compared to other modules in the network and will eventually disrupt the majority of the network function. DAVID tool was used for gene ontology analysis and KEGG pathway analysis.
Fig. 4.
a Module1 with COVID-19 target genes (in red). b Biological functions and KEGG pathways related to module-1. c Dot plot of module-1 related disease with their gene counts
Fig. 5.
a Module-2 with COVID-19 target genes (in red). b Biological functions and KEGG pathways related to module-2. c Dot plot of module-2 related disease with their gene counts
Fig. 6.
a Module-3 with COVID-19 target genes (in red). b Biological functions and KEGG pathways related to module-3. c Dot plot of module-3-related disease with their gene counts
Fig. 7.
a Module4 with COVID-19 target genes (in red). b Biological functions and KEGG pathways related to module-4. c Dot plot of module-4-related disease with their gene counts
Fig. 8.
a Module-5 with COVID-19 target genes (in red). b Biological functions and KEGG pathways related to module-5. c Dot plot of module-5-related disease with their gene counts
Biological functional enrichment analysis of module-1 revealed its role in RNA splicing, mRNA processing, protein complex biogenesis and assembly, regulation of programmed cell death and pathway analysis reveal module-1 role in proteolysis, ribosome pathway, spliceosome pathway, in Huntington’s disease pathway, and cancer pathways (Fig. 4b). The nodes of module1 were associated with disorders like ataxia, dysarthria, spasticity, encephalopathy, coma, and delayed speech and language development along with many other disorders (Fig. 4c).
Similarly, biological functional analysis indicates that module-2 plays a role in macromolecular complex assembly, negative regulation of gene expression, RNA transport, and localization, and play role in pathways like spliceosome, ribosome, cell cycle, gliomas, pathways in cancer, and RIG-I like receptor signaling pathways (Fig. 5b). The nodes are associated with disorders like hydrocephalus, dementia, autism, muscular dystrophy, language impairment, and frontotemporal dementia (Fig. 5c).
Enrichment analysis of module-3 reveals its role in RNA splicing, cell division, mRNA processing, spindle assembly, and nuclear division-related biological process, whereas pathway analysis reveals roles in Notch signaling pathways, Toll-like receptor signaling pathways, and SNARE interaction in vesicular transport (Fig. 6b). Nodes of the module-3 are associated with diseases like ataxia, sleep apnea, leukodystrophy, cerebral ataxia, and Joubert syndrome (Fig. 6c).
Module-4 plays a role in biological functions like RNA processing, translational elongation, ncRNA processing, and in the viral infection cycle, and also plays a role in pathways like NOD-like receptor signaling pathways, Parkinson’s disease, Huntington’s disease, and Cytosolic DNA sensing pathways (Fig. 7b). Module-4 is associated with diseases like ataxia, spasticity, amyotrophic lateral sclerosis, chorea, and auditory neuropathy (Fig. 7c).
Module-5 biological functional analysis reveals its role in the regulation of transcription from RNA polymerase II promoter, chromatin modification, regulation of transcription, and in cellular protein catabolic process, pathways enrichment analysis reveal a role in Wnt signaling pathways, viral reproduction, MAPK signaling pathway, neurotrophin signaling pathway, TGF-beta signaling pathway, and ErbB signaling pathway (Fig. 8b). Module 5 is associated with diseases such as ataxia, myopathy, Chiari malformation, GAIT ataxia, limb ataxia, and febrile seizures (Fig. 8c).
Interestingly, during the early stage of infection, SARS-CoV-2 hijack these modules and the concerned biological processes and synthesize its RNA. Many clinical conditions such as ataxia, spasticity, encephalopathy, and dementia are associated with all modules, revealing a greater risk of the severe illness of COVID-19 patients. The presence of common disease, the pathway, and the biological functions in the modules indicated that these modules are connected with each other to some extent (Supplementary Table 7). Besides, we also observed a different spectrum of disorders like cancer, myopia, dysarthria, sleep apnea, and thromboembolic stroke that were associated with these modules. Therefore, besides neurologic comorbidity, disorders in various other organs can also be a potential threat for COVID-19 patients as also demonstrated by Gysi et al. [54]. Our network-based results will thus add to strengthen these observations.
Core Regulatory Hubs of the Protein Network
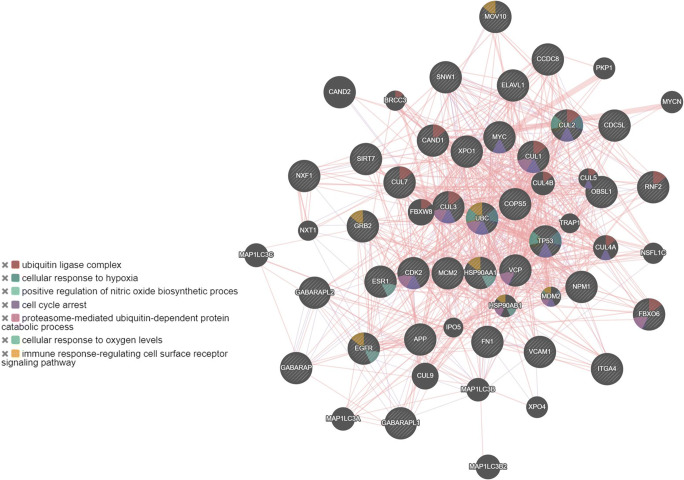
Several virus-host network studies have indicated that viral proteins target the hub proteins which have a high degree of connectivity in the network [22, 24–26, 55]. Here, we have identified 35 candidate hub proteins that exhibit a high degree of connectivity and betweenness value, using the Cytoscape’s Network analyzer tool (Supplementary Table 8). Out of these 35 hub proteins, 16 proteins bind to SARS-CoV-2 [27]. Biological process enrichment analysis of these 35 hub proteins using GeneMania webserver and GO biological process analysis indicated their role in proteasome-mediated ubiquitin-dependent protein catabolic process, G1/S transition of the mitotic cell cycle, Notch signaling pathway, ERBB signaling pathway, and response to hypoxia (Fig. 9 and Supplementary Table 9). The GO function analysis reveals the roles of hub proteins in nitric-oxide synthase regulator activity, ubiquitin-protein ligase binding, transcription regulator activity, RNA binding, and cytoskeletal protein binding (Supplementary Table 9).
Fig. 9.
