Abstract
Adjuvant chemotherapy for solid tumors based on platinum-derived compounds such as cisplatin is the treatment of choice in most cases. Cisplatin triggers signaling pathways that lead to cell death, but it also induces changes in tumor cells that modify the therapeutic response, thereby leading to cisplatin resistance. We have recently reported that microRNA-7 is silenced by DNA methylation and is involved in the resistance to platinum in cancer cells through the action of the musculoaponeurotic fibrosarcoma oncogene family, protein G (MAFG). In the present study, we first confirm the miR-7 epigenetic regulation of MAFG in 44 normal- and/or tumor-paired samples in non small-cell lung cancer (NSCLC). We also provide translational evidence of the role of MAFG and the clinical outcome in NSCLC by the interrogation of two extensive in silico databases of 2019 patients. Moreover, we propose that MAFG-mediated resistance could be conferred due to lower reactive oxygen species production after cisplatin exposure. We developed specifically selected aptamers against MAFG, with high sensitivity to detect the protein at a nuclear level probed by aptacytochemistry and histochemistry analyses. The inhibition of MAFG activity through the action of the specific aptamer apMAFG6F increased the levels of reactive oxygen species production and the sensitivity to cisplatin. We report first the specific nuclear identification of MAFG as a novel detection method for diagnosis in NSCLC, and then we report that MAFG modulates the redox response and confers cell protection against free radicals generated after platinum administration, thus also being a promising therapeutic target.
INTRODUCTION
Cisplatin (CDDP) is the current and most widely used chemotherapeutic agent in solid malignancies, including lung and ovarian cancers. CDDP is a platinum compound that binds to and crosslinks DNA, thus inducing apoptosis in cancer cells.1,2 CDDP has also been reported to generate an increase in oxidative stress by increasing levels of the superoxide anion, H2O2, and hydroxyl radicals that can lead to cell death, enhancing CDDP action.3,4 Despite a reasonable rate of initial response, however, CDDP treatment frequently results in chemoresistance development, leading to therapeutic failure in these tumor types. Many studies and reviews in the last 30 years have centered on gaining insight into the molecular mechanisms that account for the CDDP-resistant phenotype of tumor cells, which could provide crucial information for treatment resensitizing.4,5 A number of events have been proposed to underlie this phenomenon. One such event consists of the ability of cancer cells to overcome the reactive oxygen species (ROS) generated by CDDP, thus avoiding apoptosis. The complex cellular response against ROS is mainly mediated through the antioxidant response element (ARE), nuclear factor (erythroid-derived 2)-like 2 (NRF2), and the small MAF proteins (sMAFs) family of transcription fac-tors.6 The accumulation of reactive oxygen or nitrogen species in the cell triggers the translocation of NRF2 to the nucleus, where it interacts with sMAFs. NRF2-sMAF heterodimers are the predominant complexes that bind to the ARE and promote the transcription of detoxification genes.7–12 An overexpression of these elements could be responsible for acquired resistance to CDDP. In fact, we recently reported that miR-7 hypermethylation-derived deficiency mediates CDDP resistance and identified MAFG as a miR-7-direct target gene that is upregulated in CDDP-resistant tumor cells. Furthermore, miR-7 overexpression resulted in a repression of MAFG expression in lung and ovarian human cancer cells.13 Our previous results suggest a relevant role for MAFG conferring resistance to ROS-inducing compounds such as CDDP via the increased detoxification of free radicals. In addition, despite its important role in ROS detoxification, little is known about the involvement of MAFG in human diseases.
In the present study, we have studied in depth the role of the miR-7 and/or MAFG axis in acquired resistance to CDDP in lung cancer and explored MAFG as a possible diagnostic and therapeutic target by developing aptamers that show high affinity for the protein as a novel approach for chemotherapy resensitization. Aptamers are a recently developed alternative to specific biomolecule recognition; they are single-stranded nucleic acid molecules with high affinity for a particular target due to their ability to fold into tertiary shapes. These oligonucleotides are generated from an in vitro selection process and can be considered a novel alternative in therapy, diagnosis, and in the development of new biotechnological tools.14–16
MATERIALS AND METHODS
Cell cultures and treatments.
The human cancer cells H23 and A2780, and the HEK-293T epithelial cells were purchased from ATCC (Manassas, USA) and ECACC (Sigma, Spain) and cultured as recommended. The CDDP-resistant variants H23R and A2780R were established in our laboratory as previously described,13,17 using CDDP (Farma Ferrer, Spain) for cell viability assays. To validate the results obtained from the resistant cell lines established in our laboratory, we also used the CDDP-resistant lung cancer cell line H1299, with IC50 over 7, purchased from ATCC and maintained as recommended. Cell authentication is shown in Supplementary Table 3.
Clinical sample and data collection.
Fresh-frozen tumors (T) and adjacent-tumor tissue (ATT)-paired samples were obtained from 22 patients with non-small-cell lung cancer (NSCLC) from La Paz University Hospital. All patients had both a perioperative positron emission tomography-computed tomography scan showing localized disease and a pathological confirmation of stages after having undergone a complete resection for a histologically confirmed early NSCLC. Follow-up was conducted according to the criteria of the medical oncology division from the institution. In addition, five saliva samples from healthy donors were used as controls. All samples were collected after the approval of the appropriate Human Research Ethics Committee at the contributing center, including an informed consent within the context of research. Clinical, pathological, and therapeutic data were recorded by an independent observer, and a blind statistical analysis was performed on the data.
RNA isolation, RT-PCR, and quantitative RT-PCR.
Total RNA from surgical samples was isolated, reverse transcribed and quantitative real-time polymerase chain reaction (qRT-PCR) analysis was performed as previously described.17,18 Samples were analyzed in triplicate using the HT7900Real-Time PCR system (Applied Biosystems, USA). Relative expression levels were calculated according to the comparative threshold cycle method (2−ΔΔCt) using GAPDH as an endogenous control gene and RNU48 as endogenous control miRNA. Primers and probes for expression analysis were purchased from Applied Biosystems (MAFG:Hs 01034678_g1; GAPDH: Hs03929097_g1; miRNA-7: 000268; and RNU48: 001006).
For semiquantitative RT-PCR, 500 ng of total RNA isolated from cell lines was used for RT reaction using Prime-Script RT Master (Clontech-Takara, USA) and subsequent semiquantitative PCR using Promega PCR Mix (Promega, USA) as previously described.19 Relative quantification was performed by measuring the intensity of band amplified using ImageJ software. Primers for HMOX1 (F: 50'-TGAGTTTCAAGTATCCTTGTTGACAC-3'; R: 5'-CTT GGTCTAACTTTTGTGTGAAATAA-3'), MAFG (F: 5'-TCAGATTTCAGAGGAATACCCAGCAG-3'; R:5'-TG ATCACCAGTCAGAAGTGTACACAC-3'), and GAP DH (F: 5'-GAGAGACCCTCACTGCTG-3'; R: 5'GATGGTACATGACAAGGTGC-3') were designed tO include the probe from the array in order to assure the correct transcript identification.
DNA extraction, bisulfite modification, and quantitative methylation-specific PCR.
DNA from a total of 44 surgical samples from patients with NSCLC was isolated and bisulfite modified as previously described18 and was used to analyze miRNA-7 methylation status. For quantitative methylation-specific PCR, we used the primer and/or probe set to detect levels of either methylation (F: 5'-GGGTGGGGTTTTTTAAGAATC-3'; R: 50-ACATTCTCCTCCTTCGATCG-3'; Probe: 5'-FAM-ACCCCTCTTCGTTCTCGAT-3') or unmethy-lation (F: 5'-GGGGTGGGGTTTTTTAAGAATT-3'; R: 5'-ATAACATTCTCCTCCTTCAATCA-3'; Probe: 5'-VIC-ACCCCTCTTCATTCTCAAT-3'). All assays were performed in duplicate using the QuantiTect Multiplex PCR Kit (Qiagen, USA) and the HT7900 Applied Biosystems. The percentage of methylation of each sample was calculated according to previously published reports.20
Western blot analysis.
Proteins (30 μg) from cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Immobilon-P membranes (Millipore Ibérica SA, Madrid, Spain). Membranes were incubated with anti-Nrf2 at 1:10,000, anti-HO-1 at 1:10,000 (Abcam), and anti-β -actin at 1:100,000 (Sigma, Madrid, Spain). Peroxidase-conjugated secondary antibodies (1:10,000) were used to detect proteins by enhanced chemiluminescence detected by Advance Western-blotting Detection Kit (GE Healthcare, Barcelona, Spain).
In silico databases: The Cancer Genome Atlas and Total Cancer Care.
The Cancer Genome Atlas (TCGA) data: We obtained RNA sequencing data for the MAFG of 984 NSCLC tumors from the TCGA. The raw reads were quantified by RSEM21 in order to determine the read counts for each gene and miRNA (calculated separately). Then, we filtered out genes and miRNAs having less than one count-per-million reads in all samples. The normalization process was performed with trimmed mean of M values22 to obtain the MAFG sequence count data in all patients.
