Abstract
Oropharyngeal cancer (OPC) incidence is increasing significantly among men and often requires intensive therapy causing significant morbidity. Early detection of OPC is needed, when monotherapy can be safely delivered with less treatment-associated morbidity, while maintaining high cure rates. We conducted a study of 101 pretreatment male OPC cases matched 1:1 to 101 disease-free controls for age and smoking history. Oral gargles were collected from cases and controls with additional biopsies or aspirates from cases. The HPV SPF10-LiPA25 PCR assay was utilized for HPV genotyping. Methylation of three CpG sites (438, 427 and 425) in the EPB41L3 gene and methylation status of the L1 (6,367, 6,389), L2 (4,257, 4,262, 4,266, 4,269, 4,275, 4,282) and E2 (3,412, 3,415, 3,417, 3,433, 3,436) CpG sites of HPV 16 positive specimens was assessed by pyrosequencing. Significant correlations were observed between tumor and oral specimens for all methylation biomarkers (p < 0.01). EPB41L3 and HPV 16 L1, L2 and E2 methylation were significantly (p < 0.0001) higher among cases than controls, regardless of early vs. late disease. When HPV 16 genes and EPB41L3 methylation status were combined in a logistic regression analysis, a sensitivity of 70.3% and a specificity of 90.9% were observed for the detection of OPC from an oral gargle. Our data suggest that methylation biomarkers measured in oral gargles may have utility in identifying OPC early. Future studies are needed to replicate these findings and to inform additional biomarkers that can maximize specificity and sensitivity for early OPC detection.
Keywords: HPV, oropharyngeal cancer, methylation, screening, early detection
Introduction
In the US, approximately 70% of oropharyngeal cancers (OPCs) are attributable to human papillomavirus (HPV) infection, predominantly HPV 16.1,2 HPV-related OPC incidence is four to fivefold higher in men compared to women and is increasing rapidly among males worldwide.2,3 OPC incidence among US men is now higher than cervical cancer incidence in women,4 with a significant increase in the proportion of cases attributable to HPV in recent decades.2 No screening tests are available for OPC, nor have current HPV vaccines been proven to protect against these cancers. As a result, most OPC tumors are diagnosed with advanced disease, with multiple bilateral positive neck nodes. Although treatment outcomes of HPV-related OPC are superior to those of HPV negative cases, treatment may cause significant morbidity, and rates of recurrence are still 10–30%. Detection of cancers earlier (T1–2 N0–1 [small tumors with only a single ipsilateral positive node <3 cm]) when tumors can be effectively and safely treated with a single modality provides an opportunity to achieve a cure while limiting adverse consequences.5 To increase patient survival and quality of life, biomarkers are needed to diagnose OPCs earlier.
Multiple biomarkers have been evaluated for head and neck cancer detection,6–8 each with drawbacks that limit their utility.7,9 Marketed imaging devices and screening tests have not been effective to date,9 and an oral cytology test has been attempted with poor results.10 In contrast, oral gargles are recognized as the standard method to assess HPV status, preferable to other more direct sampling methods.11,12 However, agreement between oral rinses and tumor HPV 16 DNA has been variable, ranging between 5913 and 96%,14 with most studies reporting values of approximately 70–80%.13,15–20 In addition, we noted the performance of the oral HPV 16 test was significantly lower among younger cases and those diagnosed with earlier disease that can be treated with monotherapy.20 Hence, additional biomarkers are needed to improve oral HPV 16 test characteristics to identify OPC early.
After HPV acquisition, the biological mechanisms leading to carcinogenesis appear similar at each anatomic site where HPV is known to cause cancer (e.g., cervix, oropharynx and anus).21 Like cervical and anal cancer, HPV-related OPC is primarily caused by HPV type 16,22 although detection of HPV 16 is insufficient as a biomarker at any anatomic site as only a subset of HPV 16 infections progress to cancer. Methylation of the HPV 16 L1, L2 and E2 regions and the host EPB41L3 gene have been validated as biomarkers for the detection of high-grade cervical intraepithelial lesions and cancers and for differentiating these from lesions that are likely to clear.23,24 HPV L1 methylation has also been detected in oral squamous cell carcinomas,25 and elevated levels of methylation of HPV 16 genes may be useful in predicting or detecting oral HPV infections at risk of progression to OPC. Thus, we evaluated methylation of the L1, L2 and E2 regions of HPV 16, and methylation levels of the tumor suppressor gene EPB41L3 as an indicator of epigenetic changes that have occurred in the host genome in a sample set of 101 OPC cases and 101 disease free controls. As a proof of principle study, we assessed these biomarkers among OPC cases and controls by first examining the correlation of the markers in oral gargle and tumor biopsy and/or fine needle aspirated specimens among cases, and then compared oral gargle biomarker levels among cases and controls, overall and by clinical disease presentation at diagnosis.
Materials and Methods
From May 2014 to December 2016, newly diagnosed first primary OPC cases were recruited from the Head and Neck Cancer, Radiation Oncology and the Senior Adult Oncology Clinics at the Moffitt Cancer Center. Patients who were male, ≥18 years, had histologically confirmed squamous cell carcinoma of the oropharynx stages I–IV (C01.9 base of tongue; C05.1 soft palate, not otherwise specified [NOS]; C05.2 uvula; C09.0 tonsillar fossa; C09.1 tonsillar pillar; C09.8 overlapping lesion of the tonsil; C09.9 tonsil, NOS; C10.0 vallecula; C10.2 lateral wall of epiglottis; C10.3 posterior wall of epiglottis; C10.8 overlapping lesion of oropharynx and C10.9 oropharynx, NOS), had not yet initiated treatment (radiation or chemoradiation or surgery), and were able to provide an oral gargle specimen were eligible for study. A clinical trial coordinator approached potential study participants, described the study aims and answered questions. Men who were interested and met study eligibility criteria were invited to enroll by signing an informed consent form, escorted to a private consultation room where they completed a health and risk factor questionnaire and provided an oral gargle specimen. Our study was approved by the Liberty IRB, Chesapeake IRB and Moffitt’s Scientific Review Committee, and written informed consent was obtained from all enrolled participants.
The number of cases approached for study participation, exclusions and study participation rate and reasons for nonparticipation are shown in Figure 1a. Briefly, 600 patients were reviewed for participation, 225 met study criteria of which 57 refused study participation. Of the 168 cases that signed consent forms, 18 were subsequently found ineligible for study. Of the 150 enrolled cases, the first 101 were included in the current methylation biomarker study.
Figure 1.
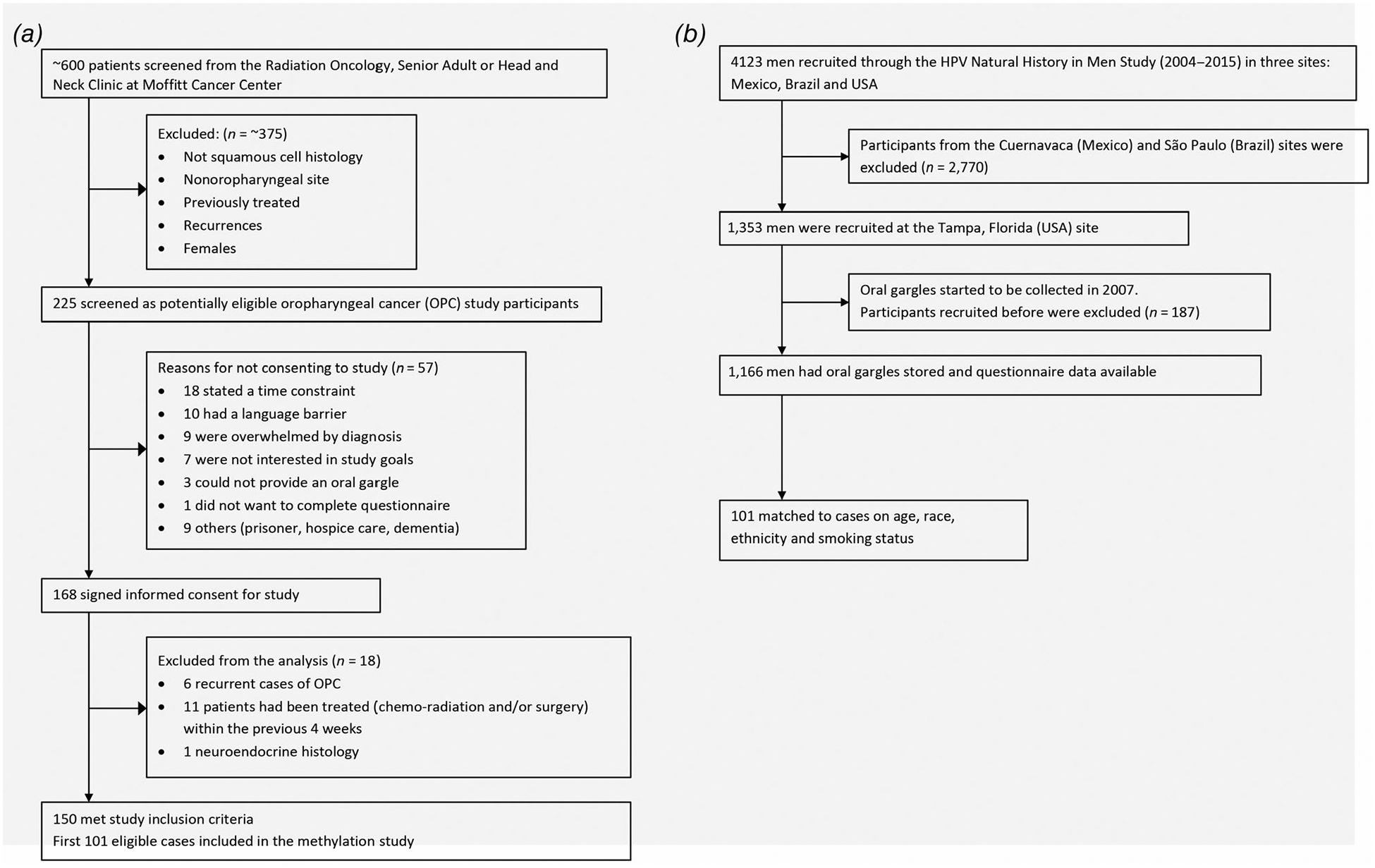
(a) Recruitment and enrollment of oropharyngeal cancer cases. (b) Selection of HIM Study comparison population.
