Abstract
The incidence of esophageal adenocarcinoma (EAC) and other gastrointestinal (GI) cancers have risen dramatically, thus defining the oncogenic drivers to develop effective therapies are necessary. Patients with Barrett’s Esophagus (BE), have an elevated risk of developing EAC. Around 70%–80% of BE cases that progress to dysplasia and cancer have detectable TP53 mutations. Similarly, in other GI cancers higher rates of TP53 mutation are reported, which provide a significant survival advantage to dysplastic/cancer cells. Targeting molecular chaperones that mediate mutant p53 stability may effectively induce mutant p53 degradation and improve cancer outcomes. Statins can achieve this via disrupting the interaction between mutant p53 and the chaperone DNAJA1, promoting CHIP-mediated degradation of mutant p53, and statins are reported to significantly reduce the risk of BE progression to EAC. However, statins demonstrated sub-optimal efficacy depending on cancer types and TP53 mutation specificity. Besides the well-established role of MDM2 in p53 stability, we reported that individual isoforms of the E3 ubiquitin ligase GRAIL (RNF128) are critical, tissue-specific regulators of mutant p53 stability in BE progression to EAC, and targeting the interaction of mutant p53 with these isoforms may help mitigate EAC development. In this review, we discuss the critical ubiquitin-proteasome and chaperone regulation of mutant p53 stability in EAC and other GI cancers with future insights as to how to affect mutant p53 stability, further noting how the precise p53 mutation may influence the efficacy of treatment strategies and identifying necessary directions for further research in this field.
Keywords: Ubiquitin Proteasome System, UPS, RNF128/GRAIL, Heat Shock Protein, HSP, p53
Abbreviations used in this paper: BE, Barrett’s esophagus; EAC, esophageal adenocarcinoma; GI, gastrointestinal; GOF, gain-of-function; HCC, hepatocellular carcinoma; HGD, high-grade dysplasia; HOP, Hsp70/Hsp90 organizing protein; HSP, heat shock protein; LGD, low-grade dysplasia; LOH, loss of heterozygosity; MVP, mevalonate-5-phosphate; RE, responsive element; UPS, ubiquitin proteasome system; WT, wild-type
Summary.
TP53 mutations stabilize its protein and are frequently observed in Barrett’s adenocarcinoma and other gastrointestinal cancers. We examine mechanisms of the ubiquitin proteasome system and chaperone machinery as key regulators of mutant p53 stability and as potential therapeutic opportunities.
Esophageal adenocarcinoma (EAC) incidence has increased over 700% in the last 4 decades.1 Unfortunately, most patients experience poor prognosis when EAC is diagnosed beyond stage I.2,3 The typical precursor tissue for EAC is Barrett’s esophagus (BE), which is generally restricted to the lower esophagus, close to or overlapping the gastroesophageal junction. This condition is characterized by a change in the esophageal epithelium composition, in which columnar epithelial cells replace the squamous epithelial mucosa. BE is highly correlated with repeated acid exposure and prolonged gastroesophageal reflux disease. Patients with BE may progress to dysplastic states, termed low-grade dysplasia (LGD) and high-grade dysplasia (HGD), and finally to EAC.4 BE affects a significant number of people (about 1.6%–11% of adults); however, only a small fraction of these patients (∼1.1%) will progress to EAC.5 It is, therefore, important to use reliable and efficient methods of detecting progression risk and treating progressors early. Currently, detection relies on collection of biopsies subject to histological screening. Patients who are deemed higher risk based on dysplasia grading may then undergo more intense screenings or procedures such as ablation or surgical esophagectomy. This approach is not ideal, as histological grading is subjective, and intensive screenings and interventions can be expensive and invasive.6 Understanding additional reliable markers of risk may lead to improvements in screening as well as inform effective therapeutic strategies.
Significance of TP53 mutation in BE progression
Mutation in TP53 is a viable contender for this purpose. TP53 mutation often occurs early in the development of esophageal and gastric cancers and is associated with increased likelihood of progression from BE to EAC.3,7 Stachler et al6 compared tissue samples from BE progressors who eventually developed HGD or EAC with those of BE nonprogressors. TP53 mutation was detected in the nondysplastic BE tissue of progressors vs nonprogressors at frequencies of 46% and 3.4%, respectively, and overall, BE patients with TP53 mutation were more likely to progress by a factor of 13.8-fold.6
Additionally, loss of heterozygosity (LOH) on chromosome 17p allows for the loss of wild-type (WT) TP53 allele (located on chromosome 17p13) after acquiring an initial mutation. Studies suggest that 17p LOH is an early event in BE progression, which contributes to the selection of genetically aberrant cells that drive neoplastic transformation.8,9 In a study using endoscopic biopsies from BE patients, 17p LOH was detected in 6% of nondysplastic samples and 15% of HGD biopsies. Furthermore, in BE patients with negative dysplasia, indefinite dysplasia, or LGD, 17p LOH correlated with significantly increased likelihood of progression to HGD and EAC. Moreover, 17p LOH also correlated with increased incidence of 4N and aneuploidy, suggesting that this event leaves cells more vulnerable to genetic instability and consequent cancer progression.9 This is in line with the fact that WT TP53 loss impairs checkpoint and DNA repair mechanisms that usually help preserve the integrity of the genome (reviewed by Williams and Schumacher).10 The deleterious effects of TP53 mutation and subsequent LOH on genomic stability may extend beyond the loss of protective WT p53 function, including the potential for mutant p53 gain-of-function (GOF) activities to exacerbate genomic instability.11
The significance of TP53 alteration in driving progression in this context is confirmed in mouse models assessing gastric cancer development. Scientists have been unable to develop an ideal rodent model of BE due to the significant differences between the human and rodent esophagus12; however, gastric adenocarcinomas resemble EACs in their molecular and cellular properties, making this a useful model system to examine the role of TP53 alterations. In a study by Sethi et al,7 mice with Trp53 deletion in Lgr5+ gastric cells were significantly more likely to develop dysplasia compared with their Trp53 WT counterparts following each group’s exposure to the carcinogens DCA/MNU. Organoids developed from Trp53 knockout dysplastic lesions exhibited upregulation in pathways related to inflammation, WNT, stem cell renewal, and cell cycle signaling.7 Furthermore, in another study, TP53 mutation in gastric adenocarcinoma and EAC cell lines was found to contribute to primary tumor growth through GOF activity causing elevated hypoxia signaling. Supporting this, mice with conditionally expressed Trp53R270H in gastric tissue developed dysplastic lesions with increased hypoxia signaling compared with their Trp53+/– counterparts after exposure to DCA and MNU.13 These studies suggest that TP53 status is a promising predictor of BE progression risk, and a better understanding of the biological consequences of TP53 mutation may provide insights relevant to effective treatment strategies.
TP53 mutation among gastrointestinal cancers
The tumor suppressor gene TP53 is of critical importance in many human cancers, as it is the most commonly mutated gene. WT p53 functions as a transcription factor that usually helps keep cancer at bay through regulating programs such as DNA repair, cell cycle arrest, senescence, and apoptosis in response to cellular stress.14,15 TP53 is often the most commonly altered gene in gastrointestinal cancer studies.16 TP53 mutation rates are subject to variations in clinical selection methodology, cohort populations, and detection strategies; however, frequencies of 80% in EAC,17 35%–55% in gastric cancer,18 70% in pancreatic cancer,19 60% in colorectal cancer,20 30%–70% in gallbladder carcinoma,21 10%–50% in hepatocellular carcinoma,22 and 50% in duodenal carcinoma23 have been reported. Several studies, including in vivo mouse models, assess the role of TP53 mutation in gastrointestinal (GI) cancer development, and highlight the impact of loss of WT p53 function as well as mutant p53 GOF activities.24
As mentioned, TP53 mutation in gastric adenocarcinoma and EAC is associated with promoting tumor growth through increasing hypoxia signaling. In the context of colorectal cancer, TP53 mutation is associated with functions that drive the evolution of adenoma to adenocarcinoma.25 In mice harboring APCΔ716 mutation, the introduction of Trp53R270H mutation in the intestinal epithelia caused mice to exhibit higher rates of invasive tumors and shorter lifespan compared with Trp53-null mice.26 Mutant TP53 was also associated with the evolution of pancreatic intraepithelial neoplasia to aggressive pancreatic ductal adenocarcinoma.27 In a pancreatic ductal adenocarcinoma mouse model, Morton et al28 compared the outcomes of mice with the driver mutation KrasG12D expressed in the pancreas alone (KC mice), alongside Trp53R172H/+ mutation (KPC mice) or alongside Trp53R172H/R172H mutation (KPPC mice). Complete loss of p53 through homozygous Trp53R172H/R172H mutation provided a significant survival advantage to the pancreatic cells of KPPC mice and led to rapid tumorigenesis, likely through enabling circumvention of KrasG12D-induced senescence and growth arrest. In addition, the Trp53R172H mutation exhibited a GOF prometastatic effect, as 65% of KPC mice experienced metastatic spread to the liver, in contrast to 0% of their counterparts expressing heterozygous Trp53 knockout (Trp53loxP/+) rather than the Trp53 mutation in pancreatic cells.28 In hepatocellular carcinoma (HCC), it is hypothesized that p53 mutation is a critical event due to the ability of certain p53 mutants (including p53R175H and p53R248W) to bind to and compromise the proapoptotic activity of the transcription factors p63 and p73.29 In a mouse model of HCC, however, p53 deficiency and Trp53R172H mutation led to similar phenotypic outcomes, including rates of survival, tumor incidence, and metastasis. Additionally, both conditions were associated with similar levels of p63 and p73 target genes. This study therefore suggests that in the context of HCC, p53 mutation may not carry tumorigenic properties beyond those of p53 deficiency. However, p53 mutation and p53 deficiency may impair p63 and p73 through different mechanisms, and p53 mutations other than human TP53R175H (mouse Trp53R172H) may possess GOF activities.30 The significance of mutant p53 in these varying gastrointestinal cancers suggests that targeting mutant p53 stability or its activity may serve as effective therapeutic approaches in many of these contexts.
Effects of TP53 mutation
Mutation in TP53 results in a variety of deleterious effects contingent on several factors, such as mutation type and cell context. These effects can be grouped into 3 categories: (1) loss of WT p53 function, (2) dominant negative effects, and (3) GOF activities. However, these effects are nuanced and there is much to be learned about each.
Loss of WT p53 function involves the inability to activate transcription of p53 target genes.31 Reduced transactivation of WT p53 target genes has been linked to effects on biological outcomes such as clonal survival and induction of apoptosis in human osteosarcoma cell lines.32 However, studies show that p53 mutants vary in their impacts on the spectrum and magnitude of transcriptional activation of p53 responsive elements (REs). For example, in a yeast model by Resnick and Inga,33 the p53T123A mutant caused enhanced transactivation at the p53 REs for GADD45, CCNG1, CDKN1A and BAX, while the p53 hotspot mutant p53R282Q exhibited reduced activity at GADD45 and CCNG1 REs and failed to induce transcription at P21-3′ and BAX-B REs.
