Abstract
Objective
This study describes the occurrence of a silent mutation in the RNA binding domain of nucleocapsid phosphoprotein (N protein) coding gene from SARS-CoV-2 that may consequence to a missense mutation by onset of another single nucleotide mutation.
Results
In the DNA sequence isolated from severe acute respiratory syndrome (SARS-CoV-2) in Iran, a coding sequence for the RNA binding domain of N protein was detected. The comparison of Chinese and Iranian DNA sequences displayed that a thymine (T) was mutated to cytosine (C), so “TTG” from China was changed to “CTG” in Iran. Both DNA sequences from Iran and China have been encoded for leucine. In addition, the second T in “CTG” in the DNA or uracil (U) in “CUG” in the RNA sequences from Iran can be mutated to another C by a missense mutation resulting from thymine DNA glycosylase (TDG) of human and base excision repair mechanism to produce “CCG” encoding for proline, which consequently may increase the affinity of the RNA binding domain of N protein to viral RNA and improve the transcription rate, pathogenicity, evasion from human immunity system, spreading in the human body, and risk of human-to-human transmission rate of SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, RNA binding domain, N protein, Mutation, Zinc, Thymine DNA glycosylase (TDG), Base excision repair
Introduction
Coronaviruses (CoVs) are the causative agent of diversity of infections in birds and mammals such as upper respiratory infection of chickens and enteritis in pigs and cows [1]. In addition to animals, human can be infected with different isolates of CoVs such as severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) with common cold, bronchitis, and pneumonia symptoms [2]. CoVs contain a positive-sense RNA strand with the genome size between 26.2 and 31.7 kb. The genomes of CoVs contain 3′ poly (A) and 5′ cap structures play as a mRNA in the translation activity of replicase enzyme [3]. It is specified that about 70% of the total size of the genome is occupied by replicase gene (20 kb) encoding for nonstructural proteins (nsp), while about 10 kb of the genome encodes for structural and accessory proteins. The studies have shown that the accessory proteins are not necessary for the viral replication, while are required in the CoVs pathogenesis and morphogenesis [4]. The CoVs contain four main structural proteins including spike (S), membrane (M), nucleocapsid (N), and envelope (E), which are expressed by the 3′ end of RNA genome [5]. S protein (150 kDa) contains a signal peptide sequence at the N-terminal with the ability to lead the S proteins to endoplasmic reticulum (ER) for glycosylation [6]. M protein (25–30 kDa) is the most abundant structural protein containing three transmembrane domains, which play a role in the virion formation. M proteins contain a small glycosylated N-terminal with the capability to bind to nucleocapsid so the M–N proteins binding may stabilize the complexes in the assembly of CoVs [7]. N protein (50–60 kDa) has the RNA binding capability by both N-terminal domain (NTD) and C-terminal domain (CTD) with two different mechanisms [8]. Three main factors have the crucial roles in the binding of N protein to RNA including phosphorylation of N protein, transcription regulatory signal (TRS), and genomic packaging signal (GPS). N protein binds to RNA, M protein, nsp3, replicase-transcriptase complex (RTC), which finally packages the RNA genome into CoVs. E protein (8–12 kDa) is a transmembrane protein with ion channel activity so that the lipid membrane charges affect the cation preference of these ion channels [9]. E protein facilitates the assemblage and release of virions from the infected cells. E protein is not necessary for CoV replication, while it is required for the CoV pathogenesis [10]. The studies revealed that SARS-CoV enters to the host cell using angiotensin converting enzyme 2 (ACE2) as the host receptor, while the host cell receptor for MERS-CoV is Dipeptidyl peptidase 4 (DPP4) [11].
Since December 2019, a novel member of CoVs have emerged from an outbreak in Wuhan, China, later on the causative agent was nominated as SARS-CoV-2 and the infection was nominated as coronavirus disease 2019 (COVID-19) [12]. Thereupon, the World Health Organization declared COVID-19 as a pandemic infection in March 2020 [13–15].
