Abstract
An exponential rise in studies regarding the association among human gut microbial communities, human health, and diseases is currently attracting the attention of researchers to focus on human gut microbiome research. However, even with the ever-growing number of studies on the human gut microbiome, translation into improved health is progressing slowly. This hampering is due to the complexities of the human gut microbiome, which is composed of >1,000 species of microorganisms, such as bacteria, archaea, viruses, and fungi. To overcome this complexity, it is necessary to reduce the gut microbiome, which can help simplify experimental variables to an extent, such that they can be deliberately manipulated and controlled. Reconstruction of synthetic or established gut microbial communities would make it easier to understand the structure, stability, and functional activities of the complex microbial community of the human gut. Here, we provide an overview of the developments and challenges of the synthetic human gut microbiome, and propose the incorporation of multi-omics and mathematical methods in a better synthetic gut ecosystem design, for easy translation of microbiome information to therapies.
Keywords: Synthetic microbiota, Gut ecosystem, Omics, Mathematical modelling
1. Introduction
The human gut microbiome is composed of numerous microbes performing functions that affect human health. The majority of human-inhabiting microorganisms reside inside the intestines and are affected by the host’s mode of birth, life style, and genetics. In the training of host immunity, controlling gut endocrine function and neurological signalling, digesting food, altering drug action and metabolism, removing toxins, and producing various compounds that affect the host, the gut microbiome has important roles as a desirable target for therapeutic applications [1]. However, unravelling human gut microbiome therapeutic potential has been hampered due to its complexity, which includes > 1,000 species of bacteria, archaea, and fungi, which are mostly unculturable [2]. Although numerous efforts have been made to understand the gut microbiome through genome sequencing platforms, microbial phenotypes and their roles in microbial populations have not been completely predicted by sequencing data alone. Recently, sophisticated culturomic approaches have increased the number of cultivable gut microbial species [2], [3], [4]. The challenge of investigating the human gut microbiome can be solved by using a combination of genome sequence and culturomics in an experimental system where variables can be tightly controlled and intentionally manipulated. Therefore, synthetic microbial communities have been proposed.
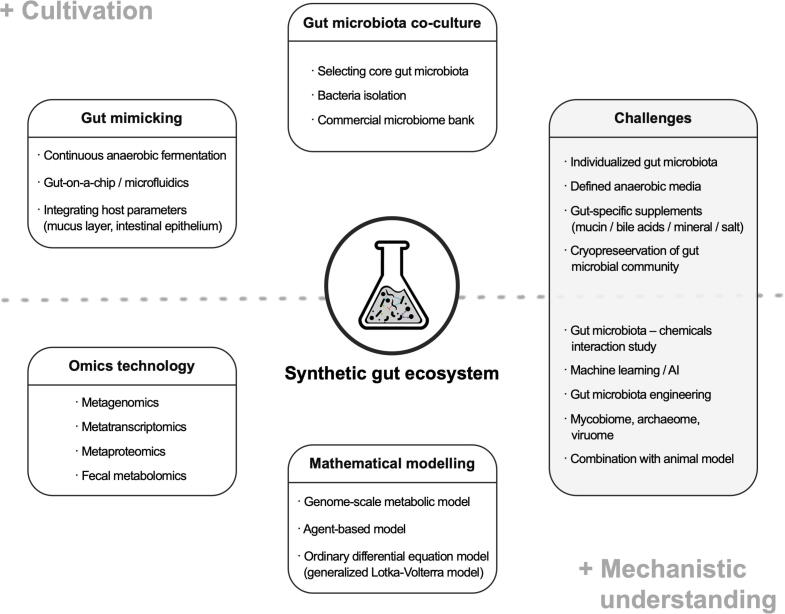
These communities are designed to represent the normal human intestinal microbiota, and are characterised by reduced microbial complexity that is amenable to experimental intervention [5]. Following the use of small-scale reduced interactions to unravel the complex behaviours in other systems, the synthetic microbiome has great potential for understanding the complicated human gut microbiome [6]. For the effective reconstruction of synthetic microbial communities, firstly, focus should be on the interaction between biologically relevant core bacterial strains in a controlled environment. Thereafter, to make reliable predictions, sufficient information should be provided through a useful omics approach, even if it is difficult to distinguish which variables are important modulators of group dynamics. Lastly, high-throughput data from complex gut microbiome studies should be integrated, and mathematical models and artificial intelligence (AI) should be involved in their simulation and testing. In this minireview, we address the advances and challenges in the design of synthetic intestinal microbiota. Our ultimate aim is to emphasise the importance of combining culturomics and multi-omics in synthetic gut microbiome design with mathematical modelling and AI to build therapies against human gut microbiome alterations (Fig. 1).
Fig. 1.
Overview of the current applications and challenges of synthetic gut microbiome research.
2. Systemic features of gut ecosystems
2.1. Gut mimicking using faecal samples
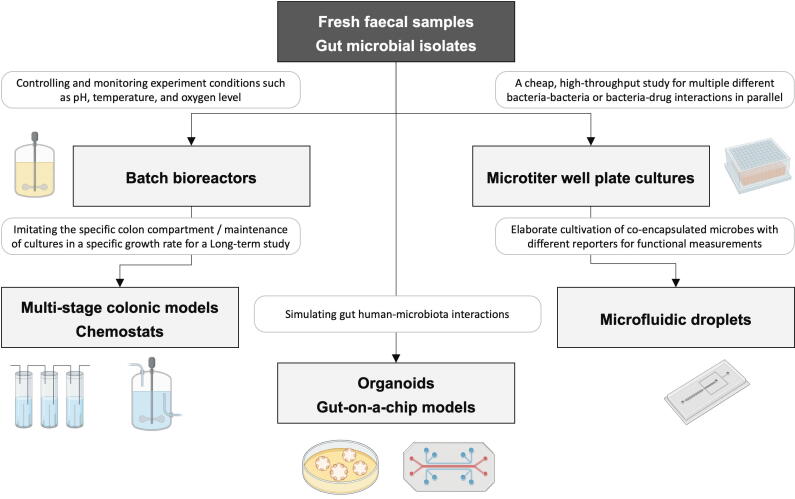
Currently, synthetic microbiomes are constructed using two approaches: bottom-up or top-down [7]. Top-down approaches primarily involve metagenomic studies that promote looking at the composition of the gut population, whereas bottom-up approaches are instrumental in understanding how these diverse bacterial communities form, communicate, and impact their host's health [8]. Hundreds of bacterial species are composed of faecal microbiota, most of which include Firmicutes, Bacteroidetes, and Actinobacteria [9]. Owing to their ability to imitate microbial composition in the natural system, faecal samples are well suited for the design of synthetic microbial communities. Most microbial data from both in vivo and in vitro systems have been collected through metagenomic studies of faecal samples [10]. Additionally, faecal samples are progressively used to examine microbiome responses to drugs and dietary fibre in batch and continuous fermentation systems [11]. To design the synthetic gut microbial community with human stools, it is important to study bacterial populations as communities in a well-controlled gut environment of temperature, pH, and other conditions, such as residence time, with gut mimicking systems using bioreactors (Fig. 2). Zihler et al revealed that a pH reduction of 0.2 units increased the relative abundance of butyrate producers and decreased Bacteroidetes phylum [12]. Additionally, the diversity of gut microbiota in the transverse and distal compartments of an in vitro gut model was shown to be affected by residence time [13]. Other factors such as temperature, anaerobic conditions, and dissolved oxygen could also affect microbial community and diversity [14].
