SUMMARY
Understanding the effects of microgravity on human organs is crucial to exploration of low-earth orbit, the moon, and beyond. Drosophila can be sent to space in large numbers to examine the effects of microgravity on heart structure and function, which is fundamentally conserved from flies to humans. Flies reared in microgravity exhibit cardiac constriction with myofibrillar remodeling and diminished output. RNA sequencing (RNA-seq) in isolated hearts revealed reduced expression of sarcomeric/extracellular matrix (ECM) genes and dramatically increased proteasomal gene expression, consistent with the observed compromised, smaller hearts and suggesting abnormal proteostasis. This was examined further on a second flight in which we found dramatically elevated proteasome aggregates co-localizing with increased amyloid and polyQ deposits. Remarkably, in long-QT causing sei/hERG mutants, proteasomal gene expression at 1g, although less than the wild-type expression, was nevertheless increased in microgravity. Therefore, cardiac remodeling and proteostatic stress may be a fundamental response of heart muscle to microgravity.
Graphical Abstract
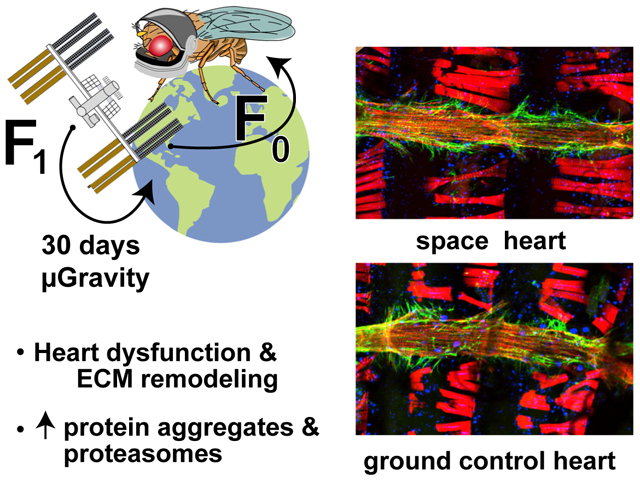
In Brief
Walls et al. find that exposure to microgravity aboard the ISS causes heart dysfunction in a fly cardiac model. Hearts are less contractile and exhibit changes in genes and proteins that maintain heart structure and function. Effects are seen in several lines of flies, suggesting a common response to microgravity.
INTRODUCTION
Throughout history, humans have explored the earth, so much so, that few regions outside of the ocean depths still remain unexplored. This drive to go “where no man has gone before,” coupled with increasing pollution, climate change, and competition for scarcer resources, has brought humans to the point at which exploration and exploitation of other celestial bodies, such as the moon, asteroids, and other planets, is inevitable. However, our organ systems have evolved to work under 1g, and it is not clear what the effect of long-term exposure to lower gravity environments might be and how those effects are exerted at the cellular and molecular levels.
Cardiac health has been a major concern of the National Aeronautics and Space Administration (NASA) for many years, but the clinical evidence for decreased function and unmasking of asymptomatic cardiovascular disease is limited. Early studies performed on astronauts who had been in microgravity for only a few weeks to a few months failed to uncover any cardiac effects (Hamilton et al., 2011), and some studies suggested that changes in function could be explained by dehydration (Summers et al., 2005). However, the number of subjects examined in those studies was low, with sometimes conflicting results, and they were without discernable (cardiac) health risks, so it is difficult to draw conclusions. Experiments designed to mimic longer-term exposure to microgravity (bed rest in humans and hind limb unloading in mice) showed reductions in cardiac contractility (Platts et al., 2009) and increases in arrhythmia (Respress et al., 2014). Increases in ventricular contraction (QT) intervals have also been reported in astronauts after longer-duration flights (D’Aunno et al., 2003), suggesting reductions in the cardiac repolarization reserve (Akoum et al., 2011), but those conclusions are again based on few subjects with differing genetic backgrounds.
Dysfunction in several cardiac ion channels are known to cause long QT syndrome (LQTS) in humans, which can lead to ventricular arrhythmias and sudden cardiac arrest. The human ether-a-go-go-related gene, also known to code for the hERG channel, is one of the ion channels that contribute to LQTS. hERG channel dysfunction can be caused by alterations in the genes encoding this channel as well as by some commonly used medications (Sanguinetti and Tristani-Firouzi, 2006).
To systematically explore the effects of spaceflight on the heart, we used the Drosophila cardiac model to examine how prolonged exposure to microgravity affects cardiac health. We report the effects of microgravity on heart function in wild-type (WT) fruit flies (Drosophila Canton-S [CS]) that were born, developed, and spent 1–3 weeks as adults aboard the International Space Station (ISS) compared with their ground-based controls (Figure 1A). We also examined the effects of microgravity on flies with mutations in seizure (sei), the fly homolog of the human hERG gene. Our results show that structural and functional cardiac remodeling occurs in response to microgravity and that remodeling is more severe in the channel mutants. Cardiac RNA sequencing (RNA-seq) of WT and sei/hERG mutant flies and follow-up immunohistochemistry studies suggest that alterations in proteostasis likely contribute to the observed changes in cardiac function in microgravity.
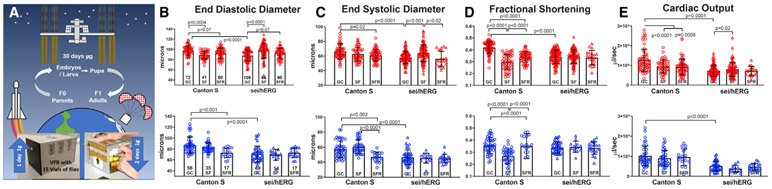
Figure 1. Heart Size and Contractility Are Reduced under Microgravity.
(A) Drosophila flies were reared aboard the ISS in a customized Nanoracks vented fly box (VFB) containing 15 vials of flies and food (bottom, left). Each vial contained 10 virgin females and 5 males when launched. Vials remained in the VFB for the duration of the microgravity exposure.
(B) End-diastolic diameters (EDDs) in hearts from females (red, top) and males (blue, bottom) were significantly decreased in female, but not male, space flown (SF) Canton-S flies compared with ground controls (GCs). EDDs recovered in hearts from female flies born in space but cultured as adults in 1g (space flight recovery [SFR]). EDDs in all sei/hERG mutant flies were reduced relative to Canton-S flies; genetic background controls were not significantly affected by microgravity.
(C) End systolic diameters (ESDs) were also reduced in sei/hERG/hERG mutant flies under both gravity conditions relative to Canton-S flies.
(D and E) Fractional shortening (D), a measure of contractility, and cardiac output (E) were significantly reduced in hearts from both male and female Canton-S flies under microgravity and showed partial recovery in flies born in space and reared as adults in 1g (SFR). Heart function parameters were analyzed using a two-way ANOVA with Tukey’s multiple comparison post hoc test. p values are shown. Fly numbers are indicated in the bars in (B).
RESULTS
Lifelong Microgravity Exposure Caused Cardiac Dysfunction and Remodeling
We used vented fly boxes (VFBs) to carry multiple vials of adult Drosophila, each with 10 females and 5 males, to the ISS (see protocol schematic; Figure 1A). Eggs laid on the food in the bottom of the vials hatched into larva that then metamorphosed into adults in about 10–12 days, all under microgravity. VFBs and vials containing live adult flies were returned to earth gravity, and flies were processed within ~2 days of exposure to 1g (space-flown [SF] flies). We also collected flies that emerged from those same vials at 1g (days 3–5 after splash down) and aged them for 2 weeks at 1g before processing (space flight recovery [SFR]).
For cardiac function assays, flies were dissected under artificial hemolymph and allowed to equilibrate for 15–30 min in fresh, aerated, artificial hemolymph. The contracting hearts were filmed with high-speed digital cameras (140–160 frames/s [fps]) for subsequent heart-function analysis (Fink et al., 2009; Ocorr et al., 2007). Measurements of heart diameters were made at the widest portion of the second abdominal chamber during systole and diastole. Exposure to microgravity resulted in significant reductions in end-diastolic diameters (EDDs) of SF CS female fly hearts compared with hearts from ground controls (GCs) (Figure 1B, top left). This reduction was not the result of changes in the overall size of female flies because there was no significant difference in body length in response to microgravity (Figure S1A). There was a trend toward reduced diastolic diameters in male flies, which did not reach statistical significance, as well as a small reduction in the overall body length of SF CS males (Figures 1B, bottom left, and S1A). No significant effects of microgravity were observed on end-systolic diameters (ESDs) under microgravity in either sex (Figure 1C). Importantly, both sexes showed significant decreases in fractional shortening ([EDD – ESD]/[EDD]), which is an indirect measure of cardiac contractility (Figure 1D). The functional consequence of reduced diameters in females was a highly significant reduction in cardiac output (Figure 1E).
