Label-free stimulated Raman scattering microscopy resolves amphotericin B orientation in fungal cell membrane.
Abstract
Ergosterol-targeting amphotericin B (AmB) is the first line of defense for life-threatening fungal infections. Two models have been proposed to illustrate AmB assembly in the cell membrane; one is the classical ion channel model in which AmB vertically forms transmembrane tunnel and the other is a recently proposed sterol sponge model where AmB is laterally adsorbed onto the membrane surface. To address this controversy, we use polarization-sensitive stimulated Raman scattering from fingerprint C═C stretching vibration to visualize AmB, ergosterol, and lipid in single fungal cells. Intracellular lipid droplet accumulation in response to AmB treatment is found. AmB is located in membrane and intracellular droplets. In the 16 strains studied, AmB residing inside cell membrane was highly ordered, and its orientation is primarily parallel to phospholipid acyl chains, supporting the ion channel model. Label-free imaging of AmB and chemical contents offers an analytical platform for developing low-toxicity, resistance-refractory antifungal agents.
INTRODUCTION
It is estimated that nearly a billion people have skin, nail, and fungal infections ranging from asymptomatic, mild mucocutaneous, to potentially life-threatening systemic infections (1). Among the pathogenic fungi, Aspergillus, Candida, and Cryptococcus species remain the main fungal species responsible for the majority causes of severe fungal infections (2). Moreover, Candida spp. are the fourth most common cause of nosocomial bloodstream infections in the United States (3). In particular, Candida auris is an emerging and notable Candida species that has been associated with recent nosocomial outbreaks on all five continents (4). Notably, C. auris–related invasive bloodstream infection has been reported to result in 30 to 60% mortality (5).
For more than 50 years, amphotericin B (AmB), an amphipathic polyene macrolide (Fig. 1A) derived from Streptomyces nodosus (6), has remained a powerful but highly toxic first line of defense against clinically severe invasive fungal infections (7) with minimal development of microbial resistance (8). Its selective toxicity against fungi compared to mammalian cells lies in the higher binding affinity of AmB for ergosterol (Fig. 1B) than for cholesterol (9, 10). However, adverse side effects such as nephrotoxicity and electrolyte disturbances still occur with use of AmB (9, 11). Thus, understanding how AmB interacts with fungal membrane is essential to guide the development of novel polyene derivatives with low toxicity or repurposing of existing drugs. Extensive research has been conducted to understand AmB’s working mechanism. The most prevailing one is the ion channel model, also called the barrel-stave model (Fig. 1C), where AmB and ergosterol aggregates remain vertical inside a lipid bilayer and subsequently cause membrane permeabilization and metabolite leakage (12–20). Recently, Anderson et al. (21) proposed a sterol sponge model, in which AmB sequesters ergosterol from lipid bilayers and stacks likely in a way parallel to the cell membrane plane (Fig. 1D). These two models contradict each other on the alignment of AmB, as AmB assembly is parallel to the phospholipid acyl chains in the ion channel model, whereas AmB stays in an orthogonal direction against the phospholipid in the sterol sponge model.
Fig. 1. Major models of AmB’s mechanism of action in a lipid membrane.
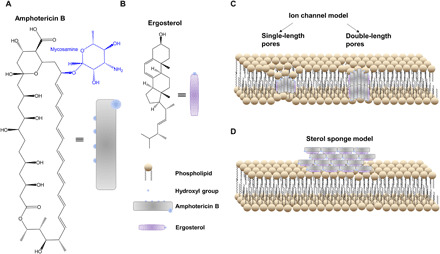
(A and B) Chemical structures of AmB (A) and ergosterol (B), respectively. (C) Classic ion channel model. (D) Sterol sponge model where large extramembranous aggregates sequester ergosterol from lipid bilayers.
Extensive studies have been conducted to understand these models, including circular dichroism spectroscopy (16, 17, 22), Raman spectroscopy (23), atomic force microscopy (19), freeze-etch electron microscopy (24), solution and solid-state nuclear magnetic resonance spectroscopy (13, 14, 17), and theoretical simulation (18). Fluorescence-based approaches have also been developed to modify the hydroxyl and carboxylic acid groups of AmB, making this molecule fluorescent for visualization (17, 25). However, these modifications usually conjugate AmB with a bulky fluorophore, thus likely affecting its interaction with ergosterol. In addition, many of the abovementioned methods used an artificial cell membrane but were not able to depict AmB orientation in situ in a fungal cell membrane. Therefore, an imaging approach allowing for direct visualization of AmB orientation in situ in fungal cell membrane is highly desired.
Here, grounded on the intrinsic Raman signal from fingerprint C═C stretching vibration mode, we report polarization-sensitive stimulated Raman scattering (SRS) microscopy (26–28) as an advanced imaging approach for in situ visualization of AmB in fungal cell membrane. Using parallel-polarized pump and Stokes fields, the SRS signal intensity is anticipated to be maximized when the laser fields are polarized along the backbone of the molecule and minimized when the laser polarization is rotated to be perpendicular to the backbone of the molecule. With this principle, we studied a total of 16 clinically isolated fungal species, including seven clinical C. auris isolates. On the basis of the strong signal dependence on laser polarization, we confirm that AmB is concentrated at the cell membrane in a highly ordered manner. With the symmetric CH2 stretching vibration signal from the acyl chains in phospholipids as reference, we further confirm that AmB molecules are oriented parallel to the phospholipid’s acyl chains. These findings support the classical ion channel model and offer guidance for developing low-toxicity polyene-based antifungal drugs.
