Abstract
The pandemic of COVID-19 is still ongoing, and many studies on serum antibodies have been reported, however, there are few studies about asymptomatic and mild patients. In this study, we enrolled 44 COVID-19 patients with relatively mild disease and 48 pre-pandemic controls. We measured serum antibodies against extracellular domain, S1 domain, and receptor-binding domain of Spike and N protein, examined neutralization titers by authentic virus neutralization assay and newly-developed bead/cell-based Spike-ACE2 inhibition assay, and compared them with clinical features. Most of these antibodies, including neutralizing titers, were mutually correlated, and the production of antibodies were associated with low Ct values of PCR test, disease severity, symptoms especially pneumonia, lymphopenia, and serological test including CRP, LD, D-dimer, and procalcitonin. Notably, 87.5% of asymptomatic and 23.5% of mild patients did not have antibody against SARS-CoV-2. Our results revealed the inadequate acquisition of humoral immunity in patients with asymptomatic and mild COVID-19 patients.
Keywords: COVID-19, SARS-CoV-2, Antibody, Neutralizing titer, Neutralizing assay, Clinical features, Disease severity, Asymptomatic, Serological test, Immune response
1. Introduction
Although the SARS-CoV-2 pandemic continues worldwide, in some countries, especially those in East Asia, the number of patients tends to be lower than in other areas. Several hypotheses, such as the difference in genetic backgrounds, socio-behavioral differences, mutations in SARS-CoV-2 genomes, and the possibility of having cross-reactive immunity to similar viruses, are inferred as causes of this difference but has not been identified (Yamamoto et al., 2020; Ng et al., 2020; Forster et al., 2020). In Japan, the number of infected patients has started to increase; however, massive outbreaks have been avoided without using forced policies.
Antibody titers against various SARS-CoV-2-derived antigens have been measured and reported to have high specificity for the Spike protein and Nucleocapsid protein (N)(Jiang et al., 2020), and many of the antibody tests have been developed using Spike and/or N as antigens. The serum antibody titers were reported to rise from 10 to 14 days after onset and peak at the third week (Wellinghausen et al., 2020; Liu et al., 2020; Lau et al., 2020; Shaw et al., 2020; Caturegli et al., 2020). To measure the neutralizing titer, several procedures using live virus or pseudovirus have been developed, and it is also known that the titer of neutralizing antibody correlates to some extent with the antibodies against Spike and the N protein (Wu et al., 2020; Robbiani et al., 2020; Okba et al., 2020; Wang et al., 2020). In Japan, although two groups reported that seroconversion was observed in 87% and 96% of patients using a lateral flow kit two weeks after onset (Kaneko et al., 2020; Imai et al., 2020), there are few detailed studies about the association between serum antibodies and clinical parameters.
Our hospital has cared for various patients, including those with symptoms from asymptomatic to severe, and reported their clinical symptoms (Nakagawara et al., 2020). In this study, we aimed to investigate their serological features. Here, we established a bead-based assay for measuring serum antibodies against Spike-derived proteins and N, and bead-based and cell-based procedures for examining the serum Spike-ACE2 inhibition rate, in addition to authentic virus neutralization assay. This study reported the humoral immune responses in Japanese patients with relatively mild disease and their association with detailed clinical features.
2. Material and methods
2.1. Clinical samples
Patients were recruited at Keio University Hospital from April to July 2020. PCR tests were performed on admission and once every 3–5 days thereafter. Serum samples from patients were collected at discharge or at the first outpatient visit. Serum samples from healthy controls were collected and preserved before November 2019. The following parameters were collected from medical charts: signs and symptoms; neutrophil and lymphocyte counts; serum parameters of lactate dehydrogenase (LD), C-reactive protein (CRP), ferritin, D-dimer, procalcitonin, sialylated carbohydrate antigen KL-6, and estimated glomerular filtration rate (eGFR); and medication history. The pneumonia was diagnosed based on the lung CT. This study was approved by the Ethics Committee of our University School of Medicine and conducted in compliance with the tenets of the Declaration of Helsinki. Informed consent was obtained from all participating individuals.
2.2. PCR test for SARS-CoV-2
SARS-CoV-2 detection by real-time RT-PCR from nasopharyngeal swabs was performed with routine diagnostics for examination of potentially infected personnel using BD-MAX (BD, NJ, USA) with the primers N_Sarbeco_F1: CACATTGGCACCCGCAATC, N_Sarbeco_R1: GAGGAACGAGAAGAGGCTTG, N_Sarbeco_P1: ACTTCCTCAAGGAACAACATTGCCA, NIID_2019-nCOV_N_F2: AAATTTTGGGGACCAGGAAC, NIID_2019-nCOV_N_R2: TGGCAGCTGTGTAGGTCAAC, and NIID_2019-nCOV_N_P2: ATGTCGCGCATTGGCATGGA(Shirato et al., 2020).
2.3. Production of recombinant ACE2 and SARS-CoV-2 proteins
The extracellular domain of ACE2 (NCBI, NM_021804.3, 1–708 AA) was cloned from cDNA of human PBMCs into the pcDNA3.4 expression vector (Thermo Fisher Scientific, MA, USA) with a streptavidin-binding peptide (SBP) tag or FLAG tag at the C-terminus. The codon-optimized double-stranded DNA fragments coding SARS-COV-2 Spike and N (NCBI, MN908947.3) were purchased from Genewiz (NJ, USA). The extracellular domain (SECD), S1 domain, and receptor-binding domain (RBD) of the Spike protein and N were inserted into pcDNA3.4 with an SBP tag at the C-terminus. The Spike-derived proteins and ACE2 were produced using the Expi293 Expression System (Thermo) according to manufacturer's instruction. The supernatants were concentrated and buffer exchanged into PBS using Amicon Ultra filters (Merck, Darmstadt, Germany) and incubated with Streptavidin Sepharose High Performance beads (Cytiva, Tokyo, Japan) overnight at 4 °C with shaking. Then, the streptavidin beads were washed 5 times with PBS, and proteins were eluted with 1x Buffer BXT (IBA, Goettingen, Germany).
