Abstract
Genetically defined amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), collectively named c9ALS/FTD, are triggered by hexanucleotide GGGGCC repeat expansions [r(G4C2)exp] within the C9orf 72 gene. In these diseases, neuronal loss occurs through an interplay of deleterious phenotypes, including r(G4C2)exp RNA gain-of-function mechanisms. Herein, we identified a benzimidazole derivative, CB096, that specifically binds to a repeating 1×1 GG internal loop structure, 5′CGG/3′GGC, that is formed when r(G4C2)exp folds. Structure−activity relationship (SAR) studies and molecular dynamics (MD) simulations were used to define the molecular interactions formed between CB096 and r(G4C2)exp that results in the rescue of disease-associated pathways. Overall, this study reveals a unique structural feature within r(G4C2)exp that can be exploited for the development of lead medicines and chemical probes.
Graphical Abstract
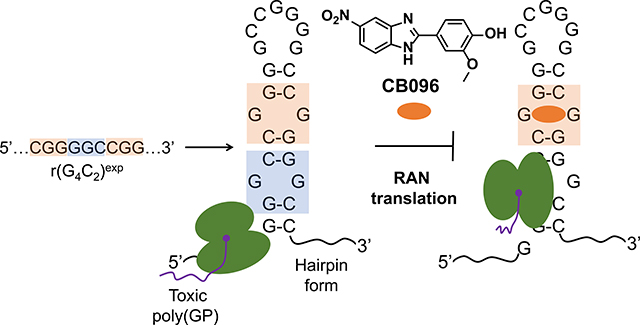
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are fatal neurodegenerative diseases characterized by a loss of motor function and cognitive impairment.1 The lack of effective treatments, despite multiple clinical trials,2 can be attributed to heterogeneous manifestation of the disorders3 and an incomplete understanding of their etiology and development.4,5 Recently, long stretches of hexanucleotide GGGGCC repeat expansions [hereafter r(G4C2)exp] in intron 1 of chromosome 9 open reading frame 72 (C9orf72) were identified as the main cause of genetically defined ALS and FTD, collectively named c9ALS/FTD.6 Whereas neurologically healthy individuals have up to 30 r(G4C2) repeats,1 affected individuals house hundreds to thousands of such consecutive repeats in C9orf72. Neuronal death in c9ALS/FTD patients is triggered by an interplay of pathological mechanisms7 originating from both C9orf72 protein loss-of-function8 and r(G4C2)exp-mediated RNA gain-of-function.9 In particular, r(G4C2)exp accumulates in the nucleus of a subset of central nervous system (CNS) cells in nuclear foci by sequestering various RNA binding proteins (RBPs) such as heterogeneous nuclear ribonucleoprotein H1 (hnRNP H1).10 The depletion of the RBP pool triggers RNA splicing defects, as well as dysfunction in RNA metabolism and nucleocytoplasmic transport.11 Moreover, when exported to the cytoplasm, r(G4C2)exp undergoes an unconventional translation process, named repeat associated non-ATG (RAN) translation, which occurs in the absence of an AUG start codon.12 RAN translation generates toxic dipeptide repeats (DPRs), which can cause formation of stress granules, defects in autophagosome formation, and induction of endoplasmic reticulum (ER) stress.13 Thus, an efficient therapeutic approach to ameliorate the RNA-mediated pathological mechanisms is to directly target r(G4C2)exp.
Thus far, antisense oligonucleotides (ASOs) and small molecules have been proposed as promising therapeutic solutions.14–16 However, there are challenges associated with ASOs, such as limited cellular and tissue penetrance, off-target effects, suboptimal physicochemical properties, and toxicity in clinical settings.17 Alternatively, small molecules offer several advantages including (i) synthetic ease from commercially available starting materials, (ii) tunable physicochemical properties, and (ii) temporal control of biological activity via structure–activity relationships (SAR).18,19 Indeed, recent studies showed that small molecules avidly bind two structures adopted by r(G4C2)exp that are in equilibrium, a G-quadruplex20 (stabilized in the presence of K+ ions) and a hairpin that forms a periodic array of 1 × 1 GG internal loops21 (stabilized in the presence of Li+ or Na+ ions), and alleviate c9ALS/FTD pathologies.21–23 Great effort has been expended to determine how each of these two structures contributes to r(G4C2)exp-mediated molecular defects. For example, hnRNPH1 has been shown to bind the G-quadruplex form in vitro and nuclear foci sequestering hnRNP H1 have been observed in patient-derived cells.10 We have previously shown that the r(G4C2)exp hairpin, not the G-quadruplex, undergoes RAN translation in vitro and that this hairpin structure forms in cells.21 Moreover, we previously reported a selective small molecule that targets the hairpin form of r(G4C2)exp and inhibits the production of poly(GP) DPR by stabilizing the 1 × 1 GG internal loops within r(G4C2)exp.21 Interestingly, under pathogenically relevant lengths (≥30 repeats), the formation of the G-quadruplex comprises a minor population of the folding landscape, and the hairpin form predominates.24 Additionally, a structural transition from the hairpin form to the G-quadruplex form is unlikely at pathogenic lengths due to its high activation energy barrier.21,24 Collectively, these studies suggest that both structural forms contribute to c9ALS/FTD pathology, and small molecules selectively targeting each will allow these contributions to be teased out. Herein, we studied the ligandability of the r(G4C2)exp hairpin form, identifying chemotypes that potently and specifically bind to structural features uniquely present within this structure and inhibit disease-relevant pathologies.
Results and Discussion
High-Throughput Screening Identifies CB096 As a Specific r(G4C2)8 Binder
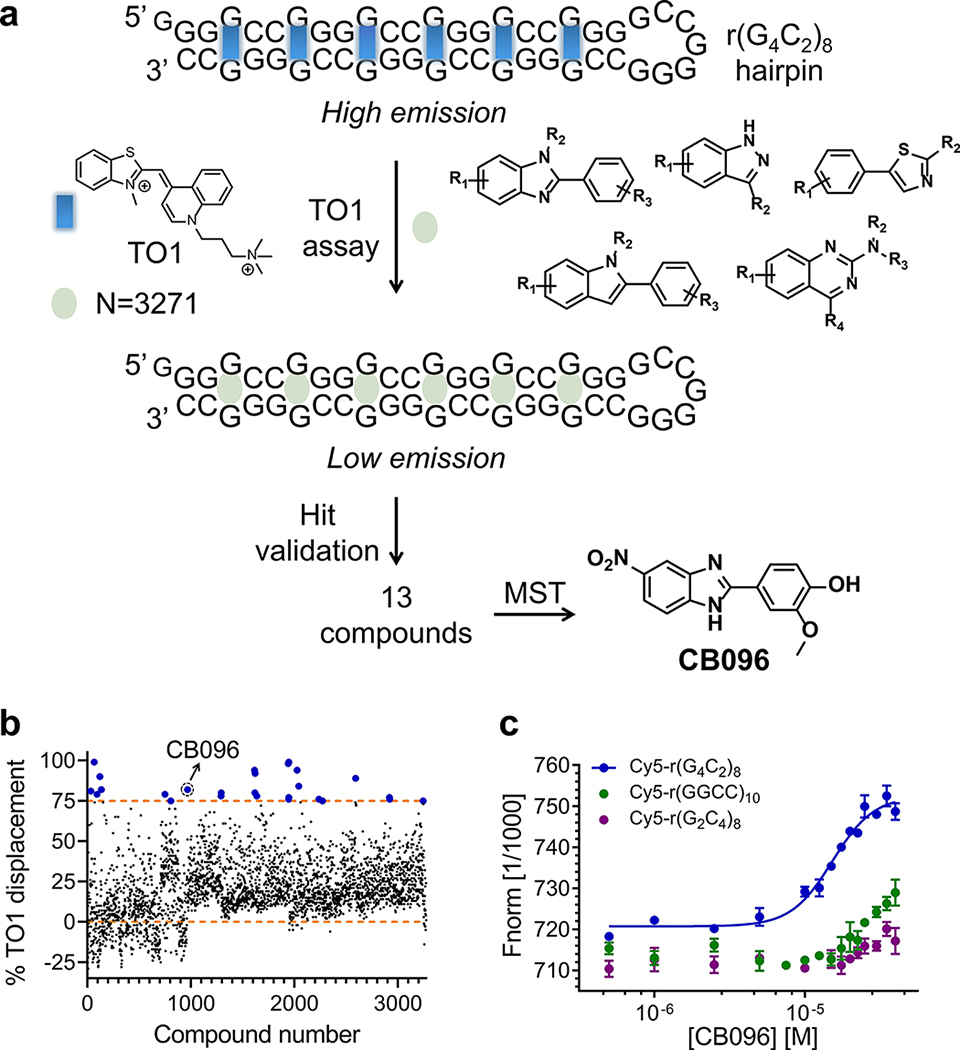
The ligandability of the r(G4C2)exp hairpin form was explored using a two-pronged screening strategy (Figure 1A). First, a TO-PRO-1 (TO1) fluorescent dye displacement assay22,25,26 was used to identify small molecules within an RNA-focused library that bind a r(G4C2)8 hairpin. The library comprises 3271 nitrogen-rich heterocyclic compounds featuring drug-like properties.27 By monitoring the decrease in fluorescence upon incubation with the small molecules (tested at 100 μM), 26 derivatives were identified that induced ≥75% displacement of TO1 from r(G4C2)8 (0.8% hit rate; Z-factor28 = 0.6; Figure 1B). False positives were removed by applying stringent criteria: (i) compounds must lack interference with the signal readout due to intrinsic fluorescence; (ii) compounds must not aggregate at concentrations up to 100 μM; and (iii) compounds must exhibit dose-dependent displacement of TO1 (Supplementary Figure 1). A total of 13 hits were validated, seven of which were substituted benzimidazoles (Supplementary Figure 1).
Figure 1.
Identification of CB096, a lead molecule that binds to the r(G4C2)8 hairpin. (a) Graphical representation of the two-pronged screening strategy leading to the identification of CB096. TO-PRO-1 (TO1) dye displacement assay was performed on 3271 compounds in 384-well format at 100 μM (single dose). Derivatives showing ≥75% TO1 quenching of the initial fluorescence (blue dots) were selected and further validated. A total of 13 derivatives met the criteria and were dose dependently evaluated for binding the hairpin form of Cy5-r(G4C2)8 via microscale thermophoresis (MST), of which CB096 is the best binder. (b) Results of the high throughput TO1 dye displacement assay. Compounds that exhibited ≥75% TO1 displacement were considered hits (upper orange dashed line). (c) CB096 specifically binds r(G4C2)8 (sense) over r(G2C4)8(antisense) and r(GGCC)10 (base pair) control RNAs. EC50 values were determined by plotting the changes in normalized fluorescence upon dose-dependent addition of CB096. The data points are reported as the mean values ± SD and are representative of two independent experiments each performed in duplicate.
