Abstract
Background
The sequencing and annotations of cotton genomes provide powerful theoretical support to unravel more physiological and functional information. Plant homeodomain (PHD) protein family has been reported to be involved in regulating various biological processes in plants. However, their functional studies have not yet been carried out in cotton.
Results
In this study, 108, 55, and 52 PHD genes were identified in G. hirsutum, G. raimondii, and G. arboreum, respectively. A total of 297 PHD genes from three cotton species, Arabidopsis, and rice were divided into five groups. We performed chromosomal location, phylogenetic relationship, gene structure, and conserved domain analysis for GhPHD genes. GhPHD genes were unevenly distributed on each chromosome. However, more GhPHD genes were distributed on At_05, Dt_05, and At_07 chromosomes. GhPHD proteins depicted conserved domains, and GhPHD genes exhibiting similar gene structure were clustered together. Further, whole genome duplication (WGD) analysis indicated that purification selection greatly contributed to the functional maintenance of GhPHD gene family. Expression pattern analysis based on RNA-seq data showed that most GhPHD genes showed clear tissue-specific spatiotemporal expression patterns elucidating the multiple functions of GhPHDs in plant growth and development. Moreover, analysis of cis-acting elements revealed that GhPHDs may respond to a variety of abiotic and phytohormonal stresses. In this regard, some GhPHD genes showed good response against abiotic and phytohormonal stresses. Additionally, co-expression network analysis indicated that GhPHDs are essential for plant growth and development, while GhPHD genes response against abiotic and phytohormonal stresses may help to improve plant tolerance in adverse environmental conditions.
Conclusion
This study will provide useful information to facilitate further research related to the vital roles of GhPHD gene family in plant growth and development.
Keywords: Cotton, PHD, Transcription factor, Phytohormone, Stress tolerance, Co-expression network, Transcriptome analysis
Background
Plants often face various abiotic and biotic stress conditions. Abiotic stresses include heat, cold, drought, and salinity, whereas biotic stresses mainly come from bacteria, fungi, viruses, and insects. These abiotic and biotic stresses significantly reduce crop quality and productivity world-wide [1, 2]. In order to adapt such unfavorable environment, plants have established a comprehensive mechanism to combat stress signals and mitigate their effects on plant growth and development [3]. Phytohormones play significant roles in regulating developmental processes and signal transduction networks, which respond to various abiotic stresses. Brassinosteroid (BR), jasmonate (JA), gibberellin (GA), salicylic acid (SA), auxin, and abscisic acid (ABA) regulate plant growth, development, stress, and defense responses [4–11], but how phytohormones mediate the growth and stress trade-off is unclear.
Zinc finger protein motifs are part of many protein families and widely distributed in eukaryotic organisms. The term “zinc finger” represents the sequence motif in which cysteines and/or histidines coordinate the zinc atom(s) to form the local peptide structure that are required for their specific functions. The “finger” structural motif has been divided into different types, such as TFIIIA-type zinc finger (EPF1, SUPERMAN) [12, 13], WRKY family (WRKY1, 2, and 3), GATA1-type protein (NTL1) [14, 15], Dof family (Dof1) [16, 17], RING-finger type (COP1) [18], PHD-finger family (AtHAT3.1 and ZmHOX1a) [19, 20], LIM family (SF3) [21, 22], and other uncategorized types. Plant homeodomain (PHD) zinc fingers are small reader domains found in several chromatin-binding proteins. In plants, PHD proteins are usually zinc finger proteins with one or more PHD domains, which have a Cys4-His-Cys3 zinc-binding motif consisting of about 60 amino acids [23]. It is worth noting that the number of amino acids between cysteine and histidine or between cysteine residues in the PHD domain are conserved, while second amino acid (before the penultimate cysteine residue) is usually an aromatic amino acid, such as tryptophan [24].
Since the discovery of the first PHD protein HAT3.1 (Histone acetyltransferase 3.1) in Arabidopsis, more PHD proteins have been identified to participate in many physiological and biochemical processes involved in the structure and transcription of chromatin [25]. In Arabidopsis, PHD protein MMD1 (Male meiocyte death 1)/DUET is specifically expressed in male meiocytes and involved in regulating gene expression during meiosis, mutations of mmd1 gene leads to the death of male meiotic cells [26–28]. Epigenetic regulation in eukaryotes is performed through complex signal interactions between chromatin markers and small RNA species. AtVIM1 (Variant in methylation 1) functions in DNA methylation-histone interface to maintain the centromeric heterochromation in Arabidopsis [29]. In addition, PHD proteins are involved in regulating plant response to abiotic stresses and altering plant growth and development [30, 31]. In soybean, six Alfin1-type PHD proteins were identified to respond against salt, cold, drought, and ABA treatment. For instance, GmPHD2 improve salt tolerance in transgenic Arabidopsis plants compared with the wild type plants [32]. In Arabidopsis, AtVIN3 (Vernalization insensitive 3) protein binds to modified histone in vitro to change the binding specificity of PHD-finger domain and accelerate the vernalization reaction in vivo [33]. During seed germination, the AL PHD-PRC1 complex affect seed developmental genes from the active state associated with H3K4me3 to the repressive transcriptional state associated with H3K27me3, thereby promote seed germination [34]. PHD protein GSR1 (Germostatin resistance locus 1) is a member of auxin-mediated genetic network for seed germination and form a corepressor with ARF16 (Auxin response factor 16) to regulate seed germination [35]. Therefore, PHD proteins play irreplaceable roles in the biological processes of life.