Protein-protein interaction network of 35 hub genes derived from GeneMANIA along with functional enrichment
Strikingly, 26 of the SARS-COV-2 viral proteins interact with the above-identified 16 hub genes (Table 1). As can be seen, SARS-CoV-2’s NSP8, NSP13, ORF3a, and ORF9c target the most of these hub proteins (i.e., eight in total), SARS-CoV-2 NSP2 and ORF7a targets seven hub proteins, while SARS-CoV-2 N, M, NSP6, and NSP7 have six hub protein targets (Table S3). Other SARS-CoV-2 proteins like NSP’s (4, 9, 12) and SARS-CoV-2 ORFs’ (3b, 8, 10) have five targets. Intriguingly, all the 16 hub genes are targets of more than one SARS-CoV-2 protein (Table 1), out of which NPM1, SNW1, GRB2, HSP90AA1, and ELAVL1 are the target of several SARS-CoV-2 proteins. These findings are consistent with previous findings which indicates that an individual viral protein can target multiple host proteins and several viral proteins can interact with the same host protein [25, 26, 55–57].
Table 1.
Interaction of 16 hub genes of the CTN with SARS-CoV-2 proteins
| Proteins | The hub genes of the CTN |
|---|---|
| SARS-COV2 NSP1 | NPM1, SNW1 |
| SARS-COV2 NSP9 | ELAVL1, HSP90AA1, NPM1, SNW1, TP53 |
| SARS-COV2 NSP4 | HSP90AA1, NPM1, SNW1, VCP, XPO1 |
| SARS-COV2 SPIKE | GRB2, NPM1, SNW1 |
| SARS-COV2 ORF6 | CDK2, ELAVL1, NPM1, SNW1, VCP |
| SARS-COV2 NSP14 | GRB2, SNW1 |
| SARS-COV2 NSP7 | CAND1, CDK2, GRB2, HSP90AA1, NPM1, XPO1 |
| SARS-COV2 ORF9C | CAND1, CCDC8, ELAVL1, GRB2, HSP90AA1, NPM1, VCP, XPO1 |
| SARS-COV2 ORF9B | ELAVL1, NPM1, SNW1, XPO1 |
| SARS-COV2 ORF3B | ELAVL1, GRB2, HSP90AA1, NPM1, SNW1 |
| SARS-COV2 M | CAND1, CDK2, HSP90AA1, NPM1, SNW1, XPO1 |
| SARS-COV2 ORF10 | CUL2, NPM1, SNW1, VCP, XPO1 |
| SARS-COV2 ORF8 | APP, CCDC8, NPM1, SNW1, XPO1 |
| SARS-COV2 N | CDC5L, ELAVL1, HSP90AA1, MOV10, NPM1, VCP |
| SARS-COV2 NSP11 | CUL2, ELAVL1, HSP90AA1, NPM1, SNW1 |
| SARS-COV2 NSP15 | NPM1, SNW1 |
| SARS-COV2 NSP2 | CDC5L, CDK2, ELAVL1, GRB2, HSP90AA1, NPM1, SNW1 |
| SARS-COV2 NSP5 | GRB2, NPM1, SNW1 |
| SARS-COV2 NSP8 | CCDC8, CDC5L, ELAVL1, GRB2, MOV10, NPM1, RNF2, SNW1 |
| SARS-COV2 NSP13 | CDC5L, CDK2, GRB2, HSP90AA1, NPM1, SNW1, TP53, VCP |
| SARS-COV2 E | CDC5L, ELAVL1, NPM1, SNW1 |
| SARS-COV2 NSP6 | CAND1, GRB2, NPM1, SNW1, VCP, XPO1 |
| SARS-COV2 ORF7A | CAND1, CDC5L, HSP90AA1, NPM1, SNW1, VCP, XPO1 |
| SARS-COV2 NSP12 | APP, GRB2, HSP90AA1, NPM1, SNW1 |
| SARS-COV2 NSP10 | ELAVL1, NPM1, SNW1 |
| SARS-COV2 ORF3A | CDC5L, ELAVL1, GRB2, HSP90AA1, NPM1, RNF2, SNW1, TP53 |
Moreover, the spatio-temporal expression analysis of these 35 hub genes was studied through the BEST (Brain Expression Spatio-Temporal) webserver (http://best.psych.ac.cn/#) [58]. This server utilizes eight human brain expression datasets obtained from BrainSpan Atlas, Allen brainmap, GTEx, and other sources. The expression pattern of selected genes in different brain regions (spatial pattern) and age stages (temporal pattern) were analyzed using RNA-seq Data from Brainspan and RNA-seq Data from GTEx and has been shown in Supplementary Fig. 5. As can be seen in the figure that most of the genes were upregulated from the neonatal stages to adulthood, these genes were downregulated in the hypothalamus, olfactory bulb, substantia nigra, insula, and parahippocampal region (Supplementary Fig. 5A). However, in older age, the genes were moderately upregulated in the hypothalamus and substantia nigra, whereas they largely get downregulated in the olfactory bulb, thalamus, and in the cortical region (Supplementary Fig. 5B). The downregulation of these genes in the olfactory bulb, thalamus, substantia nigra, and the cortical regions of the brain indicates the impairment of sensory systems, memory, and cognition.
Network-Based Drug Repurposing
Because of high connectivity and the ability to rapidly transfer information in the network, hub genes could be the most appropriate target in any network for drug identification [26]. We propose to target these hub proteins, hijacked by SARS-CoV-2, through drug repositioning. Initially, we identified drugs for the 35 hub genes by screening multiple databases related to drug-genes interaction [42, 59, 60]. A total of 3286 drug-gene interactions for 35 hub genes were identified (Supplementary Table 10). For 16 hub genes showing interaction with COVID-19 viral protein, 1477 drug-gene interactions were identified. Out of these 16 genes, 12 genes showed significant interactions with 41 drugs having a combined score > 0.7 in the STITCH database (Supplementary Fig. 6).
Considering the severity and complexity of COVID-19, we also target key genes connecting the functional modules using drug repurposing. Targeting these key genes makes much more sense, as information transmitted by these key genes will further provide instructions to other modules controlling the network. We have identified five key genes (ESR1, TP53, UBC, HSP90AA1, and MYC) that were linking the modules in-between and can act as a suitable candidate for drug repurposing (Supplementary Fig. 7A). A total of 138 number of FDA approved drugs were identified for the 5 key genes. A drug-gene interaction network was prepared, and the STITCH database was used for the final categorization of interactions. Interactions having a combined score value > 0.7 were considered as high confidence interactions (Supplementary Fig. 7B). Moreover, earlier clinical studies indicated that many of the drugs used for the treatment of viral CNS infections have poor CNS penetration abilities and thus were unable to inhibit viral RNA replication [61]. These drugs were shown to have lower log P values (CNS penetration ability) and hence were unable to cross the blood-brain barrier. Ideally, according to Lipinski’s rule of 5, a log P value less than 5 has been considered as a pharmacologically effective drug [62]. The log P value of the identified drugs has been shown in parentheses and this information would be highly useful for selecting the drugs for drug repurposing against COVID-19 induced CNS infection.