N was produced by 293T cells by transient expression using polyethyleneimine (Polysciences, PA, USA). Two days after transfection, N-expressing cells were lysed in Tris-buffered saline containing 1% Triton X-100 (TBSTx) with protease inhibitor cocktail (FUJIFILM Wako, Osaka, Japan). Cell lysates were cleared by centrifugation at 16,000 g for 15 min at 4 °C, and the supernatant was purified using streptavidin beads as described above.
These eluates were concentrated and buffer exchanged into PBS again using Amicon Ultra filters. The protein purity was determined by SDS-PAGE and Coomassie Brilliant Blue (CBB) staining using a 12.5% Supersep precast gel and Quick CBB Plus (FUJIFILM Wako), and the concentration was determined by a BCA Protein Assay Kit (Thermo). The purity of the protein are shown in Supplementary Fig. 1A
2.4. Measurement of antibodies against viral proteins by antigen-binding bead assay
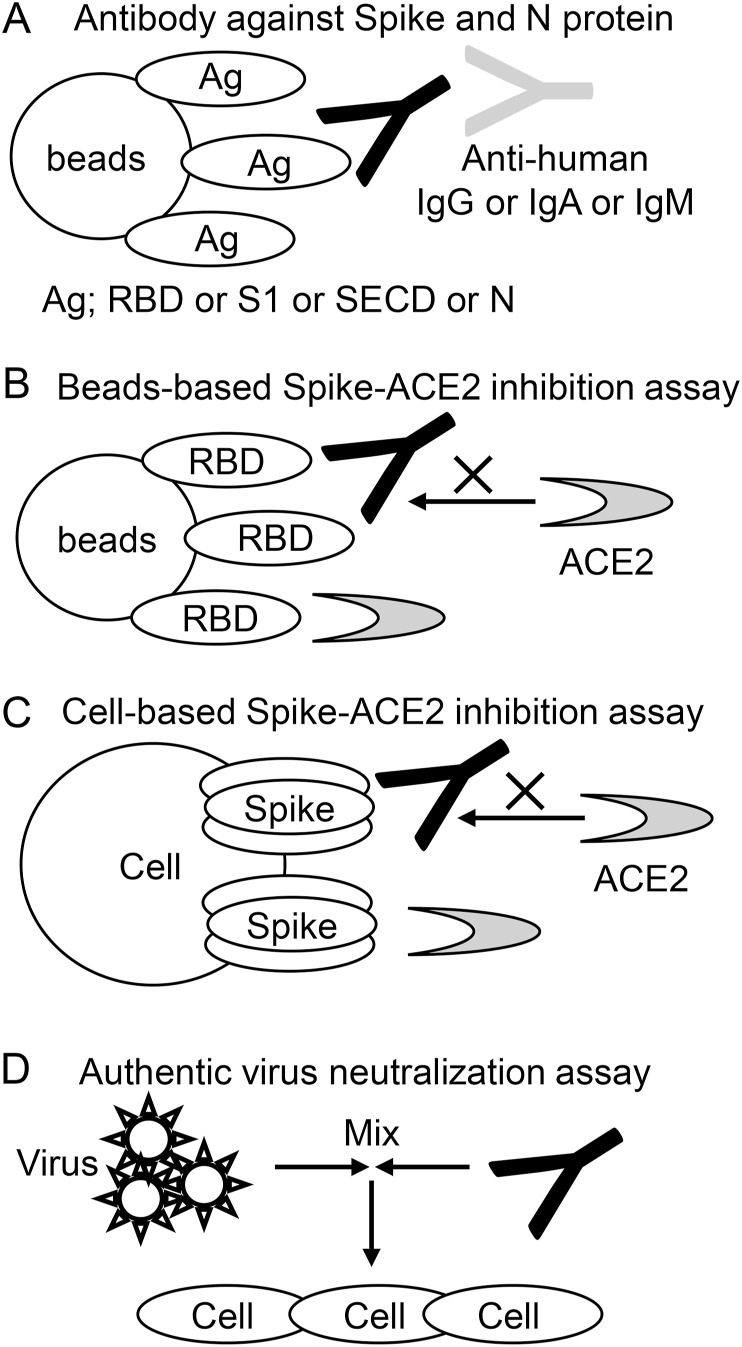
The antibodies against SECD, S1, RBD, and N were measured by antigen-binding bead assays as previously described (Takeshita et al., 2020). In detail, 20 μl of Dynabeads M280 streptavidin (Thermo) were incubated with 4 μg of RBD-SBP, 16 μg of S1-SBP, 32 μg of SECD-SBP, or 10 μg of N-SBP in 100 μl of TBSTx overnight at 4 °C with shaking. After washing with PBS with 0.05% Tween-20 (PBS-T), RBD-, S1-, SECD-, and N-binding beads were transferred into 96 well plate and incubated with 50 μl of 1:100-diluted serum for 20 min at 4 °C. After washing, beads were incubated with 30 μl of an allophycocyanin (APC)-conjugated antibody against human IgG-Fc, IgA-Fc, or IgM-Fc. After the final wash, the beads were analyzed by a FACS Verse (BD). The mean fluorescent intensity (MFI) of APC among singlet-gated beads was used as an indicator of the titer of anti-RBD, S1, SECD, or N antibodies. Representative FACS plot is shown in Supplementary Fig. 1B. The representative results of optimization of serum dilution factor are shown in Supplementary Fig. 1C. The measurement procedures are illustrated in Fig. 1A.
Fig. 1.