RESULTS
Raman spectroscopy uncovers different levels of AmB in stationary versus log phase Candida albicans
Considering the heptaene structure inside AmB (9) and the fact that conjugated C═C bonds normally yield strong Raman scattering signal (29), we reasoned that Raman-based microscopic techniques can be a viable imaging tool to study AmB. To prove this point, we first measured the Raman spectra of ergosterol and AmB to obtain their specific vibrational fingerprints. As shown in Fig. 2A, ergosterol demonstrates strong Raman peaks in the fingerprint (800 to 1800 cm−1) region, where the peak at 1602 cm−1 originates from the two conjugated C═C bonds (30) in a six-member ring and the peak at 1666-cm−1 peak is from the isolated C═C bond in the alkyl tail (30). In the case of AmB dissolved inside dimethyl sulfoxide (DMSO), the prototypical Raman peak at 1560 cm−1 arises from the conjugated heptaene (Fig. 2B) (23). Moreover, under the resonance excitation wavelength of 532 nm, we could reach detection sensitivity of ~100 μM in DMSO (Fig. 2B). To further test AmB sensitivity in situ on fungal cells, we measured the Raman spectra from untreated and AmB-treated C. albicans SC5314 (CASC5314) (at a sublethal concentration) aggregates dried onto an aluminum wafer. We observed distinguishable Raman signal from AmB at 1556 cm−1 (fig. S1), which has around 4-cm−1 shift compared to solution-form AmB. Next, we recorded Raman spectra from AmB-treated single CASC5314 cells sandwiched between a cover glass and a polylysine-coated cover slide. Notably, AmB from AmB-treated single log-phase CASC5314 cells was clearly detectable (Fig. 2C), whereas the untreated ones did not exhibit this signal at all (fig. S2A). However, under the same treatment schemes as that of log-phase cells, we did not obtain discernible AmB signals from AmB-treated and untreated stationary-phase CASC5314 cells (Fig. 2D and figs. S2B and S3). These results suggest that AmB shows high affinity to the cell membrane in the metabolically active phases. We used log-phase fungal cells in our follow-up imaging experiments with detailed procedure depicted in fig. S4.
Fig. 2. Raman spectra of ergosterol, AmB, and AmB-treated single log-phase and stationary-phase CASC5314 cells.
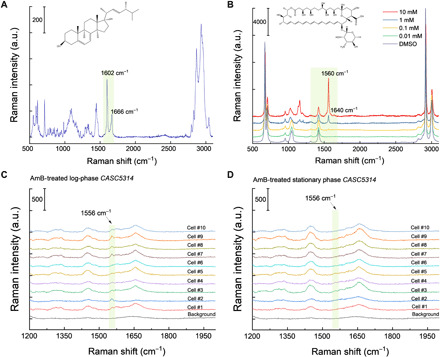
(A and B) Raman spectra of pure ergosterol (A) and AmB (B) dissolved in DMSO at a series of concentrations, respectively. a.u., arbitrary unit. (C) Raman spectra of AmB-treated single log-phase CASC5314 cells. (D) Raman spectra of AmB-treated single stationary-phase CASC5314 cells. Regions of interest are highlighted by green boxes.
SRS imaging of single fungal cells in fingerprint region reveals distribution of AmB, ergosterol, and accumulation of lipids
The pronounced Raman signal of AmB from single yeast cells prompted us to deploy Raman scattering to visualize AmB in situ. However, spontaneous Raman scattering is a weak process, hindering the feasibility of imaging AmB in single fungal cells, especially because AmB is sensitive to long-time light irradiance (31). In recent years, SRS microscopy has emerged as an advanced nonlinear imaging technique to achieve high-sensitivity fast chemical mapping of biological analytes (32–34). Femtosecond pulse–based hyperspectral SRS (hSRS), which can be achieved either through spectral focusing method (27) or pulse shaping approach (28), allows chemical imaging of multiple analytes using their representative fingerprint Raman bands. In our laboratory-built SRS microscope (Fig. 3A), we set the wavelength of femtosecond pump laser at 895 nm and femtosecond Stokes laser at 1040 nm (in this case, the center beating frequency between pump and Stokes is 1556 cm−1) and then chirped the two beams with NSF57 rods to obtain hSRS imaging capability.
Fig. 3. SRS microscope, performance characterization, and SRS imaging of untreated CASC5314 and CAC15 at 1556, 1602, and 1650 cm−1.
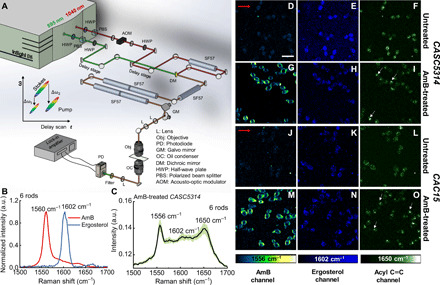
(A) Schematic of the laboratory-built polarization-sensitive SRS microscope. Two half-wave plates (HWPs) were used to control the polarization of each beam. (B) SRS spectra of AmB (10 mg/ml) and pure ergosterol, with optimal Raman peaks highlighted. Data acquired under six-rod chirping. (C) SRS spectra of single AmB-treated CASC5314 cells. Data: means ± SD from at least 10 yeasts. AmB: 3.2 μg/ml for 1 hour. Data acquired under six-rod chirping condition. (D to F) SRS images of untreated CASC5314 cells at 1556, 1602, and 1650 cm−1, respectively. (G to I) SRS images of AmB-treated CASC5314 cells at 1556, 1602, and 1650 cm−1, respectively. (J to L) SRS images of untreated CAC15 cells at 1556, 1602, and 1650 cm−1, respectively. (M to O) SRS images of AmB-treated CAC15 cells at 1556, 1602, and 1650 cm−1, respectively. Pixel dwell time: 50 μs. Scale bar, 10 μm. Laser polarization direction is indicated by red arrows. Lipid droplet is indicated by white arrows.
For imaging in the fingerprint region, we initially chirped the pump and Stokes beams with three rods and could separate the SRS spectra between AmB and ergosterol crystal (fig. S5A). However, for the AmB-treated CASC5314 cells, the AmB signal was indiscernible and suffered from a large background (fig. S5B). To improve the spectral resolution, we then chirped the two beams with six rods in total, with five of them chirping after the dichroic mirror combiner. AmB, ergosterol, and glyceryl trioleate were then used to calibrate this six-rod chirping SRS system (fig. S5, C and D). Under the same circumstances, the spectral resolution is estimated to be 10 cm−1 with six-rod chirping based on the 1560-cm−1 peak, increased by around two times from that of three-rod chirping (Fig. 3B). Moreover, peaks at 1556, 1602, and 1650 cm−1 were clearly discernible from AmB-treated CASC5314 cell (Fig. 3C). In particular, the AmB signal is allocated at 1556 cm−1, around 4-cm−1 shift from the solution-form AmB. As these three peaks are pronounced compared to the background under the six-rod chirping condition, we recorded SRS images at these wave numbers. We imaged untreated and AmB-treated CASC5314 and CAC15 cells at 1556, 1602, and 1650 cm−1 (Fig. 3, D to O), respectively.
