Abstract
A novel coronavirus disease (COVID-19) caused by SARS-CoV2 has now spread globally. Replication/transcription machinery of this virus consists of RNA-dependent RNA polymerase (nsp12 or RdRp) and its two cofactors nsp7 and nsp8 proteins. Hence, RdRp has emerged as a promising target to control COVID-19. In the present study, we are reporting a novel inhibitor VTRM1.1 against the RdRp protein of SARS CoV2. A series of antivirals were tested for binding to the catalytic residues of the active site of RdRp protein. In-silico screening, molecular mechanics, molecular dynamics simulation (MDS) analysis suggest ribavirin, and remdesivir have good interaction with the binding site of the RdRp protein as compared to other antiviral investigated. Hence, ribavirin and remdesivir were used for the denovo fragments based antiviral design. This design, along with docking and MDS analysis, identified a novel inhibitor VTRM1 that has better interaction with RdRp as compared to their parent molecules. Further, to produce a lead-like compound, retrosynthetic analysis, and combinatorial synthesis were performed, which produces 1000 analogs of VTRM1. These analogs were analysed by docking and MDS analysis that identified VTRM1.1 as a possible lead to inhibit RdRp protein. This lead has a good docking score, favourable binding energy and bind at catalytic residues of the active site of RdRp. The VTRM1.1 also interacts with RdRp in the presence of RNA primer and other cofactors. It was also seen that, VTRM1.1 do not have off-target in human. Therefore, the present study suggests a hybrid inhibitor VTRM1.1 for the RNA-dependent RNA polymerase of SARS CoV2 that may be useful to control infection caused by COVID-19.
Keywords: Denovo designed antiviral, SARS-CoV2, RNA-dependent RNA polymerase (RdRp), Retrosynthetic analysis, Molecular dynamics simulation
1. Introduction
A novel coronavirus disease (Covid-19) caused by SARS-CoV2 [1] has now spread globally [2]. It has shifted its epicenter at different places across the globe and has now spread in more than 200 countries. On March 11, 2020, the world health organization (WHO) declared this outbreak a pandemic. On Apr 15, 2020 there were > 15,00,00 cumulative cases globally and > 90,000 death with ~5% mortality rate in outcome cases. The main symptom of this disease includes fever, shortness of breath, cough, and gastrostomy complications. Coronavirus employs a multi-subunit replication/transcription machinery. Different non-structural proteins (nsp) produced as a cleavage product of ORF1a and ORF1b viral polyproteins [3] that are assemble to facilitate viral replication and transcription. A key component of this machinery is nsp12 also known as RNA-dependent RNA polymerase (RdRp), and responsible for RNA synthesis. RNA synthesis of nsp12 in SARS CoV is activated by two other non-structural proteins nsp7 and nsp8 [4]. In addition to that, this complex (nsp12/nsp7/nsp8) associates with nsp14 and has shown 3′-5′ exoribonuclease (involved in replication fidelity) and RNA cap N-7-guanine methyltransferase activities (associated with 5’-RNA capping) in SARS-CoV [4]. This shows the importance of RdRp in the biology of SARS-CoV2. Hence, to control COVID-19 infection, there is an urgent need to find an inhibitor for RdRp. It is also reported that nsp12 (or RdRp) was considered as a primary target to control the infection of SARS-CoV by different inhibitors [5]. RdRp can be a potential therapeutic target to control the infection caused by SARS CoV2 as its human homolog of this nsp12 has not reported [6] therefore, therapeutic against nsp12 may be significant to control the COVID-19. The attempt has made to evaluate the FDA approved molecule like Ribavirin, Remdesivir, etc. as an inhibitor of RdRp [7]. Remdesivir, Ritonavir, and Lopinavir have been used to control other coronaviruses [8]. Remdesivir (GS-5734) is a broad-spectrum antiviral nucleotide prodrug with potent antiviral activity against diverse RNA viruses like SARS-CoV, MERS-CoV [9]. Designing of a new molecule takes a long time to develop it as effective antiviral, hence in the present study, we have designed a denovo synthesized VTRM1.1 from FDA approved antiviral molecules against SARS-CoV2 RNA-depended RNA polymerase using in-silico drug design, molecular mechanics, retrosynthetic analysis, combinatorial synthesis, and molecular dynamics simulation analysis.
