Abstract
Background
Argiope bruennichi, the European wasp spider, has been investigated intensively as a focal species for studies on sexual selection, chemical communication, and the dynamics of rapid range expansion at a behavioral and genetic level. However, the lack of a reference genome has limited insights into the genetic basis for these phenomena. Therefore, we assembled a high-quality chromosome-level reference genome of the European wasp spider as a tool for more in-depth future studies.
Findings
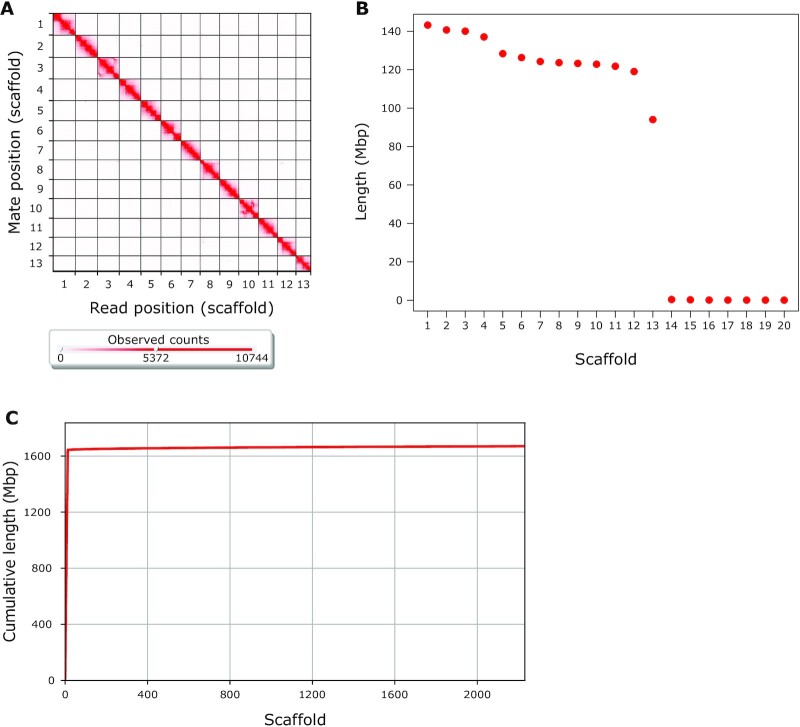
We generated, de novo, a 1.67 Gb genome assembly of A. bruennichi using 21.8× Pacific Biosciences sequencing, polished with 19.8× Illumina paired-end sequencing data, and proximity ligation (Hi-C)-based scaffolding. This resulted in an N50 scaffold size of 124 Mb and an N50 contig size of 288 kb. We found 98.4% of the genome to be contained in 13 scaffolds, fitting the expected number of chromosomes (n = 13). Analyses showed the presence of 91.1% of complete arthropod BUSCOs, indicating a high-quality assembly.
Conclusions
We present the first chromosome-level genome assembly in the order Araneae. With this genomic resource, we open the door for more precise and informative studies on evolution and adaptation not only in A. bruennichi but also in arachnids overall, shedding light on questions such as the genomic architecture of traits, whole-genome duplication, and the genomic mechanisms behind silk and venom evolution.
Keywords: Argiope bruennichi, genome assembly, Araneae, spider, PacBio, Hi-C, chromosome-level, Hox duplication, silk, venom
Data description
Context
Spider genomes are of great interest, e.g., in the context of silk and venom evolution and biomedical and technical applications. In addition, spiders are fascinating from ecological and evolutionary perspectives. As the most important predators of terrestrial arthropods, they play a key role in terrestrial food webs [1–4]. Spiders are distributed on every continent except Antarctica, and diverse habitats can be occupied by single species or multiple close relatives [5, 6], making them ideal for studies on environmental plasticity, adaptation, and speciation. With regards to adaptation, work on cobweb spiders (Theridiidae) has revealed a whole-genome duplication (WGD) that may have facilitated diversification [7], with other studies highlighting a key role of tandem duplication and neofunctionalization of genes in the diversification and specialization of spider silks [8] and venoms [9]. A key aspect that has been missing from studies to date is the role of genome organization in promoting or impeding adaptation because there have been no studies on spiders that have provided a chromosomal framework for the genome.
Understanding the chromosomal organization of a genome is critical for identification of processes underlying divergence between populations, adaptation, and speciation. Indeed, the potential role of chromosomal reorganization in species formation has long been the subject of debate, in particular in Drosophila species, where polytene chromosomes allowed early visualization of chromosomal rearrangements [10]. For spiders, karyotype data are still used to identify changes in chromosomes associated with speciation [11]. With the advent of detailed genomic data, there has been renewed focus on the role that structural variants in the genome can play as drivers of adaptation and speciation, associated with translocations, fusions, and inversions [12], as well as with admixture and associated demographic changes [13]. Recent data from sister species of the genus Drosophila suggest that the establishment of inversion polymorphisms within isolated and/or heterogeneous environments may well set the stage for species formation [14]. To develop a broader understanding of the role of structural variation in adaptation and speciation [15–22], we need chromosome-level genomes that provide the ability to map the order of genes, define chromosomal gene neighborhoods, and identify potential genomic islands of differentiation [23–26].
To the best of our knowledge, 10 draft spider genomes have been published to date [7, 27–33], most of which focus on silk and venom genes, while one discusses WGD [7], and the publication of the most recent two focuses on gene content evolution across arthropods [33]. There is one additional, as yet unpublished, spider genome assembly available on NCBI (Anelosimus studiosus, accession No. GCA_008297655.1). Spider genomes are considered notoriously difficult to sequence, assemble, and annotate for a number of reasons, including their relatively high repeat content, low guanine cytosine (GC) content, high levels of heterozygosity in the wild [27], and owing to the fact that they possess some extremely long coding genes in the spidroin gene families [28, 29, 34, 35]. As a result of these challenges, the completeness of the available spider genomes varies greatly between assemblies (Supplementary Table S1). All of them are incomplete and there is no chromosome-level assembly published for any spider to date. While this does not diminish the conclusions of the aforementioned studies, a chromosome-level assembly would open doors for more detailed studies on the genomic architecture of gene families, such as silk and venom genes, providing greater understanding of the evolutionary mechanisms driving the diversification of these gene families and genome evolution, in addition to the aforementioned applications in understanding adaptation and speciation.
The European wasp spider, Argiope bruennichi (Scopoli, 1772), is an orb-weaving spider in the family Araneidae (Fig. 1). Despite the lack of a reference genome, A. bruennichi has been the focal species for studies on local adaptation, range expansion, admixture, and biogeography [5, 36–38]. These studies have suggested that the range expansion and subsequent local adaptation of A. bruennichi from southern to northern Europe was caused by genetic admixture. However, it is not yet known which regions of the genome are admixed and whether these regions are truly responsible for adaptation to colder climates. A. bruennichi has also been well studied in the context of dispersal and life history traits [39], as well as sexual selection and chemical communication (e.g., [40–44]). A high-quality reference genome would allow new insights into our understanding of the genetic basis of these phenomena. Considering this background, a chromosome-level reference genome would be desirable for the species.
Figure 1:
Female Argiope bruennichi spider in orb web from Loulé (Faro, Portugal). Photograph by Monica M. Sheffer.
Sampling, DNA extraction, and sequencing
Adult female Argiope bruennichi individuals (NCBI:txid94029) were collected in the south of Portugal in 2013 and 2019 (37° 44.34' N, 7° 51.18' W). Because inbred lines of the species do not exist, we selected a population that was previously found to have low heterozygosity in the wild, likely due to naturally high levels of inbreeding [5].