Schematic representation of the measurement procedure.
(A) The antibodies against SECD, S1, RBD, and N protein were measured. Antigen-binding beads were incubated with serum, and antibodies bound to the beads (Black) were detected by secondary antibodies (Gray) against IgG, IgA, or IgM. (B) Bead-based Spike-ACE2 inhibition assay. RBD-binding beads were incubated with serum including inhibitory antibody (Black) and then incubated with soluble ACE2 (Gray). The amount of ACE2 bound to beads was measured. (C) Cell-based Spike-ACE2 inhibition assay. Spike-transfected cells were incubated with serum including inhibitory antibody (Black) and then incubated with soluble ACE2 (Gray). The amount of ACE2 bound to beads was measured. (D) Authentic virus neutralization assay. SARS-CoV-2 virus were mixed with serially diluted sera. The mixtures were placed on VeroE6/TMRRSS2 cells and cultured. The highest sera dilution factor with 100% CPE inhibition was defined as authentic virus neutralization titer.
2.5. Bead-based Spike-ACE2 inhibition assay
RBD-binding beads were prepared as described above, and were incubated with 1:20-, 1:100-, or 1:500-diluted serum for 20 min at 4 °C, washed, incubated with ACE2-FLAG for 20 min at 4 °C, washed, and incubated with an anti-DYKDDDDK antibody for 20 min at 4 °C. After the final wash, the beads were analyzed by a FACS Verse. The MFI of beads incubated without serum was used as a negative control (0% inhibition), and beads incubated without ACE2-FLAG were used as a positive control (100% inhibition). The inhibition rate was calculated as follows: inhibition rate = 1 - (MFI of sample – MFI of positive control)/(MFI of negative control – MFI of positive control). The measurement procedures are illustrated in Fig. 1B.
2.6. Cell-based Spike-ACE2 inhibition assay
The expression vectors (12 μg of full-length spike protein and 6 μg of pMX-GFP) were cotransfected into 293T cells on 100 mm dish using 43.2 μg of Polyethylenimine Max (Polyscience, PA, USA). After two days, cells were washed with PBS supplemented with 0.5% BSA and 2 mM EDTA (staining buffer), incubated with diluted serum samples for 20 min at 4 °C, washed again, and incubated with premixed ACE2-SBP and APC-conjugated streptavidin (0.6 μg/ml and 0.4 μg/ml, respectively) for 20 min at 4 °C. After the final wash, the cells were analyzed by a FACS Verse. The MFI among GFP+ cells was calculated using FlowJo (BD) and used as an indicator of Spike-ACE2 binding. Cells without serum and without ACE2-SBP were used as negative and positive controls, respectively. The representative plots are shown in Supplementary Fig. 1D. The measurement procedures are illustrated in Fig. 1C.
2.7. Authentic virus neutralization assay
Heat-inactivated sera were serially diluted with Dulbecco modified Eagle medium (FUJIFILM Wako) supplemented with 2% fetal bovine serum (Biowest, Nuaillé, France) and 100 unit/ml penicillin, 100 μg/ml streptomycin (Thermo). Diluted sera were mixed with 100 TCID50 SARS-CoV-2 JPN/TY/WK-521 strain(Matsuyama et al., 2020) and incubated at 37 °C for 1 h. The mixtures were placed on VeroE6/TMRRSS2 cells (JCRB1819, JCRB Cell Bank, Osaka, Japan) and cultured for 5 days at 37 °C with 5% CO2. For the evaluation of cytopathic effect (CPE), plates were fixed with 20% formalin (FUJIFILM Wako) and stained with crystal violet solution (Sigma-Aldrich, MO, USA). The highest sera dilution factor with 100% CPE inhibition was defined as authentic virus neutralization titer. The measurement procedures are illustrated in Fig. 1D.
2.8. Antibodies
The following antibodies were used: anti-human IgG-Fc (APC, goat-F(ab’)2 fragment), anti-human IgM-Fc (APC, goat-F(ab’)2 fragment), and anti-human IgA-Fc (APC, goat-F(ab’)2 fragment) from Jackson ImmunoResearch (PA, USA); anti-human IgG1 hinge (AF647, 4E3), anti-human IgG2-Fc (AF647, HP6002), anti-human IgG3 hinge (AF647, HP6050), and anti-human IgG4-Fc (AF647, HP6025) from Southern Biotech (AL, USA); and anti-DYKDDDDK antibody (AF647, FLA1) from MBL (Nagoya, Japan).
2.9. Statistics
Continuous data are presented as the median and interquartile range (IQR) or as a number with the percentage value, as appropriate. The chi-squared test was used to examine the categorical variables, and the Wilcoxon rank sum test was used to examine the continuous variables. Correlations between two continuous variables were analyzed using Spearman's rank correlation coefficient. A model for the authentic virus neutralization titer was made using multiple linear regression analysis. The variables that were associated with the neutralizing titer, other than highest LD level and lowest lymphocyte count, which vary even in healthy subjects, were entered into the model using forward selection with a threshold p-value of 0.05. P-value < 0.05 were considered to be statistically significant. All statistical analyses were performed with JMP 15 (SAS Institute, NC, USA).
3. Results
3.1. Cohort of patients
Our hospital experienced nosocomial infections from asymptomatic patients in April 2020 and started to perform PCR tests extensively on potentially infected personnel. Therefore, we found and treated many patients who had mild or no symptoms but were positive on PCR for SARS-CoV-2, in addition to patients who had symptoms and were diagnosed with COVID-19. Because antibodies against SARS-CoV-2 have been reported to appear 10–14 days after onset and most seroconversion develops by the third week, we mainly collected serum samples cross-sectionally three weeks after the positive PCR test. The characteristics of participants are shown in Table 1 .
Table 1.
