Abstract
Background
Peritoneal metastasis from pancreatic cancer (PM-PC) may be treated with repeated pressurised intraperitoneal aerosol chemotherapy (PIPAC). Utility of next-generation sequencing (NGS) to detect cancer-related mutations in peritoneal quadrant biopsies (QBs) and peritoneal fluid (PF) after systemic and PIPAC treatment has not been evaluated. Around 90% of pancreatic cancers (PCs) harbour a KRAS mutation, making PC ideal for the evaluation of this aspect.
Aims
Evaluation of PM-PC in terms of (1) histological response to PIPAC using Peritoneal Regression Grading Score (PRGS), (2) clinical characteristics and (3) frequency of mutations in QBs and PF before and after PIPAC.
Methods
Peritoneal QBs and PF were obtained prior to each PIPAC. NGS for 22 cancer-related genes was performed on primary tumours, QBs and PFs. Response was assessed by the four-tiered PRGS.
Results
Sixteen patients treated with a median of three PIPAC procedures were included. The mean PRGS was reduced from 1.91 to 1.58 (p=0.02). Fifty-seven specimens (13 primary tumours, 2 metastatic lymph nodes, 16 PFs and 26 QB sets) were analysed with NGS. KRAS mutation was found in 14/16 patients (87.50%) and in QBs, primary tumours and PF in 8/12 (66.67%), 8/13 (61.53%) and 6/9 (66.67%). The median overall survival was 9.9 months (SE 1.5, 95% CI 4.9 to 13.9).
Conclusion
PIPAC induces histological response in the majority of patients with PM-PC. KRAS mutation can be found in PM-PC after PIPAC at a frequency similar to the primaries. NGS may be used to detect predictive mutations in PM-PC of various origins, also when only post-PIPAC QBs or PFs are available.
Keywords: pancreatic neoplasms, pancreas, peritoneum
Introduction
The incidence of pancreatic cancer (PC) is rising, but the prognosis for the majority of these patients remains poor, especially when the disease has metastasised to the peritoneum. Even in selected patients with peritoneal metastasis from pancreatic cancer (PM-PC), the median survival after systemic combination chemotherapy leads to a median overall survival of only 7–8 months.1 Pressurised intraperitoneal aerosol chemotherapy (PIPAC) is a treatment alternative to systemic chemotherapy, where studies in patients with peritoneal metastasis (PM) from ovarian, gastric and colorectal cancers and PC show a high safety profile and promising survival data.2–6 Currently, PIPAC is an experimental treatment, and randomised controlled trials are lacking.
The choice of drugs used for PIPAC has depended more on PIPAC tradition and safety rather than disease profile. However, based on the preliminary but promising results of PIPAC-directed treatment in PM-PC, the next obvious step is to look at individual patients with PC that may harbour rare molecular alterations that may be targeted by specific agents.7 8 The mutational profile of PM of various origin before and after PIPAC may provide important prognostic and predictive information. Studies have shown that chemotherapy treatment in patients with, for example, acute myeloid leukaemia could ‘breed’ new mutations in different genes, so-called subclones.9 10 It is unknown whether PIPAC can have similar effects regarding the mutational profile of PM. Knowledge regarding if and to what extent there is discordance between the molecular alterations in the primary PC tumour and its PM, and particularly between PM before and after PIPAC, is sparse.
It remains to be elucidated whether treatment may influence the utility of biopsy specimens from the peritoneum for next-generation DNA sequencing (NGS) analyses. As more than 90% of PCs harbour a KRAS mutation, this type of cancer is ideal for the evaluation of the latter aspect. The aims of the present study were to evaluate a series of patients with PM-PC treated with PIPAC regarding the following aspects: (1) evaluation of the histological response of PM-PC to PIPAC by means of the Peritoneal Regression Grading Score (PRGS), (2) description of the clinical characteristics and (3) the frequency of mutations in cancer-related genes in peritoneal quadrant biopsies (QBs) and peritoneal fluid (PF) with PM-PC before and after PIPAC.