For the baseline assembly, DNA was extracted from a female collected in 2013 using the ArchivePure blood and tissue kit (5 PRIME, Hamburg, Germany), according to the manufacturer's protocol. An RNA digestion step was included using RNAse A solution (7,000 U mL−1; 5 PRIME). The DNA was stored at −80°C until library preparation in 2017. The DNA extract was cleaned using a salt: phenol chloroform isoamyl alcohol cleaning step and had a fragment size distribution of 1,300–165,500 bp (peak at 14,002 bp) before size selection. The library was size selected to 15 kb using Pippin prep and subsequently sequenced in 2018 at the QB3 Genomics facility at the University of California Berkeley on a Pacific Biosciences Sequel I platform (PacBio, Menlo Park, CA, USA) on 10 cells.
The specimen collected in 2019 was used to build a proximity-ligation-based short-read library (Hi-C). Four Hi-C libraries were prepared from a single individual using a DovetailTM Hi-C library preparation kit according to the manufacturer's protocol (Dovetail Genomics, Santa Cruz, CA). The specimen was anesthetized with CO2 before preparation. In brief, the legs were removed from the body and stored in liquid nitrogen, and the leg tissue was disrupted in liquid nitrogen using a mortar and pestle. Chromatin was fixed with formaldehyde, then extracted. Fixed chromatin was digested with DpnII, the 5′ overhangs filled in with biotinylated nucleotides, and the free blunt ends were ligated. After ligation, cross-links were reversed and the DNA was purified to remove proteins. Purified DNA was treated to remove biotin that was not internal to ligated fragments. The DNA was then sheared to ∼350 bp mean fragment size using a Covaris S2 Focused-ultrasonicator. A typical Illumina library preparation protocol followed, with end repair and Illumina adapter ligation. Biotinylated fragments were captured with streptavidin beads before PCR amplification (12 cycles), and size selection was performed using SPRI-select beads (Beckman Coulter GmbH, Germany) for a final library size distribution centered at ∼450 bp. The library was sequenced to ∼440 million paired-end reads on 1 Flowcell of an Illumina NextSeq 550 with a High Output v2 kit (150 cycles).
Genome size estimation and coverage
We estimated the genome size of Argiope bruennichi on the basis of data for closely related species, and bioinformatically on the basis of previously published Illumina paired-end data derived from a single female individual from a population in Madeira (SRA accession No. ERX533198) [5], which we later used for polishing the assembly.
The closely related species Argiope aurantia and Argiope trifasciata have genome size estimates based on Feulgen densitometry data of 1.620 Gb [45] or 1.650 Gb [46] for A. aurantia and 1.690 Gb for A. trifasciata [45, 47]. Using the backmap.pl (v. 0.3) pipeline [48–55] on the Illumina data from A. bruennichi [5], we generated a genome size estimate of 1.740 Gb. Averaging these 4 genome size measurements yields an estimate of 1.675 Gb.
Given this estimate, the PacBio sequencing yielded 21.8× coverage (∼36.65 Gb sequenced, with an estimated genome size of 1.67 Gb). The previously published Illumina data [5] have a coverage of 19.8× (33.05 Gb sequenced).
De novo genome assembly
First, we generated a baseline assembly using 21.8× long-read PacBio Sequel I sequencing data and the wtdbg2 assembler (v. 2.3) (WTDBG, RRID:SCR_017225) [56]. Next, we polished the assembly by applying 3 rounds of Pilon (v. 1.23) (Pilon, RRID:SCR_014731) [57] using the 19.8× of previously published Illumina paired-end data [5]. Mapping for the 3 rounds of polishing resulted in a mapping rate ranging from 92.55% to 93.69%. The polishing resulted in 13,843 contigs with an N50 of 288.4 kb, and an overall assembly size of 1.67 Gb. Analysis of BUSCO (v. 3.1.0) scores, using the arthropod dataset (BUSCO, RRID:SCR_015008) [58], showed the presence of 90.2% of complete BUSCOs, with 86.4% complete and single-copy BUSCOs, 3.8% complete and duplicated BUSCOs, 3.3% fragmented BUSCOs, and 6.5% missing BUSCOs (Table 1). Next, we scaffolded the contigs using a proximity-ligation-based short-read library [59]. The sequences from this library had a 94.71% mapping rate against the polished assembly. Scaffolding using HiRise v. 2.1.7, a software pipeline designed specifically for using proximity ligation data to scaffold genome assemblies [59], resulted in 12 scaffolds >1 Mb in size and 1 scaffold just slightly less than 1 Mb in size. These 13 scaffolds comprise 98.4% of the assembly, with a genome assembly scaffold N50 of 124 Mb and BUSCO scores of 91.1% complete genes (Fig. 2, Table 1). Genome assembly statistics were calculated using QUAST v. 5.0.2 (QUAST, RRID:SCR_001228) [60] applying default parameters, except setting the minimum contig length (–min-contig parameter) to 0. Previous studies have inferred the chromosome number of A. bruennichi to be 13, indicating that our genome assembly achieved full-chromosome level [61, 62]. As an additional assessment of assembly quality, we ran the K-mer Analysis Toolkit v. 2.4.2 (KAT, RRID:SCR_016741) [63] “comp" tool, comparing the k-mer content in the Illumina sequencing data to the k-mer content in the final assembly. Different values of the parameter k (k = 17, 27, 29, 30, and 37) yielded k-mer completeness estimates ranging from 86.55% to 90.43% (Supplementary Fig. S1). The missing k-mer content in the final assembly may be attributed to errors remaining in the assembly, likely within repeat regions. This could be attributed to the moderate 19.8× coverage Illumina reads used for polishing and their short read length, which may have been insufficient to correct the more error-prone PacBio reads.
Table 1:
Argiope bruennichi genome assembly completeness
| Genome assembly statistic | Unscaffolded | Scaffolded |
|---|---|---|
| Assembly size (bp) | 1,669,116,561 | 1,670,285,661 |
| AT/GC/N content (%) | 70.7/29.3/0 | 70.6/29.3/0.1 |
| No. of contigs/scaffolds | 13,843 | 2,231 |
| Longest contig/scaffold (bp) | 2,039,454 | 143,171,375 |
| Contig/scaffold N50 (bp) | 288,395 | 124,235,998 |
| Contig/scaffold N90 (bp) | 67,231 | 119,022,586 |
| % Repetitive | 34.66 | 34.64 |
| BUSCO analysisa | ||
| Complete (%) | 90.2 | 91.1 |
| Complete and single-copy (%) | 86.4 | 87.8 |
| Complete and duplicated (%) | 3.8 | 3.3 |
| Fragmented (%) | 3.3 | 2.8 |
| Missing (%) | 6.5 | 6.1 |
Genome assembly statistics were calculated using QUAST v. 5.0.2 (QUAST, RRID:SCR_001228) [60] using default parameters, except –min-contig 0. AT: adenine thymine.
BUSCO analysis using default parameters against the arthropod dataset.
