Abstract
Embedded in an extracellular matrix, biofilm-residing bacteria are protected from diverse physicochemical insults. In accordance, in the human host the general recalcitrance of biofilm-grown bacteria hinders successful eradication of chronic, biofilm-associated infections. In this study, we demonstrate that upon addition of promethazine, an FDA approved drug, antibiotic tolerance of in vitro biofilm-grown bacteria can be abolished. We show that following the addition of promethazine, diverse antibiotics are capable of efficiently killing biofilm-residing cells at minimal inhibitory concentrations. Synergistic effects could also be observed in a murine in vivo model system. PMZ was shown to increase membrane potential and interfere with bacterial respiration. Of note, antibiotic killing activity was elevated when PMZ was added to cells grown under environmental conditions that induce low intracellular proton levels. Our results imply that biofilm-grown bacteria avoid antibiotic killing and become tolerant by counteracting intracellular alkalization through the adaptation of metabolic and transport functions. Abrogation of antibiotic tolerance by interfering with the cell’s bioenergetics promises to pave the way for successful eradication of biofilm-associated infections. Repurposing promethazine as a biofilm-sensitizing drug has the potential to accelerate the introduction of new treatments for recalcitrant, biofilm-associated infections into the clinic.
Author summary
At sub-minimal inhibitory concentrations, phenothiazines have been shown to inhibit virulence as well as the formation of biofilms in a wide range of different bacterial pathogens. In this study, we analyzed the anti-bacterial effect of the FDA-approved drug, promethazine, on biofilm-grown Pseudomonas aeruginosa. We demonstrate that PMZ interferes with bacterial bioenergetics and sensitizes biofilm-grown P. aeruginosa cells to bactericidal activity of several different classes of antibiotics by several orders of magnitude. This effect was most pronounced when cells were grown under environmental conditions that induce low intracellular proton levels. Thus, it seems that a reduced proton efflux in cells that exhibit decreased respiratory activity due to their biofilm mode of growth might explain their general antimicrobial tolerance. The use of PMZ as an antibiotic sensitizer holds promise that targeting tolerance mechanisms of biofilm-grown bacteria could become a practicable way to change the way physicians treat biofilm-associated infections.
Introduction
Biofilms are widely found in natural habitats and are considered an adaptation of microbes to hostile environments [1]. Bacteria in biofilms are embedded in a self-produced extracellular matrix and exhibit recalcitrance to a wide range of adverse conditions [2–4]. In the human host, biofilm-associated bacteria efficiently withstand antibiotic treatment and effectors of the immune system. Even in the absence of genotypic drug-resistance, antimicrobials have been demonstrated to be unable to clear biofilm infections [5–7]. Several factors, individually, or combinatorially, help to explain biofilm survival [8–12]. These include protection by extracellular polymeric substances (EPSs) [13,14], decreased antibiotic target activity due to decreased growth rate [15], biofilm-specific expression of possible resistance genes or gene products [16–18], and profound metabolic changes of biofilm-associated bacteria [19,20].
Pseudomonas aeruginosa is an exceedingly problematic gram-negative, opportunistic pathogen that has emerged as one of the most important human pathogens due to its role in acute nosocomial infections. Furthermore, chronic biofilm-associated P. aeruginosa infections, like those of the respiratory tract of cystic fibrosis patients, largely determine morbidity and mortality in the affected patients [21,22]. The urgent medical need for new therapy options spurred global biofilm research. However, despite a large body of research into biofilms, much remains to be learned about the mechanisms of biofilm formation and antimicrobial tolerance. Several anti-biofilm compounds have been described, with most interfering with structural biofilm components [6,23,24]. Potential anti-biofilm targets include adhesion molecules and biofilm matrix components such as extracellular DNA [25], lipopolysaccharides and exopolysaccharides [26]. Additionally, therapeutics that interfere with bacterial quorum sensing pathways [27,28] or secondary messengers involved in various signaling pathways [29–31] seem promising. Nevertheless, although there are numerous different biofilm targets, there are currently no anti-biofilm compounds in clinical use.
In this study, we analyzed the anti-bacterial effect of the phenothiazine promethazine (PMZ) on biofilm-grown P. aeruginosa. Phenothiazines are a class of highly versatile biological compounds and are well-known for their antipsychotic, sedative and antihistaminic effects [32]. Research conducted in the last couple of decades has indicated that phenothiazine compounds could play an important role in other fields of medicine as well, such as in the treatment of tumorous, neurodegenerative, or infectious diseases [33–35]. At sub-minimal inhibitory concentrations (MIC), phenothiazines have been shown to inhibit virulence as well as the formation of biofilms in a wide range of different bacterial pathogens [36–45].
Among the phenothiazines, the structure of promethazine (PMZ) is most similar to that of histamine. PMZ inhibits histamine H1 receptors and has an additional anticholinergic and a mild anti-dopaminergic activity. In this study, we show that PMZ interferes with bacterial bioenergetics and abrogates biofilm tolerance. PMZ sensitizes biofilm-grown P. aeruginosa cells to bactericidal activity of several different classes of antibiotics by several orders of magnitude. We also provide evidence for in vivo activity. The use of PMZ as an antibiotic sensitizer that targets microbial bioenergetics could change the way physicians treat chronically infected patients and pave the way for successful eradication of biofilm-associated infections.
Materials and methods
Ethics statement
All animal experiments were performed according to the guidelines of the German Recommendation of the Society for Laboratory Animal Science (GV-SOLAS) and the European Health Recommendations of the Federation of Laboratory Animal Science Associations. The animal protocol was approved by the local ethics committee and the “Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit (LAVES)”, permission number 33.9-42502-04-12/0713.
Bacterial strains, media and growth conditions
All P. aeruginosa strains and mutants used in this study, including the two reference strains PA14 and PAO1, are listed in S1 Table. Bacteria were cultivated in standard Lysogenic broth (LB) medium at 37°C with shaking (180 rpm) unless otherwise stated. For the cultivation of transposon mutants, 15 μg/ml gentamicin was added to the medium. Respiratory activity was measured following cultivation of the bacteria in a modified M9 minimal medium adapted from Abril et al. [46]. This medium contained 1 mM MgSO4, 0.1 mM CaCl2, 20 mM glucose, 0.01 mM FeSO4, 22 mM KCl, 5 mM HEPES, 2 mM Na2HPO4, 18.7 mM NH4Cl, 8.6 mM NaCl, and trace metals [H3BO3 9.6 μM, ZnSo4 0.7 μM, MnSO4 0.45 μM, CoCl2 2.1 μM, CuSO4 0.15 μM, Na2MoO4 0.3 μM]. Promethazine (PMZ) was used as a ready-to-use Promethazine hydrochloride injection solution (25 mg/ml, Promethazin-neuraxpharm, Neuraxpharm, Langenfeld, Germany).
To maintain the pH of the LB medium at 5.5, 7.2, and 8.5, respectively, we used the following buffer solutions: MES (2-(N-morpholino)ethanesulfonic acid), MOPS (3-(N-morpholino)propanesulfonic acid) and TAPS (N-[Tris(hydroxymethyl)methyl]-3-aminopropanesulfonic acid) at a final concentration of 50 mM. The buffers were prepared as 0.5 M stock solutions, adjusted to pH with NaOH/KOH at 37°C and filter-sterilized before use.
Planktonic growth
Growth of planktonic P. aeruginosa PA14 cells with and without 1 μM, 10 μM, 25 μM and 100 μM PMZ was monitored using an automated growth analysis system (Bioscreen C MBR plate reader; Oy Growth Curves Ab, Helsinki, Finland). Overnight grown pre-cultures were diluted to a starting OD600 of 0.02 in 200 μl and the OD600 was measured every 30 minutes for up to 20 h.
Bacterial respiration
Oxygen consumption rates (OCRs) and extracellular acidification rates (ECARs) of P. aeruginosa PAO1 and PA14 wild type strains were quantified using the Seahorse XFe96 Analyzer (Agilent, Santa Clara, California, USA). The protocol previously described by Dwyer et al. [47] was adopted and slightly modified. Briefly, overnight cultures of bacteria were grown in LB, diluted 1:100 in fresh modified M9 medium and grown to an OD600 of ~0.5 with shaking at 300 rpm. The cells were diluted to two times the final OD600 (OD600 of 0.05 for PA14 and OD600 of 0.04 for PAO1) in modified M9 medium. 90 μl were seeded onto poly-D-lysine-coated 96-well XF cell culture microplates. Cells were centrifuged for 10 min at 4,000 rpm in Heraeus Megafuge 40R centrifuge (ThermoFisher Scientific, Waltham, Massachusetts, USA), followed by increasing the well volume to 180 μl using modified M9 medium. Basal OCR and ECAR were measured for an initial 16 minutes, followed by automatic injection of 20 μl PMZ from 10x stock solutions (final concentrations of 200, 300 and 400 μM PMZ, respectively). OCR and ECAR measurements were then acquired every 4 minutes for up to 90 min. PMZ added to cell-free medium served as a control to account for background values upon PMZ addition; these OCR and ECAR values were subtracted to obtain final OCR and ECAR cell response profiles.
Membrane potential
The BacLight Bacterial Membrane potential kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA) was used according to the manufacturer’s instructions. In brief, overnight grown cells were inoculated in 10 ml of LB medium at OD600 of ~0.05 and cultivated at 37°C until they reached early logarithmic phase (OD600 ~ 0.3–0.4). PMZ was added to the cultures at final concentrations of 100 μM, 250 μM and/or 400 μM, and the samples were incubated for an additional 10 minutes at 37°C. After PMZ addition, sample aliquots were directly diluted into 0.5 ml of filtered PBS (0.22 nm pore size) to approximately 106 cells ml-1. The samples were exposed to 30 μM DiOC2(3) (3,3'-Diethyloxacarbocyanine Iodide) prepared from a 3 mM stock solution in DMSO, followed by further incubation for 30 min at room temperature in the absence of light. Stained bacteria were assayed in an LSR Fortessa flow cytometer (BD Biosciences, San Jose, California, USA) equipped with a 488 nm laser excitation source and both green (525/50 nm bandpass filter) and red (685/35 bandpass filter) fluorescence emission channels. Forward and side scatter density plots were used to identify the bacterial cell population of interest and to exclude debris. Fluorescence was recorded for at least 10,000 bacteria in both green and red emission channels and the red/green fluorescence ratios were subsequently calculated using population mean fluorescence intensities. The experiments were performed in independent triplicates.