Mutant p53 dominant negative activity describes the ability of mutant p53 to compromise the activity of WT p53 protein and is associated with hetero-oligomerization. WT p53 functions as a tetramer, and when mutant p53 replaces 1 or more WT p53 molecules, this can impede the productivity of the complex. The extent to which p53 mutants hinder or prevent WT p53 activity in this way requires further investigation. For example, in studies using MEFs, heterozygous expression of p53 mutants Trp53R172H/+ or Trp53R270H/+ disrupted regulation of cell proliferation but did not hinder DNA damage–induced cell cycle arrest when compared with Trp53+/– MEFs.34
Mutant p53 GOF activities represent novel, tumor-promoting functions differing from those of WT p53. These are attributed to the ability of p53 mutants to bind novel protein partners, including transcription factors such as Sp1, Yap1, NF-Y, CBP, NF-κB, VDR, SREBP, E2Fs, Smad2/Smad3, and p63/p73, thereby affecting regulation of novel target genes.34,35 Phenotypic consequences of this GOF activity include faster rate of cell division in cell culture, higher tumorigenic potential in nude mice, increased cellular invasion and migration, and drug resistance.36 However, there is still much to be learned regarding the mechanisms and biological outcomes of mutant p53 GOF,37 and it is unclear if all p53 mutants are capable of GOF activity as well as how this activity is impacted by the exact point mutation or cellular environment.36
The specific consequences of TP53 mutation in the context of BE are therefore complex and may involve different aspects of loss of WT p53 function and the dominant negative and GOF effects of mutant p53. Furthermore, these effects may differ between BE patients based on factors such as the exact p53 mutation.
Targeting the stability of mutant TP53
Studies in human lung cancer cells and mouse models show that mutant p53 knockdown, inactivation or destabilization can impair or prevent tumor formation. This suggests that cancers may display oncogenic addiction toward mutant p53, meaning the formation and maintenance of these tumor cells depends on the gain of function activity of mutant p53.38 Mutant p53 protein is greatly stabilized in cancers including EAC. Our studies using siRNA reveal BE cells with mutant p53 are dependent on this protein for their survival,39 suggesting oncogenic addiction toward mutant p53 in this context.39 Therefore, targeting mutant p53 may serve as a promising therapeutic strategy. While WT p53 is maintained at low levels in the cell under normal conditions, mutant p53 accumulates to high levels in tumors, suggesting that specific mechanisms for stabilizing mutant p53 are at play.40 Fully elucidating these mechanisms of stabilization may allow scientists to develop translational approaches to effectively targeting mutant p53 in cancers or precancerous states.
While most TP53 mutations found in cancers are missense mutations that occur in the DNA-binding domain, the precise amino acid change affects the mutant p53 protein properties, such as its ability to interact with DNA, GOF capabilities, and stability.40 On the one hand, "contact mutants" (eg, p53R248Q and p53R273H) involve changes to sites that normally participate in direct DNA contact; this affects DNA binding capability but has little impact on protein folding. “Conformational mutants,” such as p53R175H and p53R249S, on the other hand, carry amino acid changes that result in local or global unfolding.41 These mutants lie on a spectrum of different folding tendencies and stability. While exploring mechanisms governing mutant p53 stability (Figure 1), greater understanding of how these mutants are differentially regulated will inform on the efficacy and applicability of translatable strategies targeting mutant p53 in the context of EAC and beyond. TP53 mutations observed in BE and EAC tissues include both contact and conformational mutants. Figure 2 (upper panel) includes a list of common p53 mutations observed in BE, as well as information on their folding and functional statuses (Table 1), drawn from the International Agency for Research on Cancer p53 mutation database (version R20).42 For comparison, we also show the prevalence of these same p53 mutations in other gastrointestinal cancers (Figure 2, lower panel). As with other molecular characterizations, based on overall gene expression or mutation profile, there are differences between cancers arising in squamous or mucosal tissue. Squamous cell types show a lower frequency of hot-spot mutations in Figure 2, as a fraction of all TP53 mutations (17% head and neck and 19% for esophageal squamous cell carcinoma compared with 39%, 32%, and 43% across esophageal, gastric, and colon adenocarcinoma, respectively) and are characterized by a broader spectrum of mutations, yet still a high rate of overall p53 mutation. Other GI mucosal cancers show similar hotspot mutant frequencies to EAC/HGD (Figure 2, lower panel), with a reduction in overall p53 mutation rate down the alimentary canal, which may relate to local environment or shifts in cancer subtype frequencies.43
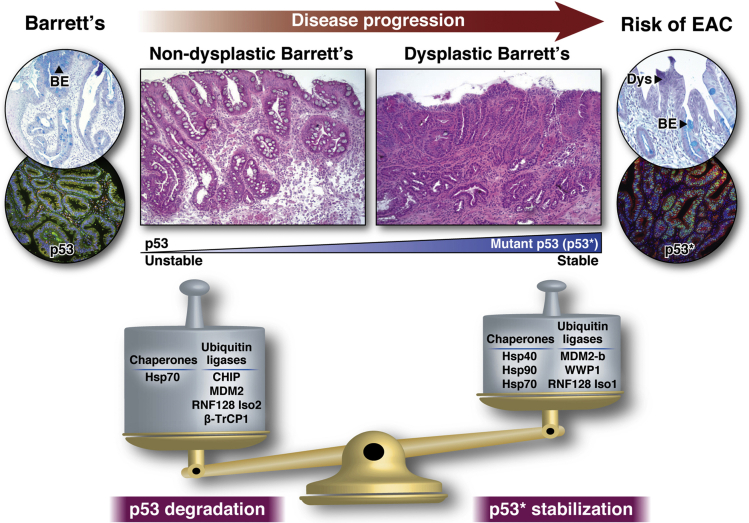
Figure 1.
Key ubiquitin ligases (E3) and chaperones involved in tilting the balance toward increased stabilization of mutant p53 (p53∗) to promote BE progression to EAC. Critical ligases include MDM2, CHIP, β-TrCP1, WWP1, RNF128, etc, and various HSP family members including Hsp40, Hsp70, and Hsp90 are the chaperones involved those determine the p53∗ protein levels tilting the balance toward increased stability during BE progression.
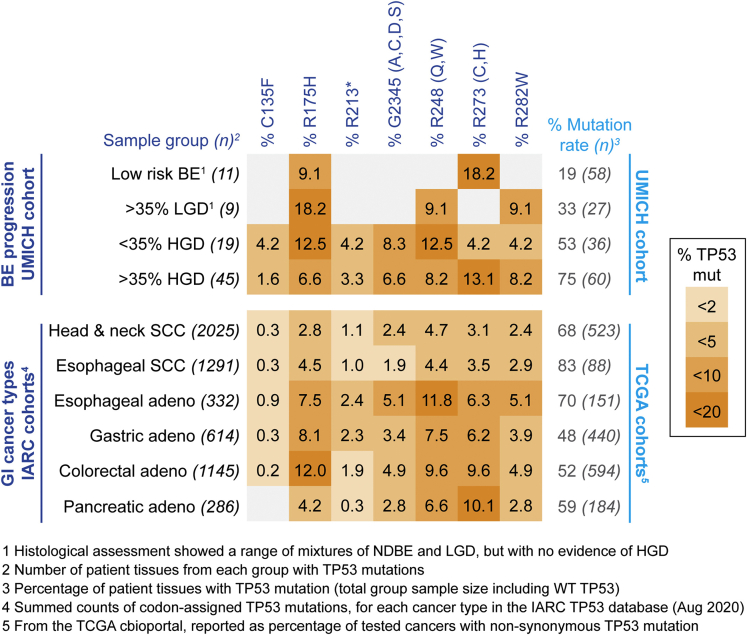
Figure 2.
Common TP53 mutations in BE with frequency data across common GI cancers. (Upper panel) Using our extended progression-related cohort of dysplasia-enriched BE tissues (n = 181) from HGD/EAC resection patients, we present frequencies for the most commonly targeted amino acids across BE dysplasia groups, based on histological assessment39: BE tissue with a low-risk of progression based on absence or low levels of dysplasia (<50% of BE tissue as LGD, with regional no evidence of HGD); high proportion LGD (50% or greater, with no evidence of HGD); regions with <50% HGD; regions with more than 50% HGD and EAC. Numbers given on the left represent the number of cases with TP53 mutations, used as a base line for % AA targeted mutations, while numbers of the right show the overall TP53 mutation percentage, with total number of samples for each group (in brackets) used as a baseline. (Lower panel) Comparative mutation rates for common GI cancers, using the International Agency for Research on Cancer (IARC) TP53 database42 of curated TP53 mutations associated with human cancers either from peer-reviewed literature or curated from other genomic databases. Baseline totals (shown on the left) represent the number of records from each cancer group with p53 mutations reported with designated amino acid localizations. The rightmost columns in the lower section shows the overall percentage of TP53 mutations for each cancer type, drawn from The Cancer Genome Atlas (TCGA)–based cohorts (from cBioportal interface) used in comparable GI cancer–type studies, with the total number of samples for each cancer type (shown in brackets) used as a baseline. Color coding key on the far right is a categorical visual guide to the quoted amino acid specific percentages, with frequencies rare (>2%) to common (10%–20%) shaded from light to dark. NDBE, nondysplastic Barrett’s esophagous; SCC, squamous cell carcinoma.
Table 1.
Features for Common TP53 Mutations in BE Progression Samples
| Codon | Exon | Protein Foldinga |
Residue Functionb |
Dominant Negativeb |
Structural Motifb |
Mutation Typec |
|---|---|---|---|---|---|---|
| C135 | 5-exon | NA | Buried | Moderate | L1/S/H2 | Loss |
| R175 | 5-exon | mostly unfolded | Buried | Yes | L2/L3 | NA |
| R213 | 6-exon | nonsense | Buried | NA | NDBL/beta-sheets | NA |
| G245 | 7-exon | NA | Buried | Yes | L2/L3 | Loss |
| R248 | 7-exon | mostly folded | DNA binding | Yes | L2/L3 | Loss |
| R273 | 8-exon | mostly folded | DNA binding | Yes | L1/S/H2 | Loss |
| R282 | 8-exon | mostly unfolded | Buried | Moderate | L1/S/H2 | NA |
NA, not available.