Total Cancer Care (TCC): We obtained MAFG gene expression data for 1035 lung cancer samples from the Moffitt Cancer Center Total Cancer Care Bioreposi-tory23 that were assayed on a custom Affymetrix 2.0 microarray. Normalized intensity values for MAFG probe sets were obtained and the probe with highest average intensity was retained for gene expression analysis.
ROS measurement.
H23 and/or A2780 cells were cultured in 96-well black plates at density of 10,000 cells/well. Cells were treated with 6 different doses of CDDP for 24, 48, and 72 hours. Then treatments were removed and cells were incubated with the fluorescent probe 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA) (10 μM) for 45 minutes in Roswell Park Memorial Institute medium (FBS-free). Cells were washed twice with Roswell Park Memorial Institute (10% FBS), and fluorescence was recorded in a Fluostar Optima at 520 nm after excitation at 485 nm. At the end of the experiment, solutions were replaced for fresh media containing MTT (0.5 mg/mL concentration) in order to determinate viability. ROS production was calculated dividing the mean H2DCFDA fluorescence by the mean viability. Data were normalized with respect to basal conditions that were considered as 100%.
Aptamer selection for MAFG.
Selection of DNA aptamers for MAFG was performed using the systematic evolution of ligands by exponential enrichment (SELEX) method with several modifications. First, MAFG-Myc overexpressed in HEK293T cells were bound to Myc-agarose beads (Thermo Scientific 20168), following the supplier’s instructions. The same amount of lysates from HEK293T cells transfected with empty vector was immunoprecipitated in parallel and used for the contraselection step. Synthetic random ssDNA (IBA Life Sciences, Germany), containing a central randomized region of 40 nucleotides flanked by two conserved 18-nucleotide regions in each end (RND40, 50-GCGGATGAAGACTGGTGT-40N-GCCCTAAATAC-GAGCAAC-30) was denatured at 95°C for 10 minutes and then cooled on ice for 10 minutes. Further steps of the selection were followed as previously described with minor modifications.16 Finally, contraselection prepared as above was performed after round 2 and 5.
Analysis of aptamer-MAFG complexes by RT-PCR: The aptamer populations obtained after 3 and 6 rounds of selection (SEL3MAFG and SEL6MAFG) or RND40 were incubated at 0.4 nM with 20 pmoles of MAFG bound to Myc-agarose for 1 hour at 37°C. In parallel, the same amount of each aptamer was incubated with the same volume of Myc-agarose resin without MAFG. After centrifugation at 12,000 g for 10 minutes, complexes were washed four times with 250 μL of selection buffer and confirmation the aptamers presence was performed as described.16
Enzyme-linked oligonucleotide assay (ELONA): Aptamers were labeled by PCR using 5' digoxigenin-labeled F3/5' digoxigenin-labeled R3 primers (IBA Life Sciences). To assess the enrichment of the selected population and the affinity of the individual aptamers for the target, MAFG-Myc (50 ng) was incubated with the rounds 3, 6 of aptamers or the RND40 library, as previously described with minor modifications.16 OD405nm values were determined using a SpectraFluor microplate reader (TECAN, Barcelona, Spain).
Aptacytochemistry: HEK293T cells were seeded on glass coverslips pretreated with poly-L-lysine (Sigma-Aldrich). After 16–24 hours, the cells were transfected with 0.4 μg of MAFG-Myc plasmid using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Twenty-four hours post-transfection, the cells were fixed with cold methanol for 20 minutes at −20°C and followed incubation with aptamer populations SEL3MAFG and SEL6-MAFG, as described previously.16 Colocalization was assessed by confocal microscopy using a Nikon ECLIPSE Ti-e inverted fluorescence microscope equipped with a Nikon C1 laser scanning confocal microscope system (Nikon, Tokyo, Japan) and a 60x oil immersion objective.
Aptamer cloning, sequencing, and secondary structure prediction: The dsDNA products with “A”-overhangs from SEL6MAFG were cloned onto pGEM-T Easy-cloning vector (Promega), following the manufacturer’s instructions. Individual clones were sequenced using T7 (5'-TAATACGACTCACTA-TAGGG-3') and Sp6 primers (5'-ATTTAGGTGA-CACTATAGAA-3') provided by the Sequencing core. Selected ssDNA molecules were subjected to secondary structure prediction using mFold software (http://mfold.rna.albany.edu/?q=mfold/DNA-Folding-Form) 41 at 37°C in 150 mmol/L [Na+] and 1 mmol/L [Mg+2].
Aptacytochemistry and aptahistochemistry with selected dig-labeled aptamers.
Following the methodology described above, H23R cells were incubated with 6 pmol of digoxigenin-labeled aptamers (IBA Life Sciences) in selection buffer for 1 hour at room temperature. Subsequently, cells were washed three times with PBS and incubated with anti-digoxigenin conjugated with 5' AlexaFluor 488 (R&D systems, USA) at 1/500 dilution in blocking buffer for 1 hour at room temperature, as secondary antibody. Finally, the cells were mounted on glass slides using glycerol-buffer containing p-phenylenediamine and 1/750 dilution of Dapi for nuclear staining. Controls were made by omitting the aptamer. Colocalization was assessed by confocal microcopy as described in (iii).
The formalin-fixed and paraffin-embedded tissue sections of patients with lung cancer were baked 15 minutes at 60°C. Deparaffinization was performed by two washes of xylene for 10 minutes each. Tissue sections were rehydrated by a sequential wash in 100%, 90%, 80%, and 70% ethanol and distilled water for 5 minutes each. Antigen retrieval was achieved by heat treatment in a pressure cooker for 2 minutes in 10 mM citrate buffer (pH 6.5). Endogenous peroxidase activity of tissues was blocked with 0.3% H2O2. The primary binding reaction was performed with 10 pmol/mL of digoxigenin-conjugated aptamers overnight at room temperature. The secondary binding reaction was performed with anti-digoxigenin conjugated with POD (Roche; 1/200 dilution in TBS) for 45 minutes. Staining was done using the Immunoperoxidase DAB kit (Master Diagnostica), according to the manufacturer’s instructions. The sections were counterstained with hematoxylin.
Cell transfection and viability assays.
HEK293 and H23S/R cells were seeded into 96-well plates then transfected with 5, 12.5, 25, 75, and 100 nM of MAFG aptamers or treated with PBS-Mg using Jet Pei Poly-Plus transfection reagent (PolyPlus Transfection, USA). The dose of 25 nM aptamer was afterward used in combination with increased doses of CDDP to assay viability and ROS production as described above. Viability to aptamers was assayed as previously described.24 To confirm the aptamer-transfection efficiency, cells were seeded in 24-well plates for aptamer isolation and confirmation of aptamer transfection by quantitative analysis as previously described.16 A Myc-DDK-tagged ORF clone of MAFG the negative control pCMV6 were used for in transient transfection (OriGene, USA) in combination with MAFG-aptamers following the same procedure.
Statistical analysis.
The data were compared using the Chi-squared test or Fisher’s exact test for qualitative variables, and Student’s t test or the Wilcoxon-Mann-Whitney test (non-normal distribution) for quantitative variables. Correlation of quantitative variables was analyzed by Pearson’s test. For the in silico databases, the data were stratified for patients with high or low expression of MAFG according to the median of the gene expression (TCGA cutoff: 2018 counts per million; TCC cutoff: 8.70 of intensity probe). Overall survival was estimated according to the Kaplan-Meier method and compared between groups by means of the Log Rank test. All the P values were two sided, and the type I error was set at 5%. Statistical analyses were performed using SPSS 20 software.
RESULTS
MAFG overexpression is associated with a poor prognosis in patients with non—small-cell lung cancer.
Previously published studies from our group showed that the epigenetic regulation of miR-7 through DNA methylation is a mechanism involved in platinum resistance in cancer cell lines directly regulating the action of MAFG, which is overexpressed in resistant pheno-types.13 In the present study, we explored the involvement of the miR-7 and/or MAFG axis in NSCLC primary tumors. We first analyzed the quantitative DNA methylation levels of miR-7 and RNA expression levels of miR-7 and MAFG in a cohort of 22 paired samples from fresh-frozen T and ATT from patients with NSCLC (Table I) and controls.
Table I.