The SRS signals of ergosterol at 1602 cm−1 were obtained for both untreated and AmB-treated CASC5314 (Fig. 3, E and H) and CAC15 cells (Fig. 3, K and N). To confirm that what we observed at 1602 cm−1 was primarily from ergosterol, we imaged CASC5314 along with its isogenic mutants CAUPC and CADBC46 at this wave number. CAUPC has a homozygous activating mutation in UPC2, which regulates the expression of genes involved in ergosterol synthesis, including lanosterol 14α-demethylase (ERG11) (35). Therefore, ERG11 overexpression inside CAUPC leads to increased ergosterol production. On the contrary, CADBC46 has a heterozygous deletion of ERG11, thus reducing the ergosterol level. After culturing the untreated and AmB-treated strains in yeast peptone dextrose (YPD) broth for 1 hour at 37°C, followed by formalin fixation, we imaged the samples at 1602 cm−1. Consistently, we found the weakest ergosterol signal in the case of CADBC46 (fig. S6C) compared to other two strains (fig. S6, A and B). Ergosterol was found to accumulate inside CAUPC as “droplets” (fig. S6B), which suggests that excess ergosterol was stored intracellularly. Moreover, ergosterol reallocation to membrane happens in response to AmB treatment. As shown in Fig. 3 (H and N), ergosterol appeared to aggregate around the cell membrane, forming a ring pattern.
Notably, we observed a significant change of SRS signal at 1650 cm−1 after AmB treatment. In the case of untreated CASC5314 and CAC15, SRS signal depicts evenly at 1650 cm−1 primarily because of the amide I C═O stretching (Fig. 3, F and L). After AmB treatment, lipid droplets (from acyl C═C bond in the unsaturated fatty acids) were found to accumulate intracellularly (Fig. 3, I and O, and fig. S7), and these droplets were mostly adjacent to the big vacuole (Fig. 3, I and O).
The SRS signals of AmB at 1556 cm−1 were clearly obtained from the cell membrane region (Fig. 3, G and M) in the case of AmB-treated CASC5314 and CAC15. As control, no discernible signal at 1556 cm−1 was observed for untreated CASC5314 and CAC15 (Fig. 3, D and J). Notably, the SRS signals of AmB are orientated in a certain direction in the fungal cell membrane. This finding triggered us to further explore AmB distribution and orientation by polarization-sensitive SRS.
Polarization-sensitive SRS imaging of ghost red blood cells and AmB-treated fungal species determines AmB orientation in cell membrane
To understand how AmB orientates in the cell membrane, we used polarization-sensitive SRS microscopy (see Materials and Methods and fig. S8) achieved by using two half-wave plates (one beam each) to control the laser polarization (Fig. 3A). By rotating both plates at 45° simultaneously, the laser polarization is changed by 90°. Using this method, we recorded the symmetric phospholipid acyl CH2 stretching signal at 2850 cm−1 and the symmetric C═C stretching signal of AmB at 1556 cm−1 under the same laser polarizations. The CH2 groups are the major constituent chemical groups of phospholipid backbone, and they lie perpendicularly to the membrane phospholipids (fig. S9A), thus providing a reference to AmB orientation in the cell membrane. Initially, we recorded the SRS images of the untreated CASC5314 at 2850 cm−1 under both polarization directions (fig. S9, B and C). However, the CH2 signal from untreated fungal cell membrane was largely imperceptible (fig. S9, B and C). It was reported that fungal cell wall has abundant glucans, glycoproteins, where CH2 groups exist extensively (36). Thus, we could not be able to clearly observe the polarized behavior of CH2 from the cell membrane only probably because of the interference from that of cell wall. Nonetheless, there were still some yeast cells with big vacuoles inside, demonstrating SRS signal difference between horizontal and vertical polarization directions. The zoom-in view of fig. S9 (B and C) showed a strong signal around the yeast edges (left/right) under horizontal polarization direction, and the top and down SRS signals indeed became dimmer under vertical polarization direction (fig. S9, D and E).
To clearly resolve the CH2 signal only from cell membrane, we used a ghost red blood cell (RBC) model (37) where hemoglobin was deprived to preclude the strong transient absorption background. As shown in Fig. 4 (A1 and A2), we found that sharply bright signal at 2850 cm−1 appeared perpendicularly against the laser polarization direction. Collectively, these two independence evidences consolidate the fact that CH2 group in the cell membrane is orthogonal to the phospholipid backbones.
Fig. 4. Polarization-sensitive SRS imaging of ghost RBCs at 2850 cm−1 and various AmB-treated fungal species at 1556 cm−1.
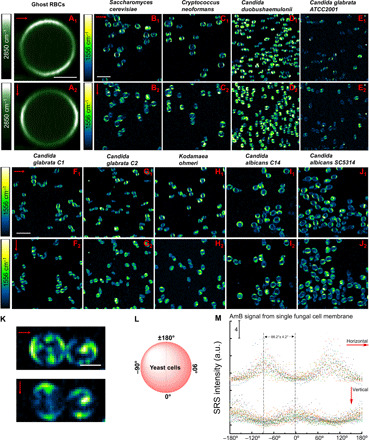
(A1 and A2) SRS images of ghost RBCs at 2850 cm−1 in the horizontal and vertical polarization directions, respectively. Pump, 802 nm; Stokes, 1040 nm. Pixel dwell time: 50 μs. Scale bar, 2.5 μm. (B1 to J1) SRS images of AmB-treated S. cerevisiae AR-399, C. neoformans, C. duobushaemulonii AR-0394, C. glabrata ATCC2001, C. glabrata C1, C. glabrata C2, K. ohmeri AR-0396, C. albicans C14, and C. albicans SC5314 at 1556 cm−1 with laser polarization in the horizontal direction. Pump, 895 nm; Stokes, 1040 nm. Pixel dwell time: 50 μs. Scale bar, 10 μm. (B2 to J2) SRS images of the same strains but with laser polarization in the vertical direction. Pump, 895 nm; Stokes, 1040 nm. Pixel dwell time: 50 μs. Scale bar, 10 μm. (K) Zoom-in view of AmB-treated C. neoformans under two laser polarization directions. Scale bar, 2.5 μm. (L) Schematic of fungal cell membrane with angles defined. (M) Quantitative analysis of SRS signal intensity of single fungal cell at 1556 cm−1 under two laser polarization directions. AmB: 3.2 μg/ml, 1-hour treatment at 30°C. Laser orientation is indicated by red arrows.