In this study and through a biodata mining approach, we evaluated a partial RNA binding domain of nucleocapsid protein (N protein) coding gene of SARS-CoV-2 isolated in Iran. In this coding gene, we found a silent thymine (T) to cytosine (C) mutation expressing leucine, which the encoded amino acid was similar to the query DNA sequence from Wuhan, China. This study alerts the incidence of second single nucleotide mutation in the subject coding sequence in Iran with the equal probability happened in the first T and changing middle T in DNA or U in RNA to C to form “CCG”. The encoded leucine will be changed to proline, which can influence the RNA binding activity of N protein and pathogenicity of SARS-CoV and induction of human immune system based on the 3D structure of newly emerged N protein.
Main text
Materials and methods
Sequence analysis
The coding gene for RNA binding domain of N protein of SARS-CoV-2 isolated in Iran was extracted from NCBI Virus database (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/) [16]. The accession number of MT186676 was identified, which was related to a DNA sequence with 363 b in length, isolated in Iran, and annotated in the NCBI Virus database on 2020-03-13. The details of this annotated DNA sequence has been described as “Severe acute respiratory syndrome coronavirus 2 isolate SARS-CoV-2/1337/human/2020/IRN nucleocapsid phosphoprotein (N) gene, partial cds (Mokhtari Azad et al.)”. The accession number of encoded protein is QIK02784, which was nominated as partial nucleocapsid phosphoprotein with 122 amino acids in length.
The Iranian CDS sequence (MT186676) and the query CDS sequence from Wuhan, China (NC_045512.2) were aligned using Clustal Omega algorithm (https://www.ebi.ac.uk/Tools/msa/clustalo/) [17] of multiple sequence alignment (MSA). Then, the partial nucleocapsid phosphoprotein (QIK02784) from Iran and the query protein sequence from Wuhan, China (QIE07458) were aligned using Clustal Omega algorithm of MSA.
Structural analysis
MT186676 related to the partial nucleocapsid phosphoprotein from Iran was analyzed in the RCSB Protein Data Bank (PDB) (https://www.rcsb.org/) [18]. In this data bank, a basic local alignment search tool (BLAST) was used to detect the most identical PDB identification number related to a crystalized protein to the Iranian protein sequence. In the next step, the NGL (WebGL) viewer [19] was employed to visualize NMR structure of the most identical protein structure to the partial nucleocapsid phosphoprotein from Iran.
Results
Sequence analysis
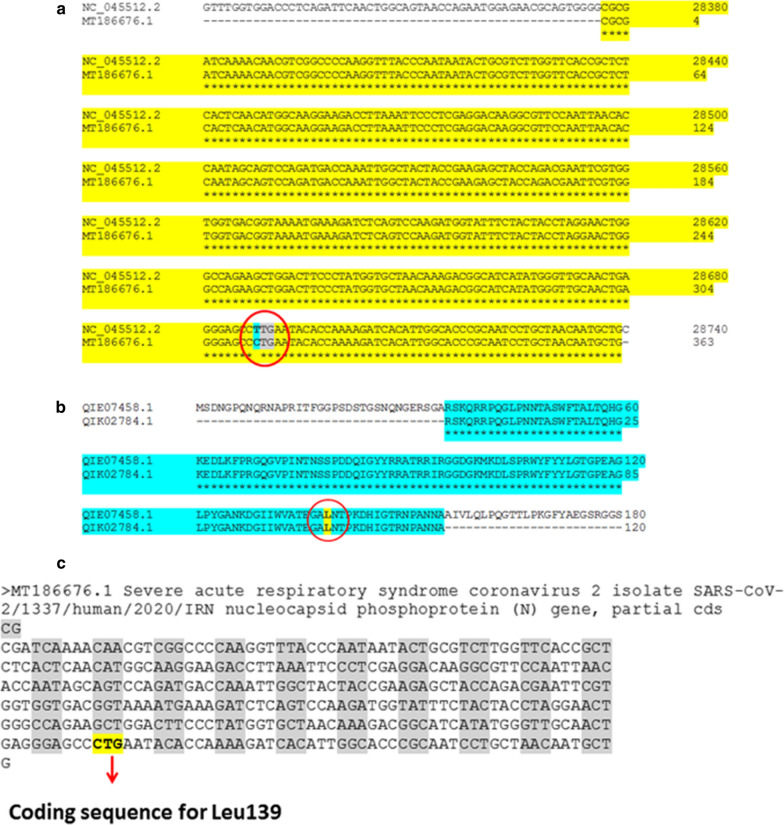
The MSA analysis of the CDS sequence coding for the partial RNA binding domain of N protein of SARS-CoV-2 isolated in Iran (MT186676) and the query CDS sequence from Wuhan, China (NC_045512.2) showed high similarity in both CDS sequences. A nucleotide mismatch was identified at nucleotide No. 312 that mutates T to C. The analysis revealed that both coding nucleotide sequences including “TTG” in the query CDS sequence from Wuhan, China and “CTG” in the subject CDS sequence from Iran encode for leucine (Fig. 1a). In addition, the MSA analysis of the query protein sequence from Wuhan, China (QIE07458) and the partial RNA binding domain of N protein from Iran (QIK02784) showed 100% similarity in both protein sequences (Fig. 1b and c) (Additional file 1: Fig. S1).
