Abstract
Background & Aims
Transient expression of Neurog3 commits intestinal secretory progenitors to become enteroendocrine-biased progenitors and hence drive enteroendocrine differentiation. Loss of Neurog3 in mouse resulted in the depletion of intestinal enteroendocrine cells (EECs) and an increase in goblet cells. Earlier studies in developing mouse pancreas identified a role of Neurog3 gene dosage in endocrine and exocrine cell fate allocation. We aimed to determine whether Neurog3 gene dosage controls fate choice of enteroendocrine progenitors.
Methods
We acquired mutant Neurog3 reporter mice carrying 2, 1, or null Neurog3 alleles to study Neurog3 gene dosage effect by lineage tracing. Cell types arising from Neurog3+ progenitors were determined by immunohistochemistry using antibodies against intestinal lineage-specific markers. RNA sequencing of sorted Neurog3+/+, Neurog3+/-, or bulk intestinal cells were performed and differentially expressed genes were analyzed.
Results
We identified 2731 genes enriched in sorted Neurog3+/+-derived cells in the Neurog3+/+EYFP mouse intestine when compared with bulk duodenum epithelial cells. In the intestine of Neurog3+/-EGFP heterozygous mouse, we observed a 63% decrease in EEC numbers. Many Neurog3-derived cells stained for goblet marker Mucin 2. RNA sequencing of sorted Neurog3+/- cells uncovered enriched expression of genes characteristic for both goblet and enteroendocrine cells, indicating the mixed lineages arose from Neurog3+ progenitors. Consistent with this hypothesis, deletion of both Neurog3 alleles resulted in the total absence of EECs. All Neurog3+-derived cells stained for Mucin 2.
Conclusions
We identified that the fate of Neurog3+ enteroendocrine progenitors is dependent on Neurog3 gene dosage. High Neurog3 gene dosage enforces the commitment of secretory progenitors to an EE lineage, while constraining their goblet cell lineage potential. Transcriptome profiling data was deposited to Gene Ontology omnibus, accession number: GSE149203.
Keywords: Enteroendocrine Progenitors, Neurog3 Gene Dosage, Goblet Cell Differentiation
Abbreviations used in this paper: Asxl3, additional sex-combs like 3; ChgA, chromogranin A; DEG, differentially expressed gene; EE, enteroendocrine; EEC, enteroendocrine cell; EGFP, enhanced green fluorescent protein; EYFP, enhanced yellow fluorescent protein; FACS, fluorescent-activated cell sorting; Gfi, growth factor independence 1; GO, Gene Ontology; Hmgn, high-mobility group nucleosome; IF, immunofluorescent; ISC, intestinal stem cell; Lyz, lysozyme; MMP, matrix metalloproteinase; mRNA, messenger RNA; Muc2, Mucin 2; Neurod, Neurod1; Neurog, Neurogenin; PBS, phosphate-buffered saline; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; RNA-seq, RNA sequencing; Spdef, SAM pointed domain-containing ETS transcription factor; TF, transcription factor
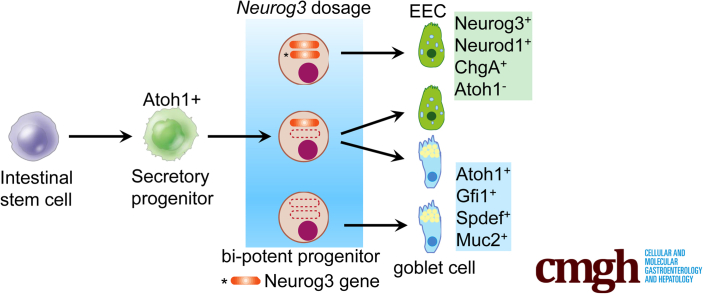
Graphical abstract
Summary.
This study reports the transcriptome profiles for Neurog3+-derived cells isolated from mouse duodenum with 2 or 1 Neurog3 alleles. Deletion of 1 Neurogenin3 allele resulted in reduced enteroendocrine cells, increased goblet cell number, and altered gene expression profile, indicating that Neurog3 gene dosage determined intestinal Neurog3+ progenitor fate specification.
Single-columnar intestinal epithelial cells fall into 2 broad classes: the absorptive enterocyte lineage (∼80%) and the secretory cell lineage, including mucus-secreting goblet cells (∼10%–15%), hormone-secreting enteroendocrine (EE) cells (EECs) (∼1%), chemosensory tuft cells (<1%), and antimicrobial Paneth cells (<5%).1, 2, 3 Other than Paneth cells that cluster at the bottom of the crypt base, EEC, goblet, and tuft cells appear as singlets scattered along the intestinal epithelium.
Intestinal stem cells (ISCs) that reside at the bottom of crypts give rise to all types of intestinal epithelial cells.4 Intestinal cell-type specification and differentiation are orchestrated by the expression of a cascade of transcription factors. The expression of Atonal bHLH transcription factor 1 (Atoh1), a basic helix-loop-helix transcription factor (TF), in the ISCs initiates ISC differentiation toward secretory cell lineages.5 Subsequent downstream lineage-specific transcription factors regulate secretory progenitors to differentiate into mature cell types: Neurogenin 3 (Neurog3) and NeuroD1 (Neurod1) for enteroendocrine,6, 7, 8, 9, 10 and Growth factor independent 1 transcriptional repressor (Gfi1) and SAM Pointed Domain Containing ETS Transcription Factor (Spdef) for goblet/Paneth cells.11,12 Gfi1, a direct target of Atoh1 that is critical for goblet cell lineage specification, was shown to repress the expression of Neurog3,13 suggesting an interplay between Neurog3 and Gfi1 that may determine the allocation of secretory cell fate among EE, goblet, and Paneth cells.
Neurog3, a basic helix-loop-helix transcription factor, is expressed transiently in endocrine progenitor cells of the pancreas and intestine.14 In pancreas, where it is known to play an essential role in controlling pancreatic islet cell development and differentiation,15 Neurog3 is expressed in endocrine progenitors, but not mature islets. Ectopic expression of Neurog3 in adult human pancreatic duct cells results in fate shifting to endocrine differentiation.16 Forced Neurog3 expression in Pdx1+ pancreatic precursor cells leads to their precocious endocrine differentiation.17 Conversely, Neurog3 deficiency results in the absence of all islet cell types and neonatal lethality from diabetes.14,18 Lowered Neurog3 expression levels in the developing mouse pancreas was shown to regulate allocation of endocrine and exocrine cell fates.19 However, it occurred only in Neurog3 hypomorphic mice in which the Neurog3 expression level was less than 10% of littermate controls. No noticeable difference was observed in islet development in Neurog3+/- heterozygous mice.
Similar to pancreatic islet development, Neurog3 in the intestinal epithelium acts as an enteroendocrine lineage-determining transcription factor.20,21 Neurog3 is expressed transiently in early secretory progenitors, where it initiates EEC differentiation by activating a cascade of downstream target genes including Neurod1, Pax4/6, Nkx2.2, IA-1/Insm1, and Pdx1.20,22, 23, 24, 25 Forced overexpression of Neurog3 in Atoh1+ early progenitor cells directs their differentiation toward EE cells21,26 at the expense of reducing goblet cell numbers,6 whereas Neurog3 knock-out in mice results in loss of most endocrine cells in the gastrointestinal tract and neonatal death because of the failure in pancreatic islet development.21,26
Transgenic overexpression of Neurog3 in the intestine expands the number of EECs at the expense of goblet cells, suggesting that Neurog3 plays a critical role in the bipotential secretory progenitor’s fate choice between EE and goblet cells.6 We hypothesize that the fate choice between EE and goblet cells may depend on Neurog3 gene dosage. To test this hypothesis, we generated mutant mice carrying 2, 1, or null Neurog3 alleles with fluorescent reporters in Neurog3+ cells as a lineage tracing marker. Neurog3+ progenitor cell fate was analyzed by immunofluorescent staining using antibodies against secretory lineage markers. Molecular signatures of those Neurog3+ derived cells were determined by RNA sequencing (RNA-seq). We found that Neurog3+ progenitor cell fate choice between EE and goblet cells is sensitive to Neurog3 gene dosage. Furthermore, RNA-seq data showed that deletion of 1 Neurog3 allele produced substantial changes in gene expression, with the reduction of transcripts related to EEC differentiation and function. In addition, a significant increase of transcripts pertaining to goblet cell differentiation and function was observed. Our results substantiated our hypothesis that Neurog3 gene dosage regulates the allocation of their fates toward EEC vs goblet cells. This hypothesis was supported further by lineage tracing in Neurog3 null mice, which showed that all Neurog3-derived cells become goblet cells.