As previously observed (Ocorr et al., 2017), cardiac size and output in sei/hERG mutant hearts were already significantly reduced at 1g compared with CS hearts (Figures 1B and 1C). Interestingly, there was a small but significant increase in diastolic diameters in response to microgravity in hearts from female sei/hERG mutants, which translated into a modest increase in the cardiac output (Figure 1C, top).
The length of time the heart spent in diastole and systole was also quantified from high-speed movies. There were no significant differences between 1g- and microgravity-exposed flies (Figures S1B and S1C). However, hearts from female SFR flies did show a significant increase in the systolic intervals, perhaps a response to increased load when returned to 1g. Male CS hearts showed significantly increased cardiac arrhythmia (Figure S1D, bottom; arrhythmia index [AI], a measure of beat-to-beat variability) (Ocorr et al., 2007), which was not seen in SFR flies. The AI trended higher in females but did not reach the level of significance (Figure S1D, top). We observed the expected bradycardia and increased arrhythmia in sei/hERG mutant flies compared with CS controls at 1g, but it was not significantly worsened in microgravity (Figures S1B-S1D). Representative high-speed videos of hearts from both lines reared under 1g and microgravity are provided (Video S1).
Transcriptome Analysis Reveals Differential Expression in Extracellular Matrix (ECM) and Proteasome Regulatory Genes under Microgravity
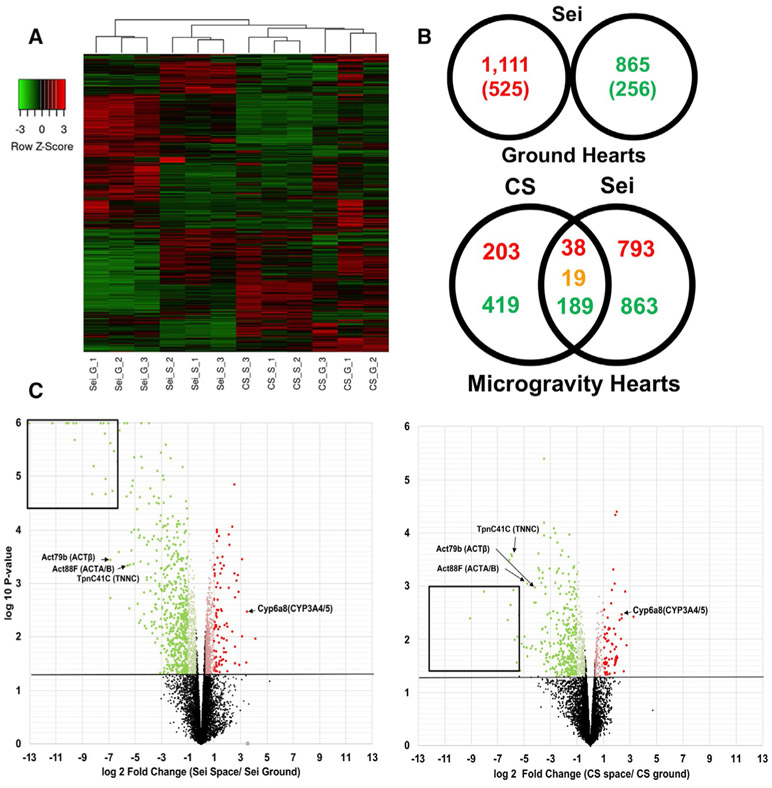
After filming for cardiac functional analyses, hearts were removed and processed for transcriptomic analysis. Hierarchical-cluster analyses from triplicate samples from CS 1g (CS_G), CS microgravity-exposed (CS_S), sei/hERG 1g (sei_G), and sei/hERG-microgravity-exposed (sei_S) flies were performed. The results showed that triplicates from each genotype and condition clustered within their respective group. Genotypes clustered at shorter distances then those with microgravity exposure.
Hearts from WT CS flies reared in microgravity compared with 1g GCs showed upregulation of 250 genes (1.25-fold; adjusted [adj.] p value ≤ 0.05); of which, 86 genes were increased ≥2.0-fold (adj. p value ≤ 0.01) (Figure 2B). Ontological analysis of the 250 upregulated genes using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) functional annotation bioinformatics microarray analysis showed an ~20-fold enrichment in proteasome-mediated ubiquitin (Ub)-dependent protein catabolism genes, a 23-fold enrichment in chromatin assembly and disassembly genes, and a 12-fold enrichment in genes with endopeptidase activity, among other ontologies (Table 1). Conversely, 618 genes were downregulated (1.25-fold; adj. p value ≤ 0.05); of which, 313 genes were decreased ≥2.0-fold (adj. p value ≤ 0.01) (Figure 2B). Here, ontological analysis identified 2-fold or greater enrichment in gene subsets enriched for proteolysis, carbohydrate metabolic processes, and the ECM (Table 1).
Figure 2. RNA-Seq Analysis of Differential Transcriptome Expression in Drosophila Cardia under Microgravity.
(A) Average linkage cluster analysis using Kendall’s Tau distance measurement method comparing Canton-S GCs (CS_G), Canton-S microgravity exposed (CS_S), sei/hERG GCs (SEI_G), and sei/hERG microgravity exposed (SEI_S) fly hearts. Column dendrograms showed samples clustered by both genetic background and (micro)gravity exposure. Genes (rows) normalized using row Z score. p < 0.05, 1.2-fold (p < 0.01, 2.0-fold in parenthesis)
(B) Differentially expressed genes in sei/hERG mutants at 1g (top). Venn Diagram of up (red) and down (green) regulated genes in CS and sei/hERG under microgravity normalized to their respective GCs (bottom). Overlap denotes shared up- and downregulated genes. Orange denotes contra-regulated genes.
(C) Volcano plots of CS (left) and sei/hERG (right) under microgravity. Black box denotes highly downregulated chitin-related fly-specific clusters. Red denotes upregulated 2.0+-fold; light red, 1.25- to 1.99-fold; green, downregulated 2.0+-fold; light green, 1.25- to 1.99-fold. Triplicate samples of n = 20 fly hearts.
Table 1.
Differentially Expressed Gene Subsets in CS Hearts under Microgravity versus CS at 1g
| Term | Dir. | Description | Count | Fold | FDR |
|---|---|---|---|---|---|
| BP_GO: 0043161 | up | proteasome-mediated Ub-dependent catabolism | 37 | 4.4 | <0.001 |
| BP_GO: 0030497 | up | fatty acid elongation | 9 | 1.9 | 0.031 |
| BP_GO: 0006974 | up | cellular response to DNA damage stimulus | 19 | 2.2 | 0.048 |
| MF_GO: 0004298 | up | threonine-type endopeptidase activity | 12 | 1.4 | <0.001 |
| MF_GO: 0004175 | up | endopeptidase activity | 12 | 1.43 | 0.042 |
| CC_GO: 0000502 | up | proteosome complex | 29 | 3.5 | <0.001 |
| CC_GO: 0005634 | up | nucleus | 159 | 18.9 | 0.002 |
| KEGG: dme03050 | up | proteasome | 28 | 3.3 | <0.001 |
| BP_GO: 0006598 | dwn | proteolysis | 41 | 2.3 | 0.001 |
| BP_GO: 0005975 | dwn | carbohydrate metabolic process | 15 | 3.8 | 0.05 |
| CC_GO: 0005616 | dwn | extracellular space | 48 | 2.2 | <0.001 |
| CC_GO: 0005887 | dwn | integral component of plasma membrane | 38 | 2.1 | 0.03 |
Table uses gene ontology (GO) terms. Dir., direction; up, upregulated; dwn, downregulated; BP, biological process; MF, molecular function; CC, cellular localization; KEGG, Kyoto Encyclopedia of Genes and Genomes pathway identification; FDR, false discovery rate.
Transcriptomic analysis of hearts from GC sei/hERG mutants compared with GC CS flies (Figures 2A and 2B) showed upregulation of 1,111 genes (1.25-fold; adj. p value ≤ 0.05), with 525 genes showing increased expression ≥2.0-fold (adj. p value ≤ 0.01). Ontological analysis of the 1,111 upregulated genes showed a 5.8-fold enrichment in chromatin assembly genes, 2-fold enrichment in proteolysis genes, and 2.9-fold increase in carbohydrate metabolic process genes, among other ontologies (Table 2). Conversely, 865 genes were downregulated (1.25-fold; adj. p value ≤ 0.05), with 256 genes ≥2.0-fold (adj. p value ≤ 0.01) (Figures 2A and 2B). Here, ontological analysis revealed a 4.4-fold enrichment in proteasome-mediated Ub-dependent catabolism genes, 1.9-fold enrichment in fatty-acid-elongation genes, and a 2.2-fold enrichment in cellular response to DNA damage genes, among other ontologies. Importantly, expression of many ECM-related genes in hearts from sei/hERG GC flies was already significantly reduced compared with WT GCs (Table 2).