The comparison group of healthy (cancer free) men was selected from the US participants of the HPV Infection in Men (HIM) Study.26 In the US, 1,353 participants were recruited in Tampa, FL. HIM Study participants selected for our study were frequency matched on age (±5 years), and smoking history (never, former and current) to OPC cases (Fig. 1b). All HIM Study participants completed a similar health and risk factor questionnaire as the cases. At the time of HIM Study enrollment, participants met the following eligibility criteria: 18–70 years of age, reported no prior diagnosis of penile or anal cancers (both related with HPV infection) nor had been diagnosed with genital or anal warts, had never participated in an HPV vaccine study, reported no prior diagnosis of HIV or AIDS, were not being treated for an STD nor had current penile discharge or dysuria, and were willing to comply with study requirements and follow-up schedule.
Oral gargle specimen collection and processing
All participants (cases and controls) provided an oral gargle sample using a previously described protocol.27 Briefly, participants were instructed to gargle with 15 ml of locally available mouthwash (Target Up&Up® [Water, alcohol (15% WT), glycerin, flavor, polysorbate 80, sodium saccharin, sodium benzoate, cetylpyridinium chloride, benzoic acid, blue 1, yellow 5]) by first swishing the mouthwash from cheek to cheek for 15 sec and then gargling at the back of the throat for another 15 sec before dispensing the sample back into a 50 ml conical tube. If the participant had difficulty gargling, he was asked to simply swish the mouthwash for the full 30 sec. With this swish and gargle method, multiple studies14,26,28,29 have demonstrated the feasibility of assessing HPV DNA genotype status.
After collection, oral gargle specimens were maintained at 4°C until processing was completed within 24 hr of collection. Specimens were centrifuged at 2,000g for 15 min. About 1.8 ml of the supernatant was pipetted into three individual 2.0 ml cryovials and archived at −80°C. The cell pellet underwent three wash procedures by resuspending the pellet in 20 ml cold phosphate buffered solution (PBS), repeatedly mixed by inversion to assure thorough homogenization, then centrifuged at 2,000g for 15 min at 4°C. The remaining cell pellet was resuspended in 1.2 ml PBS and archived at −80°C until analysis.
Health and risk factor questionnaire
Participants completed an extensive questionnaire that assessed demographic characteristics, personal and familial cancer history, personal oral health and sexual behavior history as well as alcohol, tobacco and other drug use. The questionnaire was administered using a passcode-locked tablet and the reliable Computer-Assisted Self-Interviewing (CASI) survey method.30–32 For participants who were not comfortable using a tablet, a hard copy (paper) version of the questionnaire was provided.
Biopsy/tissue block/fine needle aspirate (cases only)
Electronic medical records and pathology reports were reviewed. A surgical specimen, biopsy and/or fine needle aspirate (FNA) was identified and retrieved for study from the procedure conducted closest to the initial cancer diagnosis and prior to receiving treatment for each case. Anatomic pathology reports were reviewed to select the optimal formalin-fixed paraffin-embedded (FFPE) biopsy block representative of the tumor. Tissue blocks or slides received both from Moffitt cases and from outside facilities were centrally managed through Moffitt’s Tissue Core where blocks were cut and stained. A 4-μm section from the face of the block was removed and discarded prior to cutting. Using sterile plastic forceps, one 4-μm paraffin section was collected and floated in a water bath for the preparation of one H&E slide. Forceps were discarded and a new blade was inserted into the microtome. Nine additional 4-μm sections were cut using the clean, central part of the microtome blade and placed in individual sterile labeled tubes. One additional 4-μm section was cut and placed on another slide for H&E staining. One of the nine FFPE sections was stained for p16INK4a expression. Another 1–2 slides were used for HPV DNA extraction, and the others stored for future studies.
HPV genotyping
DNA was extracted from oral gargle cell pellets using the automated BioRobot MDx (Qiagen, Inc.) following the manufacturer’s instructions. For the extraction of FFPE tumor DNA, the QIAamp® DNA FFPE Tissue Kit (Qiagen, Inc., Germantown, MD) was used according to the manufacturer’s instructions.
The RHA Kit HPV SPF10-LiPA25 (DDL Diagnostic Laboratory, Rijswijk, The Netherlands) an in vitro reverse hybridization assay (RHA) for the qualitative identification of HPV DNA was utilized for HPV genotyping of tumor and oral gargle specimens. The LiPA25 targets a 65 base pair fragment of the L1 region of the HPV genome. This assay requires a three step process: (i) qPCR that determines sample adequacy; (ii) a DNA enzyme immunoassay (DEIA) or ELISA method that detects 65 HPV types; and (iii) a LiPA25 genotyping multiplex PCR that selectively identifies the following HPV types by reverse hybridization: 6, 11, 16, 18, 31, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68/73, 70 and 74.33 Using this method, our group has reliably identified HPV types in anogenital mucosa34,35 and oral gargle specimens.26,36
HPV and host gene methylation
All samples (tumor and oral gargle cell pellet) were tested for methylation of three CpG sites (438, 427 and 425) in the host tumor suppressor gene EPB41L3 using a previously validated pyrosequencing method (PyroMark).24,37 HPV 16 positive oral gargle (n = 67) and tumor specimens (n = 90) were selected for evaluation of viral methylation status of the CpG sites in the L1 (6,367, 6,389), L2 (4,257, 4,262, 4,266, 4,269, 4,275, 4,282) and E2 (3,412, 3,415, 3,417, 3,433, 3,436) regions by pyrosequencing.23,24,37,38 Briefly, 200 ng of DNA was used in bisulfite conversion reactions in which unmethylated cytosines were converted to uracil using the EZ DNA methylation kit (Zymo Research, Irvine, CA). Converted DNA was purified and amplified by PCR primers, with one biotin primer for each pair. Primers were designed with short amplicons (90–140 base pairs each) using the PyroMark Assay Design software (V2.0.1.15 Qiagen). PCR was performed using a converted DNA equivalent of 1,500 cells using the PyroMark PCR kit (Qiagen). PCR products were then captured by streptavidin beads (GE Healthcare, Buckinghamshire, UK) in 96-well plates and pyrosequenced using PyroGold reagents with the signal analyzed using a PyroMark TMQ96 ID (Qiagen) instrument as previously described.37 All runs included standard curves as positive controls of 0, 50 and 100% methylated human DNA and a nontemplate control.
Statistical analyses
Sociodemographic characteristics, sexual behavior, oral health and HPV status in oral gargles were compared for cases (n = 101) and controls (n = 101) using Cochran–Mantel–Haenszel (CMH) exact test. To examine the correlation between HPV 16 gene and EPB41L3 methylation in oral gargle specimens and tumor tissues among cases, the Spearman correlation coefficient was calculated. Kruskal–Wallis test was used to compare the mean value of EPB41L3 methylation in oral gargles comparing early disease (T1–2 N0–1 [small tumors with only a single ipsilateral positive node <3 cm]) and late disease cases versus controls, and post hoc analysis Dunn’s test was used for pair-wise comparisons.