Results
Expression Profiling of Intestinal EE Cells
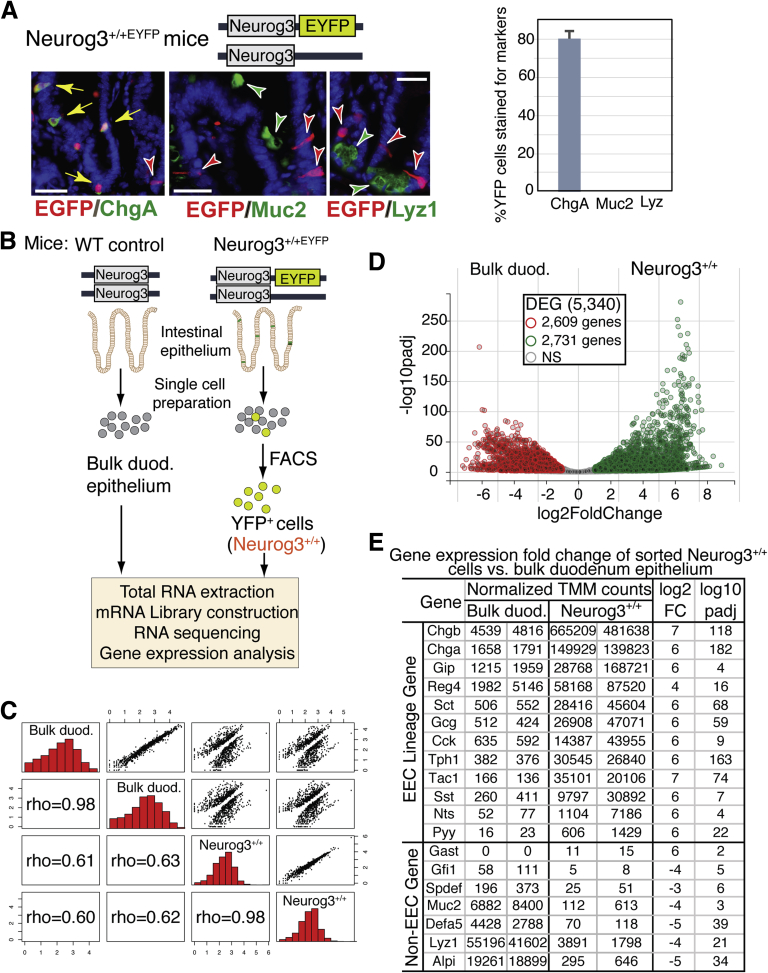
In this study, we used Neurog3+/+EYFP (Neurog3+/+) transgenic reporter mice with an internal ribosome entry site-enhanced yellow fluorescent protein (EYFP) reporter gene knock-addon at the 3’-untranslated region of the Neurog3 gene (Figure 1A, illustration). An early study showed restricted endocrine expression of EYFP cells in pancreas. In the small intestine, EYFP expression was confined to scattered cells throughout the intestinal epithelium.27 Here, we used an enhanced green fluorescent protein (EGFP) antibody (recognizing both EGFP and EYFP proteins) and a chromogranin A (ChgA) antibody to examine the EYFP-tagged cells for expression of the pan-endocrine marker ChgA using double immunofluorescent (IF) staining on small intestinal tissue sections. IF staining showed that approximately 80% (80% ± 4%) of EGFP+ cells stained with ChgA antibody (Figure 1A, bar graph on the right). Those EGFP+ cells lacking ChgA (EGFP+ChgA- cells) may represent early EEC progenitor cells that do not yet express ChgA, or express it at undetectable levels (Figure 1A, red arrow). However, none of the EGFP+ cells were seen to co-express the other intestinal secretory lineage markers Mucin 2 (Muc2) for goblet cells or lysozyme (Lyz) for Paneth cells (Figure 1A, green arrowhead). These results clearly indicate that EYFP+ cells with EYFP gene knock-addon to the Neurog3 locus faithfully tagged endogenous EE cells derived from Neurog3-expressing progenitors, similar to what was observed in pancreas.27
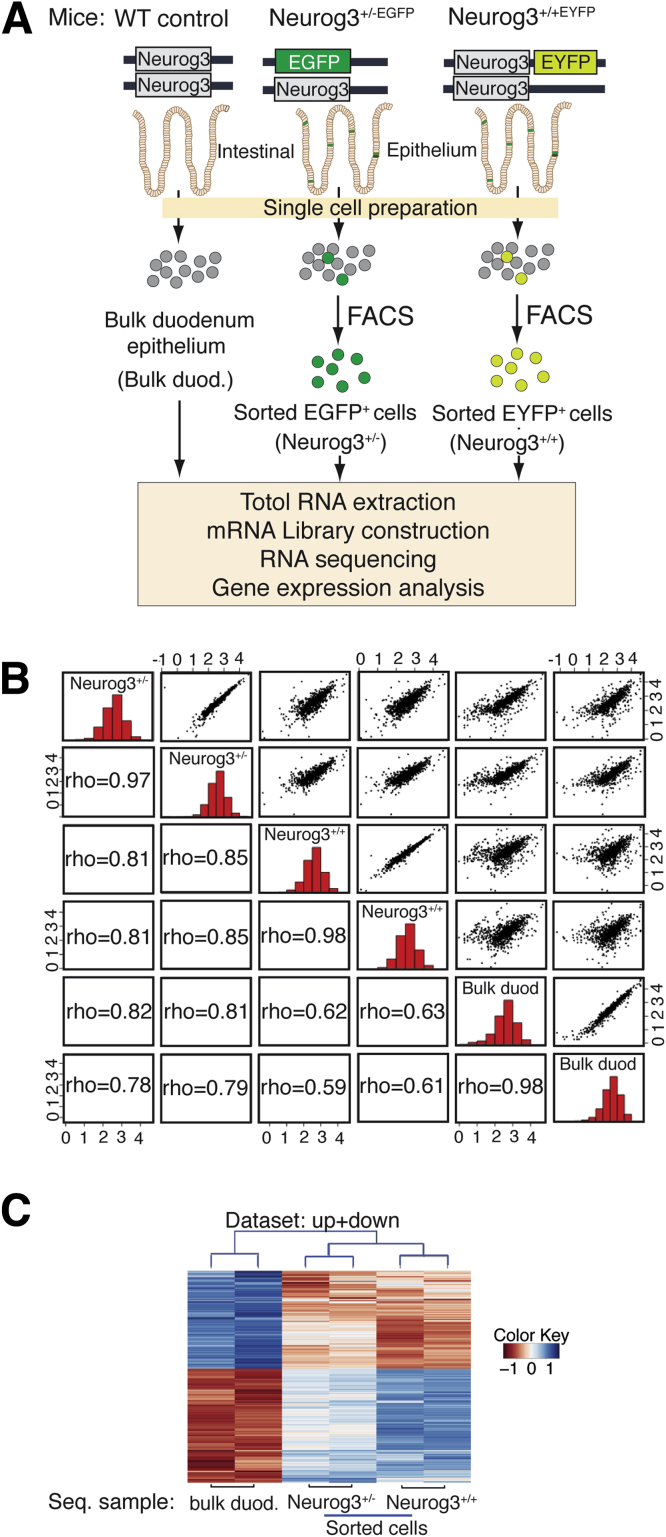
Figure 1.
EYFP expression is restricted to EECs in the small intestine of Neurog3+/+EYFP(Neurog3+/+) reporter mouse. (A) Top: Schematic diagram of Neurog3 and EYFP at the Neurog3 locus in Neurog3+/+EYFP mouse. Bottom: Adult mouse intestinal sections from Neurog3+/+ mice were co-stained with antibodies against GFP (in red) and against ChgA, Muc2, or lysozyme, markers (in green) for enteroendocrine, goblet, and Paneth cells, respectively. Yellow arrows indicate co-expression, and 4′,6-diamidino-2-phenylindole (in blue) counterstained for nuclei. Scale bars: 50 um. Percentage of GFP cells stained for ChgA, Muc2, or Lyz was summarized in the bar graph (right) (N = 3). P < .00001 as indicated. The P value was calculated by 1-tailed Student t test. (B) Experimental flow chart of cell isolation and RNA-seq procedure. (C) All2all scatter plot of all transcripts detected after batch correction: an overview of similarities and variance between samples sequenced. Each Neurog3+/+ library was made from RNAs isolated and sorted duodenum cells from 2 Neurog3+/+EYFP mice. Each bulk duodenum (Bulk duod.) library was made from RNAs from 1 wild-type (WT) control mouse duodenum epithelial cells. Two independent replicates were performed for each genotype. The upper side of the plot shows a log10 normalized scatter plot. The bottom side of the plot is the Pearson correlations of each paired sample. Diagonal shows the histogram of normalized read counts in log10 scale. (D) Volcano plots of normalized TMM counts comparing the transcriptomes of sorted Neurog3+/+ cells and control intestinal epithelial cells (Unsorted). A total of 5340 transcripts were found to be expressed differentially (|Fc| ≥2), with 2609 genes enriched in the control cell population and 2731 genes enriched in the Neurog3+/+ cell population. (E) Selected transcript levels of EEC-specific hormones and marker genes expressed in Neurog3+/+ cells vs control unsorted intestinal cells. Normalized gene expression values (TMM) were adjusted using edgeR testing methods that account for library depth, RNA composition, and gene length among samples. Fc, fold change.
To interrogate the gene expression pattern of all EE cells derived from Neurog3-expressing progenitor cells, we isolated singlet EYFP+ cells from the upper intestinal epithelium duodenum by fluorescent-activated cell sorting (FACS) and performed RNA-seq to accurately measure gene expression in those cells as shown in an experimental flowchart diagram in Figure 1B. An RNA-seq library made from extracted total RNAs from unsorted bulk duodenum epithelial cells comprising mainly enterocytes (90%) and secretory cells (goblet cells, 8%–10%; EECs, ∼1%) was sequenced and used as a control for differentially expressed gene (DEG) analysis. We conducted differential expression analysis using DEseq2 (1.26.0)28 to identify DEGs in the EECs as compared with bulk epithelial cells. Quality control of the count data is shown in an All2all scatter plot28 (Figure 1C), with coefficient rho of 0.98 between the 2 gene expression data sets from 2 sequenced libraries of sorted duodenum from Neurog3+/+EYFP mice or bulk duodenum from wild-type control mice. The rho number was less than 0.63 when compared with each library between Neurog3+/+ and bulk duodenum cells, indicating the data are suitable for differential expression analysis. From the 23,846 detected transcripts, there were 2731 genes showing highly enriched expression in Neurog3+/+ cells (>2-fold, P-adjusted value < .01), when compared with bulk duodenum epithelial cells (Figure 1D; total genes are listed in Supplementary Table 1).
The top 40 most abundant differentially expressed transcripts consist of those genes encoding intestinal peptide hormones or related key enzymes including ghrelin (Ghrl), cholecytokinin (Cck), secretin (Sct), somatostatin (Sst), gastric inhibitory polypeptide, substance P (Tac1), Glp-1 precursor (Gcg), and tryptophan hydroxylase 1 (Tph1), a rate-limiting enzyme in serotonin biosynthesis (Figure 1E). Pan-endocrine markers ChgA, ChgB, and Reg4 were the most abundant transcripts.29 Other neuroendocrine peptides including galanin (Gal), amylin (Iapp), and nucleobindin 2 (Nucb2) were found at very low levels in those sorted Neurog3+/+ cells. No enrichment was found for the stomach-specific hormone gastrin (Gast) or pancreatic-specific hormone insulin (Ins1).
The lineage specificity of Neurog3+ progenitor-derived cells were assessed further. As expected, the RNA-seq gene expression profile of Neurog3+/+ cells showed no enrichment of lineage-specific marker genes for goblet (Gfi1, Muc2, and Spdef), Paneth (Mptx2, Defa, and Lyz1), or tuft (Dmck1, Gfi1b, and Trpm5) cells (Figure 1E and Supplementary Table 2). Taken together, the EYFP+ cells derived from Neurog3+ progenitors in the intestines of Neurog3+/+EYFP mice comprised a heterogeneous population of gut hormone–producing cells.