2. Results
2.1. Receptor grid of RNA-dependent RNA polymerase (RdRp) was prepared targeted to the catalytic residue of the active site
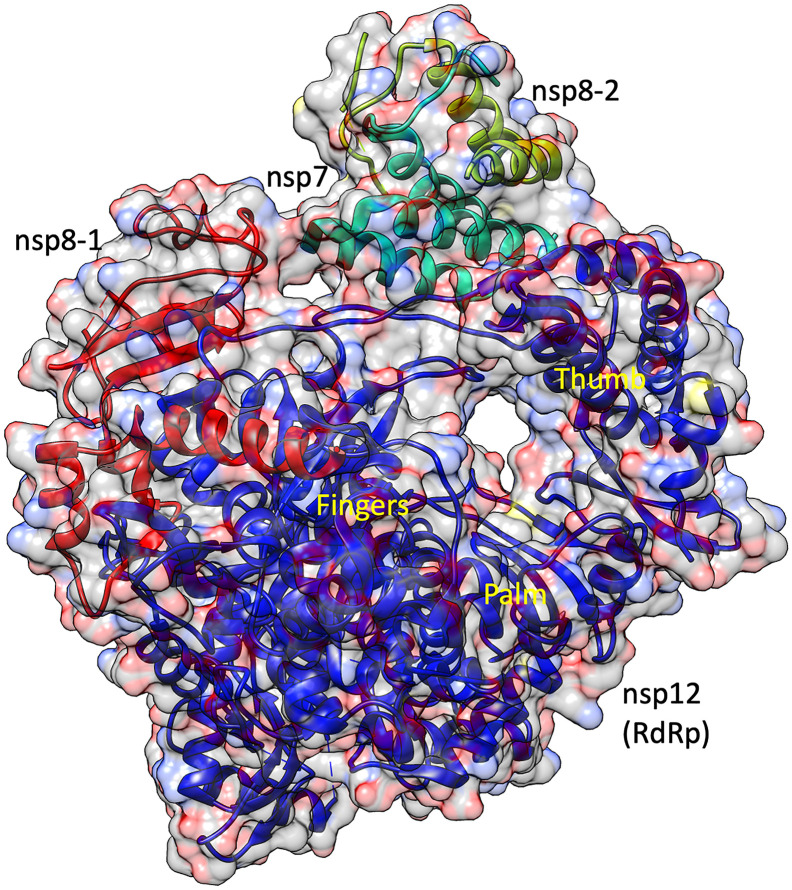
The PDB structure of RNA-dependent RNA polymerase (RdRp) was taken from RCSB (PDB number 6m71, resolution 2.9 Å). This PDB structure has four chains [10] including nsp12 (chain A), nsp7 (chain C), nsp8 (chain B and chain D). The nsp12 consists of the thumb, palm, and finger domain (Fig. 1 ). The PDB structure showed that the active site of RdRp domain involves residues like Asp618 of the motif A; Ser759, Asp760, and Asp761 of motif C. It was also seen that Asp618, Asp760 and Asp761 are involved in the binding with Mn2+; Arg555, Val557, Asp623, Thr680, Ser682, Asn691, and Asp760 are involved in binding of UDP and Remdesivir [10]. Hence, these residues Asp618, Ser759, Asp760, Asp761, Asp623, Arg555, Val557, Ser682, Asn691, Thr680 were used for receptor grid generation. This receptor gird was used to screen antiviral molecule targeting RdRp.
Fig. 1.
Binding sites of SARS-CoV2 RNA-dependent RNA polymerase (nsp12) complexed with cofactors nsp7 and nsp8. The figure shows the thumb, palm, and fingers domain of the nsp12 protein.
2.2. Selection of antiviral molecules
The antiviral molecules were selected based on the currently available literature search and their efficacy on the SARS-CoV2. The selected antiviral drugs in the present study were remdesivir, ribavirin, favipiravir, ritonavir, umifenovir, hydroxychloroquine, ascorbate, oseltamivir, tenofovir, and acetyl-serine. The ligand preparation of these antiviral molecules has resulted in the 87 tautomers, and prepared ligands were investigated for its interaction with the binding site of RdRp.
2.3. Virtual screening showed that ribavirin, and remdesivir have best docking in the active site of RdRp protein
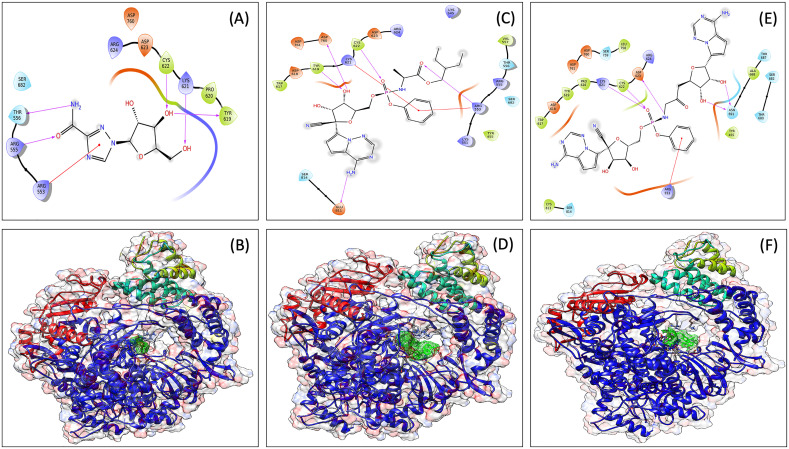
We have performed docking studied in HTVS, SP, and XP mode. Docking results suggested that among all the antiviral molecules investigated, ribavirin (G-score, −6.109 kcal/mol) and remdesivir (G-score, −6.038 kcal/mol) have shown better interaction with the active site of RdRp (Supplementary Table ST1). The docking poses are shown in Fig. 2 , and docking scores, along with binding energies, are shown in Table 1 . The residues of RdRp protein that interact with individual antiviral molecules are different for both antiviral molecules. Ribavirin involves Arg553, Arg555, Thr556, Ser682, Arg624, Asp623, Cys622, Lys621, Pro620, Tyr619 and Asp760 (Fig. 2A) while remdesivir interacts with Trp617, Ap618, Tyr619, Lys621, Cys622, Asp623, Ard624, Asp760, Asp761, Lys545, Lys551, Arg553, Arg555, Thr556, Val557, Ser682, Tyr455, Glu811, and Ser814 (Fig. 2C). It is also seen in the present study that the interaction of both the antiviral involves the crucial amino acid of RdRp that are involved in the catalysis of this enzyme.
Fig. 2.
Interaction diagram showing interacting amino acid residues and their docking pose in SARS-CoV2 RdRp. RdRp-Ribavirin complex (A and B), RdRp-Remdesivir complex (C and D), and RdRp-VTRM1 complex (E and F).