Fig. 1.
MSA analysis of RNA binding domain of N protein sequence. a MSA analysis of the CDS subject from Iran with the CDS query from Wuhan, China (yellow highlight); b MSA analysis of the partial RNA binding domain of N protein sequence from Iran with the query protein sequence from Wuhan, China (cyan highlight). Although both analyses show high similarity, a mismatch nucleotide is shown by the red circles in part A and B; and (c) The coding sequences of “TTG” in the CDS query sequence from Wuhan, China and “CTG” in the CDS subject sequence from Iran, which both encode for leucine
Structural analysis
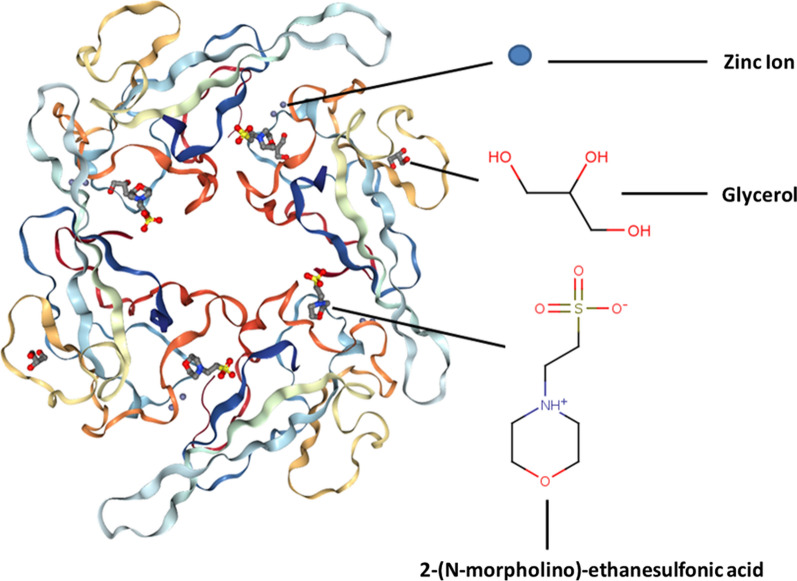
The structural analysis of the partial RNA binding domain of N protein from Iran (QIK02784) in the RCSB PDB Protein Data Bank defined that the NMR entry ID: 6VYO belongs to the crystal structure of RNA binding domain of N protein from SARS-CoV-2 with E-value: 6.19076E−76, which in comparison to each other sequences showed 100% identity. This tetrameric protein structure contains four Glu62 and Glu136 with the highest exposure rate at the surface of the protein, while Leu139 encoded by the silent mutation (CTG) showed less exposure rate (Fig. 2). The analysis of the crystal structure of 6VYO revealed that the RNA binding domain of N protein from SARS-CoV-2 is a tetrameric structure, which each monomer contains three binding sites to unique ligands including zinc ion (Zn2+), glycerol (GOL), and 2-(N-morpholino)-ethanesulfonic acid (MES) (Fig. 3).
Fig. 2.