Since AmB is located in the fungal cell membrane in a highly ordered form (Fig. 3), we asked how the SRS signal level depends on the laser polarization. Consistently, the horizontal parts of fungal cells from nine different species exhibit pronounced signals under the horizontal laser polarization direction (Fig. 4, B1 to J1). When the laser polarization direction was rotated by 90°, the AmB signal pattern changed accordingly, with the vertical part of the membrane becoming brighter (Fig. 4, B2 to J2). Notably, in the case of Saccharomyces cerevisiae AR-0399 and Cryptococcus neoformans, AmB-containing droplets formed intracellularly (Fig. 4, B1, B2, and K), which indicates that AmB might reside inside the intracellular lipid droplets besides cell membrane. According to the structure of AmB, the SRS signal intensity is maximized when the laser fields are polarized along the molecule. We then further analyzed the SRS signal intensity of single AmB-treated fungal cells at 1556 cm−1 along the peripheral of these cells (Fig. 4, L and M). The signal distribution between CH2 and AmB exhibited a ~90° difference (Fig. 4, A1, A2, and M) under the same laser polarization settings. Thus, our data indicate that AmB molecules are primarily oriented parallel to the laser polarization and perpendicular to the axis of CH2 groups in the membrane.
Polarization-sensitive SRS imaging determines AmB orientation in clinical isolates of C. auris
C. auris is an emerging multidrug-resistant pathogen that can cause invasive infections and has demonstrated high mortality in health care settings (38, 39). So far, 90% of C. auris isolates in the United States have been found resistant to fluconazole, and 30% have been resistant to AmB (38). It was believed that modification of sterol composition of cell membrane accounts for AmB and fluconazole resistance mechanism (40). To have a better understanding of how AmB works for C. auris, we used seven C. auris clinical isolates with six of them from the U.S. Centers for Disease Control and Prevention (CDC) and one from the Massachusetts General Hospital (MGH), Boston, MA. Most of the C. auris isolates were found to be highly sessile and sticky to surfaces, which is consistent with previous reports (41). Moreover, the sizes of these C. auris strains are commonly smaller than other fungal strains. As shown in Fig. 5, AmB signal from AmB-treated C. auris is patterned in a similar manner as other fungal strains shown in Fig. 4; that is, the AmB signal distribution is parallel to the laser polarization direction. In particular, some C. auris strains are prone to aggregate between cells (Fig. 5, E1 and E2), which suggests that C. auris demonstrates the propensity to form sessile biofilms to evade antifungals. Collectively, the SRS images at 1556 cm−1 from altogether 16 strains including 7 C. auris strains consolidate a universal phenomenon: AmB molecules are oriented parallel to the laser polarization. In combination with the fact that CH2 groups from the phospholipids are perpendicular to the phospholipid chain, these evidences show that AmB resides in the cell membrane along the acyl chains in the membrane (Fig. 5H). Notably, we still could observe a weak SRS signal from the top and bottom parts of membranes of AmB-treated fungal cells when the laser polarization is in the horizontal direction (Fig. 4, G to J), which indicates that the sterol sponge model might also exist. Therefore, our imaging results provide in situ evidence that AmB positions itself primarily perpendicular to the membrane plane, which supports the classical ion channel model.
Fig. 5. Polarization-sensitive SRS imaging of AmB-treated various C. auris strains at 1556 cm−1.
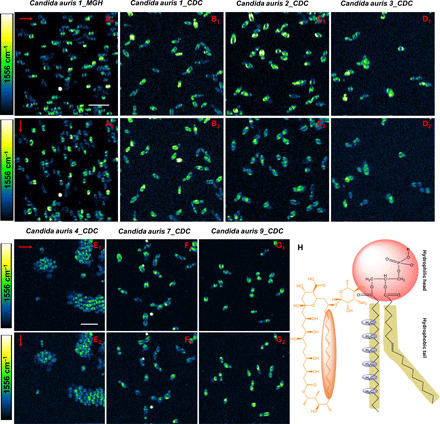
(A1 to G1) SRS images of AmB-treated C. auris 1_MGH, C. auris 1_CDC, C. auris 2_CDC, C. auris 3_CDC, C. auris 4_CDC, C. auris 7_CDC, and C. auris 9_CDC at 1556 cm−1 with the laser polarization in the horizontal direction. (A2 to G2) SRS images of the same strains on the same field of view but with laser polarization in the vertical direction. (H) Orientation of ─CH2 in phospholipid backbone versus that of AmB. AmB: 3.2 μg/ml, 1-hour treatment at 30°C. Pixel dwell time: 50 μs. Scale bar, 10 um. Pump, 895 nm; Stokes, 1040 nm. Laser polarization direction is indicated by red arrows.
Note that the level of AmB signal significantly differs among cells within the same population, as shown in Fig. 5 (A1, A2, F1, and F2), which indicates a large heterogeneity in C. auris’ responses to AmB treatment. Such heterogeneity might be due to the presence of a persister subpopulation, which accounts for AmB persistence. Another interesting phenomenon is that AmB forms “microdomains” in some cell membrane, as shown in Fig. 5 (B1 to C2). Given that AmB-treated CADBC46 exhibits AmB microdomain in the hyphae cell membrane due to the lack of ergosterol in the cell membrane (fig. S10), we postulate that some C. auris cells might have less ergosterol content in the cell membrane.
DISCUSSION
Over 50 years, AmB, the prototypic representative of membrane-targeting antifungals (9), has been the mainstay for treating clinical invasive fungal infections despite its nephrotoxicity (21). However, its working mechanism remains elusive. Better understanding of how AmB interacts with the cell membrane is essential for guiding future development of novel resistance-refractory and low-toxicity polyene or other antifungals. For decades, AmB has been generally believed to aggregate with ergosterol to from ion channel vertically in the cell membrane, subsequently causing catastrophic membrane curvature and intracellular substances leakage (20). Until recently, a sterol sponge model was proposed to serve as another mechanism (21). As for the lateral orientation of AmB in the sterol sponge model, the paper by Anderson et al. (21) demonstrated that AmB from both surface adsorption model and sterol sponge model likely exist parallel with the membrane surface. In addition, when AmB positions in the membrane surface, especially in the presence of ergosterol, most of the AmB molecules stay horizontally with respect to the cell membrane plane (42, 43). Moreover, a recent paper by Yamamoto et al. (13) also suggested that the orientation of AmB in the ion channel model is different from that of sterol sponge model. Therefore, the orientation direction of AmB inside the ion channel model is likely different from that of sterol sponge model. Given the limitation and drawbacks of current methods, a novel approach that allows for direct visualization of AmB orientation is highly needed.
