Graphical abstract
Keywords: High-density lipoprotein, HDL subpopulation, apolipoprotein A-I, Molecular dynamics simulation, Coarse-grained
Highlights
-
•
High-density lipoprotein subpopulations have unique surface profiles and dynamics.
-
•
Relative hydrophobic surface area decreases with increasing lipoprotein size.
-
•
Core lipid exposure at the lipoprotein surface decreases with increasing size.
-
•
Cholesterol molecules localise near apolipoprotein A-I central helices.
-
•
Lipid and protein interactions stabilise multifoil models of apolipoprotein A-I.
Abstract
Human serum high-density lipoproteins (HDLs) are a population of small, dense protein-lipid aggregates that are crucial for intravascular lipid trafficking and are protective against cardiovascular disease. The spheroidal HDL subfraction can be separated by size and density into five major subpopulations with distinct molecular compositions and unique biological functionalities: HDL3c, HDL3b, HDL3a, HDL2a and HDL2b. Representative molecular models of these five subpopulations were developed and characterised for the first time in the presence of multiple copies of its primary protein component apolipoprotein A-I (apoA-I) using coarse-grained molecular dynamics simulations. Each HDL model exhibited size, morphological and compositional profiles consistent with experimental observables. With increasing particle size the separation of core and surface molecules became progressively more defined, resulting in enhanced core lipid mixing, reduced core lipid exposure at the surface, and the formation of an interstitial region between core and surface molecules in HDL2b. Cholesterol molecules tended to localise around the central helix-5 of apoA-I, whilst triglyceride molecules predominantly interacted with aromatic, hydrophobic residues located within the terminal helix-10 across all subpopulation models. The three intermediate HDL models exhibited similar surface profiles despite having distinct molecular compositions. ApoA-I in trefoil, quatrefoil and pentafoil arrangements across the surface of HDL particles exhibited significant warping and twisting, but largely retained intermolecular contacts between adjacent apoA-I chains. Representative HDL subpopulations differed in particle size, morphology, intermolecular interaction profiles and lipid and protein dynamics. These findings reveal how different HDL subpopulations might exhibit distinct functional associations depending on particle size, form and composition.
1. Introduction
Plasma high-density lipoprotein (HDL) particles are highly heterogeneous polymolecular assemblies that primarily mediate intravascular lipid transport and regulate cardiovascular health (Lund-Katz and Phillips, 2010). The peripheral HDL subfraction is also associated with a number of vasoprotective properties, including anti-inflammatory, anti-oxidative, anti-thrombotic, anti-infectious, vasodilatory, and cytoprotective functionality (Camont et al., 2011). In normal human plasma, the overwhelming majority of circulating HDL particles are spheroidal in geometry, representing the mature forms of HDL. The structure–function relationship of mature HDL has nonetheless remained largely elusive, despite significant and intensive efforts across the preceding three decades.
It is worth briefly summarising the maturation process of endogenous HDL to provide context for the predominant mature species. Lipid-poor apolipoprotein A-I (apoA-I) is secreted from the liver or small intestine and is rapidly lipidated via interaction with the ATP-binding cassette transporter A1 (ABCA1) in a process involving the hydrophobic α-helices in the C-terminal domain of apoA-I (Nagao et al., 2014, Panagotopulos et al., 2002). The resulting nascent HDL particle is typically comprised of phospholipid (PL) molecules, cholesterol (CHOL) and up to two apolipoproteins. Nascent HDL readily interacts with the lecithin:cholesterolacyl transferase (LCAT) enzyme via apoA-I, which then facilitates the esterification of CHOL cargo into cholesteryl ester (CE) molecules. This process is accompanied by a profound geometric change from a discoidal particle to the prevailing spheroidal morphology. In this state, mature HDL particles undergo significant size and compositional changes in a multistage, receptor-mediated process known as remodelling (Zannis et al., 2015). Mature HDL remodelling proceeds within a non-specific, bidirectional cycle involving dynamic interconversions between particles of different sizes via the exchange of lipid/protein cargo with other circulating lipoproteins via cholesteryl ester transfer protein (CETP) and cell-surface receptors (scavenger receptor class B member 1, SRB1; ATP-binding cassette transporter G1, ABCG1) (Zannis et al., 2015).
Despite the dynamicity of HDL remodelling, within the mature HDL subfraction five major subpopulations can be separated and characterised based on increasing average particle size and decreasing average density: HDL3c (7.2–7.8 nm; 1.154–1.25 g/mL), HDL3b (7.8–8.2 nm; 1.129–1.154 g/mL), HDL3a (8.2–8.8 nm; 1.110–1.129 g/mL), HDL2a (8.8–9.7 nm; 1.088–1.110 g/mL) and HDL2b (9.7–12.9 nm; 1.063–1.087 g/mL). In addition, each major HDL subpopulation exhibits a distinct average molecular composition (Camont et al., 2013), which can inform the overall particle structure as well as its unique biological functionality (Camont et al., 2011). In this regard, HDL particle size and composition can profoundly influence the specificity of the particle to different receptors and ultimately dictate the overall progression of HDL remodelling (Asztalos et al., 2015). Furthermore, certain HDL compositional profiles and subpopulation distributions have been linked to a variety of vascular-related conditions (Boyce et al., 2017). For example, small, dense HDL3-family subtypes have been associated with favourable vasoprotective properties relative to larger HDL2 particles (Hye et al., 2014).
Developing a molecular understanding of the major endogenous HDL subpopulations is critical towards evaluating and elucidating their diverse biological functionality. Yet the structural and compositional heterogeneity of native HDL, in combination with their small size and soft nature, has historically made high-resolution structural investigations of these species via nuclear magnetic resonance (NMR) and X-ray crystallography techniques prohibitively challenging. As a consequence, high-resolution structural studies of HDL have focussed primarily on synthetic reconstituted HDL (rHDL) particles modelled on the relatively low-abundance and short-lived nascent HDL subfraction of human serum (Gogonea, 2016). Such synthetic particles can be readily prepared to exhibit considerable structural and compositional homogeneity (Cao et al., 2017), but are understandably limited in their biological relevance. Representative nascent rHDL are discoidal in structure and are typically composed of a single PL species in complex with two apoA-I molecules (Jonas et al., 2008).
ApoA-I is the major protein component of endogenous HDL and is responsible for providing considerable structural integrity to HDL and facilitating much of its functional specificity (Davidson and Thompson, 2007). Specifically, apoA-I is understood to exist in a largely exposed fashion on the HDL surface to represent one of the first points of interaction with circulating and cell-surface receptors (Jonas et al., 2008, Kontush et al., 2015). The prevailing arrangement of apoA-I molecules in discoidal rHDL is well characterised: the lipid-binding C-terminal regions of each apoA-I chain (residues 44 to 243) encircle a small PL bilayer patch in an anti-parallel arrangement centred upon helix-five (H5; residues 121–142) (Kontush et al., 2015). This arrangement is referred to as the “double-belt” model of apoA-I, and it owes much of its lipid solubilising capacity to the unique helical propensity of this lipid-binding C-terminal region (Segrest et al., 1999).
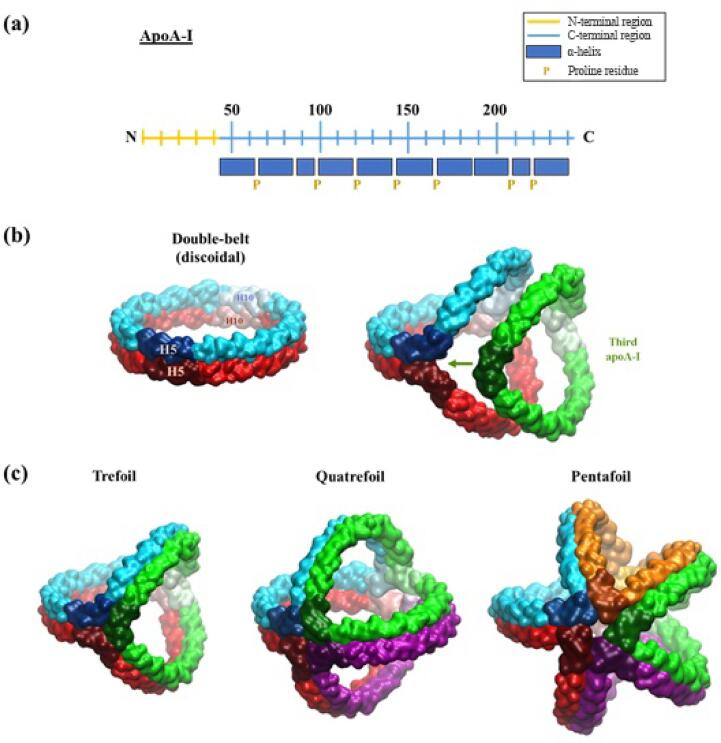
The C-terminal region of lipidated apoA-I is characterised by a series of ten 11- and 22-mer helical repeats primarily punctuated by proline residues (Fig. 1, a). For the most part, helices within this region are considered to be Class-A amphipathic α-helices (AαHs), which are characterised by opposing polar- and non-polar faces along the long axes of each helix (Phillips, 2013, Segrest et al., 1992). In addition, the outermost polar face of AαHs is generally populated by negatively charged residues, whereas positively charged residues tend to localise in the regions flanking the non-polar face. In the double-belt model, the non-polar faces of AαHs orient inwards towards PL tails, minimising solvent exposure at the edge of the bilayer patch, whilst polar faces orient outwards into solution. Charged residues within the external polar faces of apoA-I chains are understood to coordinate PL headgroups and interact with the adjacent apoA-I chain. In regards to this latter point, the stability of the double-belt model has been widely attributed to a complex network of intermolecular protein interactions including salt-bridges (Segrest et al., 1999) and, more recently, a collection of cation-π interactions within the defined “zipper” region of helix-4 (H4) and the opposing helix-6 (H6) (Bibow et al., 2017).
Fig. 1.
(a) Distribution of helices across the C-terminal region of lipidated apoA-I. (b) The transition from a double-belt model for two apoA-I molecules in discoidal HDLs, to the trefoil model for apoA-I containing three molecules in spheroidal HDLs. (c) Multifoil models for spheroidal HDLs containing three, four and five apoA-I chains. ApoA-I chains drawn in surface representation and coloured according to chain. Helix 5 (H5) and helix 10 (H10) for all chains are represented in dark and light shading respectively. ().
