Abstract
Background & Aims
The etiology of nonalcoholic fatty liver disease (NAFLD) is poorly understood, with males and certain populations exhibiting markedly increased susceptibility. Using a systems genetics approach involving multi-omic analysis of ∼100 diverse inbred strains of mice, we recently identified several candidate genes driving NAFLD. We investigated the role of one of these, liver pyruvate kinase (L-PK or Pklr), in NAFLD by using patient samples and mouse models.
Methods
We examined L-PK expression in mice of both sexes and in a cohort of bariatric surgery patients. We used liver-specific loss- and gain-of-function strategies in independent animal models of diet-induced steatosis and fibrosis. After treatment, we measured several metabolic phenotypes including obesity, insulin resistance, dyslipidemia, liver steatosis, and fibrosis. Liver tissues were used for gene expression and immunoblotting, and liver mitochondria bioenergetics was characterized.
Results
In both mice and humans, L-PK expression is up-regulated in males via testosterone and is strongly associated with NAFLD severity. In a steatosis model, L-PK silencing in male mice improved glucose tolerance, insulin sensitivity, and lactate/pyruvate tolerance compared with controls. Furthermore, these animals had reduced plasma cholesterol levels and intrahepatic triglyceride accumulation. Conversely, L-PK overexpression in male mice resulted in augmented disease phenotypes. In contrast, female mice overexpressing L-PK were unaffected. Mechanistically, L-PK altered mitochondrial pyruvate flux and its incorporation into citrate, and this, in turn, increased liver triglycerides via up-regulated de novo lipogenesis and increased PNPLA3 levels accompanied by mitochondrial dysfunction. Also, L-PK increased plasma cholesterol levels via increased PCSK9 levels. On the other hand, L-PK silencing reduced de novo lipogenesis and PNPLA3 and PCSK9 levels and improved mitochondrial function. Finally, in fibrosis model, we demonstrate that L-PK silencing in male mice reduced both liver steatosis and fibrosis, accompanied by reduced de novo lipogenesis and improved mitochondrial function.
Conclusions
L-PK acts in a male-specific manner in the development of liver steatosis and fibrosis. Because NAFLD/nonalcoholic steatohepatitis exhibit sexual dimorphism, our results have important implications for the development of personalized therapeutics.
Keywords: Liver Pyruvate Kinase, Sex Differences, NAFLD, Liver Fibrosis, Mitochondrial Dysfunction
Abbreviations used in this paper: AAV8, adeno-associated virus serotype 8; CDAHFD, choline-deficient, L-amino acid-defined, high-fat diet with 0.1% methionine; CE, cholesteryl ester; DEGs, differentially expressed genes; DNL, de novo lipogenesis; ETC, electron transport chain; FFA, free fatty acid; GGT, gamma-glutamyl transpeptidase; GTT, glucose tolerance tests; HDL, high-density lipoprotein; HF/HS diet, diet rich in fat and sucrose; HMDP, Hybrid Mouse Diversity Panel; ITT, insulin tolerance tests; LPK, liver pyruvate kinase; L/PTT, lactate/pyruvate tolerance tests; NAFLD, nonalcoholic fatty liver disease; NAS, NAFLD Activity Score; NASH, nonalcoholic steatohepatitis; OXPHOS, oxidative phosphorylation; qPCR, quantitative polymerase chain reaction; ROS, reactive oxygen species; scrRNA, scrambled RNA; SEM, standard error of the mean; shRNA, short hairpin RNA; siRNA, small interfering RNA; TBG, thyroxine binding globulin; TC, total cholesterol; TG, triglyceride
Graphical abstract
Summary.
In both mice and humans, high levels of liver pyruvate kinase (L-PK) are strongly associated with NAFLD severity only in males. Mechanistically, L-PK regulates mitochondrial pyruvate flux and its incorporation into citrate, thereby affecting de novo lipogenesis accompanied by mitochondrial dysfunction/maladaptation.
Nonalcoholic fatty liver disease (NAFLD) is the leading cause of chronic liver disease in the United States and worldwide.1, 2, 3, 4 NAFLD manifestations range from simple steatosis to complex nonalcoholic steatohepatitis (NASH), fibrosis and cirrhosis, and liver cancer.1, 2, 3 NASH-related cirrhosis is the most common non-viral cause of liver failure requiring liver transplantation worldwide.5 Despite intensive research efforts, the etiology of this disease is poorly understood; in particular, factors governing the progression from steatosis to NASH and fibrosis are largely unknown. Population studies have shown that NAFLD is strongly associated with obesity and diabetes.2,4,6,7 To understand the genetic and molecular factors underlying NAFLD, we used an integrative multi-omics approach by combining liver transcriptomics, expression quantitative trait loci, and intrahepatic triglyceride levels from our extensively phenotyped mouse cohort, the Hybrid Mouse Diversity Panel (HMDP).8 This led to the identification of the liver isoform of pyruvate kinase (L-PK or Pklr) as one of the candidate genes regulating NAFLD via mitochondrial involvement.9 However, this study involved only male HMDP strains.
Recent studies have also revealed that NAFLD is more prevalent in males than females, with males exhibiting more severe NAFLD symptoms.2,10, 11, 12, 13, 14 Sex differences in susceptibility to obesity, insulin resistance, or other metabolic phenotypes have been amply described in mice, humans, and other species, with females generally exhibiting more beneficial metabolic profiles.15, 16, 17, 18, 19, 20, 21, 22 Sex differences in experimental organisms, including the mouse, have generally been studied on a single genetic background. Our goal was to study sex differences in the context of genetic variation, so that we could examine interactions between genetic background and sex. For this, we used liver tissues from both sexes of HMDP and identified tissue-specific patterns of sex-dimorphic gene expression, with more than 3719 liver transcripts differentially expressed between the sexes.23 Follow-up gonadectomy studies revealed that testosterone plays a dominant role in regulating the sex-dimorphic gene expression in the liver.23 It is important to further understand and validate these sex-dimorphic genes in the context of disease pathogenesis because that knowledge can be used for personalized drug development.
Our current focus is to understand and delineate the potential sex-dimorphic roles of L-PK in NAFLD pathogenesis. To this end, we used liver tissues from both sexes of mouse and human patients and revealed that indeed, L-PK exhibited sex-dimorphic and testosterone-regulated associations and suggested a role in NAFLD and NASH development primarily in males. Using both loss- and gain-of-function strategies in independent mouse models of NAFLD/NASH, we demonstrate that L-PK alters both steatosis and fibrosis in males but not females. Furthermore, we show that L-PK induces mitochondrial stress in both steatosis and fibrosis and that silencing L-PK relieves this stress and promotes NAFLD/NASH resolution.
Results
Male-Specific Roles of L-PK in Both NAFLD and NASH
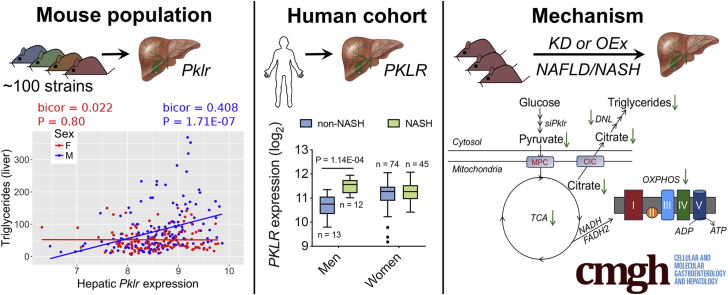
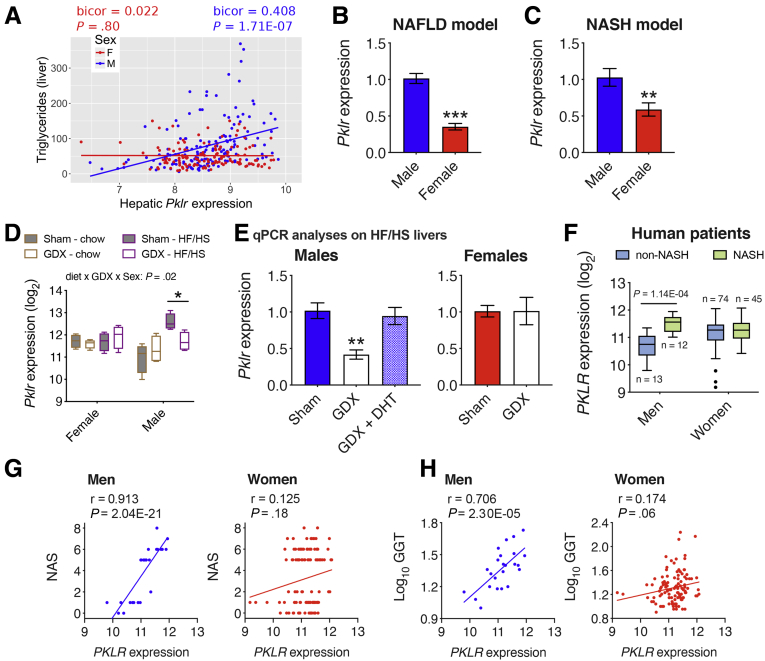
Recently, we reported L-PK as a key driver gene in potentiating liver steatosis.9 However, these studies were performed only using males, and when we investigated our mouse population of ∼98 sex-matched strains maintained on a diet rich in fat and sucrose (HF/HS diet), we observed that only male hepatic Pklr expression was positively correlated with liver triglyceride (TG) levels (bicor 0.408; P = 1.71E-07), whereas there was no correlation in females (Figure 1A). This suggested that L-PK has little or no role in developing steatosis in females.
Figure 1.
Male L-PK expression is associated with NAFLD/NASH in both mice and humans. (A) Correlation plots between hepatic Pklr expression and hepatic TG in male and female HMDP cohorts (n = 98 sex-matched strains with 2–4 mice per sex/strain). Pklr expression from (B) C57BL/6J mice fed HF/HS diet for 8 weeks (n = 3–7 mice per group), (C) C57BL/6J-APOE-Leiden mice fed a Western diet for 16 weeks (n = 6 mice per group), (D and E) gonadectomized male and female C57BL/6J mice maintained on chow or HF/HS diets (n = 4 mice per group), or (F) human patient population (n = 25 men, 119 women). Correlation plots between hepatic PKLR expression and (G) NAS or (H) plasma GGT levels in men and women cohorts. Blue and red represent males and females, respectively. DHT, dihydrotestosterone; GDX, gonadectomized. Data are presented as mean ± SEM. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001 by (A) Bicorrelation; (B and C) t test; (D) three-factor or (E) one-factor or (F) two-factor analysis of variance with Holm-Sidak’s post hoc test; (G and H) partial correlation.