Biofilm microscopy
A static microtiter plate assay combined with automated confocal laser scanning microscopy was used as previously described [48]. Briefly, P. aeruginosa PA14 pre-cultures were grown overnight for 16 h in 3 ml of LB medium. One milliliter of each pre-culture was harvested by centrifugation (5000 g, 5 min), and the bacteria were washed twice with 1 ml of fresh LB. Cells were suspended in fresh LB medium, adjusted to OD600 of 0.002 and 100 μl of the bacterial solution were added to the wells of a sterile black half-area 96-well μClear microtiter plate (Greiner Bio-One). The microtiter plate was sealed with an air-permeable BREATHseal cover foil (Greiner Bio-One) and incubated for 24 h in a humid atmosphere at 37°C. The bacteria were then treated with PMZ at a concentration of 100 μM and stained by carefully adding the fluorescence dyes Syto9 and propidium iodide [final concentrations of 2.1 μM and 12.5 μM respectively] of the LIVE/DEAD BacLight Bacterial Viability Kit (Molecular Probes, Life Technologies) and incubated for another 24 hours.
Automated confocal microscopy was performed with an inverted SP8 system (Leica Microsystems) and the Leica application suite LAS X including the Matrixscreener module. Focal planes were acquired starting from the bottom of the plate (20 focal planes, z-step size: 3 μm) by using a PL APO 40x/1.10 W water immersion objective. Two Z-stacks (overview and zoom) were recorded in parallel at the center of a well: Stack 1 (overview stack) was acquired using a zoom x0.75, while stack 2 (zoom stack) was acquired with a zoom x4 in order to visualize biofilm details with higher resolution. Syto9 was excited at 488 nm, whereas propidium iodide (PI) was excited at 561 nm. Emission was detected with hybrid photo-detectors (HyD) in the range of 500–550 nm (Syto9) and 675–725 nm (PI) respectively. For automated image acquisition of all test samples, a number of pre-defined laser/detector settings were assigned to compensate for inter-well fluctuations in fluorescence intensity, avoiding under- and over-exposed images for the different treatments.
The image stacks were subsequently processed and analyzed with the software Developer XD 64 (Definiens) with a customized programmed solution. Moreover, biofilm structures were visualized with the software Imaris (version 7.6, Bitplane).
Drug susceptibility testing and MIC determination
We performed antimicrobial susceptibility testing for tobramycin (TOB), ciprofloxacin (CIP), ceftazidime (CAZ) and gentamycin (GEN). Minimal inhibitory concentrations (MIC) were determined by a standard broth microdilution procedure [49] in LB medium using 96-well microtitre plates. Growth was evaluated after overnight incubation at 37°C with shaking at 180 rpm. The MIC was defined as the lowest antibiotic concentration for which no visible growth could be detected.
Antimicrobial susceptibility testing of biofilm grown bacterial cells was performed in a static microtiter plate assay as previously described [48] with slight modifications. In brief, overnight cultures of the cells were adjusted to an OD600 of 0.002 and 100 μl of the bacterial suspension were added to the wells of a sterile half-area, black 96-well μClear microtiter plate (Greiner Bio-One). The plate was sealed with an air-permeable BREATHseal cover foil (Greiner Bio-One) and incubated without shaking at 37°C in a humid atmosphere. After 24 h, the biofilms were treated with the antibiotics (at the given concentrations) either alone or in combination with PMZ (100μM) and in selected experiments also with 300 mM KCl, 20 μM CCCP (Carbonyl cyanide 3-chlorophenylhydrazone; Sigma Aldrich dissolved in DSMO) or 100 μM PQS (2-Heptyl-3-hydroxy-4(1H)-quinolone; Sigma Aldrich dissolved in DMSO).
The biofilms were incubated for another 24 h before the wells were resuspended and tenfold serial dilutions (10−1 to 10−6) were prepared using a 96-channel pipetting device (Platemaster, Gilson). Dilutions were spot-plated onto rectangular LB agar plates and incubated at 37°C overnight. Growth of surviving bacteria was evaluated after 16 h and colony-forming units (CFU) per ml were determined.
Susceptibility on ex vivo lung tissue
In this study, tissue samples were obtained from three explanted lungs of CF patients undergoing double lung transplantation at Hannover Medical School (MHH). Procession of the tissue was done immediately after explantation at room temperature in the pathology department of the MHH. The dissected tissue was then cooled on ice and further kept at 4°C until its subsequent use for the determination of an ex vivo antibacterial activity.
Destruction of the tissue by homogenization processes, such as vortexing, was avoided. Instead, sampling of the mucopurulent secretion areas of the chronically P. aeruginosa infected CF lungs was achieved by collecting the secretions with the help of a sterile inoculation loop into 0.8 ml of PBS in a 2 ml eppendorf reaction tube. The weight of each of the collected mucopurulent secretion mass was recorded in order to adjust CFU determination relative to the weight of the patient´s material. Multiple experimental replicates were prepared for each of the tested treatment condition (PBS control, tobramycin 16 μg ml-1, PMZ 100 μM, and the combination of the two (PMZ and TOB)). The ex vivo mucus biofilms were incubated in the treatment solutions for 24 h at 37°C without shaking, and the material was then resuspended by vortexing the tubes for 30–60 sec. Serial dilutions of the resuspended biofilm were prepared in PBS and 100 μl of the dilutions were plated onto LB agar and selective Pseudomonas Cetrimide Agar. The plates were then incubated for at least 24 h at 37°C to obtain CFU/mg mucus.
In vivo murine tumor model
Experiments were performed as described previously [50]. In brief, seven to eight-week old female BALB/c mice (Janvier, Germany) were injected intradermally into the left flank with 5x105 of CT26 colon carcinoma cells or F1A11 fibro-sarcoma cells in 100 μl PBS. After roughly 10 days the tumors had grown to a volume of 150–200 mm3. The mice were then infected with 5x106 P. aeruginosa PA14 intravenously (i.v.). Under these conditions, the bacteria colonize preferentially the tumors while spleen and liver will bear 100-to 1000-fold less bacteria. Systemic application of such bacteria induces rapid release of cytokines with TNF-α being most dominant. This results in a severe hemorrhage in the tumor and the formation of a large necrotic region in the center of the tumor. P. aeruginosa proliferate in this necrotic area but especially in the region between necrosis and the remaining viable, hypoxic interface. We could show that in this region the bacteria form biofilms [51,52]. Interestingly, from transcriptional profiling we conclude that for the bacteria this microenvironment very closely resembles the environment they encounter in the cystic fibrotic lung [50]. Experimentally, robust bacterial biofilms are established within the tumor by 48 h. Therefore, colonized tumor bearing mice were treated locally intra-tumorally (i.t.) with 50mg kg -1 PMZ once or twice with 24 h intervals. Starting at the same time, tobramycin (2mg kg-1 in 100μl PBS) was given i.v. three or five times at 12 h intervals as specified. Six h after the last application, the mice were euthanized by CO2 asphyxiation and tumors isolated. The tumors were homogenized in 0.1% (v/v) Triton X-100/PBS using gentle MACS M-tubes and a dissociator from Miltenyi Biotec. The samples were serially diluted and plated on LB agar plates containing ampicillin (0.1 mg ml-1).
Statistical analysis
All experiments were performed at least in biological triplicates, and with three technical replicates per experiment. Statistical analysis was made using GraphPad Prism version 5 (GraphPad Software, La Jolla, California, USA, www.graphpad.com).
Results
PMZ sensitizes biofilm-grown bacteria to antibiotic killing
In order to test the anti-biofilm activity of PMZ, the P. aeruginosa type strain PA14 was grown in 96-well plates for 24 h to establish biofilms prior to the addition of 100 μM PMZ with and without additional antibiotics. Following live/dead staining, the bactericidal activity of the compounds on biofilm-grown PA14 was monitored by acquiring confocal images at two positions in duplicate wells. Despite modest effects on the structure of P. aeruginosa biofilms, no profound PMZ-mediated effects on the viability of the biofilm bacteria were observed (Fig 1A and 1B). PMZ did not exhibit an antibiotic activity on planktonic bacteria, and growth was not altered at PMZ concentrations as high as 100 μM (Fig 1C).
Fig 1. PMZ does not affect bacterial growth nor viability.