ESR1 gene showed high confidence interactions with FDA approved drugs, fulvestrant (logP, 6.54), tamoxifen (logP, 5.93), raloxifene (logP, 5.45), estradiol (logP, 3.57), and ethinylestradiol (logP, 3.63). TP53 gene showed interaction with etoposide (logP, 0.73), 5-fluorouracil (logP, − 0.89), mitomycin-c (logP, − 0.40), bortezomib (logP, 0.89), and doxorubicin (logP, 1.41) drugs. MYC gene showed interaction with imatinib (logP, 3.47), calcitriol (logP, 5.51), and amifostine (logP, − 1.4) drugs and HSP90AA1 gene showed interaction with rifabutin (logP, 4.25) drug.
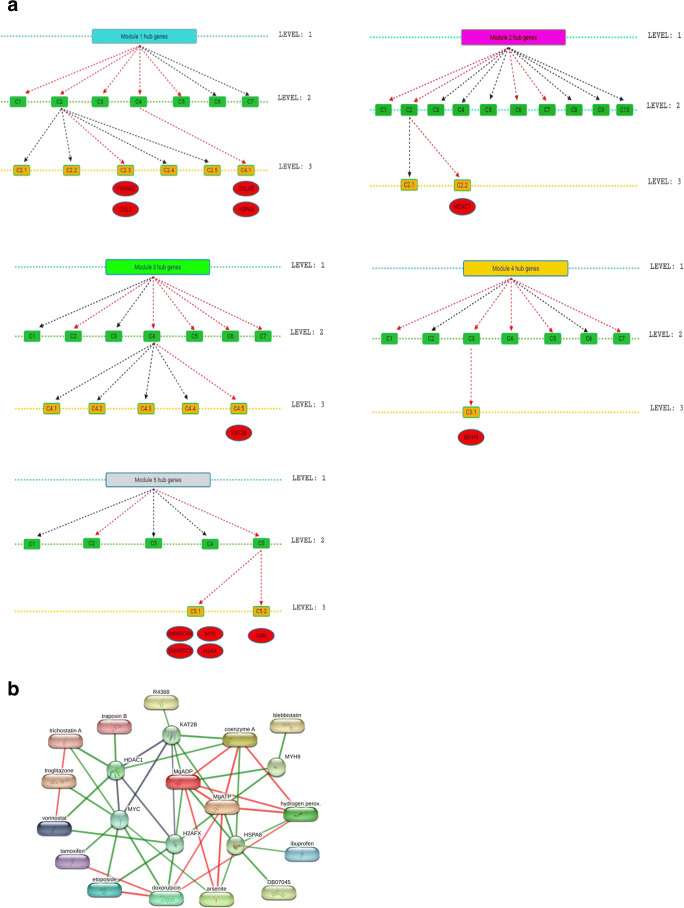
Key regulators in a network are the proteins that are deeply rooted in the network/modules and serve as the backbone of the network. Since networks serve as hierarchical characteristics, removal of key regulators from the network may cause alarming changes in the whole network and especially in the deeper levels of the network [63]. Thus, these key regulators will be proved as a good target gene in the modules. A total of 12 key regulators in 5 modules were identified by tracing the hub proteins of each module up to the motif level (Fig. 10 a). For module 1, YWHAQ, CUL4B, CUL2, and HSPA8 genes were identified as key regulators. Similarly, for module 2, the HDAC1 gene was identified, for module 3, the KAT2B gene, for module 4, the MYH9 gene, and for module 5, SMARCA4, SMARCC2, MYC, H2AX, and GAN genes were identified as key regulators. Drug targets against these key regulators were identified. A total of 107 FDA approved drugs were identified for these key regulators. Out of these 12 key regulators, 6 key genes showed significant interaction with drugs (combined value of > 0.7) using the STITCH database (Fig. 10b). For the HSPA8 protein of module-1, significant interactions were obtained with compounds such as hydrogen peroxide (logP, − 0.45), ibuprofen (logP, 3.97), arsenite, and DB07045 (logP, 1.39). Similarly, the HDAC1 protein of module-2 showed interactions with trapoxin B (logP, 3.1), trichostatin A (logP, 2.36), and vorinostat (logP, 1.88) drug molecules. Module-3 protein, KAT2B showed interaction with R4368 (N-(3-AMINOPROPYL)-N-(2-NITROPHENYL)) (logP, 1.6) drug molecule. MYH9 protein of module-4 showed interaction with blebbistatin (logP, 2) and module 5 protein, MYC showed interactions with trichostatin A (logP, 2.36), troglitazone, tamoxifen, etoposide, doxorubicin, and arsenite. Another protein of module-5, H2AX showed interaction with vorinostat, etoposide, and doxorubicin. Many of the drugs targeting these key regulators can be used either individually or in combination.
Fig. 10.
Identification of key regulators of CTN. a Tracing of hub genes through different levels in the modules. The red arrow represents the transfer of hub genes to the next level. b Interaction network of key regulators with FDA-approved drugs. The green color interaction between the drug and genes was a significant interaction with a combined score > 0.7
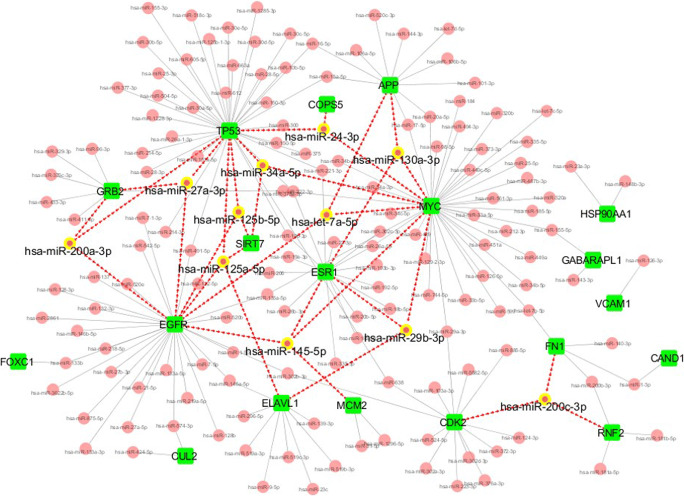
Finally, we searched the miRNAs target for the 35 hub proteins that may be considered as a potential drug to target the comorbidity. Among these 35 proteins, 19 proteins were targeted by 157 miRNAs. Of which, 11 miRNAs including, miR-125a-5p, miR-125b-5p, let-7a-5p, miR-130a-3p, miR-34a-5p, miR-29b-3p, miR-27a-3p, miR-24-3p, miR-145-5p, miR-200a-3p, and miR-200c-3p were noticed to target more than three interconnected proteins (Fig. 11). These miRNAs thus may show its utility as a drug for therapeutic intervention.