To further determine the sex-specific roles of L-PK in NAFLD and NASH, Pklr expression in livers from males and females of either C57BL/6J mice maintained on a HF/HS diet (NAFLD model) or C57BL/6J-APOE-Leiden mice maintained on a “Western” diet (NASH model) were measured. Hepatic Pklr expression was significantly lower in both female NAFLD mice (2.9-fold) (Figure 1B) and female NASH mice (1.7-fold) (Figure 1C), compared with their respective male counterparts. To further characterize the role of sex hormones in hepatic Pklr expression, liver tissues from intact and gonadectomized C57BL/6J mice maintained on a chow or HF/HS diet were analyzed. We observed that gonadectomy in males maintained on HF/HS diet significantly lowered hepatic Pklr expression (Figure 1D and E), and that this was rescued by testosterone replacement (Figure 1E). In contrast, ovariectomy in females had no impact on hepatic Pklr expression in either diets (Figure 1D and E).
Finally, to understand the clinical relevance of hepatic PKLR in human NASH, we sequenced liver biopsies from a total of 144 morbidly obese Mexican patients who underwent bariatric surgery (Table 1). We found that hepatic PKLR expression was higher only in biopsy proven NASH men compared with non-NASH men (Figure 1F). In contrast, women had no difference in PKLR expression between their NASH status (Figure 1F). Furthermore, we observed strong positive associations between hepatic PKLR expression and NASH-related phenotypes including NAFLD Activity Score (NAS) (Figure 1G) and plasma gamma-glutamyl transpeptidase (GGT) levels (Figure 1H) only in men. In contrast, only weak or no correlations were observed in women (Figure 1G and H). It should be noted that there were only 25 men compared with 119 women, yet we observed distinct male-specific hepatic PKLR associations with NASH phenotypes. Taken together, these results suggest that L-PK plays an important sex-specific role in the pathophysiology of both NAFLD and NASH.
Table 1.
Clinical and Biochemical Characteristics of Subjects With Gene Expression Data (N = 144) Shown in Figure 1
| Characteristics | Non-NASH (n = 87) | NASH (n = 57) | P value |
|---|---|---|---|
| Age (y) | 37.0 (30.0–46.0) | 39.0 (34.0–47.0) | .472 |
| Sex (male %) | 13 (14.9) | 12 (21.1) | .344 |
| BMI (kg/m2) | 41.0 (37.9–45.9) | 43.7 (38.9–48.8) | .319 |
| Fasting glucose (mg/dL) | 89.0 (80.0–99.0) | 94.0 (86.0–100.0) | .172 |
| Fasting insulin (μU/L) | 9.6 (6.1–12.7) | 14.5 (11.3–18.5) | 2.2E-04 |
| HOMA-IR | 2.1 (1.5–3.1) | 3.3 (2.5–4.2) | .001 |
| Triglycerides (mg/dL) | 120.8 (89.5–153.0) | 161.5 (110.0–219.0) | 2.7E-04 |
| Total cholesterol (mg/dL) | 168.5 (147.5–194.8) | 172.0 (148.0–204.8) | .799 |
| HDL-C (mg/dL) | 36.0 (32.0–44.0) | 34.5 (31.0–41.0) | .140 |
| AST (IU/mL) | 26.0 (20.3–31.0) | 29.5 (25.0–36.3) | .016 |
| ALT (IU/mL) | 26.0 (18.0–35.0) | 28.5 (23.0–41.8) | .024 |
| GGT (IU/mL) | 16.5 (13.0–24.8) | 23.0 (18.0–32.8) | 2.4E-04 |
NOTE. Data are shown as median (interquartile range) or n (%) according to distribution of variables. Data were analyzed using Mann-Whitney U or χ2 test. P values in bold indicate significant differences.
ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; HDL-C, high-density lipoprotein cholesterol; HOMA-IR, homeostatic model assessment for insulin resistance.
L-PK Overexpression Exacerbates Liver Steatosis Only in Males
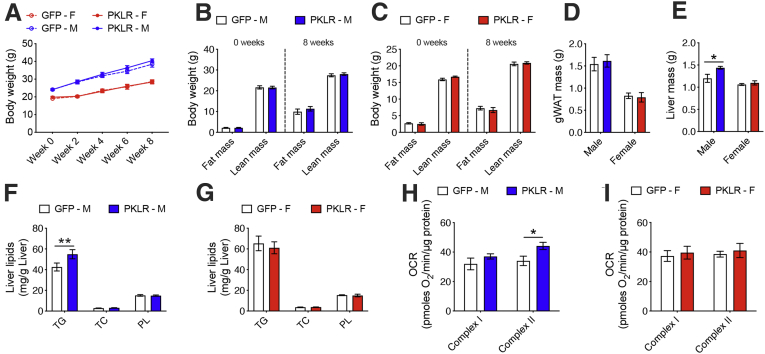
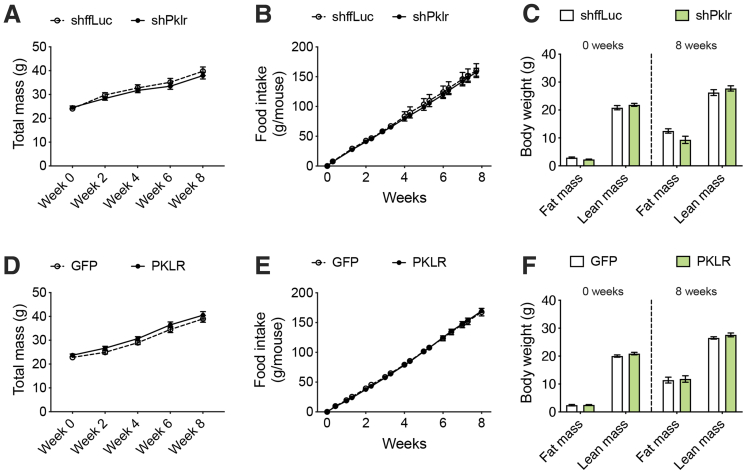
Next, to functionally validate the male-specific role of L-PK in liver steatosis, we specifically overexpressed L-PK or green fluorescent protein in a liver-specific manner using recombinant adeno-associated virus serotype 8 (AAV8) under the control of thyroxine binding globulin (TBG) promoter in both sexes of C57BL/6J mice. These animals were then placed on a HF/HS diet for 8 weeks to develop liver steatosis. The quantitation of AAV8-mediated hepatic L-PK overexpression is shown in Figure 2. After 8 weeks, we observed that hepatic L-PK overexpression did not affect obesity phenotypes in either sex as measured by body weight, body composition, and gonadal white adipose tissue mass (Figure 3A–D). However, the liver mass and the liver TG levels were increased only in males overexpressing L-PK, whereas the females had no effect (Figure 3E–g). We have earlier reported that altered liver mitochondrial respiration affects hepatic TG accumulation.9 Supporting this concept, we noted that only males overexpressing L-PK had higher complex-II mediated respiration, whereas females had no change (Figure 3H and I). Taken together, we conclude that L-PK has a male-specific role in exacerbating liver steatosis via altered mitochondrial respiration.
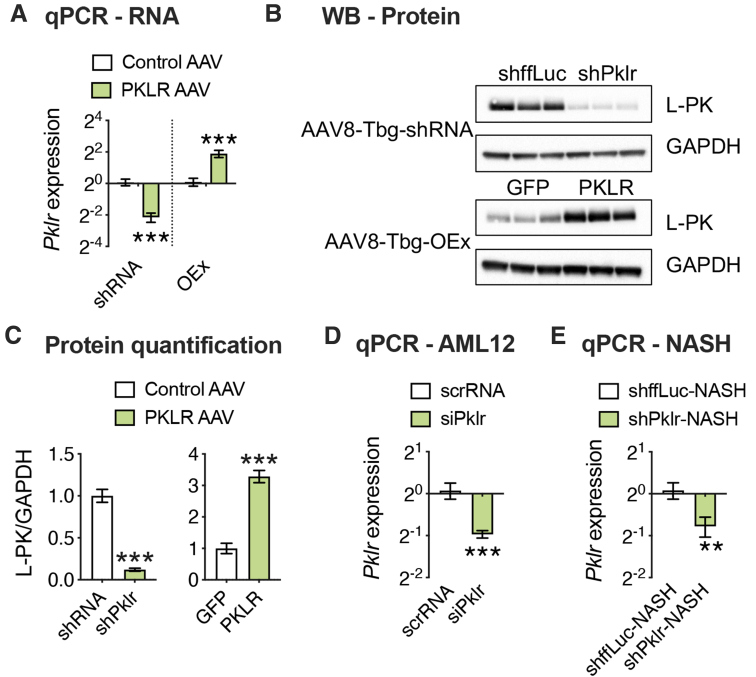
Figure 2.
AAV-mediated modulation of hepatic L-PK expression. Eight-week-old male C57BL/6J mice were injected with either loss-of-function (shffLuc or shPklr) or gain-of-function (green fluorescent protein or PKLR) AAV vectors under the control of Tbg promoter and fed HF/HS diet for 17 additional weeks. (A) qPCR analyses of hepatic Pklr expression and (B) immunoblot analyses of hepatic PKLR in L-PK KD and OEx mice. GAPDH was used as a loading control. (C) Quantification of average PKLR levels normalized to GAPDH for each group. qPCR analyses of Pklr expression (D) 48 hours after siRNA transfection of AML12 cells (experiment was repeated 2 independent times with n = 3 wells per group each time) and (E) 12 weeks after NASH diet in L-PK KD mice. Data are presented as mean ± SEM (n = 6–8 livers for RNA and 3 for protein analyses per group). P values were calculated by unpaired Student t test. ∗∗∗P < .001.
Figure 3.
L-PK is causal in developing liver steatosis but only in males. Eight-week-old males and females of C57BL/6J mice were injected with AAV8-TBG vectors carrying either green fluorescent protein or PKLR and fed HF/HS diet for 8 weeks. (A) Body weight monitored every 2 weeks and (B and C) body composition monitored before and after the study are shown. Comparisons of (D) gWAT and (E) liver weights in males and females. Similarly, comparisons of liver lipids in (F) males and (G) females, respectively. State 3 liver mitochondrial respiration from (H) males and (I) females, respectively. Blue and red represent male and female, respectively. Data are presented as mean ± SEM (n = 6 mice or 3 mitochondria per group). ∗P < .05; ∗∗P < .01 by two-factor analysis of variance with Holm-Sidak’s post hoc test. gWAT, gonadal white adipose tissue.
