Abstract
Background and objective
Little is yet known whether pathogenesis of COVID-19 is different between young and elder patients. Our study aimed to investigate the clinical characteristics and provide predictors of mortality for young adults with severe COVID-19.
Methods
A total of 77 young adults with confirmed severe COVID-19 were recruited retrospectively at Tongji Hospital. Clinical characteristics, laboratory findings, treatment and outcomes were obtained from electronic medical records. The prognostic effects of variables were analyzed using logistic regression model.
Results
In this retrospective cohort, non-survivors showed higher incidence of dyspnea and co-existing laboratory abnormalities, compared with young survivals in severe COVID-19. Multivariate logistic regression analysis showed that lymphopenia, elevated level of d-dimer, hypersensitive cardiac troponin I (hs-CTnI) and high sensitivity C-reactive protein (hs-CRP) were independent predictors of mortality in young adults with severe COVID-19. Further analysis showed that severely young adults with two or more factors abnormalities above would be more prone to death. The similar predictive effect of above four factors had been observed in all-age patients with severe COVID-19.
Conclusion
Lymphopenia, elevated level of d-dimer, hs-CTnI and hs-CRP predicted clinical outcomes of young adults with severe COVID-19.
Keywords: COVID-19, Predictors, SARS-CoV-2, Severe, Young adults
Background
The newly emergent human severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19), resulting in epidemics and pandemics [1, 2]. As of April 18th 2020, SARS-CoV-2 has caused more than 2,000,000 infections and 100,000 deaths worldwide [3].
Previous studies have focused on general epidemiological findings, clinical presentations, and clinical outcomes of patients of COVID-19 [4, 5]. Accumulating studies have suggested that all ages people are susceptible to SARS-CoV-2 infection, which can result in severe and even fatal respiratory diseases [6–8]. As reported by Liu et al., clinical features of elderly patients with COVID-19 were significantly different from that of younger patients [6]. It has to be noted that elderly patients were with more comorbidities, leading to more complicated pathogenesis in COVID-19 [8]. Would the pathogenesis of COVID-19 be different in young adults, with less comorbidities and more strong host immune? Indeed, there are plenty of severe COVID-19 cases in young adults. However, the characteristics and associated risk factors of non-elderly patients, especially with severe COVID-19, have not been fully elucidated so far. Furthermore, young people as the main social labor force, it is urgent to identify the risk factors associated with mortality of young adults in severe COVID-19.
We intended to investigate the clinical characteristics and provide predictors of mortality for young adults with severe COVID-19.
Methods
Study participants and data collection
For this retrospective, non-interventional study, a total of 376 patients with COVID-19 were recruited retrospectively at Tongji Hospital from January 25 to February 15, 2020, and 299 cases were excluded due to ineligible. According to final outcome, we classified 77 young adults with severe COVID-19 into survivor group (37patients) and non-survivor group (40 patients).
The study was performed in accordance with Tongji Hospital Ethics Committee (TJ-IRB20200353). Written informed consent was waived by the Ethics Commission owing to the rapid emergence of this infectious disease.
We prospectively collected information of all patients including demographic data, clinical characteristics, laboratory findings, treatment and outcomes from reviewing medical records. Three researchers individually checked the collected data. To investigate the risk of in-hospital death, all patients were followed from admission to discharge or death (1–58 days). The primary outcome was death during hospitalization.
Definitions
The diagnosis of COVID-19 was created according to the definition established by World Health Organization (WHO) interim guidance [9]. The clinical classifications of patients as having severe or not COVID-19 are established based on the 2019 American Thoracic Society / Infectious Disease Society of America guideline, taking into account its global acceptance for severity stratification of community-acquired pneumonia although lacking of validation in patients with viral pneumonia [10]. The young adults were defined as people under age of 65 years old.
Primary variables selection in logistic regression model
Univariate and multivariate logistic regression were performed to make out the association of clinical characteristics and laboratory parameters for the risk of death. Taking the total deaths events (n = 40) of our study into account and to avoid overfitting in multivariate logistic regression model, four factors were chosen for multivariable logistic analysis on the basis of previous results and clinical constraints. Original researches have shown plasma levels of d-dimer and high sensitivity cardiac troponin I (hs-CTnl) to be higher in severe or critical ill cases, whereas lymphopenia has been less observed in surviving or moderate ill patients [11–13]. Therefore, we chose lymphocyte count, d-dimer, hs-CTnl, and other variable as the four variables in our multivariable logistic regression model. According to the AIC level of each model, we chose the best regression model.
We ruled out variables from the multivariable analysis if the differences between-group were not significant, if they had collinearity, if the accuracy was not confirmed (eg, exposure, which was self-reported), and if the number of incidences was too small to calculate odds ratios [11].
