Abstract
Chronic myelomonocytic leukemia (CMML) is a clinically heterogeneous neoplasm in which JAK2 inhibition has demonstrated reductions in inflammatory cytokines and promising clinical activity. We hypothesize that annotation of inflammatory cytokines may uncover mutation-independent cytokine subsets associated with novel CMML prognostic features. A Luminex cytokine profiling assay was utilized to profile cryopreserved peripheral blood plasma from 215 CMML cases from three academic centers, along with center-specific, age-matched plasma controls. Significant differences were observed between CMML patients and healthy controls in 23 out of 45 cytokines including increased cytokine levels in IL-8, IP-10, IL-1RA, TNF-α, IL-6, MCP-1/CCL2, hepatocyte growth factor (HGF), M-CSF, VEGF, IL-4, and IL-2RA. Cytokine associations were identified with clinical and genetic features, and Euclidian cluster analysis identified three distinct cluster groups associated with important clinical and genetic features in CMML. CMML patients with decreased IL-10 expression had a poor overall survival when compared to CMML patients with elevated expression of IL-10 (P = 0.017), even when adjusted for ASXL1 mutation and other prognostic features. Incorporating IL-10 with the Mayo Molecular Model statistically improved the prognostic ability of the model. These established cytokines, such as IL-10, as prognostically relevant and represent the first comprehensive study exploring the clinical implications of the CMML inflammatory state.
Introduction
Chronic myelomonocytic leukemia (CMML) is a hematopoietic neoplasm with features of both myelodysplastic (MDS) and myeloproliferative (MPN) diseases hallmarked by a peripheral blood monocytosis (≥1 × 109/L) that constitutes greater than 10% of white blood cells [1]. CMML is often accompanied by cytopenias and/or the presence of constitutional symptoms such as fatigue, weight loss, night sweats, symptoms from organomegaly (early satiety or abdominal pain) [1]. Previous French-American-British (FAB) classifications separate CMML based on white blood cell count into MPN-CMML and MDS-CMML [2]. World Health Organization (WHO) classifications separate CMML based on bone marrow blast count in three prognostically relevant groups [1]. However, these classification systems do not fully exemplify the clinical heterogeneity observed in CMML because many clinically relevant symptoms such as constitutional symptoms, splenomegaly, and peripheral cytopenias are not captured. Although recurrently mutated genes in CMML have been rigorously annotated, the spectrum of mutations is homogeneous relative to other adult cancers and also does not reflect the clinical diversity seen in patients with CMML [3–6].
To address the wide clinical heterogeneity, many investigators have devised prognostic scoring systems that predict overall and leukemia-free survival [7–11]. We have demonstrated that, although valid and reproducible, these prognostic scores have only modest predictive capacity and are vulnerable to up-staging via competing models [12]. Numerous studies have also explored the prognostic capacity of somatic gene mutations in CMML [3, 13, 14]. Despite several independent studies, only ASXL1 frameshift or nonsense mutations have demonstrated to be independently and reproducibly associated with survival [3, 15, 16]. This has led to at least three new prognostic scoring systems that incorporate ASXL1 and clinical features [3, 12, 15–18]. Given these efforts, we reason that identifying non-mutational molecular characteristics may further refine prognostication in CMML.
We have demonstrated that ruxolitinib, a JAK1/2 inhibitor FDA approved for treatment of primary myelofibrosis (PMF), is an effective therapeutic in CMML [19, 20]. Similar to that seen in MPNs, CMML patients treated with ruxolitinib were found to have marked reduction in circulating inflammatory cytokines and improvement in constitutional symptoms and spleen volume [19, 21, 22]. Previous literature investigating JAK inhibition in MPNs has demonstrated that inflammatory cytokines are reduced or normalized after treatment correlating with symptom improvement [19, 22, 23]. This suggests that inflammatory cytokines may associate with clinical features not captured by current models or genetic mutations. However, the spectrum of cytokine secretion in CMML has not been previously comprehensively evaluated in the literature.
To understand the non-mutational molecular correlates driving CMML clinical diversity, we comprehensively profiled the inflammatory secretome in CMML patients. We hypothesize that annotation of inflammatory cytokines may uncover cytokine subsets associated with CMML-specific clinical and genetic features and refine existing prognostic models.
Methods
Patient samples and controls
The study was approved by the Institutional Review Board (IRB) for each respective institution including Moffitt Cancer Center (Tampa, Florida), Institute Gustave-Roussy (Paris, France), and the University of Florence (Florence, Italy). All patients provided informed written consent for the collection of peripheral blood and bone marrow samples and required a confirmed WHO diagnosis of CMML [1]. Age-matched, center-specific controls (n = 35) were also obtained under institute-specific IRB-approved protocols. Fifteen single donor healthy control plasma samples were purchased from Innovative Research (Innovative Research, Inc., Novi, MI) with an average age 66 (range 60–80) and a total of nine men and six women. These 15 samples from Innovative Research, Inc. were used as comparison for samples from Moffitt Cancer Center and University of Florence. All healthy control plasma samples were obtained at an FDA licensed commercial donor center or facility within the United States and each sample was tested and found negative for Hepatitis B, Hepatitis C, HIV, and syphilis as well as negative for active diagnosis of cancer. Twenty single donor healthy control plasma samples were obtained by the Institute Gustave-Roussy as comparison for their samples. Pretreatment plasma was isolated from whole blood and bone marrow samples obtained from patients at each individual institution (Fig. 1).
Fig. 1.
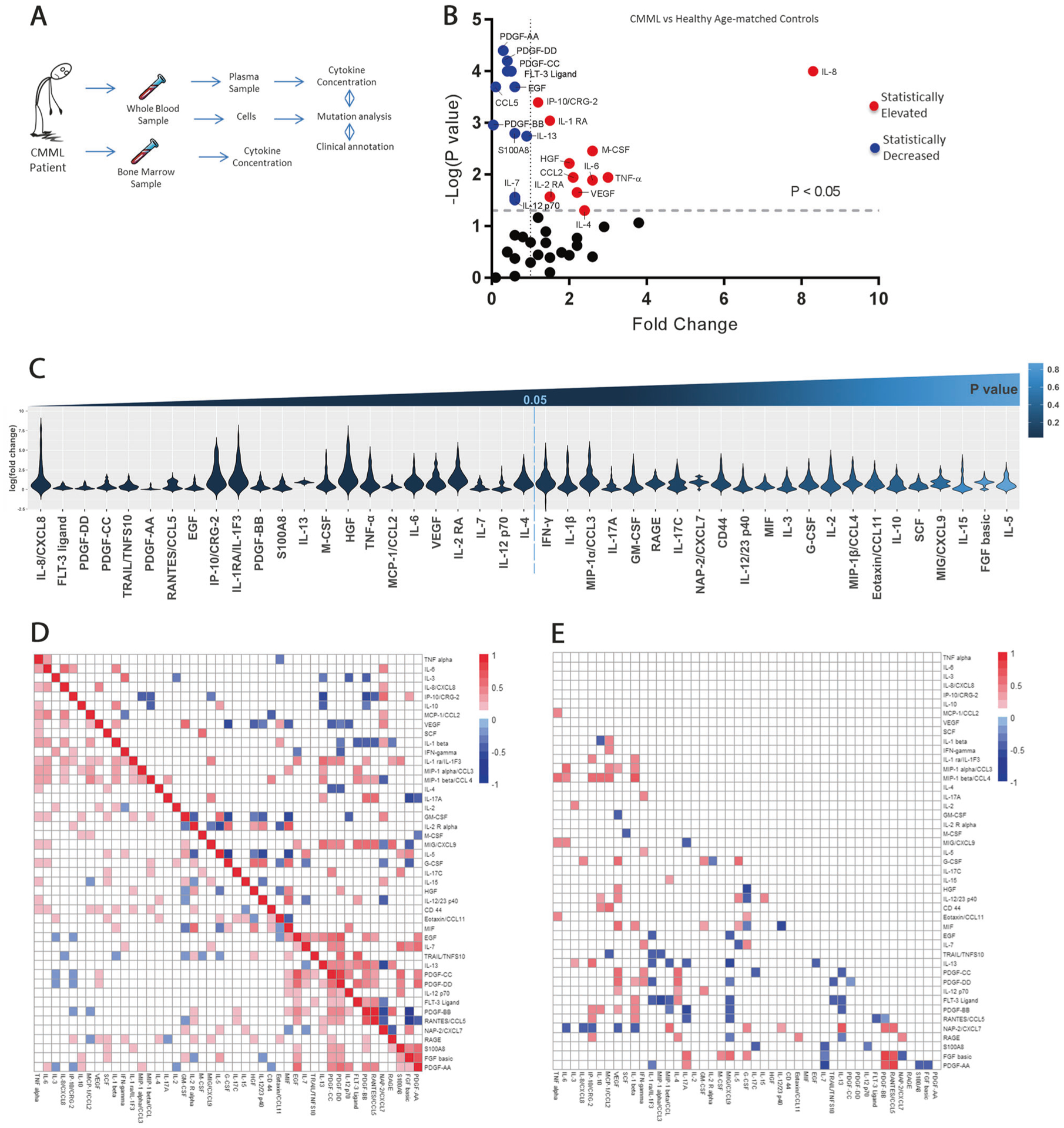
