Abstract
Fibroblast growth factor 21 (FGF21) is a regulator of glucose and lipid metabolism. It has been widely considered as a promising candidate for the treatment of type 2 diabetes mellitus (T2DM) and other related metabolic disorders. However, lack of structural and dynamic information has limited FGF21‐based drug development. Here, using nuclear magnetic resonance (NMR) spectroscopy, we determine the structure of FGF21 and find that its non‐canonical flexible β‐trefoil conformation affects the folding of β2‐β3 hairpin and further overall protein stability. To modulate folding dynamics, we designed an FGF21‐FGF19 chimera, FGF21SS. As expected, FGF21SS shows better thermostability without inducing hepatocyte proliferation. Functional characterization of FGF21SS shows its better insulin sensitivity, reduced inflammation in 3T3‐L1 adipocytes, and lower blood glucose and insulin levels in ob/ob mice compared with wild type. Our dynamics‐based rational design provides a promising approach for FGF21‐based therapeutic development against T2DM.
Keywords: anti‐diabetes, disulfide bond mutation, FGF21, folding dynamics, NMR
Subject Categories: Metabolism, Structural Biology
NMR structural analysis of FGF21 reveals a flexible conformation that affects heparan sulfate binding and β2‐β3 hairpin folding dynamics. A folding dynamic modulated variant, FGF21SS, improves protein thermostability and anti‐diabetic activity.

Introduction
FGF21, a member of the FGF family, exerts diverse pharmacological effects on the regulation of glucose homeostasis, lipid metabolism, and insulin sensitivity. Administration of FGF21 leads to reduction of circulating glucose, triglycerides, insulin levels and body weight, as well as improvement of insulin sensitivity, energy metabolism, and amelioration of hepatic steatosis in diabetic rodents and non‐human primate (Kharitonenkov et al, 2005; Badman et al, 2007; Kharitonenkov et al, 2007; Dutchak et al, 2012; Schlein et al, 2016; BonDurant et al, 2017; Li et al, 2018). Besides, FGF21 prevents the apoptosis of insulin‐producing INS1E cells induced by glucolipotoxicity and cytokine and promotes the survival of pancreatic β‐cell from diabetic rodents (Wente et al, 2006). Extensive researches have suggested FGF21 as a promising therapy for T2DM, obesity, non‐alcoholic steatohepatitis (NASH), and related metabolic disorders (Degirolamo et al, 2016; Potthoff, 2017; Tucker et al, 2019; Geng et al, 2020).
The human FGF21 is a 181‐amino‐acid secreted FGF19 subfamily protein. Unlike classical paracrine‐acting FGFs, which signal by interacting with FGF receptors (FGFRs) in the presence of heparan sulfate (HS) proteoglycan, the FGF19 subfamily members achieve their functions in an endocrine fashion. FGF21 requires β‐klotho, a single transmembrane glycoprotein, as a scaffold for its binding to FGFR (Chen et al, 2018; Lee et al, 2018). Therefore, FGF21 performs its metabolic activities in liver, pancreas, adipocytes, and neurons, where the expression level of β‐klotho is high (Fon Tacer et al, 2010).
The native human FGF21 protein exhibits poor pharmacokinetics (PK) properties, resulting in a great challenge for clinical application (Degirolamo et al, 2016; Potthoff, 2017; Tucker et al, 2019; Geng et al, 2020). To improve the protein activity, stability, and solubility, different engineering approaches have been developed. For example, Xu et al described an Fc tag fused variant, Fc‐FGF21(RG), to improve PK, in which they mutated Leu98Arg to resist aggregation and Pro171Gly to impede the C‐terminal proteolytic cleavage at Pro171 (Hecht et al, 2012). Kharitonenkov et al designed LY2405319, in which an additional disulfide bond (SS bond) was added at Leu118Cys‐Ala134Cys to stabilize the predicted loop between β10 and β12 (Kharitonenkov et al, 2013). Lee et al designed Arg185Trp/Leu166Phe double mutation based on the structure of β‐klotho and FGF21 C‐terminus complex, to enhance the binding of FGF21 with β‐klotho but no in vivo activity result was reported (Lee et al, 2018). Besides, a variety of modifications have been utilized to extend the FGF21 half‐life, including Fc fusion (Hecht et al, 2012; Stanislaus et al, 2017), scaffold antibody conjugation (Talukdar et al, 2016), pegylation (Huang et al, 2011; Mu et al, 2012; Xu et al, 2013; Song et al, 2014; Ye et al, 2014), and glycosylation (Weng et al, 2018).
However, most of the designing strategies of FGF21 variants so far were limited in protein fusion or modification combined with empirical mutations. Although some variants enhance the protein PK property, they showed limited anti‐diabetic therapeutic improvement in vivo. The main obstacle of FGF21 drug development is the lack of structural information of FGF21. Here, a FGF21 structure was determined using NMR spectroscopy and the dynamic property was also characterized. We found that the flexible non‐canonical β‐trefoil conformation in the β10‐β12 region of FGF21 severely affects the folding stability of the neighboring β2‐β3 hairpin. The understanding of structural and folding dynamic property of FGF21 enables a rational design of a novel FGF21 variant, with significantly increased anti‐diabetic activities.
Results
Solution structure of FGF21 core region
FGF21 belongs to the endocrine FGF19 subfamily. Structural prediction based on homologues reveals that FGF21 contains about 13 N‐terminal and 40 C‐terminal residues extended random coil regions (FGF21CT) flanking a β‐trefoil core domain (FGF21core, residues 14–141), which shares only 38% and 33% sequence identity with other subfamily members FGF19 and FGF23, respectively (Fig EV1). NMR spectral comparison shows that FGF21core has a similar overall fold compared with full‐length protein, but less spectral overlap and better T2 relaxation (Fig EV2). Therefore, we determined FGF21core structure using standard multidimensional heteronuclear solution NMR spectroscopy (Fig 1A and B, and Table EV1). The FGF21core structure ensemble shows a converged non‐canonical β‐trefoil fold, which contains 11 β‐strands (residues 17–22 (β1), 32–36 (β2), 40–44 (β3), 53–58 (β4), 62–67 (β5), 72–77 (β6), 81–85 (β7), 95–100 (β8), 104–109 (β9), 114–117 (β10), and 136–139 (β12)). Strand β11 of canonical β‐trefoil conformation is missing in FGF21, and β10 and β12 are linked by an 18‐residue proline‐rich disordered random coil (residues 118–135), for which few inter‐residue Nuclear Overhauser Effect (NOE) restraints are available.
Figure EV1. Structural‐based sequence alignment of FGF21, FGF19, FGF23, and selected paracrine‐acting FGFs.

Predicted signal peptide sequences were omitted. Residue numbers indicates the sequence of mature FGF21. The locations and lengths of the secondary structure elements are indicated by boxes in the sequences. Cysteine residues that form disulfide bonds are highlighted on a yellow background. Glycine and threonine residues of the GXXXXGXX(T/S) motif are highlighted on an orange background. Residues strictly conserved are highlighted on a red background. Similar residues are rendered as red characters with blue frames.
Figure EV2. 2D 1H‐15N Heteronuclear Single Quantum Correlation (HSQC) spectra of FGF21 (blue) and FGF21core (red).