Fig. 11.
miRNA network. The network shows the miRNA (red node) targeted 19 hub genes (green node). The miRNA (yellow highlighted node) that binds to three or more than three hub genes are shown only
Discussion
The most alarming situation in COVID-19 infection is the associated comorbidity in the aged patients which increases the severe health risks worldwide. The present work has shown the risk of COVID-19 infection on the onset of various neurological disorders and the molecular basis of this comorbidity through the principle of system-based network biology. We here showed the complexity of COVID-19 and wide range of SARS-CoV-2 targets in the host cell, which establishes the molecular connection with various brain-related disorders. The disease-gene and disease-disease network map has revealed the overlapping molecular connections and high clustering of diseases within the vicinity of the same network, thus demonstrating a close pathobiological similarity. Moreover, the formation of a scale-free network among brain disorders and COVID-19 indicates the presence of a high-density comorbidity cluster. These results increase our understanding of the molecular basis of the neurological comorbidity associated with COVID-19 patients.
Another significant finding is the existence of functional modules that exhibit greater connectivity among nodes within a module (Figs. 4, 5, 6, 7, 8). Therefore, targeting the functional protein modules which are primarily hijacked by SARS-CoV-2 and are the origin of many brain disorders will prevent the virus replication and growth. The most highly connected module relates to the ubiquitin-proteasome system (UPS), ribosomal proteins, cell signaling, and cellular export proteins that can be considered as critical targets for reducing the SARS-CoV-2 infection. Furthermore, in-depth network analyses revealed 35 hub proteins present in different functional modules and are involved in more than one function (Supplementary Table 8). To provide the system-wide importance of these hub proteins in COVID-19 and associated comorbidity, we categorized these hub proteins into three groups based on their functionality. The proteins in the first group have been largely relevant to the UPS, cell cycle, and cell death. Similarly, the proteins in the other two groups are involved in transcription regulator activity, RNA metabolic pathway, signaling pathway, nuclear transport, and cytoskeletal protein binding.
The group 1 proteins are generally multifunctional and contain genes responsible for highly represented UPS. These are CUL1, CUL2, CUL3, CUL7, CAND1, OBSL1, VCP, FBXO6, UBC, and RNF2. Four of these genes (CUL1, CUL2, CUL3, and CUL7) belong to the core component of E3 ubiquitin-protein ligase complexes, which mediate the ubiquitination of target proteins. Three other UPS-related proteins, VCP, FBXO6, and UBC are the key mediators of the ER-associated degradation (ERAD) pathway. The UPS is largely involved in the maintenance of cellular homeostasis and plays a fundamental role in viral replication [64, 65]. Several viruses, including coronaviruses, often modulate ubiquitin and ubiquitin-related pathways for their survival [66, 67]. In this study, we find that module 1 and module 2 includes several members of ubiquitin-ligase and ER-associated proteins (Fig. 4a and 5a). Since the loss of ubiquitin-proteasome activity results in aggregation of proteins and disturbs cellular functions, we suggested that SARS-CoV-2 targets these modules to hinder the UPS-mediated cellular responses. The other genes of this group are involved in the regulation of cell cycle and cell death. It was recently suggested that COVID-19 infection may cause neurodegenerative disease and leads to neuronal death. The study revealed that SARS-CoV-2 infection-induced pathological effects closely resembling tauopathies and neuronal cell death [18]. In our study, we found genes like APP, TP53, MYC1, VCP, and UBC as hub genes that are involved in protein misfolding and aggregation, which ultimately leads to cell death.
Moreover, the second largest group of hub genes is involved in transcription regulation and RNA binding. The genes in this group include MOV10, XPO1, ESR1, SIRT7, NPM1, ELAVL1, NXF1, TP53, CDC5L, SNW1, RNF2, and COPS5. A recent network study identified XPO1, NPM1, HNRNPA1, and JUN, as “hub proteins” with the highest number of functional connections within 119 host proteins that interact with the human SARS-CoV-2 [56]. Of note, both NPM1 and HNRNPA1 interact directly with XPO1 in normal cells, and JUN is involved in inflammatory responses, including those associated with viral infections.
A recent single-cell RNA sequencing of the SARS-CoV-2 infected human brain organoid showed that SARS-CoV-2 infection induces a locally hypoxic environment in neurons [19]. Also, post-mortem studies of the brain of COVID-19 patients indicated the acute hypoxic-ischemic damage in the brain [14]. Consistent with these results, our findings indicate the enrichment of the hypoxia-inducing genes in the BDGN, including CUL2, TP53, UBC, and MDM2, suggesting a possible mechanism of SARS-CoV-2 induced neuropathology.
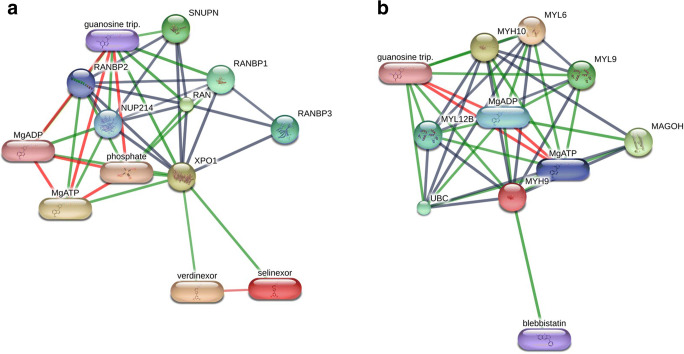
We then mapped the known drug-target network to search for druggable, cellular targets. We identified 41 approved targets for the 16 hub genes. For example, TP53, CDK2, HSP90AA, HDAC1, NPM1, and ESR1 were the most targetable proteins. One of the important drug targets is the export protein, XPO1, which enables the transport of viral proteins from the nucleus to the cytoplasm. The FDA-approved drug, Selinexor (logP, 2.85), (Fig. 12a) is a selective inhibitor of nuclear export (SINE) compound which blocks the cellular protein XPO1 and is now entered into a randomized clinical trial in COVID-19 patients as announced by Karyopharm Therapeutics Inc. [68]. Selinexor is FDA-approved drug against multiple myeloma. SINE compounds have been used to block the replication of several viruses and act as anti-inflammatory drugs [69]. In a study, verdinexor (logP, 3.7), which is highly similar in structure and biological activity to selinexor, was identified as a candidate drug inhibiting the interactions of several of the SARS-CoV-2 proteins to their human targets and was recommended for further evaluation to target COVID-19 [27].