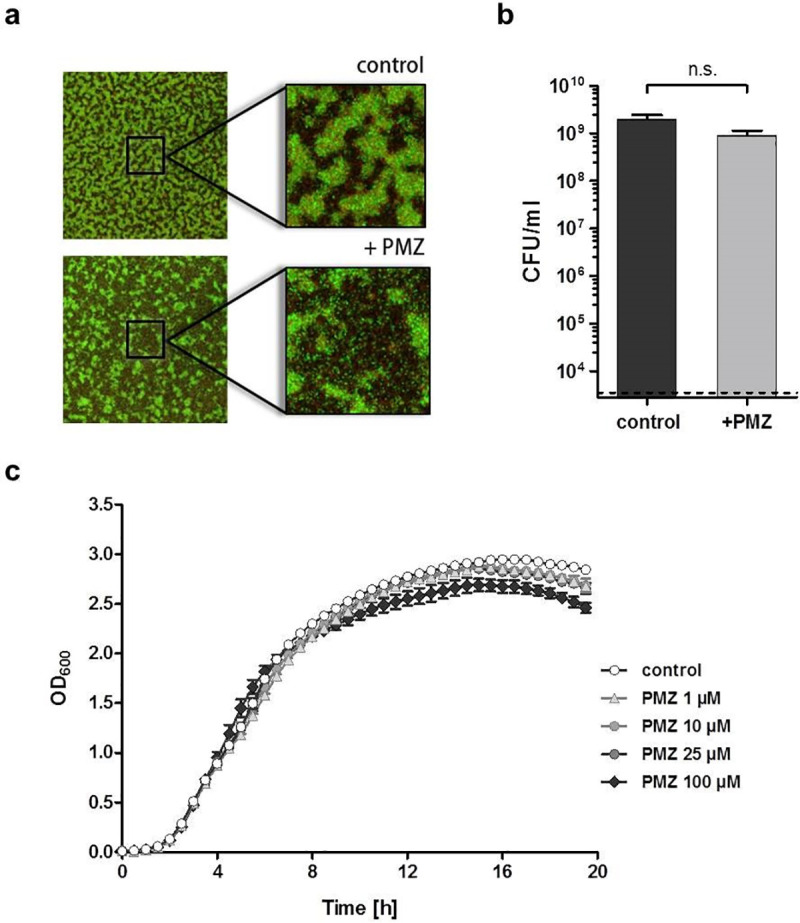
100 μM promethazine (PMZ) slightly influenced the structure, but not the viability of biofilm-grown P. aeruginosa cells. (a) Representative maximum intensity projections of PA14 biofilms treated with PMZ for 24 h in comparison to untreated control biofilms. The biofilms were stained with the BacLight Bacterial Viability Kit, visualizing dead cells in red (propidium iodide) and living cells in green (Syto9). (b) Determination of colony forming units (CFU) from the untreated (dark grey) and PMZ-treated (light grey) biofilms revealed no significant differences in CFU/viability (Significance was calculated using the t- test). Data show mean CFU counts from overall 6 independent experiments. (c) Increasing concentrations of PMZ (1 μM, 10 μM, 25 μM and 100 μM) marginally affected planktonic growth of P. aeruginosa PA14 cells. Experiments were performed three times with at least three biological replicates each; here an exemplary data set from one experiment is shown.
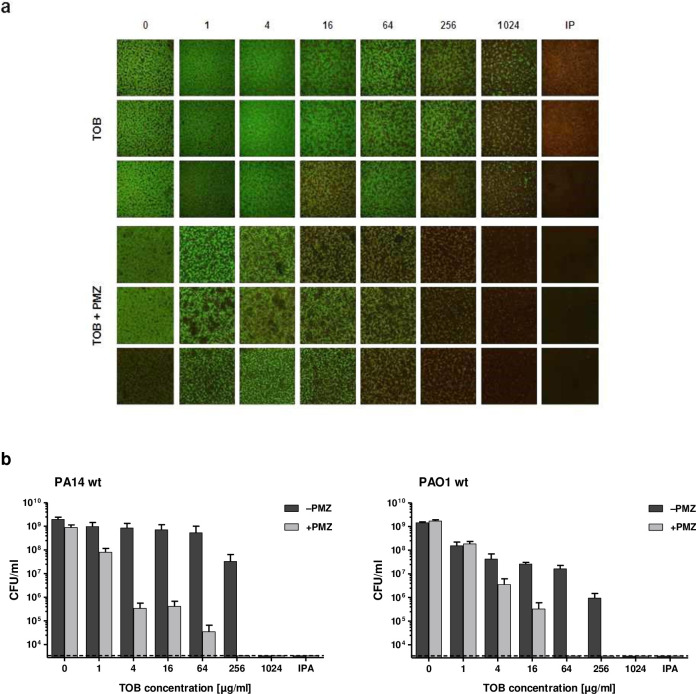
However, treatment of P. aeruginosa PA14 and PAO1 biofilms with PMZ, in combination with tobramycin, strongly enhanced the killing activity of tobramycin (Fig 2). Up to a four-log reduction of viable bacteria were observed in biofilms treated with a combination of PMZ and tobramycin, in comparison to tobramycin alone. The sensitizing activity of PMZ was not restricted to tobramycin, but was also observed for ciprofloxacin, ceftazidime and gentamicin (S1A, S1B and S1C Fig). Furthermore, we tested the sensitizing effect of PMZ on Escherichia coli K12 cultures. We observed a synergistic, bactericidal effect when E. coli biofilm-grown cells were treated with PMZ and tobramycin; albeit, the effect was less pronounced than in P. aeruginosa cultures (S1D Fig).
Fig 2. Synergistic anti-biofilm activity of PMZ in combination with tobramycin.
P. aeruginosa static biofilms grown in 96-well plates were treated with increasing concentrations of tobramycin (TOB) with or without the addition of 100 μM promethazine (PMZ). (a) Representative maximum intensity projections of PA14 biofilms (triplicates) treated with TOB alone (upper panel), or in combination with PMZ (bottom panel). The biofilms were stained with the BacLight Bacterial Viability Kit, visualizing dead cells in red (propidium iodide) and living cells in green (Syto9). Isopropanol treatment (IPA) was used as a killing control. Biofilms were assessed in triplicate wells, and at least three independent experiments were performed. (b) To assess the viability of the bacteria upon synergistic treatment, colony forming units (CFU) were determined from the tobramycin treated (dark grey) and tobramycin plus PMZ treated (light grey) biofilms of P. aeruginosa PA14 as well as the PAO1 reference strain. Mean CFU counts were performed on pools of three wells in at least three independent experiments. Error bars represent the standard error of the mean, while the dashed line indicates the lower limit of detection of the assay.
PMZ sensitized biofilm-grown cells can be efficiently killed at MIC concentrations
Since the combined use of antibiotics and PMZ killed the majority of P. aeruginosa biofilm-associated bacteria, when the antibiotics were added at MICs concentrations, we tested how an increase in the MIC would affect killing of biofilm-associated cells. We generated two ciprofloxacin resistant strains by introducing a target mutation into the gyrA gene individually (MIC of 2 μg/ml), and in combination with a target mutation in parC (MIC of 32 μg/ml). The introduction of these two target mutations drastically elevated the MIC of ciprofloxacin from that measured for WT cultures (0.125 μg/ml). Additionally, we used a PA14 transposon mutant that harbored a gentamicin resistance cassette, which increased the MIC of gentamicin from 2 μg/ml to >2048 μg/ml. The three resistant strains were grown under biofilm conditions and killing by the respective antibiotics was monitored in PMZ-sensitized cells. Both, ciprofloxacin and gentamicin, exhibited synergistic activities on biofilm-grown bacteria with PMZ, even in the resistant PA14 strains. For bacteria of such strains exhibiting increased MIC values, at least MIC concentrations were required to sufficiently kill the biofilm-grown PA14 variants (S2 Fig). In conclusion, PMZ reverts antimicrobial tolerance in all the strains tested under biofilm growth conditions. Thus, biofilm-grown cells can be effectively killed at MIC concentrations. Differences in antimicrobial killing of planktonic and biofilm-grown bacteria level off.
PMZ–mediated synergistic activity is enhanced in PQS producing cells
We observed that clinical P. aeruginosa isolates and mutant strains that exhibited low or abolished expression of the Pseudomonas quinolone signaling (PQS) system were less prone to sensitization towards antimicrobial killing upon PMZ treatment. Interestingly, it has previously been demonstrated that increases in the levels of PQS, or decreases in the ability to induce an oxidative stress response, influence the antibiotic susceptibility of biofilm-grown P. aeruginosa isolates [53]. Here, the treatment of biofilms of a pqsA negative mutant, which is not able to produce PQS signaling molecules, revealed that there is significantly diminished sensitizing activity of PMZ towards the bactericidal activity of tobramycin (S3 Fig; compare to results of Fig 2B). In line with this, we observed that E. coli biofilms were sensitized to a lower extent by PMZ towards tobramycin killing activity (S1 Fig). The exogenous addition of PQS to the non-producing P. aeruginosa mutant restored the sensitizing activity of PMZ in pqsA mutant biofilms (S3 Fig). Furthermore, also in E. coli biofilms more cells were killed by PMZ/tobramycin if the biofilms were simultaneously treated with PQS. Thus, it seems that strong synergistic PMZ/tobramycin anti-biofilm activity in P. aeruginosa is dependent on the presence of PQS. Since PQS enhanced PMZ/tobramycin killing not only in P. aeruginosa but also in E. coli biofilm cells, this implicates that the activity of PQS per se and not its impact on bacterial signaling contributes to the killing activity.
Synergistic activity of PMZ is dependent on the pH of the medium
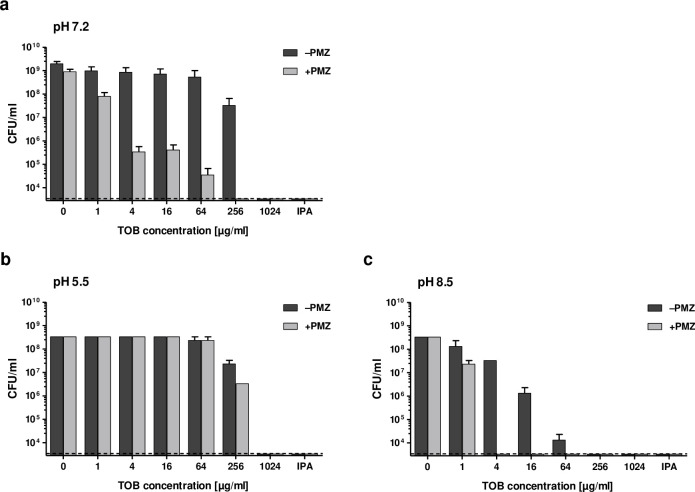
The extracellular pH has previously been demonstrated to have an impact on the activity of various antibiotics [54,55]. Therefore, we repeated the biofilm killing experiments with PA14 biofilms grown in pH buffered medium to stabilize the pH at 5.5, 7.2, and 8.5, respectively. An increase or decrease in the pH of the medium strongly influenced the susceptibility of PA14 biofilms to the synergistic effects of PMZ and tobramycin. In accordance to previous studies, the killing activity of tobramycin was more effective under higher pH medium conditions [54,55]. At a pH of 8.5, the combination of PMZ with tobramycin reduced the CFU counts below the detection limit at tobramycin concentration levels as low as 4 μg/ml, whereas 256 μg/ml was needed at pH of 7.2 and even 1024 μg/ml at pH of 5.5 (Fig 3).