In the second step of the screening funnel, binding of the 13 hits to the r(G4C2)8 hairpin was assessed via microscale thermophoresis (MST).29–31 Of these, the most promising was benzimidazole derivative CB096, which had an EC50 of 33(±9) μM in the TO1 displacement assay and an EC50 of 19(±3) μM from binding studies (Figure 1C and Supplementary Figure 2A). Circular dichroism (CD) spectroscopy was performed to determine the binding stoichiometry of CB096 to the r(G4C2)8 hairpin. The addition of CB096 triggered a dose-dependent increase of signals at 235 and 298 nm that reached saturation upon the addition of 4 equiv of CB096 (Supplementary Figure 4).
Interestingly, the RNA-focused library houses 262 benzimidazoles (8%), of which three have a nitro group (−NO2) in position 5 (CB096, CB141, and compound 11) and two were hits in the primary screen (CB096 and CB141). Of the 47 benzimidazoles with a −NO2 in a different position, none were hits in the TO-1 displacement primary screen nor were different scaffolds such as thiazoles, quinazolines, and indazoles that contain −NO2 groups (n = 12).
The binding selectivity of CB096 was then assessed relative to r(G2C4)8 (antisense) and the base paired construct r(GGCC)10, for which only a minor change in thermophoresis and no saturation were detected, respectively (Figure 1C and Supplementary Figure 2B and C). Benzimidazoles are a privileged chemotype for binding RNA26,27,32 and have been previously shown to bind 1 × 1 nucleotide internal loops.33 We therefore studied the binding of CB096 to other 1 × 1 nucleotide internal loops, including 1 × 1 AA loops in Cy5-r(CAG)12 and 1 × 1 UU internal loops in Cy5-r(CUG)12. Indeed, no saturable binding was observed for either loop (Supplementary Figure 3). Collectively, these data indicate that CB096 is indeed specific for r(G4C2)exp.
CB096 Alters the Dynamics of the 5′CGG/3′GGC Site within r(G4C2)exp
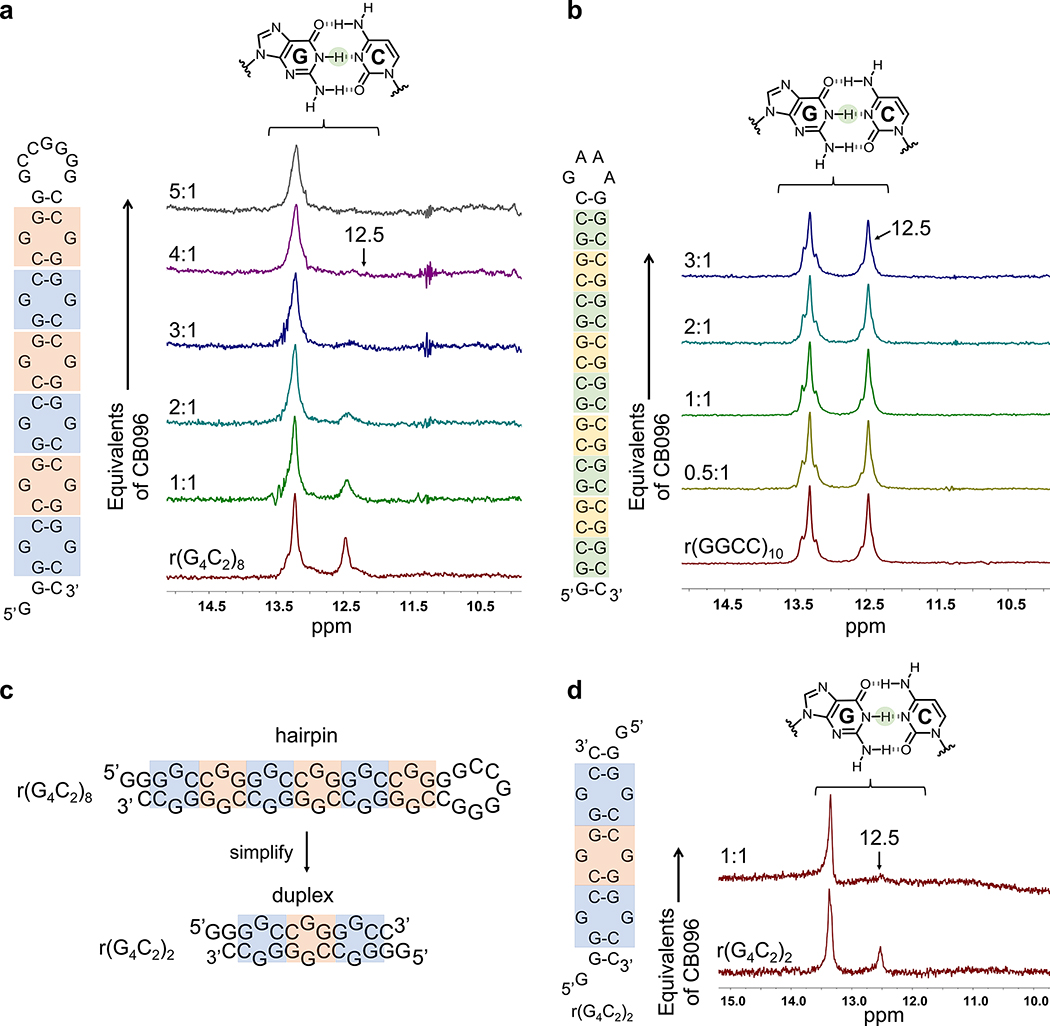
To investigate the exact residues involved in CB096 binding to the r(G4C2)8 hairpin, 1D imino NMR spectroscopy was employed. The r(G4C2)8 RNA displays a total of six 1 × 1 GG internal loops containing two types of binding sites with different closing base pairs: three 5′GGC/3′CGG loops (highlighted in blue; G highlights the 1 × 1 GG non canonical base pair; Figure 2A) and three 5′CGG/3′GGC loops (highlighted in orange, Figure 2A). Each type of loop alternates to form a periodic array of 5′GGCCGG/3′CGGGGC. The imino 1H NMR spectrum recorded at 25 °C exhibits a peak at 13.4 ppm, indicating a stable GC/CG base pair, and a less intense peak at 12.5 ppm, which could indicate that the base pair is more dynamic and hence less stable (i.e., faster exchange rate with H2O; Figure 2A). The closing pairs of the two types of 1 × 1 GG internal loops within 5′GGCCGG/3′CGGGGC are not identical, and these differences could be distinguished by small molecule ligands. Indeed, the addition of CB096 only broadened the GC/CG base pair at 12.5 ppm dose-dependently, reaching saturation at 4 equiv of CB096, in agreement with the CD spectroscopy data (Figure 2A). Further, resonances at 13.4 and 12.5 ppm were also observed for the r(G4C2)4 hairpin, and the resonance at 12.5 ppm also broadened upon the addition of CB096, saturating at 1 equiv of CB096 (Supplementary Figure 5). To confirm that the GG loop is required for binding, we studied the binding of CB096 to a fully paired RNA construct, r(G2C2)10, and to a model of the C9orf72 antisense repeat, r(G2C4)8. This RNA, complemented with a GAAA tetraloop for loop stability, adopts a rigid hairpin structure that maintains a similar nucleotide composition to r(G4C2)4. No changes were observed in the 1D imino proton spectra of either RNA upon the addition of up to 3 equiv of CB096 (Figure 2B and Supplementary Figure 6).
Figure 2.
CB096 mode of binding investigated via 1D imino proton NMR. (a) CB096 broadens the imino proton signal of GC/CG base pairs at 12.5 ppm within the r(G4C2)8 hairpin. Blue labels indicate 5′GGC/3′CGG internal loops, whereas the orange labels indicate 5′CGG/3′GGC loops (G represents the G residues within the 1 × 1 GG internal loops). (b) CB096 does not affect the 1D imino proton signal of GC/CG base pairs at 12.5 ppm within the base pair control r(GGCC)10 RNA. Unique GC/CG base pairs within r(GGCC)10 are highlighted in green and yellow. The 1D NMR spectra are representative of two independent experiments. (c) Simplification of r(G4C2)8 secondary structure to the r(G4C2)2 duplex featuring two 5′GGC/3′CGG (blue squares) and one 5′CGG/3′GGC (orange square). (d) The 1D imino proton spectra of r(G4C2)2 duplex recapitulates the 1D imino proton peak pattern of r(G4C2)4 and r(G4C2)8. CB096 (1 equiv) broadens the peak at 12.5 ppm. 1D NMR spectra are representative of two independent experiments.
Interestingly, imino proton signals of equal intensities were observed for r(GGCC)10 at 13.4 and 12.5 ppm, but the addition of CB096 did not lead to the broadening effect observed for r(G4C2)4 and r(G4C2)8. Since the secondary structures of r(G4C2)8 and r(GGCC)10 share the same array of GC/CG base pairs and vary solely by the 1 × 1 GG noncanonical base pairs in the former, the loops may trigger unique conformational features within the r(G4C2)8 hairpin. These data provide structural support for specific targeting of the c9ALS/FTD r(G4C2)exp RNA hairpin by CB096 observed in binding studies. Moreover, the substoichiometric ratio between CB096 and the number of 1 × 1 GG internal loops in r(G4C2)4 and r(G4C2)8 indicates that this compound might specifically bind to one type of GG loop and its closing pairs (5′GGC/3′CGG vs 5′CGG/3′GGC) and/or exhibit multiple binding modes. Indeed, previous studies have shown that both the loop nucleotides and closing base pairs influence conformation.34,35
We therefore designed and studied a simplified duplex derived from r(G4C2)4 and r(G4C2)8, r(G4C2)2, that features two 5′GGC/3′CGG and one 5′CGG/3′GGC and thus has two full r(G4C2) repeat sequences on each strand (Figure 2C). The imino-proton spectrum of r(G4C2)2 was similar to r(G4C2)4 and r(G4C2)8 exhibiting peaks at both 13.4 and 12.5 ppm, with the latter completely broadened out upon the addition of 1 equiv of CB096 (Figure 2D). Notably, free energy minimization using the RNAstructure software36 predicted that r(G4C2)2 can also form a 2 × 2 GG internal loop containing structure with the same folding energy as the 1 × 1 GG internal loop (Supplementary Figure 7A). However, the 1D imino proton spectrum of a 2 × 2 GG containing duplex model did not recapitulate either the peak pattern or the specific signal broadening upon the addition of CB096 (Supplementary Figure 7B).