Clinicopathological and experimental data obtained from patients with NSCLC from La Paz University Hospital
| miR-7 methylation | miR-7 expression | MAFG expression | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Histology | Sex | Stage | Chemotherapy | ATT (%) | T (%) | ATT (2−ΔCt) | T (2−ΔCt) | ATT (2−ΔCt) | T (2−ΔCt) | apMAFG3F (% of PC) | apMAFG6F (% of PC) | apMAFG11F (% of PC) | Status | OS, days |
| 1 | Adenocarcinoma | Female | IA | No | 58.49 | NA | 0.50 | 0.19 | 0.62 | 0.43 | 70.81 | NA | 79.26 | Alive | 2220 |
| 2 | Epidermoid | Male | IB | No | 5.63 | 5.35 | 1.71 | 2.61 | 0.65 | 0.53 | 84.14 | 90.93 | 27.83 | Exitus | 1022 |
| 3 | Adenocarcinoma | Male | IB | No | 14.98 | 13.12 | 1.16 | 0.31 | 0.59 | 0.28 | NA | 77.11 | NA | Exitus | 3 |
| 4 | Adenocarcinoma | Male | NA | No | 12.47 | 26.06 | 1.11 | 0.51 | 0.43 | 4.37 | 90.61 | 91.08 | 87.18 | Exitus | 626 |
| 5 | Large Cell | Male | IIB | No | 9.20 | 12.69 | 0.80 | 0.34 | 0.28 | 0.29 | 44.69 | 25.55 | 21.72 | Exitus | 62 |
| 6 | Adenocarcinoma | Male | IIIA | Other | 7.28 | 3.74 | 2.33 | 0.38 | 0.31 | 2.12 | 84.77 | 79.60 | 40.41 | Exitus | 228 |
| 7 | Epidermoid | Female | IIIB | CDDP + Other | 8.47 | 5.69 | 1.65 | 1.77 | 0.58 | 0.05 | 67.48 | 87.21 | 22.58 | Exitus | 109 |
| 8 | Adenocarcinoma | Female | IIA | CDDP + Other | 8.37 | 28.94 | 0.70 | 2.42 | 0.91 | 0.13 | 77.15 | 79.55 | 59.50 | Alive | 2260 |
| 9 | Epidermoid | Male | IB | No | 9.41 | 8.09 | 1.07 | 1.17 | 5.59 | 0.29 | 74.31 | 73.12 | NA | Alive | 1853 |
| 10 | Adenocarcinoma | Male | IA | No | 8.19 | 38.79 | 1.00 | 0.09 | 1.00 | 0.04 | 75.39 | 69.14 | 74.06 | Exitus | 216 |
| 11 | Adenocarcinoma | Female | IIIA | CBDCA + Other | 6.51 | 6.08 | 0.99 | 5.83 | 1.91 | 0.20 | 53.02 | 35.66 | 68.15 | Alive | 2192 |
| 12 | Epidermoid | Male | IB | CDDP + Other | 7.59 | 30.28 | 0.92 | 0.75 | 2.24 | 0.81 | 77.27 | 92.45 | NA | Alive | 2341 |
| 13 | Epidermoid | Male | IIA | No | 12.44 | NA | 0.77 | 1.51 | 0.70 | 0.09 | 88.89 | 84.07 | 75.52 | Exitus | 289 |
| 14 | Epidermoid | Male | IIA | No | 12.66 | 10.98 | 2.21 | 2.07 | 0.73 | 0.84 | 90.76 | 96.88 | NA | NA | 109 |
| 15 | Adenocarcinoma | Female | IIIA | CDDP + Other | 4.72 | 3.57 | 0.73 | 0.32 | 0.76 | 0.49 | NA | NA | NA | Alive | 2228 |
| 16 | Adenocarcinoma | Male | IIB | Other | 35.35 | 10.00 | 0.80 | 0.31 | 1.03 | 0.78 | 75.77 | 95.65 | 94.51 | Exitus | 888 |
| 17 | Epidermoid | Male | IIB | CBDCA + Other | 8.24 | 53.19 | 1.35 | 1.05 | 0.57 | 0.03 | 74.61 | 28.90 | 44.30 | Exitus | 259 |
| 18 | Adenocarcinoma | Female | IB | CDDP + Other | 10.29 | 9.42 | 1.19 | 2.86 | 0.38 | 1.52 | 95.72 | 93.75 | 87.23 | Exitus | 936 |
| 19 | Epidermoid | Male | IIB | CDDP + Other | 13.67 | 9.52 | 1.50 | 2.87 | 0.99 | 0.50 | NA | NA | NA | Exitus | 1224 |
| 20 | Epidermoid | Male | IIIB | CBDCA + Other | 4.06 | 13.45 | 0.80 | 1.33 | 0.22 | 0.43 | 85.99 | NA | 2.24 | Exitus | 603 |
| 21 | Adenocarcinoma | Male | IIIA | CDDP + Other | 9.55 | 63.07 | 1.14 | 0.65 | 1.11 | 2.57 | NA | 67.21 | 71.08 | NA | 421 |
| 22 | Adenocarcinoma | Female | IIB | CDDP + Other | 4.43 | 6.84 | 0.50 | 2.07 | 0.26 | 2.35 | 80.30 | 92.55 | 78.10 | NA | 184 |
Abbreviations: ATT: adjacent tumor tissue; CBDCA: carboplatin; CDDP: cisplatin; NA: not available; NSCLC: non-small cell lung cancer; OS, Overall Survival; % of PC: % of Positive Cells; T: tumor.
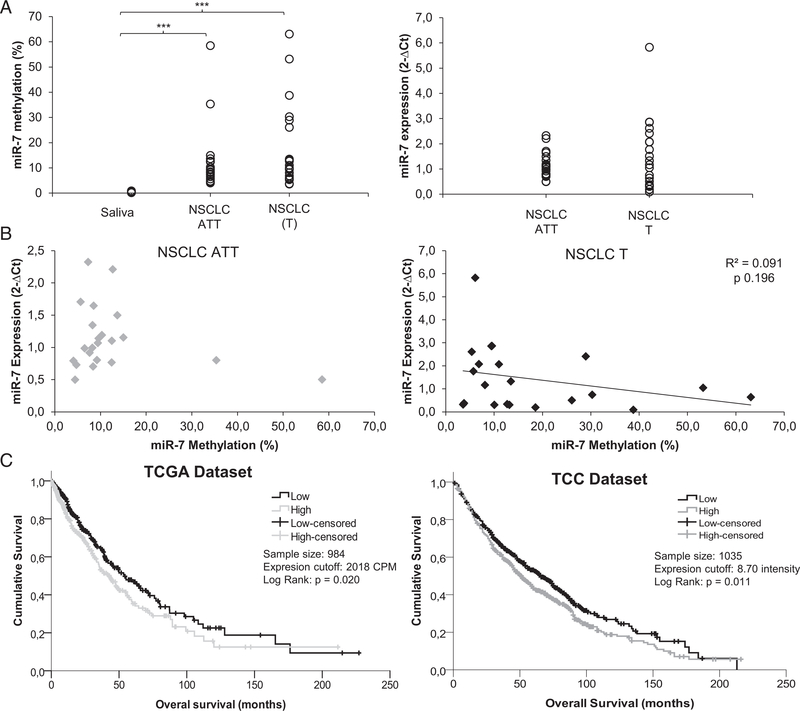
We observed an increase in the data dispersion in the NSCLC T samples versus the ATTs in miR-7 methylation and expression levels (Supplementary Table 1). The higher dispersion was statistically significant in both groups only when compared with saliva samples from healthy donors (P =0.0002;Fig 1A and Supplementary Table 1). A negative tendency in the correlation between the percentage of methylation and expression levels of miR-7 was found only in the T samples (Fig 1B). The opposite tendency was found, toward more expression of MAFG in T samples, when the percentage of miR-7 methylation increased (Supplementary Figure 1). Neither tendency was statistically significant, probably due to the limited sample numbers; however, it is interesting to note that the two samples that showed the highest dispersion for MAFG expression presented a miR-7 methylation level over 20%.
Fig 1.
Involvement of miR-7 and MAFG in patients with NSCLC. (A-B) Assessment of miR-7 methylation levels measured by qMSP and expression levels of miR-7 measured by qRT-PCR in fresh samples from a cohort of 22 patients with NSCLC; (A) Left, quantitative methylation percentage of miR-7; right, miR-7 expression levels in 44 paired samples (ATT and T); (B) Correlation between the methylation percentage of miR-7 and expression levels of miR-7. For all the analyses, data represent the percentage of methylation according to a previous report20 and expression levels in 2−ΔCt; (C) Analysis of mRNA expression of MAFG in two in silico cohorts of patients with NSCLC. Survival analysis in 984 (left panel) and 1035 (right panel) NSCLC samples from the TCGA and TCC, respectively. LogRank test was used for comparisons and P < 0.05 was considered as a significant change in OS. NSCLC, non-small cell lung cancer; ATT, adjacent tumor tissue; T, tumor; CPM, counts per million; TCGA, The Cancer Genome Atlas; TCC, Total Cancer Care. *** P < 0.001
To determine whether MAFG expression correlated with clinical outcome in patients with NSCLC, we investigated the gene expression levels in 984 patients with NSCLC from the TCGA database and 1035 patients with NSCLC from the Total Cancer Care Bio-repository at the Moffitt Cancer Center. When comparing data from both data sets, we observed statistical significance according to the median level of MAFG expression; patients with low expression levels had a clearly increased overall survival compared with the group of patients with high MAFG expression levels, with P values of 0.020 and 0.011 for both cohorts, respectively (Fig 1C).