Oral HPV 16 alone does not accurately reflect case status, and its performance is worse among younger cases and cases with early disease, the group of patients we seek to identify with screening.20 In the screening setting, some who test oral HPV 16 positive will not have cancer, and some testing oral HPV 16 negative will have cancer. However, oral HPV 16 methylation is rarely detected among those without cancer. As multiple biomarkers in a panel typically improve test characteristics, and EPB41L3 is measurable in all cases and controls, we performed a logistic regression analysis to combine the two measures (oral HPV 16 methylation and EPB41L3 methylation). All CpG sites were given equal weight. Cases and controls that had detectable oral HPV 16 methylation of one or more of the three HPV genes assessed (L1, L2 and E2) were assigned a dummy variable value of 1, those that did not were assigned a variable value of 0. Receiver operating characteristic (ROC) curves were generated combining HPV 16 gene (L1, L2 or E2) methylation and EPB41L3 methylation levels. The performance of continuous risk scores was measured by area under the curve (AUC) with the Wilcoxon test. Youden’s J (J statistic) was calculated to identify the cut-point yielding the highest sum value for sensitivity and specificity. Cut points were selected to maximize specificity while maintaining sensitivity at 70% or higher. Internal validation was performed by 0.632 bootstrap resampling (n = 300) using the validate procedure in the rms software package in R,39 as recommended by Steyerberg et al.40 All other analyses were performed in SAS 9.3.
Data availability
We make the data collected as part of this protocol available to outside investigators, but within the limitations of preserving the anonymity of individuals since these data could theoretically identify a study participant. In order to maintain compliance with HIPAA regulations, we execute a data sharing agreement with the requestor for a limited use dataset as defined by the US Department of Health and Human Services (DHHS). We also encourage, but not require, collaborative use of the data. As with all datasets, there are many subtle nuances to the coding and interpretation of the data that cannot be completely documented in coding books or comment fields. Through a collaborative agreement, we feel that the quality of secondary analyses derived from these data can be greatly enhanced.
Data will be made available no later than 120 days after the publications of the main findings related to that dataset. We have established an internal Data Access Committee to review requests for the use of data according to these stipulations. Requests for data will be handled on a case-by-case basis, and will be evaluated by this Committee for scientific merit and ethical issues. For consortium use, our data are posted on a secure internal network drive accessed by passwords released on an individual basis. To make data available to outside investigators we employ Moffitt’s ELF (Exchange of Large Files) technology which allows users to access our data files from a secure website with a password and set of instructions sent by the IT department at Moffitt. Final datasets available to outside investigators will include reported demographic, diagnostic and risk factor information. Limited genotypic data will be made available to outside investigators to maintain compliance with confidentiality commitments. Outside investigators access to the secure ELF website is limited to the dataset prepared for that investigator. No biological samples will be released under this mechanism due to the limited nature of these samples. Outside investigators desiring access to such samples must set up collaboration with our consortium and also must agree that any data generated through their work will be governed by the data access rules that apply to the rest of this collaborative effort.
Because all of the collaborating sites have obligations to the participants to return important new information, we have collected and maintained personal identifying information. Although the final dataset will be stripped of identifiers prior to release for sharing, we believe that there remains the possibility of deductive disclosure of subjects. Thus, we will follow the data enclave model that make data available to users only under a data sharing agreement that provides for (i) a detailed summary of the proposed research project, including a complete list of data requested and limited specific aims, (ii) a commitment to using the data only for research purposes, (iii) a commitment to maintaining confidentiality of the data and not to identify any individual participant and (iv) a commitment to securing the data summary statistics using appropriate computer technology, (v) a commitment to destroying or returning the user data after analyses are completed, (vi) a commitment to publication of all results of these analyses and (vii) a commitment to reporting all results to the consortium in a timely manner (at least annually). The internal Data Access Committee is also responsible for evaluating the scientific validity, projected power and ethical aspects of the proposed use of our data prior to approval of release to any requesting investigator(s).
Results
Characteristics of OPC cases and controls are presented in Table 1. The median age of cases and controls was 60.0 years with 40.6% reporting never smoking. The majority of cases (93.1%) and controls (76.2%) were White, married/cohabiting (74.3 and 63.4%) and consumed 1–4 drinks per occasion (52.5 and 60.4%). Cases were significantly more likely to have reported having had a tonsillectomy than controls (39.6% vs. 10.9%), with the surgery occurring most often 30 years in the past compared to controls who underwent tonsillectomy <2 years in the past. Cases also reported a greater number of teeth extracted prior to diagnosis with 18.8% reporting ≥10 teeth removed compared to 7.9% among controls. Among cases, 45.5% of tumors were at the tonsils and 49.5% at the base of tongue, with the minority (18.8%) diagnosed with early disease defined as T1–2 and N0–1 only if there was a single ipsilateral node <3 cm. The majority of tumors (95.0%) were HPV positive with 89.1 and 3.0% HPV 16 and HPV 18 positive, respectively. Oral gargle HPV status differed significantly by case status with 66.3% of cases and 3.0% of controls oral HPV 16 positive.