Adapted from Kontush et al., 2015, Huang et al., 2011, Segrest et al., 2015
Accumulating evidence suggests that the double-belt model of apoA-I in discoidal rHDL is the prevailing structural arrangement found within the majority of circulating nascent HDL (Huang et al., 2011). However, a double-belt model of apoA-I quickly becomes inadequate within the context of mature HDL subpopulations given their relatively large diameters, spheroidal geometries (ranging from spherical to ellipsoidal particles) with curved surfaces, as well as complex and diverse molecular compositions including multiple additional apoA-I molecules (Huang et al., 2011). Remarkably, despite these profound structural differences and diverse physicochemical properties, cross-linking studies of synthetic spherical particles as well as plasma-derived HDL subpopulations have unequivocally demonstrated that intermolecular apoA-I distances, consistent with a double-belt arrangement, remained largely conserved across all HDL species including mature HDL subpopulations (Huang et al., 2011, Silva et al., 2008).
In an effort to rationalise the apparent conservation of intermolecular apoA-I contacts across all HDL subpopulations, whilst also accounting for the profound and dynamic structural differences between nascent HDL and mature species, the “multifoil” arrangement of apoA-I was proposed (Huang et al., 2011, Silva et al., 2008). The multifoil arrangement is a variation of the double-belt model that describes the opening of each anti-parallel apoA-I chain via opposing lateral bending within the relatively flexible H5 regions and corresponding helix-10 (H10) regions. In this way, a bend angle of 120° at these points can allow for the incorporation of one additional apoA-I chain with the same geometry, whilst at the same time preserving the H5/H5 registry and intermolecular contacts found in the double-belt model (Fig. 1, b). Expansion of the HDL and further bending of the aforementioned angles within each apoA-I chain to 90° and 72° can allow for the incorporation of four and five apoA-I molecules per particle in the same registry (Fig. 1, c). Multifoil arrangements composed of three, four and five apoA-I chains are known as trefoil, quatrefoil and pentafoil models, respectively.
The multifoil arrangement of apoA-I in mature HDL is a promising model for how apoA-I chains may be intrinsically organised across the surface of endogenous particles. Firstly, all apoA-I contacts are consistent with the double-belt model and thus with cross-linking data by virtue of each chain retaining the same local intermolecular protein interactions. Secondly, multifoil models satisfy the geometries of mature HDL subpopulations given that the resulting arrangements resemble a spheroidal cage-like structure that is presumed to spread across the particle surface. Thirdly, progressive bending of apoA-I chains to accommodate the incorporation of additional molecules can theoretically continue beyond five chains, rationalising the existence of large HDL2b subpopulations containing seven (or more) apoA-I molecules (Huang et al., 2011). On the other hand, multifoil arrangements are also capable of accommodating smaller HDL particles with reduced surface areas via condensation and twisting of the entire protein model along one axis in a torsional fashion (Huang et al., 2011). Finally, the roughly symmetrical distribution of proteins in multifoil models allows for the division of the lipid surface area into approximately equivalent segments, in a configuration that is theoretically consistent with the lowest free energy state possible (Pan and Segrest, 2016).
It is one matter that the conceptual multifoil arrangement of apoA-I is compatible with the available structural data for mature endogenous HDL, but it is an altogether different matter as to whether such a model is biologically viable and stable within the context of the diverse molecular compositions and functionalities of mature HDL subpopulations. Such validation would ideally require detailed molecular descriptions of (1) the dynamic folding and inter-chain interactions between multifoil arrangements of different sizes in complex with well-defined mature HDL particles, and (2) the interplay between protein chains and constituent HDL lipid molecules. However, in the absence of compositionally and morphologically homogenous populations of mature endogenous HDL subpopulations, the required high-resolution structural characterisations of such particles are unfeasible using the currently available experimental techniques. To this end, sufficient isolation of well-defined HDL subpopulations from human plasma is impractical due to the intrinsic dynamicity and heterogeneity of endogenous HDL. Whilst the emergence and development of synthetic spherical HDL mimics (Thaxton et al., 2009, Luthi et al., 2015, Luthi et al., 2010, Cormode et al., 2008) could represent a promising strategy towards the structural characterisation of spherical HDL-like particles (in much the same way as synthetic discoidal rHDL have been used as a proxy for the nascent HDL population), it remains an ongoing challenge to synthesise spherical HDL-like mimics with the wide-ranging form and functionality of endogenous HDL subpopulation species (Henrich and Thaxton, 2019).
Since the early 1990s, advanced computational approaches such as molecular dynamics (MD) simulations have been utilised as a complementary strategy to characterise simplified, well-defined HDL particle models (Pan and Segrest, 2016). In addition to developing and exploring the structure of single HDL particles at a molecular and atomistic scale, MD simulations can be presently used to investigate the biophysical molecular properties of representative particles across timescales approaching tens of microseconds. The majority of HDL simulations have focussed on discoidal rHDL models due in large part to their smaller relative size and simple molecular composition, which are two qualities conducive to conducting HDL simulations across timescales long enough to generate sufficient statistics for comparison with, and validation against, the wealth of experimental structural data pertaining to rHDL. Some notable findings from MD simulations of discoidal HDL include spontaneous particle self-assembly (Shih et al., 2007), conformation, registry and stability of the double-belt model for apoA-I (Klon et al., 2002), as well as the gradual maturation of an rHDL particle to a spheroidal model following the progressive addition of CE molecules (Shih et al., 2009).
To date, a small number of MD simulations exploring the biophysical properties of representative spheroidal HDL systems have been reported (Shih et al., 2009, Vuorela et al., 2010, Koivuniemi et al., 2009, Jones et al., 2009, Yetukuri et al., 2010), owing to their relatively more complex compositions, larger sizes and limited experimental data for cross-validation purposes. Of the reported simulation studies of spheroidal HDL, most systems suffer from a combination of inadequate simulation times, randomly arranged or an insufficient number of apolipoprotein chains, and/or very simplified lipid compositions not especially representative of any particular endogenous HDL subpopulation. Indeed, in all reported cases simulated mature HDL models were composed of a neutral lipid core (typically comprised of CE, and/or triacylglycerides, TG) surrounded by a surface PL monolayer intercalated between embedded apoA-I chains. Vuorela et al. reported one of the most compositionally complex MD simulation studies of a representative spherical HDL2b-sized particle (Vuorela et al., 2010), comprised of experimentally relevant fractions of the four major HDL lipid groups – PL, CE, TG and CHOL, as well as a small quantity of curvature-promoting lysophosphotidylcholine (LysoPC) molecules. The resulting lipid droplet was complexed with two apoA-I chains in an arrangement resembling the double-belt model of apoA-I, with a third chain introduced semi-randomly across the HDL surface following particle equilibration (Vuorela et al., 2010). However, in a subsequent experimental study Huang et al. identified that plasma-derived HDL2b species typically contain between five and seven apoA-I chains on average (Huang et al., 2011). In light of this information, it remains unclear as to how realistic quantities of apoA-I chains complexed to HDL2b influence the structure, arrangement and dynamics of particles within this subpopulation and their constituent lipid and protein molecules.
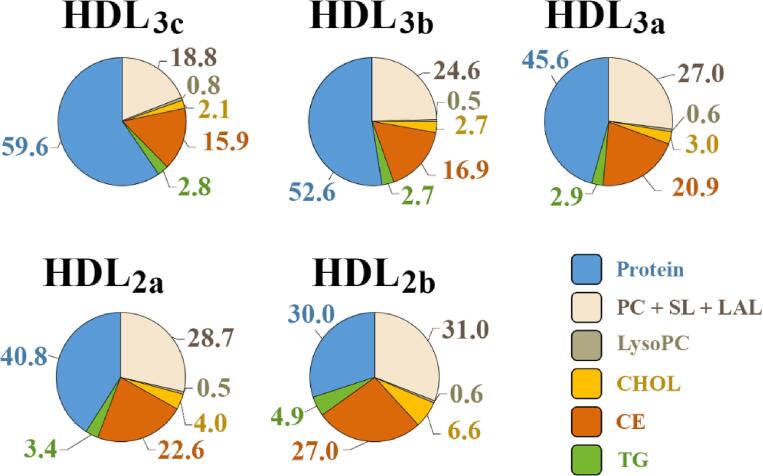
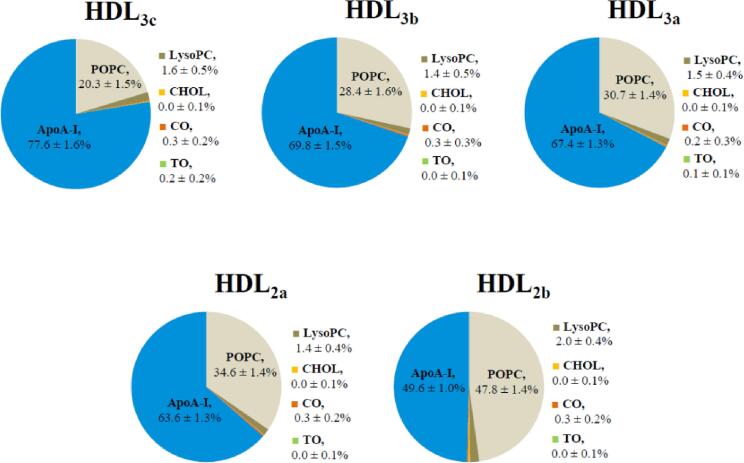
Throughout the preceding decade significant progress has indeed been made towards defining the average proteome and lipidome for the five major endogenous HDL subpopulations. Whilst the proteome of endogenous HDL has ballooned out to currently include between 85 and 204 different HDL proteins (Mangé et al., 2012, Shah et al., 2013), apoA-I remains the most abundant protein in periphery HDL fractions and is believed to be found in almost all circulating particles (Asztalos and Schaefer, 2003, Schaefer et al., 2010). However, as alluded to previously, the average distribution of total apoA-I chains across HDL subpopulations varies significantly (HDL3c – three to five; HDL3b – three to five; HDL3a – four to six; HDL2a – four to six; HDL2b – five to seven) (Huang et al., 2011), whilst the average relative proportion of protein to lipid is also subpopulation-specific (Camont et al., 2013) (Fig. 2). In terms of the HDL lipidome, Camont et al. utilised a combined ultra-performance liquid chromatography and mass spectrometry approach to determine that 162 unique lipid species can be found within the circulating HDL fraction in normolipidemic individuals (Camont et al., 2013). Importantly, endogenous HDL lipids are non-uniformly distributed across HDL subpopulations giving rise to distinct average molecular profiles (Camont et al., 2013) (Fig. 2). Phosphatidylcholine (PC) accounts for the overwhelming majority of PL at the surface of all mature HDL subpopulations (32–35 mol % of total lipids), CE forms the major core lipid component (15.9–27.0 mol %), whilst relatively small but significant concentrations of CHOL and TG are found in low and varying abundance across the five major HDL subpopulations (Camont et al., 2013). Sphingolipids (SL) including sphingomyelin and low-abundance lipid (LAL) species like LysoPC account for the remaining fractions of the HDL lipidome.
