Abstract
Novel functional coding sequences (altORFs) are camouflaged within annotated ones (CDS) in a different reading frame. We show here that an altORF is nested in the FUS CDS, encoding a conserved 170 amino acid protein, altFUS. AltFUS is endogenously expressed in human tissues, notably in the motor cortex and motor neurons. Over‐expression of wild‐type FUS and/or amyotrophic lateral sclerosis‐linked FUS mutants is known to trigger toxic mechanisms in different models. These include inhibition of autophagy, loss of mitochondrial potential and accumulation of cytoplasmic aggregates. We find that altFUS, not FUS, is responsible for the inhibition of autophagy, and pivotal in mitochondrial potential loss and accumulation of cytoplasmic aggregates. Suppression of altFUS expression in a Drosophila model of FUS‐related toxicity protects against neurodegeneration. Some mutations found in ALS patients are overlooked because of their synonymous effect on the FUS protein. Yet, we show they exert a deleterious effect causing missense mutations in the overlapping altFUS protein. These findings demonstrate that FUS is a bicistronic gene and suggests that both proteins, FUS and altFUS, cooperate in toxic mechanisms.
Keywords: alternative ORF, amyotrophic lateral sclerosis, dual coding, FUS, polycistronic
Subject Categories: Molecular Biology of Disease, Neuroscience
The FUS gene encodes two proteins: one large and nuclear, FUS, and another small and mitochondrial, altFUS. AltFUS is expressed in human brains and cooperates with FUS in FUS‐mediated cellular toxicity.
Introduction
FUS is a nuclear RNA‐binding protein, with a C‐terminal nuclear localization signal (NLS) (Vance et al, 2009; Deng et al, 2014). The protein is involved in RNA processing, DNA repair and cellular proliferation, and although some of its functions and mechanisms are described, our understanding of FUS‐related pathological mechanisms is still incomplete (Deng et al, 2014). Mutations in FUS gene associate with amyotrophic lateral sclerosis (ALS), frontotemporal lobar dementia (FUS‐FTLD) and essential tremor, all characterized by FUS cytoplasmic inclusions in neurons and glial cells (Deng et al, 2014). Such cytoplasmic FUS aggregates are pathological features in patients with FUS mutations or sporadic disease.
Recently, mutations in FUS 3’UTR were described in ALS patients and linked to an increased level of FUS mRNA and protein (Zou et al, 2012; Sabatelli et al, 2013; Dini Modigliani et al, 2014). Surprisingly, over‐expression of wild‐type FUS provokes an aggressive ALS phenotype in mice and fruit flies, in accordance with findings in yeast and mammalian cells (Ju et al, 2011; Chen et al, 2011; Miguel et al, 2012; Ajmone‐Cat et al, 2019; Ling et al, 2019). The mechanism of the wild‐type or ALS‐linked mutated FUS toxicity remains unclear (Deng et al, 2014; Taylor et al, 2016; Nolan et al, 2016).
With currently non‐annotated proteins being increasingly reported (Samandi et al, 2017; Saghatelian & Couso, 2015; Delcourt et al, 2017; Brunet et al, 2018), we hypothesized that the toxicity resulting from wild‐type FUS over‐expression may come from another, unseen, actor (Brunet et al, 2018). These novel proteins are coded by alternative open reading frames (altORFs) that are located within “non‐coding” RNAs (ncRNA), within the 5’ or 3’ “untranslated” regions (UTR) of mRNAs, or overlapping a known coding sequence (CDS) within a different frame of an mRNA (Delcourt et al, 2017; Brunet et al, 2018, 2019). Serendipitous discoveries and ribosome profiling have recently highlighted the distribution of altORFs throughout the human genome, and the consequences of their absence from current databases (Brunet et al, 2018). For example, mass spectrometry‐based proteomics has become the gold standard for protein identification and has been extensively used in ALS studies (Collins et al, 2015; Umoh et al, 2017). However, if a protein is not annotated, it is not included in the protein database (e.g. UniProtKB) and thus cannot be detected by mass spectrometry. An estimated 50% of mass spectra from a proteomics experiment are unmatched at the end of the analysis (Chick et al, 2015; Brunet et al, 2018).
Genome annotations must avoid spurious ORF annotations. Thus, unless functional characterization has been published, they rely upon 2 arbitrary criteria: a minimum length of 100 codons and a single ORF per transcript. Several groups developed tools to challenge such criteria, such as the sORF repository (Olexiouk et al, 2018) and the OpenProt database (Brunet et al, 2019), which offer a data‐driven broader view of eukaryotic proteomes. The OpenProt database is based on a polycistronic model of ORF annotation (Brunet et al, 2019) and reports any ORF longer than 30 codons within any frame of an mRNA or ncRNA. It contains currently annotated proteins (RefProts), novel isoforms and novel alternative proteins (altProts). Here, we used the OpenProt database (www.openprot.org) to ask whether FUS may encode additional proteins that could explain the toxicity of the wild‐type protein over‐expression. In support of this hypothesis, FUS displays an N‐terminal prion‐like domain. These low‐complexity domains are known to harbour overlapping ORFs (Kovacs et al, 2010; Pancsa & Tompa, 2016).
Results
AltFUS is a novel 170 amino acid protein, endogenously expressed in cell lines and tissues
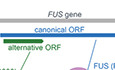
We began by querying OpenProt (Brunet et al, 2019) predictions for FUS canonical mRNA (ENST00000254108 or NM_004960), which led to 8 predicted altORFs, either overlapping the coding sequence (CDS) or within the 3’UTR (Table EV1). Amongst these, IP_243680 or altFUS, a 170 codon altORF overlapping FUS N‐terminal prion‐like domain, presents convincing experimental evidence of expression (OpenProt v1.3). AltFUS overlaps the FUS CDS in an open reading frame shifted by one nucleotide (Figs 1Aand EV1A). FUS is a complex gene with 13 annotated transcripts resulting from alternative splicing. Based on the GTEx expression data in brain tissues and nerves, five transcripts are more abundant and represent 85% of all transcripts (Fig EV1B). Three of them (FUS‐206, FUS‐211 and FUS‐203) are non‐coding according to Ensembl, while only two (FUS‐211 and FUS‐203) are non‐coding according to OpenProt (Figs 1B and EV1C). Ensembl (Zerbino et al, 2018) annotates two transcripts as coding (FUS‐201 and FUS‐202), for either the 526 amino acid FUS protein or its 525 amino acid isoform (Fig EV1D). From OpenProt prediction, these two transcripts also encode altFUS (IP_243680), or its 169 amino acid isoform (IP_243691), respectively (Fig EV1E). Moreover, the second most abundant transcript in brain tissues and nerves (FUS‐206), representing about 20% of all transcripts, is non‐coding according to Ensembl, but OpenProt predicts it contains the altFUS CDS (Figs 1B and EV1C). Thus, of the five most abundant transcripts in brain tissues and nerves, two code for both FUS and altFUS proteins, one codes for altFUS alone and the remaining two are non‐coding.
Figure 1. FUS is a bicistronic gene.
-
AFUS gene bicistronic annotation, with the canonical FUS CDS (in blue, +1 frame) and altFUS CDS (in green, +2 frame), represented on the FUS canonical transcript (ENST00000254108 or NM_004960). Sequence length proportions are respected, and the scale bar corresponds to 300 nucleotides.
-
BGenome browser view of FUS gene. The five most abundant transcripts in the brain are shown in the “Transcript” track. Transcripts predicted coding by the OpenProt resource are coloured in blue, and in grey if predicted non‐coding. The “Protein” track contains all predicted protein products. The known FUS protein (ENSP00000254108) and its isoform (ENSP00000369594) are coloured in green. The novel‐predicted altFUS protein (IP_243680) and its isoform (IP_243691) are coloured in red.
-
CPhyloP nucleotidic conservation scores are represented in grey across the FUS mRNA (ENST00000254108). The noise reduction after FFT (fast Fourier transformation) is outlined in blue. The average PhyloP score on the bicistronic and the monocistronic region are represented as dotted red lines. The position of the FUS CDS is represented by a blue rectangle and that of altFUS CDS by a green rectangle.
-
DAlignment (Clustalω) of altFUS protein sequences in human (Homo sapiens), chimpanzee (Chimp.—Pan troglodytes), rat (Rattus norvegicus), mouse (Mus musculus) and dog (Canis lupus familiaris). Residues are coloured based on their identity across species, from white (not conserved) to red (conserved in all species).
-
ERibo‐seq data over the FUS gene from the GWIPS portal. Initiating ribosome reads are indicated by blue bars, and elongating ribosomes footprints are indicated by the blue curve. The graph captures the beginning of the FUS gene with FUS and altFUS methionines indicated by blue arrows. The genomic positions are indicated relative to the start of exon 1.
-
F, GGenome browser view of FUS gene, centred on altFUS. The “Transcript” track contains the beginning of the canonical FUS transcript (ENST00000254108) in blue. The “Protein” track contains the beginning of the FUS protein (green) and the whole altFUS protein (red). In F, the “Peptide” track contains all the peptides identified by the OpenProt resource using the classical spectrum‐centric approach. The peptides sequences are indicated and are unique to altFUS or its isoform (see Fig EV2B for an example spectrum). In G, the “Peptide” track contains all the peptides identified by a peptide‐centric approach. The peptides indicated matched better to at least one spectrum than any known protein and are coloured in yellow if they matched better than any known protein with any PTM (see Fig EV2C for an example spectrum). The peptides sequences are indicated and are unique to altFUS or its isoform.
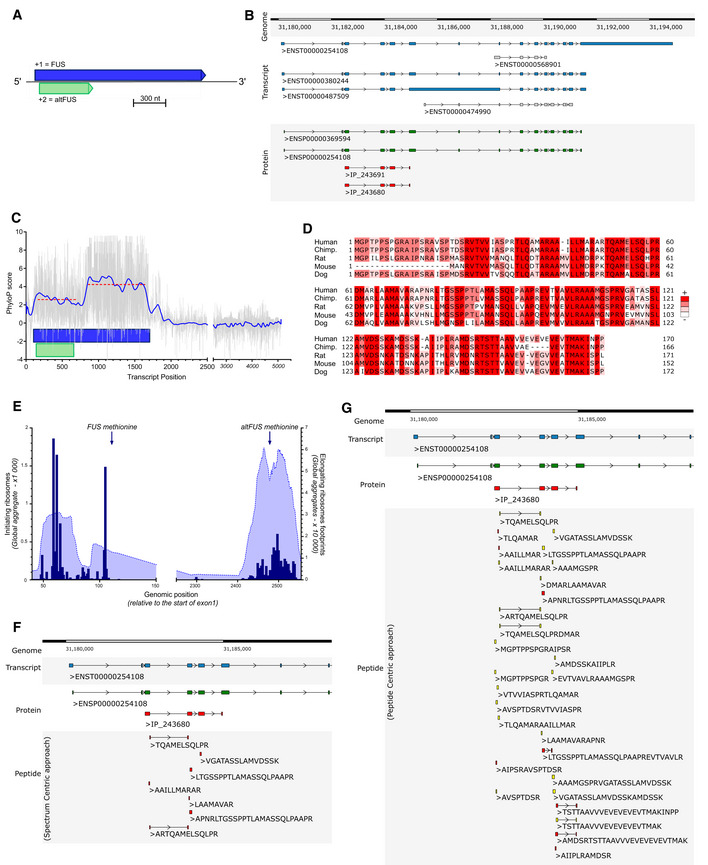
Figure EV1. FUS gene protein diversity.
-
AAltFUS is encoded within an alternative open reading frame (ORF) overlapping the FUS canonical CDS. When read in the + 1 frame, the FUS mRNA (here ENST00000254108) codes for a 526 amino acid protein (highlighted in blue), named FUS. In the + 2 frame, the FUS mRNA contains a second ORF (highlighted in green) that codes for a novel 170 amino acid protein, named altFUS. The two proteins are not isoforms.
-
BGTEx portal data on FUS mRNA expression in the brain and nerves are shown in blue coloured scale (TPM = transcripts per million). Transcripts are identified with Ensembl accessions (the number after the dot corresponds to the version used in the analysis) and are quantified across 14 tissues. The five transcripts framed in red share 85% of all mRNA expression level.
-
CTable compiling the protein information relayed by Ensembl and OpenProt resources for the five transcripts highlighted in panel B. The FUS protein is highlighted in blue. The altFUS protein is highlighted in green.
-
D–EAlignment (Clustalω) of protein sequences for FUS (ENSP00000254108) and its isoform (ENSP00000369594) in panel D (blue) and altFUS (IP_243680) and its isoform (IP_243691) in panel E (green). The residues are coloured based on their degree of identity.
- F
We retrieved nucleotide conservation scores (PhyloP) for FUS transcripts over 100 vertebrates. PhyloP scores range from −10 (highly variable) to 10 (highly conserved). Scores over the FUS CDS are under a constraint at the altFUS CDS locus (average score of 2.6 instead of 4 elsewhere on the FUS CDS), which is consistent with a selection pressure across 2 overlapping frames (Fig 1C) (Pavesi, 2019). We then retrieved altFUS protein sequences over 84 species and observed a strong protein conservation across mammals, and primates notably (75 to 99.4% of sequence identity—Table EV2, Fig EV1F). Thus, AltFUS is well conserved, with domains showing little to no sequence variations (Fig 1D). No functional domain nor clear secondary structures could be inferred from bioinformatics predictions.
