Graphical abstract
Keywords: DFT method, Structural analysis, HOMO-LUMO, MEP, Molecular docking calculations
Abstract
Structure−activity relationships for hydroxychloroquine compound and its derivatives resulted in a potent antiviral activity. Where hydroxychloroquine derivatives showed an apparent efficacy against coronavirus related pneumonia. For this reason, the current study is focused on the structural properties of hydroxychloroquine and hydroxychloroquine sulfate. Optimized structures of these molecules have been reported by using DFT method at B3LYP/6-31G* level of theory. The geometric were determined and compared with the experimental crystal structure. The intra and intermolecular interactions which exist within these compounds are analyzed by different methods namely the topological analysis AIM, ELF and the reduced gradient of the density. These approaches make it possible in particular to study the properties of hydrogen bonds. The highest occupied molecular orbital and the lowest unoccupied molecular orbital energy levels are constructed and the corresponding frontier energy gaps are determined to realize the charge transfer within the molecule. The densities of state diagrams were determined to calculate contributions to the molecular orbitals. The molecular electrostatic potential surfaces are determined to give a visual representation of charge distribution of these ligands and to provide information linked to electrophilic and nucleophilic sites localization. Finally, these derivatives were evaluated for the inhibition of COVID-19 activity by using the molecular docking method.
1. Rationale
Over the past two decades, several infectious diseases caused by various viruses have occurred frequently, what affecting not only the life of human beings, but also affecting national security and stability. At the beginning of the year, Coronavirus disease 2019 namely COVID-19 is a contagious disease caused by a novel strain of beta-coronavirus called SARS-CoV-2 or 2019-nCoV (Huang et al., 2019) that had never been known in humans before. This new coronavirus pneumonia (COVID-19) has swept the world causing enormous loss of life and property to mankind. So far, there is no specific treatment for COVID-19. Coronavirus are enclosed RNA viruses originating from Coronaviridae family. This beta-Coronavirus has flu-like symptoms as fever but it causes respiratory illness which can tend to pneumonia. This virus generally transmits for the duration of coughs, sneezes, saliva droplets or nasal secretions of infected people. This emerging infectious disease appeared in November 2019 in Wuhan, central China. The China National Health Commission studied the first 17 deaths cases in order to fix the median period from the first symptom to death. It founded that this time interval were 14 days and tended to be shorter for people whose exceeds 70 years. Therefore, there is an urgent need to find effective antiviral drugs to combat this disease.
Transmissions ways and treatment results of COVID-19 paid much attention; consequently this latter was the subject of several papers (Romano et al., 2020, Sagaama et al., 2020). In this context, we can mention our previous work (Romani et al., 2020) in which we did a complete study on the properties of Niclosamide molecule using DFT calculations and molecular docking. Molecular docking is a fast and efficient method of drug discovery by testing whether existing antiviral molecules can effectively treat associated viral infections. Results reveal that the niclosamide could bind to the COVID-19 protein with high affinity and it can be a potent inhibitor against coronavirus (Fischer et al., 2020). Numerous clinical tests in China have exposed chloroquine phosphate, an aminoquinoline used in malaria treatment, to be efficient against COVID-19. In addition, hydroxychloroquine was less toxic and more soluble than chloroquine. Both chloroquine and hydroxychloroquine increase the pH and award antiviral effects. Besides, hydroxychloroquine is favored over chloroquine due to its lower ocular toxicity.
Several works were made to utilize the computational methods to screen molecules to find inhibitors against SARS-CoV-2 main protease (Wang et al., 2019). In this context, the present work reports on the geometric optimization of hydroxychloroquine and hydroxychloroquine sulfate by using the hybrid B3LYP/6-31G* level of theory. Then, their reactivity and behaviours were predicted at the same level of theory by using the frontier molecular orbitals. This analysis allows us to find the most reactive ligands against the COVID-19 virus. Moreover, the sites of the nucleophilic and electrophilic regions of chloroquine derivatives can be clearly predicted by means of the different colors discovered on the molecular electrostatic potential surfaces mapped. All these calculations help us a lot in the biological part since the use of optimized structures of ligands is more precise in molecular docking calculations, which makes the program more reliable to be used in structure-based drug design. We proposed three crystal structures of COVID-19 viruses and we will test the antiviral activity of hydroxychloroquine and hydroxychloroquine sulfate compounds against these proteins via molecular docking calculations. Docking studies survey was made to examine and to compare the inhibition mechanism of these chloroquine derivatives against the novel coronavirus disease. So to conclude, this work aims to treat the activities of chloroquine derivatives against COVID-19 virus on the basis of certain important properties predicted by DFT method and specific molecular docking calculations.
2. Methodology calculations
Theoretical results were determined by GaussView (GaussView) and Gaussian programs (Gaussian 09 et al., 2009). The molecular geometries structures of chloroquine derivatives were optimised with the density functional theory (DFT). The quantum chemical calculations were done applying the functional hybrid B3LYP with 6-31G* basis set. Currently this functional correlation exchange is the most used in DFT theory. It is based on a linear combination between fractions of the exchange energy of LDA, HF and B (Becke1988). The reactivity of the chloroquine derivatives are predicted by using the differences between the HOMO and LUMO orbitals via TD-DFT approach. Density of states (DOS) plots was obtained by using TD-DFT Gaussian output via Gauss-Sum software (O'Boyle et al., 2008). The mapped molecular electrostatic potentials (MEPs) are determined at the B3LYP/6-31G* optimized geometry.
The choice of our ligands was mainly based on their anti-viral activities. So, to test the biological activity of the hydroxychloroquine and the hydroxychloroquine sulfate against coronavirus diseases (Protein Data Bank (http) code: 5R7Y, 6M03 and 6LU7), partial flexible docking calculations (rigid protein-flexible ligand) analysis have been carried out using iGEMDOCK program (Yang and Chen, 2004). Molecular docking analysis is a powerful tool for the detection of specific Covid-19 inhibitors, their binding requirements and the mode of ligand–protein interactions. It is a procedure that predicts the orientation of a molecule to another; it allows finding the orientation that maximizes the interaction even as minimizing the total energy score of the studied complex. In all the conditions of docking calculation, we used the settings as follows: population size is 800, the number 10 of generations is 80, and the number of solutions is 10. The docking results were viewed with the help of Discovery studio visualizer interface (Visualizer, 2005).