Table 1.
Sociodemographic characteristics of oropharyngeal cancer cases and disease-free controls
| Cases (n = 101) | Controls (n = 101) | CMH (Exact) p-value | |||
|---|---|---|---|---|---|
| Characteristics | n | % | n | % | |
| Race | |||||
| White | 94 | 93.1 | 77 | 76.2 | 0.0011 |
| Black | 5 | 5.0 | 19 | 18.8 | |
| Other | 2 | 1.9 | 5 | 5.0 | |
| Ethnicity | |||||
| Hispanic | 6 | 5.9 | 9 | 8.9 | 0.5929 |
| Non-Hispanic | 95 | 94.1 | 92 | 91.1 | |
| Age (years) | |||||
| Median (SD) | 60.0 (45) | 60.0 (45) | 1.0000 | ||
| 35–49 | 12 | 11.9 | 12 | 11.9 | |
| 50–59 | 29 | 28.7 | 29 | 28.7 | |
| 60–69 | 35 | 34.7 | 35 | 34.7 | |
| ≥70 | 25 | 24.7 | 25 | 24.7 | |
| Marital status | |||||
| Single/divorced/separated/widowed | 25 | 24.8 | 37 | 36.6 | 0.0929 |
| Married/cohabiting | 75 | 74.3 | 64 | 63.4 | |
| Refused or N/A | 1 | 0.9 | 0 | 0.0 | |
| Education | |||||
| High school (<12 years) | 27 | 26.7 | 13 | 12.9 | 0.1169 |
| Some college/vocational school | 32 | 31.7 | 37 | 36.6 | |
| College graduate | 20 | 19.8 | 30 | 29.7 | |
| Postgraduate/professional school | 21 | 20.8 | 21 | 20.8 | |
| Refused or N/A | 1 | 1.0 | 0 | 0.0 | |
| Smoking status | |||||
| Never | 41 | 40.6 | 41 | 40.6 | 1.0000 |
| Current | 9 | 8.9 | 10 | 9.9 | |
| Former | 51 | 50.5 | 50 | 49.5 | |
| Pack-years smoking | |||||
| Never | 41 | 40.6 | 41 | 40.6 | 0.0950 |
| ≤5 | 9 | 8.9 | 22 | 21.8 | |
| 6–29 | 22 | 21.8 | 26 | 25.7 | |
| ≥30 | 28 | 27.7 | 12 | 11.9 | |
| N/A or refused | 1 | 1.0 | 0 | 0.0 | |
| Alcohol drinks per occasion in the past month | |||||
| None | 41 | 40.6 | 28 | 27.7 | 0.0505 |
| 1–4 | 53 | 52.5 | 61 | 60.4 | |
| ≥5 | 7 | 6.9 | 12 | 11.9 | |
| Lifetime number of people kissed with tongue | |||||
| None | 6 | 5.9 | 9 | 8.9 | 0.1629 |
| 1–9 | 28 | 27.7 | 33 | 32.7 | |
| 10–24 | 26 | 25.7 | 23 | 22.8 | |
| 25–49 | 15 | 14.9 | 19 | 18.8 | |
| ≥50 | 22 | 21.8 | 13 | 12.9 | |
| N/A or refused | 4 | 4.0 | 4 | 3.9 | |
| Gave oral sex in past 6 months | |||||
| No | 52 | 51.5 | 50 | 49.5 | 0.8844 |
| Yes | 43 | 42.6 | 45 | 44.6 | |
| N/A or refused | 6 | 5.9 | 6 | 5.9 | |
| Tonsillectomy | |||||
| No | 60 | 59.4 | 90 | 89.1 | <0.0001 |
| Yes | 40 | 39.6 | 11 | 10.9 | |
| N/A or refused | 1 | 1.0 | 0 | 0.0 | |
| Time since tonsillectomy (years ago) | |||||
| Never removed | 60 | 59.4 | 90 | 89.1 | <0.0001 |
| <2 years | 5 | 5.0 | 11 | 10.9 | |
| 2–29 years | 2 | 1.9 | 0 | 0.0 | |
| 30+ years | 32 | 31.7 | 0 | 0.0 | |
| N/A or Refused | 2 | 2.0 | 0 | 0.0 | |
| Gingivitis | |||||
| No | 78 | 77.2 | 71 | 70.3 | 0.3304 |
| Yes | 22 | 21.8 | 28 | 27.7 | |
| N/A or refused | 1 | 1.0 | 2 | 1.9 | |
| Teeth extracted prior to diagnosis | |||||
| 0 | 44 | 43.6 | 54 | 53.5 | 0.0558 |
| 1–9 | 31 | 30.7 | 35 | 34.7 | |
| ≥10 | 19 | 18.8 | 8 | 7.9 | |
| N/A or refused | 7 | 6.9 | 4 | 3.9 | |
| Tumor location | |||||
| Tonsil | 46 | 45.5 | |||
| Base of tongue (BOT) | 50 | 49.5 | |||
| Other oropharynx | 5 | 5.0 | |||
| Early or late disease presentation | |||||
| Early (T1–2 N0–1 [small tumors with only a single ipsilateral positive node <3 cm]) | 19 | 18.8 | |||
| Late | 82 | 81.2 | |||
| P16INK4a (IHC) | |||||
| Positive | 88 | 87.1 | |||
| Negative | 8 | 7.9 | |||
| N/A1 | 5 | 5.0 | |||
| HPV status in oral gargle (PCR) | |||||
| Any HPV type—positive | 92 | 91.1 | 33 | 32.7 | <0.001 |
| HPV 16—positive | 67 | 66.3 | 3 | 3.0 | <0.001 |
| HPV 18—positive | 3 | 3.0 | 0 | 0.0 | 0.2463 |
| HPV status in tumor (PCR) | |||||
| Any HPV type—positive | 96 | 95.0 | |||
| HPV 16—positive | 90 | 89.1 | |||
| HPV 18—positive | 3 | 3.0 | |||
| HR HPV (other than 16/18)—positive | 3 | 3.0 | |||
p16 staining was missing for five cases.
Figure 2 presents the distribution of methylation values among cases for each of the measured biomarkers, comparing values in the oral gargle and the tumor biopsy specimens, and comparing early vs. late disease presentation within the oral gargle and the tumor biopsy specimen. EPB41L3, HPV 16 L1, HPV 16 L2 and HPV 16 E2 methylation values were significantly higher in the tumor biopsy compared to the oral gargle specimen (p < 0.001). No significant differences in EPB41L3, HPV 16 L1 or HPV 16 E2 methylation levels were observed by extent of disease in either the oral gargle or tumor biopsy specimen (all p > 0.05). HPV 16 L2 oral gargle methylation levels were significantly lower for early vs. late disease, but were not different when measured in the tumor biopsy specimen. Tumor biopsy EPB41L3, HPV 16 L1, L2 and E2 methylation levels were similar regardless of tumor location (p > 0.05); however, oral gargle HPV 16 L1, L2 and E2 methylation levels were significantly lower for base of tongue compared to tumors diagnosed at the tonsils (p < 0.01; Supporting Information Fig. S1).
Figure 2.
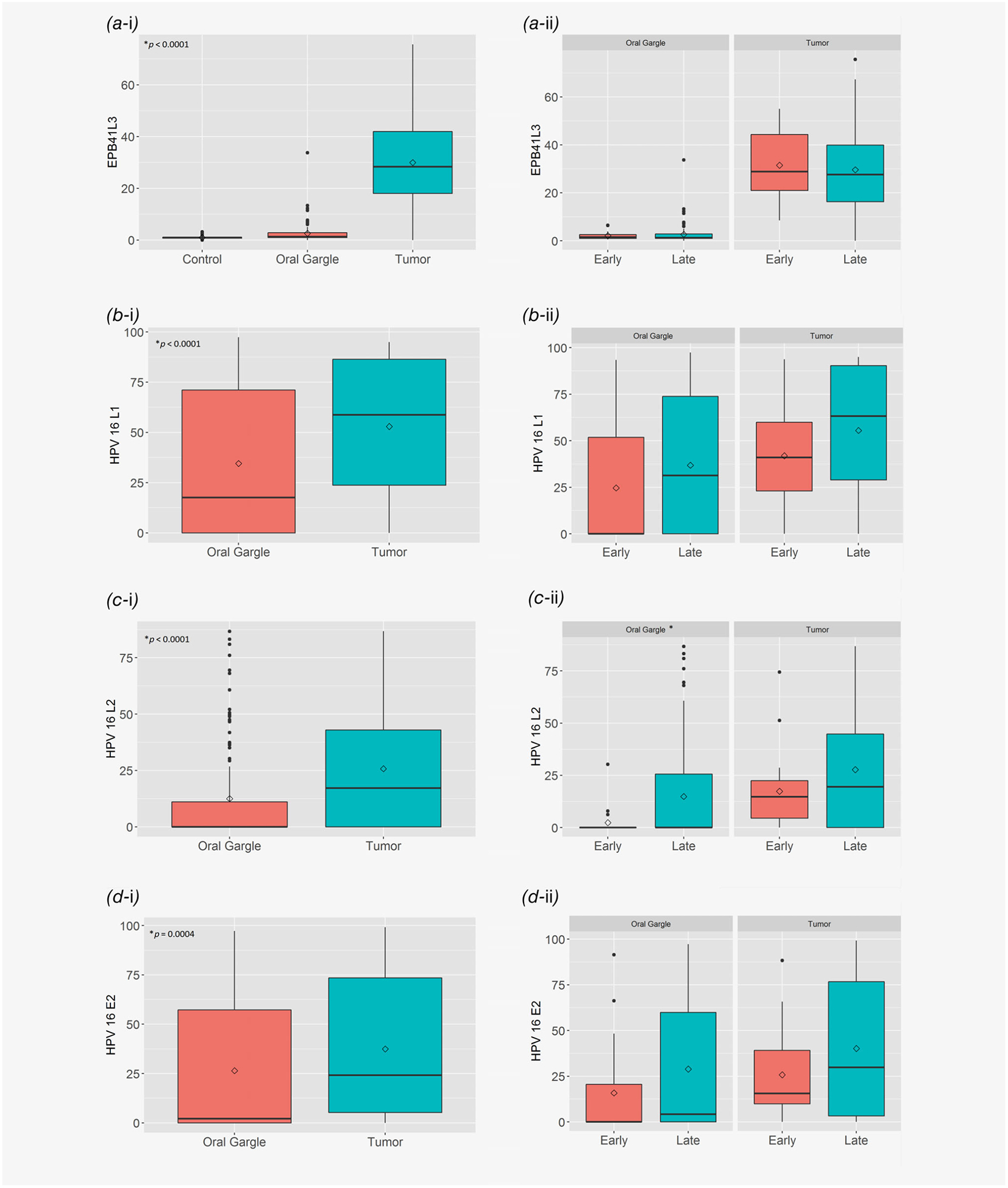
Distribution of oral gargle and tumor biopsy. (a) EPB41L3, (b) HPV 16 L1, (c) HPV 16 L2, and (d) HPV 16 E2 methylation levels among oropharyngeal cancer cases comparing oral gargle vs. tumor biopsy and early vs. late disease presentation: (a-i) EPB41L3—controls and cases (*Wilcoxon signed rank test, oral gargle vs. tumor; **p < 0.0001, Wilcoxon rank sum test, control vs. oral gargle or control vs. tumor); (a-ii) EPB41L3—early vs. late cases (*all comparisons p > 0.05, Wilcoxon rank sum test); (b-i) HPV 16 L1—cases only (*Wilcoxon signed rank test); (b-ii) HPV 16 L1—early vs. late cases (*all comparisons p > 0.05, Wilcoxon rank sum test); (c-i) HPV 16 L2—cases only (*Wilcoxon signed rank test); (c-ii) HPV 16 L2—early vs. late cases (*p < 0.05, Wilcoxon rank sum test); (d-i) HPV 16 E2—cases only (*Wilcoxon signed rank test). (d-ii) HPV 16 E2—early vs. late cases (*all comparisons p > 0.05, Wilcoxon rank sum test).