Fig. 2.
Average molecular profile for mature HDL subpopulations consisting of the major component groups, according to (Camont et al., 2013).
A defined average molecular profile for each HDL subpopulation provides a basis for developing more realistic conceptual and MD simulation models for the molecular arrangement and structure of all five major HDL fractions. Of course, the substantially heterogeneous and soft nature of endogenous HDL still underpins the immense challenges associated with effectively validating theoretical computational models of representative HDL particles. However, a number of broad biophysical averages for equilibrated representative HDL models (particle geometry and size, as well as molecular distributions) can be determined across long, multi-microsecond timescales by utilising a coarse-grained (CG) force-field model, and these properties can then be assessed and compared with measured experimental quantities. Additionally, particle surface profiles, quaternary protein arrangements, and intra- and inter-molecular contacts and residue distances can easily be assessed for consistency with known physicochemical properties of endogenous HDL as well as with other computational studies and theoretical models for apoA-I in spheroidal HDL.
The present study has set out to elucidate the structure and dynamics of representative models of spheroidal HDL subpopulations containing realistic average protein and lipid compositions. In this study, five HDL models representing the five human plasma spheroidal HDL subpopulations (HDL3c, HDL3b, HDL3a, HDL2a and HDL2b) have been developed and characterised for the first time. Each model was composed with lipid and protein quantities reflective of experimentally-determined averages for serum-derived HDL subpopulations.
2. Material and methods
Five HDL particle models representing the major subpopulations of HDL (HDL3c, HDL3b, HDL3a, HDL2a, and HDL2b) were developed using long-scale CG MD simulations. HDL compositions were calculated to accurately represent the average primary lipid and protein compositions of the major HDL subpopulations as derived experimentally for normolipidic HDL from human plasma by Camont et al. (Camont et al., 2013). The major HDL lipid groups PC, CHOL, CE and TG were represented by 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), unesterified free cholesterol molecules (CHOL), cholesteryl oleate (CO) and glycerine trioleate (TO), respectively. For this study, the sphingolipid (SL) contribution (<6 wt%) was combined into the POPC lipid group. A small but representative LysoPC portion represented by 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine was included in each model due to its known ability to enhance lipid monolayer curvature. The total HDL protein fraction of each model was represented by multiple apoA-I chains. The lowest predicted average number of apoA-I molecules per HDL-AI subpopulation (as reported in (Huang et al., 2011) was used for each system in an attempt to limit the total simulation system size, whilst also allowing for the investigation of three separate protein arrangements in complex with correspondingly proportionate lipid droplets. Thus, in HDL3c and HDL3b three apoA-I molecules were included: the larger HDL3a and HDL2a contained four apoA-I molecules, whilst the HDL2b system was developed with five apoA-I molecules. Each multifoil model was developed to exhibit a helical content consistent with experimentally determined helicity ranges for plasma-derived HDL-AI subpopulations (Huang et al., 2011). A full description of the HDL model preparation including multifoil development can be found in the ‘Model preparation’ section of Appendix A.
CG MD simulations were performed with the GROMACS 4.6.7 package using a time step of 20 fs for integrating the equations of motion for the lipid droplet (LD) systems and 10 fs for the full HDL particles. LD simulations and subsequent HDL simulations each totalled 25.0 µs (50.0 µs combined) to allow for sufficient lipid diffusion and particle equilibration (Vuorela et al., 2010). The standard components of the coarse-grained Martini force-field v.2.2 were used to describe POPC, LysoPC, CHOL, ions and water. The Martini model parameters of CO and TO, as parameterised by Vuorela et al. (Vuorela et al., 2010), were taken from the Martini website (http://md.chem.rug.nl/). Multifoil protein models were also described by the Martini v.2.2 force-field. The Nosé-Hoover thermostat (Hoover, 1985, Nosé, 1984) was used to maintain the temperature at 310 K and the Parrinello-Rahman barostat (Parrinello and Rahman, 1981) was used to maintain pressure at 1.0 bar using coupling constants of 1.0 ps and 4.0 ps, respectively. Lipids, proteins, and solvent were coupled to separate thermostats, and the whole system was coupled to the barostat isotropically. Cut-offs for Lennard-Jones and electrostatic interactions were set to 1.2 nm, consistent with the parameterisation of the Martini force-field (Monticelli et al., 2008). Additionally, for Lennard-Jones interactions the Gromacs shift function was introduced at 0.9 nm. Further details pertaining to all simulations presented in this study are outlined in the ‘Simulation details’ section of Appendix A.
The final 5.0 µs of HDL model simulation time was used for analysis. Analysis of trajectories was conducted using numerous in-house Python scripts utilising the MDAnalysis package (Gowers et al., 2019), as well as a variety of Gromacs 5.1.1 analysis programs. HDL model validation as well as structural and biophysical analyses included: spheroidal radial density profiles (RDPs); solvent accessible surface area (SASA); effective particle radius (RS); 2-dimensional (2D) and 3D lipid diffusion coefficients; inter- and intramolecular lipid-lipid contacts, inter- and intramolecular lipid-protein contacts including annular lipid identification, contact lifetimes, localisation maps, and residue-wise average intermolecular contacts; key inter- and intramolecular protein–protein interactions; and minimum geodesic lysine-lysine distances. A detailed description for each analysis method as well as additional analyses not presented in the main text of this manuscript can be found in section ‘Analysis methods’ within Appendix A.
Visualisation and figure preparation were conducted via Visual Molecular Dynamics (VMD) v. 1.9.3 (Humphrey et al., 1996). The Python plotting package Matplotlib v. 2.2.4 (Hunter, 2007) as well as Microsoft Excel (2016) were utilised for plotting. For all reported numerical results, the standard deviation from the mean is reported unless otherwise stated.
3. Results and discussion
3.1. Particle morphology, size and molecular distribution
Five representative HDL subpopulation models containing multiple chains of apoA-I and five lipid groups were simulated using the described methodology. Visual inspection of the final HDL structures shows that apoA-I chains form a distorted cage-like arrangement surrounding the lipid molecules (Fig. 3, a). In this arrangement, lipids molecules are bound at the surface by encircling apoA-I chains forming discrete lunes. Each lipid lune is composed primarily of POPC and LysoPC molecules oriented with polar headgroups facing the solvent.
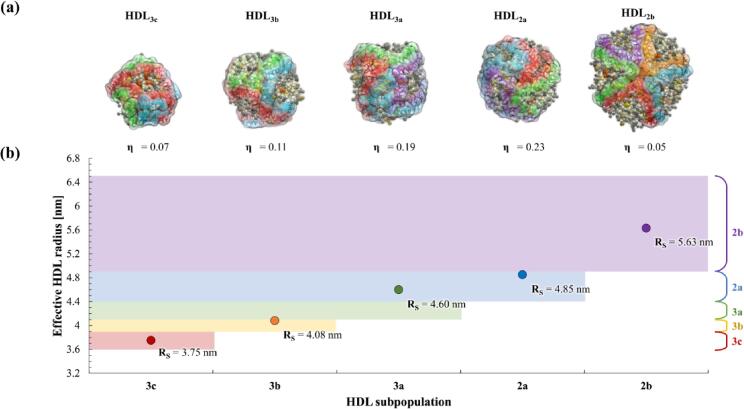
Fig. 3.
(a) Final snapshots of mature HDL subpopulation models centred on the apoA-I H5/H5 junction. Corresponding particle eccentricity values (η) are reported below each snapshot. (b) Effective HDL particle radii (RS) relative to experimentally determined HDL subpopulation particle size ranges as reported in (Blanche et al., 1981).
As expected, calculated Rs values for the five representative HDL models increase with increasing molecular composition (Fig. 3, b). Predicted Rs values for the mature HDL models are in very good agreement with experimental size ranges derived via native polyacrylamide gel electrophoresis (PAGE) (Blanche et al., 1981), with four of the five predicted values falling within their respective endogenous HDL subpopulation range. The exception to this is the predicted Rs value for HDL3a, which is 0.2 nm larger than the upper limit of its corresponding range, representing a small but significant deviation away from the predicted HDL3a particle size.
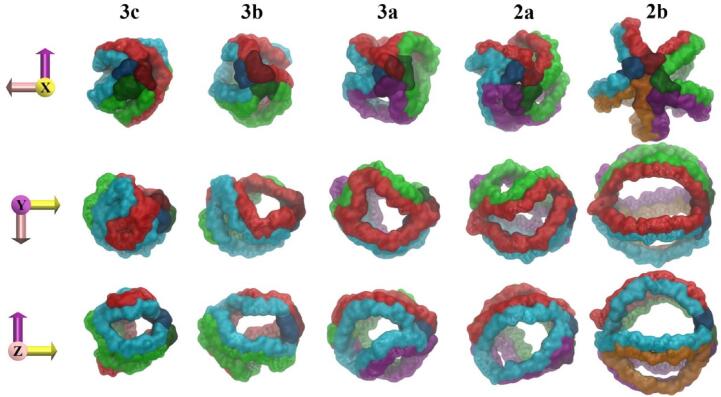
All HDL models display stable spheroidal geometries. HDL3c, HDL2b and to a lesser extent HDL3b are effectively spherical in geometry as indicated by η values approaching zero (Fig. 3, a). On the other hand, η values and corresponding axial radii of gyration (see Figure S2 in Appendix A) for HDL3a and HDL2a models are indicative of a prolate ellipsoid geometry. Comparison of the axial semi-radii for overall HDL particles with their respective core lipid regions (defined as all CO and TO molecules) follow similar trends, as illustrated in Fig. 4. This relationship suggests that the average geometry of the dynamic hydrophobic core is correlated with the overall average particle geometry. In this regard, it is interesting to consider that constituent LD and quatrefoil models used in the development of HDL3a and HDL2a models exhibited spherical forms (η < 0.05 ± 0.01 for each component; illustrated in Figure S3 in Appendix A) prior to association and preceding the formation of ellipsoidal particles. Hence, for these two systems the interaction between the quatrefoil model and each corresponding LD led to a profound geometric change in the HDL lipid phase (including the core region), the protein arrangement, and thus the overall HDL particle.
