Abstract
Objective
This study evaluated the degree to which recommendations for demographic data standardization improve patient matching accuracy using real-world datasets.
Materials and Methods
We used 4 manually reviewed datasets, containing a random selection of matches and nonmatches. Matching datasets included health information exchange (HIE) records, public health registry records, Social Security Death Master File records, and newborn screening records. Standardized fields including last name, telephone number, social security number, date of birth, and address. Matching performance was evaluated using 4 metrics: sensitivity, specificity, positive predictive value, and accuracy.
Results
Standardizing address was independently associated with improved matching sensitivities for both the public health and HIE datasets of approximately 0.6% and 4.5%. Overall accuracy was unchanged for both datasets due to reduced match specificity. We observed no similar impact for address standardization in the death master file dataset. Standardizing last name yielded improved matching sensitivity of 0.6% for the HIE dataset, while overall accuracy remained the same due to a decrease in match specificity. We noted no similar impact for other datasets. Standardizing other individual fields (telephone, date of birth, or social security number) showed no matching improvements. As standardizing address and last name improved matching sensitivity, we examined the combined effect of address and last name standardization, which showed that standardization improved sensitivity from 81.3% to 91.6% for the HIE dataset.
Conclusions
Data standardization can improve match rates, thus ensuring that patients and clinicians have better data on which to make decisions to enhance care quality and safety.
Keywords: record linkage, patient matching, data standards, interoperability, patient identification
INTRODUCTION
Every time a patient visits a hospital, health system, outpatient provider, clinic, pharmacy, long-term care provider, or public health agency, new information is generated. This information is stored in independent clinical repositories and, critically, no single unique identifier exists to easily and definitively combine this disparate information into a single comprehensive patient record.1,2 Even within single large institutions, like a health system or hospital, the internal billing systems, laboratory information systems, and electronic health records may effectively function as independent data silos relying on different patient identifiers to manage information (some of which may or may not exist in other systems). Fragmented patient information risks patient safety, hinders data aggregation for clinical decision support, prevents physicians from having comprehensive medical information, deters effective population health approaches, creates inefficiencies by delaying care, limits public health reporting, and severely reduces the utility of electronic data for clinical research.3,4 Using transaction volumes among health systems exchanging data within the Indiana Health Information Exchange (personal communication, Keith Kelly, April 18, 2018), we extrapolate a lower-bound estimate of 30 billion Health Level 7 messages transmitted (and requiring matching to a patient’s record) for the U.S. healthcare system annually. Consequently, even a small improvement in matching accuracy can potentially improve integration of a significant volume of clinical data into the appropriate patient record nationally.
The United States is the last industrialized nation without a national unique identification system.5 As a result, healthcare organizations must rely on patient matching algorithms driven by varying combinations of patient demographics and other identifiers. Matching algorithms can be effective, achieving match rates above 90% when implemented properly.6 However, algorithms must be paired with high quality, standardized data elements to optimize matching accuracy. Problematically, patient demographic data are captured in varying formats by healthcare organizations and health information technology systems. This lack of consistent approaches to data standardization has generated multiple best-practice recommendations, but no consensus on specific standardization approaches. Several organizations, including the Agency for Healthcare Research and Quality,7 the Health Information Management Systems Society,8 the Bipartisan Policy Center,9 the eHealth Initiative,10 the Markle Foundation,11 the Sequoia Project,12 and the Office of the National Coordinator for Health Information Technology (ONC),13–15 have either promoted or published best-practice approaches for optimizing patient matching, including data standardization.
While patient record linkage in the United States requires effective patient matching algorithms, recommendations for patient demographic data standardization have not been formally evaluated using real-world demographic data from diverse settings. Patient data generated through actual clinical care and business processes contains numerous inherent limitations, errors, and variations. Additionally, data generation and collection processes can be idiosyncratic by organization or by type of healthcare provider. Thus, this study seeks to evaluate the degree to which recommendations for demographic data standardization improve patient matching accuracy in real-world datasets.
MATERIALS AND METHODS
Using patient demographic data from 4 health datasets, we compared baseline matching accuracy to matching results after implementing best-practice recommendations. Matching represented 4 distinct use cases: hospital-to-hospital linkage, deduplicating a public health registration file, linking death records to clinical data, and matching newborn screening laboratory data to health information exchange (HIE) registration data.
Datasets and use cases
This analysis used 4 manually curated gold-standard analytic datasets, which contained a random selection of true-positive and true-negative matches.16–22 Through the manual curation process, we know the true match status for each record, which is a significant advantage over databases where the true matches are unknown.
HIE for hospital-to-hospital record matching
This dataset reflected demographic records from 2 geographically proximal hospital systems participating in an HIE. The data contained 50 000 sampled gold-standard pairs with 35 152 (70.3%) true positives and 14 858 (29.7%) true negatives.18,19 Patients from hospitals in close proximity cross over to other nearby institutions at significant rates,2 thereby creating the need to identify common records. The need to identify and capture information on patients seeking care from other institutions is dramatically increased by new value-based purchasing models like Accountable Care Organizations.2,23–25
Public health registry for de-duplicating
This dataset comes from the Marion County Health Department, Indiana’s largest public health department. The registry contains a master list of demographic information for clients who receive public health services such as immunizations, Women, Infants, and Children nutrition support, and laboratory testing.20,21 The registry also tracks health trends of populations and supports other public health activities, and duplicate patients can be unintentionally added. This dataset contained 33 005 sample pairs with 1950 (5.9%) true positives and 31 055 (94.1%) true negatives. We de-duplicated the complete patient registry. De-duplication is a process for identifying multiple copies of the same person in a single patient registry.