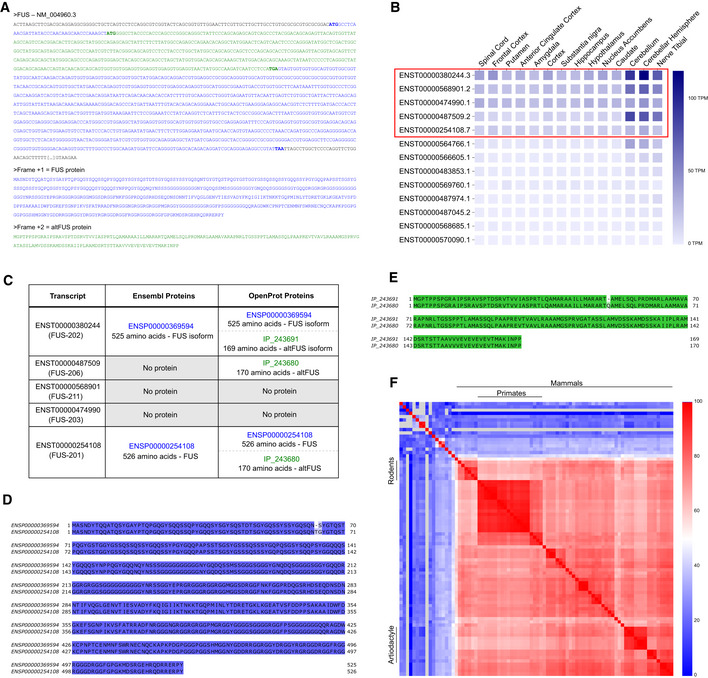
Published Ribo‐seq data in human, retrieved from the Gwips portal (Michel et al., 2018), revealed an accumulation of initiating ribosomes around the altFUS initiating methionine, in association with an increase in the density of elongating ribosomes over altFUS CDS (Fig 1E). These results suggest that altFUS is translated. Similar results were observed in mouse (Fig EV2A).
Figure EV2. AltFUS is endogenously expressed.
- Ribo‐seq data over the mouse FUS gene from the GWIPS portal (Mus musculus). Initiating ribosome reads are indicated by green bars, and elongating ribosomes footprints are indicated by the green curve. The graph captures the beginning of the FUS gene with FUS and altFUS methionines indicated by green arrows. The genomic positions are indicated relative to the start of exon 1.
- MS/MS spectrum confidently mapped to a peptide unique to altFUS by the OpenProt resource. Peaks are represented by their mass over charge ratios (m/z) and their intensity relative to the highest (relative intensity). The y ions are coloured in blue, the b ions in red and the unannotated peaks appear in grey. The charge state and m/z error of the matched peptide are indicated on the top right corner of the spectrum. The original study, file and spectrum number are indicated below the graph.
- Comparison of one MS/MS spectrum confidently mapped to a peptide unique to altFUS (left graph) and its best possible annotation with any known protein with any post‐translational modification (PTM—right graph). Peaks are represented by their mass over charge ratios (m/z) and their intensity relative to the highest (relative intensity). The y ions are coloured in blue, the b ions in red and the unannotated peaks appear in grey. The charge state and m/z error for each of the matched peptides are indicated on the top right corner of each panel. The original study, file and spectrum number are indicated below the graph. On the top of each panel is indicated the peptide matched to the spectrum and the corresponding protein and PTMs.
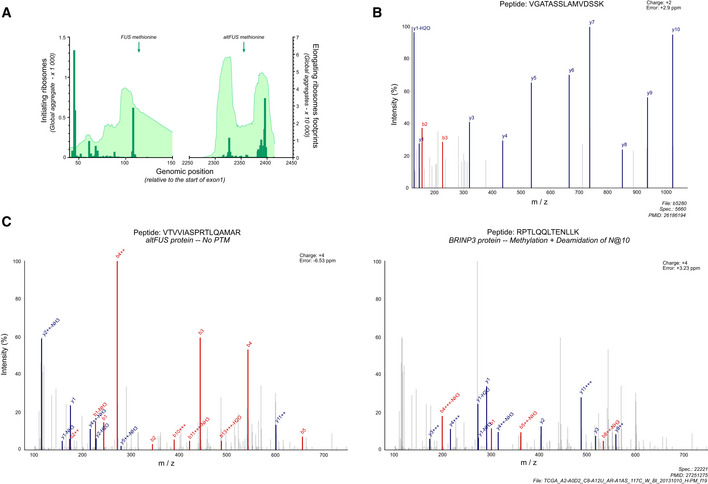
Based on the OpenProt database, AltFUS was identified in multiple proteomics data sets, with up to 7 confident peptides (Fig 1F, Table EV1). These peptides are unique to altFUS (Fig EV2B), or shared with its isoform (IP_243691), and represent a 41% sequence coverage. Furthermore, we used a peptide‐centric approach to query the TCGA data sets for altFUS expression with the PepQuery algorithm. This approach allowed us to identify 28 peptides unique to altFUS, or shared with its isoform (IP_243691), confidently mapped to mass spectra that could not be better explained by any known protein (hg38_Ensembl database; Table EV3). These peptides span through the entire altFUS sequence, representing a full sequence coverage. Out of these, 20 peptides were confidently mapped to mass spectra that could not be better explained by any known protein with any post‐translational modification and/or chemical artefact (Fig 1G, Fig EV2C).
To further validate altFUS protein expression, we developed a custom antibody targeting two unique altFUS peptides (Appendix Fig S1A and B) and tested it using three constructs: FUS, altFUS and FUS(Ø). The latter is a monocistronic FUS version, where all altFUS methionines are mutated for threonines in a manner synonymous for FUS (Appendix Fig S1C–E). Thus, the FUS protein sequence is unchanged, but the altFUS sequence does not contain any methionines. Transfection of HEK293 cells revealed expression of both proteins, FUS and altFUS, from the FUS nucleotide sequence (Fig 2A). A slight decrease in endogenous FUS expression could be observed although not significant (P value = 0.255, Appendix Fig S1F). As expected, altFUS expression was lost with the monocistronic FUS(Ø) construct. HEK293 cells transfected with a siRNA targeting FUS mRNA showed a significant knockdown of both proteins, FUS and altFUS, whereas altFUS endogenous expression was visible in scrambled control siRNA and mock‐transfected cells (Fig 2B and Appendix Fig S1B). These results validate the specificity of the custom antibody for altFUS protein detection by Western blot and demonstrate altFUS endogenous expression in HEK293‐cultured cells.
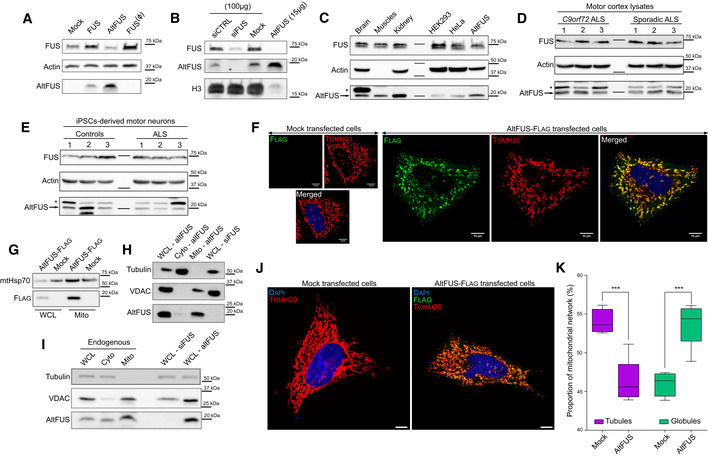
Figure 2. altFUS, a novel endogenous mitochondrial protein.
-
AExpression of untagged versions of both FUS and altFUS proteins from transfection of the FUS cDNA in HEK293 cells by Western blot, and expression of FUS with the monocistronic construct FUS(Ø) (representative image from n = 3). The slight decrease in endogenous FUS expression upon altFUS over‐expression (third lane) was not significant (see Appendix Fig S1F for quantification, P = 0.255, n = 3).
-
BAltFUS endogenous expression in HEK293 cells using a siRNA targeting FUS mRNA as negative control and over‐expression of altFUS CDS as positive control (representative image from n = 3). For the mock, siCTRL and siFUS conditions, 100 μg of total protein was loaded, when only 15 μg of total proteins was loaded for the altFUS over‐expression condition.
-
CAltFUS (arrow) endogenous expression in human tissues (brain, muscles and kidney—100 μg), in HEK293 and HeLa‐cultured cells (100 μg) and using the over‐expression of altFUS CDS in HEK293 cells (50 μg) as positive control (representative image from n = 3). The asterisk indicates a protein species detected with the anti‐altFUS antibody specifically in the brain.
-
D, EAltFUS (arrow) endogenous expression in the motor cortex of three C9orf72 and three sporadic ALS patients (D) or in iPSC‐derived motor neurons of three lines from controls and from ALS patients (E) (representative image from n = 3). The asterisk indicates a protein species detected with the anti‐altFUS antibody specifically in the brain.
-
FImages by confocal microscopy of altFUS‐flag (green) in HeLa cells, using Tomm20 (red) as a mitochondrial marker (representative image from n = 3, Pearson’s correlation r = 0.92). Mock‐transfected cells were identically stained, highlighting the specificity of the observed altFUS signal (Flag). The white scale bar corresponds to 10 μm.
-
GAltFUS‐flag enrichment in mitochondrial extracts from transfected HEK293 cells (representative image from n = 3) with mtHsp70 used as a mitochondrial marker (WCL = whole cell lysate, Mito = mitochondrial extract).
-
HAltFUS mitochondrial expression in untagged altFUS‐transfected HEK293 cells following fractionation (representative image from n = 3), with Tubulin as a marker of the cytosolic fraction and VDAC as a marker of the mitochondrial fraction (WCL = whole cell lysate, Cyto = cytosol fraction, Mito = mitochondrial fraction).
-
IEndogenous altFUS mitochondrial expression in HEK293 cells following fractionation (representative image from n = 3), with Tubulin as a marker of the cytosolic fraction and VDAC as a marker of the mitochondrial fraction. We used siFUS‐transfected cells as a negative control and altFUS‐transfected cells as a positive control for altFUS expression (WCL = whole cell lysate, Cyto = cytosol fraction, Mito = mitochondrial fraction).
-
JRepresentative images of the mitochondrial network (Tomm20 in red) in mock and altFUS‐Flag (green)‐transfected HeLa cells (n = 3). The white scale bar corresponds to 10 μm.
-
KProportion of tubules and globules in the mitochondrial network of mock HeLa cells and HeLa cells transfected with altFUS‐flag (see Appendix Fig S2A). Quantification was done over a minimum of 100 cells across a technical duplicate per independent experiments (n = 3, i.e. a minimum of 300 cells per biological conditions, p‐value < 0.001, Mann–Whitney U test). The boxes extend to the 25th and 75th percentiles, with the median marked. The whiskers correspond to the 5th and 95th percentiles.
AltFUS endogenous expression was visible in control human tissues, HEK293 and HeLa cell lines (Fig 2C). In order to test altFUS expression in pathological tissues, we retrieved motor cortex lysates from 3 ALS patients with a C9orf72 mutation (most common genetic cause) and 3 sporadic ALS patients (most common aetiology). AltFUS endogenous expression was detected in all cases (Fig 2D). Furthermore, as ALS is a motor neuron disease, we derived functional ventral spinal motor neurons from induced pluripotent stem cells (iPSCs; Hall et al, 2017; Luisier et al, 2018) from healthy controls and ALS patients carrying valosin‐containing protein mutations (3 lines per group). AltFUS endogenous expression was detected in all samples (Fig 2E). We noticed that brain (Fig 2C) and motor cortex lysates (Fig 2D), as well as iPSC‐derived motor neurons (Fig 2E), from healthy controls and ALS patients, presented a higher band detected with the custom altFUS antibody. This band is not present in cultured cell lines or other tissues. It could come from a non‐specific signal or a post‐translational modification on altFUS that is specific to the motor cortex and spinal cord motor neurons (Fig 2C–E). Deep learning predictions of post‐translational modifications on altFUS revealed an extensive propensity to phosphorylation and O‐GlcNAcylation (O‐linked β‐N‐acetylglucosamine glycosylation), with up to 19 and 18 sites, respectively, under a high stringency model (Table EV4). These two post‐translational modifications are abundant in the eukaryotic brain and known to be dysregulated in neurodegenerative diseases or ageing (Hart & Akimoto, 2009; Didonna & Benetti, 2015; Santos & Lindner, 2017; Thompson et al, 2018). We could also observe a lower band in the line 2 of controls motor neurons, which may correspond to a degradation product or an initiation at a downstream methionine in altFUS sequence. This band has never been observed in other samples so far. We demonstrate that the FUS gene encodes two proteins, FUS and altFUS, both endogenously expressed in human tissues, iPSCs and cell lines. Furthermore, we spiked GFP‐FUS or GST‐altFUS recombinant proteins in human brain lysates to evaluate the stoichiometry of the FUS gene proteins. The ratio of altFUS to FUS in brain lysates is 0.29 (1/3.5) (Fig EV3A–D).
Figure EV3. AltFUS/ FUS stoichiometry in human brain lysates.
