Abstract
The task of eukaryotic genome annotation remains challenging. Only a few genomes could serve as standards of annotation achieved through a tremendous investment of human curation efforts. Still, the correctness of all alternative isoforms, even in the best-annotated genomes, could be a good subject for further investigation. The new BRAKER2 pipeline generates and integrates external protein support into the iterative process of training and gene prediction by GeneMark-EP+ and AUGUSTUS. BRAKER2 continues the line started by BRAKER1 where self-training GeneMark-ET and AUGUSTUS made gene predictions supported by transcriptomic data. Among the challenges addressed by the new pipeline was a generation of reliable hints to protein-coding exon boundaries from likely homologous but evolutionarily distant proteins. In comparison with other pipelines for eukaryotic genome annotation, BRAKER2 is fully automatic. It is favorably compared under equal conditions with other pipelines, e.g. MAKER2, in terms of accuracy and performance. Development of BRAKER2 should facilitate solving the task of harmonization of annotation of protein-coding genes in genomes of different eukaryotic species. However, we fully understand that several more innovations are needed in transcriptomic and proteomic technologies as well as in algorithmic development to reach the goal of highly accurate annotation of eukaryotic genomes.
INTRODUCTION
Constantly improving next generation sequencing (NGS) technology makes it now possible to finish sequencing of a complete eukaryotic genome within several days. Not surprisingly, the computational methods reducing the time of the genome annotation stage were in high demand since the dawn of the NGS era. A self-training algorithm for ab initio gene prediction in eukaryotic genomes, GeneMark-ES (1), has accelerated the process of structural annotation for a number of genome projects, e.g. (2–7). Application of NGS to transcript sequencing (RNA-seq) motivated active development of methods integrating genomic and transcriptomic information. A new self-training algorithm, GeneMark-ET (8), integrated data on spliced aligned RNA-seq reads into GeneMark-ES.
On a parallel avenue, yet another algorithm, AUGUSTUS (9–14), was demonstrated to be one of the most accurate gene prediction tools (15–17). AUGUSTUS carried a flexible mechanism for integration of external evidence generated by spliced-aligned RNA-seq reads or homologous proteins into gene prediction. AUGUSTUS also used this evidence to predict alternative isoforms. Still, for model parameter estimation, AUGUSTUS required an expert curated training set of genes.
It was apparent that a useful automatic tool could be created by combining strong features of GeneMark-ET and AUGUSTUS. A pipeline BRAKER1 was developed and released in 2015 (18) to become a frequently used tool in genome annotation projects, e.g. (19–24). BRAKER1 requires availability of RNA-seq data, however, not all novel genomes are sequenced along with the species’ transcriptomes, e.g. within the Earth BioGenome Project (25). Moreover, for various reasons, a significant fraction of genes may not be covered by transcripts even if the transcriptome data are generated in the project.
Here, we introduce BRAKER2, for which the sequences of known proteins, readily available for any genome project, are used as external evidence. Mapping cross-species proteins to a novel genome presents a challenge, due to the protein divergence and uneven speed of evolution among protein families. Nonetheless, information contained in large numbers of homologous proteins, particularly proteins from remotely related species, has a potential to improve genome annotation. Recently developed GeneMark-EP+ (26) was able to improve its self-training process by hints originating from multiple spliced alignments of cross-species proteins. It was logical to integrate GeneMark-EP+ and AUGUSTUS in a pipeline using already known protein sequences as external evidence.
Several computational tools created earlier addressed the task of identification of a eukaryotic gene structure via spliced alignment of a protein to the genomic locus encoding homologous protein, e.g. (27–32). However, it was observed that the accuracy of the spliced alignments deteriorated quickly with increase of the evolutionary distance between two species. In GeneMark-EP+ this trend was neutralized by search for consensus in spliced alignments of multiple proteins including ones from remotely related species. Particular role was played by evolutionary conserved protein domains that would sustain the accuracy of intron mapping on larger evolutionary distances.
Salient features of BRAKER2 are (i) fully automatic run (ii) massive database search for proteins homologous to proteins encoded in the new genome (yet unknown ones) (iii) processing of millions of protein to genome spliced alignments to generate hints to exon-intron structures, (iv) integration of genomic sequence patterns and protein hints to the gene structure at all iterative steps of model training and gene prediction.
We assessed gene prediction accuracy of BRAKER2 on well-studied and, arguably, well-annotated as a whole, genomes of Arabidopsis thaliana, Caenorhabditis elegans and Drosophila melanogaster. For the tests on nine more genomes we selected sets of genes whose annotation was corroborated by RNA-seq evidence. Besides performance and accuracy of BRAKER2 we determined and compared the ones of MAKER2 (33), run with two distinct execution protocols, as well as of BRAKER1 (18).
MATERIALS AND METHODS
Materials
For testing BRAKER2, we used genomic sequences and gene annotations of 12 species. Among them were the early sequenced model organisms: A. thaliana, C. elegans and D. melanogaster. The other nine species were: the plants Populus trichocarpa, Medicago truncatula, Solanum lycopersicum, the arthropods Bombus terrestris, Rhodnius prolixus, Parasteatoda tepidariorum and the vertebrates Tetraodon nigroviridis, Danio rerio and Xenopus tropicalis (Table 1). We used the OrthoDB database (34) as a source of protein data. RNA-seq data used in runs of BRAKER1 was sampled from the Sequence Read Archive (35) by VARUS (36). To determine to which degree both predicted and annotated genes covered the sets of universal single copy genes identified by the BUSCO protein families, we used the BUSCO database v4 (37).
Table 1.