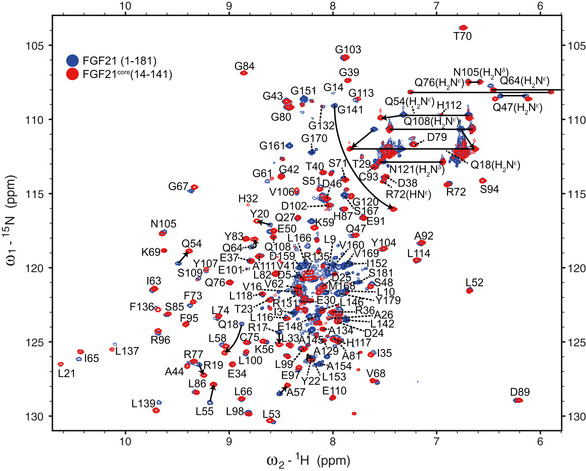
Backbone amide resonance assignments were indicated with one‐letter amino acid code and sequence number. Arrows indicate the peak shifts upon the truncation of N‐ and C‐termini in FGF21core.
Figure 1. Solution structure of FGF21core .
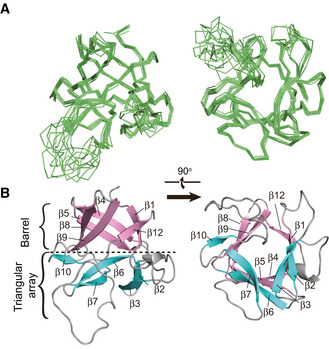
- Ribbon representation of 10 energy‐refined structure ensemble of FGF21core.
- Cartoon representation of a representative structure of FGF21core. β sheets for the barrel and triangular array regions were colored pink and cyan, respectively.
Structural features of FGF21 for low HS‐binding affinity
Unlike classical paracrine‐acting FGFs, which interact with FGF receptors (FGFRs) in the presence of HS, FGF19 subfamily members have weak binding affinity for both HS and their cognate FGFRs. For FGF21, which diffuses from the HS‐rich secretory tissues and performs its function as an endocrine hormone, it interacts with cognate receptor in the assistance of β‐klotho (Goetz et al, 2007; Ogawa et al, 2007; Kharitonenkov et al, 2008; Suzuki et al, 2008; Beenken & Mohammadi, 2012; Ding et al, 2012; Yie et al, 2012; Chen et al, 2018; Lee et al, 2018). The low HS‐binding affinity of FGF21 results from the aberrant conformation of FGF21 β10‐β12 region. For details, a typical FGF molecule like FGF2 consists of a six‐stranded antiparallel β‐barrel closed off on one end by a triangular array, where the arrangement of three β‐hairpins (the edges are named clock‐wisely as β2‐β3 hairpin, β6‐β7 hairpin and β10‐β11 hairpin) gives the molecule a pseudo 3‐fold symmetry (Fig 2A, left panel). HS binds to the positively charged groove formed by β10‐β11 hairpin and β1‐β2 loop (Fig 2A, left panel, and Fig 3). A conserved GXXXXGXX(T/S) motif termed glycine box within β10‐β11 hairpin (Fig EV1) interacts with HS by forming tight hydrogen bonds using the sidechains of Arg/Lys and backbone atoms (Luo et al, 1998; Plotnikov et al, 2000). In the case of FGF21, instead of a β‐hairpin and/or glycine box, it forms a disordered loop stretching out of the triangular array (Fig 2A, middle panel). The steric hindrance therefore reduces the HS binding to the disrupted FGF21 β10‐β11 segment (similar to the cases in FGF19 and FGF23; Goetz et al, 2007; Fig EV3). Moreover, the HS‐binding region of FGF21 is primarily more negatively charged than other FGFs; thus, electrostatic force will further repel HS containing highly acidic sulfate groups (Fig 3). The low receptor and HS‐binding affinity of FGF21 are of great significance to understand the endocrine properties of FGF21 and the β‐klotho‐dependent signal transduction mechanism.
Figure 2. Dynamics caused folding instability of FGF21 in the triangular array region.
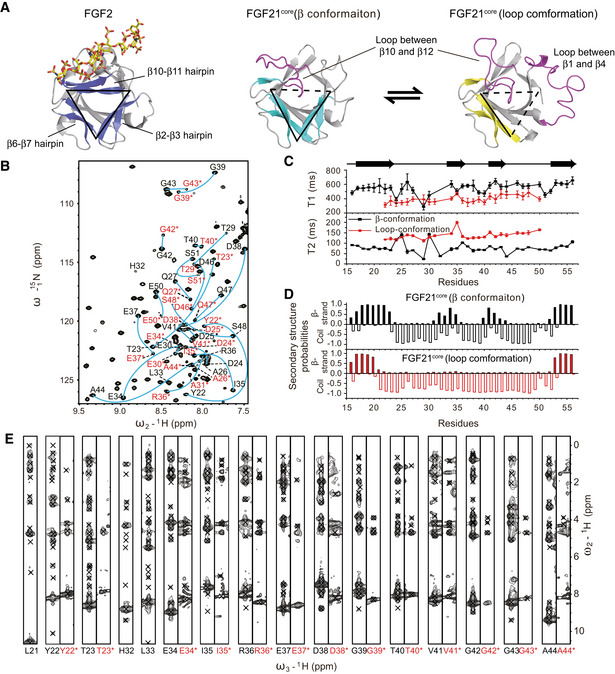
- Triangular array region of FGF2 (PDB ID: 2FGF), FGF21core (β conformation) and FGF21core (loop conformation). The edges of the triangular formed by hairpins or missed (colored magenta) are indicated by solid or dashed lines, respectively.
- 2D 1H‐15N Heteronuclear Single Quantum Correlation (HSQC) spectrum showing the two sets of assignments for residue Tyr22 to Glu50 (labeled by one‐letter amino acid code and sequence number). Labels for the loop conformation are colored red and marked with asterisks.
- 15N T1 and T2 relaxation times of the residues in β2‐β3 region. The secondary structure of FGF21core (β conformation) is shown on the top. Error bars show the fitting SDs of ten different experiments.
- Secondary structure prediction of residues in β2‐β3 region using TALOS + program. The probability of β‐strand and random coil of the residues are shown by histogram.
- 1H‐1H slices of 15N‐edited 3D NOESY‐HSQC spectrum for β2‐β3 residues. Residue codes are labeled as in (B).
Figure 3. Heparin binding sites of FGF21 differ from that of paracrine‐acting and other endocrine‐acting FGFs.
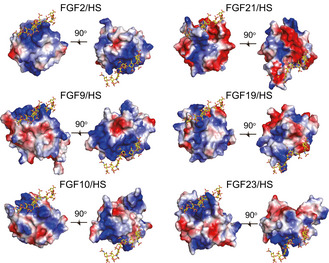
Electrostatic potential surface of different FGFs for representing the (HS) binding sites. HS is placed by superimposition of FGF21core, FGF9 (PDB ID: 1IHK), FGF10 (PDB ID: 1NUN), FGF19 (PDB ID: 2P23), and FGF23 (PDB ID: 2P39) onto the FGF2/FGFR1c/HS ternary complex structure (PDB ID: 1FQ9). HS is shown as a stick representation. The positive and negative charges on the protein surface are colored blue and red, respectively.
Figure EV3. Superimposition of the FGF21core structure (cyan) with that of FGF2 (blue, PDB ID: 2FGF), FGF19 (orange, PDB ID: 2P23), and FGF23 (yellow, PDB ID: 2P39) shown as a ribbon representation.