On the other hand, the clinical settings have witnessed infections caused by increasing number of multidrug-resistant fungal pathogens such as C. albicans and Candida luasitaniae (44). The recent outbreak caused by the newly emerging C. auris severely poses alarming threat to immunocompromised patients and hospital settings (38) because of its high mortality and transmission (45). Three chronically ill patients in New York were identified as colonizing pan-resistant (resistant to all three classes of antifungals) C. auris according to CDC in 2020. As the second line of defense toward C. auris infections (46), AmB has been found ineffective to cope with 33% of C. auris–caused infections in the United States (45). Unveiling the molecular mechanism of AmB is desperately essential for future antifungal development or repurposing existing drugs to treat C. auris–caused infections. Here, through polarization-sensitive SRS microscopy, we were able to visualize AmB in its natural state at a single-cell level. Meanwhile, by imaging AmB and CH2 from ghost RBCs under the same laser polarization, we found that AmB molecules are orientated parallel to the phospholipids inside the cell membrane, thus supporting the traditional ion channel model.
In support of the ion channel model, we find that ergosterol reallocation uniformly occurs in the cell membrane region in response to AmB treatment, and lipid droplets are formed after AmB treatment in C. albicans (fig. S7) and C. auris. It is known that AmB could cause membrane leakage, induce accumulation of intracellular reactive oxygen species, and subsequently cause a plethora of deleterious cytotoxic substances (47, 48). Given that lipid droplet accumulation is a marker of cellular response to oxidative stress (49–51) and some fungal cells used lipid droplets to trap lipophilic toxins (52), we thus postulate that lipid droplet accumulation acts as a defensive response to AmB-induced cellular stress. Further lipidomics and metabolism pathway analysis of how lipid droplets are synthesized in response to AmB are needed. In this way, we could potentially combat drug-resistant fungal pathogens by combining low-dose AmB with agents that inhibit the biogenesis of lipid droplet under stress.
We note that AmB exhibits pronounced Raman signal (Fig. 2B) at 1556 cm−1 due to the seven conjugated C═C bonds, which has around 100-cm−1 shift compared to the Raman shift of single C═C bond. Meanwhile, there is a 4-cm−1 shift between solution-form AmB that is dimeric in DMSO and AmB from fungal cell membrane where AmB accumulates in a molecular aggregate state (53). Moreover, we also found a much stronger AmB signal in AmB-treated log-phase C. albicans compared to the stationary-phase one under the same settings (Fig. 2, C and D). It was reported that stationary-phase C. albicans exhibits a resistance two or three orders of magnitude greater than AmB compared to the log-phase one (54). Cell wall thickening plays an essential role in inducing this resistance by blocking the membrane insertion of AmB (54). Our data provide visual evidence that stationary-phase C. albicans is more resilient to AmB compared to the log-phase one.
Another interesting finding is that AmB molecules reside as droplets intracellularly (Fig. 4, B1 and C1). Given the fact that sterol esters also reside inside the neutral “lipid droplets” in the fungal cells (55), we make the assumption that AmB stays inside these droplets apart from cell membrane. Further study is needed to understand the mechanism underlying this phenomenon. Since polyene antimycotic agents also include other drugs, such as nystatin, natamycin, or AmB liposomal, the same microscopic approach can be used to unravel the working mechanisms of these drugs. Azoles, another major class of antifungals, are used to target ERG11 to inhibit the biosynthesis of ergosterol. As our technique also allows the detection of ergosterol in situ from single fungal cells, further research can be pursued through this approach to understand the function of ergosterol following phagocytosis of C. albicans by host innate immune cells such as macrophages or neutrophils. Collectively, our discovery paves the foundation for unraveling the mechanism of antifungal agents and future therapeutic development.
MATERIALS AND METHODS
Polarization-sensitive SRS microscope
A femtosecond solid-state laser provides two synchronized outputs as 80-MHz pulsed laser trains (InSight DeepSee, Spectra-Physics). The 1040-nm beam serves as the Stokes beam. The other beam is wavelength tunable (680 to 1300 nm) and serves as the pump beam (here, we chose 895 nm as the pump). An acousto-optic modulator is used to modulate the Stokes beam at 2.2 MHz with an efficiency of ~75%. The pump and Stokes beams were spatially aligned and sent to an upright microscope with two-dimensional galvo system for laser scanning. Half-wave plates at specific wavelength were used to control the polarization directions of these two beams. A spectral-focusing approach was used to obtain spectral domain information. In spectral focusing, the pump and Stokes pulses (stretched by one NSF50 before combiner) were stretched in time by five glass rods (NSF57) to achieve a constant instantaneous frequency difference that drives a single Raman coherence. By delaying the pump pulses controlled by a motorized stage (each step is 10 μm), a series of Raman shifts (120 data points) were generated. At a certain time delay, all the laser energy was spectrally focused to excite a narrow Raman band. The laser powers (pump ~10 mW and Stokes ~50 mW at the sample) were used to excite AmB and ergosterol from the fungal cells. A 60× water immersion objective [Olympus, numerical aperture (NA) 1.2] was used to focus pump and Stokes laser on a sample. An oil condenser (Nikon, NA 1.4) was used to collect the laser light in the forward direction. A 980-nm short-pass filter (Thorlabs) blocked the Stokes laser before a photodiode with a laboratory-built resonant amplifier. A lock-in amplifier demodulated the stimulated Raman loss signal from the pump beam detected by the photodiode. The image size for this paper was 200 × 200 pixels or 400 × 400 pixels with pixel dwell time at 50 μs unless notified, and the pixel size was 250 nm (for 200 × 200 pixels) and 125 nm (for 400 × 400 pixels). The pump power on the sample is around 10 to 15 mW, and the Stokes is around 30 to 50 mW. Images were analyzed using ImageJ (National Institutes of Health), and Raman or SRS spectra were analyzed using OriginPro 2020 (OriginLab Corporation) and MATLAB (MathWorks).
Polarization-sensitive SRS theory
The SRS signal comes from an induced third-order nonlinear polarization expressed by
| (1) |
where D = 3 for ω0 = ωp or ωs (56). χ(3) is the third-order susceptibility, which includes both an electronic and a vibrational contribution. Using the polarization of the Ep field as reference, denoted as 1, the signal field (SRL for pump) under our polarization-sensitive SRS microscope can be written as
| (2) |
Here, ES1 = EScosφ and ES2 = ESsinφ, where φ stands for the angle between the polarization of pump beam and that of Stokes beam.