Table 2.
Differential Gene Subset Expression in SEI Mutants at 1g Relative to CS at 1g
| Term | Dir. | Description | Count | Fold | FDR |
|---|---|---|---|---|---|
| BP_GO:0006333 | up | chromatin assembly or disassembly | 23 | 5.8 | <0.001 |
| BP_GO:0006508 | up | proteolysis | 74 | 2 | <0.001 |
| BP_GO:0055114 | up | oxidation-reduction process | 62 | 2 | <0.001 |
| BP_GO:0045087 | up | innate immune response | 25 | 3.2 | 0.001 |
| BP_GO:0006120 | up | mitochondrial electron transport | 13 | 4.6 | 0.017 |
| BP_GO:0005975 | up | carbohydrate metabolic process | 23 | 2.9 | 0.019 |
| MF_GO:0046982 | up | protein heterodimerization activity | 48 | 2.5 | <0.001 |
| MF_GO:0003954 | up | NADH dehydrogenase activity | 13 | 4.9 | 0.007 |
| CC_GO:0005811 | up | lipid particle | 87 | 4 | <0.001 |
| CC_GO:0000788 | up | nuclear nucleosome | 41 | 7.9 | <0.001 |
| CC_GO:0005739 | up | mitochondrion | 66 | 1.8 | 0.0065 |
| BP_GO:0043161 | dwn | proteasome-mediated Ub-dependent catabolism | 37 | 4.4 | <0.001 |
| BP_GO:0030497 | dwn | fatty acid elongation | 9 | 1.9 | 0.031 |
| BP_GO:0006974 | dwn | cellular response to DNA damage stimulus | 19 | 2.2 | 0.048 |
| MF_GO:0004298 | dwn | threonine-type endopeptidase activity | 12 | 1.4 | <0.001 |
| MF_GO:0004175 | dwn | endopeptidase activity | 12 | 1.4 | 0.042 |
| CC_GO:0000502 | dwn | proteosome complex | 29 | 3.5 | <0.001 |
| CC_GO:0005634 | dwn | nucleus | 159 | 19 | 0.002 |
| KEGG:dme03050 | dwn | proteasome | 28 | 3.3 | <0.001 |
Table uses gene ontology (GO) terms. Dir., direction; up, upregulated; dwn, downregulated; BP, biological process; MF, molecular function; CC, cellular localization; KEGG, Kyoto Encyclopedia of Genes and Genomes pathway identification; FDR, false discovery rate.
Transcriptomic analysis of hearts from sei/hERG mutants showed exposure to microgravity resulted in the upregulation of 840 genes (1.25-fold; adj. p value ≤ 0.05), with 96 genes increased ≥2.0-fold (adj. p value ≤ 0.01) (Figure 2B). Ontological analysis of the 840 upregulated genes showed a 5.4-fold enrichment in rRNA processing genes and a 4.8-fold enrichment in histone acetyltransferase genes, among other ontologies (Table 3). Conversely, 1,063 genes were downregulated (1.25-fold; adj. p value ≤ 0.05), with 582 genes decreased ≥2.0-fold (adj. p value ≤ 0.01) (Figure 2B). Here, ontological analysis revealed a 2.4-fold enrichment in oxidative-reduction, a 2.2-fold enrichment in proteolysis, and a 5.8-fold enrichment in cell adhesion via plasma membrane genes among other ontologies (Table 3).
Table 3.
Differentially Expressed Genes in SEI Hearts under Microgravity
| TERM | Dir. | Description | Count | Fold | FDR |
|---|---|---|---|---|---|
| BP_GO:0022008 | up | neurogenesis | 83 | 2.6 | <0.001 |
| BP_GO:0009267 | up | cellular response to starvation | 21 | 3.9 | <0.001 |
| BP_GO:0006364 | up | rRNA processing | 14 | 5.4 | 0.001 |
| BP_GO:0000462 | up | SSU-rRNA maturation from tricistronic rRNA |
11 | 6.4 | 0.005 |
| MF_GO:0003676 | up | nucleic acid binding | 56 | 2.2 | <0.001 |
| MF_GO:0004004 | up | ATP-dependent RNA helicase activity | 13 | 4.5 | 0.03 |
| MF_GO:0004402 | up | histone acetyltransferase activity | 11 | 4.8 | 0.08 |
| CC_GO:0032040 | up | small subunit processome | 17 | 8.7 | <0.001 |
| CC_GO:0005739 | up | nucleolus | 36 | 3.6 | <0.001 |
| CG_GO:0005634 | up | nucleus | 171 | 1.6 | <0.001 |
| BP_GO:0055114 | dwn | oxidative-reduction process | 64 | 2.4 | <0.001 |
| BP_GO:0006508 | dwn | proteolysis | 68 | 2.2 | <0.001 |
| BP_GO:0007156 | dwn | homophilic cell adhesion via plasma membrane | 13 | 5.8 | 0.001 |
| MF_GO:0020037 | dwn | heme binding | 26 | 2.8 | 0.007 |
| MF_GO:0004252 | dwn | serine-type endopeptidase | 41 | 2 | 0.02 |
| MF_GO:0005509 | dwn | calcium ion binding | 33 | 2.2 | 0.04 |
| CC_GO:0005615 | dwn | extracellular space | 99 | 2.5 | <0.001 |
| CC_GO:0005576 | dwn | extracellular region | 80 | 2.2 | <0.001 |
| CC_GO:0016021 | dwn | integral component of membrane | 255 | 1.4 | <0.001 |
| CC_GO:0005887 | dwn | integral component of plasma membrane | 59 | 1.8 | 0.01 |
| CC_GO:0031012 | dwn | extracellular matrix | 25 | 2.6 | 0.03 |
| CC_GO:0005604 | dwn | basement membrane | 7 | 9.5 | 0.04 |
| KEGG:dme01100 | dwn | metabolic pathways | 79 | 1.5 | 0.01 |
Table uses gene ontology (GO) terms. Dir., direction; up, upregulated; dwn, downregulated; BP, biological process; MF, molecular function; CC, cellular localization; KEGG, Kyoto Encyclopedia of Genes and Genomes pathway identification; FDR, false discovery rate.
We identified 38 genes that were commonly upregulated and 189 genes that were commonly downregulated in both WT and mutant hearts under microgravity (Figure 2B). Within the 38 commonly upregulated genes, we identified multiple proteasomal/Ub-dependent protein catabolic genes, including ProsAlpha6, Rpn13, Npl4, and CG2046 (Table S1). Within the 189 downregulated genes, there was a 7.9-fold enrichment in carbohydrate metabolic process genes (Table S2; Figure S2). Half of those genes mapped specifically to the Kyoto Encyclopedia of Genes and Genomes (KEGG) glycerophospholipid metabolic pathway and included key conserved enzymes, such as Agat4, Gpdh1, Gpd2, and Gapdh1 (Figure S2). Among downregulated genes, there was also a 3.2-fold enrichment in genes encoding proteins found in the ECM. In addition, oxidation-reduction and proteolysis genes were enriched in commonly downregulated genes (Table S2; see also Figure S2). Nineteen genes were contra-regulated. Volcano plots (Figure 2C) show the relative distribution of differentially expressed genes in CS and sei/hERG, normalized to their respective GCs. In general, more genes were downregulated than were upregulated under microgravity, with more genes differentially expressed at a higher magnitude in sei compared with CS. Adjusted read-count data for all samples have been provided (Data S1). It should be noted that fruit fly hearts are very small, and the material isolated was insufficient to validate our RNA-seq analyses, which were run in triplicate, with qPCR.
Microgravity Exposure Caused Myofibrillar and ECM Remodeling of the Heart
We examined the cardiac morphology of GC and microgravity-reared flies in a subset of hearts used for the functional assays. Phalloidin staining of filamentous actin (F-actin) revealed the myofibrillar structure within the myocardial cells that make up the heart tube (Figures 3A-3D). In CS GC hearts, the myofibrils are typically tightly packed and circumferentially arranged within those cells (Figure 3A), such that shortening of the sarcomeres results in a squeezing of the heart tube and ejection of the hemolymph present within the heart lumen (Video S1). Hearts from flies exposed to microgravity have myofibrils that are much more loosely arranged, with gaps, and were organized in a more-longitudinal orientation compared with 1g controls (Figures 3A and 3B). Myofibrillar reorganization or remodeling has previously been documented in hearts from sei/hERG mutants (Ocorr et al., 2017) and was again observed in hearts from 1g sei/hERG GCs (Figure 3C). However, hearts from sei/hERG mutants reared in microgravity showed even-more-extensive myofibrillar reorganization and disarray than GCs, and F-actin staining was diminished compared with GCs (Figure 3D).