Fig. 4.
Front, side and top-down snapshots of HDL subpopulation models oriented along their principal axes of inertia. HDL particles represented through a transparent surface with CO and TO core molecules drawn in a surface representation and coloured orange and green, respectively.
In all cases, it should be emphasised that the two prevailing geometries described for HDL models in the present study are entirely consistent with experimental characterisations of native HDL subpopulations (Wu et al., 2011, Matz and Jonas, 1982, Davidson et al., 1994, Zhang et al., 2011). More specifically, in vitro preparations of HDL3a-sized spheroidal rHDL have been characterised with a similarly ellipsoidal form according to low-resolution SANS techniques (Wu et al., 2011). However, to the author’s knowledge the relative prevalence of spherical and ellipsoidal particles throughout mature HDL subpopulations has not been quantified experimentally - and it would be presumptuous to assume that clear morphological preferences can be inferred for HDL subpopulations from the single representative models presented in this study alone. Nevertheless, it is interesting to consider that two previous MD simulations have also described an ellipsoidal preference in intermediate-sized HDL models containing multiple apoA-I chains. A study conducted by Shih et al. described the formation and development of ellipsoidal HDL particles throughout the transition from HDL discs to an ultimately spherical HDL2b-sized particle following the progressive incorporation and sequestration of CE molecules (Shih et al., 2009). Additionally, Catte et al. conducted simulations of a HDL3b-sized model which adopted a prolate ellipsoidal shape following equilibration in atomistic and CG representations (Catte et al., 2008). In each case, two apoA-I chains warped and twisted across each HDL surface to accommodate the overall particle shape whilst remaining fully complexed with the lipid phase of the particle.
Of additional interest is whether a genuine relationship exists between HDL particles containing four apoA-I chains in a quatrefoil arrangement and the development of an overall ellipsoidal shape. This is ostensibly intriguing when considering that the apo Δ(1–43)A-I crystal structure (PDB: 1AV1) also describes four similarly truncated molecules arranged in an overall elliptical geometry (Borhani et al., 1997). However, it should be emphasised that apoA-I molecules in this crystal structure are arranged as two sets of antiparallel dimers, as opposed to the quatrefoil arrangement investigated in the present study. Notwithstanding the antiparallel registry of apoA-I dimers and multifoil models alike, some additional structural consistencies can be observed between the 1AV1 crystal structure and the predominant arrangement of apoA-I chains in the present HDL3a and HDL2b models. Firstly, the 1AV1 crystal structure describes mostly amphipathic helices arranged such that they encircle an elliptical volume with hydrophobic faces mostly oriented inwards. Secondly, the elliptical poles in the apo Δ(1–43)A-I crystal structure are located at H5 and H10 respectively, consistent with the arrangement of all apoA-I chains in the quatrefoil arrangement following HDL simulations. Thirdly, and perhaps most significantly, the quatrefoil model contains the highest helical content of all multifoil models used in the present study and thus exhibits the highest secondary structure identity with the predominantly helical apo Δ(1–43)A-I crystal structure. It should be emphasised that the overall helical percentages are consistent with CD-derived ranges for apoA-I in these subpopulations (Huang et al., 2011), however these values are average totals not specific to any particular region. In this regard, the given mapping scheme for individual apoA-I molecules within the quatrefoil model contains a high quantity of continuous helical domains which may not reflect the native distribution of secondary structure. Moreover, such domains could be limited in their ability to bend and twist in a way that can fully accommodate the corresponding HDL3a and HDL2a lipid volumes when arranged in a spherical geometry. The inability of the Martini protein model to allow for dynamic secondary structure rearrangements could further emphasise this effect to overestimate the ellipsoidal geometry of these models. Additional simulations using an ensemble of secondary structure mapping regimes for apoA-I chains could help to confirm this.
In view of the geometric findings for the present HDL models, it may be worth reconsidering the size discrepancy between the HDL3a model in the present study and PAGE estimates within the context of an ellipsoidal HDL geometry. HDL subpopulation size ranges are derived using native PAGE techniques that not only separate HDL particles according to their size and electrophoretic mobility, but also their geometry. Simulations of rigid ellipsoid particles in a turbulent channel flow have described a tendency for such geometries to orient their smallest cross-sectional area in the mean flow direction (Mortensen et al., 2008, Zhang et al., 2001). Hence, it is conceivable that an ellipsoidal HDL3a particle could migrate through a polyacrylamide gel in a similar orientation to ultimately deposit at a point reflecting its shortest semi-axis. Taken together with the fact that HDL are soft and thus relatively plastic in nature, it is plausible that RS as a measure of size for ellipsoidal HDLs is susceptible to overestimation when comparing with experimental quantities.
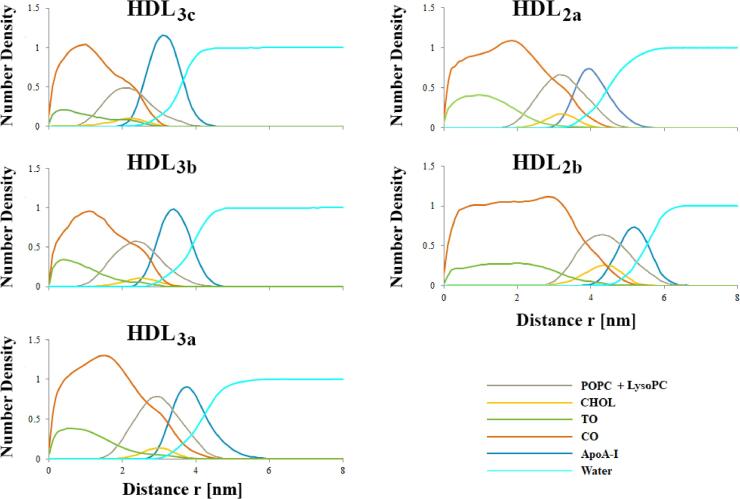
Considering the geometric findings for the present HDL models, RDP calculations assumed a prolate ellipsoidal volume for HDL3a and HDL2a whilst a conventional spherical volume was used for HDL3c, HDL3b and HDL2b. By calculating RDPs in this way, the relative distribution of molecular components, especially at the particle surface, can be appropriately assessed and compared between models. RDPs for all representative HDL models are shown in Fig. 5 and demonstrate several consistent distribution patterns for each molecular component. Specifically: (1) apoA-I curves in all models represent the outer extremity of HDL models, display considerable overlap with water curves and, along with water curves, do not penetrate significantly into the core region of HDL particles; (2) CO and TO curves overlap with each other, are predominantly located towards the particle interior in order of CO then TO at the centre, and exhibit relatively minor overlap with apoA-I and water curves, (3) combined surface lipid (POPC + LysoPC) profiles remain roughly of the same width irrespective of subpopulation model and consistent with the “Shen” structural model for lipoproteins (Shen et al., 1977), wherein the interfacial region of HDL is composed of a lipid monolayer of roughly 2 nm, and; (4) surface lipid and CHOL curves overlap considerably with one another and are located in all models between the apoA-I peak and the TO and CO maxima (with considerable overlap across both regions). Overall, the distributions for HDL components within these models are generally consistent with predicted HDL structures (Kontush et al., 2015, Shen et al., 1977, Mazer et al., 2013) as well as previous MD simulation studies (Shih et al., 2009, Vuorela et al., 2010, Koivuniemi et al., 2013, Koivuniemi et al., 2012, Ollila et al., 2012).
Fig. 5.
Combined component-wise RDPs for HDL subpopulation models. All RDPs are normalised relative to bulk water for ease of visual comparison between plots. For HDL3a and HDL2a, r is also normalised as the relative distance along all principal axes. Note: LysoPC profiles were not distinguishable in these plots, so for clarity they were combined with POPC distributions where they exhibited considerable overlap.
Amongst HDL models, the two distinct maxima observed for CO and TO within the core volume are most profound for smaller HDL. With increasing particle size these profiles progressively flatten and expand into their corresponding core volume. Ultimately for HDL2b both core lipid profiles resemble a plateau throughout the core region indicative of a volume approaching compositional heterogeneity. The migration of TO lipids towards the central inner core domain in smaller HDL subpopulation models could be interpreted as the initial stages of a microphase separation of HDL core components, as had been previously characterised via MD simulations of spheroidal LDs containing fixed levels of core lipids and a varying quantity of surface lipids (Ollila et al., 2012). In that study, higher quantities of surface lipids reduced the separation of core components and lowered their solvent accessibility. Whilst the effect of CHOL was not tested in that study, the authors postulated that CHOL enrichment in the HDL interfacial layer may further reduce core lipid solubility, consistent with the behaviour of neutral lipids in phospholipid bilayers (Spooner et al., 1986, Spooner and Small, 1987).
In addition to the overall expansion and plateauing of core lipid RDPs with increasing HDL size, the overlap of core lipid profiles with the HDL surface components progressively decreased such that the HDL2b model exhibited a clear separation between surface and core lipids. This separation of PC and CHOL from core lipids in larger HDL2 particles is consistent with the development of an interstitial region, as has been previously described for a similarly large HDL model (Vuorela et al., 2010). Alternatively, the extent of overlap between core and surface components in small HDL suggests that not only do these particles lack an interstitial region, but also that a somewhat nebulous intersection of core and interfacial regions exists. Intriguingly, a recent atomic force microscopy study of HDL subclasses identified that HDL3-sized particles contain a surprising quantity of core lipids within the interfacial region (Amigó et al., 2016). In that study the authors attributed this effect to the spatial restrictions imposed on small HDL due to the proportional volumetric differences between the interfacial shell and the HDL core. Specifically, the interfacial volume of mature HDL is defined by a relatively constant shell width of roughly 2 nm equating to the thickness of a phospholipid monolayer irrespective of particle size (Shen et al., 1977). On the other hand, the core volume in spherical HDL is proportional to the third power of the HDL radius minus the shell width. Hence, for small HDL the inner core volume is particularly sensitive to small changes in the overall particle radii, and thus core lipids could be subjected to significant spatial restrictions resulting in enhanced exposure at the surface. It is also interesting to consider this effect in relation to the resulting geometries of HDL3a and HDL2a in this study, given that a prolate spheroid HDL has a proportionally larger interfacial shell volume relative to a spherical HDL of equivalent volume. Thus, it is conceivable that the relative compositions of core and surface components used for HDL3a and HDL2a in this study are inherently conducive to the formation of an ellipsoidal geometry as opposed to this resulting geometry representing an artefact of the system (such as the quatrefoil model).