3. Results and discussion
3.1. Structural analysis
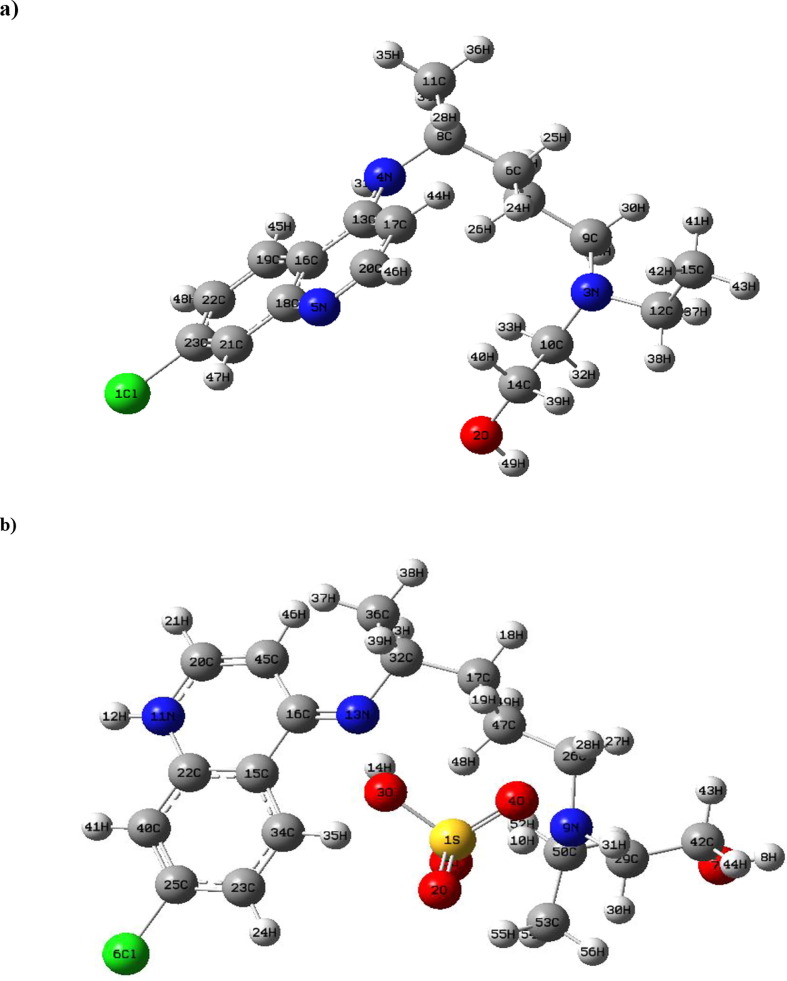
The experimental and theoretical optimized bond distances and bond angles of chloroquine derivatives forms at B3LYP/6-31G* level of theory are collected in Tables S1 and S2 with their Root Mean Square Deviation (RMSD) values. RMSD is considered one of the best tools for structural comparison between the observed and the calculated values. The experimental results are reported on the solid phase, while the theoretical results are determined in the gaseous and in water state. The calculated values reproduce reasonably well the experimentally bond distances and bond angles. The optimized geometrical structures with atom numbering are represented in Fig. 1 . Table S3 gives the geometrical parameters such as the calculated total energies (E), the RMS Cartesian force, the dipole moments (µ) and the maximum Cartesian force of chloroquine derivatives. The C1 point group symmetry is associated with the two compounds. From the data quantum calculations, the hydroxychloroquine sulfate is found to have minimum energy compared with the hydroxychloroquine (as illustrated in Table S3). In the gas phase, the global minimum energy of hydroxychloroquine molecule is equal to −1401.2376 Hartree (≈ −38129 eV) and it decreases by adding the sulfate group (−2101.4778 Hartree ≈ −57184 eV). Weaker energy values are observed for chloroquine derivatives in water solution: −1401.2620 Hartree for hydroxychloroquine and −2101.5608 Hartree (≈ −57186 eV) for hydroxychloroquine sulfate. Evidently, the adding of other atoms or groups in the structures of the hydroxychloroquine compound has an influence on their stability. This stability of the optimized structure was verified by the absence of any imaginary frequencies. In gas phase, the RMS Cartesian force values are found to be 0.0090 a.u and 0.0721 a.u for hydroxychloroquine and hydroxychloroquine sulfate, respectively. These values are weaker in water, 0.0020 a.u for the hydroxychloroquine and 0.0015 for hydroxychloroquine sulfate. The maximum Cartesian forces values of hydroxychloroquine and hydroxychloroquine sulfate are equal to 0.0325 a.u and 0.2606 a.u, while the values decrease in water respectively to 0.0079 u.a and 0.0112 u.a. In this way, it can be concluded that the calculated values in water are slightly low compared to the values in the gas phase.
Fig. 1.
Optimized structures of the hydroxychloroquine (a) and the hydroxychloroquine sulfate by using DFT/B3LYP/6-31G* method.
The hydroxychloroquine sulfate possesses the strongest dipole moment (µ) compared to the hydroxychloroquine molecule. In gas, the dipole moment is of the order of 7.33 D and 11.04 D for hydroxychloroquine and hydroxychloroquine sulfate, respectively. Whereas, in water, it is equal to 11.037 Debye and 29.853 Debye. These high values of the dipole moment allow the formation of the hydrogen bond interactions. Also, the dipole moment of the molecule can affect the interaction between the ligand and the receptor. Therefore, the analysis of the dipole moment gives significant information to find the suitable drug. The size of the dipole moment can reflect the polarity of the molecule. The greater dipole moment, the stronger polarity of the molecule, this promotes the diffusion and absorption of drug molecules.
3.1.1. Optimization of hydroxychloroquine and hydroxychloroquine sulfate
Optimized structures and numbering of atoms of the hydroxychloroquine and the hydroxychloroquine sulfate computed molecules are shown graphically in Fig. 1, obtained at B3LYP/6-31G* basis set. Their global minimum energies are found to be −1401.2376 a.u and −2101.4778 a.u in gas phase and −1401.2620 a.u and −2101.5608 a.u in water. Note that adding the sulfate group; the hydroxychloroquine molecule becomes more stable since the energy decreases. Their optimized geometrical parameters (bond distances and angles) of minimum energies have been determined by the above method and they are given in Tables S1 and S2 with the experimental bond angles and bond lengths. DFT selected structural parameters of hydroxychloroquine are found to be in agreement with experimental data (Ben-Zvi et al., 2012). The low RMSD values are noticed for the bond lengths (0.009 Å) and 1.700° for the bond angles. It can be seen that the low values of the RMSD justify the correct choice of the method used DFT/B3LYP/6-31G*. Also, the comparison of the experimental results of hydroxychloroquine sulfate (Semeniuk et al., 2008) with the calculated data shows a good agreement. The RMSD (bond lengths) = 0.111 Å and the RMSD (bond angles) = 2.523°, but the hydroxychloroquine sulfate is found to be −2101.4778 a.u more stable than hydroxychloroquine. In the solvent, the results show that the RMSD value decreases for the calculated bond lengths and angles. Moreover, it is detected that some of bond distances and angles obtained are greater than the experiment values. This overestimation can be justified that the theoretical results pertain to an isolated molecule in the gas phase while the experimental measurements are obtained in the solid state.
For the hydroxychloroquine, the optimized C–C bond distances in the benzene ring fall in the ranges 1.389–1.410 Å and between 1.380 and 1.418 Å in the pyridine cycle. Whereas, for hydroxychloroquine sulfate, it varies between 1.385 and 1.415 Å in the benzene ring and between 1.357 and 1.485 Å in the pyridine. This last molecule has seventeen C–C bond lengths, six C-N bond lengths, four S-O bond lengths, one C-O and one C-Cl bond lengths. What are left are hydrogen bonds. Theoretically, the intermolecular bond lengths between sulfate and oxygen (S−O) are: 1.459 Å for S–O2 bond, 1.630 Å for S–O3 bond, 1.490 Å for S–O4 bond and 1.528 Å for S–O5 bond. Moreover, the predicted bond distances of N9-C26 (1.510), N9-C50 (1.508), N11-C20 (1.364 Å), N11-C22 (1.388 Å), N13-C16 (1.301 Å) and N13-C32 (1.465 Å) are in accord with the experimental data reported in Table S2. Concerning the N–C bonds of hydroxychloroquine, the calculated distances are: 1.458 Å for N3-C9; 1.464 Å for N3-C10; 1.464 Å for N3-C12; 1.448 Å for N4-C8; 1.383 Å for N4-C13; 1.012 Å for N4-H31, 1.346 Å for N5-C18 and 1.353 Å for N5-C20. The lengthening of Cl-C23 bond is about 1.721 Å. As for the B3LYP optimized C26-N9-C50, C20-N11-C22 and C16-N13-C32 bond angles of hydroxychloroquine sulfate are 111.976°, 121.371° and 120.621° and these compare satisfactorily with the experimental bond angles 109.278°, 120.518° and 124.221°. Concerning the hydrogen-carbon interactions, it varies in the range 1.084–1.103 Å.
3.1.2. Hybridization effect
In fact, the stability comparison of the chloroquine derivatives is necessarily based on the minimum energy value found and the value of RMSD calculated. The hydroxychloroquine sulfate is initially made up of hydroxychloroquine. Of course, the adding of other atoms or groups in the structure of the hydroxychloroquine will change system properties and has an influence on their stability. Analysis of the results above has shown that adding the sulfate group makes the hydroxychloroquine more stable since the energy decreases. Also, the comparison clearly shows the increase in the dipole moment value which promotes the formation of the hydrogen bond interactions.