From International Agency for Research on Cancer TP53 database (https://p53.iarc.fr).106
From Jordan et al.107
The stability of WT p53 is primarily regulated at the protein level, and mutant p53 follows similar mechanisms of regulation. Under normal conditions, WT p53 is maintained at low levels in the cell, and its degradation is mostly associated with ubiquitination by E3 ligases, principally by MDM2, followed by proteasomal degradation. Accumulation of p53 occurs upon cellular stresses such as DNA damage, aberrant cell proliferation, and lack of growth factors or nutrients.43,44 In this way, robust activation of p53 drives pathways that help prevent cell survival and proliferation in the presence of genetic lesions and environmental stresses that may otherwise promote tumorigenesis. Multiple mechanisms contribute to p53 accumulation, including posttranslational modifications of both p53 and MDM2, which act to prevent p53 ubiquitination. ATM kinase is a crucial activator of p53 in response to DNA damage, driving the direct and indirect phosphorylation of p53, MDM2, and the MDM2 homolog, MDMX. Other pathways driving p53 stabilization include the ARF-dependent response triggered by oncogene activation and the ribosomal protein-mediated response.44, 45, 46 Regulation and posttranslational modifications of WT p53 have been extensively reviewed elsewhere.43,47,48 Unlike WT p53, the majority of different p53 mutants are stable proteins, whose stability is regulated via a multitude of posttranslational modifications. Chronic mutant p53 stabilization in the context of cancers is largely attributed to differences in MDM2-mediated regulation as well as additional layers of regulation, notably protection by molecular chaperones, which register mutant p53 as a misfolded protein.45 Many tumors exhibit upregulation of molecular chaperones, leading to a buffering effect, whereby molecular chaperones play a critical role in protecting oncoproteins with destabilizing mutations that would otherwise leave them vulnerable to degradation.49 In this fashion, mutant p53 is specifically recognized by several molecular chaperones that help promote its accumulation. Mutant p53 expression in cancers is therefore regulated by 2 major opposing forces - degradation via the ubiquitin-proteasome system (UPS) and stabilization by chaperone machinery. In this review, we identify critical regulators of mutant p53 in these systems, then describe mechanisms by which the balance of the UPS and chaperone activity shift to promote mutant p53 stabilization in different cancer contexts, which may also be applicable to EAC. We will also introduce a recently discovered major regulator of mutant p53 stability in the context of BE progression specifically. Finally, we will draw from this information to provide insight into the potential for utilizing the UPS and chaperone regulation of mutant p53 to mediate EAC development and progression with “ying/yang” style overview in Figure 1 and more detailed molecular interaction representation in Figure 3.
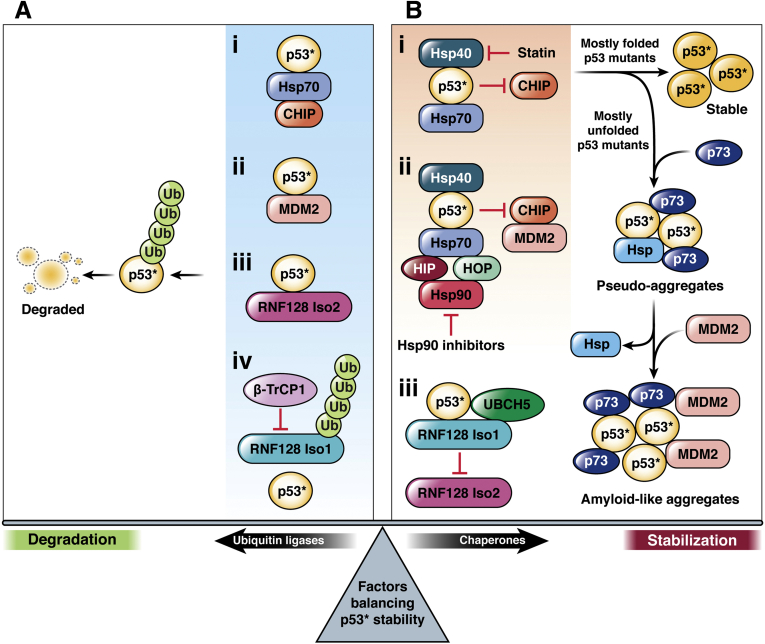
Figure 3.
Mechanisms that regulate the balance between mutant p53 (p53∗) degradation and stabilization. Misfolded/unfolded mutant p53 (p53∗) may be recognized by ubiquitin ligases or molecular chaperones. (Panel A, i-iii) Ubiquitin ligases including CHIP, MDM2 and RNF128 Iso2 are able to efficiently polyubiquitinate p53∗, leading to its subsequent proteasomal degradation. (Panel A, iv) The ubiquitin ligase β-TrCP1 may also indirectly contribute to p53∗ degradation, through promoting degradation of RNF128 Iso1. Molecular chaperones compete with the action of these ubiquitin ligases on p53∗ to instead promote stabilization. (Panel B, i-ii) Molecular chaperone activity drives p53∗ refolding and exhibits a protective effect against the activity of ubiquitin ligases such as CHIP and MDM2. p53∗ may be recognized by the molecular co-chaperone Hsp40, which is known to coordinate with Hsp70. Statins have been shown to disrupt the Hsp40 (DNAJA1)-p53∗ interaction, in turn promoting CHIP-mediated degradation. p53∗ is also observed in complexes with both HSP70 and HSP90 machinery; this may be counteracted by Hsp90 inhibitors. The refolding activity of these chaperones help p53∗ reach a more stable conformation that is less vulnerable to degradation. The ability of p53∗ to reach this stable conformation varies based on the exact mutation and its effect on the protein’s properties and structure. Mutants that are found predominantly folded in the cell reach this final conformation with more ease. The folding intermediates of other p53 mutants, most often found in an unfolded conformation in cells, are more likely to instead participate in aggregation, forming pseudo-aggregates with chaperones and other proteins such as p73 that resist degradation. The addition of MDM2 to these complexes promotes the formation of stable amyloid-like fibrils. (Panel B, iii) Additionally, RNF128 Iso1 promotes p53∗ stability through mitigating RNF128 Iso2-mediated p53∗ ubiquitination.
Key ubiquitin-proteasome and chaperone regulators of p53 stability
MDM2 and MDMX can target WT and mutant p53
MDM2 and MDMX are regarded as the 2 major negative regulators of WT p53. Through interacting with the transactivation domain of p53, MDM2 can prevent the ability of p53 to drive transcription of its target genes and can also escort p53 out of the nucleus. Additionally, MDM2 is an E3 ligase that ubiquitinates p53, targeting it for nuclear export and proteasomal degradation. These inhibitory interactions of MDM2 on p53 are reinforced through a negative feedback loop, in which active p53 drives transcription of the MDM2 gene, thus ultimately promoting p53 inactivation and degradation. Like MDM2, MDMX can bind with p53 and in turn inhibit its transactivation activity. Unlike MDM2, MDMX does not possess functional E3 ligase activity and therefore cannot directly target p53 for degradation. MDMX can, however, form heterodimers with MDM2 that are more stable than MDM2 homodimers. Through this, MDMX promotes MDM2 stability and therefore also contributes to MDM2-induced p53 degradation. In the context of cellular stress, the expression and activity of MDM2 and MDMX are suppressed through various mechanisms in order to allow for the accumulation of p53. This enables robust activation of p53 target genes such as cell cycle arrest and cell death, helping to prevent tumor development.50
MDM2 can also target degradation of mutant p53.45 However, various mechanisms can protect mutant p53 from this negative regulation. Mutant p53 is unable to drive transcription of the MDM2 gene, breaking the negative feedback loop that functions to minimize WT p53 expression. This was once regarded as the major mechanism through which mutant p53 attains hyperstabilization40; however, the work of Lang et al51 demonstrated that other modes of regulation are necessary. When the tissues of mice expressing mutant p53 were analyzed, mutant p53 accumulated to elevated levels in tumor tissue but not in normal tissue.51 This suggests that there must be additional mechanisms of mutant p53 stabilization that are unique to the tumor environment, providing justification for the importance of molecular chaperones in combating mutant p53 degradation.
Chaperone machinery stabilizes mutant oncoproteins
In the crowded environment of a cell, newly formed polypeptide chains are vulnerable to interaction with many other surfaces, which can lead to improper folding and aggregation, both of which can be toxic to the cell.52,53 Chaperones help alleviate this risk through interacting with and stabilizing unfolded and misfolded polypeptides.54 In addition to helping with the folding of de novo proteins, chaperones may also assist with refolding proteins denatured under conditions of stress, protein transport, and formation of oligomers.53 In case a protein fails to refold in its native conformation, chaperones can also promote degradation via the endoplasmic reticulum associated degradation pathway.55 Most chaperones belong to families of heat-shock proteins (HSPs), which were originally discovered as agents that help manage proteostasis under conditions of stress via mitigating protein unfolding and aggregation.49,54 Chaperones in the Hsp70 family (DnaK in prokaryotes) help with protein folding and refolding via ATP-dependent binding and release cycles. Hsp70-substrate binding helps shield hydrophobic residues of the substrate from interaction with other proteins, passively preventing aggregation. Substrate release allows for the continuation of spontaneous protein folding. Over the course of binding and release cycles, the protein substrate may progress toward its native state. HSP70 machinery also includes Hsp70 co-chaperones, such as those in the Hsp40 family (deemed DnaJ in prokaryotes), which help promote ATP hydrolysis and also recruit specific unfolded proteins to Hsp70.54 The HSP70 system may also coordinate with HSP90 machinery, linked through the co-chaperone Hsp70/Hsp90 organizing protein (HOP) (gene STIP1). A substrate may be transferred from Hsp70 to Hsp90, with the latter further regulating structural and conformational changes through its own binding and release cycles. Hsp90 is known to mediate the conformation and prevent the degradation of many mutant proteins, which can often harbor changes that may otherwise act as a hindrance to stable folding. By offering this protective function to mutant oncoproteins, Hsp90 activity can promote signaling pathways that drive tumorigenesis.53 Hsp90 machinery has been shown to coordinate with mutant p53 and contribute to its stabilization, which will be described in further depth subsequently.