Table 1.
Result showing outcome of GLIDE molecular docking in XP mode and Binding free energies result from Prime analysis using MMGBSA approach. The docking was performed using Grid of core residues of active site of RNA-dependent RNA polymerase.
| Antiviral molecules | Docking score (in Kcal/mol) |
Glide E-model (in kcal/mol) |
Binding gibbs free energy change (in kcal/mol) |
|---|---|---|---|
| Ribavarin | −6.11 | −39.97 | −24.87 |
| Remesdivir | −6.04 | −72.07 | −41.04 |
| VTRM1 | −7.95 | −99.57 | −34.44 |
| VTRM1.1 | −7.24 | −79.89 | −53.39 |
| VTRM1.2 | −5.28 | −48.47 | −36.63 |
| GTP | −6.42 | −116.24 | −36.12 |
| ATP | −6.12 | −98.25 | −19.99 |
2.4. Remdesivir and ribavirin have favourable binding free energy with RdRp
All the antivirals were undergone molecular mechanics analysis with generalized born and surface area solvation (MM-GBSA) methods to calculate the Gibbs free energy of binding. The result of this analysis is listed in Table 1. It was found that all the molecules investigated, have favourable Gibbs free energy change for binding to RdRp. Antivirals were first filtered by docking scores, followed by Gibbs free energy if their docking scores were similar. Based on docking and binding free energy results, remdesivir and ribavirin are selected for the further study. As these two molecules are nucleotide analog, hence the interaction of different NTP and dNTPs with the RdRp was also monitored. The result showed that Remdesivir has better binding energy as compared to native nucleotides (ATP, GTP, CTP, UTP), while ribavirin has better binding energy than NTPs like ATP (Supplementary Table ST2).
2.5. Denovo fragment-based designing and MDS analysis identified VTRM1 as possible lead targeting the active site of RdRp
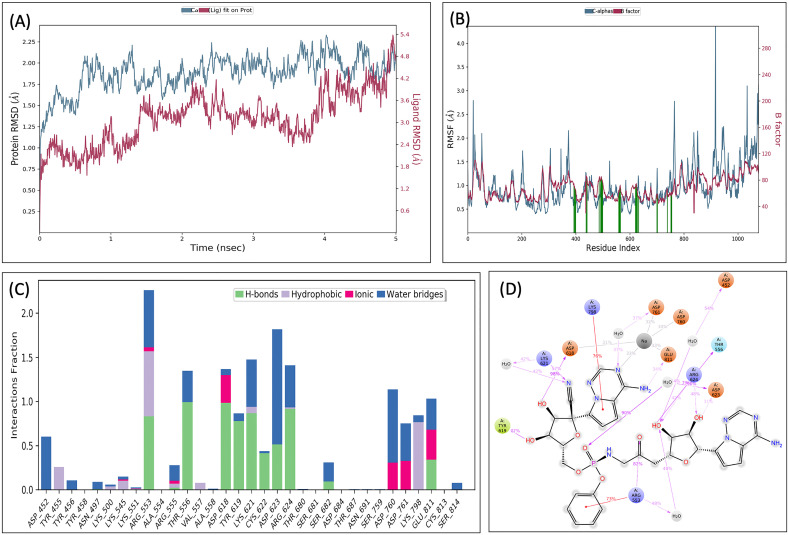
Denovo fragment-based drug designing was started with the fragmentation of the selected molecules, i.e., ribavirin, and remdesivir. A total of 93 fragments were produced that includes 5 fragments of ribavirin, and 88 fragments of remdesivir. All the 93 fragments were docked (in SP mode) to the grid of binding site of RdRp protein. Based on manual analysis of docked complexes, different fragments (fragment 4 and 5 of ribavirin; fragment 60, 61, 62, 63, 68, 71, 73, 75, 76, 86, and 87 of remdesivir) were selected for denovo fragment-based design using Breed. The breed produces 14 combinations. These 14 combinations, along with two-parent molecules (remdesivir and ribavirin), were docked (in XP mode) to the binding site of RdRp protein. That selects the denovo hybrid Breed of fragment 60 and 73 of remdesivir, which has the best docking (G-score, −7.953 kcal/mol). This docking score was found to be higher than their parent molecules (Table 1) and named as ‘VTRM1’, and chemically (2R,3R,4S,5R)-2-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-({[(S)-({3-[(2R,3S,4R,5S)-5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-3,4-dihydroxyoxolan-2-yl]-2-oxopropyl}amino)(phenoxy)phosphoryl]oxy}methyl)-3,4-dihydroxyoxolane-2‑carbonitrile (Fig. 2E and F). To further confirm the interaction, the MDS analysis of the VTRM1-RdRp protein complex was performed. The result showed that RMSD of RdRp protein (in ligand complex state) was found to be <2 Å, and ligand's RMSD is less than <4.8 Å, that showed stable complex (Fig. 3A). RMSF analysis showed that most of the protein (except some part) has the RMSF <2 Å that showed stable protein conformation (Fig. 3B). Further, the interaction between VTRM1 and RdRp protein was investigated and found that it involves Asp452, Arg553, Thr556, Asp618, Tyr619, Lys621, Cys622, Asp623, Arg624, Asp760, Asp761, Lys798, Glu811, and at least nine contacts always exist more than 30% simulation time (Fig. 3D).
Fig. 3.
Root-mean-square deviation (A) and Root mean square fluctuations (B), Interacting residues (C), and the interacting fraction (D) during molecular dynamics simulation analysis of RdRp-VTRM1 complex.