Fig 3. The pH of the growth medium affects the synergistic anti-biofilm activity of PMZ in combination with tobramycin.
P. aeruginosa static biofilms grown in 96-well plates were treated with increasing concentrations of tobramycin (TOB) with or without the addition of 100 μM promethazine (PMZ) in buffered LB. The pH of the medium was buffered to 7.2 (a) 5.5 (b) or 8.5 (c), respectively. Colony forming units (CFU) were determined from the tobramycin treated (dark grey) and tobramycin plus PMZ treated (light grey) biofilms. Isopropanol treatment (IPA) was used as a killing control. Mean CFU counts were performed on pools of three wells in at least three independent experiments. Error bars represent the standard error of the mean, while the dashed line indicates the lower limit of detection of the assay.
PMZ increases the membrane potential and results in a decreased oxygen consumption and extracellular acidification rate of P. aeruginosa cells
PMZ is known to interfere with the transport of ions across the membrane [56,57]. Thus, we measured the membrane potential of P. aeruginosa cells (PA14 and PAO1) following the addition of PMZ. An increase in the membrane potential in a concentration dependent manner was observed (Fig 4).
Fig 4. PMZ increases the membrane potential of P. aeruginosa.
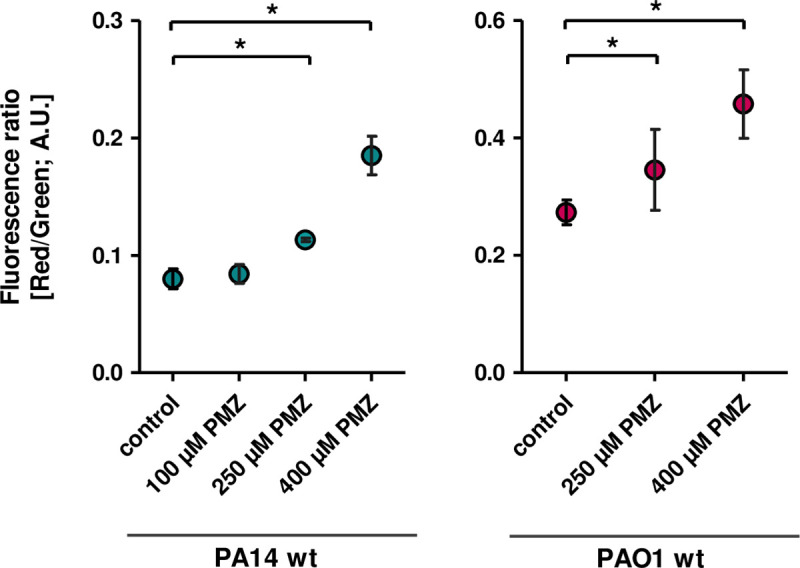
Exponentially growing cells of PA14 (left) and PAO1 (right) were challenged with increasing concentrations of PMZ. Cells were stained with the membrane potential indicator DiOC2(3) and analyzed by flow cytometry. The red/green fluorescence ratio was calculated using population mean intensities. Experiments were carried out in triplicates. Data analysis by Student’s t-test demonstrates significant difference (* p < 0.05) between the treated samples and the control.
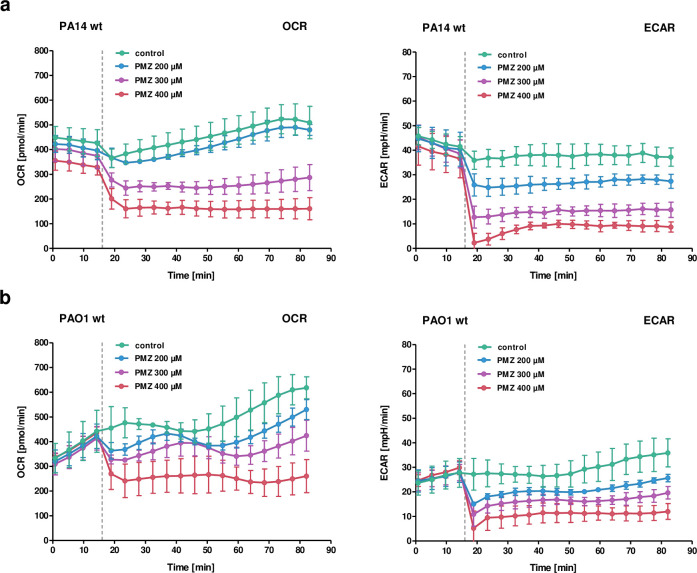
Membrane hyperpolarization can create “backpressure” on the proton pumping complexes of the electron transport chain, thereby slowing their function [58]. To measure the oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) of P. aeruginosa treated with increasing concentrations of PMZ, we used the Seahorse XFe96 Analyzer (Agilent). Immediately following PMZ exposure, both the OCR and the ECAR dropped in a concentration dependent manner Fig 5.
Fig 5. Bioenergetic analysis of PA14 upon addition of PMZ.
Oxygen consumption rate (OCR; left) and extracellular acidification rate (ECAR; right) of P. aeruginosa PA14 (a) and PAO1 (b) treated with increasing concentrations of PMZ. The dotted line indicates the addition of PMZ after four cycles of basal OCR and ECAR measurements (16 minutes). Data show the mean and standard deviation of three technical replicates, calculated by the Seahorse XF Wave software. One representative experiment is shown of at least three independent experiments. The data were corrected for background artifacts from sole addition of PMZ to cell-free buffer medium (see Experimental Procedure).
CCCP reduces the synergistic killing activity of PMZ in combination with tobramycin, whereas extracellular potassium addition increases it
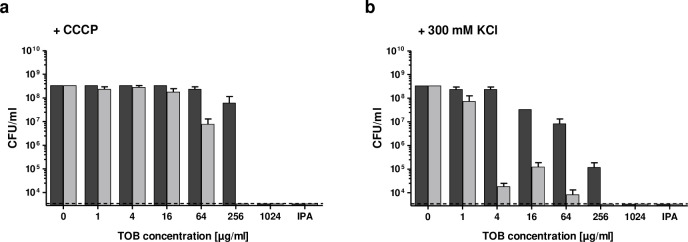
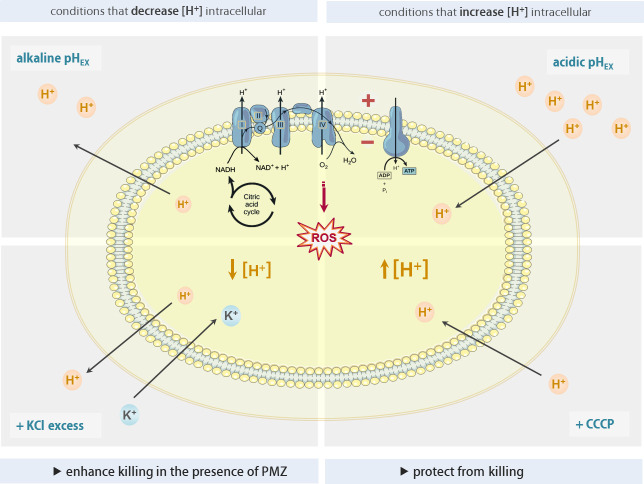
Changes in extracellular pH, and the addition of PMZ, affect the electrochemical gradient across the membrane. Therefore, we wondered whether P. aeruginosa becomes sensitive to bactericidal antibiotics upon an increase in membrane potential. To test this, we repeated the biofilm killing experiments in medium supplemented with 300mM KCl as well as the uncoupler/ionophore, CCCP (carbonyl cyanide-m-chlorophenyhydrazon), both of which decrease the membrane potential.
The addition of CCCP reduced the synergistic killing activity of PMZ in combination with tobramycin. However, high extracellular potassium concentrations increased the synergistic activity of tobramycin and PMZ (Fig 6). Interestingly, CCCP increases the membrane permeability for protons, leading to a net flow of protons into the cell; whereas under high extracellular potassium concentrations, the flow of protons is reversed, and there is a net flow of protons out of the cell. Our results indicate that a change in proton concentration, rather than a change in membrane potential, enhances the synergistic activity of PMZ and antibiotics on biofilm-associated bacteria. Decreasing the intracellular levels of protons by increasing the extracellular pH, or by increasing the extracellular potassium concentrations, increases the bactericidal effect of antibiotics. In contrast, a decrease in the extracellular pH, or an increase in membrane permeability, increases intracellular proton levels, thereby reducing the efficacy of antibiotics (Fig 7).
Fig 6. High extracellular potassium and the addition of CCCP affect the synergistic anti-biofilm activity of PMZ in combination with tobramycin.
P. aeruginosa PA14 biofilms were treated with increasing concentrations of tobramycin (TOB) alone or in combination with 100 μM PMZ and further challenged with the protonophore CCCP (a), or with high extracellular potassium (300 mM KCl) (b). Colony forming units (CFU) were determined from the tobramycin treated (dark grey) and tobramycin plus PMZ treated (light grey) biofilms. Isopropanol treatment (IPA) was used as a killing control. Mean CFU counts were performed on pools of three wells in at least three independent experiments. Error bars represent the standard error of the mean, while the dashed line indicates the lower limit of detection of the assay.
Fig 7. The levels of intracellular protons modulate the synergistic killing activity of PMZ and antibiotics.