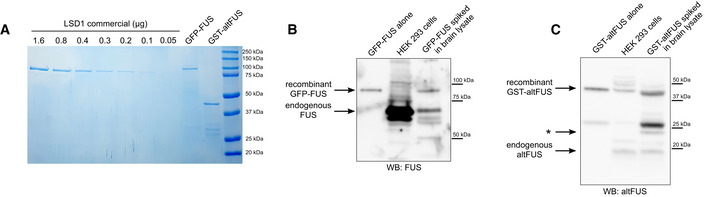
- Estimation of the concentration of GFP‐FUS and GST‐altFUS recombinant proteins (10 μL loaded each). A commercial LSD1 recombinant protein diluted at 0.2 μg/μL was used as standard. The concentration for GFP‐FUS was evaluated to 0.06 μg/μL and that of GST‐altFUS was evaluated to 0.07 μg/μl.
- Western blot of human brain lysates spiked with GFP‐FUS (last lane). The recombinant protein alone (first lane) and a whole cell lysate of HEK293 cells were used as size controls. The bands corresponding to the recombinant GFP‐FUS and to the endogenous FUS proteins are indicated on the left. Immunoblot: anti‐FUS (Abcam, ab84078).
- Western blot of human brain lysates spiked with GST‐altFUS (last lane). The recombinant protein alone (first lane) and a whole cell lysate of HEK293 cells were used as size controls. The bands corresponding to the recombinant GST‐altFUS and to the endogenous altFUS proteins are indicated on the left. The asterisk indicates a protein species detected with the anti‐altFUS antibody specifically in the brain (see Fig 2C–E). Immunoblot: custom anti‐altFUS.
AltFUS is a mitochondrial protein
Flag‐tagged altFUS (altFUS‐Flag) displayed a strong colocalization with a common mitochondrial marker, TOMM20 (Fig 2F). Additionally, mitochondrial extracts showed an enrichment in altFUS‐Flag (Fig 2G). Cellular fractionation of cells over‐expressing untagged altFUS further validated altFUS mitochondrial localization (Fig 2H). The endogenous altFUS protein was found in the mitochondrial fraction, although it displayed a weak cytoplasmic signal as well (Fig 2I), consistent with the immunofluorescence data (Fig 2F). Furthermore, cells over‐expressing altFUS showed an altered mitochondrial network, with a significant increase in fragmented mitochondria (globular) compared with mock cells that displayed more tubular structures (Fig 2J–K and Appendix Fig S2A).
Mitochondrial fragmentation is observed in models of over‐expression of FUS mutants (Deng et al, 2015; Carrì et al, 2017; Nakaya & Maragkakis, 2018), and we reproduced here a similar effect when over‐expressing altFUS alone. Thus, we wondered whether altFUS played a role in other mitochondrial dysfunctions observed in FUS‐linked toxicity models. To this end, we reproduced an ALS‐associated FUS mutant: FUS‐R495x (Deng et al, 2015; Nakaya & Maragkakis, 2018). This mutant leads to a premature stop codon before FUS NLS and is linked to severe fALS and sALS cases (Deng et al, 2014, 495). In this construct, altFUS is still present and not affected by the mutation (Appendix Fig S2B). Similar to FUS(Ø), we also generated the monocistronic construct FUS(Ø)‐R495x, which contains synonymous mutations for FUS‐R495x, but prevents altFUS expression. V5‐FUS(Ø‐F lag ) and V5‐FUS(Ø‐F lag )‐R495x did not express altFUS, but only the FUS protein, wild‐type or ALS‐linked mutant R495x, respectively (Appendix Fig S2C). We first investigated the effect of altFUS on the mitochondrial membrane potential using the potential sensitive dye TMRE (Fig 3A and B, Appendix Fig S2D). As previously described (Deng et al, 2015), over‐expression of bicistronic FUS or FUS‐R495x constructs (i.e. expressing both altFUS and FUS or FUS‐R495x proteins) led to a decrease in mitochondrial membrane potential. The mitochondrial membrane potential remained normal when over‐expressing monocistronic FUS(Ø) or FUS(Ø)‐R495x, underlining the role of altFUS. However, over‐expression of altFUS alone did not alter the mitochondrial membrane potential, which suggests both proteins cooperate for this FUS‐associated toxicity hallmark.
Figure 3. altFUS is involved in mitochondrial dysfunction and autophagy processes.
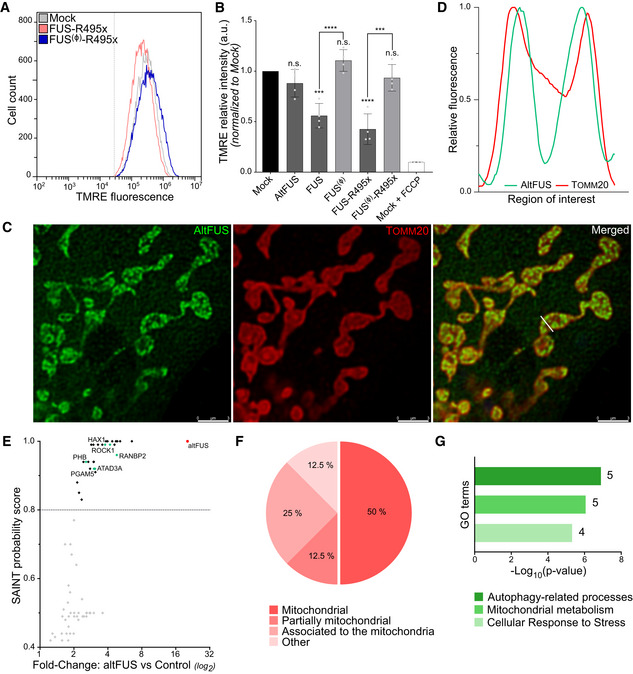
- Representative traces of TMRE fluorescence measured by flow cytometry in mock‐transfected cells and cells over‐expressing the bicistronic FUS‐R495x or the monocistronic FUS(Ø)‐R495x constructs (n = 4, minimum of 50 000 live cells per independent replicates). Mean fluorescence intensity of mock‐transfected cells treated with a decoupling agent, FCCP, is indicated by a grey dotted line.
- Mean TMRE fluorescence intensity measures in mock‐transfected cells, cells over‐expressing altFUS, FUS, FUS(Ø), FUS‐R495x or FUS(Ø)‐R495x, or mock‐transfected cells treated with FCCP across 4 independent experiments (n = 4, also see Appendix Fig S2D, mean ± SD). Statistical significance is relative to the mock condition unless otherwise indicated (***P < 0.001, ****P < 0.0001, n.s. = non‐significant, two‐way ANOVA with Tukey’s multiple comparison correction).
- Representative image by stimulated emission depletion microscopy (STED) of altFUS‐Flag (green) localization within mitochondria (Tomm20 marker in red). The white bar across the mitochondria represents the region of interest quantified in panel D. The white scale bar corresponds to 3 μm.
- Relative fluorescence histogram for altFUS‐Flag and Tomm20 across the region of interest highlighted by a white line on panel C.
- Scatter plot of the proteins identified by AP‐MS (see Appendix Fig S3) indicating their enrichment (fold change over control) and their SAINT probability score. Proteins above the 0.8 threshold (grey line) are indicated in black, others in grey. AltFUS is indicated in red (bait), and preys known to regulate the autophagy or the cellular stress response are indicated in green.
- Subcellular localizations of proteins identified by AP‐MS from panel E (see Appendix Fig S3).
- Enrichment of biological processes in altFUS‐interacting proteins compared with the human mitochondrial proteome (Fisher’s exact test with FDR < 0.1%). The number of proteins identified in each GO term is indicated next to the corresponding bar.
To further characterize altFUS, we investigated its protein interactors. Using stimulated emission depletion microscopy (STED), we observed that altFUS localized in puncta following a cristae‐like pattern inside the mitochondria, delimited using an outer‐membrane mitochondrial marker, TOMM20 (Fig 3C and D). We then used size‐exclusion chromatography on mitochondrial extracts to isolate altFUS‐Flag macromolecular complexes (Appendix Fig S3A and B). Following a Flag affinity purification and mass spectrometry (AP‐MS) analysis, using a no‐bait control for quantitative comparison, we confidently identified 12 interacting proteins (Fig 3E, Appendix Fig S3C and D, Table EV5). These proteins were identified with a minimum of 2 unique peptides and displayed over a twofold enrichment to the control. A gene enrichment analysis to the human proteome on subcellular localization showed that the identified interactors are in majority proteins known to localize at the mitochondria (Fig 3F). Amongst the identified interactors is the heat shock protein HSPA9, a chaperone crucial in the mitochondrial iron–sulphur cluster biogenesis (Shan & Cortopassi, 2016) (Appendix Fig S3D, Table EV5). Although HSPA9 was identified with 6 unique peptides and a fold change of 6.53 to the control, this protein is commonly detected in AP‐MS data sets and thus likely non‐specific nor functionally relevant (Mellacheruvu et al, 2013). Several altFUS‐interacting proteins are known to interact together, such as PHB, ATAD3A, ERLIN2, RANBP2 and EMD (Appendix Fig S3D). A refined gene enrichment analysis to human mitochondrial proteome identified three significantly enriched biological processes: autophagy‐related pathways, mitochondrial metabolism and cellular response to stress (Fig 3G). Disruptions within these pathways are pathological hallmarks of FUS‐linked toxicity and ALS (Taylor et al, 2016; Monahan et al, 2016; Nakaya & Maragkakis, 2018; Ling et al, 2019).
AltFUS inhibits autophagy and drives the accumulation of FUS‐ and TDP43‐positive cytoplasmic aggregates
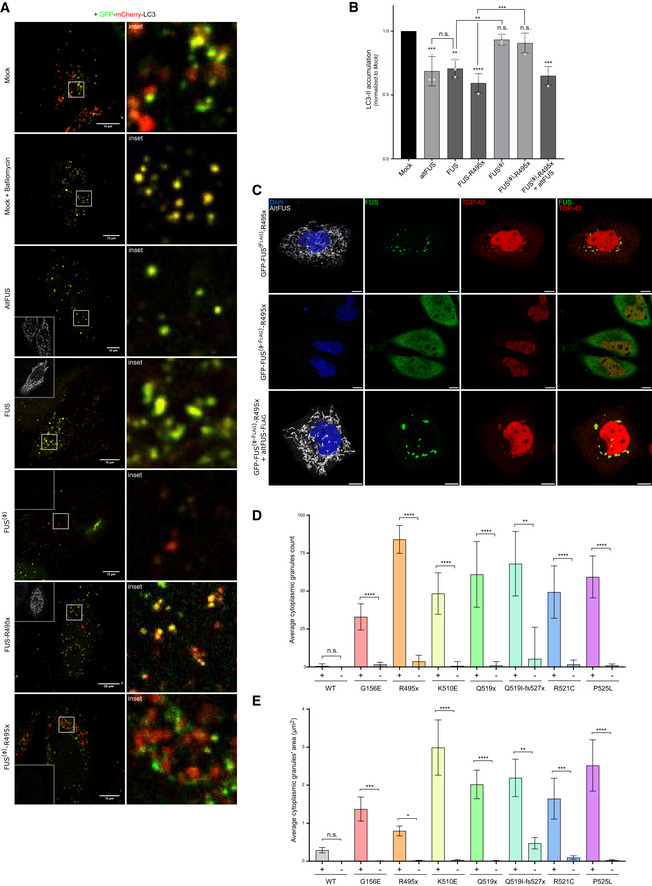
Following on these results, we hypothesized that the inhibition of autophagy observed with ALS‐associated FUS mutants may instead be attributed to altFUS. We used the mCherry‐GFP‐LC3 reporter to track the autophagic flux by confocal microscopy (Appendix Fig S4A). Under basal conditions, cells displayed red and yellow foci as expected (Fig 4A). An accumulation of yellow foci was observed when cells were treated with bafilomycin, an inhibitor of autophagy. Similarly, cells over‐expressing altFUS displayed a significant accumulation of yellow foci (Fig 4A). Furthermore, our results were consistent with previously published data (Marrone et al, 2019) as cells transfected with FUS or FUS‐R495x displayed a decreased autophagic flux (Fig 4A). This accumulation of yellow foci was absent in cells that express monocistronic FUS constructs, thus lacking altFUS expression (FUS(Ø) or FUS(Ø)‐R495x). We used bafilomycin followed by LC3 probing to further validate the impact of altFUS on autophagy (Fig 4B, Appendix Fig S4B). Similarly, an inhibition of autophagy was observed only in cells over‐expressing altFUS. Furthermore, in cells over‐expressing monocistronic FUS(Ø)‐R495x, the inhibition of autophagy could be restored by co‐transfecting altFUS (Fig 4B, Appendix Fig S4B). These results establish altFUS, rather than FUS, as the protein responsible of the inhibition of autophagy.
Figure 4. AltFUS is necessary for FUS‐associated inhibition of autophagy and accumulation of FUS/TDP‐43 cytoplasmic aggregates.
-
AImages by confocal microscopy of mCherry‐GFP‐LC3 signal in HeLa cells across biological conditions: untreated mock, bafilomycin‐treated mock, altFUS, FUS, FUS(Ø), FUS‐R495x and FUS(Ø)‐R495x (representative images of n = 3). The altFUS signal (white) is shown as an inset in the top or bottom left of the left panels. The white scale bar corresponds to 10 μm, and the zoomed in region (right panels) is delimited by a white box.
-
BLC3‐II accumulation after bafilomycin treatment from mock, altFUS, FUS, FUS(Ø), FUS‐R495x, FUS(Ø)‐R495x transfected cells and FUS(Ø)‐R495x and altFUS co‐transfected cells across 3 independent experiments (n = 3, mean ± SD). The quantification corresponds to the treated/untreated ratio of LC3‐II abundance (see Appendix Fig S4B for a representative image). Statistical significance is relative to the mock condition unless otherwise indicated (****P < 0.0001, ***P < 0.001, **P < 0.01, n.s. = non‐significant, two‐way ANOVA with Tukey’s multiple comparison correction).