Fig. 2.
The current in vitro gut mimicking systems for gut microbiota cultivation.
The simplest gut bioreactor systems are batch bioreactors and microtitre well plate systems, which are effective for investigating the simple interaction of prebiotics and drugs with single gut microbiota [15], [16], [17]. Continuous bioreactor systems such as chemostats, mini-bioreactor arrays, and multi-stage colonic models enable the inflow and outflow of materials. They provide useful resources for investigating the long-term effects of nutraceuticals on gut microbiota [18]. As some bioreactors are combined with simulated digestion systems which include the addition of digestive enzymes and bile salts, alterations to pH and residence time, and the peristaltic motility of the human intestine [19], they can better mimic the intestinal physiochemistry and gut ecology. While the effects of the host epithelium have not been taken into account by any of the previously mentioned bioreactor systems, gut-on-a-chip is an example of a bioreactor that mimics the host epithelium structure [20]. However, faecal samples are not preferred for designing bottom-up synthetic microbial communities owing to their large number of bacteria. The unintentional contribution of diverse food components and microorganisms, including bacteria, fungi, and viruses with high variability of inocula from different donors, might be a major setback in deciphering microbial interactions with entire faecal samples. Additionally, faecal samples comprise a meaningful percentage of uncultured, unannotated microbes with unknown interactions, thus making predictive modelling prior to synthetic microbial community reconstruction a challenging activity.
2.2. Synthetic ecosystems using isolated gut microbiota
To better understand the human microbiome, reconstructing defined microbial communities starting with the most prevalent microbes in the faecal samples under well-controlled conditions could be a reasonable approach. There are several benefits to using these isolates for the reconstruction of synthetic communities over the whole faecal specimen: First, the bacterial composition in the synthetic consortium would be known, controllable, and reproducible. Second, due to the absence of viruses and pathogens, a pure consortium is more stable than stool, and the formulation will be safe. Finally, isolates can be selected on the basis of their sensitivity to the testing environment, such as oxygen requirement, which further increases the success of synthetic microbial communities. Venturelli et al. conducted experiments with a simulated ecosystem utilising isolated gut microbiota with a bottom-up approach [21]. They used representatives of gut microbiota isolated from human faeces and discovered that pairwise interactions are necessary to model the microbial community consortium of multiple species, and that certain pairwise characteristics may be important over time to ensure resilience of a stable microbial community. Although their model population was less complex and diverse than the populations of real gut microbiota, the twelve species included in the experiment could span the key phyla of human-associated intestinal gut microbiota. Petrof successfully designed a 33-member synthetic microbiome, using faecal isolates also representing Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria. Their research shows that a synthetic microbial environment is capable of curing antibiotic-resistant Clostridium difficile colitis [22]. The 15-member synthetic ecosystem developed from human faeces isolates by the Welch group was applied to investigate spatial organisation at different scales of the main gut microbiota [23].
The availability of well-documented cultivable strain collections, particularly the core human gut microbiome, is essential to establish a good synthetic gut ecosystem. These core strains include keystone species important for the structure and function of the microbial ecosystem, such as the breakdown of complex carbon sources to support the growth of other members [24], [25]. Examples of keystone microbial species in the human gut include Bacteroides thetaiotaomicron, Bacteroides ovatus, and Bacteroides caccae, which are capable of breaking down complex dietary polysaccharides; Akkermansia muciniphila enables the degradation of mucin glycans as well as other gut members, such as Desulfovibrio piger, that are capable of eliminating the end products of fermentation by reducing the sulphate [26], [27], [28]. This could increase our understanding of gut ecology and promote the design of a synthetic microbial community representative of balanced human gut microbiota by mapping the keystone species into a stable microbiota and comparing it with diseased microbiota. Experiments involving co-cultures of core gut microbes have shown fascinating cross-feeding pathways, which involve the use of short chain fatty acids (SCFAs) [29] and have identified properties associated with major metabolic pathways for amino acid and vitamin metabolism, in combination with molecular analysis techniques [30], [31]. There are several routes for obtaining reliable core gut microbiota. Public microbial resource centres, such as the German Collection of Microorganisms and Cell Cultures, the University of Gothenburg Culture Collection, the Human Gastrointestinal Bacteria Culture Collection, and the Cultivable Genome Reference, are the current suppliers of a variety of isolated microbial strains that could be used for synthetic microbial community reconstruction [32]. New requirements for the isolation of keystone species necessary for the design of synthetic microbial communities may be provided through increased insights into the human gut microbiota metagenome and functional core microbes. On the other hand, fungal synthetic communities have been successfully reconstructed previously. Hu et al designed a synthetic fungal-bacterial microbial consortium to improve lingo-cellulolytic enzyme activity [33]. Other researchers used synthetic Saccharomyces cerevisiae consortium for the production of cellulosic ethanol or isobutanol [34], [35]. Similarly, more fungi-fungi microbial consortia systems for bioethanol production have been designed [36], [37], [34].
2.3. Challenges for a better synthetic gut ecosystem
Synthetic microbiomes are hampered by their inability to completely imitate the natural gut microbiome, considering their capacity and broad applicability. Appropriate indicators such as the relative abundance of keystone gut microbiota, the microbial diversity index which can show microbial richness and evenness of the sample, and the content and ratio of major gut metabolites like short chain fatty acids are required to evaluate the availability of synthetic gut ecosystem. However, selection of core gut microbiota and their metabolic functions may be difficult as there are approximately 10 to 100 trillion microorganisms that make up the human gut microbiota, and the microbiome contains 100 to 150 times more genes than our genome [38]. It is thus cumbersome to classify specific microbes integrated into synthetic microbiome reconstruction. Most existing studies have depended solely on a phylogeny for species selection [21], [39]. As metabolic interactions occur between distant and closely related species, selection of species based on the functional population should be considered. Moreover, for customisation and specialisation of the synthetic microbiome, some disease-related or age-related gut microbes may be included to establish a personalised synthetic gut ecosystem [40], [41].