Figure 2:
Argiope bruennichi genome assembly completeness. (A) Contact heat map of Hi-C scaffolding shows long-range contacts of paired-end Hi-C reads. Gray gridlines denote scaffold (chromosome) boundaries. Visualized with Juicebox (v. 1.11.08) [64]. (B) The length of the 20 longest scaffolds in the assembly shows that the 13 putative chromosome scaffolds are much larger than the next largest. Red points represent individual scaffolds, ordered from largest to smallest. (C) Cumulative length of assembly contained within scaffolds. Note that almost all (98.4%) of the genome is contained within very few scaffolds. Visualized with QUAST v. 5.0.2 [60] using default parameters, except –min-contig 0.
The 13 largest scaffolds are henceforth referred to as Chromosomes 1–13, ordered according to size (Fig. 2B). The 14th-largest scaffold (Scaffold 839) contained the 16S sequence of a recently discovered, as yet unnamed, bacterial symbiont of A. bruennichi [48]. The remaining 2,217 scaffolds are much smaller, ranging from 1,747 to 258,743 bp in length (Supplementary Fig. S2) and will henceforth be referred to as “lesser scaffolds.”
Repeat masking and removal of contaminants
The assembly was repeat-masked using a combination of the de novo repeat finder RepeatModeler (v. open-1.0.11) (RepeatModeler, RRID:SCR_015027) [65] and the homology-based repeat finder RepeatMasker (v. open-4.0.9) (RepeatMasker, RRID:SCR_012954) [66]. Repetitive regions accounted for 34.64% of the genome assembly, of which the majority (20.52% of the genome) consisted of unclassified repeats, meaning that they have not been classified in previous studies. The remaining repetitive elements were made up of DNA elements (i.e., transposable elements: 6.27%), long interspersed nuclear elements (LINEs: 1.60%), simple repeats (i.e., duplications of 1–5 bp: 1.58%), long terminal repeat (LTR) elements (0.76%), satellites (0.63%), low-complexity repeats (i.e., polypurine or polypyrimidine stretches: 0.42%), and short interspersed nuclear elements (SINEs: 0.08%) (Table 2). BlobTools (v. 1.0) (Blobtools, RRID:SCR_017618) [67] was used to search for contamination by bacterial or mitochondrial sequences, finding none.
Table 2:
Argiope bruennichi repetitive DNA elements
| Type of element | No. of elements | Length (bp) | Proportion of assembly (%) |
|---|---|---|---|
| SINEs | 4,643 | 1,314,740 | 0.08 |
| LINEs | 52,648 | 26,768,096 | 1.60 |
| LTR elements | 21,649 | 12,683,330 | 0.76 |
| DNA elements | 282,019 | 104,785,665 | 6.27 |
| Unclassified | 1,359,138 | 342,727,030 | 20.52 |
| Small RNA | 0 | 0 | 0 |
| Satellites | 28,474 | 10,495,658 | 0.63 |
| Simple repeats | 595,962 | 26,379,486 | 1.58 |
| Low complexity | 137,182 | 6,952,634 | 0.42 |
| Total | 34.64 |
Genome annotation
Raw reads from previously published transcriptome sequencing data of different life stages: 20 pooled eggs (accession No. SRR11861505), 20 pooled first instar spiderlings (accession No. SRR11861504), 1 whole body of an adult female (accession No. SRR11861502), and 1 whole body of an adult male (accession No. SRR11861503) [5] were mapped against the repeat-masked assembly using HISAT2 (v. 2.1.0) (HISAT2, RRID:SCR_015530) [68]. After conversion of the resulting SAM file into a BAM file and subsequent sorting using SAMtools (v. 1.7) (SAMTOOLS, RRID:SCR_002105) [49], the sorted BAM file was converted to intron-hints for AUGUSTUS (v. 3.3.2) (Augustus, RRID:SCR_008417) [69] using AUGUSTUS scripts. AUGUSTUS was run on the soft-masked genome with the Parasteatoda parameter set. The resulting gff file containing predicted genes was converted into a gtf file using the AUGUSTUS script gtf2gff.pl. Additional AUGUSTUS scripts (getAnnoFastaFromJoinGenes.py and fix_in_frame_stop_codon_genes.py) were used to find and replace predicted protein-coding genes containing in-frame stop codons with newly predicted genes. The resulting gtf file containing 23,270 predicted genes (26,318 transcripts) was converted to gff3 format using gtf2gff.pl and protein sequences of predicted genes were extracted with getAnnoFastaFromJoinGenes.py. Finally, functional annotation was performed using InterProScan (v. 5.39–77.0) (InterProScan, RRID:SCR_005829) [70, 71] (Table 3). The majority of annotated genes fall on the 13 chromosome scaffolds, although 272 transcripts were predicted on the lesser scaffolds. The annotation gff3 file and the files containing predicted transcripts and proteins are available on GigaDB [72].
Table 3:
Argiope bruennichi genome annotation statistics
| Statistic | Value |
|---|---|
| No. of protein-coding genes | 23,270 |
| Functionally annotated genes (%) | 81.0 |
| Mean exon length (bp) | 200 |
| Mean intron length (bp) | 4,035 |
| BUSCO analysisa | |
| Complete (%) | 89.3 |
| Complete and single-copy (%) | 76.7 |
| Complete and duplicated (%) | 12.6 |
| Fragmented (%) | 7.0 |
| Missing (%) | 3.7 |
BUSCO analysis using default parameters against the arthropod dataset.
Comparative genomic analysis of repeat content
High repetitiveness is characteristic of spider genomes [27]. To compare the repeat content of A. bruennichi with that of other spiders, we downloaded the genome assemblies of several other spider species from NCBI and DDBJ (accession numbers in Table 4), then treated them in the same manner as the A. bruennichi genome, masking the repeats using RepeatModeler (v. open-1.0.11) [65] and RepeatMasker (v. open-4.0.9) [66]. Acanthoscurria geniculata was excluded from this analysis owing to the relatively poorly assembled genome. The A. bruennichi genome has a slightly lower percentage of repetitive element content (34.64%) compared to most other spiders (Table 4). Some species, such as Loxosceles reclusa, Trichonephila clavipes (formerly Nephila clavipes), Anelosimus studiosus, and Parasteatoda tepidariorum, have similar repetitive content (36.51%, 36.61%, 35.98%, and 36.79%, respectively); other species have much higher repetitive content, such as Araneus ventricosus, Dysdera silvatica, Stegodyphus dumicola, Stegodyphus mimosarum, and Pardosa pseudoannulata (55.96%, 60.03%, 58.98%, 56.91%, and 48.61%, respectively). Only Latrodectus hesperus has lower repetitive content (20.97%). The classification and relative percentage of these repeats can be found in Supplementary Table S2 and Supplementary Fig. S3. It is often asserted that the repeat content in spiders is higher in general than in other arthropod groups (i.e., [27]). To test this assertion, we looked into the repeat content in genomes of additional arthropod species. We obtained repeat content estimates, for which the repeats were masked using RepeatModeler and RepeatMasker, for 3 insect species (Bombus terrestris, Drosophila melanogaster, and Rhodnius prolixus [73]) and 7 tick and mite species (Ixodes persulcatus, Haemaphysalis longicornis, Dermacentor silvarum, Hyalomma asiaticum, Rhipicephalus sanguineus,Rhipicephalus microplus, andIxodes scapularus [74]). We additionally downloaded the genomes of 4 more arthropod species, generated custom species-specific repeat libraries with RepeatModeler, and masked the genomes with RepeatMasker to avoid any issues of under- or overmasking using other repeat-masking programs: a butterfly, Heliconius melpomene [75]; a beetle, Tribolium castaneum [76]; a millipede, Helicorthomorpha holstii [77]; and a scorpion, Centruroides sculpturatus [7, 33]. The percentage of total repetitive content for all of these species is presented in Table 4. In general, spiders do have a higher repetitive content than insects, but there is a large range of repetitive content in spiders, compared to which the repetitive content in A. bruennichi is relatively low. All of the selected spider species, aside from L. hesperus, have higher repetitive content than all other investigated groups, with the exception of ticks and mites, which have very high repetitive content overall (range: 52.6–64.4% repetitive). We conclude from this preliminary investigation that spider genomes, and arachnid genomes generally, do indeed have a higher repeat content than other arthropods.
