Abstract
Disulfide rebridging methods have recently emerged as a route to hinge region-specific antibody modification, and there exist numerous examples of successful rebridging chemistry applied to clinically relevant human IgG1 antibodies. Here, dibromopyridazinedione disulfide rebridging is adapted to fast trans-cyclooctene/tetrazine (TCO/Tz) bioorthogonal ligations and extended beyond therapeutic human IgG1 antibodies for the first time to include mouse and rat monoclonal antibodies integral to multiplexed analytical diagnostics. In spite of a common architecture, only a subset of antibody host species and IgG isotype subclasses can be rebridged, highlighting the intricate relationship between hinge region sequence, structure, biological activity, and the conjugation chemistry of IgG antibodies.
Keywords: bioorthogonal chemistry, antibodies, site-specific bioconjugation
Graphical Abstract
INTRODUCTION
The therapeutic potential of antibody-drug conjugates has inspired ongoing efforts to develop robust chemical methods to functionalize antibodies, prioritizing reproducible labeling, linkage stability, and minimal perturbation of binding affinity.1 Site-selective modifications that minimize functional interference are particularly desirable for all of these agendas.2–4 Simultaneously, a range of multiplexed single-cell analytics have emerged that require primary antibodies to be conjugated to fluorophores,5 mass tags,6–8 or DNA-barcodes9–11 for detection of protein biomarkers. While antibody re-engineering for site-selective conjugation is occasionally performed for therapeutic clinical applications,12 versatile chemical linkers that can directly modify native commercial antibodies are needed for reliable diagnostics and facile multiplexing techniques.
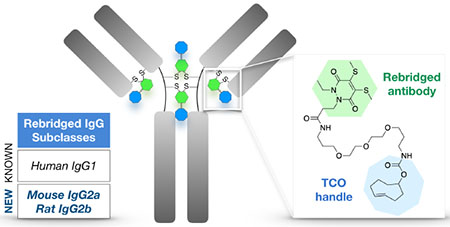
Routine labeling of abundant protein-surface lysine residues (e.g. via NHS ester chemistry) relies on the random distribution of linkers/tags, which creates unpredictable molecular heterogeneity and risks impairing antibody binding affinity and specificity. Moreover, this can alter pharmacokinetics and therapeutic efficacy with antibody-drug conjugates13 and result in errant or diminished signal in antibody-based diagnostic assays. As an alternative to lysine conjugation, cysteine-selective chemistries have been developed to modify comparatively scarce nucleophilic cysteine residues following reduction of an antibody’s inter-chain disulfide bonds.14 However, this too can lead to heterogeneously labeled products (due to incomplete labeling) as well as more deleterious effects such as disulfide scrambling. In this context, reagents that both label and functionally rebridge the reduced disulfides of native antibodies are desirable, conserving the covalent inter-chain linkage and site-specifically attaching tags to the conformationally flexible antibody hinge region, away from the epitope recognition and Fc receptor binding sites (Figure 1a). This permits a degree of labeling (DOL) of up to four tags per antibody, well within the desired stoichiometry for most applications.
Figure 1.
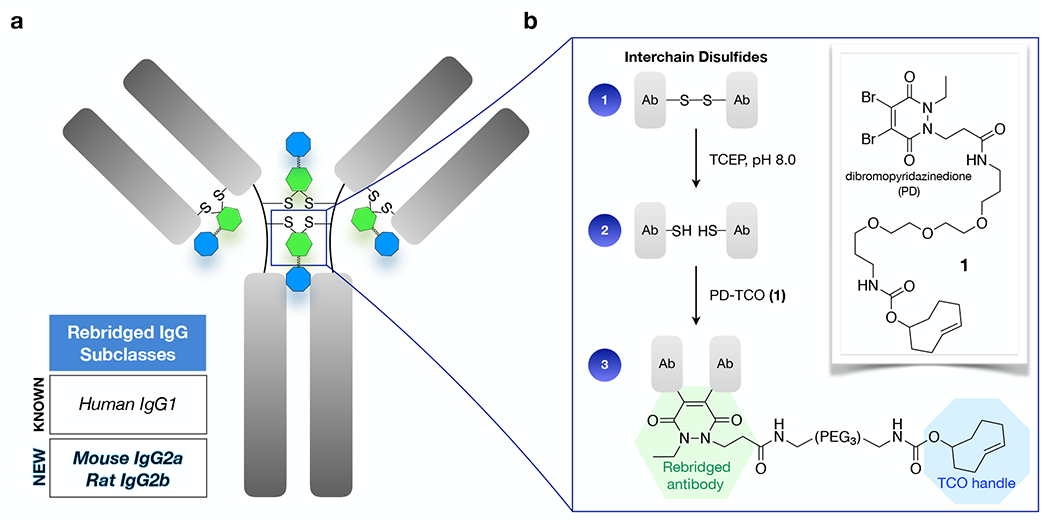
Scheme of rebridging with PD-TCO 1. (a) interchain disulfide bonds (up to four for human IgG1, as depicted) in the antibody hinge region are functionalized with a TCO for ultra-rapid/quantitative Tz ligation. (b) Reduction with TCEP and sequential conjugation with 1 results in a rebridged disulfide bond and embedded bioorthogonal tag.