Inflammatory cytokines are heterogeneously expressed in CMML patients. a Study design. b Significant differences observed in measured inflammatory cytokines in patients with CMML compared to healthy aged-matched controls. Y-axis is negative log(P-value). X-axis is fold change of CMML compared to healthy aged-matched controls. c Violin plot displaying the variable distribution of cytokine expression for each cytokine in CMML patients. On the y-axis is the log of the fold change for CMML patients. On the x-axis is each of the 45 cytokines evaluated. Cytokines to left of vertical dotted line were statistically significant when comparing expression in CMML patients to normal controls (P < 0.05). d Pearson correlation displaying cytokine–cytokine interaction in 45 cytokines measured in CMML patients. Top figure displays positive and negative cytokine interactions in normal patients and bottom section of figure displays cytokine interactions in CMML patients. Blue notes negative R coefficient correlation while red notes positive R coefficient correlation. e The difference of correlation between CMML patients and normal patients. The colored squares represent all statistically significant interactions. The more blue present in the square denotes more negative R coefficient correlation between two cytokines while the more red in the square denotes more positive R coefficient correlation. Interactions between cytokines that were not statistically significant are shaded white
Cytokine secretion profiling
Cytokine expression levels were measured, in duplicate, from plasma samples using a custom 45-plex Luminex-based inflammatory cytokine screening assay kit and Luminex 100™ according to the manufacturer’s protocol (R&D Systems, Minneapolis, MN). Forty-five cytokines (Supplemental Table 1) were analyzed, which included 19 cytokines previously implicated in the pathogenesis of myeloid neoplasms [24]. Cytokine measurements for the CMML patients were normalized to the site-specific healthy control for each batch and a fold change was calculated for each cytokine measured for every CMML patient and healthy control enrolled in the study. Cytokine expression levels with a fold change above 1 in the CMML patients were deemed elevated compared to healthy normal controls, and cytokine expression levels with a fold change less than 1 in the CMML patients were deemed to be decreased compared to healthy normal controls.
Somatic mutation profiling
Gene mutation testing was achieved via specific, in-house, targeted next-generation sequencing platform. Both full gene and hotspot sequencing were generated on the following genes: TET2, ASXL1, SRSF2, CSF3R, KRAS, NRAS CBL, DNMT3A, SF3B1, EZH2, JAK2, IDH1/2, FLT3, SETBP1, U2AF1, and ZRSR2. All variants reported were manually reviewed and validated to be variants of known significance as previously described [25].
Colony formation assay and gene expression analysis
Colony formation assays from cryopreserved primary CMML bone marrow mononuclear cells were conducted in duplicate using Methocult substrate (Stemcell, Vancouver, Canada) as previously described by our group in the presence or absence of human recombinant interleukin-10 (IL-10) or IL-8 (Stemcell) at a concentration of 10 μg/mL [26]. Cryopreserved primary CMML bone marrow mononuclear cells were thawed from seven CMML patients enrolled in our study for the assay. Colonies were counted using the StemVision automated colony-counting instrument and software (Stemcell). A paired T-test was used to compare colony output between patient samples. Multiplex gene expression analysis was performed using Nanostring nCounter PanCancer Pathway Panel and nSolver™ Analysis software (NanoString Technologies, Inc., Seattle, WA) on four CMML patients enrolled in our study.
Statistical methods
Cytokine expression levels were derived using Luminex software MasterPlex 2010 (MiraiBio Group of Hitachi Solutions America, Ltd). All analyses involving cytokines were completed after applying a log-transformation to reduce the impact of outliers. Principal component analysis (PCA) was completed with the first two principal components (PCs) plots to investigate potential batch effects. Cytokine expression levels were adjusted for batch effect (i.e., day the sample was run) using an ANOVA model for each cytokine (see Supplemental Methods). PCA demonstrated significant batch effect that was corrected by using an ANOVA model that considered the timing of experiments (Supplemental Fig. 1). Comparison of peripheral blood cytokines in CMML patients to each respective healthy age-matched and center-matched control was completed using Wilcoxon rank sum test. Cytokine levels in the peripheral blood of 20 CMML patients were compared to matched bone marrow samples using Pearson’s correlation. Log-rank tests were used to determine if individual cytokines (high/low) or clusters of cytokines were independently associated with overall survival (OS). Cox proportional hazards models were used to incorporate clinical and gene variables into a multivariate survival model. Model development was completed by first including any clinical, genetic, or cytokine variable with P < 0.05 into an initial model, followed by backward elimination to remove variables with P > 0.05 from the final model. Proportional hazards assumptions were confirmed with Schoenfeld residual plots. Cluster analysis was completed on the log-transformed batch-adjusted cytokine data with Gaussian model-based clustering using the R package mclust [27, 28] and Recursively Partitioned Mixture Model with R package RPMM [29]. Determination of difference in OS between the clusters was completed using log-rank tests. Statistical analysis was completed in collaboration with the Department of Biostatistics and Bioinformatics at Moffitt Cancer Center.
Results
Patient characteristics
A total of 215 patients with a diagnosis of CMML and median age of 74 (range 29–91, 65% males) had diagnostic pretreatment peripheral blood plasma samples available for the study. Of the 215 CMML patients with available clinical and genetic information, 105 (61% of evaluable patients) patients were classified in the CMML-0 subgroup, 43 (25% of evaluable patients) patients in the CMML-1, 25 (14% of evaluable patients) patients in the CMML-2 subgroup by 2016 WHO classification(1), and 42 patients did not have clinical data available (Table 1). Overall, 122 (68% of evaluable patients) CMML patients had a hemoglobin level greater than 10 g/dL (median 11.2, range 5.9–16.7 g/dL), 58 (32% of evaluable patients) patients with hemoglobin less than 10 g/dL, and 35 patients did not have clinical data. TET2 was the most common mutation seen in our cohort with 87 patients (58% of evaluated patients) with mutated TET2. Our patient demographics and clinical characteristics are summarized in Table 1 and Supplemental Table 2.
Table 1.