The total intensity is the mixing of the local oscillator and the signal
| (3) |
Considering that our lock-in amplifier extracts the interference term through heterodyne detection
| (4) |
By introducing , the Raman depolarization ratio, 0 < ρ < 0.75, then we have
| (5) |
Here, we denote I∥ as the SRL signal when the polarization direction of pump beam is parallel to that of the Stokes beam (φ = 0) and I⊥ as the SRL signal with orthogonal pump and Stokes polarization . Then, ; for totally polarized Raman modes, SRL signal drops to zero (ρ = 0) when switching from parallel to orthogonal polarization scheme, and for depolarized modes, SRL signal drops by ¼ times (ρ = 0.75). Totally polarized Raman mode happens when there is symmetric vibration and depolarization occurs in the case of asymmetric vibration.
In the case of AmB-treated fungal cells, when changing the polarization direction of pump beam with respect to the Stokes beam at 90°, we found that the SRS signal at 1556 cm−1 nearly drops to zero (fig. S8), which suggests that AmB at 1556 cm−1 behaves in a totally polarized Raman mode. These data further indicate that SRS signal of AmB in the fungal membrane at 1556 cm−1 comes from the symmetric vibrations of conjugated C═C bonds.
Spontaneous Raman measurement
Sample (prepared in the following method session) was sandwiched between a polylysine-coated cover slide and a cover glass. A HORIBA Raman microscope was used to acquire the Raman spectra of analytes. Excitation laser was 532 nm with a power on the sample ~8 mW. Spectrum acquisition time for each Raman spectrum was 30 s.
Fungal strains and chemicals
Fungal strains
C. albicans SC5314 (wild type) was purchased from American Type Culture Collection (ATCC). C. albicans UPC and C. albicans DBC46 are from M. N. Seleem’s laboratory at Purdue University. All the other strains are from M. K. Mansour’s laboratory (clinical strains from MGH, Boston, MA) including all the C. auris strains: C. auris 1_MGH, C. auris 1_CDC (AR-0381), C. auris 2_CDC (AR-0382), C. auris 3_CDC (AR-0383), C. auris 4_CDC (AR-0384), C. auris 7_CDC (AR-0387), and C. auris 9_CDC (AR-0389); C. albicans C15; S. cerevisiae (AR-0399); C. neoformans; Candida glabrata C1, C2, and ATCC 2001; C. albicans C14; Candida duobushaemulonii (AR-0394), Kodamaea ohmeri (AR-0396).
Chemicals
Amphotericin B (A9528-100MG), glyceryl trioleate (T1740-1G), DMSO (D8418-500ML), YPD broth (Y1375-250G), dibasic potassium phosphate (P3786), monobasic potassium phosphate (P0662), hydrogen chloride (345547-100ML), sodium chloride (S9888), sodium hydroxide (221465), and 10% formalin (HT501128-4L) were purchased from Sigma-Aldrich. Ergosterol (AC117810050) and phosphate buffered saline (BP399500) were purchased from Thermo Fisher Scientific.
RBC ghosts
The erythrocyte ghosts were prepared according to established protocol (37). Briefly, the fresh healthy human blood (from Boston Children’s Hospital Blood Donor Center) was first diluted with 1× phosphate buffered saline at a ratio of 1:20. After washing three times (3000× rpm centrifuge), the erythrocytes were then exposed to ice-cold 5 mM phosphate buffer (lysing buffer, pH 8.0), which caused the erythrocytes to lyse. Then, samples were kept close to 0°C for 2 min (found that the supernatant was deeply red), after which the solution was centrifuged for 15 min at a speed of 13,000g at 4°C, and then, the supernatant was removed from the pellet. Thereafter, the resealing buffer [50 mM NaCl and 5 mM phosphate buffer (pH 8.0)] was added to pellet to allow for resealing of the membranes. The resealing process lasted for 1 hour at room temperature. To fully get rid of all hemoglobin from the cells, we repeated the lysis and resealing procedures twice. When imaged by SRS microscopy, the formed erythrocyte ghost appeared as a spherical vesicle.
AmB-treated fungal strains
As illustrated in fig. S4, pathogens were initially cultured overnight by inoculating one to two single colonies streaked from frozen stock (at −80°C) in sterile YPD broth at 30°C with the shaking speed of 200 rpm at a tilted angle of 45°. The resulting pathogen solution was designated as stationary-phase pathogen. Log-phase pathogen was prepared by 1:20 dilution of overnight-cultured fungal pathogens into fresh prewarm YPD and cultured for another 2 to 3 hours at 30°C with the shaking speed of 200 rpm. After that, pathogen specimens were centrifuged, and supernatant was discarded and then suspended with fresh PBS. The resulting solution was vortexed homogenously, further diluted into 1× PBS with a concentration of 106 cells/ml. AmB stock (3.2 μl; 1 mg/ml in DMSO) was added into 1 ml of the above solution to make a final working concentration of 3.2 μg/ml. Then, the resulting solution was incubated at 30°C with the shaking speed of 200 rpm for 1 hour. After that, AmB-treated fungi were fixed in 10% formalin overnight to ensure the total inactivation of C. auris strains. Formalin was washed away twice by 1× PBS before imaging. Fungal specimen was sandwiched between a polylysine-coated glass slide (P0425, Sigma-Aldrich) and a cover glass (16004-348, VWR international).
Statistical analysis
Student’s unpaired t test and one-way analysis of variance (ANOVA) were used to determine whether there is any statistically significant difference between groups (*P < 0.05, **P < 0.01, and ***P < 0.001).