Clinical characteristics of the patients.
| HC (N = 48) | PT (N = 44) | |
|---|---|---|
| Sex (male) | 6 (40) | 22 (50) |
| Age (year) | 36 (30–60) | 30 (26–46) |
| Smoking: never/ex/current | 34/7/3 | |
| Days after first positive PCR | 55 (34–69) | |
| Previous coexisting disease | 30 (68) | |
| <Signs and symptoms> | ||
| Disease severitya | ||
| Asymptomatic | 16 | |
| Mild | 17 | |
| Moderate | 9 | |
| Severe | 1 | |
| Critical | 1 | |
| Fever (≥37.5°) | 22 (50) | |
| Pneumonia | 11 (25) | |
| Upper respiratory symptomsb | 21 (48) | |
| Lower respiratory symptomsc | 15 (34) | |
| <Laboratory data> | ||
| Lowest PCR Ct valued | 30.0 (19.0–40.6) | |
| Neutrophil count (/μl)e | 2905 (2018–4935) | |
| Lowest lymphocyte count (/μl) | 1464 (968–1961) | |
| Highest LD level (U/l) | 189 (158–234) | |
| Highest CRP level (mg/dl) | 0.18 (0.03–0.74) | |
| Highest ferritin level (ng/ml)f | 172 (69–255) | |
| Highest D-dimer level (μg/ml)g | <0.5 (<0.5–0.8) | |
| Highest procalcitonin level (ng/ml)f | <0.02 (<0.02–0.04) | |
| Highest KL-6 level (U/ml)h | 277 (169–67) | |
| eGFR (ml/min)g | 84 (76–97) | |
| <Treatment> | ||
| Systemic corticosteroids | 3 (7) | |
| Inhaled corticosteroids | 5 (11) | |
| Favipiravir | 7 (16) | |
| Hydroxychloroquine | 4 (9) | |
Data are shown as numbers, numbers (%), or medians (interquartile ranges), as appropriate.
Disease severity is based on “Clinical management of COVID-19” edited by the World Health Organization (May 27, 2020).
Upper respiratory symptoms include rhinorrhea, sore throat, loss of smell, and loss of taste.
Lower respiratory symptoms include cough and sputum production.
N = 40, within 10 days from disease onset.
Neutrophil count at the time of lowest lymphocyte count.
N = 38.
N = 40.
The eGFR on admission is shown. CRP, C-reactive protein; Ct, threshold cycle; PCR, polymerase chain reaction; LD, lactate dehydrogenase; CRP, C-reactive protein; KL-6, Krebs von den Lungen-6; eGFR, estimated glomerular filtration rate.
3.2. Serum antibodies against SECD, S1, RBD, and N
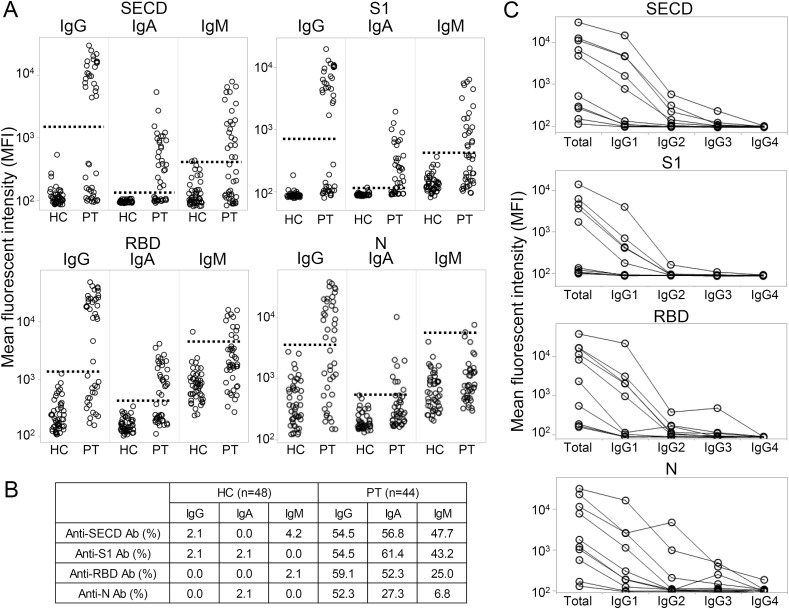
We first measured the serum antibodies against SECD, S1, RBD, and N for each isotype. As shown in Fig. 2 A, some of the patients had antibodies against each antigen, while healthy individuals rarely responded to any antigen. Among the three isotypes, IgG was most clearly separated into positive and negative results, followed by IgA and IgM, and among the four tested antigens, the results for the SECD and S1 were most clearly separated. IgA and IgM against N were less positive than others in the patient group. The overall positive rate is shown in Fig. 2B, which was significantly lower than the reported 80–100% (Wellinghausen et al., 2020; Liu et al., 2020; Lau et al., 2020; Shaw et al., 2020; Caturegli et al., 2020). We also checked IgG subclass against these antigens using 10 representative serum samples. As shown in Fig. 2C, IgG1 was the most abundant for all antigens, followed by IgG2, IgG3, and IgG4.
Fig. 2.
The antibodies against SECD, S1, RBD, and N of each isotype.
(A) The serum IgG, IgA, and IgM antibodies against each protein were measured in 44 patients (PT) and 48 healthy controls (HC). The dashed line indicates the cut-off that was determined as the center of two distinct groups (IgG for the SECD and S1) or 75% quantile + 5 × (75% quantile - 25% quantile) in others. (B) The positive rates of each antibody in the HC and PT are shown. (C) The serum antibodies of each IgG subclass from 10 representative patients were measured. Total indicates total IgG, and the line indicates each individual.