3.2. Non-covalent interactions
3.2.1. Topological analysis (AIM)
The quantum theory AIM (Atoms-in-Molecules) is a model of quantum chemistry characterizing the chemical bond of a system based on the study of the topology of the electron density. This latter characterizes the distribution of electrons in space. The AIM approach allows us to better understand the nature of the non-covalent interactions revealed within the molecular system and precisely the strengths of the hydrogen bonds from the different topological and energetic properties. Among these properties, we can cite: the electron density ρ(r), the Laplacian of the electron density ∇2 ρ (r), the eigenvalues of the Hessian matrix (λ1, λ2, λ3), density total energy H (r), local electron potential V (r) and bond energy Ebond = V(r)/2) (Ben Issa et al., 2020). This approach makes it possible to locate critical points of density that correspond to a topological feature. According to Bader's theory, the points where the gradient of the electron density is zero (∇ρ = 0) correspond to the critical points. The Laplacian of electron density is the sum of three eigenvalues of the Hessian matrix (λ 1, λ2, λ3). It is a powerful tool for the characterization of binding zones. From the Rozas criterion (Rozas et al., 2000), hydrogen bonds can be classified into 3 categories: a strong hydrogen bond if the Laplacian of the electron density ∇2 ρ (r) < 0 and the total energy density H (r) < 0; a moderate hydrogen bond if ϖ2 ρ (r) > 0 and H (r) < 0 and a weak hydrogen bond if ∇2 ρ (r) > 0 and H (r) > 0.
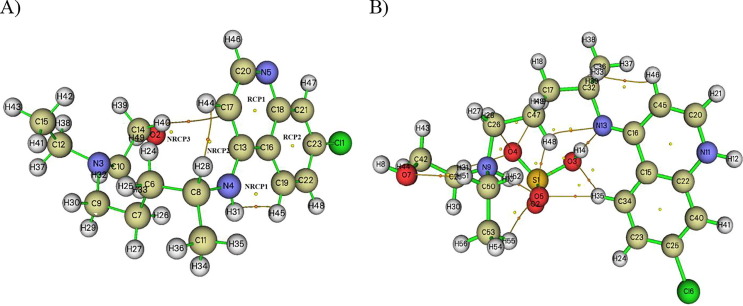
Graphical representations of the AIM analysis of hydroxychloroquine and hydroxychloroquine sulfate compounds are shown in Fig. 2 using Multiwfn (Lu and Chen, 2012). The topological parameters of the non-covalent interactions are grouped together in Table 1 . AIM results shows that the hydroxychloroquine complex is characterized by three BCPs: two describing an N-H…H and C-H…H type interaction and one characterizing a C-H…C type hydrogen bond. The BCP located at the level of the hydrogen bond H…C is characterized by a low electron density (0.0011 a.u), and a positive Laplacian value (0.0035 a.u). The positive Laplacian value indicates depletion of electronic charge along the binding path. In the case of hydroxychloroquine sulfate, AIM analysis showed nine BCPs: two located at the H…N hydrogen bonds and seven at the H…O bonds. From the topological parameters presented in Table 1, the electron density and its Laplacian values are slightly larger than those found for the hydroxychloroquine molecule. The hydrogen bond energy values are also greater. Results show the presence of weak hydrogen bonds: C47-H48…N13, C47-H48…O5, S1-H55…O5, C26-H28…O4, C17-H19…O4 and C34-H35…O3, since ∇2 ρ (r) > 0, and H (r) > 0 and their energy Ebond < 15 kJ/mol. Both C32-H14…N13 (39.119 kJ/mol) and C29-H31…O4 (15.490 kJ/mol) bonds are considered moderate interactions. N9-H 10… O5 is the only strong hydrogen bond whose energy is in the order of 73.907 kJ/mol.
Fig. 2.
Graphical representations of the AIM analysis of the hydroxychloroquine (A) and hydroxychloroquine sulfate (B).
Table 1.
Topological properties of chloroquine derivatives.
| Interactions | ρ (r) | ∇2 ρ (r) | H(r) | G(r) | V(r) | λ 1 | λ 2 | λ 3 | Ebond kJ.mol−1 | ELF |
|---|---|---|---|---|---|---|---|---|---|---|
| Hydroxychloroquine | ||||||||||
| N4-H31…H45 | 0.0126 | 0.0542 | 0.0025 | 0.01106 | −0.0085 | 0.0766 | −0.0136 | −0.0087 | 11.158 | 0.0308 |
| C8-H28…H44 | 0.0105 | 0.0467 | 0.0028 | 0.0089 | −0.0060 | −0.0080 | −0.0021 | 0.0569 | 7.876 | 0.0259 |
| C14-H40…C17 | 0.0011 | 0.0035 | 0.0003 | 0.0006 | −0.0003 | −0.0006 | 0.0046 | −0.0004 | 0.393 | 0.0025 |
| Hydroxychloroquine sulfate | ||||||||||
| C47-H48…N13 | 0.0141 | 0.0583 | 0.0023 | 0.0122 | −0.0099 | 0.0723 | −0.0019 | −0.0121 | 12.996 | 0.0360 |
| C32-H14…N13 | 0.0396 | 0.0994 | −0.0024 | 0.0273 | −0.0298 | −0.0605 | −0.0582 | 0.2181 | 39.119 | 0.1892 |
| C47-H48…O5 | 0.0082 | 0.0310 | 0.0011 | 0.0066 | −0.0054 | −0.0032 | 0.0415 | −0.7288 | 7.088 | 0.0213 |
| N9-H10…O5 | 0.0635 | 0.1772 | −0.0060 | 0.0503 | −0.0563 | 0.4135 | −0.1155 | −0.1208 | 73.907 | 0.2500 |
| S1-H55…O5 | 0.0062 | 0.0245 | 0.0010 | 0.0051 | −0.0040 | −0.0056 | −0.0027 | 0.0329 | 0.525 | 0.0141 |
| C29-H31…O4 | 0.0154 | 0.0497 | 0.0002 | 0.0121 | −0.0118 | 0.0815 | −0.0167 | −0.0151 | 15.490 | 0.0488 |
| C26-H28…O4 | 0.0103 | 0.0389 | 0.0011 | 0.0085 | −0.0073 | −0.0086 | −0.0075 | 0.0550 | 9.583 | 0.0267 |
| C17-H19…O4 | 0.0107 | 0.0350 | 0.0006 | 0.0081 | −0.0074 | −0.0104 | 0.0553 | −0.0099 | 9.714 | 0.0326 |
| C34-H35…O3 | 0.0091 | 0.0394 | 0.0016 | 0.0081 | −0.0065 | −0.0074 | −0.0047 | 0.0516 | 8.532 | 0.0192 |
3.2.2. Reduced gradient of density (RDG)
To study the different non-covalent interactions existing within the molecules, the reduced gradient of the RDG density is used.
It is a dimensionless quantity and is written in the following form:
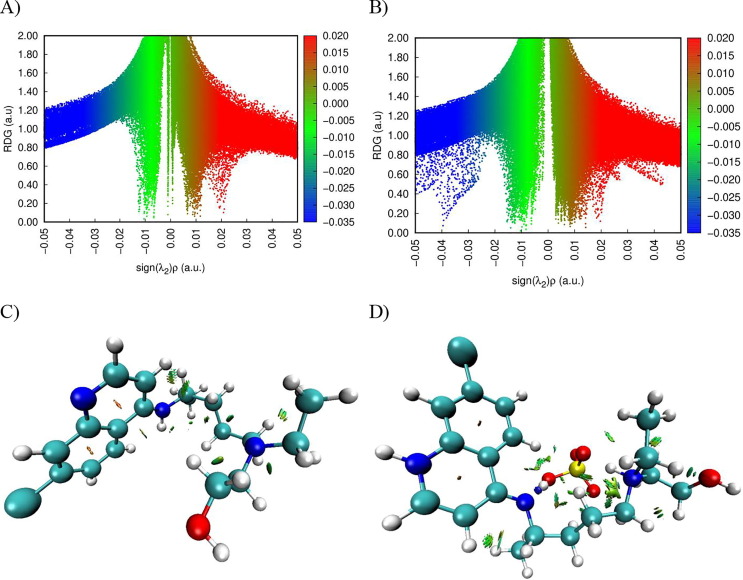
We have represented, in Fig. 3 (A, B), the reduced gradient of the density as a function of the electron density multiplied by the sign of the second eigenvalue of the matrix of Hessian. λ2 is the greatest eigenvalue of the Hessian matrix; it characterizes the fluctuation of the density in the vicinity of the critical point. From the value of the sign (λ2)*ρ, we can distinguish between the different types of non-covalent interactions. Fig. 3(C, D) shows zone of the different interactions in hydroxychloroquine and hydroxychloroquine sulfate. As clearly seen, there are three regions of interaction. The regions marked in green represent the Van der Waals interactions. The red color corresponds to repulsive interactions (steric effect) mainly localized at the cycle level. These interactions are based on the properties of electron density. The blue spots are indicators of the presence of hydrogen bonds. Generally, the VDW interactions have very low electron densities, while the regions of interactions corresponding to the hydrogen bond and the steric effect have a high density.