Genomes used in the tests; asterisks indicate model organisms
| Species | Annotation version | Genome size (Mb) | # Genes in annotation | # Introns per gene | % Non-canonical or incomplete genes |
|---|---|---|---|---|---|
| Species with early sequenced genomes | |||||
| A. thaliana* | Tair Araport 11 (Jun 2016) | 119 | 27 445 | 4.9 | 0.3 |
| C. elegans* | WormBase WS271 (May 2019) | 100 | 20 172 | 5.7 | 0.2 |
| D. melanogaster* | FlyBase R6.18 (Jun 2019) | 138 | 13 929 | 4.3 | 0.3 |
| Other species | |||||
| Plantae | |||||
| P. trichocarpa* | JGI Ptrichocarpa_533_v4.1 (Nov 2019) | 389 | 34 488 | 4.9 | 0.3 |
| M. truncatula* | MtrunA17r5.0-ANR-EGN-r1.6 (Feb 2019) | 430 | 44 464 | 2.9 | 0.0 |
| S. lycopersicum | Consortium ITAG4.0 (May 2019) | 773 | 33 562 | 3.5 | 14.5 |
| Arthropoda | |||||
| B. terrestris | NCBI Annotation Release 102 (Apr 2017) | 249 | 10 581 | 7.1 | 4.7 |
| R. prolixus | VectorBase RproC3.3 (Oct 2017) | 707 | 15 061 | 4.8 | 34.7 |
| P. tepidariorum | NCBI Annotation Release 101 (May 2017) | 1445 | 18 602 | 7.3 | 18.2 |
| Vertebrata | |||||
| T. nigroviridis | TETRAODON8.99 (Nov 2019) | 359 | 19 589 | 10.4 | 63.8 |
| D. rerio* | Ensembl GRCz11.96 (May 2019) | 1345 | 25 254 | 8.2 | 11.8 |
| X. tropicalis* | NCBI Annotation Release 104 (Apr 2019) | 1449 | 21 821 | 12.1 | 2.4 |
An average number of introns per gene was determined with respect to the number of all annotated genes in the genome. For a gene to be considered complete and canonical, at least one of the gene's transcripts had to be annotated with ATG starting the initial coding exon and the terminal coding exon ending with TAA, TAG or TGA.
Methods
Description of BRAKER2, step-by-step
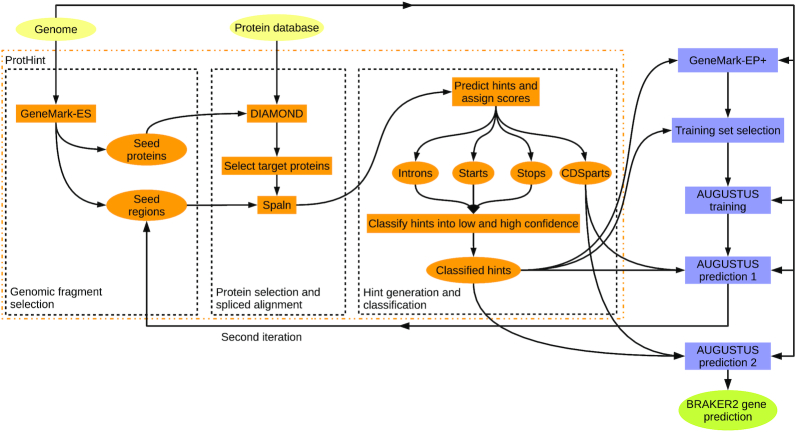
At the first step, the ab initio gene finder GeneMark-ES (1) runs self-training on a eukaryotic genome of interest and generates a set of initial gene predictions, the set of seed genes. This first step is a part of the described earlier protein hint generating pipeline ProtHint (26) that executes GeneMark-ES, DIAMOND (38) and Spaln (31) (Figure 1). The link between translated seed genes (seed proteins) and the genomic loci where the seed genes are residing (seed regions) is important for the subsequent improvement of the initial gene predictions. The seed proteins are used as queries in the DIAMOND similarity search to identify potentially homologous cross-species (target) proteins in a database of reference proteins (26). The selected target proteins are spliced aligned by Spaln back to the seed regions where the queries were encoded. From a set of alignments to the same seed region we infer hints to introns and translation initiation (start codon) and termination site (stop codon) with the scores characterizing hints reliability. The protein hints to the exon borders may coincide with the exon borders predicted ab initio by GeneMark-ES. These sites predicted by the two independent methods, called anchored sites (26) define complete and incomplete anchored gene structures used for iterative model training in GeneMark-EP+ (26). We also define a set of high confidence protein hints (see Supplementary Materials and (26)). At the gene prediction step, the high confidence hints to exon borders are enforced in GeneMark-EP+ (26).
Figure 1.
Flowchart of the BRAKER2 pipeline. Input, intermediate and output data are shown by ovals. The tools and processes of the ProtHint pipeline are shown in orange; other components of BRAKER2 are shown in blue.
In BRAKER2, in addition to the hint generation scheme implemented in GeneMark-EP+, ProtHint makes hints called CDSpart chains. This type of hints helps to combine exons predicted by AUGUSTUS into a single transcript. The CDSpart chain is defined by a spliced alignment of the highest scoring target protein to the seed region.
From the whole complement of genes predicted by GeneMark-EP+, we select a set of anchored genes that contain (i) the multi-exon genes that have all GeneMark-EP+ predicted introns either matching protein hints or enforced by high confidence hints and (ii) the single-exon genes with protein hints matching predicted ab initio start- and stop-codons (Figure 2). These anchored genes make a set for AUGUSTUS training (13). Note that at the stage of gene prediction, AUGUSTUS, in turn, enforces the high confidence hints. The CDSpart chain hints and non-high-confidence hints are integrated into the AUGUSTUS gene prediction as well (Supplementary Materials, Sections 1.2 and 1.3). In the genomic regions lacking hints from cross-species protein alignments, GeneMark-EP+ and AUGUSTUS predict genes in an ab initio mode.
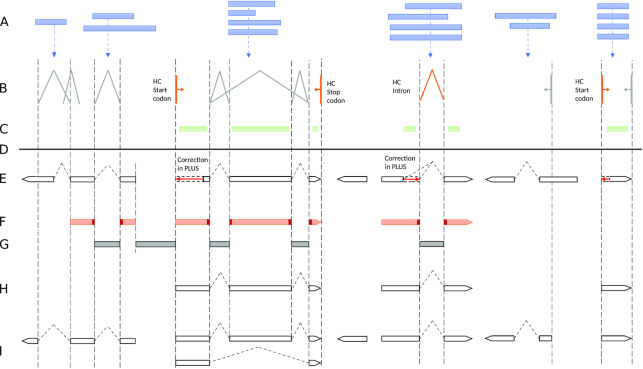
Figure 2.
Evidence integration in BRAKER2. (A) Target proteins; (B) Introns, gene start and stop sites defined by spliced alignments of target proteins to genome; (C) CDSpart chains; (D) Genome sequence; (E) Genes predicted by GeneMark-EP+ at a given iteration. The high confidence hints are enforced (red arrows); (F) Anchored sites, the splice sites and gene ends predicted ab initio and corroborated by protein hints; (G) Anchored introns and intergenic sequences bounded by anchored gene ends are selected into training of non-coding sequence model for GeneMark-EP+; (H) Anchored multi-exon and single exon genes predicted by GeneMark-EP+ and selected for training AUGUSTUS; (I) Transcripts predicted by AUGUSTUS with support of an external evidence.