For each methylation biomarker evaluated (EPB41L3, HPV 16 L1, HPV 16 L2 and HPV 16 E2) among OPC cases, significant correlations (p < 0.01) were observed between the levels in tumor and levels measured in oral gargles (Table 2). In stratified analyses by extent of disease, p16 status and tumor location, significant correlations (p < .05) were observed for all four methylation biomarkers among late presentation cases, p16 cases and tonsillar tumors. In addition, oral gargle and tumor HPV 16 L1 and E2 methylation levels were significantly correlated for base of tongue tumors. Significant oral gargle-tumor biopsy correlation among early presentation cases was only observed for HPV 16 L1 (p = 0.01).
Table 2.
Oral gargle–tumor biopsy HPV status correlation, overall and by clinical disease presentation, p16 status and tumor location—cases only
| n | Oral gargle median | Tumor biopsy median | Correlation1 | p-value2 | |
|---|---|---|---|---|---|
| EPB41L3 | |||||
| Overall | 101 | 1.30 | 28.33 | 0.289 | 0.003 |
| Early disease3 | 19 | 1.53 | 28.80 | 0.376 | 0.113 |
| Late disease | 82 | 1.25 | 27.57 | 0.268 | 0.015 |
| p16 positive | 88 | 1.32 | 28.15 | 0.279 | 0.008 |
| p16 negative | 8 | 1.03 | 32.18 | 0.355 | 0.388 |
| Base of tongue | 50 | 1.08 | 23.27 | 0.094 | 0.516 |
| Tonsil | 46 | 1.80 | 30.22 | 0.426 | 0.003 |
| HPV 16 L1 methylation | |||||
| Overall | 101 | 17.60 | 58.75 | 0.430 | <0.0001 |
| Early disease3 | 19 | 0 | 41.00 | 0.575 | 0.010 |
| Late disease | 82 | 31.30 | 63.18 | 0.390 | 0.0003 |
| p16 positive | 88 | 30.83 | 60.60 | 0.435 | <0.0001 |
| p16 negative | 8 | 0 | 0 | 0.472 | 0.237 |
| Base of tongue | 50 | 0 | 54.03 | 0.465 | 0.0007 |
| Tonsil | 46 | 51.43 | 60.60 | 0.459 | 0.001 |
| HPV 16 L2 methylation | |||||
| Overall | 101 | 0 | 17.16 | 0.278 | 0.005 |
| Early disease3 | 19 | 0 | 14.71 | 0.237 | 0.329 |
| Late disease | 82 | 0 | 19.46 | 0.276 | 0.012 |
| p16 positive | 88 | 0 | 19.97 | 0.288 | 0.006 |
| p16 negative | 8 | 0 | 0 | - | - |
| Base of tongue | 50 | 0 | 16.46 | 0.235 | 0.101 |
| Tonsil | 46 | 3.87 | 18.34 | 0.303 | 0.041 |
| HPV 16 E2 methylation | |||||
| Overall | 101 | 2.12 | 24.10 | 0.454 | <0.0001 |
| Early disease3 | 19 | 0 | 15.60 | 0.451 | 0.053 |
| Late disease | 82 | 4.20 | 29.84 | 0.444 | <0.0001 |
| p16 positive | 88 | 2.90 | 28.67 | 0.471 | <0.0001 |
| p16 negative | 8 | 0 | 15.52 | 0.571 | 0.131 |
| Base of tongue | 50 | 0 | 19.45 | 0.358 | 0.011 |
| Tonsil | 46 | 28.13 | 31.54 | 0.524 | 0.0002 |
p16 was evaluated in 96/101 cases. Tumors originating at sites in the oropharynx other than base of tongue and tonsil were excluded due to sparse sampling (n = 5).
Spearman’s correlation coefficient.
p-value for Spearman’s correlation coefficient.
Early Disease = T1–2 with only a single ipsilateral positive node <3 cm, or less.
Oral gargle EPB41L3 methylation was significantly (p < 0.0001) higher among cases compared to controls (Table 3). The mean (SD) EPB41L3 methylation level was 2.59 (3.93) among all cases combined compared to 0.97 (0.63) among controls. In addition, significant differences (p < 0.001) in oral gargle EPB41L3 methylation levels were observed when comparing early disease cases (2.14 ± 1.75) with controls (0.97 ± 0.63). Oral gargle methylation of HPV 16 L1 and L2 was not detected among the three controls that were oral HPV 16 positive, and methylation of HPV 16 E2 was detected in one of three controls only. In contrast, oral gargle HPV 16 methylation was detected in all HPV 16 positive OPC cases (n = 63) for one or more of the three HPV genes evaluated (data not shown).
Table 3.
Oral gargle EPB41L3 methylation status among controls and oropharyngeal cancer cases stratified by early (T1–2 N0–1 [small tumors with only a single ipsilateral positive node <3 cm]) and late disease presentation
| Methylation | Controls (n = 101) | All cases (n = 101) | Early disease (n = 19) | Late disease (n = 82) | p-value1 |
|---|---|---|---|---|---|
| EPB41L3 | |||||
| Median | 0.90 | 1.30 | 1.53 | 1.25 | <0.0001 |
| Mean (SD) | 0.97 (0.63) | 2.59 (3.93) | 2.14 (1.75) | 2.69 (4.28) | 0.0003 |
| Interquartile range (Q1-Q3) | 0.73–1.13 | 0.97–2.87 | 1.00–3.07 | 0.93–2.87 | <0.0001 |
p values are from Wilcoxon Rank Sum tests comparing across two groups, that is, control vs. case, control vs. early disease and control vs. late disease.
Figures 3a–3c present the combined oral gargle HPV 16 gene methylation and EPB41L3 methylation receiver operator (ROC) curves for all cases (a), early disease cases (b) and late disease cases (c). Figure 3d (Supporting Information Table 1) presents the ROC for EPB41L3 methylation alone indicating an area under the curve (AUC) of 0.738. In comparison, the AUC for the combined EPB41L3 and HPV 16 methylation panel was 0.885 when all cases were evaluated, 0.866 when restricting to early disease cases and controls and 0.890 when restricting to late disease cases and controls. The bootstrap interval validation AUCs were 0.884, 0.864 and 0.890, respectively; thus, the AUC values are quite robust.
Figure 3.