To model synthetic microbiomes, a controlled culture-based system with suitable media where all bacteria can grow optimally is required. However, most gut microbiota have thus far been cultivated in complex media of unknown chemical composition, and only a handful of species have been identified in specified or minimal media [42]. Anaerobic cultivation is fastidious for several gut bacteria owing to their challenging nutritional and physiological requirements [43]. As media, such as modified Gifu anaerobic media, brain heart infusion media, reinforced clostridial media, and tryptic soy media, which are generally used for anaerobic culture, have special features, optimal media selection strongly restricts reconstruction attempts of microbial communities. As useful information and roles regarding certain ingredients in anaerobic media for the growth or function of specific bacteria continue to accumulate, big data and AI will be combined to predict and utilise the optimised media composition for cultivation of specific gut microbial communities in advance [44]. In addition, it is proposed that more potentially universal growth media be tested with growth supplements such as mucin, bile acid, minerals, and salt concentration, in an attempt to mirror the actual gut [45]. Along with the selection of an appropriate medium, technology to keep the synthetic microbial community well-preserved is required so that it can be implemented in a reproducible manner. Cryopreservation and bio-banking approaches are currently being developed for the preservation of viable synthetic microbial communities [46]. Glycerol and dimethyl sulfoxide are the most widely used cryoprotectant additives, as their addition decreases the adverse effects of freezing [47]. Importantly, the rate of freezing and thawing, cryoprotective medium composition, cryoprotectant concentration, growth-enhancing additives, and cell properties at the time of freezing are crucial factors to consider in the cryopreservation of bacterial cells [48].
Once the medium is established, different in vitro configurations that imitate colonic environments [5], [49] can be applied to predict and evaluate the behaviour of the synthetic microbial community and their functions, in a more realistic manner. More sophisticated in vitro colon models that integrate host parameters, such as mucus layer and human cell lines, for study questions involving host-microbe interaction should be considered [50]. To better mimic host factors, the enterocyte, mucin-secreting cell lines, and antimicrobial peptides with immune cells can be included in gut microbial culture [51], [52], [53]. Also, the existing sophisticated colon models such as Host-Microbiota Interaction (HMI) model, organoids, and 3D-culture models need to be further developed [54], [55], [56].
Although these in vitro colon models that reflect the host effect could allow for some control in the production of diverse and metabolically active gut microbiota, the composition of the end product and microbial community might be different from that of human faeces [57]. Therefore, the evaluation of synthetic microbial communities in animal models, such as Caenorhabditis elegans and mice, are essential. Most recent in vivo gut microbiota studies have only used mice, but certain host-specific variations still exist between mice and humans, which should not be overlooked. As these variations include immunological differences, transit time, intestinal structure, and functional caecum presence [58], [59], current in vivo studies are also limited in their ability to predict the actions of synthetic microbial communities directly. Therefore, the interrelationship between intestinal microbes and other gut microbiota and the host should be studied through a combination of synthetic gut ecosystems and in vivo animal models.
3. Mechanistic insights into the synthetic gut microbiome
3.1. Omics approach beyond DNA in the synthetic microbiome
Detailed and reliable functional and compositional knowledge is crucial before and after the reconstruction of synthetic communities, to create an efficient synthetic microbiome and to understand their output. Accurate confirmation of unique characteristics of the synthetic microbial community consortium depends on omics-driven analysis. The available standard is 16S rRNA amplicon sequencing, which is the most widely used and efficient method for identifying intestinal microbiota [60]. However, meta-taxonomic studies are limited to genus level resolution and cannot expose the functional role of the human gut microbiome. In contrast, shotgun sequencing metagenomics provide strain-level resolution, from which it is possible to classify various bacterial strains and genes as putative biomarkers discriminating the diseased and non-diseased human gut microbiome [61].
To understand the active functional contributions of each bacterial species forming the natural human gut ecosystem, further complementation with metatranscriptomics and metaproteomics is useful [62]. Several synthetic microbiome studies primarily focus on metagenomics, and only a few studies have focussed on the metaproteomics of synthetic microbiota [63], [64]. In modelling human synthetic gut microbiome communities, metaproteomic studies can add functional, taxonomic, and biomass dimensions. Daniel Figeys’ lab developed a tool called iMetaLab for the analysis of metaproteomics data [65] and carried out meta-analysis using in vitro microbial cultivation [66]. The benefit of the combination of metaproteomics and a synthetic microbiome model is that, compared to in vivo studies, researchers can easily control the spatiotemporal variables during microbiota culture and unveil their functionalities. By using the model of in vitro metaproteomics, Hao et al demonstrated that metformin can lead to phase-dependent growth responses [66]. Regarding their growth curve, taxonomy, and functional profiles, metformin was applied at various growth phases, including lag, log, and stationary stages, resulting in various effects.
Alternatively, faecal metabolomics has been used to monitor metabolites produced by gut microbes, and to investigate the components of the host and diet. Thus, the metabolomic approach in the gut ecosystem is an extremely valuable method for understanding the complex metabolic interactions between the gut and host’s microbes [67]. Metabolomics can be applied through a targeted and/or non-targeted method using a high-throughput analytical platform, such as nuclear magnetic resonance spectroscopy, gas chromatography-mass spectrometry, and liquid chromatography-mass spectrometry [68]. Although the human metabolome database has recently grown to > 40,000 chemicals, accurate identification of microbial-derived compounds is still limited. In particular, optimisation of sample handling and preparation for metabolomic analysis of faeces should be developed more [69]. Recently, some researchers have used commercial kits for targeted faecal metabolic profiling and quantification by covering hundreds of metabolites, including short chain fatty acids, bile acids, amino acids and their derivatives, and lipids [70], [71].
However, omics profiling alone cannot completely predict bacterial interactions critical for the reconstruction of synthetic microbiomes or interactions between host bacteria [72], and the accuracy of functional gene assignments, which depends on these approximations, also depends on manual refining of genome annotations [73], [74]. In addition, statistical association between the microbial community and other omics data is required, and several omics data should be integrated to classify main interactions, by identifying gut microbes and bacterial populations that can have a positive or negative effect on gut ecology and the host [75]. Nevertheless, omics data-based modelling can predict and provide essential cross-feeding and competition mechanisms for the design of multi-strain formulations [68], [73], and facilitates the design of the optimal bacterial community for the desired metabolite, such as the production of butyrate and functional exopolysaccharides [44].
3.2. Mathematical modelling
Mathematical modelling has become valuable in understanding gut microbial community dynamics and emergent behaviours [5], [76]. By describing the system with mathematical concepts and languages, it has been possible to quantify and predict the behaviour of the system under different conditions, such as different diet, disease, or treatment conditions [77], [78], [79], [80], [81]. Combined with a detailed understanding of gut microbiota biology, mathematical models can provide new insights into the mechanisms and first principles of community dynamics.