Patient demographics and clinical characteristics
| Characteristics | Median (range) CMML | CMML (%) (N = 215) | Control (N = 35) |
|---|---|---|---|
| Age, years | 74 (29–91) | 66 (60–80) | |
| <65 | 32 (14.9%) | 4 (11.4%) | |
| ≥65 | 176 (81.8%) | 11 (31.4) | |
| Data not available | 7 (3.3%) | 20 (57.1%) | |
| Gender | |||
| Male | 139 (64.7%) | 9 (25.7%) | |
| Female | 76 (35.3%) | 6 (17.1) | |
| Data not available | 20 (57.1%) | ||
| WHO subtype | |||
| CMML-0 | 105 (48.9%) | ||
| CMML-1 | 43 (20%) | ||
| CMML-2 | 25 (11.6%) | ||
| Data not available | 42 (19.5%) | ||
| Hemoglobin level, g/dL | 11.2 (5.9–16.7) | ||
| <10g/dL | 58 (27.0%) | ||
| ≥10 g/dL | 122 (56.7%) | ||
| Data not available | 35 (16.3%) | ||
| Leukocyte count, ×109/L | 19.2 (1.0–235) | ||
| FAB-MDS | 107 (49.8%) | ||
| FAB-MPN | 83 (38.6%) | ||
| Data not available | 25 (11.6%) | ||
| Platelet count, ×109/uL | 101 (8.0–812.0) | ||
| <100,000/uL | 89 (41.4%) | ||
| ≥100,000/uL | 90 (41.9%) | ||
| Data not available | 36 (16.7%) | ||
| Blasts in BM, % | |||
| <10% | 148 (68.8%) | ||
| ≥10% | 25 (11.6%) | ||
| Data not available | 42 (19.5%) | ||
| Presence splenomeg; | |||
| Yes | 61 (28.4%) | ||
| No | 111 (51.6%) | ||
| Data not available | 43 (20.0%) | ||
| Presence of circulating immature cells | |||
| Yes | 88 (40.9%) | ||
| No | 74 (34.4%) | ||
| Data not available | 53 (24.7%) | ||
| ASXL1 status | |||
| Mutated | 49 (22.8%) | ||
| Wild type | 121 (56.3%) | ||
| Data not available | 45 (20.9%) | ||
| TET2 status | |||
| Mutation | 87 (40.5%) | ||
| Wild type | 62 (28.8%) | ||
| Data not available | 66 (30.7%) |
Concordance between bone marrow and peripheral blood samples
Because our study focused on annotating peripheral blood cytokine levels, we explored the concordance between peripheral blood and bone marrow plasma samples in 20 CMML patients with blood and bone marrow obtained concomitantly. When all data were aggregated across patients, we found a positive concordance between bone marrow plasma samples and peripheral blood samples of the majority of the cytokines evaluated (r = 0.7891, P < 0.0001, 95% CI 0.592–0.8971), suggesting that peripheral blood is a reasonable representation of the bone marrow cytokine milieu. However, certain cytokines appeared to be discordant suggesting enrichment or de-enrichment in the bone marrow compared to peripheral blood. These discordant cytokines included Granulocyte-macrophage colony-stimulating factor (GM-CSF) (r = −0.1395), vascular endothelial growth factor (VEGF) (r = −0.1415), and tumor necrosis factor-α (TNF-α) (r = −0.1561) (Supplemental Fig. 2).
Cytokine expression in CMML patients compared to matched controls
We evaluated the peripheral inflammatory secretome of CMML by annotating 45 chemokine/cytokine levels and compared these levels to age- and center-matched normal controls (n = 35) (Fig. 1a, b; Supplemental Table 3). After data normalization and quality control, significant differences in 23 of 46 cytokine levels between CMML and healthy controls were measured. We observed significantly downregulated FLT-3 ligand (fold change (FC) = 0.4), PDGF-AA (FC = 0.3), PDGF-BB (FC = 0.04), PDGF-CC (FC = 0.5), PDGF-DD (FC = 0.4), EGF (FC = 0.6), TRAIL/TNFS10 (FC = 0.5), and CCL5/RANTES (FC = 0.1) when comparing CMML patients to healthy age-matched controls (P ≤ 0.001). IL-8 (FC = 8.3), IP-10 (FC = 1.2), and IL-1RA (FC = 1.5) were found to be significantly elevated in CMML patients compared to healthy age-matched controls (P ≤ 0.001) (Fig. 1b, Supplemental Table 3). Proinflammatory cytokines including TNF-α (FC = 3, P = 0.011), IL-6 (FC = 2.6, P = 0.0129), MCP- 1/CCL2 (FC = 2.1, P = 0.0114), macrophage colony-stimulating factor (M-CSF) (FC = 2.6, P = 0.0035), HGF (FC = 2, P = 0.0061), VEGF (FC = 2.2, P = 0.0223), IL-4 (FC = 2.4, P = 0.0499), and IL-2RA (FC = 1.5, P = 0.0269) were found to be significantly elevated in CMML patients. We observed significant CMML cytokine/chemokine heterogeneity within our cohort, suggesting that annotating the secretome may reveal relevant disease subsets. We therefore reasoned that cytokines/chemokines that were not observed to be statistically up- or downregulated should be considered in our downstream analysis (Fig. 1c).
Inflammatory cytokine signals are an established feature of stress-induced hematopoiesis and the secreted inflammatory response [30]. We hypothesized that CMML patients may exhibit distinct cytokine expression patterns due to underlying somatic mutations. To evaluate this, we used Pearson’s correlation to determine pairwise correlations between cytokines measured in CMML and normal age-matched controls, respectively (Fig. 1d, e, Supplemental Tables 4–6). Cytokine associations were identified in CMML patients not seen in normal controls between IL-1β, IL-6, IL-8, and TNF-α previously identified in the literature in other proinflammatory diseases such as osteoarthritis [31–35]. Only in normal control patients, GM-CSF was positively correlated with IL-5 (r = 0.577, P = 0.0002) and CXCL7 (r = 0.432, P = 0.009), but negatively correlated with IL-2RA (r = −0.449, P = 0.006), HGF (r = −0.341, P = 0.044), and MIF (r = −0.628, P < 0.0001). A statistically significant, positive correlation was also observed between IL-10 and GM-CSF (r = 0.151, P = 0.025), M-CSF (r = 0.138, P = 0.0417), CCL3 (r = 0.169, P = 0.0125), FGF-basic (r = 0.146, P = 0.032), and TNF-α (r = 0.197, P = 0.0035) that was present only in CMML patients.
Clinical and genetic associations with the inflammatory secretome
When comparing the CMML secretome with clinical parameters, cytokine and chemokine secretion was associated with a variety of clinically relevant features. For example, patients who were classified as MPN-CMML by the FAB had significantly elevated inflammatory cytokine and chemokine secretion that included S100A8 (FC = 1.8, P = 0.005), HGF (FC = 1.3, P = 0.004), MIF (FC = 1.5, P = 0.006), and IL-1RA (FC = 1.3, P = 0.001). CCL3 was found to be downregulated in MPN-CMML patients compared to MDS-CMML patients (FC = 0.3, P = 0.012) (Fig. 2a; Supplemental Table 7). Other relevant clinical associations included IL-2RA (FC = 1.9, P < 0.001), which statistically associated with higher risk CMML (Fig. 2b; Supplemental Table 8) and IL-8 (FC = 2.1, P = 0.041) and IL-2RA (FC = 0.045, P = 1.5), which statistically associated with constitutional symptoms (Fig. 2c; Supplemental Table 9). IL-8 (FC = 6.1, P = 0.04) was found to be profoundly elevated in CMML patients classified in the CMML-2 subcategory by 2016 WHO classification system [1]. Interestingly, the association with constitutional symptoms and IL-8 (FC = 2.1, P = 0.041) is consistent with the clinical efficacy of ruxolitinib, an FDA approved JAK1/2 inhibitor, that potently down-regulates IL-8 and improves constitutional symptoms in patients with myelofibrosis and CMML [22]. Patients with enlarged spleens by physical exam were observed to have elevated CXCL9 levels compared to those without enlarged spleen (FC = 1.6, P = 0.042) (Fig. 2d; Supplemental Table 10). Finally, CMML patients with anemia had elevated levels of IL-5 (FC = 1.2, P = 0.014) while those with thrombocytopenias had elevated levels in IP-10/CXCL10 (FC = 1.9, P = 0.005) and IL2-RA (FC = 1.5, P = 0.008) (Fig. 2e, f; Supplemental Tables 11 and 12).
Fig. 2.
