Abstract
SARS-CoV-2 is a new strain of coronavirus that appeared in China in December 2019, in recent years, great progress has been made in developing new antiviral drugs, and natural products, are important sources of potential and new antiviral drugs. The present study aimed to assess some biologically active compounds present in medicinal plants as potential COVID-19 inhibitors, using molecular docking methods. The Docking study was performed by Molecular Operating Environment software (MOE). About 20 Compounds were screened in this study; these compounds were selected based on classification of their chemical origin and their antiviral activity from literature. These compounds might be used to inhibit COVID-19 infection. The results demonstrate the effectiveness of this screening strategy, which can lead to rapid drug discovery in response to new infectious diseases. The results showed that many compounds isolated from medicinal plants such as; Gallic acid (− 17.45), Quercetin (− 15.81), Naringin (− 14.50), Capsaicin (− 13.90), and Psychotrine (− 13.5) are important sources for novel antiviral drugs targeting COVID-19.
Keywords: COVID-19, Anti-viral, Medicinal plants, Docking study, Sudan
Introduction
The world faces a severe and acute public health emergency due to the ongoing global pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Wu and McGoogan 2020). SARS-CoV-2 is a new strain of coronavirus that appeared in China in December 2019 (Guo et al. 2020). Symptoms of COVID-19 can be relatively non-specific and infected people may be asymptomatic. People infected with COVID-19 generally develop signs and symptoms including mild respiratory symptoms and fever, dry cough, fatigue, shortness of breath, and loss of smell on an average of 5–6 days after infection but may range from two to fourteen days. Complications may include pneumonia and acute respiratory distress syndrome which may lead to death (Singhal 2020; Pal et al. 2020). There has been no noticeable progress in managing this disease so far and the patient is being given treatment based on his observable and diagnosed symptom (Organization et al. 2009). Until now, there is no known vaccine or specific antiviral treatment (Sheahan et al. 2020). The Human Genome Project helped to increase the number of new therapeutic targets for drug discovery. Meanwhile, high-throughput protein purification techniques, crystallography, and nuclear magnetic resonance spectroscopy techniques have contributed to the structural details of proteins and protein complexes. These developments allow computational strategies to penetrate in all its aspects (Jorgensen 2004; Bajorath 2002; Langer and Hoffmann 2001; Walters et al. 1998 and Kitchen et al. 2004), such as virtual screening techniques (VS) (Gohlke and Klebe 2002) to determine results and methods for improving molecules. Molecular docking, a computational technique used to estimate the affinity of association between two molecules such as ligand–protein and protein–protein. The use of molecular docking became an essential component in drug discovery. These techniques also help in predicting the side and toxic effects of compounds (Bacha et al. 2008). A number of conventional drugs such as chloroquine, hydroxychloroquine, remediffer have been tested and found with a specific therapeutic effect in the laboratory. However, the clinical pharmacological response is not very encouraging and toxicity remains an inevitable problem that causes serious adverse effects (Otitoloju et al. 2020). The lack of a specific treatment for COVID-19 led most researchers in many regions of the world, to use medicinal herbs known in the science of ethnopharmacology as an antiviral (Rajabian and Hosseinzadeh 2020). The development of viral resistance to current antiviral agents reinforces the need for new effective compounds against viral infections (Chattopadhyay et al. 2009). In this study, some of the active compounds present in some medicinal plants used in Sudan traditional medicine to treat viral diseases were selected based on literature review for docking studies and the interactions of these compounds have been clarified to determine the preferred molecules for the treatment of COVID-19 via the inhibition of SARS-CoV-2 main protease enzyme, and compare the results with the proposed drugs such as chloroquine and hydroxychloroquine. Furthermore, the toxicity of these compounds was performed in order to assess their safety for human use. The study of the crystal structure of therapeutic targets is crucial for knowing how the drugs will be discovered in the future. Alkaloids are a rich source of important chemical compounds which can be used for development of new anti-viral agents. Alkaloids have a wide distribution in the plant kingdom and are mainly found in higher plants, some viral diseases can be treated with approved antiviral, but for others they still have no any vaccines or drugs available. Most of the approved antiviral are directly or indirectly related to side effects, which ultimately leads to an increased need to develop antiviral based on natural phytochemicals. Globally, antiviral are shifting toward plant-derived products because they are less toxic and have less chance of developing resistance (Ghildiyal et al. 2020). All selected compounds from different groups were selected based on their chemical origin were used to treat many diseases such as antiviral activity. The present paper aimed to evaluate the antiviral activity of some compounds from medicinal plants against COVID-19 using Molecular Docking Analysis.