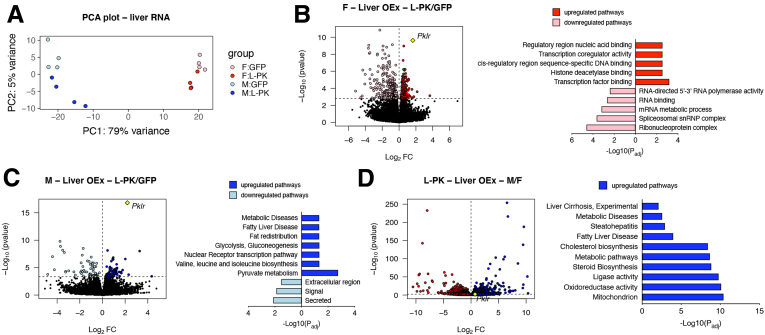
To further explore sex-specific mechanistic differences exerted by L-PK overexpression, we performed whole genome RNA sequencing on livers extracted from males and females that were expressing green fluorescent protein or L-PK. Principal component analyses revealed that the majority of the variances were explained, as expected, by sex (79%) and to a minor extent by L-PK (5%) (Figure 4A). Differential expression analyses followed by DAVID24 and ToppGene25 enrichment analyses revealed that females overexpressing L-PK had differentially regulated nucleus-related pathways such as DNA binding, histone deacetylase binding, and RNA binding among others (Figure 4B). In contrast, L-PK overexpressing males had up-regulated NAFLD-related metabolic pathways such as glycolysis, gluconeogenesis, fatty liver disease, and pyruvate metabolism among others (Figure 4C). Finally, when we compared male vs female livers (both overexpressing L-PK), we observed that males had slightly higher Pklr expression (log2FC = 0.73, P = .005) and were strongly enriched for mitochondria, cholesterol biosynthesis, and NAFLD/NASH pathways (Figure 4D). Taken together, our data clearly show that mechanistically, L-PK functions in a male-specific manner in up-regulating mitochondrial and metabolic pathways and thus accelerating NAFLD/NASH.
Figure 4.
L-PK alters mitochondrial and metabolic pathways in a male-specific manner. (A) Principal component analyses, a data reduction method, summarize total gene expression in livers from males and females expressing green fluorescent protein or L-PK in 2 dimensions. (B–D) Global genome-wide transcriptomics of livers from females or males that were expressing green fluorescent protein or L-PK (n = 4 per group) along with their respective pathway enrichments by DAVID and ToppGene analyses. Males are in blue, and females are in red; green fluorescent protein expression is in light color, and L-PK expression is in dark color. P values were calculated by DESeq function of Bioconductor R-package. OEx, overexpressing.
L-PK Silencing Attenuates Insulin Resistance, Plasma Cholesterol Levels, and Liver Steatosis While Overexpression Exacerbates Them via Altered Liver Mitochondrial Respiration
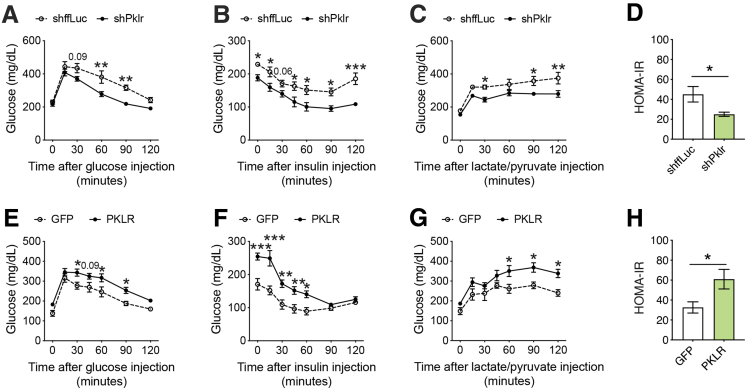
To exclude any possibility of off-target effects on the observed phenotypes and to fully characterize the role of L-PK, we repeated our NAFLD studies in male C57BL/6J mice using both gene silencing and overexpression strategies via AAV8 vectors. For gene silencing, the animals received liver-specific AAV harboring short hairpin RNA (shRNA) against ffLuc or Pklr, and for gene overexpression, the animals received liver-specific AAV harboring green fluorescent protein or L-PK. The validity of both these systems is shown in Figure 2. All these animals were placed on a HF/HS diet for a total of 17 weeks without any interventions for the first 8 weeks of diet, with careful monitoring of body weight, food intake, and body composition. We noted that both our silencing and overexpression models for hepatic Pklr expression did not affect any of these obesity phenotypes (Figure 5). In contrast, our previous study involving adenoviral vectors for 14 days had changes in adipose tissue mass.9 Because both our silencing and overexpression AAV strategies reproducibly show that food intake and/or obesity phenotypes were not affected by L-PK (Figures 3, and 5), we believe adipose tissue differences seen in our original study were mostly consequences due to different viral strategies used between the 2 studies. After 8 weeks of diet, all the animals underwent a series of metabolic tests including glucose tolerance tests (GTT), insulin tolerance tests (ITT), and lactate/pyruvate tolerance tests (L/PTT) with 2-week rest time between each test. We observed that L-PK silencing promoted enhanced glucose tolerance and insulin sensitivity (Figure 6A–C). We also noted that these animals had improved insulin sensitivity as measured by endpoint homeostatic model assessment of insulin resistance (Figure 6D). Consistent with these observations, our L-PK overexpression promoted glucose intolerance and insulin resistance (Figure 6E–H).
Figure 5.
Hepatic L-PK does not alter obesity or food intake. Comparisons of (A and D) body weight measurements, (B and E) food intake, and (C and F) body composition (fat and lean mass) during, before, and after the diet challenge are shown. Data are presented as mean ± SEM (n = 7–8 mice per group).
Figure 6.
L-PK is causal in developing insulin resistance. Eight-week-old male C57BL/6J mice were injected with either loss-of-function (shffLuc or shPklr) or gain-of-function (green fluorescent protein or PKLR) AAV8-TBG vectors and then fed HF/HS diet for 17 weeks. Comparisons of (A and E) GTT, (B and F) ITT, (C and G) L/PTT, and (D and H) homeostatic model assessment of insulin resistance measures of L-PK silencing (KD) or overexpressing (OEx) mice, respectively. Data are presented as mean ± SEM (n = 7–8 mice per group). ∗P < .05; ∗∗P < .01 by (A–C and E–G) two-factor analysis of variance with Holm-Sidak’s post hoc test; (D and H) t test. ∗∗∗P < .001.
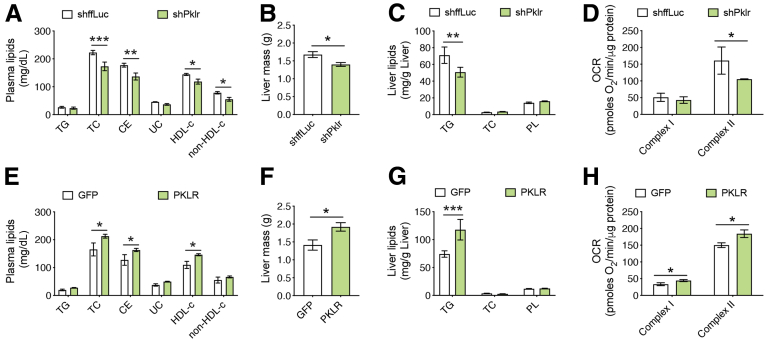
Analysis of plasma lipids revealed that L-PK silencing reduced plasma levels of total cholesterol (TC), cholesteryl ester (CE), high-density lipoprotein (HDL), and non-HDL cholesterol (Figure 7A). Also, we noted that both the liver mass and liver TG levels were reduced with L-PK silencing (Figure 7B and C). As we have seen earlier, we found that L-PK silencing reduced complex-II mediated liver mitochondrial respiration (Figure 7D). Consistent with these data, we observed increased plasma lipid levels, liver mass, liver TG levels, and a corresponding increase in liver mitochondrial respiration in L-PK overexpressing mice (Figure 8E–H). Taken together, we conclude that L-PK is a causal gene for the development of insulin resistance, steatosis, and altered plasma cholesterol levels via altering liver mitochondrial respiration.
Figure 7.
L-PK alters plasma cholesterol levels and liver steatosis via altered liver mitochondrial respiration. Comparisons of (A and E) plasma lipid levels, (B and F) liver weights, (C and G) hepatic lipid levels, and (D and H) State 3 liver mitochondrial respiration from L-PK silencing (KD) or overexpressing (OEx) mice, respectively. Data are presented as mean ± SEM (n = 7–8 mice or 4 mitochondria per group). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001 by t test for liver weights; two-factor analysis of variance with Holm-Sidak’s post hoc test for the rest. OCR, oxygen consumption rate; PL, phospholipid; UC, unesterified cholesterol.
Figure 8.
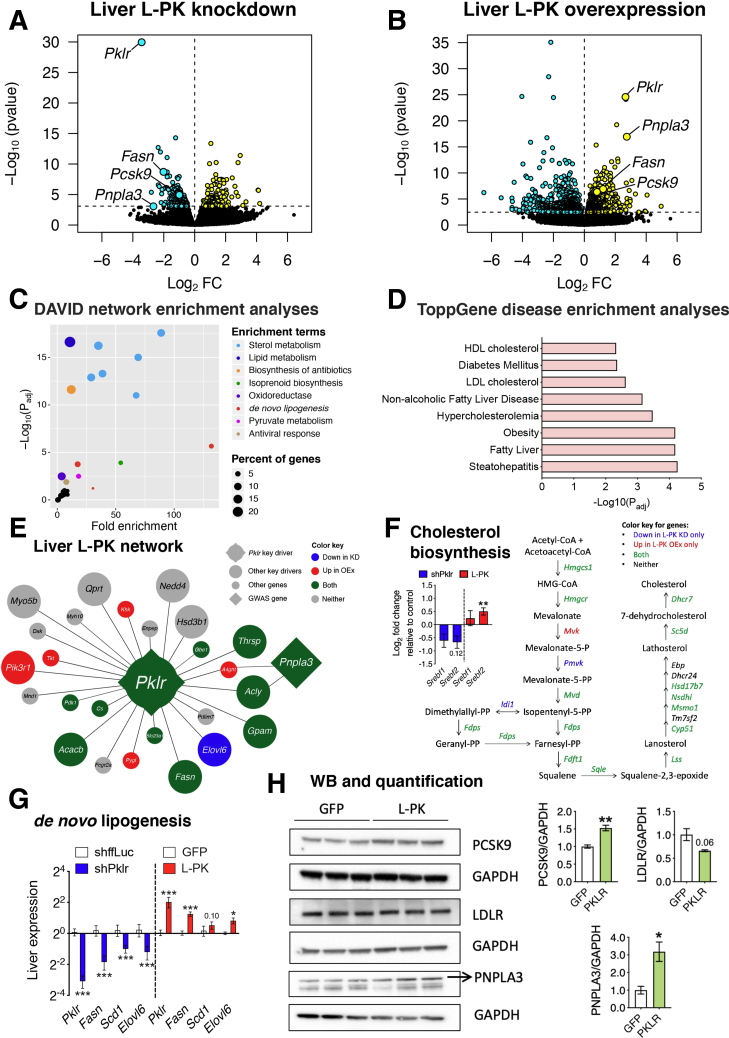
L-PK alters liver steatosis via de novo lipogenesis and PNPLA3 and plasma cholesterol levels via PCSK9. Global genome-wide transcriptomics of livers extracted from L-PK (A) KD or (B) OEx mice. Turquoise and yellow represent down-regulated and up-regulated genes, respectively. (C) DAVID pathway and (D) ToppGene disease enrichment analyses of 124 common DEGs (Supplementary Table 1) between L-PK KD and OEx mice. Overlay of DEGs on (E) L-PK key driver network from our previous study9 or (F) cholesterol biosynthesis pathway. Blue represents genes going down in KD mice, red represents genes going up in OEx mice, and green represents changing in both mice (down in KD, up in OEx). Inset shows log2 fold change of Srebf1 and Srebf2 in L-PK KD and OEx mice. Follow-up (G) qPCR analyses of liver DNL genes such as Pklr, Fasn, Scd1, and Elovl6 and (H) immunoblot analyses of liver proteins such as PCSK9, LDLR, and PNPLA3 in L-PK OEx mice. Data are presented as mean ± SEM (n = 5 livers for RNA and 3 for protein analyses per group). ∗P < .05; ∗∗P < 0.1; ∗∗∗P < .001 by t test. KD, knockdown; LDL, low-density lipoprotein; OEx, overexpressing.
