Dear Editor,
Recent articles in this Journal have suggested the potential of antigen-based rapid diagnostic tests (Ag-RDT) as low-cost and ease-of-use tools for massive screening and epidemiological surveillance of SARS-CoV-2 spread.1 , 2 Based on a pre-screening of four Ag-RDT on 40 frozen specimens from nasopharyngeal swabs with known PCR results (Table S1, Appendix), we selected the Panbio COVID-19 Ag Test (Abbott) for investigating its analytical and clinical performance.
The analysis of serial dilutions of a SARS-CoV-2 isolate, propagated in Vero E6 cells, yielded a limit of detection (LoD) of 6.5 × 105 genome copies/reaction (Table S2). According to this value, the test would not detect SARS-CoV-2 infection in respiratory specimens with very low viral load. Still, the LoD was one logarithmic unit below the 106 copies/mL threshold necessary for successful virus isolate from respiratory samples.3
The clinical performance was analysed on frozen swabs from 1406 individuals (mean age 40.4 years; SD 24.5) with an RT-qPCR result available: 951 (67.6%) positive and 455 (32.4%) negative. Overall, 446 (31.7%) and 473 (33.6%) samples were nasopharyngeal swabs from symptomatic individuals and contacts exposed to symptomatic cases, respectively, and 487 (34.6%) were nasal mid-turbinate swabs from asymptomatic individuals collected in screening campaigns. The cycle threshold (Ct) of PCR-positive samples was <20, 20–24, 25–29, and >30 in 258 (17.1%), 305 (32.1%), 285 (30.0%), and 103 (10.8%), respectively (median Ct 23.6; interquartile range 19.7–27.3).
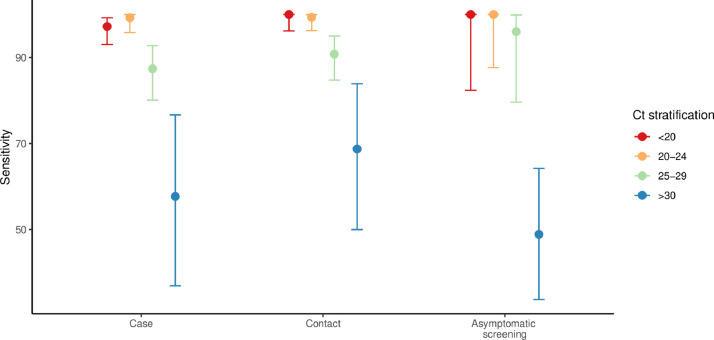
Overall, the Ag-RDT identified the presence of SARS-CoV-2 in 872 of 951 PCR-positive samples (sensitivity 91.7%; 95% CI 89.8–93.4) and ruled out its presence in 450 of 455 PCR-negative samples (specificity 98.9%; 97.5–99.6)(Table S3). In line with previous reports of clinical performance of this test,2 sensitivity increased with lower Ct values (Ct <25, 98.2%; Ct<30, 94.9%) and was higher among samples collected in the setting of case identification (92.6%) and contact tracing (94.2%) than asymptomatic screening (79.5%). The increasing trend of sensitivity with lower Ct values was maintained in the setting of asymptomatic screening: sensitivity of samples with Ct <25 and <30 were 100% and 98.6%, respectively (Fig. 1 ). The high sensitivity of the Ag-RDT in samples with low Ct values, which was consistent with the LoD, has important implications for using this test as an epidemiological surveillance tool. A growing body of evidence indicates that PCR-positive respiratory specimens with low viral load (i.e., Ct >25 or <106 copies/mL) have a limited capacity for effective transmission.3 , 4 Also, studies based on contact tracing strategies showed that the secondary attack rate significantly increases among index cases with Ct values <25.5 The higher sensitivity of the Ag-RDT in samples with low Ct, irrespective of the presence of symptoms, indicates that the test is particularly suitable for identifying individuals who are contagious.
Fig. 1.
SARS-CoV-2 detection using the Ag-RDT Panbio COVID-19 Ag-Test on PCR-positive samples according to rt-qPCR Ct value. Sensitivity (95CI) of the Ag-RDT according to the disease status and RT-qPCR Ct value.
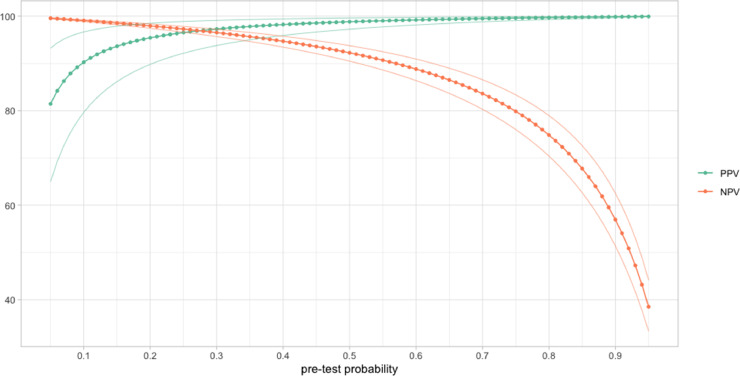
Like most studies investigating the clinical performance of RDTs,6 our assessment was retrospectively on frozen samples from three different settings. Our internal validation showed no relevant differences between tests performed on fresh samples using the Abbot test Kit buffer and 1:3 dilutions of the Kit buffer and frozen specimens stored on transport media. Although we encourage internal validation before using this approach, our experience suggests the suitability of parallel sampling for Ag-RDT and PCR tests. Another consequence of not sampling in the intended setting was the impossibility of direct estimates of the positive predictive value (PPV) and negative predictive value (NPV). Alternatively, we modelized the PPV and NPV assuming a wide range of prevalence (i.e., pre-test probability) in the target population (Fig. 2 ). At a pre-test probability of 5%, consistent with the prevalence observed in asymptomatic screening campaigns in high-risk settings,7 the NPV was 99.6% (99.5–99.7) (Table S4) and increased as the pre-test probability dropped. Correspondingly, the PPV at 5% pre-test probability was 81.5% (65.0–93.2) and decreased as pre-test probability decreased. At 5% pre-test probability, the estimated number of false-negative and false-positive values per thousand tests were 4 (3–5) and 12 (4–27), respectively (TableS5).