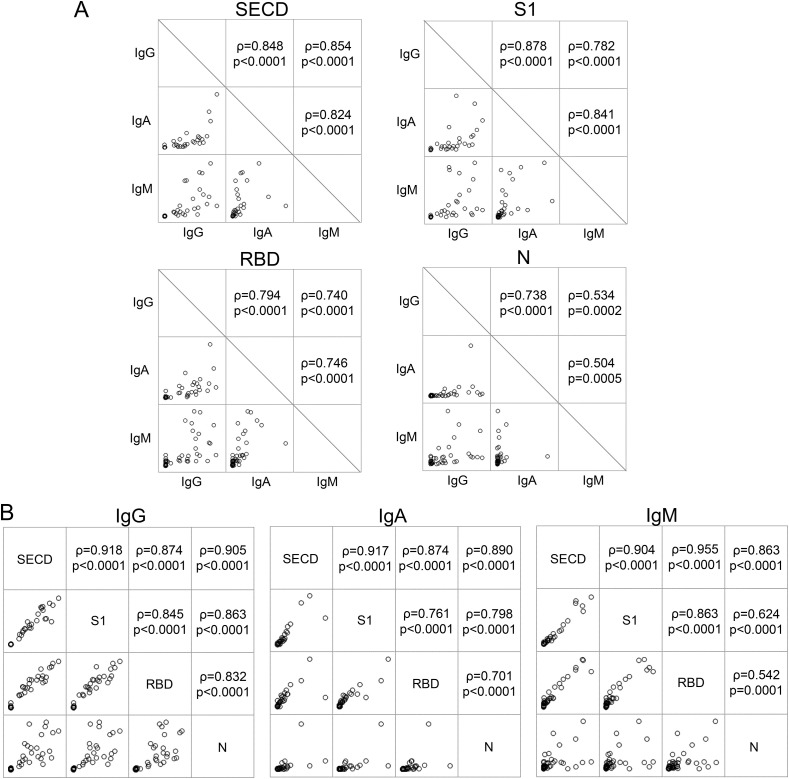
Next, we examined the correlation of the amount of antibodies against each antigen and each isotype. As shown in Fig. 3 A, the antibodies against SECD and S1 had the strongest correlation between each isotype, followed by RBD and N. The correlation of antibodies against various antigens was high for IgG, followed by for IgA and IgM, as shown in Fig. 3B. High correlations were observed among the Spike-derived antigens (SECD, S1, and RBD) for all isotypes, whereas some variations were shown between the antibodies against Spike-derived antigens and antibodies against N.
Fig. 3.
Correlations of antibodies against different targets and different isotypes.
(A) The correlations of the amount of each isotype against SECD, S1, RBD, and N are shown. (B) The correlations of IgG-, IgA-, and IgM-antibodies against each antigen are shown. Spearman's test.
3.3. Serum neutralizing antibody titers
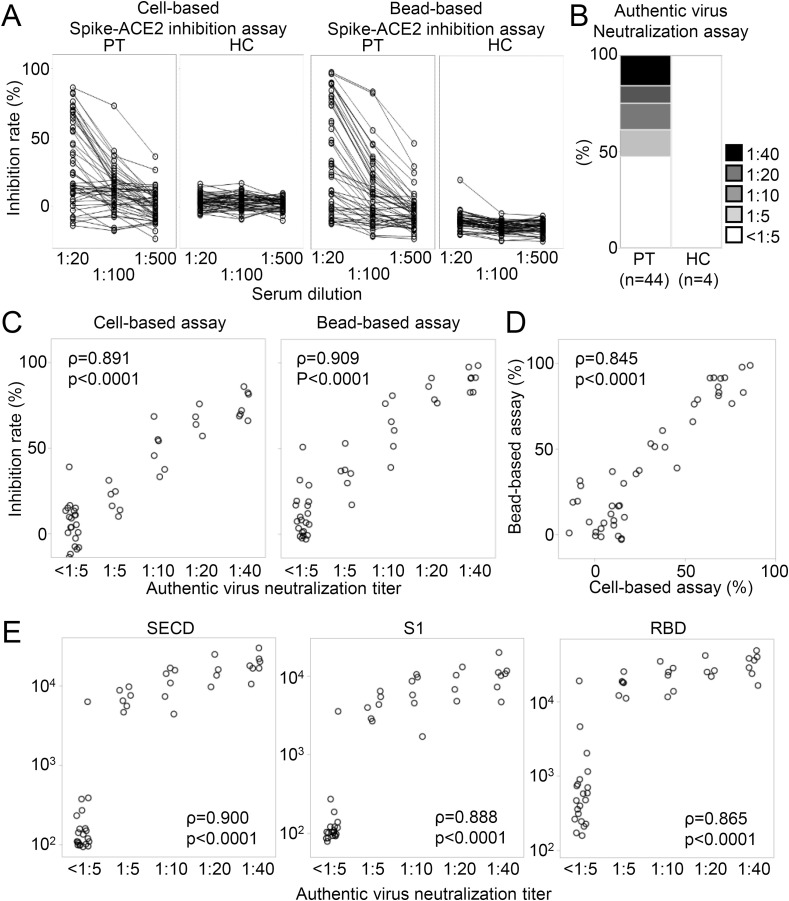
Next, we measured the serum antibodies that inhibit binding of Spike and ACE2 by two procedures. We prepared full-length spike-expressing cells and RBD-binding beads and incubated them with 20-fold-, 100-fold-, or 500-fold-diluted serum to examine how much subsequent binding of soluble ACE2 could be inhibited. As shown in Fig. 4 A, a concentration-dependent inhibition effect was observed with sera from patients but not with most of the control serum. Only one sample from healthy control reacted weakly with RBD beads. Although the IC50 is often used as an index of the these measurements, there were many samples that did not reach 50% inhibition even with a 20-fold dilution in our cohort, so we used the inhibition rate at a 20-fold dilution as an index. In addition to these, to directly measure the virus neutralizing ability of serum, a neutralization test using authentic SARS-CoV-2 was performed for all patients' samples and some healthy control samples (Fig. 4B). The neutralizing titers could not be detected in about half of patients’ samples and all healthy control samples, similar to the results of antibodies against Spike and N.