CHIP: An E3 ubiquitin ligase that interacts with chaperone machinery
CHIP (gene STUB1) is another E3 ligase that ubiquitinates substrates and sends them for proteasomal degradation. A distinct feature of CHIP is its tight coordination with chaperone machinery, as indicated by its name, which stands for carboxyl terminus of Hsc70-interacting protein. CHIP is capable of binding to molecular chaperones, notably Hsp70 (3 protein-forming human genes HSPA1A, HSPA1B, and HSPA1L) and its constitutively expressed homolog, Hsc70 (gene HSPA8), and targeting its clients for degradation.56 CHIP is also capable of binding directly to Hsp90 (6 protein-forming human genes HSP90AA1, HSP90AA2, HSP90N, HSP90AB1, HSP90B1, and TRAP1; mitochondrial), but it is mainly found directly bound to Hsp70, which has a higher binding affinity for CHIP. CHIP acts as a co-chaperone that competes with HOP for direct binding to Hsp70 to either form the Hsp70-CHIP complex driving substrate degradation or the Hsp70-HOP-Hsp90 complex driving substrate folding. This competitive binding, depicted in Figure 3, thus acts as an essential inflection point in protein quality control, regulating the balance between chaperone machinery and the UPS. One of the factors that modulates this balance is the concentration of Hsp70 cofactors in the cell.54 Under normal conditions, the concentration of the Hsp70-HOP-Hsp90 complex far exceeds that of the Hsp70-CHIP complex, such that folding of a client protein is favored over degradation.57 This balance, however, may be shifted through relative changes in HOP or CHIP concentration. In addition, posttranslational modification of chaperone proteins can further regulate their interactions. Muller et al58 found that CHIP interaction is inhibited when Hsp70 and Hsp90 are phosphorylated, while HOP binding is enhanced. Correspondingly, this group found elevated levels of phosphorylated chaperones and HOP expression in primary human cancers. The activity of molecular chaperones is also critical in moderating this balance between folding and degradation. The Hsp70-HOP-Hsp90 complex can be functionally compromised by Hsp90 inhibitors such as tanespimycin (17-AAG). In this case, the unfolded client protein will be released from the complex, but may subsequently rebind to a Hsp70 molecule. Eventually, the client may be complexed with Hsp70 and CHIP, through which it will be targeted for degradation.57 CHIP exhibits specificity for degrading unfolded proteins, including its specificity for unfolded mutant p53, while having little to no effect on folded p53 or WT p53.40
WWP1: An E3 ligase that stabilizes p53
While ubiquitin ligases are mostly associated with p53 degradation, WWP1 (WW domain containing E3 ubiquitin protein ligase 1) was the first E3 ubiquitin ligase found to stabilize and reduce the transcriptional activity of WT p53. This ligase associates with p53 within the proline-rich region and ubiquitination results in p53 nuclear export and stabilization; conversely, compromising WWP1 activity induces p53 degradation. The stabilizing effect of WWP1 is possibly due to WWP1’s capacity to monoubiquitinate p53.59 The exact consequences of substrate monoubiquitination have yet to be clarified, but evidence shows that p53 undergoes nuclear export and generally retains its stability after monoubiquitination by low levels of MDM2, as opposed to polyubiquitination mediated by high levels of MDM2, which results in p53 nuclear export and degradation.60 The activity of WWP1 on mutant p53 has yet to be studied; however, this may be a topic worth investigating, as it may represent a mechanism through which ubiquitination contributes to mutant p53 stabilization in cancers.
Expression of p53 regulators in gastrointestinal cancers
Regulators of p53 are often aberrantly expressed in gastrointestinal cancers and linked to clinicopathologic outcomes. WWP1 is overexpressed in gastric cancer, hepatocellular carcinoma, and colorectal cancer and correlates with tumor advancement.61, 62, 63 Similarly, MDM2 overexpression has been observed in gastric cancer, hepatocellular carcinoma, the primary site of colorectal cancers, and pancreatic cancer, and is associated with tumor progression, poor prognosis, and chemoresistance.64, 65, 66, 67 CHIP exhibits a tumor suppressor role in several gastrointestinal cancers, including colorectal, gastric, pancreatic, and gallbladder cancers, in which lower expression correlates to poorer prognosis.68, 69, 70 Due to their role in stabilizing oncoproteins, overexpression of HSPs benefits cancer progression and is therefore a common feature in cancers.71 Colorectal cancer, gastric cancer, hepatocellular carcinoma, and pancreatic cancer exhibit high levels of HSPs, including Hsp70, Hsp90, and small HSPs. In many of these cases, HSP overexpression correlates to worse prognosis.72, 73, 74, 75, 76, 77 The significance of p53 mutation in various gastrointestinal cancers coupled with perturbation of p53 regulators in the UPS and chaperone systems suggests that inducing p53 degradation through targeting ubiquitin ligases or molecular chaperones may help mitigate EAC as well as several related cancers.
Chaperone machinery protects mutant p53 from the UPS to promote stabilization
HSP90 protects mutant p53 from MDM2 and CHIP
Several mechanisms involving competition between molecular chaperones and E3 ligases have been discovered to assist with the stabilization of mutant p53 in cancers. HSP machinery can recognize mutant p53 as a misfolded protein, and Hsp70 and Hsp90, linked through the co-chaperone HOP have been shown to form complexes with mutant p53. These complexes help stabilize both conformational and contact mutant p53, through inhibiting the ubiquitination activity of MDM2 and CHIP on mutant p53.40 Li et al78 suggested that this may occur because MDM2 and CHIP may be locked in these complexes in an inactive state, though it has been found that CHIP and HOP compete for binding at the C-terminal EEVD site on Hsp70.54
Additionally, the binding and release cycles of Hsp70 and Hsp90 can drive spontaneous folding of mutant p53 toward its native conformation.49 Predominantly well-folded p53 mutants are more likely to reach the WT-like, native conformation of p53, which is far less susceptible to CHIP-mediated degradation compared with unfolded mutant p53. Accordingly, mutants of p53 that are predominantly well folded in the cell (eg, p53R273H) exhibit greater stability than p53 mutants that are predominantly denatured in the cell (eg, p53R175H).40 Therefore, in cells with mutant p53 that is predominantly well folded, the refolding activity of Hsp70 and Hsp90 machinery helps sustain the high ratio of folded to unfolded mutant p53, acting as an additional means of reducing mutant p53 degradation. Muller et al40 demonstrated this concept using MDA-MB-468 cells, which express the predominantly well-folded p53R273H mutant. Treatment of the cells with the Hsp90 inhibitor 17-AAG resulted in a higher ratio of unfolded to folded p53R273H. Furthermore, treatment of the cells with the proteasomal inhibitor MG132 in the absence or presence of 17-AAG revealed that Hsp90 inhibition caused increased ubiquitination of mutant p53. Immunoprecipitation also revealed that following Hsp90 inhibition, p53R273H interacts with Hsp70 and CHIP.40 A correlative study in EAC suggested that Hsp90 promotes the stability and activity of the oncoprotein, Her2,79 and therefore it would be reasonable to hypothesize that Hsp90 may protect mutant p53 in a similar manner in the EAC context, but confirmation would require specific investigation.
Chaperone-mediated mutant p53 aggregation
It is unlikely that many unfolded p53 mutants will reach their native conformation upon interaction with Hsp70 and Hsp90 machinery. However, this interaction may lead to the formation of mutant p53 folding intermediates which expose sites that are vulnerable to aggregation to other proteins, including the tumor suppressor, TAp73ɑ.49 The interaction between mutant p53 and TAp73ɑ can be stabilized by interacting chaperones, including Hsp70, Hsp90, and the Hsp40 isoform, DNAJB1.80 These mutant p53-TAp73ɑ-HSP complexes can form pseudo-aggregates, which tend to evade degradation mechanisms, therefore contributing to the stability of mutant p53 in the cell. Moreover, TAp73ɑ is a transcription factor that promotes apoptosis. Sequestering TAp73ɑ to these pseudo-aggregates compromises its proapoptotic activity, contributing to cancer cell survival and chemoresistance. Conformational mutants have a higher affinity toward TAp73ɑ compared with contact mutants, suggesting that this mode of mutant p53 stabilization is most relevant for conformational mutant p53. Additionally, overexpressed MDM2 can replace HSPs in these pseudo-aggregates, leading to mutant p53-TAp73ɑ-MDM2 complexes, which form amyloid-like fibrils, enhancing mutant p53 aggregation in the cell and leading to augmented chemoresistance. Using the MDM2 inhibitor Nutlin-3, investigators were able to disrupt this complex and induce mutant p53 degradation.49 The work of Tracz-Gaszewska et al80 suggests that cancer patients with TP53 mutation paired with MDM2 overexpression manifested worse prognosis than patients with only 1 of these conditions. Furthermore, this study shows that in breast invasive carcinoma patients, poor survival rate of patients with TP53 mutation and MDM2 overexpression correlated with high levels of chaperones in the Hsp40, Hsp70, and Hsp90 families, suggesting that the potential to form chaperone-mediated mutant p53-TAp73ɑ-MDM2 complexes worsens prognosis. We have not yet studied whether BE and EAC tissue contains mutant p53 aggregates. However, if this is the case, it would be worthwhile to also analyze the composition of these aggregates, giving attention to the presence of TAp73ɑ, molecular chaperones, MDM2, and other ubiquitin ligases.
Hsp40-mediated mutant p53 stabilization competes with CHIP-mediated degradation
The work of Parrales et al81,82 demonstrates that the Hsp40 isoform DNAJA1 can also help protect conformational p53 mutants from CHIP-mediated degradation. These studies showed that the level of mevalonate-5-phosphate (MVP), a metabolite in the mevalonate pathway, regulates the interaction between conformational mutant p53 and DNAJA1. Decreasing MVP levels through treatment with statins impaired the interaction between mutant p53 and DNAJA1 and in turn promoted CHIP-mediated ubiquitination and degradation of conformational mutant p53, including p53R156P, p53V157F, p53R175H, and p53Y220C. While statins were able to induce degradation of conformational mutant p53 in this way, these drugs had minimal effects on WT p53 or the well-folded p53 contact mutants p53R273H and p53R280K. Reduction of mevalonate kinase, an enzyme that converts mevalonic acid to MVP, or DNAJA1 knockdown also enhanced mutant p53 degradation.
This study also suggests that this mechanism of DNAJA1-mediated protection of mutant p53 stability acts independently of Hsp70 and Hsp90 activity. The steady-state levels of conformational mutant p53 were not affected by Hsc70 knockdown in CAL33 cells (squamous cell carcinoma derived from tongue), and furthermore, the effects of lovastatin treatment or DNAJA1 knockdown on conformational mutant p53 degradation were not augmented by additionally knocking down Hsc70. Additionally, statins did not affect the levels of Hsp90 or its client proteins, demonstrating that the mode of action is unlikely via Hsp90. Overall these findings suggest that the stability of p53 conformational mutants is governed by the opposing actions of DNAJA1-mediated stabilization and CHIP-mediated degradation. This is similar to the competitive forces of Hsp90-mediated stabilization and CHIP-mediated degradation of contact and conformational mutant p53 described previously. As depicted in Figure 3, studies81,83 suggest that this competition between DNAJA1 (Hsp40) and CHIP acts as a significant layer of regulation of mutant p53 stability in BE progression.
Individual RNF128 isoforms regulate mutant p53 stability during BE progression
While MDM2 is regarded as the predominant regulator of p5384 and its activity has been well studied, we have focused on the E3 ligase RNF128, or GRAIL (gene related to anergy in lymphocytes). As indicated by its name, this protein is mostly attributed to its role in inciting T cell anergy through ubiquitinating the TCR-CD3 complex.85 Chen et al86 found that RNF128 is also capable of targeting p53 for ubiquitination and proteasomal degradation. We then identified RNF128 as a major regulator of p53 in the context of BE to EAC progression and demonstrated that regulation occurs in an isoform-specific manner.39 RNF128 has 2 isoforms, which we refer to as Iso1 and Iso2, that differ in their first exon while their remaining exons 2–7 are identical. Interestingly, the differences within exon 1 lead to opposing effects on mutant p53 regulation. On the one hand, Iso2 acts as a negative regulator, capable of effectively ubiquitinating both WT and mutant p53 and decreasing its steady state levels. Iso1 on the other hand, exhibits limited ubiquitin ligase activity on WT and mutant p53, and is capable of increasing mutant p53 steady state levels and its half-life. Furthermore, Iso1 knockdown reduced the clonogenic survival of mutant p53 bearing BE cells (CpD cells). Correspondingly, over the course of BE to EAC progression, the stabilization of mutant p53 coincides with a change in the ratio of RNF128 isoforms, in which a significant decrease in Iso2 is observed, leading to a lower Iso2 to Iso1 ratio. According to RNA sequencing data, RNF128 is expressed at a level 3 times higher than MDM2 in BE samples. In addition, expression of MDM2 and MDMX remained relatively constant over the course of progression, contrasting with the shift in the Iso2 to Iso1 ratio which helps favor mutant p53 stabilization over degradation.39 Together, this data suggests that in BE tissues, RNF128 isoforms exert significant control over mutant p53 levels and likely take precedence over the activity of MDM2 and MDMX.