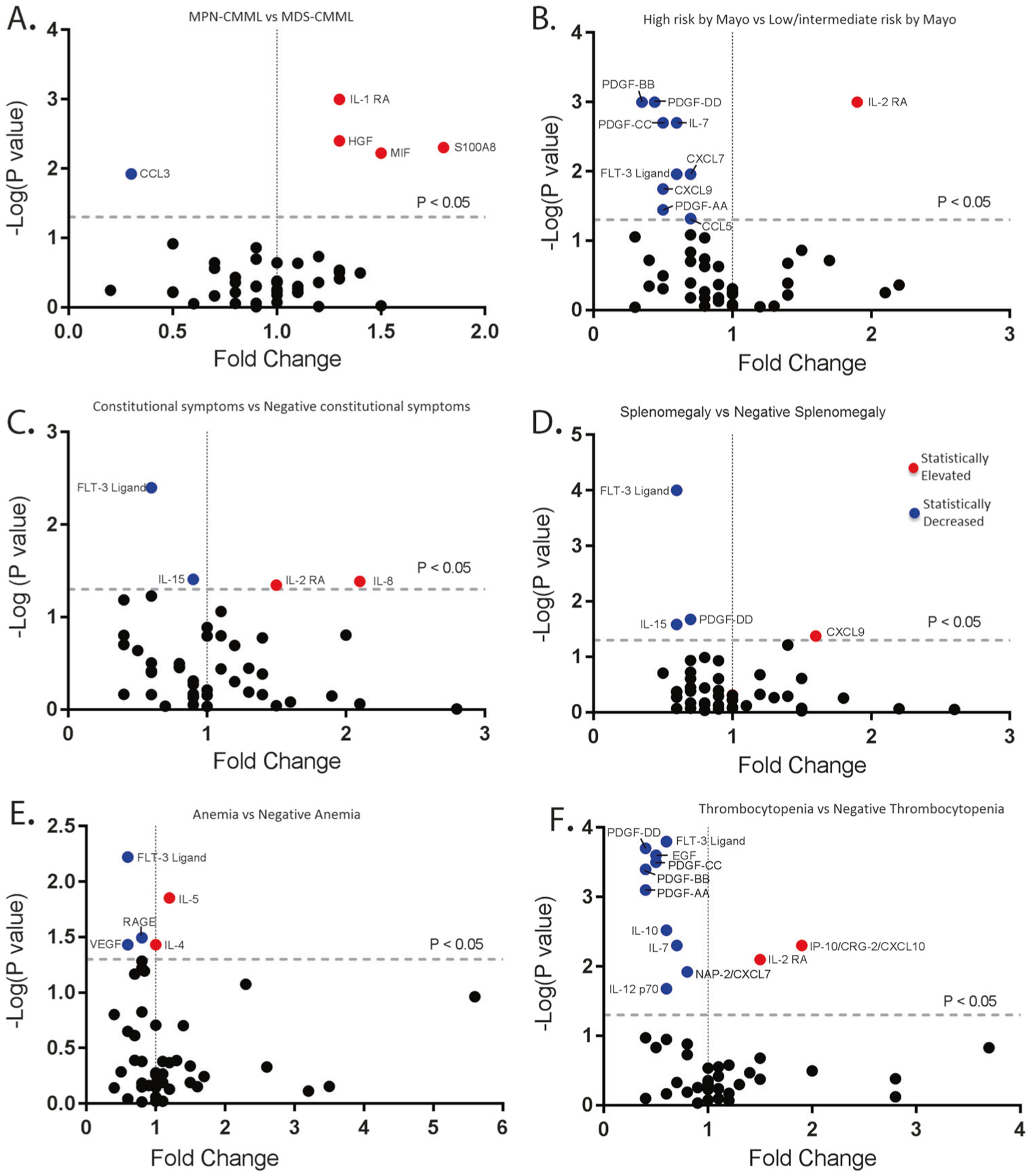
Cytokine–phenotype associations in CMML. a Cytokine expression in patients with MPN-CMML compared to patients categorized in MDS-CMML. Y-axis is negative log(P-value). X-axis is the calculated fold change from the mean of the batch-adjusted cytokine levels normalized to healthy controls. b Cytokine expression in patients with high-risk CMML by Mayo Prognostic Model compared to patients categorized to low-risk CMML by Mayo Prognostic Model. c Cytokine expression in CMML patients with constitutional symptoms compared to CMML patients without constitutional symptoms. d Cytokine expression in CMML patients with splenomegaly compared to CMML patients without the presence of splenomegaly. e Cytokine expression in CMML patients with hemoglobin <10 g/dL compared to CMML patients with hemoglobin >10 g/dL. f Cytokine expression in CMML patients with platelet count <100,000/μL compared to CMML patients with platelet count >100,000/μL
When comparing inflammatory cytokine secretion with the presence or absence of commonly mutated genes, we observed TET2 mutant patients had statistically decreased levels of IL-6 (FC = 0.2, P = 0.005), PDGF-AA (FC = 0.4, P < 0.001), and PDGF-DD (FC = 0.5, P = 0.007) (Fig. 3a, Supplemental Table 13). Combining all mutated epigenetic regulator genes tested, significant increases in IL-1β (FC = 1.2, P = 0.029) secretion were observed (Fig. 3b, Supplemental Table 14). CMML patients with mutations in splicing regulators had statistically elevated levels of IL-2 (FC = 1.7, P = 0.043) and CD44 (FC = 1.6, P = 0.041) (Fig. 3c, Supplemental Table 15). However, other than PDGF-AA, no cytokines were found to be significantly different among patients with ASXL1 mutation compared to those with wild-type ASXL1 (Fig. 3d, Supplemental Table 16). CMML patients with signaling mutations, alternatively, were found to have the greatest increases in inflammatory cytokines to include IL-10 (FC = 3.8, P = 0.006), CCL2/MCP-1 (FC = 1.5, P = 0.033), CD44 (FC = 2.2, P = 0.002), IL-1RA (FC = 1.3, P = 0.027), IL-12 23p40 (FC = 6.6, P = 0.019), and CXCL7 (FC = 1.5, P = 0.012) (Fig. 3e, Supplemental Table 17). CXCL7 has been previously reported to be differentially methylated and overexpressed in CMML patients resistant to decitabine supporting its clinical relevance [36]. GM-CSF was observed to be elevated in patients with signaling mutations, with a fold change of 3.8 with P = 0.073, but it did not reach statistical significance. Overall, distinct cytokine signatures were associated with somatic mutations, but the greatest number and increase were in those with signaling mutations (Fig. 3e).
Fig. 3.