Fig. 3.
Graphical representation of the reduced density gradient versus the electron density (A, B) and the different types of interactions (C, D) of hydroxychloroquine and hydroxychloroquine sulfate compounds.
3.2.3. Electron localization function (ELF)
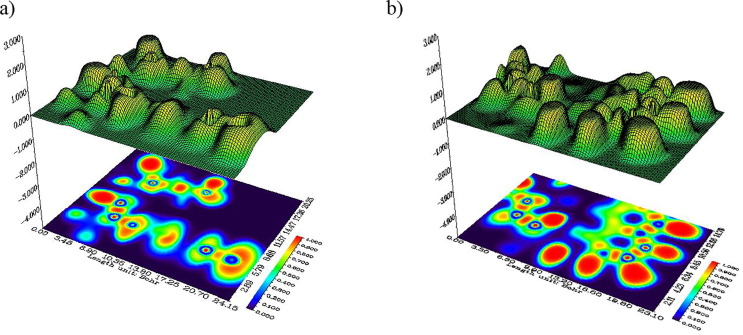
The topological analysis of the ELF function, published in 1990, was subsequently analyzed by Silvi and Savin (Silvi and Savin, 1994). It allows a partition of the molecular space not in atomic basin as in Bader's theory, or in regions of charge concentration, but in basins of electronic localization within which the excess of kinetic energy due to the repulsion of Pauli is minimum. The spatial position of these attractors makes it possible to differentiate the core basins and the valence basins. Heart basins are located around nuclei (except for the hydrogen atom). The valence basins are classified according to their connectivity with the core basins. Topological analysis of the localization function of electrons constitutes the suitable mathematical model for the characterization of chemical bonds. The 2D representation of the ELF isosurface for chloroquine derivatives was determined by Multiwfn program. In Fig. 4 , we present the shaded surface maps with projection effect of electron localization function (ELF). Other ELF surface maps of hydroxychloroquine (a) on the plane of C23, C8, N3 atoms and of hydroxychloroquine sulfate (b) on the plane of N11, N13, N9 atoms, are plotted as shown in supplementary Fig. S1. As it shown, several colors are represented on this surface. The red and orange colors represent a strong electronic localization. The blue color circle represents a depletion region between inner shell and valence shell. The hydrogen and the carbon regions have the minimum values of the localized orbital locator.
Fig. 4.
Shaded surface maps with projection effect of electron localization function (ELF) of hydroxychloroquine (a) and hydroxychloroquine sulfate (b).
3.3. Electronic properties
3.3.1. Frontier orbitals and global descriptors studies
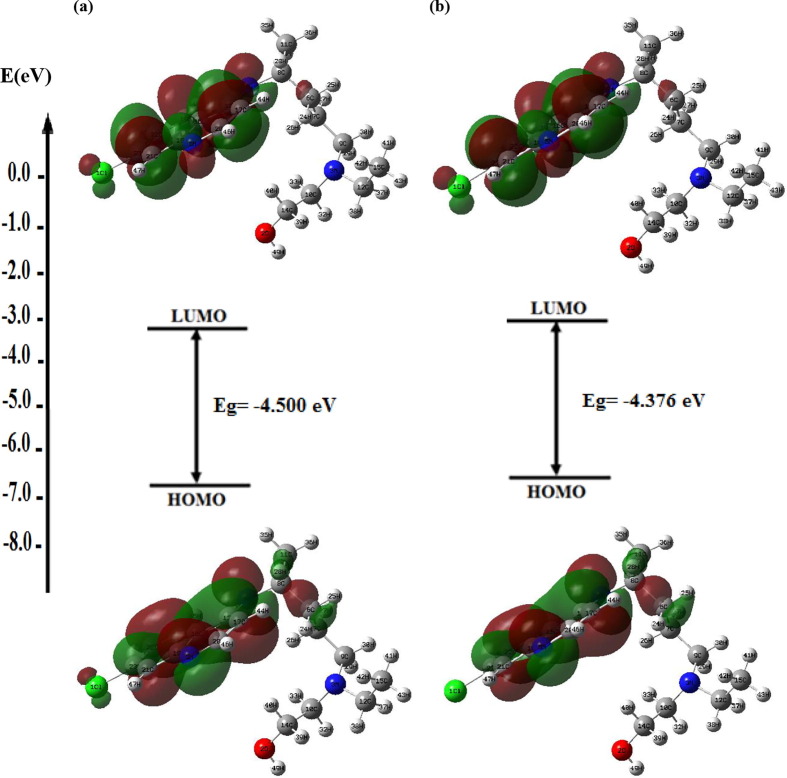
Our goal in this work is to find the most reactive ligands against the COVID-19 virus. For these reasons, in this part, we have to determine the frontier molecular orbitals (FMOs) and their properties like energy gap since they are used to provide information on the transfer of charges (Gatfaoui et al., 2019, Renug and Muthu, 2014). Lowest unoccupied molecular orbital (LUMO) and highest occupied molecular orbital (HOMO) are very important parameters for quantum chemistry (Renug and Muthu, 2014). These orbitals allow determining the manner the molecule interacts with other species. LUMO, related with the ionization potential, can be thought the most frontiers orbital having free places for accepting electrons. On the other hand HOMO, related with the electron affinity of the molecule, represents the capability to donate electrons. Therefore, studying frontiers orbitals of molecules can provide important information to explore the mechanism of action and determine the active sites (Tahenti et al., 2020). Hence, the reactivity of the chloroquine derivatives is predicted by using the differences between the HOMO and LUMO orbitals. Those orbitals were determined via TD-DFT approach and the pictorial illustrations are given in Figs. 5 and S2. The green color corresponds to the negative phase, while the positive phase is represented by the red color. The distributions of these orbitals and energies are very useful indicators of reactivity (Noureddine et al., 2021). Concerning the hydroxychloroquine molecule, the energy level of the HOMO orbital is equal to −5.553 eV and the energy level of the LUMO orbital ELUMO = −1.052 eV (in gas phase). The energy separation value between HOMO and LUMO is called energy gap and it is found to be 4.500 eV (in ultimate value), while it is equal to 4.376 eV in water
Fig. 5.
Plots of the frontier molecular orbitals of hydroxychloroquine in gas phase (a) and in water (b) by using TD-DFT calculations.
As everyone knows, the quantum chemical parameters of the molecule are very important for its practical application. Therefore, in this paper we calculated several quanta chemical reactivity using FMOs energies. Table 2 represents the calculated reactivity descriptors such as the energies of frontier molecular orbitals (EHOMO, ELUMO), energy band gap, ionization potential, electron affinity, chemical hardness, chemical softness, electronegativity, chemical potential, electrophilicity index and the maximum charge transfer index. All these parameters are allied to the energies of FMOs by the corresponding equations mentioned in the literature: the electron affinity A = − ELUMO, the ionization potential I = − EHOMO and μ = 1/2(I + A) which represent the electronic chemical potential. The chemical hardness (η) (ELUMO–EHOMO= (I − A)) is equal to 4.501 eV and 2.188 eV in gas and water, respectively. The chemical softness can be estimated as ζ = 1/2η and found to be 2.2505 eV−1 and 1.094 eV−1. Moreover, the mentioned molecule has an electronegativity (χ = (I + A)/2) about χ gas = 3.3025 eV and χ water = 3.327 eV. The electrophilicity index (ω = μ2/2 η) has been computed and found to be ω gas = 1.211 eV and ω water = 2.529 eV. Lastly, the maximum charge transfer index (ΔNmax = –μ/ η) of hydroxychloroquine is equal to 0.733 in the gas and 1.520 in water. This index represents the proportion of maximum charge that a system can acquire from its environment, or ΔNmax is maximum in water, which means that water retains more electrons in its environment than in gas.
Table 2.