According to the level of lymphocyte, d-dimer, hs-CTnI and high sensitivity C-reactive protein (hs-CRP), we classified of young adults with severe COVID-19 to subgroups. For each factor, cut points used to define a high level were as following: Lymphocyte < 0.5 × 109/L, d-dimer > 21 μg/mL, hs-CTnI > 15.6 pg/ml and hs-CRP > 100 mg/L. High-risk group indicated elevation in two or more factors, while low-risk group indicated elevation in one or no factors.
Statistical analysis
We described the categorical variables as frequency rates and percentages, and continuous variables median and interquartile range (IQR) values. Unpaired 2-sided Student’s t test was used for continuous variables if the data were normally distributed; if not, Mann–Whitney test was used. The frequencies of categorical variables were compared using χ2 test or Fisher’s exact test as appropriate.
All statistical analyses and graphs were generated and plotted using SPSS (version 22.0) and GraphPad Prism version 7.0 software (GraphPad Software Inc). P < 0.05 was considered statistically significant.
Results
Demographics and baseline characteristic of young adults with severe COVID-19
From 25 Jan 2020 to 15 Feb 2020, 376 patients were admitted to Tongji hospital with confirmed COVID-19, of whom 299 were considered ineligible. 77 young adults with severe COVID-19 were included in this study (Additional file 1: Figure S1). Baseline characteristics of patients were divided into subgroups by survival or non-survival (Table 1). Different from all-age populations, there were no significant difference in age and sex among young adults with severe COVID-19 (Additional file 1: Table S1). Patients in non-survivor group were with faster heart rate than survival group. Other characteristics such as exposure history, smoking history, comorbidities, respiratory rate, percutaneous oxygen saturation, blood pressure showed no significance between two groups.
Table 1.
Demographics and baseline characteristic of young adults with severe COVID-19
| Total n = 77 | Survivor n = 37 | Non-survivor n = 40 | P value | |
|---|---|---|---|---|
| Age, years | 59 (54–63) | 58 (50–62) | 60 (57–64) | 0.077 |
| Sex (male) | 50 (65%) | 21 (57%) | 29 (73%) | 0.148* |
| Exposure history | 10 (13%) | 5 (14%) | 5 (13%) | 0.895* |
| Smoker | 3 (4%) | 2 (6%) | 1 (3%) | 0.981* |
| Comorbidity | 72 (94%) | 35 (95%) | 97 (93%) | 1* |
| Hypertension | 26 (34%) | 12 (33%) | 414 (35%) | 0.812* |
| Diabetes | 9 (12%) | 5 (14%) | 4 (10%) | 0.901* |
| Coronary heart disease | 3 (4%) | 0 (0) | 3 (8%) | 0.241* |
| Malignancy | 2 (3%) | 2 (6%) | 0 (0) | 0.228* |
| Chronic kidney disease | 2 (3%) | 0 (0) | 2 (5%) | 0.494* |
| Tuberculosis | 1 (1%) | 0 (0) | 1 (3%) | 1* |
| Chronic hepatitis B | 4 (5%) | 2 (5%) | 2 (5%) | 1* |
| Others | 28 (36%) | 14 (38%) | 14 (35%) | 0.796* |
| Respiratory rate, > 30 breath per min | 13/75 (17%) | 4/37 (11%) | 9/40 (24%) | 0.141* |
| Heart rate, ≥ 125 beats per min | 7/76 (9%) | 0/36 (0) | 7/40 (18%) | 0.012* |
| Percutaneous oxygen saturation, ≤ 93% | 55 (71%) | 25 (68%) | 30 (75%) | 0.471 |
| Systolic pressure, mmH | 131 (119–146) | 130 (114–142) | 132 (121–151) | 0.582 |
| Diastolic pressure, mmHg | 79 (67–85) | 76 (62–85) | 80 (71–88) | 0.185 |
| Fever | 72 (94%) | 35 (95%) | 37 (93%) | 1* |
| Sore throat | 4 (5%) | 2 (5%) | 2 (5%) | 1* |
| Cough | 59 (77%) | 27 (73%) | 32 (80%) | 0.467* |
| Chest pain | 3 (4%) | 1 (3%) | 2 (5%) | 1* |
| Dyspnea | 51 (66%) | 18 (49%) | 33 (83%) | 0.002* |
| Fatigue | 42 (55%) | 20 (54%) | 22 (55%) | 0.834* |
| Myalgia | 14 (18%) | 8 (22%) | 6 (15%) | 0.452* |
| Nausea or vomiting | 5 (7%) | 1 (3%) | 4 (10%) | 0.403* |
| Diarrhea | 25 (33%) | 12 (33%) | 13 (33%) | 0.995* |
| Stomachache | 5 (5%) | 1 (3%) | 4 (10%) | 0.403* |
| Headache | 7 (9%) | 3 (8%) | 5 (10%) | 1* |
| Unconscious | 2 (3%) | 0 (0) | 2 (5%) | 0.494* |
| Dizziness | 3 (4%) | 0 (0) | 3 (8%) | 0.241* |
Data are median (IQR), n (%), or n/N (%). p values were calculated by Mann–Whitney U test, χ2 test, or Fisher’s exact test, as appropriate
*χ2 test comparing all subcategories
Similar to the results reported in previous researches, we pointed out that the top four symptoms included fever (94%), cough (77%), dyspnea (66%), fatigue (55%) in hospital among all-age population (Table 1, Additional file 1: Table S2) [1, 12]. Except for dyspnea that were more often present in non-survivor group than survivor group (83 vs. 49%), other symptoms were comparable in two groups. But in all-age patients, incidence of unconscious and dizziness were higher in non-survivors than that of survivors.