HIE and vital records for ascertaining death status
These data reflect a combination of the Social Security Death Master File and HIE data. This dataset contained 20 000 pairs with 16 873 (84.3%) true positives and 3127 (15.7%) true negatives. Accurately and comprehensively updating health records with patients’ accurate death status is critical to robust clinical quality measurement, public health reporting requirements, and high-quality clinical research.6,22
Laboratory results and HIE for newborn screening
This dataset included demographic data for newborns screened for congenital diseases (eg, sickle cell anemia, congenital hypothyroidism, etc.) as reported by multiple hospitals and private laboratories across these state and clinical records from the HIE. These data are limited to patients less than 1 month of age.16,17 This dataset contained 15 000 sampled gold-standard pairs with 13 456 (89.7%) true positives and 1544 (10.3%) true negatives. Not all infants are appropriately screened for harmful or potentially fatal disorders that are otherwise unapparent at birth.26 Although public health authorities can link vital records data with newborn screening results to identify unscreened infants, such processes may be delayed and some cases may remain undetected by this process.27
All 4 datasets contained subsets of the following fields: medical record number (MRN), social security number (SSN), last name (LN), first name (FN), middle name (MN), gender (G), month of birth (MB), day of birth (DB), year of birth (YB), full date of birth (DOB), street name, zip code (ZIP), city, state (ST), telephone number (TEL).
Data preparation and standardization
We standardized patient demographic data following the recommendations outlined in a 2014 report to the ONC.15 We selected the recommendations from the report to the ONC given the comprehensiveness of the evaluation as well as the agency’s responsibility to advance health data interoperability and ability to enforce or encourage standardization of data.
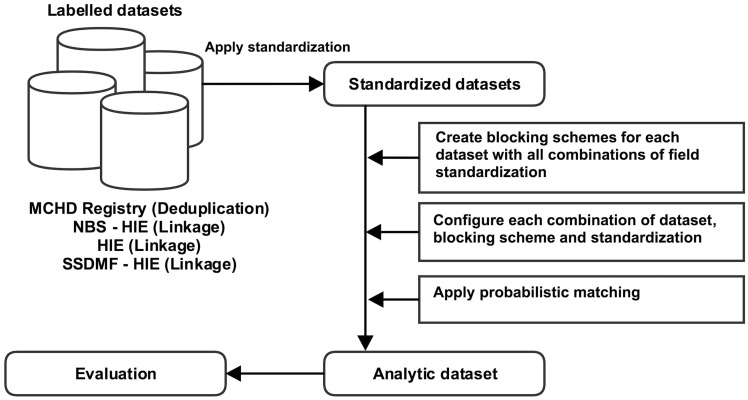
Figure 1 illustrates the data preparation process. First, we created a uniform analytical format for all datasets, with each dataset containing record pairs representing potentially matching patient records. To prevent an unmanageable and unnecessary number of record pairs from being created, we used a commonly applied technique in record linkage called “blocking.”28 Blocking refers to the process of grouping similar records together to form candidate matches. It is analogous to sorting socks by color before pairing them. Candidate matches are then evaluated to identify true matches. Fields suitable for use as blocking fields ideally have high variety of values and a low missing value rate. Blocking increases the proportion of true matches among possible pairs while decreasing the number of pairs to be evaluated.
Figure 1.
Overview of data analysis. We applied standardization methods to labelled datasets. We then applied blocking schemes to each dataset, applied probabilistic matching, and measured performance characteristics (e.g., sensitivity, specificity, etc.)
We standardized fields from across each dataset as follows15:
Last Name (LN): We applied the LN normalization rule defined by CAQH,29, which requires the removal of special characters such as apostrophes, hyphens, etc., and suffixes such as Jr, III, MD, etc. Examples include O’BRIEN→OBRIEN, SMITH JR.→SMITH, and JONES-THOMAS→JONESTHOMAS.
Telephone Number (TEL): Telephone numbers were standardized in adherence to International Telecommunications Union Recommendation E.123.30 This required converting raw telephone numbers into the standard format (123) 456–7890. Examples include 232 832–5555→(232) 832 5555, 2328325555→(232) 832 5555, 0002863866→286 3866, and 832–5555→832 5555.
Social Security Number (SSN): Invalid SSN numbers were identified based on rules provided by the Social Security Administration and replaced with null values.31 For example, if the first 3 digits were “000” or if the last 4 digits were “0000”, or the SSN lacked 9 digits, then the SSN was deemed invalid. Examples include 000004197→(null), 111220000→(null). The standardization method also removed hyphens: 123–45–6789→123456789. Additionally, SSNs that appeared in advertisements were deemed invalid and were thus replaced with null values. For example, 078051120→(null).
Full Date of Birth (DOB): All dates were converted to the format MM/DD/YYYY. Incorrect date values such as February 30 and September 31 were replaced with blank values. Month values greater than 12 and day of month values greater than 31 were nullified. Dates earlier than January 1, 1850, were also invalidated because our data source records do not include patients born before such a date. Examples include 2/15/197→(null), 9/28/→(null).
Address (ADD): We applied U.S. Postal Service certified address standardization rules,32 to correct and standardize address data including individual components (eg, standardizing Boulevard to Blvd, Drive to Dr) as well other formatting errors that would render the addresses undeliverable by the postal service. Examples include 6275 E WILSON CRK DR→6725 WILSON CREEK DR E, and 1902 N MARKET #312→1902 MARKET ST APT 312. Because street name and ZIP fields are strongly correlated with city and ST fields, we used only street and ZIP in our analysis.