L-PK Alters Liver TG Levels via De Novo Lipogenesis and PNPLA3 and Plasma Cholesterol Levels via PCSK9
To further explore the mechanistic roles of L-PK in the observed metabolic alterations, we performed whole genome RNA sequencing on the extracted liver tissues from both L-PK silencing and overexpression groups. We found 509 differentially expressed genes (DEGs) in L-PK knockdown and 1731 DEGs in L-PK overexpression animals, with a common 124 DEGs (significant overlap by hypergeometric distribution, P = 2.38E-40) between the 2 groups (Figure 8A and B, Supplementary Table 1). Key NAFLD-related genes that were significantly changed in both the groups include Fasn, Pnpla3, and Pcsk9 (Figure 8A and B). Notably, Pnpla3 was the third most down-regulated gene in L-PK knockdown mice and second most up-regulated gene in L-PK overexpression mice (Figure 8A and B, Supplementary Table 1). Network enrichment analyses of the common 124 DEGs between the 2 groups by DAVID24 revealed NAFLD-related pathways such as lipid metabolism, sterol metabolism, acetyl-CoA metabolic process, oxidoreductase, de novo lipogenesis (DNL), and pyruvate metabolism (Figure 8C). Also, we noted that antiviral defense was also nominally enriched, possibly because of AAV usage (Figure 8C). Furthermore, disease enrichments by ToppGene25 revealed metabolic syndrome-related disease traits including obesity, diabetes, and NAFLD/NASH (Figure 8D). When we reanalyzed the L-PK key driver network from our previous study,9 we found that 15 of 25 genes directly connected to Pklr hub through a single edge were affected in either L-PK silencing or overexpression models (Figure 8E). In particular, the well-known human genome-wide association studies candidate PNPLA3, which was connected to Pklr hub through 2 edges (via Acly or A4gnt), is affected in both genetic models. Other key network genes affected by L-PK were the DNL-related genes Pdk1, Cs, Thrsp, Acly, Acacb, Fasn, and Elovl6 (Figure 8E). Moreover, 16 of 19 genes involved in cholesterol biosynthesis pathway were affected in at least one of the genetic models (Figure 8F). Because Srebf1 and Srebf2 control DNL and cholesterol biosynthesis, respectively, we inspected our transcriptomic data for both these genes. We found that only Srebf2 was significantly altered in either L-PK silencing or overexpression models (Figure 8F). Follow-up quantitative polymerase chain reaction (qPCR) analyses revealed that both L-PK silencing and overexpression indeed affected DNL genes including Fasn, Scd1, and Elovl6 (Figure 8G). Finally, Western blot analyses revealed that L-PK overexpression increased liver protein levels of both PNPLA3 and PCSK9 and decreased LDLR protein levels (Figure 8H). We also observed strong correlations between human PKLR expression and these lipogenic genes in our Mexican cohort, thus corroborating our results (Table 2).
Table 2.
Correlations of Human Liver PKLR Expression With Target Genes Shown in Figure 8
| Genes | All subjects (n = 144) |
Men (n = 25) |
Women (n = 119) |
|||
|---|---|---|---|---|---|---|
| R | P value | r | P value | r | P value | |
| FASN | 0.437 | 4.5E-08 | 0.775 | 5.4E-06 | 0.389 | 1.2E-05 |
| SCD | 0.306 | 1.9E-04 | 0.541 | .005 | 0.249 | .006 |
| ELOVL6 | 0.347 | 2.1E-05 | 0.314 | .126 | 0.355 | 7.3E-05 |
| PNPLA3 | –0.044 | .597 | 0.175 | .403 | –0.078 | .402 |
| PCSK9 | 0.144 | .085 | 0.472 | .017 | 0.091 | .324 |
NOTE. Data were analyzed by Pearson correlations using normalized liver expression data. P values in bold indicate significant differences.
Stable Isotope Tracing Revealed That L-PK Silencing Attenuates Mitochondrial Glucose Oxidation
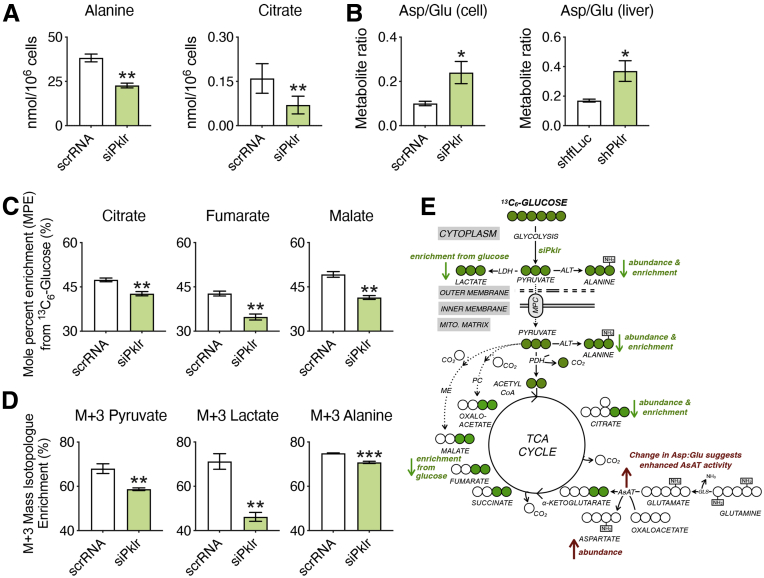
Because transcriptomic data established that L-PK silencing lowers DNL, we hypothesized that this would also be reflected in corresponding changes in TCA cycle metabolism. We therefore measured abundances of TCA cycle intermediates after transfection of AML12 liver cells with either Pklr small interfering RNA (siRNA) or scrambled RNA (scrRNA). The knockdown efficiency is shown in Figure 2. We observed significant reductions in the abundance of alanine and citrate levels by L-PK silencing (Figure 9A), both of which mimic the well-documented consequences of reduced mitochondrial uptake or oxidation of glucose-derived pyruvate.26 In addition, we also observed an increase in the aspartate to glutamate ratio in response to L-PK silencing (Figure 9B). This phenocopies the metabolic rewiring observed on chemical or genetic ablation of the mitochondrial pyruvate carrier, as cells increase glutamine oxidation and flux through the aspartate aminotransferase to maintain energetics and anaplerosis.27, 28, 29 Consistent with cell culture experiments, metabolomic analysis of hepatic extracts from L-PK knockdown mice revealed a significantly increased aspartate to glutamate ratio as well (Figure 9B). Taken together, our data show that L-PK silencing exhibits metabolic hallmarks consistent with decreased mitochondrial pyruvate uptake and oxidation.
Figure 9.
L-PK silencing attenuates mitochondrial glucose oxidation. Metabolomic analyses of AML12 cells transfected with either scrRNA or siPklr and grown in media containing [U-13C6]-glucose and hepatic extracts from L-PK KD mice. (A) Abundance levels of alanine and citrate; (B) metabolite ratios of aspartate to glutamate in AML12 cells and their corresponding levels in liver extracts of L-PK KD mice. (C) Mole percent enrichment of labeled carbon into citrate, fumarate, and malate. (D) Enrichment of glucose-derived carbons into the M+3 isotopomers of pyruvate, lactate, and alanine in AML12 cells. (E) Schematic of changes in pyruvate flux into TCA cycle using (U)13C glucose tracing in AML12 cells. ALT, alanine aminotransferase; AsAT, aspartate aminotransferase; GLS, glutaminase; KD, knockdown; LDH, lactate dehydrogenase; ME, malic enzyme; MPC, mitochondrial pyruvate carrier; PC, pyruvate carboxylase; PDH, pyruvate dehydrogenase. Data are presented as mean ± SEM (n = 3 cells or 5 livers per group). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001 by t test.