Integrating mathematical models with controlled lab experiments is key to making progress in understanding microbial communities. It is not realistic to capture the full interplay between all reactions from every individual or all interactions between all species or individuals simultaneously. Thus, simplifying the system with known or reduced complexity is required for mathematical model development and microbial experiments. For integration, experimental data are fed into the building model assumption, and mathematical model parameters that minimise model prediction and experimental observation are estimated. Calibrated model simulations make predictions that can be tested in the laboratory. Iteration of mathematical model refinement and experiments leads to a better understanding of the system observed in different biological systems, such as cancer [82], [83], [84]. The same process has been applied to the human gut microbial community, as described below.
Various integrated approaches of mathematical modelling and experiments have been applied to gut microbiota at different resolutions, different times, and spatial scales, including genes, individuals, and populations. Genome-scale metabolic models (GEMs) describe the metabolism of the microbial community [81], [85], [86], [87], [88]. The modelling approach reconstructs the metabolic network based on high-throughput omics data and known biochemical reaction databases, such as KEGG (Kyoto Encyclopaedia of Genes and Genomes). A system of mass balance equations is sub developed to describe the flow of metabolites. This modelling approach has been applied to simulate substrate uptake rates and to predict short chain fatty acid secretion profiles validated with in vitro experiments [89], [90]. Agent-based models (ABMs) simulate the emergent behaviour of communities assuming individuals as discrete autonomous agents. This modelling approach is useful for describing detailed microbial behaviour (e.g. active motion like chemotaxis) and interactions between individual microorganisms or species [91], [92], [93]. A GEM was integrated with ABMs to simulate the interactions of a synthetic gut microbial community of seven species [94]. This study demonstrates the importance of the spatial gradient of mucus glycans in the gut microbial community structure. As ABMs can readily incorporate various individual types and spatial components, these approaches can be used to understand the interaction between the microbial community and the host. For example, ABMs coupled with continuous microenvironmental factors prove to be efficient tools for investigating the role of interactions between the tumour and microenvironment in driving tumour progression systems [82].
At the population level, ordinary differential equation models, such as the generalised Lotka-Volterra model, have been applied to human gut microbiota. This approach uses the intrinsic growth rate of individual populations and growth rates modulated by the interaction between microbial populations, to explain community dynamics. The models can be tightly integrated with synthetic microbe experiments as the modelling assumes fewer parameters than others [79], [80], [95]. For example, a generalised Lotka-Volterra model trained with time-series data of 12 different microbial species predicted that pairwise interactions drive microbial community dynamics, and revealed multiple positive and negative interactions that allow the community to coexist [95]. Although not discussed here, several different modelling approaches, such as the Boolean network model, partial differential equation model, and game theory model, were developed to understand various microbial communities (see the comprehensive methodology review in [96]).
3.3. Considering possible modulators of the gut ecosystem
The current application of mathematical modelling to the synthetic gut ecosystem has primarily been conducted with the aim of understanding the interactions between gut microbial individuals and the population. However, recent studies have indicated the influence of xenobiotic compounds, including medications, phytochemicals, food components, environmental chemicals, and metals, on gut microbiota [97], [98], [99]. Some gut microbiota play key roles in the digestion and metabolism of xenobiotic compounds, and the metabolites of gut microbiota may be accompanied by a change in gut microbial composition, induction of unique bacterial genes, and microbiome-derived chemical transformation [100]. Such interactions between the gut microbiome and the metabolites of drugs or drugs themselves have also been documented in a well-designed research paper, indicating that compound changes induced by gut microbes may lead to either activation, inactivation, or toxification of these compounds [101]. Additionally, several studies have shown that exposure to environmental pollutants or heavy metals may change the composition of the gut microbiome, resulting in disorders of energy metabolism, absorption of nutrients, and functioning of the immune system [102], [103]. Some useful phytochemicals, such as catechins, procyanidins, and lignans, have been reported to ameliorate gut microbial dysbiosis by inducing growth of beneficial bacteria as essential growth factors, inhibiting the growth of harmful microbes through quorum sensing, and reducing intestinal oxidative stress as antioxidants [98], [104], [105]. Figeys and co-workers showed a high degree of correlation between gut microbiome responses to metformin in the luminal colon bioreactor model and those in mice fed a high-fat [106]. The authors also tested the effect of 43 drugs on individual faecal microbiomes with the meta-proteomics approach and showed that drugs affect functions of different members of the microbiome without even a shift in the abundance of the bacteria [107]. Therefore, it is necessary to study the interaction between gut microbiota and these chemicals or drugs or food components, and determine the accurate mechanism using the synthetic microbiome research platform to develop mathematical modelling techniques accordingly.
Microbial biotechnology has been steadily developing owing to its capability to produce valuable pharmaceuticals and drugs. Most recent achievements in engineering co-cultures for biotechnological applications have been performed using Escherichia coli to improve the functionality via specialisation, and reduce the metabolic burden [108], [109]. To improve co-operation or reduce competition between microbial populations, numerous strategies, such as resource partitioning, chemical symbiosis, horizontal gene transfer, and spatial organisation, have been employed. Such applications can be extended to diverse and complex gut environments. Certain probiotics have been engineered to identify quorum sensing molecules and to use them as live biotherapeutics to prevent viral infections [110], [111]. Some studies indicated that the engineered E. coli as probiotics could inhibit the infection of Vibrio cholerae through the increase of quorum sensing chemicals and the engineered B. ovatus could secrete human growth factors against inflammation [110], [111]. Additionally, there are specific tools for engineering Bacteroides that exhibit stable abundance and long-term colonisation in the gut environment to control the expression of their reporter genes [112]. In a recent study, the intentional shaping of microbial consortia by genetic modifications decreased the antagonistic interactions in a synthetic ecosystem using four commensal gut microbes and induced beneficial interactions, showing an increase in population evenness and ecological diversity by engineered cross-feeding [113]. Therefore, the application of gut microbe engineering to synthetic microbiomes will greatly assist in understanding the basic principles of microbial interactions, which have not yet been revealed.
4. Outlook and future prospects
Human gut microbiome analysis has primarily focussed on the analysis of high-throughput multi-omics systems of in vivo faecal samples. However, owing to the generation of big data and noise from unknown components of microbes within faecal samples, the research using diverse in vivo samples has limitations. Thus, to analyse complex and physiologically important human gut microbiome relationships, there is a great need for experimental models involving synthetic gut ecosystems with an advanced in vitro method. Advances in cultural techniques, particularly in the field of microfluidics, such as organ-on-chip technology, have the potential to fundamentally change analytical techniques of gut microbiota. Organ-on-chip technology is low cost owing to its micrometre-sized chamber and feasible microfluidic perfusion. The use of human cells in microfluidics technology can better replicate human physiology and also model the complex disease-specific conditions using individual patient-derived cells. These individualised gut microbiota approaches would be highly important for personalised medicine.