Table 4:
Total repetitive content in the genomes of spiders and selected other arthropods
| Class | Order | Species | % Repetitive | Accession No. [reference] |
|---|---|---|---|---|
| Arachnida | Araneae | Argiope bruennichi | 34.64 | |
| Araneus ventricosus | 55.96 | BGPR01000001-BGPR01300721a [29] | ||
| Trichonephila clavipes | 36.61 | GCA_002102615.1b [28] | ||
| Dysdera silvatica | 60.03 | GCA_006491805.1b [32] | ||
| Stegodyphus dumicola | 58.98 | GCA_010614865.1b [31] | ||
| Stegodyphus mimosarum | 56.91 | GCA_000611955.2b [27] | ||
| Pardosa pseudoannulata | 48.61 | GCA_008065355.1b [30] | ||
| Loxosceles reclusa | 36.51 | GCA_001188405.1b [33] | ||
| Anelosimus studiosus | 35.98 | GCA_008297655.1b,c | ||
| Latrodectus hesperus | 20.97 | GCA_000697925.2b [33] | ||
| Parasteatoda tepidariorum | 36.79 | GCA_000365465.3b [7] | ||
| Scorpiones | Centruroides sculpturatus | 34.40 | GCA_000671375.2b [7, 33] | |
| Acari | Ixodes persulcatus | 64.40 | GCA_013358835.1b [74] | |
| Haemaphysalis longicornis | 59.30 | GCA_013339765.1b [74] | ||
| Dermacentor silvarum | 60.20 | GCA_013339745.1b [74] | ||
| Hyalomma asiaticum | 52.60 | GCA_01333685.1b [74] | ||
| Rhipicephalus sanguineus | 61.60 | GCA_013339695.1b [74] | ||
| Rhipicephalus microplus | 63.10 | GCA_013339725.1b [74] | ||
| Ixodes scapularis | 63.50 | GCF_002892825.2b [74, 78] | ||
| Diplopoda | Helminthomorpha | Helicorthomorpha holstii | 23.50 | GCA_013389785.1b [77] |
| Insecta | Hemiptera | Rhodnius prolixus | 29.25 | GCA_000181055.3b [73] |
| Hymenoptera | Bombus terrestris | 12.51 | GCA_000214255.1b [73] | |
| Coleoptera | Tribolium castaneum | 28.50 | GCA_000002335.3b [76] | |
| Lepidoptera | Heliconius melpomene | 32.40 | GCA_000313835.2b [75] | |
| Diptera | Drosophila melanogaster | 19.31 | GCA_000001215.4b [73] |
Genome architecture of Hox, spidroin, and venom genes
Previous studies on spider genomes have focused on WGD, silk gene evolution, and venom gene evolution [7, 27–30]. Therefore, to place the A. bruennichi genome into the same context, we manually curated 3 gene sets from publicly available protein sequences: Hox, spidroin (silk), and venom genes. Because Hox genes are highly conserved across taxa [79], we chose the most complete sequences for the 10 arthropod Hox gene classes from spiders without regard to the relatedness of the species to A. bruennichi (Supplementary File S1). In contrast to Hox genes, spidroin and venom genes are highly polymorphic and species specific [80–83]. For the spidroin gene set, we downloaded protein sequences of the 7 spidroin gene classes exclusively from 5 species of the genus Argiope (Supplementary File S2). Venom genes are best studied in spiders that are medically significant to humans, which are very distant relatives to A. bruennichi [84–87]. To allow comparison, we focused on venom gene sequences available for araneid spiders (2 species, Supplementary File S3); however, the function and classification of these genes is poorly understood. With these 3 gene sets (Hox, spidroin, and venom), we performed a TBLASTN search against our genome assembly (v. 2.10.0+) (TBLASTN, RRID:SCR_011822) [88, 89]. We recorded the genomic position of the best matches and compared them with the AUGUSTUS gene predictions for those locations. We used a conservative E-value cut-off of <1.00 × 10−20 and only included results with an identity >60%. If hits overlapped on a scaffold or mapped to the same gene, only the hit with the highest identity and lowest E-value was retained. In cases where these metrics conflicted, the hit with the longest match length was retained. The manually curated FASTA files of each gene set used for the TBLASTN search are available in Supplementary Files S1–S3 and on GigaDB [72]. A table of the best matches with accession numbers for each gene set is available in Supplementary Tables S3–S5.
Hox cluster duplication
In 2017, Schwager et al. revealed that a WGD event occurred in the ancestor of scorpions and spiders, as evidenced by a high number of duplicated genes, including 2 clusters of Hox genes in the common house spider P. tepidariorum and the bark scorpion C. sculpturatus [7]. They found 1 nearly complete cluster of Hox genes on a single scaffold, lacking the fushi tarazu (ftz) gene, which they argued may be the case for this cluster in all spiders. The second set of Hox genes was distributed across 2 scaffolds, which the authors attributed to incompleteness of the assembly due to patchy sequencing coverage [7]. For consistency, we use the same nomenclature for Hox genes as used in [7] (Abdominal-B: AbdB, Abdominal-A: AbdA, Ultrabithorax: Ubx, Antennapedia: Antp, fushi tarazu: ftz, sex combs reduced: scr, Deformed: Dfd, Hox3, proboscipedia: pb, labial: lab). Corresponding with the results from P. tepidariorum, we found 2 clusters of Hox genes in A. bruennichi, with no evidence of tandem duplication. The 2 clusters occurred on 2 chromosomes (Chromosomes 6 and 9). In these locations, InterProScan generally annotated the genes as Hox genes but did not identify the specific type. On Chromosome 9, the Hox genes were in reverse colinear order (ordered according to their expression in development), with no overlapping regions. Because the cluster on Chromosome 9 is complete, we refer to it as “Cluster A.” On Chromosome 6 (“Cluster B”) the genes were out of colinear order, with the position of AbdA and Ubx switched, and the coordinates for Dfd, Hox3, and pb from the blast search overlapping (Fig. 3A). The hits for Antp and ftz in Cluster B fell onto a single predicted gene in the annotation. Thus, it is unclear whether A. bruennichi lacks 1 copy of ftz, as in P. tepidariorum, or whether the annotation incorrectly fused the 2 genes in this cluster. In the study by Schwager et al. [7], low sequencing coverage of Cluster B downstream of Dfd limited their inference. In our genome assembly, by mapping the PacBio reads against the final assembly, we calculated that we have an average of >12× coverage across the length of both clusters, suggesting that Cluster B is not out of order due to problems arising from low coverage. It is possible that Hox Cluster B in spiders has changed or lost functionality following the proposed ancestral WGD event. To check whether the 2 Hox-containing chromosomes show evidence of duplication, we performed an analysis of conserved synteny using the tool SatsumaSynteny2 [90]. “Synteny” here refers to loci occurring on the same chromosome; chromosomes with conserved synteny will have a high degree of syntenic blocks in common. In the genome of A. bruennichi, Chromosomes 6 and 9 show a high level of conserved synteny (Fig. 3B). The presence of 2 Hox clusters on highly syntenic chromosomes in our assembly is suggestive, but not evidence, of WGD in A. bruennichibecause it could also have arisen from duplication of only the ancestral Hox-containing chromosome; future studies will be able to capitalize on the now-available chromosome-level assemblies for several groups (e.g., horseshoe crabs, ticks, and our spider) [74, 91] to perform more detailed analyses of duplication across chelicerates.