-
CImages by confocal microscopy of altFUS (flag‐tagged—white), FUS (GFP‐tagged—green) and TDP‐43 (red) signals in HeLa cells transfected with the bicistronic GFP‐FUS(F lag )‐R495x or the monocistronic GFP‐FUS(Ø‐ flag )‐R4955x constructs, or co‐transfected with the monocistronic GFP‐FUS(Ø‐F lag )‐R495x and altFUS‐Flag constructs (representative images from n = 3). The white scale bar corresponds to 10 μm.
-
D, EQuantification of FUS cytoplasmic granules, number (D) and area (μm2) (E) in cells over‐expressing the bicistronic (+) or monocistronic (−) construct for FUS, FUS‐G156E, FUS‐R495x, FUS‐K510E, FUS‐Q519x, FUS‐Q519I‐fs527x, FUS‐R521C and FUS‐P525L. Statistical comparisons are made between bicistronic and monocistronic versions of each construct (n = 3—biological replicates, mean ± SD, ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, n.s. = non‐significant, one‐way ANOVA test with Sidak’s multiple comparison).
Furthermore, altFUS interactome analysis suggested a role in the cellular stress response, which is known to be altered in ALS with a TDP‐43 cytoplasmic accumulation in 98% of patients (Aulas & Vande Velde, 2015, 43; Hergesheimer et al, 2019). In FUS‐linked ALS and some sALS cases, FUS cytoplasmic aggregates or mislocalization is also observed (Deng et al, 2010; Farrawell et al, 2015; Tyzack et al., 2019). In our hands, cells over‐expressing FUS‐R495x displayed cytoplasmic aggregates that were positive for both FUS‐R495x and TDP‐43 (Fig 4C). Although TDP‐43 aggregates are not a common observation with FUS‐associated mutants, co‐aggregation has already been reported in patients, animal models and cultured cell lines across multiple studies (Deng et al, 2010; Shan et al, 2010; Cohen et al, 2011; Kryndushkin et al, 2011; Keller et al, 2012; Farrawell et al, 2015; Wiesner et al, 2018). In cells over‐expressing the monocistronic FUS(Ø)‐R495x construct, thus lacking altFUS expression, FUS‐R495x displayed a more diffuse cytoplasmic localization, and TDP‐43 remained in the nucleus (Fig 4C). FUS cytoplasmic aggregates were significantly more numerous and larger when altFUS was co‐expressed (Fig 4D and E). Accumulation of FUS‐R495x and TDP‐43 in cytoplasmic aggregates could be reconstituted by co‐transfecting altFUS and the monocistronic FUS(Ø)‐R495x construct (Fig 4C). These observations were repeated across all 7 ALS‐associated FUS mutations tested, where all observed FUS aggregates were also TDP‐43‐positive (Appendix Fig S4C–E). The cytoplasmic aggregates were also TIA‐1‐positive as observed with ALS‐linked FUS mutants in previous work (Aulas & Vande Velde, 2015; Fig EV4A–D). These results suggest that altFUS enhances the assembly of cytoplasmic FUS mutants aggregates and is responsible for the recruitment of TDP‐43 in these aggregates.
Figure EV4. AltFUS potentiate FUS recruitment to stress granules.
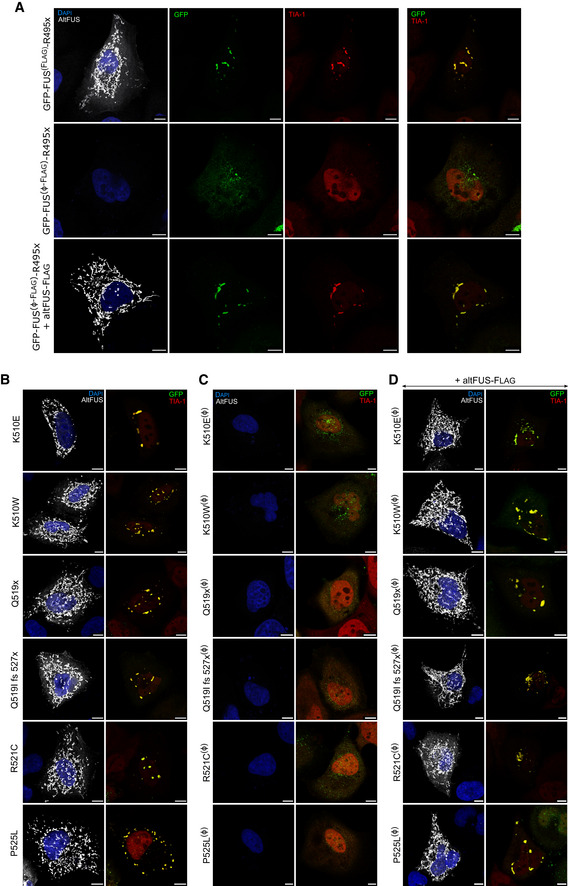
-
AImages by confocal microscopy of altFUS (Flag‐tagged—white), FUS (GFP‐tagged—green) and TIA‐1 (red) signals in HeLa cells transfected with the bicistronic GFP‐FUS(F lag )‐R495x or the monocistronic GFP‐FUS(Ø‐ flag )‐R495x constructs, or co‐transfected with the monocistronic GFP‐FUS(Ø‐F lag )‐R495x and altFUS‐Flag constructs (representative images from n = 3). The white scale bar corresponds to 10 μm.
-
B–DImages by confocal microscopy of altFUS(Flag‐tagged—white), FUS (GFP‐tagged—green) and TIA‐1 (red) signals in HeLa cells transfected with the bicistronic constructs (B), monocistronic constructs (C) or co‐transfected with altFUS‐Flag and the monocistronic constructs (D) of 6 ALS‐associated mutants: FUS‐G156E, FUS‐K510E, FUS‐Q519x, FUS‐Q519I‐fs527x, FUS‐R521C and FUS‐P525L (representative images from n = 3). The white scale bar corresponds to 10 μm.
AltFUS protects against neurodegeneration in FUS‐associated Drosophila models
In order to investigate the role of altFUS in an already established in vivo model of FUS‐related neurodegeneration, we generated Drosophila models expressing either the bicistronic, FUS and FUS‐R495x constructs, or the monocistronic, FUS(Ø) and FUS(Ø)‐R495x, constructs. We used the Elav‐GeneSwitch‐GAL4 driver strain, as previously described (Lanson et al, 2011), as it allows for an inducible over‐expression in motor neurons and avoids lethality at the larval stage from FUS over‐expression in the central nervous system (Lanson et al, 2011; Bogaert et al, 2018). First, we generated flies containing the sequences for UASt‐altFUS, UASt‐FUS, UASt‐FUS(Ø), UASt‐FUS‐R495x or UASt‐FUS(Ø)‐R495x. These flies were then crossed with the Elav‐GeneSwitch‐GAL4 driver strain (Fig 5A). UASt‐mCherry flies were used as controls. Selected F1 individuals were then divided into 2 groups with equal proportions of males/females. The first group received standard food, while the other received RU‐486‐treated food. The treatment induces a conformational change in the Elav‐GeneSwitch driver, which allows activation of the UAS promoter and thus expression of the target protein. We retrieved flies at selected time points to validate protein expression in the RU‐486‐treated population through time, while the controls showed no expression (Fig 5B).
Figure 5. AltFUS expression is necessary for the full FUS‐linked ALS phenotype in Drosophila .
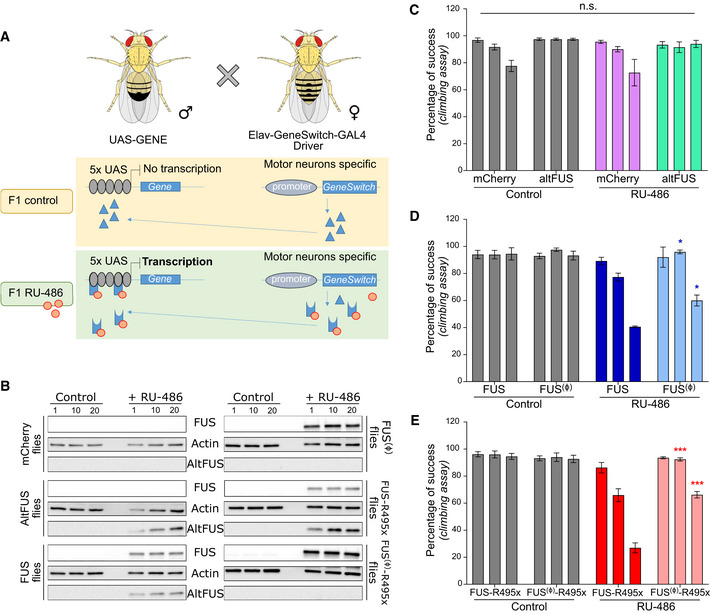
-
ACrossbreeding strategy for Drosophila generation using the Elav‐GeneSwitch‐GAL driver as an inducible expression system specific to the motor neurons.
-
BFUS and altFUS expression in mCherry (control), altFUS, FUS, FUS(Ø), FUS‐R495x or FUS(Ø)‐R495x expressing Drosophila from the control F1 and the RU‐486‐treated F1 (see panel A) at 1, 10 or 20 days post‐induction (representative image from n = 3).
-
C–ELocomotion assay represented by the percentage of climbing success in control and RU‐486‐treated transgenic Drosophila expressing mCherry or altFUS (C), the bicistronic FUS or the monocistronic FUS(Ø) (D), and the bicistronic FUS‐R495x or the monocistronic FUS(Ø)‐R495x (E) at days 1, 10 and 20 post‐induction. Statistical comparisons were made between each population (n = 4—biological replicates). Indicated significance is between the monocistronic and the bicistronic transgenic flies of the RU‐486‐treated population (mean ± SD, n.s. = non‐significant, *P < 0.05, ***P < 0.001, two‐way ANOVA with Tukey’s multiple comparison correction).
The motor neuron degeneration linked to ALS provokes a progressive locomotion loss measurable with a well‐described climbing assay (Chambers et al, 2013). The control populations did not show any significant locomotion loss at day 1, day 10 nor day 20 (Fig 5C–E). Similarly, the RU‐486‐treated control group did not show a significant effect, although a decrease in climbing ability could be observed as previously reported (Mawhinney & Staveley, 2011; Robles‐Murguia et al, 2019) (mCherry transgenic flies—Fig 5C). AltFUS flies did not show any significant locomotion loss through time (Fig 5C). This result is consistent with the in cellulo data showing altFUS alone is not sufficient to provoke pathological hallmarks. As previously shown with this model (Chen et al, 2011), the bicistronic FUS flies, which express both FUS and altFUS proteins, displayed a significant locomotion loss (Fig 5D). Bicistronic ALS‐linked FUS‐R495x flies showed an even greater motor neuron degeneration through time compared with FUS (Fig 5E). Monocistronic FUS(Ø) (Fig 5D) and FUS(Ø)‐R495x (Fig 5E) flies, which do not express altFUS, displayed both a delay in the onset of motor neuron degeneration and a reduced drop in climbing success at 20 days post‐induction (40% vs. 60% for FUS(Ø), 30% vs. 70% for FUS(Ø)‐R495x). These results in Drosophila confirm a role for altFUS in FUS‐related neurodegeneration in vivo, are consistent with our in cellulo observations and highlight the toxic cooperation between FUS and altFUS.
ALS‐associated mutations, synonymous for FUS, alter AltFUS and lead to TDP‐43 cytoplasmic aggregates
As of today, over 50 mutations in the FUS gene have been associated with ALS (Deng et al, 2014). However, most of these locate at the carboxyl end of the protein and as such have no effect on altFUS (Fig 1A). We wondered whether mutations altering altFUS might have been overlooked as non‐consequential in the FUS reading frame. We retrieved FUS synonymous mutations found in ALS patients, with an allelic frequency below 0.01%, from previous studies and the ALS Variant Server (http://als.umassmed.edu/ Table EV6). The retrieved mutations clustered on the altFUS locus (Fig 6A), with 60% of FUS synonymous mutations found in sALS patients and 50% of FUS synonymous mutations found in fALS patients, which is significantly higher than expected by chance (34%) (Table EV6). We selected 4 mutations for further analysis based on the residue conservation: altFUS‐P31L, altFUS‐A38V, altFUS‐A46V and altFUS‐R64P. We generated them in GFP‐FUS constructs: GFP‐FUS(P31L‐ flag )‐S44=; GFP‐FUS(A38V‐ flag )‐G51=; GFP‐FUS(A46V‐ flag )‐G59=; and GFP‐FUS(R64P‐ flag )‐S77=. All altFUS mutants still localized to the mitochondria (Fig EV5A). To investigate whether these mutations may provoke an ALS‐like phenotype, we quantified the number of cells presenting TDP‐43 aggregates. All 4 altFUS mutants displayed clear TDP‐43 aggregates and showed a 1.8‐ to 2.4‐fold increase compared with wild‐type altFUS (Fig 6B and C). This result indicates that altFUS mutations potentiate TDP‐43 cytoplasmic aggregation, a pathological hallmark in 98% of ALS cases. No aggregates of endogenous or GFP‐tagged FUS could be seen upon over‐expression of the altFUS constructs alone (Fig EV5B). Hence, some FUS mutations, synonymous for the FUS protein, exert a deleterious effect through their missense consequence on the altFUS protein.