Fig. 4.
Serum cell- and bead-based Spike-ACE2 inhibition rates and authentic virus neutralization titers.
(A) Serum Spike-ACE2 inhibition rates were measured by cell- and bead-based inhibition assays. The inhibition rates of sera from patients (PT) and healthy controls (HC) at 1:20, 1:100, and 1:500 dilution are shown. The line indicates each individual. (B) Serum authentic virus neutralization titers among all patients (PT) and some healthy controls (HC) are shown. (C) The correlations between the authentic virus neutralization titers and the cell- and bead-based Spike-ACE2 inhibition assays are shown. (D) The correlation between the cell-based and bead-based inhibition assay is shown. (E) The correlations between the neutralization titers and antibodies against the SECD, S1, and RBD are shown. Spearman's test.
We next compared the results of Spike-ACE2 inhibition assay and authentic virus neutralization assay. As shown in Fig. 4C, the inhibition rates of both procedures were highly correlated with the authentic virus neutralization titers, and in addition, the results of the two inhibition assays were also highly correlated (Fig. 4D). These multiple measurements confirmed that our cohort included patients with no serum neutralizing antibodies against SARS-CoV-2. Next, we examined the relationships between the serum antibodies against Spike-derived proteins and authentic virus neutralization titers. As expected, the neutralizing titer strongly correlated with the antibodies against Spike-derived proteins (Fig. 4E). These results means that about half of the patients in this study did not have serum antibodies to SARS-CoV-2.
3.4. Clinical parameters highly correlated with neutralization titers
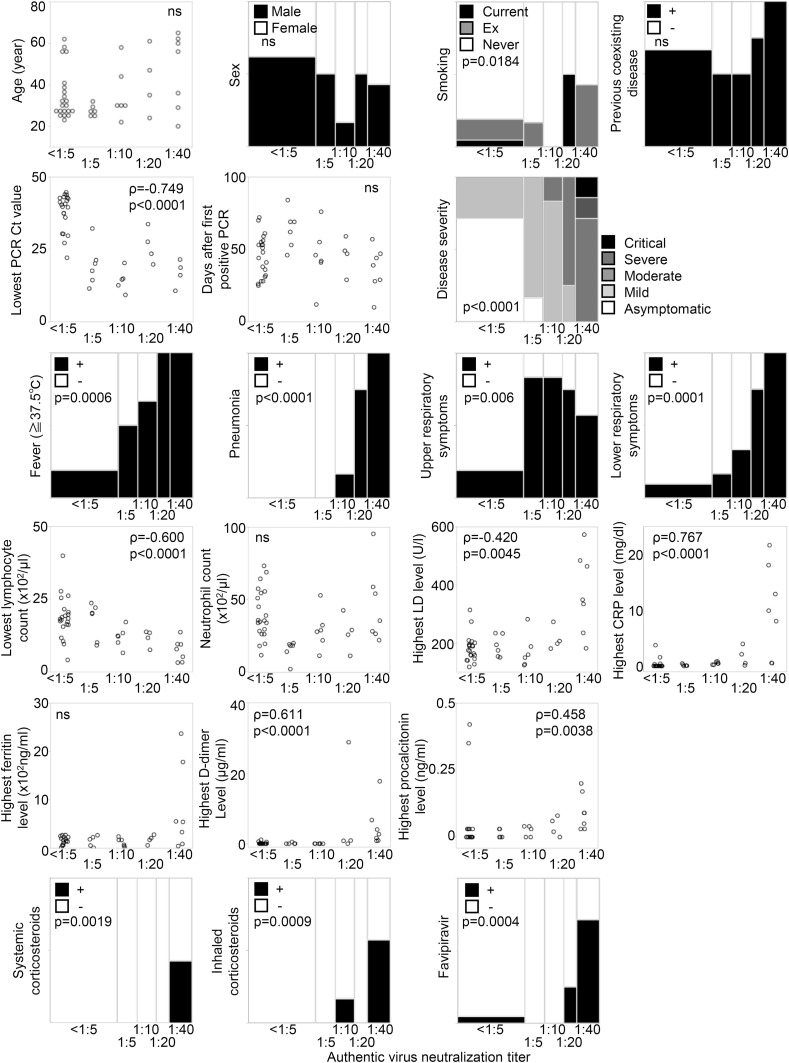
Finally, we examined the relationship between serum antibodies and clinical information/parameters to investigate what factors influence the production of antibodies. Fig. 5 shows correlations between clinical features and authentic virus neutralization titers. Many parameters including Lowest PCR Ct value, disease severity, pneumonia, lymphocytopenia, highest CRP level, and highest D-dimer level were clearly and significantly correlated with neutralizing titers, and notably, neutralizing titers were undetectable among 15/16 (93.8%) of asymptomatic patients and 6/17 (35.3%) of mild patients. Together with the results of serum antibodies against Spike and N, 87.5% of asymptomatic patients and 23.5% of mild patients did not have any antibody against SARS-CoV-2.
Fig. 5.
The association between clinical parameters and the authentic virus neutralization titers.
The associations between various clinical parameters and neutralization titers are shown. For categorical parameters, neutralization titers were used as ordinal variables, and for continuous variables, neutralization titers were used as continuous variables (<1:5 = 0). Spearman's test and Wilcoxon rank sum test.
To examine which of these factors had a strong effect on acquisition of humoral immunity, we next performed multiple linear regression analysis. As shown in Table 2 , three variables, highest CRP level, existence of pneumonia, and lowest PCR Ct value, had statistically significant effects on neutralizing titer with an adjusted R2 of 0.90.