BRAKER2 runs in two major iterations (Figure 1). The first one starts with the seed genes predicted by GeneMark-ES (1). Seeds for some true genes might be missed at this stage; however, they could be recovered in the second BRAKER2 iteration that uses the genes predicted in the first iteration as seed genes. In the second iteration, ProtHint runs the database search only for the newly added seed genes and merges the newly defined hints with the hints from the first iteration. Then, AUGUSTUS uses the models trained in the first iteration along with the updated protein hints to predict the final set of genes. The second BRAKER2 iteration has fewer steps and runs faster than the first iteration.
Accuracy assessment
Selection of protein data sets and test sets of annotated genes
A test of BRAKER2 on a well-studied genome should utilize a set of cross-species proteins that imitates a protein set available for running BRAKER2 on a newly sequenced genome. Proteins that originate from the most evolutionarily close species are expected to be most informative for the BRAKER2 algorithm. Therefore, a meaningful characteristic of a selected set of reference proteins is the least evolutionary distance from the reference genomes to the genome in the test.
To make these selections for A. thaliana, C. elegans and D. melanogaster, we have started from large clades (Plantae, Metazoa and Arthropoda, respectively) and have created three sets of proteins for each species by excluding either (i) proteins from the given species per se, (ii) proteins from all species of the same family, (iii) proteins from all species of the same order. For the other nine species, we also have defined large clades and then have only used partitions of type (iii) (Table 2).
Table 2.
Composition of the clades of OrthoDB v10 used by BRAKER2
| # of species in the OrthoDB clade | ||||||||
|---|---|---|---|---|---|---|---|---|
| Species | Genus | Family | Order | Class | Phylum | Kingdom | Name of the largest OrthoDB segment | # of proteins in the OrthoDB segment |
| A. thaliana | 2 | 8 | 10 | 100 | 117 | Plantae | 3 510 742 | |
| C. elegans | 3 | 3 | 5 | 6 | 7 | 448 | Metazoa | 8 266 016 |
| D. melanogaster | 20 | 20 | 56 | 148 | 170 | Arthropoda | 2 601 995 | |
| P. trichocarpa* | 1 | 5 | 5 | 100 | 117 | Plantae | 3 510 742 | |
| M. truncatula | 1 | 10 | 10 | 100 | 117 | Plantae | 3 510 742 | |
| S. lycopersicum | 2 | 10 | 11 | 100 | 117 | Plantae | 3 510 742 | |
| B. terrestris* | 1 | 7 | 40 | 148 | 170 | Arthropoda | 2 601 995 | |
| R. prolixus | 1 | 1 | 16 | 148 | 170 | Arthropoda | 2 601 995 | |
| P. tepidariorum | 1 | 1 | 2 | 10 | 170 | Arthropoda | 2 601 995 | |
| T. nigroviridis* | 0 | 1 | 1 | 50 | 246 | Chordata | 5 003 104 | |
| D. rerio | 1 | 5 | 5 | 50 | 246 | Chordata | 5 003 104 | |
| X. tropicalis | 2 | 2 | 3 | 3 | 246 | Chordata | 5 003 104 | |
Numbers in black bold show the largest numbers of species used to support gene predictions for a given species (left column). The numbers of species removed from the largest OrthoDB segment in the tests described below are shown in blue. Species whose proteins are not present in OrthoDB v10 are marked with asterisks.
In the tests done with 12 eukaryotic genomes (Table 1), we used as ‘gold standards’ either whole genome annotation (A. thaliana, C. elegans and D. melanogaster) or annotation of sets of complete multi-exon genes with all introns fully supported by mapped RNA-seq reads.
Protein-coding gene prediction accuracy was defined at exon and gene levels by values of sensitivity (Sn), specificity (Sp) as well as their harmonic mean (F1), with the definitions given in the Supplementary Materials. At the gene level, in presence of alternative splicing, a gene was considered to be predicted correctly if the predicted CDS matches precisely a CDS of one of the annotated transcript isoforms.
Use of universal single copy genes from BUSCO families
The BUSCO metrics is supposed to evaluate the completeness of a genome assembly and annotation; it is based on collections of single copy genes expected to be present in a particular lineage (37). The ‘BUSCO genes’ may constitute <5% of genes in the genome, nonetheless, this approach is practical for novel genomes given its relatively easy application. We used the BUSCO metrics to characterize gene or protein sets predicted by BRAKER2 in several genomes.
While, the BUSCO metrics give an idea on the gene prediction algorithm's Sn value, it does not quantify the algorithm's tendency to predict false positives (the Sp value). Moreover, since the BUSCO based method relies on HMMER3 (39) search for detecting homologs of the BUSCO proteins, it does not discriminate between precisely and approximately predicted exon–intron structures. Therefore, the BUSCO metrics are less precise in assessment of accuracy of gene prediction than the methods comparing coordinates of predicted and annotated genes and computing Sn, Sp and F1 values.
Testing MAKER2
The MAKER2 genome annotation pipeline can combine information from several sources, such as ab initio gene predictions, mapped RNA-seq reads as well as alignments of proteins to the genome (33,40,41).
For our tests, we have chosen genomes of A. thaliana, C. elegans and D. melanogaster, arguably the best annotated genomes among the genomes that we have considered. Also, for each species, we have used the relevant segment of the OrthoDB database described above, with exclusion of species of the same taxonomic order.
All the components of the MAKER2 pipeline, e.g. repeat annotation or training of gene finders, have been executed in de novo mode, i.e. each of the three genomes was considered to be a ‘novel’ one.
By design, the protein mapping in MAKER2 is much slower than protein mapping done by ProtHint in BRAKER2, therefore, we have further limited each of the three OrthoDB partitions to randomly selected ten species (Supplementary Table S3). We have used two MAKER2 execution protocols (described in detail in Supplementary Materials). In the first protocol recommended by the authors (41), the protein spliced alignments have been used to create training sets for AUGUSTUS and SNAP (42). The final gene predictions have been made by combining predictions of self-training GeneMark-ES with ones from AUGUSTUS and SNAP both using the protein derived hints (Supplementary Figure S5A). We have introduced the second training protocol, somewhat similar to the one of BRAKER2, in which protein spliced alignments and GeneMark-ES predictions have been used to create a training set for AUGUSTUS. The final gene predictions have been made by only two gene finders, GeneMark-ES, and AUGUSTUS with hints (Supplementary Figure S5B).
MAKER2 offers two modes of gene prediction: to only get predictions supported by external evidence or to add predictions generated without support. Given that the set of proteins has provided support for a limited number of genes (Supplementary Table S7), we have executed MAKER2 in the second mode, the one producing higher Sn values.