Calculated of some global reactivity descriptors of chloroquine derivatives.
| Parameters | Hydroxychloroquine | Hydroxychloroquine sulfate | ||
|---|---|---|---|---|
| In gas | In Water | In gas | In Water | |
| ELUMO | −1.052 | −1.139 | −1.271 | −1.843 |
| EHOMO | −5.553 | −5.515 | −5.618 | −6.094 |
| Energy band gap | −4.500 | −4.376 | −4.347 | −4.250 |
| Reactivity descriptors | ||||
| Ionization potential (I) | 5.553 | 5.515 | 5.618 | 6.094 |
| Electron affinity (A) | 1.052 | 1.139 | 1.271 | 1.843 |
| Chemical hardness (η) | 4.501 | 2.188 | 4.347 | 2.125 |
| Chemical softness (ζ) | 2.250 | 1.094 | 2.173 | 1.062 |
| Electronegativity (χ) | 3.302 | 3.327 | 3.444 | 3.968 |
| Chemical potential (μ) | −3.302 | −3.327 | −3.444 | −3.968 |
| Electrophilicity index (ω) | 1.211 | 2.529 | 1.364 | 3.704 |
| Maximum charge transfer index (ΔNmax) | 0.733 | 1.520 | 1.012 | 1.867 |
I = –EHOMO, A = –ELUMO, η = (I–A)/2, ζ = 1/2 η, χ = (I + A)/2, μ = –(I + A)/2, ω = μ2/2 η and ΔNmax. = –μ/ η.
Passing to the hydroxychloroquine sulfate molecule, in gas phase the energy gap between the HOMO and LUMO was found to be 4.347 eV with a chemical hardness value of 4.347 eV. It is lower in water in the order of Eg = 4.250 eV. By comparing the values of the energy differences of different phases (gas and solvent), we can conclude that the two ligands are more reactive and more stable in water. Based on the dipole moment values, we see that the dipole moment is greater in water, knowing that the larger the dipole moment, the stronger the intermolecular interactions. We also notice that the gap energy decreases by adding the sulfate group. In that case, the flow of electrons in the higher energy state is easy; therefore the molecule becomes soft and more reactive. So, from these results, we find that hydroxychloroquine sulfate is the most reactive antiviral compound compared to the other antiviral one since it has the highest transfer index and the weaker energy gap. The density of states (DOS) plots of chloroquine derivatives was also obtained in Fig. S3 by using Gauss-Sum software. These spectra describe the energy levels per unit energy increment and its composition in energy between −20 and 0 eV. The displaying analysis per orbital indicates that the red and green lines in the DOS spectrum represent the LUMO (ELUMO) and HOMO (EHOMO) levels, respectively. As expected, the energy difference between HOMO and LUMO for the two ligands is compatible with the values determined by the DOS spectra.
3.3.2. Molecular electrostatic potential (MEP)
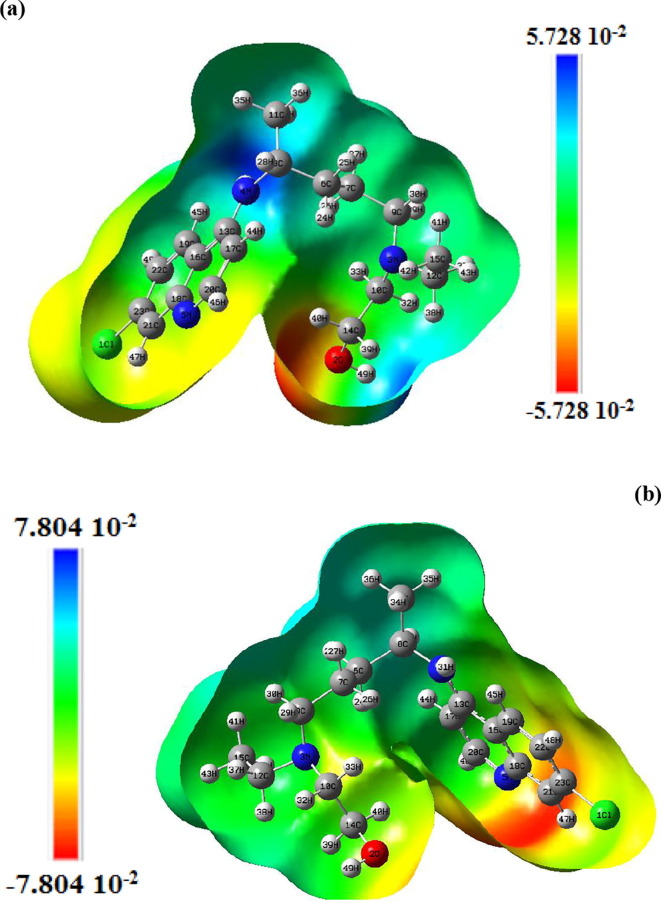
Interactions between bio-molecules and drugs mainly include hydrogen and halogen bonds dominated by electrostatic interactions. Molecular electrostatic potential (MEP) is the real spatial function most closely related to electrostatic effects, which is very suitable for dominance analysis electrostatic. Therefore, the analysis of MEP is useful for understanding the charge related properties in the molecular structure of the complex. The mapped molecular electrostatic potentials (MEPs) of the chloroquine derivatives are theoretically predicted to benefit a better comprehension at a molecular level. This analysis allows us to determine the most reactive electrophilic and nucleophilic regions of the molecule against the reactive biologic potentials. The mapped MEP surfaces are graphed in Figs. 6 and S4 at the B3LYP/6-31G* optimized geometries. In the gas phase, these surfaces are illustrated using a color code mapped in the range between −5.728 0.10−2 a.u (deepest red) to 5.728 0.10−2 a.u (deepest blue) for hydroxychloroquine and between −9.211 0.10−2 a.u to 9.211 0.10−2 a.u for hydroxychloroquine sulfate. In water, MEP surfaces varied between −7.804 0.10−2 a.u and 7.804 0.10−2 a.u for hydroxychloroquine and between −0.186 a.u and 0.186 a.u for hydroxychloroquine sulfate. Potential decreases in the order blue > green > yellow > orange > red. The negative regions (red colors) of MEP are related to nucleophilic reactivity and the positive regions (blue color) to electrophilic reactivity, whereas neutral regions of the molecule are identified by green colors. As can be seen from the MEP map of hydroxychloroquine molecule (in the gas phase), the positive potential sites are around the N4, H28 and C8 atoms with a maximum value of 5.728 0.10−2 a.u, as well as the negative potential sites, which are mainly localized on hydrogen H40 atom with a minimum value of −5.728 0.10−2 a.u. From the MEP surface of hydroxychloroquine sulfate, it is evident that the positive zones (blue) cover the hydrogen H12 and H8 atoms. In water, for hydroxychloroquine red colors are observed on the C21 and C23 atoms of benzene ring while the colorations on these atoms are neutral in the gas. For hydroxychloroquine sulfate, the strong red colors are observed on O2, O3 and O4 atoms. The benzene and pyridine rings are located in the green zone (neutral regions).
Fig. 6.
Molecular electrostatic potential (MEP) maps of hydroxychloroquine in gas phase (a) and in water (b).
3.4. Biological activities
3.4.1. Molecular docking analysis
Chloroquine derivatives have a wide scope in the medicinal field as an anti-viral drug (Hobbs et al., 1959, Ben-Zvi et al., 2012, Canadian Hydroxychloroquine Study Group, 1991, Murray et al., 2010). For many years, these compounds are a drug utilized in the treatment of rheumatic diseases and especially in the treatment of malaria Brocks and Mehvar, 2003, Borden and Parke, 2001. They have a very important role in respiratory viral loads. To appraise the antiviral activity of chloroquine derivatives, molecular docking analysis was expanded out against the protein of the antiviral strains. In this part, our goal is to test the hydroxychloroquine and hydroxychloroquine sulfate as coronavirus inhibitors and to dock at binding sites of COVID-19 (PDB ID: 5R7Y, 6M03 and 6LU7) for determining their potential binding modes. These proteins are exported from Protein Data Bank of the Structural Bioinformatics Research Laboratory (RCSB). Thereafter, the water molecules and the co-constituent in the target structure were removed using the Discovery Studio visualizer software. Molecular docking calculations have been performed by iGEMDOCK program through the generic evolutionary method (GA) and an empirical scoring function. It is a method that illustrates the potential binding posture of ligands flexibly against the ligand-binding cavities of receptor protein determination. Generally, the docking studies are utilized to explore the different interactions between ligand and receptor Seeliger and de Groot, 2010, Sagaama et al., 2020, Noureddine et al., 2021, Noureddine et al., 2020, Noureddine et al., 2020, Jomaa et al., 2020, Ghalla et al., 2018. Experimental results have shown that this analysis is extremely trustworthy for drug design (Cox et al., 2000, Kuntz, 1992); also it is a time-saving and cost-effective method. During this calculation, all the feasible conformations of the ligand and their best-docked poses have been determined. After each run, the top-ranked pose was selected from the ten runs. The best position is who has the lowest energy which corresponds to the most stable complex.