Figure 6. FUS mutations, synonymous for FUS but missense for altFUS, potentiate TDP‐43 cytoplasmic aggregation.
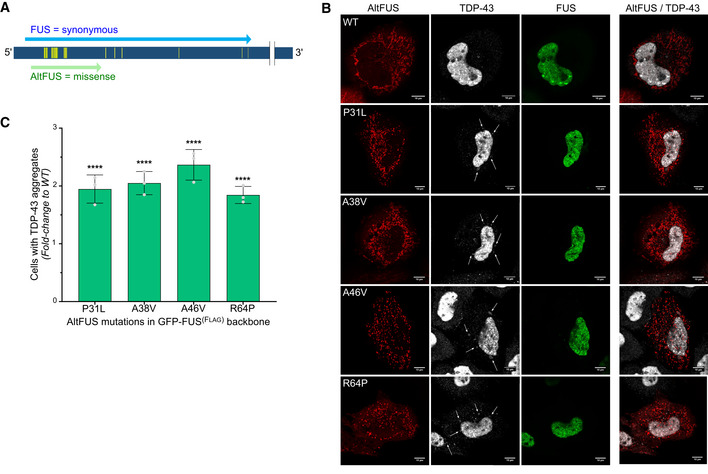
- Graphical representation of FUS synonymous mutations (yellow) found in ALS patients. The canonical FUS mRNA is represented in dark blue (ENST00000254108 or NM_004960). The FUS protein‐coding sequence is indicated in light blue, and the altFUS protein‐coding sequence is indicated in green.
- Images by confocal microscopy of TDP‐43 (white), FUS (GFP‐tagged—green) and altFUS (Flag‐tagged—red) in HeLa cells over‐expressing GFP‐FUS(F lag), GFP‐FUS(P31L‐F lag)‐s44=, GFP‐FUS(A38V‐F lag)‐g51=, GFP‐FUS(A46V‐F lag)‐g59 = and GFP‐FUS(R64P‐F lag)‐s77= (representative images from n = 3). White arrows indicate some TDP‐43 aggregates. The white scale bar corresponds to 10 μm.
- Quantification of cells with TDP‐43 aggregates in HeLa cells (see panel B). The data are represented as the fold change compared with the GFP‐FUS(F lag ) expressing cells. Statistical significance is indicated above the bars (n = 3—biological replicates, mean ± SD, ****P < 0.0001, two‐way ANOVA with Tukey’s multiple comparison correction).
Figure EV5. Mitochondrial altFUS mutants potentiates TDP‐43 cytoplasmic aggregates.
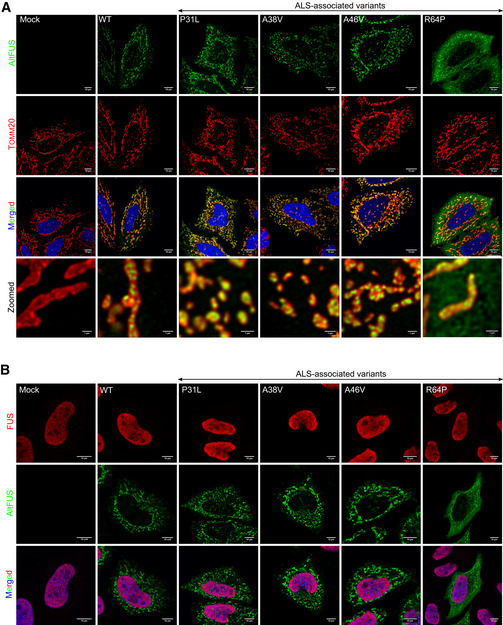
- Images by confocal microscopy of altFUS (FLAG‐tagged—green) and mitochondria (Tomm20 marker, red) in HeLa cells over‐expressing the empty vector (mock), altFUS‐Flag, altFUS‐P31L‐Flag, altFUS‐A38V‐Flag, altFUS‐A46V‐Flag or altFUS‐R64P‐Flag constructs (representative images from n = 3). The white scale bar corresponds to 10 μm. Deconvolution over a maximum of 30 iterations on the green and red channels was performed for the zoomed in pictures. The white scale bar corresponds to 1 μm.
- Images by confocal microscopy of endogenous FUS (red) and altFUS (Flag‐tagged—green) in HeLa cells over‐expressing the empty vector (mock), altFUS‐Flag, altFUS‐P31L‐Flag, altFUS‐A38V‐Flag, altFUS‐A46V‐Flag or altFUS‐R64P‐Flag constructs (representative images from n = 3). The white scale bar corresponds to 10 μm.
Discussion
Despite considerable advances in the field, current genome annotations still uphold arbitrary assumptions, such as the monocistronic nature of eukaryotic genes (Brunet et al, 2018). Here, we demonstrate FUS is a bicistronic gene. We discovered FUS CDS contains a second protein‐coding sequence in a shifted frame overlapping its prion‐like intrinsically disordered domain, regions known to host dual‐coding events (Kovacs et al, 2010; Pancsa & Tompa, 2016). This novel protein, named altFUS, is not an isoform but an entirely new sequence of 170 amino acids. AltFUS is endogenously expressed in human tissues and cultured cell lines, as demonstrated by ribosome profiling, mass spectrometry and with a custom antibody. AltFUS is notably expressed in the motor cortex and iPSCs‐derived motor neurons of healthy controls and ALS patients. Because altFUS is embedded within the FUS CDS, this discovery is of crucial importance to the field. Indeed, over‐expression studies on FUS actually implicate two proteins: FUS and altFUS. Similarly, FUS knockdown or knockout studies actually inhibit expression of both proteins (Scekic‐Zahirovic et al, 2016). Moreover, previous work has shown that gene‐editing techniques targeting a specific CDS do not necessarily result in knockout of the gene in case of dual‐coding gene (Delcourt et al, 2018). Our discovery thus suggests that FUS‐edited cells or models, notably targeting its last exons (Hicks et al, 2000; Kino et al, 2015; An et al, 2019), might only impair FUS protein expression but not altFUS, thus not resulting in a true FUS knockout. Our work provides a more accurate view of FUS coding potential to better understand its physiological function and models of FUS‐related neurodegeneration.
Following the discovery of altFUS, we developed tools in order to differentiate the specific roles and phenotypes of FUS and altFUS. Our study demonstrates that altFUS is necessary for three toxic molecular hallmarks previously attributed to FUS: mitochondrial fragmentation and loss of mitochondrial membrane potential, inhibition of autophagy and cytoplasmic aggregation of FUS. The inhibition of autophagy was observed when over‐expressing altFUS alone, absent when over‐expressing FUS alone (wild‐type or ALS‐associated R495x mutant) and reconstituted when co‐expressing FUS and altFUS. This demonstrates that the inhibition of autophagy, previously described in FUS‐ALS (Marrone et al, 2019; Ling et al, 2019), has been incorrectly associated with the FUS protein. AltFUS inhibits autophagy. Moreover, altFUS is necessary but not sufficient for the mitochondrial membrane potential loss and cytoplasmic aggregation of FUS and TDP‐43. Although TDP‐43 aggregates are not commonly seen with FUS mutations (Farrawell et al, 2015), it has already been reported (Deng et al, 2010; Shan et al, 2010; Cohen et al, 2011; Kryndushkin et al, 2011; Keller et al, 2012; Farrawell et al, 2015; Wiesner et al, 2018) and this study lends support for altFUS conspiring with FUS and TDP‐43 to lead to this molecular hallmark. Our data suggest the stoichiometry between FUS and altFUS, evaluated to 3.5 to 1 in human brain lysates, may be important for the development of cytoplasmic aggregates. Both proteins are required to observe the phenotype, highlighting a functional alliance between FUS and altFUS. This pathological synergy was also observed in the Drosophila model. The cooperation between FUS and altFUS may be orchestrated by the regulation of post‐translational modifications (PTMs) on each protein. PTMs on FUS are known to regulate its subcellular localization and/or propensity to aggregate (Rhoads et al, 2018). With preliminary evidence of altFUS carrying PTMs, a likely hypothesis is that both proteins are linked by a signalling cascade.
The physiological function of altFUS is still unclear, although our work provides evidence for its role in mitochondrial dynamics and the cellular response to stress. One mechanism put forward in ALS is that the disease originates from a sub‐optimal resolution of cellular stresses, which can come from environmental sources or mutated proteins (Al‐Chalabi & Hardiman, 2013; Monahan et al, 2016). We have shown that altFUS, not FUS, inhibits autophagy, most likely via its interaction partners (Table EV5). Since an inhibition of autophagy inhibits the dissociation of stress granules (Monahan et al, 2016), we suggest altFUS potentiates stress granule accumulation under stress conditions and that FUS phase separation properties then lead to the formation of solid and toxic aggregates (Qamar et al, 2018). Further work is needed to fully understand the role of altFUS in physiological and pathological conditions, yet our study shows this novel protein plays a crucial role in FUS‐linked gain‐of‐toxic dysfunctions in models of FUS‐related neurodegeneration. It is to note that altFUS alone did not have any effect in the Drosophila model. One likely explanation is that the expression level of altFUS in this model is not comparable to that in cultured cells. Secondly, some of altFUS partners may be absent in fruit flies, one evident cooperating protein absent in fruit flies in FUS (the fruit fly homolog being very different to the human FUS).
Recent studies have addressed the toxicity resulting from over‐expression of wild‐type FUS. Bogaert and colleagues used FUS domain truncation mutants to investigate wild‐type FUS toxicity (Bogaert et al, 2018). A FUS mutant lacking its N‐terminal intrinsically disordered domain, thus lacking altFUS, displayed reduced toxicity. This study corroborates our findings that the absence of altFUS reduces the toxicity. Furthermore, Bogaert and colleagues concluded that FUS N‐terminal synergizes with the C‐terminal domain to mediate toxicity in Drosophila (Bogaert et al, 2018). Here, we showed that altFUS synergizes with FUS to mediate ALS‐like toxic features in cultured cells and toxicity in Drosophila. Despite different laboratories and different techniques, our data are in agreement with theirs. Yet, we point to an alternative (not necessarily mutually exclusive) explanation whereby altFUS, not the FUS N‐terminal domain, synergizes for toxicity. Additionally, this discussion shows that not being aware of overlapping CDSs, especially in deletion studies, precludes alternative interpretations.
Current genome annotations guide the interpretation of data and the design of studies; however, they also affect the way we screen for pathological mutations (Brunet et al, 2018). Most of the ALS‐linked FUS mutations affect the carboxyl end of the protein, as shown with the example used in this study, the R495x mutation (Naumann et al, 2018). These mutations do not alter the altFUS protein, which is embedded at the beginning of the FUS CDS and span on exons 3 to 6. How could altFUS be important for the disease if most pathological mutations do not alter it? To answer this question, one needs to grasp how much genome annotations shape today’s research. For example, when screening for pathological mutations, those that are synonymous for FUS are discarded early in analyses as insignificant (Richards et al, 2015). Yet, a synonymous mutation for FUS may not be for altFUS. Our work shows that FUS synonymous mutations found in patients cluster on altFUS genomic locus. We tested 4 of these mutations, synonymous for the FUS protein and missense for the altFUS protein, and we found that each of them potentiated TDP‐43 cytoplasmic aggregation, a pathological hallmark of ALS (Hergesheimer et al, 2019).
Overall, we have shown that FUS is a bicistronic gene and our results indicate that altFUS interferes with mitochondrial homeostasis and autophagy in cell culture and induces motor neuron toxicity in Drosophila models. It will be important to further characterize the function of altFUS and determine whether it has a role in ALS and/or FTLD.
Materials and Methods
FUS constructs
FUS and altFUS sequences were obtained from Bio Basic Gene Synthesis service. All FUS constructs were subcloned into pcDNA3.1‐ (Invitrogen) using Gibson assembly (New England Biolabs, E26115). FUS and altFUS wild‐type sequences correspond to that of the human FUS canonical transcript (ENST00000254108 or NM_004960). FUS and altFUS proteins were either untagged or tagged with V5 (GKPIPNPLLGLDST) and 2 Flag (dykddddkdykddddk), respectively. When tagged, FUS was tagged on the N‐terminal, and altFUS was tagged on the C‐terminal. For immunofluorescence assays, N‐terminal GFP‐tagged FUS was also cloned into pcDNA3.1‐ by Gibson assembly. The necessary gBlocks were purchased from IDT. The monocistronic constructs FUS(Ø) and FUS(Ø)‐R495x were generated by mutating all altFUS methionines (ATG) to threonines (ACG). These mutations are synonymous in the FUS CDS (TAT > TAC, both coding for tyrosine). The altFUS‐mutated sequence was obtained from Bio Basic Gene Synthesis service and then subcloned in FUS sequences in pcDNA3.1‐ using Gibson assembly. The bicistronic constructs are named as follows throughout the article: FUS, FUS‐R495x, or FUS(F lag ) and FUS( flag )‐R495x when altFUS is Flag‐tagged in the + 2 reading frame. The monocistronic constructs are named as follows throughout the article: FUS(Ø) or FUS(Ø)‐R495x to indicate altFUS absence.