Fig. 2.
Modelling of positive predictive value (PPV) and negative predictive value (NPV) assuming different pre-test probabilities. Dots represent the PPV and NPV at sequential increment of 0.01; lines are the 95% confidence interval . (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Our analytical and clinical performance findings suggest that the Ag-RDT cannot replace nucleic acid amplification tests (NAAT) as a tool to confirm or rule out the presence of SARS-CoV-2. However, the high sensitivity of NAAT has raised questions as to the clinical and epidemiological meaning of being positive for SARS-CoV-2 infection. In some patients, low levels of viral RNA can remain detectable by RT-qPCR for months, with doubtful transmission capacity. The Ag-RDT reliably identifies people with high viral loads, and therefore it could be useful for screening strategies to identify and isolate asymptomatic COVID-19 people while they are still infectious. Although the kinetics of SARS-CoV-2 have not been well established, current evidence suggests that this time window may begin approximately four days after exposure and last for nearly ten days.8 Of note, during the exponential phase, viral RNA may rise from undetectable levels (i.e., Ct >40) to millions of RNA copies/mL (i.e., Ct < 25) in the order of a day,9 thus limiting the temporal validity of a negative result.
Currently, the WHO recommends using Ag-RDTs to support the diagnosis of cases and contacts during outbreak investigations and monitor trends in disease incidence, particularly in remote settings or closed groups (e.g., schools, care homes, or prisons), but not to screen asymptomatic populations.10 However, our findings suggest that Ag-RDT might be useful for screening asymptomatic individuals, particularly in communities with high prevalence. Furthermore, the high sensitivity for detecting infected individuals with transmission capacity makes the test suitable for creating safe environments in time-limited social activities with high-risk of transmission, including―but not limited to―visiting relatives at nursing homes, playing sports, going to a crowded place like movie theatres, music concerts, and airports.
Contributors
OM, AA, BB, CGB, IB, JV designed the study. AA, BB, MU, MCM, JR, LR performed the laboratory procedures, and organized the data. DO did statistical analysis. OM wrote the first draft with revisions and input from JR, JS, CE, GF, QB, BC, JA, MVM, CGB. All authors approved the final version.
Declaration of Competing Interest
We declare no conflicts of interest.
Acknowledgments
Acknowledgements
The authors would like to thank Gerard Carot-Sans (PhD) for providing professional medical writing support during the preparation of the manuscript. We thank Andrea Tiburcio Lara and Elisabeth Bascuñana Prieto for technical support
Bárbara Baró is a Beatriu de Pinós postdoctoral fellow granted by the Government of Catalonia's Secretariat for Universities and Research, and by Marie Sklodowska-Curie Actions COFUND Programme (BP3, 801370).
Funding
Blueberry diagnostics, Fundació Institut d'Investigació en Ciències de la Salut Germans Trias i Pujol, and #YoMeCorono.org crowfunding campaing.
References
- 1.Azzi L., Baj A., Alberio T., Lualdi M., Veronesi G., Carcano G., et al. Rapid Salivary Test suitable for a mass screening program to detect SARS-CoV-2: a diagnostic accuracy study. J Infect. 2020;81(3) doi: 10.1016/j.jinf.2020.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agulló V., Fernández-González M., Ortiz de la Tabla V., Gonzalo-Jiménez N., García J.A., Masiá M., et al. Evaluation of the rapid antigen test Panbio COVID-19 in saliva and nasal swabs in a population-based point-of-care study. J Infect. 2020;0(0) doi: 10.1016/j.jinf.2020.12.007. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. May 28. [DOI] [PubMed] [Google Scholar]
- 4.Singanayagam A., Patel M., Charlett A., Bernal J.L., Saliba V., Ellis J., et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance. 2020;25(32) doi: 10.2807/1560-7917.ES.2020.25.32.2001483. Aug 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marc M., Millat P., Ouchi D., Roberts C., Alemany A., Corbacho-Monne M., et al. Transmission of Covid-19 in 282 clusters in Catalonia, Spain: a cohort study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30985-3. Manuscript accepted (in Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deeks J.J., Dinnes J., Takwoingi Y., Davenport C., Spijker R., Taylor-Phillips S., et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. 2020;(6) doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oran D.P., Topol E.J. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann Intern Med. 2020 doi: 10.7326/M20-3012. doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larremore D.B., Wilder B., Lester E., Shehata S., Burke J.M., Hay J.A., et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv. 2020:eabd5393. doi: 10.1126/sciadv.abd5393. Nov 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith A.M., Perelson A.S. Influenza A virus infection kinetics: quantitative data and models. Wiley Interdiscip Rev Syst Biol Med. 2011;3(4):429–445. doi: 10.1002/wsbm.129. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization (WHO) 2020. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays [Internet]https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays [cited 2020 Sep 29]. Available from: [Google Scholar]