Materials and methods
Clinical data and inclusion of specimens
The patients with PM-PC included in this study are also included in the PIPAC-OPC-1 or PIPAC-OPC-2 studies at Odense PIPAC Center (OPC), Odense University Hospital, Denmark.11 12 We ensured that patients had not advocated against the use of their tissue in the Danish registry for the use of tissue in research (‘Vævsanvendelsesregisteret’). Patients with PM-PC were included if they had biopsy-proven malignancy, clinically proven PM and a maximum of one extraperitoneal metastasis. Five of our 16 patients were included in a previous publication.13
Application of PIPAC and sampling of PF and histological peritoneal biopsies
Using a standard laparoscopic two-trocar approach, a pressure injector and a nebuliser, aerosolised chemotherapeutics (cisplatin and doxorubicin) were distributed within the peritoneal cavity, leading to a deeper and more uniform penetration into the peritoneum compared with traditional chemotherapy.4 14 The PIPAC procedure was repeated every 4–6 weeks. Prior to each treatment, saline was irrigated into the peritoneal cavity and 150 mL of peritoneal lavage fluid (PF) was collected for further analyses, while the remaining PF was disposed. Quadrant biopsies (QBs) were taken from the peritoneum for assessment of histological response to treatment. The PFs were processed as follows: if a spontaneous coagulum was present, it was fixed in formalin and embedded in paraffin (formalin-fixed paraffin-embedded (FFPE)). Three tubes were filled with 50 mL and centrifuged. From the sediment of the first tube, two smears were produced and stained with Papanicolaou and May-Giemsa Grünwald. From the sediment of the second tube, a cell block was prepared and FFPE. From each of the two FFPE blocks, a thick section of 4–5 µm was cut with a microtome and stained with H&E for microscopic analysis. The sediment of the third tube was stored at −80°C in MagNa Pure LC Lysis Buffer (Roche) for subsequent NGS analysis. For cytological evaluation, a five-tiered score was used: malignant cells, suspicious cells, atypical cells, no malignant cells and other.
Histology and immunochemistry
The histological QBs were FFPE. Four-micron sections were cut and mounted on FLEX IHC Microscope slides. Three step sections were cut and stained with H&E, followed by a section immunostained for epithelial cell adhesion molecule and a final series of three H&E-stained step sections.13 15 Sections were dried at room temperature and baked at 60°C for 60 min before immunostaining, which was automated at the Dako Omnis immunostainer using the EnVision FLEX+DAB detection with mouse linker (Dako/Agilent, Glostrup, Denmark). The primary antibody was the clone BS14-Epithelial Specific Antigen/ CD326 (code: BSH-7402–1) (Nordic Biosite, Copenhagen, Denmark). Dilution was 1:600. For retrieval, target retrieval solution buffer with pH 9.0 was used for 30 min at 97°C. Incubation at 32°C was done for 20 min. Nuclear counterstaining was performed using Hematoxylin FLEX at the Dako Omnis platform. Slides were washed, dehydrated and cover slipped using an automated Dako cover slipper (Dako/Agilent, Glostrup, Denmark).
Peritoneal Regression Grading Score
The four-tiered PRGS for the histological assessment of response to therapy in PM was assessed for each quadrant biopsy and as a mean score for all quadrant biopsies obtained prior to a given PIPAC treatment.15 The scoring was performed by the same pathologist with special interest for peritoneal pathology (SD) in all cases. It was recently found that the interobserver variability of the PRGS is moderate to good.16 The PRGS defines four categories, based on the presence of residual tumour cells and the extent of regressive features. Major histological features of regression are fibrosis, inflammation, hyalinosis, acellular mucin pools, ischaemic necrosis, accumulation of macrophages, multinucleated giant cells and granulomas. PRGS 1 corresponds to a complete regression with absence of tumour cells; PRGS 2 corresponds to a major histological response with regressive features predominant over residual tumour cells; PRGS 3 corresponds to a minor histological response with predominance of residual tumour cells over regressive features; and PRGS 4 corresponds to a lack of histological response to therapy where the tumour cells were not accompanied by any regressive features.15
DNA extraction
For genomic DNA purification from FFPE tissue, GeneRead DNA FFPE Kit (QIAGEN, Hilden, Germany) was used according to the manufacturer’s recommendations. When possible, macrodissection was performed to increase the relative amount of tumour DNA, aiming at 20% or more. The GeneRead DNA FFPE procedure was used to remove paraffin and reverses formalin cross-links from the DNA before it was bound to QIAamp MinElute column (QIAGEN). After heating to remove cross-links, the DNA was accessible for removal of deaminated cytosine residues. After binding of the DNA to the spin column, residual contaminants such as salts were washed away with two wash buffers and ethanol. Any residual ethanol was removed by an additional centrifugation step. Finally, DNA was eluted. DNA extraction from PFs was performed using the MagNA pure LC Instrument (Roche Applied Science, Mannheim, Germany). DNA was extracted in a volume of 100 µL of elution buffer using the MagNa Pure LC DNA Isolation Kit I (Roche Applied Science).
Specimens for NGS
The inclusion criteria for the specimens used for NGS analyses were availability of cancer tissue in biopsy or resection specimen. For histological PM biopsy before and after PIPAC, only QB sets containing cancer cells in at least one of the QBs were included, regardless of the amount of cancer cells. The QB with the highest amount of tumour tissue was chosen. NGS was performed on all PFs with either ‘malignant tumour cells’ or ‘cells suspicious of malignancy’.