To both identify as many true positive matches as possible and eliminate many obvious nonmatches, we created multiple blocking schemes,33 for each dataset with up to 32 (25) possible field standardization combinations. We used the expectation–maximization algorithm,34 to configure the matching algorithm for each combination of (1) dataset, (2) blocking fields, and (3) standardization fields. Finally, we used the Fellegi-Sunter (FS) probabilistic matching algorithm to identify matches for all datasets. The FS algorithm assigns a match score to each record pair based on the number and type of agreeing fields, producing algorithm-determined matches and nonmatches.
Analyses
The matching performance was evaluated using 4 metrics: sensitivity, specificity, positive predictive value, and overall accuracy, as indicated by the percent correctly classified as either true matches (sensitivity) or nonmatches (specificity), along with accurately identified pairs. These metrics were established based on the selection of match-score thresholds, defined in the algorithm as the likelihood of a correct match, where record pairs with a match score at or above the threshold were classified as matches and those with a matching score below the threshold were classified as nonmatches. Because match accuracy measurements will vary by the choice of matching score threshold, we elected to use 2 different match thresholds based on independent criteria: (1) optimizing Youden’s J statistic,35 which is the sum of sensitivity and specificity minus 1, and (2) optimizing the F score, which is a weighted harmonic mean of the sensitivity and positive predictive value. The use of these measures to optimize algorithm performance is well documented in the literature.36,37
Using both criteria for selecting matching score thresholds, we evaluated whether standardizing fields individually or collectively resulted in differences in each matching accuracy metric using the generalized estimating equations approach with logistic regression. Estimated mean differences as well as 95% confidence intervals (CIs) were calculated based on the model to evaluate the changes. This evaluation was performed for every blocking scheme and dataset because the samples of record pairs were randomly selected within each blocking scheme. Data from multiple blocking schemes were then pooled together for a dataset to examine the overall effect of field standardization.
RESULTS
HIE for hospital-to-hospital record matching
The HIE record pairs were created using 5 blocking combinations, including SSN; LN FN DOB; TEL; LN G DB YB ZIP; and FN G DB YB ZIP. Blocking fields are not used as matching fields. Consequently, when day and birth year are used as blocking fields, only birth month is used as a matching field in that particular blocking combination. Similarly, when the ZIP is used as a blocking field, street address will be used as a matching field in that particular blocking combination.
Table 1 lists the proportion of records standardized for demographic fields in each dataset. Note that address and TEL fields exhibited the highest proportion of transformed records, while LN exhibited a small amount of change within the newborn dataset. Standardization had little impact on DOB and SSN. While TEL underwent significant transformation, the changes largely altered formatting rather than content, and thus did not result in improved match accuracy.
Table 1.
Proportion of records standardized for demographic fields in each dataset
| Demographic Field | HIE | Public Health | Newborn | Death |
|---|---|---|---|---|
| address | 33.8% | 26.5% | 0.9% | 24.1% |
| dob | 0.0% | 0.1% | 0.0% | 0.0% |
| last name | 0.7% | 0.2% | 5.4% | 0.1% |
| ssn | 0.2% | 0.0% | 0.0% | 0.5% |
| tel | 88.6% | 70.6% | 92.3% | 35.2% |
HIE: health information exchange.
The SSN block contains 27 083 record pairs with 26 591 (98.18%) true matches. In this block, the only noticeable difference in matching accuracy occurs with address standardization. When thresholds are chosen to maximize Youden’s J statistic, there is a 1.3% (95% CI, 1.2%-1.5%) increase in sensitivity and a 3.5% (95% CI, 1.9%-5.1%) decrease in specificity. This results in a 1.3% (95% CI, 1.1%-1.4%) improvement in the overall accuracy, in which the percent of record pairs correctly classified increases from 90.7% without address standardization to 92% with address standardization.
The LN FN DOB block contains 32 171 record pairs with 64 778 (88.20%) true matches. Again, in this block, address standardization makes the only noticeable difference in matching accuracy when thresholds are chosen to maximize Youden’s J statistic. There is a 3.3% (95% CI, 3.1%-3.5%) increase in sensitivity and an 8.2% (95% CI, 6.9%-9.4%) decrease in specificity. The overall accuracy is improved from 84.9% without address standardization to 87.6% with address standardization, indicating a 2.7% (95% CI, 2.5%-2.9%) absolute improvement.
The TEL block captured 21 415 record pairs with 11 604 (54.19%) true matches. Field standardization does not result in meaningful changes in any matching accuracy metrics, regardless of how the matching score thresholds are selected.
The LN G DB YB ZIP block contains 24 406 record pairs with 23 064 (94.5%) true matches. Again, in this block, address standardization has the only noticeable effect when thresholds are chosen to maximize Youden’s J statistic. Sensitivity increases by 3.2% (95% CI, 2.9%-3.5%) and specificity decreases by 9.5% (95% CI, 8%-11.1%) after address is standardized. The overall accuracy is improved from 77.7% without address standardization to 80.2% with address standardization, indicating a 2.5% (95% CI, 2.2%-2.8%) improvement.