Methodology
Ligands preparation and optimization
Ligands for this study were drawn using ChemDraw Professional 15.0, then three- dimensional structures of ligands were generated by Open babel (O’Boyle et al. 2011) and saved as SDF format for further preparation and molecular docking study.
The COVID-19 receptor preparation and optimization
The crystallographic structure of SARS-CoV-2 main protease (Mpro) (PDB ID: 5R80) in complex with Z18197050, was retrieved from protein data bank with a resolution: 1.93 Å. Interactive visualization and analysis of molecular structures were done using MOE and PyMol software for better understanding of protein-ligands interactions.
Geometry optimization and pre-docking procedure
Hydrogens and Gasteiger charges were added and all the hetero-atoms and water molecules were removed from protein structure. The protonated protein initially optimized in order to remove all the bad steric clashes using AMBER99 force field (Cornell et al. 1996), while MMFF94s force field parameters (Halgren 1996) and (Halgren 1999) were performed for small molecules. The protonated and optimized structures were saved for further preparation and analysis. All the minimizations were performed with MOE (grade < 0.001).
Docking strategy and setup
Docking studies were performed by MOE with 10 poses for each ligand (other parameters were kept default). The unnecessary chain was deleted. Ligand interactions were computed for the X-rayco-crystallized protein to reveal the different types of interaction as a validation for the coming docking procedure. The docking was done with the default settings of the MOE–DOCK as following: The scoring function was London d G. & 10 conformers of the ligand were retained with highest and best score by default. The scoring configuration of the ligand–Target complexes was selected on energetic grounds (MM/GBVI); best poses with the lowest binding energy was chosen for each compound. The docking scores, docking binding energy, and chemical structures of selected ligands were then presented. ProTox (Drwal et al. 2014) a webserver was used for acute oral toxicity prediction based on chemical similarities between compounds with known toxic effects and the presence of toxic fragments.Toxic doses are often given as LD50values in mg/kg body weight, whereas toxicity classes are defined according to the globally harmonized system of classification of labelling of chemicals (Cheng et al. 2012).
Results and discussion
Natural products are among the most important major resources for drug research and development in the past decades, especially for treating infectious diseases. Contagious disease treatment and control is widely demonstrated by effectiveness of medicinal herbs (Bouchentouf and Missoum 2020; Asadbeigi et al. 2014; Bouredja et al. 2020). A large number of diverse chemical structure compounds isolated from medicinal plants possessing antiviral activity and in this study only highly effective compounds were registered (Table 1). Twenty-one compounds were screened for in silico analysis using MOE Docking software. The best compounds interacting with SARS-CoV-2 Mpro are shown in Table 2.
Table 1.