The repeat masking for both BRAKER2 and MAKER2 has been done with the same genome specific repeat library (generated by RepeatModeler). Training and predictions have been done on a repeat-masked sequence. However, BRAKER2 and MAKER2 have different methods for processing repeat-masked sequences (see Supplementary Materials).
Testing BRAKER1
BRAKER1 is a genome annotation pipeline that combines self-training GeneMark-ET with AUGUSTUS (18). External evidence in a form of short RNA-seq reads to genome alignments is used to generate hints to intron borders. BRAKER1 and BRAKER2 use conceptually similar features, such as anchored elements of exon-intron structure.
BRAKER1 has been run on the genomes of A. thaliana, C. elegans and D. melanogaster with hints originating from RNA-seq reads sampled by VARUS (36) from the NCBI Sequence Read Archive (35). VARUS used HISAT2 (43) for mapping RNA-seq reads to genomic sequences (Supplementary Materials section 1.10).
RESULTS
Assessment of gene prediction accuracy of BRAKER2 and comparison with BRAKER1
Genomes of A. thaliana, C. elegans and D. melanogaster
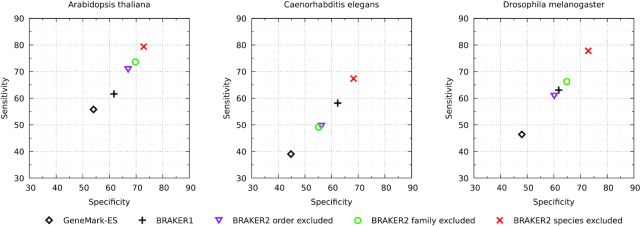
The accuracy of BRAKER2 was determined at exon and gene level (Figures 3 and 4). The exon level Sn and Sp were determined in comparison with annotated exons of all genes of A. thaliana, C. elegans and D. melanogaster, including exons of alternative isoforms, and showed the following patterns (Figure 3). BRAKER1 clearly improved both Sn and Sp values of the ab initio GeneMark-ES. In turn, BRAKER2 performed better than BRAKER1 when BRAKER2 used the largest for each genome set of reference proteins (these largest sets excluded proteins known for the same species). With the smaller protein sets: those excluding proteins from all species of same family or of the same order, the results were mixed. BRAKER2 performed better on A. thaliana, but not on D. melanogaster, and especially not on C. elegans (Figure 3).
Figure 3.
Exon level Sn and Sp determined for each genome in the three runs of BRAKER2 with protein support, the run of BRAKER1 with RNA-seq support and the run of GeneMark-ES. BRAKER2 was run with support of proteins from OrthoDB excluding proteins (i) of the same species, (ii) of all species of the same taxonomic family, (iii) of all species of the same taxonomic order.
Figure 4.
Gene level Sn and Sp determined in the tests described in the legend for Figure 3.
The pattern of accuracy change on exon level was mainly translated into the pattern observed at the gene level (Figure 4 and Supplementary Tables S4–6). In this case, vectors (Sp, Sn) were lined up along bisectors (Figure 4) thus making the ranking of the gene prediction tools unambiguous. Contrary to the exon level, at the gene level, BRAKER2 outperformed BRAKER1 on D. melanogaster when the OrthoDB reference proteins from the phylum Arthropoda excluded the proteins from the same taxonomic family. As expected, the behavior of the F1 values for BRAKER1 and BRAKER2 did correlate with the patterns shown in Figures 3 and 4 (Supplementary Tables S4–6).
Additional set of test genomes
Model organisms A. thaliana, C. elegans and D. melanogaster were subjects of the pilot genome sequencing projects, therefore, we used their longtime curated genome annotations as whole genome test sets.
In conducting tests on genomes of the other nine species (the blue color names in Table 3) we used a different approach motivated by the following example. Upon comparison of the gene predictions made by BRAKER2 in the R. prolixus genome with its current annotation (Table 1) the gene level Sn value appeared to be 13.2% (Table 3). However, the Sn value was 45.5% when it was computed against a set of multi-exon R. prolixus genes with all introns supported by at least one mapped RNA-seq read (a 26.4% subset of all multi-exon genes). In seven out of nine genomes (except for P. trichocarpa and X. tropicalis), large improvements of the gene level Sn values were observed when the base for comparison was changed from a whole set of genes annotated in genomes to a narrower (verified) set. Such an effect was not observed for A. thaliana, C. elegans and D. melanogaster (Table 3).
Table 3.
Gene prediction sensitivity of BRAKER2 at the gene and exon levels
| Gene Sn | Exon Sn | ||||
|---|---|---|---|---|---|
| Species | All | Reliable | All | Reliable | % Reliable genes |
| A. thaliana | 70.2 | 78.8 | 81.5 | 87.9 | 83.5 |
| C. elegans | 49.8 | 57.8 | 75.7 | 81.0 | 81.1 |
| D. melanogaster | 59.5 | 61.6 | 71.9 | 74.4 | 93.2 |
| P. trichocarpa | 69.3 | 76.4 | 86.2 | 90.4 | 84.6 |
| M. truncatula | 48.3 | 63.2 | 82.7 | 90.0 | 69.6 |
| S. lycopersicum | 40.7 | 68.0 | 78.5 | 92.1 | 54.4 |
| B. terrestris | 45.7 | 56.7 | 74.6 | 79.5 | 75.1 |
| R. prolixus | 13.2 | 45.5 | 61.4 | 80.2 | 26.4 |
| P. tepidariorum | 24.6 | 40.2 | 67.9 | 79.9 | 50.6 |
| T. nigroviridis | 10.4 | 67.7 | 60.6 | 89.5 | 11.2 |
| D. rerio | 39.1 | 50.3 | 75.6 | 86.3 | 70.8 |
| X. tropicalis | 38.9 | 46.3 | 75.3 | 80.0 | 74.8 |
The test sets were (All) all annotated multi-exon genes and (Reliable) all annotated complete multi-exon genes having all introns supported by mapped RNA-seq reads, the ones sampled by VARUS (36).
Therefore, we used the test sets of genes supported by the mapped RNA-seq reads. We observed exon Sn values near 80% for the three arthropods, between 80 and 87% for the three vertebrates, and the highest, close to 90%, for the three plants (Table 3).
Among the genes predicted by BRAKER2 in each of the nine genomes, we identified genes encoding proteins from the species-specific BUSCO protein families (44). For a given genome, a percentage of such recognized ‘BUSCO members’ among the full species specific BUSCO set provided an estimate of the sensitivity of gene prediction method (assuming no errors in assembly) and could be compared with the similar figures determined for the reference genome annotation (Supplementary Figure S2).