RNF128 isoforms parallel the roles of MDM2 and MDMX
It is worth noting that the 2 isoforms of RNF128 bear significant parallels to MDM2 and MDMX. RNF128 Iso2 and MDM2 are both capable of efficient ubiquitin ligase activity upon WT p53, which is mediated through their RING finger domains. Despite possessing RING finger domains, RNF128 Iso1 and MDMX exhibit little to no ubiquitin ligase activity on WT p53. A study by Iyappan et al87 revealed that substituting 2 regions of MDMX with the corresponding parts of MDM2 can impart MDMX with functional ubiquitin ligase activity on p53. Because Iso1 and Iso2 differ only in their first exon, there may be a particular region(s) within exon 1 of Iso1 which prevents functional RING finger–mediated ubiquitination. Furthermore, MDM2 and MDMX can interact with each other via their RING finger domains, creating a heterodimer that has both enhanced stability and ubiquitination activity on WT p53 compared with MDM2 or MDMX homodimers.50 Our experiments reveal that RNF128 Iso1 and Iso2 also form heterodimers, which have greater stability than Iso2 homodimers. However, our results suggest that the presence of Iso1 reduces the ubiquitination activity of Iso2 on WT or mutant p53.39 Iso1 may therefore promote mutant p53 stabilization by interacting with mutant p53 and preventing contact with Iso2 (Figure 3B, iii), as well as through a dominant negative effect in which Iso1 heterodimerizes with Iso2, compromising the efficiency of Iso2-mediated ubiquitination of mutant p53.
Work by Zheng et al,88 exploring the role of MDM2-B, showed this MDM2 isoform interacted with full-length MDM2 and exerted a dominant negative effect on MDM2-induced mutant p53 degradation. This interaction was hypothesized to reduce mutant p53 degradation by increasing the cytoplasmic fraction of MDM2, leaving less of the full-length ligase in the nucleus where mutant p53 tends to accumulate. Additionally, MDM2-B may contribute to this dominant negative effect by disrupting the formation of full-length MDM2 oligomers capable of efficient E3 ligase activity. In this way, RNF128 Iso1 also mimics the inhibitory activity of MDM2-B upon MDM2 function.
Another layer of complexity to this regulation is the involvement of an F-box family ubiquitin ligase SCFβ–TrCP1. Prior studies identified β-TrCP1 as an E3 ligase for MDM289 as well as for WT p53.90 Our data identified RNF128 as a novel substrate of β-TrCP1,39 suggesting conservation of regulatory factors involved in controlling MDM2 and RNF128 protein stabilities. In the context of BE progression in which Iso1 becomes the dominant RNF128 isoform and promotes mutant p53 stability, β-TrCP1–mediated Iso1 degradation can therefore help tilt the balance toward mutant p53 degradation, an area needing further investigation. We have included such thoughts in Figure 3.
RNF128 isoforms may represent another form of competition between chaperone activity and the UPS
Through ATP-dependent transient interaction with p53, Hsp90 helps p53 reach a conformation competent for promoter binding.91 Wawrzynow et al49 showed that MDM2 is capable of substituting for Hsp90 in this role. Furthermore, MDM2 also mimics chaperone activity through its role in nascent p53 protein synthesis, regulation of other transcription factors, interaction with Hsp90, and ability to bind ATP. These findings add diversity to the possible functions of MDM2, complicating the role of MDM2 in protein quality control by demonstrating roles in protein regulation beyond promoting proteasomal degradation. The mechanism through which RNF128 Iso1 is able to stabilize mutant p53 is an open question. Given that MDM2 exhibits chaperone-like activity on WT p53, we hypothesize that Iso1 is also capable of chaperone-like activity that promotes mutant p53 stability. The opposing roles of RNF128 Iso1 and Iso2 may therefore act as another example of competition between chaperone and E3 ligase activity.
The effect of RNF128 isoforms on specific p53 mutations
As discussed, the effect of the precise TP53 mutation on protein folding results in differential regulation by various molecular chaperones and E3 ligases. In our lab’s studies of RNF128 isoforms,39 we carried out experiments using the 3 most common p53 mutations found in EAC (p53R175H, p53R248Q, and p53R273H); the most common indel site p53R213; and the rare p53C135S mutation found in the commonly used EAC cell line (OE33). This list includes both contact and conformational mutants. Figure 2 lists these and other common p53 mutations found in BE and EAC samples, along with information on their folding status. It is worth noting that when assessing the impact of Iso1 on the steady state levels of p53R175H, p53R248Q, p53R213Q, and p53C135S, the conformational mutant p53R175H exhibited the greatest increase in expression (∼10-fold), followed by p53R213Q (∼4-fold). Iso2 was shown to effectively downregulate contact mutants, p53R273H and p53R248Q, but the steady-state levels of the other p53 mutants in the presence of Iso2 were not investigated. The impact of each RNF128 isoform on different p53 mutations is therefore a topic for further investigation. A more thorough understanding of how chaperone and UPS machinery impacts specific p53 mutants may inform the development of personalized treatment strategies for Barrett’s-associated cancers according to molecular signature.
Targeting mutant p53 in BE progression via regulating the UPS and chaperone machinery
Statins mitigate BE progression
Singh et al83 carried out a systematic analysis investigating the effect of statins on the risk of developing esophageal cancers, including tracking the outcomes of 2125 BE patients across 5 studies. It was found that overall, statin treatment led to a 41% reduced risk of developing EAC. While statins are anti-cholesterol drugs typically used for cardiovascular disease prevention, they exhibit pleiotropic effects including impacts on cell proliferation, apoptosis, angiogenesis, and immune activity.92 Given that mutant p53 is a common occurrence in the development of EAC, it is plausible that statin-induced degradation of mutant p53 may provide therapeutic advantage in mitigating BE progression. Further, our studies show that TP53 knockdown in mutant-p53 driven BE cells (CpD cells) significantly reduces clonogenic survival.39 Congruently, treating CpD cells with simvastatin resulted in a reduction of mutant p53 levels and clonogenic cell survival. This likely occurs, at least in part, due to the mechanism proposed by Parrales et al,82 in which statins disrupt the interaction between mutant p53 and DNAJA1, promoting mutant p53 proteasomal degradation. Statin treatment therefore may represent a therapeutic approach to reduce EAC development that utilizes the interplay of mutant p53 with chaperone and ubiquitin-proteasome machinery.
The therapeutic potential of statins has generated mixed results across cancer types. Studies suggest statins may not be effective against breast, lung, bladder, pancreatic cancers, or melanoma. However, evidence suggests these drugs may help prevent prostate, esophageal, gastric, liver, and colorectal cancers.93 These varied outcomes have given statins a controversial reputation as a potential effective agent in cancer prevention and treatment. While cancer type is important to take into account, Abdullah et al94 also attributed this variation to confounding factors that are often ignored when designing clinical trials. For example, the inhibitory effect of statins on isoprenoid synthesis may contribute to cancer prevention. However, exogenous isoprenoids introduced in standard mouse chow may mask this effect. Additionally, variables such as the use of lipophilic vs hydrophilic statins, dosage, and dose frequency may substantially alter outcomes.94,95
Several studies suggest that statins can play an antitumorigenic role through counteracting mutant p53 stabilization.41,81,96, 97, 98 As previously mentioned, statins interfere with the mevalonate pathway through reducing the production of MVP and downstream products. Through an unknown mechanism, reducing MVP levels in this way disrupts the mutant p53 and Hsp40 interaction, permitting mutant p53 degradation by CHIP.81 Additionally, statins can impair Hsp90 function, resulting in elevated mutant p53 degradation by MDM2. Hsp90 is activated through deacetylation by HDAC6. Statins can block HDAC6 activity directly or indirectly via interference in the mevalonate pathway.95,96 In the latter case, statin-induced reduction of MVP impairs the function of RhoA-GTPases, which usually stimulate HDAC6 activity through cytoskeletal cues.99
While many studies demonstrate the ability of statins to destabilize mutant p53 and impair growth of p53-driven cancer cells, these are mostly restricted to in vitro systems. However, a recent study by Tutuska et al95 demonstrates the effect of statins in several mouse models, including mice with xenografts of various mutant p53 human cancer cell lines, mice with allografts of mutant p53 lymphoma lines, and an autochthonous model of mice with mutant p53-driven T lymphomas. Across these groups, rosuvastatin treatment demonstrated modest success in reducing tumor growth and final size. In an experiment using mice with MDA-MB-231 xenografts treated with pitavastatin and zoledronic acid, the 2 smallest tumors at endpoint had the lowest levels of mutant p53, justifying the connection between the antagonistic effect of statins on mutant p53 stabilization and reduction in tumor growth. In the allograft and autochthonous models, rosuvastatin only exhibited a considerable therapeutic effect against p53R248Q/– (a DNA contact mutant) tumors, as opposed to p53R172H/R172H (conformational mutant) or p53–/– tumors. These results emphasize the importance of mutant p53 genotype specificity in the potential efficacy of statin treatment.95
However, when comparing the results of several similar studies, the efficacy of statins does not seem to strictly depend on the exact p53 mutation, or its classification as a contact vs conformational mutant. The results by Tutuska et al95 suggest that statins are more effective in mitigating tumors driven by p53R248Q (a DNA contact mutant) compared with p53R172H (conformational mutant). This matches data by Turrell et al97 demonstrating that murine KrasG12D lung tumors expressing contact mutant p53R270H were more sensitive to statins compared with their counterparts harboring conformational mutant p53R172H. However, in the aforementioned study by Parrales et al,81 statin treatment in various human cancer cell lines lowered levels of conformational p53 mutants including p53R156P, p53V157F, p53R175H (corresponding to murine p53R172H), and p53Y220C but did not significantly reduce levels of contact mutants including p53R273H and p53R280K. Statins also lowered cell viability and colony formation more effectively in cancer cells driven by conformational mutant p53R175H compared with contact mutant p53R273H. Another study using murine and human pancreatic cell lines follows this trend, in which statins effectively reduced the levels of conformational p53 mutants (p53R172H, p53I255N, p53G245S, and p53Y220C) but not contact mutants (p53R273H and p53R248W).98 Overall, studies analyzing anticancer effects of statins demonstrate considerable success, and it is worthwhile to continue investigating statin usage in mutant p53–driven cancers, including elucidating factors that affect its efficacy and whether statins may exhibit synergistic effects when combined with chemotherapeutics or other targeted therapies.95
Hsp90 inhibitors can induce mutant p53 degradation
As described previously, HSP90 machinery plays roles in preventing MDM2 and CHIP-mediated ubiquitination of mutant p53 as well as facilitating the formation of aggregate structures that resist proteolysis. In these ways, Hsp90 counters the degradative forces of MDM2 and CHIP, promoting mutant p53 stabilization. Many Hsp90 inhibitors have shown promising results in inducing mutant p53 degradation, mitigating tumor progression, and prolonging the survival of p53R175H and p53R248Q knock-in mice. However, no Hsp90 inhibitor has fulfilled the criteria to become an Food and Drug Administration–approved drug due to their insufficient single-agent efficacy and dose-limiting toxicity.100, 101, 102
Therapeutic strategies for reducing BE progression utilizing key UPS and chaperone players
Through continuing to study RNF128 isoforms and associated proteins that ultimately regulate mutant p53 levels, we aim to uncover insightful ways to effectively mitigate EAC development. It is important to explore a number of therapeutic approaches given the prevalence of intertumor heterogeneity and development of resistance to targeted therapy. Figures 1 and 2 depict the balance between mutant p53 stabilization and degradation as described in this review, including key players that influence this regulation.