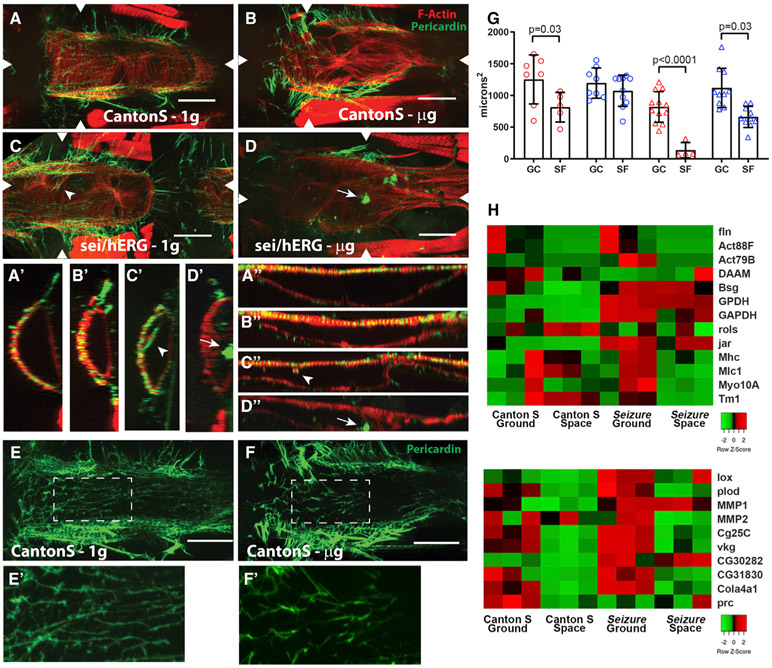
Figure 3. Microgravity Causes Myofibrillar and Fibrotic Remodeling.
(A) z stack projection of one chamber of a heart from a GC Canton S fly showing circumferentially arranged myofibrils stained for F-actin (red, phalloidin) and the supporting collagen IV extracellular matrix (green, anti-pericardin antibody [Ab]) that interacts extensively with the myofibrils. (A′) Transverse optical cross section at arrowheads reveals close association of collagen with actin-containing myofibrils. (A″) Longitudinal optical cross section. For all images, anterior is left; scale bars are 50 μm.
(B) Cardiac z stack projection from a fly raised in microgravity. Note the disorganized, non-circumferential myofibrils. Transverse (B′) and longitudinal (B″) optical cross sections.
(C) Heart from a GC sei/hERG mutant showing some myofibrillar disarray and gaps. Transverse (C′) and longitudinal (C″) cross sections show aberrant myofibrils surrounded by collagen (arrow).
(D) Heart from a microgravity-raised sei/hERG mutant shows large collagen aggregates (arrowhead). Transverse (D′) and longitudinal (D″) cross sections show that aggregates (arrowheads) appear to be located within the gaps between myofibrils and extend into the chamber.
(E) The collagen IV network from the GC heart in (A). (E′) an enlarged view of the region of interest (ROI) in GC heart (E).
(F) The effect of microgravity on the collagen IV network from the heart in (B). (F′) An enlarged view of the ROI in microgravity-exposed heart (E) reveals a reduced myofibrillar interacting network compared with GCs and SF CS.
(G) Quantification of myocardial-cell-associated collagen IV staining from z stack images. Significance was determined by a two-way ANOVA with Sidak’s multiple comparison post hoc test.
(H) Heatmap of significantly altered mRNA expression of sarcomere-associated genes (top) and ECM genes (bottom). Data were normalized to the average expression across row for each gene (Z score).
To further quantify myofibrillar structure, we calculated relative sarcomere lengths from our F-actin-stained z stack images (Figure S3) by quantifying the number of sarcomeres per 30-μm myofilament length. Sarcomere number was significantly reduced under microgravity for both female and male Canton-S flies (Figure S3), suggesting that hearts from SF flies were capable of generating less force than their GC siblings. Sarcomere counts in Canton-S SFR hearts were not different from GC hearts and, for heart diameters, there were no significant differences among any of the hearts from sei/hERG mutants (Figure S3).
In conjunction with the myofibrillar remodeling, we also observed a significant reorganization of the ECM surrounding the heart. In the Drosophila heart, this extracellular network consists primarily of the collagen IV homolog pericardin. Immunohistochemical staining revealed that network of collagen IV fibers surrounding the heart (Figure 3), and we show that it partially co-localizes with the z-band protein α-actinin (Figures S4A-S4H). In GC CS flies, those collagen fibers run orthogonally over the myofibrils of myocardial cells (Figures 3A and 3E) to form what appears to be a stabilizing network. Hearts from female CS flies reared in microgravity showed noticeable reductions in that myofiber-associated network (Figures 3B and 3F), consistent with the myofibrillar remodeling. Quantification of the cardiac muscle associated networks showed that this reduction was significant in SF female hearts (Figure 3G), although the decrease seen for CS male flies did not reach significance (Figure 3G). This result was unexpected because cardiomyopathy from compromised heart function is usually accompanied by increased collagen deposition, i.e., fibrosis.
This myofibril-associated collagen network in sei/hERG mutant GCs (Figures 3C and 3G) was already significantly reduced compared with that of CS GCs (Figures 3B and 3G), also likely a reflection of the previously observed myofibrillar remodeling in these mutants at 1g (Ocorr et al., 2017). Interestingly, we did observe intra-chamber fibrosis in a large percentage of hearts analyzed from sei mutants on the ground (arrows in Figures 2C″,3C, and 3C′), something we rarely observed in CS flies at 1g or microgravity. At 1g, 60% of sei/hERG versus 1% of CS females and 65% of sei/hERG versus 16% CS males showed evidence of intra-chamber collagen IV localization (p < 0.005, Mann Whitney unpaired t test for both males and females). Unexpectedly, most hearts from SF sei/hERG females exhibited only faint collagen fibers, and there were large globular aggregates of collagen in the cardiac lumen as well as deposits that appeared to fill-in gaps between myofibrils (Figures 3D, 3D′, and 3D″). Hearts from SF sei/hERG males exhibited more-extensive pericardin networks compared with SF sei/hERG females, but those networks were also reduced in microgravity compared with sei/hERG male GCs (Figure 3G).
The observed structural and morphological changes (Figures 3A-3D and 3G) correlate with a complex misexpression of various cardiac sarcomeric genes, (Figure 3H, top). Control and mutant flies exposed to microgravity exhibit downregulation of human actin orthologs, including Actin88F (hACTB) and Actin79b (hACTA/B). However, unique to CS, expression of the sarcomeric organizational gene basigin (hBSG), z-band localized metabolic genes GAPDH1 and GPDH, and unconventional myosin jaguar (jar) (hMYO6) were reduced, whereas the later three were upregulated in sei/hERG flies under 1g. Conversely, sarcomeric organizational gene rolling pebbles (rols) (hTANC2) was upregulated in CS in microgravity. Interestingly, reduced expression of a variety of myosin-related genes was observed exclusively in sei/hERG mutants under microgravity, including myosin heavy chain (Mhc) (hMHC7), myosin light chain1 (Mlh1) (hMYL1), Myo10A (hMYO15A), and Tropomyosin 1 (Tm1) (hTPM1).
Additionally, the observed changes in ECM content and morphology also correlated with misexpression of collagen/ECM regulatory genes (Figure 3H, bottom). Both control and mutant flies exposed to microgravity exhibit downregulation of procollagen lysyl hydroxylase (plod) (hPLOD1-3) and lysyl oxidase-1 (lox) (hLOXL2). These collagen maturation genes promote collagen cross-linking and ECM formation. However, in SF CS flies, the collagen network degradative gene, matrix metalloproteinase 1 (MMP1) (hMMP14/24), was also downregulated. In sei/hERG mutants, cardiac MMP1 expression was elevated under both gravity conditions, relative to CS. However, for CS, MMP2 (hMMP15) expression was reduced in microgravity. CS flies exhibited a reduction in multiple collagen-type-IV-encoding genes, including Cg25C (hCOL4A1), viking (vkg) (hCOL4A1), Col4a1(hCOL4A1/2/6), and pericardin (prc), as well as collagen and fibrinogen domain containing CG30280 (hFIBCD1) and CG31832 (hFCN1). Interestingly, most of these genes are upregulated in sei/hERG at 1g compared with CS, as would be expected under pathological cardiac remodeling conditions. Nevertheless, their expression shows a similar reduction under microgravity. The notable exceptions are the elevated CG30282 (see Figure 3H) expression in sei/hERG mutants, which remains high in space. Importantly, this misexpression of cell adhesion genes may underlie the observed misregulation of the pericardin network under microgravity (Figure S5).