In the plant and arthropod genomes, BRAKER2 missed ∼3% or less of the BUSCO genes. Moreover, fewer BUSCO genes were missed by BRAKER2 than by the current annotations of genomes of M. truncatula, S. lycopersicum, P. tepidariorum and R. prolixus
The percentage of BUSCO genes missed by BRAKER2 In the vertebrate genomes were: ∼12% in T. nigroviridis, ∼5% in D. rerio, ∼9% in X. tropicalis while the genome annotations missed ∼12, 3 and 3%, respectively.
Prediction accuracy change within the BRAKER2 pipeline
For D. melanogaster, A. thaliana and C. elegans genomes we observed a steady increase of the prediction accuracy upon moving from one to another step of the BRAKER2 pipeline (Supplementary Table S12). For instance, at gene level the F1 value for D. melanogaster increased from GeneMark-ES to GeneMark-EP+ by 17.1 percentage points. Runs of AUGUSTUS with hints added 8.2 percentage points in the first iteration, and 1.1 percentage points in the second iteration.
For the F1 value at the exon level, the numbers of increase were 8.8, 4.6 and 0.4 percentage points, respectively.
Effect of the selection of training genes on gene prediction accuracy
As described in ‘Materials and Methods’ section, for training AUGUSTUS we use fully anchored genes predicted by GeneMark-EP+. The use of these narrower sets improved the gene level F1 values of the ab initio gene prediction by AUGUSTUS in A. thaliana, C. elegans and D. melanogaster genomes by two to five percentage points (Supplementary Table S8). We cite here the accuracy of ab initio gene prediction since the full BRAKER2 could get further improvement from the external protein hints that could overshadow the effects of training.
The use of anchored genes for the AUGUSTUS training had an even stronger effect for the large genomes where the difference in F1 value at exon level for D. rerio reached ∼10 percentage points (Supplementary Table S8).
Effects of the repeat masking on gene prediction accuracy
To identify repetitive sequences (interspersed repeats and low complexity sequences) we used RepeatModeler along with RepeatMasker (www.repeatmasker.org). A run of RepeatModeler on a whole genome produced a repeat library. Next, the locations of repeats were identified and soft-masked by RepeatMasker.
Repeat masking by RepeatModeler and RepeatMasker with default settings was sufficient to achieve high gene prediction accuracy in all the tested genomes except for X. tropicalis. That genome contained a large number of long tandem repeats (∼60 Mb in total) identified by a run of Tandem Repeats Finder (TRF) (45) with maximum repeat period size = 500. The presence of the tandem repeats with elevated GC content (Supplementary Figure S4), when left unmasked, caused GeneMark-ES, running in the initial step of the pipeline, to converge to an incorrect statistical model of a protein-coding region and to make incorrect gene predictions. Particularly, GeneMark-ES would predict a majority of coding exons (93%) in the GC-rich regions of long tandem repeats and would poorly predict the true X. tropicalis genes.
When we applied the non-standard mode of masking by TRF to genomes other than X. tropicalis, no significant change in the BRAKER2 prediction accuracy was observed (data not shown). This could be expected, since the additional repeats found by TRF in genomes other than X. tropicalis were short in size and could be caught by RepeatModeler.
On a general note, RepeatModeler and RepeatMasker may possibly mask over parts of true protein-coding genes, an effect that may decrease gene prediction sensitivity in such regions.
Assessment of accuracy of MAKER2; comparison with BRAKER2
The coordinates of genes predicted by MAKER2 in genomes of A. thaliana, C. elegans and D. melanogaster were compared to the annotations of the three genomes (Table 4). When we used the recommended MAKER2 protocol (Supplementary Figure S5a), the accuracy was significantly lower than the accuracy of BRAKER2 that was run with support of the same reference proteins. Particularly, the exon F1 values were lower for A. thaliana, C. elegans and D. melanogaster by 10.1, 15.7 and 16.1 percentage points, respectively (Table 4). Run of MAKER2 with the second, ‘BRAKER2-like’ protocol (Supplementary Figure S5b) helped to reduce the gap in F1 between MAKER2 and BRAKER2 to 4.5, 8.0 and 7.5 percentage points, respectively (Table 4). An improvement in the Sp values obtained as a result of using the second protocol was likely to be related to the absence of SNAP (42); a separate test did show that SNAP generated an elevated number of false positive predictions (Supplementary Tables S9 and 10).
Table 4.
Prediction accuracy of MAKER2 and BRAKER2
| A. thaliana | C. elegans | D. melanogaster | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MAKER2 with recommended protocol | MAKER2 with BRAKER2-like protocol | BRAKER2 | MAKER2 with recommended protocol | MAKER2 with BRAKER2-like protocol | BRAKER2 | MAKER2 with recommended protocol | MAKER2 with BRAKER2-like protocol | BRAKER2 | |
| Gene Sn | 49.3 | 53.9 | 70.6 | 25.5 | 30.4 | 43.7 | 42.6 | 48.0 | 60.0 |
| Gene Sp | 42.1 | 55.6 | 65.8 | 22.1 | 38.9 | 51.3 | 31.1 | 50.3 | 59.5 |
| Gene F1 | 45.4 | 54.7 | 68.1 | 23.7 | 34.1 | 47.2 | 35.9 | 49.2 | 59.7 |
| Exon Sn | 73.5 | 74.7 | 80.6 | 61.7 | 62.6 | 71.9 | 62.9 | 63.7 | 71.3 |
| Exon Sp | 72.6 | 83.0 | 85.8 | 64.5 | 81.4 | 87.1 | 58.7 | 76.0 | 83.2 |
| Exon F1 | 73.0 | 78.6 | 83.1 | 63.1 | 70.8 | 78.8 | 60.7 | 69.3 | 76.8 |
The runtimes of BRAKER2 and MAKER2 in our experiments were difficult to compare directly. We executed MAKER2 in the MPI mode on a computational cluster with 96 CPUs. The runtime of MAKER2 (∼10 h) using proteins from 10 species was comparable to a time needed for a run of BRAKER2 with proteins from 443 species executed on a single node with 8 CPUs.
DISCUSSION
Genome length and composition
We evaluated accuracy of BRAKER2 on genomes that varied in length from 100 Mbp (C. elegans) to 1.4 Gbp (X. tropicalis). Notably, the exon level Sn computed on test sets of ‘reliable genes’ remained at 80–90% for both shorter and longer genomes (Table 3). However, the gene level Sn, determined on the same test sets, showed noticeable negative correlation with genome length. All the genomes used in this study had relatively homogeneous nucleotide compositions. Current versions of the algorithms used in BRAKER2 employed a single set of species-specific models. Accuracy of BRAKER2 would drop down on genomes with heterogeneous composition, such as human (mammalian) or rice (grasses) where several models reflecting heterogeneous genome composition are necessary.