Laboratory findings
The non-survivors had more white blood cells and neutrophils counts than that of the survivors, may result from the presence of secondary bacterial infection as indicated by higher concentrations of hs-CRP and procalcitonin (Table 2, Additional file 1: Table S3). As expected, the non-survivors had reduced lymphocytes. Compared with survivors, those in non-survivor group underwent susceptible to abnormalities of liver, kidney and coagulation function, suggested by elevation of albumin or creatinine, and dysregulation of d-dimer. The non-survivors had experienced more frequently and severe heart injury, as all laboratory heart function parameters including hs-CTnl, myoglobin, and N-terminal pro-brain natriuretic peptide (NT-proBNP), were all significantly increased. The similar results had been shown in all-age patients.
Table 2.
Laboratory examinations of young adults with severe COVID-19
| Findings (normal range) | Total n = 77 | Survivor n = 37 | Non-survivor n = 40 | P value |
|---|---|---|---|---|
| Blood routine test | ||||
| White blood cell, × 109/L (3.5–9.5) | 7.5 (5.9–10.6) | 6.5 (4.7–8.4) | 9.7 (7.1–13.0) | < 0.001 |
| Neutrophil granulocyte, × 109/L (1.8–6.3) | 6.6 (4.2–9.7) | 5.0 (3.6–6.7) | 8.8 (5.9–12.0) | < 0.001 |
| Lymphocyte, × 109/L (1.1–3.2) | 0.7 (0.5–1.0) | 0.9 (0.7–1.2) | 0.6 (0.4–0.7) | < 0.001 |
| Red blood cell, × 109/L (3.8–5.1) | 4.2 (3.7–4.6) | 4.2 (3.7–4.4) | 4.2 (3.7–4.8) | 0.383 |
| Haemoglobin, g/L (130–175) | 129 (115–140) | 129 (118–136) | 130 (111–143) | 0.665 |
| Platelet, × 109/L (125–350) | 194 (148–128) | 201 (162–279) | 153 (127–244) | 0.092 |
| Coagulation function | ||||
| PT, s (11.5–14.5) | 14.9 (13.8–16.3) | 13.8 (13.3–14.6) | 15.7 (15.1–17.2) | < 0.001 |
| APTT, s (29.0–42.0) | 39.9 (36.2–44.9) | 40.5 (37.1–44.8) | 39.4 (34.4–45.0) | 0.468 |
| D-dimer, μg/ml (< 0.5) | 2.4 (1.0–21.0) | 1.3 (0.7–2.2) | 18.2 (3.0–21.0) | < 0.001 |
| Biochemical test | ||||
| Albumin, g/L (35.0–52.0) | 31.1 (28.5–35.0) | 32.8 (20.2–36.3) | 30.0 (27.2–33.7) | 0.001 |
| Globulin, g/L (20.0–35.0) | 34.6 (31.5–37.9) | 33.6 (31.1–36.8) | 35.5 (31.6–39.2) | 0.172 |
| Aspartate aminotransferase, U/L (≤ 40) | 34 (23–52) | 30 (20–46) | 37 (28–57) | 0.296 |
| Alanine aminotransferase, U/L (≤ 41) | 29 (20–50) | 28 (20–57) | 29 (18–48) | 0.721 |
| Total-bilirubin, umol/L (≤ 26) | 10.1 (7.5–14.9) | 8.6 (6.7–11.7) | 12.2 (8.4–19.2) | 0.002 |
| Direct-bilirubin, umol/L (≤ 8) | 4.9 (3.5–7.4) | 3.6 (3.3–5.1) | 6.4 (4.5–10.2) | < 0.001 |
| Creatinine, umol/L (59–104) | 70 (55–87) | 64 (50–82) | 78 (59–99) | 0.024 |
| Urea nitrogen, mmol/L (3.1–8.0) | 5.3 (3.5–7.2) | 3.7 (2.9–5.2) | 7.0 (5.3–9.4) | < 0.001 |
| LDH, U/L (135–225) | 460 (347–585) | 359 (268–456) | 567 (475–663) | 0.442 |
| Infection-related biomarkers | ||||
| Procalcitonin, ng/mL (0.02–0.05) | 0.13 (0.04–0.29) | 0.04 (0.02–0.13) | 0.23 (0.14–0.60) | < 0.001 |
| ERS, mm/h (0–15) | 35 (20–64) | 34 (20–71) | 37 (20–54) | 0.586 |
| Ferritin, ng (30–400) | 1336 (685–2020) | 1113 (374–1605) | 1701 (995–3113) | 0.032 |
| hs-CRP, mg/L (< 1) | 72.5 (38.3–140.2) | 52.1 (28.4–88.4) | 120 (55.9–183.0) | < 0.001 |
| Myocardial enzymes | ||||