Proteasome Numbers Are Significantly Increased in Hearts from SF Flies
Remarkably, nearly all the genes encoding proteasome subunits were significantly upregulated under microgravity in hearts from CS WT flies (Figure 4A). In contrast, at 1g, proteasome gene expression was already reduced in the hearts from sei/hERG mutants relative to CS. Nevertheless, microgravity exposure also resulted in upregulation of cardiac proteasome genes in sei/hERG mutants relative to their sei/hERG GCs (Figure 4A), albeit to levels that were still reduced relative to CS GCs. Our observation that cardiac proteasome gene expression was significantly upregulated in response to microgravity suggested that protein folding had been compromised and/or degradation was elevated. We had a fortunate and unique opportunity to test that hypothesis on a subsequent mission launched to the ISS aboard SpaceX commercial resupply service (CRS)-11 using the same protocol as for CRS-03. For this study, we generated flies that expressed UAS-polyQ46 GFP only in the myocardial cells using a tinCΔ4-Gal4 driver. This GFP-tagged 46-glutamine repeat polypeptide (PolyQ46) is an intermediate length between disease-causing polyQ repeat lengths and the WT polyQ containing proteins (Melkani et al., 2013). Previous studies have shown that treatment with agents that increase reactive oxygen species in the heart increase the number and size of GFP-positive plaques (Melkani et al., 2013) in UAS-polyQ46 GFP-expressing hearts. In this respect, polyQ46 acts as a marker for protein misfolding and aggregation under cardiac proteostatic stress.
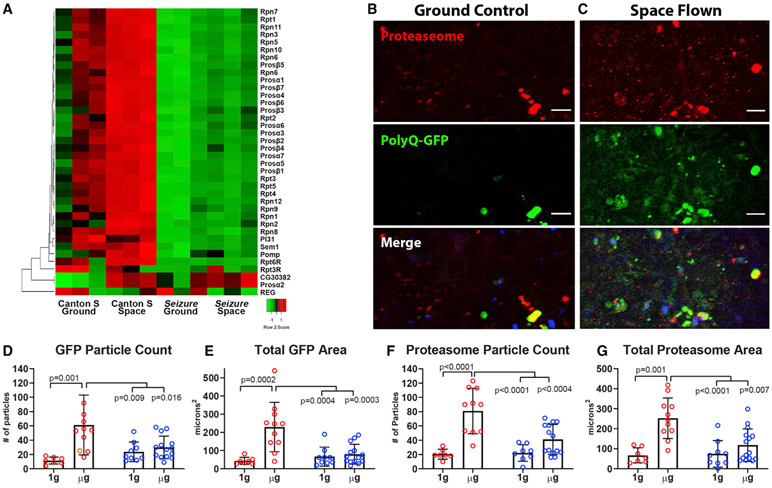
Figure 4. Proteasome mRNA and Protein Expression Is Decreased in Heart in Microgravity.
(A) Expression of mRNA encoding many proteasome subunits is dramatically increased by exposure to microgravity in hearts from CS SF flies compared with CS GCs. Hearts from sei/hERG mutant GCs have much lower expression of these same subunits, and the expression is not altered by microgravity. Each box is the average of triplicate samples normalized to the overall averaged expression for all hearts.
(B and C) Images from a single cardiac chamber in GC (B) and SF (C) CS female flies homozygous for Hand-gal4 > UAS polyQ46 GFP. Immunohistochemical staining showed proteasome immunoreactivity (red) and PolyQ-GFP (green); scale bar, 10 μm. Merged images show co-localization of some GFP-positive plaques and proteasome aggregates. Myocardial cell nuclei are stained with DAPI (blue).
(D and E) GFP particle count (D) and total area (E) were increased in hearts from females (red symbols) but not males (blue symbols) under microgravity.
(F) Proteasome particle count was significantly higher in females compared with males and was significantly increased in female hearts in response to microgravity.
(G) Proteasome aggregate total area was also significantly increased in females but not in males.
For all graphs, significance was determined by two-way ANOVA and Tukey’s multiple comparisons post hoc test. *p < 0.05, **p < 0.01, ***p < 0.005.
Immunohistochemical analysis (Figure S6F) of hearts from GC flies exhibited GFP-positive plaques and anti-proteasome puncta (Figure 4C; GC), which were typically found near myocardial cell nuclei. Hearts from microgravity-reared flies had significantly more GFP-positive polyQ plaques (Figure 4D) covering a larger area of the heart (Figure 4E) compared with 1g controls. Proteasome puncta were also more abundant (Figure 4F), covering a larger total area (Figure 4G) of the heart. In addition, in myocardial cells, a significant amount of α-proteasome “haze” was observed, which could represent less-highly aggregated proteasomes (Figure 4C; SF). Taken together, these data suggest that, in the heart in space, proteasomal gene and protein expression is dramatically upregulated, although proteosomes are likely dysfunctional, as manifest by co-localized proteasomal and polyQ aggregates.
DISCUSSION
As humans increasingly venture into our interplanetary space, it will be important to understand the different ways in which prolonged exposure to altered g forces affect organ systems that have evolved under 1g. Muscle function is known to be affected by prolonged weightlessness, but most studies address the “bulk” properties of muscle fibers, leaving the cellular and mechanistic causes relatively unexplored. Knowledge of the biomechanical properties of filaments is important because those filaments reveal how molecular properties scale up and how bulk properties of muscle fibers are dictated by nanoscale phenomena. The current work, using the genetically tractable Drosophila model, provides new and unique insights into microgravity effects on muscle cells at the molecular level and also facilitates insightful structural and functional cardiac phenotyping at the organismal level. Similar to humans, exposure to microgravity has profound effects on fly muscle function, as has been previously reported for flies exposed to microgravity for short durations (2 weeks) (Miller and Keller, 1999).
Heart function analysis in our denervated, semi-intact preparations confirmed a cardiac-specific effect of space flight, with heart chamber size and cardiac contractility and output being significantly reduced under microgravity (Figures 1B, 1D, and 1E). These findings were consistent with observed myofibrillar remodeling (Figures 3A-3D), reduced sarcomere count (Figure S3), and misregulation of sarcomeric genes (Figure 3H), including actins, myosins and tropomyosin, and sarcomere organizational genes. Troponin mutations that interfere with tropomyosin binding have already been documented in the fly heart to cause cardiac dysfunction, cardiac restriction, and myofibrillar disarray (Madan et al., 2020; Viswanathan et al., 2014), similar to our observation in space flies. In addition, we previously observed that specific myosin-transducer mutations that affect rates of ATP hydrolysis have differential effects on myofibrillar structure, including restrictive (when rates are faster) and dilatory (when rates are slower) cardiomyopathies (Cammarato et al., 2008). We have also shown that mutations in unc-45, a chaperone involved in myosin protein folding, significantly alter myofibrillar structure in the fly heart (Melkani et al., 2011). Although a role for Myo10A (Sisyphus) has not yet been described in the heart, it has been shown to promote transport of cell-adhesion cargo and to bind integrins, which we have previously shown to be critical to cardiac structure and function (Nishimura et al., 2014), thus suggesting a possible role for this unconventional myosin in the heart. Sarcomeric contractile and structural proteins provide the fundamental molecular basis for cardiac function and have well-established roles in the etiology of a variety of cardiac diseases (Marston, 2018). Our studies show that prolonged exposure to microgravity is associated with transcriptional misexpression of sarcomeric genes, cardiomyocyte myofibrillar disorganization, and ultimately, cardiac contractile dysfunction. Lack of an effect on overall chronotropy, along with an increased incidence of arrhythmia in male flies is consistent with findings in human induced pluripotent stem cell-derived cardiomyocytes (Wnorowski et al., 2019).
Under pathological cardiomyopathy conditions, collagen deposition is usually increased leading to fibrosis, which contributes to impaired contractility. Surprisingly, the collagen IV network that normally surrounds and attaches to the cardiomyocytes was decreased under microgravity. Remodeling occurred in both CS and, to a greater extent, in sei/hERG mutant hearts, which also exhibited intra-lumen fibrosis and collagen aggregation. These observations were consistent with misexpression of collagen and ECM maturation and degradative genes, as well as collagen and cell-adhesion-encoding genes. Although fibrosis is generally associated with cardiac disease, ECM degradation is an observed feature in both the volume-overloaded heart and in end-stage diastolic remodeling from pressure overload (Frangogiannis, 2019). ECM degradation is associated with features of contractile dysfunction, inflammation, and cardiomyocyte apoptosis, potentially due in part to loss of matrix-dependent signals that are required for preservation of function. Additionally, misregulation of ECM composition has also been implicated in cardiac disease. Taken together, ECM remodeling is a potential maladaptive effect of prolonged exposure to microgravity in our model and may be a contributing factor to the observed cardiac dysfunction.