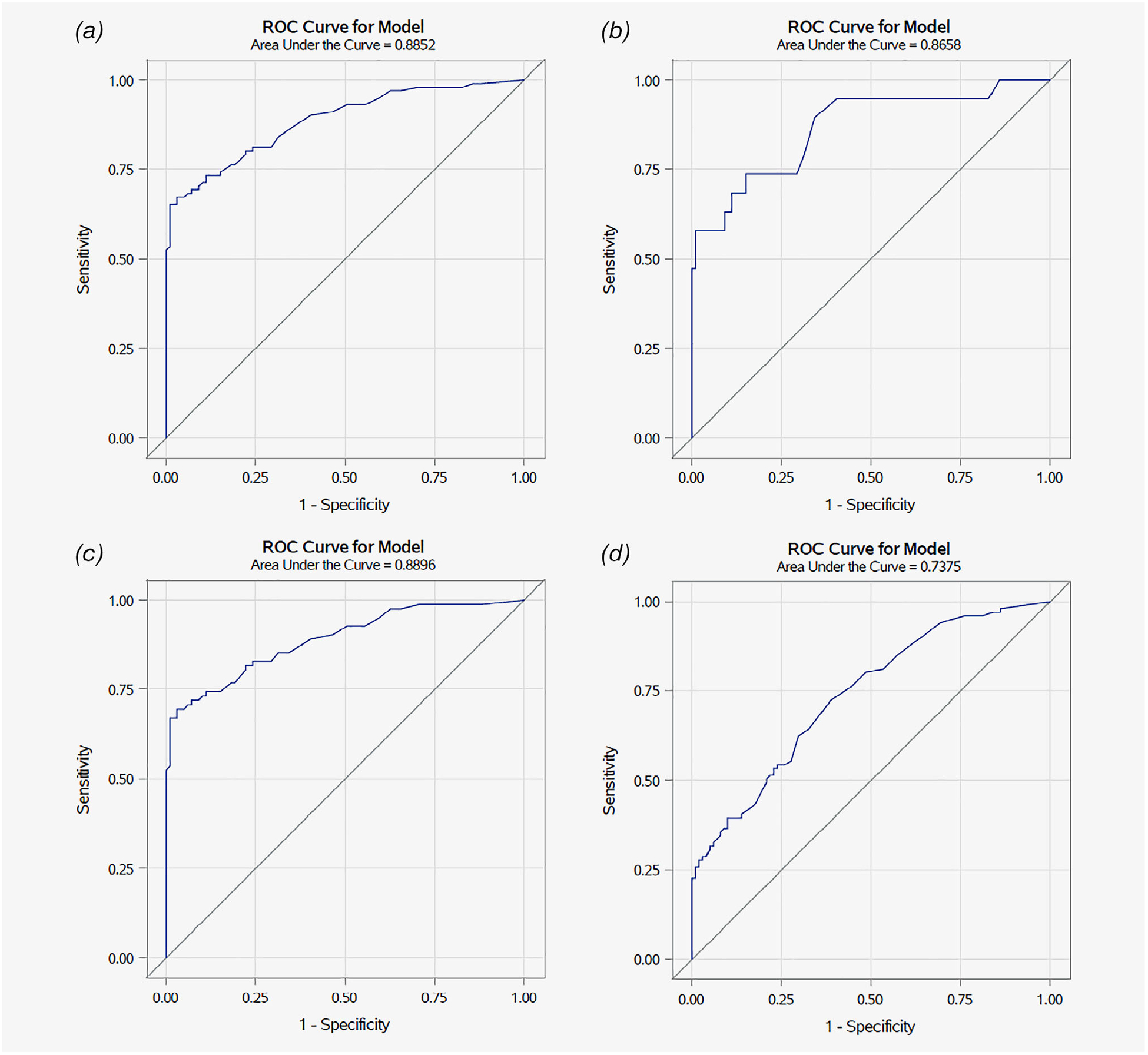
Receiver operating curve showing relationship of sensitivity and 1-specificity at various logistic regression model cut-points based on HPV 16 methylation and EPB41L3 methylation status to detect oropharyngeal cancer. Note: HPV16 methylation status was lacking for two controls. (a) All cases—ROC was calculated using 101 cases and 99 controls. (b) Early disease cases (T1–2 and N0–1, only if there is a single ipsilateral node <3 cm)—ROC was calculated using 19 cases and 99 controls. (c) Late disease cases—ROC was calculated using 82 cases and 99 controls. (d) ROC using EPB41L3 methylation alone—ROC was calculated using 101 cases and 101 controls.
As specificity is important for relatively rare outcomes such as OPC, we selected a biomarker cut point that optimized specificity while retaining >70% sensitivity (Fig. 2a; Supporting Information Table S1). Calling a score of 1.93 or higher positive gives a sensitivity of 70.3% and specificity of 90.9%. There were two Youden splits for the data; the first, at 2.70, had a sensitivity of 67.3% and specificity of 97.0%, while the second, at 3.27, had values of 65.3 and 99.0%, sensitivity and specificity, respectively. Thus, scores ≥3.27 have very high specificity, while those between 1.7 and 3.27 are highly suggestive of true disease, but for which repeat testing might be useful to reduce the false positive rate. Among early disease cases (Fig. 2b; Supporting Information Table S2) a score of 1.53 gives a sensitivity of 74% and specificity of 85%; however, these estimates are unstable due to the small sample size of early disease cases. Considering EPB41L3 methylation alone an AUC of 0.7378 was observed with specificity of 61.4% at a cut point that retained sensitivity at 70% or higher (72.3%; Supporting Information Table S3).
Discussion
OPC is one of only a few cancers whose incidence is significantly on the rise in the US41 and is the sixth most common cancer worldwide.42 While the rate of head and neck cancers (HNC) is decreasing in the US, OPC (a subset of HNCs) incidence is rapidly increasing among men and expected to continue to increase.2,43,44 Until recently, OPC was a rare cancer, hence screening was not considered a practical approach to cancer control. However, as OPC rates have significantly increased in the US, an early disease detection approach to prevent the morbidity and mortality associated with this cancer is now reasonable. Nationally, OPC incidence is 23.7–35.2/100,000 among men ≥50 years, a rate significantly higher than the incidence of cervical cancer among women of a similar age (7.9–11.8/100,000), due to effective screening for this cancer.45,46 Over the past two decades, there has been a shift in the primary cause of OPC, from tobacco to HPV, as smoking prevalence has declined. In the US, 70–90% of OPC cases are attributable to HPV.47 Two different etiologies of OPC are conceptualized—one promoted by tobacco and the other by HPV.48–52 Response to treatment differs significantly by tumor HPV status with poorer outcomes observed among HPV negative OPC cases. Despite better outcomes among HPV related OPC, ~10% of cases recur, hence the need for biomarkers that can detect cases at earlier stages when treatment is more efficacious. Our approach in our study was to evaluate two types of biomarkers, methylation of the HPV 16 viral genes, the HPV type responsible for >90% of HPV-related OPC in our study, and methylation of a tumor suppressor gene EPB41L3.
This is the first study to report levels of methylation of HPV 16 genes and EPB41L3 measured in tumors of OPC cases and oral gargle specimens from cases and controls with sufficient effect size and attributable fraction as to suggest a primary epigenetic association, a reasonable assumption given that to date no meaningful predisposing mutations have been found that may account for a majority of OPC cases. In prior studies of the cervix53 and anal canal54 methylation of HPV 16 genes and EPB41L3 were significantly positively associated with increasing grade of disease and methylation levels increased with time of progression to cancer.55 In our study, we report three important observations: (i) among OPC cases, methylation levels detected in the oral gargle and tumors were correlated, indicating that the measurement of these biomarkers in the oral gargle is representative of the tumor; (ii) HPV 16 methylation (L1, L2 and E2 genes) was predominantly observed among OPC cases and not detectable among disease-free men who were oral HPV 16 positive, except for methylation of E2 in one HPV 16 positive control; (iii) oral EPB41L3 methylation combined with HPV 16 gene methylation status had good sensitivity and high specificity for the detection of OPC. Oral gargle HPV 16 L1 and E2 methylation in p16 positive cases was observed although HPV 16 L2 methylation was not detected. These data indicate that methylation does not appear to occur across all HPV 16 genes simultaneously. It is also possible that in some samples the late region of the HPV is deleted due to genomic instability.
In prior studies, HPV 16 L1, L2 and E2 methylation was associated with prevalent cervical intraepithelial neoplasia (CIN3) as well as progression of HPV 16 to CIN3 and progression of HPV infection to invasive cancer.53,55 At the anal canal, a combined methylation score across CpG sites of these HPV 16 genes was linearly associated with anal disease severity with the lowest methylation levels observed in benign disease and highest levels among cases of high grade anal intraepithelial neoplasia (AIN).54 This pattern of HPV gene methylation associated with disease was also observed for HPV 18, HPV 31 and HPV 45 at the cervix.56,57 The consistency of these biomarkers across anatomic sites where HPV 16 and other carcinogenic HPV types cause cancer suggests that methylation of these particular HPV genes may be a common effect in HPV carcinogenesis regardless of epithelial site. Similarly, level of EPB41L3 methylation has been shown to be strongly associated with prevalent cervical and anal disease.53,54 Data from our study indicate that a similar epigenetic phenomenon to that observed at the cervix and anal canal may also mark carcinogenesis at the oropharynx.
Unlike the cervix or anal canal, it is challenging to obtain a representative cytological sample of the oropharynx and there are no reliably detected precancerous lesions of the oropharynx to utilize in screening studies. Therefore, OPC early detection will be necessity rely on measurement of informative biomarkers. The oral gargle is a convenient specimen to obtain in screening studies. In prior studies of the cervix and anal canal, HPV 16 gene and EPB41L3 methylation were measured directly from the target tissue of normal epithelium or lesions.53,54,56 As it is not feasible to reliably obtain cells from the oropharynx in routine examinations, we assessed the performance of the oral gargle as a proxy of the target tissue. To evaluate this, we measured the correlation between the oral gargle and tumor HPV 16 individual gene and EPB41L3 methylation levels. As shown, the correlation between the markers in these two specimens was high. Our data support the concept that the oral gargle may indeed be a suitable specimen to move forward with in developing early detection OPC biomarkers.