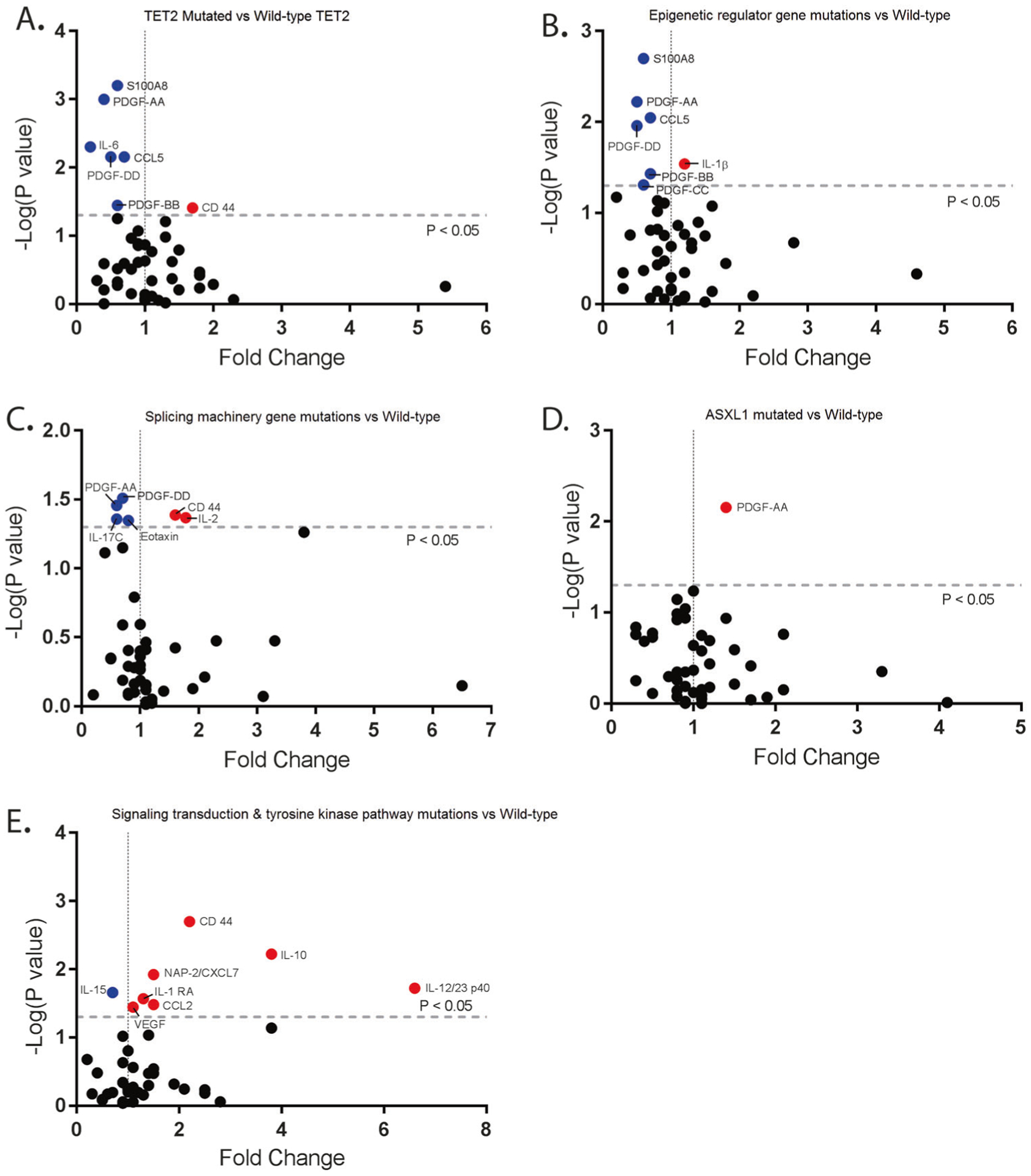
Cytokine genotype associations in CMML. a Cytokine expression in patients in CMML patients with the TET2 mutations compared to CMML patients without TET2 mutations. Y-axis is negative log (P-value). X-axis is the calculated fold change from the mean of the batch-adjusted cytokine levels normalized to healthy controls. b Cytokine expression in patients in CMML patients with the mutations in epigenetic regular genes compared to CMML patients without those mutations. c Cytokine expression in patients in CMML patients with mutations involving splicing machinery compared to CMML patients without those mutations. d Cytokine expression in patients in CMML patients with ASXL1 mutations compared to CMML patients without ASXL1 mutations. e Cytokine expression in patients in CMML patients with mutations involving signal transduction and tyrosine kinase pathway compared to CMML patients without those mutations
Unsupervised clustering analysis
Given these clinical and genetic associations with subsets of cytokine/chemokine secretion, we next tested whether unsupervised clustering of cytokines could identify clinical subtypes of CMML using two different methods, RPMM and Mclust. Using RPMM clustering, we found three distinct cluster groups when analyzed in 213 evaluable CMML patients (Fig. 4a, Supplemental Tables 18 and 19). A total of 69 patients (32%) were classified in cluster 1 driven by statistically significant increases in M-CSF (Supplemental Table 15). A total of 42 patients (20%) were classified in cluster 2 driven by statistically significant increases in 17 cytokines including IL-6, IL-8, and IL-12p70 cytokines (Supplemental Table 20). A total of 102 patients (48%) were classified in cluster 3 driven by statistically significant increases in IL-2RA (Fig. 4, Supplemental Table 18). We observed that a larger proportion of CMML patients with anemia with hemoglobin <10 (32 of 58 patients, 55.2%, P = 0.058) and thrombocytopenia with platelet count <100,000 (55 of 88 patients, 62.5%, P < 0.001) were categorized into cluster 3. Additionally, we observed that patients were significantly more likely to be lower risk by the Mayo score (P = 0.003) [17] and the Global MD Anderson score (P = 0.08) [10, 37] when assigned to cluster 2. Last, the presence of immature myeloid cells, an adverse clinical factor in CMML, was associated with Clusters 1 and 3 (P = 0.012). When comparing OS between our three distinct cluster groups, no statistical difference was observed (Supplemental Fig. 3). All tested variables and their statistical significance are presented in Supplemental Tables 20 and 21.
Fig. 4.
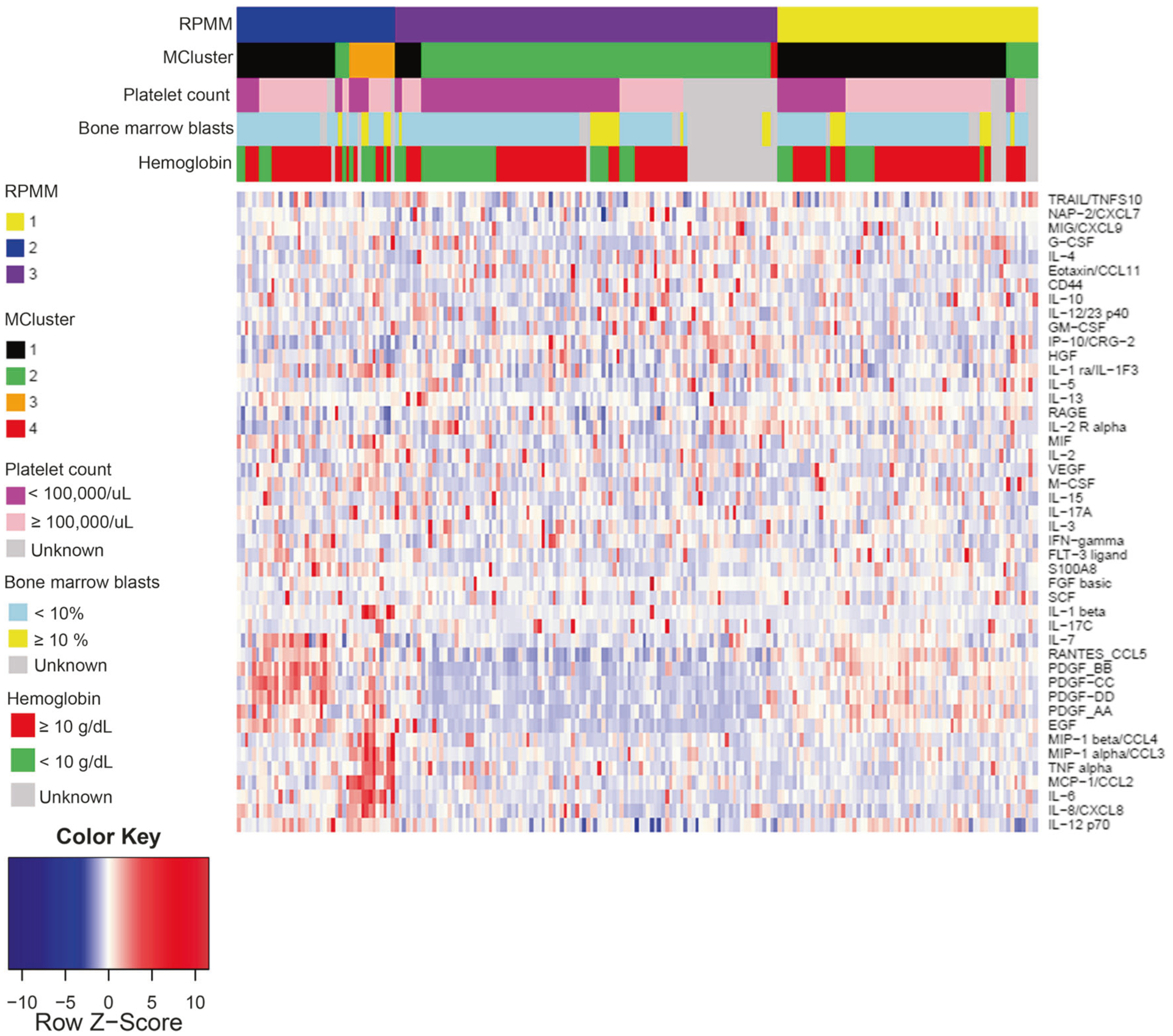
Unsupervised cluster analysis identifies three groups driven by cytokine signatures. Heatmap of batch-adjusted cytokine levels with cluster assignments denoted on the legend based on either RPMM (3 clusters) or Gaussian mixed-model clustering (4 clusters) when cytokine expression was analyzed in CMML patients (N = 213)
Impact of cytokines on survival
Using log-rank tests with all cytokines (measured as binary variable high/low), we identified M-CSF (P = 0.012, HR 0.74, 95% CI 0.58–0.94), IL-13 (P = 0.035, HR 5.59, 95% CI 1.34–23.3), and IL-10 (P = 0.037, HR 0.8, 95% CI 0.65–0.98) to be associated with OS in CMML. Confirming this, we observed that CMML patients with decreased IL-10 expression (median IL-10 = 0.895939) had an inferior OS when compared to CMML patients with elevated expression of IL-10 by Kaplan–Meier analysis (median OS of 46.98 vs 79.84 months, P = 0.0166) (Supplemental Figs. 4 and 5A). Despite small numbers of patients in subgroup analysis, investigating IL-10 expression across Mayo prognostic risk groups in CMML patients consistently demonstrated that those with elevated IL-10 secretion had a favorable OS [17] (Supplemental Fig. 5B–I).