| Creatine kinase, U/L (≤ 190) | 116 (54–308) | 100 (42–212) | 119 (61–397) | 0.442 |
| NT-BNP, pg/mL (< 285) | 292 (87–852) | 87 (39–205) | 709 (300–1773) | < 0.001 |
| hs-CTnl, pg/mL (≤ 15.6) | 13.0 (3.4–111.3) | 3.6 (2.1–10.1) | 41.5 (12.1–308.6) | < 0.001 |
| Myoglobin, ng/mL (≤ 106) | 131 (29–324) | 29 (21–123) | 258 (130–470) | 0.001 |
PT prothrombin time, APTT activated partial thromboplastin time, LDH lactate dehydrogenase, ERS erythrocyte sedimentation rate, hs-CRP high sensitivity C-reactive protein, NT-proBNP N-terminal pro-brain natriuretic peptide, hs-CtnI hypersensitive cardiac troponin I. Data are median (IQR), n (%), or n/N (%). p values were calculated by Mann–Whitney U test, χ2 test, or Fisher’s exact test, as appropriate
*χ2 test comparing all subcategories
Treatment and outcomes
More than half non-survivors experienced mechanical ventilation and ICU admission (Table 3, Additional file 1: Table S4). The median time from illness onset to death was 24 days (IQR 6–17), whereas the median time from illness onset to discharge was 38 days (IQR 21–33). The similar trend was shown in hospital length of stay (11 [IQR 6–17] vs. 25 [IQR 21–33]). Consistent with discharge standers, viral shedding of survivors was happened during treatment course.
Table 3.
Treatment and outcomes of young adults with severe COVID-19
| Total n = 77 | Survivor n = 37 | Non-survivor n = 40 | P value | |
|---|---|---|---|---|
| Treatment | ||||
| High-flow nasal cannula oxygen therapy | 15 (20%) | 6 (16%) | 9 (23%) | 0.487* |
| Non-invasive mechanical ventilation | 28 (36%) | 2 (5%) | 26 (65%) | < 0.001* |
| Invasive mechanical ventilation | 27 (35%) | 0 (0) | 27 (68%) | < 0.001* |
| ECMO | 2 (3%) | 0 (0) | 2 (5%) | 0.494* |
| Outcomes | ||||
| ICU admission | 29 (38%) | 1 (3%) | 28 (70%) | < 0.001* |
| ICU length of stay, days | 10 (4–19) | . | 10 (4–17) | . |
| Hospital length of stay, days | 19 (11–27) | 25 (21–33) | 11 (6–17) | < 0.001 |
| Time from illness onset to ICU admission, days | 16 (13–20) | . | 16 (13–21) | . |
| Time from illness onset to death or discharge, days | 30 (22–39) | 38 (32–45) | 24 (19–29) | < 0.001 |
| SARS-CoV-2 RT-PCR test (-) | 43/77 (56%) | 35/37 (95%) | 8/40 (20%) | < 0.001* |
| Duration of viral shedding after COVID-19 onset, days | 22 (18–27) | 23 nnnn | 19 n | 0.054 |
Data are median (IQR), n (%), or n/N (%). P values were calculated by χ2 test, or Fisher’s exact test, as appropriate
ECMO extracorporeal membrane oxygenation, ICU intensive care unit, RT-PCR real-time polymerase chain reaction
*χ2 test comparing all subcategories
Predictors of mortality
All demographic data, clinical symptoms, laboratory findings, treatments and outcomes were shown in Tables 1 and 2, we evaluated every variable that demonstrated statistical significance with p < 0.05 between non-survivor and survivor groups by univariate logistic regression analysis (Table 4). Slightly different from all-age population, white blood cell, neutrophil granulocyte, lymphocyte, prothrombin time, d-dimer, albumin, direct-bilirubin, urea nitrogen, procalcitonin, hs-CRP, NT-proBNP and hs-CTnl were associated with the risk of mortality (Additional file 1: Table S5).
Table 4.