It is conceivable that multi-omics big data may be a requirement for the design of synthetic microbial communities. Incidentally, to obtain as much knowledge as possible from multi-omics big data interaction datasets, new analytical methods are required to provide insights into the interactions yet to be measured and possible biological mechanisms mediating the relevant microbial interaction. Machine learning, AI, and mathematical modelling can complement high-throughput omics data by identifying complex patterns and transforming them into more useable and interpretable knowledge. This involves careful experimental design to test a hypothesis adequately, and it is necessary to recreate or combine practical synthetic microbiomes for disease diagnosis predictions and effective treatments.
The most insightful synthetic gut microbiomes can be produced by using recombinant bacterial strains genetically modified with desired characteristics. Synthetic genetic circuits in recombinant bacterial strains should be built to reduce the burden of foreign genetic material on their bacterial hosts to reduce compensatory genetic mutations, loss of engineered function, or lack of growth of engineered strains in the synthetic microbiome consortium. Additionally, a broad approach beyond bacteria-bacteria contact should also be considered in potential applications. Synthetic microbiome studies can be carried out in terms of mycobiome and viruome coupled with chemicals and other component interactions. This is possible as there has been an increase in studies evaluating the role of viral and fungal microbiome components with tailored methods and reference databases for identification and classification. In conclusion, the synthetic gut microbiome will broaden our knowledge of gut ecology and their mechanistic functions, and will be of great assistance to design effective and accurate microbiome modulation strategies for health benefits.
CRediT authorship contribution statement
Humphrey A. Mabwi: Writing - original draft. Eunjung Kim: Writing - original draft. Dae-Geun Song: Writing - original draft. Hyo Shin Yoon: Visualization, Writing - review & editing. Cheol-Ho Pan: Conceptualization. Erick.V.G. Komba: Project administration. GwangPyo Ko: Conceptualization, Supervision. Kwang Hyun Cha: Supervision, Funding acquisition, Writing - review & editing. : .
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research received funding from an intramural grant (2Z06220) from the Korea Institute of Science and Technology (KIST), Republic of Korea, in collaboration with the Regional Scholarship and Innovation Fund of the Partnership for Skills in Applied Sciences, Engineering and Technology (RSIF-PASET).
Contributor Information
GwangPyo Ko, Email: gko@snu.ac.kr.
Kwang Hyun Cha, Email: chakh79@kist.re.kr.
References
- 1.Schmidt T.S.B., Raes J., Bork P. The human gut microbiome: from association to modulation. Cell. 2018;172(6):1198–1215. doi: 10.1016/j.cell.2018.02.044. [DOI] [PubMed] [Google Scholar]
- 2.Rajilić-Stojanović M., de Vos W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38(5):996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Browne H.P., Forster S.C., Anonye B.O., Kumar N., Neville B.A., Stares M.D. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533(7604):543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Vrancken G., Gregory A.C., Huys G.R.B., Faust K., Raes J. Synthetic ecology of the human gut microbiota. Nat Rev Microbiol. 2019;17(12):754–763. doi: 10.1038/s41579-019-0264-8. [DOI] [PubMed] [Google Scholar]
- 6.Strogatz SH. Sync: how order emerges from chaos in the universe, nature, and daily life. Hachette UK; 2012.
- 7.Lawson C.E., Harcombe W.R., Hatzenpichler R., Lindemann S.R., Löffler F.E., O'Malley M.A. Common principles and best practices for engineering microbiomes. Nat Rev Microbiol. 2019;17(12):725–741. doi: 10.1038/s41579-019-0255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature 2012;486(7402):207. [DOI] [PMC free article] [PubMed]
- 9.Ley R.E., Peterson D.A., Gordon J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Wang W.L., Xu S.Y., Ren Z.G., Tao L., Jiang J.W., Zheng S.S. Application of metagenomics in the human gut microbiome. World J Gastroenterol. 2015;21(3):803–814. doi: 10.3748/wjg.v21.i3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zihler Berner A., Fuentes S., Dostal A., Payne A.N., Vazquez Gutierrez P., Chassard C. Novel Polyfermentor intestinal model (PolyFermS) for controlled ecological studies: validation and effect of pH. PLoS ONE. 2013;8(10):e77772. doi: 10.1371/journal.pone.0077772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tottey W., Feria-Gervasio D., Gaci N., Laillet B., Pujos E., Martin J.F. Colonic transit time is a driven force of the gut microbiota composition and metabolism, vitro evidence. J Neurogastroenterol Motil. 2017;23(1):124–134. doi: 10.5056/jnm16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C.-G., Xue C., Lin Y.-H., Bai F.-W. Redox potential control and applications in microaerobic and anaerobic fermentations. Biotechnol Adv. 2013;31(2):257–265. doi: 10.1016/j.biotechadv.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Likotrafiti E., Tuohy K.M., Gibson G.R., Rastall R.A. An in vitro study of the effect of probiotics, prebiotics and synbiotics on the elderly faecal microbiota. Anaerobe. 2014;27:50–55. doi: 10.1016/j.anaerobe.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Jin J.B., Cha J.W., Shin I.S., Jeon J.Y., Cha K.H., Pan C.H. Supplementation with Chlorella vulgaris, Chlorella protothecoides, and Schizochytrium sp. increases propionate-producing bacteria in in vitro human gut fermentation. J Sci Food Agric. 2020;100(7):2938–2945. doi: 10.1002/jsfa.10321. [DOI] [PubMed] [Google Scholar]
- 17.Cha K.H., Lee E.H., Yoon H.S., Lee J.H., Kim J.Y., Kang K. Effects of fermented milk treatment on microbial population and metabolomic outcomes in a three-stage semi-continuous culture system. Food Chem. 2018;263:216–224. doi: 10.1016/j.foodchem.2018.04.095. [DOI] [PubMed] [Google Scholar]
- 18.Paul W., Marta C., Tom V.W. Resolving host–microbe interactions in the gut: the promise of in vitro models to complement in vivo research. Curr Opin Microbiol. 2018;44:28–33. doi: 10.1016/j.mib.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Williams C.F., Walton G.E., Jiang L., Plummer S., Garaiova I., Gibson G.R. Comparative analysis of intestinal tract models. Ann Rev Food Sci Technol. 2015;6(1):329–350. doi: 10.1146/annurev-food-022814-015429. [DOI] [PubMed] [Google Scholar]
- 20.Kim H.J., Huh D., Hamilton G., Ingber D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12(12):2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 21.Venturelli OS, Carr AC, Fisher G, Hsu RH, Lau R, Bowen BP, et al. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol 2018;14(6):e8157-e. [DOI] [PMC free article] [PubMed]