2.6. Retrosynthetic analysis, combinatorial synthesis, and MDS analysis identified VTRM1.1 as an antiviral targeted to the binding site of RdRp protein
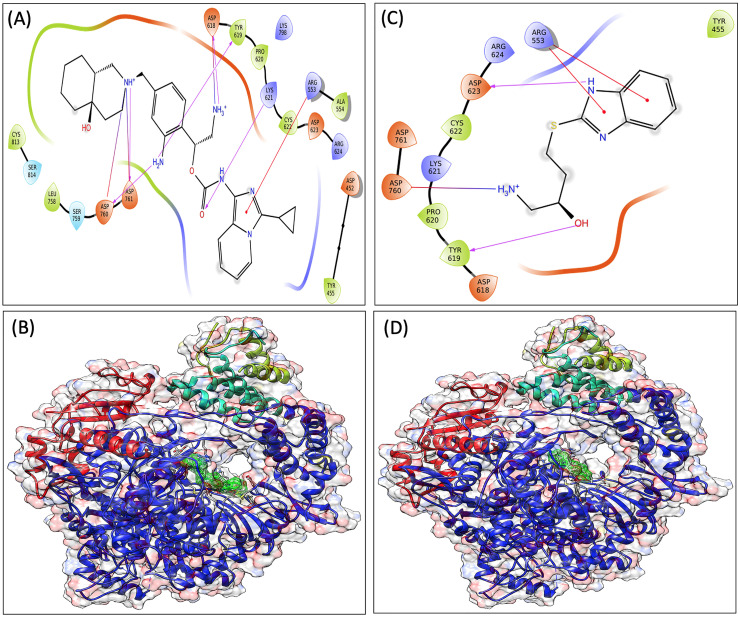
To produce lead-like compounds, retrosynthetic analysis, and combinatorial synthesis was performed that produced 1000 analogs of VTRM1 by more than 100 modifications. The 1000 products were prepared and docked to the active site of RdRp protein. Docking and binding energy calculation identified VTRM1.1 as best docked molecule (G-score, −7.239 kcal/mol) among different modified products, and identified as (1R)-1-(4-{[(4aR,8aR)-4a-hydroxy-decahydroisoquinolin-2-yl]methyl}-2-aminophenyl)-2-aminoethyl-N-{3-cyclopropylimidazo[1,5-a]pyridin-1-yl}carbamate (Fig. 4A and B). The second molecule has been named VTRM1.2 and chemically; it is (2R)-1-amino-4-(1H-1,3-benzodiazol-2-ylsulfanyl)butan-2-ol (Fig. 4C and D).
Fig. 4.
Interaction diagram showing interacting amino acid residues and their docking pose in SARS-CoV2 RdRp with denovo synthesized antiviral showing RdRp-VTRM1.1 complex (A and B), RdRp-VTRM1.2 complex (C and D).
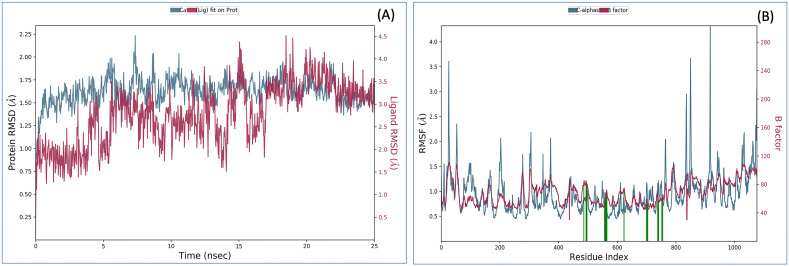
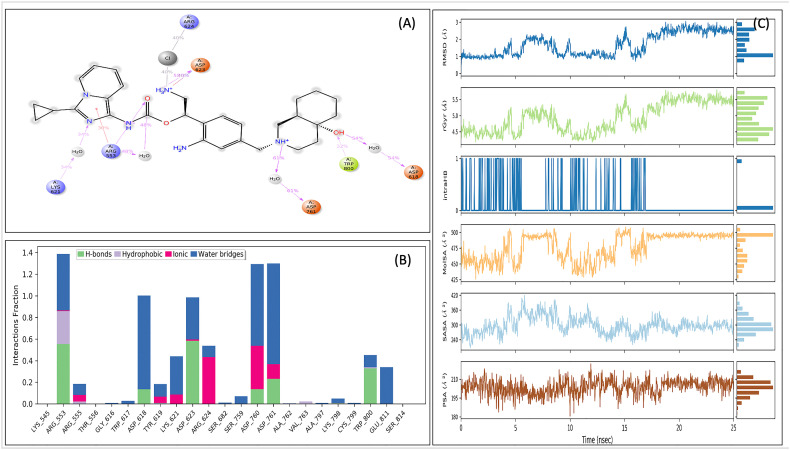
To further confirm the interaction, the MDS analysis of the VTRM1.1-RdRp (Fig. 5 ) and VTRM1.2-RdRp complex (Supplementary figure SF1) was performed. The MDS result of VTRM1.1-RdRp complex showed that RMSD of protein (in ligand complex state) is <2 Å and ligand (in protein complex state) have RMSD <4 Å that showed a very stable complex (Fig. 5A). RMSF analysis showed that most of the protein has the RMSF <2.5 Å that showed stable protein conformation (Fig. 5B). Interaction between VTRM1.1 and RdRp protein involves at least nine contacts that always exist more than 30% simulation time (Fig. 6A) and involves Arg553, Asp618, Lys621, Asp623, Arg624, Asp760, Asp761, Trp800, Glu811 amino acid residues (Fig. 6B). The ligand has a small deviation (RMSD <2.5 Å) during the simulation with reference to its conformation at the start of the simulation (Fig. 6C). Ligand has only one intramolecular hydrogen bonding till 18 ns after that there is no intramolecular bonding, which further suggests stable conformation of ligands. The van der Waals surface area (MolSA), solvent accessible surface area (SASA), polar surface area (PSA) also supported the good interaction between ligand and RdRp (Fig. 6C).