Neurog3+/+ Cells Represent EE Cell Type
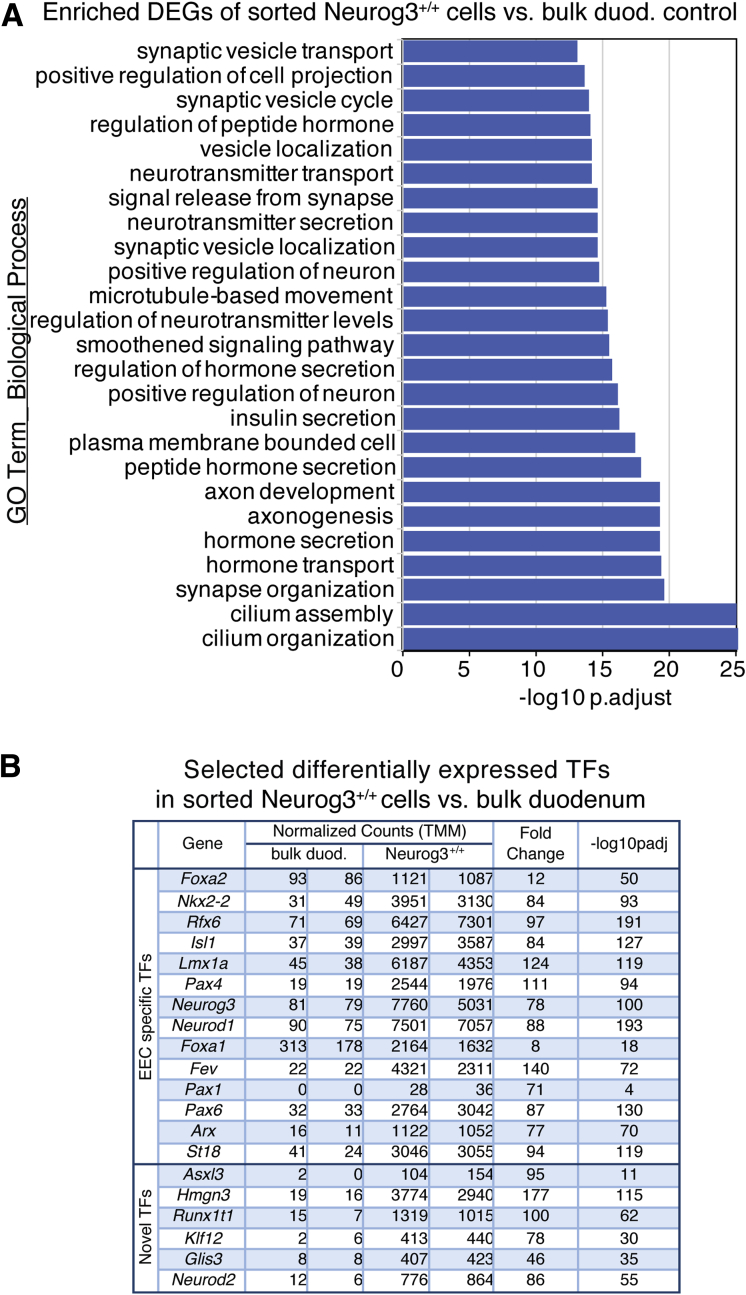
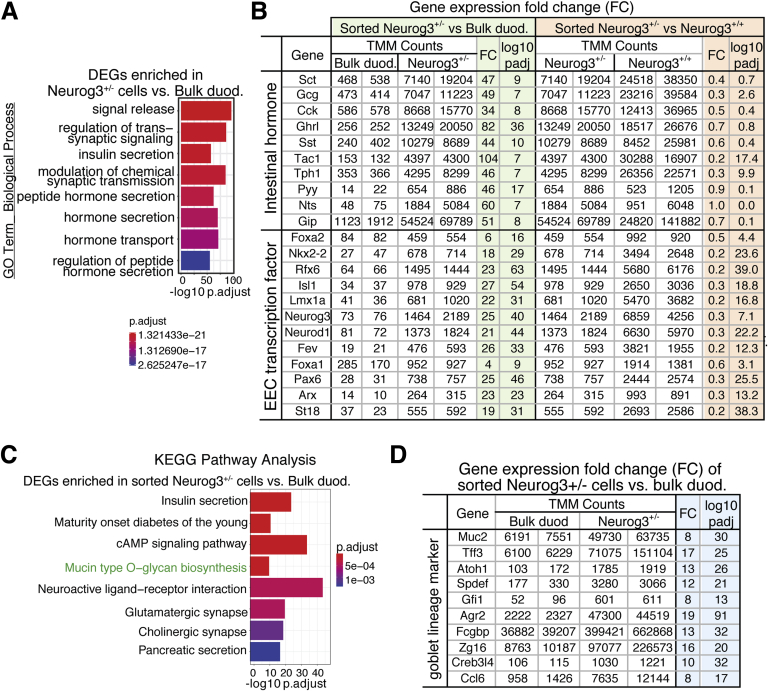
To further characterize the biological functions of those enriched genes, we selected 1399 genes that showed 8-fold enrichment in sorted Neurog3+/+ cells over bulk duodenum cells and undertook a Gene Ontology (GO) enrichment analysis using Panther (pantherdb.org/tools). The top-ranked GO term classes for biological function (false-discovery rate, <0.0001; P < .01) were associated with hormone secretory vesicles, neuron projections, vesical-mediated transporters, hormone activity and metabolism, and glucose homeostasis corresponding to the known functions of the EECs including synthesis, maturation, transport, storage, and secretion of hormones (Figure 2A). GO terms representing EEC differentiation and endocrine pancreatic development also were identified (Supplementary Table 3).
Figure 2.
(A) The top 25 most significantly enriched GO terms for biological processes of Neurog3+/+EYFP-enriched genes from 2731 DEGs enriched in intestinal Neurog3+/+cells vs bulk duodenum epithelia control. All are consistent with EEC function. The GOrilla program68 is used to identify enriched GO terms of 2731 genes with more than 2-fold enrichment and a P adjusted value < .01 in Neurog3+/+ cells. Graphs were generated by the DEBrowser (1.14.1). (B) Selected transcription factors from the top 40 highly enriched TFs listed in Neurog3+/+ cells.
Of note, the gene signature for the smoothened signaling pathway was enriched significantly. However, no transcript of Gli family genes, key effectors of canonical hedgehog signaling pathway, was detected in the transcriptomes of intestinal epithelial cells, either sorted EECs or bulk intestinal epithelium. This suggested a possible involvement of Gli-independent noncanonic hedgehog pathway30 in EEC development or function. In addition, the expression of genes representing G-protein–coupled receptors except Vipr1 also were highly enriched in those Neurog3+/+EYFP cells, reflecting their roles in luminal nutrient sensing (Supplementary Table 1). Notably, we found high EEC enrichment of tubulin α1a (Tuba1a), a microtubule component that is found predominantly in morphologically differentiated neurologic cells. Slit1, a secreted protein ligand in Slit-Robo signaling that mediates axon guidance,31 showed more than 25-fold enrichment in EECs, indicating a connection between EECs and neurons. Therefore, our data validate that those EYFP+ cells present in the Neurog3+/+EYFP mouse intestine possess characteristic features of EECs. Understanding the whole transcriptome of EECs should unravel their potential functions further.
Cell fate specification, maturation, and function are controlled by a cascade of TFs. To identify EEC-specific TFs at a global level, we used DESeq to analyze our data sets and identified 282 TFs that showed expression levels greater than 2-fold in EECs (Supplementary Table 4). As expected, TFs that have been shown to control endocrine differentiation were highly enriched (Figure 2B). These include Neurog3, Neurod1, Insm1, Isl1, Nkx2-2, Pax1/4/6, Fev, Foxa1, St18, Rfx6, and Arx.14,20, 21, 22, 23, 24,32, 33, 34, 35, 36, 37 In line with a recent report on the identification and validation of 7 novel TFs (Sox4, Rfx6, Zcchc12, Tox3, Myt1, Pbx1, and Runx1t1) as molecular regulators in EEC differentiation,37,38 we found high enrichment of those gene transcripts (3.7- to 140-fold) in Neurog3+/+ cells as well (Supplementary Table 4, highlighted in yellow). In addition, we identified more than 250 TFs including Asxl3, Hmgn3, Glis3, Runx1t1, Klf12, and Neurod2 that show cell-type–specific expression in EECs. These TFs have not been linked previously to EEC development or homeostasis, suggesting that they may contribute to EEC development, subtype differentiation, and function as well.
Reduced Expression of Neurog3 Alters Lineage Allocation of Neurog3+ Progenitors
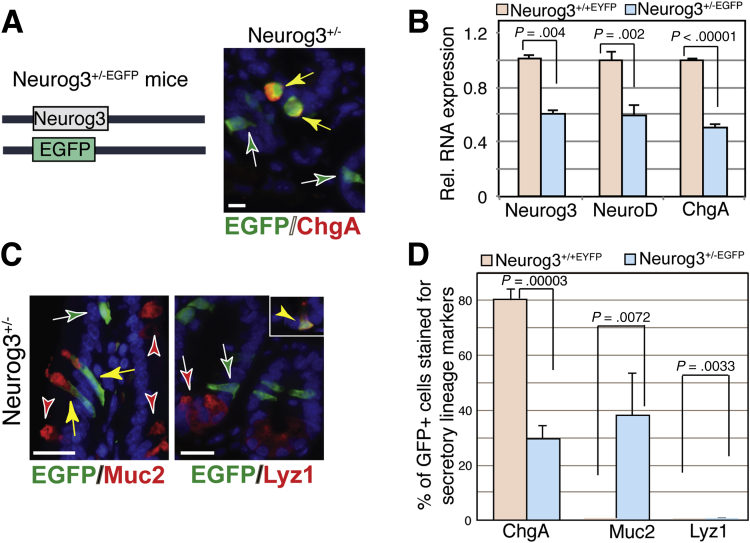
To study the cell fate of Neurog3+ progenitors, we also used another Neurog3 EGFP reporter mouse with an EGFP gene knocked-in at the Neurog3 locus to replace 1 functional Neurog3 allele (Neurog3+/-), as illustrated in Figure 3A.26 This model allows us to define Neurog3 gene activation (via reporter EGFP expression) without Neurog3 being expressed. In contrast from what we observed in the Neurog3+/+EYFP mouse intestine, the fate of Neurog3 progenitors in Neurog3+/-EGFP intestine was altered as seen by a substantial loss of ChgA+EGFP+ cells (29.3% ± 4.8%) (Figure 3A, summarized in Figure 3D). Furthermore, quantitative reverse-transcription polymerase chain reaction (qRT-PCR) showed a reduction in the transcript abundances of endocrine marker genes such as Neurog3, Neurod1, and ChgA in the intestines of heterozygous Neurog3+/-EGFP mice as compared with Neurog3+/+EYFP mice (Figure 3B). These data suggest that the reduction of Neurog3 gene dosage in the small intestine leads to its reduced expression, and therefore affects the Neurog3+ progenitor commitment to EECs.