Univariate logistic regression analysis of mortality risk factors in young adults with severe COVID-19
| Factor | Univariable OR (95% CI) | P value |
|---|---|---|
| White blood cell > 9.5 × 109/L, % | 5.167 (1.716–15.087) | 0.003 |
| Neutrophil granulocyte > 6.3 × 109/L, % | 7.118 (2.608–19.428) | < 0.001 |
| Lymphocyte < 0.5 × 109/L, % | 9.273 (2.441–35.227) | 0.001 |
| PT > 14.5 s, % | 21.778 (6.554–72.367) | < 0.001 |
| D-dimer > 21 μg/mL, % | 34.200 (4.256–274.808) | 0.001 |
| Albumin < 30 g/L, % | 7.143 (2.519–20.257) | < 0.001 |
| Direct-bilirubin > 8umol/L | 5.457 (1.410–21.117) | 0.014 |
| Urea nitrogen > 8 mmol/L | 4.857 (1.246–18.933) | 0.023 |
| Procalcitonin > 0.05 ng/mL, % | 19.559 (4.077–93.825) | < 0.001 |
| hs-CRP > 100 mg/L, % | 5.955 (2.098–16.904) | 0.001 |
| NT-proBNP ≥ 285 pg/ml, % | 18.000 (5.211–62.176) | < 0.001 |
| hs-CTnI > 15.6 pg/ml, % | 16.667 (4.234–65.601) | < 0.001 |
OR odds ratio, PT prothrombin time, hs-CRP high sensitivity C-reactive protein, NT-proBNP N-terminal pro-brain natriuretic peptide, hs-CtnI hypersensitive cardiac troponin I
Multivariate logistic regression analyses were applied to assess the independent prognostic effect of related factors (Table 5). Before adjusting other variables, odds ratio (OR) of lymphocyte < 0.5 × 109/L, d-dimer > 21 μg/mL and hs-CTnI > 15.6 pg/ml were 7.03 (95% CI 1.435–40.275), 11.012 (95% CI 1.092–111.100) and 13.876 (95% CI 2.888–66.673). When adjusting for other variables, the OR of above three fixed variables changed slightly. According to the AIC level of each model (Additional file 1: Table S6), we chose the lowest AIC level as the best. The multivariate logistic regression analyses model 5 including lymphocyte < 0.5 × 109/L, d-dimer > 21 μg/mL, hs-CTnI > 15.6 pg/ml and hs-CRP > 100 mg/L (Table 5, mode 5), were the best predictors of mortality.
Table 5.
Multivariate logistic regression analysis of mortality risk factors in young adults with severe COVID-19
| Mode | Multivariable OR (95% CI) | P value |
|---|---|---|
| Mode 1 | ||
| Lymphocyte < 0.5 × 109/L, % | 7.03 (1.435–40.275) | 0.017 |
| D-dimer > 21 μg/mL, % | 11.012 (1.092–111.100) | 0.042 |
| hs-CTnI > 15.6 pg/ml, % | 13.876 (2.888–66.673) | 0.001 |
| Mode 2 | ||
| Lymphocyte < 0.5 × 109/L, % | 6.496 (1.182–35.709) | 0.031 |
| D-dimer > 21 μg/mL, % | 8.382 (0.770–91.279) | 0.081 |
| hs-CTnI > 15.6 pg/ml, % | 14.140 (2.864–69.817) | 0.001 |
| White blood cell > 9.5 × 109/L, % | 2.883 (0.611–13.604) | 0.181 |
| Mode 3 | ||
| Lymphocyte < 0.5 × 109/L, % | 5.639 (1.027–30.958) | 0.047 |
| D-dimer > 21 μg/mL, % | 6.556 (0.624–68.908) | 0.117 |
| hs-CTnI > 15.6 pg/ml, % | 14.228 (2.841–71.252) | 0.001 |
| Neutrophil granulocyte > 6.3 × 109/L, % | 3.173 (0.712–14.129) | 0.130 |
| Mode 4 | ||
| Lymphocyte < 0.5 × 109/L, % | 6.560 (1.161–37.067) | 0.033 |
| D-dimer > 21 μg/mL, % | 5.306 (0.490–57.501) | 0.170 |
| hs-CTnI > 15.6 pg/ml, % | 8.860 (1.730–45.366) | 0.009 |
| PT > 14.5 s, % | 4.719 (0.997–22.326) | 0.050 |
| Mode 5 | ||