To further test that flux from glucose-derived pyruvate was reduced, we tracked the incorporation of uniformly labeled [U-13C6]-glucose into metabolites. Consistent with the transcriptomic and metabolite analysis, flux from glucose-derived carbon into citrate, fumarate, and malate was reduced on L-PK knockdown (Figure 9C). We also observed reduced enrichment of glucose-derived carbons into the M+3 isotopomers of pyruvate, lactate, and alanine (Figure 9D), consistent with L-PK silencing slowing the glycolytic production of pyruvate. The findings are consistent with a decreased flux of pyruvate into the TCA cycle, reducing steady-state citrate levels and, in turn, reducing DNL (Figure 9E)
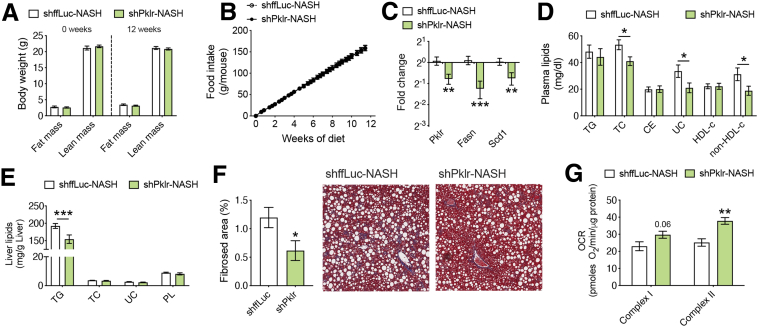
L-PK Silencing Attenuates Both Liver Steatosis and Fibrosis
Next, to verify whether L-PK silencing could attenuate severe NAFLD phenotypes such as fibrosis, we repeated our study using a diet-induced mouse model for liver fibrosis.30 We injected our AAV containing shRNA against ffLuc or Pklr in 8-week-old C57BL/6J male mice and then fed them a choline-deficient, L-amino acid-defined, high-fat diet with 0.1% methionine (CDAHFD) for an additional 12 weeks to induce steatosis and fibrosis. The quantitation of the knockdown is shown in Figure 2. As expected, we did not see any changes in body composition or food intake (Figure 10A and B), but both the plasma cholesterol levels and liver TG levels were lowered in L-PK knockdown animals, accompanied by down-regulation of DNL genes (Figure 10C–E). Furthermore, we also noted that L-PK silencing reduced liver fibrosis as measured by trichrome blue staining area (Figure 10F). We previously validated this method in a mouse model of NASH, showing that fibrosed area % strongly correlated with both hydroxyproline content and a pathologist’s fibrosis score.31 Interestingly, we also noted that reduced fibrosis in L-PK knockdown animals was accompanied by increased hepatic mitochondrial respiration (Figure 10G). It has been proposed that mitochondrial function is attenuated during NASH/fibrosis because of chronic maladaptive changes in the mitochondria caused by increased fuel oxidation (glucose and fat) and the resulting reactive oxygen species (ROS) production during steatosis.32,33 In our current study, L-PK silencing reduced pyruvate flux into the mitochondria, thereby reducing both free fatty acid (FFA) production by DNL and their eventual oxidation by mitochondria. Taken together, we interpret the increased mitochondrial function in the fibrosis model as a sign of better functional or relatively “unstressed” mitochondria.
Figure 10.
L-PK silencing protects from both liver steatosis and fibrosis. Eight-week-old male C57BL/6J mice were injected with AAV8-TBG vectors containing either shffLuc or shPklr and fed CDAHFD for 12 weeks. Comparisons of (A) body composition and (B) food intake before, after, and during the diet challenge are shown. Follow-up (C) qPCR analyses of liver DNL genes such as Pklr, Fasn, and Scd1; (D) plasma lipid levels; (E) liver lipid levels; (F) percent liver fibrosis area and representative images of trichrome blue staining and (G) State 3 liver mitochondrial respiration from L-PK KD NASH mice. Data are presented as mean ± SEM (n = 5–6 mice or 3 mitochondria per group). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001 by t test for RNA and liver fibrosis analyses; two-factor analysis of variance with Holm-Sidak’s post hoc test for the rest. OCR, oxygen consumption rate; PL, phospholipid; UC, unesterified cholesterol.
Discussion
We previously applied a systems genetics approach to identify L-PK as a strong candidate for susceptibility to steatosis in male mice fed a HF/HS diet. We subsequently validated L-PK in an acute model of steatosis (2 weeks of HF/HS diet) using in vivo knockdown in liver.9 A separate study based on human data also identified L-PK as a candidate for NAFLD.34 In the current study, we have examined the mechanism(s) underlying the role of L-PK in NAFLD. Several conclusions have emerged. First, we found that L-PK is a sexually dimorphic gene in both mice and humans and that this contributed to the greater prevalence of NAFLD in males. Second, our results have provided mechanistic information about mechanisms perturbed by L-PK, particularly relating to the role of mitochondrial metabolism such as pyruvate oxidation, generation of TCA intermediates, mitochondrial respiration, and DNL. They also validated novel interactions predicted from our network modeling, including key NAFLD genes such as PNPLA3 and PCSK9. We discuss these points in turn below. Third, using both loss- and gain-of-function studies, we show that L-PK regulates hepatic lipogenic genes and mitochondrial stress, and that it contributes to not only steatosis but also fibrosis.
Because of the greater susceptibly of males to NAFLD, our previous studies were performed in male mice. In the current study, on analysis of both sexes in the HMDP population fed a HF/HS diet, we noted that L-PK was strongly associated with liver TG in males but not females, and that L-PK was expressed at higher levels in liver in the HMDP models of steatosis and NASH. This prompted us to examine the hormonal dependence of L-PK expression, the results of which indicated a dependence on testosterone but not estrogen. The relationship between L-PK and NAFLD appears to be similar in humans. Thus, in a cohort of bariatric surgery patients, L-PK expression was associated with NASH and NASH-related phenotypes in men but not women. The fact that L-PK is sexually dimorphic, at least in mice, does entirely explain the sex-specific effect on NAFLD traits. This was clearly observed when L-PK was overexpressed in C67BL/6J mice, resulting in increased liver TG levels and liver mass in males but not females.
The explanation for the sex-dependent effect of L-PK appears to be mediated by sex-dependent mitochondrial functions, because only males overexpressing L-PK had higher complex-I and -II mediated respiration, as well as up-regulated mitochondrial-related pathways compared with females. We have observed that male mice tend to have significantly higher levels of liver mitochondrial DNA than females, suggesting differing underlying metabolic functions (data not shown). Previous studies have shown that during NAFLD/NASH, when the liver is overwhelmed with FFA flux because of (1) increased dietary fat, (2) increased hepatic fatty acid transport, and (3) augmented hepatic DNL, several mitochondrial abnormalities including ultrastructural lesions, depletion of mitochondrial DNA, decreased activity of electron transport chain (ETC) complexes, and impaired mitochondrial β-oxidation occur.32,33,35,36 This results in extra-mitochondrial fatty acid oxidation occurring in peroxisomes and endoplasmic reticulum, leading to increased ROS generation and oxidative stress as seen in NAFLD.37 Indeed, we had previously demonstrated that mitochondrial dysfunction in liver can result in liver lipid accumulation.9 Taken together, this explains how L-PK could mechanistically lead to steatosis in males; however, why overexpression of L-PK did not affect mitochondrial metabolism in females is an open question. We propose that premenopausal females have alternative nutrient preference and/or liver metabolic processes specifically pertaining to mitochondrial metabolism under conditions of excess calorie intake that keep them protected from abnormal liver lipid accumulation. Such a fundamental mechanistic difference is supported by the fact that there were 3719 sex-dimorphic mouse liver transcripts differentially expressed between the sexes, most of which are controlled by testosterone.23 Nevertheless, further investigations are warranted.
In our current study, we observed that hepatic L-PK silencing reduced mitochondrial respiration levels in a steatosis model, whereas it increased it in a fibrosis model. This apparently paradoxical finding may be related to the acute versus chronic adaptive responses of mitochondria. In a normal setting, liver mitochondria oxidize both glucose and fat, producing reducing equivalents that transfer electrons through different complexes of ETC to finally produce adenosine triphosphate (through adenosine triphosphate synthase) in a process known as oxidative phosphorylation (OXPHOS). Nevertheless, a small percentage of electron spillage occurs naturally and results in the generation of ROS. During the initial benign stages of NAFLD, such as insulin resistance and steatosis, there is an acute adaptive mitochondrial response initiated within the hepatocytes to defend against the augmented nutrient supply (such as fat and carbohydrates) to the liver. As a result, there is an increased production of reducing equivalents resulting in a hyperactive OXPHOS phenotype. Moreover, as an unavoidable consequence, this hyperactive OXPHOS also results in increased ROS generation that can directly damage mitochondrial DNA and ETC components among other things. Both of these impair electron transfer across the ETC, further resulting in electron spillage and ROS generation, thus creating a vicious cycle.32,33 Thus, the acute adaptive mitochondrial response in a chronic setting becomes maladaptive and has been proposed to be associated with the progression to later severe stages of NAFLD, such as NASH and fibrosis.32,33,35,36 In other words, in a steatosis setting, one could observe increased mitochondrial function (acute adaptive response), whereas in a NASH/fibrosis setting, there will be decreased mitochondrial function (chronic maladaptive changes). Because L-PK silencing decreased OXPHOS in steatosis and increased OXPHOS in fibrosis, the opposite of what is perceived in a pathologic scenario, our study demonstrates that L-PK silencing reverses both steatosis and fibrosis via regulating nutrient flux into mitochondria. This is important because some of the current therapeutic avenues targeting NAFLD aim at increasing mitochondrial function, with the notion that increasing fat oxidation will reduce NAFLD progression. However, this could be counterintuitive because steatosis is already accompanied by a hyperactive OXPHOS phenotype (acute adaptive response), which in long term results in excessive ROS generation and mitochondrial damage (chronic maladaptive changes).
Our studies also demonstrate that L-PK expression influences not only liver lipids and mitochondrial respiration but also systemic functions such as glucose metabolism, insulin sensitivity, and plasma cholesterol levels. Global gene expression analyses identified a surprisingly large number of hepatic genes affected by L-PK knockdown or overexpression (509 and 1731, respectively), including many lipogenic genes that were induced by L-PK overexpression in male mice. Indeed, follow-up stable isotope tracing analyses revealed that L-PK specifically affected the carbon flux from glucose/pyruvate oxidation into citrate. When high levels of citrate are produced (because of nutrient excess during steatosis), it is transported to the cytosol via mitochondrial citrate carrier resulting in FFA production via DNL.38 Citrate also allosterically activates the enzyme acetyl-CoA carboxylase that converts acetyl-coA into malonyl-coA, the committed step in DNL.39 Indeed, inhibiting mitochondrial pyruvate import40,41 or mitochondrial citrate export42 has been shown to reduce DNL and/or revert NAFLD/NASH. In addition, several investigators have shown that increased DNL is a hallmark of NAFLD in both mouse43,44 and human studies.45, 46, 47 Taken together, we believe L-PK–mediated regulation of pyruvate oxidation and DNL is causal in developing steatosis and eventually fibrosis, and limiting this could have a therapeutic potential against NAFLD.
A final important finding from our studies is the mechanistic validation of our previous network modeling involving integration of the steatosis phenotype with gene expression data across ∼100 diverse inbred strains of mice. In particular, we had observed that PNPLA3 gene, the strongest known genetic factor contributing to human NAFLD especially in the Hispanic population,48 was connected to L-PK through 2 edges via nodes Acyl or A4gnt. Also, we observed changes in 15 of 25 genes predicted to have edges to Pklr in our knockdown or overexpression studies. More interestingly, PCSK9, which was not part of our original network but was identified using human data,34 was found to be a direct target of L-PK through our loss- and gain-of-function mouse models. We also demonstrate here that L-PK not only affects the transcriptional status but also the protein levels of both PNPLA3 and PCSK9.