Figure 3.
A subset of Neurog3+/-EGFP(Neurog3+/-) cells expressed in the goblet cell marker Muc2. (A) Left: Schematic diagram of Neurog3 and EGFP at the Neurog3 locus in the Neurog3+/-EGFP mouse. (A and C) Adult mouse intestinal sections from Neurog3+/- mice were co-stained with antibodies against GFP (in red) and ChgA, Muc2, or lysozyme (in green) markers for enteroendocrine, goblet, and Paneth cells, respectively. Yellow arrow indicates co-expression. 4′,6-diamidino-2-phenylindole (in blue) counterstained for nuclei. Scale bar: 50 um. (B) qRT-PCR analysis of mRNA levels for enteroendocrine-specific genes Neurog3, Neurod1, and ChgA in adult intestinal epithelium of Neurog3+/+ or Neurog3+/- mice. Values shown are means ± SD (n = 3). Bar graph shown is representative of at least 3 independent experiments that showed similar results. The P value was calculated by 1-tailed Student t test, as indicated on the graph. (D) Bar graph summarizes the percentage of EGFP cells expressing ChgA, Muc2, and Lyz in mice intestinal tissues indicated (N = 3). The P value was calculated by 1-tailed Student t test. The P value for the percentage of ChgA, Muc2, and Lyz in EGFP+ cells between mice with Neurog3+/+EYFP and Neurog3+/-EGFP genotypes are .00003, .00717, and .0033, respectively.
To investigate which cell populations were affected by reduced Neurog3 expression levels, we examined goblet (Muc2) and Paneth (Lyz) lineage marker expression in EGFP+ cells using co-IF staining. We found that EGFP+ cells co-stained with Muc2 and ChgA, respectively (Figure 3C). The heterogeneity of different cell populations detected are summarized in Figure 3D, as follows: ChgA+EGFP+ (29.3% ± 4.8%), EGFP+Muc2+ (38.1% ± 15.2%), and, rarely, EGFP+Lyz+ cells (0.6% ± 0.16%). These results indicate that EEC differentiation is sensitive to Neurog3 gene dosage. Lower Neurog3 gene dosage results in reduced EEC cell numbers and a redirection of Neurog3+ EE progenitors to the goblet cell lineage.
Altered Neurog3+ Gene Lineage Expression Pattern With Reduced Neurog3 Gene Dosage
To understand the whole spectrum of the effect of Neurog3 gene dosage on the transcriptome of Neurog3+ cells, we performed RNA-seq using RNAs from EGFP+ cells isolated by FACS from upper intestinal epithelium of Neurog3+/-EGFP mice (Figure 4A). Quality control of the count data is shown in an All2all scatter plot (Figure 4B) with coefficient rho of 0.97 between the 2 replicates of gene expression data sets from Neurog3+/-EGFP mice and 0.98 between those replicates from Neurog3+/+ EYFP mice. The rho is 0.98 between those replicates from duodenums of wild-type control mice. The rho number was less than 0.85 when compared with individual data set pairs between different mouse lines, indicating the data are suitable for differential expression analysis. DEG cluster analysis of transcriptomes from Neurog3+/+, Neurog3+/-, and bulk duodenum cells grouped Neurog3+/+ and Neurog3+/- cells in the same cluster, showing similarities between them, and significant differences from bulk control epithelial cells (Figure 4C). DESeq analysis identified a total of 3119 differentially regulated genes when the gene expression profiles were compared between EGFP+ cells in Neurog3+/-EGFP intestines and bulk duodenum cells, with 1495 genes being differentially enriched in Neurog3+/- cells (|fold change| ≥2) (Supplementary Table 5). A heat map also showed a significant difference in transcriptomes of Neurog3+/+ cells and Neurog3+/- cells. These data indicate that deletion of 1 Neurog3 allele has a profound effect on the transcriptome of Neurog3-derived cells.
Figure 4.
(A) Experimental diagram shows the sample preparation procedure from respective mouse lines for RNA-seq analysis. (B) All2all scatter plot of all detected transcripts after batch correction: an overview of similarities and variance between samples sequenced. Each Neurog3+/- or Neurog3+/+ library replicate was made from RNAs isolated from sorted duodenum cells from 1 Neurog3+/-EGFP mice or 2 Neurog3+/+EYFP mice. Each bulk duodenum (Bulk duod.) library was made from RNAs from epithelial cells from 1 wild-type control mouse duodenum. Two independent RNA replicates were performed for each mouse genotype. The upper side of the plot shows a log10-normalized scatter plot. The bottom side of the plot shows the Pearson correlations of each paired sample. Diagonal shows the histogram of normalized read counts in log10 scale. (C) Heat map representing color-coded expression levels of differentially expressed genes (|Fc| >2 and P adjusted ≤ .01, 2-way analysis of variance) from transcriptomic sequencing of sorted Neurog3+ cells from intestines of Neurog3+/+EYFP, Neurog3+/-EGFP, and bulk duodenum epithelium control mice. Two independent transcriptome sequencing were performed for each genotype. Color key: blue, high; red, low expression. Fc, fold change.
GO term analysis of the described 1495 DEGs in Neurog3+/- cells vs bulk duodenum epithelial cells detected annotation clusters present in GO terms of DEGs from sequencing data sets of sorted Neurog3+/+ cells vs bulk duodenum epithelial cells with a false discovery rate of less than 0.05 (Database for Annotation, Visualization, and Integration Discovery, GO term) (Supplementary Table 6). Among the significantly enriched GO terms of biological function are genes involved in hormone secretion, neuron differentiation, ion transport, and primary cilium organization (Figure 5A). However, the absolute expression levels of transcripts encoding hormones and EEC-specific TFs were lower in Neurog3+/- than in Neurog3+/+ mice (Figure 5B, right).
Figure 5.
Neurog3+/-cells express genes representing cell types for both enteroendocrine and goblet cells. (A) Bar graph representing selective high-enriched GO terms in biological processes that are related to endocrine features including hormone secretion, neuron differentiation, ion transport, and synaptic signaling, after false-discovery rate correction for multiple testing. There were 1495 DEGs with 2-fold enrichment and a P adjusted value < .01 in Neurog3+/- cells vs bulk duod cells used as inputs in GO term analysis. Graphs were generated by DEBrowser (1.14.1). (B) Enteroendocrine-specific hormone and transcription factor expression levels detected by RNA-seq analysis of indicated mouse samples. Note the significant reduction in gene expression levels in sorted Neurog3+/- cells compared with that of sorted Neurog3+/+ cells. Fold change and P adjusted values between sorted Neurog3+/- vs bulk duod cells or Neurog3+/- and Neurog3+/+ are listed in the table. (C) Kyoto Encyclopedia of Genes and Genomes enrichment analysis identified a unique pathway of mucin-type O-glycan biosynthesis present in 1495 DEG genes enriched in sorted Neurog3+/- cells vs bulk duodenum epithelia cells. Graphs were generated by DEBrowser (1.14.1). (D) Normalized expression counts of goblet cell–specific genes in Neurog3+/- vs bulk duodenum epithelia cells. Fold change and P adjusted values are listed in the table. cAMP, cyclic adenosine monophosphate.
Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that those DEGs were enriched in insulin secretion, neuroactive ligand-receptor interaction, neurotransmitter synapses, and Mucin-type O-glycan biosynthesis (Figure 5C). In line with this, we found significant enrichment of goblet cell marker transcripts present in the sorted Neurog3+/- transcriptome (Figure 5D), suggesting that the Neurog3-derived cells in Neurog3+/-EGFP intestine were lineage-heterogeneous, consisting of EEC and goblet cell populations.
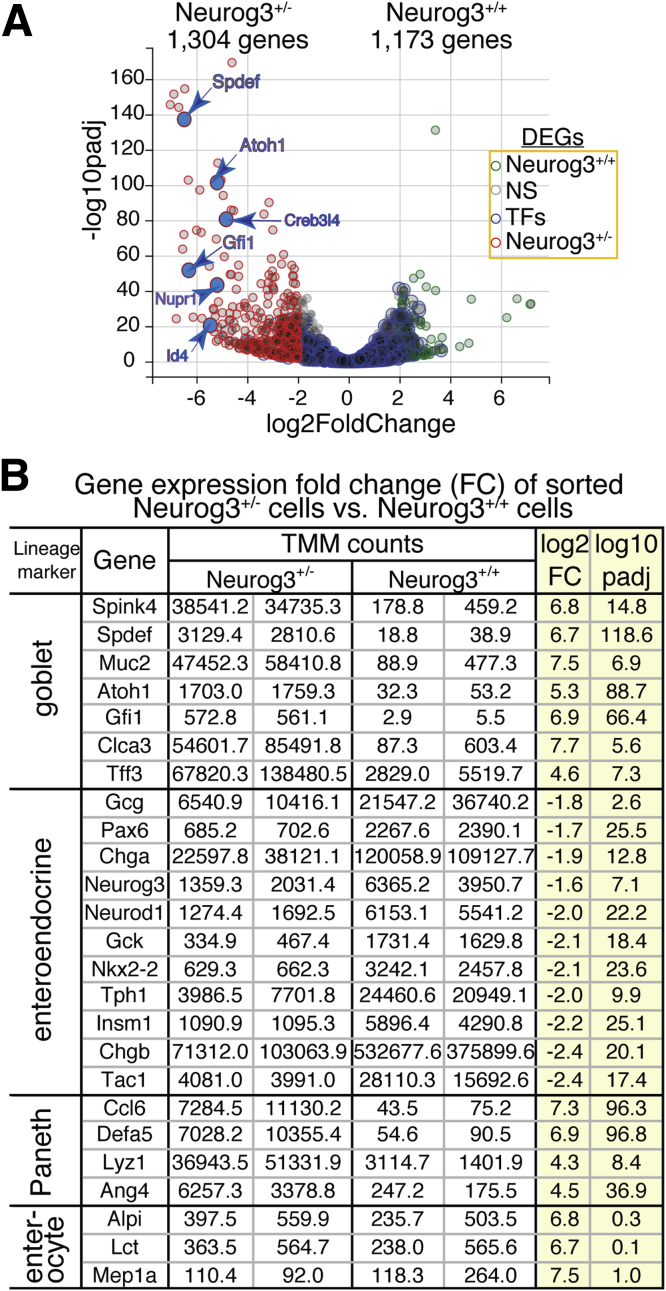
We next compared gene expression profiles of Neurog3+-derived cells isolated from Neurog3+/+ and Neurog3+/- intestines using volcano plot analysis. Here, we averaged gene expression levels over replicates for each genotype after normalization and compared the 2 genotypes by calculating the fold change as the ratio between the averages. This analysis led us to identify a total of 2477 genes with significantly altered expression levels (|FC| >2, P adjusted < .01) between sorted Neurog3+/+ and Neurog3+/- cells, of which 1304 were enriched in Neurog3+/- cells and 1173 genes in Neurog3+/+ cells (Figure 6A).