Recently developed antibody rebridging technologies include bis-sulfone reagents,15,16 divinyl pyrimidines,17 isobutylene,18 and next-generation maleimides.19,20 Chudasama, Caddick, and coworkers have developed dibromopyridazinedione (PD) reagents as a class of disulfide-bridging molecules that have yielded homogenous antibody conjugates with various payloads,21–24 including a general protocol for the chemical functionalization and rebridging of the clinically relevant human IgG1 antibody trastuzumab.25 We chose to investigate the PD rebridging method as a tool for modification of commercial monoclonal antibodies, where the heterogeneity of routine NHS-ester antibody labeling creates barriers to scaling up multiplexed testing: i) a subset of antibodies are prone to aggregation with lysine-modification, causing unacceptably poor recovery; ii) antibody-specific and dye-specific protocols are required to achieve optimal fluorophore brightness; iii) inconsistent labeling yields create normalization issues for quantitative analyses, whether for DNA-barcoded molecular targets in tissue/biopsy samples10,26 or for single molecule PCR/sequencing based readouts.27–30
While techniques for Fc modification of antibodies of different species/isotype origin have recently begun to be explored,31–33 a preponderance of the work in site-selective antibody modification and all of the published work on disulfide rebridging has been done on therapeutic human IgG1 antibodies,18 especially the anti-HER2 antibody trastuzumab.15,17,32,34,35 Here, we expand the functionalization of the PD scaffold to include trans-cyclooctene/tetrazine (TCO/Tz) tags with ultra-fast labeling kinetics, and demonstrate markedly improved labeling/recovery of NHS-intolerant antibodies. We extend that investigation to explore the generalizability of PD-rebridging methods and observe that both the IgG isotype subclass and host species play a critical role in determining the synthetic outcome, even within substantially homologous global architectures. Mouse IgG2a and rat IgG2b antibodies can be rebridged in yields that parallel trastuzumab, but other isotype subclasses cannot, highlighting connections between biological function, chemical reactivity, and sequence-dependent antibody structure.
RESULTS AND DISCUSSION
Rebridging of human IgG1 antibodies with PD-TCO 1.
The combination of site-specificity and rapid TCO/Tz kinetics makes rebridged TCO-antibodies attractive for applications that require very fast reaction kinetics such as live-cell imaging,36,37 fluorogenic imaging,38–41 or in vivo pretargeting.42–44 As we sought to label broad panels of “off-the-shelf” commercial antibodies for highly multiplexed analytical diagnostic assays, we speculated that the structural assets of PD chemistry would minimize perturbations caused by labeling and obviate the need for optimization of each individual antibody. Furthermore, TCO/Tz click kinetics provide a route to prepare DNA barcoded antibodies without onerous purification steps, enabled by quantitative ligation reactions at even submicromolar concentrations. We thus set out to test the compatibility of PD rebridging with TCO ligation by extending a PD-carboxylic acid scaffold25 (Scheme S1) to prepare PD-TCO reagent 1 for disulfide rebridging (Figure 1b). An initial panel of therapeutic antibodies was selected for testing, including the reference standard trastuzumab (anti-HER2) as well as cetuximab (anti-EGFR) and rituximab (anti-CD20), all three of which are human IgG1 isotypes. UV-vis analysis of the antibody labeling efficiency takes advantage of a characteristic 340 nm absorbance for the rebridged pyridazinedione21 and revealed that PD-TCO achieves a near-quantitative DOL for trastuzumab (Table 1), similar to other PD scaffolds previously tested.25 Cetuximab and rituximab treated with the same protocol, although of identical isotype, consistently achieved a lower ~80% labeling efficiency (DOL ~3.2).
Table 1.
Rebridging efficiency of human IgG1 antibodies with PD-TCO 1
| antibody | target | # disulfides | PD DOL | labeling % |
|---|---|---|---|---|
| Trastuzumab | HER2 | 4 | 3.7 ± 0.4 | 93 |
| Cetuximab | EGFR | 4 | 3.2 ± 0.2 | 80 |
| Rituximab | CD20 | 4 | 3.2 ± 0.3 | 80 |
As an initial validation of the rebridged antibodies, we assessed the performance of TCO-cetuximab and TCO-rituximab by immunofluorescence imaging. After reaction with Cy3-methyltetrazine and routine purification by gel-filtration spin column, the resulting Cy3-cetuximab was used to stain fixed A-431 cells, an epidermoid cancer cell line with characteristic high EGFR expression. We observed bright, membrane-specific staining for the rebridged antibody (Figure 2a), matching the morphology and relative signal intensity of unmodified cetuximab detected with an Alexa Fluor 488 anti-human secondary antibody. TCO-rituximab was reacted in parallel with AF488-tetrazine to prepare a fluorescent conjugate; simultaneous staining of DB cells with the rebridged AF488-rituximab and a validated anti-CD20/Alexa Fluor 647 secondary antibody pair displayed bright on-target staining and excellent colocalization (Figure S1). DNA barcoding also proved highly efficient, with complete ligation of the Tz-DNA at substoichiometric concentrations; rebridged TCO-cetuximab was treated with a 65 nt tetrazine-DNA barcode to produce site-specific antibody-barcode conjugates in high yield within 1 hour, completely eliminating the need to remove excess DNA (Figure S2). By contrast, traditional ligation chemistries (e.g. thiol/maleimide) require lengthy incubation times and serial precipitation and/or ultrafiltration steps for conjugate purification, leading to significant sample loss and/or mixtures containing both bound and unbound DNA.10
Figure 2.