Expression of basigin, a transmembrane protein (Suzuki et al., 2016) involved in sarcomeric organization, was significantly downregulated in sei/hERG mutant hearts relative to WT at 1g, but exposure to microgravity resulted in an increased expression in sei/hERG mutants. Basigin has been shown to promote cardiac fibrosis in pressure overload models and to activate matrix metalloproteinases that degrade ECM components and thus promote ECM remodeling. This is consistent with the expression of MMP1 maintained in sei/hERG mutants compared with CS and may have a role in the dramatic collagen IV remodeling in SF sei/hERG mutant hearts. This sarcomeric modification may serve to stiffen the heart wall, allowing cardiac output to be maintained in mutants at microgravity, despite the myofibrillar remodeling and overall reduced expression of muscle structural proteins. Interestingly, basigin (CD147) has recently emerged as a potential receptor for SARS-CoV-2 (Zhou et al., 2020).
Gap junctions are important intercellular connections among cardiomyocytes, allowing the organ to work as a syncytium. They are composed of connexins in vertebrates and innexins in flies. Innexin 2 and 3 have both been shown to be present in the fly heart (Cammarato et al., 2011). Under microgravity conditions innexin 2 is significantly downregulated in both CS (−1.76-fold; p = 0.002) and in sei/hERG mutant hearts (−1.59-fold; p = 0.045), whereas innexin 3 expression is altered only in sei/hERG mutants (−1.8-fold; p = 0.002). Together with the reduction in genes encoding cell-adhesion proteins, it is likely that electrotonic coupling of the myocardial cells was impaired.
Another surprising finding was the significant upregulation of genes encoding nearly every subunit of the proteasome. We were able to confirm that microgravity exposure resulted in an increase in protein aggregates and associated proteasome particles on a subsequent flight. The increase in polyQ-GFP aggregates and proteasome puncta suggests that microgravity induces dramatic changes in protein turnover and/or protein misfolding. Recent evidence indicates that oligomers, such as huntingtin/PolyQ proteins, bind to and inhibit proteasomes by preventing injection of proteins into the degradation chamber (Thibaudeau et al., 2018). Additionally, proteasome dysfunction has been suggested as a causative factor and is observed in a variety of cardiomyopathies (Gilda and Gomes, 2017), heart failure, and cardiac amyloidosis (Pattison et al., 2008; Sanbe et al., 2005). We have also shown in flies that cardiac amyloidosis caused structural remodeling reminiscent of the remodeling observed in flies reared in microgravity (Melkani et al., 2013). The increase in proteosome gene and protein expression in space may reflect increased turnover of sarcomeric proteins or may be a compensatory response to the likely muted proteosome function by misfolded and aggregated proteins. This interpretation is consistent with smaller hearts and less muscle mass, as in astronauts, and could be contributing to the observed cardiac remodeling and functional defects. Interestingly, in hearts from K+ channel mutants, expression of nearly all proteasome genes was already downregulated at 1g compared with WT controls. Nevertheless, proteasome gene expression was modestly upregulated in hearts from space-reared mutants suggesting that this may be a fundamental response of cardiac muscle to lowered gravity. It is likely that this generalized increase in proteasome content underlies cardiac muscle atrophy.
In contrast to the increase in proteasomes, expression of proteolytic genes was generally downregulated in microgravity. Recent quantitative analyses of human cardiac fuel usage showed that non-failing hearts undergo proteolysis and actively secrete amino acids. This process was nearly doubled in failing versus non-failing hearts (Murashige et al., 2020). It was unclear whether upregulation of proteolysis was adaptive or maladaptive. In this context, it is interesting that SF fly hearts showed decreased expression, perhaps as a reflection of an overall reduction or shift in fuel usage by the heart under microgravity.
The current studies examined the effects of prolonged exposure to microgravity, whereas the exposure of astronauts is more temporally limited. However, future space exploration efforts by government and commercial ventures are focused on missions to the moon, asteroids, and Mars, and those explorations will require astronauts to spend many months and perhaps years in altered gravity. Thus, information on how prolonged microgravity exposure affects different organ systems will be critical to the success of those efforts (Criscuolo et al., 2020; Prasad et al., 2020). It is likely that, at the cellular level, there are underlying and conserved responses to altered gravity that are shared between organisms. Approximately 75% of disease-causing genes are conserved between flies and humans. Thus, the fly model can have a key role in elucidating the effects of microgravity during prolonged space flight, with significant advantages over other model systems because of its small size, its well defined and conserved genetics, and its relatively short lifespan.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Additional information and/or requests for resources or reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Karen Ocorr (kocorr@sbpdiscovery.org).
Materials Availability
This study did not generate unique antibodies, genetic lines, reagents, software, or equipment.
Data and Code Availability
Included in Supplemental data is post-analysis normalized counts from DSeq2 for all identified genes across all samples sets in triplicate. Raw read counts and FASTQ files have been made available at the community-endorsed public repository at NASA Gene Lab https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-347.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly rearing and μg exposure –
Fly lines used in this study were Canton S laboratory WT lines (Bloomington Stock Center) and a K+ channel mutant seizure (seizurets1, a gift from Dr. Barry Ganetsky). The sei/hERG mutants were previously isolated in a screen for ethylmethane sulfonate (EMS)-induced paralytic mutations on the second chromosome (Titus et al., 1997). Flies launched to the International Space Station (space flown, SF) and the corresponding ground controls (GC) were housed in polystyrene vials (75 x24 mm) with 10 mL of standard cornmeal-based fly food containing double the usual amount of 10% Tegosept and cellulose acetate plugs. Approximately 24 hours prior to launch, 15 vials were each loaded with 10 virgin females and 5 male adult flies. Loaded vials were placed in a Plexiglas tray and inserted into a vented fly box (VFB; Figure 1A), a modified version of a Nanoracks box. The VFB was placed into a canvas case for loading into the Dragon capsule. Launch of SpaceX CRS-3 occurred at 19:25 UTC on April 18, 2014. Flies were kept in the VFB throughout the 30-day mission and were stored aboard the docked Dragon capsule that shared atmosphere with the ISS. The Dragon capsule unberthed at 13:26 UTC (Co-ordinated Universal Time) on May 18, 2014 and re-entry into the Earth’s atmosphere occurred the same day at 19:05 UTC. Following retrieval of the Dragon capsule off the coast of Mexico, the VFB was offloaded at Long Beach Port, California and transported by car to La Jolla, California in a climate-controlled vehicle. Surviving adult flies were collected from the vials at 23:30 UTC on May 20, 2014. Data collection, which included heart function studies, tissue collection for RNaseq, and tissue preservation for immunohistochemistry, was completed by 07:30 UTC on May 21, 2014 on all μg space flown and corresponding ground control flies (specific methods described below). The protocol and associated timeline are outlined schematically in Figure 1. The VFB was oriented such that the g-load was directed down into the food supply located at the bottom of the vials. This orientation was maintained throughout the entirety of the mission, from point of transfer at the lab at NASA Kennedy Space Center through stowage on the Dragon capsule, and again from Dragon un-berthing to opening of the VFB in the lab at SBP Medical Discovery Institute in La Jolla, California. Ground control flies were prepared and reared under identical conditions in ground-based incubators with temperature and humidity controlled to levels recorded on the ISS.
Following initial adult fly collections, vials were maintained for three days at 25°C and adults that emerged were collected and aged as a mixed sex cohort for two weeks at 1g (Space Flight Recovery, SFR). These flies were then analyzed as for SF and GC flies.
Identical protocols were followed for the launch of the PolyQ46 flies aboard SpaceX CRS-11 on June 03, 2017 at 21:07 UTC until splashdown which occurred at 12:12 UTC on July 03, 2017. VFBs were again returned to Long Beach Port, California and transported via a climate-controlled vehicle to La Jolla, California on the same day, arriving at approximately 03:00 UTC on July 5, 2017. All dissections and fixation of heart tissue (described below) were performed within 48 hours of arrival in La Jolla.
METHOD DETAILS
Heart Function Assays –
Flies were anaesthetized with FlyNap® (Carolina Biological Supply Co, Burlington, NC) and dissected in artificial hemolymph as previously described (Vogler and Ocorr, 2009). Cardiac function was assayed using high speed digital imaging (100-150fps, 9300 EM-CCD cameras, Hamamatsu Corp.). Heart function was quantified from movies using the Semi-automatic Optical Heartbeat Analysis software (SOHA; sohasoftware.com; Ocorr, 2009). Contractility was estimated based on end diastolic (EDD) and end systolic (ESD) diameters, measured from moves using SOHA. (i.e., fractional shortening = (EDD-ESD)/EDD). Cardiac output (CO) was calculated assuming the heart is a cylindrical tube, where volume was estimated using V = π(r2)x h, for a 1 micron portion of the tube (h = 1). Thus, Stroke Volume (SV) = (π(1/2(EDD2)−(π(1/2(ESD2) and cardiac output ©) = mean heart rate x SV.