MAFG overexpression might induce CDDP resistance, targeting ROS.
MAFG is a transcription factor involved in the detoxification of ROS, whose expression is increased in the resistant cell phenotypes H23R and A2780R, which mediates their resistance to CDDP.13 Thus, we explored whether MAFG influenced oxidative stress in our experimental model of paired sensitive and/or resistant cells by analyzing ROS production in cells after CDDP exposure.
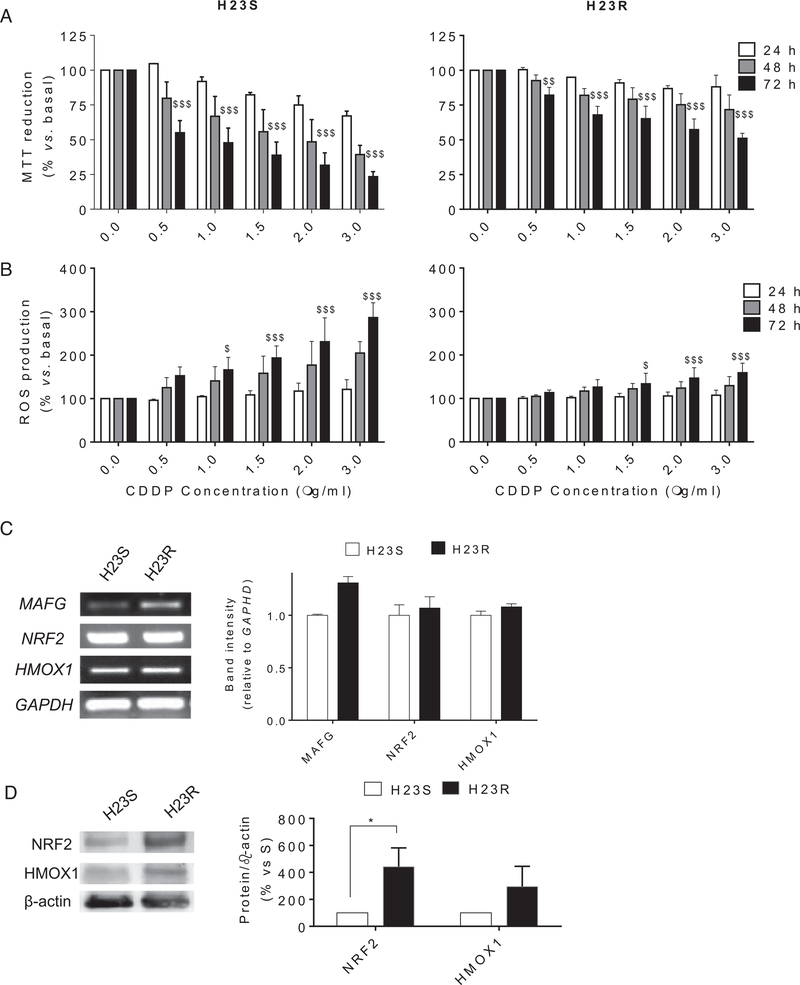
First, we confirmed the response to CDDP at 24, 48, and 72 hours after exposure to the drug for both cell lines H23S and H23R. H23R cells present a resistance index >3, in accordance with our previous results25 (Fig 2A). ROS levels were increased in sensitive cells after CDDP treatment compared with resistant cells, reaching 300% and 159% ROS production at 3.00 μg/mL CDDP, respectively, versus basal untreated cells, P < 0.001 (Fig 2B). The response to CDDP and the differences in ROS production between sensitive and resistant cells were also confirmed in A2780S/R ovarian cancer cell lines (Supplementary Figure 2A), in which we observed an effect of CDDP treatment after 48 hour of exposure, consistent with previous reports.13 Moreover, we found that the expression of ROS-detoxifying genes, such as HMOX1, NQO1, GSTO2, or GPX7 (Supplementary Table 2), was upregulated in these resistant cell lines (P < 0.05), according to our reported arrays under the same previously defined bioinformatics and statistical criteria13 (GEO GSE84201). We selected HMOX1 and NRF2 for semiquantitative RT-PCR expression pattern validation, because of their close relationship with MAFG action in the detoxification process. NRF2 expression did not changed after CDDP treatment; however, the resistant cells showed higher protein levels of NRF2, and it was also observed a slight increase in HMOX1 levels (Fig 2C, D and Supplementary Figure 2B). In addition, we observed a direct relationship between the expression of MAFG and the Resistance Index in all our lung cancer cell lines (Supplementary Figure 2C).
Fig 2.
Viability assays and reactive oxygen species detection after CDDP treatment in H23 cell lines. (A) Viability to CDDP in H23S (left) and H23R (right) at 24, 48, and 72 hours and 6 concentrations of CDDP measured by MTT assay; (B) Increment of ROS production in H23S (left) and H23R (right) after 24, 48, and 72 hours of CDDP treatment; For (A) and (B), bars represent the mean of at least two independent experiments measured by duplicate ± SD. $: P ≤ 0.05 versus basal; $$: P ≤ 0.01 versus basal; $$$: P ≤ 0.001 were considered as significant change in CDDP resistance and ROS production; (C) Semiquantitative mRNA expression analysis of downstream genes involved in ROS detoxification regulated by MAFG, which showed expression changes for H23 in the microarray data (GSE84201,13) (left panel). Representative images of MAFG and HMOX1 RT-PCR comparing sensitive and resistant subtypes. Each assay was performed at least three times to confirm the results. (Right panel) Relative intensity quantification of the amplified band for each gene measured by ImageJ Software. Bars represent the mean of the three independent experiments using the intensity of GAPDH as endogenous control and the sensitive subtype of each cell line as calibrator. (D) Immunoblots of HMOX1, NRF2, and -actin. Images correspond to a representative experiment that was repeated four times with similar results. Scion Image program was used to quantify band intensities corresponding to immunoblot detection of protein samples. CDDP, cisplatin; MAFG, musculoaponeurotic fibrosarcoma oncogene family, protein G.
Identification of aptamers binding MAFG.
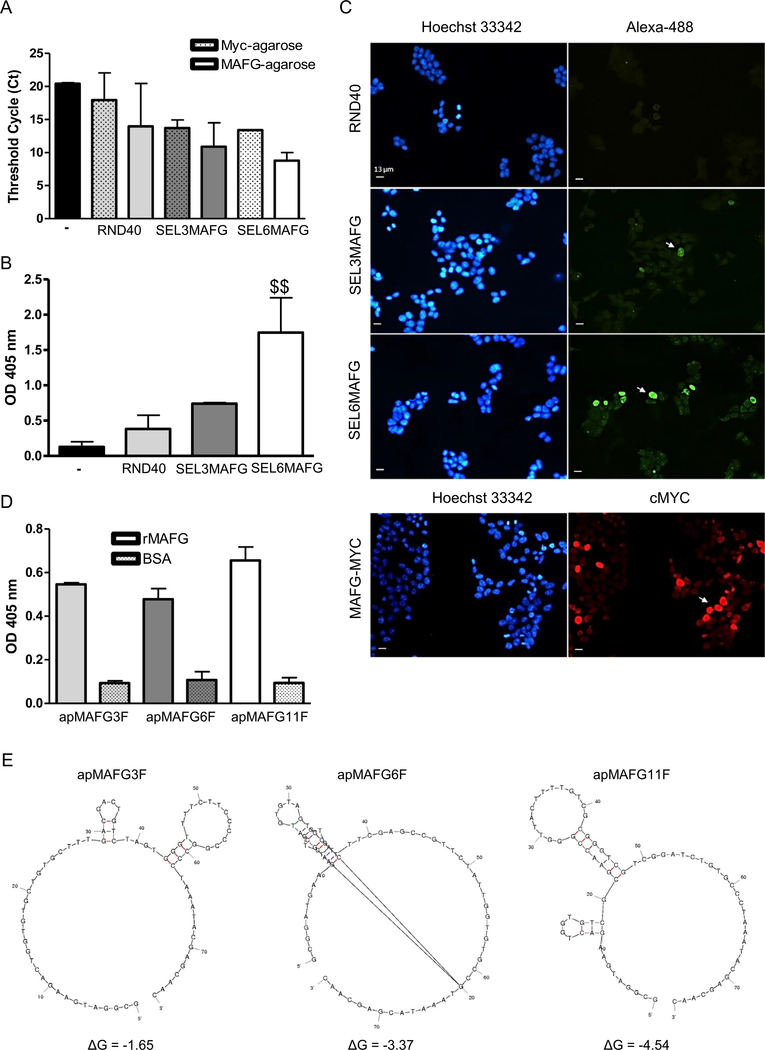
Aptamers were selected from libraries of oligonucleotides by iterative cycles of selection (SELEX methodology). We performed 6 rounds of selection and 2 contraselection rounds (after rounds 2 and 5), using a Myc agarose resin that binds the MAFG-Myc protein overexpressed in HEK293T cells. To assess the enrichment and the affinity of the population obtained after successive rounds of selection, we performed three types of assays: (i) analysis of aptamer-MAFG complexes by RT-PCR, (ii) ELONA assays, and (iii) aptacytochemistry. Quantitative PCR assays were performed using aptamers that bind to MAFG-resin complexes as a template. These assays allowed us to confirm that the selected aptamers were specific against the target and not the Myc-agarose resin. The amount of aptamers recovered in the presence of MAFG was 3 Cts lower than in that of the RND40 and SEL3MAFG population, whereas SEL6MAFG was 4.5 Cts lower in the presence of MAFG (Fig 3A), indicating a higher amount of aptamers bound in this round. In addition, ELONA assays showed a statistically significant increase in the signal of the population obtained after round 6 relative to round 3 or to the initial RND40 population (Fig 3B). Finally, we studied the capacity of the aptamer population of each round to recognize overexpressed MAFG in the cells by aptacytochemistry. For this purpose, HEK293T cells transiently transfected with MAFG-Myc were incubated with the initial RND40 population or with that of round 3 or round 6, labeled with Alexa-488. The results obtained by fluorescence microscopy indicated that RND40 showed a very low signal with a diffuse cytoplasmic pattern, whereas SEL3MAFG and most prominently SEL6MAFG aptamers showed a high nuclear signal in several cells. This pattern was very similar to that obtained by incubation with the Myc antibody (Fig 3C). In view of these results, we cloned and sequenced the SEL6MAFG population and identified three sequences (apMAFG3F, apMAFG6F, and apMAFG11F) with high binding capacity to the target in the nanomolar range, reaching values of four times that of the BSA protein (Fig 3D and Supplementary Table 4). To predict the most stable secondary structures of the MAFG aptamers, we performed a bio-informatic analysis of their sequences using mFold software. Fig 3E illustrates the most probable secondary structures of these aptamers, taking into account the lower free energy (ΔG).