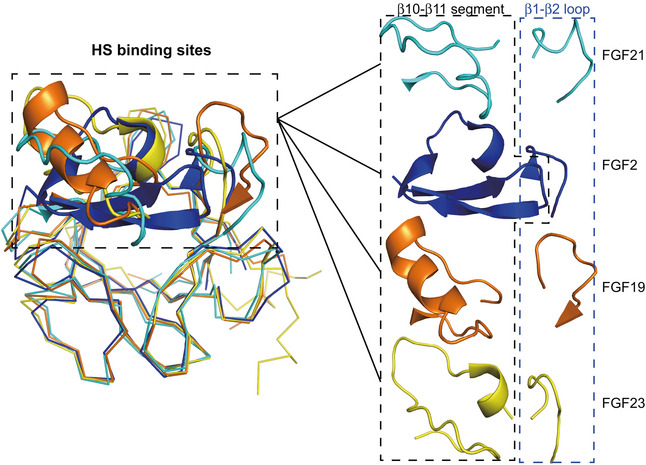
The predicted heparin binding domain is illustrated as a cartoon representation and is enlarged in the right panel separately.
Highly dynamic folding property of β2‐β3 hairpin
The untypical structural feature of FGF21 is not limited in the β10‐β11 region. Our data also revealed that adjacent β2‐β3 hairpin and the neighboring loop segments (residues Tyr22 to Glu50) exhibit highly dynamic folding characteristic. For both FGF21core and full‐length FGF21, a second set of resonances were observed for the residues in this region, whose 1H chemical shifts distribute mostly in the range of 7.7–8.6 ppm (Fig 2B and Appendix Fig S1), indicating the existence of a second conformation that is dynamically transformed on a slow time scale. Structural modeling based on the TALOS + derived dihedral angles (Fig 2D) and NOE restraints (Fig 2E) yielded a disordered loop conformation (Fig 2A, right panel). Compared with β conformation, the loop conformation displays shorter 15N T1 and longer 15N T2 relaxation times, indicating its higher flexibility (Fig 2C). The destruction of β2‐β3 hairpin will further decrease the stability of the triangular array architecture of FGF21 and obviously has a significant impact on the overall protein stability in vitro and in vivo. More importantly, the binding of FGF21 to receptor would be disturbed because the loop conformation affects the β1‐β2 loop, which directly contacts with D2 domain of FGFR1c and HS as shown in the receptor interaction model (Fig 4). Therefore, we anticipate that this highly dynamic folding property of FGF21 is unfavorable for the interaction with receptor and would help its escape from local extracellular matrix as an endocrine hormone.
Figure 4. Interaction model of 2:2:2:2 FGF21/FGFR1c/β‐klotho/HS complex.

This model is obtained by superimposition of FGF21core structure with the crystal structures of β‐klotho/FGF21‐CT (PDB ID: 5VAQ), FGF23/FGFR1c/α‐klotho (PDB ID: 5W21) and FGF2/FGFR1c/HS dimer (PDB ID: 1FQ9), and is shown as surface representation. FGF21core is colored orange. β1‐β2 loop of FGF21 (Asp24 to Ala31) directly interacting with FGFR1c is colored magenta. FGF21CT binding to β‐klotho is colored blue. FGFR1c is colored green. D1, D2 domain, and the receptor binding arm of β‐klotho are colored cyan, lightblue, and pink, respectively.
Design and structural characterization of a stable FGF21 variant
The dynamic folding property and associated protein instability of FGF21 may be important for the regulation of hormone release and degradation in the body; nevertheless, the resulted poor PK property of native human FGF21 leads to a great challenge for its clinical application (Degirolamo et al, 2016; Potthoff, 2017; Tucker et al, 2019; Geng et al, 2020). Among protein engineering strategies, SS bonds between strands have turned out to be effective to stabilize β hairpin conformation (Zhang et al, 1994; Lee & Blaber, 2009; Moore et al, 2017). SS bonds appear naturally in the triangular array region of endocrine FGFs. For example, a conserved SS bond in FGF19 subfamily (formed by Cys75 and Cys93 in FGF21) helps the tether of β6‐β7 hairpin to the barrel, and loss of this SS bond greatly impaired the activity of FGF21 (Luo et al, 2019). FGF19 has an additional SS bond between strands of β2‐β3 hairpin (Goetz et al, 2007). Attempts to introduce SS bond to FGF21 in the β10‐β12 loop were also carried out by other researchers. Unfortunately, these efforts did not significantly improve FGF21 stability and function (Kharitonenkov et al, 2013).
The availability of FGF21 structure makes it more feasible to design a better therapeutic FGF21 variant. Here, we constructed a novel FGF21‐FGF19 chimera—FGF21SS to enhance the protein stability while preserving its major binding feature to FGFR, β‐Klotho and HS. First, to stabilize β2‐β3 hairpin, Gly43 in β3 was mutated to cysteine (Cys43#, residues in FGF21SS with different sequence compared with that in FGF21 are marked with #) to form SS bond with Ala31Cys (Cys31#) in β2 strand. Secondly, considering that the conformation of HS/receptor binding β1‐β2 loop would be disturbed upon the SS bond formation, β1‐β2 loop (DDAQQTEA) was replaced by the longer FGF19 loop (SGPHGLSSC) to retain its binding to FGFR (Fig 5A).
Figure 5. Dynamic folding modulation design and structural analysis of FGF21SS variant.
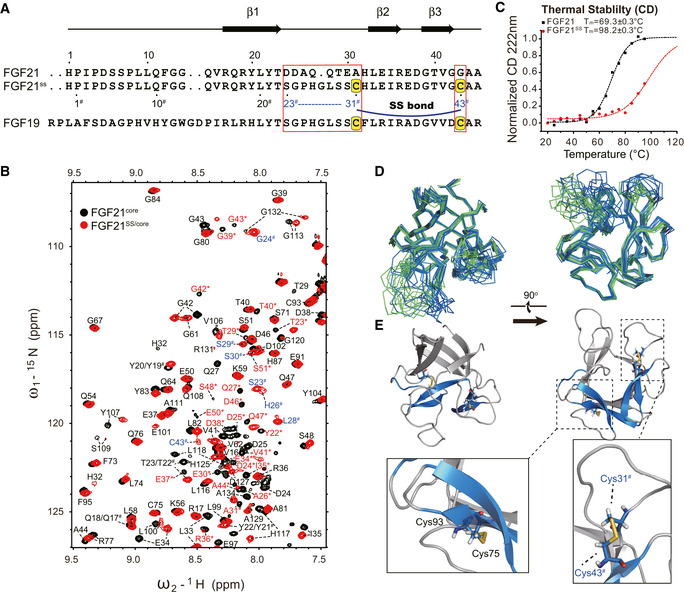
- Sequence alignment of FGF21, FGF21SS, and FGF19 in the mutated region. The mutation sites are indicated by boxes. Cysteines for disulfide bond formation are highlighted on a yellow background. The secondary structure of FGF21 is shown on the top. The N‐terminal residue IDs of FGF21SS are indicated under the sequence. # is used to identify the mutated residues (codes are colored blue) and the same residues having different sequence number in FGF21SS from wild type.
- Temperature denaturation CD experiments indicating the significant increase of thermostability. CD value was recorded and normalized at 222 nm. Tm value of the proteins is fitted using Boltzmann function.