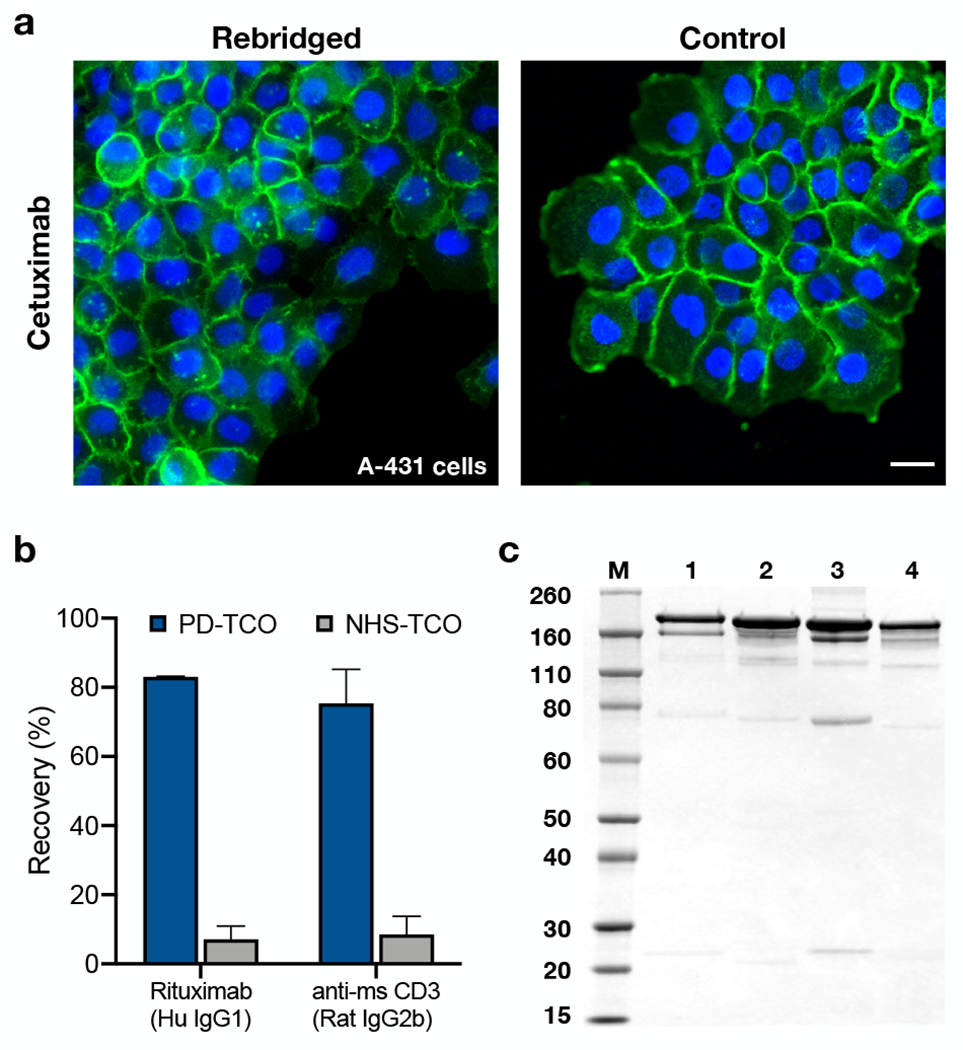
Characterization of rebridged antibodies. (a) Staining comparison of rebridged TCO-cetuximab and unmodified cetuximab (+AlexaFluor488 2° Ab) with A-431 cells. Scale bar = 25 μm. (b) TCO-antibody recovery after labeling and purification via rebridging with PD-TCO 1 (blue) or traditional NHS labeling (gray) for TCO-labeled rituximab and TCO-labeled anti-CD3 (p < 0.001, one-way ANOVA). (c) SDS-PAGE: M: molecular weight marker in kDa; 1: cetuximab rebridged with 1 (DOL = 3.2); 2: native cetuximab; 3: rituximab rebridged with 1 (DOL = 3.1); 4: native rituximab.
Antibody recovery and characterization.
In spite of the subtle differences in DOL, all three of the PD-TCO antibodies were recovered in good yield. We and others have noted in prior work that some monoclonal antibodies are especially sensitive to lysine modification, resulting in poor affinity and specificity relative to the unmodified antibody.45 A subset of these susceptible antibodies, including rituximab, are prone to aggregate in solution upon minimal labeling, i) limiting the DOL that can be achieved, and ii) resulting in marginal protein recovery. Emblematic of these issues, rituximab and a commercial anti-mouse CD3 antibody (Table S1) consistently exhibited >90% protein loss after TCO labeling and purification using traditional NHS-ester chemistry (Figure 2b), requiring much higher input quantities of antibody and generating uncertainty about the utility of recovered conjugates. We were pleased to find that protein recovery was greatly improved to 83% and 75% for rituximab and anti-CD3, respectively, when labeled with PD-TCO 1 (Figure 2b), indicating the potential usefulness of PD rebridging for dealing with compatible aggregation-prone antibodies.
In parallel to the functional imaging experiments, characterization of the rebridged antibodies by SDS-PAGE showed predominantly the intact full antibody (Figure 2c and Figure S3a), confirming that the antibody chains were productively rebridged with 1 after reduction by TCEP. Rebridged rituximab (Lane 3) showed a small band at ~75 kDa not present in native rituximab, consistent with intra-chain rebridging between cysteines on the same heavy chain. This half-bridged antibody species has been observed with other disulfide rebridging methods as well18,19 and highlights the intrinsic challenge of discriminating between the diverse inter-and intramolecular reaction pathways possible for bivalent/multi-valent reactants. This appears particularly relevant with a broader range of IgG molecules, as subtle structural and kinetic effects can dramatically alter outcomes (vide infra).
Rebridging of other species and isotype subclasses.
Having broadly recapitulated the rebridging outcomes for PD-TCO 1 with an initial panel of clinical therapeutics, we proceeded to expand testing to the broader range of monoclonal antibodies in multiplexed assay panels, where we encountered unexpectedly heterogenous results. A subset of labeling reactions worked well, as for the anti-mouse CD3 antibody tested above; in others, however, we observed poor labeling efficiencies (DOL ~1-2). Accumulated experiments suggested that the ability of an antibody to be successfully rebridged was dependent on not only the host species but also the IgG isotype subclass. We thus assembled a more systematic set of commercially-sourced mouse, rat and rabbit antibodies from a variety of vendors, spanning the range of prevalent species/isotypes46 (Table S1). In the therapeutic space, a majority of the antibodies currently in clinical use are human IgG1 isotype,47 as above; more recently, a selection of human IgG4 antibodies (e.g. nivolumab, pembrolizumab) have also become important with the success of immune checkpoint inhibition in oncology. Testing the PD-labeling protocol on this panel of animal/human antibodies allowed us to define an initial map of the scope of PD rebridging (Figure 3 and Figure S4), in which the antibodies tested are aligned by species and arranged in alphanumerical order by subclass.
Figure 3.