Fig. 5.
Root-mean-square deviation (A) and Root mean square fluctuations (B), during molecular dynamics simulation analysis of RdRp-VTRM1.1 complex.
Fig. 6.
Interacting fraction (A), interacting residues (B), properties of ligand (C), during molecular dynamics simulation analysis of RdRp-VTRM1.1 complex.
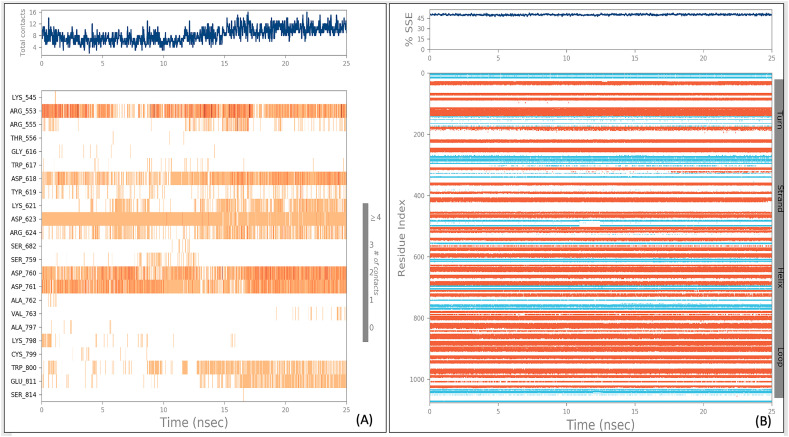
In Fig. 7A, the top panel shows the total number of specific contacts of protein with ligand throughout the trajectory. The bottom panel shows interacting residues of protein with ligand in each trajectory frame (Fig. 7A). This confirms a good interaction of VTRM1.1 with catalytically active amino acid residues like Asp760, Asp761, Asp623 of RdRp. In Fig. 7B, the secondary structure element (SSE) composition for each trajectory frame throughout the simulation are shown, and the plot at the bottom monitors each residue and its SSE assignment over time (Fig. 7). The result confirms that the SSE remains very similar throughout the simulation that confirm the stable interaction.
Fig. 7.
Representation of the interactions and contacts (H-bonds, Hydrophobic, Ionic, Water bridges) between VTRM1.1 with RdRp protein (A), and protein secondary structure elements (SSE) during simulation of RdRp-VTRM1.1 complex (B).
2.7. VTRM1.1 has better ADMET properties than VTRM1
In addition to this, ADMET analysis of all the selected lead molecules, i.e., VTRM1, VTRM1.1, and VTRM1.2 were performed, and their results are shown in Table 2 . The comparative analysis showed that although VTRM1 has the best docking score but it has bad ADMET properties that make it a weak lead. The VTRM1.1 has better ADMET parameters and has the docking and binding energy very close to VTRM1; hence VTRM1.1 is suggested as a possible inhibitor for RdRp of SARS CoV2.
Table 2.
Result showing outcome of ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity)analysis. The prediction was performed using QikProp analysis.
| ADMAT properties (normal range) |
VTRM1 | VTRM1.1 | VTRM1.2 |
|---|---|---|---|
| mol MW (130 to 725) | 736.636 | 518.658 | 237.319 |
| donorHB (1 to 6) | 9 | 5.5 | 4 |
| accptHB (2 to 20) | 24.75 | 8.75 | 4.2 |
| QPpolrz (13–70) | 63.451 | 53.612 | 63.451 |
| QPlogPC16 (4 to 18) | 23.978 | 17.059 | 8.978 |
| QPlogPoct (8 to 35) | 50.513 | 31.124 | 15.621 |
| QPlogPw (4 to 45) | 41.519 | 19.223 | 11.593 |
| QPlogPo/w (−2 to 6.5) | −1.965 | 2.812 | 1.008 |
| QPlogS (−6.5 to 0.5) | −3.941 | −3.492 | −1.817 |
| QPlogBB (−3 to 1.2) | −5.156 | −1.006 | −0.596 |
| QPlogKp (−8 to −1) | −6.038 | −7.192 | −4.7 |
| QPlogKhsa | −1.524 | 0.557 | −0.486 |
| Human Oral Absorption (2 to 3) | 1 | 2 | 3 |
| Percent Human Oral Absorption | 0 | 37.366 | 70.8 |
| Rule Of Five | 3 | 2 | 0 |
| Rule Of Three | 2 | 2 | 0 |
| QPlogHERG | −7.305 | −7.303 | −5.61 |
| QPPCaco | 1.189 | 12.888 | 131.943 |
2.8. VTRM1.1 interacts with RdRp protein in the presence of RNA primer and other cofactors
To further validate the biological relevance of the designed lead, the interaction of the VTRM1.1 with RdRp was monitored in the presence of RNA primer and other cofactors. The result showed that the binding of VTRM1.1 with RdRp is favourable (−51.67 kcal/mol) in the presence of RNA and other metal cofactors (Supplementary Table ST3) that further supports the biological significance of the lead molecule.