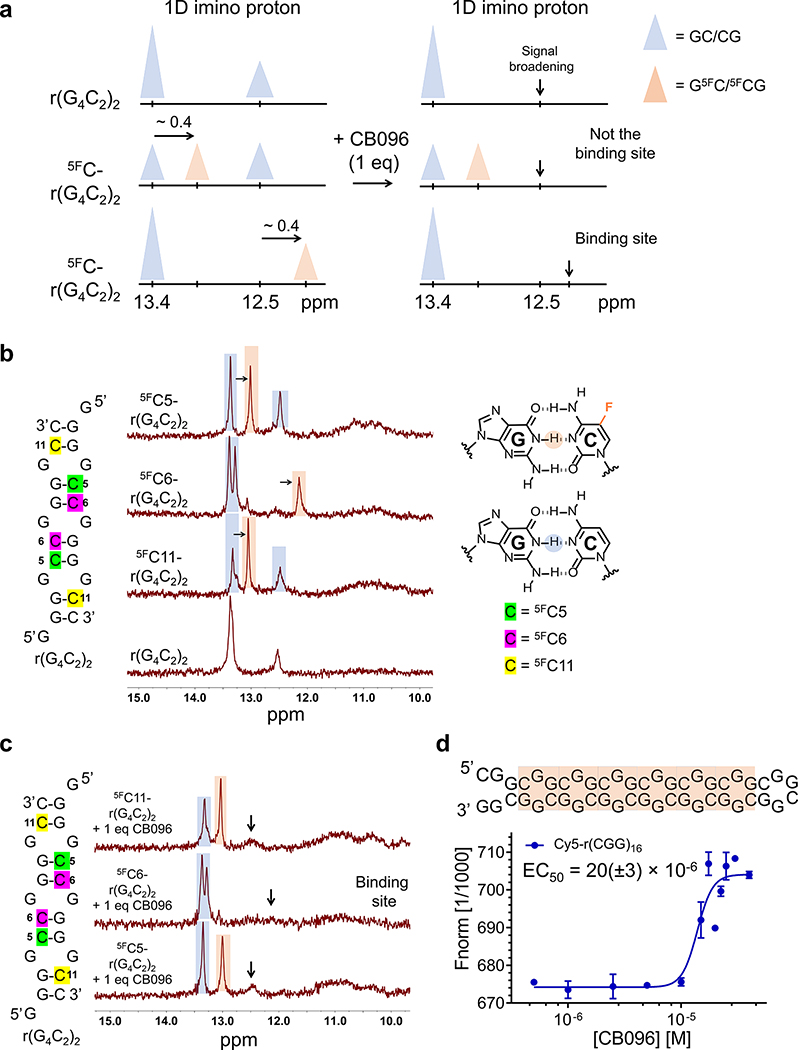
To precisely assign the two imino proton signals to their corresponding GC/CG base pairs within r(G4C2)2, cytosine (C) residues were exchanged for 5-fluoro C (5FC). Indeed, 19F has been used as a label in numerous 1D and 2D NMR spectroscopic studies of nucleic acids37,38 due to its comparable size to 1H, high detection sensitivity, and straightforward incorporation into oligonucleotides.39–41 The 5FC substitution (i) decreases the stability of GC/CG base pairs due to diminished electron density at the N(3) of C, which reduces the strength of the hydrogen bond with the N(1)–H of the paired G, and (ii) shifts the imino proton signal of the G’s N(1)–H to which it is paired upfield by ∼0.4 ppm (Figure 3A, left).37,40 By systematically substituting nonequivalent C residues, the 1D imino proton peaks at 13.4 and 12.5 ppm can be deconvoluted, allowing for identification of CB096’s binding site (Figure 3A, right).
Figure 3.
CB096 affects the dynamics of 5′CGG/3′GGC, but not 5′GGC/3′CGG within r(G4C2)2. (a) Graphical representation of the strategy to assign the imino proton signals emanating from guanine (G) residues in base pairing interactions within r(G4C2)2. Briefly, replacing cytosine (C) by 5-fluorocytosine (5FC) shifts imino proton signals of G residues in GC base pairs upfield by ∼0.4 ppm (orange triangle), compared to unlabeled base pair peaks (blue triangle). (b) 1D imino proton spectra of 5FC labeled r(G4C2)2 duplex constructs. Blue color highlights the imino proton peak corresponding to the GC base pairs while those labeled in orange are from 5FC-labeled pairs. The 1D NMR spectra are representative of two independent experiments. (c) 1D imino proton spectra of 5FC labeled r(G4C2)2 duplex constructs in complex with CB096 (1 equiv). Black arrows indicate the imino proton peaks that broadened out upon CB096 treatment. The data indicate that CB096 binds 5′CGG/3′GGC and increases the dynamics of the base pairs (likely do not form or form transiently). The 1D NMR spectra are representative of two independent experiments. (d) CB096 binds to 5′CGG/3′GGC within Cy5-r(CGG)16 (orange squares) with an EC50 of 20(±3) μM, as measured by MST. The EC50 value was determined by plotting the changes in normalized fluorescence upon dose-dependent addition of CB096. The data points are reported as the mean values ± SD and are representative of two independent experiments performed in duplicate.
Following this strategy, C5, C6, or C11 within the r(G4C2)2 duplex were replaced by 5FCs (Figure 3B). The 5FC5 (green label, Figure 3B) and 5FC11 (yellow label, Figure 3B) substitutions triggered the emergence of new imino proton peaks at 13.0 ppm, whereas the introduction of 5FC6 (purple label, Figure 3B) shifted the 12.5 ppm peak to 12.1 ppm. Thus, the imino proton signal at 13.4 ppm in r(G4C2)2 was from the guanine (G) residues of the base pairs formed between C5–G9 and C11–G3, both forming closing pairs for the 5′GGC/3′CGG loops (bold lettering indicates the base pair referred to in the motif). Likewise, the peak at 12.5 ppm was assigned to G8, which forms a base pair with C6, closing the 5′CGG/3′GGC loop (bold lettering indicates the base pair referred to in the motif).
We next determined which peaks changed upon the addition of CB096 to each construct. For the 5FC5– and 5FC11–r(G4C2)2 duplexes, the addition of 1 equiv of CB096 broadened the signal at 12.5 ppm, assigned to G8, which forms the closing base pairs of 5′CGG/3′GGC, while having no effect on the signals at 13.0 and 13.4 ppm (Figure 3C). Likewise, the addition of CB096 to 5FC6-r(G4C2)2 causes broadening of the G8 resonance (12.1 ppm), without affecting the other peaks (Figure 3C). Collectively, these data suggest that CB096 binds and selectively affects the dynamics of the 5′CGG/3′GGC but not those of the 5′GGC/3′CGG. Interestingly, nearly complete broadening of G8’s resonance occurred with 1 equiv of CB096, consistent with the construct containing only one 5′CGG/3′GGC and two 5′GGC/3′CGG motifs. This observation was further validated by binding studies of CB096 to Cy5-r(CGG)16 and Cy5-r(GGC)16 using MST. Experimental results indicated that CB096 binds Cy5-r(CGG)16 with an EC50 of 20(±3) μM (Figure 3D), whereas no change in thermophoresis was observed for Cy5-r(GGC)16 (Supplementary Figure 8). Thus, binding of CB096 to r(G4C2)exp is exclusively due to its interaction with the 5′CGG/3′GGC motif within the r(G4C2)exp hairpin.
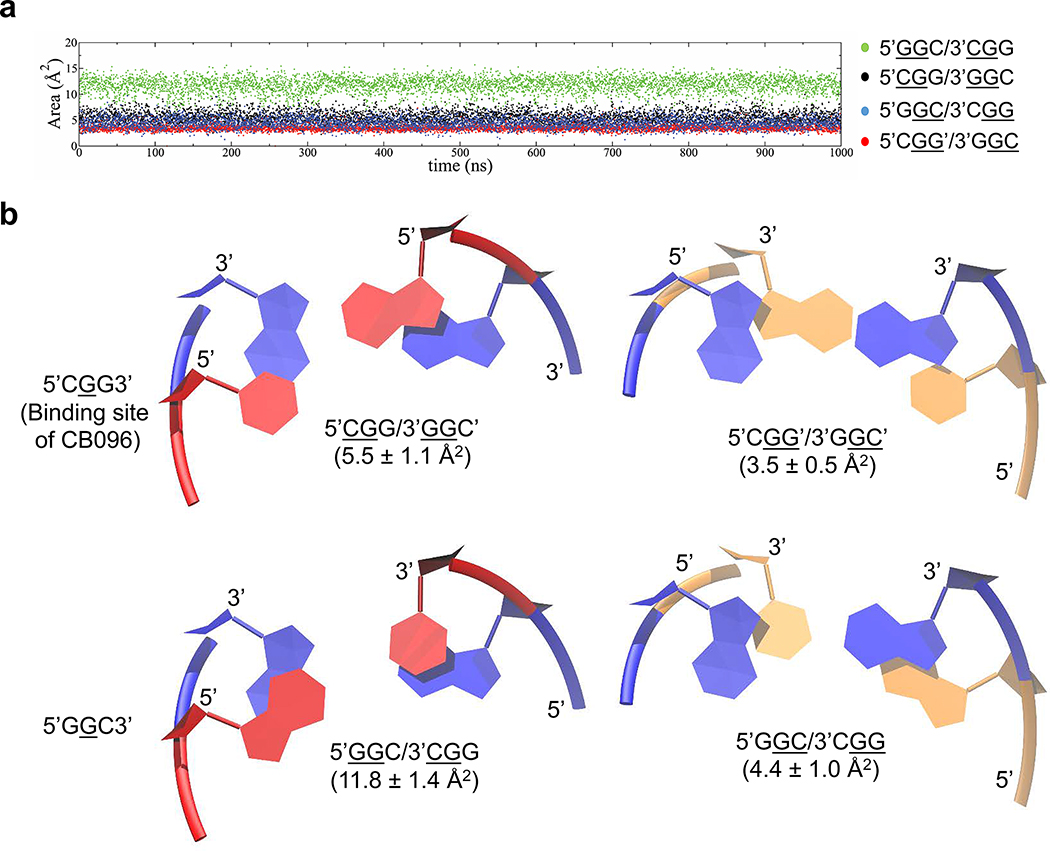
Previously reported experimental data42,43 indicate that the 5′GGC/3′CGG is more stable than 5′CGG/3′GGC, which could make the latter more amendable to structural rearrangement required for CB096 binding. The free energies, ΔG°37, of 5′GGC/3′CGG and 5′CGG/3′GGC are −1.89 and −1.40 kcal/mol, respectively, calculated from a model duplex of each loop and a duplex lacking the loop.42,43 This ∼0.5 kcal/mol difference, or 3.6-fold in an equilibrium constant, can likely be traced to more favorable stacking interactions of the 1 × 1 GG internal loop on its closing base pairs. Indeed, our molecular dynamics (MD) simulations44 confirmed more optimal stacking of loop nucleotides on their closing pairs in 5′GGC/3′CGG than in 5′CGG/3′GGC. In 5′CGG/3′GGC, the loop nucleotides stacked on the 5′ and 3′ closing pairs with 5.5 ± 1.1 Å2 (5′CGG/3′GGC′) and 3.5 ± 0.5 Å2 (5′CGG′/3′GGC′) of surface area, respectively (Figure 4A and B, black and red, respectively). In contrast, the stacking surface area of the GG loops on the 5′ and 3′ closing pairs within 5′GGC/3′CGG was 4.4 ± 1.0 Å2 (5′GGC/3′CGG) and 11.8 ± 1.4 Å2 (5′GGC/3′CGG) of surface area, respectively (Figure 4A and B, blue and green, respectively). Therefore, the computational study highlights a ∼2-fold increase of the stacking surface within 5′GGC/3′CGG compared to 5′CGG/3′GGC′. Consequently, this might affect the conformational ensemble of the 5′GGC/3′CGG motif and the ability to accommodate small molecule CB096.