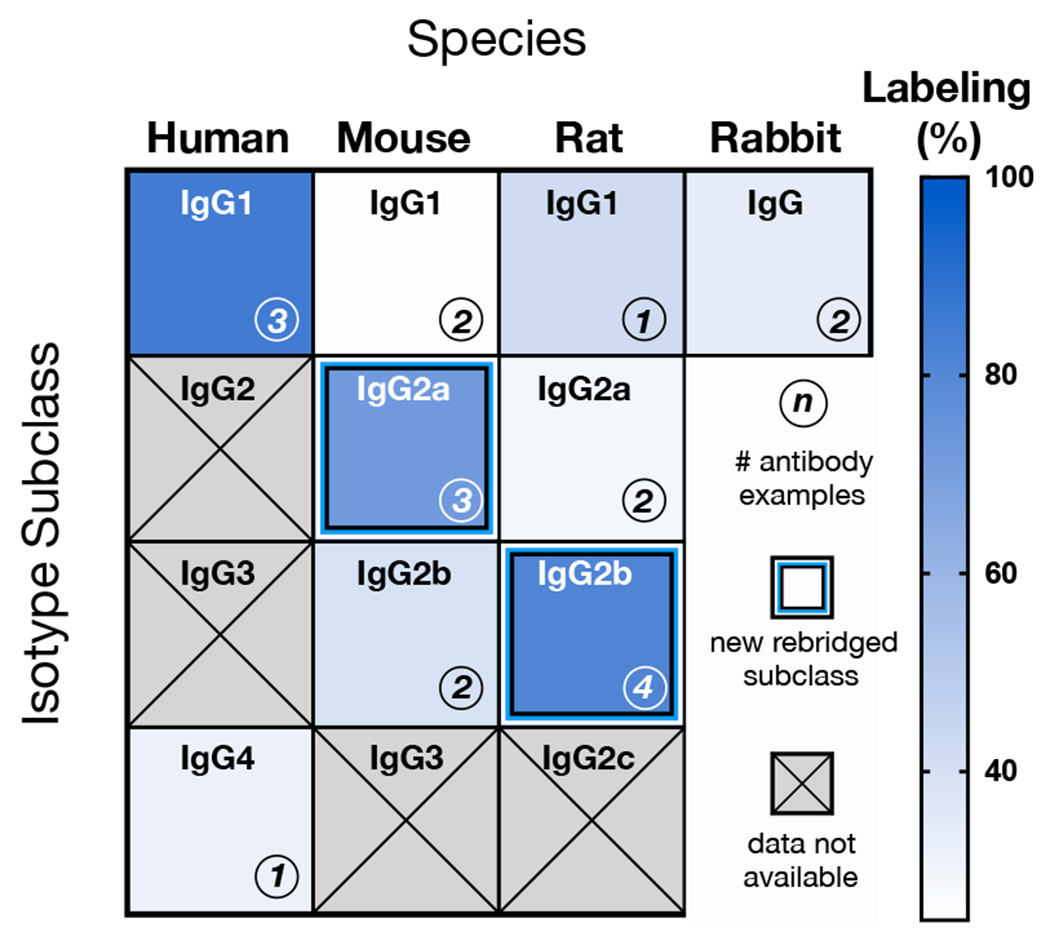
Labeling efficiency by antibody host species and isotype, organized alphanumerically by subclass nomenclature; only mouse IgG2a and rat IgG2b are labeled with yields paralleling human IgG1. Data are not available for human IgG2/3, as they are not represented in the current repertoire of therapeutic antibodies. Likewise, mouse IgG3 and rat IgG2c were not tested, as they make up a minor fraction (<2-3%) of the commercially available monoclonal antibody catalog due to their low physiologic abundance.
In contrast to the IgG1 therapeutics, the human IgG4 pembrolizumab (anti-programmed cell death-protein 1) yielded a PD DOL of just 1.3 ± 0.4 conjugates per antibody. Mouse IgG2a and rat IgG2b monoclonals were consistently rebridged with PD-TCO 1, expanding disulfide rebridging chemistry to mouse and rat antibodies for the first time. Characterization by SDS-PAGE confirmed predominantly intact full antibody, similar to rebridged human IgG1 (Figure S3b). Rebridged mouse IgG2a and rat IgG2b antibodies were also clicked with Tz-fluorophores and tested in cellular immunofluorescence assays, where they exhibited bright and target-specific staining (Figure S5 and Figure S6). Somewhat surprisingly, the remainder of the mouse, rat and rabbit isotypes tested all appear to be incompatible.
To further investigate the isotype subclasses that showed poor labeling with PD-TCO 1 under standard rebridging conditions, we selected a mouse IgG1 antibody and systematically varied the time, temperature, and equivalents of TCEP and PD-TCO 1. Under standard conditions (10 equivalents of TCEP), SDS-PAGE suggested that the low DOL was principally due to a failure to productively reduce the disulfide linkages, as we observed poor PD labeling (as determined by UV-Vis). However, increasing the amount of TCEP to facilitate the reduction step resulted in a combination of mis-rebridged species and complete reduction into heavy and light chains, as evidenced by multiple bands on SDS-PAGE (Figure S7), irrespective of the number of PD-TCO equivalents used (Figure S8); all conditions yielded poor PD labeling and/or mis-rebridged/reduced species. It is possible that an alternative approach to rebridging, either in terms of the reaction conditions or the nature of the rebridging ligand itself, would prove successful; nevertheless, our results suggest that these chemistries may be harder to generalize than expected.
Hinge region sequence comparison.
While the universality of interchain disulfide bonds and the general consistency of the immunoglobulin fold suggested to us that rebridging chemistry should be similar for all antibodies, IgG subtypes are well known to possess important diversity in their biological activities.48 And in spite of high homology of the constant region across different species and isotypes, the hinge region, where PD rebridging chemistry occurs, contains the greatest discrepancies between subclasses (Figure S9). Variation in hinge structural flexibility has been directly implicated in functional differences as well, including complement fixation.48 Comparing the aligned hinge region sequences48–51 of the antibodies tested reveals potentially informative distinctions (Figure 4). The antibody subtypes that can be successfully rebridged (human IgG1, mouse IgG2a, and rat IgG2b, shaded in blue) display the greatest sequence identity, and share functional similarities in their effectiveness in fixing complement.52 In contrast, mouse IgG2b (not rebridged) has two non-conserved sequence differences in the upper hinge and carries a substitution of glutamic acid for leucine at a key residue in the lower hinge. The specific mechanistic impact of these changes on PD rebridging is unknown, but the latter Le→Glu substitution has been shown to be a determinant of FcRI binding,53 supporting a broader functional role for this residue. Rabbit IgG (not rebridged) has just a single interheavy chain disulfide as well as three non-conserved sequence changes in the upper hinge. Finally, relative to the subclasses that can be rebridged, human IgG4, mouse IgG1, rat IgG2a, and rat IgG1 have significant truncations in the upper hinge region (Figure 4). The restricted hinge in these subclasses is known to bring the Fc and Fab segments closer together, restricting the flexibility of the Fab arms, which has been implicated in the ability of an antibody to fix complement.50,51,54 Cumulatively, the relationship between hinge region sequence, structure, biological activity, and PD chemistry is complex, with multiple factors that ultimately dictate the ability of an antibody to be rebridged.