Tobramycin and PMZ combination therapy reduces bacterial burden in an in vivo biofilm model
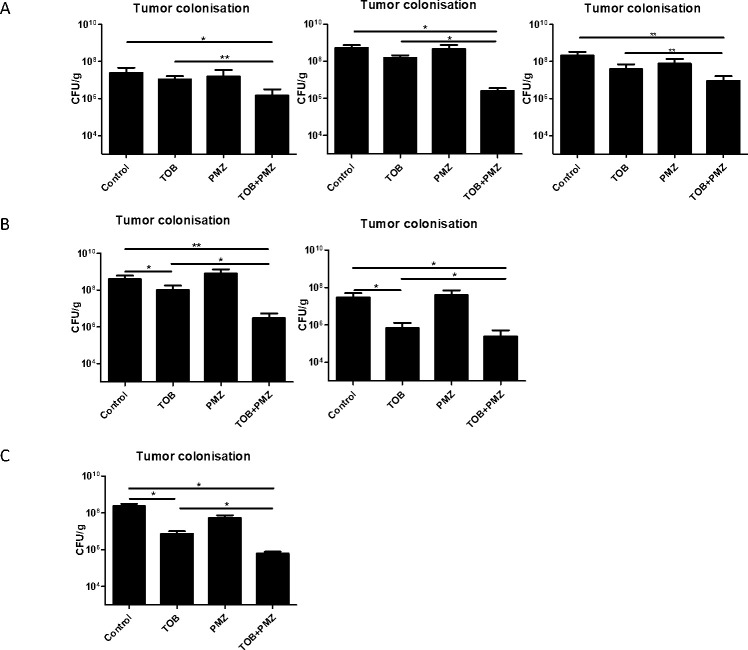
We next evaluated the synergistic activity of PMZ and tobramycin on the killing of biofilm-associated P. aeruginosa in an in vivo system. We therefore used our previously established murine tumor model for biofilm infections. According to the genetic profile of the bacteria, the tumor model very closely simulates the environment of the cystic fibrosis lung for the bacteria (50). Mice bearing CT26 tumors were infected i.v. with P. aeruginosa. This leads to stable biofilm formation within two days post infection within the tumor. The mice were then treated with a combination of PMZ and tobramycin. In the first experiment, mice bearing CT26 tumors colonized with biofilm-residing P. aeruginosa were treated once i.t. with PMZ and three times i.v. with tobramycin at 12 h intervals. Mice were sacrificed and CFU numbers in the tumors and the liver were determined by plating tissue homogenates. A reduction of bacterial load was observed at 2 mg kg-1 tobramycin in the tumor (Fig 8A). The reduction in CFU counts was drastically more pronounced in the tumor of mice that had been treated with tobramycin in combination with PMZ.
Fig 8. In vivo synergistic activity of tobramycin and PMZ on biofilm forming P. aeruginosa.
(A) PA14 colonized CT26 tumor bearing mice were treated with 50 mg kg-1 PMZ by single i.t. injection followed by three i.v. injections of 2mg kg-1 tobramycin at 12 h intervals. (B) PA14 colonized CT26 tumor bearing mice were treated twice with 50 mg kg-1 PMZ at an interval of 24 h via i.t. injections. Starting at the same time, five i.v. injections of 2mg kg-1 tobramycin at 12 h intervals were applied. (C) PA14 colonized F1A11 tumor bearing mice were treated twice with 50 mg kg-1 PMZ at an interval of 24 h via i.t. injections combined with five i.v. injections of 2mg kg-1 tobramycin at 12 h intervals. The panels represent individual experiments. At least 5 mice per group were tested. Tumors were harvested 6 h after final administration, homogenized and plated for CFU quantitation. *, P < 0.05; **, P < 0.005 (P value were determined by unpaired t test).
In an attempt to optimize the application regimen, i.t. application of PMZ was performed twice within a 24 h interval with mice bearing colonized CT26 tumors. Tobramycin was applied five times i.v. with 12 h hour intervals. Under these circumstances already an antibiotic effect could be observed (Fig 8B) as described before (51). Nevertheless, an enhanced effect of the combination therapy could still be observed (Fig 8B). To test this therapy in a tumor system less prone to bacterial toxicity, we switched to the aggressive fibrosarcoma F1A11 keeping the other experimental condition constant. Again, an effect of the antibiotics alone could be observed (Fig 8C). However, the enhanced effect of the combination of PMZ and tobramycin became much more obvious (Fig 8C). Thus, as predicted from the in vitro data when tobramycin was administered in combination with PMZ, the efficacy of tobramycin against biofilm residing bacteria in the tumor model can be strongly enhanced when combined with PMZ.
Treatment of P. aeruginosa microcolonies from chronic CF lung infections with PMZ/tobramycin
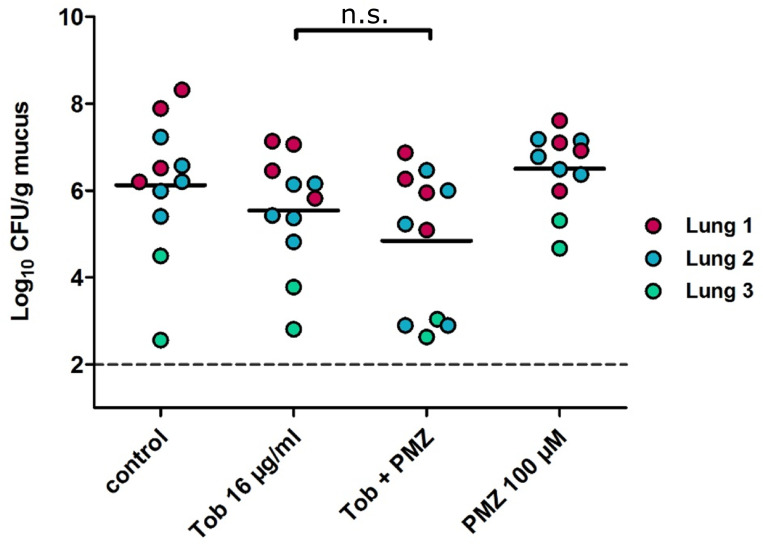
To further evaluate the synergistic activity of PMZ and tobramycin on the killing of biofilm-associated P. aeruginosa, we tested their bactericidal activity on human ex vivo samples. We collected tissue samples from three chronically infected explanted cystic fibrosis lungs and exposed the samples to tobramycin, or tobramycin in combination with PMZ. Despite a trend, the combined use of PMZ and tobramycin did not show a significant synergistic bactericidal activity on the ex vivo P. aeruginosa biofilm samples (Fig 9). More work needs to be done, in order to evaluate the optimal synergistic concentrations of PMZ and tobramycin in an in vivo/ex vivo setting, to improve delivery of the drug and to adjust environmental conditions (such as pH and efficient delivery of the drug) to promote the synergistic killing activity.
Fig 9. PMZ and tobramycin show a synergistic killing in P. aeruginosa infected ex vivo lung tissue samples.
Tissue samples were collected from three explanted CF lungs which were found to be chronically infected by P. aeruginosa. Tobramycin was added to the infected tissue at a final concentration of 16 μg/ml with and without 100 μM PMZ. Colony forming units (CFU) were determined in the PMZ, tobramycin and tobramycin plus PMZ treated samples. Each colored dot represents a different technical replicate of tissue samples (coming from the same lung) whereas the different colors are biological replicates (different lungs). The dashed line indicates the lower limit of detection of the assay. Horizontal bars represent the mean. No significant differences were observed between the tobramycin treated samples and those treated with tobramycin in combination with PMZ (Mann Whitney U test).
Discussion
The general recalcitrance of biofilm-associated bacteria diminishes the efficacy of antimicrobials. Accordingly, present antimicrobial treatment protocols that serve well for the eradication of acute infections fail to clear biofilm-associated chronic infections. As a result, morbidity and mortality due to chronic infections have remained unchanged over the past few decades. Alternative therapeutic strategies to eradicate biofilm-associated infections are desperately needed.
Many efforts to decrease the burden of chronic infections have been made; however, despite the initial enthusiasm and the vast amount of literature on the identification of novel anti-biofilm compounds, no antimicrobials are currently in clinical use to specifically treat biofilm-associated infections. An alternative way to meet the clinical need could be to concentrate on enhancing the activity of our current antibiotics [35,59]. It seems that the successful eradication of biofilm-associated bacterial infections relies on our ability to break the antibiotic tolerance of the biofilm-grown bacteria.
In this study, we show that PMZ targets antimicrobial tolerance under biofilm growth conditions. We found that in the opportunistic pathogen P. aeruginosa, PMZ fully restores the bactericidal activity of common antibiotics in in vitro grown biofilm bacteria and to some extent in vivo. The finding that PMZ breaks tolerance against several classes of antibiotics seems to be consistent with a shared underlying molecular tolerance mechanism.
It is well established that hypoxia [60], low nutrient availability [53,61], and low pH [55,62] have the potential to shift a pathogen into a drug-tolerant state [63]. The effectiveness of antibiotics appears to depend on bacterial growth and metabolism and there is considerable evidence that dysregulation of bacterial bioenergetics is important in modulating drug lethality [64–66]. Extensive screens for genes that contribute to antibiotic resistance in P. aeruginosa [67, 68] and other bacterial pathogens [69,70] have revealed that mutations influencing the production of NADH (by e.g. interrupting TCA cycle genes) or the subsequent NADH oxidation in the electron transport chain are responsible for a tolerant phenotype. Previous work also identified a link between antibiotic-induced cellular respiration and bactericidal lethality. Bactericidal activity can be arrested by attenuated respiration and potentiated by accelerated respiration [71], and has been proposed to be connected to enhanced oxidative stress generated from increased aerobic respiration [47,72]. Accordingly, killing of biofilm-associated bacteria can be counteracted by molecular mechanisms that reduce oxidative stress within the cells [53].