Next-generation DNA sequencing (NGS)
NGS was performed on the Ion Torrent PGM (Life Technologies, Carlsbad, California, USA) instrument, using the Ion AmpliSeq Colon and Lung Cancer NGS Panel v2, containing hotspot regions in 22 target genes: ALK, AKT1, BRAF, CTNNB1, DDR2, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, KRAS, MAP2K1, MET, NRAS, NOTCH1, PIK3CA, PTEN, SMAD4, STK11 and TP53. Ion Reporter software was used for variant calling. All variants were visualised using Golden Helix GenomeBrowse V.3.0.0 software (GoldenHelix, Montana, USA). A wild-type control was included in each run. A 1% cut-off was used for known KRAS hotspot mutations and a 10% cut-off was used for other variants.17 18 Only variants with coverage of >2000 were included. Samples with a DNA input of <0.1 ng/µL were discarded.
Statistics
Values were given as means or percentages. Comparisons were performed using Pearson’s χ2 test for categorical data and Wilcoxon matched pairs signed-rank test was used where appropriate. P values were two-tailed, and a p value of 0.05 was considered statistically significant. The statistical software STATA V.15.0 was used for statistical analysis.
Results
Clinical data
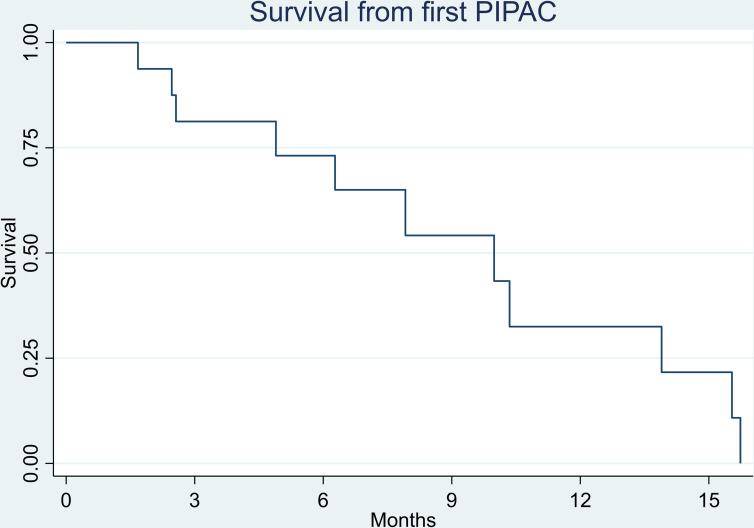
Sixteen patients with PM-PC were treated with at least one PIPAC at OPC from January 2016 to October 2019. The median number of PIPAC treatments was 3 (range 1–6). All patients underwent palliative systemic chemotherapy, and six patients underwent synchronous chemotherapy with gemcitabine+S1, gemcitabine+nab-paclitaxel or Folfirinox (folinic acid, fluorouracil, irinotecan and oxaliplatin) (table 1). The median overall survival from the first PIPAC was 9.9 months (SE 1.5, 95% CI 4.9 to 13.9). Kaplan-Meier survival analysis is shown in figure 1.
Table 1.
Baseline characteristics of patients with peritoneal metastasis from pancreatic cancer treated with PIPAC
| Demographic variables | Value |
| Number of patients | 16 |
| Age (years), mean (range) | 59 (46–72) |
| Sex, male/female | 10/6 |
| ECOG performance status 0, number of patients | 5 |
| ECOG performance status 1, number of patients | 11 |
| Number of PIPAC procedures, median (range) | 3 (1–6) |
| Previous treatment | |
| One-line palliative SC, number of patients | 12 |
| Two-line palliative SC, number of patients | 4 |
ECOG, Eastern Cooperative Oncology Group; PIPAC, Pressurised IntraPeritoneal Aerosol Chemotherapy; SC, systemic chemotherapy.
Figure 1.
Kaplan-Meier survival analysis of 16 patients with peritoneal metastasis from pancreatic cancer treated with PIPAC. Survival is shown as the time period from the first PIPAC treatment. The median overall survival from first PIPAC was 9.9 months (SE 1.5, 95% CI 4.9 to 13.9). PIPAC, pressurised intraperitoneal aerosol chemotherapy.
Histological response assessment using the PRGS
A total of 44 QB sets were collected, consisting of 111 QBs. Sixteen, 13, 6, 5, 3 and 1 QB sets were taken prior to PIPAC treatments 1, 2, 3, 4, 5 and 6. The remaining biopsies were from the epigastric peritoneum (n=4), the liver capsule (n=1), the right flank (n=3), the falciform ligament and the peritoneum without known precise location (each n=1). Thirteen patients underwent at least two PIPACs and were eligible for response assessment using the PRGS. Seven patients underwent two PIPAC treatments, while six patients underwent three PIPAC treatments or more. Based on the PRGS prior to PIPAC treatment 1 and PIPAC treatment 2 or 3, 61.50% of patients had a reduction of the mean PRGS. The overall mean PRGS was reduced from 1.91 at baseline to 1.58 prior to PIPAC 2 or 3 (p=0.02). The overall findings regarding PRGS are shown in tables 2 and 3.