- 22.Petrof E.O., Gloor G.B., Vanner S.J., Weese S.J., Carter D., Daigneault M.C. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: 'RePOOPulating' the gut. Microbiome. 2013;1(1):3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mark Welch J.L., Hasegawa Y., McNulty N.P., Gordon J.I., Borisy G.G. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc Natl Acad Sci U S A. 2017;114(43):E9105–E9114. doi: 10.1073/pnas.1711596114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ze X., Le Mougen F., Duncan S.H., Louis P., Flint H.J. Some are more equal than others: the role of “keystone” species in the degradation of recalcitrant substrates. Gut Microb. 2013;4(3):236–240. doi: 10.4161/gmic.23998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trosvik P., de Muinck E.J. Ecology of bacteria in the human gastrointestinal tract–identification of keystone and foundation taxa. Microbiome. 2015;3:44. doi: 10.1186/s40168-015-0107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faith J.J., McNulty N.P., Rey F.E., Gordon J.I. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science. 2011;333(6038):101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 2004;54(5):1469–76. [DOI] [PubMed]
- 28.Ravcheev D.A., Godzik A., Osterman A.L., Rodionov D.A. Polysaccharides utilization in human gut bacterium Bacteroides thetaiotaomicron: comparative genomics reconstruction of metabolic and regulatory networks. BMC Genomics. 2013;14(1):873. doi: 10.1186/1471-2164-14-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19(1):29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 30.Laverde Gomez J.A., Mukhopadhya I., Duncan S.H., Louis P., Shaw S., Collie-Duguid E. Formate cross-feeding and cooperative metabolic interactions revealed by transcriptomics in co-cultures of acetogenic and amylolytic human colonic bacteria. Environ Microbiol. 2019;21(1):259–271. doi: 10.1111/1462-2920.14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chia L.W., Hornung B.V., Aalvink S., Schaap P.J., de Vos W.M., Knol J. Deciphering the trophic interaction between Akkermansia muciniphila and the butyrogenic gut commensal Anaerostipes caccae using a metatranscriptomic approach. Antonie Van Leeuwenhoek. 2018;111(6):859–873. doi: 10.1007/s10482-018-1040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H.J., Li H., Collins J.J., Ingber D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A. 2016;113(1):E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu J., Xue Y., Guo H., Gao M.-t., Li J., Zhang S. Design and composition of synthetic fungal-bacterial microbial consortia that improve lignocellulolytic enzyme activity. Bioresour Technol. 2017;227:247–255. doi: 10.1016/j.biortech.2016.12.058. [DOI] [PubMed] [Google Scholar]
- 34.Zuroff T.R., Xiques S.B., Curtis W.R. Consortia-mediated bioprocessing of cellulose to ethanol with a symbiotic Clostridium phytofermentans/yeast co-culture. Biotechnol Biofuels. 2013;6(1):59. doi: 10.1186/1754-6834-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minty J.J., Singer M.E., Scholz S.A., Bae C.-H., Ahn J.-H., Foster C.E. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. P Natl Acad Sci USA. 2013;110(36):14592–14597. doi: 10.1073/pnas.1218447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patle S., Lal B. Ethanol production from hydrolysed agricultural wastes using mixed culture of Zymomonas mobilis and Candida tropicalis. Biotechnol Lett. 2007;29(12):1839–1843. doi: 10.1007/s10529-007-9493-4. [DOI] [PubMed] [Google Scholar]
- 37.Suriyachai N., Weerasaia K., Laosiripojana N., Champreda V., Unrean P. Optimized simultaneous saccharification and co-fermentation of rice straw for ethanol production by Saccharomyces cerevisiae and Scheffersomyces stipitis co-culture using design of experiments. Bioresour Technol. 2013;142:171–178. doi: 10.1016/j.biortech.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutiérrez N., Garrido D. Species Deletions from microbiome consortia reveal key metabolic interactions between gut microbes. mSystems. 2019;4(4) doi: 10.1128/mSystems.00185-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013;2:e01202. [DOI] [PMC free article] [PubMed]
- 41.Chen J., Wright K., Davis J.M., Jeraldo P., Marietta E.V., Murray J. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8(1):1–14. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tramontano M., Andrejev S., Pruteanu M., Klünemann M., Kuhn M., Galardini M. Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat Microbiol. 2018;3(4):514–522. doi: 10.1038/s41564-018-0123-9. [DOI] [PubMed] [Google Scholar]
- 43.Rettedal E.A., Gumpert H., Sommer M.O. Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat Commun. 2014;5(1):1–9. doi: 10.1038/ncomms5714. [DOI] [PubMed] [Google Scholar]
- 44.Magnúsdóttir S., Heinken A., Kutt L., Ravcheev D.A., Bauer E., Noronha A. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol. 2017;35(1):81. doi: 10.1038/nbt.3703. [DOI] [PubMed] [Google Scholar]
- 45.Yang K., Xu M., Zhu J. Evaluating the impact of four major nutrients on gut microbial metabolism by a targeted metabolomics approach. J Proteome Res. 2020;19(5):1991–1998. doi: 10.1021/acs.jproteome.9b00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terveer E.M., van Beurden Y.H., Goorhuis A., Seegers J., Bauer M., van Nood E. How to: establish and run a stool bank. Clin Microbiol Infect. 2017;23(12):924–930. doi: 10.1016/j.cmi.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 47.Hubalek Z. Protectants used in the cryopreservation of microorganisms. Cryobiology. 2003;46(3):205–229. doi: 10.1016/s0011-2240(03)00046-4. [DOI] [PubMed] [Google Scholar]
- 48.Smith D., Ryan M. Implementing best practices and validation of cryopreservation techniques for microorganisms. Scientific World J. 2012;2012 doi: 10.1100/2012/805659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pham V.T., Mohajeri M.H. The application of in vitro human intestinal models on the screening and development of pre- and probiotics. Benef Microb. 2018;9(5):725–742. doi: 10.3920/BM2017.0164. [DOI] [PubMed] [Google Scholar]
- 50.Williams C.F., Walton G.E., Jiang L., Plummer S., Garaiova I., Gibson G.R. Comparative analysis of intestinal tract models. Annu Rev Food Sci Technol. 2015;6:329–350. doi: 10.1146/annurev-food-022814-015429. [DOI] [PubMed] [Google Scholar]
- 51.Van den Abbeele P., Roos S., Eeckhaut V., MacKenzie D.A., Derde M., Verstraete W. Incorporating a mucosal environment in a dynamic gut model results in a more representative colonization by lactobacilli. Microb Biotechnol. 2012;5(1):106–115. doi: 10.1111/j.1751-7915.2011.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marzorati M., Pinheiro I., Van den Abbeele P., Van de Wiele T., Possemiers S. An in vitro technology platform to assess host-microbiota interactions in the gastrointestinal tract. Agro Food Ind Hi-Tech. 2012;23:8–11. [Google Scholar]