In the FN G DB YB ZIP block, there are 25 569 record pairs with 23 158 (90.6%) true matches. Standardizing the address again shows a large effect when thresholds are chosen to maximize Youden’s J statistic. Sensitivity increases by 8.3% (95% CI, 8%-8.6%) and specificity decreases by 11.6% (95% CI, 10.5%-12.6%) after address is standardized. The overall accuracy is improved from 82% without address standardization to 88.3% with address standardization, indicating a 6.3% (95% CI, 6.1%-6.6%) improvement. In addition, LN standardization increases the sensitivity by 2.9% (95% CI, 2.8%-3%) and decreases the specificity by 1.9% (95% CI, 1.7%-2.1%), leading to a 2.2% (95% CI, 2.1%-2.3%) improvement of the overall accuracy.
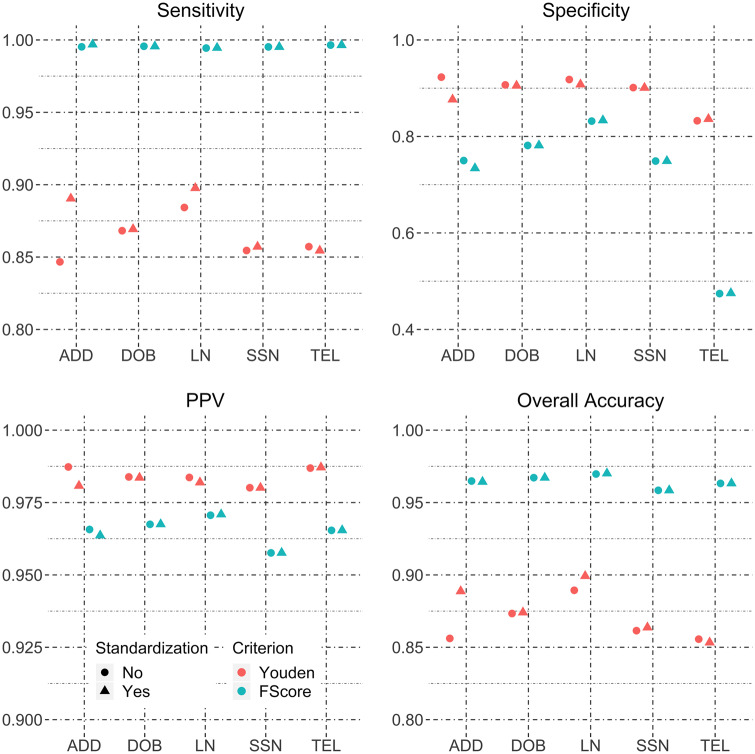
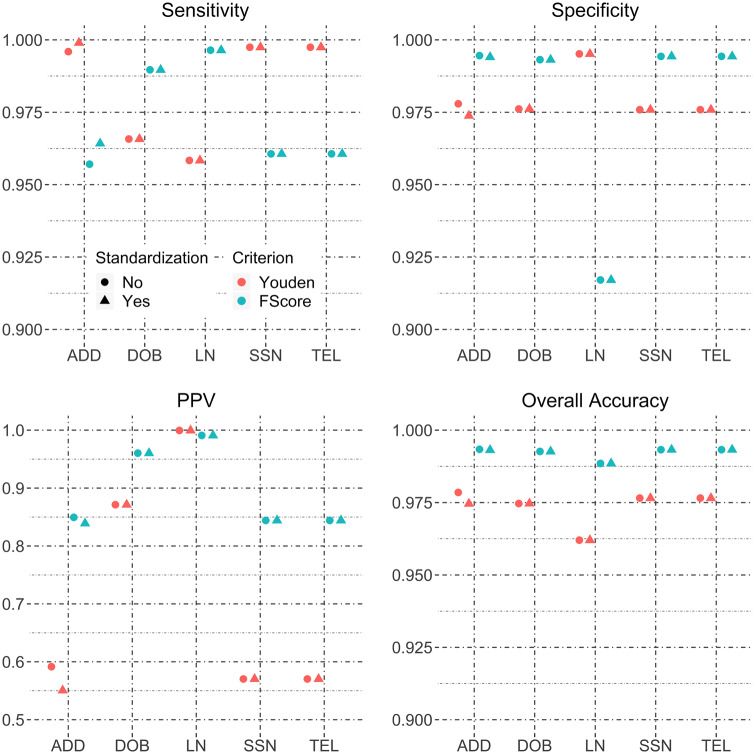
When data from all 5 blocking schemes are pooled, the estimated matching accuracies are shown in Figure 2 . It is obvious that standardizing DOB, SSN, or TEL does not improve any matching accuracy metrics. Standardizing the address appears to have a larger effect, while LN standardization shows a relatively smaller effect when matching thresholds are chosen to maximize Youden’s J statistic. When using the F score to select matching score thresholds, no field standardizations show a difference in accuracy metrics. These findings suggest that address and LN standardization improve matching performance, but the magnitude of improvement will be influenced by the choice of score threshold.
Figure 2.
Sensitivity, specificity, positive predictive value, and overall accuracy for field standardization of the HIE hospital-to-hospital data pooling data from all blocking schemes.