To evaluate whether cytokines can improve our current prognostic systems in CMML, we conducted log-rank tests on clinical and genetic variables, and found significant risk factors for survival that included anemia, absolute monocyte count, elevated WBC count, the presence of ASXL1 mutation, and FAB-MPN status (Supplemental Table 22). To create a final model that could predict OS and leukemia-free survival, we used a backward selection to eliminate non-significant variables when considering the above clinical and genetic variables, including age, hemoglobin, platelet count, monocyte count, bone marrow blast percentage, presence of circulating immature cells, ASXL1, FAB-MPN, and CMML-specific cytogenetic risk score. (Tables 2 and 3, Supplemental Tables 22 and 23). When considering this, IL-10, hemoglobin <10 g/dL, ASXL1 mutation status, and FAB-MPN status remained statistically significant (Table 2). Because the vast majority of these clinical and genetic variables were previously identified by the Mayo Molecular Model [17], we tested the additive prognostic power of IL-10 expression (including ASXL1 mutation status) and found that IL-10 statistically improved the Mayo CMML prognostic score model [17] (C-Index of 0.6736317 (model without IL-10) vs 0.7367075 (model with IL-10), P = 0.01871) in our patient cohort (Supplemental Table 22).
Table 2.
Significant variables associated with overall survival (OS)
| Covariate | Level | N | Hazard ratio (95% CI) | HR P-value | Overall P-value | Log-rank test P-value |
|---|---|---|---|---|---|---|
| IL-10 | High | 84 | 0.48 (0.26–0.89) | 0.019 | 0.019 | 0.017 |
| Low | 90 | - | - | |||
| Mayo CMML prognostic model | High | 43 | 4.20 (1.71–10.31) | 0.002 | 0.002 | <0.001 |
| Low | 66 | - | - | |||
| ASXL1 | Positive | 44 | 3.07 (1.52–6.21) | 0.002 | 0.002 | 0.001 |
| Negative | 97 | - | - |
Table 3.
Overall survival according to the defined risk categories in the training cohort determined using Mayo Molecular Model with addition of IL-10
| Number of patients (%) | Mean (months) | Proportion alive at 5 year (%) | Log- rank test (P-value) | |
|---|---|---|---|---|
| <0.001 | ||||
| Low | 39 (22.4) | 68.0 | 23.1 | |
| Intermediate | 122 (70.1) | 56.0 | 8.2 | |
| High | 13 (7.5) | 13.6 | 0 | |
| Pairwise comparisons | Low vs Intermediate, P = 0.031 Low vs High, P<0.001 Intermediate vs High, P = 0.003 |
Impact of IL-10 and IL-8 on colony formation in primary CMML patient samples
CFAs were performed in duplicate from cryopreserved bone marrow mononuclear cells of seven CMML patients. Four of the patients were found to have elevated IL-10 expression (above median IL-10 = 0.895939) using our custom luminex assay and three of the patients were found to have low IL-10 expression. We compared colony output between IL-10 treated patient sample, IL-8 treated patient sample, and untreated patient sample. We observed lower colony formation output in our CMML patients with decreased IL-10 expression (P = 0.054) (Supplementary Figure 6A). In CMML patients with elevated IL-10 expression, again we observed decreased colony formation potential compared to our untreated sample which was statistically significant (P = 0.025) (Supplementary Figure 6B). When all seven samples in duplicated treated with IL-10 was compared to the untreated samples, we persistently found decreased colony formation in IL-10-treated samples (P = 0.0007) (Supplementary Figure 5C). We, however, observed increased colony formation in IL-8-treated samples compared to our untreated samples which was statistically significant (P = 0.017) (Supplementary Figure 6D–F). RNA gene expression analysis comparing untreated cultured primary CMML bone marrow mononuclear cells to those treated with IL-10 found upregulation in cell cycle apoptotic pathways using Nanostring PanCancer Pathway (Supplementary Figure 6G, H, I).
Discussion
CMML is characterized by clinical heterogeneity that cannot be molecularly explained by known recurrent genetic mutations alone. Previous studies have demonstrated that elevated inflammatory cytokines in MPNs, such as PMF, correlate with clinical phenotypes and are associated with OS [24]. However, cytokine expression has never been evaluated in patients with CMML. Given the clinical responses to ruxolitinib, we reasoned that the inflammatory secretome may play an important role in the pathogenesis of CMML. To explore this, we profiled 45 inflammatory cytokines and chemokines in CMML patients, demonstrating that proinflammatory cytokines were heterogeneously expressed in CMML patients with 23 of 45 cytokines that were significantly discordant to matched healthy controls. Of interest, approximately 50% of these cytokine/chemokines were increased in CMML and 50% were decreased. Within clinical CMML subgroups, this heterogeneity appeared to coincide with clinically relevant phenotypes. As an illustration, proliferative CMML, which diversely combine leukocytosis, splenomegaly, and constitutional symptoms, was typically associated with a strong inflammatory cytokine profile. Cytokine heterogeneity also aligned with mutational signatures. For example, the largest number of inflammatory cytokines were, not surprisingly, associated with those patients that harbored a cytokine signaling mutation. However other genetic associations identified were unexpected. For example, it has been previously reported that TET2 function is associated with suppression of IL-6 and increased production of IL-6 in TET2-deficient mice; however, our results found the opposite effect [38, 39]. In our cohort, we observed decreased IL-6 expression in CMML patients with mutated TET2, suggesting that there are complex molecular interactions in human, polygenetic disease not predicted by single gene models (Fig. 3a).
Using unbiased cluster analysis, we were able to further validate the capacity of cytokine secretion to account for CMML clinical diversity. Surprisingly, while the greatest number of elevated individual cytokines was found in those CMML patients with proliferative symptomatology, cluster analysis was able to identify associations with higher risk disease and cytopenias. These are important clinical variables, albeit not associated with proliferative disease, suggesting that complex networks of cytokines may also govern ‘dysplastic’ symptomatology. In support of this, cytokine association studies demonstrated that CMML patients have, in addition to discordance in individual cytokine secretion, loss of normal cytokine associations.