Figure 4.
The 5′CGG/3′GGC loop is thermodynamically less stable than 5′GGC/3′CGG. (a) Overlap areas between 5′CGG/3′GGC and 5′GGC/3′CGG (G represents the G residue within the 1 × 1 GG internal loop) GG internal loops and the closing GC base pairs as a function of time, obtained by molecular dynamics (MD) simulations. Overlap areas are represented by underlining of two residues, which highlights the G residue part of the 1 × 1 GG internal loops and the G or C residue of the closing base pair. (b) Stacking observed for 1 × 1 GG internal loops on 5′ and 3′ closing base pairs in 5′CGG/3′GGC and 5′GGC/3′CGG, extracted from MD simulations. Figures display averaged structures calculated from the MD trajectories. Blue colored base pairs represent the 1 × 1 GG internal loops in syn-anti orientations. Red and orange colored base pairs highlight the closing Watson–Crick GC base pairs.
Structure–Activity Relationships Reveal Functional Groups Critical for RNA Binding
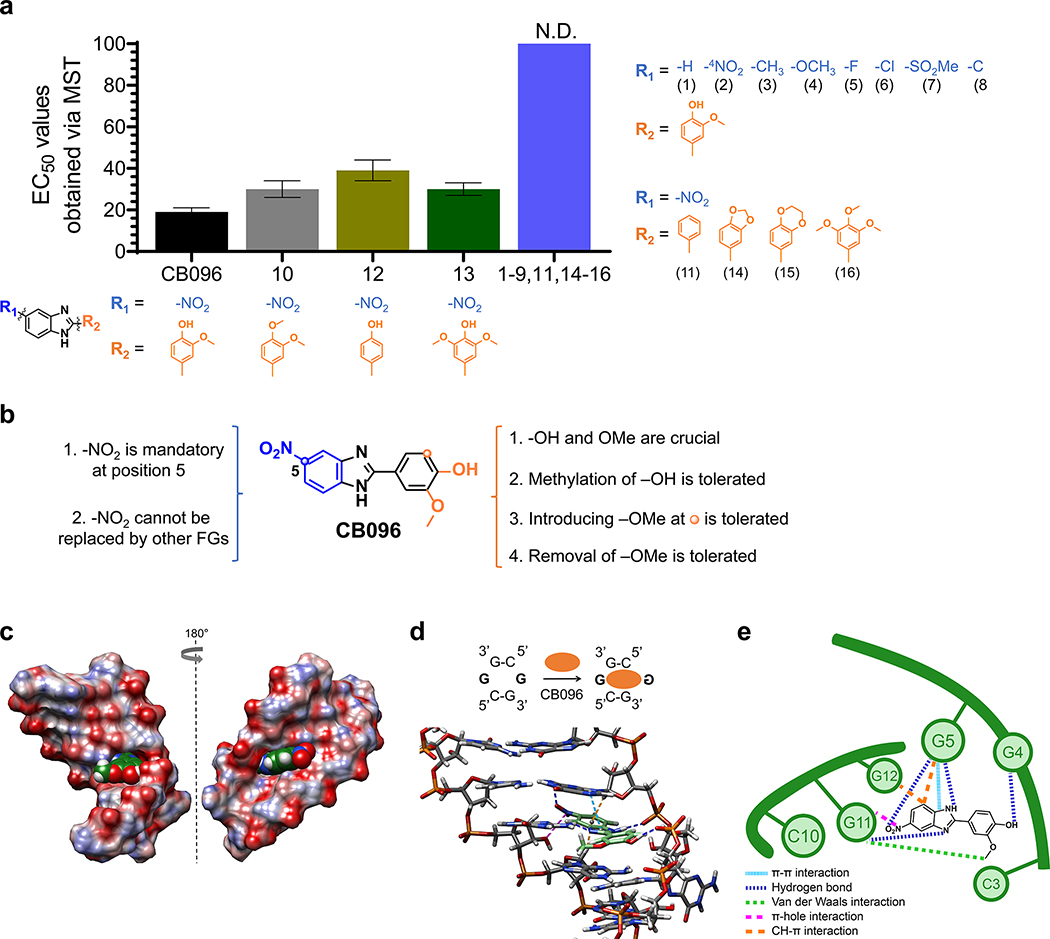
A SAR study was performed to identify the structural features that are responsible for CB096 binding to the r(G4C2)8 hairpin (Figure 5A and B). First, the importance of the nitro (−NO2) group at position 5 in CB096 was investigated (Figure 5A). Ten CB096 derivatives were synthesized according to literature procedures, maintaining the 2-methoxyphenyl unit of the parent compound, and binding to Cy5-r(G4C2)8 was assessed via MST (Supplementary Figure 9). The removal of the −NO2 group (derivative 1), or placement of the −NO2 group at position 4 (derivative 2), completely abolished binding to Cy5-r(G4C2)8 hairpin, suggesting this functional group’s placement at position 5, i.e., a directional noncovalent interaction, is key to the small molecule–RNA interaction (Figure 5A). Moreover, replacing the −NO2 group with electron-donating functional groups such as methyl (−CH3) and methoxy (−OCH3; derivatives 3 and 4, respectively) and electron-withdrawing substitutions such as fluoro (−F), chloro (−Cl), methanesulfonyl (−SO2CH3), trifluoromethyl (−CF3), or carboxyl (−COOH; derivatives 5–9, respectively) led to either no change in thermophoresis signal or no saturable binding to Cy5-r(G4C2)8 (Figure 5A and Supplementary Figure 9). Replacement of −NO2 with its isostere −CN (nitrile) as in derivative 10, yielded a ∼2-fold lower binding affinity compared to CB096. These results suggest that neither the electronic properties of the functional group nor the steric size at position 5 dramatically contribute to r(G4C2)8 hairpin binding (further investigated by MD calculations, vide infra). In conclusion, the −NO2 group at position 5 of the benzimidazole core is both unique and essential for CB096’s interaction with r(G4C2)8 (Figure 5B).
Figure 5.
Mode of binding of CB096 to 5′CGG/3′GGC motif within r(G4C2)exp hairpin revealed via SAR and MD simulations. (a) The SAR study included 17 small molecules (1–17; shown below each structure), closely related derivatives of CB096. N.D. indicates not determined. (b) Summary of the SAR study performed with 17 structurally similar derivatives of CB096. (c) Structure of CB096 in complex with the 5′CGG/3′GGC motif within the r(G4C2)-model duplex, where G represents the G residue within the 1 × 1 GG internal loop, as determined by MD simulations. Atoms of O, N, C, and H are represented in red, blue, green, and white, respectively. (d) Structure of CB096 in complex with the 5′CGG/3′GGC motif within the r(G4C2)–model duplex, highlighting the noncovalent interactions that are responsible for molecular recognition. Hydrogen (H) bonds are represented with dark blue dashed lines, π–π stacking interactions with light blue, C–H−π interactions with orange, π-hole interactions with purple, and van der Waals interactions with green dashed lines. (e) Schematic diagram of the CB096–5′CGG/3′GGC complex summarizing the noncovalent interactions that stabilize the small molecule-RNA complex with colors as indicated in panel d.
Next, derivatives that varied the 2-methoxyphenyl moiety (derivatives 11–17, Figure 5A and Supplementary Figure 10) were synthesized, while maintaining the −NO2 functionality at position 5. Binding studies of the seven derivatives showed that (i) methylation of the −OH group led to a ∼1.5-fold lower binding affinity compared to the parent compound (derivative 11); (ii) removal of both −OH and −OCH3 completely ablated binding to r(G4C2)8 (derivative 12); (iii) removal of −OCH3 reduced binding affinity by ∼2-fold (derivative 13); (iv) addition of −OCH3 at position 6 reduced affinity by ∼1.5 fold (derivative 14); (v) methylation of derivative 14 at the phenol moiety ablated binding (derivative 17); and (vi) sterically constraining the 2-methoxyphenol moiety, as in 1,3-dioxolane (derivative 15) and 1,4-dioxane (derivative 16), also ablated binding (Figure 5B).
MD Simulations to Further Interrogate CB096’s Binding Mode
In addition, we investigated the molecular recognition of r(G4C2)exp by CB096 using MD simulations. Given the rapid exchange of tautomeric forms within benzimidazoles,45–46 computational studies considered both tautomers of CB096 (CB096 and CB096T in Supplementary Figure 11 and Supplementary Tables 1 and 2). The tautomers are structurally different because of the −NO2 substitution at position 5. To determine the most preferred bound state of CB096 to 5′CGG/3′GGC, we employed an approach previously utilized to study small molecule-RNA binding.21,47–49 MD simulations combined with Molecular Mechanics Poisson–Boltzmann Surface Area (MM-PBSA)50 calculations revealed that the 5′CGG/3′GGC RNA motif prefers one of the tautomeric forms (CB096) over the other (CB096T) by ∼5 kcal/mol in binding energy (38.31 vs −32.93 kcal/mol; Supplementary Tables 3 and 4).
We therefore focused our attention on CB096’s (not CB096T’s) interaction with the 5′CGG/3′GGC motif (Figure 5C–E). The computational calculations predicted that CB096 directly interacts with one of the loop’s G residues while the other loop nucleotide is flipped out of the helix (Figure 5C and D). Various noncovalent interactions stabilize the complex including: (i) the −OH group within the 2-methoxyphenol moiety forms a hydrogen bond (H-bond) with the phosphate group of the flipped-out G4 residue (highlighted in blue in Figure 5D and E); (ii) the methoxy group forms a van der Waals interaction with G11 base in the major groove; (iii) the imidazole’s NH group in CB096 forms a H bond with the phosphate group of G5 while the imidazole nitrogen acts as a H bond acceptor for G11’s amino (−NH2) group; (iv) CB096’s imidazole and G5 form a π–π stacking interaction; (v) the oxygen atom within the nitro group at position 5 H-bonds with the −NH2 group of the G5. Interestingly, the −NO2 group could also serve as an excellent π-hole acceptor51–53 for the electron rich hydroxyl (−OH) group at the C2′ position of G11, which is favorably located on top of the nitrogen (N) atom within the −NO2 group. Indeed, the N atom of nitro-aromatics has recently been validated as an excellent π-hole acceptor;53,54 and (vi) C–H−π interactions occur between the nitrobenzene moiety and the H 1s of G5 and G12. Collectively, these MD simulations support our SAR studies, both of which highlighted the key importance of the −NO2 group at position 5 and the 2-methoxyphenol moiety.