- Superimposition of FGF21SS/core (skyblue) and FGF21core (green) structure ensembles (10 energy‐refined structures each) in ribbon representation showing the structure conservation upon SS bond mutation.
- Cartoon representation of FGF21SS/core. Cys31#‐Cys43# (between β2 and β3) and Cys75‐Cys93 (between β6 and β9) SS bonds are highlighted and shown as stick representations.
The conformation of FGF21SS after mutagenesis was analyzed using NMR spectroscopy. 2D 1H‐15N HSQC spectra and backbone atom assignments showed that FGF21SS exists in a single conformation (Figs 5B and EV4). 13Cβ chemical shifts of Cys31# and Cys43# were identified to 44.266 ppm and 44.427 ppm, respectively, indicating the oxidized form of cysteines. Liquid chromatography‐mass spectrometry (LC‐MS) also demonstrated the existence of SS bond between Cys31# and Cys43# (Appendix Fig S2). We further determined the solution structure of FGF21SS/core and found that the structure of FGF21SS/core is virtually identical to that of wild‐type FGF21core, with an overall Cα root mean square deviation (r.m.s.d.) value of 0.198 Å excluding the disordered regions (β1‐β2 loop and β10‐β12 loop) (Fig 5D and E, and Table EV1). The sidechain orientation and inter‐thiol distance of Cys31# and Cys43# support the SS bond formation (Fig 5E). As expected, FGF21SS showed significantly higher melting temperature (Tm) than wild type in temperature denaturation circular dichroism (CD) experiments (98.2 ± 0.3°C vs. 69.3 ± 0.3°C), indicating higher protein thermostability (Fig 5C).
Figure EV4. Backbone assignments of FGF21SS/core specifying one set of resonances.
# is used to identify the mutated residues (whose labels are colored blue) and the same residues having different sequence number in FGF21SS compared with FGF21.
Functional characterization of FGF21SS
The carcinogenic risk of the chimera protein needs to be evaluated first, as administration of FGF19 has been reported to increase hepatocyte proliferation (Nicholes et al, 2002). After a 6‐day injection of vehicle or 0.6 mg/kg/day proteins in ICR mice, the hepatic cell proliferation was detected by immunohistochemical analysis using BrdU assay. As shown in Fig EV5, while FGF19 obviously induced the mitosis of hepatocyte, both FGF21 and FGF21SS did not show any mitogenic activity, indicating excellent safety profile.
Figure EV5. Representative histological analysis of livers indicating the nontumorigenic feature of FGF21SS and FGF21.
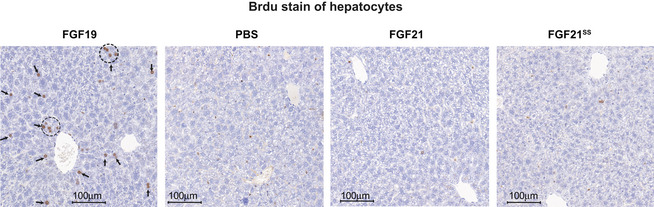
ICR mice were injected every day with vehicle or 0.6 mg/kg FGF21, FGF21SS and FGF19 for 6 days, and the hepatocyte proliferation was estimated using nuclear labeling with 5‐bromo‐2‐deoxyuridine (BrdU) (arrows and dotted circles), which was constantly infused simultaneously by osmotic minipump. Tests were repeated as five independent experiments.
Recent reports revealed that the anti‐inflammatory effect of FGF21 is involved in the improvement of insulin resistance (Li et al, 2018; Wang et al, 2018). Thus, we next tested the anti‐insulin‐resistant activity of FGF21SS in 3T3‐L1 adipocytes. Cells were treated for 24 h with vehicle, FGF21 or FGF21SS in RAW264.7 conditioned medium (RAW‐CM, containing several macrophages secreted cytokines, e.g., TNF‐α and IL‐6, which are responsible for the development of adipocytes inflammation and insulin resistance (Dandona et al, 2004; Permana et al, 2006; Zatterale et al, 2019)). AKT phosphorylation upon insulin stimulation through PI3K/Akt/mTOR signaling pathway was used here as a marker of insulin sensitivity (Haeusler et al, 2018). As shown in Fig 6A, treatment of RAW‐CM greatly inhibited insulin sensitivity in 3T3‐L1 adipocytes, while the addition of FGF21s effectively antagonized the effect of macrophage‐secreted inflammatory cytokines and significantly alleviated insulin resistance, and FGF21SS showed more significant activity than FGF21.
Figure 6. Anti‐diabetic activity characterization of FGF21SS .
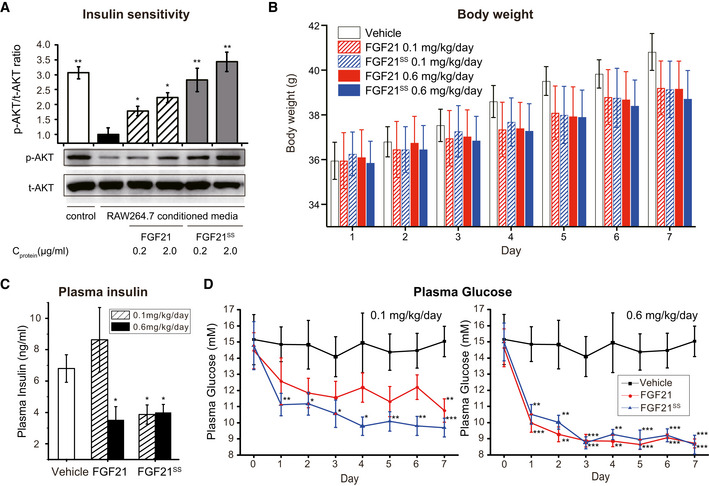
-
AFGF21SS and FGF21 improve insulin resistance by countering the inflammation in 3T3‐L1 adipocytes. Cells were cultured in RAW‐CM to induce inflammation and insulin resistance, while vehicle, FGF21 or FGF21SS were added simultaneously. The insulin sensitivity was indicated by insulin‐induced AKT phosphorylation using Western blotting analysis. Error bars show the SEM of four independent experiments. *P < 0.05, **P < 0.01 (Student's t‐test) vs. RAW‐CM treated only group.
-
B–DAnti‐diabetic effect evaluation in ob/ob mice. Mice were injected subcutaneously with vehicle, FGF21 or FGF21SS at indicated doses every day. Body weight (B) was measured, and plasma glucose (D) was measured every day. Plasma insulin (C) was analyzed at day 7 using insulin ELISA kit. Error bars show the SEM of ten independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 (Student's t‐test) vs. vehicle.
To further investigate the anti‐diabetic effect of FGF21SS in vivo, the diabetic and obese ob/ob mice were injected subcutaneously with vehicle, FGF21 or FGF21SS at doses of 0.1 mg/kg and 0.6 mg/kg once daily. As a result, both FGF21 and FGF21SS showed dose‐dependent anti‐diabetic activity. At the higher dose, the protein administration toward ob/ob mice achieved rapid and significant blood glucose‐lowering effect, and the body weight and plasma insulin level were also significantly reduced after 7‐day treatment (Fig 6B–D). At the lower dose of 0.1 mg/kg, FGF21SS decreased the blood glucose more significantly than FGF21 (1 day vs. 7 days) and finally achieved similar body weight and plasma insulin‐lowering effects with the 0.6 mg/kg dose, while 0.1mg/kg FGF21 showed only similar body weight decrease but weaker blood glucose‐lowering effect and little plasma insulin‐lowering effect (Fig 6B–D). The data show that the stability improved FGF21SS can potentially provide consistent and better efficacy against diabetes.