Table 2.
Multiple linear regression model for authentic virus neutralizing titer (%).
| Characteristics | Coefficient (95% confidence interval) | t-value | P-value |
|---|---|---|---|
| Highest CRP level (per 1 mg/dl) | 0.97 (0.57–1.37) | 4.97 | <0.0001 |
| Pneumonia | 8.12 (5.60–10.64) | 6.54 | <0.0001 |
| Lowest PCR Ct value (per cycle) | −0.28 (−0.40–−0.16) | −4.84 | <0.0001 |
CRP, C-reactive protein; PCR, polymerase chain reaction; Ct, threshold cycle; PCR, polymerase chain reaction.
4. Discussion
In this study, we examined the serum antibodies in Japanese COVID-19 patients with relatively mild severities. In our cohort, the amount of antibodies against viral proteins, Spike-ACE2 inhibition rates, and authentic virus neutralization titers were mutually correlated. The production of antibodies was associated with clinical symptoms such as severity and pneumonia, low PCR Ct values, lymphopenia, and serological test results including CRP, LD, D-dimer, and procalcitonin levels. Multiple linear regression analysis showed that highest CRP level, existence of pneumonia, and lowest PCR Ct value had a strong effect on neutralizing titer. We also revealed that approximately 87.5% of asymptomatic patients and 23.5% of mild patients did not acquire humoral immunity. Our results suggest that careful consideration should be applied when conducting epidemiological studies using sera.
Many previous studies reported a cohort that was more symptomatic and severe than our cohort, with 80–100% antibody prevalence three weeks after onset (Robbiani et al., 2020) (Wolfel et al., 2020). Even in a study with mild cases in China, neutralizing antibodies were detected in 94% of patients(Wu et al., 2020). These antibodies were reported to correlate with age, sex, presence of symptoms, severity, lymphopenia, and serum CRP levels (Wu et al., 2020; Robbiani et al., 2020; Yan et al., 2020); however, the prevalence of antibodies was much lower in our results. This difference is considered to be due to the many asymptomatic patients who do not have any antibody. Although there are few studies including many asymptomatic individuals, a German report (Wellinghausen et al., 2020) examining asymptomatic patients with a positive PCR test showed that 85% of these patients were antibody negative, and seropositivity were negatively correlated to Ct values, consistent with our results. These results indicate that a high viral load, pneumonia, and strong systemic response have important roles in the antibody production, similar to findings in Japan and Western countries. One problem is what happens in asymptomatic or mild patients without humoral immunity. Because PCR is repetitively positive in most patients, it is certain that the virus had existed these patients. Recently, T cell responses were reported to be observed in some antibody-negative patients (Sekine et al., 2020). How the immune reaction occurs in such patients and whether such patients are at high risk of reinfection is an important issue for future studies.
The strength of our study is that we used multiple assays to detect antibodies against viral antigens and the neutralizing ability. The results correlated well, indicating that the assay system was reliable. This study also has limitations. One is that the number of patient is relatively small, especially severe patients. Another is that asymptomatic and mild patients showed high PCR Ct values, and the possibility of false positive or contamination remains. Among them, however, 42 out of 44 patients were found to be positive by at least twice PCR tests. Two patients were diagnosed by only single PCR test, and we could not re-test due to lack of samples.
5. Conclusions
In summary, we showed that the viral load, pneumonia, and systemic inflammatory response are important for the production of antibodies against SARS-CoV-2. Our results revealed that the memory of the humoral immunity differ greatly in patients with different degrees of severity. Further comprehensive studies including the clinical significance of these antibodies are desired in the future.
Funding
This work was supported by intramural research funds of Keio University, and by AMED under Grant Number JP20fk0108283.
CRediT authorship contribution statement
Masaru Takeshita: Conceptualization, Methodology, Formal analysis, Writing - original draft, Funding acquisition. Naoshi Nishina: Resources, Data curation, Formal analysis, Writing - original draft. Saya Moriyama: Methodology, Investigation. Yoshimasa Takahashi: Methodology, Investigation. Yoshifumi Uwamino: Investigation. Mika Nagata: Investigation. Wataru Aoki: Investigation. Katsunori Masaki: Resources. Makoto Ishii: Resources. Hideyuki Saya: Supervision. Yasushi Kondo: Resources. Yuko Kaneko: Conceptualization. Katsuya Suzuki: Supervision, Writing - review & editing. Koichi Fukunaga: Supervision. Tsutomu Takeuchi: Supervision, Funding acquisition.
Declaration of competing interest
K.F. reports personal fees from Boehringer Ingelheim and AstraZeneca, outside the submitted work. The remaining authors declare no conflicts of interest.