Supplementary Material
Acknowledgments
We acknowledge M. N. Seleem for providing the C. albicans UPC and C. albicans DBC46 strains. Funding: This work was supported by R35GM136223 and R01AI141439 to J.-X.C. and, in part, by R01AI132638 to M.K.M. Author contributions: M.K.M. provided the clinical fungal strains including the C. auris strains and experimental discussions. P.-T.D., Z.D., and Y.Z. prepared the fungal samples. P.-T.D. and C.Z conducted the experiments. P.-T.D., J.H., J.L., C.Z., and M.Z. did data analysis. P.-T.D. and J.-X.C. co-wrote the manuscript. All authors discussed the results and contributed to the writing of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/2/eabd5230/DC1
REFERENCES AND NOTES
- 1.Brown G. D., Denning D. W., Gow N. A. R., Levitz S. M., Netea M. G., White T. C., Hidden killers: Human fungal infections. Sci. Transl. Med. 4, 165rv13 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Bongomin F., Gago S., Oladele R., Denning D., Global and multi-national prevalence of fungal diseases—Estimate precision. J. Fungi 3, 57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfaller M., Diekema D., Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 20, 133–163 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeffery-Smith A., Taori S. K., Schelenz S., Jeffery K., Johnson E. M., Borman A.; Candida auris Incident Management Team, Manuel R., Brown C. S., Candida auris: A review of the literature. Clin. Microbiol. Rev. 31, e00029-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowdhary A., Sharma C., Meis J. F., Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLOS Pathog. 13, e1006290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caffrey P., Lynch S., Flood E., Finnan S., Oliynyk M., Amphotericin biosynthesis in Streptomyces nodosus: Deductions from analysis of polyketide synthase and late genes. Chem. Biol. 8, 713–723 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Mora-Duarte J., Betts R., Rotstein C., Colombo A. L., Thompson-Moya L., Smietana J., Lupinacci R., Sable C., Kartsonis N., Perfect J.; Caspofungin Invasive Candidiasis Study Group , Comparison of caspofungin and amphotericin B for invasive candidiasis. N. Engl. J. Med. 347, 2020–2029 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Cannon R. D., Lamping E., Holmes A. R., Niimi K., Tanabe K., Niimi M., Monk B. C., Candida albicans drug resistance—Another way to cope with stress. Microbiology 153, 3211–3217 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Lemke A., Kiderlen A. F., Kayser O., Amphotericin B. Appl. Microbiol. Biotechnol. 68, 151–162 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Neumann A., Baginski M., Czub J., How do sterols determine the antifungal activity of amphotericin B? Free energy of binding between the drug and its membrane targets. J. Am. Chem. Soc. 132, 18266–18272 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Hartsel S., Bolard J., Amphotericin B: New life for an old drug. Trends Pharmacol. Sci. 17, 445–449 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Ermishkin L. N., Kasumov K. M., Potzeluyev V. M., Single ionic channels induced in lipid bilayers by polyene antibiotics amphotericin B and nystatine. Nature 262, 698–699 (1976). [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto T., Umegawa Y., Yamagami M., Suzuki T., Tsuchikawa H., Hanashima S., Matsumori N., Murata M., The perpendicular orientation of amphotericin B methyl ester in hydrated lipid bilayers supports the barrel-stave model. Biochemistry 58, 2282–2291 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto T., Umegawa Y., Tsuchikawa H., Hanashima S., Matsumori N., Funahashi K., Seo S., Shinoda W., Murata M., The amphotericin B–ergosterol complex spans a lipid bilayer as a single-length assembly. Biochemistry 58, 5188–5196 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Andreoli T. E., The structure and function of amphotericin B-cholesterol pores in lipid bilayer membranes. Ann. N. Y. Acad. Sci. 235, 448–468 (1974). [DOI] [PubMed] [Google Scholar]
- 16.Bolard J., How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim. Biophys. Acta Rev. Biomembr. 864, 257–304 (1986). [DOI] [PubMed] [Google Scholar]
- 17.Volmer A. A., Szpilman A. M., Carreira E. M., Synthesis and biological evaluation of amphotericin B derivatives. Nat. Prod. Rep. 27, 1329–1349 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Baginski M., Resat H., Borowski E., Comparative molecular dynamics simulations of amphotericin B–cholesterol/ergosterol membrane channels. BBA-Biomembranes 1567, 63–78 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Milhaud J., Ponsinet V., Takashi M., Michels B., Interactions of the drug amphotericin B with phospholipid membranes containing or not ergosterol: New insight into the role of ergosterol. BBA-Biomembranes 1558, 95–108 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Gray K. C., Palacios D. S., Dailey I., Endo M. M., Uno B. E., Wilcock B. C., Burke M. D., Amphotericin primarily kills yeast by simply binding ergosterol. Proc. Natl. Acad. Sci. U.S.A. 109, 2234–2239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson T. M., Clay M. C., Cioffi A. G., Diaz K. A., Hisao G. S., Tuttle M. D., Nieuwkoop A. J., Comellas G., Maryum N., Wang S., Uno B. E., Wildeman E. L., Gonen T., Rienstra C. M., Burke M. D., Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat. Chem. Biol. 10, 400–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chéron M., Cybulska B., Mazerski J., Grzybowska J., CzerwiŃski A., Borowski E., Quantitative structure-activity relationships in amphotericin B derivatives. Biochem. Pharmacol. 37, 827–836 (1988). [DOI] [PubMed] [Google Scholar]
- 23.Gagos M., Arczewska M., Gruszecki W. I., Raman spectroscopic study of aggregation process of antibiotic amphotericin B induced by H+, Na+, and K+ ions. J. Phys. Chem. B 115, 5032–5036 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Verkleij A., de Kruijff B., Gerritsen W. F., Demel R. A., van Deenen L. L. M., Ververgaert P. H. J., Freeze-etch electron microscopy of erythrocytes, Acholeplasma laidlawii cells and liposomal membranes after the action of filipin and amphotericin B. Biochim. Biophys. Acta Biomembr. 291, 577–581 (1973). [DOI] [PubMed] [Google Scholar]
- 25.Wu W., Wieckowski S., Pastorin G., Benincasa M., Klumpp C., Briand J.-P., Gennaro R., Prato M., Bianco A., Targeted delivery of amphotericin B to cells by using functionalized carbon nanotubes. Angew. Chem. Int. Ed. Engl. 44, 6358–6362 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Cheng J.-X., Xie X. S., Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 350, aaa8870 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Fu D., Holtom G., Freudiger C., Zhang X., Xie X. S., Hyperspectral imaging with stimulated Raman scattering by chirped femtosecond lasers. J. Phys. Chem. B 117, 4634–4640 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang D., Wang P., Slipchenko M. N., Ben-Amotz D., Weiner A. M., Cheng J.-X., Quantitative vibrational imaging by hyperspectral stimulated Raman scattering microscopy and multivariate curve resolution analysis. Anal. Chem. 85, 98–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaffer H., Chance R., Silbey R., Knoll K., Schrock R., Conjugation length dependence of Raman scattering in a series of linear polyenes: Implications for polyacetylene. J. Chem. Phys. 94, 4161–4170 (1991). [Google Scholar]