Discussion
Paracrine FGFs have relatively conserved β10‐β11 region and β1‐β2 loop responsible for high‐level HS‐binding affinity, which will promote their interaction with cognate FGFRs. Huang et al indicated that the tight binding induces robust and long‐lived FGFR dimerization, which is essential for FGF1 mitogenic response (Huang et al, 2017). Although HS serves as a universal requirement for all FGFs signaling, its binding affinity is reduced to confer endocrine ability to FGF19 subfamily members. Our data show that the low HS‐binding affinity of FGF21 is due to the flexible and disordered β10‐β11 region and the adjacent negative charges. This structural feature impacts FGF21 binding with cognate FGFR and explains why FGF21 cannot trigger signaling in the case of HS alone (Kharitonenkov et al, 2008). It is well documented that the interaction between FGF21 and FGFR requires β‐klotho as a co‐receptor for signaling (Goetz et al, 2007; Ogawa et al, 2007; Kharitonenkov et al, 2008; Suzuki et al, 2008; Beenken & Mohammadi, 2012; Ding et al, 2012; Yie et al, 2012). The crystal structures of FGF21CT/β‐klotho and its homologue FGF23/FGFR1c/α‐klotho ternary complex indicated that β‐klotho directly binds to FGF21CT and acts as a scaffold for FGF21 binding to FGFR (Chen et al, 2018; Lee et al, 2018; Shi et al, 2018). According to the dimeric FGF21/FGFR1c/β‐klotho/HS complex model (Fig 4), however, β‐klotho does not participate in the dimerization of FGFR. Therefore, although the interaction between FGF21 and HS is weak and transient, HS is still necessary and sufficient for the weak receptor dimerization enough for metabolic effects but not too strong to induce proliferative response.
The maintenance of protein’s original structural integrity and activity is critical for successful protein drug development. In the case of FGF21, many efforts were carried out to enhance its in vivo anti‐diabetic activity, but achieved only limited improvement (Huang et al, 2011; Hecht et al, 2012; Mu et al, 2012; Kharitonenkov et al, 2013; Xu et al, 2013; Song et al, 2014; Ye et al, 2014; Talukdar et al, 2016; Stanislaus et al, 2017; Weng et al, 2018). As one of the most versatile biophysical techniques, NMR spectroscopy is a useful tool to determine protein high‐resolution structure under near‐physiological conditions and provide folding dynamics and homogeneity information. We determined the structure of FGF21 and observed the inherent dynamic folding property and instability in the region of β2‐β3 hairpin, which is essential for the interaction with FGFR and HS. This conformational instability may be closely related to the regulation of hormone release and degradation in vivo, but has negatively impacts on laboratory research (e.g., protein crystallization) and clinical application.
The structural and dynamic information guided us to design next‐generation FGF21 variant. The native SS bond between β2 and β3 strands of FGF19 serves as a good template. However, the introduction of Ala31Cys and Gly43Cys double‐cysteine mutation to FGF21 did not enhance FGF21 activity (Song et al, 2014). We supposed the flexibility of β1‐β2 loop is critical for FGF21 interacting with receptor and then replaced β1‐β2 loop in FGF21SS with the sequence of FGF19, as FGF19 shows highly similar receptor binding feature and physiological function compared with FGF21 in previous study (Beenken & Mohammadi, 2012; Degirolamo et al, 2016). Despite FGF19 induces hepatocyte proliferation through a FGFR4‐dependent pathway, Wu et al suggested that β1‐β2 loop is not the critical region for the hepatocyte mitogenic activity of FGF19 (Wu et al, 2010). Our mouse hepatocyte proliferation experiment also showed high safety profile of FGF21SS. In all, the structural and dynamic study of FGF21 provides very useful guidance for a novel rational design of therapeutic variant FGF21SS. The development of FGF21SS not only significantly enhances the protein stability but also improves its anti‐diabetic activities, making it a potentially and more attractive clinical candidate drug.
Materials and Methods
Cloning, expression, and purification
A gene encoding FGF21 was optimized according to the Escherichia coli (E. coli) preferred codons and synthesized by GenScript Company. The FGF21 gene was amplified and connected with the N‐terminal 6 × His tagged SUMO encoding gene by polymerase chain reaction (PCR). Then, the SUMO‐FGF21 DNA fragments were subcloned into the protein expression vector pET22b(+) using NdeI and XhoI restriction sites and transformed into the E. coli strain BL21(DE3). For FGF21 truncations and mutations, the corresponding genes were cloned using the same method. The cells were cultured until OD600 reached 0.6‐0.8 in LB broth and then induced by 0.5 mM IPTG at 37ºC for 4 h. For the expression of 15N‐, or 15N/13C‐labeled proteins, the cells were grown in M9 minimal medium supplemented with 15NH4Cl or/and 13C6‐glucose, respectively. After centrifugation, the cell pellet was lysed in 50 mM Tris buffer (pH 8.0) containing 300 mM NaCl, 10 mM imidazole by ultrasonication. The protein was purified by Ni‐Sepharose affinity chromatography (Qiagen). After the cleavage of SUMO fusion protein by SUMO‐protease, gel filtration (Superdex‐75, GE Healthcare) and anion‐exchange chromatography (Resource Q, GE Healthcare) using an ÄKTA FPLC system (Amersham Biosciences) were taken for further purification.
NMR spectroscopy
The proteins were concentrated to ~0.5 mM for triple‐resonance experiments and 0.2 mM for 2D 1H‐15N HSQC experiments in a 10% D2O/90% H2O buffer containing 20 mM Na2HPO4/NaH2PO4, 100 mM NaCl, 50 mM arginine, 50 mM glutamic acid, and 100 mM imidazole at pH 7.0. All the NMR spectra were acquired at 298 K on Bruker AVANCE III 850 or 600 MHz spectrometers. Each spectrometer is equipped with a triple‐resonance cryoprobe and pulsed field gradients. The spectra were processed with NMRPipe (Delaglio et al, 1995) and analyzed with NMRFAM‐Sparky (Lee et al, 2015). Backbone and sidechain resonances were assigned by the analysis of HNCA, HN(CO)CA, HNCO, HN(CA)CO, CBCANH, CBCA(CO)NH, HBHA(CO)NH, 13C‐edited TOCSY, and 13C‐edited COSY experiments. 15N longitudinal relaxation time T1 and transverse relaxation time T2 were measured using 15N‐labled sample. For T1 measurements, the relaxation delays of 2, 50, 100, 200, 500, 800, 1000, 1200, 1500, and 2000 ms were performed. For T2 measurements, the data were acquired with delays of 0, 17, 34, 51, 68, 85, 102, 119, 136, and 153 ms.
NMR structure calculation
The structures of FGF21core and FGF21core/SS were calculated using the XPLOR‐NIH program (version 2.28) (Schwieters et al, 2006). The distance restraints for structure determination were derived from 15N‐ and 13C‐edited 3D NOESY‐HSQC (mixing time 120 ms) spectra. The hydrogen bonds restraints were obtained by hydrogen–deuterium exchange experiments. The protein backbone dihedral angle ψ and φ restrains were derived by the TALOS + program using backbone chemical shift assignments (Shen et al, 2009). Finally, 10 out of 200 energy refined structures were selected as the representative structures of FGF21core and FGF21core/SS.