Fig. 12.
Drug-gene interaction network of two important key genes and its drugs using the STITCH database. a The network shows selinexor, verdinexor, and guanosine triphosphate as drug candidates for XPO1. b The network shows blebbistatin as a candidate drug that binds to MYH9 and could be considered for drug repurposing
The other key drug target gene is MYH9 which has been traced to a candidate gene of module- 4 largely involved in viral infection. The MYH9 gene is involved in synthesizing a protein called myosin-9 which is one subunit of the myosin IIA protein. The MYH9-related disorder includes thrombocytopenia, hearing loss, kidney disease, and cataracts [70]. Mutations in the MYH9 gene lead to a genetic condition known as MYH9-related thrombocytopenia (MYH9RD) which is characterized by the presence of large but lesser platelets, results in bleeding in the body or the skin [71]. Thrombocytopenia is now considered as a risk factor for increased morbidity and mortality in COVID-19 patients. According to a recent study, COVID-19 patients showed blood clots in small vessels, clots in the lungs, and stroke-causing clots in cerebral arteries [72]. In some cases, a stroke may also lead to a ruptured blood vessel, and cause bleeding in the brain [73]. We here found that the chemical compounds bind to MYH9 includes guanosine triphosphate, MgADP, MgATP, and blebbistatin (logP, 2) (Fig. 12 b). The drug blebbistatin is a myosin-2 inhibitor and has been commonly used drugs in several research disciplines including neuroscience, muscle physiology, cell migration, cell differentiation, cell death, and cancer research [74]. It has also been shown to reduce the viral infection in many of the viruses by inhibiting its entry [75] and reducing the viral loads [76].Here, we also suggests eleven micro-RNAs (miRNAs) which shows the binding with hub proteins and also regulates hub proteins. The miRNA has been considered as drug molecules for targeting the SARS-CoV-2 proteins [77]. It is largely accepted that host cellular miRNA can directly target both the coding region of the viral genome and 3’UTR to induce the antiviral effect. Very recently, Fulzele et al. [78] identified over 800 miRNAs that target the COVID-19 genome. Interestingly, many of the miRNAs identified in this study (e.g., miR-130a-3p, miR-29b-3p, miR-27a-3p, miR-145-5p, and miR-200a-3p) are found to be unique to the SARS-CoV-2 genome according to the results of the Fulzele et al. [78]. Moreover, miRNAs including miR-34a-5p, miR-200a-3p, and let-7a-5p have been shown to strongly interact with the SARS-CoV-2 [79]. Besides, polyphenols are also considered as a potential and valuable natural compound for designing new drugs that could be used effectively in the combat against COVID-19 [80–82]. Overall, our BDGN analysis opens up avenues for multiple candidate repurposable drugs that target diverse cellular pathways involved in COVID-19 and associated neurologic comorbidity.
Conclusions
In conclusion, we constructed a human-SARS-CoV-2 BDGN interactome, demonstrating the risk of COVID-19 infection on multiple brain-related disorders. The study of disease-gene and disease-disease association networks reveals COVID-19-related structural and functional modules that establish the molecular connection with brain disorders. Also, the highly clustered network of the diseases suggests close pathobiological similarity and large comorbidity. We also identified crucial functional hubs in the network that showed close links with brain-related disorders including dementia, ataxia, encephalopathy, stroke, and many other disorders that indicate a high degree of comorbidity and the severity of COVID-19. We here also propose candidate drugs and miRNAs, targeting the hub proteins interconnecting the molecular pathways linked to virus infection that can be used to treat SARS-CoV-2 infection. We believe our results will help to understand the molecular pathobiology of neurological comorbidities linked to COVID-19. Given the severity and complexity of SARS-CoV-2 infection, we further suggest that drugs in combinations or drugs targeting multiple proteins will be the choice to improve clinical outcomes.
Supplementary Information
(PDF 2356 kb)
(XLSX 353 kb)
Acknowledgments
S.Y.A is grateful to the Deanship of Scientific Research, King Saud University, for funding through Vice Deanship of Scientific Research Chairs. This project was supported by the health ministry, Project number 895, Saudi Arabia. K.P. and V.K. sincerely thank the Amity University, Noida, for providing facilities. MZM is financially supported by the Department of Health and Research, Ministry of Health and Family Welfare, Government of India under young scientist FTS No. 3146887.
Authors’ Contributions
V.K. designed the study. K.P. and M.Z.M performed the experiments and calculations. K.P., M.Z.M, and V. K prepared figures for the results. V.K., K.P., S.Y.AO, and S.A.M.A analyzed the data and wrote the main text.
Funding
This work was supported by the funding from Department of Health and Research, Ministry of Health and Family Welfare, Government of India, under young scientist FTS No. 3146887. This project was supported by the health ministry, Project number 895, Saudi Arabia.
Data Availability
The data generated and analyzed during this study are included in this article and its supplementary information files.
Compliance with Ethical Standards
Not applicable.
Conflict of Interest
The authors declare that they have no conflict of interest.
Consent to Participate
Not applicable.
Consent for Publication
An informed consent and a consent to publish were obtained from each of the participants.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Md. Zubbair Malik, Email: zubbairmalik@jnu.ac.in.
Vijay Kumar, Email: vkumar33@amity.edu.