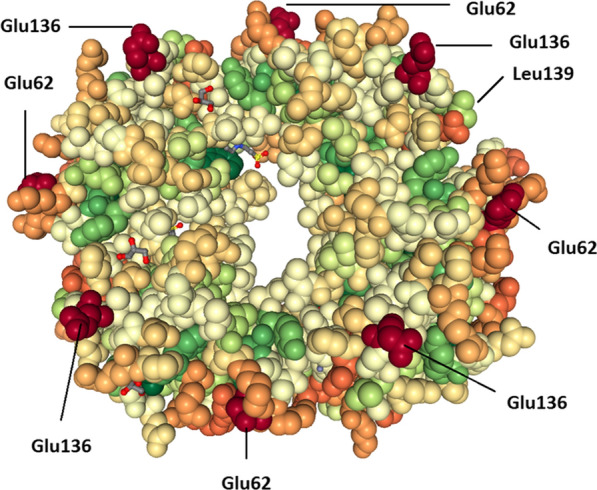
Protein structure of the RNA binding domain of N protein. The NMR structure entry ID: 6VYO was visualized by the NGL (WebGL) viewer. The exposure rate of eight glutamines including four Glu62 and four Glu136 in the tetrameric protein structure is higher than Leu139 encoded by the silent mutated sequence
Fig. 3.
Tetrameric structure of the RNA binding domain of N protein from SARS-CoV-2. Each monomer contain one binding site for unique ligands including zinc ion (Zn2+), glycerol (GOL), and 2-(N-morpholino)-ethanesulfonic acid (MES)
Discussion
The comparison of both CDS and protein sequences for subject from Iran and query from Wuhan, China by the MSA method revealed that the subject sequence from Iran is the partial RNA binding domain of N protein from SARS-CoV-2. The studies revealed that N protein is phosphorylated, which triggers the binding affinity of N protein to the RNA genome of SARS-CoV-2 [5, 20]. In addition, both TRS and GPS are considered as the main factors of binding of N protein to the viral RNA. The further analysis revealed that the CDS subject sequence from Iran contains a silent mutation, which changed “TTG” in the CDS query sequence to “CTG”. Although both “TTG” and “CTG” encode for leucine, the occurrence of mutation in the middle T in “CTG” with the equal probability will change the encoded leucine (Leu139) to proline (Pro139).
In SARS-CoV-2, the mutations can occur under the pressure of the human immune system, which can be associated with increasing the transmission rate and pathogenicity of the virus. It is speculated that single mutation of first T in “TTG” to C to form “CTG” can be one of the changes occurred under the pressure of immune system. Changing the middle T to C to form “CCG” for encoding proline instead of leucine can be one of the evolutionary strategies of SARS-CoV-2. It is speculated that the middle T in “CTG” or middle U in “CUG” have undergo a G:T (in DNA) or G:U (in RNA) mismatch, which was repaired by employing thymine DNA glycosylase (TDG) of human and base excision repair system with replacing the middle T to C. Although TDG is a crucial enzyme in the human DNA repair system, it seems that TDG can be employed within the SARS-CoV-2 genome organization during the COVID-19 infection in human as well. Further biochemical analysis of constituent amino acids of the RNA binding domain of N protein from SARS-CoV-2 defined that not only the exposure rate of the RNA binding domain of N protein is increased with replacement of leucine with proline, but also increase the chance of immune evasion of this protein [21]. Therefore, this missense mutation may increase the binding rate of N protein to the viral RNA; improving the transcription rate of the viral RNA, escaping from the human immune system, and consequently raise incidence of COVID-19 cases with higher pathogenicity rate in Iran.
The probability of emerge of next generation of SARS-CoV-2 in Iran through this missense mutation has the potential to be transmitted worldwide by human-to-human contacts as well. This happening may prepare more human-to-human transmission cases, which consequently rise the risk of evolution of SARS-CoV-2 by accumulation of silent mutations leading to a missense mutation [22]. The study of binding sites of unique ligands including Zn2+, GOL, and MES to the RNA binding domain of N protein demonstrated Zn2+ coordination has a major role in the geometry of tetrahedral structure of protein; GOL plays the role in the stability of protein, and MES is associated with the preparation of buffering condition for the protein [23]. Therefore, the ligand binding information can provide the invaluable opportunities to chelate Zn2+ by appropriate inhibitors as an antiviral therapy. The chelation of Zn2+ and the early stage inhibition of replication had been studied in dengue virus [24]. On the other hand, another study showed that RNA-dependent RNA polymerase (RdRp) of nidovirus, a CoV member, is inhibited by Zn2+ [25]. Hence, focusing on Zn2+, Zn2+ chelating agents, and inhibition of Zn2+-associated enzymes and proteins [24–26] potentially may be a pharmaceutical measure to treat COVID-19.