Using the oral gargle, our study demonstrated significant differences in methylation levels by case status. Important to the development of an early detection biomarker, significant differences in methylation levels were observed when comparing healthy controls and cases diagnosed with early disease. However, as only 3% of controls were oral HPV 16 positive, we were limited in further evaluating the HPV 16 gene methylation markers. In contrast, EPB41L3 methylation was measurable in all cases and controls. Using EPB41L3 combined with HPV 16 gene methylation status the ROC analysis indicated a relatively good sensitivity and high specificity for the detection of early disease OPC (AUC = 0.885 and 0.866 for all cases combined and early disease cases, respectively). Alone, EPB41L3 methylation yielded an AUC of 0.738. Combining methylation of HPV 16 and EPB41L3, we achieved a specificity of 99% at a sensitivity of 65% with a cutoff of 3.7. Given a relatively high cutoff, screening performance would be reasonable, with an ability to detect men most in need of expert attention while indicating very few as false positives. A methylation assay for HPV 16 and EPB41L3 could be no more expensive than an HPV screening test since it would involve only two targets as compared to 14 or more HPV types. While research progresses in identifying biomarkers that can identify OPC early, clinical research to develop methods to locate and/or detect primary tumors and small metastases is simultaneously needed.
There are several strengths to our study including the enrollment of cases pretreatment with carefully annotated disease characteristics, multiple specimens for evaluation, well characterized controls matched to cases on two key factors (tobacco use history and age) and robust assessment of HPV status and targeted gene methylation status. There are also study limitations that need to be considered in interpreting the results of our study. The relatively small sample size of early disease cases limited the statistical power to examine test sensitivity and specificity by a series of characteristics, such as HPV tumor status and smoking history. We evaluated OPC tumor-oral gargle biomarker concordance but did not have tissue from noncancer cases with which to compare tumor vs. normal tissue biomarker levels. In addition, we did not assess cellularity of the tumor specimen; however, the pathology panel confirmed presence of tumor cells in the specimens that was tested for biomarker status. In the evaluation of sensitivity and specificity, we only examined two biomarkers. It is likely that the test characteristics can be improved if a panel of biomarkers were to be evaluated. In the future, additional methylation biomarkers should be evaluated, including those identified by Ren et al. in their study of OPC tumors.58 Finally, as this is the first study to report results associating oral gargle HPV 16 and EPB41L3 methylation in OPC cases and controls, further studies are needed to replicate these findings in separate case–control sets and ultimately in large screening trials.
In conclusion, our study adds to the growing literature pointing to differential methylation of HPV genes and EPB41L3 as biomarkers of malignancy in diseases associated with HPV. To increase survival and reduce morbidity after an OPC diagnosis, an early detection-screening test that can be implemented as part of routine care is needed. The data presented here suggest that it may be possible to utilize biomarkers measured in an oral gargle as a screening specimen; however, additional test development is needed to refine the current test and move toward an early detection screening trial.
Supplementary Material
What’s new?
Oropharyngeal cancer is on the rise among men in the US, largely due to HPV infection, but it’s not typically screened for. Here, the authors investigated whether biomarkers in oral gargle specimens could provide a useful screening tool. They compared 101 OPC cases with controls matched for age and smoking history, looking at methylation in various regions known to be involved in cervical cancer. Among cases, they showed that methylation status in the oral sample correlated with the tumor sample. Methylation was significantly higher in cases than controls, suggesting that these biomarkers could be useful for early detection of OPC.
Acknowledgments
Conflict of interest: Dr Anna Giuliano is a member of the Merck Advisory Board and her institution has received funding for research through a Merck Investigator Initiated Studies Program. Dr Laura Martin-Gomez is currently an employee at GlaxoSmithKline. The rest of the authors report no conflicts.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Human papillomaviruses. IARC Monogr Eval Carcinog Risks Hum 2007;90:1–636. [PMC free article] [PubMed] [Google Scholar]
- 2.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 2011;29:4294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol 2013;31:4550–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marur S, D’Souza G, Westra WH, et al. HPVassociated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol 2010;11:781–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisbruch A, Harris J, Garden AS, et al. Multiinstitutional trial of accelerated hypofractionated intensity-modulated radiation therapy for earlystage oropharyngeal cancer (RTOG 00–22). Int J Radiat Oncol Biol Phys 2010;76:1333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minor J, Wang X, Zhang F, et al. Methylation of microRNA-9 is a specific and sensitive biomarker for oral and oropharyngeal squamous cell carcinomas. Oral Oncol 2012;48:73–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong DT. Salivary diagnostics for oral cancer. J Calif Dent Assoc 2006;34:303–8. [PubMed] [Google Scholar]
- 8.Franzmann EJ, Reategui EP, Pedroso F, et al. Soluble CD44 is a potential marker for the early detection of head and neck cancer. Cancer Epidemiol Biomarkers Prev 2007;16:1348–55. [DOI] [PubMed] [Google Scholar]
- 9.Lingen MW. Screening for oral premalignancy and cancer: what platform and which biomarkers? Cancer Prev Res 2010;3:1056–9. [DOI] [PubMed] [Google Scholar]
- 10.Fakhry C, Rosenthal BT, Clark DP, et al. Associations between oral HPV16 infection and cytopathology: evaluation of an oropharyngeal “pap-test equivalent” in high-risk populations. Cancer Prev Res 2011;4:1378–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Souza G, Sugar E, Ruby W, et al. Analysis of the effect of DNA purification on detection of human papillomavirus in oral rinse samples by PCR. J Clin Microbiol 2005;43:5526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillison ML, Broutian T, Pickard RK, et al. Prevalence of oral HPV infection in the United States, 2009–2010. J Am Med Assoc 2012;307: 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn SM, Chan JY, Zhang Z, et al. Saliva and plasma quantitative polymerase chain reactionbased detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol Head Neck Surg 2014;140: 846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chai RC, Lim Y, Frazer IH, et al. A pilot study to compare the detection of HPV-16 biomarkers in salivary oral rinses with tumour p16(INK4a) expression in head and neck squamous cell carcinoma patients. BMC Cancer 2016;16:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao M, Rosenbaum E, Carvalho AL, et al. Feasibility of quantitative PCR-based saliva rinse screening of HPV for head and neck cancer. Int J Cancer 2005;117:605–10. [DOI] [PubMed] [Google Scholar]
- 16.Agrawal Y, Koch WM, Xiao W, et al. Oral human papillomavirus infection before and after treatment for human papillomavirus 16-positive and human papillomavirus 16-negative head and neck squamous cell carcinoma. Clin Cancer Res 2008; 14:7143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koslabova E, Hamsikova E, Salakova M, et al. Markers of HPV infection and survival in patients with head and neck tumors. Int J Cancer 2013; 133:1832–9. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida H, Murono S, Ueno T, et al. Usefulness of human papillomavirus detection in oral rinse as a biomarker of oropharyngeal cancer. Acta Otolaryngol 2017;137:773–7. [DOI] [PubMed] [Google Scholar]
- 19.Gillison ML, D’Souza G, Westra W, et al. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst 2008;100:407–20. [DOI] [PubMed] [Google Scholar]
- 20.Martin-Gomez L, Fulp WJ, Schell MJ, et al. Oral gargle-tumor biopsy human papillomavirus (HPV) agreement and associated factors among Oropharyngeal squamous cell carcinoma (OPSCC) cases. Oral Oncol 2019;92:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillison ML, Castellsague X, Chaturvedi A, et al. Eurogin roadmap: comparative epidemiology of HPV infection and associated cancers of the head and neck and cervix. Int J Cancer 2014;134: 497–507. [DOI] [PubMed] [Google Scholar]
- 22.Ritchie JM, Smith EM, Summersgill KF, et al. Human papillomavirus infection as a prognostic factor in carcinomas of the oral cavity and oropharynx. Int J Cancer 2003;104:336–44. [DOI] [PubMed] [Google Scholar]
- 23.Louvanto K, Franco EL, Ramanakumar AV, et al. Methylation of viral and host genes and severity of cervical lesions associated with human papillomavirus type 16. Int J Cancer 2015;136: E638–45. [DOI] [PubMed] [Google Scholar]
- 24.Vasiljevic N, Scibior-Bentkowska D, Brentnall AR, et al. Credentialing of DNA methylation assays for human genes as diagnostic biomarkers of cervical intraepithelial neoplasia in high-risk HPV positive women. Gynecol Oncol 2014;132:709–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balderas-Loaeza A, Anaya-Saavedra G, RamirezAmador VA, et al. Human papillomavirus-16 DNA methylation patterns support a causal association of the virus with oral squamous cell carcinomas. Int J Cancer 2007;120:2165–9. [DOI] [PubMed] [Google Scholar]