Immunohistochemistry –
Dissected hearts previously used for the heart function assays were subsequently relaxed with EGTA (10mM in artificial hemolymph) and then immediately fixed for 20 minutes in a solution of hemolymph containing 4% paraformaldehyde. All hearts were stained as described in Alayari et al. (2009). Collagen IV was visualized using a monoclonal antibody against Pericardin, obtained from the Developmental Studies Hybridoma Bank and developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. Proteasomes were visualized using an antibody against 20S proteasome subunits a1,2,3,5,6 & 7 (BML-PW8195, Enzo Life Sciences, Farmingdale, NY). PolyQ-GFP plaques were identified using Mouse anti-GFP (Invitrogen, Carlsbad, CA, USA) The following secondary antibodies were used: anti-rabbit IgG conjugated with Alexa Fluor 488 and anti-mouse IgG conjugated with Alexa Fluor 594 (Invitrogen, Carlsbad, CA, USA) F-actin was detected using Phalloidin-Alexa Fluor 594 (Molecular Probes, Eugene, OR). Stained hearts were visualized with a Zeiss Apotome microscope; Z stack images were captured and processed with Axiovision software (Zeiss).
Pericardin, proteosome and polyQ46-GFP staining was quantified using the Weka Trainable Segmentation plugin in ImageJ (Schneider et al., 2012). Individual slices from Z stack images of heart chambers were constructed by optical sectioning of the heart tube starting at a point below the non-cardiac longitudinal fibers. This optical sectioning revealed the pericardin fibers that interacted with the myofibrils in the dorsal myocardial cells. We selected a 350 × 150-pixel (90x 30 um) region of interest (ROI) centered over a heart chamber (Figures 3E and 3F) and a representative image (i.e., Figure S6, left panel) was used for training; the model generated by the training was then applied to all images. Particle counts and area measurements were performed on the segmentation output images (Figure S6, middle panel) using ImageJ analysis tools (Figure S6, right panel).
Sarcomere numbers were quantified from Z stacks of hearts stained for F-actin(with phalloidin). Two to three 30um segments of single myofilaments were identified in myocardial cells from each heart (Figure S3) and the number of sarcomeres were counted and averaged for each heart.
Cardiac RNaseq Analysis
Gene expression analysis was performed on a subset of adult fly hearts from both 1g ground control flies and flies that spent their entire existence in μg. For each genotype, adult Drosophila were dissected under artificial hemolymph as described in Vogler and Ocorr (2009), on ice to expose the heart. These hearts were then carefully cleaned of surrounding fat cells, extracted, and pooled in groups of 6-15 hearts. Hearts were placed into tubes containing 300 μL of Qiazol lysis reagent, homogenized per the QIAGEN protocol, flash frozen in liquid nitrogen, and transferred to −80°C for storage. Total RNA from each sample was extracted and purified using the miRNeasy Mini kit from QIAGEN (Cat. No. 217004), yielding 173~551 ng of RNA eluted in 30 μL of RNase free water. The integrity of the extracted RNA was assessed by Agilent 2200 Tapestation system (Agilent, Santa Clara, CA), indicating RIN numbers in the range of 8.3 to 9.6. RNA samples of high quality and sufficient RNA quantities were selected for further processing.
Libraries and sequencing
Libraries of extracted RNA samples were built using the Illumina TruSeq™ RNA sample preparation kit and yielded fragments with 200~500 base pairs. The qualified fragments were ligated with indexed adapters, amplified and subjected for sequencing by Illumina HiSeq 2000 (Illumina, San Diego, CA) to generate paired end reads with a length of 100 bases.
Pre-processing of heart RNA-seq Data
A total of 8.02 billion reads were generated in FASTQ format for 24 samples, yielding coverage in the range of 35-152 bases for each sample. The quality of reads was evaluated by FASTX-Toolkit (Hannon Lab, Cold Spring Harbor, NY, USA), where all reads indicated excellent quality with an average quality score of 37. The reads were then aligned to the Drosophila melanogaster genome Ensembl.BDGP5 using TopHat2 and Bowtie2 .The aligned reads were then assembled using BEDTools (Quinlan and Hall, 2010). The samples with low alignment rate and biased expression patterns were excluded from further analyses. Expression of mRNAsin qualified samples were then normalized as Reads Per Kilobase per Million reads (RPKM) by AltAnalyze (Salomonis et al., 2009). Genes with insufficient copy numbers were filtered out before conducting further statistical analyses.
Differential expression and functional annotation clustering analysis of RNA-seq data
Differentially expressed genes were determined using a one-way ANOVA analysis considering a p value of 0.05. Pathway enrichment analysis was performed by comparing differentially expressed genes with Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using a Fisher’s exact test-based method (Evangelou et al., 2012) where p < 0.05 was considered to be statistically significant.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification methods for the various parameters are described in the relevant Methods Section. Statistical analyses were performed using Prism (Graph Pad, v.9). The specific tests used are indicated in the Figure legends. P values for significant differences are indicated in the graphs. All graphs show the individual data points used in the analysis.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| monoclonal antibody against Pericardin | Developmental Studies Hybridoma Bank Iowa City, IA 52242. | DSHB Cat # EC11 RRID:AB_528431 |
| 0S proteasome subunits a1,2,3,5,6 & 7 | Enzo Life Sciences, Farmingdale, NY | Cat # BML-PW8195 RRID:AB_10541045 |
| Mouse anti-GFP | Invitrogen, Carlsbad, CA, USA | Cat # 33-2600 RRID:AB_2533111 |
| anti-rabbit IgG conjugated with Alexa Fluor 488 | Invitrogen, Carlsbad, CA, USA | Cat # Cat # R37116 RRID:AB_2556544 |
| anti-mouse IgG conjugated with Alexa Fluor 594 | Invitrogen, Carlsbad, CA, USA | R37121 RRID:AB_2556549 |
| Phalloidin-Alexa Fluor 594 | Molecular Probes, Eugene, OR | Cat # A12381 RRID:AB_2315633 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Flynap | Carolina Biological Supply Co, Burlington, NC | #173010 |
| Critical Commercial Assays | ||
| miRNeasy Mini kit | QIAGEN | Cat. No. 217004 |
| Illumina TruSeq™ RNA sample preparation kit | Illumina, San Diego, CA) | RS-122-2001 |
| Deposited Data | ||
| Drosophila melanogaster genome Ensembl.BDGP5 | ensembl | http://uswest.ensembl.org/Drosophila_melanogaster/Info/Index |
| Kyoto Encyclopedia of Genes and Genomes | https://www.kegg.jp/ | RRID:SCR_012773 |
| NASA Gene Labs | This paper | https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-347 |
| Experimental Models: Organisms/Strains | ||
| Canton S Wildtype Strain | Bloomington Drosophila Stock Center | BDSC Cat# 64349, RRID:BDSC_64349 |
| Sei/hERG mutant | Dr. Barry Ganetsky | N/A |
| PolyQ46-GFP line | Dr. Girish Melkani | N/A |
| Software and Algorithms | ||
| SOHA Semi Automated Optical Heartbeat Analysis | http://www.sohasoftware.com | N/A |
| Axiovision software | Zeiss | RRID:SCR_002677 |
| Weka Trainable Segmentation plugin in ImageJ | https://www.cs.waikato.ac.nz/ml/weka/ | RRID:SCR_001214 RRID:SCR_001935 |
| FASTX-Toolkit | Hannon Lab, Cold Spring Harbor, NY, USA) | RRID:SCR_005534 |
| BEDTools | https://github.com/arq5x/bedtools2 | RRID:SCR_006646 |
| AltAnalyze | http://www.altanalyze.org/ | RRID:SCR_002951 |
| Top Hat | http://ccb.jhu.edu/software/tophat/index.shtml | RRID:SCR_013035 |
| Bow Tie | http://bowtie-bio.sourceforge.net/index.shtml | RRID:SCR_005476 |
| Other | ||
| 9300 EM-CCD cameras | Hamamatsu Corp. | https://www.hamamatsu.com/eu/en/product/cameras/emccd-cameras/index.html |
| Zeiss Apotome microscope | Zeiss | https://www.zeiss.com/microscopy/us/products/imaging-systems/apotome-for-biology.html |
| Agilent 2200 Tapestation system | Agilent, Santa Clara, CA | https://www.agilent.com/en/product/automated-electrophoresis/tapestation-systems |
| Illumina HiSeq 2000 | Illumina, San Diego, CA) | https://www.illumina.com |
Highlights.