Molecular chaperones and co-chaperones including Hsp90, HOP, DNAJA1, and DNAJB1 as well as RNF128 Iso1 are associated with the accumulation of mutant p53. Additionally, WWP1 is known to stabilize WT p53 and thus could also help stabilize mutant p53 in the tumor context. Reducing expression or activity of these regulators can serve as potential ways of shifting the balance of mutant p53 outcomes toward degradation. In the context of BE progression, statins may achieve this through compromising DNAJA1 interaction with mutant p53. Targeting Hsp90 presents a challenge, as this molecular chaperone has ubiquitous, housekeeping roles in preserving proteostasis. As we learn more about RNF128 Iso1, we may discover effective methods to target its expression or compromise its stabilizing activity on mutant p53.
Other possible therapeutic approaches may rely on bolstering machinery that drives mutant p53 degradation, including proteasomal degradation by CHIP and RNF128 Iso2 and the yet unexplored role of β-TrCP1 on mutant p53 protein stability. Dai et al103 reported that reduced levels of CHIP among gastric cancer tissues correlated with lower patient survival. BE is characterized by the presence of columnar epithelial cells in the esophagus which take on properties of gastric and intestinal tissue3,104; thus, it is within reason to compare BE and EAC development to gastric cancers. Elevating CHIP expression or activity may therefore represent a helpful strategy in preventing or managing EAC. Further research into RNF128 isoform regulation may also reveal promising methods of restoring Iso2 levels or activity.
Several chaperone and UPS players display specificity for recognizing and interacting with mutant p53 over WT p53. Hsp70 and Hsp90 machinery associates with unfolded mutant p53 in productive folding complexes or aggregates. CHIP also exhibits specificity for targeting unfolded mutant p53 over folded p53 or WT p53. These features are advantageous for therapeutic approaches, as they provide potential to achieve a therapeutic window that targets cancerous cells harboring unfolded or misfolded mutant proteins while sparing healthy tissue. Through investigating these approaches and exploring their effects in combination, we may be able to devise effective treatments against EAC development and advancement centered around robust degradation of mutant p53.
Conclusion
Mutation in p53 and its subsequent accumulation are critical events that drive the progression of BE to EAC. Similar observations were noted in many other GI cancers. Potential approaches for targeted therapy therefore revolve around strategies directed toward mutant p53 degradation. Among the numerous regulators of mutant p53 levels, there lies a competition between the UPS and chaperone machinery, driving mutant p53 degradation or stabilization, respectively. Impacting these systems therefore provides opportunity for tipping the balance of mutant p53 regulation toward degradation. Work with statins acts as a prototype of this concept, as these drugs impair DNAJA1-mediated stabilization and bolster CHIP-mediated degradation of conformational mutant p53, although we recognize therapeutic limitation of such an approach based on cancer type and p53 mutation specificity. Further studies involving repressing chaperone activity or elevating ubiquitin-proteasome activity may generate similar outcomes that could potentially be used to counter development and progression of EAC and other GI cancers. Important candidates for these studies include players in Hsp70 and Hsp90 machinery, CHIP, and RNF128 isoforms. Our lab is currently investigating regulation of RNF128 isoforms during BE progression to EAC, which is critical to understanding how their functions may be utilized to induce mutant p53 degradation.
When working with the various models, it is important to keep track of the specific p53 mutations used. Different amino acid changes vary in their impact on the folding of mutant p53 and its affinity to other proteins. Hsp90 machinery, in coordination with Hsp70 machinery, can protect both conformational and contact p53 mutants from CHIP and MDM2-mediated degradation. However, mutant p53 stabilization through chaperone-mediated aggregation with other proteins like TAp73ɑ and MDM2 exhibits specificity for conformational p53 mutants. In addition, DNAJA1 protection of mutant p53 from CHIP-mediated degradation is effective for conformational mutant p53. Consistent with this, CHIP exhibits specificity for unfolded mutant p53 while having little to no impact on folded mutant or WT p53. Our lab has seen that RNF128 Iso1 was most effective in upregulating the conformational mutant, p53R175H, of the 4 mutants investigated. Iso2 meanwhile, was shown to effectively downregulate contact mutants p53R248Q and p53R273H. Further research into how different p53 mutants are impacted by chaperone and UPS machinery will inform optimal approaches to mutant p53 degradation based on lesion specific TP53 genotypes. As we continue to gain greater understanding of mutant p53 regulation by the UPS and chaperone systems in the context of EAC, which may also be applicable in certain GI cancers, we may develop promising methods of challenging these cancers' onset and advancement.
Acknowledgments
We thank Steven Kronenberg for graphic assistance.
CRediT Authorship Contributions
May San Martinho, (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Lead)
Derek J. Nancarrow, (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Validation: Supporting; Writing – original draft: Supporting)
Theodore S. Lawrence (Conceptualization: Supporting; Supervision: Supporting; Writing – original draft: Supporting)
David G. Beer (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Project administration: Lead; Resources: Lead; Supervision: Supporting; Writing – original draft: Supporting)
Dipankar Ray (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Lead)
Footnotes
Conflicts of Interest The authors disclose no conflicts.
Funding This work was supported by grants nos. R01CA215596, U54CA163059, P30CA046592, and F31A200113 and the John and Carla Klein Family Research Fund.
References
- 1.Kroep S., Lansdorp-Vogelaar I., Rubenstein J.H., de Koning H.J., Meester R., Inadomi J.M., van Ballegooijen M. An accurate cancer incidence in Barrett's esophagus: a best estimate using published data and modeling. Gastroenterology. 2015;149:577–585.e4. doi: 10.1053/j.gastro.2015.04.045. quiz e14–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spechler S.J., Goyal R.K. Barrett's esophagus. N Engl J Med. 1986;315:362–371. doi: 10.1056/NEJM198608073150605. [DOI] [PubMed] [Google Scholar]
- 3.Bresalier R.S. Barrett's Esophagus and esophageal adenocarcinoma. Annu Rev Med. 2009;60:221–231. doi: 10.1146/annurev.med.59.061206.112706. [DOI] [PubMed] [Google Scholar]
- 4.Kambhampati S., Tieu A.H., Luber B., Wang H., Meltzer S.J. Risk Factors for progression of Barrett's esophagus to high grade dysplasia and esophageal adenocarcinoma. Sci Rep. 2020;10:4899. doi: 10.1038/s41598-020-61874-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishnamoorthi R., Borah B., Heien H., Das A., Chak A., Iyer P.G. Rates and predictors of progression to esophageal carcinoma in a large population-based Barrett's esophagus cohort. Gastrointest Endosc. 2016;84:40–46.e7. doi: 10.1016/j.gie.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stachler M.D., Camarda N.D., Deitrick C., Deitrick C., Kim A., Agoston A.T., Odze R.D., Hornick J.L., Nag A., Thorner A.R., Ducar M., Noffsinger A., Lash R.H., Redston M., Carter S.L., Davison J.M., Bass A.J. Detection of mutations in Barrett's esophagus before progression to high-grade dysplasia or adenocarcinoma. Gastroenterology. 2018;155:156–167. doi: 10.1053/j.gastro.2018.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sethi N.S., Kikuchi O., Duronio G.N., Stachler M.D., McFarland J.M., Ferrer-Luna R., Zhang Y., Bao C., Bronson R., Patil D., Sanchez-Vega F., Liu J.B., Sicinska E., Lazaro J.B., Ligon K.L., Beroukhim R., Bass A.J. Early TP53 alterations engage environmental exposures to promote gastric premalignancy in an integrative mouse model. Nat Genet. 2020;52:219–230. doi: 10.1038/s41588-019-0574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merola E., Claudio P.P., Giordano A. p53 and the malignant progression of Barrett's esophagus. J Cell Physiol. 2006;206:574–577. doi: 10.1002/jcp.20475. [DOI] [PubMed] [Google Scholar]
- 9.Reid B.J., Prevo L.J., Galipeau P.C., Sanchez C.A., Longton G., Levine D.S., Blount P.L., Rabinovitch P.S. Predictors of progression in Barrett's esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol. 2001;96:2839–2848. doi: 10.1111/j.1572-0241.2001.04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams A.B., Schumacher B. p53 in the DNA-damage-repair process. Cold Spring Harb Perspect Med. 2016;6:a026070. doi: 10.1101/cshperspect.a026070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanel W., Moll U.M. Links between mutant p53 and genomic instability. J Cell Biochem. 2012;113:433–439. doi: 10.1002/jcb.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapoor H., Lohani K.R., Lee T.H., Agrawal D.K., Mittal S.K. Animal models of Barrett's esophagus and esophageal adenocarcinoma-past, present, and future. Clin Transl Sci. 2015;8:841–847. doi: 10.1111/cts.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sethi N., Kikuchi O., McFarland J., Zhang Y., Chung M., Kafker N., Islam M., Lampson B., Chakraborty A., Kaelin W.G., Jr., Bass A.J. Mutant p53 induces a hypoxia transcriptional program in gastric and esophageal adenocarcinoma. JCI Insight. 2019;4 doi: 10.1172/jci.insight.128439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 15.Yue X., Zhao Y., Xu Y., Zheng M., Feng Z., Hu W. Mutant p53 in cancer: accumulation, gain-of-function, and therapy. J Mol Biol. 2017;429:1595–1606. doi: 10.1016/j.jmb.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fesler A., Zhang N., Ju J. The expanding regulatory universe of p53 in gastrointestinal cancer. F1000Res. 2016;5:756. doi: 10.12688/f1000research.8363.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steccanella F., Costanzo A., Petrelli F. Editorial on role of p53 in esophageal cancer from a meta-analysis of 16 studies by Fisher et al. J Thorac Dis. 2017;9:1450–1452. doi: 10.21037/jtd.2017.05.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan X., Ji X., Zhang R., Zhou Z., Zhong Y., Peng W., Sun N., Xu X., Xia L., Li P., Lu J., Tu J. Landscape of somatic mutations in gastric cancer assessed using next-generation sequencing analysis. Oncol Lett. 2018;16:4863–4870. doi: 10.3892/ol.2018.9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cicenas J., Kvederaviciute K., Meskinyte I., Meskinyte-Kausiliene E., Skeberdyte A., Cicenas J., Jr. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer. Cancers (Basel) 2017;9:42. doi: 10.3390/cancers9050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama M., Oshima M. Mutant p53 in colon cancer. J Mol Cell Biol. 2019;11:267–276. doi: 10.1093/jmcb/mjy075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asai T., Loza E., Roig G.V., Ajioka Y., Tsuchiya Y., Yamamoto M., Nakamura K. High frequency of TP53 but not K-ras gene mutations in Bolivian patients with gallbladder cancer. Asian Pac J Cancer Prev. 2014;15:5449–5454. doi: 10.7314/apjcp.2014.15.13.5449. [DOI] [PubMed] [Google Scholar]