Table 2.
PRGS ranging from 1 to 4 in 16 patients with peritoneal metastasis from pancreatic cancer who underwent at least one PIPAC treatment
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | |
| PRGS before PIPAC 1 (highest/mean) | 3/2.25 | 3/2 | 3/2.25 | 4/2.75 | 3/3 | 1/1 |
| PRGS before PIPAC 2 (highest/mean) | 2/1.25 | 2/2 | 3/1.75 | 3/2.25 | 2/2 | |
| Histological regression | + | U | + | + | + | |
| PRGS before PIPAC 3 (highest/mean) | 2/1.25 | 3/1.75 | 2/1.5 | 3/2 | ||
| Histological regression | U | + | + | – | ||
| PRGS before PIPAC 4 (highest/mean) | 2/1.75 | 3/2 | 3/2.67 | |||
| Histological regression | – | – | – | |||
| PRGS before PIPAC 5 (highest/mean) | 3/2.5 | 3/2.33 | ||||
| Histological regression | – | + |
| Patient 7 | Patient 8 | Patient 9 | Patient 10 | Patient 11 | Patient 12 | |
| PRGS before PIPAC 1 (highest/mean) | 1/1 | 3/3 | 1/1 | 3/2.25 | 2/1.33 | 1/1 |
| PRGS before PIPAC 2 (highest/mean) | 1/1 | 3/2 | 1/1 | 2/1,5 | ||
| Histological response | U | + | + | – | ||
| PRGS before PIPAC 3 (highest/mean) | 1/1 | 1/1 | ||||
| Histological response | U | + | ||||
| PRGS before PIPAC 4 (highest/mean) | 1/1 | 3/2.66 | ||||
| Histological response | U | – | ||||
| PRGS before PIPAC 5 (highest/mean) | 1/1 | |||||
| Histological response | U | |||||
| PRGS before PIPAC 6 (highest/mean) | 1/1 | |||||
| Histological response | U |
| Patient 13 | Patient 14 | Patient 15 | Patient 16 | |
| PRGS before PIPAC 1 (highest/mean) | 1/1 | 1/1 | 3/2 | 3/3 |
| PRGS before PIPAC 2 (highest/mean) | 1/1 | 1/1 | 3/2.33 | 2/2 |
| Histological response | U | U | – | + |
– indicates histological progression (according to mean PRGS); + indicates histological regression (according to mean PRGS).
PIPAC, Pressurised IntraPeritoneal Aerosol Chemotherapy; PRGS, Peritoneal Regression Grading Score; U, unchanged mean Peritoneal Regression Grading Score.
Table 3.
PRGS ranging from 1 to 4 in 13 patients with peritoneal metastasis from pancreatic cancer who underwent at least two PIPAC treatments
| Patient 1 | Patient 2 | Patient 3 | Patient 4* | Patient 5 | Patient 7* | Patient 10* | Patient 11 | Patient 12 | |
| PRGS before PIPAC 1 (highest/mean) | 3/2.25 | 3/2 | 3/2.25 | 4/2.75 | 3/3 | 1/1 | 3/2.25 | 2/1.33 | 1/1 |
| PRGS before PIPAC 3 (or 2*) (highest/mean) | 2/1.75 | 3/1.75 | 3/1.5 | 3/2.25 | 3/2 | 1/1 | 3/2 | 1/1 | 1/1 |
| Histological response | + | + | + | + | + | U | + | + | U |
| Patient 13* | Patient 14* | Patient 15* | Patient 16* | |
| PRGS before PIPAC 1 (highest/mean) | 1/1 | 1/1 | 3/2 | 3/3 |
| PRGS before PIPAC 3 (or 2*) (highest/mean) | 1/1 | 1/1 | 3/2.33 | 2/2 |
| Histological response | U | U | – | + |
– indicates histological progression (according to mean PRGS); + indicates histological regression (according to mean PRGS).
*Did only undergo PIPAC twice, data from PIPAC 2 instead of PIPAC 3.
PIPAC, Pressurised IntraPeritoneal Aerosol Chemotherapy; PRGS, Peritoneal Regression Grading Score; U, unchanged mean PRGS.