Fig 3.
Analysis of the affinity of the aptamers against MAFG. (A) Quantification of aptamer-MAFG complexes by qPCR comparing absence (dot pattern) or presence (filled pattern) of MAFG protein. Figure shows the differences in Threshold Cycle (Ct) between the library RND40 and the selection rounds 3 (SEL3MAFG) and 6 (SEL6MAFG). Bars represent the mean ± SEM of 2–3 experiments; (B) Specificity binding assay of aptamers to MAFG performed by ELONA. MAFG-Myc was plated at 50 ng/well in 96-well plates followed by incubation with 5' digoxigenin-labeled aptamers from RND40, SEL3MAFG, and SEL6MAFG. The figure represents the mean ± SEM of three independent experiments. $$: P < 0.01; (C) Aptacytochemistry in HEK-293T cells transiently transfected with MAFG-Myc. Cells overexpressing MAFG were incubated with 5' Alexafluor 488-conju-gated aptamers from RND40, SEL3MAFG, or SEL6MAFG (upper panel) or Myc antibody (lower panel). Fluorescence microscopy images corresponding to the staining of nuclei with Hoechst (blue), antibodies (red), and aptamers (green) are shown. Arrowhead indicates positive cells. Bar = 13 μm; (D) Study of individual aptamer affinity performed by ELONA. Recombinant MAFG was plated at 50 ng/well in 96-well plates followed by incubation with 5' digoxigenin-labeled individual aptamers apMAGF3F, apMAFG6F, and apMAFG11F. The figure represents the mean ± SEM of two independent experiments; (E) Secondary structures of aptamers apMAGF3F, apMAFG6F, and apMAFG11F obtained by bioinformatics sequence analysis using the mFold software taking into account the lower free energy (ΔG). MAFG, musculoaponeurotic fibrosarcoma oncogene family, protein G; ELONA, enzyme-linked oligonucleotide assay.
MAFG aptamers can be used to detect MAFG levels in NSCLC samples.
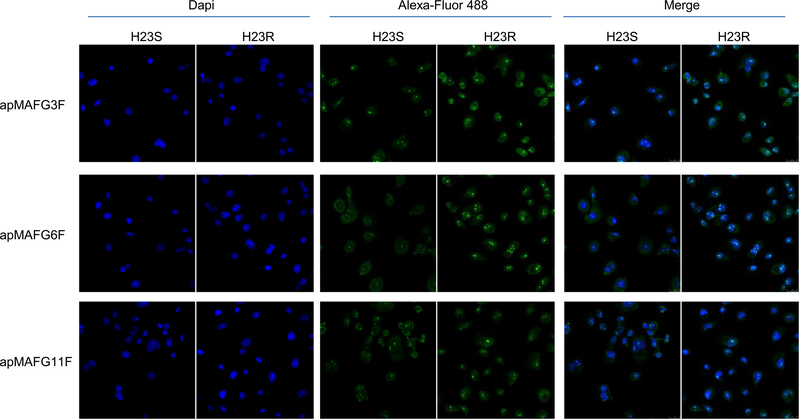
We next performed aptacytochemistry on H23S and H23R cells with the individual digoxigenin-labeled aptamers apMAFG3F, apMAFG6F, and apMAFG11F. We observed that individual aptamers were more present in H23R cells than in H23S, mostly in the nuclei of the cells and specifically accumulated in quantifiable dots (Fig 4), according to the cellular localization of MAFG. Quantification of the number of dots per nucleus showed remarkable differences between H23R and H23S for apMAFG3F and apMAFG6F, whereas no differences were observed for apMAFG11F (Supplementary Figure 3). Interestingly, apMAFG3F was also present in the cytoplasm, whereas apMAFG6F was preferentially located at the nuclei.
Fig 4.
MAFG localization pattern in H23S and H23R cells measured by aptacytochemistry. Cells were incubated using digoxigenin-labeled aptamers apMAFG3F, apMAFG6F, and apMAFG11F as primary recognition molecule and Alexafluor 488-conjugated anti-digoxigenin as secondary antibody. Confocal microscopy images corresponding to the staining of nuclei with Dapi (blue), aptamers (green) and merge are shown. Bar = 25 μm. MAFG, musculoaponeurotic fibrosarcoma oncogene family, protein G.
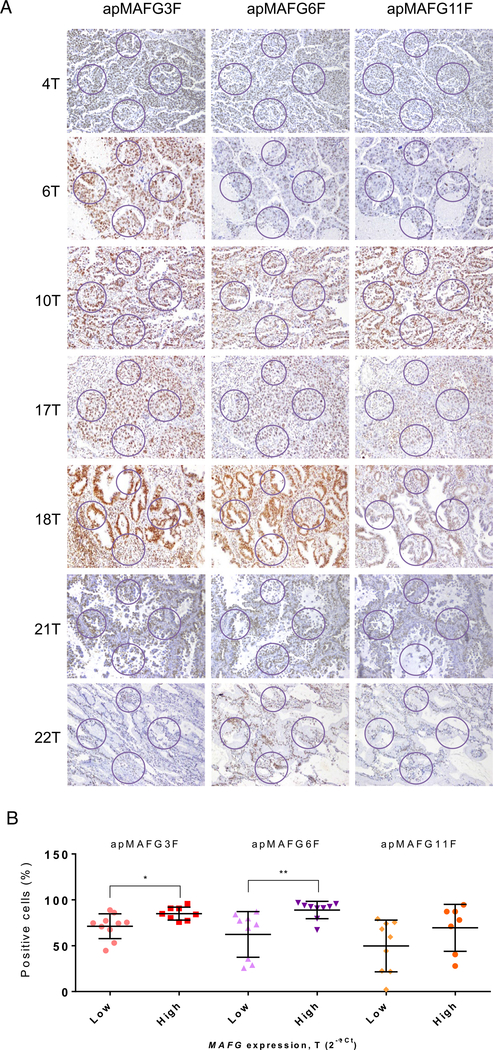
We then took advantage of the aptacytochemistry to perform an aptamer-based histochemistry in 20 tumor samples from our cohort of patients with known mRNA levels of MAFG. We found that aptamers were able to identify MAFG in the nucleus of various cell types with differential staining intensity between the tumors (Fig 5A). To determine the correlation between the aptahistochemistry and the mRNA levels of MAFG, we calculated the percentage of cells with positive staining from the total number of cells computed at four different areas randomly selected for each sample. We observed that classifying the patients according to their MAFG expression levels, those samples with higher MAFG levels presented a significant increment in the percentage of positive cells after aptahistochemistry staining with both apMAFG3F and apMAFG6F (P < 0.05 and P < 0.01, T test, respectively; Fig 5B).
Fig 5.