2.9. VTRM1.1 has no human off-targets
Presence of lead's off-targets in human may reduce the efficacy and produce the side effect. Hence, human off-targets of VTRM1.1 was predicted using Swiss Target Prediction. The result showed that the designed lead VTRM1.1 do not possess any off-targets in human. This further enhances the significance of VTRM1.1 as a possible lead against the RdRp.
3. Discussion
SARS-CoV2 has emerged as a lethal pathogen causing a global pandemic. No specific FDA approved treatment is available for COVID-19. Some non-specific treatments like remdesivir, ribavirin, etc. [11] are used. Ribavirin is a guanosine analog that has been reported to have multiple mechanisms of action [12] and has also shown to have an inhibitory effect against SARS-CoV2 [13]. Remdesivir (GS-5734) is currently used with lopinavir against COVID-19 without knowing its specific targets in SARS-CoV2 [14]. The essential components of the replication/transcription machinery of SARS CoV2 are RNA-dependent RNA polymerase (nsp12 or RdRp) along with its two cofactors nsp7 and nsp8. In addition to this, this complex nsp12/nsp7/nsp8 binds with nsp14, which then modulates replication fidelity and 5’-RNA capping in SARS-CoV [4]. Human homologs of nsp12 have not reported [6], hence, RdRp can be a potential therapeutic target to control the infection caused by SARS CoV2. In the present study, a novel inhibitor VTRM1.1 targeting to RdRp was designed using denovo designing, retrosynthetic analysis, and combinatorial synthesis, and molecular dynamic simulation.
PDB structure of RdRp was retrieved from the RCSB, and the grid was prepared at an active site that binds with metal and nucleoside-diphosphate (NDP). The docking, binding energy calculation, and molecular dynamic simulation calculation have shown that ribavirin and remdesivir have good interactions with the active site of RdRp. Various in-vitro experimental data suggest that the ribavirin has shown an inhibitory effect on SARS-CoV2 [13]. Similarly, remdesivir effectivity inhibits the 2019-nCoV in-vitro [15] by delayed chain termination and inhibits RNA synthesis [16]. Recently, it is also seen that it incorporate into the primer stand at the first replicated base pair and terminates chain elongation [17]. An in-silico study on the modeled RdRp has also shown that the ribavirin and remdesivir has the best interaction with RdRp among the approved antiviral investigated [18].
To find a novel inhibitor, we have used denovo fragment-based drug design using FDA approved antiviral molecules ribavirin and remdesivir that showed direct interaction with the active site of RdRp of SARS CoV2. A total of 93 fragments were generated from these two molecules. They were docked to the binding site of RdRp, and best-docked fragments were manually analysed of its interaction with RdRp. The selected fragments were joined via denovo fragment-based drug design. This approach joined the different fragments based on the structural similarity to the active molecules and their scaffolds, hence used to navigate a considerable chemical space [19]. The hybrid antiviral was further confirmed for its interaction with the active site of RdRp using the XP-mode of docking and molecular dynamics simulations. This select VTRM-1 as a possible denovo synthesized hybrid molecule that can interact with RdRp and involves Asp618, Asp760, Asp761 amino acid residue that is important for the catalysis of this protein [10]. VTRM1 has many ADMET parameters outside its permissible limit; hence, it was modified.
Optimization of lead required the synthesis and validation of thousands of lead's analogs before clinical candidate nomination, which takes a longer time. Further, to explore possible improvement in the VTRM1, we have used in-silico based retrosynthetic analysis, combinatorial synthesis, and MDS analysis to design an improved new antiviral molecule. Retrosynthetic analysis followed by combinatorial synthesis generates 1000 analogs of the denovo designed hybrid molecules (VTRM1) and analysed for its interaction with the active site of RdRp. ADMET properties is very important for the selection of drug candidates. VTRM1.1 has shown docking, and binding free energy value close to VTRM1, but its ADMET properties are better than VTRM1; hence VTRM1.1 is identified as better lead than VTRM1. MDS analysis confirms the interaction of VTRM1.1 with the active site of RdRp. It involves the catalytically critical residue (Asp760 and Asp761) of the active site of this protein that is important for the binding metal ion and catalysis. This showed that modification by retro-combinatorial synthesis does not alter the docking and binding energy of the denovo synthesized molecule VTRM1.1 but improves ADMET properties that enhance the drug-likeness nature of the designed units. The in-vivo condition involves the presence of RNA in the active site of RdRp hence we have further validated hybrid leads for its interaction with RdRp in the presence of RNA and metal cofactors. The result confirms the interaction between VTRM1.1 and RdRp in the presence of an RNA molecule. Hence, the present study reports denovo designing, retrosynthetic analysis, and combinatorial synthesis, molecular dynamic simulation based design of an novel antiviral VTRM1.1 that may be a possible inhibitor of RNA dependent RNA polymerase of SARS CoV2. This molecule VTRM1.1 need to be synthesized and experimentally tested against SARS-CoV, in animal and humans before been use as therapeutics.
4. Methods
4.1. Retrieval of the structure of RNA-depended RNA polymerase
The PDB structure of SARS-CoV2 RNA-depended RNA polymerase (RdRp) is available hence retrieved from RCSB (PDB number 6M71, resolution 2.9 Å). The structure was pre-processed by assign bond orders, add hydrogens, create zero-order bonds for metals, create disulfide bonds, optimise for water orientation and pH, and reduce for converge heavy atoms to 0.3RMSD using OPLS_2005 force field. The residues Asp618, Ser759, Asp760, Asp761, Asp623, Arg555, Val557, Ser682, Asn691, Thr680 are used for receptor grid generation. This receptor gird was used to screen several antiviral molecules.