Cell culture, transfections, Western blots and immunofluorescence
HEK293 and HeLa cells cultures tested negative for mycoplasma contamination (ATCC 30–1012K). Transfections, immunofluorescence, confocal analyses and Western blots were carried out as previously described (Vanderperre et al, 2011). For FUS knockdown, 150,000 HEK293 cells in a 6‐well plate were transfected with 25 nM FUS SMARTpool: siGENOME siRNA (Dharmacon, Canada, L‐009497‐00‐0005) or ON‐TARGET plus Nontargeting pool siRNAs (Dharmacon, D‐001810‐10‐05) with DharmaFECT one transfection reagent (Dharmacon, T‐2001–02) according to the manufacturer’s protocol. Cell media were changed every 24 h, and cells were processed 72 h after transfection. For immunofluorescence, primary antibodies were diluted as follows: anti‐Flag (Sigma, F1804) 1/1,000, anti‐TOMM20 (Abcam, ab186734) 1/500, anti‐V5 (Cell Signalling Technologies, #13202) 1/1,000, anti‐TDP‐43 (ProteinTech, 10782‐2‐AP) 1/500 and anti‐TIAR (Cell Signalling Technologies, #8611) 1/1,600. For Western blots, primary antibodies were diluted as follows: anti‐Flag (Sigma, F1804) 1/8,000, anti‐V5 (Sigma, V8012) 1/8,000, anti‐actin (Sigma, A5441) 1/40,000, anti‐FUS (Abcam, ab84078) 1/500, anti‐altFUS (Abcam, custom antibody) 1/3,000, anti‐H3 (Cell Signalling Technologies, 9715S) 1/6,000, anti‐LC3 (Cell Signalling Technologies, #2775) 1/1,000, anti‐Hsp70 (Thermo Fisher Scientific, MA3‐028) 1/1,000, anti‐Tubulin (Thermo Fisher Scientific, a11126) 1/2,000 and anti‐VDAC (Abcam, ab15895) 1/10,000. The altFUS antibody was generated by injection two rabbits, each with 2 unique altFUS peptide (Appendix Fig S1A). The purified antibody from rabbit 2 was used in this study at a 1/2000 dilution. As our custom antibody is a polyclonal one raised against 2 peptides, it sadly did not recognize the native form of the protein. Mitochondrial morphology was evaluated using the microP tool (Peng et al, 2011). A minimum of 100 cells per replicate were counted across 3 independent experiments (n = 3, i.e. minimum 300 cells for each experimental condition). Colocalization analyses were performed using the JACoP plugin (Just Another Colocalization Plugin) implemented in ImageJ software, as previously described (Samandi et al, 2017). When specified, images obtained by confocal microscopy on the Leica TCS SP8 STED 3X were deconvolved using the Huygens software (Scientific Volume Imaging B.V., Hilversum, Netherlands). The software uses a signal reassignment algorithm for deconvolution, and identical deconvolution parameters were applied to all images. The default parameters were used, including the classic maximum‐likelihood estimation (CMLE) algorithm, signal‐to‐noise ratio and background estimation radius. The maximum iteration number was set at 30. Human tissue lysates for altFUS endogenous expression were purchased from Zyagen Laboratories (San Diego, California, USA).
Ribo‐seq data and conservation analyses
Global aggregate reads for initiating ribosomes and elongating ribosomes footprints across all available studies were downloaded from the Gwips portal (https://gwips.ucc.ie/), for Homo sapiens and for Mus musculus. For altFUS protein conservation analysis, all FUS mRNAs with at least EST evidence were retrieved across all available species from NCBI RefSeq. We performed an in silico 3‐frame translation and retrieved the best matching protein sequence per species that displayed a minimum of 20% sequence identity with the human altFUS sequence over 25% of human altFUS length. AltFUS homologous sequences were found in 83 species, and we manually added that of Drosophila melanogaster that displayed a 37.5% sequence identity over 19% of the human altFUS length. All retrieved altFUS sequences were then aligned using Clustalω with default parameters.
Peptide‐centric analysis of proteomics data sets
The stand‐alone PepQuery tool (v1.0) (Wen et al, 2019) was downloaded from the PepQuery website (http://www.pepquery.org/). The tool was run on the following data sets from the TCGA consortium: colon cancer (COCA) proteome, ovarian cancer (OVCA) proteome, phosphoproteome and glycoproteome, and the breast cancer (BRCA) proteome and phosphoproteome. The reference database was set to the Ensembl database (hg38_Ensembl_20190910). The following parameters were set for all runs unless specified: carbamidomethylation of cysteine as fixed modification (as well as iTRAQ 4‐plex of K, iTRAQ 4‐plex of peptide N‐term for BRCA and OVCA); oxidation of methionine as variable modification (as well as iTRAQ 4‐plex of Y for BRCA and OVCA); a maximum of 3 modifications per peptides; trypsin digestion with maximum of 1 miscleavage; precursor tolerance of 10 ppm (20 ppm for COCA); fragment mass tolerance of 0.05 Da (0.6 Da for COCA); and the hyperscore was used as a scoring metric, and 10 000 randoms. For phosphoproteomes, phosphorylation of Y, T and S was added as variable modifications. For the glycoproteome, deamidation of Q and N was added as variable modifications. PepQuery was run in the protein mode, with altFUS (IP_243680) whole sequence as input. Spectra were visualized and drawn using an in‐house python script.
Human induced pluripotent stem cell differentiation into motor neurons
Directed differentiation to human iPSC‐motor neurons was performed as previously reported (Hall et al, 2017). Briefly, iPSCs were maintained on Geltrex (Life Technologies) with Essential 8 Medium Media (Life Technologies) and passaged using EDTA (Life Technologies, 0.5 mM). All cell cultures were maintained at 37°C and 5% carbon dioxide. For motor neuron differentiation, iPSCs were differentiated to neuroepithelium by plating to 100% confluency in chemically defined medium consisting of DMEM/F12 GlutaMAX, Neurobasal, L‐Glutamine, N2 supplement, non‐essential amino acids, B27 supplement, β‐mercaptoethanol (Life Technologies) and insulin (Sigma). Treatment with the following small molecules from day 0–7: 1 µM dorsomorphin (Millipore), 2 µM SB431542 (Tocris Bioscience) and 3.3 µM CHIR99021 (Miltenyi Biotec). At day 8, cells patterned for 7 days with 0.5 µM retinoic acid and 1 µM purmorphamine. At day 14, spinal cord motor neuron precursors were treated with 0.1 µM purmorphamine for a further 4 days before being terminally differentiated for > 10 days in 0.1 µM Compound E (Enzo Life Sciences) to promote cell cycle exit. Throughout the neural conversion and patterning phase (D0‐18), the neuroepithelial layer was enzymatically dissociated twice (at D4‐5 and D10‐12) using dispase (GIBCO, 1 mg/ml).
Preparation of tissue lysates of the motor cortex of ALS patients
Approximately 100mg of motor cortex from 4 sporadic ALS and 4 C9orf72‐ALS cases was lysed in 10× RIPA (50 mM Tris–HCl pH 7.8, 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP40; supplemented with protease inhibitors and EDTA) volume using TissueLyser equipment (Qiagen). Lysates were incubated on ice 20 min followed by centrifugation at 20,000 g for 20 min at 4°C. Supernatant was taken as “RIPA fraction”, and pellets were resuspended in RIPA and SDS (final concentration of 2%). 3 sporadic ALS and 3 C9orf72‐ALS samples were subsequently used as they were sufficiently concentrated to load 100 ug of proteins onto SDS–PAGE gels.
Recombinant proteins purification
GFP‐FUS recombinant protein was purified from HEK293 cells. Briefly, the GFP‐FUS construct was transfected in HEK293 cells. Cells were grown up to 80% confluence, rinsed twice with PBS, pelleted and snap‐frozen at −80°C. Cells were then resuspended in lysis buffer (10 mM HEPES‐NaOH pH 7.6, 3 mM MgCl2, 300 mM KCl, 5% glycerol, 0.5% NP‐40) complemented with phosphatase and protease inhibitors, and incubated on ice for 30 mins. The lysates were centrifuged at 16,000 g for 10 mins at 4°C. The supernatant was added to GFP‐trap sepharose beads (ChromoTek, Germany) and incubated at 4°C for 3 h on rotation. The beads were then washed 5 times with the washing buffer (10 mM HEPES‐NaOH pH 7.6, 3 mM MgCl2, 5% glycerol, 0.5% NP‐40) at increasing concentrations of KCl: twice with 100 mM KCl, once with 200 mM KCl and twice with 400 mM KCl. The GFP‐FUS protein was then eluted from the beads using 50 μl of acidic glycine solution (0.1 M, pH 3). After centrifugation at 2,500 g for 3 min at 4°C, the elution was transferred to a new Eppendorf tube and equilibrated with 50 μl of Tris solution (1 M ph 8). The elution step was repeated three times for a final total volume of 300 μl.
GST‐altFUS recombinant protein was purified from RosettaTM competent cells. AltFUS sequence was subcloned in the pGEX‐4T1. Briefly, GST‐altFUS expression was induced in competent cells at a OD600 of 0.5 with 0.5 mM of IPTG and 2% ethanol. Cells were then left to grow for 1.5 h at 37°C. Cells were then centrifuged at 5,000 rpm for 10 mins at 4°C and resuspended in lysis buffer (1 mM EDTA, 250 mM NaCl, 10% glycerol, 1 mM DTT, 25 mg/ml lysozyme, 1% Triton X‐100, in PBS) and sonicated prior to incubation on rotation for 30 mins at 4°C. Both the soluble and insoluble fractions were then collected and loaded on an acrylamide gel. After Coomassie coloration, GST‐altFUS protein was found mostly in the insoluble fraction. Thus, the pellet was resuspended in 8M Urea lysis buffer to ensure resuspension and then dialysed using a 2K 0.5ML 10/PK Slide‐A‐Lyser cassette (#PI66205, Thermo Fisher) in the usual lysis buffer (no urea). The dialysed lysate was then incubated with GST‐trap sepharose beads (Glutathione Sepharose 4B—GE Healthcare, 17‐0756‐01) for 3 h on rotation at 4°C. The beads were then washed 5 times: twice with lysis buffer, once with PBS and twice with washing buffer (50 mM Tris pH 8.0, 250 mM NaCl, 0.1% Triton X‐100). The GST‐altFUS protein was then eluted by incubating the beads in the elution buffer (10 mM reduced glutathione, 50 mM Tris pH 8.0, 250 mM NaCl, 10% glycerol and 0.1% Triton X‐100) on rotation for 15 mins at 4 °C. After centrifugation at 150 g for 3 min at 4°C, the elution was transferred to a new Eppendorf tube. The elution step was repeated twice.
The GFP‐FUS and GST‐alFUS recombinant proteins were dosed using a commercial LSD1 recombinant protein (BML‐SE544, Enzo Life Sciences) of known concentration. The concentration evaluated by this standard curve was then confirmed by NanoDrop quantification of total protein.
Mitochondrial extracts and cellular fractionation
Mitochondrial extracts were prepared as previously described (Delcourt et al, 2018). Briefly, HEK293 cells grown up to 80% confluence, were rinsed twice with PBS and gathered using a cell scraper. Cells were pelleted by centrifugation at 500 g for 10 min at 4°C. Supernatant was discarded, and cells were suspended in mitochondrial buffer (210 mM mannitol, 70 mM sucrose, 1 mM EDTA, 10 mM HEPES‐NaOH, pH 7.5, 0.5 mM PMSF and EDTA‐free protease inhibitor (Thermo Fisher Scientific)). Cells were disrupted by 15 consecutive passages through a 25G1 0.5 × 25 needle syringe on ice, followed by a 3‐min centrifugation at 2,000 g at 4°C. Supernatant was collected, and the pellet was suspended in mitochondrial buffer. The cell disruption was repeated four times, and all retrieved supernatants containing mitochondria were again passed through syringe needle in mitochondrial buffer and cleared by centrifugation for 3 min at 2,000 g at 4°C. Supernatants were pooled and centrifuged for 10 min at 13,000 g at 4°C to pellet mitochondria. The pellet was suspended in 200 μl of mitochondrial buffer until further processing. Cellular fractionation was performed using the Cell Fractionation Kit (#9038S, Cell Signaling Technology). Briefly, HEK293 cells were grown up to 80% confluence, washed twice with PBS and gathered using a cell scraper. Cells were spun at 350 g for 5 min at 4°C, and 2.5 × 106 cells were suspended in 500 μl of ice‐cold PBS. An aliquot of 100 μl was spun at 350 g for 5 min at 4°C and resuspended in SDS buffer (4% SDS, Tris–HCl 100 mM pH 7.6) and kept as WCL (whole cell lysate). The rest of the collected cells (remaining 400 μl) were spun at 500 g for 5 min at 4°C. Supernatant was discarded, and pellet was resuspended in 500 μl of CIB (cytoplasmic isolation buffer) from the kit, vortexed for 5 sec and incubated on ice for 5 min. After centrifugation at 500 g for 5 min at 4°C, the supernatant was collected as the cytosplasmic fraction. The pellet was resuspended in 500 μl of MIB buffer (membrane isolation buffer) from the kit, vortexed for 15 sec and incubated on ice for 5 min. After centrifugation at 8,000 g for 5 min at 4°C, the supernatant was collected as the membrane and organelles fraction. To each 100 μl of fraction was added 60 μl of loading buffer 1× (from Cold Spring Laemmli sample buffer: 50 mM Tris pH 6.8, 2% SDS, 10% glycerol, 5% β‐mercaptoethanol) before processing for Western blot.