A final observation from the RPDs is related to the CO “shoulder” peak coinciding with the inner-side of the apoA-I maxima. With increasing HDL size, this shoulder peak progressively declines until it is almost imperceptible in HDL2b. Given its location, this shoulder could be suggestive of CO infiltration into the interfacial lipid layer as a consequence of the aforementioned spatial restrictions in small HDL. Alternatively, considering that TO exhibits lower hydrophobicity relative to CO, it is more likely that this shoulder represents a favourable interaction between CO and the hydrophobic underside of apoA-I. Increases in HDL size and surface lipid number, as well as CHOL enrichment in the interfacial layer and presumably at the hydrophobic underside of apoA-I, may combine to ultimately redistribute apoA-I-bound CO towards the HDL core and contribute to the overall softening of this CO shoulder in large HDL subpopulations. Given that the rate of disassociation of apoA-I from HDL decreases with increasing particle size (Liang et al., 1994), is tempting to speculate that a reduction in CO molecules within the interfacial region, in combination with an increase in PC and CHOL lipidation of apoA-I, enhances the overall stability of the protein across the particle surface.
3.2. HDL surface profiles
The approximate average proportional component-wise surface profiles for the five HDL subpopulation models are shown in Fig. 6. ApoA-I and PC fractions contribute the highest proportional exposed surface areas across all HDL models, notionally consistent with the relative distribution of these components according to their respective RDP curves in Fig. 5. Additionally, the calculated average apoA-I surface coverage for HDL3c is in reasonable agreement with predicted estimates based on theoretical models of HDL3c (∼85%) (Huang et al., 2011). With increasing HDL size, the apoA-I fraction gradually reduced from 77.6% total surface coverage in HDL3c to 49.6% in HDL2b, whilst the proportional lipid exposed surface increased with increasing particle size. This progressive increase in total HDL lipid surface area with increasing HDL size is consistent with the enhanced capacity for larger HDL2 particles to interact with CETP, which binds at the HDL lipid surface to mediate the shuttling of core lipid molecules between circulating lipoproteins (Zhang et al., 2015).
Fig. 6.
Average proportional component-wise solvent-accessible surface area for HDL models.
Interestingly, for HDL3b, HDL3a and, to a lesser extent, HDL2a the proportional exposed lipid and protein surface areas were roughly equivalent despite each particle having different compositions and significantly different core to surface component ratios. It is conceivable that the transition from a spherical geometry to an ellipsoid between the HDL3b model and HDL3a and HDL2a may be somewhat responsible for preserving this proportional surface profile for the reasons discussed previously. Considering this preservation across the three intermediate HDL subpopulation models, it is also tempting to speculate that such an interfacial composition holds some biological relevance. For example, the relatively uniform proportional interaction profile may begin to explain how HDL3 and HDL2 subpopulations can be equally effective acceptors of ABCG1-derived CHOL via a passive diffusion pathway that is independent of apoA-I content and conformation (Rothblat and Phillips, 2010).
For all models, POPC is the overwhelmingly prevalent solvent-exposed lipid fraction making up 20.3% of the total solvent-accessible surface area (SASA) in HDL3c and increasing up to almost half of the total SASA (47.8%) in HDL2b. As expected, LysoPC contributes the next largest lipid fraction, accounting for 1.4–2.0% of the SASA across HDL models. On average, all other lipids represent<0.6% of the total SASA across the simulation times investigated. Somewhat surprisingly, the calculated SASA for CHOL is virtually zero in all HDL models despite occupying a similar region to surface PCs and having some overlap with water curves in all HDL RDPs. This finding may suggest that CHOL molecules are predominantly restricted to the surface lipid tail region consistent with a similar localisation of CHOL (and indeed other sterols) in MD simulations of PC/CHOL bilayers (Hughes et al., 2013, Hughes and Mancera, 2013), as well as the underside of apoA-I as was previously characterised by Vuorele et al. in their simulation study of a large HDL model (Vuorela et al., 2010), to ultimately be protected from solvent exposure across the simulations. Consistent with the notion that smaller HDL experience a higher quantity of core lipid exposure at the surface, HDL3c exhibits the highest average quantity of TO at the surface of all HDL subpopulation models in this study. However, it should be noted that the high deviation associated with this measurement in HDL3c is reflective of transient exposures of TO for this model. This level of TO exposure is noteworthy given that HDL3c also has the lowest proportional lipid exposure of all HDL models, such that TO accounts for almost 1% of the total exposed lipid content in HDL3c relative to < 0.2% for all other HDL species.
The overall average hydrophilic SASA increases significantly with increasing particle size, whilst the hydrophobic SASA moderately increases (Fig. 7, a). The former observation can be largely attributed to an increase in POPC fraction at the HDL surface, whist the latter observation is consistent with a previous MD simulation study of the progressive maturation of HDL by Shih et al. (Shih et al., 2009). It is worth noting that as a relative fraction of the total surface area, the hydrophobic contribution decreases with increasing particle size: HDL3c – 32.1%, HDL3b – 30.6%, HDL3a – 28.3%, HDL2a – 26.0% and HDL2b – 20.0%. This is consistent with the notion that a higher surface curvature in smaller HDL results in a greater degree of hydrophobicity, as has been demonstrated for liposomes (Zhang et al., 2015). In that study, increased relative surface hydrophobicity correlated with a higher CETP binding capacity (Zhang et al., 2015). However, for HDL CETP-binding increases with increasing HDL size as discussed previously. This can be explained by the fact that CETP-binding proceeds via interactions with the lipid surface of HDL, and in the present models of HDL apoA-I accounts for the overwhelming contribution to the average total hydrophobic SASA (Fig. 7, b), with this proportion slightly decreasing from 97.5% for HDL3c to roughly 92.0% for HDL2b. The next highest contributor to average total hydrophobic SASA in the HDL models is POPC, accounting for 1.5% in HDL3c and up to 6.3% in HDL2b (data not shown). Considering that apoA-I hydrophobicity mediates the binding of lipids and additional apolipoproteins during HDL biogenesis (Nagao et al., 2014), the relatively high proportional hydrophobic SASA attributed to apoA-I in the five present models may also be indicative of a similar mechanism of binding of hydrophobic species for mature HDL.
Fig. 7.
Average hydrophobic and hydrophilic solvent-accessible surface area (SASA) for (a) HDL (b) all apoA-I chains, and (c) average solvent-accessible surface area per apoA-I chain (note the different scale used for this particular plot).
The average per-chain contribution to hydrophobic SASA trends downwards for apoA-I with increasing HDL size (Fig. 7, c), indicative of fewer exposed hydrophobic regions. This effect may be explained by the larger total particle surface area allowing for a more complete incorporation of multifoil models within the surface lipid monolayer. In addition, an increase in the total number of apoA-I chains in larger particles may also be contributing towards the masking of hydrophobic lipid and protein sites. This trend in apoA-I hydrophobicity suggests that despite larger HDL containing higher quantities of apoA-I, on average these chains exhibit fewer exposed hydrophobic regions at the surface interface. To the author’s knowledge, no studies have examined the effects of HDL subpopulation size on the overall lipid binding kinetics. However, in a somewhat related fashion the effectiveness of HDL to bind the hydrophobic peptide beta-amyloid has been shown to decrease in HDL2-sized particles relative to HDL3 despite having a larger available total surface area for interaction (Koudinov et al., 1994). Indeed, the proposed beta-amyloid binding site of apoA-I is a highly conserved seven residue region containing four hydrophobic Leu residues with a proclivity to form an exposed loop (Paula-Lima et al., 2009). Taken together, it could be speculated that HDL particle size and composition modulates the overall capacity of apoA-I to bind additional lipids as well as other hydrophobic species in a self-regulating fashion.
3.3. Lipid diffusion
Long-scale lipid dynamics were determined for each species according to its diffusion coefficient, D, and characterised according to its prevalent dimension of diffusion: two-dimensions (2D) or three-dimensions (3D). Lipid-wise diffusion trends are plotted in Fig. 8 relative to the average distance of each lipid species from the HDL centre of mass (corresponding calculated D and best fitting diffusion dimensionality for each lipid group across HDL models can be found in Table S2 in Appendix A). The dimensionality of lipid diffusion for all HDL models in the present study were consistent with the findings of Vuorela et al. (Vuorela et al., 2010), whereby POPC, LysoPC and CHOL diffused primarily along the HDL interfacial layer in 2D, whilst core lipids (CO and TO) diffused in 3D within the particle core. Between HDL models, calculated D for each lipid type roughly increased with increasing HDL particle size up to HDL2a, which shared statistically equivalent values with HDL2b. Calculated D values for core lipids in HDL2a and HDL2b were roughly equivalent to those reported by Vuorela et al. for a similarly sized HDL particle (DCO = 0.36 10-7cm2 s−1, DTO = 0.27 10-7cm2 s−1 for a HDL with Rg = 4.079 nm) (Vuorela et al., 2010). On the other hand, lipid diffusion for LysoPC, POPC and CHOL proceeded roughly two to three times slower for HDL2b in the present study relative to the corresponding values from the Vuorela et al. study. This discrepancy in D for interfacial lipids across the two studies may be best explained by the differences in molecular composition, with the HDL2b model in the present study containing five apoA-I chains across the HDL surface compared with only two chains in the previous study. The total quantity of interfacial lipids is also higher in the Vuorela et al. study compared with HDL2b in the present study (319 and 277 molecules, respectively). Taken together, interfacial lipid motion in the presence of high proportions of protein is expected to be substantially more impeded on a HDL surface due to the resulting volumetric exclusion.
Fig. 8.
(a) Lipid diffusion coefficients for each lipid species across HDL models (dashed lines) relative to the average distance of each lipid from the centre of mass of the lipoprotein particle. In order to compare 2D and 3D diffusion, all diffusion coefficient values are scaled by a factor 2d, where d is the dimension of the fit. (b) Cut-away view of HDL2b model depicting the distribution of lipid species at the conclusion of the simulation, with all lipids coloured according to their respective average lipid diffusion coefficient values. ApoA-I protein chains have been drawn as a transparent outlined surface to provide visual context.