References
- 1.Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, Bikdeli B, Ahluwalia N, Ausiello JC, Wan EY, Freedberg DE, Kirtane AJ, Parikh SA, Maurer MS, Nordvig AS, Accili D, Bathon JM, Mohan S, Bauer KA, Leon MB, Krumholz HM, Uriel N, Mehra MR, Elkind MSV, Stone GW, Schwartz A, Ho DD, Bilezikian JP, Landry DW. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26(7):1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khatoon F, Prasad K, Kumar V. Neurological manifestations of COVID-19: available evidences and a new paradigm. J NeuroVirol. 2020;26(5):619–630. doi: 10.1007/s13365-020-00895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paterson RW, Brown RL, Benjamin L, Nortley R, Wiethoff S, Bharucha T, Jayaseelan DL, Kumar G, Raftopoulos RE, Zambreanu L, Vivekanandam V, Khoo A, Geraldes R, Chinthapalli K, Boyd E, Tuzlali H, Price G, Christofi G, Morrow J, McNamara P, McLoughlin B, Lim ST, Mehta PR, Levee V, Keddie S, Yong W, Trip SA, Foulkes AJM, Hotton G, Miller TD, Everitt AD, Carswell C, Davies NWS, Yoong M, Attwell D, Sreedharan J, Silber E, Schott JM, Chandratheva A, Perry RJ, Simister R, Checkley A, Longley N, Farmer SF, Carletti F, Houlihan C, Thom M, Lunn MP, Spillane J, Howard R, Vincent A, Werring DJ, Hoskote C, Jager HR, Manji H, Zandi MS. The emerging spectrum of COVID-19 neurology: clinical, radiological and laboratory findings. Brain. 2020;143:3104–3120. doi: 10.1093/brain/awaa240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, Chang J, Hong C et al (2020) Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 10.1001/jamaneurol.2020.1127 [DOI] [PMC free article] [PubMed]
- 8.Poyiadji N, Shahin G, Noujaim D, Stone M, Patel S, Griffith B. COVID-19–associated acute hemorrhagic necrotizing encephalopathy: CT and MRI features. Radiology. 2020;296(2):E119–E120. doi: 10.1148/radiol.2020201187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, Xu X, Chen Z, Duan J, Hashimoto K, Yang L, Liu C, Yang C. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav Immun. 2020;87:18–22. doi: 10.1016/j.bbi.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Q, Yang Y, Gao J. Infectivity of human coronavirus in the brain. EBioMedicine. 2020;56:102799. doi: 10.1016/j.ebiom.2020.102799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herman C, Mayer K, Sarwal A. Scoping review of prevalence of neurologic comorbidities in patients hospitalized for COVID-19. Neurology. 2020;95(2):77–84. doi: 10.1212/WNL.0000000000009673. [DOI] [PubMed] [Google Scholar]
- 12.Beyrouti R, Adams ME, Benjamin L, Cohen H, Farmer SF, Goh YY, Humphries F, Jager HR, Losseff NA, Perry RJ, Shah S, Simister RJ, Turner D, Chandratheva A, Werring DJ. Characteristics of ischaemic stroke associated with COVID-19. J Neurol Neurosurg Psychiatry. 2020;91(8):889–891. doi: 10.1136/jnnp-2020-323586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toscano G, Palmerini F, Ravaglia S, Ruiz L, Invernizzi P, Cuzzoni MG, Franciotta D, Baldanti F, Daturi R, Postorino P, Cavallini A, Micieli G. Guillain-Barre syndrome associated with SARS-CoV-2. N Engl J Med. 2020;382(26):2574–2576. doi: 10.1056/NEJMc2009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solomon IH, Normandin E, Bhattacharyya S, Mukerji SS, Keller K, Ali AS, Adams G, Hornick JL, Padera RF, Jr, Sabeti P. Neuropathological features of Covid-19. N Engl J Med. 2020;383:989–992. doi: 10.1056/NEJMc2019373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puelles VG, Lutgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, Chilla S, Heinemann A, Wanner N, Liu S, Braun F, Lu S, Pfefferle S, Schroder AS, Edler C, Gross O, Glatzel M, Wichmann D, Wiech T, Kluge S, Pueschel K, Aepfelbacher M, Huber TB. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zubair AS, McAlpine LS, Gardin T, Farhadian S, Kuruvilla DE, Spudich S (2020) Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: a review. JAMA Neurol. 10.1001/jamaneurol.2020.2065 [DOI] [PMC free article] [PubMed]
- 17.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramani A, Müller L, Ostermann PN, Gabriel E, Abida-Islam P, Müller-Schiffmann A, Mariappan A, Goureau O et al (2020) SARS-CoV-2 targets cortical neurons of 3D human brain organoids and shows neurodegeneration-like effects. bioRxiv. 10.1101/2020.05.20.106575 [DOI] [PMC free article] [PubMed]
- 19.Song E, Zhang C, Israelow B, Lu P, Weizman O-E, Liu F, Dai Y, Louvi A et al (2020) Neuroinvasive potential of SARS-CoV-2 revealed in a human brain organoid model. bioRxiv. 10.1101/2020.06.25.169946
- 20.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104(21):8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng C, Xu R. Large-scale mining disease comorbidity relationships from post-market drug adverse events surveillance data. BMC Bioinformatics. 2018;19(Suppl 17):500. doi: 10.1186/s12859-018-2468-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfefferle S, Schopf J, Kogl M, Friedel CC, Muller MA, Carbajo-Lozoya J, Stellberger T, von Dall'Armi E, Herzog P, Kallies S, Niemeyer D, Ditt V, Kuri T, Zust R, Pumpor K, Hilgenfeld R, Schwarz F, Zimmer R, Steffen I, Weber F, Thiel V, Herrler G, Thiel HJ, Schwegmann-Wessels C, Pohlmann S, Haas J, Drosten C, von Brunn A. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7(10):e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulbahce N, Yan H, Dricot A, Padi M, Byrdsong D, Franchi R, Lee DS, Rozenblatt-Rosen O, Mar JC, Calderwood MA, Baldwin A, Zhao B, Santhanam B, Braun P, Simonis N, Huh KW, Hellner K, Grace M, Chen A, Rubio R, Marto JA, Christakis NA, Kieff E, Roth FP, Roecklein-Canfield J, Decaprio JA, Cusick ME, Quackenbush J, Hill DE, Munger K, Vidal M, Barabasi AL. Viral perturbations of host networks reflect disease etiology. PLoS Comput Biol. 2012;8(6):e1002531. doi: 10.1371/journal.pcbi.1002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139(7):1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ackerman EE, Kawakami E, Katoh M, Watanabe T, Watanabe S, Tomita Y, Lopes TJ, Matsuoka Y, Kitano H, Shoemaker JE, Kawaoka Y. Network-guided discovery of influenza virus replication host factors. mBio. 2018;9(6):e02002–e02018. doi: 10.1128/mBio.02002-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasad K, Khatoon F, Rashid S, Ali N, AlAsmari AF, Ahmed MZ, Alqahtani AS, Alqahtani MS et al (2020) Targeting hub genes and pathways of innate immune response in COVID-19: a network biology perspective. Int J Biol Macromol. 10.1016/j.ijbiomac.2020.06.228 [DOI] [PMC free article] [PubMed]