4.2. Ligand preparation
The antiviral currently prescribed for COVID-19 [8,14,11,5] were selected in the present study. The selected antiviral drugs were remdesivir, ribavirin, favipiravir, ritonavir, umifenovir, hydroxychloroquine, ascorbate, oseltamivir, tenofovir, acetyl-serine. The SDF structure of the antiviral molecules was downloaded from the PubChem database. These SDF structures, along with denovo design hybrid antiviral leads, were prepared by using LigPrep modules of the Schrodinger as per our published protocol [20].
4.3. Virtual screening of antiviral on the RNA-depended RNA polymerase
All the prepared ligands were used for virtual testing of antiviral that docked to the grid of RNA-depended RNA polymerase as per our published protocol [21]. The docking score predicts the binding affinity between the target protein and particular pose of ligand docked. E-model is used to compare conformers of same ligand based on electrostatic and van der Waals energies. The virtual screening was performed in three modes of docking i.e., HTVS, SP, and XP mode [22]. HTVS and SP docking use a series of hierarchical filters to search for possible locations of the ligand in the binding site of the receptor. HTVS and SP use the same scoring function, but HTVS reduces the number of intermediate conformations in docking and also reduces the thoroughness of final torsional refinement and sampling. In contrast, Extra Precision (XP) mode employs an anchor and grow sampling approach for the docking.
4.4. Molecular mechanics/generalized born surface area (MM-GBSA) calculations of selected library compounds
Binding free energy is the sum of total intermolecular interactions that is present between ligand and protein. To get more accurate interaction, XP docked antiviral molecules with RNA-dependent RNA polymerase were further subjected to molecular mechanics with generalized born surface area (MM-GBSA) calculations using the Prime module of Schrodinger as per our published methods [23].
4.5. Molecular dynamics simulation (MDS) analysis
MDS was performed using Desmond modules of the Schrodinger 2019–4 as per published methods [24] using the OPLS3e force field. The system was built for the protein-ligand complex using the TIP3P solvent model; sodium ion was added to make charge-neutral, 0.15 M NaCl was added to make the system close to the natural system. The simulation was run for 5 ns (or 25 ns for VTRM1.1.-RdRp complex), with 5 ps trajectory recording intervals. System energy was set to be 1.2, and the ensemble class used was NPT. The simulation was set to run at 300 k at 1.01325 bar. The option to relax the system before simulation was selected. The simulated system was analysed for the simulation interaction diagram.
4.6. Denovo fragment-based drug design
De-novo fragment-based drug designing is a newly emerged approach [19]. The different fragments of selected antiviral molecules were generated. The various fragments of ligands were docked into the binding site of the RdRp. The docked complex was manually analysed for its position of docking in the binding site. Different fragments (fragments 4 and 5 of ribavirin; fragment 60, 61, 62, 63, 68, 71, 73, 75, 76, 86, and 87 of remdesivir were selected based on their interaction to the binding site of the RdRp. The selected fragments were joined using the Breed module to generate newly denovo designed antiviral molecules. The denovo designed antiviral molecules were further confirmed for its interaction with the binding site of the RdRp using XP-docking and molecular dynamics simulations analysis.
4.7. Retrosynthetic analysis and combinatorial synthesis
To produce synthetically tractable lead-like compounds, retrosynthetic analysis, and combinatorial synthesis were performed for the denovo synthesized VTRM.1 as per the published method using PathFinder [25]. PathFinder can incorporate more than 100 reactions like C—C bond formation that are required for the molecular scaffolds and drug discovery [26]. A total of 100 number of pathways are investigated to produce 1000 products. The top 10% product that is similar to the input molecule VTRM.1 are selected for further analysis. The selected 1000 products were analysed by docking, and molecular dynamics simulations analysis to select the antiviral lead.
4.8. ADMET analysis of the selected lead
The ADMET analysis was performed that identified absorption distribution, metabolism, excretion, and toxicity of the leads molecule using QikProp as per published protocol [27].
4.9. Validation of lead for its interaction with the RdRp-RNA complex
The interaction of lead with RdRp in the presence of cofactors has a biological relevance, hence the PDB structure of RdRp complexed with primer RNA, cofactors, and remdesivir was retrieved from RCSB (PDB number 7BV2, resolution 2.5 Å). The remdesivir was removed from the complex, and the structure was pre-processed, optimised, minimised to 0.3RMSD using the OPLS_2005 force field. The residues Asp618, Ser759, Asp760, Asp761, Asp623, Arg555, Val557, Ser682, Asn691, Thr680 are used for receptor grid generation. This receptor gird was used to validate the selected leads.
4.10. Identification of human off-targets of designed lead VTRM1.1
The human off-targets of the designed lead was predicted using Swiss Target Prediction [28] using the published protocol [29].
Author Contributions
Conceived and designed the experiments: V.T., Performed the experiments: V.T., Analysed the data: V.T., Wrote the manuscript: V.T., Proofread of the final version: V.T.
Declaration of competing interest
The author have declared that no competing interests exist. The experiment was performed in the absence of any financial support.
Acknowledgments
Acknowledgments
I would like to thank the Central University of Rajasthan for providing the Schrodinger suite. I would like to thank Monalisa Tiwari for proofreading the manuscript.
Ethical approval
The present study does not involve human and animal samples.