| Lymphocyte < 0.5 × 109/L, % | 9.191 (1.190–70.996) | 0.033 |
| D-dimer > 21 μg/mL, % | 24.142 (1.622–359.302) | 0.021 |
| hs-CTnI > 15.6 pg/ml, % | 10.358 (1.711–63.036) | 0.011 |
| hs-CRP > 100 mg/L, % | 19.528 (3.068–124.288) | 0.022 |
| Mode 6 | ||
| Lymphocyte < 0.5 × 109/L, % | 6.790 (1.242–37.129) | 0.027 |
| D-dimer > 21 μg/mL, % | 7.964 (0.702–90.402) | 0.094 |
| hs-CTnI > 15.6 pg/ml, % | 13.129 (2.668–64.599) | 0.002 |
| Albumin < 30 g/L, % | 2.900 (0.660–12.746) | 0.159 |
| Mode 7 | ||
| Lymphocyte < 0.5 × 109/L, % | 7.598 (1.434–40.249) | 0.017 |
| D-dimer > 21 μg/mL, % | 10.575 (0.752–148.749) | 0.080 |
| hs-CTnI > 15.6 pg/ml, % | 13.942 (2.878–67.526) | 0.001 |
| Direct-bilirubin > 8umol/L | 1.070 (0.121–9.442) | 0.952 |
| Mode 8 | ||
| Lymphocyte < 0.5 × 109/L, % | 8.684 (1.507–50.054) | 0.016 |
| D-dimer > 21 μg/mL, % | 11.719 (1.086–126.460) | 0.043 |
| hs-CTnI > 15.6 pg/ml, % | 13.209 (2.609–66.883) | 0.002 |
| Urea nitrogen > 8 mmol/L | 4.233 (0.479–37.407) | 0.194 |
| Mode 9 | ||
| Lymphocyte < 0.5 × 109/L, % | 3.796 (0.671–21.481) | 0.131 |
| D-dimer > 21 μg/mL, % | 16.923 (0.990–289.353) | 0.051 |
| hs-CTnI > 15.6 pg/ml, % | 9.509 (1.833–49.323) | 0.007 |
| Procalcitonin > 0.05 ng/mL, % | 9.876 (0.926–105.342) | 0.058 |
| Mode 10 | ||
| Lymphocyte < 0.5 × 109/L, % | 7.412 (1.205–45.602) | 0.031 |
| D-dimer > 21 μg/mL, % | 4.625 (0.446–47.978) | 0.199 |
| hs-CTnI > 15.6 pg/ml, % | 7.589 (1.360–42.335) | 0.021 |
| NT-proBNP ≥ 285 pg/ml, % | 7.011 (1.452–33.843) | 0.015 |
OR odds ratio, PT prothrombin time, hs-CRP high sensitivity C-reactive protein, NT-proBNP N-terminal pro-brain natriuretic peptide, hs-CtnI hypersensitive cardiac troponin I
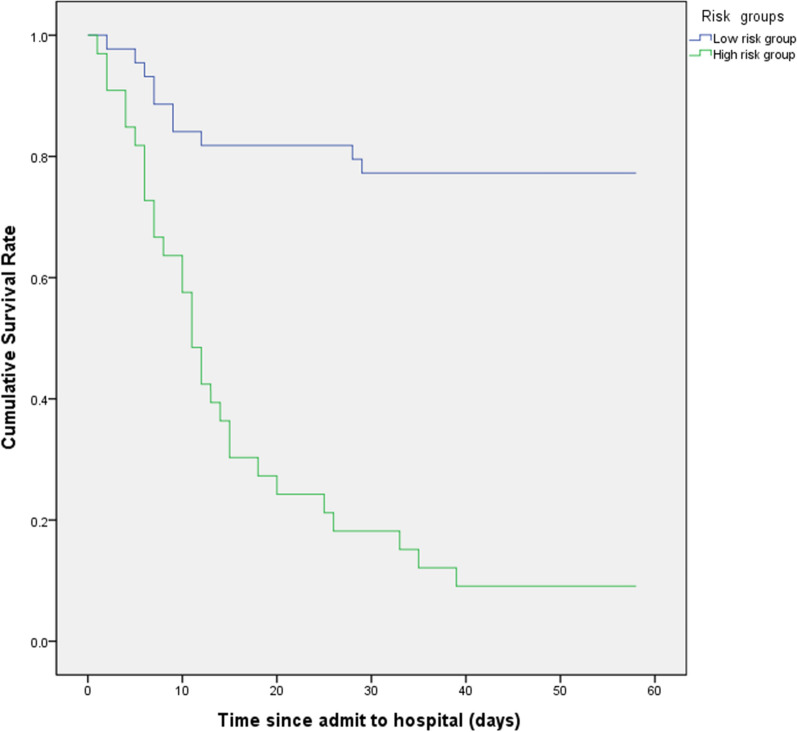
According to the level of four variables in mode 5, we classified of young adults with severe COVID-19 to low-risk and high-risk subgroups. The cumulative survival rate of low-risk group was much higher than that of high-risk group (Fig. 1). The same predictive effect of four factors were shown in all-age participants with severe COVID-19.
Fig. 1.
Survival of young adults with severe COVID-19. According to the level of lymphocytes, d-dimer, hs-CTnI and high sensitivity C-reactive protein (hs-CRP), we classified of young adults with severe COVID-19 into subgroups. Kaplan–Meier survival curves for survival rate during the time from admission to follow-up. The log-rank test was used to compare the Kaplan–Meier survival curves (Log-rank, P < 0.001)
Discussion
This retrospective study reported clinical characteristics and identified several predictors for mortality of young adults with severe COVID-19. In particular, lymphocyte count less than 0.5 × 109/L, d-dimer level greater than 21 μg/mL, hs-CTnI degree higher than 15.6 pg/ml and hs-CRP level higher than 100 mg/L were correlated with higher odds of on-admission mortality. Furthermore, we confirmed the markedly reduction of survival probability during the course of disease in severely ill young patients with high risk.