Analysis of MAFG localization pattern in paraffin samples of patients with known levels of MAFG by aptahistochemistry. Samples were incubated with digoxigenin-labeled aptamers apMAFG3F, apMAFG6F, and apMAFG11F as primary recognition molecules, and afterward with anti-digoxigenin antibodies. Staining was performed using DAB staining solution. (A) Representative aptahistochemistry pictures showing the areas of analysis in NSCLC tumor samples; (B) T test comparison between low and high MAFG mRNA levels (2−ΔCt) and the % mean ratio of positive cells analyzed in the areas defined on (A). Positive cells were counted using the Cell Counter on ImageJ software. NSCLC: non-small cell lung cancer. MAFG, musculoaponeurotic fibrosarcoma oncogene family, protein G
apMAFG6F increases ROS production and restores cell sensitivity to CDDP.
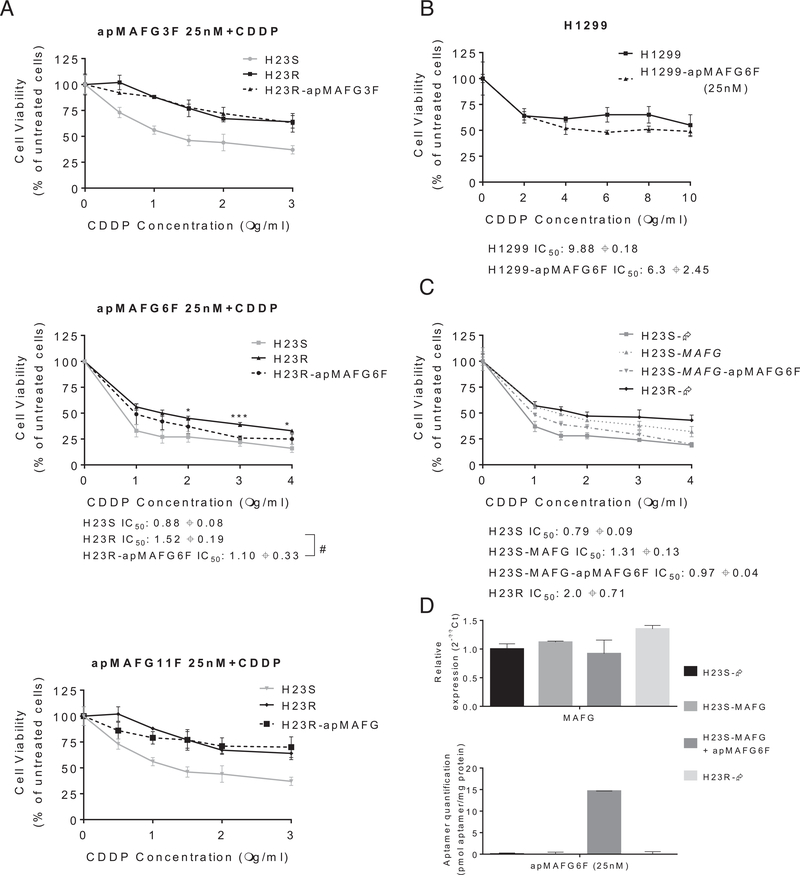
To gain insight into the biological effect of the identified aptamers and to optimize the dose for a functional analysis, we worked with the normal cell line HEK293T and the sensitive and resistant phenotypes of the lung cancer cell lines H23S/R. MAFG aptamer transfection induced similar mortality curves in the cancer cell lines, while it appeared to induce less toxicity in HEK293T, which showed better viability curves for the three aptamers tested (Supplementary Figure 4A). Concentrations higher than 25 nM induced a high mortality rate in all the cells tested, and the same effect was also observed when two intermediate doses selected between the 25 nM and 75 nM concentrations were tested in H23R (Supplementary Figure 4B). The 25 nM concentration was chosen for functional analysis because it is the minimal dose that exerts an effect on viability of tumor cells but not normal cells. The efficiency of aptamer transfection at this dose was confirmed by quantitative PCR (Supplementary Figure 4C). We next tested the specificity of the aptamers for rescuing the sensitivity to CDDP. Aptamer apMAFG6F decreased the CDDP resistance at 25 nM (resistance index 2.63 vs 2.0), whereas no changes were observed for aptamers apMAFG3F or apMAFG11F (Fig 6A). We confirmed these results in H1299, an additional lung cancer tumor cell line highly resistant to CDDP with an IC50 of 10 mg/mL (Fig 6B). In fact, we verified the ability of apMAFG6F to increase CDDP sensitivity in H1299, as we observed a decrease in the IC50 to 4.5 μg/mL platinum in comparison with the untransfected cell line. This increase in sensitivity to the drug was accompanied by a decrease in MAFG and HMOX1 expression in the cells carrying apMAFG6F (Supplementary Figure 4D). In addition, we overexpressed MAFG in H23S cells as we have done previously13 and combine the overexpression with apMAFG6F, which rescued the phenotype and resensitized the cells (Fig 6C). We also confirmed the success of the overexpression and aptamer transfection by qRT-PCR (Fig 6D).
Fig 6.
Effect of aptamers apMAFG3F, apMAFG6F, and apMAFG11F on cell viability, response to CDDP. (A) Viability curves combining the selected dose of aptamers (25 nM) and increasing doses of CDDP in H23S/R cells. #P < 0.05 (Student’s t test). The treatment with aptamer significantly increased the sensitivity to CDDP (* P < 0.05 and ***< 0.001); (B) Viability curves combining apMAFG6F at 25 nM with increasing doses of CDDP in H1299 lung cancer cells. (C) Viability curves of H23 cell line transfected with pCMV6 (S-Ø and R-), with the overexpression vectors (S-MAFG) and the combination of MAFG overexpressing plasmids together with apMAFG6F at 25 nM. For (A), (B), and (C), each experimental group was exposed for 72 h to 6 different CDDP concentrations after aptamer transfection; data were normalized to each untreated control, set to 100% and represent the mean ± SD of at least three independent experiments performed in quadruplicate at each drug concentration for each cell line analyzed. The CDDP-RI (Resistant Index to CDDP) was calculated as “IC50 from the Resistant/ IC50 from the Sensitive" and “IC50 from the transfected with aptamers or MAFG/ IC50 from the Sensitive” ± SD. (D) Validation of MAFG overexpression and aptamer transfection in the H23S cells after 72 h of transfection. Top, Relative expression levels of MAFG measured by quantitative RT-PCR, in the cell line H23, represented in 2−ΔΔCt scale; In each experimental group, the sensitive cell line transfected with pCMV6 plasmid was used as a calibrator. Each bar represents the combined relative expression of two independent experiments measured in triplicate. Bottom, quantification of apMAFG6F in the cell line H23. Quantification was performed according to the absolute quantification method by using a standard curve of known concentration of aptamers. The amount of protein per sample was used as a control. Data represent the mean of at least three independent experiments measured by triplicate ± SD. CDDP, cisplatin; RT-PCR, real-time polymerase chain reaction.
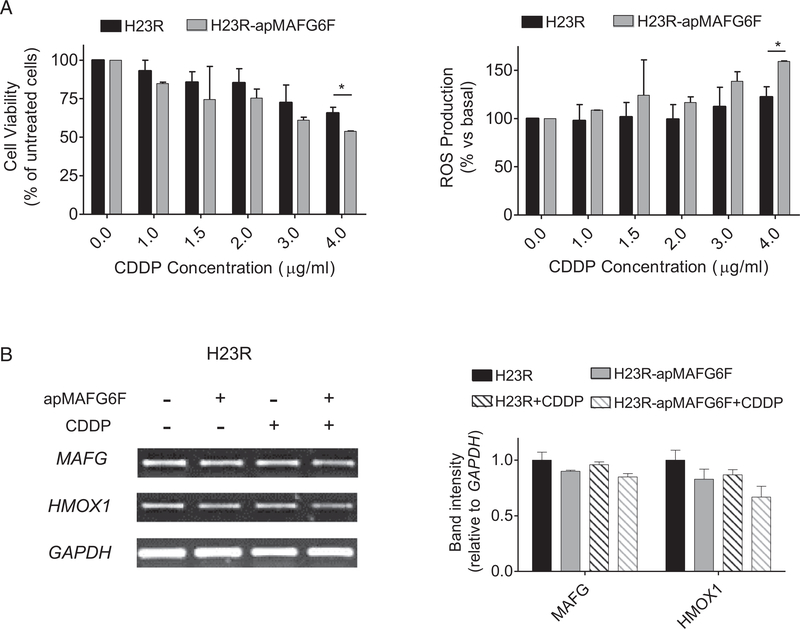
To ultimately confirm that apMAFG6F was restoring CDDP sensitivity in H23R cells by acting on the ROS, we measured ROS production after combining aptamer transfection and CDDP treatment. We observed a linear dose-response relationship between increasing amounts of CDDP and the production of ROS at 4 μg/mL of CDDP (P < 0.005) in the resistant cells carrying the apMAFG6F compared with the untransfected cells. This result was also consistent with the decrease in cell viability that we also measured in parallel in this experiment (Fig 7A). Semiquantitative RT-PCR of MAFG and HMOX1 showed a decrease in their expression in aptamer-transfected H23R cells after platinum treatment compared with H23R (Fig 7B).