Data availability
All the data are available in the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2020.12.223.
Appendix A. Supplementary data
Supplementary material
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L., Samborskiy D., Sidorov I.A., Sola I., Ziebuhr J. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. 2014;111(37) doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheahan T.P., Sims A.C., Leist S.R., Schafer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11(1):222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9(396) doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh A.K., Singh A., Shaikh A., Singh R., Misra A. Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: a systematic search and a narrative review with a special reference to India and other developing countries. Diabetes Metab Syndr. 2020;14(3):241–246. doi: 10.1016/j.dsx.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loustaud-Ratti V., Debette-Gratien M., Jacques J., Alain S., Marquet P., Sautereau D., Rousseau A., Carrier P. Ribavirin: past, present and future. World J. Hepatol. 2016;8(2):123–130. doi: 10.4254/wjh.v8.i2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khalili J.S., Zhu H., Mak N.S.A., Yan Y., Zhu Y. Novel coronavirus treatment with ribavirin: groundwork for an evaluation concerning COVID-19. J. Med. Virol. 2020;92(7):740–746. doi: 10.1002/jmv.25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choy K.T., Yin-Lam Wong A., Kaewpreedee P., Sia S.F., Chen D., Yan Hui K.P., Wing Chu D.K., Wai Chan M.C., Pak-Hang Cheung P., Huang X., Peiris M., Yen H.L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;104786 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295(20):6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.W. Yin, C. Mao, X. Luan, D.-D. Shen, Q. Shen, H. Su, X. Wang, F. Zhou, W. Zhao, M. Gao, S. Chang, Y.-C. Xie, G. Tian, H.-W. Jiang, S.-C. Tao, J. Shen, Y. Jiang, H. Jiang, Y. Xu, S. Zhang, Y. Zhang, H.E. Xu, Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir, Science (New York, N.Y.) 368(6498) (2020) 1499–1504. [DOI] [PMC free article] [PubMed]
- 18.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai K., Nagata N., Takahashi Y. De novo design of drug-like molecules by a fragment-based molecular evolutionary approach. J. Chem. Inf. Model. 2014;54(1):49–56. doi: 10.1021/ci400418c. [DOI] [PubMed] [Google Scholar]
- 20.V. Tiwari, V. Patel, M. Tiwari, In-silico screening and experimental validation reveal L-adrenaline as anti-biofilm molecule against biofilm-associated protein (Bap) producing Acinetobacter baumannii, Int J Biol Macromol 107(Pt A) (2018) 1242–1252. [DOI] [PubMed]
- 21.Verma P., Tiwari V. Targeting outer membrane protein component AdeC for the discovery of efflux pump inhibitor against AdeABC efflux pump of multidrug resistant Acinetobacter baumannii. Cell Biochem. Biophys. 2018;76(3):391–400. doi: 10.1007/s12013-018-0846-5. [DOI] [PubMed] [Google Scholar]
- 22.R.A. Friesner, J.L. Banks, R.B. Murphy, T.A. Halgren, J.J. Klicic, D.T. Mainz, M.P. Repasky, E.H. Knoll, M. Shelley, J.K. Perry, D.E. Shaw, P. Francis, P.S. Shenkin, Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy, Journal of Medicinal Chemistry 47(7) (2004) 1739-1749. [DOI] [PubMed]
- 23.Tiwari V., Rajeswari M.R., Tiwari M. Proteomic analysis of iron-regulated membrane proteins identify FhuE receptor as a target to inhibit siderophore-mediated iron acquisition in Acinetobacter baumannii. Int. J. Biol. Macromol. 2019;125:1156–1167. doi: 10.1016/j.ijbiomac.2018.12.173. [DOI] [PubMed] [Google Scholar]
- 24.Wright W.C., Chenge J., Wang J., Girvan H.M., Yang L., Chai S.C., Huber A.D., Wu J., Oladimeji P.O., Munro A.W., Chen T. Clobetasol propionate is a heme-mediated selective inhibitor of human cytochrome P450 3A5. J. Med. Chem. 2020;63(3):1415–1433. doi: 10.1021/acs.jmedchem.9b02067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konze K.D., Bos P.H., Dahlgren M.K., Leswing K., Tubert-Brohman I., Bortolato A., Robbason B., Abel R., Bhat S. Reaction-based enumeration, active learning, and free energy calculations to rapidly explore synthetically tractable chemical space and optimize potency of cyclin-dependent kinase 2 inhibitors. J. Chem. Inf. Model. 2019;59(9):3782–3793. doi: 10.1021/acs.jcim.9b00367. [DOI] [PubMed] [Google Scholar]
- 26.G.W. Bemis, M.A. Murcko, The properties of known drugs. 1. Molecular frameworks, J Med Chem 39(15) (1996) 2887–93. [DOI] [PubMed]
- 27.Tiwari M., Panwar S., Kothidar A., Tiwari V. Rational targeting of Wzb phosphatase and Wzc kinase interaction inhibits extracellular polysaccharides synthesis and biofilm formation in Acinetobacter baumannii. Carbohydr. Res. 2020;492:108025. doi: 10.1016/j.carres.2020.108025. [DOI] [PubMed] [Google Scholar]
- 28.Daina A., Michielin O., Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W364. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiwari V., Meena K., Tiwari M. Differential anti-microbial secondary metabolites in different ESKAPE pathogens explain their adaptation in the hospital setup. Infect. Genet. Evol. 2018;66:57–65. doi: 10.1016/j.meegid.2018.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
All the data are available in the manuscript.