Flies in microgravity exhibit cardiac constriction, remodeling, and diminished output
Heart defects correlate with reduced sarcomeric and extracellular matrix gene expression
Proteosome gene or protein expression is upregulated, suggesting proteostasis imbalance
ACKNOWLEDGMENTS
We would like to thank Nanoracks and Space Florida for initiating these studies with their award to us of the vented fly box. This work was supported in part by the following grants: K.O., NASA NN13AN38694, AHA SDG 0835243N, NIH HL132241, and NIA AG061598; R.B., NASA NN13AN38694, NIA AG058075, and NIH HL05432 and HL149992; and S.B., NASA NN13AN38694 and A.M., R01 HL137100.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108445.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Akoum NW, Sanders NA, Wasmund SL, and Hamdan MH (2011). Irregular ventricular activation results in QT prolongation and increased QT dispersion: a new insight into the mechanism of AF-induced ventricular arrhythmogenesis. J. Cardiovasc. Electrophysiol 22, 1249–1252. [DOI] [PubMed] [Google Scholar]
- Alayari NN, Vogler G, Taghli-Lamallem O, Ocorr K, Bodmer R, and Cammarato A (2009). Fluorescent labeling of Drosophila heart structures. J. Vis. Exp (32), 1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarato A, Dambacher CM, Knowles AF, Kronert WA, Bodmer R, Ocorr K, and Bernstein SI (2008). Myosin transducer mutations differentially affect motor function, myofibril structure, and the performance of skeletal and cardiac muscles. Mol. Biol. Cell 19, 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarato A, Ahrens CH, Alayari NN, Qeli E, Rucker J, Reedy MC, Zmasek CM, Gucek M, Cole RN, Van Eyk JE, et al. (2011). A mighty small heart: the cardiac proteome of adult Drosophila melanogaster. PLoS ONE 6, e18497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo F, Sueur C, and Bergouignan A (2020). Human adaptation to deep space environment: an evolutionary perspective of the foreseen interplanetary exploration. Front. Public Health 8, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aunno DS, Dougherty AH, DeBlock HF, and Meck JV (2003). Effect of short- and long-duration spaceflight on QTc intervals in healthy astronauts. Am. J. Cardiol 91, 494–497. [DOI] [PubMed] [Google Scholar]
- Evangelou M, Rendon A, Ouwehand WH, Wernisch L, and Dudbridge F (2012). Comparison of methods for competitive tests of pathway analysis. PLoS ONE 7, e41018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink M, Callol-Massot C, Chu A, Ruiz-Lozano P, Izpisua Belmonte JC, Giles W, Bodmer R, and Ocorr K (2009). A new method for detection and quantification of heartbeat parameters in Drosophila, zebrafish, and embryonic mouse hearts. Biotechniques 46, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangogiannis NG (2019). The extracellular matrix in ischemic and nonischemic heart failure. Circ. Res 125, 117–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilda JE, and Gomes AV (2017). Proteasome dysfunction in cardiomyopathies. J. Physiol 595, 4051–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DR, Sargsyan AE, Martin DS, Garcia KM, Melton SL, Feiveson A, and Dulchavsky SA (2011). On-orbit prospective echocardiography on International Space Station crew. Echocardiography 28, 491–501. [DOI] [PubMed] [Google Scholar]
- Madan A, Viswanathan MC, Woulfe KC, Schmidt W, Sidor A, Liu T, Nguyen TH, Trinh B, Wilson C, Madathil S, et al. (2020). TNNT2 mutations in the tropomyosin binding region of TNT1 disrupt its role in contractile inhibition and stimulate cardiac dysfunction. Proc. Natl. Acad. Sci. USA 117, 18822–18831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston S (2018). The molecular mechanisms of mutations in actin and myosin that cause inherited myopathy. Int. J. Mol. Sci 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkani GC, Bodmer R, Ocorr K, and Bernstein SI (2011). The UNC-45 chaperone is critical for establishing myosin-based myofibrillar organization and cardiac contractility in the Drosophila heart model. PLoS ONE 6, e22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkani GC, Trujillo AS, Ramos R, Bodmer R, Bernstein SI, and Ocorr K (2013). Huntington’s disease induced cardiac amyloidosis is reversed by modulating protein folding and oxidative stress pathways in the Drosophila heart. PLoS Genet. 9, e1004024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MS, and Keller TS (1999). Measuring Drosophila (fruit fly) activity during microgravity exposure. J. Gravit. Physiol 6, P99–P100. [PubMed] [Google Scholar]
- Murashige D, Jang C, Neinast M, Edwards JJ, Cowan A, Hyman MC, Rabinowitz JD, Frankel DS, and Arany Z (2020). Comprehensive quantification of fuel use by the failing and nonfailing human heart. Science 370, 364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M, Kumsta C, Kaushik G, Diop SB, Ding Y, Bisharat-Kernizan J, Catan H, Cammarato A, Ross RS, Engler AJ, et al. (2014). A dual role for integrin-linked kinase and β1-integrin in modulating cardiac aging. Aging Cell 13, 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr Karen, et al. (2009). Semi-automated Optical Heartbeat Analysis of Small Hearts. JoVE. 10.3791/1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Reeves NL, Wessells RJ, Fink M, Chen HS, Akasaka T, Yasuda S, Metzger JM, Giles W, Posakony JW, and Bodmer R (2007). KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc. Natl. Acad. Sci. USA 104, 3943–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Zambon A, Nudell Y, Pineda S, Diop S, Tang M, Akasaka T, and Taylor E (2017). Age-dependent electrical and morphological remodeling of the Drosophila heart caused by hERG/seizure mutations. PLoS Genet. 13, e1006786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison JS, Sanbe A, Maloyan A, Osinska H, Klevitsky R, and Robbins J (2008). Cardiomyocyte expression of a polyglutamine preamyloid oligomer causes heart failure. Circulation 117, 2743–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platts SH, Martin DS, Stenger MB, Perez SA, Ribeiro LC, Summers R, and Meck JV (2009). Cardiovascular adaptations to long-duration head-down bed rest. Aviat. Space Environ. Med 80 (5, Suppl), A29–A36. [DOI] [PubMed] [Google Scholar]
- Prasad B, Richter P, Vadakedath N, Mancinelli R, Krüger M, Strauch SM, Grimm D, Darriet P, Chapel JP, Cohen J, and Lebert M (2020). Exploration of space to achieve scientific breakthroughs. Biotechnol. Adv 43, 107572. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Respress JL, Gershovich PM, Wang T, Reynolds JO, Skapura DG, Sutton JP, Miyake CY, and Wehrens XH (2014). Long-term simulated microgravity causes cardiac RyR2 phosphorylation and arrhythmias in mice. Int. J. Cardiol 176, 994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N, Nelson B, Vranizan K, Pico AR, Hanspers K, Kuchinsky A, Ta L, Mercola M, and Conklin BR (2009). Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput. Biol 5, e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanbe A, Osinska H, Villa C, Gulick J, Klevitsky R, Glabe CG, Kayed R, and Robbins J (2005). Reversal of amyloid-induced heart disease in desmin-related cardiomyopathy. Proc. Natl. Acad. Sci. USA 102, 13592–13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti MC, and Tristani-Firouzi M (2006). hERG potassium channels and cardiac arrhythmia. Nature 440, 463–469. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers RL, Martin DS, Meck JV, and Coleman TG (2005). Mechanism of spaceflight-induced changes in left ventricular mass. Am. J. Cardiol 95, 1128–1130. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Satoh K, Ikeda S, Sunamura S, Otsuki T, Satoh T, Kikuchi N, Omura J, Kurosawa R, Nogi M, et al. (2016). Basigin promotes cardiac fibrosis and failure in response to chronic pressure overload in mice. Arterioscler. Thromb. Vasc. Biol 36, 636–646. [DOI] [PubMed] [Google Scholar]
- Thibaudeau TA, Anderson RT, and Smith DM (2018). A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun 9, 1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus SA, Warmke JW, and Ganetzky B (1997). The Drosophila erg K+ channel polypeptide is encoded by the seizure locus. J. Neurosci 17, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan MC, Kaushik G, Engler AJ, Lehman W, and Cammarato A (2014). A Drosophila melanogaster model of diastolic dysfunction and cardiomyopathy based on impaired troponin-T function. Circ. Res 114, e6–e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler G, and Ocorr K (2009). Visualizing the beating heart in Drosophila. J. Vis. Exp (31), 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wnorowski A, Sharma A, Chen H, Wu H, Shao NY, Sayed N, Liu C, Countryman S, Stodieck LS, Rubins KH, et al. (2019). Effects of spaceflight on human induced pluripotent stem cell-derived cardiomyocyte structure and function. Stem Cell Reports 13, 960–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Fang Y, Xu T, Ni WJ, Shen AZ, and Meng XM (2020). Potential therapeutic targets and promising drugs for combating SARS-CoV-2. Br. J. Pharmacol 177, 3147–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Included in Supplemental data is post-analysis normalized counts from DSeq2 for all identified genes across all samples sets in triplicate. Raw read counts and FASTQ files have been made available at the community-endorsed public repository at NASA Gene Lab https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-347.