CB096 Inhibits RAN Translation in r(G4C2)66-Expressing HEK293T Cells, a Model of c9ALS/FTD
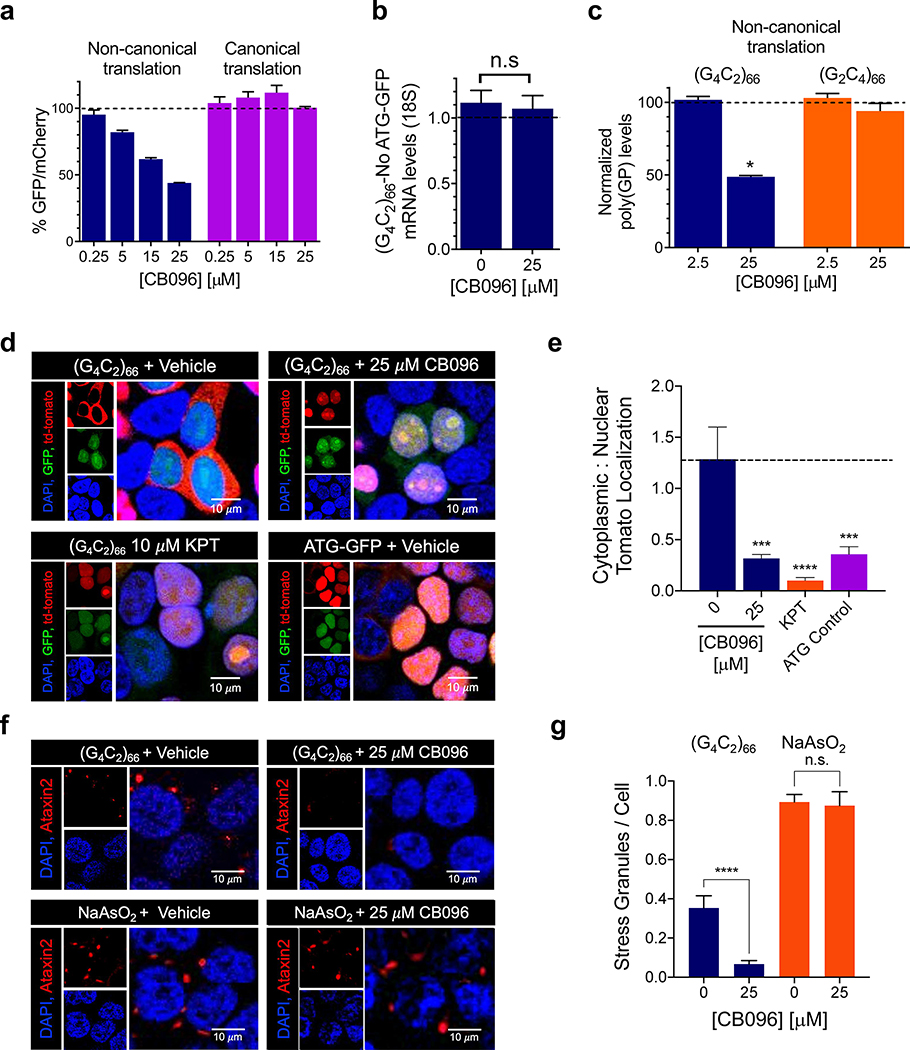
The inhibition of RAN translation by CB096 was assessed in HEK293T cells cotransfected with plasmids that encode (G4C2)66-No ATG-GFP and ATG-mCherry (the latter used for normalization).21 We first defined the therapeutic window for CB096 by using a cell viability assay, which showed that CB096 is not cytotoxic up to 25 μM upon 24 h of treatment (Supplementary Figure 12A). Further, CB096 did not alter the proliferation of r(G4C2)66-transfected HEK293T cells (Supplementary Figure 12B). Treatment with CB096 for 24 h elicited dose-dependent inhibition of RAN translation with an IC50 of ∼20 μM, as assessed by measuring GFP and mCherry fluorescence (Figure 6A). The reduced GFP signal caused by RAN translation of the repeat expansion was not due to transcriptional inhibition as GFP mRNA levels were not altered upon CB096 treatment, as measured by RT-qPCR (Figure 6B). In agreement with these observations, CB096 had no effect on the canonical translation of GFP, assessed by transfecting HEK293T cells with plasmid encoding GFP with an ATG start codon (Figure 6A). These results were further confirmed by measuring levels of a dipeptide repeat protein generated by RAN translation of r(G4C2)exp, poly(GP) (Figure 6C). Notably, CB096 did not inhibit RAN translation of r(G2C4)66, the C9orf72 antisense strand (Figure 6C). As expected, compounds 1, 2, 15, and 16 were unable to inhibit RAN translation of r(G4C2)66 as they did not bind to the Cy5-r(G4C2)8 (Supplementary Figure 13). Collectively, these in cellulis results indicate that CB096 binds r(G4C2)exp and thereby inhibited RAN translation.
Figure 6.
Biological activity of CB096 in a HEK293T cellular model of c9ALS/FTD. (a) CB096 selectively inhibits repeat-associated non-ATG (RAN) translation of r(G4C2)exp. Data indicated with blue bars were collected from HEK293T cells cotransfected with plasmids encoding r(G4C2)66-No ATG-GFP and ATG-mCherry, while data indicated with purple bars were collected from HEK293T cells cotransfected with plasmids encoding ATG-GFP and ATG-mCherry. Data are reported as the mean ± SD from three independent measurements each with three technical replicates. (b) Effect of CB096 on r(G4C2)66 mRNA levels, as measured by RT-qPCR. Data are reported as the mean ± SD from three independent measurements with three technical replicates each. “n.s.” indicates not significant. (c) Effect of CB096 on the levels of toxic dipeptide repeats of poly(GP) upon 24 h of treatment. Poly(GP) levels were measured by an electroluminescent sandwich immunoassay. Data are reported as the mean ± SD and are representative of two independent measurements with two technical replicates each. (d) Rescue of nucleocytoplasmic transport dysfunction by CB096. Representative images of HEK293T cells stably expressing Lentiviral-S-tdTomato cotransfected with plasmid encoding a (G4C2)66-No ATG-firefly luciferase (RAN translation), SV40-Renilla (canonical translation), and ATG-GFP (for distinguishing transfected from non-transfected cells in imaging studies). Cells were treated with vehicle, KPT-350 (KPT at 10 μM; a selective inhibitor of Exportin-1-mediated nuclear export), or CB096 (25 μM). (e) Quantification of the percentage of tdTomato localized to the nucleus or cytoplasm (n = 3 biological samples; 200 cells evaluated per biological sample). (f) Effect of CB096 on r(G4C2)66-induced or chemically induced stress granules. For the former, HEK293T cells stably expressing Lentiviral-S-td-Tomato were transfected with (G4C2)66-No ATG-GFP and treated with CB096 or vehicle (25 μM). For chemically induced stress granules, HEK293T cells stably expressing Lentiviral-S-td-Tomato were transfected with ATG-GFP and treated with 0.5 mM sodium arsenite (NaAsO2). Stress granules were visualized by Ataxin-2 (ATXN2) staining and imaged by confocal microscopy. (g) Quantification of average number of stress granules per nuclei upon treatment with vehicle or CB096 (n = 3 biological samples; 200 cells counted per biological sample). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001, as determined by one-way ANOVA. Data are reported as mean ± SD.
CB096 Improves Nucleocytoplasmic Transport and Reduces Stress Granule Formation in r(G4C2)66-Expressing HEK293T Cells
Accumulation of toxic DPRs can cause impaired nucleocytoplasmic transport.55 To study whether CB096 alleviates nucleocytoplasmic transport dysfunction, we used a cellular model in which HEK293T cells stably express NLS-NES-tagged tdTomato [where NLS denotes nuclear localization signal (N-terminus) and NES denotes a nuclear export sequence (C-terminus)] or Lentiviral-S-tdTomato.56 These cells were cotransfected with plasmids encoding (G4C2)66-No ATG-firefly luciferase, which solely undergoes RAN translation, Renilla firefly, used to measure canonical translation, and a plasmid encoding ATG-GFP (used to distinguish transfected cells for imaging studies).56 As expected, expression of r(G4C2)66 disrupted the nuclear localization of tdTomato (Figure 6D), which was rescued upon treatment with CB096 (Figure 6D,E). In particular, 25 μM of CB096 rescued ∼75% of tdTomato mislocalization. Rescue can be traced to inhibition of RAN translation, as a dose-dependent decrease in firefly luciferase was also observed, with no effect on Renilla luciferase levels (canonical translation; Supplementary Figure 14).
Studies by other groups have shown a link between defects in nucleocytoplasmic transport, formation of nuclear foci, and the presence of stress granules in ALS/FTD,57 toxic depositions of membrane-less condensates containing RNA/protein assemblies caused by cellular stress. Indeed, stress granules are observed in an adeno-associated virus (AAV) mouse of model c9ALS/FTD,58 and they are distinct from those caused by heat or chemical stress.59,60 We thus sought to determine if CB096 could decrease the abundance of r(G4C2)66-induced stress granules using the HEK293T stably expressing Lentiviral-S-tdTomato and forced expression of r(G4C2)66-No ATG-GFP. Upon treatment with 25 μM of CB096, the number of stress granules was reduced by 81 (±11)%, as assessed by staining of the stress granule marker Ataxin-2 (ATXN2; Figure 6F,G). To verify that this effect is specific to r(G4C2)exp, we chemically induced stress granule formation with sodium arsenite (NaAsO2), a commonly used chemical stressor that causes both cytoplasmic mislocalization and stress granule formation.61 Notably, CB096 (25 μM) was unable to reduce the number of stress granules induced by NaAsO2, indicating that the compound’s effect is r(G4C2)exp-dependent (Figure 6F,G).
Summary and Conclusions
Collectively, the ligandability of the r(G4C2)exp hairpin responsible for RAN translation in c9ALS/FTD was assessed by screening an RNA-focused library containing nitrogen-rich heterocycles featuring drug-like molecules. High throughput screening using a TO1 dye displacement assay identified 26 hit compounds that were further validated via MST. This two-step procedure yielded the benzimidazole derivative CB096 as a selective binder to the r(G4C2)8 hairpin over r(G2C4)8 (antisense), r(GGCC)10 (base pair) RNAs, and other 1 × 1 internal loops. NMR structural studies showed that CB096 selectively binds the 5′CGG/3′GGC loops (and not the 5′GGC/3′CGG loops) within r(G4C2)exp hairpin and disrupts its closing base pairs. Computational investigations revealed that 5′CGG/3′GGC has less optimal stacking interactions than 5′GGC/3′CGG and is thus less stable. These data indicate that 5′CGG/3′GGC may be more prone to structural rearrangement required for small molecule binding.
Additionally, SAR studies showed that a rather unique −NO2 group at position 5 and the 2-methoxyphenyl moiety are crucial for binding the r(G4C2)exp hairpin, which were confirmed by MD simulations. Interestingly, the contribution of the −NO2 group does not appear to be electronic in nature, as various electron-donating and -withdrawing functional groups of various sizes did not promote binding to the r(G4C2)8 hairpin. A known isostere of the −NO2 group, −CN, yielded a less avid binder compared to CB096, further highlighting the uniqueness of the −NO2. Rather, the −NO2 group acts as a H-bond acceptor and has the potential to serve as a π-hole acceptor. The 2-methoxyphenyl moiety also plays a key role in molecular recognition, where the −OH group establishes a H bond with the phosphate group of the flipped-out G4 residue and the −OCH3 group forms van der Waals interactions with the nearby G11 residue.