Figure 6.
Deletion of 1 Neurog3 allele resulted in significant alteration on the global transcriptome. (A) Volcano plot shows transcript TMM values comparing the transcriptomes of sorted Neurog3+/- cells vs Neurog3+/+ cell populations, showing 1304 transcripts enriched in Neurog3+/- cells (Fc > 2, P adjust ed< .01). The negative log of the adjusted P values (P-adjust) for each expressed gene was plotted on the y-axis and the log of the fold change between the 2 genotypes was plotted on the x-axis. Marker genes specific for goblet cells are indicated. (B) Table summarizes the fold change (log2 FC) of major lineage marker gene expression in Neurog3+/- cells compared with Neurog3+/+ cells. P adjusted values between Neurog3+/- and Neurog3+/+ are listed on the right. Fc, fold change.
In Neurog3+/- cells, several top differentially expressed genes including Muc2, Gfi1, Spdef, and Tff3 represent the gene expression signature for intestinal goblet cells, consistent with our earlier observation by immunostaining (Figure 3C). Interestingly, we found that Atoh1 expression was significantly higher in Neurog3+/- cells while no Atoh1 expression was detected in Neurog3+/+ cells. Furthermore, deletion of 1 Neurog3 allele significantly reduced the expression of genes encoding hormones important for intestinal functions, genes encoding transcription factors associated with EEC development, and genes otherwise linked to secretory function (Figure 6B). The expression of transcription factors that are critical for endocrine cell-type differentiation and function also were reduced (Supplementary Table 7). Those changes in the expression levels of lineage-specific transcription factors may redirect Neurog3+ EEC progenitors toward the goblet cell lineage when Neurog3 gene expression is reduced.
Neurog3-/- Cells Express Goblet Cell Lineage-Specific Genes
It is well established that Neurog3 is required and sufficient for EEC specification from Atoh1+ secretory progenitors.6,14,21,39,40 Our results showed that lower Neurog3 gene dosage seen in Neurog3+/- cells results in a reduction of EEC cell number associated with an increase in goblet cells. These findings suggest that the level of Neurog3 expression determines to what extent Neurog3+ progenitors adopt an EEC or a goblet cell fate.
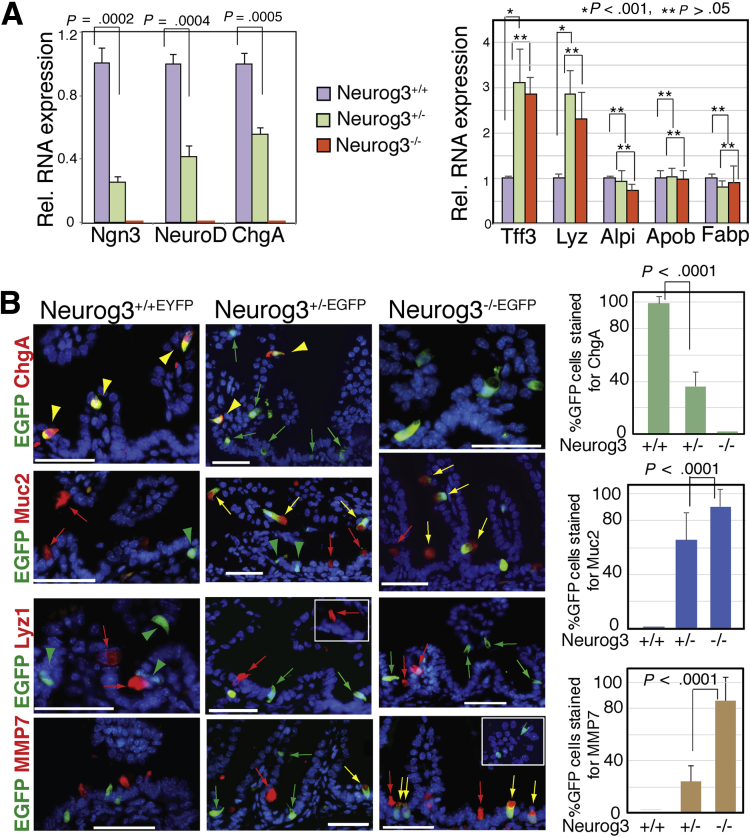
To further substantiate this hypothesis, we generated a global Neurog3 null mouse (Neurog3-/-) by in-breeding Neurog3+/-EGFP mice and determined to what extent that Neurog3+-derived cells adopted a goblet fate. As previously reported, Neurog3-/- mice died 2 days after birth (2 days post coitum) with severe neonatal diabetes.14 The expression levels of lineage-specific genes in neonate intestinal epithelia were determined by qRT-PCR. As shown in Figure 7A, the messenger RNA (mRNA) level of EEC markers Neurog3, Neurod1, and ChgA in the neonatal intestinal epithelial were reduced significantly (70%, 60%, and 55%, respectively) in 2-day-old Neurog3+/- mice in comparison with Neurog3+/+ mice. A similar comparison showed undetectable expression of the earlier-described EEC markers in Neurog3-/- mice. Conversely, a significant increase in mRNAs of goblet and Paneth cell marker genes was observed with little change in transcript abundance for enterocyte marker genes in Neurog3-/- mice when compared with that in Neurog3+/+ mice.
Figure 7.
Decreased Ngn3 expression level redirects Neurog3+ cells toward goblet cell fate. (A) Left: qRT-PCR analysis of intestinal epithelial cells isolated from P2 Neurog3+/+, Neurog3+/-, and Neurog3-/- mice shows reduced expression levels of Neurog3, Neurod1, and ChgA in Neurog3+/- mice, and undetectable expression in Neurog3 null intestine. Right: The expression of Tff3 and Lyz, but not of Alpi, Apob, and Fabp were increased significantly in Nng3+/- and Ngn3 null mice. Values shown are means ± SD (N ≥ 3). Bar graph shown on the left is representative of at least 3 independent experiments that showed similar results. The P value was calculated by 1-tailed Student t test, as indicated in the graph. Bar graph on the right is the mean value of 3 independent experiments. ∗P < .001, indicating the difference is significant. ∗∗P > .05, indicating the difference is not significant. (B) Immunofluorescent co-staining of intestinal tissue sections from 2-day-old mice indicated with antibodies against EGFP (green), and ChgA, Muc2, Lyz1, and MMP7 (red), respectively. Yellow arrow or arrowhead indicate EGFP+ cells co-expressed with protein detected. Scale bars: 50 um. Bar graph on the right summarizes the percentage of GFP+ intestinal epithelial cells co-stained for ChgA, Muc2, and MMP7. Error bars represent the SD from at least 3 experimental replicates. P values were determined by 1-tailed Student t test. Rel., relative.
We next characterized cell types that might arise from Neurog3+ progenitors using immunohistochemistry with lineage-specific antibodies (ChgA, Muc2, Lyz1, or Matrix metalloproteinase 7 [MMP7]) on intestinal tissue sections from 2-day-old postnatal Neurog3+/+, Neurog3+/-, and Neurog3-/- mice (Figure 7B). Similar to what we observed in adult mice, in the intestine of Neurog3+/+ 2-day postnatal mice, nearly all EGFP+ cells stained for ChgA (98.7% ± 5.4%), with no detection of EGFP+Muc2+, EGFP+Lyz+, or EGFP+MMP7+ cells. Likewise, in 2-day postnatal Neurog3+/- mice, in addition to EGFP+ChgA+ cells (yellow arrowhead, 36.2% ± 10.6%), we detected EGFP+Muc2+ (65.5% ± 20.7%) or EGFP+MMP7+ (24.1% ± 11%) cells, but no EGFP+Lyz+ cells. In Neurog3-/- mice, no EGFP+ChgA+ cells were detected. Instead, all EGFP+ were stained with Muc2 (90.4% ± 12.9%) or MMP7 (85.6% ± 18.3%) (yellow arrow). However, we did not detect any EGFP+Lyz+ cells. Of note, we also detected Muc2+EGFP- cells, indicating that those Muc2+ cells were not derived from Neurog3+ progenitor cells. Taken together, our data suggest that Neurog3-expressing intestinal secretory progenitor cells retain plasticity to differentiate into either an endocrine or a goblet cell fate, depending on the levels of Neurog3 expression.
Discussion
In pancreatic islet development, Neurog3+ progenitor cells give rise to mature islet cells. When Neurog3 expression levels reduced to 90% in Neurog3 hypomorphic mutant mice, those Neurog3+ progenitor cells switched their cell fate to exocrine lineage.19 In this study, we examined the possible impact of reduced Neurog3 gene dosage in EEC progenitor cell fate in the intestine. We found that EEC differentiation is more sensitive to Neurog3 expression levels. We provide evidence at the transcriptomic, protein, and genetic levels that EEC differentiation from intestinal secretory progenitors is dependent on Neurog3 levels. We found that reduced Neurog3 gene dosage in the intestinal endocrine cells alters EEC progenitor cell differentiation from endocrine to goblet cells, resulting in loss of EECs and an increase in the number of goblet cells.