Conclusion
In conclusion, the more opportunities for human-to-human transmission,the more occurrence of missense mutation risk, improving the replication and transcription rates, and raising the virulence and pathogenicity of SARS-CoV-2. In addition, Zn2+ and Zn2+ inhibition studies have the potentials to become a medication compound and strategy against COVID-19. Moreover, our results revealed that TDG of human can be employed by SARS-CoV-2 during the genome organization and repairing leading to emerge of another mutated SARS-CoV-2 strain, which this mechanism has not reported yet by molecular virology studies of SARS-CoV-2 and needs to be examined by further experimental studies in the future.
Limitations
This study was performed based on the bioinformatics and computational analyses. In addition, further mutations may be occurred in the genome of SARS-CoV-2 to increase the transcription rate of viral RNA, pathogenicity of virus, evasion from the human immune system, spreading in the human body, binding of virus to cell receptors, and human-to-human transmission rate.
Supplementary Information
Additional file 1: Fig. S1 (A) MSA between the CDS coding for the RNA binding domain of N protein from Wuhan, China, and the CDS coding for the RNA binding domain of N protein from Iran. (B) MSA between the protein sequence of the RNA binding domain of N protein from Wuhan, China, and the protein sequence of the RNA binding domain of N protein from Iran.
Acknowledgements
We thank the Deputy of Research and COVID-19 National Committee from NIGEB for preparing the condition to perform this study. No funding organizations hadn't any role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; and in the decision to publish the results.
Abbreviations
- ACE2
Angiotensin converting enzyme 2
- BLAST
Basic Local Alignment Search Tool
- C
Cytosine
- CDS
Coding sequence
- COVID-19
Coronavirus disease 2019
- DPP4
Dipeptidyl peptidase 4
- E
Envelope
- ER
Endoplasmic reticulum
- Glu
Glutamine
- GOL
Glycerol
- Leu
Leucine
- M
Membrane
- MERS-CoV
Middle Eastern Respiratory Syndrome Coronavirus
- MES
2-(N-morpholino)-ethanesulfonic acid
- MSA
Multiple sequence alignment
- N
Nucleocapsid
- NCBI
National Center for Biotechnology Information
- NMR
Nuclear magnetic resonance
- nsp
Nonstructural protein
- ORF
Open reading frame
- PDB
Protein data bank
- Pro
Proline
- RdRP
RNA-dependent RNA polymerase
- RNA
Ribonucleic acid
- RTC
Replication-transcription complexes
- S
Spike
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- T
Thymine
- TDG
Thymine DNA glycosylase
- U
Uracil
- Zn2+
Zinc ion
Authors’ contributions
RZE: Design of the study, Search for gene/protein sequences of SARS-CoVs-2, Biodata mining, Bioinformatics and computational modelling analyses, Drafting the first version of the manuscript, Review and editing. ME: English editing, graphic works, preparing the manuscript based on the guidelines of the journal. HN: Biodata mining, Review and editing. JH: Editing the medical and clinical statements, Review and editing. RF: Biodata mining, Review and editing. All authors read and approved the final manuscript.
Funding
To perform this study, RZE received a research grant support from NIGEB, the Islamic Republic of Iran.
Availability of data and materials
All data analyzed in this study were prepared from databases including NCBI Virus and RCSB PDB Protein Data Bank were included in this article. These IDs are including MT186676, QIK02784, NC_045512.2, and QIE07458 from NCBI database and 6VYO from RCSB PDB Protein Data Bank.
Ethics approval and consent to participate
The data of the RNA binding domain of N protein of SARS-CoV-2 isolated in Iran was available online in the NCBI Virus database with the following accession numbers: MT186676 and QIK02784. There is no human or animal evaluation in this study and consequently no need to ethics committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Reza Zolfaghari Emameh, Email: zolfaghari@nigeb.ac.ir.
Mahyar Eftekhari, Email: eftekhari@nigeb.ac.ir.
Hassan Nosrati, Email: h.nosrati@clin.au.dk.
Jalal Heshmatnia, Email: jalalheshmatnia@gmail.com.
Reza Falak, Email: falak.r@iums.ac.ir.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-020-05439-x.