Notably, CB096 selectively inhibited RAN translation and decreased the production of poly(GP) DPRs without affecting r(G4C2)66 mRNA levels in an HEK293T cellular model of c9ALS/FTD, alleviated dysfunctional nucleocytoplasmic transport, and selectively reduced the abundance of r(G4C2)66-induced stress granules. In conclusion, the r(G4C2)exp hairpin adopts multiple structural features that can accommodate or can be stabilized by small molecules, further substantiating the potential of r(G4C2)exp ligandability for therapeutic benefit.
Supplementary Material
Figure S1. Hit validation of from r(G4C2)8 binders via TO1 displacement in vitro. EC50 values were determined by plotting the change in normalized fluorescence as a function of compound concentration. Data points are the mean ± SD of two independent experiments in which each concentration was measured in triplicate. The binding curve was fitted using a four-parameter nonlinear regression equation of the GraphPad Prism 8.1.2 suite.
Figure S2. Representative thermophoresis curves of CB096 binding to Cy5-labeled RNAs obtained. Dose dependent addition of CB096: i) increases the thermophoresis of the hairpin form of Cy5-r(G4C2)8 (a); ii) leads to minor change of the Cy5-r(G2C4)8 (antisense) thermophoresis (b); iii) triggers no saturable change of the Cy5-r(GGCC)10 (base pair control) thermophoresis (c). The normalized fluorescence upon dose-dependent addition of CB096 was recorded in the Thermophoresis and T-jump channels. Data points are representative of two independent experiments each performed in duplicate.
Figure S3. CB096 specifically interacts with the 1×1 GG internal loops within the r(G4C2)exp hairpin RNA. a) No saturable binding was observed in thermophoresis measurement for CB096 and RNAs featuring 1×1 AA and 1×1 UU internal loops within Cy5-r(CAG)12 and Cy5-r(CUG)12 RNAs, respectively, as assessed via MST. b-c) Representative thermophoresis curves of CB096 interacting with Cy5-r(CAG)12 (b) and Cy5-r(CUG)12 (c) RNAs. Dose dependent addition of CB096: i) triggers no saturable change of the Cy5-r(CAG)12 thermophoresis (b); and ii) leads to minor change of the Cy5-r(CUG)12 thermophoresis (c). The normalized fluorescence at each CB096 concentration was recorded in the Thermophoresis and T-jump channels. Data points in panel a were generated from two independent experiments each performed in duplicate.
Figure S4. Representative CD spectrum recorded upon dose dependent addition of CB096 to r(G4C2)8 hairpin. Compound addition triggers an increase of CD signals at 235 and 298 nm (a) that reached saturation at 4 equivalents CB096, i.e. a binding stoichiometry of CB096 to r(G4C2)8 of 4:1 (b). The spectra are representative of two independent experiments.
Figure S5. CB096 mode of binding to the r(G4C2)4 hairpin. CB096 broadens the imino proton peak of GC/CG base pairs at 12.5 ppm. Blue labels indicate 5’GGC/3’CGG (G represents the G residues within the 1×1 GG internal loops) internal loops, whereas the orange labels indicate 5’CGG/3’GGC loops. Further addition of CB096 triggered precipitation. The 1D NMR spectra are representative of two independent experiments.
Figure S6. Dose dependent addition of CB096 does not change the imino proton signals of the 5’GG/3’CC (blue squares) and 5’CC/3’GG (orange squares) base pairs at 13.1 ppm and 12.8 ppm within r(G2C4)8 (antisense). The 1D NMR spectra are representative of two independent experiments.
Figure S7. CB096 does not interact with a 2×2 GG internal loop-containing duplex. a) Free energy minimization (RNAstructure software) of the r(G4C2)2 duplex predicts secondary structures with 1×1 GG and 2×2 GG loops have the same free energy of folding at 37 oC (ΔGo37). b) Dose dependent addition of CB096 does not affect the imino proton signal of GC/CG base pairs at 12.5 ppm within the 2×2 GG internal loop containing model construct. The 1D NMR spectra are representative of two independent experiments.
Figure S8. Binding of CB096 to 5’GGC/3’CGG sites (highlighted in blue) within Cy5-r(GGC)16 via MST. No change in thermophoresis, i.e. no binding, was observed upon dose dependent addition of CB096 to Cy5-r(GGC)16. The secondary structure was predicted by RNAstructure software. The normalized fluorescence was recorded in the Thermophoresis and T jump channels. Data points indicate mean values ± SD from two independent experiments each performed in duplicate.
Figure S9. In vitro binding of compounds 1–10 to Cy5-r(G4C2)8 hairpin, as measured by MST. No change in thermophoresis or no saturation, i.e. no binding, was observed upon dose dependent addition of derivatives 1–10 to Cy5-r(G4C2)8 as shown by the normalized fluorescence signal recorded in the Thermophoresis and T jump channels. Saturable binding was observed for derivative 10 only, with EC50,total value of 32(±6) μM, calculated by plotting the changes in normalized fluorescence as function of compound concentration. EC50,total considered the EC50 values for the two observed binding modes and was generated by calculating the square root of the first binding mode (EC50,1) multiplied by the value of the second binding mode (EC50,2). Data points are reported as the mean ± SD from two independent experiments each performed in duplicate.
Figure S10. In vitro binding of compounds 11–17 to Cy5-r(G4C2)8 hairpin, as measured by MST. Saturable binding was observed for derivatives 11, 13, and 14 only, with EC50 values of 30(±4) μM, 39(±5) μM, and 30(±3) μM, respectively. EC50 values were determined by plotting the changes in normalized fluorescence as function of compound concentration. No change in thermophoresis or no saturation, i.e. no binding, was observed addition of derivatives 12,15–17 to Cy5-r(G4C2)8. Data points are reported as mean ± SD from two independent experiments each performed in duplicate. Binding curves were fitted using a four-parameter nonlinear regression equation of the GraphPad Prism 8.1.2 suite.
Figure S11. Atom numbering of CB096 (a) and its tautomer CB096T (b).
Figure S12. CB096 is not cytotoxic in r(G4C2)66-transfected HEK293T cells nor affects cell proliferation. a) Cellular viability of r(G4C2)66-transfected HEK293T cells upon incubation with CB096 for 24 h assessed via CellTiter-Glo. Data points are reported as the mean ± SD from two independent experiments each performed in triplicate. b) CB096 (25 μM) does not alter the cell proliferation of r(G4C2)66-transfected HEK293T cells. Data points are reported as the mean ± SD relative to vehicle (0.5% DMSO) from two independent experiments each performed in triplicate. n.s. = not significant, as determined by a two-tailed Student t test.
Figure S13. Inhibition of RAN translation by CB096 and compounds 1, 2, 15 and 16. CB096 binds r(G4C2)8 hairpin in vitro and inhibits RAN translation in cells in a dose dependent manner. Conversely, derivatives 1, 2, 15 and 16 do not bind r(G4C2)8 and do not block RAN translation. Data are reported as the mean ± SD from three independent experiments each performed in duplicate.
Figure S14. CB096 selectively inhibits RAN translation of r(G4C2)exp in HEK293T cells that stably express Lentiviral-S-tdTomato co-transfected with plasmind encoding a (G4C2)66-No ATG-firefly luciferase (RAN translation), Renilla luciferase (canonical translation), and ATG-GFP. Relative translation was measured by normalizing the signal from firefly luciferase to Renilla luciferase (non-canonical translation) or the signal from Renilla luciferase to GFP (canonical translation). Data are reported as the mean ± SD (n = 4).
Acknowledgments
This work was funded by the NIH (DP1 NS096898 and R35 NS116846 to M.D.D.; P01 NS099114 to M.D.D. and L.P.; and R35 NS097273 to L.P.), Target ALS (to M.D.D.), and the Nelson Family Fund (to M.D.D). A.U. acknowledges the Deutsche Forschungsgemeinschaft (DFG) for the DFG Postdoctoral Fellowship and the ALS Association (ALSA) for the Milton Safenowitz Postdoctoral Fellowship. We thank T. Bailey for the preliminary TO1 high throughput screening efforts. We acknowledge C. Williams, S. Meyer, H. Haniff, Y. Li, H. Aikawa, and the members of the Disney lab for the helpful discussions and valuable input throughout the study. We thank J. Childs-Disney for assistance with writing and editing the manuscript and X. Kong for his assistance in acquiring NMR spectra. Purchase of the 600 MHz NMR instrument was supported in part by the NIH (S10OD021550).
Footnotes
The authors declare the following competing financial interest(s): M.D.D. is a founder of Expansion Therapeutics.