The intestinal enteroendocrine cells are the largest hormone-secreting organ in the body, scattered as single cells along the intestinal epithelium. Even though much effort has been committed to the study of EEC development and differentiation, many studies have identified a number of key transcription factors and signaling pathways as contributors to EEC development. Low cell density and a dispersed distribution of EECs have slowed research progress on their function and homeostasis regulation. Recent studies using a fluorescent transgenic reporter and single-cell sequencing have advanced EEC studies.37,38,41,42 Unlike the present work reported here, previous studies used fluorescence-tagged cells derived from a Neurog3cre;tdTomfloxed mouse as intestinal EECs in their bulk RNA-seq.41 Cre-mediated lineage tracing does not always reflect the endogenous EEC gene expression pattern. Previous studies have shown that Neurog3cre:ROSAEGFP progenitor cells give rise not only to endocrine cells, but also ectopically to goblet and Paneth cells.8 Therefore, the transcriptome profiled in the Neurog3cre;tdTomfloxed studies was not for EECs only, rather a mixed population of EEC, goblet, and Paneth cells. Here, we purified and analyzed transcriptomes of fluorescent-tagged Neurog3+ cells from the intestines of an EYFP knocked-addon (Neurog3+/+EYFP) reporter mouse as endogenous EE cells.
A recent study using single-cell sequencing technology defined the gene expression signature for each intestinal cell type, each of which contained a list of genes including EE, goblet, Paneth, and tuft cells.42 We therefore examined the presence of those cell-type signatures in our data sets. In our Neurog3+/+ transcriptome, all genes listed in EE signatures were highly enriched. The expression levels of genes listed in goblet, tuft, and Paneth cells were low to none. Therefore, our data not only fortify the reported EE signature genes, but also expanded the gene list from 36 to 2731. Furthermore, we identified novel EE-specific transcription factors, ion channels, and primary cilia genes.
Lineage specification and cell differentiation and function are governed by a complex regulatory network of transcription factors. Even though more than 20 transcription factors including Neurog3, Neurod1, and Nkx2-2 have been identified for their crucial roles in EEC differentiation and function, the mechanisms underlying some differentiation processes such as enteroendocrine cell subtyping and EEC cellular functions remain elusive. Here, we sequenced duodenum EEC transcriptome. Our bulk sequencing provided much higher sequencing depth than that of single-cell sequencing. High sequencing depth covers more transcripts, especially genes with low expression. We uncovered 282 transcription factors that enriched more than 2-fold in EECs, many of them (>250) have not been linked to EEC differentiation and function.
For instance, we found high expression of Additional sex-combs like 3 (Asxl3) and Hmgn3 (95-fold and 177-fold enrichment compared with control bulk epithelial cells, respectively) in Neurog3+/+ EE cells (Figure 2B, table). Asxl3 is a component of the polycomb repressive de-ubiquitination complex that functions as the epigenetic regulators. It has been linked to human neurodevelopmental disorders.43 Its knockdown highly perturbs neural cell fate specification in xenopus embryo development.44 Hmgns, a family of high-mobility group nucleosome–binding proteins, have been shown to play a critical role in stabilizing cell identify through binding to cell-specific enhancers.45 High tissue-specific expression of Hmgn3 up-regulates glycine transporter 1 gene expression in glia cells and eye,46 glut2 in pancreas, and hormone-dependent thyroid hormone–receptor transcription.47 The loss of Hmgn3 in pancreas β cells blunt glucose-stimulated insulin secretion.48 Asxl3 and Hmgn are epigenetic regulators, suggesting the involvement of dynamic chromatin modification in EEC development, differentiation, and function. Therefore, those EEC-enriched TFs we identified that have not been linked to EEC development and function such as Asxl3 and Hmgn3 should provide novel tools to study their roles in EEC cell fate restriction, subtyping, and EEC identity.
Ion channels conduct electrical currents, which have been shown to be important for controlling cell cycle, hormone release, and motility.49 It has been reported that ion channels play an important role in the development as well as tumor biology, and may contribute to tumor cell invasion and metastasis.50,51 Our findings of EE-specific ion channels may provide a new avenue to study EE differentiation and neuroendocrine tumor biology and to develop novel therapeutic strategies.
Primary cilia are antenna-like cellular protrusions that present in many types of vertebrate cells.52,53 They act as sensory organelles to transduce extracellular cues by regulating many signaling pathways to control developmental programs, cell proliferation, cell fate determination, and metabolic homeostasis.54 Even though all primary cilia have a similar structure in many cell types, their protein components are tightly regulated and not identical among different cell types. Defects in primary cilia cause ciliopathy and have been implicated in metabolic disorders and cancer progression. In the endocrine system, primary cilia in islet and gastric endocrine cells have been detected and have been linked to islet function and development in pancreas55, 56, 57 and fat regulation of gastrin secretion and gastric acidity in the stomach.58 No primary cilia structure in intestinal enteroendocrine cells has been detected. Here, we reported high enrichment of primary cilia structure proteins in endogenous enteroendocrine cells. Although deletion of 1 Neurog3 allele resulted in reduced EECs and increased Neurog3+-derived goblet cells, we observed decreased expression of cilia-annotated genes in Neurog3+-derived intestinal cells, suggesting a link between cilia genes and EE progenitor cell fate choice.
Gli-independent noncanonic hedgehog pathway has been linked to cellular responses ranging from Ca2+ signaling and cytoskeletal rearrangement, axon guidance, and metabolic rewiring in adipose tissue and skeletal muscle (for a review, see Teperino et al59). In cell line 3T3L1 adipocytes adipocytes, primary cilia-dependent Smoothened gene activation is coupled to a rapid L-type–dependent Ca2+ influx,60 which leads to the activation of calcium/calmodulin dependent protein kinase kinase 2 and AMP-activated protein kinase phosphorylation and activation. Ampk is a nutrient and energy sensor of cellular metabolic state that maintains energy homeostasis.61 Its activation results in a rapid, insulin-independent glucose uptake and aerobic glycolysis. L-type–dependent Ca2+ influx is responsible for peptide hormone secretion in enteroendocrine cells. Our RNA-seq showed high enrichment of transcripts of L-type calcium channel genes (Supplementary Table 1). Thus, the Gli-independent noncanonic hedgehog pathway may serve as a novel mechanism to regulate hormone release in intestinal EECs and to maintain their energy homeostasis.
In the Neurog3+/- transcriptome, we found high enrichment (>13-fold) of potential signature genes not only for EEC, but also for goblet, Paneth, and tuft cells. However, our immunostaining confirmed the presence of ChgA and Muc2 in EGFP+ cells in adult Neurog3+/- intestinal epithelium. No Doublecortin Like Kinase 1 or lysozyme, characteristic markers for tuft and Paneth cells, respectively, was found to co-express in EGFP+ cells. Because the Paneth and tuft cell signatures were identified by single-cell sequencing, the expression of most of the cell-type–specific signature genes have not been validated. It also is possible that those signature genes are not expressed at the protein level.
The high depth of our RNA-seq detects both known and novel transcripts enriched in EECs compared with previous single-cell sequencing studies. However, we would like to draw attention to the potential limitation of our RNA-seq studies owing to the low biological replicates (n = 2) used. Additional replicates or qRT-PCR are needed to make a firm conclusion for future validation studies.
In the intestine of 2-day-old Neurog3-/- mice, all GFP+ cells become Muc2-expressing goblet cells, with a small subset of cells in the intervillus zone co-expressing MMP7, located in the intervillus zone. Our findings were supported by a previous observation of an increased number of goblet cells in the newborn small intestine of Neurog3 knockout mice.21 The intervillus zone present in embryonic and neonatal intestines consists of undifferentiated precursor cells that eventually invaginate into the mucosa to form crypts and establish a crypt–villus axis in the first few days after birth in the rodent.62 Those MMP7+/Muc2+ double-positive cells may represent Paneth/goblet intermediate cells. It is conceivable that the prospective secretory progenitor cells, generated in the absence of Neurogenin3, are fated to goblet cells via these Paneth/goblet intermediate cells. The predominant expression of Jag2 in EECs and of Delta-like protein 1 (Dll1) in goblet cells suggest that goblet cells and EECs use different Notch ligands to communicate with neighboring cells for their singlet appearance in the intestinal epithelia.
Gene dosage–dependent regulation of cell differentiation by lineage-restricted transcription factors has been well recognized in several developmental systems including sex determination in drosophila and hematopoietic differentiation in mice.63,64 EECs, goblet, and Paneth cells are derived from Atoh1-expressing common secretory progenitors. Subsequent expression of Neurog3, Gfi1, and Spdef direct secretory progenitor differentiation into EEC, goblet, and Paneth cells. Our results suggest a reciprocal interaction between Neurog3 and Gfi1/Spdef in secretory progenitors. This may explain our observation that reduced Neurog3 gene dosage in progenitor cells results in an increased goblet cell population at the expense of the EEC population.
Methods
All authors had access to the study data and reviewed and approved the final manuscript.
Mouse Models
All procedures using mice were approved by the Institutional Animal Care and Use Committee at the University of Massachusetts Medical School and in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals: Reporting of In Vivo Experiments Standards. The Neurog3+/+EYFP and Neurog3+/-EGFP mice were kindly provided by Drs G. Gradwohl27 and K. Kaestner, respectively.26 All mice were bred in-house and were kept on a 12-hour light-dark cycle. Neurog3-/-EGFP mice were obtained by intercrossing Neurog3+/-EGFP mice.
Intestinal Epithelial Cell Isolation and FACS Sorting of EGFP+ and EYFP+ Cells
Two adult Neurog3+/+EYFP or Neurog3+/-EGFP mice were used in each sorting, and sorted cells were collected for RNA-seq. Two replicates were performed for each mouse line. Wild-type bulk duodenum epithelia of C57Bl6 adult mice were isolated, sequenced, and used as controls. In brief, duodenum was removed from 10- to 12-week-old euthanized mice and was inverted and rinsed in ice-cold phosphate-buffered saline (PBS) 4 times before cutting into 1- to 2-cm fragments. The small duodenal pieces were placed in dissociation buffer (1 mmol/L EDTA, 1.5 mmol/L dithiothreitol in PBS) for 20 minutes before transferring to 1 mmol/L EDTA in PBS buffer and followed by incubating at 37°C for 8 minutes. Epithelial cells were dissociated from intestinal mucosa by gentle rotating at 37°C for 20 minutes. The intestinal epithelial cells were collected by centrifugation at 4°C at 1000 rpm for 5 minutes. The pellet was dissociated further into single cells by incubation in a pronase buffer (12% pronase, 1 mmol/L EDTA, 10 mmol/L HEPES, pH 7.4 in MEM) for 30 minutes at 37°C. Single cells were collected by passing through a 20-umol/L cell strainer (Thermo Fisher) to remove cell clumps and mucus followed by centrifugation at 4°C at 1500 rpm for 5 minutes.