- 26.Kreimer AR, Pierce Campbell CM, Lin HY, et al. Incidence and clearance of oral human papillomavirus infection in men: the HIM cohort study. Lancet 2013;382:877–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreimer AR, Villa A, Nyitray AG, et al. The epidemiology of oral HPV infection among a multinational sample of healthy men. Cancer Epidemiol Biomarkers Prev 2011;20:172–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bottalico D, Chen Z, Dunne A, et al. The oral cavity contains abundant known and novel human papillomaviruses from the Betapapillomavirus and Gammapapillomavirus genera. J Infect Dis 2011; 204:787–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Souza G, Gross ND, Pai SI, et al. Oral human papillomavirus (HPV) infection in HPV-positive patients with oropharyngeal cancer and their partners. J Clin Oncol 2014;32:2408–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown JL, Vanable PA, Eriksen MD. Computerassisted self-interviews: a cost effectiveness analysis. Behav Res Methods 2008;40:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson AM, Copas AJ, Erens B, et al. Effect of computer-assisted self-interviews on reporting of sexual HIV risk behaviours in a general population sample: a methodological experiment. AIDS 2001;15:111–5. [DOI] [PubMed] [Google Scholar]
- 32.Fairley CK, Sze JK, Vodstrcil LA, et al. Computerassisted self interviewing in sexual health clinics. Sex Transm Dis 2010;37:665–8. [DOI] [PubMed] [Google Scholar]
- 33.Geraets DT, Struijk L, Kleter B, et al. The original SPF10 LiPA25 algorithm is more sensitive and suitable for epidemiologic HPV research than the SPF10 INNO-LiPA extra. J Virol Methods 2015; 215–216:22–9. [DOI] [PubMed] [Google Scholar]
- 34.Giuliano AR, Lee JH, Fulp W, et al. Incidence and clearance of genital human papillomavirus infection in men (HIM): a cohort study. Lancet 2011; 377:932–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sudenga SL, Ingles DJ, Pierce Campbell CM, et al. Genital HPV infection progression to external genital lesions: the HIM study. Eur Urol 2016;69:166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierce Campbell CM, Kreimer AR, Lin HY, et al. Long-term persistence of oral human papillomavirus type 16: the HPV infection in men (HIM) study. Cancer Prev Res 2015;8:190–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasiljevic N, Wu K, Brentnall AR, et al. Absolute quantitation of DNA methylation of 28 candidate genes in prostate cancer using pyrosequencing. Dis Markers 2011;30:151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorincz AT, Brentnall AR, Scibior-Bentkowska D, et al. Validation of a DNA methylation HPV triage classifier in a screening sample. Int J Cancer 2016;138:2745–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrel FE Jr. Regression Modeling Strategies. New York, NY: Springer, 2019. [Google Scholar]
- 40.Steyerberg EW, Harrell FE Jr, Borsboom GJ, et al. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J Clin Epidemiol 2001;54:774–81. [DOI] [PubMed] [Google Scholar]
- 41.Ryerson AB, Eheman CR, Altekruse SF, et al. Annual report to the nation on the status of cancer, 1975–2012, featuring the increasing incidence of liver cancer. Cancer 2016;122:1312–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- 43.Gillison ML, Chaturvedi AK, Anderson WF, et al. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J Clin Oncol 2015;33:3235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viens LJ, Henley SJ, Watson M, et al. Human papillomavirus-associated cancers—United States, 2008–2012. MMWR Morb Mortal Wkly Rep 2016; 65:661–6. [DOI] [PubMed] [Google Scholar]
- 45.Van Dyne EA, Henley SJ, Saraiya M, et al. Trends in human papillomavirus-associated cancers—United States, 1999–2015. MMWR Morb Mortal Wkly Rep 2018;67:918–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jemal A, Simard EP, Dorell C, et al. Annual report to the nation on the status of cancer, 1975–2009, featuring the burden and trends in human papillomavirus(HPV)-associated cancers and HPV vaccination coverage levels. J Natl Cancer Inst 2013;105:175–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young D, Xiao CC, Murphy B, et al. Increase in head and neck cancer in younger patients due to human papillomavirus (HPV). Oral Oncol 2015; 51:727–30. [DOI] [PubMed] [Google Scholar]
- 48.D’Souza G, Fakhry C, Sugar EA, et al. Six-month natural history of oral versus cervical human papillomavirus infection. Int J Cancer 2007;121:143–50. [DOI] [PubMed] [Google Scholar]
- 49.Cook RL, Thompson EL, Kelso NE, et al. Sexual behaviors and other risk factors for oral human papillomavirus infections in young women. Sex Transm Dis 2014;41:486–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schabath MB, Villa LL, Lazcano-Ponce E, et al. Smoking and human papillomavirus (HPV) infection in the HPV in men (HIM) study. Cancer Epidemiol Biomarkers Prev 2012;21:102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schabath MB, Villa LL, Lin HY, et al. A prospective analysis of smoking and human papillomavirus infection among men in the HPV in men study. Int J Cancer 2014;134:2448–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giuliano AR, Sedjo RL, Roe DJ, et al. Clearance of oncogenic human papillomavirus (HPV) infection: effect of smoking (United States). Cancer Causes Control 2002;13:839–46. [DOI] [PubMed] [Google Scholar]
- 53.Mirabello L, Sun C, Ghosh A, et al. Methylation of human papillomavirus type 16 genome and risk of cervical precancer in a costa Rican population. J Natl Cancer Inst 2012;104:556–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorincz AT, Nathan M, Reuter C, et al. Methylation of HPV and a tumor suppressor gene reveals anal cancer and precursor lesions. Oncotarget 2017;8:50510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cook DA, Krajden M, Brentnall AR, et al. Evaluation of a validated methylation triage signature for human papillomavirus positive women in the HPV FOCAL cervical cancer screening trial. Int J Cancer 2019;144:2587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wentzensen N, Sun C, Ghosh A, et al. Methylation of HPV18, HPV31, and HPV45 genomes and cervical intraepithelial neoplasia grade 3. J Natl Cancer Inst 2012;104:1738–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasiljevic N, Scibior-Bentkowska D, Brentnall A, et al. A comparison of methylation levels in HPV18, HPV31 and HPV33 genomes reveals similar associations with cervical precancers. J Clin Virol 2014;59:161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren S, Gaykalova D, Wang J, et al. Discovery and development of differentially methylated regions in human papillomavirus-related oropharyngeal squamous cell carcinoma. Int J Cancer 2018;143: 2425–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We make the data collected as part of this protocol available to outside investigators, but within the limitations of preserving the anonymity of individuals since these data could theoretically identify a study participant. In order to maintain compliance with HIPAA regulations, we execute a data sharing agreement with the requestor for a limited use dataset as defined by the US Department of Health and Human Services (DHHS). We also encourage, but not require, collaborative use of the data. As with all datasets, there are many subtle nuances to the coding and interpretation of the data that cannot be completely documented in coding books or comment fields. Through a collaborative agreement, we feel that the quality of secondary analyses derived from these data can be greatly enhanced.
Data will be made available no later than 120 days after the publications of the main findings related to that dataset. We have established an internal Data Access Committee to review requests for the use of data according to these stipulations. Requests for data will be handled on a case-by-case basis, and will be evaluated by this Committee for scientific merit and ethical issues. For consortium use, our data are posted on a secure internal network drive accessed by passwords released on an individual basis. To make data available to outside investigators we employ Moffitt’s ELF (Exchange of Large Files) technology which allows users to access our data files from a secure website with a password and set of instructions sent by the IT department at Moffitt. Final datasets available to outside investigators will include reported demographic, diagnostic and risk factor information. Limited genotypic data will be made available to outside investigators to maintain compliance with confidentiality commitments. Outside investigators access to the secure ELF website is limited to the dataset prepared for that investigator. No biological samples will be released under this mechanism due to the limited nature of these samples. Outside investigators desiring access to such samples must set up collaboration with our consortium and also must agree that any data generated through their work will be governed by the data access rules that apply to the rest of this collaborative effort.
Because all of the collaborating sites have obligations to the participants to return important new information, we have collected and maintained personal identifying information. Although the final dataset will be stripped of identifiers prior to release for sharing, we believe that there remains the possibility of deductive disclosure of subjects. Thus, we will follow the data enclave model that make data available to users only under a data sharing agreement that provides for (i) a detailed summary of the proposed research project, including a complete list of data requested and limited specific aims, (ii) a commitment to using the data only for research purposes, (iii) a commitment to maintaining confidentiality of the data and not to identify any individual participant and (iv) a commitment to securing the data summary statistics using appropriate computer technology, (v) a commitment to destroying or returning the user data after analyses are completed, (vi) a commitment to publication of all results of these analyses and (vii) a commitment to reporting all results to the consortium in a timely manner (at least annually). The internal Data Access Committee is also responsible for evaluating the scientific validity, projected power and ethical aspects of the proposed use of our data prior to approval of release to any requesting investigator(s).