At present, the PHD protein family has been studied in several plants, such as Arabidopsis thaliana, poplar (Populus trichocarpa) [36], maize (Zea mays) [30], moso bamboo (Phyllostachys edulis) [37], carrot (Daucus carota L.) [38], potato (Solanum tuberosum) [39], and pear (Pyrus bretschneideri) [40]. However, comprehensive identification and characterization of cotton PHD protein family has not been carried out till date. Upland cotton (Gossypium hirsutum) is the most important natural fiber crop in the world. Recently, the availability of the complete genome sequence and annotations of G. hirsutum [41], G. arboreum [42], and G. raimondii [43] provided an excellent opportunity to identify and characterize PHD transcription factors in cotton. In this study, we performed the whole genome-wide analysis, tissue expression pattern analysis, relative expression level analysis under different stresses and phytohormones treatment, and co-expression network analysis of GhPHD genes in upland cotton. Our results indicated that GhPHD genes are involved in various processes of plant growth and development, and phytohormones mediate responses of GhPHD genes against abiotic stresses.
Results
Genome-wide identification of PHD proteins in cotton
Based on the homology of protein sequences, 108, 52, and 55 PHD proteins were identified in three cotton species G. hirsutum, G. arboreum, and G. raimondii, respectively. In addition, 39 and 43 PHD proteins were identified in Arabidopsis and rice, respectively (Table S1). Among 108 GhPHD proteins, 56 members belong to the At subgenome and 52 members belong to the Dt subgenome. The predicted biophysical characteristic of GhPHDs (Table 1) indicates that the length of GhPHD proteins ranges from 159 aa (GhPHD28) to 2231 aa (GhPHD39) with an average length of 741 aa. Moreover, the molecular weight of GhPHD proteins ranges from 17.76 kD (GhPHD28) to 247.42 kD (GhPHD39) with an average value of 93.09 kD. The isoelectric point (pI) of GhPHD proteins ranges from 4.58 (GhPHD38) to 10.41 (GhPHD103) with an average value of 6.89. Furthermore, the predicted subcellular localization indicated that 93 GhPHD proteins are located in nucleus, ten in cytoplasm, and five are extracellular.
Table 1.
Physicochemical parameters of 108 GhPHD genes in G. hirsutum
| Name | Protein length (aa) | Molecular weight (kDa) | Charge | Isoelectric point | Grand average of hydropathy | Subcellular localization |
|---|---|---|---|---|---|---|
| GhPHD1 | 217 | 24.915 | 5 | 7.895 | − 0.694 | Nuclear |
| GhPHD2 | 1033 | 114.441 | 32.5 | 8.49 | −0.274 | Nuclear |
| GhPHD3 | 1030 | 114.182 | 36 | 8.594 | −0.306 | Nuclear |
| GhPHD4 | 815 | 90.024 | 5 | 6.895 | −0.323 | Nuclear |
| GhPHD5 | 1303 | 144.878 | −9.5 | 6.002 | −0.713 | Nuclear |
| GhPHD6 | 700 | 79.363 | −4.5 | 6.21 | −0.308 | Nuclear |
| GhPHD7 | 700 | 79.363 | −4.5 | 6.21 | −0.308 | Nuclear |
| GhPHD8 | 345 | 39.474 | 12 | 8.648 | −0.573 | Nuclear |