Next-generation sequencing
NGS analysis was performed on 57 specimens (13 primary tumours, 2 metastatic lymph nodes, 16 PFs and 26 QBs) (table 4). A mutation was detected in 15 of 16 patients (93.75%), and a KRAS mutation (p.Gly12Asp, p.Gly12Val, p.Gly12Ala or p.Gln61Arg) in 14 of 16 patients (87.50%). A KRAS mutation was found in QBs, primary tumours and PFs in 8/12 (66.67%), 8/13 (61.53%) and 6/9 (66.67%) patients. A KRAS mutation was found in the primary tumour in 8/13 patients (61.53%) and in at least one of the metastatic biopsies/PFs in 10/13 patients (76.92 %). A KRAS mutation was found in QBs prior to PIPAC 1, in PFs prior to PIPAC 1, QBs after PIPAC and PFs after PIPAC in 8/11 (61.54%), 4/6 (66.67%), 5/8 (62.50%) and 4/7 (57.14%) patients. A KRAS mutation was found in both the primary tumour and at least one metastasis in 4/11 patients (36.36%). In three patients, a KRAS mutation was detected in a metastasis but not in the primary tumour. In one patient, a KRAS mutation was detected in the primary tumour but not in the metastases.
Table 4.
Results of NGS analyses of cytological and histological specimens from primary tumour and metastases from 16 patients with PM from pancreatic cancer
| Primary tumour | Histological biopsy prior to PIPAC* | PF before PIPAC | Histological peritoneal biopsy after PIPAC | PF after PIPAC | |
| Patient 1 | KRAS (p.Gly12Asp) | KRAS (p.Gly12Asp) | KRAS (p.Gly12Asp) | ND (PIPAC 3 and 4) | NA |
| Patient 2 |
KRAS (p.Gly12Val) TP53 (p.Cys275Tyr) |
KRAS (p.Gly12Val) | NA | KRAS (p.Gly12Val) (PIPAC 2 and 3) | NA |
| Patient 3 | ND | ND | NA | ND (PIPAC 2, 3, 4 and 5) |
ND (PIPAC 5) |
| Patient 4 | ND | KRAS (p.Gly12Val) | ND | KRAS (p.Gly12Val) (PIPAC2) | KRAS (p.Gly12Val) (PIPAC 2) |
| Patient 5 |
KRAS (p.Gly12Asp) SMAD4 (p.Arg135Ter) |
NA | NA |
KRAS (p.Gly12Asp) (PIPAC 2) SMAD4 (p.Arg135Ter) (PIPAC 2) ND (PIPAC 3) KRAS (p.Gly12Asp) (PIPAC 4) |
KRAS (p.Gly12Asp) (PIPAC 5) |
| Patient 6 | KRAS (Gly12Asp) | NA | NA | NA | NA |
| Patient 7 | KRAS (p.Gly12Val) | KRAS (p.Gly12Val) (lymph node) | NA | NA | NA |
| Patient 8 | NA | ND | KRAS (p.Gly12Asp) | NA | NA |
| Patient 9 | MET (p.Arg988Cys) | MET (p.Arg988Cys) (found in PM biopsy and lymph node biopsy) | NA | NA | NA |
| Patient 10 | ND |
KRAS (p.Gln61Arg) SMAD4 (p.Arg361Cys) |
KRAS (p.Gln61Arg) | KRAS (p.Gln61Arg) (PIPAC 2) | KRAS (p.Gln61Arg) (PIPAC 2) |
| Patient 11 | FGFR2 (p.Asn549Lys) | KRAS (p.Gly12Asp) | NA | NA | NA |
| Patient 12 |
KRAS
(p.Gly12Asp) |
NA | NA | NA | ND (PIPAC 3 and 4) |
| Patient 13 | KRAS (p.Gly12Val) | NA | NA | NA | NA |
| Patient 14 |
KRAS (p.Gly12Val) TP53 (p.Arg273His) |
NA | NA | NA | NA |
| Patient 15 | NA | KRAS (p.Gly12Arg) | KRAS (p.Gly12Arg) | KRAS (p.Gly12Arg) | KRAS (p.Gly12Arg) (PIPAC 2) |
| Patient 16 | NA | KRAS (p.Gly12Ala) | ND | ND | ND (PIPAC 2) |
*NGS of histological biopsy from peritoneum, unless something else is stated.
NA, not available; ND, not detected; NGS, next-generation sequencing; PF, peritoneal fluid; PM, peritoneal metastasis.
We also found mutations in TP53 in the primary tumour in two patients (patient 2 and patient 14), a mutation in MET in both primary tumour and two metastases (patient 9), a SMAD4 mutation in the primary tumour and PM (patient 5), a SMAD4 mutation in QB (patient 10) and a FGFR2 mutation in the primary tumour (patient 11).
Discussion
To our knowledge, this is the first study evaluating the utility of NGS for detection of mutations in cancer-related genes in QBs and PFs from PM treated with PIPAC. PIPAC induced a histological response, according to the mean PRGS, in 8/13 patients (61.50%), while 4/13 (30.77%) had stable disease. Using NGS, we found the KRAS mutation in peritoneal QBs and PF, even after PIPAC treatment, at a frequency similar to that in the primary tumour biopsies.