Recently, it was also proposed that in antibiotic-treated bacterial cells in the absence of reduced respiration, protons would be drained from the cytoplasm and not replaced through proton re-entry via ATP synthesis as the cells switch from high to low ATP states [54,58]. It was suggested that the effects of antibiotics on pH homeostasis should be considered a potential mechanism that contributes to antibiotic lethality. However, as the chemistries of protons and electrons are closely linked, it may be not possible to decipher if the bactericidal effect of an antibiotic is due to oxidative stress, or the disruption of pH homeostasis.
In accordance with the pH homeostatic model, we demonstrate that the antibiotic tolerance of biofilm-grown cells can be reversed under conditions that induce low concentrations of intracellular protons by increasing the extracellular pH, or extracellular potassium concentrations. Antibiotic activity was counteracted by increasing the intracellular levels of protons via decreasing the extracellular pH or increasing the membrane permeability for protons (addition of CCCP). Thus, changes in environmental conditions, which lead to decreased intracellular proton levels, promoted antibiotic killing of biofilm-grown bacteria. Of note, this killing could be clearly enforced by the addition of PMZ. We demonstrate that the simultaneous treatment of in vitro grown biofilm bacteria with PMZ and tobramycin was sufficient to fully revert the biofilm-growth mediated tolerance phenotype and tobramycin killed biofilm-grown cells at MIC concentrations.
This is remarkable because the PMZ-mediated inhibitory effect on cellular respiration curtails cellular alkalization. However, PMZ induced membrane hyperpolarization also leads to conditions that are energetically less favorable for proton-pumping electron transport chain complexes. While this hyperpolarization counteracts alkalization of the cytoplasm, it may also increase the formation of ROS due to the stalling of electrons on the electron transport chain complexes. Our finding that the activity of PMZ sensitization was diminished in P. aeruginosa strains with low or abolished production of PQS supports the finding that ROS production contributes to effective antibiotic killing. PQS is an iron scavenger and has been demonstrated to have anti- and pro-oxidant activities [73]. Its presence was determined to be essential for antibiotic killing of starved biofilm-grown cells [53].
It is also conceivable that the PQS governed 4-quinolone derivate 2-heptyl-4-hydroxyquinoline n-oxide (HQNO) contributes to the sensitization of the biofilm cells to the synergistic activity of PMZ and tobramycin. HQNO has previously been shown to be a cytochrome bc1 complex inhibitor and to promote stationary phase killing of P. aeruginosa populations due to an inhibition of the electron transfer chain leading to enhanced ROS production [74]. Of note, an interference of phenothiazines with the respiratory chain of Mycobacterium tuberculosis has been demonstrated previously [75].
In addition, the phenothiazines were shown to inhibit the activity of multi-drug efflux pumps and to disrupt biofilms [34,76–79]. Multi-drug efflux pumps play an essential role in adapted antibiotic resistance [80–82] and are specifically upregulated under biofilm growth conditions [83–86]. Thus, an accumulation of antibiotics in PMZ treated bacterial cells could contribute to the synergistic activity observed between PMZ and tobramycin.
In conclusion, our data suggest that reduced proton efflux in cells that exhibit decreased respiratory activity (due to their growth in a biofilm) contributes to general antimicrobial tolerance. This tolerance phenotype can be overcome if cells are subjected to environmental conditions that lead to lower intracellular proton levels. Under these conditions, PMZ acts synergistically with antibiotics to fully revert the tolerance phenotype of biofilm-associated bacteria by inducing hyperpolarization of the cellular membrane. In this study, the simultaneous addition of PMZ and tobramycin was not sufficient to eradicate the bacteria from the in vivo biofilms in mouse tumor tissue or within microcolonies that were recovered from chronically infected explanted CF lungs. Clearly, more work is needed to evaluate whether and how the in vivo environment could be influenced to promote low intracellular proton levels in biofilm-associated cells to maximize the synergistic activity of PMZ and antibiotics.
Our findings open new avenues, and hold promise that targeting tolerance mechanisms of biofilm-associated bacteria could become a practicable way to change the way physicians treat biofilm-associated infections. Furthermore, since PMZ is already used in the clinic, bioavailability and safety profiles for this drug are already available. Proven formulation and manufacturing routes and reasonably characterized pharmacology for this drug would also aid in repurposing this drug, thereby enabling the entry into clinical phases more rapidly, and at a lower cost than novel compounds. Repurposing of PMZ as an antibiotic sensitizer of biofilm-grown bacterial cells may foster its fast introduction into clinical practice to enhance the activity of existing antibiotics so that chronic biofilm-associated infections can be targeted more effectively.
Supporting information
(DOCX)
P. aeruginosa static biofilms grown in 96-well plates were treated with increasing concentrations of ciprofloxacin (CIP) (a), ceftazidime (CAZ) (b) or gentamicin (GEN) (c) with or without the addition of 100 μM promethazine (PMZ). Colony forming units (CFU) were determined from the antibiotics treated (dark grey) and antibiotics plus PMZ treated (light grey) biofilms. Isopropanol treatment (IPA) was used as killing control. Mean CFU counts were performed on pools of three wells in at least three independent experiments. Error bars represent the standard error of the mean, while the dashed line indicates the lower limit of detection of the assay. (d) Biofilms of Escherichia coli K12 were treated in the same manner with increasing concentrations of either tobramycin alone or in combination with promethazine, and CFU were determined accordingly.
(TIF)
P. aeruginosa PA14 isogenic mutants, which exhibited an increase in the minimal inhibitory concentration (MIC indicated by the arrow) against ciprofloxacin due to a target mutation in gyrA (a), gyrA and parC (b) and gentamicin due to a gentamicin harboring resistance cassette transposon insertion (c), were grown in 96-well plates and treated with increasing concentrations of ciprofloxacin (a, b) and gentamicin (c) with or without the addition of 100 μM promethazine (PMZ). Colony forming units (CFU) were determined from the antibiotics treated (dark grey) and antibiotics plus PMZ treated (light grey) biofilms. Isopropanol treatment (IPA) was used as a killing control. Mean CFU counts were performed on pools of three wells in at least three independent experiments. Error bars represent the standard error of the mean, while the dashed line indicates the lower limit of detection of the assay.
(TIF)
(a) A P. aeruginosa PA14 pqsA transposon mutant was grown in 96-well plates and treated with increasing concentrations of tobramycin with or without the addition of 100 μM promethazine (PMZ). Colony forming units (CFU) were determined from the tobramycin treated (dark grey) and tobramycin plus PMZ treated (light grey) biofilms. Isopropanol treatment (IPA) was used as a killing control. External addition of PQS (100 μM) restored the killing activity of tobramycin/PMZ in the P. aeruginosa PA14 pqsA transposon mutant (b) and enhanced the killing activity of tobramycin/PMZ in E. coli K12 (c). Mean CFU counts were performed on pools of three wells in at least three independent experiments. Error bars represent the standard error of the mean, while the dashed line indicates the lower limit of detection of the assay.
(TIF)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
S.H. was funded by the EU (ERC Consolidator Grant COMBAT 724290) and received funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC 2155 “RESIST” – Project ID 39087428. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hall-Stoodley L, Stoodley P : Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol 2005, 13(1):7–10. 10.1016/j.tim.2004.11.004 [DOI] [PubMed] [Google Scholar]
- 2.Maunders E, Welch M : Matrix exopolysaccharides; the sticky side of biofilm formation. FEMS Microbiol Lett 2017, 364(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fong JNC, Yildiz FH : Biofilm Matrix Proteins. Microbiol Spectr 2015, 3(2). 10.1128/microbiolspec.MB-0004-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teschler JK, Zamorano-Sanchez D, Utada AS, Warner CJ, Wong GC, Linington RG, Yildiz FH : Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol 2015, 13(5):255–268. 10.1038/nrmicro3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costerton JW, Stewart PS, Greenberg EP: Bacterial biofilms: a common cause of persistent infections. Science 1999, 284(5418):1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- 6.de la Fuente-Nunez C, Reffuveille F, Fernandez L, Hancock RE: Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol 2013, 16(5):580–589. 10.1016/j.mib.2013.06.013 [DOI] [PubMed] [Google Scholar]
- 7.Romling U, Balsalobre C: Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med 2012, 272(6):541–561. 10.1111/joim.12004 [DOI] [PubMed] [Google Scholar]
- 8.Stewart PS: Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol 2002, 292(2):107–113. 10.1078/1438-4221-00196 [DOI] [PubMed] [Google Scholar]
- 9.Stewart PS: Antimicrobial Tolerance in Biofilms. Microbiol Spectr 2015, 3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart PS, Franklin MJ, Williamson KS, Folsom JP, Boegli L, James GA: Contribution of stress responses to antibiotic tolerance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 2015, 59(7):3838–3847. 10.1128/AAC.00433-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh S, Singh SK, Chowdhury I, Singh R: Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol J 2017, 11:53–62. 10.2174/1874285801711010053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall CW, Mah TF: Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev 2017, 41(3):276–301. 10.1093/femsre/fux010 [DOI] [PubMed] [Google Scholar]