Liquid chromatography‐mass spectrometry (LC‐MS)
To identify the disulfide bond formed by Cys31# and Cys43# for FGF21SS, protein was subjected to LC‐MS/MS analysis. After digestion by trypsin, the peptides were analyzed using an Easy‐nLC1200 system (Thermo Fisher Scientific) interfaced with a Q‐Exactive mass spectrometer (Thermo Fisher Scientific). A 60‐min reverse‐phase gradient was used to separate peptides. The top 10 most intense precursor ions from each full scan were isolated for Electrospray Ionization (ESI) MS2. The MS data were analyzed using pLink‐SS, a software tool for identification of disulfide bridges as described in detail previously by Dong and co‐workers (Lu et al, 2015).
Circular dichroism spectroscopy
Far‐UV CD spectra were recorded at wavelengths between 190 and 240 nm using 200 μg/ml proteins in a 0.1 cm path length cell on a Jasco‐810 spectropolarimeter equipped with the Julabo temperature controller F25‐HD. The temperature dependence of the CD spectra of FGF21 and FGF21SS was determined by a rate of 1 K/min continuous heating in the temperature range of 20‐95ºC. All the CD spectra were shown by the average of three independent experiments.
Cell culture and adipocyte differentiation
The 3T3‐L1 preadipocytes were plated in six‐well plates at a density of 5 × 104 cells in DMEM containing 10% calf serum (CS) (Gibco). Cultures were maintained at 37ºC in a humidified 5% CO2 incubator. For differentiation to adipocytes, the post‐confluent cells were cultured in DMEM supplemented with 10% Fetal Bovine Serum (FBS) (Hyclone), 0.5 mM 3‐isobutyl‐1‐methylxanthine (IBMX), 1 μM dexamethasone and 1 μM insulin (IBMX, dexamethasone, and insulin were purchased from Sigma‐Aldrich) for 2 days, and then cultured in DMEM supplemented with 10% FBS and 1 μM insulin for another 2 days. After that, the media was changed every 2 days with DMEM supplemented with 10% FBS for 4‐8 days before used for experiments. The lipid droplets were observed in > 95% of cells by Oil Red (Sangon) staining.
Insulin sensitivity analysis of 3T3‐L1 adipocytes
For the collection of media conditioned by RAW264.7 macrophages (RAW‐CM), RAW264.7 macrophages were grown to 90% confluency in DMEM containing 10% FBS. Then, the cells were stimulated with 100 ng/ml lipopolysaccharide (LPS) (Sigma‐Aldrich) for 3 h. After stimulation, the cells were cultured in new serum‐free DMEM for 24 h. The media was collected, filtrated through a 0.22 μm filter, and used as RAW‐CM.
Insulin resistance of 3T3‐L1 adipocytes could be induced by incubation with RAW‐CM (Permana et al, 2006). To assess the insulin resistance improvement effects of FGF21 and FGF21SS, RAW‐CM containing vehicle, 200 μg/ml or 2000 μg/ml proteins were added into 3T3‐L1 adipocytes. After treatment for 24 h, 1 nM insulin was added to activate the insulin‐induced PI3K/Akt/mTOR signaling pathway (Haeusler et al, 2018). The insulin sensitivity was detected by the AKT (Ser473) phosphorylation, which was qualified using western blotting: The cells were collected and lysed in RIPA lysis buffer (50 mM tris, pH 7.4, 150 mM NaCl, 0.1% SDS, 1% NP‐40, and 0.5% sodium deoxycholate) containing protease inhibitor cocktail (Roche) and phosphatase inhibitor (Sangon). Then, the lysate was centrifuged at 12,000 g and 4 °C for 10 min, and the protein concentration was determined using BCA assay kit (Thermo Fisher Scientific). The total cellular proteins were resolved on SDS–PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). The protein blots were probed with antibodies to phosphorylated AKT (1:1000; #4060, Cell Signaling Technology). Antibodies to AKT (1:1000; #4685, Cell Signaling Technology) were used as the control of total AKT expression.
Animals
Male ob/ob mice (aged 6–7 weeks) and male ICR mice (aged 8–10 weeks) were purchased from the Model Animal Research Center of Nanjing University. Animals were maintained in a temperature‐controlled (20°C‐26°C) room with a 12 h light/dark cycle and had free access to food and water. Animal experiments in this study were approved and carried out in accordance with the protocol provided by the Institutional Animal Care and Use Committee (IACUC) at Hefei University of Technology. IACUC uses Guidelines for the ethical review of laboratory animal welfare, People’s Republic of China National Standard GB/T 35892‐2018 for the Care and Use of Laboratory Animals.
In vivo hepatocyte BrdU labeling
To determine whether FGF21 and FGF21SS would induce the mitosis of hepatocyte, ICR mice were subcutaneously injected with vehicle, FGF19, FGF21 or FGF21SS at a dose of 0.6 mg/kg/day (n = 4 for each group). For in vivo labeling of S‐phase hepatocytes, an osmotic minipump (ALZET, model 1007D) containing 5‐bromo‐2‐deoxyuridine (BrdU, Sigma) (16 mg/ml) was implanted subcutaneously into each mouse. Samples of liver were collected from each mouse on the day 6 following the last protein injection and placed in 10% neutral‐buffered formalin in preparation for paraffin‐embedding, sectioning, and light microscopic evaluation.
Immunohistochemical analysis
Hepatocyte proliferation was evaluated using a monoclonal antibody to BrdU (#5292, Cell Signaling Technology). After deparaffinization, sections were treated with 2 N HCl for 30 min and digested in preheated 0.01% trypsin (Sigma) for 3 min at 37°C. Endogenous proteins were blocked with 10% normal goat serum (Gibco) in 3% bovine serum albumin (BSA, Sigma) and PBS for 30 min. Then, the sections were incubated with anti‐BrdU antibody for 60 min, followed by incubation with SignalStain® Boost Detection Reagent (#8125, Cell Signaling Technology) for 30 min at room temperature, and then stained with the Signal Stain DAB substrate kit (#8059, Cell Signaling Technology) for visualization. Digital images were obtained using Pannoramic MIDI II and analyzed using CaseViewer 2.2 software (3DHISTECH Ltd).
In vivo anti‐diabetic activity evaluation
For anti‐diabetic effects’ evaluation experiments of FGF21 and FGF21SS, 6–7‐week‐old male ob/ob mice were randomized into different groups (n = 10) based on blood glucose levels and body weight. Mice were weighed and subcutaneous administered with vehicle, FGF21 or FGF21SS at doses of 0.1 mg/kg or 0.6 mg/kg every day. The blood glucose levels were measured by Omnitest® plus blood glucose monitor (B|BRAUN) every day. After 7‐day treatment, the blood insulin levels were measured using Human Insulin ELISA Kit (Crystal Chem).
Statistical analysis
All experiments were repeated at least three times. Results were expressed as mean ± standard error of the mean (SEM) (n = 3–10) unless otherwise indicated. Statistical analysis was performed using one‐way ANOVA, followed by Student’s t‐tests for each pair of comparisons. Furthermore, P < 0.05, P < 0.01 and P < 0.001 were considered statistically significant.