Figure 4.
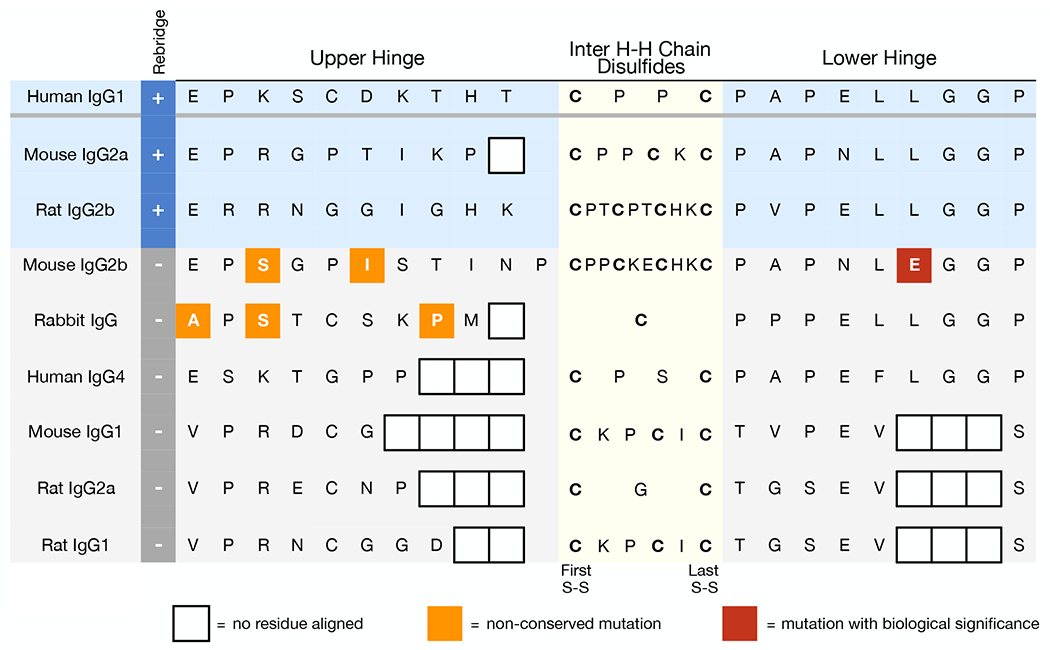
Comparison of IgG hinge region sequences. Non-conserved mutations (highlighted in orange/red) and truncated hinge regions are present in isotype subclasses that cannot be rebridged (shaded in gray) compared to isotype subclasses that were successfully rebridged (blue).
CONCLUSIONS
While disulfide rebridging remains an attractive site-selective conjugation method for human IgG1 therapeutics and may advance applications such as antibody-drug conjugates, this work brings to light a multifactorial and underappreciated link between hinge-region structure and the feasibility of rebridging. Within the subset of monoclonals congruent with pyridazinedione rebridging chemistry, PD-TCO 1 conjugates exhibit excellent staining performance, can improve recovery of sensitive antibodies, and enable efficient DNA barcoding via fast and quantitative ligation reactions. However, because the prevalence of distinct IgG subclasses in mouse and rat monoclonal antibody collections mirrors their natural physiological abundance,55 a significant fraction of monoclonal antibodies available in commercial catalogs are currently incompatible with this chemistry. For mouse, the most common subtypes available are IgG1 (~70%), IgG2a (~20%, rebridging-compatible), and IgG2b (~10%).46,56 Similarly for rat, IgG2a and IgGlare more widely available than the PD-rebridged IgG2b, consistent with the proportion of these antibody subclasses produced by rat hybridomas.57 Further work to develop a potential one-size-fits-all disulfide modification reagent will require careful study of monoclonal antibodies beyond human IgG1. The increasing utility58 and relative simplicity of rabbit antibodies (only one subclass, only three inter-chain disulfides) suggests one option for further stress-testing of disulfide rebridging reagents. Simultaneously, a rebridging reagent optimized for antibodies with restricted hinge regions would encompass commercially abundant subclasses (mouse IgG1, rat IgG2a, and rat IgG1) and facilitate site-specific hinge-region labeling of human IgG4 therapeutics. Pursuit of these goals may require further creative strategies to prepare tractable reagents that synchronize disulfide reduction with rebridging ligation22 or ultimately highlight a need for other chemical/biological approaches to versatile, broadly-compatible antibody linkers.
MATERIALS AND METHODS
General Methods.
All reagents were obtained from commercial sources and used without further purification. TCO-PEG3-Amine, TCO-PEG4-NHS, Methyltetrazine-PEG4-NHS, and tetrazine-fluorophores (AF488, Cy3, AF594) were purchased from Click Chemistry Tools, LLC. Flash column chromatography was performed using Sorbtech purity flash cartridges or Biotage SNAP Bio C18 columns for reversed phase chromatography. NMR spectra were recorded on a Bruker Avance UltraShield 400 MHz spectrometer. Chemical shifts are reported in parts per million (δ) and referenced to the residual solvent. Reactions were monitored via liquid chromatography-mass spectrometry (LC-MS) on a Waters instrument equipped with a Waters 2424 ELS Detector, Waters 2998 UV-Vis Diode array Detector, and a Waters 3100 Mass Detector. UV-Vis analysis of antibodies was performed on a NanoDrop 1000 spectrophotometer.
Rebridging with PD-TCO 1.