- 13.Flemming HC, Wingender J: The biofilm matrix. Nat Rev Microbiol 2010, 8(9):623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 14.Limoli DH, Jones CJ, Wozniak DJ: Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol Spectr 2015, 3(3). 10.1128/microbiolspec.MB-0011-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pontes MH, Groisman EA: Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci Signal 2019, 12(592). 10.1126/scisignal.aax3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA: A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003, 426(6964):306–310. 10.1038/nature02122 [DOI] [PubMed] [Google Scholar]
- 17.Sauer K: The genomics and proteomics of biofilm formation. Genome Biol 2003, 4(6):219 10.1186/gb-2003-4-6-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beloin C, Ghigo JM: Finding gene-expression patterns in bacterial biofilms. Trends Microbiol 2005, 13(1):16–19. 10.1016/j.tim.2004.11.008 [DOI] [PubMed] [Google Scholar]
- 19.Crabbe A, Jensen PO, Bjarnsholt T, Coenye T: Antimicrobial Tolerance and Metabolic Adaptations in Microbial Biofilms. Trends Microbiol 2019. 10.1016/j.tim.2019.05.003 [DOI] [PubMed] [Google Scholar]
- 20.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T: Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 2008, 68(1):223–240. 10.1111/j.1365-2958.2008.06152.x [DOI] [PubMed] [Google Scholar]
- 21.Moradali MF, Ghods S, Rehm BH: Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front Cell Infect Microbiol 2017, 7:39 10.3389/fcimb.2017.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoiby N, Ciofu O, Johansen HK, Song ZJ, Moser C, Jensen PO, Molin S, Givskov M, Tolker-Nielsen T, Bjarnsholt T: The clinical impact of bacterial biofilms. Int J Oral Sci 2011, 3(2):55–65. 10.4248/IJOS11026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjarnsholt T, Ciofu O, Molin S, Givskov M, Hoiby N: Applying insights from biofilm biology to drug development—can a new approach be developed? Nat Rev Drug Discov 2013, 12(10):791–808. [DOI] [PubMed] [Google Scholar]
- 24.Roy R, Tiwari M, Donelli G, Tiwari V: Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9(1):522–554. 10.1080/21505594.2017.1313372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS: Extracellular DNA required for bacterial biofilm formation. Science 2002, 295(5559):1487 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- 26.Powell LC, Pritchard MF, Ferguson EL, Powell KA, Patel SU, Rye PD, Sakellakou SM, Buurma NJ, Brilliant CD, Copping JM et al. : Targeted disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate oligosaccharides. NPJ Biofilms Microbiomes 2018, 4:13 10.1038/s41522-018-0056-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen TB, Bjarnsholt T, Skindersoe ME, Hentzer M, Kristoffersen P, Kote M, Nielsen J, Eberl L, Givskov M: Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J Bacteriol 2005, 187(5):1799–1814. 10.1128/JB.187.5.1799-1814.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu C, Kirsch B, Zimmer C, de Jong JC, Henn C, Maurer CK, Musken M, Haussler S, Steinbach A, Hartmann RW: Discovery of antagonists of PqsR, a key player in 2-alkyl-4-quinolone-dependent quorum sensing in Pseudomonas aeruginosa. Chem Biol 2012, 19(3):381–390. 10.1016/j.chembiol.2012.01.015 [DOI] [PubMed] [Google Scholar]
- 29.Dingemans J, Al-Feghali RE, Lau GW, Sauer K: Controlling chronic Pseudomonas aeruginosa infections by strategically interfering with the sensory function of SagS. Mol Microbiol 2019, 111(5):1211–1228. 10.1111/mmi.14215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valentini M, Filloux A: Biofilms and Cyclic di-GMP (c-di-GMP) Signaling: Lessons from Pseudomonas aeruginosa and Other Bacteria. J Biol Chem 2016, 291(24):12547–12555. 10.1074/jbc.R115.711507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta K, Liao J, Petrova OE, Cherny KE, Sauer K: Elevated levels of the second messenger c-di-GMP contribute to antimicrobial resistance of Pseudomonas aeruginosa. Mol Microbiol 2014, 92(3):488–506. 10.1111/mmi.12587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varga B, Csonka A, Csonka A, Molnar J, Amaral L, Spengler G: Possible Biological and Clinical Applications of Phenothiazines. Anticancer Res 2017, 37(11):5983–5993. 10.21873/anticanres.12045 [DOI] [PubMed] [Google Scholar]
- 33.Jaszczyszyn A, Gasiorowski K, Swiatek P, Malinka W, Cieslik-Boczula K, Petrus J, Czarnik-Matusewicz B: Chemical structure of phenothiazines and their biological activity. Pharmacol Rep 2012, 64(1):16–23. 10.1016/s1734-1140(12)70726-0 [DOI] [PubMed] [Google Scholar]
- 34.Grimsey EM, Piddock LJV: Do phenothiazines possess antimicrobial and efflux inhibitory properties? FEMS Microbiol Rev 2019. 10.1093/femsre/fuz017 [DOI] [PubMed] [Google Scholar]
- 35.Melander RJ, Melander C: The Challenge of Overcoming Antibiotic Resistance: An Adjuvant Approach? ACS Infect Dis 2017, 3(8):559–563. 10.1021/acsinfecdis.7b00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baugh S, Phillips CR, Ekanayaka AS, Piddock LJ, Webber MA: Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J Antimicrob Chemother 2014, 69(3):673–681. 10.1093/jac/dkt420 [DOI] [PubMed] [Google Scholar]
- 37.Nzakizwanayo J, Scavone P, Jamshidi S, Hawthorne JA, Pelling H, Dedi C, Salvage JP, Hind CK, Guppy FM, Barnes LM et al. : Fluoxetine and thioridazine inhibit efflux and attenuate crystalline biofilm formation by Proteus mirabilis. Sci Rep 2017, 7(1):12222 10.1038/s41598-017-12445-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denis K, Le Bris M, Le Guennec L, Barnier JP, Faure C, Gouge A, Bouzinba-Segard H, Jamet A, Euphrasie D, Durel B et al. : Targeting Type IV pili as an antivirulence strategy against invasive meningococcal disease. Nat Microbiol 2019, 4(6):972–984. 10.1038/s41564-019-0395-8 [DOI] [PubMed] [Google Scholar]
- 39.Kristiansen JE, Mortensen I: Antibacterial effect of four phenothiazines. Pharmacol Toxicol 1987, 60(2):100–103. 10.1111/j.1600-0773.1987.tb01504.x [DOI] [PubMed] [Google Scholar]
- 40.Bettencourt MV, Bosne-David S, Amaral L: Comparative in vitro activity of phenothiazines against multidrug- resistant Mycobacterium tuberculosis. Int J Antimicrob Agents 2000, 16(1):69–71. 10.1016/s0924-8579(00)00199-0 [DOI] [PubMed] [Google Scholar]
- 41.El-Nakeeb MA, Abou-Shleib HM, Khalil AM, Omar HG, El-Halfawy OM: I n vitro antibacterial activity of some antihistaminics belonging to different groups against multi-drug resistant clinical isolates. Braz J Microbiol 2011, 42(3):980–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakrabarty AN, Acharya DP, Neogi D, Dastidar SG: Drug interaction of promethazine & other non-conventional antimicrobial chemotherapeutic agents. Indian J Med Res 1989, 89:233–237. [PubMed] [Google Scholar]
- 43.Molnar J, Batho N, Csik V, Chevalier J, Cremieux A: Interaction between tricyclic psychopharmacons and some antibiotics. Acta Microbiol Immunol Hung 1995, 42(3):277–285. [PubMed] [Google Scholar]
- 44.Gunics G, Motohashi N, Amaral L, Farkas S, Molnar J: Interaction between antibiotics and non-conventional antibiotics on bacteria. Int J Antimicrob Agents 2000, 14(3):239–242. 10.1016/s0924-8579(00)00131-x [DOI] [PubMed] [Google Scholar]
- 45.Lehtinen J, Lilius EM: Promethazine renders Escherichia coli susceptible to penicillin G: real- time measurement of bacterial susceptibility by fluoro-luminometry. Int J Antimicrob Agents 2007, 30(1):44–51. 10.1016/j.ijantimicag.2007.02.019 [DOI] [PubMed] [Google Scholar]
- 46.Abril MA, Michan C, Timmis KN, Ramos JL: Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol 1989, 171(12):6782–6790. 10.1128/jb.171.12.6782-6790.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG et al. : Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 2014, 111(20):E2100–2109. 10.1073/pnas.1401876111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Müsken M, Di Fiore S, Romling U, Haussler S: A 96-well-plate-based optical method for the quantitative and qualitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nat Protoc 2010, 5(8):1460–1469. 10.1038/nprot.2010.110 [DOI] [PubMed] [Google Scholar]
- 49.Wiegand I, Hilpert K, Hancock RE: Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 2008, 3(2):163–175. 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- 50.Pawar V, Komor U, Kasnitz N, Bielecki P, Pils MC, Gocht B, Moter A, Rohde M, Weiss S, Häussler S: In Vivo Efficacy of Antimicrobials against Biofilm-Producing Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy 2015, 59(8):4974–4981. 10.1128/AAC.00194-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pawar V, Crull K, Komor U, Kasnitz N, Frahm M, Kocijancic D, Westphal K, Leschner S, Wolf K, Loessner H et al. : Murine solid tumours as a novel model to study bacterial biofilm formation in vivo. J Intern Med 2014, 276(2):130–139. 10.1111/joim.12258 [DOI] [PubMed] [Google Scholar]
- 52.Crull K, Rohde M, Westphal K, Loessner H, Wolf K, Felipe-López A, Hensel M, Weiss S: Biofilm formation by Salmonella enterica serovar Typhimurium colonizing solid tumours. Cellular microbiology 2011, 13(8):1223–1233. 10.1111/j.1462-5822.2011.01612.x [DOI] [PubMed] [Google Scholar]
- 53.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y et al. : Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 2011, 334(6058):982–986. 10.1126/science.1211037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bartek IL, Reichlen MJ, Honaker RW, Leistikow RL, Clambey ET, Scobey MS, Hinds AB, Born SE, Covey CR, Schurr MJ et al. : Antibiotic Bactericidal Activity Is Countered by Maintaining pH Homeostasis in Mycobacterium smegmatis. mSphere 2016, 1(4). 10.1128/mSphere.00176-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lebeaux D, Chauhan A, Letoffe S, Fischer F, de Reuse H, Beloin C, Ghigo JM: pH-mediated potentiation of aminoglycosides kills bacterial persisters and eradicates in vivo biofilms. J Infect Dis 2014, 210(9):1357–1366. 10.1093/infdis/jiu286 [DOI] [PubMed] [Google Scholar]