References
- 1.Balendra R and Isaacs AM (2018) C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol. 14, 544–558, DOI: 10.1038/s41582-018-0047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrov D, Mansfield C, Moussy A, and Hermine O (2017) ALS clinical trials review: 20 years of failure. Are we any closer to registering a new treatment?. Front. Aging Neurosci. 9, 68, DOI: 10.3389/fnagi.2017.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardiman O, Al-Chalabi A, Chio A, Corr EM, Logroscino G, Robberecht W, Shaw PJ, Simmons Z, and van den Berg LH (2017) Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 3, 17071, DOI: 10.1038/nrdp.2017.71 [DOI] [PubMed] [Google Scholar]
- 4.Swinnen B and Robberecht W (2014) The phenotypic variability of amyotrophic lateral sclerosis. Nat. Rev. Neurol. 10, 661–670, DOI: 10.1038/nrneurol.2014.184 [DOI] [PubMed] [Google Scholar]
- 5.Taylor JP, Brown RH Jr., and Cleveland DW (2016) Decoding ALS: from genes to mechanism. Nature 539, 197–206, DOI: 10.1038/nature20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung G-YR, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, and Rademakers R (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256, DOI: 10.1016/j.neuron.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vatsavayai SC, Nana AL, Yokoyama JS, and Seeley WW (2019) C9orf72-FTD/ALS pathogenesis: evidence from human neuropathological studies. Acta Neuropathol. 137, 1–26, DOI: 10.1007/s00401-018-1921-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizielinska S and Isaacs AM (2014) C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia: gain or loss of function?. Curr. Opin. Neurol. 27, 515–523, DOI: 10.1097/WCO.0000000000000130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen X, Westergard T, Pasinelli P, and Trotti D (2017) Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene. Neurosci. Lett. 636, 16–26, DOI: 10.1016/j.neulet.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conlon EG, Lu L, Sharma A, Yamazaki T, Tang T, Shneider NA, and Manley JL (2016) The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. eLife 5, e17820 DOI: 10.7554/eLife.17820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barker HV, Niblock M, Lee Y-B, Shaw CE, and Gallo J-M (2017) RNA misprocessing in C9orf72-linked neurodegeneration. Front. Cell. Neurosci 11, 195–195, DOI: 10.3389/fncel.2017.00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green KM, Linsalata AE, and Todd PK (2016) RAN translation—What makes it run?. Brain Res. 1647, 30–42, DOI: 10.1016/j.brainres.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freibaum BD and Taylor JP (2017) The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front. Mol. Neurosci. 10, 35, DOI: 10.3389/fnmol.2017.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Hu C, Moufawad El Achkar C, Black LE, Douville J, Larson A, Pendergast MK, Goldkind SF, Lee EA, Kuniholm A, Soucy A, Vaze J, Belur NR, Fredriksen K, Stojkovska I, Tsytsykova A, Armant M, DiDonato RL, Choi J, Cornelissen L, Pereira LM, Augustine EF, Genetti CA, Dies K, Barton B, Williams L, Goodlett BD, Riley BL, Pasternak A, Berry ER, Pflock KA, Chu S, Reed C, Tyndall K, Agrawal PB, Beggs AH, Grant PE, Urion DK, Snyder RO, Waisbren SE, Poduri A, Park PJ, Patterson A, Biffi A, Mazzulli JR, Bodamer O, Berde CB, and Yu TW (2019) Patient-customized oligonucleotide therapy for a rare genetic disease. N. Engl. J. Med. 381, 1644–1652, DOI: 10.1056/NEJMoa1813279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mis MSC, Brajkovic S, Tafuri F, Bresolin N, Comi GP, and Corti S (2017) Development of therapeutics for C9ORF72 ALS/FTD-related disorders. Mol. Neurobiol. 54, 4466–4476, DOI: 10.1007/s12035-016-9993-0 [DOI] [PubMed] [Google Scholar]
- 16.Yin W and Rogge M (2019) Targeting RNA: a transformative therapeutic strategy. Clin. Transl. Sci. 12, 98–112, DOI: 10.1111/cts.12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khorkova O and Wahlestedt C (2017) Oligonucleotide therapies for disorders of the nervous system. Nat. Biotechnol. 35, 249–263, DOI: 10.1038/nbt.3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connor CJ, Laraia L, and Spring DR (2011) Chemical genetics. Chem. Soc. Rev. 40, 4332–4345, DOI: 10.1039/c1cs15053g [DOI] [PubMed] [Google Scholar]
- 19.Eggert US, Field CM, and Mitchison TJ (2006) Small molecules in an RNAi world. Mol. BioSyst. 2, 93–96, DOI: 10.1039/B515335B [DOI] [PubMed] [Google Scholar]
- 20.Zamiri B, Reddy K, Macgregor RB, and Pearson CE (2014) TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J. Biol. Chem. 289, 4653–4659, DOI: 10.1074/jbc.C113.502336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z-F, Ursu A, Childs-Disney JL, Guertler R, Yang W-Y, Bernat V, Rzuczek SG, Fuerst R, Zhang Y-J, Gendron TF, Yildirim I, Dwyer BG, Rice JE, Petrucelli L, and Disney MD (2019) The hairpin form of r(G4C2)exp in c9ALS/FTD is repeat-associated non-ATG translated and a target for bioactive small molecules. Cell Chem. Biol. 26, 179–190, DOI: 10.1016/j.chembiol.2018.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang W-Y, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, Johnston A, Overstreet K, Oh S-Y, Todd PK, Berry JD, Cudkowicz ME, Boeve BF, Dickson D, Floeter MK, Traynor BJ, Morelli C, Ratti A, Silani V, Rademakers R, Brown RH, Rothstein JD, Boylan KB, Petrucelli L, and Disney MD (2014) Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 83, 1043–1050, DOI: 10.1016/j.neuron.2014.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simone R, Balendra R, Moens TG, Preza E, Wilson KM, Heslegrave A, Woodling NS, Niccoli T, Gilbert-Jaramillo J, Abdelkarim S, Clayton EL, Clarke M, Konrad MT, Nicoll AJ, Mitchell JS, Calvo A, Chio A, Houlden H, Polke JM, Ismail MA, Stephens CE, Vo T, Farahat AA, Wilson WD, Boykin DW, Zetterberg H, Partridge L, Wray S, Parkinson G, Neidle S, Patani R, Fratta P, and Isaacs AM (2018) G-quadruplex-binding small molecules ameliorate c9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol. Med. 10, 22–31, DOI: 10.15252/emmm.201707850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Goodrich KJ, Conlon EG, Gao J, Erbse AH, Manley JL, and Cech TR (2019) C9orf72 and triplet repeat disorder RNAs: G-quadruplex formation, binding to PRC2 and implications for disease mechanisms. RNA 25, 935–947, DOI: 10.1261/rna.071191.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asare-Okai PN and Chow CS (2011) A modified fluorescent intercalator displacement assay for RNA ligand discovery. Anal. Biochem. 408, 269–276, DOI: 10.1016/j.ab.2010.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran T and Disney MD (2012) Identifying the preferred RNA motifs and chemotypes that interact by probing millions of combinations. Nat. Commun. 3, 1125, DOI: 10.1038/ncomms2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haniff HS, Graves A, and Disney MD (2018) Selective small molecule recognition of RNA base pairs. ACS Comb. Sci. 20, 482–491, DOI: 10.1021/acscombsci.8b00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J-H, Chung TDY, and Oldenburg KR (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screening 4, 67–73, DOI: 10.1177/108705719900400206 [DOI] [PubMed] [Google Scholar]
- 29.Jerabek-Willemsen M, André T, Wanner R, Roth HM, Duhr S, Baaske P, and Breitsprecher D (2014) Microscale thermophoresis: interaction analysis and beyond. J. Mol. Struct. 1077, 101–113, DOI: 10.1016/j.molstruc.2014.03.009 [DOI] [Google Scholar]
- 30.Scheuermann TH, Padrick SB, Gardner KH, and Brautigam CA (2016) On the acquisition and analysis of microscale thermophoresis data. Anal. Biochem. 496, 79–93, DOI: 10.1016/j.ab.2015.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon MH, Hilimire TA, Sanders AM, and Schneekloth JS (2018) Measuring RNA-ligand interactions with microscale thermophoresis. Biochemistry 57, 4638–4643, DOI: 10.1021/acs.biochem.7b01141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Velagapudi SP, Seedhouse SJ, French J, and Disney MD (2011) Defining the RNA internal loops preferred by benzimidazole derivatives via 2D combinatorial screening and computational analysis. J. Am. Chem. Soc. 133, 10111–10118, DOI: 10.1021/ja200212b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, and Disney MD (2009) Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J. Am. Chem. Soc. 131, 9767–9779, DOI: 10.1021/ja9020149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.SantaLucia J Jr. and Turner DH (1993) Structure of (rGGCGAGCC)2 in solution from NMR and restrained molecular dynamics. Biochemistry 32, 12612–12623, DOI: 10.1021/bi00210a009 [DOI] [PubMed] [Google Scholar]
- 35.Wu M and Turner DH (1996) Solution structure of (rGCGGACGC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry 35, 9677–9689, DOI: 10.1021/bi960133q [DOI] [PubMed] [Google Scholar]
- 36.Mathews DH (2014) RNA secondary structure analysis using RNAstructure. Curr. Protoc. Bioinformatics 46, 1–25, DOI: 10.1002/0471250953.bi1206s46 [DOI] [PubMed] [Google Scholar]
- 37.Hennig M, Scott LG, Sperling E, Bermel W, and Williamson JR (2007) Synthesis of 5-fluoropyrimidine nucleotides as sensitive NMR probes of RNA structure. J. Am. Chem. Soc. 129, 14911–14921, DOI: 10.1021/ja073825i [DOI] [PubMed] [Google Scholar]
- 38.Puffer B, Kreutz C, Rieder U, Ebert M-O, Konrat R, and Micura R (2009) 5-Fluoro pyrimidines: labels to probe DNA and RNA secondary structures by 1D 19F NMR spectroscopy. Nucleic Acids Res. 37, 7728–7740, DOI: 10.1093/nar/gkp862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graber D, Moroder H, and Micura R (2008) 19F NMR spectroscopy for the analysis of RNA secondary structure populations. J. Am. Chem. Soc. 130, 17230–17231, DOI: 10.1021/ja806716s [DOI] [PubMed] [Google Scholar]
- 40.Scott LG, and Hennig M (2016) 19F-site-specific-labeled nucleotides for nucleic acid structural analysis by NMR, Methods Enzymol. (Kelman Z, Ed.), pp 59–87, Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- 41.Zhao C, Anklin C, and Greenbaum NL (2014) Use of 19F NMR methods to pobe conformational heterogeneity and dynamics of exchange in functional RNA molecules, Methods Enzymol. (Burke-Aguero DH, Ed.), pp 267–285, Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- 42.Kierzek R, Burkard ME, and Turner DH (1999) Thermodynamics of single mismatches in RNA duplexes. Biochemistry 38, 14214–14223, DOI: 10.1021/bi991186l [DOI] [PubMed] [Google Scholar]
- 43.Burkard ME and Turner DH (2000) NMR structures of r(GCAGGCGUGC)2 and determinants of stability for single guanosine-guanosine base pairs. Biochemistry 39, 11748–11762, DOI: 10.1021/bi000720i [DOI] [PubMed] [Google Scholar]
- 44.Šponer J, Bussi G, Krepl M, Banáš P, Bottaro S, Cunha RA, Gil-Ley A, Pinamonti G, Poblete S, Jurečka P, Walter NG, and Otyepka M (2018) RNA structural dynamics as captured by molecular simulations: a comprehensive overview. Chem. Rev. 118, 4177–4338, DOI: 10.1021/acs.chemrev.7b00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dall’Oglio E, Caro MB, Gesser JC, Zucco C, and Rezende MC (2002) The influence of substituents on the tautomerism of symmetrically substituted 2,2′-bis-benzimidazoles. J. Braz. Chem. Soc. 13, 251–259, DOI: 10.1590/S0103-50532002000200018 [DOI] [Google Scholar]