Formerly, elder age has been announced as an independent predictor of mortality in COVID-19 (Additional file 1: Table S1) [13]. More attention had been paid to older patients [14]. What’s more, clinical features of elderly patients with COVID-19 were significantly different from that of younger patients. Indeed, there are plenty of severe COVID-19 cases in young adults. However, clinical characters and risk factors of young adults with severe COVID-19 had not been fully understood.
Comparing with all-age patients, a lot of clinical factors such as sex, comorbidities and some clinical symptoms, showed no significant difference between non-survivors and survivors in severely young adults, indicating that characters of young adults were far more different from older patients in severe COVID-19 [12, 15]. It has to be noted that elderly patients were with more comorbidities, leading to more complicated pathogenesis in COVID-19 [8]. In severely young adults, comorbidities of non-survivors were similar to that of survivors. Alternatively, in all-age patients, the presence of unconscious and dizziness were higher in non-survivors than that of survivors, which were not observed in young adults of severe COVID-19. In the current study, dyspnea related to hypoxemia were more frequent in non-survivors, consistent with previous studies [16]. Correspondingly, non-survivors more often required mechanical ventilation and admission to Intensive Care Unit (ICU), indicating more prone to experience lung injury.
The pathogenesis of highly pathogenic human coronavirus is still not completely understood. Viral evasion of cellular immune responses and cytokine storm are thought to play important roles in disease severity [17]. Lymphopenia (< 0.5 × 109/L) was found 45% in non-survivors, while 8% in survivors. The SARS-CoV-2 infection may impact on lymphocytes particularly, CD4 + T and CD8 + T cells, causing decrease in viral shedding [18, 19]. Increased levels of d-dimer (21 μg/mL), hs-CTnI (> 15.6 pg/mL) and hs-CRP (> 100 mg/L) were also correlated with a higher risk for death in multivariate logistic regression analysis. Increased levels of d-dimer, fibrin degradation products, and prolonged prothrombin time (PT) have been related with poor prognosis of COVID-19 patients [20]. Multiple pathogenetic mechanisms are included, such as Toll-like receptor activation, endothelial dysfunction, and tissue-factor pathway activation [21–23]. Myocardial insult is significantly associated with mortality of COVID-19, while the prognosis of patients with underlying coronary artery disease but without myocardial insult is relatively beneficial [24]. On the basis of recent studies, angiotensin-converting enzyme 2 (ACE2) is the human cell receptor with strong binding affinity to the Spike protein of SARS-CoV-2, and ACE2 is also highly expressed in heart [25, 26]. Thus, it is rational to hypothesize that COVID-19 induced cardiac injury might be mediated by ACE2. High level of hs-CRP, maker of systemic inflammation, was observed between severe and non-severe patients [15, 27]. As a classic acute phase protein, hs-CRP levels rise quickly responding to inflammation and might adjust the innate immune response by activating complement, stimulating the production of excessive inflammatory cytokines, or bonding fragment crystallizable region (Fc) receptors to activate phagocytosis [28, 29].
Among plenty of factors, lymphopenia, elevated level of d-dimer, hs-CTnT and hs-CRP were independently associated with COVID-19 mortality, suggesting multi-organ dysfunction in young adults of severe COVID-19. Further analysis showed that severely young adults with two or more factors abnormalities above would be more prone to death. Earlier identification of high-risk subgroup and subsequent timely appropriate therapy might improve outcomes. Though predictors of severely young adults showed great influence on mortality of COVID-19, the same effect of four predictors was comparable to that of all-age patients with severe COVID-19.
Tis study had some notable limitations too. Firstly, not all laboratory tests were done in all patients, due to the retrospective study design. Secondly, because the clinical observation of patients is still ongoing, some have not reached clinical end points. Thirdly, as an observational and retrospective study, we currently could not establish a validation cohort to evaluate the predictive effect due to the imperative timeline under this special situation.
Conclusion
In conclusion, lymphopenia, elevated level of d-dimer, hs-CTnI and hs-CRP were independent predictors of mortality in young adults with severe COVID-19. Earlier confirmation, more intensive observation and appropriate treatment should be considered in high-risk young patients.
Supplementary Information
Additional file 1. Additional Figure and Tables.
Acknowledgements
We thank all the patients and their families involved in this study, as well as numerous doctors, nurses and civilians working together to fight against SARS-CoV-2.