- 56.Eilam Y: Membrane effects of phenothiazines in yeasts. I. Stimulation of calcium and potassium fluxes. Biochim Biophys Acta 1983, 733(2):242–248. 10.1016/0005-2736(83)90528-x [DOI] [PubMed] [Google Scholar]
- 57.Eilam Y: Effects of phenothiazines on inhibition of plasma membrane ATPase and hyperpolarization of cell membranes in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1984, 769(3):601–610. 10.1016/0005-2736(84)90059-2 [DOI] [PubMed] [Google Scholar]
- 58.Voskuil MI, Covey CR, Walter ND: Antibiotic Lethality and Membrane Bioenergetics. Adv Microb Physiol 2018, 73:77–122. 10.1016/bs.ampbs.2018.06.002 [DOI] [PubMed] [Google Scholar]
- 59.Ejim L, Farha MA, Falconer SB, Wildenhain J, Coombes BK, Tyers M, Brown ED, Wright GD: Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat Chem Biol 2011, 7(6):348–350. 10.1038/nchembio.559 [DOI] [PubMed] [Google Scholar]
- 60.Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS: Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother 2004, 48(7):2659–2664. 10.1128/AAC.48.7.2659-2664.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernier SP, Lebeaux D, DeFrancesco AS, Valomon A, Soubigou G, Coppee JY, Ghigo JM, Beloin C: Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet 2013, 9(1):e1003144 10.1371/journal.pgen.1003144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leyer GJ, Johnson EA: Acid adaptation induces cross-protection against environmental stresses in Salmonella typhimurium. Appl Environ Microbiol 1993, 59(6):1842–1847. 10.1128/AEM.59.6.1842-1847.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deb C, Lee CM, Dubey VS, Daniel J, Abomoelak B, Sirakova TD, Pawar S, Rogers L, Kolattukudy PE: A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One 2009, 4(6):e6077 10.1371/journal.pone.0006077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amato SM, Fazen CH, Henry TC, Mok WW, Orman MA, Sandvik EL, Volzing KG, Brynildsen MP: The role of metabolism in bacterial persistence. Front Microbiol 2014, 5:70 10.3389/fmicb.2014.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baek SH, Li AH, Sassetti CM: Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol 2011, 9(5):e1001065 10.1371/journal.pbio.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Castaneda-Garcia A, Do TT, Blazquez J: The K+ uptake regulator TrkA controls membrane potential, pH homeostasis and multidrug susceptibility in Mycobacterium smegmatis. J Antimicrob Chemother 2011, 66(7):1489–1498. 10.1093/jac/dkr165 [DOI] [PubMed] [Google Scholar]
- 67.Dotsch A, Becker T, Pommerenke C, Magnowska Z, Jansch L, Haussler S: Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009, 53(6):2522–2531. 10.1128/AAC.00035-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schurek KN, Marr AK, Taylor PK, Wiegand I, Semenec L, Khaira BK, Hancock RE: Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2008, 52(12):4213–4219. 10.1128/AAC.00507-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Bojer MS, George SE, Wang Z, Jensen PR, Wolz C, Ingmer H: Inactivation of TCA cycle enhances Staphylococcus aureus persister cell formation in stationary phase. Sci Rep 2018, 8(1):10849 10.1038/s41598-018-29123-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas VC, Kinkead LC, Janssen A, Schaeffer CR, Woods KM, Lindgren JK, Peaster JM, Chaudhari SS, Sadykov M, Jones J et al. : A dysfunctional tricarboxylic acid cycle enhances fitness of Staphylococcus epidermidis during beta-lactam stress. MBio 2013, 4(4). 10.1128/mBio.00437-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lobritz MA, Belenky P, Porter CB, Gutierrez A, Yang JH, Schwarz EG, Dwyer DJ, Khalil AS, Collins JJ: Antibiotic efficacy is linked to bacterial cellular respiration. Proc Natl Acad Sci U S A 2015, 112(27):8173–8180. 10.1073/pnas.1509743112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ: A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130(5):797–810. 10.1016/j.cell.2007.06.049 [DOI] [PubMed] [Google Scholar]
- 73.Haussler S, Becker T: The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog 2008, 4(9):e1000166 10.1371/journal.ppat.1000166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hazan R, Que YA, Maura D, Strobel B, Majcherczyk PA, Hopper LR, Wilbur DJ, Hreha TN, Barquera B, Rahme LG: Auto Poisoning of the Respiratory Chain by a Quorum-Sensing-Regulated Molecule Favors Biofilm Formation and Antibiotic Tolerance. Curr Biol 2016, 26(2):195–206. 10.1016/j.cub.2015.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weinstein EA, Yano T, Li LS, Avarbock D, Avarbock A, Helm D, McColm AA, Duncan K, Lonsdale JT, Rubin H: Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc Natl Acad Sci U S A 2005, 102(12):4548–4553. 10.1073/pnas.0500469102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amaral L, Cerca P, Spengler G, Machado L, Martins A, Couto I, Viveiros M, Fanning S, Pages JM: Ethidium bromide efflux by Salmonella: modulation by metabolic energy, pH, ions and phenothiazines. Int J Antimicrob Agents 2011, 38(2):140–145. 10.1016/j.ijantimicag.2011.03.014 [DOI] [PubMed] [Google Scholar]
- 77.Kaatz GW, Moudgal VV, Seo SM, Kristiansen JE: Phenothiazines and thioxanthenes inhibit multidrug efflux pump activity in Staphylococcus aureus. Antimicrob Agents Chemother 2003, 47(2):719–726. 10.1128/aac.47.2.719-726.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bailey AM, Paulsen IT, Piddock LJ: RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob Agents Chemother 2008, 52(10):3604–3611. 10.1128/AAC.00661-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sidrim JJ, Vasconcelos DC, Riello GB, Guedes GM, Serpa R, Bandeira TJ, Monteiro AJ, Cordeiro RA, Castelo-Branco DS, Rocha MF et al. : Promethazine improves antibiotic efficacy and disrupts biofilms of Burkholderia pseudomallei. Biofouling 2017, 33(1):88–97. 10.1080/08927014.2016.1262846 [DOI] [PubMed] [Google Scholar]
- 80.Lowrence RC, Subramaniapillai SG, Ulaganathan V, Nagarajan S: Tackling drug resistance with efflux pump inhibitors: from bacteria to cancerous cells. Crit Rev Microbiol 2019, 45(3):334–353. 10.1080/1040841X.2019.1607248 [DOI] [PubMed] [Google Scholar]
- 81.Spengler G, Kincses A, Gajdacs M, Amaral L: New Roads Leading to Old Destinations: Efflux Pumps as Targets to Reverse Multidrug Resistance in Bacteria. Molecules 2017, 22(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Du D, Wang-Kan X, Neuberger A, van Veen HW, Pos KM, Piddock LJV, Luisi BF: Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 2018, 16(9):523–539. 10.1038/s41579-018-0048-6 [DOI] [PubMed] [Google Scholar]
- 83.Van Acker H, Coenye T: The Role of Efflux and Physiological Adaptation in Biofilm Tolerance and Resistance. J Biol Chem 2016, 291(24):12565–12572. 10.1074/jbc.R115.707257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zaborskyte G, Andersen JB, Kragh KN, Ciofu O: Real-Time Monitoring of nfxB Mutant Occurrence and Dynamics in Pseudomonas aeruginosa Biofilm Exposed to Subinhibitory Concentrations of Ciprofloxacin. Antimicrob Agents Chemother 2017, 61(3). 10.1128/AAC.02292-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kvist M, Hancock V, Klemm P: Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl Environ Microbiol 2008, 74(23):7376–7382. 10.1128/AEM.01310-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alav I, Sutton JM, Rahman KM: Role of bacterial efflux pumps in biofilm formation. J Antimicrob Chemother 2018, 73(8):2003–2020. 10.1093/jac/dky042 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
P. aeruginosa static biofilms grown in 96-well plates were treated with increasing concentrations of ciprofloxacin (CIP) (a), ceftazidime (CAZ) (b) or gentamicin (GEN) (c) with or without the addition of 100 μM promethazine (PMZ). Colony forming units (CFU) were determined from the antibiotics treated (dark grey) and antibiotics plus PMZ treated (light grey) biofilms. Isopropanol treatment (IPA) was used as killing control. Mean CFU counts were performed on pools of three wells in at least three independent experiments. Error bars represent the standard error of the mean, while the dashed line indicates the lower limit of detection of the assay. (d) Biofilms of Escherichia coli K12 were treated in the same manner with increasing concentrations of either tobramycin alone or in combination with promethazine, and CFU were determined accordingly.
(TIF)
P. aeruginosa PA14 isogenic mutants, which exhibited an increase in the minimal inhibitory concentration (MIC indicated by the arrow) against ciprofloxacin due to a target mutation in gyrA (a), gyrA and parC (b) and gentamicin due to a gentamicin harboring resistance cassette transposon insertion (c), were grown in 96-well plates and treated with increasing concentrations of ciprofloxacin (a, b) and gentamicin (c) with or without the addition of 100 μM promethazine (PMZ). Colony forming units (CFU) were determined from the antibiotics treated (dark grey) and antibiotics plus PMZ treated (light grey) biofilms. Isopropanol treatment (IPA) was used as a killing control. Mean CFU counts were performed on pools of three wells in at least three independent experiments. Error bars represent the standard error of the mean, while the dashed line indicates the lower limit of detection of the assay.
(TIF)
(a) A P. aeruginosa PA14 pqsA transposon mutant was grown in 96-well plates and treated with increasing concentrations of tobramycin with or without the addition of 100 μM promethazine (PMZ). Colony forming units (CFU) were determined from the tobramycin treated (dark grey) and tobramycin plus PMZ treated (light grey) biofilms. Isopropanol treatment (IPA) was used as a killing control. External addition of PQS (100 μM) restored the killing activity of tobramycin/PMZ in the P. aeruginosa PA14 pqsA transposon mutant (b) and enhanced the killing activity of tobramycin/PMZ in E. coli K12 (c). Mean CFU counts were performed on pools of three wells in at least three independent experiments. Error bars represent the standard error of the mean, while the dashed line indicates the lower limit of detection of the assay.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.