Compounds of medicinal plants screened for antiviral activity targeting COVID-19
| No. | Compound | Formula | M.W | Type of compound |
|---|---|---|---|---|
| 1 | Gallic acid | C7H6O5 | 170.12 g/mol | Phenol |
| 2 | Quercetin | C15H10O7 | 302.23 g/mol | Flavonoid glycoside |
| 3 | Naringin | C27H32O14 | 580.5 g/mol | Flavonoid glycoside |
| 4 | Capsaicin | C18H27NO3 | 305.4 g/mol | Pyridine Alkaloid |
| 5 | Psychotrine | C28H36N2O4 | 464.6 g/mol | Phenyl and phenylpropyl Alkaloid |
| 6 | Curcumin | C21H20O6 | 368.4 g/mol | Phenol |
| 7 | Plicamine | C26H26N2O6 | 462.5 g/mol | Amaryllidaceae Alkaloid |
| 8 | Narciclasine | C14H13NO7 | 307.25 g/mol | Amaryllidaceae Alkaloid |
| 9 | Catechin | C15H14O6 | 290.27 g/mol | Flavonoid glycoside |
| 10 | Lycoricidine | C14H13NO6 | 291.26 g/mol | Amaryllidaceae Alkaloid |
| 11 | Cryptopleurine | C24H27NO3 | 377.5 g/mol | Phenanthroindolizidine and phenanthroquinolizidine Alkaloid |
| 12 | Oliverine | C20H22CINO4 | 375.8 g/mol | Aporphinoid Alkaloid |
| 13 | Papaverine | C20H21NO4 | 339.4 g/mol | Benzylisoquinoline Alkaloid |
| 14 | Hyoscyamine | C17H23NO3 | 289.4 g/mol | Tropane Alkaloid |
| 15 | Lycorenine | C18H23NO4 | 317.4 g/mol | Amaryllidaceae Alkaloid |
| 16 | Galantamine | C17H21NO3 | 287.35 g/mol | Amaryllidaceae Alkaloid |
| 17 | Mesembrine | C17H23NO3 | 289.4 g/mol | Tropane ester Alkaloid |
| 18 | Cysteine | C3H7NO2S | 121.16 g/mol | Opium Alkaloid |
| 19 | Hydroxychloroquine | C18H26CIN3O | 335.9 g/mol | Standard (Alkaloid) |
| 20 | Chloroquine | C18H26CIN3 | 319.9 g/mol | Standard (Alkaloid) |
Table 2.
Molecular docking analysis of several compounds against SARS-CoV-2 main protease
| No. | Name | Energy (kcal/mol) | LD50 class | Predicted LD50 mg/kg |
|---|---|---|---|---|
| 1 | Gallic acid | − 17.45 | 5 | 2260 |
| 2 | Quercetin | − 15.81 | 3 | 159 |
| 3 | Naringin | − 14.50 | 5 | 2300 |
| 4 | Capsaicin | − 13.90 | 2 | 47 |
| 5 | Psychotrine | − 13.50 | 4 | 480 |
| 6 | Curcumin | − 12.80 | 5 | 4000 |
| 7 | Plicamine | − 12.75 | 3 | 230 |