- 22.Lombardo D., Saitta C., Giosa D., Casuscelli Di Tocco F., Musolino C., Caminiti G., Chines V., Franzè M.S., Alibrandi A., Navarra G., Raimondo G., Pollicino T. Frequency of somatic mutations in TERT promoter, TP53 and CTNNB1 genes in patients with hepatocellular carcinoma from Southern Italy. Oncol Lett. 2020;19:2368–2374. doi: 10.3892/ol.2020.11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu K., Zhang X., Cui R., Liu C. Meta-signature of mutated genes in gallbladder cancer: evidence based high throughput screening assays. Ann Transl Med. 2016;4:229. doi: 10.21037/atm.2016.05.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia P.B., Attardi L.D. Illuminating p53 function in cancer with genetically engineered mouse models. Semin Cell Dev Biol. 2014;27:74–85. doi: 10.1016/j.semcdb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H., Zhang J., Tong J.H.M., Chan A.W.H., Yu J., Kang W., To K.F. Targeting the oncogenic p53 mutants in colorectal cancer and other solid tumors. Int J Mol Sci. 2019;20:5999. doi: 10.3390/ijms20235999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakayama M., Sakai E., Echizen K., Yamada Y., Oshima H., Han T.S., Ohki R., Fujii S., Ochiai A., Robine S., Voon D.C., Tanaka T., Taketo M.M., Oshima M. Intestinal cancer progression by mutant p53 through the acquisition of invasiveness associated with complex glandular formation. Oncogene. 2017;36:5885–5896. doi: 10.1038/onc.2017.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weissmueller S., Manchado E., Saborowski M., Morris J.P., 4th, Wagenblast E., Davis C.A., Moon S.H., Pfister N.T., Tschaharganeh D.J., Kitzing T., Aust D., Markert E.K., Wu J., Grimmond S.M., Pilarsky C., Prives C., Biankin A.V., Lowe S.W. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell. 2014;157:382–394. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morton J.P., Timpson P., Karim S.A., Ridgway R.A., Athineos D., Doyle B., Jamieson N.B., Oien K.A., Lowy A.M., Brunton V.G., Frame M.C., Evans T.R.J., Sanson O.J. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U S A. 2010;107:246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaiddon C., Lokshin M., Ahn J., Zhang T., Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahronian L.G., Driscoll D.R., Klimstra D.S., Lewis B.C. The p53R172H mutant does not enhance hepatocellular carcinoma development and progression. PLoS One. 2015;10 doi: 10.1371/journal.pone.0123816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36:3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menendez D., Inga A., Resnick M.A. The biological impact of the human master regulator p53 can be altered by mutations that change the spectrum and expression of its target genes. Mol Cell Biol. 2006;26:2297–2308. doi: 10.1128/MCB.26.6.2297-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resnick M.A., Inga A. Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity. Proc Natl Acad Sci U S A. 2003;100:9934–9939. doi: 10.1073/pnas.1633803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Kim M.P., Lozano G. Mutant p53 partners in crime. Cell Death Differ. 2018;25:161–168. doi: 10.1038/cdd.2017.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baugh E.H., Ke H., Levine A.J., Bonneau R.A., Chan C.S. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018;25:154–160. doi: 10.1038/cdd.2017.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine A.J. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20:471–480. doi: 10.1038/s41568-020-0262-1. [DOI] [PubMed] [Google Scholar]
- 38.Singh S., Vaughan C.A., Frum R.A., Grossman S.R., Deb S., Deb S.P. Mutant p53 establishes targetable tumor dependency by promoting unscheduled replication. J Clin Invest. 2017;127:1839–1855. doi: 10.1172/JCI87724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray D., Ray P., Ferrer-Torres D., Wang Z., Nancarrow D., Yoon H.W., San Martinho M., Hinton T., Owens S., Thomas D., Jiang H., Lawrence T.S., Lin J., Lagisetty K., Chang A.C., Beer D.G. Isoforms of RNF128 regulate the stability of mutant P53 in Barrett's esophageal cells. Gastroenterology. 2020;158:583–597.e1. doi: 10.1053/j.gastro.2019.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller P., Hrstka R., Coomber D., Lane D.P., Vojtesek B. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene. 2008;27:3371–3383. doi: 10.1038/sj.onc.1211010. [DOI] [PubMed] [Google Scholar]
- 41.Freed-Pastor W.A., Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouaoun L., Sonkin D., Ardin M., Hollstein M., Byrnes G., Zavadil J., Olivier M. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum Mutat. 2016;37:865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 43.Kruse J.P., Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng Q., Chen J. Mechanism of p53 stabilization by ATM after DNA damage. Cell Cycle. 2010;9:472–478. doi: 10.4161/cc.9.3.10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vijayakumaran R., Tan K.H., Miranda P.J., Haupt S., Haupt Y. Regulation of mutant p53 protein expression. Front Oncol. 2015;5:284. doi: 10.3389/fonc.2015.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meek D.W., Anderson C.W. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol. 2009;1 doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y., Tavana O., Gu W. p53 modifications: exquisite decorations of the powerful guardian. J Mol Cell Biol. 2019;11:564–577. doi: 10.1093/jmcb/mjz060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lavin M.F., Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 49.Wawrzynow B., Zylicz A., Zylicz M. Chaperoning the guardian of the genome. The two-faced role of molecular chaperones in p53 tumor suppressor action. Biochim Biophys Acta Rev Cancer. 2018;1869:161–174. doi: 10.1016/j.bbcan.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Shadfan M., Lopez-Pajares V., Yuan Z.M. MDM2 and MDMX: alone and together in regulation of p53. Transl Cancer Res. 2012;1:88–89. [PMC free article] [PubMed] [Google Scholar]
- 51.Lang G.A., Iwakuma T., Suh Y.A., Liu G., Rao V.A., Parant J.M., Valentin-Vega Y.A., Terzian T., Caldwell L.C., Strong L.C., El-Naggar A.K., Lozano G. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Ellis R.J. Revisiting the Anfinsen cage. Fold Des. 1996;1:R9–R15. [PubMed] [Google Scholar]
- 53.Hartl F.U. Chaperone-assisted protein folding: the path to discovery from a personal perspective. Nat Med. 2011;17:1206–1210. doi: 10.1038/nm.2467. [DOI] [PubMed] [Google Scholar]
- 54.Esser C., Alberti S., Hohfeld J. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim Biophys Acta. 2004;1695:171–188. doi: 10.1016/j.bbamcr.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 55.Stolz A., Wolf D.H. Endoplasmic reticulum associated protein degradation: a chaperone assisted journey to hell. Biochim Biophys Acta. 2010;1803:694–705. doi: 10.1016/j.bbamcr.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 56.Theodoraki M.A., Caplan A.J. Quality control and fate determination of Hsp90 client proteins. Biochim Biophys Acta. 2012;1823:683–688. doi: 10.1016/j.bbamcr.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kundrat L., Regan L. Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry. 2010;49:7428–7438. doi: 10.1021/bi100386w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller P., Ruckova E., Halada P., Coates P.J., Hrstka R., Lane D.P., Vojtesek B. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene. 2013;32:3101–3110. doi: 10.1038/onc.2012.314. [DOI] [PubMed] [Google Scholar]
- 59.Laine A., Ronai Z. Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene. 2007;26:1477–1483. doi: 10.1038/sj.onc.1209924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li M., Brooks C.L., Wu-Baer F., Chen D., Baer R., Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 61.Zhang L., Wu Z., Ma Z., Liu H., Wu Y., Zhang Q. WWP1 as a potential tumor oncogene regulates PTEN-Akt signaling pathway in human gastric carcinoma. Tumour Biol. 2015;36:787–798. doi: 10.1007/s13277-014-2696-0. [DOI] [PubMed] [Google Scholar]
- 62.Zhang X.F., Chao J., Pan Q.Z., Pan K., Weng D.S., Wang Q.J., Zhao J.J., He J., Liu Q., Jiang S.S., Chen C.L., Zhang H.X., Xia J.C. Overexpression of WWP1 promotes tumorigenesis and predicts unfavorable prognosis in patients with hepatocellular carcinoma. Oncotarget. 2015;6:40920–40933. doi: 10.18632/oncotarget.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J.J., Zhang W. High expression of WWP1 predicts poor prognosis and associates with tumor progression in human colorectal cancer. Am J Cancer Res. 2018;8:256–265. [PMC free article] [PubMed] [Google Scholar]
- 64.Hou Y.C., Deng J.Y. Role of E3 ubiquitin ligases in gastric cancer. World J Gastroenterol. 2015;21:786–793. doi: 10.3748/wjg.v21.i3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang W., Qin J.J., Voruganti S., Nijampatnam B., Velu S.E., Ruan K.H., Hu M., Zhou J., Zhang R. Discovery and characterization of dual inhibitors of MDM2 and NFAT1 for pancreatic cancer therapy. Cancer Res. 2018;78:5656–5667. doi: 10.1158/0008-5472.CAN-17-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugano N., Suda T., Godai T.I., Tsuchida K., Shiozawa M., Sekiguchi H., Yoshihara M., Matsukuma S., Sakuma Y., Tsuchiya E., Kameda Y., Akaike M., Miyagi Y. MDM2 gene amplification in colorectal cancer is associated with disease progression at the primary site, but inversely correlated with distant metastasis. Genes Chromosomes Cancer. 2010;49:620–629. doi: 10.1002/gcc.20774. [DOI] [PubMed] [Google Scholar]