Author contributions
HZ and Juanjuan L purified the proteins. LZ, HZ, and ZL performed the NMR experiment and analyzed the NMR data. LZ and BW determined the NMR structure. Juanjuan L, HC, QL, BB, and Jian L designed, conducted, and analyzed cell‐based experiments. LZ, HZ, and Juanjuan L designed, conducted, and analyzed in vivo experiments. LZ, SZ, XL, HD, and JW designed experiments, analyzed data, and wrote the manuscript. All of the authors contributed to the discussion and editing of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Review Process File
Acknowledgements
We thank Dr. Y. Bi for critically reading and editing the manuscript. All NMR experiments were performed at High Magnetic Field Laboratory of the Chinese Academy of Sciences. This work was primarily supported by the National Natural Science Foundation of China (Grant No. U1532269 to J.W., 31800645 to L.Z., 21673244 to B.W. and 31100539 to Z.L.) and the Ministry of Science and Technology of China (Grant No. 2016YFA0400900 to J.W.). A portion of this work was supported by the High Magnetic Field Laboratory of Anhui Province.
EMBO Reports (2021) 22: e51352.
Contributor Information
Han Dai, Email: daihan@hmfl.ac.cn.
Junfeng Wang, Email: junfeng@hmfl.ac.cn.
Data availability
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession code 6M6E (FGF21core) and 6M6F (FGF21SS/core). Assigned 1H, 15N, and 13C chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) under accession number 36323 (FGF21core) and 36324 (FGF21SS/core).
References
- Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos‐Flier E (2007) Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab 5: 426–437 [DOI] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M (2012) The structural biology of the FGF19 subfamily. Adv Exp Med Biol 728: 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BonDurant LD, Ameka M, Naber MC, Markan KR, Idiga SO, Acevedo MR, Walsh SA, Ornitz DM, Potthoff MJ (2017) FGF21 regulates metabolism through adipose‐dependent and ‐independent mechanisms. Cell Metab 25: 935‐+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GZ, Liu Y, Goetz R, Fu LL, Jayaraman S, Hu MC, Moe OW, Liang G, Li XK, Mohammadi M (2018) alpha‐Klotho is a non‐enzymatic molecular scaffold for FGF23 hormone signalling. Nature 553: 461–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandona P, Aljada A, Bandyopadhyay A (2004) Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol 25: 4–7 [DOI] [PubMed] [Google Scholar]
- Degirolamo C, Sabba C, Moschetta A (2016) Therapeutic potential of the endocrine fibroblast growth factors FGF19, FGF21 and FGF23. Nat Rev Drug Discov 15: 51–69 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Ding XS, Boney‐Montoya J, Owen BM, Bookout AL, Coate KC, Mangelsdorf DJ, Kliewer SA (2012) beta Klotho Is required for fibroblast growth factor 21 effects on growth and metabolism. Cell Metab 16: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutchak PA, Katafuchi T, Bookout AL, Choi JH, Yu RT, Mangelsdorf DJ, Kliewer SA (2012) Fibroblast growth factor‐21 regulates PPARgamma activity and the antidiabetic actions of thiazolidinediones. Cell 148: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fon Tacer K, Bookout AL, Ding X, Kurosu H, John GB, Wang L, Goetz R, Mohammadi M, Kuro‐o M, Mangelsdorf DJ et al (2010) Research resource: Comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol Endocrinol 24: 2050–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L, Lam KSL, Xu A (2020) The therapeutic potential of FGF21 in metabolic diseases: from bench to clinic. Nat Rev Endocrinol 16: 654–667. [DOI] [PubMed] [Google Scholar]
- Goetz R, Beenken A, Ibrahimi OA, Kalinina J, Olsen SK, Eliseenkova AV, Xu C, Neubert TA, Zhang F, Linhardt RJ et al (2007) Molecular insights into the klotho‐dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol Cell Biol 27: 3417–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, McGraw TE, Accili D (2018) Biochemical and cellular properties of insulin receptor signalling. Nat Rev Mol Cell Biol 19: 31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht R, Li YS, Sun J, Belouski E, Hall M, Hager T, Yie J, Wang W, Winters D, Smith S et al (2012) Rationale‐based engineering of a potent long‐acting FGF21 analog for the treatment of type 2 diabetes. PLoS One 7: e49345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Tan Y, Gu J, Liu Y, Song L, Niu J, Zhao L, Srinivasan L, Lin Q, Deng J et al (2017) Uncoupling the mitogenic and metabolic functions of FGF1 by tuning FGF1‐FGF receptor dimer stability. Cell Rep 20: 1717–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Wang H, Lu M, Sun C, Wu X, Tan Y, Ye C, Zhu G, Wang X, Cai L et al (2011) A better anti‐diabetic recombinant human fibroblast growth factor 21 (rhFGF21) modified with polyethylene glycol. PLoS One 6: e20669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A, Beals JM, Micanovic R, Strifler BA, Rathnachalam R, Wroblewski VJ, Li S, Koester A, Ford AM, Coskun T et al (2013) Rational design of a fibroblast growth factor 21‐based clinical candidate, LY2405319. PLoS One 8: e58575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A, Dunbar JD, Bina HA, Bright S, Moyers JS, Zhang C, Ding L, Micanovic R, Mehrbod SF, Knierman MD et al (2008) FGF‐21/FGF‐21 receptor interaction and activation is determined by beta Klotho. J Cell Physiol 215: 1–7 [DOI] [PubMed] [Google Scholar]
- Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, Galbreath EJ, Sandusky GE, Hammond LJ, Moyers JS, Owens RA et al (2005) FGF‐21 as a novel metabolic regulator. J Clin Invest 115: 1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A, Wroblewski VJ, Koester A, Chen YF, Clutinger CK, Tigno XT, Hansen BC, Shanafelt AB, Etgen GJ (2007) The metabolic state of diabetic monkeys is regulated by fibroblast growth factor‐21. Endocrinology 148: 774–781 [DOI] [PubMed] [Google Scholar]
- Lee J, Blaber M (2009) Structural basis of conserved cysteine in the fibroblast growth factor family: evidence for a vestigial half‐cystine. J Mol Biol 393: 128–139 [DOI] [PubMed] [Google Scholar]
- Lee S, Choi J, Mohanty J, Sousa LP, Tome F, Pardon E, Steyaert J, Lemmon MA, Lax I, Schlessinger J (2018) Structures of beta‐klotho reveal a 'zip code'‐like mechanism for endocrine FGF signalling. Nature 553: 501–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tonelli M, Markley JL (2015) NMRFAM‐SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31: 1325–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HT, Wu GY, Fang QC, Zhang ML, Hui XY, Sheng B, Wu L, Bao YQ, Li P, Xu AM et al (2018) Fibroblast growth factor 21 increases insulin sensitivity through specific expansion of subcutaneous fat. Nat Commun 9: 272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Fan SB, Yang B, Li YX, Meng JM, Wu L, Li P, Zhang K, Zhang MJ, Fu Y et al (2015) Mapping native disulfide bonds at a proteome scale. Nat Methods 12: 329–331 [DOI] [PubMed] [Google Scholar]