| 8 | Narciclasine | − 12.67 | 3 | 80 |
| 9 | Catechin | − 11.48 | 6 | 10,000 |
| 10 | Lycoricidine | − 11.47 | 4 | 460 |
| 11 | Cryptopleurine | − 11.28 | 3 | 100 |
| 12 | Oliverine | − 11.06 | 4 | 450 |
| 13 | Papaverine | − 10.43 | 3 | 69 |
| 14 | Hyoscyamine | − 10.10 | 3 | 75 |
| 15 | Lycorenine | − 9.99 | 4 | 795 |
| 16 | Galantamine | − 9.44 | 2 | 19 |
| 17 | Mesembrine | − 9.43 | 4 | 369 |
| 18 | Cystin | − 8.20 | 3 | 156 |
| 19 | Hydroxychloroquine | − 12.25 | 4 | 1240 |
| 20 | Chloroquine | − 10.04 | 4 | 311 |
| RZG (standard) | − 10.30 |
Class I: fatal if swallowed (LD50 ≤ 5 mg/kg), Class II: fatal if swallowed (5 < LD50 ≤ 50 mg/kg)
Class III: toxic if swallowed (50 < LD50 ≤ 300 mg/kg), Class IV: harmful if swallowed (300 < LD50 ≤ 2000 mg/kg), Class V: may be harmful if swallowed (2000 < LD50 ≤ 5000 mg/kg), Class VI: non-toxic (LD50 > 5000 mg/kg)
RZG: methyl 4-sulfamoylbenzoate
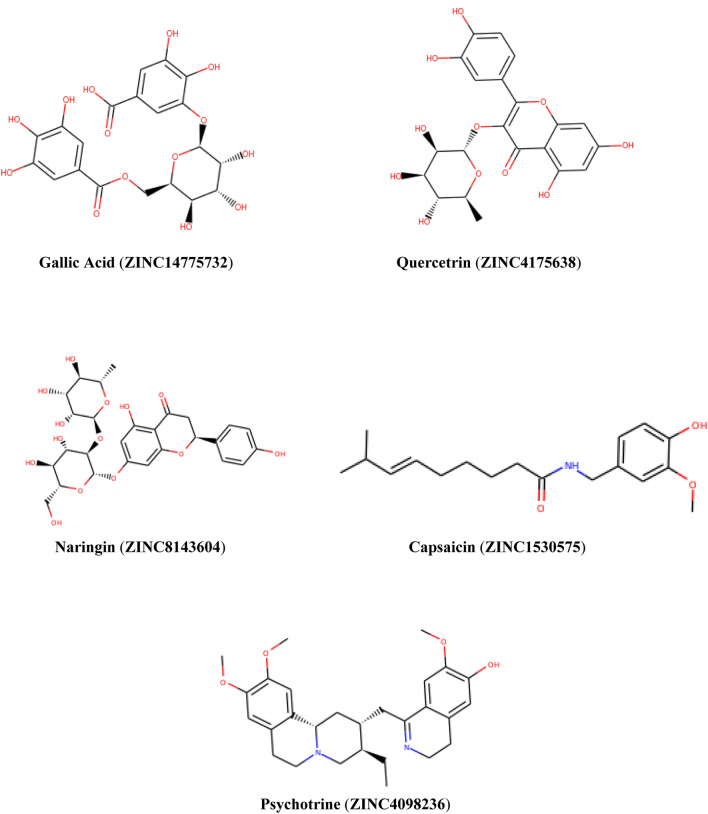
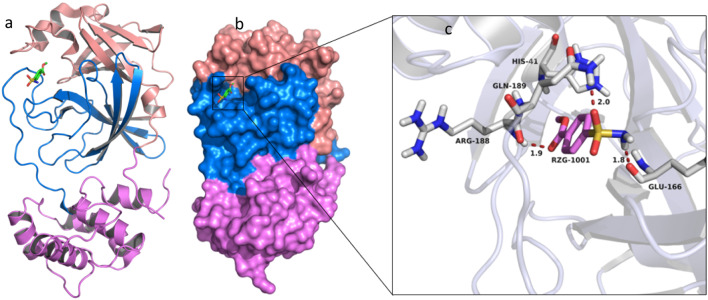
The results showed that the compounds have a rank scores ranged from − 17.45 to − 8.20 kcal/mol. The docking simulations resulted in five compounds that were assigned as the most potential candidate among the docked ones; the compounds’ poses were ranked with respect to their binding energy values. We noted that the inhibition potentials of the Gallic acid (1) are very encouraging. The compounds including Quercetin (2), Naringin (3), Capsaicin (4), and Psychotrine (5) (Fig. 1), have larger inhibition potentials than that of chloroquine as a reference too. The standard was docked to protomer A of the dimeric COVID-19 Mpro (Fig. 2a). The co-crystallized ligand (RZG; methyl 4-sulfamoylbenzoate) is interacting with the enzyme residues in the substrate-binding site located in a cleft between domain I and II (Fig. 2b). Figure 2c shows the interaction of RZG. The other compounds also possess certain inhibition properties against SARS-COV2 Mpro.
Fig. 1.
Chemical structure of the top ranked compounds
Fig. 2.