Mitochondrial membrane potential measurements
Mitochondrial membrane potential was measured by flow cytometry in HEK293 cells using TMRE (tetramethylrhodamine ethyl ester, Abcam, ab113852). FCCP was used as a positive control to validate each independent experiment. Cells were grown up to 80% confluence and washed twice with PBS. The cells were then incubated for 5 mins at 37°C, 5% CO2 with PBS/A (0.2% BSA in PBS) solution (experimental) or 3 μM FCCP in PBS/A solution (positive control). Then, 100 nM of TMRE was added and cells were incubated 15 min at 37°C, 5% CO2. After incubation, cells were trypsinized and centrifuged at 800 g for 5 min at 4°C and resuspended in 500 μl of PBS and kept on ice. Cells were immediately analysed by flow cytometry. A gate for living cells was set, as well as a second gate to filter out cell doublets. TMRE fluorescence (PE‐A) was recorded over a minimum of 50,000 gated cells for each experimental condition. The mean TMRE fluorescence intensity was measured over 3 independent experiments for each experimental condition.
Stimulated Emission Depletion (STED) microscopy
Samples were prepared as described above for confocal microscopy. A Leica TCS SP8 STED 3X was used with a 100x objective lens and immersion oil for Dual Color STED images. Images were obtained by sequential scanning of a given area. The combination of Alexa Fluor 488 (Thermo Fisher Scientific, A‐11017) and Alexa Fluor 568 (Thermo Fisher Scientific, A‐21069) dyes was chosen for STED imaging. Alexa Fluor 488 dye was excited with a white light laser (WLL) at 488 nm and was depleted using the 660 nm STED laser. Alexa Fluor 568 dye was excited with a WLL at 561 nm and was depleted using the 660 nm STED laser. The STED laser (660 nm) was applied at 80% of maximum power.
Fast Protein Liquid Chromatography (FPLC) and affinity purification–mass spectrometry (AP‐MS)
Mitochondrial extracts of HEK293 cells were centrifuged at 13,000 g for 10 min at 4°C to remove the supernatant and were resuspended in FPLC buffer (50 mM Tris–HCl, 1 mM EDTA, 150 mM NaCl, 1% Triton X‐100, pH 7.5, filtered with 0.2‐μm filters) at 2 mg/ml for a total of 4 mg of mitochondrial proteins. Samples were incubated on ice for 15 min and then centrifuged at 10,000 g for 5 min at 4°C, and the supernatant was loaded in the injection syringe without disrupting the pellet. The FPLC was performed on a HiLoad 16/60 Superdex 200 pg Column (GE Healthcare, Chicago, USA) at 4°C. The column was pre‐equilibrated with the FPLC buffer for up to 0.2 CV (column volume), and the sample was applied at a flow rate of 0.5 ml/min with a pressure alarm set at 0.5 MPa. The elution was performed over 72 fractions of 1.5 ml for a maximum of 1.1 CV. For altFUS probing by Western blot, proteins were precipitated from 150 μl of each 4 fractions in technical duplicates. First, 600 μl of methanol was added to each tube and mixed gently, before adding 150 μl of chloroform. Tubes were gently inverted 10 times before adding 450 μl of milli‐Q H2O and vortexing briefly. After centrifugation at 12,000 g for 3 min, the upper phase was discarded, and 400 μl of methanol was added. Tubes are centrifuged at 16,000 g for 4 min, and the pellet was resuspended in loading buffer. For interactome analysis by mass spectrometry, fractions of interest (8–14) were pooled together and incubated at 4°C overnight with magnetic Flag beads (Sigma, M8823) pre‐conditioned with FPLC buffer. The beads were then washed 3 times with 5 ml of FPLC buffer and 5 times with 5 ml of 20 mM NH4HCO3 (ABC). Proteins were eluted and reduced from the beads using 10 mM DTT (15 min at 55°C) and then treated with 20 mM IAA (1 h at room temperature in the dark). Proteins were digested overnight by adding 1 μg of Trypsin (Promega, Madison, Wisconsin) in 100 μl ABC at 37°C overnight. Digestion was quenched using 1% formic acid, and supernatant was collected. Beads were washed once with acetonitrile/water/formic acid (1/1/0.01 v/v) and pooled with supernatant. Peptides were dried with a SpeedVac, desalted using a C18 Zip‐Tip (Millipore Sigma, Etobicoke, Ontario, Canada) and resuspended into 30 μl of 1% formic acid in water prior to MS analysis.
Mass spectrometry analysis
Peptides were separated in a PepMap C18 Nano Column (75 μm × 50 cm, Thermo Fisher Scientific). The setup used a 0–35% gradient (0–215 min) of 90% acetonitrile, 0.1% formic acid at a flow rate of 200 nl/min followed by acetonitrile wash and column re‐equilibration for a total gradient duration of 4 h with a RSLC Ultimate 3000 (Thermo Fisher Scientific, Dionex). Peptides were sprayed using an EASY‐Spray source (Thermo Fisher Scientific) at 2 kV coupled to a Quadrupole‐Orbitrap (Q Exactive, Thermo Fisher Scientific) mass spectrometer. Full‐MS spectra within a m/z 350–1,600 mass range at 70,000 resolution were acquired with an automatic gain control (AGC) target of 1e6 and a maximum accumulation time (maximum IT) of 20 ms. Fragmentation (MS/MS) of the top ten ions detected in the Full‐MS scan at 17,500 resolution, AGC target of 5e5, a maximum IT of 60 ms with a fixed first mass of 50 within a 3 m/z isolation window at a normalized collision energy (NCE) of 25. Dynamic exclusion was set to 40 s. Mass spectrometry RAW files were searched with Andromeda search engine implemented in MaxQuant 1.5.5.1. The digestion mode was set at Trypsin/P with a maximum of two missed cleavages per peptides. Oxidation of methionine and acetylation of N‐terminal were set as variable modifications, and carbamidomethylation of cysteine was set as fixed modification. Precursor and fragment tolerances were set at 4.5 and 20 ppm, respectively. Files were searched using a target‐decoy approach against UniprotKB (Homo sapiens 03/2017 release) with the addition of altFUS sequence for a total of 92,949 entries. The false discovery rate (FDR) was set at 1% for peptide spectrum match, peptide and protein levels. Protein interactions were then scored using the SAINT algorithm, with Mock cells as control and the magnetic Flag beads in HEK293 cell CRAPome (Choi et al, 2011). Proteins with a SAINT score above 0.99 were considered, as well as those presenting a SAINT score above 0.88 with a minimum of two unique peptides.
Biological processes and cellular compartment enrichment analysis
Proteins identified in altFUS interactome were screened for cellular compartment and biological processes enrichment using Gene Ontology (GO) enrichment. Proteins were queried against the whole human proteome for cellular compartment and against the human mitochondrial proteome (MitoCarta 2.0) for biological processes. The statistical analysis used Fisher’s exact test with a FDR set at 1%.
Autophagic flux measurements
The mCherry‐GFP‐LC3 was used to evaluate the autophagic vesicles within HeLa cells by confocal microscopy. Before fusion with the lysosome, the LC3 molecules on the autophagosome display a yellow fluorescence (combined mCherry and GFP fluorescence). After fusion, the GFP fluorescence is quenched by the lysosomal pH, and as such, the LC3 molecules display a red signal (mCherry alone). This allows a visual representation of the autophagic flux in a given cell. Cells treated with 50 nM bafilomycin for 4 h were used as a positive control to validate each independent experiment. Observations were made across 2 technical duplicates for each biological condition, across 3 independent experiments (n = 3). Alternatively, the autophagic flux was also evaluated by LC3 probing before and after bafilomycin treatment (50 nM for 4 h). The quantification corresponds to the treated/ untreated ratio of LC3‐II abundance.
Cytoplasmic aggregates measurements
Images of HeLa cells were taken by confocal microscopy and then processed using the Image J 3D Objects Counter plugin. FUS cytoplasmic aggregates were then quantified in number and size (μm2) for each cell. A total of 100 cells across two technical replicates were taken for each independent experiment (n = 3, i.e. a minimum of 300 cells per biological conditions).
Transgenic Drosophila and climbing assay
The bicistronic constructs, FUS and FUS‐R495x, and the monocistronic constructs, altFUS, FUS(Ø) and FUS(Ø)‐R495x, were subcloned in the pUASTattB expression vector for site‐specific insertion into attP2 on chromosome 3. Transgenic flies were generated by Best Gene (Best Gene Inc., California, USA). The Elav‐GeneSwitch‐GAL4 driver (stock number: 43642, genotype: y[1] w[*]; P{w[+mC]=elav‐Switch.O}GSG301) and the UAS‐mCherry flies (stock number: 35787, genotype: y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=UAS‐mCherry.VALIUM10}attP2) was purchased from Bloomington (Bloomington Drosophila Stock Center, Indiana, USA). All stocks were in a w1118 background and were cultured on standard medium at 25°C or room temperature. Transgenic flies were crossed with the Elav‐GeneSwitch‐GAL4 driver strain. The F1 was equally divided into two groups with equal proportion of males and females: one group will feed on standard food supplemented with ethanol (0.2%—control flies) and the other on standard food supplemented with RU‐486 at 10 μM diluted in ethanol (induced flies). The climbing assay was performed as previously described (Chambers et al, 2013). Briefly, flies were transferred into an empty vial and tapped to the bottom. After 18 s, the number of flies at the top of the tube was considered successful. The assay was done at days 1, 10 and 20 post‐induction, across 4 independent F1. Five flies were taken at days 1, 10 and 20 post‐induction to validate expression of the proteins of interest.
Statistical analyses and representation
Unless otherwise stated, the statistical analysis carried was a two‐way ANOVA with Tukey’s multiple comparison correction. The box plots represent the mean with the 5–95% percentile. The bar graphs represent the mean, and error bars correspond to the standard deviation. When using parametric tests, normality of data distribution was verified beforehand using the Shapiro–Wilk test.
Author contributions
MAB and XR designed and wrote the study. MAB and J‐FJ did the experiments, and MAB did the analyses and figures. SN and SJ assisted with the Drosophila experiments. PM, LZ and JR provided the motor cortex tissue lysates. GET and RP provided the iPSC‐derived motor neurons. All authors proofread the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Review Process File
Acknowledgements
This research was supported by CIHR grants MOP‐137056 and MOP‐136962, by an ALS Canada Project Grant and by a Canada Research Chair in Functional Proteomics and Discovery of Novel Proteins to X.R. X.R and S.J are members of the Fonds de Recherche du Québec Santé (FRQS)‐supported Centre de Recherche du Centre Hospitalier Universitaire de Sherbrooke. P.M. is supported by the ALS Double Play Christopher Chiu Postdoctoral Fellowship. We thank J. Ule, M‐J. Boucher and D. Hunting for helpful discussions. The autopsy programme is supported by the James Hunter and Family ALS Initiative. The Genotype‐Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal in April 2019.
EMBO Reports (2021) 22: e50640.
Contributor Information
Marie A Brunet, Email: marie.brunet@usherbrooke.ca.
Xavier Roucou, Email: xavier.roucou@usherbrooke.ca.
Data availability
The OpenProt database is available at www.openprot.org. The GTEx portal is available via www.gtexportal.org. The GWIPS portal is available at www.gwips.ucc.ie. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/projects/PXD021320) and assigned the identifier PXD021320. AltFUS coding sequence was submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and assigned the identifier BK012000. Any other relevant data are available from the corresponding authors upon reasonable request.