- 27.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O'Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Huettenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy-Lugo J, Lin Y, Huang XP, Liu Y, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Tran QD, Shengjuler D, Fletcher SJ, O'Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu D, Wang HY, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d'Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, Garcia-Sastre A, Shokat KM, Shoichet BK, Krogan NJ. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basha O, Barshir R, Sharon M, Lerman E, Kirson BF, Hekselman I, Yeger-Lotem E. The TissueNet v.2 database: a quantitative view of protein-protein interactions across human tissues. Nucleic Acids Res. 2017;45(D1):D427–D431. doi: 10.1093/nar/gkw1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gokhman D, Kelman G, Amartely A, Gershon G, Tsur S, Carmel L. Gene ORGANizer: linking genes to the organs they affect. Nucleic Acids Res. 2017;45(W1):W138–W145. doi: 10.1093/nar/gkx302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doncheva NT, Assenov Y, Domingues FS, Albrecht M. Topological analysis and interactive visualization of biological networks and protein structures. Nat Protoc. 2012;7(4):670–685. doi: 10.1038/nprot.2012.004. [DOI] [PubMed] [Google Scholar]
- 31.Vella D, Marini S, Vitali F, Di Silvestre D, Mauri G, Bellazzi R. MTGO: PPI network analysis via topological and functional module identification. Sci Rep. 2018;8(1):1–13. doi: 10.1038/s41598-018-23672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barabasi A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 33.Pinkert S, Schultz J, Reichardt J. Protein interaction networks—more than mere modules. PLoS Comput Biol. 2010;6(1):e1000659. doi: 10.1371/journal.pcbi.1000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci. 2003;100(21):12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terentiev A, Moldogazieva N, Shaitan K. Dynamic proteomics in modeling of the living cell. Protein-protein interactions. Biochem Mosc. 2009;74(13):1586–1607. doi: 10.1134/S0006297909130112. [DOI] [PubMed] [Google Scholar]
- 36.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC bioinformatics. 2003;4(1):2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dam S, Vosa U, van der Graaf A, Franke L, de Magalhaes JP. Gene co-expression analysis for functional classification and gene–disease predictions. Brief Bioinform. 2018;19(4):575–592. doi: 10.1093/bib/bbw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 40.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, Lahr DL, Hirschman JE, Liu Z, Donahue M, Julian B, Khan M, Wadden D, Smith IC, Lam D, Liberzon A, Toder C, Bagul M, Orzechowski M, Enache OM, Piccioni F, Johnson SA, Lyons NJ, Berger AH, Shamji AF, Brooks AN, Vrcic A, Flynn C, Rosains J, Takeda DY, Hu R, Davison D, Lamb J, Ardlie K, Hogstrom L, Greenside P, Gray NS, Clemons PA, Silver S, Wu X, Zhao WN, Read-Button W, Haggarty SJ, Ronco LV, Boehm JS, Schreiber SL, Doench JG, Bittker JA, Root DE, Wong B, Golub TR. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171(6):1437–1452 e1417. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(Database issue):D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G, Wollam A, Spies NC, Griffith OL, Griffith M. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46(D1):D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36(Database issue):D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang HY, Lin YC, Li J, Huang KY, Shrestha S, Hong HC, Tang Y, Chen YG, Jin CN, Yu Y, Xu JT, Li YM, Cai XX, Zhou ZY, Chen XH, Pei YY, Hu L, Su JJ, Cui SD, Wang F, Xie YY, Ding SY, Luo MF, Chou CH, Chang NW, Chen KW, Cheng YH, Wan XH, Hsu WL, Lee TY, Wei FX, Huang HD. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48(D1):D148–D154. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’networks. Nature. 1998;393(6684):440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 46.Groningen Uo Scale-free networks.
- 47.Hu T, Andrew AS, Karagas MR, Moore JH. Functional dyadicity and heterophilicity of gene-gene interactions in statistical epistasis networks. BioData mining. 2015;8(1):1–11. doi: 10.1186/s13040-015-0062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park J, Barabási A-L. Distribution of node characteristics in complex networks. Proc Natl Acad Sci. 2007;104(46):17916–17920. doi: 10.1073/pnas.0705081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, Liu W, Ning S, Ye J, Huang T, Li Y, Wang P, Shi H, Li X. Community of protein complexes impacts disease association. Eur J Hum Genet. 2012;20(11):1162–1167. doi: 10.1038/ejhg.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hage P, Harary F. Eccentricity and centrality in networks. Soc Networks. 1995;17(1):57–63. doi: 10.1016/0378-8733(94)00248-9. [DOI] [Google Scholar]
- 51.Tripathi S, Moutari S, Dehmer M, Emmert-Streib F. Comparison of module detection algorithms in protein networks and investigation of the biological meaning of predicted modules. BMC bioinformatics. 2016;17(1):129. doi: 10.1186/s12859-016-0979-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z, Zhang J. In search of the biological significance of modular structures in protein networks. PLoS Comput Biol. 2007;3(6):e107. doi: 10.1371/journal.pcbi.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gysi DM, Do Valle I, Zitnik M, Ameli A, Gan X, Varol O, Sanchez H, Baron RM, Ghiassian D, Loscalzo J, Barabasi AL. Network medicine framework for identifying drug repurposing opportunities for COVID-19. ArXiv. 2020;arXiv:2004.07229v1. doi: 10.1073/pnas.2025581118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bosl K, Ianevski A, Than TT, Andersen PI, Kuivanen S, Teppor M, Zusinaite E, Dumpis U, Vitkauskiene A, Cox RJ, Kallio-Kokko H, Bergqvist A, Tenson T, Merits A, Oksenych V, Bjoras M, Anthonsen MW, Shum D, Kaarbo M, Vapalahti O, Windisch MP, Superti-Furga G, Snijder B, Kainov D, Kandasamy RK. Common nodes of virus-host interaction revealed through an integrated network analysis. Front Immunol. 