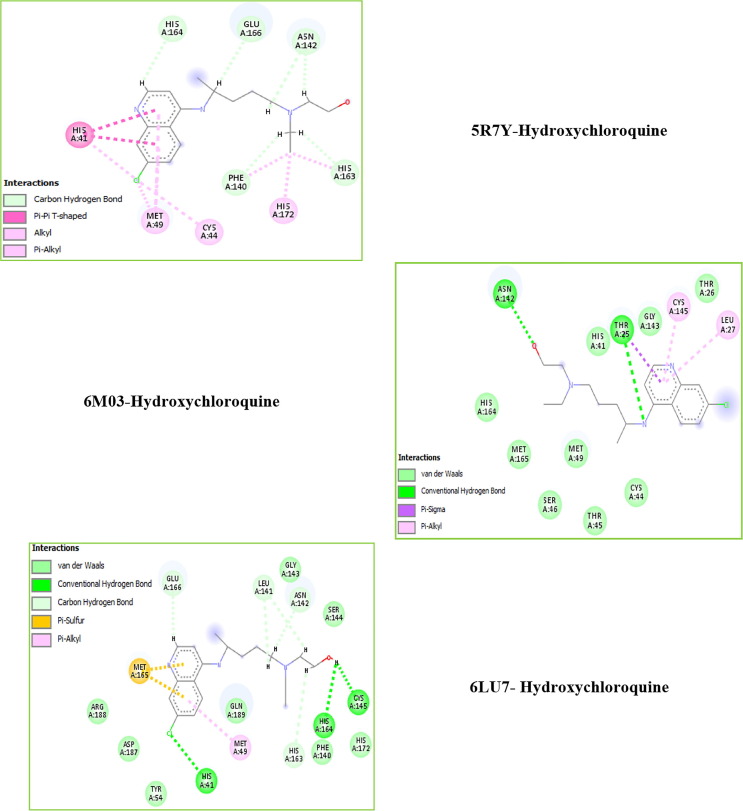
The chloroquine derivatives molecules suggest a strong prophylactic and therapeutic capacity in the fight against the COVID-19 virus, where they were found to have powerful antiviral effects infection. These inhibitory effects were discovered when they were used in the treatment before or after exposure to the coronavirus (Ng et al., 2003). For that, in the instant study the antiviral activity of the chloroquine derivatives compounds were screened against three crystal structures of COVID-19 viruses: 5R7Y, 6M03 and 6LU7 via molecular docking calculations. These compounds were docked in a functional place of selected proteins. Between all the molecular docking conformations, the only one which confines totally atone which linked perfectly at the active place was extracted through discovery studio program for the elaborated interactions. The docking pictures of our ligands against 5R7Y, 6M03 and 6LU7 targets are given in Fig. S5 and their 2D plots are mapped in Fig. 7 . Also, we have plotted in Fig. S6 the representation of diverse interactions present in the ligand-proteins complex. As you can see below, the ligands are linked to the protein by several interactions such as alkyl, pi-alkyl, pi-donor, pi-Anion, carbon-hydrogen bond, conventional hydrogen bonding, donor-hydrogen bonding interaction, etc… These interactions were pictured by colored lines. For example, the van der Waals (VDW) and H-bond interactions are represented by a green color (one is darker than the other). Orange and yellow color corresponds to Pi-Anion and Pi-Sulfur interactions, respectively. Whereas, the purple color degradation shows the Alkyl, Pi-Alkyl and Pi-Sigma interactions. The molecular docking binding energies of the docked compounds in the various viruses were enrolled in Table 3 . The implications of these results are discussed below.
Fig. 7.
2D visual representation of hydroxychloroquine ligand-COVID-19 virus.
Table 3.
Docking results of hydroxychloroquine and hydroxychloroquine sulfate in COVID-19 protein.
| Ligands | Hydroxychloroquine |
Hydroxychloroquine sulfate |
||||
|---|---|---|---|---|---|---|
| Target protein | 5R7Y | 6M03 | 6LU7 | 5R7Y | 6LU7 | 6M03 |
| Total energy | −76.987 | −73.430 | −72.878 | −117.738 | −105.153 | −98.412 |
| VDW | −67.392 | −64.115 | −57.505 | −94.484 | −93.360 | −95.386 |
| H-bond | −9.595 | −9.315 | −15.373 | −18.717 | −12.361 | −3.026 |
| Electronic | 0 | 0 | 0 | −4.536 | 0.578 | 0 |
| Binding affinities | −7.1 | −6.6 | −6.1 | −2.8 | −2.8 | −2.4 |
3.5. Hydroxychloroquine
In this part, we explore the docking calculations of the hydroxychloroquine ligand that is predicted to be relevant for antiviral activity. For obtaining insight into the binding mode and predicting the potential molecules, docking studies were carried out for hydroxychloroquine molecule against three structures of COVID-19 (5R7Y, 6M03 and 6LU7). Molecular calculations of these protein–ligand complexes show that the interaction between the inhibitor and protein surface is playing an important role, and some active site residues present a few flexibilities. The results for 5R7Y, 6M03 and 6LU7 are given in Figs. 7 and S5, while the calculated binding energies are tabulated in Table 3. Calculations reveal an interesting comparison between 5R7Y, 6M03 and 6LU7 receptors, in which differ only in their configurations. Their total energy scores are equal to −76.987, −73.430 and −72.878 kcal/mol, respectively. Docking results showed that 5R7Y has an inhibition value which is even better than other proteins. It is seen there that the predominant interaction is of VDW type. As shown in Table 3, it is characterized by the highest total energy score and the biggest van der Waals interactions (−67.392 kcal/mol). The hydrogen-bond interaction of hydroxychloroquine in 5R7Y protein is equal to −9.595 kcal/mol. Then, the docking of 6M03 protein posses a docked energy score of −73.430 kcal/mol with a range of docked interactions energies: EVDW = −64.115 kcal/mol and EH-bond = −9.315 kcal/mol. Thereafter, 6LU7 protein exhibits total energy values of −72.878 kcal/mol and van der Waals interactions of −57.505 kcal/mol which are a little weaker than the two other proteins, besides it has the highest hydrogen bonding interaction (equal to −15.373 kcal/mol). It can be seen that the totality of interaction energy is van der Waals and H-bond type. All docked hydroxychloroquine structures don’t have electronic interaction. Visual inspection of the 2D interaction representations showed that for 5R7Y protein, the following atoms: H46, H27, (H32, H38), C14, (C14, H32) and (C14, H33) are linked to HIS164, GLU166, ASN142, HIS172, HIS163 and PHE140 residues respectively via carbon-hydrogen bond interactions, with a range of distances varies between 2.15 and 3.25 Å. The binding residues MET49, CYS44 and HIS41 form, respectively, with Cl atom an alkyl, Pi- Alkyl and Pi-Pi T-shaped interactions. Their bond lengths are equal to 4.06, 5.05 and 4.75 Å, as indicated in Table S4. For the protein of PDB ID: 6M03: The oxygen atom O2 as well as the N4 atom forms two conventional H-bond interactions with ASN142 and THR25 residues, having bond distances 2.75 and 3.27 Å, respectively. Subsequently, CYS145 and LEU27 binding residues have Pi-Alkyl interactions, indicating distance 4.93 and 5.05 Å. A Pi-Sigma interaction exists formed by THR25 residue. Finally, the other amino acids HIS164, MET165, MET49, SER46, THR45, CYS44, HIS41 and THR26 forms VDW interactions for PDB ID: 6M03. Concerning 6LU7-hydroxychloroquine complex, MET165 and MET49 amino residues making a Pi-Sulfur and Pi-Alkyl interactions with the studied compound marking bond lengths equal to 5.26 and 5.20 Å, respectively. The H39, H46, (H33, H38) and H37 atoms have a carbon H-bond interactions with HIS163, GLU166, LEU141 and ASN142 residues with distances values range between 1.92 and 2.98 Å. In addition, three conventional H-bond interactions are performed between CYS145, HIS41, HIS164 amino residues and H49, Cl and H49 atoms, respectively. Their bond distances vary between 1.93 and 2.98 Å. Finally, the van der Waals interactions were being formed with the remaining amino acids: ARG188 (Arginine), ASP187 (AsparticAcid), TYR54 (Tyrosine), GLN189 (Glutamine), CLY143 (Glycine), SER144 (Serine), PHE140 (Phenylalanine) and HIS172 (as clearly seen in Fig. S6).