References
- 1.To KK, et al. From SARS coronavirus to novel animal and human coronaviruses. J Thorac Dis. 2013;5(Suppl 2):S103–S108. doi: 10.3978/j.issn.2072-1439.2013.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim YX, et al. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4(3):26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakagawa K, Lokugamage KG, Makino S. Viral and cellular mRNA translation in coronavirus-infected cells. Adv Virus Res. 2016;96:165–192. doi: 10.1016/bs.aivir.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang R, et al. The ns12.9 accessory protein of human coronavirus OC43 is a viroporin involved in virion morphogenesis and pathogenesis. J Virol. 2015;89(22):11383–95. doi: 10.1128/JVI.01986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belouzard S, et al. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arndt AL, Larson BJ, Hogue BG. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J Virol. 2010;84(21):11418–11428. doi: 10.1128/JVI.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuniga S, et al. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J Virol. 2010;84(4):2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdia-Baguena C, et al. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432(2):485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zolfaghari Emameh R, Falak R, Bahreini E. Application of system biology to explore the association of neprilysin, angiotensin-converting enzyme 2 (ACE2), and carbonic anhydrase (CA) in pathogenesis of SARS-CoV-2. Biol Proced Online. 2020;22:11. doi: 10.1186/s12575-020-00124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosch BJ, Smits SL, Haagmans BL. Membrane ectopeptidases targeted by human coronaviruses. Curr Opin Virol. 2014;6:55–60. doi: 10.1016/j.coviro.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang JJ, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75(7):1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 13.Mahase E. Covid-19: WHO declares pandemic because of "alarming levels" of spread, severity, and inaction. BMJ. 2020;368:m1036. doi: 10.1136/bmj.m1036. [DOI] [PubMed] [Google Scholar]
- 14.Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zolfaghari Emameh R, Nosrati H, Taheri RA. Combination of biodata mining and computational modelling in identification and characterization of ORF1ab polyprotein of SARS-CoV-2 isolated from oronasopharynx of an Iranian patient. Biol Proced Online. 2020;22:8. doi: 10.1186/s12575-020-00121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brister JR, et al. NCBI viral genomes resource. Nucleic Acids Res. 2015;43(Database issue):D571–7. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27(1):135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodsell DS, et al. The RCSB PDB "Molecule of the Month": inspiring a molecular view of biology. PLoS Biol. 2015;13(5):e1002140. doi: 10.1371/journal.pbio.1002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose AS, et al. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics. 2018;34(21):3755–3758. doi: 10.1093/bioinformatics/bty419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura T, et al. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur J Biochem. 2003;270(20):4070–4081. doi: 10.1046/j.1432-1033.2003.03796.x. [DOI] [PubMed] [Google Scholar]
- 22.Reperant LA, Grenfell BT, Osterhaus AD. Quantifying the risk of pandemic influenza virus evolution by mutation and re-assortment. Vaccine. 2015;33(49):6955–6966. doi: 10.1016/j.vaccine.2015.10.056. [DOI] [PubMed] [Google Scholar]
- 23.van Kasteren PB, et al. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc Natl Acad Sci U S A. 2013;110(9):E838–E847. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kar M, et al. Zinc chelation specifically inhibits early stages of dengue virus replication by activation of NF-kappaB and induction of antiviral response in epithelial cells. Front Immunol. 2019;10:2347. doi: 10.3389/fimmu.2019.02347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.te Velthuis AJ, et al. Zn(2+) inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6(11):e1001176. doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zolfaghari Emameh R, et al. Identification and inhibition of carbonic anhydrases from nematodes. J Enzyme Inhib Med Chem. 2016;31(sup4):176–184. doi: 10.1080/14756366.2016.1221826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1 (A) MSA between the CDS coding for the RNA binding domain of N protein from Wuhan, China, and the CDS coding for the RNA binding domain of N protein from Iran. (B) MSA between the protein sequence of the RNA binding domain of N protein from Wuhan, China, and the protein sequence of the RNA binding domain of N protein from Iran.
Data Availability Statement
All data analyzed in this study were prepared from databases including NCBI Virus and RCSB PDB Protein Data Bank were included in this article. These IDs are including MT186676, QIK02784, NC_045512.2, and QIE07458 from NCBI database and 6VYO from RCSB PDB Protein Data Bank.