The crystal structure of COVID-19 Mpro in complex with RZG (methyl 4-sulfamoylbenzoate). a Cartoon representation of protomer A of the dimeric Mpro-inhibitor complex. b Surface representation of the Protomer A. Domain I is in purple, domain II is in blue, and domain III is in salmon-red. RZG is presented as green sticks. c A close view of the substrate-binding pocket. The key residues forming the binding pocket are shown as grey sticks
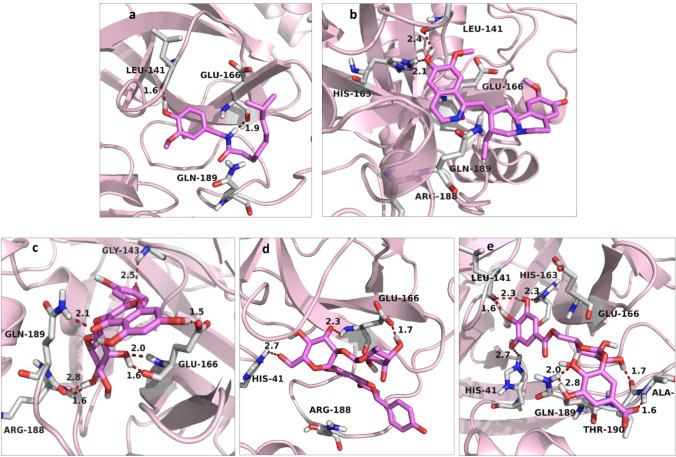
Gallic acid revealed the highest docking score − 17.45 kcal/mol, the depiction of the key interactions is displayed in Table 1 and Fig. 3d. The compound revealed an interaction profile better that displayed by HCQ. It is completely trapped in the binding site of the enzyme by a complicated network of strong hydrogen bonding generated by His41, Leu141, His163, Gln189, Thr190, and Ala191, with distances ranging between 1.6 Å to 2.8 Å as shown in Fig. 3e. Quercetin with a docking score − 15.80 kcal/mol is connected to the enzyme’s residues Gly143, Glu166, Arg188, and Glu189, through strong hydrogen bonding with distances range 1.5–2.8 Å (Fig. 3c). Naringin, Capsaicin and Psychotrine (Fig. 3a, b, and d) are interacting with most of these amino acid residues with potential hydrogen bonding, however, they revealed binding scores − 14.50, − 13.90, − 13.50 kcal/mol respectively, which are much smaller than that displayed by compounds 5.
Fig. 3.
3D view of the interaction of the top ranked compounds in the binding site of COVD-19 Mpro. The compounds shown as pink sticks, and residues as gray sticks, the rest of the enzyme structure is represented as faint pink cartoons. The red dashed lines indicate the hydrogen bonds which are labeled with their lengths
Throughout human history, thousands of biologically active plants have been discovered, identified and used in medicine. Most cultures around the world rely on medicinal plants for disease treatment (Halberstein 2005). Plants possess the ability to produce a wide range of compounds including flavonoids, alkalis, lignans and tannins, and are responsible for the primary functions of plant growth and development (Saxena et al. 2013). Molecular docking studies and explores the interaction mechanism between ligands and receptors. Interactions between the compound and the receptor play a crucial role in the discovery of drugs. All these 21 molecules were docked against the target enzyme SARS-COV2 Mpro and ranked based on their dock scores. Flavonoids are important natural compounds with diverse biologic activities. Citrus flavonoids constitute an important series of flavonoids. Naringin and its aglycone naringenin belong to this series of flavonoids and were found to display strong anti-in flammatory and antioxidant activities (Alam et al. 2014). Naringenin (flavanones), quercetin (flavonols) and Catechins (flavans) which are found in Citrus spp and different plants showed the highest result against COVID-19, flavonoids are widely distributed as secondary metabolites produced by plants and play important roles in plant physiology, and they have a variety of potential biological benefits such as antioxidants, anti-inflammatory, anti-cancer, anti-bacterial, antifungal and antiviral agents. Various flavonoids have been investigated for their potential antiviral activities and many have shown important antiviral properties in the laboratory and even in live studies (Zakaryan et al. 2017). Recent research has shown that Amaryllidaceae alkaloids represent a rich reservoir of potential small chemical molecules that display many medicinal properties through various mechanisms. Among the many Amaryllidaceae compounds, galanthamine has been given a lot of attention since it has a strong inhibitory activity of acetylcholine esterase (Habartova et al. 2016). Among the many species investigated for components of small molecules with treatment potential, the Amaryllidaceae family plants were particularly fruitful (Evidente et al. 2009). Psychotrine Occurs in ipecacuanha root Cephaelis acuminata (Rubiaceae); active against HIV-1 (Manske and Brossi 1985). Capsaicin is an alkaloid from Capsicum genus is one of the principle substances which have many important biological activities in food and medicine (Reyes-Escogido et al. 2011).
Conclusion
To combat the life-threatening corona virus infection, several studies are ongoing using antiviral drug therapies. This study will be useful in recent and future for designing novel drugs for the treatment of COVID-19. In this study, docking analyses showed that the COVID-19 protease (6LU7) may be inhibited by some compounds from herbal plants, based on the binding energy score, we suggest that these compounds such as; Naringin, Quercetin, Capsaicin, Psychotrine and Gallic acid can be tested against Corona virus and used to develop effective antiviral drugs. These molecules could be utilized for further innovation and development of antiviral compounds against Corona virus. However, further researches warranted to investigate the potential uses of the medicinal plants containing these compounds.