Although the present study represents the first interrogation of inflammatory cytokines/chemokines in CMML, circulating cytokine levels have been previously evaluated in PMF and other myeloid diseases [1, 24]. When comparing abnormal cytokine secretion between CMML and PMF, for example, several cytokines in CMML patients that were elevated compared to healthy controls were also increased in PMF patients, including IL-1RA, Il-2RA, IL-6, IL-8, TNF-α, HGF, IP-10, MCP-1, and VEGF [24]. Further, cytokine–phenotype associations discovered in our cohort were also seen in PMF, validating our findings. Specifically, statistically increased IL-8 expression in patients with constitutional symptoms was observed in our cohort and in PMF (Fig. 2c, Supplemental Table 6) [24]. However, not every cytokine association in CMML was similar to that in PMF. IFN-γ was found to be decreased in PMF, but in our cohort, IFN-γ was elevated in CMML patients compared to healthy controls, though it did not reach statistical significance [24]. Further, 50% of cytokines differentially secreted were decreased in CMML, a number far greater than in PMF. Of these cytokines, RANTES/CCL5 was among the most decreased cytokines in CMML, with similar findings previously reported in MDS and aplastic anemia, providing cytokine-level support for the phenotypic overlap between bone marrow failure and myeloproliferation in CMML [40].
Prognostic risk models are valuable tools for assessing individual disease risk and clinical decision-making. In CMML, these risks models have modest predictive capacity even after adding prognostically informative genetic mutations. We identified IL-10 as an independent prognostic factor for OS in CMML, consistent with findings in other malignancies [41, 42]. This cytokine has been observed to suppress cytokine secretion and inhibit the induction of proinflammatory cytokines such as IL-1β, TNF-α, GM-CSF, and IFN-γ [43]. Further, CMML patients with downregulated IL-10 expression were identified to have poor OS when compared to CMML patients with elevated expression of IL-10 (P = 0.0166). Importantly, the addition of IL-10 statistically improved the predictive power of the Mayo Molecular Model [17] within our cohort. Although the magnitude of improvement was small, these data provide the foundational evidence supporting the notion that inflammatory cytokines represent a tractable, clinically relevant molecular characteristic of CMML. In fact, a sequential analysis of eight CMML patients treated with 13.75 mean cycles of 5-azacitidine demonstrated that 50% (16 of 32 tested) of cytokines tested were statistically augmented by therapy suggestive that inflammatory cytokines may have therapeutic relevance (Supplemental Figure 7).
IL-10 is a multifunctional cytokine produced by several cell types including monocytes and has been described to have both immunosuppressive and proinflammatory properties [43–45]. IL-10 has been implicated in the pathogenesis of multiple tumor types including CMML, and its potential to suppress cytokine expression has made it of particular interest as a possible therapeutic target [46–49]. Geissler et al. [50] observed that IL-10 could inhibit the growth of CMML primary cells in a dose-dependent manner in vitro potentially via the suppression of endogenous GM-CSF. The group observed that treatment of exogenous GM-CSF was able to reverse IL-10 growth inhibition of CMML primary cells, and observed that GM-CSF mRNA levels and protein levels were decreased after treatment with IL-10 [50]. In our cohort, we observed that CMML patients with high IL-10 expression had a favorable OS perhaps mediated by IL-10 growth suppression of CMML leukemia cells. In support of this notion, a small study of five CMML patients treated with recombinant human IL-10 observed some clinical efficacy with improvement in one of three patients with confirmed skin infiltration by CMML suggesting that IL-10 is a potential therapeutic target [51]. Our own experiment confirmed decreased colony formation in primary CMML patient samples treated with IL-10 compared to untreated samples and IL-8-treated samples (Supplementary Figure 6).
Our study comprehensively evaluated cytokine expression in CMML patients, which has not been evaluated in the literature to understand the pathophysiology as well as evaluate the potential for new therapeutic targets. The annotation of inflammatory cytokine/chemokines in this study also raises several important questions. For example, what is the individual contribution of the stroma and the leukemic cells to the overall cytokine milieu? Additionally, can inhibition of cytokines critical to the symptomatology of CMML such as IL-8 be selectively targeted in CMML? Based on the findings from this study, further investigation is warranted into the inflammatory state of CMML, its therapeutic relevance, and its use to improve our current prognostic models. Last, our data demonstrate that IL-10 has the potential to improve existing clinical and genetic prognostic model and serve as a therapeutic target in CMML.
Supplementary Material
Acknowledgements
This research was supported in part by the USF GME Research Grant. Research was funded by EP.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Electronic supplementary material The online version of this article (https://doi.org/10.1038/s41375-018-0203-0) contains supplementary material, which is available to authorized users.
References
- 1.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 2.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol. 1976;33:451–8. [DOI] [PubMed] [Google Scholar]
- 3.Elena C, Galli A, Such E, Meggendorfer M, Germing U, Rizzo E, et al. Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood. 2016;128:1408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball M, List AF, Padron E. When clinical heterogeneity exceeds genetic heterogeneity: thinking outside the genomic box in chronic myelomonocytic leukemia. Blood. 2016;128:2381–7. [DOI] [PubMed] [Google Scholar]
- 5.Merlevede J, Droin N, Qin T, Meldi K, Yoshida K, Morabito M, et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat Commun. 2016;7:10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deininger MWN, Tyner JW, Solary E. Turning the tide in myelodysplastic/myeloproliferative neoplasms. Nat Rev Cancer. 2017; 17:425–40. [DOI] [PubMed] [Google Scholar]
- 7.Beran M, Wen S, Shen Y, Onida F, Jelinek J, Cortes J, et al. Prognostic factors and risk assessment in chronic myelomonocytic leukemia: validation study of the M.D. Anderson Prognostic Scoring System. Leuk Lymphoma. 2007;48:1150–60. [DOI] [PubMed] [Google Scholar]
- 8.Aul C, Gattermann N, Heyll A, Germing U, Derigs G, Schneider W. Primary myelodysplastic syndromes: analysis of prognostic factors in 235 patients and proposals for an improved scoring system. Leukemia. 1992;6:52–9. [PubMed] [Google Scholar]
- 9.Worsley A, Oscier DG, Stevens J, Darlow S, Figes A, Mufti GJ, et al. Prognostic features of chronic myelomonocytic leukaemia: a modified Bournemouth score gives the best prediction of survival. Br J Haematol. 1988;68:17–21. [DOI] [PubMed] [Google Scholar]
- 10.Onida F, Kantarjian HM, Smith TL, Ball G, Keating MJ, Estey EH, et al. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002;99:840–9. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Sole F, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padron E, Garcia-Manero G, Patnaik MM, Itzykson R, Lasho T, Nazha A, et al. An international data set for CMML validates prognostic scoring systems and demonstrates a need for novel prognostication strategies. Blood Cancer J. 2015;5:e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B, et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010; 28:3858–65. [DOI] [PubMed] [Google Scholar]
- 14.Palomo L, Garcia O, Arnan M, Xicoy B, Fuster F, Cabezon M, et al. Targeted deep sequencing improves outcome stratification in chronic myelomonocytic leukemia with low risk cytogenetic features. Oncotarget. 2016;7:57021–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelsi-Boyer V, Trouplin V, Roquain J, Adelaide J, Carbuccia N, Esterni B, et al. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol. 2010;151:365–75. [DOI] [PubMed] [Google Scholar]