Fig 7.
Effect of aptamer apMAFG6F on response to CDDP and ROS production. (A) Viability and ROS production assay in H23R cells after apMAFG6F transfection. Each experimental group was exposed for 72 h to 6 different CDDP concentrations, and data were normalized to each untreated control, set to 100%. The data represent the mean ± SD of at least four independent experiments performed in duplicate at each drug concentration for each cell line analyzed. * P < 0.05 was considered as significant change in CDDP resistance and ROS production; (B) Expression analysis of downstream genes regulated by MAFG involved in ROS detoxification in H23R and H23R transfected with apMAFG6F. Left, representative images of MAFG, HMOX1, and GAPDH RT-PCR. Each assay was performed at least three times to confirm the results. Right, relative intensity quantification of the amplified band for each gene measured by ImageJ Software. Bars represent the mean of three independent experiments using the intensity of GAPDH as endogenous control and the resistant subtype of each cell line as calibrator ± SD. CDDP, cisplatin; ROS, reactive oxygen species; MAFG, musculoaponeurotic fibrosarcoma oncogene family, protein G.
DISCUSSION
The study of epigenetic, transcriptional, and posttranscriptional regulatory mechanisms of genes and miRNAs will help to elucidate key players involved in resistance to chemotherapy drugs. Current treatments may be improved and new ones developed based on the genetic and epigenetic profiles of various cancer patients, thus leading to personalized therapies. The search for new biomarkers for survival, prognosis, and drug-resistance in cancer is a wide-open field for exploration and investigation. In fact, despite significant scientific effort in recent decades, we have not yet overcome the strategic challenge of restoring sensitivity to platinum-derived drugs that are still the paradigm treatment for many malignancies, including lung cancer.
For NSCLC, differential expressions have been reported for several miRNAs, such as miR-200c in NSCLC cell lines,26 or miR-138 in the A549 CDDP-resistant subtype cell line, in comparison with the parental A549 and how their enhanced expression increased the response to CDDP.27 However, there are very few studies linking the role of miRNA methylation with tumor development or clinical outcomes such as miR-34a.28,29 We have previously reported that hypermethylation of regulatory regions of miR-7s may predict relapse in platinum-treated patients with ovarian cancer. We intended first to deepen our understanding of this aspect in a cohort of 22 paired samples from patients with NSCLC. We found similar methylation levels of miR-7 in tumor samples and in normal adjacent tissues, and no association of methylation levels with therapy response or overall survival. We observed, however, a higher dispersion of methylation and expression of miR-7 and MAFG in the T than in the ATT and in the control samples, suggesting that different molecular changes in miR-7 and MAFG could be mediating the carcinogenic process, as has been observed for other markers.30 We have not specifically observed clear differences either in methylation or expression of any candidate in NSCLC T versus ATT samples, which could indicate that the samples adjacent to the tumor have already acquired molecular changes probably associated with tumor features as has been described for other miRNAs.31
Moreover, the methylation analysis of the ATT compared with the five normal saliva samples indicates that the methylation of this miRNA is an event that occurs in the early stages of the disease. This fact suggests that molecular changes such as miR-7 methylation within the tumor tissue and surrounding tissue generate the microenvironment necessary for tumor cells to grow, and this event could be involved not only in tumorigenesis but also in other malignancies associated with smoking.32,33 We could not find a clear correlation in either methylation or miR-7 and MAFG expressions in NSCLC T samples. Although these results are not as compelling as previously observed with our in vitro models (ref theranostics), we cannot dismiss the potential coregulation of epigenetic factors that can interact with miR-7 and MAFG, like lncRNAs that regulate miR-7 (CDR1AS), and MAFG expression (MAF-GAS1) in a more complex dynamic environment like the human body. A strength of the current study is that the results are based on an analysis of fresh tissues from patients with NSCLC, whereas most of the data in the literature report the expression of miRNAs and candidate genes from culture cell lines. In accordance with our “in house” results, we observed a significant relationship between the expression of MAFG and survival in our additional large cohort of 1035 patients analyzed from Moffitt Center and in the public database with 984 from TCGA with a clear trend toward higher overall survival when MAFG expression is low. This suggests an oncogenic role of MAFG in lung cancer, supporting its potential as a prognostic biomarker.
The mechanistic approach we used in this study was aimed at directly linking the development of in vitro CDDP resistance with the overexpression of MAFG through the decrease of ROS production in our model of cancer cell lines. It has previously been shown that the resistance to CDDP in the A2780 cell line is an event caused by overactivation of the redox-detoxifying path-way.34 In line with these, the results obtained with the ROS production assay in our experimental models indicate that CDDP-resistant subtypes have a lesser increase in ROS after CDDP exposure, probably due to its ability to detoxify the oxidative stress produced by the drug, as a consequence of an overexpression of MAFG. W ealso found a significant overexpression of reported redox detoxifying-related gene35 in the resistant cell lines harboring an increased expression of MAFG,rein-forcing the idea that MAFG mediates the resistance to CDDP through the modulation of ROS. Indeed, our results also indicate that MAFG could regulate this process in response to CDDP by increasing NRF2 stabilization in the nucleus. Therefore, targeting MAFG may lower NRF2 activity and potentially restore the sensitivity to the drug.
We then sought to explore new methodologies to detect and inhibit the action of MAFG in order to resensitize cancer cells to CDDP. Aptamers constitute a novel field of research, which has led to important achievements in therapeutics and diagnosis.14,36 Therefore, in this study, we aimed to select and characterize aptamers that are highly specific against MAFG and that could show potential therapeutic and/or diagnostic properties in cancer, as has been shown for other aptamers.15,37 The SELEX technology allowed us to select and identify three aptamers with high affinity to MAFG after 6 rounds of selection. Although the three selected aptamers (apMAFG3F, apMAFG6F, and apMAFG11F) showed high affinity to the protein with Kd values in the nanomolar range, apMAFG3F and apMAFG6F showed the lower Kd and the highest signal in resistant cells in aptacytochemistry. The fact that apMAFG6F staining remains preferentially at the nuclei could indicate that it is, in essence, more specific for nuclear MAFG. In addition, we observed a greater presence of the three aptamers in H23R cells, suggesting greater expression of the protein MAFG in resistant cells; indeed, this result is in accordance with the high mRNA levels of MAFG observed in the resistant cancer cells.13 We were also able to detect MAFG in human formalin-fixed paraffin-embedded tumor samples with known levels of MAFG mRNA. Specifically, we found the strongest difference between low and high expression levels of MAFG when using apMAFG6F staining in the group of samples analyzed. These results, although preliminary because of the limited number of samples tested, are promising, indicating that these aptamers, particularly apMAFG6F, could be used as potential diagnostic tools to identify patients with higher expression of MAFG. It is a robust starting point for future studies involving larger cohorts of patients to analyze the correlation between aptamer intensity and patients’ clinical outcome.
As expected, the combination of aptamers apMAFG3F, apMAFG6F, and apMAFG11F and CDDP treatment in resistant lung cancer cells showed that only apMAFG6F can change the response to CDDP, partially restoring the sensitivity to the drug, without causing cell mortality in normal cells. In fact, even when MAFG was overexpressed, inducing an increase of platinum resistance in H23 sensitive cells, apMAFG6F was able to revert the resistance to platinum. This resensitization was promoted by an increase in ROS production, probably mediated by a decrease in the expression of antioxidant genes such as HMOX1, as previously shown.38–40 We also observed a decrease in MAFG expression levels when transfecting apMAFGF6, which suggests an auto-regulation of MAFG transcription and is consistent with the fact that MAFG shows ARE motifs on its regulatory region.10,41 Therefore, our results present a new mechanism for restoring CDDP sensitivity in lung cancer through the use of aptamer apMAFG6F.
Taken together, our experimental results strongly support the role of MAFG in the development of CDDP resistance through ROS detoxification. To the best of our knowledge, this is the first report describing the possible role of MAFG expression as a biomarker of poor prognosis in NSCLC and its potential as a therapeutic and diagnostic target in cancer and other pathologies through the use of aptamers.
Supplementary Material
ATA GLANCE COMMENTARY.
Vera-Puente O et al.
Background
We have recently reported that microRNA-7 is silenced by DNA methylation and is involved in the resistance to platinum in cancer cells through the action of the musculoaponeurotic fibrosarcoma oncogene family, protein G(MAFG).
Translational Significance
In the present manuscript, we study the molecular mechanism underlying cisplatin (CDDP)-resis-tance mediated by MAFG in human cancer cell lines by analyzing the reactive oxygen species production after CDDP treatment. We have also identified highly-specific aptamers against MAFG that modulate its activity. The final intention of our work is to provide new tools for the diagnosis and therapeutic treatment of CDDP-resistant tumors by targeting MAFG with aptamers.