Across all HDL particles lipid diffusion was slowest within the core, with TO molecules diffusing slower than CO (Fig. 8). This finding is likely to reflect the larger size and geometry of TO relative to CO, such that a large molecular morphology can significantly impede diffusion in a crowded and restricted volume such as the HDL core. Similarly, LysoPC diffusion was faster than POPC in the interfacial region for all models except HDL3b, where POPC and LysoPC shared statistically equivalent values of D. In the case of CHOL, which straddles the region between core and interfacial volumes according to RDPs, predicted D is marginally slower than PCs but substantially higher than core lipids across all HDL models. In this local environment beneath the lipid headgroups, it is conceivable that CHOL motion is somewhat inhibited by entanglement within acyl chains.
3.4. Inter- and intramolecular lipid-protein interactions
The residue-wise average number of molecular contacts between lipid groups and apoA-I are shown in Fig. 9. These plots highlight which lipids exhibit a preferential affinity for the different apoA-I residue types throughout each simulation. Across all HDL models, aromatic hydrophobic residues (Trp, Tyr and Phe) exhibited the greatest affinity for each lipid species. Unsurprisingly, other hydrophobic residues make up the majority of additional interactions with all lipid molecules, whilst aromatic His residues also feature prominently for the five lipid types. Interactions with Arg residues are well represented for POPC molecules, perhaps suggestive of an electrostatic interaction involving POPC headgroups and these residues. On the other hand, as expected, hydrophilic residues tended to exhibit the least preference for lipid molecules.
Fig. 9.
Average number of lipid contacts with apoA-I residue types, normalised per number of residues for the five HDL subpopulation models. Corresponding average values are labelled for each residue. Note: Average number of protein residue contacts with TO have been plotted in separate plots for each HDL on a different scale to POPC/LysoPC and CHOL/CO plots to enhance readability.
The distribution of minimum average intermolecular distances between lipid types CHOL and TO and apoA-I residues from all chains are represented for the five HDL models in Fig. 10, providing an indication of the preferential distribution of these lipid types relative to regions of apoA-I. Corresponding fingerprint distribution plots for POPC and CO, as well as all chain-wise plots can be seen in Figure S8 and Figure S9, respectively, in Appendix A. For all HDL models, the H5 region corresponding to the multifoil junction (residues 133–138) was largely lipidated with CHOL molecules. The localisation of CHOL around H5 is interesting to consider within the context of LCAT-mediated esterification of HDL-CHOL, which proceeds following binding of apoA-I at H5, helix 6 and to a lesser extent helix 7 (Maiorano et al., 2004, Frank and Marcel, 2000). In this respect, it is conceivable that this region of HDL-bound apoA-I can act as a CHOL reservoir in order to enhance the LCAT-mediated conversion of CHOL to CE and thus HDL maturation. With increasing HDL size and thus increasing number of CHOL molecules, CHOL distributions tended to broaden from H5 towards both termini. The exception to this trend is for H4, which notably lacked CHOL interactions across all HDL models. This is a curious finding given that H4 contains four aromatic hydrophobic residues (Y100, F104, W108, Y115), which have previously been shown to preferentially interact with CHOL in a study of a HDL2b-sized particle (Vuorela et al., 2010). Indeed, in the present study the preference for CHOL to interact with aromatic, hydrophobic residues was only notable in the HDL2b model (see Fig. 9).
Fig. 10.
Fingerprint plots showing the distribution of minimum average intermolecular contact distances between apoA-I residues and (a) cholesterol and (b) trioleate, for all apoA-I chains per HDL model. ApoA-I helices are coloured as alternating slate-grey and pale-blue rectangles at the base of each fingerprint plot.
Across all HDL models TO interactions with apoA-I were typically facilitated via hydrophobic and/or aromatic residues and focused towards the termini of apoA-I (Fig. 9 and Fig. 10, b, respectively). Specifically, TO molecules tended to localise around H10 in all models except HDL2a, where TO was predominantly located within H9. In smaller HDL3-family models TO was also found to interact at H1, which could be due to the smaller particle size and the anti-parallel H10-H1 interaction facilitating TO interactions with both AαHs. Given that hydrophobic amino acids represent the majority of interacting residues with TO across the five HDL models, it is likely that the preferential localisation of TO molecules towards H10 is related to this region exhibiting the highest total hydrophobicity and most hydrophobic helical face of all 22-mer AαH repeats in apoA-I (Palgunachari et al., 1996). Additionally, the capacity to directly interact with core lipids may be related to H10 having the highest relative capacity to deeply penetrate lipid surfaces according to AαH modelling (Palgunachari et al., 1996). Furthermore, the localisation of TO molecules towards the C-termini of apoA-I chains is consistent with in vitro studies of apoA-I and apoA-I-derivatives adsorbing at TO/water and POPC/TO/water interfaces (Wang et al., 2007, Mitsche and Small, 2011). In these studies, H10 was identified to exhibit high affinity for TO molecules and regulate apoA-I adsorption at each lipid interface (Wang et al., 2007, Mitsche and Small, 2011). Moreover, these studies revealed that the desorption of apoA-I from such interfaces was significantly higher for C-terminal truncated apoA-I to suggest that H10 interactions with TG stabilise apoA-I on lipid interfaces and could thus represent a critical modulating factor for apoA-I exchange between lipoproteins (Wang et al., 2007, Mitsche and Small, 2011).
These studies highlight the importance of both region- and lipid-specific protein-lipid interactions towards modulating and regulating apoA-I stability at the HDL surface. With respect to the present models, for which all apoA-I molecules remained complexed to each HDL particle throughout the simulation time periods, it could be extrapolated that specific apoA-I lipidation profiles including CHOL accumulation centred around H5 as well as C-terminal lipidation via TO molecules could play important structural and/or functional roles towards facilitating apoA-I retention, moderating protein conformation and stability, and more broadly helping to preserve and stabilise the overall structures and morphologies of spheroidal HDL particles.
4. Protein arrangement and conformation
The extent of quaternary structure distortion of the multifoil arrangements following HDL simulations is shown in Fig. 11. The warping of multifoil models following incorporation within LD surfaces appears to be most pronounced for the smallest HDL3c model and becomes progressively less noticeable with increasing HDL size. Despite this rearrangement, intermolecular interactions between neighbouring apoA-I chains across the three multifoil arrangements appear to be largely preserved across all HDL models, and specifically all chains retain the H5/H5 registry in all HDL simulations. All transitional HDL models up to HDL2b demonstrate structural distortion somewhat resembling a torsional rearrangement, as predicted by Huang et al. as a way for the multifoil arrangement to accommodate smaller surface areas in transitional HDL (Huang et al., 2011). This is best illustrated from the x-axis perspective of HDL2a in Fig. 11 where a twisting of the quatrefoil model can be seen. Incidentally, the observed warping of multifoil arrangements is centred along the axis passing through the H5/H5 junction. Considering that the H5/H5 junction serves as one of the fixed points from which a torsional rearrangement proceeds within multifoil models, it suggests that this region exhibits relative rigidity and stability following the incorporation of each multifoil model into a HDL surface.
Fig. 11.
Final snapshots of all multifoil apoA-I arrangements for the five HDL models (lipids not shown). ApoA-I chains are drawn as a surface and coloured individually. Helix 5 sections of apoA-I are centred in the x-axis perspective and shaded in a darker hue.
As a way to assess the spatial validity of multifoil models complexed to spheroidal HDL, relevant Lys-Lys cross-linking geodesic distances were monitored. In each HDL model simulated, all 19 cross-linking distances relevant to the apo Δ(1–43)A-I chains used in this study were found to be satisfied via intra- and/or intermolecular geodesic distances during the simulation time period (Table S4 in Appendix A). Moreover, at least one average geodesic distance remained within the cut-off throughout the final 5 µs of simulation time for each cross-link evaluated across all HDL models investigated, with the exception of three cross-links within the HDL2b model (Lys182-Lys226, Lys182-Lys238 and Lys182-Lys239). For all HDL models, the shortest average intermolecular cross-linking distances tended to be associated with the H5/H5 intersection (Lys133-Lys140) as well as the C-termini of apoA-I chains (Lys226-Lys238, Lys238-Lys239 and Lys239-Lys239). This is illustrated in Fig. 12 corresponding to the HDL3c model, whilst Lys-Lys backbone bead distance wheels for each HDL model can be seen in Figure S14 in Appendix A.
Fig. 12.
(a) Intra- and intermolecular apoA-I lysine-lysine backbone bead distance wheel for HDL3c model containing three apoA-I chains. (b) Visual representation of all satisfied geodesic lysine-lysine backbone bead distances for HDL3c. In each representation, coloured lines indicate average minimum geodesic distances below the 2.6 nm cut-off and greyscale lines in the background indicate all global minimum geodesic distances that satisfy the cut-off. Lysine residues are coloured as yellow nodes in (a), and lysine backbone beads are drawn as yellow spheres in (b). Standard error estimates for all average intra- and intermolecular distances were measured to be < 0.15 nm.
5. Conclusions
In this study, five computational models of the major HDL subpopulations in human plasma have been developed using representative protein and lipid compositional profiles and investigated using long-scale CG MD simulations. The simulated models exhibited discrete size and form profiles largely consistent with experimental data for their respective subpopulations. Moreover, the five HDL models were found to share similarities regarding the general distribution of constituent molecules. Each subpopulation particle could nonetheless be defined by a distinct molecular arrangement, surface profile and a number of biophysical properties. Across all HDL models, lipid molecules were found to separate into a core region composed predominantly of TO and CO, whilst POPC, LysoPC and CHOL molecules arranged to form a roughly 2 nm thick HDL surface monolayer bound between apoA-I chains in lipid lunes. Increasing HDL size correlated with a reduction in core lipid infiltration at the HDL surface layer, enhanced core lipid mixing, faster lipid diffusion and ultimately the emergence of an interstitial region between core and surface layers in the largest HDL2b model. The distribution of lipid molecules with respect to different regions of apoA-I revealed that in all HDL models, CHOL molecules localised around H5 – a site critical for the LCAT-mediated esterification of CHOL. Moreover, the aromatic and/or hydrophobic residues within the C-terminal region of apoA-I in all HDL models were found to preferentially interact with TO in a mode consistent with the penetration of this region into the HDL core, and more broadly implicated in apoA-I retention and stability on the HDL surface.