- 16.Cui Y, Tong H, Du X, Li B, Gale RP, Qin T, et al. Impact of TET2, SRSF2, ASXL1 and SETBP1 mutations on survival of patients with chronic myelomonocytic leukemia. Exp Hematol Oncol. 2015;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patnaik MM, Padron E, LaBorde RR, Lasho TL, Finke CM, Hanson CA, et al. Mayo prognostic model for WHO-defined chronic myelomonocytic leukemia: ASXL1 and spliceosome component mutations and outcomes. Leukemia. 2013;27:1504–10. [DOI] [PubMed] [Google Scholar]
- 18.Lin Y, Zheng Y, Wang ZC, Wang SY. Prognostic significance of ASXL1 mutations in myelodysplastic syndromes and chronic myelomonocytic leukemia: a meta-analysis. Hematology. 2016; 21:454–61. [DOI] [PubMed] [Google Scholar]
- 19.Padron E, Dezern A, Andrade-Campos M, Vaddi K, Scherle P, Zhang Q, et al. A multi-institution phase I trial of ruxolitinib in patients with chronic myelomonocytic leukemia (CMML). Clin Cancer Res. 2016;22:3746–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geissler K, Jager E, Barna A, Sliwa T, Knobl P, Schwarzinger I, et al. In vitro and in vivo effects of JAK2 inhibition in chronic myelomonocytic leukemia. Eur J Haematol. 2016;97:562–7. [DOI] [PubMed] [Google Scholar]
- 21.Mascarenhas J, Mughal TI, Verstovsek S. Biology and clinical management of myeloproliferative neoplasms and development of the JAK inhibitor ruxolitinib. Curr Med Chem. 2012;19: 4399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med. 2010;363:1117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyner JW, Bumm TG, Deininger J, Wood L, Aichberger KJ, Loriaux MM, et al. CYT387, a novel JAK2 inhibitor, induces hematologic responses and normalizes inflammatory cytokines in murine myeloproliferative neoplasms. Blood. 2010;115:5232–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tefferi A, Vaidya R, Caramazza D, Finke C, Lasho T, Pardanani A. Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. J Clin Oncol. 2011;29: 1356–63. [DOI] [PubMed] [Google Scholar]
- 25.Gillis NK, Ball M, Zhang Q, Ma Z, Zhao Y, Yoder SJ, et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. Lancet Oncol. 2017;18:112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padron E, Painter JS, Kunigal S, Mailloux AW, McGraw K, McDaniel JM, et al. GM-CSF-dependent pSTAT5 sensitivity is a feature with therapeutic potential in chronic myelomonocytic leukemia. Blood. 2013;121:5068–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraley CRA, Murphy TB, Scrucca L. mclust version 4 for R: normal mixture modeling for model-based clustering, classification, and density estimation. Technical Report No. 597, Department of Statistics, University of Washington. 2012. [Google Scholar]
- 28.Raftery CFaAE. Model-based clustering, discriminant analysis and density estimation. J Am Stat Assoc. 2002;97:611–31. [Google Scholar]
- 29.HousemanEA CB, Yeh RF, Marsit CJ, Karagas MR, Wrensch M, Nelson HH, Wiemels J, Zheng S, Wiencke JK, Kelsey KT. Model-based clustering of DNA methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics. 2008; 9:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirantes C, Passegue E, Pietras EM. Pro-inflammatory cytokines: emerging players regulating HSC function in normal and diseased hematopoiesis. Exp Cell Res. 2014;329:248–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bender S, Haubeck HD, Van de Leur E, Dufhues G, Schiel X, Lauwerijns J, et al. Interleukin-1 beta induces synthesis and secretion of interleukin-6 in human chondrocytes. FEBS Lett. 1990;263:321–4. [DOI] [PubMed] [Google Scholar]
- 32.Guerne PA, Carson DA, Lotz M. IL-6 production by human articular chondrocytes. Modulation of its synthesis by cytokines, growth factors, and hormones in vitro. J Immunol. 1990;144: 499–505. [PubMed] [Google Scholar]
- 33.Lotz M, Terkeltaub R, Villiger PM. Cartilage and joint inflammation. Regulation of IL-8 expression by human articular chondrocytes. J Immunol. 1992;148:466–73. [PubMed] [Google Scholar]
- 34.Pulsatelli L, Dolzani P, Piacentini A, Silvestri T, Ruggeri R, Gualtieri G, et al. Chemokine production by human chondrocytes. J Rheumatol. 1999;26:1992–2001. [PubMed] [Google Scholar]
- 35.Honorati MC, Bovara M, Cattini L, Piacentini A, Facchini A. Contribution of interleukin 17 to human cartilage degradation and synovial inflammation in osteoarthritis. Osteoarthr Cartil. 2002;10:799–807. [DOI] [PubMed] [Google Scholar]
- 36.Meldi K, Qin T, Buchi F, Droin N, Sotzen J, Micol JB, et al. Specific molecular signatures predict decitabine response in chronic myelomonocytic leukemia. J Clin Invest. 2015;125: 1857–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Germing U, Strupp C, Aivado M, Gattermann N. New prognostic parameters for chronic myelomonocytic leukemia. Blood. 2002;100:731–2. author reply 2–3 [DOI] [PubMed] [Google Scholar]
- 38.Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bird L Inflammation: TET2: the terminator. Nat Rev Immunol. 2015;15:598. [DOI] [PubMed] [Google Scholar]
- 40.Feng X, Scheinberg P, Wu CO, Samsel L, Nunez O, Prince C, et al. Cytokine signature profiles in acquired aplastic anemia and myelodysplastic syndromes. Haematologica. 2011; 96:602–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boulland ML, Meignin V, Leroy-Viard K, Copie-Bergman C, Briere J, Touitou R, et al. Human interleukin-10 expression in T/natural killer-cell lymphomas: association with anaplastic large cell lymphomas and nasal natural killer-cell lymphomas. Am J Pathol. 1998;153:1229–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nemunaitis J, Fong T, Shabe P, Martineau D, Ando D. Comparison of serum interleukin-10 (IL-10) levels between normal volunteers and patients with advanced melanoma. Cancer Invest. 2001;19:239–47. [DOI] [PubMed] [Google Scholar]
- 43.Mannino MH, Zhu Z, Xiao H, Bai Q, Wakefield MR, Fang Y. The paradoxical role of IL-10 in immunity and cancer. Cancer Lett. 2015;367:103–7. [DOI] [PubMed] [Google Scholar]
- 44.Mocellin S, Marincola F, Rossi CR, Nitti D, Lise M. The multi-faceted relationship between IL-10 and adaptive immunity: putting together the pieces of a puzzle. Cytokine Growth Factor Rev. 2004;15:61–76. [DOI] [PubMed] [Google Scholar]
- 45.Kasamatsu T, Saitoh T, Minato Y, Shimizu H, Yokohama A, Tsukamoto N, et al. Polymorphisms of IL-10 affect the severity and prognosis of myelodysplastic syndrome. Eur J Haematol. 2016;96:245–51. [DOI] [PubMed] [Google Scholar]
- 46.Torisu-Itakura H, Lee JH, Huynh Y, Ye X, Essner R, Morton DL. Monocyte-derived IL-10 expression predicts prognosis of stage IV melanoma patients. J Immunother. 2007;30:831–8. [DOI] [PubMed] [Google Scholar]
- 47.Vahl JM, Friedrich J, Mittler S, Trump S, Heim L, Kachler K, et al. Interleukin-10-regulated tumour tolerance in non-small cell lung cancer. Br J Cancer. 2017;117:1644–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clerici M, Merola M, Ferrario E, Trabattoni D, Villa ML, Stefanon B, et al. Cytokine production patterns in cervical intraepithelial neoplasia: association with human papillomavirus infection. J Natl Cancer Inst. 1997;89:245–50. [DOI] [PubMed] [Google Scholar]
- 49.Emmerich J, Mumm JB, Chan IH, LaFace D, Truong H, McClanahan T, et al. IL-10 directly activates and expands tumor-resident CD8(+) T cells without de novo infiltration from secondary lymphoid organs. Cancer Res. 2012;72:3570–81. [DOI] [PubMed] [Google Scholar]
- 50.Geissler K, Ohler L, Fodinger M, Virgolini I, Leimer M, Kabrna E, et al. Interleukin 10 inhibits growth and granulocyte/macrophage colony-stimulating factor production in chronic myelomonocytic leukemia cells. J Exp Med. 1996;184: 1377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pochlauer S, Jager E, Jager U, Geissler K. Recombinant human interleukin-10 in patients with chronic myelomonocytic leukemia. Ann Hematol. 2014;93:1775–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