In conclusion, we originally identified L-PK as a causal candidate for hepatic steatosis in male mice fed a HF/HS diet,9 but our present findings strongly suggest that L-PK acts in a sex-specific manner and that it also contributed to the more advanced stages of NAFLD including fibrosis. A “two hit model” has been proposed for NASH, with one hit mediating steatosis and a second inflammation/fibrosis. This is based on the fact that only a subset of patients with steatosis progress to NASH. However, the underlying factors involved in NASH progression are poorly understood, and genetic variants of PNPLA3 clearly promote both steatosis and NASH.48 Because in our study, among other functions, L-PK also modulated both the RNA and protein levels PNPLA3, we believe L-PK could be a strong candidate added to the growing repertoire of potential NAFLD drugs. One important advantage of L-PK over the other candidates is that L-PK affects a mitochondrial metabolic process (conversion of glucose/pyruvate into adenosine triphosphate, ROS, and FFA) that has strong associations with both NAFLD development and progression.
Methods
Animals
All animals were purchased from the Jackson Laboratory and bred at UCLA according to approved IACUC protocols with daily monitoring by vivarium personnel. We have previously described both the HMDP and gonadectomy study design in detail.17 Briefly, 8-week-old males and females of ∼98 inbred strains were fed a HF/HS diet (Research Diets-D12266B) for 8 weeks. For the gonadectomy studies, at 6 weeks of age, both sexes of C57BL/6J mice were either gonadectomized or sham-operated under isoflurane anesthesia and placed on a HF/HS diet around 8 weeks of age. For NASH models, 8-week-old C57BL/6J-APOE-Leiden were fed a Western diet (Research Diets-D10042101) for 16 weeks,31 or C57BL/6J mice were fed CDAHFD (Research Diets-A06071302) for 12 weeks.30
HMDP Liver Gene Expression Analysis
Isolated RNA from HMDP liver tissues were used for global gene expression analysis using Affymetrix HT_MG430A arrays, and microarray data were filtered as previously described.17 We used ComBat method from the SVA Bioconductor package49 to remove known batch effects and bicorAndPvalue function of the Weighted Correlation Network Analysis package50 to calculate biweight midcorrelation between liver gene expression and phenotypes.
Human Study Population
The study detail and its characteristics have been reported elsewhere (Table 1).51,52 The study included a total of 144 Mexican subjects who underwent bariatric surgery for morbid obesity (body mass index ≥35 kg/m2). Blood samples were taken after a 10-hour overnight fast for biochemical measurements. During surgery, liver biopsies were collected and stored in RNAlater (Sigma-Aldrich, St Louis, MO) and later processed for RNA sequencing analyses as described previously.31 Liver biopsy specimens were also fixed in 10% formaldehyde, embedded in paraffin, stained with hematoxylin-eosin and Masson’s trichrome, and evaluated by an experienced pathologist according to the Kleiner scoring system.53 On the basis of the NAS, participants were classified as non-NASH (including subjects with normal liver histology and simple steatosis without inflammation) and NASH (NAS ≥5).53 This study was performed according to the principles of the Declaration of Helsinki and was approved by the institutional review boards of the National Institute of Genomic Medicine (Mexico) and the Hospital Dr. General Ruben Leñero. All participants provided written informed consent before their inclusion. Correlations of normalized values of RNA sequencing data with NAS or GGT levels were evaluated by using partial correlations adjusting for age, body mass index, and type 2 diabetes status. Because GGT levels were not normally distributed, the values were log-transformed before performing statistical analyses.
AAV Expression System
We modulated liver gene expression using AAV-mediated gene transfer, a technique successfully applied in our previous studies.54 For AAV overexpression of Pklr, the cDNA sequence (NM_001099779.1) was cloned into an AAV8 expression plasmid under a TBG promoter. To generate deficiency models, we knockdown Pklr, using the AAV8-TBG vector expressing shRNA sequences (ACCGCCTCAAGGAGATGATCAA) as described previously.54 AAV syntheses were carried out by the University of Pennsylvania vector core.
L-PK Gain- and Loss-of-Function Studies
AAVs were intravenously or intraperitoneally injected into 8-week-old male or female C57BL/6J mice (∼1 × 1012 genome copies diluted in 0.2 mL saline). Control mice were injected with either AAV8 expressing green fluorescent protein gene (for overexpression) or AAV8 expressing shRNA against firefly luciferase gene (for knockdown). Because AAV8-mediated gene manipulation provides long-term changes in expression, metabolic traits were evaluated multiple times over a period of several months. Body composition was determined by using NMR (Brüker Biospin Corp, Billerica, MA). On the day of death, mice were fasted for 4 hours, followed by their death and tissue extraction. Retro-orbital blood was collected to isolate plasma for analyzing glucose, insulin, and lipids; liver and gonadal fat tissues were collected for weight and/or lipid measurements.
Glucose, Insulin, and Lactate/Pyruvate Tolerance Tests
For GTT and ITT, animals were fasted for 6 hours before being injected intraperitoneally with glucose (1 g/kg) or insulin (1 U/kg), respectively. For L/PTT, animals were fasted for 16 hours before being injected intraperitoneally with 9:1 lactate to pyruvate ratio (2 g/kg). Glucometer measures were taken at regular time intervals.
Bioenergetics of Isolated Mitochondria
Isolated mitochondrial respiration from liver tissue was measured as described previously.55 Briefly, mitochondria were obtained by dual centrifugation, resuspended in respiration buffer, and kept on ice. Mitochondrial respiration was obtained with XF96 Seahorse bioanalyzer (Agilent Technologies, Santa Clara, CA). For the complex I respiration, the measures were collected in presence of 10 mmol/L pyruvate (Complex I substrate), 2 mmol/L malate, and 4 μmol/L FCCP. For the complex II respiration, the measures were collected in presence of 10 mmol/L succinate (Complex II substrate) and 2 mmol/L rotenone (Complex I inhibitor).
RNA Isolation, Library Preparation, and Sequencing
Flash-frozen liver samples on death were weighed and homogenized in QIAzol (Qiagen, Germantown, MD), and after chloroform phase separation, RNA was isolated according to the manufacturer’s protocol using miRNeasy columns (Qiagen). Libraries were prepared from these extracted liver RNA (Agilent 2200 Tapestation eRIN >7) using Illumina Stranded Total RNA Kit (Illumina, San Diego, CA) per the manufacturers’ instructions. The pooled libraries were sequenced with an Illumina NovaSeq 6000 instrument, PE50 bp reads (Illumina). Reads were aligned to the mouse genome mm10 using HISAT256 aligner and quantfiied using the Bioconductor R packages as described in the RNA-Seq workflow.57 P values were adjusted by using the Benjamini-Hochberg procedure of multiple hypothesis testing.58
RNA Isolation for qPCR
Total RNA was isolated from frozen liver tissues or AML12 cells using QIAzol (Qiagen) following manufacturer’s RNA isolation protocol. Next, first-strand complementary DNA (cDNA) was made from 2 μg total RNA according to the manufacturer’s protocol using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA). Relative quantitative gene expression levels were measured by qPCR using Kapa SYBR Fast qPCR kit (Kapa Biosystems, Inc, Wilmington, MA) on a LightCycler 480 II (Roche, Basel, Switzerland) and analyzed using the Roche LightCycler1.5.0 Software. The geometric mean of B2m and Tbp was used to normalize all qPCR targets.59 Relative normalized expression was measured using the equation 2-ΔΔCt. All qPCR primer sequences are listed here: B2m, forward: TACGTAACACAGTTCCACCCGCCTC, reverse: GCAGGTTCAAATGAATCTTCAGAGCATC; Tbp, forward: CAAACCCAGAATTGTTCTCCTT, reverse: ATGTGGTCTTCCTGAATCCCT; Pklr, forward: AGATGCAACATGCGATTGCC, reverse: GCACAGCACTTGAAGGAAGC; Fasn, forward: TGCACCTCACAGGCATCAAT, reverse: GTCCCACTTGATGTGAGGGG; Scd1, forward: TTCCCTCCTGCAAGCTCTAC, reverse: CAGAGCGCTGGTCATGTAGT; Elovl6, forward: ACACGTAGCGACTCCGAAGAT, reverse: CGTACAGCGCAGAAAACAGG.
Immunoblotting Analyses
Immunoblotting of liver samples was performed as described previously.60 Briefly, liver samples were lysed, and proteins were separated by using 4%-12% Bis-Tris gels (Invitrogen, Carlsbad, CA). Protein was then transferred to polyvinylidene difluoride membranes (Immobilon; Milllipore Sigma, Burlington, MA) and probed by using rabbit polyclonal PKLR (ThermoFisher #PA5-79824; Waltham, MA), goat polyclonal PCSK9 (R&D Systems #AF3985; Minneapolis, MN), goat polyclonal LDLR (R&D Systems #AF2255), mouse polyclonal PNPLA3 (Abcam # ab69170; Cambridge, United Kingdom), and rabbit monoclonal GAPDH (Cell Signaling #5174S; Danvers, MA) and their respective secondary antibodies. Bands were quantified by using ImageJ.
Metabolomics and Stable Isotope Tracing
All experiments were conducted using an Agilent 7890 gas chromatograph coupled to a 5977 mass spectrometer and collected using Agilent MassHunter software. AML12 cells were seeded on 6-well plates and transfected with either scr or si RNA against Pklr using TransIT-X2 Dynamic Delivery System (Mirus Bio LLC, Madison, WI). The next day medium was removed; cells were washed and replaced with medium containing [U-13C6]-glucose (Sigma-Aldrich) and grown for 24 hours before being analyzed. Samples were derivatized and analyzed as described previously.61
Data Availability
PKLR RNA sequencing raw data can be accessed at the Gene Expression Omnibus under accession GSE157203.
Statistical Analysis
Statistical analyses were performed using Prism v8.4.2 (GraphPad Software, San Diego, CA). Errors bars plotted on graphs are presented as the mean ± standard error of the mean (SEM) unless reported otherwise. The critical significance value (α) was set at .05, and if the P values were less than α, we reported that by rejecting the null hypothesis, the observed differences were statistically significant.
Acknowledgments
The authors thank Zhiqiang Zhou and Yonghong Meng for assistance in animal experiments; Sarada Charugundla for plasma metabolite and liver lipid analyses; Calvin Pan for informatics; Yeram Hong, Inshirah Kawsar, Sara Fardeheb, and Norma Marshall for their help in constructing the AAV plasmids; and Sereena Nand and Diana Anum for their help in mouse NMR analyses.