Figure 3:
Duplication of the Hox-containing chromosomes. (A) Hox gene clusters. Genes connected by a black line occur on the same scaffold, in the order depicted. Cluster A occurs on Chromosome 9, and Cluster B occurs on Chromosome 6. (B) A synteny plot of the results of SatsumaSynteny2 [90] visualized in Circos [92] shows chromosome-scale conservation of synteny for the Hox-containing chromosomes (Chromosomes 6 and 9). The 2 curved rectangles represent Chromosomes 6 and 9, and the tick marks represent the position on the chromosome, in megabase pairs. Lines between the 2 rectangles show the shared syntenic blocks between the chromosomes, based on sequence homology. The presence of 2 Hox gene clusters on 2 highly syntenic chromosomes is suggestive of whole-genome duplication in Argiope bruennichi, as was found previously for Parasteatoda tepidariorum [7].
Spidroin genes
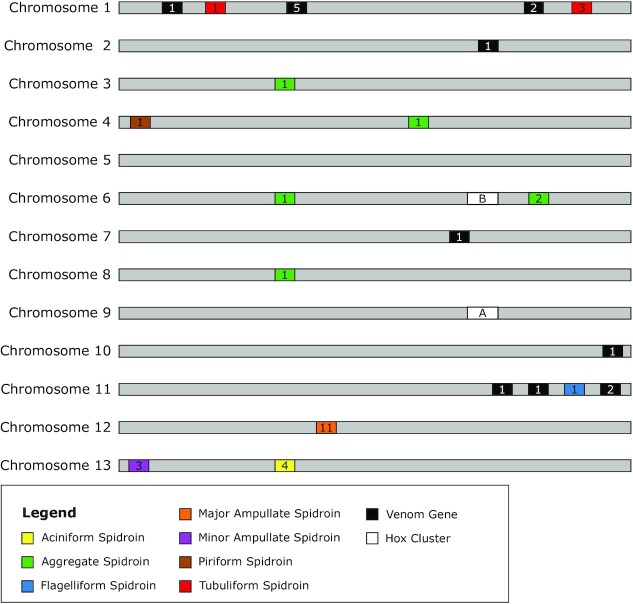
There are 7 classes of silk produced by araneomorph spiders, each with 1 or more unique uses; it is important to note that the uses of these silk types are best understood for spiders in the family Araneidae, and the number and uses of silk types can vary widely between families [28, 29, 93, 94]. The classes of silk are major ampullate (MaSp), minor ampullate (MiSp), piriform (PiSp), aggregate (AgSp), aciniform (AcSp), tubuliform (also referred to as cylindrical) (TuSp), and flagelliform (Flag). In A. bruennichi, spidroin genes occur on 8 of the 13 chromosome scaffolds (Chromosomes 1, 3, 4, 6, 8, 11, 12, and 13) (Fig. 4). There were no hits on the lesser scaffolds. We found 4 unique hits for AcSp, 6 hits for AgSp, 1 hit for Flag, 11 hits for MaSp, 3 hits for MiSp, 1 hit for PiSp, and 4 hits for TuSp. In the majority of cases, all blast hits for a single spidroin type occurred on a single chromosome; the only exception was for AgSp, which had hits on 4 different chromosomes. However, these were not all annotated as spidroins; on Chromosome 6 there were 2 AgSp hits that were annotated as spidroins and 1 hit that was annotated as a chitin-binding domain, while on Chromosome 4 the AgSp hit was annotated as tropoelastin, on Chromosome 3 the hit was annotated as a chitin-binding domain, and on Chromosome 8 the hit was annotated as a serine protease. All hits for TuSp occurred on Chromosome 1, but there were hits in 2 physically separated areas of the chromosome; in 1 region there were hits on 3 annotated genes, and only 1 hit in the other region. There are more sequences available on NCBI for MaSp than any of the other spidroin types in the genus Argiope, which allowed us to find matches for several unique MaSp genes in the A. bruennichi assembly. These occur in a small region of Chromosome 12, in close proximity to one another, suggesting that the spidroin genes in this group may have diversified via tandem duplication, as has been suggested in previous studies [95].
Figure 4:
Schematic representation of the location of gene families on the 13 chromosomes. The light grey bars represent chromosomes, the coloured rectangles represent the 7 different spidroin gene families, the black rectangles represent venom genes, and the white rectangles represent Hox gene clusters. The numbers inside of the rectangles represent the number of genes found within that cluster.
Venom genes
We found high identity matches for venom toxins on 5 of the chromosome scaffolds (Chromosomes 1, 2, 7, 10, and 11) (Fig. 4), but the majority of hits were on Chromosome 1. In most cases, each region containing venom gene matches contained only 1 gene, with the exception of a region on Chromosome 1, which contained 5 genes in very close proximity to one another, and 2 other regions (on Chromosome 1 and Chromosome 11), which contained matches to 2 genes. Babb et al. 2017 [28] conducted a study on silk genes in T. clavipes, in which they found a novel flagelliform-type gene (FLAG-b), which was expressed most highly in the venom glands, not the flagelliform silk glands. This added to previous findings in the S. mimosarum genome, where spidroin-like proteins are also found in the venom glands [27]. Interestingly, in the A. bruennichi genome assembly, there are several venom genes on Chromosome 11 in close proximity to the flagelliform spidroin gene.
Conclusions
We have assembled and annotated the first chromosome-level genome for a spider. The assembly approach of combining long-read, short-read, and proximity ligation data overcame the challenges of assembling arachnid genomes, namely, large genome size, high repetitiveness, and low GC content. In our study, we made a preliminary analysis of the location of certain gene families of interest in the context of spider genomics, which hinted at several interesting directions for future studies on the evolution of silk and venom genes. Furthermore, because this species has undergone a recent and rapid range expansion, the well-resolved genome assembly will be useful for studies on the genomic underpinnings of range expansion and evolutionary adaptation to novel climates.
Data Availability
The final genome assembly and raw data from the PacBio and Hi-C libraries, as well as the annotation, have been deposited at NCBI under BioProject PRJNA629526. A publicly accessible genome browser hub with the annotation, raw transcriptome, and PacBio read coverage can be found on the UCSC Genome Browser server (under “My Data” > “Track Hubs” > “My Hubs” enter the cited URL [96]). Supporting data are available via the GigaScience data repository, GigaDB, including the softmasked assembly in FASTA format, the output file from RepeatMasker, predicted coding genes and their functional annotation in GFF3 formats, predicted coding gene nucleotide and translated sequences in FASTA formats, functional annotation from InterProScan in TSV format, the blast query results for Hox, spidroin, and venom genes in FASTA format, and the BUSCO output files in a zip folder [72].