Role of the evolutionary distances and the total number of species in the reference set
It has been generally assumed that the accuracy of a gene finding algorithm using external protein support would be higher if the evolutionary distance to the closest relative providing reference proteins would be smaller (10,26,46). Indeed, for A. thaliana, C. elegans and D. melanogaster we saw that the accuracy increased step by step when the closest relative in the supporting protein set was outside the order then the family, then the same species (Figure 4). However, another factor improving the accuracy of BRAKER2 is the number of species whose proteins were used for generating protein hints. For instance, the gene prediction accuracy observed for A. thaliana and D. melanogaster that had more species involved in hints generation (Table 2) was higher than the accuracy for C. elegans that had fewer number of species in each instance of the protein reference set (Figure 4, species, family and order excluded).
Demonstration that the increase of the number of species in the protein reference set is a positive factor for the accuracy of predictions was the goal of additional experiments (Supplementary Figure S6). Moreover, we did show that the increase of the total number of species in the protein set of BRAKER2 could compensate the benefit of having closer relatives. For instance, the use of a number of species outside of the D. melanogaster order (Supplementary Figure S7) delivered better accuracy than the use of several Anopheles species within the taxonomic order (Supplementary Table S15).
Of course, it could be that a species with sequenced genome exists at a very close distance, e.g., when the average nucleotide identity (ANI) computed for two genomes is close to 100%. In this case, gene annotation transfer from one genome to another could be the most efficient approach assuming that the reference annotation is of high quality. Otherwise, if the reference annotation is not trusted, use of BRAKER2 would be a reasonable choice; BRAKER2 mechanism of hints generation is insensitive to presence of random errors in the reference protein annotations.
General issues with making gene sets for model training
Ab initio self-training algorithms were shown to deliver high gene annotation accuracy. Still, arguably, training on a sufficiently large set of manually curated gene structures (a supervised training) could outperform, albeit slightly, a self-training algorithm. For instance, AUGUSTUS trained on a randomly selected set of genes annotated in a well-studied genome slightly outperforms AUGUSTUS trained by BRAKER2 in an ab initio mode. Nonetheless, such an idealistic condition (a random sampling from a 100% correct annotation) is an unlikely case when working with a novel genome.
Attempts to create a large enough training set were made by approaches centered around mapping of highly conserved cross-species proteins. Nonetheless, attempts to get an unbiased set of parameters by this approach have not been successful (47). To make one more attempt of this kind, we mapped the BUSCO protein families to genomes of A. thaliana, C. elegans and D. melanogaster to generate training sets of genes. We observed still, that the ‘BUSCO genes’ based models produced lower prediction accuracy than BRAKER2 (Supplementary Table S11).
The new approach used in BRAKER2 is using cross-species protein conservation to predict introns and ab initio gene prediction to connect introns and exons into gene structures. This new method has led to a significant increase in the size of the gene set supported by protein evidence. For almost all the genomes selected for our tests, more than four thousand gene structures were selected into training sets.
Improvement in the method for generation of external evidence
BRAKER2 is using a new approach for creating protein hints. The protein mapping pipeline, ProtHint, makes sets of hints with higher and lower confidence. All hints contribute to generation of anchored genes used in training. GeneMark-EP+ is enforcing high confidence hints in the prediction step. In turn, AUGUSTUS utilizes low and high confidence hints at the prediction step along with information about the hints’ connections within a putative transcript (CDSpart chain). The flexible use of hints leads to an increase in accuracy of BRAKER2 (Supplementary Table S13). Particularly, BRAKER2 appears to be a useful tool for annotation of genomes of deep branching species, since BRAKER2 is tuned up to generate accurate hints upon the use of proteins from remotely related species.
BRAKER2 iterations
The number of hints generated by the ProtHint pipeline depends on the number of genes predicted by GeneMark-ES. A solid performance of GeneMark-ES was demonstrated (8,26), but any ab initio gene finder may miss genes. Missed genes would translate in BRAKER2 into missed protein hints to the corresponding genomic loci. The second iteration of BRAKER2 recovers hundreds of missed genes and leads to an increase of gene prediction accuracy (Supplementary Table S12). Still, the effect of the second iteration on the overall accuracy of BRAKER2 is relatively small. Another albeit computationally expensive way to recover some missed genes is to align proteins from a protein database to the 6-frame translations of the genomic DNA (33). Use of this approach did not produce a better Sn value than the iterative procedure of BRAKER2 (data not shown).
The ‘BUSCO genes’ in BRAKER2 predictions
As a part of the accuracy assessment we selected a set of the BRAKER2 predicted genes identified by the search with the BUSCO tools (37) as ones that belong to the species-specific BUSCO family (Supplementary Figure S2). The completeness of such set was determined as percentage of the whole BUSCO set. In all cases, but X. tropicalis, the selected sets of genes were comparable to, or, even more complete than the sets of ‘BUSCO genes’ identified in the reference genome annotations. We should note that some BUSCO families included species within the same taxonomic order as the species of interest (e.g. Hemiptera order of R. prolixus or Solanales order for S. lycopersicum). On the other hand, the input to BRAKER2 were proteins of the species outside of the corresponding taxonomic order.
A lower level of accuracy of BRAKER2 for X. tropicalis could be related to the insufficient number of external proteins. Removing the Anura taxonomic order from the OrthoDB partition left no proteins from the Amphibia taxonomic class among input proteins (Table 2). Also, a reason for missing some ‘BUSCO genes’ could be inaccurate de novo repeat masking. For example, among annotated genes missed by BRAKER2 in the P. trichocarpa genome, more than half were genes partially masked by long repeats (>1000 nt).
Comparison with MAKER2
Differences in accuracy of MAKER2 and BRAKER2 observed in our experiments were quite large despite the attempts to find a way to improve the MAKER2 protocol (Table 4). The differences in the outcomes could be caused by the differences in the methods of data preparation, processing repeats, ways of generating and selecting external evidence, connecting main elements of the pipelines as well as combining the gene predictions into the final annotation. Therefore, we presented detailed descriptions of the protocols used for running MAKER2 (Supplementary Materials).