CRediT Authorship Contributions
Karthickeyan Chella Krishnan, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Supporting; Investigation: Lead; Methodology: Lead; Project administration: Lead; Supervision: Supporting; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead; Co-corresponding author: Equal)
Raquel R. Floyd, BS (Investigation: Equal)
Simon Sabir, BS (Investigation: Equal)
Dulshan W. Jayasekera, BS (Investigation: Equal)
Paola V. Leon-Mimila, PhD (Investigation: Supporting)
Anthony E. Jones, BS (Funding acquisition: Supporting; Investigation: Supporting)
Angel A. Cortez, BS (Investigation: Supporting)
Varun Shravah, BS (Investigation: Supporting)
Miklós Péterfy, PhD (Resources: Supporting)
Linsey Stiles, PhD (Investigation: Supporting)
Samuel Canizales-Quinteros, PhD (Resources: Supporting)
Ajit S. Divakaruni, PhD (Investigation: Supporting; Resources: Supporting; Supervision: Supporting; Visualization: Supporting)
Adriana Huertas-Vazquez, PhD (Funding acquisition: Supporting; Investigation: Supporting; Resources: Supporting; Supervision: Supporting; Visualization: Supporting)
Aldons J. Lusis, PhD (Conceptualization: Equal; Funding acquisition: Lead; Investigation: Supporting; Resources: Lead; Supervision: Lead; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Footnotes
Conflicts of interest This author discloses the following: A.S.D. has previously served as a paid consultant for Agilent Technologies. The remaining authors disclose no conflicts.
Funding Supported by NIH-P01HL028481 (A.J.L.), NIH-R01DK117850 (A.J.L.), NIH-K99DK120875 (K.C.K), AHA fellowship 18POST33990256 (K.C.K.), UCLA Seed grant (S.C.Q., A.H.V., A.J.L.), NIH-R35GM138003 (A.S.D.), NIH-P30DK063491 (A.S.D.), and UCLA Tumor Cell Biology Training grant T32CA009056 (A.E.J.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Karthickeyan Chella Krishnan, Email: kchellakrishnan@mednet.ucla.edu.
Aldons J. Lusis, Email: jlusis@mednet.ucla.edu.
Supplementary Material
References
- 1.Adams L.A., Lymp J.F., St Sauver J., Sanderson S.O., Lindor K.D., Feldstein A., Angulo P. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–121. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Browning J.D., Szczepaniak L.S., Dobbins R., Nuremberg P., Horton J.D., Cohen J.C., Grundy S.M., Hobbs H.H. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 3.Kopec K.L., Burns D. Nonalcoholic fatty liver disease: a review of the spectrum of disease, diagnosis, and therapy. Nutr Clin Pract. 2011;26:565–576. doi: 10.1177/0884533611419668. [DOI] [PubMed] [Google Scholar]
- 4.Marchesini G., Bugianesi E., Forlani G., Cerrelli F., Lenzi M., Manini R., Natale S., Vanni E., Villanova N., Melchionda N., Rizzetto M. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology. 2003;37:917–923. doi: 10.1053/jhep.2003.50161. [DOI] [PubMed] [Google Scholar]
- 5.Ratziu V., Bellentani S., Cortez-Pinto H., Day C., Marchesini G. A position statement on NAFLD/NASH based on the EASL 2009 special conference. J Hepatol. 2010;53:372–384. doi: 10.1016/j.jhep.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 6.de Alwis N.M., Day C.P. Non-alcoholic fatty liver disease: the mist gradually clears. J Hepatol. 2008;48(Suppl 1):S104–S112. doi: 10.1016/j.jhep.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 7.McCullough A.J. The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin Liver Dis. 2004;8:521–533. doi: 10.1016/j.cld.2004.04.004. viii. [DOI] [PubMed] [Google Scholar]
- 8.Bennett B.J., Farber C.R., Orozco L., Kang H.M., Ghazalpour A., Siemers N., Neubauer M., Neuhaus I., Yordanova R., Guan B., Truong A., Yang W.P., He A., Kayne P., Gargalovic P., Kirchgessner T., Pan C., Castellani L.W., Kostem E., Furlotte N., Drake T.A., Eskin E., Lusis A.J. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chella Krishnan K., Kurt Z., Barrere-Cain R., Sabir S., Das A., Floyd R., Vergnes L., Zhao Y., Che N., Charugundla S., Qi H., Zhou Z., Meng Y., Pan C., Seldin M.M., Norheim F., Hui S., Reue K., Lusis A.J., Yang X. Integration of multi-omics data from mouse diversity panel highlights mitochondrial dysfunction in non-alcoholic fatty liver disease. Cell Syst. 2018;6:103–115 e7. doi: 10.1016/j.cels.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan J.J., Fallon M.B. Gender and racial differences in nonalcoholic fatty liver disease. World J Hepatol. 2014;6:274–283. doi: 10.4254/wjh.v6.i5.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruhl C.E., Everhart J.E. Determinants of the association of overweight with elevated serum alanine aminotransferase activity in the United States. Gastroenterology. 2003;124:71–79. doi: 10.1053/gast.2003.50004. [DOI] [PubMed] [Google Scholar]
- 12.Clark J.M., Brancati F.L., Diehl A.M. The prevalence and etiology of elevated aminotransferase levels in the United States. Am J Gastroenterol. 2003;98:960–967. doi: 10.1111/j.1572-0241.2003.07486.x. [DOI] [PubMed] [Google Scholar]
- 13.Ioannou G.N., Boyko E.J., Lee S.P. The prevalence and predictors of elevated serum aminotransferase activity in the United States in 1999-2002. Am J Gastroenterol. 2006;101:76–82. doi: 10.1111/j.1572-0241.2005.00341.x. [DOI] [PubMed] [Google Scholar]
- 14.Lazo M., Hernaez R., Eberhardt M.S., Bonekamp S., Kamel I., Guallar E., Koteish A., Brancati F.L., Clark J.M. Prevalence of nonalcoholic fatty liver disease in the United States: the Third National Health and Nutrition Examination Survey, 1988-1994. Am J Epidemiol. 2013;178:38–45. doi: 10.1093/aje/kws448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenney-Hunt J.P., Wang B., Norgard E.A., Fawcett G., Falk D., Pletscher L.S., Jarvis J.P., Roseman C., Wolf J., Cheverud J.M. Pleiotropic patterns of quantitative trait loci for 70 murine skeletal traits. Genetics. 2008;178:2275–2288. doi: 10.1534/genetics.107.084434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittelstrass K., Ried J.S., Yu Z., Krumsiek J., Gieger C., Prehn C., Roemisch-Margl W., Polonikov A., Peters A., Theis F.J., Meitinger T., Kronenberg F., Weidinger S., Wichmann H.E., Suhre K., Wang-Sattler R., Adamski J., Illig T. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parks B.W., Sallam T., Mehrabian M., Psychogios N., Hui S.T., Norheim F., Castellani L.W., Rau C.D., Pan C., Phun J., Zhou Z., Yang W.P., Neuhaus I., Gargalovic P.S., Kirchgessner T.G., Graham M., Lee R., Tontonoz P., Gerszten R.E., Hevener A.L., Lusis A.J. Genetic architecture of insulin resistance in the mouse. Cell Metab. 2015;21:334–347. doi: 10.1016/j.cmet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varlamov O., Bethea C.L., Roberts C.T., Jr. Sex-specific differences in lipid and glucose metabolism. Front Endocrinol (Lausanne) 2014;5:241. doi: 10.3389/fendo.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White U.A., Tchoukalova Y.D. Sex dimorphism and depot differences in adipose tissue function. Biochim Biophys Acta. 2014;1842:377–392. doi: 10.1016/j.bbadis.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karp N.A., Mason J., Beaudet A.L., Benjamini Y., Bower L., Braun R.E., Brown S.D.M., Chesler E.J., Dickinson M.E., Flenniken A.M., Fuchs H., Angelis M.H., Gao X., Guo S., Greenaway S., Heller R., Herault Y., Justice M.J., Kurbatova N., Lelliott C.J., Lloyd K.C.K., Mallon A.M., Mank J.E., Masuya H., McKerlie C., Meehan T.F., Mott R.F., Murray S.A., Parkinson H., Ramirez-Solis R., Santos L., Seavitt J.R., Smedley D., Sorg T., Speak A.O., Steel K.P., Svenson K.L., International Mouse Phenotyping C., Wakana S., West D., Wells S., Westerberg H., Yaacoby S., White J.K. Prevalence of sexual dimorphism in mammalian phenotypic traits. Nat Commun. 2017;8:15475. doi: 10.1038/ncomms15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norheim F., Bjellaas T., Hui S.T., Chella Krishnan K., Lee J., Gupta S., Pan C., Hasin-Brumshtein Y., Parks B.W., Li D.Y., Bui H.H., Mosier M., Wu Y., Huertas-Vazquez A., Hazen S.L., Gundersen T.E., Mehrabian M., Tang W.H.W.H.W., Hevener A.L., Drevon C.A., Lusis A.J. Genetic, dietary, and sex-specific regulation of hepatic ceramides and the relationship between hepatic ceramides and IR. J Lipid Res. 2018;59:1164–1174. doi: 10.1194/jlr.M081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norheim F., Hasin-Brumshtein Y., Vergnes L., Chella Krishnan K., Pan C., Seldin M.M., Hui S.T., Mehrabian M., Zhou Z., Gupta S., Parks B.W., Walch A., Reue K., Hofmann S.M., Arnold A.P., Lusis A.J. Gene-by-sex interactions in mitochondrial functions and cardio-metabolic traits. Cell Metab. 2019;29:932–949 e4. doi: 10.1016/j.cmet.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metallo C.M., Gameiro P.A., Bell E.L., Mattaini K.R., Yang J., Hiller K., Jewell C.M., Johnson Z.R., Irvine D.J., Guarente L., Kelleher J.K., Vander Heiden M.G., Iliopoulos O., Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du J., Cleghorn W.M., Contreras L., Lindsay K., Rountree A.M., Chertov A.O., Turner S.J., Sahaboglu A., Linton J., Sadilek M., Satrustegui J., Sweet I.R., Paquet-Durand F., Hurley J.B. Inhibition of mitochondrial pyruvate transport by zaprinast causes massive accumulation of aspartate at the expense of glutamate in the retina. J Biol Chem. 2013;288:36129–36140. doi: 10.1074/jbc.M113.507285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vacanti N.M., Divakaruni A.S., Green C.R., Parker S.J., Henry R.R., Ciaraldi T.P., Murphy A.N., Metallo C.M. Regulation of substrate utilization by the mitochondrial pyruvate carrier. Mol Cell. 2014;56:425–435. doi: 10.1016/j.molcel.