ACKNOWLEDGMENTS
The authors thank Hayley Pickett ServingMed.com for the English language correction. The authors also acknowledge Biobank from HULP for sample processing.
Conflicts of Interest: All the authors have read the journal’s authorship statement and have no conflicts of interest to declare. The information provided in this study is included in a patent application process (EP17382610.8) and therefore it must be treated, solely and exclusively, based on the purposes of this paper, and should not be published if it does not respond to the purpose thereof. This application and its contents are protected by the Spanish Law on Intellectual and Industrial Property, prohibiting the distribution, reproduction, disclosure, transformation and sale of the entire document or part thereof, as well as the use, under any circumstances, of the trademarks appearing therein, without the prior express written consent of the Foundation for Biomedical Research of La Paz University Hospital-IdiPAZ (FIBHULP), which holds the ownership.
Financial support: This study was supported by the “Fondo de Investigación Sanitaria-Institutode Salud Carlos III” [PI15/00186 and CP 08/000689 to I.I.C.]; and the European Regional Development Fund/European Social Fund FIS [FEDER/FSE, Una Manera de Hacer Europa]. MINECO funds support O.V., C.R.A. and O.P. contracts through RTC-2015-4362-1 and RTC-2016-5314-1 projects.
Abbreviations
- ATT
adjacent tumor tissues
- CDDP
cisplatin
- MAFG
musculoaponeurotic fibrosarcoma oncogene family, protein G
- miR-7
microRNA-7
- NSCLC
non small-cell lung cancer
- qMSP
quantitative methylation-specific PCR
- qRT-PCR
quantitative real-time PCR
- ROS
reactive oxygen species
- TCC
Moffitt’s Total Cancer Care
- TCGA
The Cancer Genome Atlas
Footnotes
Supplementary Data
Supplementary data associated with this article can be found online at https://doi.org/10.1016/j.trsl.2018.06.005.
REFERENCES
- 1.Haslehurst AM, Koti M, Dharsee M, et al. EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer 2012;12:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karaca B, Atmaca H, Bozkurt E, et al. Combination of AT-101/ cisplatin overcomes chemoresistance by inducing apoptosis and modulating epigenetics in human ovarian cancer cells. Mol Biol Rep.40:3925–33. [DOI] [PubMed] [Google Scholar]
- 3.Deavall DG, Martin EA, Horner JM, Roberts R. Drug-induced oxidative stress and toxicity. J Toxicol 2012;2012:645460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryter SW, Kim HP, Hoetzel A, et al. Mechanisms of cell death in oxidative stress. Antioxid Redox Signal 2007;9:49–89. [DOI] [PubMed] [Google Scholar]
- 5.Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene 2012;31:1869–83. [DOI] [PubMed] [Google Scholar]
- 6.Kilic U, Kilic E, Tuzcu Z, et al. Melatonin suppresses cisplatin-induced nephrotoxicity via activation of Nrf-2/HO-1 pathway. Nutr Metab (Lond).10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katsuoka F, Motohashi H, Engel JD, Yamamoto M. Nrf2 transcriptionally activates the mafG gene through an antioxidant response element. J Biol Chem 2005;280:4483–90. [DOI] [PubMed] [Google Scholar]
- 8.Motohashi H, Katsuoka F, Miyoshi C, et al. MafG sumoylation is required for active transcriptional repression. Mol Cell Biol 2006;26:4652–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Yu S, Liu T, et al. Heterodimerization with small Maf proteins enhances nuclear retention of Nrf2 via masking the NESzip motif. Biochim Biophys Acta 2008;1783:1847–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirotsu Y, Katsuoka F, Funayama R, et al. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res 2012;40:10228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Aguiar Vallim TQ, Tarling EJ, Ahn H, et al. MAFG is a transcriptional repressor of bile acid synthesis and metabolism. Cell metabolism 2015;21:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katsuoka F, Yamamoto M. Small Maf proteins (MafF, MafG, MafK): history, structure and function. Gene 2016;586:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vera O, Jimenez J, Pernia O, et al. DNA Methylation of miR-7 is a Mechanism Involved in Platinum Response through MAFG Overexpression in Cancer Cells. Theranostics 2017;7:4118–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakravarthy U, Adamis AP, Cunningham ET Jr., et al. Year 2 efficacy results of 2 randomized controlled clinical trials of pegaptanib for neovascular age-related macular degeneration. Ophthalmology 2006;113:e1–25. [DOI] [PubMed] [Google Scholar]
- 15.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Experimental and molecular pathology 2009;86:151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Recio EM, Pinto-Diez C, Perez-Morgado MI, et al. Char-acterization of MNK1b DNA aptamers that inhibit proliferation in MDA-MB231 breast cancer cells. Molecular therapy Nucleic acids 2016;5:e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibanez de Caceres I, Cortes-Sempere M, Moratilla C, et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene.29:1681–90. [DOI] [PubMed] [Google Scholar]
- 18.Ibanez de Caceres I, Dulaimi E, Hoffman AM, Al-Saleem T, Uzzo RG, Cairns P. Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer Res 2006;66:5021–8. [DOI] [PubMed] [Google Scholar]
- 19.Vera O, Rodriguez-Antolin C, de Castro J, Karreth FA, Sellers TA, de Caceres II. An epigenomic approach to identifying differential overlapping and cis-acting lncRNAs in cisplatin-resistant cancer cells. Epigenetics 2018: 01–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000;28:E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome biol-ogy 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer journal 2011;17:528–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chattopadhyay S, Machado-Pinilla R, Manguan-Garcia C, et al. MKP1/CL100 controls tumor growth and sensitivity to cisplatin in non-small-cell lung cancer. Oncogene 2006;25:3335–45. [DOI] [PubMed] [Google Scholar]
- 25.Ibanez de Caceres I, Cortes-Sempere M, Moratilla C, et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene 2010;29:1681–90. [DOI] [PubMed] [Google Scholar]
- 26.Ceppi P, Mudduluru G, Kumarswamy R, et al. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res.8:1207–16. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Xiang S, Williams KA, et al. Depletion of HDAC6 enhances cisplatin-induced DNA damage and apoptosis in non-small cell lung cancer cells. PLoS One.7:e44265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallardo E, Navarro A, Vinolas N, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 2009;30:1903–9. [DOI] [PubMed] [Google Scholar]
- 29.Pronina IV, Loginov VI, Burdennyy AM, et al. DNA methylation contributes to deregulation of 12 cancer-associated micro-RNAs and breast cancer progression. Gene.604:1–8. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt M, Hellwig B, Hammad S, et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin kappa C as a compatible prognostic marker in human solid tumors. Clin Cancer Res.18:2695–703. [DOI] [PubMed] [Google Scholar]
- 31.Jusufovic E, Rijavec M, Keser D, et al. let-7b and miR-126 are down-regulated in tumor tissue and correlate with microvessel density and survival outcomes in non small cell lung cancer. PLoS One.7:e45577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostrow KL, Michailidi C, Guerrero-Preston R, et al. Cigarette smoke induces methylation of the tumor suppressor gene NISCH. Epigenetics.8:383–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guida F, Sandanger TM, Castagne R, et al. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum Mol Genet 2015;24:2349–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao LJ, Jaramillo MC, Zhang ZB, et al. Nrf2 induces cisplatin resistance through activation of autophagy in ovarian carcinoma. Int J Clin Exp Pathol.7:1502–13. [PMC free article] [PubMed] [Google Scholar]
- 35.Chen XL, Kunsch C. Induction of cytoprotective genes through Nrf2/antioxidant response element pathway: a new therapeutic approach for the treatment of inflammatory diseases. Current Pharmaceutical Design 2004;10:879–91. [DOI] [PubMed] [Google Scholar]
- 36.Yao C, Qi Y, Zhao Y, Xiang Y, Chen Q, Fu W. Aptamer-based piezoelectric quartz crystal microbalance biosensor array for the quantification of IgE. Biosensors and Bioelectronics 2009;24:2499–503. [DOI] [PubMed] [Google Scholar]
- 37.Mongelard F, Bouvet P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Current Opinion in Molecular Therapeutics 2010;12:107–14. [PubMed] [Google Scholar]
- 38.Cui Y, She K, Tian D, Zhang P, Xin X. miR-146a inhibits proliferation and enhances chemosensitivity in epithelial ovarian cancer via reduction of SOD2. Oncology Research 2016;23:275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HR, Kim S, Kim EJ, et al. Suppression of Nrf2-driven heme oxygenase-1 enhances the chemosensitivity of lung cancer A549 cells toward cisplatin. Lung Cancer 2008;60:47–56. [DOI] [PubMed] [Google Scholar]
- 40.Wu S, Zhang T, Du J. Ursolic acid sensitizes cisplatin-resistant HepG2/DDP cells to cisplatin via inhibiting Nrf2/ARE pathway. Drug Design, Development and Therapy 2016;10:3471–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warnatz HJ, Schmidt D, Manke T, et al. The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J Biol Chem 2011;286:23521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.