- Luo W, Lin XM, Wang TX, Cai JL, Zeng XF, Zhu CR, Li RZ, Wang H, Wu XP (2019) Identification of a crucial amino acid responsible for the loss of specifying FGFR1‐KLB affinity of the iodinated FGF21. J Cell Physiol 234: 2500–2510 [DOI] [PubMed] [Google Scholar]
- Luo Y, Lu W, Mohamedali KA, Jang JH, Jones RB, Gabriel JL, Kan M, McKeehan WL (1998) The glycine box: a determinant of specificity for fibroblast growth factor. Biochemistry 37: 16506–16515 [DOI] [PubMed] [Google Scholar]
- Moore EJ, Zorine D, Hansen WA, Khare SD, Fasan R (2017) Enzyme stabilization via computationally guided protein stapling. Proc Natl Acad Sci USA 114: 12472–12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Pinkstaff J, Li Z, Skidmore L, Li N, Myler H, Dallas‐Yang Q, Putnam AM, Yao J, Bussell S et al (2012) FGF21 analogs of sustained action enabled by orthogonal biosynthesis demonstrate enhanced antidiabetic pharmacology in rodents. Diabetes 61: 505–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholes K, Guillet S, Tomlinson E, Hillan K, Wright B, Frantz GD, Pham TA, Dillard‐Telm L, Tsai SP, Stephan JP et al (2002) A mouse model of hepatocellular carcinoma: ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am J Pathol 160: 2295–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Kurosu H, Yamamoto M, Nandi A, Rosenblatt KP, Goetz R, Eliseenkova AV, Mohammadi M, Kuro‐o M (2007) beta Klotho is required for metabolic activity of fibroblast growth factor 21. P Natl Acad Sci USA 104: 7432–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permana PA, Menge C, Reaven PD (2006) Macrophage‐secreted factors induce adipocyte inflammation and insulin resistance. Biochem Biophys Res Commun 341: 507–514 [DOI] [PubMed] [Google Scholar]
- Plotnikov AN, Hubbard SR, Schlessinger J, Mohammadi M (2000) Crystal structures of two FGF‐FGFR complexes reveal the determinants of ligand‐receptor specificity. Cell 101: 413–424 [DOI] [PubMed] [Google Scholar]
- Potthoff MJ (2017) FGF21 and metabolic disease in 2016: A new frontier in FGF21 biology. Nat Rev Endocrinol 13: 74–76 [DOI] [PubMed] [Google Scholar]
- Schlein C, Talukdar S, Heine M, Fischer AW, Krott LM, Nilsson SK, Brenner MB, Heeren J, Scheja L (2016) FGF21 lowers plasma triglycerides by accelerating lipoprotein catabolism in white and brown adipose tissues. Cell Metab 23: 441–453 [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Clore GM (2006) Using Xplor‐NIH for NMR molecular structure determination. Prog Nucl Mag Res Sp 48: 47–62 [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS plus : a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44: 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SY, Lu YW, Richardson J, Min XS, Weiszmann J, Richards WG, Wang ZL, Zhang ZQ, Zhang J, Li Y (2018) A systematic dissection of sequence elements determining beta‐Klotho and FGF interaction and signaling. Sci Rep 8: 11045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song LT, Zhu YL, Wang HY, Belov AA, Niu JL, Shi L, Xie YY, Ye CH, Li XK, Huang ZF (2014) A solid‐phase PEGylation strategy for protein therapeutics using a potent FGF21 analog. Biomaterials 35: 5206–5215 [DOI] [PubMed] [Google Scholar]
- Stanislaus S, Hecht R, Yie J, Hager T, Hall M, Spahr C, Wang W, Weiszmann J, Li Y, Deng L et al (2017) A novel Fc‐FGF21 with improved resistance to proteolysis, increased affinity toward beta‐klotho, and enhanced efficacy in mice and cynomolgus monkeys. Endocrinology 158: 1314–1327 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Uehara Y, Motomura‐Matsuzaka K, Oki J, Koyama Y, Kimura M, Asada M, Komi‐Kuramochi A, Oka S, Imamura T (2008) betaKlotho is required for fibroblast growth factor (FGF) 21 signaling through FGF receptor (FGFR) 1c and FGFR3c. Mol Endocrinol 22: 1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar S, Zhou Y, Li D, Rossulek M, Dong J, Somayaji V, Weng Y, Clark R, Lanba A, Owen BM et al (2016) A long‐acting FGF21 molecule, PF‐05231023, decreases body weight and improves lipid profile in non‐human primates and type 2 diabetic subjects. Cell Metab 23: 427–440 [DOI] [PubMed] [Google Scholar]
- Tucker B, Li H, Long X, Rye KA, Ong KL (2019) Fibroblast growth factor 21 in non‐alcoholic fatty liver disease. Metabolism 101: 153994 [DOI] [PubMed] [Google Scholar]
- Wang N, Xu TY, Zhang X, Li JY, Wang YX, Guo XC, Li SM, Wang WF, Li DS (2018) Improving hyperglycemic effect of FGF‐21 is associated with alleviating inflammatory state in diabetes. Int Immunopharmacol 56: 301–309 [DOI] [PubMed] [Google Scholar]
- Weng Y, Ishino T, Sievers A, Talukdar S, Chabot JR, Tam A, Duan W, Kerns K, Sousa E, He T et al (2018) Glyco‐engineered long acting FGF21 variant with optimal pharmaceutical and pharmacokinetic properties to enable weekly to twice monthly subcutaneous dosing. Sci Rep 8: 4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente W, Efanov AM, Brenner M, Kharitonenkov A, Koster A, Sandusky GE, Sewing S, Treinies I, Zitzer H, Gromada J (2006) Fibroblast growth factor‐21 improves pancreatic beta‐cell function and survival by activation of extracellular signal‐regulated kinase 1/2 and Akt signaling pathways. Diabetes 55: 2470–2478 [DOI] [PubMed] [Google Scholar]
- Wu X, Ge H, Lemon B, Vonderfecht S, Baribault H, Weiszmann J, Gupte J, Gardner J, Lindberg R, Wang Z et al (2010) Separating mitogenic and metabolic activities of fibroblast growth factor 19 (FGF19). Proc Natl Acad Sci USA 107: 14158–14163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bussiere J, Yie J, Sickmier A, An P, Belouski E, Stanislaus S, Walker KW (2013) Polyethylene glycol modified FGF21 engineered to maximize potency and minimize vacuole formation. Bioconjug Chem 24: 915–925 [DOI] [PubMed] [Google Scholar]
- Ye X, Qi J, Wu Y, Yu D, Xu P, Li S, Zhu S, Wu Q, Ren G, Li D (2014) Comparison of PEGylated FGF‐21 with insulin glargine for long‐lasting hypoglycaemic effect in db/db mice. Diabetes Metab 41(1): 82–90. [DOI] [PubMed] [Google Scholar]
- Yie J, Wang W, Deng L, Tam LT, Stevens J, Chen MM, Li Y, Xu J, Lindberg R, Hecht R et al (2012) Understanding the Physical Interactions in the FGF21/FGFR/beta‐Klotho Complex: Structural Requirements and Implications in FGF21 Signaling. Chem Biol Drug Des 79: 398–410 [DOI] [PubMed] [Google Scholar]
- Zatterale F, Longo M, Naderi J, Raciti GA, Desiderio A, Miele C, Beguinot F (2019) Chronic adipose tissue inflammation linking obesity to insulin resistance and type 2 diabetes. Front Physiol 10: 1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Bertelsen E, Alber T (1994) Entropic effects of disulphide bonds on protein stability. Nat Struct Biol 1: 434–438 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Review Process File
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession code 6M6E (FGF21core) and 6M6F (FGF21SS/core). Assigned 1H, 15N, and 13C chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) under accession number 36323 (FGF21core) and 36324 (FGF21SS/core).