A histological tumour response to PIPAC treatment measured by PRGS has been observed in 67% of patients with PM of various origin in a previous study.13 Our study cohort was more homogenous, and therefore, only patients with PC were included.2 13 19 In another study on PIPAC treatment of 20 patients with PM-PC, 10 of whom underwent at least two PIPACs or more, a histological tumour response was observed in 35%.20 PRGS was not used, but instead, tumour regression grade. Horvath et al reported a histological regression in 40% of their 12 PIPAC-treated patients with PM from pancreatobiliary cancer (PC (n=6) or cholangiocarcinoma (n=6)), using the PRGS.21 The histological response was evaluated in 6 of the 12 included patients who had undergone two PIPACs or more. Furthermore, Graversen et al demonstrated histological regression, measured by PRGS, in four out of five patients with PM-PC (80 %), while one patient had stable disease.13 Hence, the present study is the to date the largest study providing histological response evaluation in patients with PM-PC treated with PIPAC (n=13). The median survival from first PIPAC that we found, 9.9 months (95% CI 4.9 to 13.9), is similar to the median survival reported by Khosrawipour et al and Horvath et al.20 21
We detected a KRAS mutation and other mutations in QBs and PF before and after PIPAC treatment. The KRAS gene is mutated in 88% to 95% of patients with PC.17 18 22 These previous studies were mainly based on snap-frozen tissue from pancreatic surgery or FFPE tissue from surgical specimens from chemotherapy-naïve patients, meaning that much more tumour tissue was available than in our QB and PF specimens before and after systemic chemotherapy and PIPAC. In many of our specimens, regressive changes and only few cancer cells were present. In PF specimens before and after PIPAC, we found a mutation in around 60% of the patients. This indicates that NGS for cancer-related genes has a lower sensitivity compared with PCR for messenger RNA (mRNA) of tumour markers in this setting. Graversen et al reported a sensitivity of 0.88 and a specificity of 1.00 in patients with PM of various origins, including PC, when performing PCR for a combination of carcinoembryonic antigen (CEA)/EpCAM mRNA on PF specimens.19 In patient 9, no KRAS mutation but instead a MET mutation was found. A previous study has shown that targeting MET is possible and can be used for treatments of various cancers.23 Future studies have to elucidate whether treatment with, for example, crizotinib, targeting MET, may be of value in some patients with PM-PC.24 It has to be emphasised that our data do not support the routine use of NGS in PM treated with PIPAC. However, NGS using post-PIPAC peritoneal biopsies could be considered in the following setting: (1) the NGS panel includes predictive markers (eg, in patients with PM from colorectal cancer, where the KRAS mutation has predictive value for treatment with EGF-R antagonists); (2) cancer cells are present in the biopsy (but some histological regression (PRGS 2 or 3) may be present); and (3) an adequate specimen from the primary tumour or from a pre-PIPAC biopsy is not available. However, that being said, a peritoneal post-PIPAC biopsy may be even preferred over a biopsy from the primary tumour, particularly in patients who underwent several PIPACs; as such a biopsy may be more representative of the current mutational profile of the PM compared with a primary tumour biopsy that may have been taken many months (or years) previously.
Recent studies found the histological PRGS useful for evaluation of response to PIPAC.19 21 The PRGS has a moderate to good interobserver reproducibility and a near-perfect intraobserver reproducibility.16 The effect of PIPAC has been defined and monitored by different methods, for example, quality of life, median survival, histological tumour regression, PCI score and changes in gene expression.2 The combined progression index, based on the highest PRGS and cytology of PF, was independently associated with worse prognosis for OS and for PFS in PM.25
Four of our patients with PM-PC were treated with a combination of chemotherapy and PIPAC, and all patients underwent systemic chemotherapy. Therefore, it is difficult to assess whether the histological response was based on the systemic chemotherapy or PIPAC or both. This is particularly true for those patients who were found to have a baseline mean PRGS of 1. Both treatments may have led to improvement of overall outcome. The presence of regressive features prior to the first PIPAC indicates a systemic effect. However, an increase in some regressive features from PIPAC 1 to PIPAC 3 could indicate a PIPAC-related effect. The fact that three patients underwent only one PIPAC was the main cause of why some data were missing, which may have had an impact on the results and interpretations. However, we think that it had a very little or maybe no impact on the results regarding the utility of NGS, because these analyses were independent of how many PIPACs each patient received. A total of five patients underwent more than three PIPACs, but due to the low number of patients, the effect of more than three PIPAC procedures still remains to be investigated.