Antibodies (Table S1) were modified with 1 following a previously published general protocol for trastuzumab.25 Antibodies were concentrated to 5 mg/mL using Amicon (100 K MWCO) Ultra centrifugal filters. (Antibodies that were supplied at low concentration or in low quantity were concentrated to 1-2 mg/mL.) After centrifugal concentration, antibodies were solvent-exchanged with Zeba spin desalting columns (40 K MWCO) into a borate-buffered saline solution (25 mM sodium borate, 25 mM NaCl, 0.5 mM EDTA, 2% DMSO, pH 8.0). Antibodies were then incubated at 4 °C with 20 equivalents 1 (from a stock prepared at 50 mM in DMSO). After one hour, 10 equivalents TCEP · HCl (10 mM in MQ H2O) were added and the mixture incubated at 4 °C for a further 16 h. Excess labeling reagents were then removed using Zeba spin desalting columns (40 K MWCO) equilibrated with PBS, and TCO-antibody conjugates were analyzed by UV-Vis spectrometry. Degree of TCO-labeling (DOL) for each antibody was calculated according to the previously published formula21 where ε280 = 215,000 M−1 cm−1 for trastuzumab and 210,000 M−1 cm−1 for all other antibodies, ε345 = 9,100 M−1 cm−1 for 1, and CF345 = 0.25 as a correction factor for the absorbance at 280 nm. The nmol of antibody in solution was calculated before and after labeling and purification to determine antibody recovery.
Labeling with NHS-ester chemistry.
Antibodies (5 mg/mL) were solvent-exchanged with Zeba spin desalting columns (40 K MWCO) into a PBS-bicarbonate solution (100 mM sodium bicarbonate in PBS, pH 8.4) and incubated with TCO-PEG4-NHS (20 equivalents, Click Chemistry Tools, #A137) at room temperature for 25 min. After this, excess reagents were removed using Zeba spin desalting columns (40 K MWCO) into PBS. To determine the degree of labeling, TCO-antibodies were treated with Tz-AF488 (10 equivalents, Click Chemistry Tools, #1361) at room temperature for 20 min followed by purification with Zeba spin desalting columns (40 K MWCO) equilibrated with PBS and analyzed by UV-Vis spectrometry. The nmol of antibody in solution was calculated before and after labeling and purification to determine antibody recovery.
Statistical Analysis.
Statistical analyses were performed in GraphPad Prism 8.3. Error bars represent the standard deviation (n ≥ 2). A one-way analysis of variance (Tukey correction for multiple comparisons) was used to determine statistical significance.
SDS-PAGE.
Non-reducing SDS-PAGE was performed following standard lab procedures on NuPAGE 4-12% Bis-Tris Protein Gels (ThermoFisher). Antibody samples (2 ug in 15 uL) were mixed with 5 uL 4x NuPAGE LDS Sample Buffer (ThermoFisher) and heated at 75 °C for 5 min. Samples were then loaded onto the gel and run in 1x NuPAGE MOPS SDS Running Buffer (ThermoFisher) at constant 200 V for 50 min. Novex Sharp Pre-Stained Protein Standard (15-260 kDa, ThermoFisher) was used to estimate protein molecular weights. Gels were stained using SimplyBlue SafeStain (Invitrogen) following the manufacturer’s protocol and imaged on an Azure Biosystems Sapphire Biomolecular Imager. Antibody-DNA samples were run at constant 120 V for 75 min and stained with both SimplyBlue SafeStain and SYBR Safe DNA gel stain (Invitrogen) before imaging.
Cell Culture.
Cell lines (A-431 (epidermoid carcinoma), DB (germinal center B-cell-like DLBCL), Jurkat (T-cell leukemia)) were purchased from the American Tissue Culture Collection (ATCC). Cells were passaged in media prepared to the specifications of each cell line according to ATCC: DB and Jurkat cells were maintained in RPM1-1640 medium and A-431cells were maintained in DMEM medium. All media were supplemented with 10% heat-inactivated fetal bovine serum, 100 IU penicillin, and 100 μg/mL streptomycin. Cells were cultured at 37 °C under a humidified atmosphere of 5% CO2. Cell lines were routinely tested for mycoplasma contamination with MycoAlert Mycoplasma Detection Kit (Lonza).
Fluorescence Imaging.
Fluorescent images were acquired on an Olympus BX-63 upright automated epifluoresccence microscope. Hoechst and DAPI-stained nuclei were visualized using a DAPI filter cube. Alexa Fluor 488, Cy3, Alexa Fluor 594, and Alexa Fluor 647-conjugated markers were excited with traditional FITC, Cy3, TexasRed, and Cy5 filter cubes, respectively. Images were processed and colocalized using ImageJ.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by UH3-CA202637, UO1CA206997 and T32CA079443 (ANM). The authors would like to thank Dr. Katy Yang for assistance with SDS-PAGE, Dr. Juhyun Oh for obtaining mouse splenocytes, Dr. Peter Koch for isolating PBMCs, and Dr. Jina Ko for providing tetrazine-DNA barcodes.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Additional cellular fluorescence images, additional SDS-PAGE gels, list of antibodies tested, cellular staining conditions, cetuximab-barcode preparation, synthesis, and NMR spectra (PDF)
REFERENCES
- (1).Alves NJ (2019) Antibody conjugation and formulation. Antibody Ther. 2, 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Agarwal P and Bertozzi CR (2015) Site-specific antibody–drug conjugates: the nexus of bioorthogonal chemistry, protein engineering, and drug development. Bioconjugate Chem. 26, 176–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Akkapeddi P, Azizi S-A, Freedy AM, Cal PMSD, Gois PMP and Bernardes GJL (2016) Construction of homogeneous antibody–drug conjugates using site-selective protein chemistry. Chem. Sci 7, 2954–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hoyt EA, Cal PMSD, Oliveira BL and Bernardes GJL (2019) Contemporary approaches to site-selective protein modification. Nat. Rev. Chem 3, 147–171. [Google Scholar]