Additionally, the structure and dynamics of conceptual multifoil models for apoA-I were investigated in three configurations (trefoil, quatrefoil and pentafoil) within the context of full HDL particles. Upon partial incorporation across the surface of HDL particles, each multifoil model underwent significant warping and twisting but largely retained intermolecular contacts between adjacent apoA-I chains and relevant geodesic cross-linking distances throughout all simulations. In summary, these results provide greater insight towards understanding how different HDL subpopulations and respective apoA-I arrangements may exhibit distinct functional associations depending on particle size, form and composition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by resources provided by the Pawsey Supercomputing Centre with funding from the Australian Government and the Government of Western Australia, as well as resources and services from the National Computational Infrastructure (NCI), which is supported by the Australian Government.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.yjsbx.2020.100042.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Amigó N., Mallol R., Heras M., Martínez-Hervás S., Blanco-Vaca F., Escolà-Gil J.C., Plana N., Yanes Ó., Masana L., Correig X. Lipoprotein hydrophobic core lipids are partially extruded to surface in smaller HDL: “Herniated” HDL, a common feature in diabetes. Sci. Rep. 2016;6:19249. doi: 10.1038/srep19249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asztalos B.F., Horvath K.V., Goldfine A., Schaefer E.J., Particles H.D.L., Functions H.D.L. Arterioscl. Thrombos. Vascul. Biol. 2015;35(Suppl 1):A124. [Google Scholar]
- Asztalos B.F., Schaefer E.J. HDL in atherosclerosis: actor or bystander? Atherosclerosis Supplements. 2003;4(1):21–29. doi: 10.1016/s1567-5688(03)00006-0. [DOI] [PubMed] [Google Scholar]
- Bibow S., Polyhach Y., Eichmann C., Chi C.N., Kowal J., Albiez S., McLeod R.A., Stahlberg H., Jeschke G., Guntert P., Riek R. Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I. Nat. Struct. Mol. Biol. 2017;24(2):187–193. doi: 10.1038/nsmb.3345. [DOI] [PubMed] [Google Scholar]
- Blanche P.J., Gong E.L., Forte T.M., Nichols A.V. Characterization of human high-density lipoproteins by gradient gel electrophoresisBiochimica et Biophysica Acta (BBA) - Lipids and Lipid. Metabolism. 1981;665(3):408–419. doi: 10.1016/0005-2760(81)90253-8. [DOI] [PubMed] [Google Scholar]
- Borhani D.W., Rogers D.P., Engler J.A., Brouillette C.G. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. PNAS. 1997;94(23):12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce G., Button E., Soo S., Wellington C. The pleiotropic vasoprotective functions of high density lipoproteins (HDL) J Biomed. Res. 2017;32(3):164–182. doi: 10.7555/JBR.31.20160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camont L., Chapman M.J., Kontush A. Biological activities of HDL subpopulations and their relevance to cardiovascular disease. Trends Mol. Med. 2011;17(10):594–603. doi: 10.1016/j.molmed.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Camont L., Lhomme M., Rached F., Le Goff W., Nègre-Salvayre A., Salvayre R., Calzada C., Lagarde M., Chapman M.J., Kontush A. Small, dense high-density lipoprotein-3 particles are enriched in negatively charged phospholipids relevance to cellular cholesterol efflux. Antioxid., Antithromb., Anti-Inflammatory, Antiapopt. Functional., Arterioscl., thromb. Vascul Biol. 2013;33(12):2715–2723. doi: 10.1161/ATVBAHA.113.301468. [DOI] [PubMed] [Google Scholar]
- Cao Y.-N., Xu L., Han Y.-C., Wang Y.-N., Liu G., Qi R. Recombinant high-density lipoproteins and their use in cardiovascular diseases. Drug Discovery Today. 2017;22(1):180–185. doi: 10.1016/j.drudis.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Catte A., Patterson J.C., Bashtovyy D., Jones M.K., Gu F., Li L., Rampioni A., Sengupta D., Vuorela T., Niemelä P., Karttunen M., Marrink S.J., Vattulainen I., Segrest J.P. Structure of spheroidal HDL particles revealed by combined atomistic and coarse-grained simulations. Biophys. J. 2008;94(6):2306–2319. doi: 10.1529/biophysj.107.115857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormode D.P., Skajaa T., van Schooneveld M.M., Koole R., Jarzyna P., Lobatto M.E., Calcagno C., Barazza A., Gordon R.E., Zanzonico P., Fisher E.A., Fayad Z.A., Mulder W.J.M. Nanocrystal core high-density lipoproteins: a multimodality contrast agent platform. Nano Lett. 2008;8(11):3715–3723. doi: 10.1021/nl801958b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson W.S., Sparks D.L., Lund-Katz S., Phillips M.C. The molecular basis for the difference in charge between pre-beta- and alpha-migrating high density lipoproteins. J. Biol. Chem. 1994;269(12):8959–8965. [PubMed] [Google Scholar]
- Davidson W.S., Thompson T.B. The structure of apolipoprotein AI in high density lipoproteins. J. Biol. Chem. 2007;282(31):22249–22253. doi: 10.1074/jbc.R700014200. [DOI] [PubMed] [Google Scholar]
- Frank P.G., Marcel Y.L. Apolipoprotein A-I: structure–function relationships. J. Lipid Res. 2000;41(6):853–872. [PubMed] [Google Scholar]
- Gogonea V. Structural insights into high density lipoprotein: old models and new facts. Front. Pharmacol. 2016;6:318. doi: 10.3389/fphar.2015.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers, R.J., Linke, M., Barnoud, J., Reddy, T.J.E., Melo, M.N., Seyler, S.L., Domanski, J., Dotson, D.L., Buchoux, S., Kenney, I.M., 2019. MDAnalysis: a Python package for the rapid analysis of molecular dynamics simulations, Los Alamos National Lab.(LANL), Los Alamos, NM (United States).
- Henrich S.E., Thaxton C.S. An update on synthetic high-density lipoprotein-like nanoparticles for cancer therapy. Expert Rev. Anticancer Ther. 2019;19(6):515–528. doi: 10.1080/14737140.2019.1624529. [DOI] [PubMed] [Google Scholar]
- Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A. 1985;31(3):1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- Huang R., Silva R.G.D., Jerome W.G., Kontush A., Chapman M.J., Curtiss L.K., Hodges T.J., Davidson W.S. Apolipoprotein AI structural organization in high-density lipoproteins isolated from human plasma. Nat. Struct. Mol. Biol. 2011;18(4):416–422. doi: 10.1038/nsmb.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes Z.E., Mancera R.L. Molecular dynamics simulations of mixed DOPC–β-sitosterol bilayers and their interactions with DMSO. Soft Matter. 2013;9(10):2920–2935. [Google Scholar]
- Hughes Z.E., Malajczuk C.J., Mancera R.L. The effects of cryosolvents on DOPC−β-sitosterol bilayers determined from molecular dynamics simulations. J. Phys. Chem. B. 2013;117(12):3362–3375. doi: 10.1021/jp400975y. [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9(3):90–95. [Google Scholar]
- Hye A., Riddoch-Contreras J., Baird A.L., Ashton N.J., Bazenet C., Leung R., Westman E., Simmons A., Dobson R., Sattlecker M., Lupton M., Lunnon K., Keohane A., Ward M., Pike I., Zucht H.D., Pepin D., Zheng W., Tunnicliffe A., Richardson J., Gauthier S., Soininen H., Kłoszewska I., Mecocci P., Tsolaki M., Vellas B., Lovestone S. Plasma proteins predict conversion to dementia from prodromal disease. Alzheimer's & Dementia. 2014;10(6):799–807.e2. doi: 10.1016/j.jalz.2014.05.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas A., Phillips M.C. Lipoprotein structure. In: Vance D.E., Vance J.E., editors. Biochemistry of Lipids, Lipoproteins and Membranes (Fifth Edition) Elsevier; San Diego: 2008. pp. 485–506. [Google Scholar]
- Jones M.K., Catte A., Li L., Segrest J.P. Dynamics of activation of lecithin: cholesterol acyltransferase by apolipoprotein A-I. Biochemistry. 2009;48(47):11196–11210. doi: 10.1021/bi901242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klon A.E., Segrest J.P., Harvey S.C. Molecular dynamics simulations on discoidal HDL particles suggest a mechanism for rotation in the apo A-I belt model. J. Mol. Biol. 2002;324(4):703–721. doi: 10.1016/s0022-2836(02)01143-9. [DOI] [PubMed] [Google Scholar]
- Koivuniemi A., Heikelä M., Kovanen P.T., Vattulainen I., Hyvönen M.T. Atomistic simulations of phosphatidylcholines and cholesteryl esters in high-density lipoprotein-sized lipid droplet and trilayer: clues to cholesteryl ester transport and storage. Biophys. J. 2009;96(10):4099–4108. doi: 10.1016/j.bpj.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivuniemi A., Vuorela T., Kovanen P.T., Vattulainen I., Hyvönen M.T. Lipid exchange mechanism of the cholesteryl ester transfer protein clarified by atomistic and coarse-grained simulations. PLoS Comput. Biol. 2012;8(1) doi: 10.1371/journal.pcbi.1002299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivuniemi A., Sysi-Aho M., Orešič M., Ollila S. Interfacial properties of high-density lipoprotein-like lipid droplets with different lipid and apolipoprotein A-I compositions. Biophys. J. 2013;104(10):2193–2201. doi: 10.1016/j.bpj.2013.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontush A., Lindahl M., Lhomme M., Calabresi L., Chapman M.J., Davidson W.S. Structure of HDL: Particle Subclasses and Molecular Components. In: von Eckardstein A., Kardassis D., editors. High Density Lipoproteins. Springer International Publishing; 2015. pp. 3–51. [Google Scholar]
- Koudinov A., Matsubara E., Frangione B., Ghiso J. The soluble form of Alzheimer′s amyloid β protein is complexed to high density lipoprotein 3 and very high density lipoprotein in normal human plasma. Biochem. Biophys. Res. Commun. 1994;205(2):1164–1171. doi: 10.1006/bbrc.1994.2788. [DOI] [PubMed] [Google Scholar]
- Liang H.Q., Rye K.A., Barter P.J. Dissociation of lipid-free apolipoprotein A-I from high density lipoproteins. J. Lipid Res. 1994;35(7):1187–1199. [PubMed] [Google Scholar]
- Lund-Katz, S., Phillips, M.C., 2010. High density lipoprotein structure–function and role in reverse cholesterol transport, Cholesterol Binding and Cholesterol Transport Proteins, Springer, pp. 183–227. [DOI] [PMC free article] [PubMed]