Combined field standardization
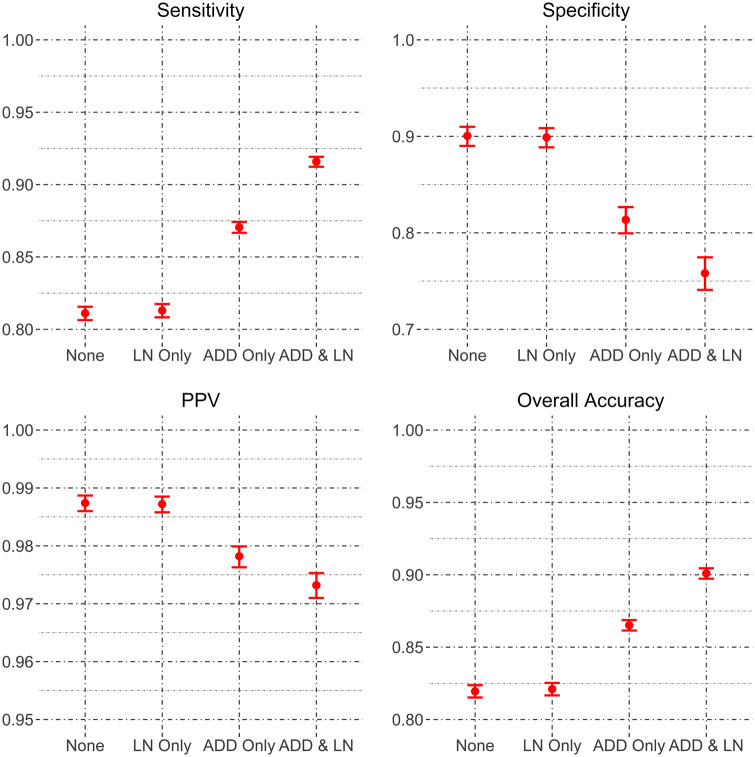
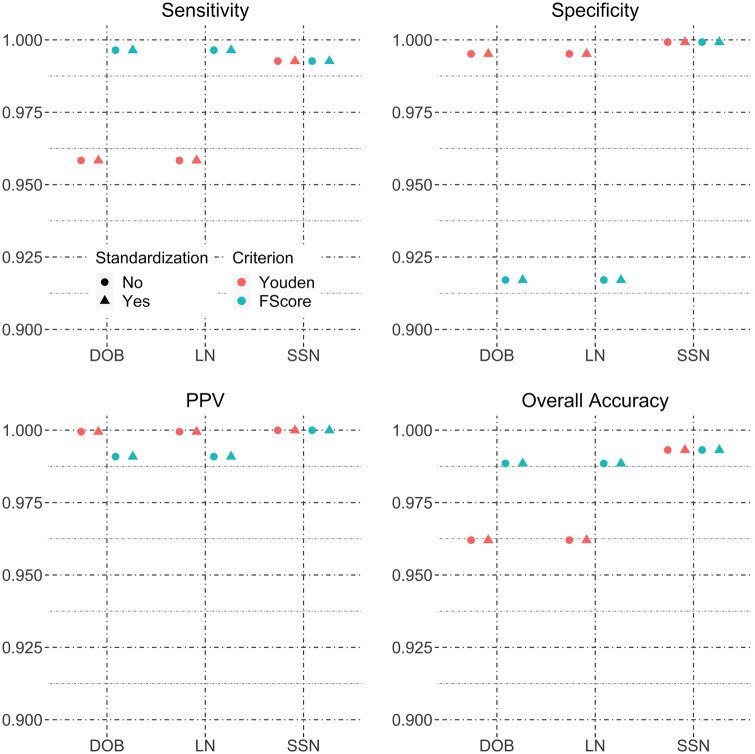
In the FN G DB YB ZIP block, because standardizing address and LN improved matching sensitivity, we further examine the combined effect of address and LN standardization. Estimated matching accuracies when both address and LN are standardized, compared with when only 1 field is standardized or no field is standardized, are shown in Figure 3 . Standardizing both address and LN improves the sensitivity to 91.6% from 81.3% (increase = 10.3%; 95% CI, 10%-10.6%) when only LN is standardized or 87.1% (increase = 4.5%; 95% CI, 4.4%-4.7%) when only address is standardized. The dual standardization decreases the specificity to 75.8% from 89.9% (decrease = 14.1%; 95% CI, 13.2%-15.6%) when only LN is standardized or 81.4% (decrease = 5.6%; 95% CI, 4.8%-5.8%) when only address is standardized. The increased sensitivity has greater importance as vast majority of the record pairs in the data are true matches. As a result, the overall accuracy improves to 90% with dual standardization from 82.1% (increase = 7.9%; 95% CI, 7.6%-8.3%) when only LN is standardized or 86.5% (increase = 3.5%; 95% CI, 3.4%-3.8%) when only address is standardized.
Figure 3.
Sensitivity, specificity, positive predictive value, and overall accuracy for standardizing address and/or last name for the HIE hospital-to-hospital FN G DB YB ZIP block.
Laboratory results and HIE for newborn screening
The newborn screening data was blocked using 3 schemes, with TEL, LN FN, and MRN as blocking variables. The TEL block contains 11 029 record pairs with 9739 (88.3%) true matches. In this block, neither LN nor DOB standardization results in changes in matching accuracy, regardless of the criteria used for the selecting the matching score thresholds. The LN FN block contained 2716 record pairs with 2583 (95.1%) true matches. None of the 4 matching accuracy measures are changed by the standardizing the fields, regardless of how matching score thresholds are selected. The MRN block contained 9179 record pairs with 9038 (98.46%) true matches. Field standardization again does not result in improved matching accuracy. The matching accuracies estimated based on pooling all 3 blocking schemes together are shown in Figure 4 . Standardizing DOB, LN, or TEL does not improve any of the 4 matching accuracy metrics.
Figure 4.
Sensitivity, specificity, positive predictive value, and overall accuracy for field standardization of the NBS data pooling data from all 3 blocking schemes.