Additional Files
Supplementary Figure S1. KAT plots
Supplementary Figure S2. Histogram of minor scaffold lengths
Supplementary Figure S3. Stacked barplot of repeat content in spiders
Supplementary File S1. Hox blast query sequences
Supplementary File S2. Spidroin blast query sequences
Supplementary File S3. Venom blast query sequences
Supplementary Table S1. Spider genome assembly statistics
Supplementary Table S2. Repetitive content in spiders
Supplementary Table S3. Hox blast results
Supplementary Table S4. Spidroin blast results
Supplementary Table S5. Venom blast results
Abbreviations
Abd-A: Abdominal-A; Abd-B: Abdominal-B; AcSp: aciniform spidroin;AgSp: aggregate spidroin;Antp: Antennapedia; AT: adenine thymine; bp: base pairs; BUSCO: Benchmarking Universal Single Copy Orthologs; DDBJ: DNA Data Bank of Japan;Dfd: Deformed; Flag: flagelliform spidroin;ftz: fushi tarazu; Gb: gigabase pairs; GC: guanine cytosine; kb: kilobase pairs;lab: labial; LINE: long interspersed nuclear element; LTR: long terminal repeat;MaSp: major ampullate spidroin; Mb: megabase pairs;MiSp: minor ampullate spidroin; NCBI: National Center for Biotechnology Information; PacBio: Pacific Biosciences;pb: proboscipedia; PiSp: piriform spidroin; scr: sex combs reduced; SINE: short interspersed nuclear element; SRA: Sequence Read Archive; TSV: tab-separated value;TuSp: tubuliform spidroin;Ubx: Ultrabithorax; UCSC: University of California Santa Cruz; WGD: whole-genome duplication.
Competing Interests
The authors declare that they have no competing interests.
Funding
Funding for this study was provided by the Deutsche Forschungsgemeinschaft (DFG) as part of the Research Training Group 2010 RESPONSE (GRK 2010) to G.U.
Authors’ Contributions
M.M.S., H.K., G.U., and S.P. conceived of the study; M.M.S., H.K., and G.U. collected the spiders. H.K. extracted DNA for the PacBio sequencing; M.M.S. prepared and submitted the DNA for PacBio sequencing, with input and infrastructure provided by R.G.G. M.M.S. and C.J. constructed and sequenced the Hi-C library, with input and infrastructure provided by L.J. and A.W.K. M.M.S., A.H., and S.P. performed the genome assembly, and A.H. and K.J.H. performed the genome annotation with input and infrastructure provided by M.M.S. and S.P. A.H. and K.J.H. analysed the repeat content of other arthropod species; M.M.S. performed the analysis of Hox duplication, spidroin genes, and venom genes. M.M.S., A.H., K.J.H., and S.P. wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Jessica Garb -- 7/7/2020 Reviewed
Jessica Garb -- 11/3/2020 Reviewed
Jean-François Flot -- 8/23/2020 Reviewed
Jean-François Flot -- 11/10/2020 Reviewed
ACKNOWLEDGEMENTS
We thank the California Academy of Sciences for allowing us access to their computing resources for the genome assembly, and Dovetail Genomics for their support in troubleshooting the Hi-C kit and running HiRise. M.M.S. thanks José Cerca for helpful ideas and discussions about the silk and venom gene analysis.
Contributor Information
Monica M Sheffer, Zoological Institute and Museum, University of Greifswald, Loitzer Str. 26, 17489 Greifswald, Germany.
Anica Hoppe, Institute of Mathematics and Computer Science, University of Greifswald, Walther-Rathenau-Str. 47, 17489 Greifswald, Germany; Center for Functional Genomics of Microbes, University of Greifswald, Felix-Hausdorf-Str. 8, 17489 Greifswald, Germany.
Henrik Krehenwinkel, Department of Biogeography, University of Trier, Universitätsring 15, 54296 Trier, Germany.
Gabriele Uhl, Zoological Institute and Museum, University of Greifswald, Loitzer Str. 26, 17489 Greifswald, Germany.
Andreas W Kuss, Center for Functional Genomics of Microbes, University of Greifswald, Felix-Hausdorf-Str. 8, 17489 Greifswald, Germany; Interfaculty Institute for Genetics and Functional Genomics, University of Greifswald, Felix-Hausdorf-Str. 8, 17489 Greifswald, Germany.
Lars Jensen, Center for Functional Genomics of Microbes, University of Greifswald, Felix-Hausdorf-Str. 8, 17489 Greifswald, Germany; Interfaculty Institute for Genetics and Functional Genomics, University of Greifswald, Felix-Hausdorf-Str. 8, 17489 Greifswald, Germany.
Corinna Jensen, Center for Functional Genomics of Microbes, University of Greifswald, Felix-Hausdorf-Str. 8, 17489 Greifswald, Germany; Interfaculty Institute for Genetics and Functional Genomics, University of Greifswald, Felix-Hausdorf-Str. 8, 17489 Greifswald, Germany.
Rosemary G Gillespie, Department of Environmental Science Policy and Management, University of California Berkeley, 130 Mulford Hall #3114, Berkeley, CA, 94720, USA.
Katharina J Hoff, Institute of Mathematics and Computer Science, University of Greifswald, Walther-Rathenau-Str. 47, 17489 Greifswald, Germany; Center for Functional Genomics of Microbes, University of Greifswald, Felix-Hausdorf-Str. 8, 17489 Greifswald, Germany.
Stefan Prost, LOEWE-Centre for Translational Biodiversity Genomics, Senckenberganlage 25, 60325 Frankfurt, Germany; South African National Biodiversity Institute, National Zoological Gardens of South Africa, 232 Boom St., Pretoria 0001, South Africa.