By using AutoDockTools (ADT) (Morris et al., 2008), we have calculated the affinities (in Kcal/mol) of theses complexes. These binding affinities characterize the strength of a non-covalent interaction between the ligand and its receptor which linking to a site on its surface. This calculation allows ameliorating the identification of the interactions existing between protein and ligand. As given in Table 3, the binding affinities values of hydroxychloroquine ligand are found to be −7.1 kcal.mol−1 (for 5R7Y), −6.6 kcal.mol−1 (for 6M03) and −6.1 kcal.mol−1 (for 6LU7).
3.6. Hydroxychloroquine sulfate
The binding mode of hydroxychloroquine sulfate is shown in Table 3. The examination of docking computation indicates that 5R7Y protein showed the highest inhibition activity among all complexes; the activity is identified by the COVID-19 inhibition activity test. This latter presents the strongest VDW energy about −94.484 kcal/mol and the strongest H-bond interaction −18.717 kcal/mol. The high inhibition constant of hydroxychloroquine sulfate ligand towards 5R7Y could be justified by these two strong interactions. The other structure of COVID-19 “6LU7”, has weaker energy score (−105.153 kcal/mol) compared to 5R7Y protein. It presents the greatest electronic interaction (−4.536 kcal/mol). It exhibited the VDW score of −93.360 kcal/mol and the H-bond interaction of −12.361 kcal/mol. Despite the corresponding total energy score of 6M03 is weaker than the total energy of the other two proteins (−98.412 kcal/mol), but it has a higher VDW interaction (EVDW = −95.386 kcal/mol). The increasing order of the electronic interaction of the present ligand-proteins is 5R7Y > 6LU7 > 6M03 with interaction energies −4.536, −0.578 and 0 kcal/mol, respectively.
The docking calculations showed that for 5R7Y-hydroxychloroquine sulfate complex, the A:HIS41 amino acid form a Pi-Sulfur interaction with S1 atom and having 4.10 Å bond lengths (as listed in Table S5). CYS145 (5.28 Å) interact with the benzene ring via a Pi-Alkyl interaction, whereas with pyridine ring (3.26 Å) via a Pi-Donor hydrogen-bond interaction. The THR25 (3.32 Å) and SER46 (1.89 Å) form two conventional H-bond interactions with O4 and H19 atoms, respectively. For 6M03 receptor, the Cl atom of hydroxychloroquine sulfate interact with LEU286 and MET276 residues via an alkyl interactions; including bond distances 4.74 and 4.91 Å. LEU287 residues interact with benzene ring forming Pi-Alkyl interaction with bond distance equal to 5.09 Å. Also, a Pi Donor H-bonding was identified between the pyridine group and TYR239 binding residues and the distance was found to be 3.71 Å. Furthermore, two carbon hydrogen bonding interactions wrapped by the amino acids GLU166, LEU287 and THR199 were formed with bond lengths 2.95 and 1.93 Å. The hydroxychloroquine sulfate is oriented with the conventional H-bond interactions surrounded by the chains of HIS41 (3.16 Å), HIS164 (3.31 Å) and CYS145 (2.62 Å) binding residues in the 6LU7 protein. Moreover, MET165 amino residues form an Alkyl interaction with Cl atom (3.71 Å), a Pi-Sulfur interaction with the benzene ring (5.48 Å) and a halogen interaction with Cl atom (3.21 Å). C18 atom made a Pi-Alkyl interaction with HIS41 residues and having distance 4.88 Å. By comparing the values of the affinities, we find that hydroxychloroquine sulfate present weaker affinities than hydroxychloroquine: −2.8 kcal.mol−1 (for 5R7Y and 6LU7 proteins) and −2.4 kcal.mol−1 for 6M03.
-
•
Molecular docking analyses of the hydroxychloroquine and the hydroxychloroquine sulfate showed a good to moderate anti-inflammatory activity. We have found that the hydroxychloroquine sulfate is the most important candidate in the treatment of COVID-19, since it has a significant total energy scores with high affinities. This result is expected because the hydroxychloroquine sulfate is considered to be the most reactive inhibitor, also having the highest dipole moment which promotes the diffusion and absorption of drug molecules.
4. Conclusion
Two stable molecules of antiviral activities were theoretically studied by using molecular docking and DFT methods. The ground state molecular geometries of hydroxychloroquine and hydroxychloroquine sulfate was obtained in the gas phase and in water using the hybrid B3LYP with 6-31G* basis set. A good accordance between the calculated geometrical parameters and the experimental data were found. The study show that the two ligands are more reactive and more stable in water. Evidently, the adding of other atoms in the structure of the hydroxychloroquine has an influence on their stability; the results have shown that adding the sulfate group makes the hydroxychloroquine more stable. Then, the frontier molecular orbitals HOMO and LUMO and their properties were determined. Results reveal that the hydroxychloroquine sulfate is the most reactive antiviral compound than the hydroxychloroquine. The molecular orbital contributions were analyzed by density of states (DOSs). Thereafter, the molecular electrostatic potentials (MEPs) surfaces are calculated to benefit better comprehension at a molecular level. The last part is to study the biological activities of our ligands by molecular docking analysis. This latter provided us with invaluable data to find out results that permitted us to estimate the total energy and the binding mode using iGEMDOCK program. The calculation is promising for the discovery of active inhibitors valuable as pharmacological agents. The fruitful results attained from the docking calculations show that the hydroxychloroquine sulfate could bind to the 5R7Y target sites with high affinity (−7.1 Kcal/mol). It has a significant total energy score −117.738 Kcal/mol. The other complexes showed also an important total energy which makes them an important candidate to study. Therefore, from our study, it is confirmed that the addition of the sulfate group makes it possible to strengthen the interactions of the binding modes, as well as strengthening the inhibitors. These compounds discover reasonable and excellent antiviral activity against COVID-19 diseases. So, we can use these compounds as antibiotics to a greater extent.
Funding
Researchers supporting project number (RSP-2020/61), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2020.101334.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with, novel coronavirus in Wuhan, China. Lancet. 2019;395(2020):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano E., Issaoui N., Manzur M.E., Brandán S.A. Properties and molecular docking of antiviral to COVID-19 chloroquine combining DFT calculations with SQMFF approach. Inter. J. Curr. Adv. Res. 2020;9(08(A)):22862–22876. [Google Scholar]
- Sagaama A., Brandan S.A., Ben Issa T., Issaoui N. Searching potential antiviral candidates for the treatment of the 2019 novel coronavirus based on DFT calculations and molecular docking. Heliyon. 2020;6(8):e04640. doi: 10.1016/j.heliyon.2020.e04640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani D., Noureddine O., Issaoui N., Brandán S.A. Properties and reactivities of niclosamide in different media, a potential antiviral to treatment of COVID-19 by using DFT calculations and molecular docking. Biointerface Res. Appl. Chem. 2020;10:7295–7328. [Google Scholar]
- Fischer A., Sellner M., Neranjan S., Lill M.A., Smieško M. Inhibitors for novel coronavirus protease identified by virtual screening of 687 million compounds. ChemRxiv. 2020 doi: 10.3390/ijms21103626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang Y., Ye D., Liu D. A review of the novel coronavirus (SARS-CoV-2) based on current evidence. Int. J. Antimicrob. Agents. 2019;2020:105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GaussView, Guassian, Inc., Carnergie Office Parck-Building6 Pittsburgh PA 151064 USA, Copyright © 2000-2003 Semichem. Inc.