2019;10:2186. doi: 10.3389/fimmu.2019.02186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar N, Mishra B, Mehmood A, Athar M, Mukhtar MS. Integrative network biology framework elucidates molecular mechanisms of SARS-CoV-2 pathogenesis. Science:SSRN. 2020;23(9):101526. doi: 10.2139/ssrn.3581857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo L, Lin W, Zhang Y, Li W, Wang J. BEST: a web server for brain expression Spatio-temporal pattern analysis. BMC Bioinformatics. 2019;20(1):632. doi: 10.1186/s12859-019-3222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ. The comparative toxicogenomics database: update 2019. Nucleic Acids Res. 2019;47(D1):D948–D954. doi: 10.1093/nar/gky868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nightingale S, Winston A, Letendre S, Michael BD, McArthur JC, Khoo S, Solomon T. Controversies in HIV-associated neurocognitive disorders. Lancet Neurol. 2014;13(11):1139–1151. doi: 10.1016/S1474-4422(14)70137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, Smoot J, Gregg AC, Daniels AD, Jervey S, Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farooqui A, Tazyeen S, Ahmed MM, Alam A, Ali S, Malik MZ, Ali S, Ishrat R. Assessment of the key regulatory genes and their Interologs for Turner syndrome employing network approach. Sci Rep. 2018;8(1):1–13. doi: 10.1038/s41598-018-28375-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raaben M, Posthuma CC, Verheije MH, te Lintelo EG, Kikkert M, Drijfhout JW, Snijder EJ, Rottier PJ, de Haan CA. The ubiquitin-proteasome system plays an important role during various stages of the coronavirus infection cycle. J Virol. 2010;84(15):7869–7879. doi: 10.1128/JVI.00485-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang Q, Wu P, Chen H, Li G. Pleiotropic roles of the ubiquitin-proteasome system during viral propagation. Life Sci. 2018;207:350–354. doi: 10.1016/j.lfs.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin D, Zhong B. Regulation of cellular innate antiviral signaling by ubiquitin modification. Acta Biochim Biophys Sin Shanghai. 2015;47(3):149–155. doi: 10.1093/abbs/gmu133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Longhitano L, Tibullo D, Giallongo C, Lazzarino G, Tartaglia N, Galimberti S, Li Volti G, Palumbo GA, Liso A. Proteasome inhibitors as a possible therapy for SARS-CoV-2. Int J Mol Sci. 2020;21(10):3622. doi: 10.3390/ijms21103622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karyopharm to evaluate low dose selinexor as a potential treatment for hospitalized patients with COVID-19. [news release]. Newton, MA: Karyopharm Therapeutics Inc.; April 7, 2020. (2020). https://bitly/2VftoVw Accessed July15, 2020
- 69.Widman DG, Gornisiewicz S, Shacham S, Tamir S. In vitro toxicity and efficacy of verdinexor, an exportin 1 inhibitor, on opportunistic viruses affecting immunocompromised individuals. PLoS One. 2018;13(10):e0200043. doi: 10.1371/journal.pone.0200043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fernandez-Prado R, Carriazo-Julio SM, Torra R, Ortiz A, Perez-Gomez MV. MYH9-related disease: it does exist, may be more frequent than you think and requires specific therapy. Clin Kidney J. 2019;12(4):488–493. doi: 10.1093/ckj/sfz103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ban BH, Shah V. MYH9-related thrombocytopenia. Mayo Clin Proc. 2017;92(7):1169–1170. doi: 10.1016/j.mayocp.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 72.Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin Chim Acta. 2020;506:145–148. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu P, Zhou Q, Xu J. Mechanism of thrombocytopenia in COVID-19 patients. Ann Hematol. 2020;99(6):1205–1208. doi: 10.1007/s00277-020-04019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rauscher AA, Gyimesi M, Kovacs M, Malnasi-Csizmadia A. Targeting myosin by Blebbistatin derivatives: optimization and pharmacological potential. Trends Biochem Sci. 2018;43(9):700–713. doi: 10.1016/j.tibs.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 75.Sanchez EG, Quintas A, Perez-Nunez D, Nogal M, Barroso S, Carrascosa AL, Revilla Y. African swine fever virus uses macropinocytosis to enter host cells. PLoS Pathog. 2012;8(6):e1002754. doi: 10.1371/journal.ppat.1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumakura M, Kawaguchi A, Nagata K. Actin-myosin network is required for proper assembly of influenza virus particles. Virology. 2015;476:141–150. doi: 10.1016/j.virol.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 77.Wang X, Dhindsa R, Povysil G, Zoghbi A, Motelow J, Hostyk J, Goldstein D (2020) Transcriptional inhibition of host viral entry proteins as a therapeutic strategy for SARS-CoV-2. Preprints. 10.20944/preprints202003.0360.v1
- 78.Fulzele S, Sahay B, Yusufu I, Lee TJ, Sharma A, Kolhe R, Isales CM. COVID-19 virulence in aged patients might be impacted by the host cellular MicroRNAs abundance/profile. Aging Dis. 2020;11(3):509–522. doi: 10.14336/AD.2020.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bartoszewski R, Dabrowski M, Jakiela B, Matalon S, Harrod KS, Sanak M, Collawn JF. SARS-CoV-2 may regulate cellular responses through depletion of specific host miRNAs. Am J Phys Lung Cell Mol Phys. 2020;319(3):L444–L455. doi: 10.1152/ajplung.00252.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fusco R, Scuto M, Cordaro M, D'Amico R, Gugliandolo E, Siracusa R, Peritore AF, Crupi R et al (2019) N-palmitoylethanolamide-oxazoline protects against middle cerebral artery occlusion injury in diabetic rats by regulating the SIRT1 pathway. Int J Mol Sci 20(19). 10.3390/ijms20194845 [DOI] [PMC free article] [PubMed]
- 81.Brunetti G, Di Rosa G, Scuto M, Leri M, Stefani M, Schmitz-Linneweber C, Calabrese V, Saul N. Healthspan maintenance and prevention of Parkinson’s-like phenotypes with hydroxytyrosol and oleuropein aglycone in C. elegans. Int J Mol Sci. 2020;21(7):2588. doi: 10.3390/ijms21072588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Calabrese EJ, Calabrese V, Tsatsakis A, Giordano JJ. Hormesis and Ginkgo biloba (GB): Numerous biological effects of GB are mediated via hormesis. Ageing Res Rev. 2020;64:101019. doi: 10.1016/j.arr.2020.101019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 2356 kb)
(XLSX 353 kb)
Data Availability Statement
The data generated and analyzed during this study are included in this article and its supplementary information files.