MAKER2 uses the ab initio self-training algorithm GeneMark-ES, while BRAKER2 uses the more recent self-training GeneMark-EP+ algorithm that integrates protein hints into training and prediction (26). Comparison of protein hints is difficult since BRAKER2 uses hints to splice sites and start/stop codons while MAKER2 uses hints to parts of exons. More efficient training of AUGUSTUS is one of the important factors for elevated accuracy of BRAKER2. In our experiments, an effective way to improve MAKER2 accuracy was a reduction of the number of gene finders from three to two (Supplementary Figure S5, Table 4 and Supplementary Tables S9-10). The difference in accuracy of BRAKER2 and MAKER2 is likely to be even larger for eukaryotic genomes with longer length, however, such a comparison is harder to make due to less accurate reference annotations. Since a comprehensive comparison of the two methods is not a goal of this paper, the comparisons are limited to the three well studied genomes.
Last but not least, the training of gene finders is not fully automated in MAKER2. Users have to execute the training steps manually, even though recommendations are given on the training protocols. On the other hand, BRAKER2 can be executed from start to finish by a single command.
Comparison with BRAKER1
As we have demonstrated, the accuracy of BRAKER2 depends on the number of reference proteins and on distribution of evolutionary distance to the reference species. The accuracy of BRAKER1 depends on the volume of the RNA-seq data. Experiments with BRAKER1 on genomes of A. thaliana, C. elegans and D. melanogaster used RNA-seq reads from SRA retrieved by VARUS, e.g. the non-redundant volumes of RNA-Seq reads from the maximum number of libraries available for each species. When we used the largest number of the proteins for each species (the species-specific OrthoDB partition excluding proteins of the same species) we observed the accuracy of BRAKER2 comparable or better than the one of BRAKER1.
In genomes of A. thaliana, C. elegans and D. melanogaster, both BRAKER1 and BRAKER2 made correct predictions of a rather low number of annotated alternative isoforms (Supplementary Table S14). This is a result of a deliberate parameter setting in AUGUSTUS to reduce the number of false positives. Particularly, AUGUSTUS ignored an RNA-seq or a protein hint contradicting another hint with 10 times larger support. On the other hand, the reference genome annotations of the three species are rather inclusive in a sense of presenting isoforms that have low support (potentially lowly expressed ones).
EuGene and other gene finders
A gene finder for eukaryotic genomes, EuGene, provides a mechanism for integration of several sources of information into the gene prediction process (48). This modular tool can integrate data derived from protein spliced alignment to genomic DNA. Unfortunately, EuGene does not provide recommendations on model training for the case when protein sequences are the only source of the external evidence; therefore, we could not immediately use this tool in the comparative study.
Several tools attempt accurate identification of gene structures in a novel genome by mapping homologous proteins (e.g. GenomeThreader (28), Scipio (30)). This approach limits the gene discovery to genes of homologs present in the input protein set; the accuracy of this method drops significantly with increase of evolutionary distance between the two species (10,46). Another significant challenge for a number of earlier developed tools is the processing of large volumes of proteins. This challenge is much smaller if a tool, like GeMoMa (49,50), is oriented on getting the protein information from closely related species. In addition, GeMoMa requires gene coordinates in reference genomes. We did not make comparisons of BRAKER2 to the above mentioned tools since these tools were not designed for the situation when the reference protein data does not contain proteins from closely related species.
CONCLUSION
BRAKER2, a fully automated pipeline for gene prediction in novel eukaryotic genomes allows to produce hints to gene structures from protein databases. BRAKER2 runs processing of millions of proteins in the course of several hours (for instance, in case of D. melanogaster, ∼2.6 millions of proteins were processed in ∼3 h on a single node with eight CPU). In the tests on genomes of plants, and animals, we observed that BRAKER2 delivered state-of-the-art annotation accuracy and was favorably compared to already existing tools.
DATA AVAILABILITY
BRAKER2 is available at https://github.com/Gaius-Augustus/BRAKER. All additional scripts and data used to generate figures and tables in this manuscript are available at https://github.com/gatech-genemark/BRAKER2-exp.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stephane Rombauts for useful comments.
Contributor Information
Tomáš Brůna, School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA 30332, USA.
Katharina J Hoff, Institute of Mathematics and Computer Science, University of Greifswald, 17489 Greifswald, Germany; Center for Functional Genomics of Microbes, University of Greifswald, 17489 Greifswald, Germany.
Alexandre Lomsadze, Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology, Atlanta, GA 30332, USA.
Mario Stanke, Institute of Mathematics and Computer Science, University of Greifswald, 17489 Greifswald, Germany; Center for Functional Genomics of Microbes, University of Greifswald, 17489 Greifswald, Germany.
Mark Borodovsky, Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology, Atlanta, GA 30332, USA; School of Computational Science and Engineering, Georgia Institute of Technology, Atlanta, GA 30332, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
National Institutes of Health (NIH) [GM128145 to M.B., M.S., in part]. Funding for open access charge: NIH [GM128145].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lomsadze A., Ter-Hovhannisyan V., Chernoff Y.O., Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005; 33:6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shulaev V., Sargent D.J., Crowhurst R.N., Mockler T.C., Folkerts O., Delcher A.L., Jaiswal P., Mockaitis K., Liston A., Mane S.P. et al. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 2011; 43:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhan S., Merlin C., Boore J.L., Reppert S.M. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011; 147:1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng H., Zhang W., Zhang L., Zhang Z., Li J., Lu G., Zhu Y., Wang Y., Huang Y., Liu J. et al. The genome of the hydatid tapeworm Echinococcus granulosus. Nat. Genet. 2013; 45:1168–1175. [DOI] [PubMed] [Google Scholar]
- 5. Suga H., Chen Z., de Mendoza A., Sebe-Pedros A., Brown M.W., Kramer E., Carr M., Kerner P., Vervoort M., Sanchez-Pons N. et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat. Commun. 2013; 4:2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chu H., Qian Q., Liang W., Yin C., Tan H., Yao X., Yuan Z., Yang J., Huang H., Luo D. et al. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 2006; 142:1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woycicki R., Witkowicz J., Gawronski P., Dabrowska J., Lomsadze A., Pawelkowicz M., Siedlecka E., Yagi K., Plader W., Seroczynska A. et al. The genome sequence of the North-European cucumber (Cucumis sativus L.) unravels evolutionary adaptation mechanisms in plants. PLoS One. 2011; 6:e22728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lomsadze A., Burns P.D., Borodovsky M. Integration of mapped RNA-seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 2014; 42:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoff K.J., Stanke M. WebAUGUSTUS–a web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res. 2013; 41:W123–W128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Konig S., Romoth L.W., Gerischer L., Stanke M. Simultaneous gene finding in multiple genomes. Bioinformatics. 2016; 32:3388–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stanke M., Diekhans M., Baertsch R., Haussler D Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008; 24:637–644. [DOI] [PubMed] [Google Scholar]