Public health registry for de-duplicating
The Marion County Health Department data was blocked using 2 schemes, 1 with SSN and the other with LN and LN FN as blocking variables. The SSN block contains 2141 record pairs with 1508 (70.43%) true matches. In this block, the only observed difference in matching accuracy occurs with address standardization when thresholds are chosen to maximize Youden’s J statistic. There is a 0.3% (95% CI, -0.1%-0.7%) increase in sensitivity and a 2.4% (95% CI, 1.1%-3.6%) decrease in specificity. This, however, does not change the overall accuracy due to the fact that very few record pairs are nonmatches. Standardizing other fields or using the F measure for threshold selection does not lead to changes in any matching accuracy metrics.
The LN FN block contained 31 420 record pairs with 978 (3.11%) true matches. Standardizing address appears to slightly increase the sensitivity and decrease the specificity by less than half-percent changes, regardless of how matching score thresholds are selected. The leads to a 4% (95% CI, 3.3%-4.9%) and 1% (95% CI, 0.4%-1.7%) reduction in positive predictive value when selecting matching score thresholds by optimizing Youden’s J statistic and F score, respectively. However, these did not change the overall percent of correctly classified pairs due to the vast majority of record pairs being nonmatches.
When data from both blocking schemes are pooled, the estimated matching accuracies are shown in Figure 5 . Standardizing DOB, LN, SSN, or TEL does not improve any of the 4 matching accuracy metrics. Standardizing address appears to have some effect, but this is negligible.
Figure 5.
Sensitivity, specificity, positive predictive value, and overall accuracy for field standardization of the public health registry data pooling data from both blocking schemes.
HIE and vital records for ascertaining death status
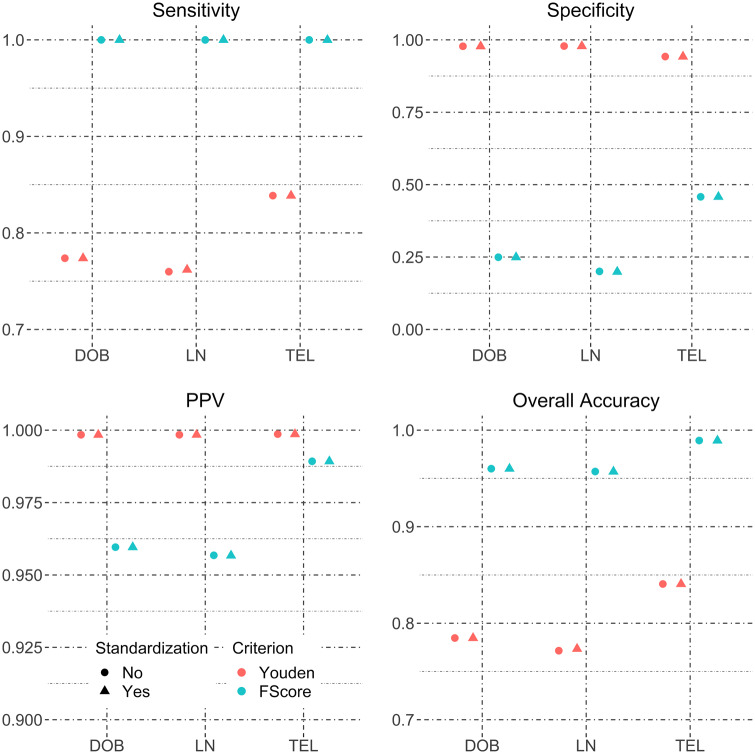
The Social Security Death Master File data were blocked using 2 schemes, 1 with SSN and the other with LN FN DOB as blocking variables. The SSN block contains 18 615 record pairs with 16 758 (90%) true matches, while the LN FN DOB block contained 16 798 record pairs with 15 527 (92.4%) true matches. Standardization results in no changes for any matching accuracy metrics, regardless of how matching score thresholds are selected. This is shown by the same matching accuracy before and after field standardization for every field and every threshold selection criterion in Figure 6 .
Figure 6.
Sensitivity, specificity, positive predictive value, and overall accuracy for field standardization of the HIE-to-social security data pooling data from both blocking schemes.
DISCUSSION
Our research found that standardizing certain individual demographic data fields yields incremental improvements in match performance among hospital-to-hospital exchange, while combined standardization can produce more meaningful improvements, which would further ensure that patient data can be linked across organizations to improve care coordination and lower costs. Specifically, using a database of 100 000 records that represents hospital to hospital data exchange, we found sensitivity increases of up to 10%, which reduces the number of unlinked records in the dataset by nearly half.
Several limitations exist. First, while we observed increased false positive matches (decreased specificity) associated with data standardization, this is not unexpected. False positive matches can be mitigated by introducing deterministic exclusion criteria specific to the offending match pattern (eg, declare a nonmatch if corresponding dates of birth disagree), an approach commonly taken when optimizing match performance for specific data sources. Second, our results are specific to the blocking schemes we selected. Other blocking schemes may yield different results. Third, while we used only 1 algorithm, the FS model, this model is a common component of many patient matching systems. Fourth, we limited our analysis to datasets specific to Indiana. However, the data included in this study represent a broad spectrum of healthcare settings from rural to urban as well as hospital and clinic settings, and therefore the findings likely are to have applicability to hospital to hospital exchange on a nationwide scale.