References
- Ajmone‐Cat MA, Onori A, Toselli C, Stronati E, Morlando M, Bozzoni I, Monni E, Kokaia Z, Lupo G, Minghetti L et al (2019) Increased FUS levels in astrocytes leads to astrocyte and microglia activation and neuronal death. Sci Rep 9: 4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Chalabi A, Hardiman O (2013) The epidemiology of ALS: a conspiracy of genes, environment and time Nat Rev Neurol 9: 617–628 [DOI] [PubMed] [Google Scholar]
- An H, Skelt L, Notaro A, Highley JR, Fox AH, La Bella V, Buchman VL, Shelkovnikova TA (2019) ALS‐linked FUS mutations confer loss and gain of function in the nucleus by promoting excessive formation of dysfunctional paraspeckles. Acta Neuropathol Commun 7: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A, Vande Velde C (2015) Alterations in stress granule dynamics driven by TDP‐43 and FUS: a link to pathological inclusions in ALS? Front Cell Neurosci 9: 423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert E, Boeynaems S, Kato M, Guo L, Caulfield TR, Steyaert J, Scheveneels W, Wilmans N, Haeck W, Hersmus N et al (2018) Molecular dissection of FUS points at synergistic effect of low‐complexity domains in toxicity. Cell Rep 24: 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet MA, Brunelle M, Lucier J‐F, Delcourt V, Levesque M, Grenier F, Samandi S, Leblanc S, Aguilar J‐D, Dufour P et al (2019) OpenProt: a more comprehensive guide to explore eukaryotic coding potential and proteomes. Nucleic Acids Res 47: D403–D410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet MA, Levesque SA, Hunting DJ, Cohen AA, Roucou X (2018) Recognition of the polycistronic nature of human genes is critical to understanding the genotype‐phenotype relationship. Genome Res 28: 609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrì MT, D’Ambrosi N, Cozzolino M (2017) Pathways to mitochondrial dysfunction in ALS pathogenesis. Biochem Biophys Res Commun 483: 1187–1193 [DOI] [PubMed] [Google Scholar]
- Chambers RP, Call GB, Meyer D, Smith J, Techau JA, Pearman K, Buhlman LM (2013) Nicotine increases lifespan and rescues olfactory and motor deficits in a Drosophila model of Parkinson’s disease. Behav Brain Res 253: 95–102 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang M, Deng J, Chen X, Ye Y, Zhu L, Liu J, Ye H, Shen Y, Li Y et al (2011) Expression of human FUS protein in Drosophila leads to progressive neurodegeneration. Protein Cell 2: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick JM, Kolippakkam D, Nusinow DP, Zhai B, Rad R, Huttlin EL, Gygi SP (2015) A mass‐tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat Biotechnol 33: 743–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin Z‐Y, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras A‐C, Nesvizhskii AI (2011) SAINT: Probabilistic Scoring of Affinity Purification ‐ Mass Spectrometry Data. Nat Methods 8: 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NR, Hammans SR, Macpherson J, Nicoll JAR (2011) New neuropathological findings in Unverricht‐Lundborg disease: neuronal intranuclear and cytoplasmic inclusions. Acta Neuropathol (Berl) 121: 421–427 [DOI] [PubMed] [Google Scholar]
- Collins MA, An J, Hood BL, Conrads TP, Bowser RP (2015) Label‐free LC–MS/MS proteomic analysis of cerebrospinal fluid identifies protein/pathway alterations and candidate biomarkers for amyotrophic lateral sclerosis. J Proteome Res 14: 4486–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcourt V, Brunelle M, Roy AV, Jacques J‐F, Salzet M, Fournier I, Roucou X (2018) The protein coded by a short open reading frame, not by the annotated coding sequence, is the main gene product of the dual‐coding gene MIEF1. Mol Cell Proteomics MCP 17: 2402–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcourt V, Staskevicius A, Salzet M, Fournier I, Roucou X (2018) Small proteins encoded by unannotated ORFs are rising stars of the proteome, confirming shortcomings in genome annotations and current vision of an mRNA. Proteomics 18: e1700058. [DOI] [PubMed] [Google Scholar]
- Deng H, Gao K, Jankovic J (2014) The role of FUS gene variants in neurodegenerative diseases. Nat Rev Neurol 10: 337–348 [DOI] [PubMed] [Google Scholar]
- Deng H‐X, Zhai H, Bigio EH, Yan J, Fecto F, Ajroud K, Mishra M, Ajroud‐Driss S, Heller S, Sufit R et al (2010) FUS‐immunoreactive inclusions are a common feature in sporadic and non‐SOD1 familial amyotrophic lateral sclerosis. Ann Neurol 67: 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Yang M, Chen Y, Chen X, Liu J, Sun S, Cheng H, Li Y, Bigio EH, Mesulam M et al (2015) FUS interacts with HSP60 to promote mitochondrial damage. PLOS Genet 11: e1005357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didonna A, Benetti F (2015) Post‐translational modifications in neurodegeneration. Biophys 3: 27–49. [Google Scholar]
- Dini Modigliani S, Morlando M, Errichelli L, Sabatelli M, Bozzoni I (2014) An ALS‐associated mutation in the FUS 3′‐UTR disrupts a microRNA–FUS regulatory circuitry. Nat Commun 5: 4335 [DOI] [PubMed] [Google Scholar]
- Farrawell NE, Lambert‐Smith IA, Warraich ST, Blair IP, Saunders DN, Hatters DM, Yerbury JJ (2015) Distinct partitioning of ALS associated TDP‐43, FUS and SOD1 mutants into cellular inclusions. Sci Rep 5: 13416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CE, Yao Z, Choi M, Tyzack GE, Serio A, Luisier R, Harley J, Preza E, Arber C, Crisp SJ et al (2017) Progressive motor neuron pathology and the role of astrocytes in a human stem cell model of VCP‐related ALS. Cell Rep 19: 1739–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Akimoto Y (2009) The O‐GlcNAc modification, In Essentials of Glycobiology, Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. (eds.), Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, Chapter 18 [Google Scholar]
- Hergesheimer RC, Chami AA, de Assis DR , Vourc’h P, Andres CR, Corcia P, Lanznaster D, Blasco H (2019) The debated toxic role of aggregated TDP‐43 in amyotrophic lateral sclerosis: a resolution in sight? Brain 142: 1176–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks GG, Singh N, Nashabi A, Mai S, Bozek G, Klewes L, Arapovic D, White EK, Koury MJ, Oltz EM et al (2000) Fus deficiency in mice results in defective B‐lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet 24: 175–179 [DOI] [PubMed] [Google Scholar]
- Ju S, Tardiff DF, Han H, Divya K, Zhong Q, Maquat LE, Bosco DA, Hayward LJ, Brown RH, Lindquist S et al (2011) A Yeast Model of FUS/TLS‐Dependent Cytotoxicity. PLoS Biol 9: e1001052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller BA, Volkening K, Droppelmann CA, Ang LC, Rademakers R, Strong MJ (2012) Co‐aggregation of RNA binding proteins in ALS spinal motor neurons: evidence of a common pathogenic mechanism. Acta Neuropathol (Berl) 124: 733–747 [DOI] [PubMed] [Google Scholar]
- Kino Y, Washizu C, Kurosawa M, Yamada M, Miyazaki H, Akagi T, Hashikawa T, Doi H, Takumi T, Hicks GG et al (2015) FUS/TLS deficiency causes behavioral and pathological abnormalities distinct from amyotrophic lateral sclerosis. Acta Neuropathol Commun 3: 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs E, Tompa P, Liliom K, Kalmar L (2010) Dual coding in alternative reading frames correlates with intrinsic protein disorder. Proc Natl Acad Sci 107: 5429–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D, Wickner RB, Shewmaker F (2011) FUS/TLS forms cytoplasmic aggregates, inhibits cell growth and interacts with TDP‐43 in a yeast model of amyotrophic lateral sclerosis. Protein Cell 2: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanson NA, Maltare A, King H, Smith R, Kim JH, Taylor JP, Lloyd TE, Pandey UB (2011) A Drosophila model of FUS‐related neurodegeneration reveals genetic interaction between FUS and TDP‐43. Hum Mol Genet 20: 2510–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S‐C, Dastidar SG, Tokunaga S, Ho WY, Lim K, Ilieva H, Parone PA, Tyan S‐H, Tse TM, Chang J‐C et al (2019) Overriding FUS autoregulation in mice triggers gain‐of‐toxic dysfunctions in RNA metabolism and autophagy‐lysosome axis. eLife 8: e40811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisier R, Tyzack GE, Hall CE, Mitchell JS, Devine H, Taha DM, Malik B, Meyer I, Greensmith L, Newcombe J et al (2018) Intron retention and nuclear loss of SFPQ are molecular hallmarks of ALS. Nat Commun 9: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone L, Drexler HCA, Wang J, Tripathi P, Distler T, Heisterkamp P, Anderson EN, Kour S, Moraiti A, Maharana S, et al (2019) FUS pathology in ALS is linked to alterations in multiple ALS‐associated proteins and rescued by drugs stimulating autophagy. Acta Neuropathologica 138: 67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawhinney RMS, Staveley BE (2011) Expression of GFP can influence aging and climbing ability in Drosophila. Genet Mol Res GMR 10: 494–505 [DOI] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert J‐P, St‐Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY et al (2013) The CRAPome: a contaminant repository for affinity purification‐mass spectrometry data. Nat Methods 10: 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Kiniry SJ, O’Connor PBF, Mullan JP, Baranov PV. GWIPS‐viz: 2018 update. Nucleic Acids Res 46: D823–D830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel L, Avequin T, Delarue M, Feuillette S, Frébourg T, Campion D, Lecourtois M (2012) Accumulation of insoluble forms of FUS protein correlates with toxicity in Drosophila. Neurobiol Aging 33: 1008.e1–1008.e15 [DOI] [PubMed] [Google Scholar]
- Monahan Z, Shewmaker F, Pandey UB (2016) Stress granules at the intersection of autophagy and ALS. Brain Res 1649: 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya T, Maragkakis M (2018) Amyotrophic Lateral Sclerosis associated FUS mutation shortens mitochondria and induces neurotoxicity. Sci Rep 8: 15575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M, Pal A, Goswami A, Lojewski X, Japtok J, Vehlow A, Naujock M, Günther R, Jin M, Stanslowsky N et al (2018) Impaired DNA damage response signaling by FUS‐NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat Commun 9: 335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan M, Talbot K, Ansorge O (2016) Pathogenesis of FUS‐associated ALS and FTD: insights from rodent models. Acta Neuropathol Commun 4: 99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olexiouk V, Van Criekinge W, Menschaert G (2018) An update on sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res 46: D497–D502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancsa R, Tompa P (2016) Coding regions of intrinsic disorder accommodate parallel functions. Trends Biochem Sci 41: 898–906 [DOI] [PubMed] [Google Scholar]
- Pavesi A (2019) Asymmetric evolution in viral overlapping genes is a source of selective protein adaptation. Virology 532: 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J‐Y, Lin C‐C, Chen Y‐J, Kao L‐S, Liu Y‐C, Chou C‐C, Huang Y‐H, Chang F‐R, Wu Y‐C, Tsai Y‐S et al (2011) Automatic Morphological Subtyping Reveals New Roles of Caspases in Mitochondrial Dynamics. PLOS Comput Biol 7: e1002212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, Phillips EC, Miyashita A, Williams D, Ströhl F et al (2018) FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation‐π interactions. Cell 173: 720–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads SN, Monahan ZT, Yee DS, Shewmaker FP (2018) The role of post‐translational modifications on prion‐like aggregation and liquid‐phase separation of FUS. Int J Mol Sci 19: 886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier‐Foster J, Grody WW, Hegde M, Lyon E, Spector E et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med Off J Am Coll Med Genet 17: 405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles‐Murguia M, Hunt LC, Finkelstein D, Fan Y, Demontis F (2019) Tissue‐specific alteration of gene expression and function by RU486 and the GeneSwitch system. Npj Aging Mech Dis 5: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatelli M, Moncada A, Conte A, Lattante S, Marangi G, Luigetti M, Lucchini M, Mirabella M, Romano A, Del Grande A et al (2013) Mutations in the 3′ untranslated region of FUS causing FUS overexpression are associated with amyotrophic lateral sclerosis. Hum Mol Genet 22: 4748–4755 [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Couso JP (2015) Discovery and characterization of smORF‐encoded bioactive polypeptides. Nat Chem Biol 11: 909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samandi S, Roy AV, Delcourt V, Lucier J‐F, Gagnon J, Beaudoin MC, Vanderperre B, Breton M‐A, Motard J, Jacques J‐F et al (2017) Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. eLife 6: e27860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AL, Lindner AB (2017) Protein posttranslational modifications: roles in aging and age‐related disease. Oxid Med Cell Longev 2017: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scekic‐Zahirovic J, Sendscheid O, El Oussini H, Jambeau M, Sun Y, Mersmann S, Wagner M, Dieterlé S, Sinniger J, Dirrig‐Grosch S et al (2016) Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. EMBO J 35: 1077–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Chiang P‐M, Price DL, Wong PC (2010) Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP‐43 transgenic mice. Proc Natl Acad Sci USA 107: 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y, Cortopassi G (2016) Mitochondrial Hspa9/Mortalin regulates erythroid differentiation via iron‐sulfur cluster assembly. Mitochondrion 26: 94–103 [DOI] [PubMed] [Google Scholar]
- Taylor JP, Brown RH Jr, Cleveland DW (2016) Decoding ALS: from genes to mechanism. Nature 539: 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JW, Sorum AW, Hsieh‐Wilson LC (2018) Deciphering the Functions of O‐GlcNAc Glycosylation in the Brain: The Role of Site‐Specific Quantitative O‐GlcNAcomics. Biochemistry 57: 4010–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyzack GE, Luisier R, Taha DM, Neeves J, Modic M, Mitchell JS, Meyer I, Greensmith L, Newcombe J, Ule J et al (2019) Widespread FUS mislocalization is a molecular hallmark of amyotrophic lateral sclerosis. Brain 142: 2572–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umoh ME, Dammer EB, Dai J, Duong DM, Lah JJ, Levey AI, Gearing M, Glass JD, Seyfried NT (2017) A proteomic network approach across the ALS‐FTD disease spectrum resolves clinical phenotypes and genetic vulnerability in human brain. EMBO Mol Med 10: 48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobágyi T, Vos KJD, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P et al (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderperre B, Staskevicius AB, Tremblay G, McCoy M, O’Neill MA, Cashman NR, Roucou X (2011) An overlapping reading frame in the PRNP gene encodes a novel polypeptide distinct from the prion protein. FASEB J 25: 2373–2386 [DOI] [PubMed] [Google Scholar]
- Wen B, Wang X, Zhang B (2019) PepQuery enables fast, accurate, and convenient proteomic validation of novel genomic alterations. Genome Res 29(3): 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner D, Tar L, Linkus B, Chandrasekar A, Olde Heuvel F, Dupuis L, Tsao W, Wong PC, Ludolph A, Roselli F (2018) Reversible induction of TDP‐43 granules in cortical neurons after traumatic injury. Exp Neurol 299: 15–25 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG et al (2018) Ensembl 2018. Nucleic Acids Res 46: D754–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z‐Y, Peng Y, Feng X‐H, Wang X‐N, Sun Q, Liu M‐S, Li X‐G, Cui L‐Y (2012) Screening of the FUS gene in familial and sporadic amyotrophic lateral sclerosis patients of Chinese origin. Eur J Neurol 19: 977–983 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Review Process File
Data Availability Statement
The OpenProt database is available at www.openprot.org. The GTEx portal is available via www.gtexportal.org. The GWIPS portal is available at www.gwips.ucc.ie. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/projects/PXD021320) and assigned the identifier PXD021320. AltFUS coding sequence was submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and assigned the identifier BK012000. Any other relevant data are available from the corresponding authors upon reasonable request.