- Luthi A.J., Patel P.C., Ko C.H., Mutharasan R.K., Mirkin C.A., Thaxton C.S. Nanotechnology for synthetic high-density lipoproteins. Trends Mol. Med. 2010;16(12):553–560. doi: 10.1016/j.molmed.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi A.J., Lyssenko N.N., Quach D., McMahon K.M., Millar J.S., Vickers K.C., Rader D.J., Phillips M.C., Mirkin C.A., Thaxton C.S. Robust passive and active efflux of cellular cholesterol to a designer functional mimic of high density lipoprotein. J. Lipid Res. 2015;56(5):972–985. doi: 10.1194/jlr.M054635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano J.N., Jandacek R.J., Horace E.M., Davidson W.S. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal high-density lipoproteins of different size. Biochemistry. 2004;43(37):11717–11726. doi: 10.1021/bi0496642. [DOI] [PubMed] [Google Scholar]
- Mangé A., Goux A., Badiou S., Patrier L., Canaud B., Maudelonde T., Cristol J.-P., Solassol J. HDL proteome in hemodialysis patients: a quantitative nanoflow liquid chromatography-tandem mass spectrometry approach. PLoS ONE. 2012;7(3) doi: 10.1371/journal.pone.0034107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C.E., Jonas A. Reaction of human lecithin cholesterol acyltransferase with synthetic micellar complexes of apolipoprotein A-I, phosphatidylcholine, and cholesterol. J. Biol. Chem. 1982;257(8):4541–4546. [PubMed] [Google Scholar]
- Mazer N.A., Giulianini F., Paynter N.P., Jordan P., Mora S. A comparison of the theoretical relationship between HDL size and the ratio of HDL cholesterol to apolipoprotein A-I with experimental results from the Women's health study. Clin. Chem. 2013;59(6):949–958. doi: 10.1373/clinchem.2012.196949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsche M.A., Small D.M. C-terminus of apolipoprotein A-I removes phospholipids from a triolein/phospholipids/water interface, but the N-terminus does not: a possible mechanism for nascent HDL assembly. Biophys. J. 2011;101(2):353–361. doi: 10.1016/j.bpj.2011.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli L., Kandasamy S.K., Periole X., Larson R.G., Tieleman D.P., Marrink S.-J. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 2008;4(5):819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- Mortensen P.H., Andersson H.I., Gillissen J.J.J., Boersma B.J. Dynamics of prolate ellipsoidal particles in a turbulent channel flow. Phys. Fluids. 2008;20(9) [Google Scholar]
- Nagao K., Hata M., Tanaka K., Takechi Y., Nguyen D., Dhanasekaran P., Lund-Katz S., Phillips M.C., Saito H. The roles of C-terminal helices of human apolipoprotein A-I in formation of high-density lipoprotein particles. Biochim. Biophys. Acta (BBA) – Mol. Cell Biol. Lipids. 2014;1841(1):80–87. doi: 10.1016/j.bbalip.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81(1):511–519. [Google Scholar]
- Ollila O.H.S., Lamberg A., Lehtivaara M., Koivuniemi A., Vattulainen I. Interfacial tension and surface pressure of high density lipoprotein, low density lipoprotein, and related lipid droplets. Biophys. J. 2012;103(6):1236–1244. doi: 10.1016/j.bpj.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palgunachari M.N., Mishra V.K., Lund-Katz S., Phillips M.C., Adeyeye S.O., Alluri S., Anantharamaiah G.M., Segrest J.P. Only the two end helixes of eight tandem amphipathic helical domains of human apo A-I have significant lipid affinity. Arterioscler. Thromb. Vasc. Biol. 1996;16(2):328–338. doi: 10.1161/01.atv.16.2.328. [DOI] [PubMed] [Google Scholar]
- Pan L., Segrest J.P. Computational studies of plasma lipoprotein lipids. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2016;1858(10):2401–2420. doi: 10.1016/j.bbamem.2016.03.010. [DOI] [PubMed] [Google Scholar]
- Panagotopulos S.E., Witting S.R., Horace E.M., Hui D.Y., Maiorano J.N., Davidson W.S. The role of apolipoprotein A-I Helix 10 in apolipoprotein-mediated cholesterol efflux via the ATP-binding cassette transporter ABCA1. J. Biol. Chem. 2002;277(42):39477–39484. doi: 10.1074/jbc.M207005200. [DOI] [PubMed] [Google Scholar]
- Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52(12):7182–7190. [Google Scholar]
- Paula-Lima A.C., Tricerri M.A., Brito-Moreira J., Bomfim T.R., Oliveira F.F., Magdesian M.H., Grinberg L.T., Panizzutti R., Ferreira S.T. Human apolipoprotein A-I binds amyloid-β and prevents Aβ-induced neurotoxicity. Int. J. Biochem. Cell Biol. 2009;41(6):1361–1370. doi: 10.1016/j.biocel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Phillips M.C. New insights into the determination of HDL structure by apolipoproteins: thematic review series: high density lipoprotein structure. Function, Metabol., J. Lipid Res. 2013;54(8):2034–2048. doi: 10.1194/jlr.R034025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothblat G.H., Phillips M.C. High-density lipoprotein heterogeneity and function in reverse cholesterol transport. Curr. Opin. Lipidol. 2010;21(3):229–238. doi: 10.1097/mol.0b013e328338472d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer E.J., Santos R.D., Asztalos B.F. Marked HDL deficiency and premature coronary heart disease. Curr. Opin. Lipidol. 2010;21(4):289–297. doi: 10.1097/MOL.0b013e32833c1ef6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrest J.P. HDL and the Amphipathic Helix. In: Anantharamaiah G.M., Goldberg D., editors. Apolipoprotein Mimetics in the Management of Human Disease. Springer International Publishing; Cham: 2015. pp. 1–13. [Google Scholar]
- Segrest J.P., Jones M.K., De Loof H., Brouillette C.G., Venkatachalapathi Y.V., Anantharamaiah G.M. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 1992;33(2):141–166. [PubMed] [Google Scholar]
- Segrest J.P., Jones M.K., Klon A.E., Sheldahl C.J., Hellinger M., De Loof H., Harvey S.C. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 1999;274(45):31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- Shah A.S., Tan L., Long J.L., Davidson W.S. Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. J. Lipid Res. 2013;54(10):2575–2585. doi: 10.1194/jlr.R035725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B.W., Scanu A.M., Kézdy F.J. Structure of human serum lipoproteins inferred from compositional analysis. Proc. Natl. Acad. Sci. 1977;74(3):837–841. doi: 10.1073/pnas.74.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih A.Y., Freddolino P.L., Arkhipov A., Schulten K. Assembly of lipoprotein particles revealed by coarse-grained molecular dynamics simulations. J. Struct. Biol. 2007;157(3):579–592. doi: 10.1016/j.jsb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Shih A.Y., Sligar S.G., Schulten K. Maturation of high-density lipoproteins. J. R. Soc. Interface. 2009 doi: 10.1098/rsif.2009.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva R.A.G.D., Huang R., Morris J., Fang J., Gracheva E.O., Ren G., Kontush A., Jerome W.G., Rye K.-A., Davidson W.S. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. 2008;105(34):12176–12181. doi: 10.1073/pnas.0803626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner P.J., Small D.M. Effect of free cholesterol on incorporation of triolein in phospholipid bilayers. Biochemistry. 1987;26(18):5820–5825. doi: 10.1021/bi00392a036. [DOI] [PubMed] [Google Scholar]
- Spooner P.J., Hamilton J.A., Gantz D.L., Small D.M. The effect of free cholesterol on the solubilization of cholesteryl oleate in phosphatidylcholine bilayers: a 13C-NMR study. Biochim. et Biophys. Acta (BBA)-Biomembr. 1986;860(2):345–353. doi: 10.1016/0005-2736(86)90531-6. [DOI] [PubMed] [Google Scholar]
- Thaxton C.S., Daniel W.L., Giljohann D.A., Thomas A.D., Mirkin C.A. Templated Spherical High Density Lipoprotein Nanoparticles. J. Am. Chem. Soc. 2009;131(4):1384–1385. doi: 10.1021/ja808856z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorela T., Catte A., Niemelä P.S., Hall A., Hyvönen M.T., Marrink S.-J., Karttunen M., Vattulainen I. Role of lipids in spheroidal high density lipoproteins. PLoS Comput. Biol. 2010;6(10) doi: 10.1371/journal.pcbi.1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hua N., Atkinson D., Small D.M. The N-terminal (1–44) and C-terminal (198–243) peptides of apolipoprotein A-I behave differently at the triolein/water interface. Biochemistry. 2007;46(43):12140–12151. doi: 10.1021/bi7010114. [DOI] [PubMed] [Google Scholar]
- Wu Z., Gogonea V., Lee X., May R.P., Pipich V., Wagner M.A., Undurti A., Tallant T.C., Baleanu-Gogonea C., Charlton F., Ioffe A., DiDonato J.A., Rye K.-A., Hazen S.L. The low resolution structure of ApoA1 in spherical high density lipoprotein revealed by small angle neutron scattering. J. Biol. Chem. 2011;286(14):12495–12508. doi: 10.1074/jbc.M110.209130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetukuri L., Söderlund S., Koivuniemi A., Seppänen-Laakso T., Niemelä P.S., Hyvönen M., Taskinen M.-R., Vattulainen I., Jauhiainen M., Orešič M. Composition and lipid spatial distribution of HDL particles in subjects with low and high HDL-cholesterol. J. Lipid Res. 2010;51(8):2341–2351. doi: 10.1194/jlr.M006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis, V.I., Fotakis, P., Koukos, G., Kardassis, D., Ehnholm, C., Jauhiainen, M., Chroni, A., 2015. HDL biogenesis, remodeling, and catabolism, High Density Lipoproteins, Springer, pp. 53–111. [DOI] [PubMed]
- Zhang H., Ahmadi G., Fan F.-G., McLaughlin J.B. Ellipsoidal particles transport and deposition in turbulent channel flows. Int. J. Multiph. Flow. 2001;27(6):971–1009. [Google Scholar]
- Zhang M., Charles R., Tong H., Zhang L., Patel M., Wang F., Rames M.J., Ren A., Rye K.-A., Qiu X., Johns D.G., Charles M.A., Ren G. HDL surface lipids mediate CETP binding as revealed by electron microscopy and molecular dynamics simulation. Sci. Rep. 2015;5(1):8741. doi: 10.1038/srep08741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Song J., Cavigiolio G., Ishida B.Y., Zhang S., Kane J.P., Weisgraber K.H., Oda M.N., Rye K.-A., Pownall H.J., Ren G. Morphology and structure of lipoproteins revealed by an optimized negative-staining protocol of electron microscopy. J. Lipid Res. 2011;52(1):175–184. doi: 10.1194/jlr.D010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.