Ultimately, we found that the recommendation by many policy organizations for enhanced standardization of certain data elements can yield matching improvements. Because LN and address information are commonly entered as unstructured text, they can exhibit meaningful variation. We hypothesize that LN and address standardization is associated with match rate improvement because it minimizes this variation. Consequently, we found utility in standardizing address and LN in combination, which can significantly reduce the number of unmatched records. Conversely, we did not find evidence for standardizing DOB and TEL. Given the limited degrees of freedom for information for these fields, we hypothesize that these fields require little standardization.
We also sought information on the costs needed to implement standardization by hospitals or health information technology vendors. While we could not find clear cost data, standardization and deployment of those standards in to their products by electronic health record developers would help consolidate costs as opposed to having every healthcare facility conduct the standardization. We also note that the costs for standardizing the data as part of this project were limited, which may provide some context for nationwide standardization among vendors.
With an incomplete evidence base to more firmly support standardization, healthcare organizations, health information technology (IT) developers and policymakers may be less inclined to pursue approaches with unclear value or they may implement methods that, upon further study, prove to be less effective and generalizable than initially perceived.
Given that accurate patient matching is essential for maximizing health data quality, where opportunities exist, health IT vendors should prioritize incorporating address standardization—which we found in this study can decrease the number of unlinked records by up to 20%—functionality into their patient registration products. Relying solely on individual vendors may result in incomplete implementation across the industry because some may elect not to implement address standardization. Little information exists on the degree to which organizations currently standardize data, though informal discussions and recommendations made by many groups suggest that there is not widespread use of the same standards. While data standardization would yield partial benefit even if only 1 party in a transaction standardizes the data, maximal benefit requires that all systems adopt the same standardization rules. Consequently, health IT policymakers, including the ONC, should explore strategies for expanding the evidence base for the value of data consistency and encouraging broad deployment of address standardization. These strategies may include updates to its certification criteria for electronic health records, issuing guidance to encourage the voluntary standardization of data, or incorporating standardization in to its plans to create a nationwide interoperability framework. Similarly, vendors and ONC should further examine the utility of LN standardization so that the increased match rates when used in conjunction with LN can be realized.
CONCLUSIONS
Standardizing certain demographic data on a broader scale can improve match rates, ensuring that patients and clinicians have better data on which to make decisions to enhance care quality and safety.
FUNDING
This work was supported by Pew Charitable Trust grant number PEW30381.
CONTRIBUTORS
SJG contributed to the conception, design, acquisition, analysis, and interpretation of data for the work. HX and NB performed analysis and interpretation of data. JV, SK, and JR provided interpretation of data. BM and RT contributed to conception and design.
Conflict of interest statement. None declared.
REFERENCES
- 1. McDonald CJ, Overhage JM, Dexter PR et al. . Canopy computing using the web in clinical practice. JAMA 1998; 280 (15): 1325–9. [DOI] [PubMed] [Google Scholar]
- 2. Finnell JT, Overhage JM, Grannis SJ. All health care is not local: an evaluation of the distribution of emergency department care delivered in Indiana. AMIA Annu Symp Proc 2011; 2011: 409–16. [PMC free article] [PubMed] [Google Scholar]
- 3. Friedman CP, Wong AK, Blumenthal D. Achieving a nationwide learning health system. Sci Transl Med 2010; 2 (57): 57cm29. [DOI] [PubMed] [Google Scholar]
- 4. Mason AR, Barton AJ. The emergence of a learning health care system. Clin Nurse Spec 2013; 27 (1): 7–9. [DOI] [PubMed] [Google Scholar]
- 5. Hillestad R, Bigelow JH, Chaudhry B et al. . Identity Crisis: An Examination of the Costs and Benefits of a Unique Patient Identifier for the US Health Care System. Santa Monica, CA: RAND Corporation, MG-753-HLTH; 2008. [Google Scholar]
- 6. Grannis SJ, Overhage JM, Hui S, McDonald CJ. Analysis of a probabilistic record linkage technique without human review. AMIA Annu Symp Proc 2003; 2003: 259–63. [PMC free article] [PubMed] [Google Scholar]
- 7. Grannis SJ, Banger A, Harris D Perspectives on Patient Matching: Approaches, Findings, and Challenges. Agency for Health Care Research and Quality and Office of the National Coordinator for Health Information Technology; 2009. http://www.healthit.gov/policy-researchers-implementers/health- information-security-privacy-collaboration-hispc. Accessed April 28, 2014.
- 8. Consistent Nationwide Patient Data Matching Strategy. Health Information Management System Society 2013 Policy Summit Congressional Ask #1 Recommendation to Congress; September 2013. http://www.himss.org/files/HIMSSorg/Congressional_Asks_1%202013_InteroperabilityPatientID.pdf. Accessed April 2, 2014.
- 9. Marchibroda J. Challenges and strategies for accurately matching patients to their health data. Bipartisan Policy Center; June 2012. http://bipartisanpolicy.org. Accessed January 2, 2014.
- 10. Health IT: Setting the Foundation to Transform Our Future. eHealth Initiative Government Affairs Retreat; February 2014. http://www.ehidc.org/resource-center/publications/doc_download/386-event-summary-ehi-government-affairs-retreat-2014-health-it-setting-the-foundation-to-transform-our-future. Accessed April 20, 2014.
- 11. Linking Health Care Information: Proposed Methods for Improving Care and Protecting Privacy. The Markle Foundation; February 2005. http://www.markle.org/publications/863-linking-health-care-information-proposed-methods-improving-care-and-protecting-priv. Accessed March 20, 2014.