Acknowledgements
The authors are grateful to the Dean of the Faculty of Pharmacy Prof. Abdelazim Elshaikh Madani and to Prof. Saad Mohamed Hussein Ayoub, the Head of Pharmacognosy Department, Faculty of Pharmacy, University of Medical Sciences and Technology (UMST) for their assistance and support.
Abbreviations
- COVID-19
Corona virus disease-2019
- MOE
Molecular Operating Environment software
- M.W
Molecular weight
- LD50
Lethal dose that inhibit 50%
Author contributions
AAA coordinated the initial idea, designed the project and drafted the manuscript. MY and TAA carried out docking analysis. TAA reviewed and corrected the manuscript. All authors have read and approved the final manuscript.
Availability of data and materials
Research data have been provided in the manuscript, and for more details, they are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alam MA, Subhan N, Rahman MM, Uddin SJ, Reza HM, Sarker SD. Effect of citrus flavonoids, naringin and naringenin, on metabolic syndrome and their mechanisms of action. Adv Nutr. 2014;5:404–417. doi: 10.3945/an.113.005603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadbeigi M, Mohammadi T, Rafieian-Kopaei M, Saki K, Bahmani M, Delfan M. Traditional effects of medicinal plants in the treatment of respiratory diseases and disorders: an ethnobotanical study in the Urmia. Asian Pac J Trop Med. 2014;7:S364–S368. doi: 10.1016/S1995-7645(14)60259-5. [DOI] [PubMed] [Google Scholar]
- Bacha U, Barrila J, Gabelli SB, Kiso Y, Mario Amzel L, Freire E. Development of broad-spectrum halomethyl ketone inhibitors against coronavirus main protease 3CLpro. Chem Biol Drug Des. 2008;72:34–49. doi: 10.1111/j.1747-0285.2008.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baildya N, Ghosh NN, Chattopadhyay AP. Inhibitory activity of hydroxychloroquine on COVID-19 main protease: an insight from MD-simulation studies. J Mol Struct. 2020;2:128595. doi: 10.1016/j.molstruc.2020.128595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorath J. Integration of virtual and high-throughput screening. Nat Rev Drug Discov. 2002;1:882–894. doi: 10.1038/nrd941. [DOI] [PubMed] [Google Scholar]
- Bouchentouf S, Missoum N (2020) Identification of compounds from nigella sativa as new potential inhibitors of 2019 novel coronavirus (COVID-19): Molecular Docking Study.
- Bouredja N, Bouthiba M, Kebir M (2020) Ethnobotanical study of medicinal plants used by herbalists for the treatment of respiratory diseases in the region of Oran, Algeria. Br J Med Health Sci (BJMHS) 2.
- Chattopadhyay D, Chawla-Sarkar M, Chatterjee T, Dey RS, Bag P, Chakraborti S, Khan MTH. Recent advancements for the evaluation of anti-viral activities of natural products. New Biotechnol. 2009;25:347–368. doi: 10.1016/j.nbt.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, Tang Y. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. Washington: ACS Publications; 2012. [DOI] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules J. Am. Chem. Soc. 1995, 117, 5179–5197. J Am Chem Soc. 1996;118:2309–2309. doi: 10.1021/ja955032e. [DOI] [Google Scholar]
- Drwal MN, Banerjee P, Dunkel M, Wettig MR, Preissner R. ProTox: a web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014;42:W53–W58. doi: 10.1093/nar/gku401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evidente A, Kireev AS, Jenkins AR, Romero AE, Steelant WF, Van Slambrouck S, Kornienko A. Biological evaluation of structurally diverse amaryllidaceae alkaloids and their synthetic derivatives: discovery of novel leads for anticancer drug design. Planta Med. 2009;75:501–507. doi: 10.1055/s-0029-1185340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon D, Powell AJ, Douangamath A, Owen CD, Wild C, Krojer T, Lukacik P, Strain-Damerell CM, Walsh MA, von Delft F (2020) PanDDA analysis of COVID-19 main protease against the DSI-poised Fragment Library. PDB ID: 5R82. 2020.