- 30.Chiu L.-d., Hullin-Matsuda F., Kobayashi T., Torii H., Hamaguchi H.-o., On the origin of the 1602 cm–1 Raman band of yeasts; contribution of ergosterol. J. Biophotonics 5, 724–728 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Shadomy S., Brummer D. L., Ingroff A. V., Light sensitivity of prepared solutions of amphotericin B. Am. Rev. Respir. Dis. 107, 303–304 (1973). [DOI] [PubMed] [Google Scholar]
- 32.Li J., Condello S., Thomes-Pepin J., Ma X., Xia Y., Hurley T. D., Matei D., Cheng J.-X., Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell Stem Cell 20, 303–314.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M. C., Min W., Freudiger C. W., Ruvkun G., Xie X. S., RNAi screening for fat regulatory genes with SRS microscopy. Nat. Methods 8, 135–138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P., Li J., Wang P., Hu C.-R., Zhang D., Sturek M., Cheng J.-X., Label-free quantitative imaging of cholesterol in intact tissues by hyperspectral stimulated Raman scattering microscopy. Angew. Chem. Int. Ed. Engl. 52, 13042–13046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasicek E. M., Berkow E. L., Flowers S. A., Barker K. S., Rogers P. D., UPC2 is universally essential for azole antifungal resistance in Candida albicans. Eukaryot. Cell 13, 933–946 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang X., Kirui A., Muszyński A., Dickwella Widanage M. C., Chen A., Azadi P., Wang P., Mentink-Vigier F., Wang T., Molecular architecture of fungal cell walls revealed by solid-state NMR. Nat. Commun. 9, 2747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potma E. O., Xie X. S., Detection of single lipid bilayers with coherent anti-Stokes Raman scattering (CARS) microscopy. J. Raman Spectrosc. 34, 642–650 (2003). [Google Scholar]
- 38.Forsberg K., Woodworth K., Walters M., Berkow E. L., Jackson B., Chiller T., Vallabhaneni S., Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 57, e7 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Lockhart S. R., Candida auris and multidrug resistance: Defining the new normal. Fungal Genet. Biol. 131, 103243 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zamith-Miranda D., Heyman H. M., Cleare L. G., Couvillion S. P., Clair G. C., Bredeweg E. L., Gacser A., Nimrichter L., Nakayasu E. S., Nosanchuk J. D., Multi-omics signature of Candida auris, an emerging and multidrug-resistant pathogen. mSystems 4, e00257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piedrahita C. T., Cadnum J. L., Jencson A. L., Shaikh A. A., Ghannoum M. A., Donskey C. J., Environmental surfaces in healthcare facilities are a potential source for transmission of Candida auris and other Candida species. Infect. Control Hosp. Epidemiol. 38, 1107–1109 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Gagoś M., Gabrielska J., Dalla Serra M., Gruszecki W. I., Binding of antibiotic amphotericin B to lipid membranes: Monomolecular layer technique and linear dichroism-FTIR studies. Mol. Membr. Biol. 22, 433–442 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Man D., Olchawa R., Two-step impact of amphotericin B (AmB) on lipid membranes: ESR experiment and computer simulations. J. Liposome Res. 23, 327–335 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Peyron F., Favel A., Michel-Nguyen A., Gilly M., Regli P., Bolmström A., Improved detection of amphotericin B-resistant isolates of Candida lusitaniae by Etest. J. Clin. Microbiol. 39, 339–342 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chow N. A., Gade L., Tsay S. V., Forsberg K., Greenko J. A., Southwick K. L., Barrett P. M., Kerins J. L., Lockhart S. R., Chiller T. M., Litvintseva A. P.; US Candida auris Investigation Team , Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: A molecular epidemiological survey. Lancet Infect. Dis. 18, 1377–1384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J. Y., Bradley N., Brooks S., Burney S., Wassner C., Management of patients with Candida auris Fungemia at Community Hospital, Brooklyn, New York, USA, 2016–2018. Emerg. Infect. Dis. 25, 601–602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mesa-Arango A. C., Trevijano-Contador N., Román E., Sánchez-Fresneda R., Casas C., Herrero E., Argüelles J. C., Pla J., Cuenca-Estrella M., Zaragoza O., The production of reactive oxygen species is a universal action mechanism of amphotericin B against pathogenic yeasts and contributes to the fungicidal effect of this drug. Antimicrob. Agents Chemother. 58, 6627–6638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belenky P., Camacho D., Collins J. J., Fungicidal drugs induce a common oxidative-damage cellular death pathway. Cell Rep. 3, 350–358 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S.-J., Zhang J., Choi A. M. K., Kim H. P., Mitochondrial dysfunction induces formation of lipid droplets as a generalized response to stress. Oxid. Med. Cell. Longev. 2013, 327167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shyu P. Jr., Fah X., Wong A., Crasta K., Thibault G., Dropping in on lipid droplets: Insights into cellular stress and cancer. Biosci. Rep. 38, BSR20180764 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi K., Gao Z., Shi T.-Q., Song P., Ren L.-J., Huang H., Ji X.-J., Reactive oxygen species-mediated cellular stress response and lipid accumulation in oleaginous microorganisms: The state of the art and future perspectives. Front. Microbiol. 8, 793 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang W., Zhang M., Zheng S., Li Y., Li X., Li W., Li G., Lin Z., Xie Z., Zhao Z., Lou H., Trapping toxins within lipid droplets is a resistance mechanism in fungi. Sci.Rep. 5, 15133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyaoka R., Hosokawa M., Ando M., Mori T., Hamaguchi H.-O., Takeyama H., In situ detection of antibiotic amphotericin B produced in Streptomyces nodosus using Raman microspectroscopy. Mar.Drugs 12, 2827–2839 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gale E. F., Ingram J., Kerridge D., Notario V., Wayman F., Reduction of amphotericin resistance in stationary phase cultures of Candida albicans by treatment with enzymes. Microbiology 117, 383–391 (1980). [DOI] [PubMed] [Google Scholar]
- 55.Lv Q.-Z., Yan L., Jiang Y.-Y., The synthesis, regulation, and functions of sterols in Candida albicans: Well-known but still lots to learn. Virulence 7, 649–659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y. R. Shen, The Principles of Nonlinear Optics (Wiley, 2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/2/eabd5230/DC1