Single-cell pellets were resuspended in cell-sorting buffer (12.5 mmol/L HEPES [pH 7.4], 2 mmol/L EDTA, 1% fetal bovine serum in 1 × minimal essential media). 7-actinomycin D was added to sorting media 20 minutes before sorting to exclude dead cells. EGFP+ or EYFP+ live cells were sorted and collected on a BD FACS Aria cell sorter (BD Biosciences, Woburn, MA) gated with fluorescent and forward/side scatter (to exclude cell debris).
RNA Extraction, Library Construction, and Real-Time PCR Analysis
Total RNAs were extracted using the RNeasy Micro kit (Qiagen, Germantown, MD) according to the manufacturer’s protocol with in-column DNase digestion. RNA quality was assessed using a Bioanalyzer (Agilent Technologies, Lexington, MA).
For qPCR analysis, 100 ng RNA was reverse-transcribed using the Maxima First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions. qPCR reactions were performed using iTaq SYBR green super mix (Bio-Rad, Hercules, CA) and primers as indicated on a Bio-Rad CFX96 real-time system. Gene expression levels were normalized to β-actin. Relative gene expression was quantified using the comparative cycle threshold method.
PolyA+ RNAs from ∼100 ng of total RNAs isolated from FACS-sorted cells with a RNA integrity number >8 were used as input for library preparation after the Illumina San Diego, CA Truseq RNA sample preparation protocol. Each library was constructed with an adapter containing a unique 6-nucleotide index sequence ligated to double-stranded complementary DNAs. Sequencing was run on an Illumina HiSeq2000 instrument at the University of Massachusetts deep sequencing core using multiplexed 50–base pair single-end reads, with an average of 20–30 million reads obtained per sample.
RNA-Seq Data Analysis
RNA-seq data analysis was performed using DESeq pipeline (DolphinNext, version 1.2.10).65 In brief, adapter sequences were removed using the program Trimmomatic 0.32. Ribosomal RNA reads were filtered out by Bowtie2. The raw reads were mapped onto the Mus musculus University of California, Santa Cruz mm10 reference genome. mRNA was annotated and quantification was calculated using RSEM v1.2.7. Differential gene expression was calculated using the DESeq2 (1.26.0) in DEBrowser (1.14.1),28 with the mean value parameter of gene-wise dispersion estimates.
GO Term Analysis
Enriched GO categories in the clusters were detected by Database for Annotation, Visualization, and Integration Discovery, and analyzed by GOnet: https://tools.dice-database.org/GOnet.66 Graphs were generated by DEBrower (1.14.1).28
Immunofluorescent Microscopy
Immunofluorescent staining on intestinal tissues was performed as previously described.67 In brief, cryopreserved mouse intestinal tissues fixed in 4% paraformaldehyde were cryostat-sectioned into 5-um thickness onto glass slides. Sections were rehydrated and permeabilized in PBS containing 0.2% Triton X-100 followed by blocking in Blocking Reagent (Vector Laboratories, Burlingame, CA). Sections then were incubated with primary antibody overnight in the cold room and detected by incubating with fluorophore-conjugated secondary antibodies for 1 hour at room temperature. Samples were mounted in 4′,6-diamidino-2-phenylindole containing anti-fade medium (ProLong Gold antifade reagent; Thermo Fisher Scientific, Frankin, MA). Images were acquired with a Nikon epifluorescence microscope.
Data Availability
RNA-seq data sets were deposited in NCBI, Gene ontology omnibus (GEO), accession number: GSE149203. All authors had access to all data and reviewed and approved the final manuscript.
Statistical Analysis
Results between samples were compared by 1-tailed Student t tests as indicated in Figure legends where applicable.
Acknowledgments
The authors thank Dr K. Kaestner for providing Neurog3þ/-EGFP reporter mice, and Dr Stephen Krasinski for critical reading and editing of the manuscript.
CRediT Authorship Contributions
Hui Joyce Joyce Li, PhD (Conceptualization: Lead; Data curation: Lead; Methodology: Lead; Project administration: Lead; Writing – original draft: Lead); Subir K. Ray, PhD (Data curation: Supporting; Writing – review & editing: Supporting); Alper Kucukural, PhD (Software: Lead); Gerard Gradwohl, PhD (Resources: Supporting); Andrew B. Leiter, MD, PhD; (Conceptualization: Equal; Funding acquisition: Lead; Methodology: Equal; Project administration: Equal; Writing – review & editing: Supporting).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported in part by National Institutes of Health grants R01 DK100223 and R01 DK110614 (A.B.L.). Also supported by funds from the National Human Genome Research Institute grant U01 HG007910–01 and the National Center for Advancing Translational Sciences grant UL1 TR001453–01 (A.K.).
Supplementary Material
References
- 1.de Santa Barbara P., van den Brink G.R., Roberts D.J. Development and differentiation of the intestinal epithelium. Cell Mol Life Sci. 2003;60:1322–1332. doi: 10.1007/s00018-003-2289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 3.Noah T.K., Donahue B., Shroyer N.F. Intestinal development and differentiation. Exp Cell Res. 2011;317:2702–2710. doi: 10.1016/j.yexcr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gehart H., Clevers H. Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol. 2019;16:19–34. doi: 10.1038/s41575-018-0081-y. [DOI] [PubMed] [Google Scholar]
- 5.Yang Q., Bermingham N.A., Finegold M.J., Zoghbi H.Y. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Diaz L., Jain R.N., Keeley T.M., VanDussen K.L., Brunkan C.S., Gumucio D.L., Samuelson L.C. Intestinal Neurogenin 3 directs differentiation of a bipotential secretory progenitor to endocrine cell rather than goblet cell fate. Dev Biol. 2007;309:298–305. doi: 10.1016/j.ydbio.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjerknes M., Cheng H. Neurogenin 3 and the enteroendocrine cell lineage in the adult mouse small intestinal epithelium. Dev Biol. 2006;300:722–735. doi: 10.1016/j.ydbio.2006.07.040. [DOI] [PubMed] [Google Scholar]
- 8.Schonhoff S.E., Giel-Moloney M., Leiter A.B. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol. 2004;270:443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Mutoh H., Fung B.P., Naya F.J., Tsai M.J., Nishitani J., Leiter A.B. The basic helix-loop-helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc Natl Acad Sci U S A. 1997;94:3560–3564. doi: 10.1073/pnas.94.8.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naya F.J., Huang H.P., Qiu Y., Mutoh H., DeMayo F.J., Leiter A.B., Tsai M.J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shroyer N.F., Wallis D., Venken K.J., Bellen H.J., Zoghbi H.Y. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noah T.K., Kazanjian A., Whitsett J., Shroyer N.F. SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp Cell Res. 2010;316:452–465. doi: 10.1016/j.yexcr.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bjerknes M., Cheng H. Cell Lineage metastability in Gfi1-deficient mouse intestinal epithelium. Dev Biol. 2010;345:49–63. doi: 10.1016/j.ydbio.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 14.Gradwohl G., Dierich A., LeMeur M., Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rukstalis J.M., Habener J.F. Neurogenin3: a master regulator of pancreatic islet differentiation and regeneration. Islets. 2009;1:177–184. doi: 10.4161/isl.1.3.9877. [DOI] [PubMed] [Google Scholar]
- 16.Heremans Y., Van De Casteele M., in't Veld P., Gradwohl G., Serup P., Madsen O., Pipeleers D., Heimberg H. Recapitulation of embryonic neuroendocrine differentiation in adult human pancreatic duct cells expressing neurogenin 3. J Cell Biol. 2002;159:303–312. doi: 10.1083/jcb.200203074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwitzgebel V.M., Scheel D.W., Conners J.R., Kalamaras J., Lee J.E., Anderson D.J., Sussel L., Johnson J.D., German M.S. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- 18.Wang S., Hecksher-Sorensen J., Xu Y., Zhao A., Dor Y., Rosenberg L., Serup P., Gu G. Myt1 and Ngn3 form a feed-forward expression loop to promote endocrine islet cell differentiation. Dev Biol. 2008;317:531–540. doi: 10.1016/j.ydbio.2008.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S., Yan J., Anderson D.A., Xu Y., Kanal M.C., Cao Z., Wright C.V., Gu G. Neurog3 gene dosage regulates allocation of endocrine and exocrine cell fates in the developing mouse pancreas. Dev Biol. 2010;339:26–37. doi: 10.1016/j.ydbio.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H.J., Ray S.K., Singh N.K., Johnston B., Leiter A.B. Basic helix-loop-helix transcription factors and enteroendocrine cell differentiation. Diabetes Obes Metab. 2011;13(Suppl 1):5–12. doi: 10.1111/j.1463-1326.2011.01438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenny M., Uhl C., Roche C., Duluc I., Guillermin V., Guillemot F., Jensen J., Kedinger M., Gradwohl G. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 2002;21:6338–6347. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsson L.I., St-Onge L., Hougaard D.M., Sosa-Pineda B., Gruss P. Pax 4 and 6 regulate gastrointestinal endocrine cell development. Mech Dev. 1998;79:153–159. doi: 10.1016/s0925-4773(98)00182-8. [DOI] [PubMed] [Google Scholar]
- 23.Gierl M.S., Karoulias N., Wende H., Strehle M., Birchmeier C. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 2006;20:2465–2478. doi: 10.1101/gad.381806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desai S., Loomis Z., Pugh-Bernard A., Schrunk J., Doyle M.J., Minic A., McCoy E., Sussel L. Nkx2.2 regulates cell fate choice in the enteroendocrine cell lineages of the intestine. Dev Biol. 2008;313:58–66. doi: 10.1016/j.ydbio.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Offield M.F., Jetton T.L., Labosky P.A., Ray M., Stein R.W., Magnuson M.A., Hogan B.L.M., Wright C.V.E. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 26.Lee C.S., Perreault N., Brestelli J.E., Kaestner K.H. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488–1497. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellitzer G., Martin M., Sidhoum-Jenny M., Orvain C., Barths J., Seymour P.A., Sander M., Gradwohl G. Pancreatic islet progenitor cells in neurogenin 3-yellow fluorescent protein knock-add-on mice. Mol Endocrinol. 2004;18:2765–2776. doi: 10.1210/me.2004-0243. [DOI] [PubMed] [Google Scholar]