- 12. Heflin E. A Framework for Cross-Organizational Patient Identity Management: Draft for Public Review and Comment. The Sequoia Project; November 10, 2015. http://sequoiaproject.org. Accessed October 1, 2016.
- 13. Morris G, et al. Patient identification and matching initial findings. HealthIT.gov: the official site for Health IT information; December 16, 2013. http://www.healthit.gov. Accessed January 22, 2016.
- 14. Tang P. Recommendations to the Department of Health and Human Services on patient matching. Health IT Policy Committee: Recommendations to the National Coordinator for Health IT; February 8, 2011. http://www.healthit.gov. Accessed March 3, 2014
- 15. Morris G, et al. Patient Identification and Matching Final Report. HealthIT.gov: the official site for Health IT information; February 7, 2014. http://www.healthit.gov. Accessed September 13, 2016.
- 16. Zhu VJ, Overhage MJ, Egg J, Downs SM, Grannis SJ. An empiric modification to the probabilistic record linkage algorithm using frequency-based weight scaling. J Am Med Inform Assoc 2009; 16 (5): 738–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Daggy J, Xu H, Hui S, Grannis S. Evaluating latent class models with conditional dependence in record linkage. Statist Med 2014; 33 (24): 4250–65. [DOI] [PubMed] [Google Scholar]
- 18. Wu J, Xu H, Finnell JT, Grannis SJ. A practical method for predicting frequent use of emergency department care using routinely available electronic registration data. AMIA Annu Symp Proc 2013; 2013: 1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grannis SJ, Overhage JM, McDonald C. Real world performance of approximate string comparators for use in patient matching. Stud Health Technol Inform 2004; 107 (Pt 1): 43–7. [PubMed] [Google Scholar]
- 20. Xu H, Hui SL, Grannis S. Optimal two-phase sampling design for comparing accuracies of two binary classification rules. Statist Med 2014; 33 (3): 500–13. [DOI] [PubMed] [Google Scholar]
- 21. Daggy JK, Xu H, Hui SL, Gamache RE, Grannis SJ. A practical approach for incorporating dependence among fields in probabilistic record linkage. BMC Med Inform Decis Mak 2013; 13: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grannis SJ, Overhage JM, McDonald CJ. Analysis of identifier performance using a deterministic linkage algorithm. Proc AMIA Symp 2002: 305–9. [PMC free article] [PubMed] [Google Scholar]
- 23. Devore S, Champion RW. Driving population health through accountable care organizations. Health Aff (Millwood) 2011; 30 (1): 41–50. [DOI] [PubMed] [Google Scholar]
- 24. Wu FM, Rundall TG, Shortell SM, Bloom JR. Using health information technology to manage a patient population in accountable care organizations. J Health Org Mgt 2016; 30 (4): 581–96. [DOI] [PubMed] [Google Scholar]
- 25. McWilliams JM, Hatfield LA, Chernew ME, Landon BE, Schwartz AL. Early performance of accountable care organizations in medicare. N Engl J Med 2016; 374 (24): 2357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rock MJ, Levy H, Zaleski C, Farrell PM. Factors accounting for a missed diagnosis of cystic fibrosis after newborn screening. Pediatr Pulmonol 2011; 46 (12): 1166–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoff T, Ayoob M, Therrell BL. Long-term follow-up data collection and use in state newborn screening programs. Arch Pediatr Adolesc Med 2007; 161 (10): 994–1000. [DOI] [PubMed] [Google Scholar]
- 28. Michelson M, Knoblock CA. Learning blocking schemes for record linkage. In: Proceedings of the 21st National Conference on Artificial Intelligence – Volume 1 (AAAI’06), Boston, MA: AAAI Press; 2006: 440–5 [Google Scholar]
- 29. Council for Affordable Quality Health Care 2011. Normalizing Patient Last Name Rule; March 2011. http://www.caqh.org. Accessed December 18, 2013
- 30. Series E: Overall Network Operation Telephone Service, Service Operation and Human Factors. ITU-T Recommendation E.123. https://www.itu.int/rec/dologin_pub.asp?lang=e&id=T-REC-E.123–200102-I!! PDF-E&type=items. Accessed May 27, 2016.
- 31. High Group List and Other Ways to Determine if an SSN is Valid. SSA.gov. https://www.ssa.gov/employer/ssnvhighgroup.htm. Accessed May 25, 2017.
- 32. Mailing Standards of the United States Postal Service Publication 28 – Postal Addressing Standards. USPS Publication 28. PSN 7610–03–000–3688; Updated 2013. Usps.gov. http://pe.usps.gov/cpim/ftp/pubs/pub28/pub28.pdf. Accessed February 20, 2014.
- 33. Christen P. A survey of indexing techniques for scalable record linkage and deduplication. IEEE Trans Knowl Data Eng 2011; 24 (9): 1537–55. June; [Google Scholar]
- 34. Moon TK. The expectation-maximization algorithm. IEEE Signal Process Mag 1996; 13 (6): 47–60. [Google Scholar]
- 35. Youden WJ. Index for rating diagnostic tests. Cancer 1950; 3 (1): 32–5. [DOI] [PubMed] [Google Scholar]
- 36. Unal I. Defining an optimal cut-point value in roc analysis: an alternative approach. Comput Math Methods Med 2017; 2017: 3762651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Z, Tan M, Jiang F. Regularized F-Measure Maximization for Feature Selection and Classification. J Biomed Biotechnol 2009; 2009: 617946. [DOI] [PMC free article] [PubMed] [Google Scholar]