- (5).Lin J-R, Fallahi-Sichani M and Sorger PK (2015) Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun 6, 8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD and Zhao S (2014) Multiplexed ion beam imaging of human breast tumors. Nat. Med 20, 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, Gherardini PF, Prestwood TR, Chabon J and Bendall SC (2017) Systemic immunity is required for effective cancer immunotherapy. Cell 168, 487–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Jackson HW, Fischer JR, Zanotelli VRT, Ali HR, Mechera R, Soysal SD, Moch H, Muenst S, Varga Z, Weber WP and Bodenmiller B (2020) The single-cell pathology landscape of breast cancer. Nature 578, 615–620. [DOI] [PubMed] [Google Scholar]
- (9).Agasti SS, Liong M, Peterson VM, Lee H and Weissleder R (2012) Photocleavable DNA barcode-antibody conjugates allow sensitive and multiplexed protein analysis in single cells. J. Am. Chem. Soc 134, 18499–18502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Giedt RJ, Pathania D, Carlson JCT, McFarland PJ, del Castillo AF, Juric D and Weissleder R (2018) Single-cell barcode analysis provides a rapid readout of cellular signaling pathways in clinical specimens. Nat. Commun 9, 4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, Black S and Nolan GP (2018) Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Beck A, Goetsch L, Dumontet C and Corvaïa N (2017) Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discovery 16, 315–337. [DOI] [PubMed] [Google Scholar]
- (13).Tsuchikama K and An Z (2018) Antibody-drug conjugates: recent advances in conjugation and linker chemistries. Protein Cell 9, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Gunnoo SB and Madder A (2016) Chemical protein modification through cysteine. ChemBioChem 17, 529–553. [DOI] [PubMed] [Google Scholar]
- (15).Badescu G, Bryant P, Bird M, Henseleit K, Swierkosz J, Parekh V, Tommasi R, Pawlisz E, Jurlewicz K and Farys M (2014) Bridging disulfides for stable and defined antibody drug conjugates. Bioconjugate Chem. 25, 1124–1136. [DOI] [PubMed] [Google Scholar]
- (16).Bryant P, Pabst M, Badescu G, Bird M, McDowell W, Jamieson E, Swierkosz J, Jurlewicz K, Tommasi R and Henseleit K (2015) In vitro and in vivo evaluation of cysteine rebridged trastuzumab–MMAE antibody drug conjugates with defined drug-to-antibody ratios. Mol. Pharmaceutics 12, 1872–1879. [DOI] [PubMed] [Google Scholar]
- (17).Walsh SJ, Omarjee S, Galloway WRJD, Kwan TT-L, Sore HF, Parker JS, Hyvönen M, Carroll JS and Spring DR (2019) A general approach for the site-selective modification of native proteins, enabling the generation of stable and functional antibody–drug conjugates. Chem. Sci 10, 694–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sun S, Akkapeddi P, Marques MC, Martïnez-Sáez N, Torres VM, Cordeiro C, Boutureira O and Bernardes GJL (2019) One-pot stapling of interchain disulfides of antibodies using an isobutylene motif. Org. Biomol. Chem 17, 2005–2012. [DOI] [PubMed] [Google Scholar]
- (19).Schumacher FF, Nunes JPM, Maruani A, Chudasama V, Smith MEB, Chester KA, Baker JR and Caddick S. (2014) Next generation maleimides enable the controlled assembly of antibody–drug conjugates via native disulfide bond bridging. Org. Biomol. Chem 12, 7261–7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Nunes JPM, Morais M, Vassileva V, Robinson E, Rajkumar VS, Smith MEB, Pedley RB, Caddick S, Baker JR and Chudasama V (2015) Functional native disulfide bridging enables delivery of a potent, stable and targeted antibody–drug conjugate (ADC). Chem. Commun 51, 10624–10627. [DOI] [PubMed] [Google Scholar]
- (21).Maruani A, Smith MEB, Miranda E, Chester KA, Chudasama V and Caddick S (2015) A plug-and-play approach to antibody-based therapeutics via a chemoselective dual click strategy. Nat. Commun 6, 6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lee MTW, Maruani A, Baker JR, Caddick S and Chudasama V (2016) Next-generation disulfide stapling: reduction and functional re-bridging all in one. Chem. Sci 7, 799–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lee MTW, Maruani A, Richards DA, Baker JR, Caddick S and Chudasama V (2017) Enabling the controlled assembly of antibody conjugates with a loading of two modules without antibody engineering. Chem. Sci 8, 2056–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Robinson E, Nunes JPM, Vassileva V, Maruani A, Nogueira JCF, Smith MEB, Pedley RB, Caddick S, Baker JR and Chudasama V (2017) Pyridazinediones deliver potent, stable, targeted and efficacious antibody–drug conjugates (ADCs) with a controlled loading of 4 drugs per antibody. RSC Adv. 7, 9073–9077. [Google Scholar]
- (25).Bahou C, Richards DA, Maruani A, Love EA, Javaid F, Caddick S, Baker JR and Chudasama V (2018) Highly homogeneous antibody modification through optimisation of the synthesis and conjugation of functionalised dibromopyridazinediones. Org. Biomol. Chem 16, 1359–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Saka SK, Wang Y, Kishi JY, Zhu A, Zeng Y, Xie W, Kirli K, Yapp C, Cicconet M, Beliveau BJ, Lapan SW, Yin S, Lin M, Boyden ES, Kaeser PS, Pihan G, Church GM and Yin P (2019) Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol 37, 1080–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Gaublomme JT, Li B, McCabe C, Knecht A, Yang Y, Drokhlyansky E, Van Wittenberghe N, Waldman J, Dionne D, Nguyen L, De Jager PL, Yeung B, Zhao X, Habib N, Rozenblatt-Rosen O and Regev A (2019) Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat. Commun 10, 2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Granja JM, Klemm S, McGinnis LM, Kathiria AS, Mezger A, Corces MR, Parks B, Gars E, Liedtke M, Zheng GXY, Chang HY, Majeti R and Greenleaf WJ (2019) Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol 37, 1458–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Wu D, Yan J, Shen X, Sun Y, Thulin M, Cai Y, Wik L, Shen Q, Oelrich J, Qian X, Dubois KL, Ronquist KG, Nilsson M, Landegren U and Kamali-Moghaddam M (2019) Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nat. Commun 10, 3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R and Smibert P (2017) Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ohata J and Ball ZT (2017) A hexa-rhodium metallopeptide catalyst for site-specific functionalization of natural antibodies. J. Am. Chem. Soc 139, 12617–12622. [DOI] [PubMed] [Google Scholar]