In conclusion, PIPAC treatment induces objective response according to the PRGS in the majority of patients with PM-PC. The PRGS seems to be a useful tool for the histological assessment of response to PIPAC. PIPAC does not seem to influence the utility or sensitivity of NGS because molecular alterations were identified at expected frequencies. Our data indicate that NGS is a useful tool also in peritoneal biopsies and PFs after PIPAC. Hence, it may be hypothesised that NGS in the future may become a tool for detection of predictive mutations in patients treated with PIPAC for PM of various origins, also when only peritoneal post-PIPAC QBs or PFs are available.
Take home messages.
Pressurised IntraPeritoneal Aerosol Chemotherapy (PIPAC) can be a treatment alternative to conventional, systemic chemotherapy in patients with peritoneal metastasis from pancreatic cancer (PM-PC).
PIPAC induces a histological response in peritoneal biopsies taken prior to PIPAC treatment 2 or 3, compared with biopsies taken prior to PIPAC treatment 1, in the majority of patients.
Next-generation sequencing (NGS) is able to identify the KRAS mutation in peritoneal biopsies and peritoneal fluids taken after systemic chemotherapy and PIPAC at a frequency similar to biopsies taken from the primary tumour prior to treatment.
NGS may in the future become a tool for detection of predictive mutations in patients treated with PIPAC for PM of various origins, also when only peritoneal post-PIPAC biopsies or peritoneal fluids are available.
Acknowledgments
Parts of this study were part of MN’s MD thesis.
Footnotes
Handling editor: Runjan Chetty.
Contributors: The study was designed by SDE and MBM. All authors contributed to the data collection and data analysis, discussed the results, and commented on and approved the final manuscript. MN and SD prepared the manuscript draft. SD was the main supervisor.
Funding: This study was supported by funding from Odense Pancreas Center.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: The study was conducted according to the Helsinki Declaration and approved by The Regional Committees on Health Research Ethics for Southern Denmark (project-ID S-20180185) and the Danish Data Protection Agency (project-ID 19/10354).
Provenance and peer review: Not commissioned; internally peer reviewed.
Data availability statement: Data are available upon reasonable request. Participant data will be retained at the study facility for five years after completion of the trial according to Danish law. After this time period, it will be de-identified and made available for other researchers upon reasonable request to the corresponding author.
References
- 1. Tabernero J, Chiorean EG, Infante JR, et al. Prognostic factors of survival in a randomized phase III trial (MPACT) of Weekly nab-paclitaxel plus gemcitabine versus gemcitabine alone in patients with metastatic pancreatic cancer. Oncologist 2015;20:143–50. 10.1634/theoncologist.2014-0394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grass F, Vuagniaux A, Teixeira-Farinha H, et al. Systematic review of pressurized intraperitoneal aerosol chemotherapy for the treatment of advanced peritoneal carcinomatosis. Br J Surg 2017;104:669–78. 10.1002/bjs.10521 [DOI] [PubMed] [Google Scholar]
- 3. Demtröder C, Solass W, Zieren J, et al. Pressurized intraperitoneal aerosol chemotherapy with oxaliplatin in colorectal peritoneal metastasis. Colorectal Dis 2016;18:364–71. 10.1111/codi.13130 [DOI] [PubMed] [Google Scholar]
- 4. Solaß W, Hetzel A, Nadiradze G, et al. Description of a novel approach for intraperitoneal drug delivery and the related device. Surg Endosc 2012;26:1849–55. 10.1007/s00464-012-2148-0 [DOI] [PubMed] [Google Scholar]
- 5. Dueckelmann AM, Fink D, Harter P, et al. The use of PIPAC (pressurized intraperitoneal aerosol chemotherapy) in gynecological oncology: a statement by the "Arbeitsgemeinschaft Gynaekologische Onkologie Studiengruppe Ovarialkarzinom (AGO-OVAR)", the Swiss and Austrian AGO, and the North-Eastern German Society of Gynaecologic Oncology. Arch Gynecol Obstet 2018;297:837–46. 10.1007/s00404-018-4673-0 [DOI] [PubMed] [Google Scholar]
- 6. Tempfer CB, Winnekendonk G, Solass W, et al. Pressurized intraperitoneal aerosol chemotherapy in women with recurrent ovarian cancer: a phase 2 study. Gynecol Oncol 2015;137:223–8. 10.1016/j.ygyno.2015.02.009 [DOI] [PubMed] [Google Scholar]
- 7. Abramson MA, Jazag A, van der Zee JA, et al. The molecular biology of pancreatic cancer. Gastrointest Cancer Res 2007;1:S7–12. [PMC free article] [PubMed] [Google Scholar]