- 67.Wang W., Hu B., Qin J.J., Cheng J.W., Li X., Rajaei M., Fan J., Yang X.R., Zhang R. A novel inhibitor of MDM2 oncogene blocks metastasis of hepatocellular carcinoma and overcomes chemoresistance. Genes Dis. 2019;6:419–430. doi: 10.1016/j.gendis.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paul I., Ghosh M.K. The E3 ligase CHIP: insights into its structure and regulation. Biomed Res Int. 2014;2014:918183. doi: 10.1155/2014/918183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang T., Yang J., Xu J., Li J., Cao Z., Zhou L., You L., Shu H., Lu Z., Li H., Li M., Zhang T., Zhao Y. CHIP is a novel tumor suppressor in pancreatic cancer through targeting EGFR. Oncotarget. 2014;5:1969–1986. doi: 10.18632/oncotarget.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y., Ren F., Wang Y., Feng Y., Wang D., Jia B., Qiu Y., Wang S., Yu J., Sung J.J., Xu J., Zeps N., Chang Z. CHIP/Stub1 functions as a tumor suppressor and represses NF-kappaB-mediated signaling in colorectal cancer. Carcinogenesis. 2014;35:983–991. doi: 10.1093/carcin/bgt393. [DOI] [PubMed] [Google Scholar]
- 71.Chatterjee S., Burns T.F. Targeting heat shock proteins in cancer: a promising therapeutic approach. Int J Mol Sci. 2017;18:1978. doi: 10.3390/ijms18091978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xia Y., Rocchi P., Iovanna J.L., Peng L. Targeting heat shock response pathways to treat pancreatic cancer. Drug Discov Today. 2012;17:35–43. doi: 10.1016/j.drudis.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 73.Zhang T., Hamza A., Cao X., Wang B., Yu S., Zhan C.G., Sun D. A novel Hsp90 inhibitor to disrupt Hsp90/Cdc37 complex against pancreatic cancer cells. Mol Cancer Ther. 2008;7:162–170. doi: 10.1158/1535-7163.MCT-07-0484. [DOI] [PubMed] [Google Scholar]
- 74.Gress T.M., Muller-Pillasch F., Weber C., Lerch M.M., Friess H., Büchler M., Beger H.G., Adler G. Differential expression of heat shock proteins in pancreatic carcinoma. Cancer Res. 1994;54:547–551. [PubMed] [Google Scholar]
- 75.Wang J., Cui S., Zhang X., Wu Y., Tang H. High expression of heat shock protein 90 is associated with tumor aggressiveness and poor prognosis in patients with advanced gastric cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ge H., Yan Y., Guo L., Tian F., Wu D. Prognostic role of HSPs in human gastrointestinal cancer: a systematic review and meta-analysis. Onco Targets Ther. 2018;11:351–359. doi: 10.2147/OTT.S155816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang C., Zhang Y., Guo K., Wang N., Jin H., Liu Y., Qin W. Heat shock proteins in hepatocellular carcinoma: molecular mechanism and therapeutic potential. Int J Cancer. 2016;138:1824–1834. doi: 10.1002/ijc.29723. [DOI] [PubMed] [Google Scholar]
- 78.Li D., Marchenko N.D., Schulz R., Fischer V., Velasco-Hernandez T., Talos F., Moll U.M. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res. 2011;9:577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Slotta-Huspenina J., Becker K.F., Feith M., Walch A., Langer R. Heat shock protein 90 (HSP90) and Her2 in adenocarcinomas of the esophagus. Cancers (Basel) 2014;6:1382–1393. doi: 10.3390/cancers6031382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tracz-Gaszewska Z., Klimczak M., Biecek P., Biecek P., Herok M., Kosinski M., Olszewski M.B., Czerwinska P., Wiech M., Wiznerowicz M., Zylicz A., Zylicz M., Wawrzynow B. Molecular chaperones in the acquisition of cancer cell chemoresistance with mutated TP53 and MDM2 up-regulation. Oncotarget. 2017;8:82123–82143. doi: 10.18632/oncotarget.18899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parrales A., Ranjan A., Iyer S.V., Padhye S., Weir S.J., Roy A., Iwakuma T. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat Cell Biol. 2016;18:1233–1243. doi: 10.1038/ncb3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parrales A., Thoenen E., Iwakuma T. The interplay between mutant p53 and the mevalonate pathway. Cell Death Differ. 2018;25:460–470. doi: 10.1038/s41418-017-0026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh S., Singh A.G., Singh P.P., Murad M.H., Iyer P.G. Statins are associated with reduced risk of esophageal cancer, particularly in patients with Barrett's esophagus: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2013;11:620–629. doi: 10.1016/j.cgh.2012.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nag S., Qin J., Srivenugopal K.S., Wang M., Zhang R. The MDM2-p53 pathway revisited. J Biomed Res. 2013;27:254–271. doi: 10.7555/JBR.27.20130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nurieva R.I., Liu X., Dong C. Molecular mechanisms of T-cell tolerance. Immunol Rev. 2011;241:133–144. doi: 10.1111/j.1600-065X.2011.01012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen Y.C., Chan J.Y., Chiu Y.L., Liu S.T., Lozano G., Wang S.L., Ho C.L., Huang S.M. Grail as a molecular determinant for the functions of the tumor suppressor p53 in tumorigenesis. Cell Death Differ. 2013;20:732–743. doi: 10.1038/cdd.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iyappan S., Wollscheid H.P., Rojas-Fernandez A., Marquardt A., Tang H.C., Singh R.K., Scheffner M. Turning the RING domain protein MdmX into an active ubiquitin-protein ligase. J Biol Chem. 2010;285:33065–33072. doi: 10.1074/jbc.M110.115113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng T., Wang J., Zhao Y., Zhang C., Lin M., Wang X., Yu H., Liu L., Feng Z., Hu W. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat Commun. 2013;4:2996. doi: 10.1038/ncomms3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Inuzuka H., Tseng A., Gao D., Zhai B., Zhang Q., Shaik S., Wan L., Ang X.L., Mock C., Yin H., Stommel J.M., Gygi S., Lahav G., Asara J., Xiao Z.X.J., Kaelin W.G., Jr., Harper J.W., Wei W. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell. 2010;18:147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xia Y., Padre R.C., De Mendoza T.H., Bottero V., Tergaonkar V.B., Verma I.M. Phosphorylation of p53 by IkappaB kinase 2 promotes its degradation by beta-TrCP. Proc Natl Acad Sci U S A. 2009;106:2629–2634. doi: 10.1073/pnas.0812256106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Walerych D., Kudla G., Gutkowska M., Wawrzynow B., Muller L., King F.W., Helwak A., Boros J., Zylicz A., Zylicz M. Hsp90 chaperones wild-type p53 tumor suppressor protein. J Biol Chem. 2004;279:48836–48845. doi: 10.1074/jbc.M407601200. [DOI] [PubMed] [Google Scholar]
- 92.Demierre M.F., Higgins P.D., Gruber S.B., Hawk E., Lippman S.M. Statins and cancer prevention. Nat Rev Cancer. 2005;5:930–942. doi: 10.1038/nrc1751. [DOI] [PubMed] [Google Scholar]
- 93.Wu X.D., Zeng K., Xue F.Q., Chen J.H., Chen Y.Q. Statins are associated with reduced risk of gastric cancer: a meta-analysis. Eur J Clin Pharmacol. 2013;69:1855–1860. doi: 10.1007/s00228-013-1547-z. [DOI] [PubMed] [Google Scholar]
- 94.Abdullah M.I., de Wolf E., Jawad M.J., Richardson A. The poor design of clinical trials of statins in oncology may explain their failure - lessons for drug repurposing. Cancer Treat Rev. 2018;69:84–89. doi: 10.1016/j.ctrv.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 95.Tutuska K., Parrilla-Monge L., Di Cesare E., Nemajerova A., Moll U.M. Statin as anti-cancer therapy in autochthonous T-lymphomas expressing stabilized gain-of-function mutant p53 proteins. Cell Death Dis. 2020;11:274. doi: 10.1038/s41419-020-2466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ingallina E., Sorrentino G., Bertolio R., Lisek K., Zannini A., Azzolin L., Severino L.U., Scaini D., Mano M., Mantovani F., Rosato A., Bicciato S., Piccolo S., Del Sal G. Mechanical cues control mutant p53 stability through a mevalonate-RhoA axis. Nat Cell Biol. 2018;20:28–35. doi: 10.1038/s41556-017-0009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Turrell F.K., Kerr E.M., Gao M., Thorpe H., Doherty G.J., Cridge J., Shorthouse D., Speed A., Samarajiwa S., Hall B.A., Griffiths M., Martins C.P. Lung tumors with distinct p53 mutations respond similarly to p53 targeted therapy but exhibit genotype-specific statin sensitivity. Genes Dev. 2017;31:1339–1353. doi: 10.1101/gad.298463.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu D., Tong X., Sun L., Li H., Jones R.D., Liao J., Yang G.Y. Inhibition of mutant Kras and p53-driven pancreatic carcinogenesis by atorvastatin: mainly via targeting of the farnesylated DNAJA1 in chaperoning mutant p53. Mol Carcinog. 2019;58:2052–2064. doi: 10.1002/mc.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sorrentino G., Mantovani F., Del Sal G. The stiff RhoAd from mevalonate to mutant p53. Cell Death Differ. 2018;25:645–647. doi: 10.1038/s41418-018-0091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shimomura A., Yamamoto N., Kondo S., Fujiwara Y., Suzuki S., Yanagitani N., Horiike A., Kitazono S., Ohyanagi F., Doi T., Kuboki Y., Kawazoe A., Shitara K., Ohno I., Banerji U., Sundar R., Ohkubo S., Calleja E.M., Nishio M. First-in-human phase I study of an oral HSP90 inhibitor, TAS-116, in patients with advanced solid tumors. Mol Cancer Ther. 2019;18:531–540. doi: 10.1158/1535-7163.MCT-18-0831. [DOI] [PubMed] [Google Scholar]
- 101.Garcia-Carbonero R., Carnero A., Paz-Ares L. Inhibition of HSP90 molecular chaperones: moving into the clinic. Lancet Oncol. 2013;14:e358–e369. doi: 10.1016/S1470-2045(13)70169-4. [DOI] [PubMed] [Google Scholar]
- 102.Park H.K., Yoon N.G., Lee J.E., Yoon N.G., Lee J.E., Hu S., Yoon S., Kim S.Y., Hong J.H., Nam D., Chae Y.C., Park J.B., Kang B.H. Unleashing the full potential of Hsp90 inhibitors as cancer therapeutics through simultaneous inactivation of Hsp90, Grp94, and TRAP1. Exp Mol Med. 2020;52:79–91. doi: 10.1038/s12276-019-0360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dai H., Chen H., Xu J., Zhou J., Shan Z., Yang H., Zhou X., Guo F. The ubiquitin ligase CHIP modulates cellular behaviors of gastric cancer cells by regulating TRAF2. Cancer Cell Int. 2019;19:132. doi: 10.1186/s12935-019-0832-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Que J., Garman K.S., Souza R.F., Spechler S.J. Pathogenesis and cells of origin of barrett's esophagus. Gastroenterology. 2019;157:349–364.e1. doi: 10.1053/j.gastro.2019.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bullock A.N., Henckel J., Fersht A.R. Altered-function p53 missense mutations identified in breast cancers can have subtle effects on transactivation. Mol Cancer Res. 2010;8:701–716. doi: 10.1158/1541-7786.MCR-09-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jordan J.J., Inga A., Conway K., Edmiston S., Carey L.A., Wu L., Resnick M.A. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene. 2000;19:1245–1256. doi: 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]