- 53.Marzorati M., Possemiers S., Van Den Abbeele P., Van De Wiele T., Vanhoecke B., Verstraete W. Technology and method to study microbial growth and adhesion to host-related surfaces and the host-microbiota interaction. Google Patents. 2014 [Google Scholar]
- 54.Marzorati M., Vanhoecke B., De Ryck T., Sadabad M.S., Pinheiro I., Possemiers S. The HMI™ module: a new tool to study the Host-Microbiota Interaction in the human gastrointestinal tract in vitro. BMC Microbiol. 2014;14(1):133. doi: 10.1186/1471-2180-14-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barrila J., Radtke A.L., Crabbé A., Sarker S.F., Herbst-Kralovetz M.M., Ott C.M. Organotypic 3D cell culture models: using the rotating wall vessel to study host–pathogen interactions. Nat Rev Microbiol. 2010;8(11):791–801. doi: 10.1038/nrmicro2423. [DOI] [PubMed] [Google Scholar]
- 56.Rubert J., Schweiger P.J., Mattivi F., Tuohy K., Jensen K.B., Lunardi A. Intestinal organoids: a tool for modelling diet–microbiome–host interactions. Trends Endocrinol Metab. 2020;31(11):848–858. doi: 10.1016/j.tem.2020.02.004. [DOI] [PubMed] [Google Scholar]
- 57.Bircher L., Schwab C., Geirnaert A., Lacroix C. Cryopreservation of artificial gut microbiota produced with in vitro fermentation technology. Microb Biotechnol. 2018;11(1):163–175. doi: 10.1111/1751-7915.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Byndloss M.X., Pernitzsch S.R., Bäumler A.J. Healthy hosts rule within: ecological forces shaping the gut microbiota. Mucosal Immunol. 2018;11(5):1299–1305. doi: 10.1038/s41385-018-0010-y. [DOI] [PubMed] [Google Scholar]
- 59.Salzman N.H., Hung K., Haribhai D., Chu H., Karlsson-Sjöberg J., Amir E. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11(1):76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knight R., Vrbanac A., Taylor B.C., Aksenov A., Callewaert C., Debelius J. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 61.Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35(9):833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 62.Bashiardes S., Zilberman-Schapira G., Elinav E. Use of metatranscriptomics in microbiome research. Bioinf Biol Insights. 2016;10:BBI.:S34610. doi: 10.4137/BBI.S34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moss E.L., Maghini D.G., Bhatt A.S. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat Biotechnol. 2020;38(6):701–707. doi: 10.1038/s41587-020-0422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eberl C., Ring D., Münch P.C., Beutler M., Basic M., Slack E.C. Reproducible colonization of germ-free mice with the oligo-mouse-microbiota in different animal facilities. Front Microbiol. 2020;10(2999) doi: 10.3389/fmicb.2019.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng K, Ning Z, Zhang X, Li L, Liao B, Mayne J, et al. MetaLab: an automated pipeline for metaproteomic data analysis. 2017;5(1):157. [DOI] [PMC free article] [PubMed]
- 66.Hao Z., Li L., Ning Z., Zhang X., Mayne J., Cheng K. Metaproteomics reveals growth phase-dependent responses of an in vitro gut microbiota to metformin. J Am Soc Mass Spectrom. 2020;31(7):1448–1458. doi: 10.1021/jasms.0c00054. [DOI] [PubMed] [Google Scholar]
- 67.Pedersen H.K., Gudmundsdottir V., Nielsen H.B., Hyotylainen T., Nielsen T., Jensen B.A. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 68.Lamichhane S., Sen P., Dickens A.M., Orešič M., Bertram H.C. Gut metabolome meets microbiome: a methodological perspective to understand the relationship between host and microbe. Methods. 2018;149:3–12. doi: 10.1016/j.ymeth.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 69.Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vazquez-Fresno R. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Connors J., Dunn K.A., Allott J., Bandsma R., Rashid M., Otley A.R. The relationship between fecal bile acids and microbiome community structure in pediatric Crohn's disease. ISME J. 2020;14(3):702–713. doi: 10.1038/s41396-019-0560-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuhring M., Eisenberger A., Schmidt V., Krankel N., Leistner D.M., Kirwan J. Concepts and software package for efficient quality control in targeted metabolomics studies: MeTaQuaC. Anal Chem. 2020;92(15):10241–10245. doi: 10.1021/acs.analchem.0c00136. [DOI] [PubMed] [Google Scholar]
- 72.Röttjers L., Faust K. From hairballs to hypotheses–biological insights from microbial networks. FEMS Microbiol Rev. 2018;42(6):761–780. doi: 10.1093/femsre/fuy030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muller E.E., Faust K., Widder S., Herold M., Arbas S.M., Wilmes P. Using metabolic networks to resolve ecological properties of microbiomes. Curr Opin Syst Biol. 2018;8:73–80. [Google Scholar]
- 74.Magnúsdóttir S., Thiele I. Modeling metabolism of the human gut microbiome. Curr Opin Biotechnol. 2018;51:90–96. doi: 10.1016/j.copbio.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 75.Hornung B, Martins Dos Santos VAP, Smidt H, Schaap PJ. Studying microbial functionality within the gut ecosystem by systems biology. Genes Nutr 2018;13:5. [DOI] [PMC free article] [PubMed]
- 76.Kumar M., Ji B., Zengler K., Nielsen J. Modelling approaches for studying the microbiome. Nat Microbiol. 2019;4(8):1253–1267. doi: 10.1038/s41564-019-0491-9. [DOI] [PubMed] [Google Scholar]
- 77.Bucci V., Bradde S., Biroli G., Xavier J.B. Social interaction, noise and antibiotic-mediated switches in the intestinal microbiota. PLoS Comput Biol. 2012;8(4):e1002497. doi: 10.1371/journal.pcbi.1002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buffie C.G., Bucci V., Stein R.R., McKenney P.T., Ling L., Gobourne A. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stein R.R., Bucci V., Toussaint N.C., Buffie C.G., Ratsch G., Pamer E.G. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput Biol. 2013;9(12):e1003388. doi: 10.1371/journal.pcbi.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stein R.R., Tanoue T., Szabady R.L., Bhattarai S.K., Olle B., Norman J.M. Computer-guided design of optimal microbial consortia for immune system modulation. Elife. 2018;7 doi: 10.7554/eLife.30916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shoaie S., Ghaffari P., Kovatcheva-Datchary P., Mardinoglu A., Sen P., Pujos-Guillot E. Quantifying Diet-induced metabolic changes of the human gut microbiome. Cell Metab. 2015;22(2):320–331. doi: 10.1016/j.cmet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 82.Anderson A.R., Weaver A.M., Cummings P.T., Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127(5):905–915. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 83.Frankenstein Z., Basanta D., Franco O.E., Gao Y., Javier R.A., Strand D.W. Stromal reactivity differentially drives tumour cell evolution and prostate cancer progression. Nat Ecol Evol. 2020;4(6):870–884. doi: 10.1038/s41559-020-1157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim E., Kim J.Y., Smith M.A., Haura E.B., Anderson A.R.A. Cell signaling heterogeneity is modulated by both cell-intrinsic and -extrinsic mechanisms: an integrated approach to understanding targeted therapy. PLoS Biol. 2018;16(3):e2002930. doi: 10.1371/journal.pbio.2002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chan S.H.J., Simons M.N., Maranas C.D. SteadyCom: predicting microbial abundances while ensuring community stability. PLoS Comput Biol. 2017;13(5):e1005539. doi: 10.1371/journal.pcbi.1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lewis N.E., Nagarajan H., Palsson B.O. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat Rev Microbiol. 2012;10(4):291–305. doi: 10.1038/nrmicro2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orth J.D., Thiele I., Palsson B.O. What is flux balance analysis? Nat Biotechnol. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zengler K., Palsson B.O. A road map for the development of community systems (CoSy) biology. Nat Rev Microbiol. 2012;10(5):366–372. doi: 10.1038/nrmicro2763. [DOI] [PubMed] [Google Scholar]
- 89.Heinken A., Sahoo S., Fleming R.M., Thiele I. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes. 2013;4(1):28–40. doi: 10.4161/gmic.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shoaie S., Karlsson F., Mardinoglu A., Nookaew I., Bordel S., Nielsen J. Understanding the interactions between bacteria in the human gut through metabolic modeling. Sci Rep. 2013;3:2532. doi: 10.1038/srep02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bonabeau E. Agent-based modeling: methods and techniques for simulating human systems. P Natl Acad Sci USA. 2002;99:7280–7287. doi: 10.1073/pnas.082080899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hellweger F.L., Bucci V. A bunch of tiny individuals-Individual-based modeling for microbes. Ecol Model. 2009;220(1):8–22. [Google Scholar]
- 93.Shashkova T., Popenko A., Tyakht A., Peskov K., Kosinsky Y., Bogolubsky L. Agent based modeling of human gut microbiome interactions and perturbations. PLoS ONE. 2016;11(2) doi: 10.1371/journal.pone.0148386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bauer E., Zimmermann J., Baldini F., Thiele I., Kaleta C. BacArena: individual-based metabolic modeling of heterogeneous microbes in complex communities. PLoS Comput Biol. 2017;13(5):e1005544. doi: 10.1371/journal.pcbi.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Venturelli O.S., Carr A.C., Fisher G., Hsu R.H., Lau R., Bowen B.P. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol. 2018;14(6):e8157. doi: 10.15252/msb.20178157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Song HS, Cannon WR, Beliaev AS, Konopka A. Mathematical modeling of microbial community dynamics: a methodological review (vol 2, pg 711, 2014). Processes 2015;3(3):699-+.
- 97.Velmurugan G. Gut microbiota in toxicological risk assessment of drugs and chemicals: the need of hour. Gut Microbes. 2018;9(5):465–468. doi: 10.1080/19490976.2018.1445955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Carrera-Quintanar L., Lopez Roa R.I., Quintero-Fabian S., Sanchez-Sanchez M.A., Vizmanos B., Ortuno-Sahagun D. Phytochemicals that influence gut microbiota as prophylactics and for the treatment of obesity and inflammatory diseases. Mediators Inflamm. 2018;2018:9734845. doi: 10.1155/2018/9734845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jin Y., Wu S., Zeng Z., Fu Z. Effects of environmental pollutants on gut microbiota. Environ Pollut. 2017;222:1–9. doi: 10.1016/j.envpol.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 100.Maurice C.F., Haiser H.J., Turnbaugh P.J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152(1–2):39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570(7762):462–467. doi: 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jin Y., Zeng Z., Wu Y., Zhang S., Fu Z. Oral exposure of mice to carbendazim induces hepatic lipid metabolism disorder and gut microbiota dysbiosis. Toxicol Sci. 2015;147(1):116–126. doi: 10.1093/toxsci/kfv115. [DOI] [PubMed] [Google Scholar]
- 103.Zhang S., Jin Y., Zeng Z., Liu Z., Fu Z. Subchronic exposure of mice to cadmium perturbs their hepatic energy metabolism and gut microbiome. Chem Res Toxicol. 2015;28(10):2000–2009. doi: 10.1021/acs.chemrestox.5b00237. [DOI] [PubMed] [Google Scholar]
- 104.Mujawdiya P.K., Kapur S. Modulation of gut microbiota through dietary phytochemicals as a novel anti-infective strategy. Curr Drug Discov Technol. 2020;17(4):498–506. doi: 10.2174/1570163816666191107124214. [DOI] [PubMed] [Google Scholar]
- 105.Yan T., Wang N., Liu B., Wu B., Xiao F., He B. Schisandra chinensis ameliorates depressive-like behaviors by regulating microbiota-gut-brain axis via its anti-inflammation activity. Phytother Res. 2020 doi: 10.1002/ptr.6799. [DOI] [PubMed] [Google Scholar]
- 106.Li L., Abou-Samra E., Ning Z., Zhang X., Mayne J., Wang J. An in vitro model maintaining taxon-specific functional activities of the gut microbiome. Nat Commun. 2019;10(1):4146. doi: 10.1038/s41467-019-12087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li L., Ning Z., Zhang X., Mayne J., Cheng K., Stintzi A. RapidAIM: a culture- and metaproteomics-based Rapid Assay of Individual Microbiome responses to drugs. Microbiome. 2020;8(1):33. doi: 10.1186/s40168-020-00806-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rubens J.R., Selvaggio G., Lu T.K. Synthetic mixed-signal computation in living cells. Nat Commun. 2016;7:11658. doi: 10.1038/ncomms11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sheth R.U., Cabral V., Chen S.P., Wang H.H. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 2016;32(4):189–200. doi: 10.1016/j.tig.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hamady Z.Z., Scott N., Farrar M.D., Lodge J.P., Holland K.T., Whitehead T. Xylan-regulated delivery of human keratinocyte growth factor-2 to the inflamed colon by the human anaerobic commensal bacterium Bacteroides ovatus. Gut. 2010;59(4):461–469. doi: 10.1136/gut.2008.176131. [DOI] [PubMed] [Google Scholar]
- 111.Jayaraman P., Holowko M.B., Yeoh J.W., Lim S., Poh C.L. Repurposing a two-component system-based biosensor for the killing of vibrio cholerae. ACS Synth Biol. 2017;6(7):1403–1415. doi: 10.1021/acssynbio.7b00058. [DOI] [PubMed] [Google Scholar]
- 112.Whitaker W.R., Shepherd E.S., Sonnenburg J.L. Tunable expression tools enable single-cell strain distinction in the gut microbiome. Cell. 2017;169(3):538–46 e12. doi: 10.1016/j.cell.2017.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ziesack M., Gibson T., Oliver J.K.W., Shumaker A.M., Hsu B.B., Riglar D.T. Engineered interspecies amino acid cross-feeding increases population evenness in a synthetic bacterial consortium. mSystems. 2019;4(4) doi: 10.1128/mSystems.00352-19. [DOI] [PMC free article] [PubMed] [Google Scholar]