- 8. Strimpakos AS, Syrigos KN, Saif MW. The molecular targets for the diagnosis and treatment of pancreatic cancer. Gut Liver 2010;4:433–49. 10.5009/gnl.2010.4.4.433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kohlmann A, Bacher U, Schnittger S, et al. Perspective on how to approach molecular diagnostics in acute myeloid leukemia and myelodysplastic syndromes in the era of next-generation sequencing. Leuk Lymphoma 2014;55:1725–34. 10.3109/10428194.2013.856427 [DOI] [PubMed] [Google Scholar]
- 10. Cheson BD, Bennett JM, Kopecky KJ, et al. Revised recommendations of the International Working group for diagnosis, standardization of response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol 2003;21:4642–9. 10.1200/JCO.2003.04.036 [DOI] [PubMed] [Google Scholar]
- 11. Graversen M, Detlefsen S, Bjerregaard JK, et al. Prospective, single-center implementation and response evaluation of pressurized intraperitoneal aerosol chemotherapy (PIPAC) for peritoneal metastasis. Ther Adv Med Oncol 2018;10:1–11. 10.1177/1758835918777036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graversen M, Detlefsen S, Asmussen J, et al. Treatment of peritoneal carcinomatosis with Pressurized IntraPeritoneal Aerosol Chemotherapy - PIPAC-OPC2. Pleura Peritoneum 2018;3:20180108. 10.1515/pp-2018-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graversen M, Detlefsen S, Bjerregaard JK, et al. Peritoneal metastasis from pancreatic cancer treated with pressurized intraperitoneal aerosol chemotherapy (PIPAC). Clin Exp Metastasis 2017;34:309–14. 10.1007/s10585-017-9849-7 [DOI] [PubMed] [Google Scholar]
- 14. Solass W, Kerb R, Mürdter T, et al. Intraperitoneal chemotherapy of peritoneal carcinomatosis using pressurized aerosol as an alternative to liquid solution: first evidence for efficacy. Ann Surg Oncol 2014;21:553–9. 10.1245/s10434-013-3213-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Solass W, Sempoux C, Detlefsen S, et al. Peritoneal sampling and histological assessment of therapeutic response in peritoneal metastasis: proposal of the peritoneal regression grading score (PRGS). Pleura Peritoneum 2016;1:99–107. 10.1515/pp-2016-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Solass W, Sempoux C, Carr NJ, et al. Reproducibility of the peritoneal regression grading score for assessment of response to therapy in peritoneal metastasis. Histopathology 2019;74:1014–24. 10.1111/his.13829 [DOI] [PubMed] [Google Scholar]
- 17. Cicenas J, Kvederaviciute K, Meskinyte I, et al. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer. Cancers 2017;9. 10.3390/cancers9050042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Connor AA, Denroche RE, Jang GH, et al. Association of distinct mutational signatures with correlates of increased immune activity in pancreatic ductal adenocarcinoma. JAMA Oncol 2017;3:774–83. 10.1001/jamaoncol.2016.3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Graversen M, Fristrup C, Kristensen TK, et al. Detection of free intraperitoneal tumour cells in peritoneal lavage fluid from patients with peritoneal metastasis before and after treatment with pressurised intraperitoneal aerosol chemotherapy (PIPAC). J Clin Pathol 2019;72:368–72. 10.1136/jclinpath-2018-205683 [DOI] [PubMed] [Google Scholar]
- 20. Khosrawipour T, Khosrawipour V, Giger-Pabst U. Pressurized intra peritoneal aerosol chemotherapy in patients suffering from peritoneal carcinomatosis of pancreatic adenocarcinoma. PLoS One 2017;12:e0186709. 10.1371/journal.pone.0186709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horvath P, Beckert S, Struller F, et al. Pressurized intraperitoneal aerosol chemotherapy (PIPAC) for peritoneal metastases of pancreas and biliary tract cancer. Clin Exp Metastasis 2018;35:635–40. 10.1007/s10585-018-9925-7 [DOI] [PubMed] [Google Scholar]
- 22. Sausen M, Phallen J, Adleff V, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 2015;6:7686. 10.1038/ncomms8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peruzzi B, Bottaro DP. Targeting the c-Met signaling pathway in cancer. Clin Cancer Res 2006;12:3657–60. 10.1158/1078-0432.CCR-06-0818 [DOI] [PubMed] [Google Scholar]
- 24. Takiguchi S, Inoue K, Matsusue K, et al. Crizotinib, a Met inhibitor, prevents peritoneal dissemination in pancreatic cancer. Int J Oncol 2017;51:184–92. 10.3892/ijo.2017.3992 [DOI] [PubMed] [Google Scholar]
- 25. Benzerdjeb N, Durieux E, Tantot J, et al. Prognostic impact of combined progression index based on peritoneal grading regression score and peritoneal cytology in peritoneal metastasis. Histopathology 2020. 10.1111/his.14092 [DOI] [PubMed] [Google Scholar]