- 46.Nieto CI, Cabildo P, Garcia MA, Claramunt RM, Alkorta I, and Elguero J (2014) An experimental and theoretical NMR study of NH-benzimidazoles in solution and in the solid state: proton transfer and tautomerism. Beilstein J. Org. Chem. 10, 1620–1629, DOI: 10.3762/bjoc.10.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costales MG, Aikawa H, Li Y, Childs-Disney JL, Abegg D, Hoch DG, Pradeep Velagapudi S, Nakai Y, Khan T, Wang KW, Yildirim I, Adibekian A, Wang ET, and Disney MD (2020) Small molecule targeted recruitment of a nucleaase to cleave an oncogenic RNA in a mouse model of metastatic cancer. Proc. Natl. Acad. Sci. U. S. A. 117, 2406–2411, DOI: 10.1073/pnas.1914286117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Childs-Disney JL, Yildirim I, Park H, Lohman JR, Guan L, Tran T, Sarkar P, Schatz GC, and Disney MD (2014) Structure of the myotonic dystrophy type 2 RNA and designed small molecules that reduce toxicity. ACS Chem. Biol. 9, 538–550, DOI: 10.1021/cb4007387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Childs-Disney JL, Stepniak-Konieczna E, Tran T, Yildirim I, Park H, Chen CZ, Hoskins J, Southall N, Marugan JJ, Patnaik S, Zheng W, Austin CP, Schatz GC, Sobczak K, Thornton CA, and Disney MD (2013) Induction and reversal of myotonic dystrophy type 1 pre-mRNA splicing defects by small molecules. Nat. Commun. 4, 2044, DOI: 10.1038/ncomms3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller BR, McGee TD, Swails JM, Homeyer N, Gohlke H, and Roitberg AE (2012) MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 8, 3314–3321, DOI: 10.1021/ct300418h [DOI] [PubMed] [Google Scholar]
- 51.Bauzá A, Frontera A, and Mooibroek TJ (2019) π-hole interactions involving nitro aromatic ligands in protein structures. Chem. - Eur. J 25, 13436–13443, DOI: 10.1002/chem.201903404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franconetti A, Frontera A, and Mooibroek TJ (2019) Intramolecular π-hole interactions with nitro aromatics. CrystEngComm 21, 5410–5417, DOI: 10.1039/C9CE01015G [DOI] [Google Scholar]
- 53.Bauzá A, Mooibroek TJ, and Frontera A (2015) Directionality of π-holes in nitro compounds. Chem. Commun. 51, 1491–1493, DOI: 10.1039/C4CC09132A [DOI] [PubMed] [Google Scholar]
- 54.Báuza A, Frontera A, and Mooibroek TJ (2016) π-hole interactions involving nitro compounds: directionality of nitrate esters. Cryst. Growth Des. 16, 5520–5524, DOI: 10.1021/acs.cgd.6b00989 [DOI] [Google Scholar]
- 55.Zhang Y-J, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu Y-F, Katzman RB, Gass J, Murray ME, Shinohara M, Lin W-L, Garrett A, Stankowski JN, Daughrity L, Tong J, Perkerson EA, Yue M, Chew J, Castanedes-Casey M, Kurti A, Wang ZS, Liesinger AM, Baker JD, Jiang J, Lagier-Tourenne C, Edbauer D, Cleveland DW, Rademakers R, Boylan KB, Bu G, Link CD, Dickey CA, Rothstein JD, Dickson DW, Fryer JD, and Petrucelli L (2016) C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat. Neurosci. 19, 668–677, DOI: 10.1038/nn.4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, Gupta S, Thomas MA, Hong I, Chiu SL, Huganir RL, Ostrow LW, Matunis MJ, Wang J, Sattler R, Lloyd TE, and Rothstein JD (2015) The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61, DOI: 10.1038/nature14973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, Bowen KE, Wadhwa H, Yang P, Rigo F, Taylor JP, Gitler AD, Rothstein JD, and Lloyd TE (2018) Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971, e917 DOI: 10.1016/j.cell.2018.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chew J, Cook C, Gendron TF, Jansen-West K, Del Rosso G, Daughrity LM, Castanedes-Casey M, Kurti A, Stankowski JN, Disney MD, Rothstein JD, Dickson DW, Fryer JD, Zhang YJ, and Petrucelli L (2019) Aberrant deposition of stress granule-resident proteins linked to C9orf72-associated TDP-43 proteinopathy. Mol. Neurodegener. 14, 9, DOI: 10.1186/s13024-019-0310-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee K-H, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, Maxwell BA, Kim NC, Temirov J, Moore J, Kolaitis R-M, Shaw TI, Bai B, Peng J, Kriwacki RW, and Taylor JP (2016) C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788, e717 DOI: 10.1016/j.cell.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, and Kaufman RJ (2005) Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280, 16925–16933, DOI: 10.1074/jbc.M412882200 [DOI] [PubMed] [Google Scholar]
- 61.Bernstam L and Nriagu J (2000) Molecular aspects of arsenic stress. J. Toxicol. Environ. Health, Part B 3, 293–322, DOI: 10.1080/109374000436355 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Hit validation of from r(G4C2)8 binders via TO1 displacement in vitro. EC50 values were determined by plotting the change in normalized fluorescence as a function of compound concentration. Data points are the mean ± SD of two independent experiments in which each concentration was measured in triplicate. The binding curve was fitted using a four-parameter nonlinear regression equation of the GraphPad Prism 8.1.2 suite.
Figure S2. Representative thermophoresis curves of CB096 binding to Cy5-labeled RNAs obtained. Dose dependent addition of CB096: i) increases the thermophoresis of the hairpin form of Cy5-r(G4C2)8 (a); ii) leads to minor change of the Cy5-r(G2C4)8 (antisense) thermophoresis (b); iii) triggers no saturable change of the Cy5-r(GGCC)10 (base pair control) thermophoresis (c). The normalized fluorescence upon dose-dependent addition of CB096 was recorded in the Thermophoresis and T-jump channels. Data points are representative of two independent experiments each performed in duplicate.
Figure S3. CB096 specifically interacts with the 1×1 GG internal loops within the r(G4C2)exp hairpin RNA. a) No saturable binding was observed in thermophoresis measurement for CB096 and RNAs featuring 1×1 AA and 1×1 UU internal loops within Cy5-r(CAG)12 and Cy5-r(CUG)12 RNAs, respectively, as assessed via MST. b-c) Representative thermophoresis curves of CB096 interacting with Cy5-r(CAG)12 (b) and Cy5-r(CUG)12 (c) RNAs. Dose dependent addition of CB096: i) triggers no saturable change of the Cy5-r(CAG)12 thermophoresis (b); and ii) leads to minor change of the Cy5-r(CUG)12 thermophoresis (c). The normalized fluorescence at each CB096 concentration was recorded in the Thermophoresis and T-jump channels. Data points in panel a were generated from two independent experiments each performed in duplicate.
Figure S4. Representative CD spectrum recorded upon dose dependent addition of CB096 to r(G4C2)8 hairpin. Compound addition triggers an increase of CD signals at 235 and 298 nm (a) that reached saturation at 4 equivalents CB096, i.e. a binding stoichiometry of CB096 to r(G4C2)8 of 4:1 (b). The spectra are representative of two independent experiments.
Figure S5. CB096 mode of binding to the r(G4C2)4 hairpin. CB096 broadens the imino proton peak of GC/CG base pairs at 12.5 ppm. Blue labels indicate 5’GGC/3’CGG (G represents the G residues within the 1×1 GG internal loops) internal loops, whereas the orange labels indicate 5’CGG/3’GGC loops. Further addition of CB096 triggered precipitation. The 1D NMR spectra are representative of two independent experiments.
Figure S6. Dose dependent addition of CB096 does not change the imino proton signals of the 5’GG/3’CC (blue squares) and 5’CC/3’GG (orange squares) base pairs at 13.1 ppm and 12.8 ppm within r(G2C4)8 (antisense). The 1D NMR spectra are representative of two independent experiments.
Figure S7. CB096 does not interact with a 2×2 GG internal loop-containing duplex. a) Free energy minimization (RNAstructure software) of the r(G4C2)2 duplex predicts secondary structures with 1×1 GG and 2×2 GG loops have the same free energy of folding at 37 oC (ΔGo37). b) Dose dependent addition of CB096 does not affect the imino proton signal of GC/CG base pairs at 12.5 ppm within the 2×2 GG internal loop containing model construct. The 1D NMR spectra are representative of two independent experiments.
Figure S8. Binding of CB096 to 5’GGC/3’CGG sites (highlighted in blue) within Cy5-r(GGC)16 via MST. No change in thermophoresis, i.e. no binding, was observed upon dose dependent addition of CB096 to Cy5-r(GGC)16. The secondary structure was predicted by RNAstructure software. The normalized fluorescence was recorded in the Thermophoresis and T jump channels. Data points indicate mean values ± SD from two independent experiments each performed in duplicate.
Figure S9. In vitro binding of compounds 1–10 to Cy5-r(G4C2)8 hairpin, as measured by MST. No change in thermophoresis or no saturation, i.e. no binding, was observed upon dose dependent addition of derivatives 1–10 to Cy5-r(G4C2)8 as shown by the normalized fluorescence signal recorded in the Thermophoresis and T jump channels. Saturable binding was observed for derivative 10 only, with EC50,total value of 32(±6) μM, calculated by plotting the changes in normalized fluorescence as function of compound concentration. EC50,total considered the EC50 values for the two observed binding modes and was generated by calculating the square root of the first binding mode (EC50,1) multiplied by the value of the second binding mode (EC50,2). Data points are reported as the mean ± SD from two independent experiments each performed in duplicate.
Figure S10. In vitro binding of compounds 11–17 to Cy5-r(G4C2)8 hairpin, as measured by MST. Saturable binding was observed for derivatives 11, 13, and 14 only, with EC50 values of 30(±4) μM, 39(±5) μM, and 30(±3) μM, respectively. EC50 values were determined by plotting the changes in normalized fluorescence as function of compound concentration. No change in thermophoresis or no saturation, i.e. no binding, was observed addition of derivatives 12,15–17 to Cy5-r(G4C2)8. Data points are reported as mean ± SD from two independent experiments each performed in duplicate. Binding curves were fitted using a four-parameter nonlinear regression equation of the GraphPad Prism 8.1.2 suite.
Figure S11. Atom numbering of CB096 (a) and its tautomer CB096T (b).
Figure S12. CB096 is not cytotoxic in r(G4C2)66-transfected HEK293T cells nor affects cell proliferation. a) Cellular viability of r(G4C2)66-transfected HEK293T cells upon incubation with CB096 for 24 h assessed via CellTiter-Glo. Data points are reported as the mean ± SD from two independent experiments each performed in triplicate. b) CB096 (25 μM) does not alter the cell proliferation of r(G4C2)66-transfected HEK293T cells. Data points are reported as the mean ± SD relative to vehicle (0.5% DMSO) from two independent experiments each performed in triplicate. n.s. = not significant, as determined by a two-tailed Student t test.
Figure S13. Inhibition of RAN translation by CB096 and compounds 1, 2, 15 and 16. CB096 binds r(G4C2)8 hairpin in vitro and inhibits RAN translation in cells in a dose dependent manner. Conversely, derivatives 1, 2, 15 and 16 do not bind r(G4C2)8 and do not block RAN translation. Data are reported as the mean ± SD from three independent experiments each performed in duplicate.
Figure S14. CB096 selectively inhibits RAN translation of r(G4C2)exp in HEK293T cells that stably express Lentiviral-S-tdTomato co-transfected with plasmind encoding a (G4C2)66-No ATG-firefly luciferase (RAN translation), Renilla luciferase (canonical translation), and ATG-GFP. Relative translation was measured by normalizing the signal from firefly luciferase to Renilla luciferase (non-canonical translation) or the signal from Renilla luciferase to GFP (canonical translation). Data are reported as the mean ± SD (n = 4).