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Divakaruni A.S., Wallace M., Buren C., Martyniuk K., Andreyev A.Y., Li E., Fields J.A., Cordes T., Reynolds I.J., Bloodgood B.L., Raymond L.A., Metallo C.M., Murphy A.N. Inhibition of the mitochondrial pyruvate carrier protects from excitotoxic neuronal death. J Cell Biol. 2017;216:1091–1105. doi: 10.1083/jcb.201612067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto M., Hada N., Sakamaki Y., Uno A., Shiga T., Tanaka C., Ito T., Katsume A., Sudoh M. An improved mouse model that rapidly develops fibrosis in non-alcoholic steatohepatitis. Int J Exp Pathol. 2013;94:93–103. doi: 10.1111/iep.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hui S.T., Kurt Z., Tuominen I., Norheim F., Davis R.C., Pan C., Dirks D.L., Magyar C.E., French S.W., Chella Krishnan K., Sabir S., Campos-Perez F., Mendez-Sanchez N., Macias-Kauffer L., Leon-Mimila P., Canizales-Quinteros S., Yang X., Beaven S.W., Huertas-Vazquez A., Lusis A.J. The genetic architecture of diet-induced hepatic fibrosis in mice. Hepatology. 2018;68:2182–2196. doi: 10.1002/hep.30113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Begriche K., Igoudjil A., Pessayre D., Fromenty B. Mitochondrial dysfunction in NASH: causes, consequences and possible means to prevent it. Mitochondrion. 2006;6:1–28. doi: 10.1016/j.mito.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Pessayre D., Fromenty B. NASH: a mitochondrial disease. J Hepatol. 2005;42:928–940. doi: 10.1016/j.jhep.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Lee S., Zhang C., Liu Z., Klevstig M., Mukhopadhyay B., Bergentall M., Cinar R., Stahlman M., Sikanic N., Park J.K., Deshmukh S., Harzandi A.M., Kuijpers T., Grotli M., Elsasser S.J., Piening B.D., Snyder M., Smith U., Nielsen J., Backhed F., Kunos G., Uhlen M., Boren J., Mardinoglu A. Network analyses identify liver-specific targets for treating liver diseases. Mol Syst Biol. 2017;13:938. doi: 10.15252/msb.20177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caldwell S.H., Swerdlow R.H., Khan E.M., Iezzoni J.C., Hespenheide E.E., Parks J.K., Parker W.D., Jr. Mitochondrial abnormalities in non-alcoholic steatohepatitis. J Hepatol. 1999;31:430–434. doi: 10.1016/s0168-8278(99)80033-6. [DOI] [PubMed] [Google Scholar]
- 36.Sanyal A.J., Campbell-Sargent C., Mirshahi F., Rizzo W.B., Contos M.J., Sterling R.K., Luketic V.A., Shiffman M.L., Clore J.N. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120:1183–1192. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]
- 37.Aubert J., Begriche K., Knockaert L., Robin M.A., Fromenty B. Increased expression of cytochrome P450 2E1 in nonalcoholic fatty liver disease: mechanisms and pathophysiological role. Clin Res Hepatol Gastroenterol. 2011;35:630–637. doi: 10.1016/j.clinre.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 38.Ferramosca A., Zara V. Dietary fat and hepatic lipogenesis: mitochondrial citrate carrier as a sensor of metabolic changes. Adv Nutr. 2014;5:217–225. doi: 10.3945/an.113.004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin D.B., Vagelos P.R. The mechanism of tricarboxylic acid cycle regulation of fatty acid synthesis. J Biol Chem. 1962;237:1787–1792. [PubMed] [Google Scholar]
- 40.Rauckhorst A.J., Gray L.R., Sheldon R.D., Fu X., Pewa A.D., Feddersen C.R., Dupuy A.J., Gibson-Corley K.N., Cox J.E., Burgess S.C., Taylor E.B. The mitochondrial pyruvate carrier mediates high fat diet-induced increases in hepatic TCA cycle capacity. Mol Metab. 2017;6:1468–1479. doi: 10.1016/j.molmet.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCommis K.S., Finck B.N. Treating hepatic steatosis and fibrosis by modulating mitochondrial pyruvate metabolism. Cell Mol Gastroenterol Hepatol. 2019;7:275–284. doi: 10.1016/j.jcmgh.2018.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan M., Mosaoa R., Graham G.T., Kasprzyk-Pawelec A., Gadre S., Parasido E., Catalina-Rodriguez O., Foley P., Giaccone G., Cheema A., Kallakury B., Albanese C., Yi C., Avantaggiati M.L. Inhibition of the mitochondrial citrate carrier, Slc25a1, reverts steatosis, glucose intolerance, and inflammation in preclinical models of NAFLD/NASH. Cell Death Differ. 2020;27:2143–2157. doi: 10.1038/s41418-020-0491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knebel B., Fahlbusch P., Dille M., Wahlers N., Hartwig S., Jacob S., Kettel U., Schiller M., Herebian D., Koellmer C., Lehr S., Muller-Wieland D., Kotzka J. Fatty liver due to increased de novo lipogenesis: alterations in the hepatic peroxisomal proteome. Front Cell Dev Biol. 2019;7:248. doi: 10.3389/fcell.2019.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim C.-W., Addy C., Kusunoki J., Anderson N.N., Deja S., Fu X., Burgess S.C., Li C., Chakravarthy M., Previs S., Milstein S., Fitzgerald K., Kelley D.E., Horton J.D. Acetyl coA carboxylase inhibition reduces hepatic steatosis but elevates plasma triglycerides in mice and humans: a bedside to bench investigation. Cell Metabolism. 2017;26:394–406.e6. doi: 10.1016/j.cmet.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lambert J.E., Ramos-Roman M.A., Browning J.D., Parks E.J. Increased de novo lipogenesis is a distinct characteristic of individuals with nonalcoholic fatty liver disease. Gastroenterology. 2014;146:726–735. doi: 10.1053/j.gastro.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umpleby A.M., Shojaee-Moradie F., Fielding B., Li X., Marino A., Alsini N., Isherwood C., Jackson N., Ahmad A., Stolinski M., Lovegrove J.A., Johnsen S., Jeewaka R.M.A.S., Wright J., Wilinska M.E., Hovorka R., Bell J.D., Thomas E.L., Frost G.S., Griffin B.A. Impact of liver fat on the differential partitioning of hepatic triacylglycerol into VLDL subclasses on high and low sugar diets. Clin Sci (Lond) 2017;131:2561–2573. doi: 10.1042/CS20171208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwarz J.-M., Clearfield M., Mulligan K. Conversion of sugar to fat: is hepatic de novo lipogenesis leading to metabolic syndrome and associated chronic diseases? J Am Osteopath Assoc. 2017;117:520. doi: 10.7556/jaoa.2017.102. [DOI] [PubMed] [Google Scholar]
- 48.Trepo E., Romeo S., Zucman-Rossi J., Nahon P. PNPLA3 gene in liver diseases. J Hepatol. 2016;65:399–412. doi: 10.1016/j.jhep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gutiérrez-Vidal R., Vega-Badillo J., Reyes-Fermín L.M., Hernández-Pérez H.A., Sánchez-Muñoz F., López-Álvarez G.S., Larrieta-Carrasco E., Fernández-Silva I., Méndez-Sánchez N., Tovar A.R., Villamil-Ramírez H., Mejía-Domínguez A.M., Villarreal-Molina T., Hernández-Pando R., Campos-Pérez F., Aguilar-Salinas C.A., Canizales-Quinteros S. SFRP5 hepatic expression is associated with non-alcoholic liver disease in morbidly obese women. Annals of Hepatology. 2015;14:666–674. [PubMed] [Google Scholar]
- 52.León-Mimila P., Vega-Badillo J., Gutiérrez-Vidal R., Villamil-Ramírez H., Villareal-Molina T., Larrieta-Carrasco E., López-Contreras B.E., Kauffer L.R.M., Maldonado-Pintado D.G., Méndez-Sánchez N., Tovar A.R., Hernández-Pando R., Velázquez-Cruz R., Campos-Pérez F., Aguilar-Salinas C.A., Canizales-Quinteros S. A genetic risk score is associated with hepatic triglyceride content and non-alcoholic steatohepatitis in Mexicans with morbid obesity. Exp Mol Pathol. 2015;98:178–183. doi: 10.1016/j.yexmp.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 53.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.C., Torbenson M.S., Unalp-Arida A., Yeh M., McCullough A.J., Sanyal A.J., Nonalcoholic Steatohepatitis Clinical Research N Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 54.Chella Krishnan K., Sabir S., Shum M., Meng Y., Acin-Perez R., Lang J.M., Floyd R.R., Vergnes L., Seldin M.M., Fuqua B.K., Jayasekera D.W., Nand S.K., Anum D.C., Pan C., Stiles L., Peterfy M., Reue K., Liesa M., Lusis A.J. Sex-specific metabolic functions of adipose lipocalin-2. Mol Metab. 2019;30:30–47. doi: 10.1016/j.molmet.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogers G.W., Brand M.D., Petrosyan S., Ashok D., Elorza A.A., Ferrick D.A., Murphy A.N. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Love M.I., Anders S., Kim V., Huber W. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000Res. 2015;4:1070. doi: 10.12688/f1000research.7035.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 59.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seldin M.M., Koplev S., Rajbhandari P., Vergnes L., Rosenberg G.M., Meng Y., Pan C., Phuong T.M.N., Gharakhanian R., Che N., Makinen S., Shih D.M., Civelek M., Parks B.W., Kim E.D., Norheim F., Chella Krishnan K., Hasin-Brumshtein Y., Mehrabian M., Laakso M., Drevon C.A., Koistinen H.A., Tontonoz P., Reue K., Cantor R.M., Bjorkegren J.L.M., Lusis A.J. A strategy for discovery of endocrine interactions with application to whole-body metabolism. Cell Metab. 2018;27:1138–1155 e6. doi: 10.1016/j.cmet.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cordes T., Metallo C.M. Quantifying intermediary metabolism and lipogenesis in cultured mammalian cells using stable isotope tracing and mass spectrometry. Methods Mol Biol. 2019;1978:219–241. doi: 10.1007/978-1-4939-9236-2_14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PKLR RNA sequencing raw data can be accessed at the Gene Expression Omnibus under accession GSE157203.
Statistical Analysis
Statistical analyses were performed using Prism v8.4.2 (GraphPad Software, San Diego, CA). Errors bars plotted on graphs are presented as the mean ± standard error of the mean (SEM) unless reported otherwise. The critical significance value (α) was set at .05, and if the P values were less than α, we reported that by rejecting the null hypothesis, the observed differences were statistically significant.