- 28.Kucukural A., Yukselen O., Ozata D.M., Moore M.J., Garber M. DEBrowser: interactive differential expression analysis and visualization tool for count data. BMC Genomics. 2019;20:6. doi: 10.1186/s12864-018-5362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grun D., Lyubimova A., Kester L., Wiebrands K., Basak O., Sasaki N., Clevers H., van Oudenaarden A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 30.Robbins D.J., Fei D.L., Riobo N.A. The Hedgehog signal transduction network. Sci Signal. 2012;5:re6. doi: 10.1126/scisignal.2002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blockus H., Chedotal A. Slit-Robo signaling. Development. 2016;143:3037–3044. doi: 10.1242/dev.132829. [DOI] [PubMed] [Google Scholar]
- 32.Beucher A., Gjernes E., Collin C., Courtney M., Meunier A., Collombat P., Gradwohl G. The homeodomain-containing transcription factors Arx and Pax4 control enteroendocrine subtype specification in mice. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y.C., Zuraek M.B., Kosaka Y., Ota Y., German M.S., Deneris E.S., Bergsland E.K., Donner D.B., Warren R.S., Nakakura E.K. The ETS oncogene family transcription factor FEV identifies serotonin-producing cells in normal and neoplastic small intestine. Endocr Relat Cancer. 2010;17:283–291. doi: 10.1677/ERC-09-0243. [DOI] [PubMed] [Google Scholar]
- 34.Jia S., Ivanov A., Blasevic D., Muller T., Purfurst B., Sun W., Chen W., Poy M.N., Rajewsky N., Birchmeier C. Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic beta-cell function. EMBO J. 2015;34:1417–1433. doi: 10.15252/embj.201490819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henry C., Close A.F., Buteau J. A critical role for the neural zinc factor ST18 in pancreatic beta-cell apoptosis. J Biol Chem. 2014;289:8413–8419. doi: 10.1074/jbc.M114.554915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki K., Harada N., Yamane S., Nakamura Y., Sasaki K., Nasteska D., Joo E., Shibue K., Harada T., Hamasaki A., Toyoda K., Nagashima K., Inagaki N. Transcriptional regulatory factor X6 (Rfx6) increases gastric inhibitory polypeptide (GIP) expression in enteroendocrine K-cells and is involved in GIP hypersecretion in high fat diet-induced obesity. J Biol Chem. 2013;288:1929–1938. doi: 10.1074/jbc.M112.423137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piccand J., Vagne C., Blot F., Meunier A., Beucher A., Strasser P., Lund M.L., Ghimire S., Nivlet L., Lapp C., Petersen N., Engelstoft M.S., Thibault-Carpentier C., Keime C., Correa S.J., Schreiber V., Molina N., Schwartz T.W., De Arcangelis A., Gradwohl G. Rfx6 promotes the differentiation of peptide-secreting enteroendocrine cells while repressing genetic programs controlling serotonin production. Mol Metab. 2019;29:24–39. doi: 10.1016/j.molmet.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gehart H., van Es J.H., Hamer K., Beumer J., Kretzschmar K., Dekkers J.F., Rios A., Clevers H. Identification of enteroendocrine regulators by real-time single-cell differentiation mapping. Cell. 2019;176:1158–1173 e16. doi: 10.1016/j.cell.2018.12.029. [DOI] [PubMed] [Google Scholar]
- 39.McGrath P.S., Watson C.L., Ingram C., Helmrath M.A., Wells J.M. The basic helix-loop-helix transcription factor NEUROG3 is required for development of the human endocrine pancreas. Diabetes. 2015;64:2497–2505. doi: 10.2337/db14-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sinagoga K.L., McCauley H.A., Munera J.O., Reynolds N.A., Enriquez J.R., Watson C., Yang H.C., Helmrath M.A., Wells J.M. Deriving functional human enteroendocrine cells from pluripotent stem cells. Development. 2018;145:dev165795. doi: 10.1242/dev.165795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommer C.A., Mostoslavsky G. RNA-Seq analysis of enteroendocrine cells reveals a role for FABP5 in the control of GIP secretion. Mol Endocrinol. 2014;28:1855–1865. doi: 10.1210/me.2014-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., Tirosh I., Beyaz S., Dionne D., Zhang M., Raychowdhury R., Garrett W.S., Rozenblatt-Rosen O., Shi H.N., Yilmaz O., Xavier R.J., Regev A. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bainbridge M.N., Hu H., Muzny D.M., Musante L., Lupski J.R., Graham B.H., Chen W., Gripp K.W., Jenny K., Wienker T.F., Yang Y., Sutton V.R., Gibbs R.A., Ropers H.H. De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring-Opitz syndrome. Genome Med. 2013;5:11. doi: 10.1186/gm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lichtig H., Artamonov A., Polevoy H., Reid C.D., Bielas S.L., Frank D. Modeling Bainbridge-Ropers syndrome in Xenopus laevis embryos. Front Physiol. 2020;11:75. doi: 10.3389/fphys.2020.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He B., Deng T., Zhu I., Furusawa T., Zhang S., Tang W., Postnikov Y., Ambs S., Li C.C., Livak F., Landsman D., Bustin M. Binding of HMGN proteins to cell specific enhancers stabilizes cell identity. Nat Commun. 2018;9:5240. doi: 10.1038/s41467-018-07687-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.West K.L., Castellini M.A., Duncan M.K., Bustin M. Chromosomal proteins HMGN3a and HMGN3b regulate the expression of glycine transporter 1. Mol Cell Biol. 2004;24:3747–3756. doi: 10.1128/MCB.24.9.3747-3756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J.W., Choi H.S., Gyuris J., Brent R., Moore D.D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 48.Ueda T., Furusawa T., Kurahashi T., Tessarollo L., Bustin M. The nucleosome binding protein HMGN3 modulates the transcription profile of pancreatic beta cells and affects insulin secretion. Mol Cell Biol. 2009;29:5264–5276. doi: 10.1128/MCB.00526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bates E. Ion channels in development and cancer. Annu Rev Cell Dev Biol. 2015;31:231–247. doi: 10.1146/annurev-cellbio-100814-125338. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y., Wang H., Qian Z., Feng B., Zhao X., Jiang X., Tao J. Low-voltage-activated T-type Ca2+ channel inhibitors as new tools in the treatment of glioblastoma: the role of endostatin. Pflugers Arch. 2014;466:811–818. doi: 10.1007/s00424-013-1427-5. [DOI] [PubMed] [Google Scholar]
- 51.Lang F., Stournaras C. Ion channels in cancer: future perspectives and clinical potential. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130108. doi: 10.1098/rstb.2013.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hua K., Ferland R.J. Primary cilia proteins: ciliary and extraciliary sites and functions. Cell Mol Life Sci. 2018;75:1521–1540. doi: 10.1007/s00018-017-2740-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anvarian Z., Mykytyn K., Mukhopadhyay S., Pedersen L.B., Christensen S.T. Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol. 2019;15:199–219. doi: 10.1038/s41581-019-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basten S.G., Giles R.H. Functional aspects of primary cilia in signaling, cell cycle and tumorigenesis. Cilia. 2013;2:6. doi: 10.1186/2046-2530-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.diIorio P., Rittenhouse A.R., Bortell R., Jurczyk A. Role of cilia in normal pancreas function and in diseased states. Birth Defects Res C Embryo Today. 2014;102:126–138. doi: 10.1002/bdrc.21064. [DOI] [PubMed] [Google Scholar]
- 56.Lodh S. Primary cilium, an unsung hero in maintaining functional beta-cell population. Yale J Biol Med. 2019;92:471–480. [PMC free article] [PubMed] [Google Scholar]
- 57.Kluth O., Stadion M., Gottmann P., Aga H., Jahnert M., Scherneck S., Vogel H., Krus U., Seelig A., Ling C., Gerdes J., Schurmann A. Decreased expression of cilia genes in pancreatic islets as a risk factor for type 2 diabetes in mice and humans. Cell Rep. 2019;26:3027–3036 e3. doi: 10.1016/j.celrep.2019.02.056. [DOI] [PubMed] [Google Scholar]
- 58.Saqui-Salces M., Dowdle W.E., Reiter J.F., Merchant J.L. A high-fat diet regulates gastrin and acid secretion through primary cilia. FASEB J. 2012;26:3127–3139. doi: 10.1096/fj.11-197426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teperino R., Aberger F., Esterbauer H., Riobo N., Pospisilik J.A. Canonical and non-canonical Hedgehog signalling and the control of metabolism. Semin Cell Dev Biol. 2014;33:81–92. doi: 10.1016/j.semcdb.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teperino R., Amann S., Bayer M., McGee S.L., Loipetzberger A., Connor T., Jaeger C., Kammerer B., Winter L., Wiche G., Dalgaard K., Selvaraj M., Gaster M., Lee-Young R.S., Febbraio M.A., Knauf C., Cani P.D., Aberger F., Penninger J.M., Pospisilik J.A., Esterbauer H. Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell. 2012;151:414–426. doi: 10.1016/j.cell.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 61.Hardie D.G., Ross F.A., Hawley S.A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt G.H., Winton D.J., Ponder B.A. Development of the pattern of cell renewal in the crypt-villus unit of chimaeric mouse small intestine. Development. 1988;103:785–790. doi: 10.1242/dev.103.4.785. [DOI] [PubMed] [Google Scholar]
- 63.Kung A.L., Rebel V.I., Bronson R.T., Ch'ng L.E., Sieff C.A., Livingston D.M., Yao T.P. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 64.Lee H., Cho D.Y., Whitworth C., Eisman R., Phelps M., Roote J., Kaufman T., Cook K., Russell S., Przytycka T., Oliver B. Effects of gene dose, chromatin, and network topology on expression in Drosophila melanogaster. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yukselen O., Turkyilmaz O., Ozturk A.R., Garber M., Kucukural A. DolphinNext: a distributed data processing platform for high throughput genomics. BMC Genomics. 2020;21:310. doi: 10.1186/s12864-020-6714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pomaznoy M., Ha B., Peters B. GOnet: a tool for interactive Gene Ontology analysis. BMC Bioinformatics. 2018;19:470. doi: 10.1186/s12859-018-2533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H.J., Johnston B., Aiello D., Caffrey D.R., Giel-Moloney M., Rindi G., Leiter A.B. Distinct cellular origins for serotonin-expressing and enterochromaffin-like cells in the gastric corpus. Gastroenterology. 2014;146:754–764 e3. doi: 10.1053/j.gastro.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data sets were deposited in NCBI, Gene ontology omnibus (GEO), accession number: GSE149203. All authors had access to all data and reviewed and approved the final manuscript.