References
- 1. Wise DH. Spiders in Ecological Webs. Cambridge: Cambridge University Press; 1993. [Google Scholar]
- 2. Spiller DA, Schoener TW. Effects of top and intermediate predators in a terrestrial food web. Ecology. 1994;75:182–96. [Google Scholar]
- 3. Moulder BC, Reichle DE. Significance of spider predation in the energy dynamics of forest-floor arthropod communities. Ecol Monogr. 1972;42:473–98. [Google Scholar]
- 4. Wirta HK, Weingartner E, Hambäck PA, et al. Extensive niche overlap among the dominant arthropod predators of the High Arctic. Basic Appl Ecol. 2015;16:86–92. [Google Scholar]
- 5. Krehenwinkel H, Rödder D, Tautz D. Eco-genomic analysis of the poleward range expansion of the wasp spider Argiope bruennichi shows rapid adaptation and genomic admixture. Glob Change Biol. 2015;21:4320–32. [DOI] [PubMed] [Google Scholar]
- 6. Garb JE, González A, Gillespie RG. The black widow spider genus Latrodectus (Araneae: Theridiidae): Phylogeny, biogeography, and invasion history. Mol Phylogenet Evol. 2004;31:1127–42. [DOI] [PubMed] [Google Scholar]
- 7. Schwager EE, Sharma PP, Clarke T, et al. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 2017;15:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clarke TH, Garb JE, Hayashi CY, et al. Spider transcriptomes identify ancient large-scale gene duplication event potentially important in silk gland evolution. Genome Biol Evol. 2015;7:1856–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gendreau KL, Haney RA, Schwager EE, et al. House spider genome uncovers evolutionary shifts in the diversity and expression of black widow venom proteins associated with extreme toxicity. BMC Genomics. 2017;18:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carson HL, Clayton FE, Stalker HD. Karyotypic stability and speciation in Hawaiian Drosophila. Proc Natl Acad Sci U S A. 1967;57:1280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Řezáč M, Arnedo MA, Opatova V, et al. Taxonomic revision and insights into the speciation mode of the spider Dysdera erythrina species-complex (Araneae: Dysderidae): Sibling species with sympatric distributions. Invertebr Syst. 2018;32:10–54. [Google Scholar]
- 12. Mérot C, Oomen RA, Tigano A, et al. A roadmap for understanding the evolutionary significance of structural genomic variation. Trends Ecol Evol. 2020;35:561–72. [DOI] [PubMed] [Google Scholar]
- 13. Shchur V, Svedberg J, Medina P, et al. On the distribution of tract lengths during adaptive introgression. G3 (Bethesda). 2020;10:3663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fuller ZL, Koury SA, Phadnis N, et al. How chromosomal rearrangements shape adaptation and speciation: Case studies in Drosophila pseudoobscura and its sibling species Drosophila persimilis. Mol Ecol. 2019;28:1283–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Faria R, Navarro A. Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends Ecol Evol. 2010;25:660–9. [DOI] [PubMed] [Google Scholar]
- 16. White MJD. Chromosomal rearrangements and speciation in animals. Annu Rev Genet. 1969;3:75–98. [Google Scholar]
- 17. Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–8. [DOI] [PubMed] [Google Scholar]
- 18. Noor MAF, Gratos KL, Bertucci LA, et al. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A. 2001;98:12084–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yannic G, Basset P, Hausser J. Chromosomal rearrangements and gene flow over time in an inter-specific hybrid zone of the Sorex araneus group. Heredity (Edinb). 2009;102:616–25. [DOI] [PubMed] [Google Scholar]
- 20. Feulner PGD, De-Kayne R. Genome evolution, structural rearrangements and speciation. J Evol Biol. 2017;30:1488–90. [DOI] [PubMed] [Google Scholar]
- 21. Castiglia R. Sympatric sister species in rodents are more chromosomally differentiated than allopatric ones: Implications for the role of chromosomal rearrangements in speciation. Mamm Rev. 2014;44, doi: 10.1111/mam.12009. [DOI] [Google Scholar]
- 22. Wellenreuther M, Mérot C, Berdan E, et al. Going beyond SNPs: The role of structural genomic variants in adaptive evolution and species diversification. Mol Ecol. 2019;28:1203–9. [DOI] [PubMed] [Google Scholar]
- 23. Vijay N, Bossu CM, Poelstra JW, et al. Evolution of heterogeneous genome differentiation across multiple contact zones in a crow species complex. Nat Commun. 2016;7, doi: 10.1038/ncomms13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hejase HA, Salman-Minkov A, Campagna L, et al. Genomic islands of differentiation in a rapid avian radiation have been driven by recent selective sweeps. Proc Natl Acad Sci U S A. 2020;117(48):30554–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duranton M, Allal F, Fraïsse C, et al. The origin and remolding of genomic islands of differentiation in the European sea bass. Nat Commun. 2018;9(1):2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanggaard KW, Bechsgaard JS, Fang X, et al. Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun. 2014;5:3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Babb PL, Lahens NF, Correa-Garhwal SM, et al. The Nephila clavipes genome highlights the diversity of spider silk genes and their complex expression. Nat Genet. 2017;49:895–903. [DOI] [PubMed] [Google Scholar]
- 29. Kono N, Nakamura H, Ohtoshi R, et al. Orb-weaving spider Araneus ventricosus genome elucidates the spidroin gene catalogue. Sci Rep. 2019;9:8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu N, Li J, Liu M, et al. Genome sequencing and neurotoxin diversity of a wandering spider Pardosa pseudoannulata (pond wolf spider). bioRxiv. 2019, doi: 10.1101/747147. [DOI] [Google Scholar]
- 31. Liu S, Aagaard A, Bechsgaard J, et al. DNA methylation patterns in the social spider, Stegodyphus dumicola. Genes (Basel). 2019;10:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sánchez-Herrero JF, Frías-López C, Escuer P, et al. The draft genome sequence of the spider Dysdera silvatica (Araneae, Dysderidae): A valuable resource for functional and evolutionary genomic studies in chelicerates. Gigascience. 2019;8:giz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thomas GWC, Dohmen E, Hughes DST, et al. Gene content evolution in the arthropods. Genome Biol. 2020;21:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stellwagen SD, Renberg RL. Toward spider glue: Long read scaffolding for extreme length and repetitious silk family genes AgSp1 and AgSp2 with insights into functional adaptation. G3 (Bethesda). 2019;9:1909–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ayoub NA, Garb JE, Kuelbs A, et al. Ancient properties of spider silks revealed by the complete gene sequence of the prey-wrapping silk protein (AcSp1). Mol Biol Evol. 2013;30:589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krehenwinkel H, Tautz D. Northern range expansion of European populations of the wasp spider Argiope bruennichiis associated with global warming-correlated genetic admixture and population-specific temperature adaptations. Mol Ecol. 2013;22:2232–48. [DOI] [PubMed] [Google Scholar]
- 37. Wawer W, Rutkowski R, Krehenwinkel H, et al. Population structure of the expansive wasp spider (Argiope bruennichi) at the edge of its range. J Arachnol. 2017;45:361–9. [Google Scholar]
- 38. Krehenwinkel H, Graze M, Rödder D, et al. A phylogeographical survey of a highly dispersive spider reveals eastern Asia as a major glacial refugium for Palaearctic fauna. J Biogeogr. 2016;43:1583–94. [Google Scholar]
- 39. Wolz M, Klockmann M, Schmitz T, et al. Dispersal and life-history traits in a spider with rapid range expansion. Mov Ecol. 2020;8, doi: 10.1186/s40462-019-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fromhage L, Uhl G, Schneider JM. Fitness consequences of sexual cannibalism in female Argiope bruennichi. Behav Ecol Sociobiol. 2003;55:60–4. [Google Scholar]
- 41. Schneider JM, Fromhage L, Uhl G. Extremely short copulations do not affect hatching success in Argiope bruennichi (Araneae, Araneidae). J Arachnol. 2005;33:663–9. [Google Scholar]
- 42. Schneider J, Uhl G, Herberstein ME. Cryptic female choice within the genus Argiope: A comparative approach. In: Peretti A, Aisenberg A, eds. Cryptic Female Choice in Arthropods. Cham: Springer; 2015:55–77. [Google Scholar]
- 43. Chinta SP, Goller S, Lux J, et al. The sex pheromone of the wasp spider Argiope bruennichi. Angew Chem Int Ed Engl. 2010;49:2033–6. [DOI] [PubMed] [Google Scholar]