- 12. Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006; 34:W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stanke M., Schoffmann O., Morgenstern B., Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006; 7:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stanke M., Steinkamp R., Waack S., Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004; 32:W309–W312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guigo R., Flicek P., Abril J.F., Reymond A., Lagarde J., Denoeud F., Antonarakis S., Ashburner M., Bajic V.B., Birney E. EGASP: the human ENCODE genome annotation assessment project. Genome Biol. 2006; 7(Suppl. 1):S21–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coghlan A., Fiedler T.J., McKay S.J., Flicek P., Harris T.W., Blasiar D. nGASP Consortium nGASP Consortium Stein L.D. nGASP–the nematode genome annotation assessment project. BMC Bioinformatics. 2008; 9:549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Steijger T., Abril J.F., Engstrom P.G., Kokocinski F., Consortium R., Hubbard T.J., Guigo R., Harrow J., Bertone P. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods. 2013; 10:1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoff K.J., Lange S., Lomsadze A., Borodovsky M., Stanke M. BRAKER1: unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 2016; 32:767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bickhart D.M., Rosen B.D., Koren S., Sayre B.L., Hastie A.R., Chan S., Lee J., Lam E.T., Liachko I., Sullivan S.T. et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 2017; 49:643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoshida Y., Koutsovoulos G., Laetsch D.R., Stevens L., Kumar S., Horikawa D.D., Ishino K., Komine S., Kunieda T., Tomita M. et al. Comparative genomics of the tardigrades Hypsibius dujardini and Ramazzottius varieornatus. PLoS Biol. 2017; 15:e2002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bowman J.L., Kohchi T., Yamato K.T., Jenkins J., Shu S., Ishizaki K., Yamaoka S., Nishihama R., Nakamura Y., Berger F. et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017; 171:287–304. [DOI] [PubMed] [Google Scholar]
- 22. Munoz J.F., Gade L., Chow N.A., Loparev V.N., Juieng P., Berkow E.L., Farrer R.A., Litvintseva A.P., Cuomo C.A. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 2018; 9:5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Bekker C., Ohm R.A., Evans H.C., Brachmann A., Hughes D.P. Ant-infecting Ophiocordyceps genomes reveal a high diversity of potential behavioral manipulation genes and a possible major role for enterotoxins. Sci. Rep. 2017; 7:12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Costa M.D., Artur M.A., Maia J., Jonkheer E., Derks M.F., Nijveen H., Williams B., Mundree S.G., Jimenez-Gomez J.M., Hesselink T. et al. A footprint of desiccation tolerance in the genome of Xerophyta viscosa. Nat. Plants. 2017; 3:17038. [DOI] [PubMed] [Google Scholar]
- 25. Lewin H.A., Robinson G.E., Kress W.J., Baker W.J., Coddington J., Crandall K.A., Durbin R., Edwards S.V., Forest F., Gilbert M.T.P. et al. Earth BioGenome Project: sequencing life for the future of life. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:4325–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bruna T., Lomsadze A., Borodovsky M. GeneMark-EP+: eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genome Bioinform. 2020; 2:lqaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gelfand M.S., Mironov A.A., Pevzner P.A. Gene recognition via spliced sequence alignment. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:9061–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gremme G., Brendel V., Sparks M.E., Kurtz S. Engineering a software tool for gene structure prediction in higher organisms. Inf. Softw. Technol. 2005; 47:965–978. [Google Scholar]
- 29. Sayers E.W., Barrett T., Benson D.A., Bolton E., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Federhen S. et al. Database resources of the National Centerfor Biotechnology Information. Nucleic Acids Res. 2011; 39:D38–D51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Keller O., Odronitz F., Stanke M., Kollmar M., Waack S. Scipio: using protein sequences to determine the precise exon/intron structures of genes and their orthologs in closely related species. BMC Bioinformatics. 2008; 9:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gotoh O. Direct mapping and alignment of protein sequences onto genomic sequence. Bioinformatics. 2008; 24:2438–2444. [DOI] [PubMed] [Google Scholar]
- 32. Rogozin I.B., Milanesi L., Kolchanov N.A. Gene structure prediction using information on homologous protein sequence. Comput. Appl. Biosci. 1996; 12:161–170. [DOI] [PubMed] [Google Scholar]
- 33. Holt C., Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011; 12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kriventseva E.V., Kuznetsov D., Tegenfeldt F., Manni M., Dias R., Simao F.A., Zdobnov E.M. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019; 47:D807–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leinonen R., Sugawara H., Shumway M., International Nucleotide Sequence Database C. The sequence read archive. Nucleic Acids Res. 2011; 39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stanke M., Bruhn W., Becker F., Hoff K.J. VARUS: sampling complementary RNA reads from the sequence read archive. BMC Bioinformatics. 2019; 20:558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seppey M., Manni M., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 2019; 1962:227–245. [DOI] [PubMed] [Google Scholar]
- 38. Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015; 12:59–60. [DOI] [PubMed] [Google Scholar]
- 39. Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cantarel B.L., Korf I., Robb S.M., Parra G., Ross E., Moore B., Holt C., Sanchez Alvarado A., Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008; 18:188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Campbell M.S., Holt C., Moore B., Yandell M. Genome annotation and curation using MAKER and MAKER-P. Curr. Protoc. Bioinform. 2014; 48:4.11.1–4.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004; 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019; 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Waterhouse R.M., Seppey M., Simao F.A., Manni M., Ioannidis P., Klioutchnikov G., Kriventseva E.V., Zdobnov E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018; 35:543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999; 27:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Birney E., Clamp M., Durbin R. GeneWise and Genomewise. Genome Res. 2004; 14:988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parra G., Bradnam K., Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007; 23:1061–1067. [DOI] [PubMed] [Google Scholar]
- 48. Sallet E., Gouzy J., Schiex T. EuGene: an automated integrative gene finder for eukaryotes and prokaryotes. Methods Mol. Biol. 2019; 1962:97–120. [DOI] [PubMed] [Google Scholar]
- 49. Keilwagen J., Wenk M., Erickson J.L., Schattat M.H., Grau J., Hartung F. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016; 44:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Keilwagen J., Hartung F., Paulini M., Twardziok S.O., Grau J. Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinformatics. 2018; 19:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BRAKER2 is available at https://github.com/Gaius-Augustus/BRAKER. All additional scripts and data used to generate figures and tables in this manuscript are available at https://github.com/gatech-genemark/BRAKER2-exp.