Acknowledgments
We thank Yukari Kaneda and Mami Yamada for helping with the experiments, the member of Department of Laboratory Medicine, Keio University School of Medicine for repeated scientific discussion, and the Collaborative Research Resources, School of Medicine, Keio University for technical assistances.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2020.12.020.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- Caturegli G., Materi J., Howard B.M., Caturegli P. Clinical validity of serum antibodies to SARS-CoV-2: a case-control study. Ann. Intern. Med. 2020:M20–M2889. doi: 10.7326/M20-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. U.S.A. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K., Tabata S., Ikeda M., Noguchi S., Kitagawa Y., Matuoka M., Miyoshi K., Tarumoto N., Sakai J., Ito T., Maesaki S., Tamura K., Maeda T. Clinical evaluation of an immunochromatographic IgM/IgG antibody assay and chest computed tomography for the diagnosis of COVID-19. J. Clin. Virol. 2020;128:104393. doi: 10.1016/j.jcv.2020.104393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.W., Li Y., Zhang H.N., Wang W., Yang X., Qi H., Li H., Men D., Zhou J., Tao S.C. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat. Commun. 2020;11:3581. doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Nukui Y., Arashiro T., Aiso Y., Sugii M., Hadano Y., Nagata K., Taki R., Ueda K., Hanada S., Suzaki S., Harada N., Yamaguchi Y., Nakanishi H., Kurosaki M., Nagasawa M., Izumi N. Clinical validation of an immunochromatographic SARS-Cov-2 IgM/IgG antibody assay with Japanese cohort. J. Med. Virol. 2020 doi: 10.1002/jmv.26363. (in press) [DOI] [PubMed] [Google Scholar]
- Lau C.S., Hoo S.P., Yew S.F., Ong S.K., Lum L.T., Heng P.Y., Tan J.G., Wong M.S., Aw T.C. Evaluation of an electrochemiluminescent SARS-CoV-2 antibody assay. J Appl Lab Med. 2020:jfaa134. doi: 10.1093/jalm/jfaa134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. Evaluation of Nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawara K., Masaki K., Uwamino Y., Kabata H., Uchida S., Uno S., Asakura T., Funakoshi T., Kanzaki S., Ishii M., Hasegawa N., Fukunaga K. Acute onset olfactory/taste disorders are associated with a high viral burden in mild or asymptomatic SARS-CoV-2 infections. Int. J. Infect. Dis. 2020;99:19–22. doi: 10.1016/j.ijid.2020.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K., Faulkner N., Cornish G., Rosa A., Earl C., Wrobel A., Benton D., Roustan C., Bolland W., Thompson R., Agua-Doce A., Hobson P., Heaney J., Rickman H., Paraskevopoulou S., Houlihan C.F., Thomson K., Sanchez E., Shin G.Y., Spyer M.J., Walker P.A., Kjaer S., Riddell A., Beale R., Swanton C., Gandhi S., Stockinger B., Gamblin S., McCoy L.E., Cherepanov P., Nastouli E., Kassiotis G. bioRxiv; 2020. Pre-existing and <em>de novo</em> humoral immunity to SARS-CoV-2 in humans; p. 2020. 2005.2014.095414. [Google Scholar]
- Okba N.M.A., Muller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C., Bosch B.J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Jr., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., Wullimann D.J., Kammann T., Emgård J., Parrot T., Folkesson E., Rooyackers O., Eriksson L.I., Henter J.-I., Sönnerborg A., Allander T., Albert J., Nielsen M., Klingström J., Gredmark-Russ S., Björkström N.K., Sandberg J.K., Price D.A., Ljunggren H.-G., Aleman S., Buggert M. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168. doi: 10.1016/j.cell.2020.08.017. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.M., Hyde C., Merrick B., James-Pemberton P., Squires B.K., Olkhov R.V., Batra R., Patel A., Bisnauthsing K., Nebbia G., MacMahon E., Douthwaite S., Malim M., Neil S., Martinez Nunez R., Doores K., Mark T.K.I., Signell A.W., Betancor G., Wilson H.D., Galao R.P., Pickering S., Edgeworth J.D. Real-world evaluation of a novel technology for quantitative simultaneous antibody detection against multiple SARS-CoV-2 antigens in a cohort of patients presenting with COVID-19 syndrome. Analyst. 2020;145:5638–5646. doi: 10.1039/d0an01066a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y., Kageyama T., Matsuyama S., Takeda M. Development of genetic diagnostic methods for detection for novel coronavirus 2019(nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020;73 doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- Takeshita M., Suzuki K., Kaneda Y., Yamane H., Ikeura K., Sato H., Kato S., Tsunoda K., Arase H., Takeuchi T. Antigen-driven selection of antibodies against SSA, SSB and the centromere 'complex', including a novel antigen, MIS12 complex, in human salivary glands. Ann. Rheum. Dis. 2020;79:150–158. doi: 10.1136/annrheumdis-2019-215862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang L., Sang L., Ye F., Ruan S., Zhong B., Song T., Alshukairi A.N., Chen R., Zhang Z., Gan M., Zhu A., Huang Y., Luo L., Mok C.K., Al Gethamy M.M., Tan H., Li Z., Huang X., Li F., Sun J., Zhang Y., Wen L., Li Y., Chen Z., Zhuang Z., Zhuo J., Chen C., Kuang L., Wang J., Lv H., Jiang Y., Li M., Lin Y., Deng Y., Tang L., Liang J., Huang J., Perlman S., Zhong N., Zhao J., Malik Peiris J.S., Li Y., Zhao J. Kinetics of viral load and antibody response in relation to COVID-19 severity. J. Clin. Invest. 2020;130(10):5235–5244. doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinghausen N., Plonne D., Voss M., Ivanova R., Frodl R., Deininger S. SARS-CoV-2-IgG response is different in COVID-19 outpatients and asymptomatic contact persons. J. Clin. Virol. 2020;130:104542. doi: 10.1016/j.jcv.2020.104542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brunink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Liu M., Wang A., Lu L., Wang Q., Gu C., Chen J., Wu Y., Xia S., Ling Y., Zhang Y., Xun J., Zhang R., Xie Y., Jiang S., Zhu T., Lu H., Wen Y., Huang J. Evaluating the association of clinical characteristics with neutralizing antibody levels in patients who have recovered from mild COVID-19 in shanghai, China. JAMA Intern Med. 2020;180(10):1356–1362. doi: 10.1001/jamainternmed.2020.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Ariumi Y., Nishida N., Yamamoto R., Bauer G., Gojobori T., Shimotohno K., Mizokami M. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene. 2020;758:144944. doi: 10.1016/j.gene.2020.144944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Zheng Y., Sun Y., Wang L., Luan L., Liu J., Tian X., Wan N. Analysis of the diagnostic value of serum specific antibody testing for coronavirus disease 2019. J. Med. Virol. 2020 doi: 10.1002/jmv.26230. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.