- 44. Uhl G, Zimmer SM, Renner D, et al. Exploiting a moment of weakness: male spiders escape sexual cannibalism by copulating with moulting females. Sci Rep. 2015;5:16928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gregory TR, Shorthouse DP. Genome sizes of spiders. J Hered. 2003;94:285–90. [DOI] [PubMed] [Google Scholar]
- 46. Rasch EM, Connelly BA. Genome size and endonuclear DNA replication in spiders. J Morphol. 2005;265:209–14. [DOI] [PubMed] [Google Scholar]
- 47. Gregory TR. Animal Genome Size Database. 2020. http://www.genomesize.com/index.php. Accessed on August 28, 2020. [Google Scholar]
- 48. Schell T, Feldmeyer B, Schmidt H, et al. An annotated draft genome for Radix auricularia (Gastropoda, Mollusca). Genome Biol Evol. 2017;9:585–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013: 1303.3997. [Google Scholar]
- 51. Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 53. Ewels P, Magnusson M, Lundin S, et al. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruan J, Li H. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 2020;17:155–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Walker BJ, Abeel T, Shea T, et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Simão FA, Waterhouse RM, Ioannidis P, et al. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2. [DOI] [PubMed] [Google Scholar]
- 59. Putnam NH, O'Connell BL, Stites JC, et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 2016;26:342–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gurevich A, Saveliev V, Vyahhi N, et al. QUAST: Quality Assessment Tool for genome assemblies. Bioinformatics. 2013;29:1072–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang YJ, Tong SJ. The routine method for preparing the chromosomes in spiders. Chinese J Zool. 1990;25:30–1. [Google Scholar]
- 62. Araujo D, Mattos VF, Giroti AM, et al. Cytogenetical characterization of six orb-weaver species and review of cytogenetical data for Araneidae. J Arachnol. 2011;39:337–44. [Google Scholar]
- 63. Mapleson D, Accinelli GG, Kettleborough G, et al. KAT: A K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics. 2017;33:574–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Durand NC, Robinson JT, Shamim MS, et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016;3:99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smit AFA, Hubley R. RepeatModeler-1.0. 2008-2015. http://www.repeatmasker.org. Accessed on March 8, 2019. [Google Scholar]
- 66. Smit AFA, Hubley R. RepeatMasker-4.0. 2013-2015. http://www.repeatmasker.org. Accessed on December 17, 2019. [Google Scholar]
- 67. Laetsch DR, Blaxter ML. BlobTools: Interrogation of genome assemblies. F1000Res. 2017;6:1287. [Google Scholar]
- 68. Kim D, Paggi JM, Park C, et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hoff KJ, Stanke M. Predicting genes in single genomes with AUGUSTUS. Curr Protoc Bioinformatics. 2019;65:e57. [DOI] [PubMed] [Google Scholar]
- 70. Jones P, Binns D, Chang HY, et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Quevillon E, Silventoinen V, Pillai S, et al. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005;33:W116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sheffer MM, Hoppe A, Krehenwinkel H, et al. Supporting data for “Chromosome-level reference genome of the European wasp spider Argiope bruennichi: a resource for studies on range expansion and evolutionary adaptation.”. GigaScience Database. 2020. 10.5524/100837. [DOI] [PMC free article] [PubMed]
- 73. Brůna T, Hoff KJ, Lomsadze A, et al. BRAKER2: Automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. bioRxiv. 2020, doi: 10.1101/2020.08.10.245134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jia N, Wang J, Shi W, et al. Large-scale comparative analyses of tick genomes elucidate their genetic diversity and vector capacities. Cell. 2020;182(5):1328–40.e13. [DOI] [PubMed] [Google Scholar]
- 75. Dasmahapatra KK, Walters JR, Briscoe AD, et al. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim HS, Murphy T, Xia J, et al. BeetleBase in 2010: Revisions to provide comprehensive genomic information for Tribolium castaneum. Nucleic Acids Res. 2010;38:D437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Qu Z, Nong W, So WL, et al. Millipede genomes reveal unique adaptations during myriapod evolution. PLoS Biol. 2020;18, doi: 10.1371/journal.pbio.3000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Miller JR, Koren S, Dilley KA, et al. A draft genome sequence for the Ixodes scapularis cell line, ISE6. F1000Res. 2018;7:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. [DOI] [PubMed] [Google Scholar]
- 80. Gatesy J, Hayashi C, Motriuk D, et al. Extreme diversity, conservation, and convergence of spider silk fibroin sequences. Science. 2001;291(5513):2603–5. [DOI] [PubMed] [Google Scholar]
- 81. Hayashi CY, Shipley NH, Lewis RV. Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int J Biol Macromol. 1999;24:271–5. [DOI] [PubMed] [Google Scholar]
- 82. Casewell NR, Wüster W, Vonk FJ, et al. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 2013;28:219–29. [DOI] [PubMed] [Google Scholar]
- 83. Fry BG, Roelants K, Champagne DE, et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 2009;10:483–511. [DOI] [PubMed] [Google Scholar]
- 84. Grishin E. Polypeptide neurotoxins from spider venoms. Eur J Biochem. 1999;264:276–80. [DOI] [PubMed] [Google Scholar]
- 85. Escoubas P. Molecular diversification in spider venoms: A web of combinatorial peptide libraries. Mol Divers. 2006;10:545–54. [DOI] [PubMed] [Google Scholar]
- 86. Escoubas P, Sollod B, King GF. Venom landscapes: Mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon. 2006;47:650–63. [DOI] [PubMed] [Google Scholar]
- 87. Diniz CR, do Nascimento Cordeiro M, Junor LR, et al. The purification and amino acid sequence of the lethal neurotoxin Tx1 from the venom of the Brazilian ‘armed’ spider Phoneutria nigriventer. FEBS Lett. 1990;263:251–3. [DOI] [PubMed] [Google Scholar]
- 88. Gerts EM, Yu YK, Agarwala R, et al. Composition-based statistics and translated nucleotide searches: Improving the TBLASTN module of BLAST. BMC Biol; 2006;4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Altschul SF, Gish W, Miller W, et al. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- 90. Satsuma2. https://github.com/bioinfologics/satsuma2. Accessed on September 8, 2020. [Google Scholar]
- 91. Shingate P, Ravi V, Prasad A, et al. Chromosome-level assembly of the horseshoe crab genome provides insights into its genome evolution. Nat Commun. 2020;11:2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Krzywinski M, Schein J, Birol I, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Vollrath F. Biology of spider silk. Int J Biol Macromol. 1999;24:81–8. [DOI] [PubMed] [Google Scholar]
- 94. Blackledge TA, Hayashi CY. Silken toolkits: Biomechanics of silk fibers spun by the orb web spider Argiope argentata (Fabricius 1775). J Exp Biol. 2006;209:2452–61. [DOI] [PubMed] [Google Scholar]
- 95. Zhao Y, Ayoub NA, Hayashi CY. Chromosome mapping of dragline silk genes in the genomes of widow spiders (Araneae, Theridiidae). PLoS One. 2010;5:e12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hoppe A. Argiope bruennichidata hub. http://bioinf.uni-greifswald.de/hubs/argiope/hub.txt. Accessed on February 15, 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Sheffer MM, Hoppe A, Krehenwinkel H, et al. Supporting data for “Chromosome-level reference genome of the European wasp spider Argiope bruennichi: a resource for studies on range expansion and evolutionary adaptation.”. GigaScience Database. 2020. 10.5524/100837. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Jessica Garb -- 7/7/2020 Reviewed
Jessica Garb -- 11/3/2020 Reviewed
Jean-François Flot -- 8/23/2020 Reviewed
Jean-François Flot -- 11/10/2020 Reviewed
Data Availability Statement
The final genome assembly and raw data from the PacBio and Hi-C libraries, as well as the annotation, have been deposited at NCBI under BioProject PRJNA629526. A publicly accessible genome browser hub with the annotation, raw transcriptome, and PacBio read coverage can be found on the UCSC Genome Browser server (under “My Data” > “Track Hubs” > “My Hubs” enter the cited URL [96]). Supporting data are available via the GigaScience data repository, GigaDB, including the softmasked assembly in FASTA format, the output file from RepeatMasker, predicted coding genes and their functional annotation in GFF3 formats, predicted coding gene nucleotide and translated sequences in FASTA formats, functional annotation from InterProScan in TSV format, the blast query results for Hox, spidroin, and venom genes in FASTA format, and the BUSCO output files in a zip folder [72].