- Gaussian 09, Revision C.01, Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G.A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H.P., Izmaylov, A.F., Bloino, J., Zheng, G., Sonnenberg, J.L., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Montgomery, J.A., Jr., Peralta, J.E., Ogliaro, F., Bearpark, M., Heyd, J.J., Brothers, E., Kudin, K.N., Staroverov, V.N., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A., Burant, J.C., Iyengar, S.S., Tomasi, J., Cossi, M., Rega, N., Millam, N.J., Klene, M., Knox, J.E., Cross, J.B., Bakken, V., Adamo, C., Jaramillo, J., Gomperts, R., Stratmann, R.E., Yazyev, O., Austin, A.J., Cammi, R., Pomelli, C., Ochterski, J.W., Martin, R.L., Morokuma, K., Zakrzewski, V.G., Voth, G.A., Salvador, P., Dannenberg, J.J., Dapprich, S., Daniels, A.D., Farkas, Ö., Foresman, J.B., Ortiz, J.V. Cioslowski, J., Fox, D.J., Gaussian, Inc., Wallingford CT, 2009.
- O'Boyle N.M., Tenderholt A.L., Langer K.M. A library for package independent computational chemistry algorithms. J. Comput. Chem. 2008;29:839–845. doi: 10.1002/jcc.20823. [DOI] [PubMed] [Google Scholar]
- http://www.rcsb.org/pdb/
- Yang J.-M., Chen C.-C. GEMDOCK: a generic evolutionary method for molecular docking proteins. Struct. Funct. Bioinform. 2004;55:288–304. doi: 10.1002/prot.20035. [DOI] [PubMed] [Google Scholar]
- Visualizer, D.S., 2005. Accelrys Software Inc. Discovery Studio Visualizer, p. 2.
- Ben-Zvi I., Kivity S., Langevitz P., Shoenfeld Y. Hydroxychloroquine: from malaria to autoimmunity. Expert. Rev. Clin. Immunol. 2012;42:145–153. doi: 10.1007/s12016-010-8243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semeniuk A., Kalinowska-Tluscik J., Nitek W., Oleksyn B.J. Intermolecular interactions in crystalline hydroxychloroquine sulfate in comparison with those in selected antimalarial drugs. J. Chem. Crystallogr. 2008;38:333–338. [Google Scholar]
- Ben Issa T., Sagaama A., Issaoui N. Computational study of 3-thiophene acetic acid: molecular docking, electronic and intermolecular interactions investigations. Comput. Biol. Chem. 2020;86:107268. doi: 10.1016/j.compbiolchem.2020.107268. [DOI] [PubMed] [Google Scholar]
- Rozas I., Alkorta I., Elguero J. Behavior of Ylides containing N, O, and C atoms as hydrogen bond acceptors. J. Am. Chem. Soc. 2000;122:11154–11161. [Google Scholar]
- Lu T., Chen F. Multiwfn: a multifunctional wavefunction analyzer. J. Comput. Chem. 2012;33:580–592. doi: 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- Silvi B., Savin A. Nature. 1994;371:683. [Google Scholar]
- Gatfaoui S., Issaoui N., Roisnel T., Marouani H. A proton transfer compound template phenylethylamine: synthesis, a collective experimental and theoretical investigations. J. Mol. Struct. 2019;1191:183–196. [Google Scholar]
- Renug S., Muthu S. Molecular structure, normal coordinate analysis, harmonic vibrational frequencies, NBO, HOMO–LUMO analysis and detonation properties of (S)-2-(2-oxopyrrolidin-1-yl) butanamide by density functional methods. Spect. Acta A: Mol. Bio. Spect. 2014;118:702–715. doi: 10.1016/j.saa.2013.09.055. [DOI] [PubMed] [Google Scholar]
- Renug S., Muthu S. Vibrational spectra and normal coordinate analysis of 2-hydroxy-3-(2-methoxyphenoxy) propyl carbamate. Spect. Acta A: Mol. Bio. Spect. 2014;132:313–325. doi: 10.1016/j.saa.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Tahenti M., Gatfaoui S., Issaoui N., Roisnel T., Marouani H. A tetrachlorocobaltate(II) salt with 2-amino-5-picolinium: synthesis, theoretical and experimental characterization. J. Mol. Struct. 2020;1207:127781. [Google Scholar]
- Noureddine O., Issaoui N., Gatfaoui S., Al-Dossary O., Marouani H. Quantum chemical calculations, spectroscopic properties and molecular docking studies of a novel piperazine derivative. J. King Saud Univ. – Sci. 2021;33 [Google Scholar]
- Hobbs H.E., Sorsby A., Freedman A. Retinopaihy following chloroquine therapy. Lancet. 1959:478–480. doi: 10.1016/s0140-6736(59)90604-x. [DOI] [PubMed] [Google Scholar]
- Canadian Hydroxychloroquine Study Group A randomized study of the effect of withdrawing hydroxychloroquine sulfate in systemic lupus erythematosus. N. Engl. J. Med. 1991;324:150–154. doi: 10.1056/NEJM199101173240303. [DOI] [PubMed] [Google Scholar]
- Murray S.M., Down C.M., Boulware D.R., Stauffer W.M., Cavert W.P., Schacker T., Douek D.C. Reduction of immune activation with chloroquine therapy during chronic HIV infection. J. Virol. 2010;84:12082–12086. doi: 10.1128/JVI.01466-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocks D.R., Mehvar R. Clin. Pharmacokinet. 2003;42:1359. doi: 10.2165/00003088-200342150-00004. [DOI] [PubMed] [Google Scholar]
- Borden M.B., Parke A.L. Drug Saf. 2001;24:1055. doi: 10.2165/00002018-200124140-00004. [DOI] [PubMed] [Google Scholar]
- Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagaama A., Noureddine O., Brandán S.A., Jarczyk-Jędryka A., Flakus H.T., Ghalla H., Issaoui N. Molecular docking studies, structural and spectroscopic properties of monomeric and dimeric species of benzofuran-carboxylic acids derivatives: DFT calculations and biological activities. Comput. Biol. Chem. 2020;87:107311. doi: 10.1016/j.compbiolchem.2020.107311. [DOI] [PubMed] [Google Scholar]
- Noureddine O., Issaoui N., Al-Dossary O. DFT and molecular docking study of chloroquine derivatives as antiviral to coronavirus COVID-19. J. King Saud Univ. – Sci. 2021;33:101248. doi: 10.1016/j.jksus.2020.101248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noureddine O., Gatfaoui S., Brandan S.A., Marouani H., Issaoui N. Structural, docking and spectroscopic studies of a new piperazine derivative, 1-phenylpiperazine-1,4-diium-bis (hydrogen sulfate) J. Mol. Struct. 2020;1202:127351. [Google Scholar]
- Jomaa I., Noureddine O., Gatfaoui S., Issaoui N., Roisnel T., Marouani H. Experimental, computational, and in silico analysis of (C8H14N2) 2 [CdCl6] compound. J. Mol. Struct. 2020;128186 [Google Scholar]
- Noureddine O., Gatfaoui S., Brandan S.A., Saagama A., Marouani H., Issaoui N. Experimental and DFT studies on the molecular structure, spectroscopic properties, and molecular docking of 4-phenylpiperazine-1-ium dihydrogen phosphate. J. Mol. Struct. 2020;1207:127762. [Google Scholar]
- Ghalla H., Issaoui N., Bardak F., Atac A. Intermolecular interactions and molecular docking investigations on 4-methoxybenzaldehyde. Comput. Mater. Sci. 2018;149:291–300. [Google Scholar]
- Cox B., Denyer J.C., Binnie A., Donnelly M.C., Evans B., Green D.V., Lewis J.A., Mander T.H., Merritt A.T., Valler M.J., Watson S.P. Application of high-throughput screening techniques to drug discovery. Prog. Med. Chem. 2000;37:83–133. doi: 10.1016/s0079-6468(08)70058-4. [DOI] [PubMed] [Google Scholar]
- Kuntz I.D. Structure-based strategies for drug design and discovery. Science. 1992;257:1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- Ng M.L., Tan S.H., See E.E., Ling A.E. Early events of SARS coronavirus infection in vero cells. J Med Virol. 2003;71:323–331. doi: 10.1002/jmv.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Arthur J.O. Using autodock for ligand-receptor docking. Curr. Protoc. Bioinform. 2008;24:8–14. doi: 10.1002/0471250953.bi0814s24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.