- Ghildiyal R, Prakash V, Chaudhary VK, Gupta V, Gabrani R. Plant-derived bioactives. Singapore: Springer; 2020. Phytochemicals as antiviral agents: recent updates; pp. 279–295. [Google Scholar]
- Gohlke H, Klebe G. Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew Chem Int Ed. 2002;41:2644–2676. doi: 10.1002/1521-3773(20020802)41:15<2644::AID-ANIE2644>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Guo Y-R, Cao Q-D, Hong Z-S, Tan Y-Y, Chen S-D, Jin H-J, Tan K-S, Wang D-Y, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil Med Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habartov K, Cahlikova L, Řezáčová M, Havelek R (2016) The biological activity of alkaloids from the Amaryllidaceae: from cholinesterases inhibition to anticancer activity. Nat Product Commun 11, 1934578X1601101038. [PubMed]
- Halberstein RA. Medicinal plants: historical and cross-cultural usage patterns. Ann Epidemiol. 2005;15:686–699. doi: 10.1016/j.annepidem.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17:490–519. doi: 10.1002/(SICI)1096-987X(199604)17:5/6<490::AID-JCC1>3.0.CO;2-P. [DOI] [Google Scholar]
- Halgren TA. MMFF VI. MMFF94s option for energy minimization studies. J Comput Chem. 1999;20:720–729. doi: 10.1002/(SICI)1096-987X(199905)20:7<720::AID-JCC7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Duan Y, Yu J (2020) Structure-based drug design, virtual screening and high-throughput screening rapidly identify antiviral leads targeting COVID-19. BioRxiv
- Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- Langer T, Hoffmann R. Virtual screening an effective tool for lead structure discovery. Curr Pharm Des. 2001;7:509–527. doi: 10.2174/1381612013397861. [DOI] [PubMed] [Google Scholar]
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization, W. H., Research, S. P. f., Diseases, T. i. T., Diseases, W. H. O. D. o. C. o. N. T., Epidemic, W. H. O., and Alert, P. (2009) Dengue: guidelines for diagnosis, treatment, prevention and control. World Health Organization. [PubMed]
- Otitoloju AA, Okafor IP, Fasona M, Bawa-Allah KA, Isanbor C, Onyeka CS, Folarin OS, Adubi TO, Sogbanmu TO, Ogbeibu AE (2020) COVID-19 pandemic: examining the faces of spatial differences in the morbidity and mortality in sub-Saharan Africa, Europe and USA. medRxiv.
- Pal M, Berhanu G, Desalegn C, Kandi V (2020) Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus 12. [DOI] [PMC free article] [PubMed]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rajabian A, Hosseinzadeh H (2020) Dermatological effects of nigella sativa and its constituent, thymoquinone: a review. In: Nuts and Seeds in Health and Disease Prevention, pp. 329–355. Elsevier.
- Reyes-Escogido MDL, Gonzalez-Mondragon EG, Vazquez-Tzompantzi E. Chemical and pharmacological aspects of capsaicin. Molecules. 2011;16:1253–1270. doi: 10.3390/molecules16021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R. Effects of chloroquine on viral infections: an old drug against today’s diseases. Lancet Infect Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M, Saxena J, Nema R, Singh D, Gupta A (2013) Phytochemistry of medicinal plants. J Pharmacogn Phytochem, p 1.
- Sheahan TP, Sims AC, Leist SR, Schäfer A, Won J, Brown AJ, Montgomery SA, Hogg A, Babusis D, Clarke MO. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11:1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T (2020) A review of coronavirus disease-2019 (COVID-19). Indian J Pediatr, pp 1–6. [DOI] [PMC free article] [PubMed]
- Walters WP, Stahl MT, Murcko MA. Virtual screening—an overview. Drug Discov Today. 1998;3:160–178. doi: 10.1016/S1359-6446(97)01163-X. [DOI] [Google Scholar]
- Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Zakaryan H, Arabyan E, Oo A, Zandi K. Flavonoids: promising natural compounds against viral infections. Adv Virol. 2017;162:2539–2551. doi: 10.1007/s00705-017-3417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data have been provided in the manuscript, and for more details, they are available from the corresponding author on reasonable request.