Abbreviations
- hs-CTnl
Hypersensitive cardiac troponin I
- hs-CRP
High sensitivity C-reactive protein
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- COVID-19
Coronavirus disease 2019
- WHO
World Health Organization
- IQR
Interquartile range
- NT-proBNP
N-terminal pro-brain natriuretic peptide
- OR
Odds ratio
- ICU
Intensive Care Unit
- ACE2
Angiotensin-converting enzyme 2
Authors’ contributions
YJL, ZLH, MJW, KT, SSW and PFG contributed equally to the study and shared first authorship. JPZ and TW designed the study, had full access to all data in the study and take responsibility for the integrity of data and the accuracy of the data analysis. YJL, ZLH, MJW, KT, SSW and PFG contributed to patient recruitment, data collection, data analysis, data interpretation, the literature search, and the writing of the manuscript. All authors contributed to data acquisition, data analysis, or data interpretation, and reviewed and approved the final version of the manuscript. The corresponding author attests that all listed authors meet the authorship criteria and that no others meeting the criteria have been omitted. JPZ is the guarantor. All authors read and approved the final manuscript.
Funding
This work was funded by the HUST COVID-19 Rapid Response Call (2020kfyXGYJ015) and The National Major Science and Technology Project for the Control and Prevention of Major Infectious Diseases of China (2017ZX10103004).
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
The study was performed in accordance with the guidelines of the Tongji Hospital Ethics Committee (TJ-IRB20200353). The requirement for written informed consent was waived by the Ethics Commission owing to the rapid emergence of this infectious disease.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tao Wang, Email: wt7636@126.com.
Jianping Zhao, Email: Zhaojp88@126.com.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12941-020-00412-9.
References
- 1.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Coronavirus disease 2019 (COVID-19) Situation report—89. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 4.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323:1488. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu K, Chen Y, Lin R, Han K. Clinical features of COVID-19 in elderly patients: A comparison with young and middle-aged patients. J Infect. 2020;80:e14. doi: 10.1016/j.jinf.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu T, Wang Y, Zhou S, Zhang N, Xia L. A comparative study of chest computed tomography features in young and older adults with Corona Virus Disease (COVID-19) J Thorac Imaging. 2020;35:97. doi: 10.1097/RTI.0000000000000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen T, Dai Z, Mo P, Li X, Ma Z, Song S, et al. Clinical characteristics and outcomes of older patients with coronavirus disease 2019 (COVID-19) in Wuhan, China (2019): a single-centered, retrospective study. J Gerontol A Biol Sci Med Sci. 2020;75:1788. doi: 10.1093/gerona/glaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organisation . Clinical management of severe acute respiratory infection when Novel coronavirus (nCoV) infection is suspected: interim guidance. Geneva: World Health Organisation; 2020. [Google Scholar]
- 10.Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, et al. Diagnosis and treatment of adults with community-acquired pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Resp Crit Care Med. 2019;200(7):45–67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu X, Sun X, Cui P, Pan H, Lin S, Han R, et al. Epidemiological and clinical characteristics of 333 confirmed cases with coronavirus disease 2019 in Shanghai, China. Transboundary Emerg Dis. 2020;67:1697. doi: 10.1111/tbed.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen R, Liang W, Jiang M, Guan W, Zhan C, Wang T, et al. Risk factors of fatal outcome in hospitalized subjects with coronavirus disease 2019 from a nationwide analysis in China. Chest. 2020;158:97. doi: 10.1016/j.chest.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J Clin Invest. 2020;130:2620. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moro-García MA, Mayo JC, Sainz RM, Alonso-Arias R. Influence of inflammation in the process of T lymphocyte differentiation: proliferative, metabolic, and oxidative changes. Front Immunol. 2018;9:339. doi: 10.3389/fimmu.2018.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramaniam S, Scharrer I. Procoagulant activity during viral infections. Front Biosci. 2018;23:1060–1081. doi: 10.2741/4633. [DOI] [PubMed] [Google Scholar]
- 22.van Gorp EC, Suharti C, ten Cate H, Dolmans WM, van der Meer JW, ten Cate JW, et al. Review: infectious diseases and coagulation disorders. J Infect Dis. 1999;180(1):176–186. doi: 10.1086/314829. [DOI] [PubMed] [Google Scholar]
- 23.Key NS, Vercellotti GM, Winkelmann JC, Moldow CF, Goodman JL, Esmon NL, et al. Infection of vascular endothelial cells with herpes simplex virus enhances tissue factor activity and reduces thrombomodulin expression. Proc Natl Acad Sci USA. 1990;87(18):7095–7099. doi: 10.1073/pnas.87.18.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo T, Fan Y, Chen M, Wu X, Zhang L, He T, et al. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:811. doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14:185. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang AP, Liu JP, Tao WQ, Li HM. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int Immunopharmacol. 2020;84:106504. doi: 10.1016/j.intimp.2020.106504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Potempa LA, El Kebir D, Filep JG. C-reactive protein and inflammation: conformational changes affect function. Biol Chem. 2015;396(11):1181–1197. doi: 10.1515/hsz-2015-0149. [DOI] [PubMed] [Google Scholar]
- 29.Marnell L, Mold C, Du Clos TW. C-reactive protein: ligands, receptors and role in inflammation. Clin Immunol. 2005;117(2):104–111. doi: 10.1016/j.clim.2005.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional Figure and Tables.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.