- (32).Yu C, Tang J, Loredo A, Chen Y, Jung SY, Jain A, Gordon A and Xiao H (2018) Proximity-induced site-specific antibody conjugation. Bioconjugate Chem. 29, 3522–3526. [DOI] [PubMed] [Google Scholar]
- (33).Wouters SFA, Vugs WJP, Arts R, de Leeuw NM, Teeuwen RWH and Merkx M (2020) Bioluminescent Antibodies through Photoconjugation of Protein G-Luciferase Fusion Proteins. Bioconjugate Chem. 10.1021/acs.bioconjchem.9b00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Vinogradova EV, Zhang C, Spokoyny AM, Pentelute BL and Buchwald SL (2015) Organometallic palladium reagents for cysteine bioconjugation. Nature 526, 687–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Adusumalli SR, Rawale DG, Singh U, Tripathi P, Paul R, Kalra N, Mishra RK, Shukla S and Rai V (2018) Single-site labeling of native proteins enabled by a chemoselective and site-selective chemical technology. J. Am. Chem. Soc 140, 15114–15123. [DOI] [PubMed] [Google Scholar]
- (36).Devaraj NK, Upadhyay R, Haun JB, Hilderbrand SA and Weissleder R (2009) Fast and sensitive pretargeted labeling of cancer cells through a tetrazine/trans–cyclooctene cycloaddition. Angew. Chem., Int. Ed 48, 7013–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Rahim MK, Kota R and Haun JB (2015) Enhancing reactivity for bioorthogonal pretargeting by unmasking antibody-conjugated trans-cyclooctenes. Bioconjugate Chem. 26, 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Carlson JC, Meimetis LG, Hilderbrand SA and Weissleder R (2013) BODIPY-tetrazine derivatives as superbright bioorthogonal turn-on probes. Angew. Chem., Int. Ed 52, 6917–6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Meimetis LG, Carlson JCT, Giedt RJ, Kohler RH and Weissleder R (2014) Ultrafluorogenic coumarin–tetrazine probes for real-time biological imaging. Angew. Chem., Int. Ed 53, 7531–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Vázquez A, Dzijak R, Dračínský M, Rampmaier R, Siegl SJ and Vrabel M (2017) Mechanism–based fluorogenic trans–cyclooctene–tetrazine cycloaddition. Angew. Chem, Int. Ed 56, 1334–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Siegl SJ, Galeta J, Dzijak R, Vázquez A, Del Rïo-Villanueva M, Dračínský M and Vrabel M (2019) An Extended Approach for the Development of Fluorogenic trans-Cyclooctene-Tetrazine Cycloadditions. ChemBioChem 20, 886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Rondon A and Degoul F (2020) Antibody Pretargeting Based on Bioorthogonal Click Chemistry for Cancer Imaging and Targeted Radionuclide Therapy. Bioconjugate Chem. 31, 159–173. [DOI] [PubMed] [Google Scholar]
- (43).Rossin R, van Duijnhoven SMJ, Läppchen T, van den Bosch SM and Robillard MS (2014) Trans-Cyclooctene tag with improved properties for tumor pretargeting with the Diels–Alder reaction. Mol. Pharmaceutics 11, 3090–3096. [DOI] [PubMed] [Google Scholar]
- (44).Läppchen T, Rossin R, van Mourik TR, Gruntz G, Hoeben FJM, Versteegen RM, Janssen HM, Lub J and Robillard MS (2017) DOTA-tetrazine probes with modified linkers for tumor pretargeting. Nucl. Med. Biol 55, 19–26. [DOI] [PubMed] [Google Scholar]
- (45).Hermanson GT (2013) Bioconjugate Techniques, 3rd Edition, Academic Press, [Google Scholar]
- (46).Andrews NP, Boeckman JX, Manning CF, Nguyen JT, Bechtold H, Dumitras C, Gong B, Nguyen K, van der List D, Murray KD, Engebrecht J and Trimmer JS (2019) A toolbox of IgG subclass-switched recombinant monoclonal antibodies for enhanced multiplex immunolabeling of brain. eLife 8, 43322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Beers SA, Glennie MJ and White AL (2016) Influence of immunoglobulin isotype on therapeutic antibody function. Blood 127, 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Burton DR (1985) Immunoglobulin G: functional sites. Mol. Immunol 22, 161–206. [DOI] [PubMed] [Google Scholar]
- (49).Clark MR (1997) Antibody Engineering. Chem. Immunol 65, 88–110. [PubMed] [Google Scholar]
- (50).Dangl JL, Wensel TG, Morrison SL, Stryer L, Herzenberg LA and Oi VT (1988) Segmental flexibility and complement fixation of genetically engineered chimeric human, rabbit and mouse antibodies. The EMBO Journal 7, 1989–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Feinstein A, Richardson N and Taussig MI (1986) Immunoglobulin flexibility in complement activation. Immunol. Today 7, 169–174. [DOI] [PubMed] [Google Scholar]
- (52).Collins AM (2016) IgG subclass co–expression brings harmony to the quartet model of murine IgG function. Immunol. Cell Biol 94, 949–954. [DOI] [PubMed] [Google Scholar]
- (53).Duncan AR, Woof JM, Partridge LJ, Burton DR and Winter G (1988) Localization of the binding site for the human high-affinity Fc receptor on IgG. Nature 332, 563–564. [DOI] [PubMed] [Google Scholar]
- (54).Brekke OH, Michaelsen TE and Sandlie I (1995) The structural requirements for complement activation by IgG: does it hinge on the hinge. Immunol. Today 16, 85–90. [DOI] [PubMed] [Google Scholar]
- (55).Natsuume-Sakai S, Motonishi K and Migita S (1977) Quantitative estimations of five classes of immunoglobulin in inbred mouse strains. Immunology 32, 861–866. [PMC free article] [PubMed] [Google Scholar]
- (56).Manning CF, Bundros AM and Trimmer JS (2012) Benefits and pitfalls of secondary antibodies: why choosing the right secondary is of primary importance. PLoS One 7, 38313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Kishiro Y, Kagawa M, Naito I and Sado Y (1995) A novel method of preparing rat-monoclonal antibody-producing hybridomas by using rat medial iliac lymph node cells. Cell Struct. Funct 20, 151–156. [DOI] [PubMed] [Google Scholar]
- (58).Weber J, Peng H and Rader C (2017) From rabbit antibody repertoires to rabbit monoclonal antibodies. Exp. Mol. Med 49, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