| GhPHD9 | 251 | 28.253 | −8 | 4.891 | −0.621 | Nuclear |
| GhPHD10 | 786 | 86.704 | −35.5 | 4.631 | −0.966 | Nuclear |
| GhPHD11 | 216 | 24.846 | 8.5 | 8.262 | −0.789 | Nuclear |
| GhPHD12 | 375 | 42.862 | 5.5 | 7.542 | −0.708 | Nuclear |
| GhPHD13 | 237 | 26.757 | −4 | 5.421 | −0.596 | Nuclear |
| GhPHD14 | 959 | 104.731 | −3 | 6.227 | −1.126 | Nuclear |
| GhPHD15 | 733 | 82.869 | 40.5 | 9.936 | −0.907 | Nuclear |
| GhPHD16 | 252 | 28.482 | −8.5 | 4.84 | −0.717 | Nuclear |
| GhPHD17 | 1680 | 189.096 | 46.5 | 8.1 | −0.669 | Nuclear |
| GhPHD18 | 252 | 28.35 | −7.5 | 4.894 | −0.661 | Nuclear |
| GhPHD19 | 238 | 27.277 | 4.5 | 7.669 | −0.648 | Cytoplasmic |
| GhPHD20 | 493 | 55.306 | 5 | 7.03 | −0.485 | Nuclear |
| GhPHD21 | 600 | 67.261 | 5.5 | 7.049 | −0.611 | Nuclear |
| GhPHD22 | 1084 | 122.987 | 34 | 8.276 | −0.574 | Nuclear |
| GhPHD23 | 253 | 28.577 | −5.5 | 5.132 | −0.736 | Nuclear |
| GhPHD24 | 259 | 29.239 | −7.5 | 4.915 | −0.708 | Nuclear |
| GhPHD25 | 224 | 25.723 | 6.5 | 8.087 | −0.682 | Cytoplasmic |
| GhPHD26 | 870 | 95.472 | 3 | 6.876 | −0.472 | Nuclear |
| GhPHD27 | 1358 | 154.506 | 35 | 7.891 | −0.677 | Nuclear |
| GhPHD28 | 159 | 17.763 | 0 | 6.496 | −0.666 | Extracellular |
| GhPHD29 | 733 | 80.954 | 22 | 8.271 | −0.664 | Nuclear |
| GhPHD30 | 1247 | 141.67 | 24 | 7.655 | −0.43 | Nuclear |
| GhPHD31 | 949 | 104.907 | 2.5 | 6.779 | −0.416 | Nuclear |
| GhPHD32 | 1618 | 180.35 | 45 | 8.404 | −0.446 | Nuclear |
| GhPHD33 | 1618 | 180.725 | 41.5 | 8.289 | −0.442 | Nuclear |
| GhPHD34 | 216 | 24.95 | 8.5 | 8.399 | −0.783 | Nuclear |
| GhPHD35 | 321 | 35.88 | −4.5 | 5.599 | −0.049 | Extracellular |
| GhPHD36 | 822 | 88.768 | 2 | 6.651 | −0.539 | Nuclear |
| GhPHD37 | 1305 | 143.316 | −33.5 | 4.951 | −0.624 | Nuclear |
| GhPHD38 | 705 | 78.949 | 22 | 8.309 | −0.315 | Extracellular |
| GhPHD39 | 2231 | 247.421 | −36 | 5.321 | −0.444 | Nuclear |
| GhPHD40 | 226 | 25.942 | 6 | 8.086 | −0.788 | Nuclear |
| GhPHD41 | 1685 | 187.671 | −4.5 | 6.321 | −0.389 | Nuclear |
| GhPHD42 | 1239 | 138.122 | −0.5 | 6.487 | −0.735 | Nuclear |
| GhPHD43 | 253 | 28.585 | −6 | 5.13 | −0.76 | Nuclear |
| GhPHD44 | 531 | 58.455 | −4 | 5.77 | −0.564 | Nuclear |
| GhPHD45 | 389 | 44.625 | 26.5 | 9.906 | −0.405 | Nuclear |
| GhPHD46 | 803 | 90.452 | −16.5 | 5.132 | −0.836 | Nuclear |
| GhPHD47 | 851 | 94.805 | −26 | 4.895 | −1.011 | Nuclear |
| GhPHD48 | 212 | 23.802 | 26 | 10.41 | −0.745 | Nuclear |
| GhPHD49 | 1019 | 116.396 | 18 | 7.609 | −0.584 | Nuclear |
| GhPHD50 | 655 | 74.308 | 11 | 7.433 | −0.207 | Cytoplasmic |
| GhPHD51 | 1091 | 124.448 | 45 | 8.516 | −0.59 | Nuclear |
| GhPHD52 | 237 | 26.997 | −3.5 | 5.244 | −0.666 | Nuclear |
| GhPHD53 | 216 | 24.723 | 6.5 | 8.049 | −0.785 | Nuclear |
| GhPHD54 | 716 | 81.912 | −38.5 | 4.581 | −1.1 | Nuclear |
| GhPHD55 | 1367 | 152.049 | 3 | 6.689 | −0.451 | Nuclear |
| GhPHD56 | 254 | 28.492 | −6.5 | 5.136 | −0.576 | Cytoplasmic |
| GhPHD57 | 217 | 24.929 | 5 | 7.895 | −0.684 | Nuclear |
| GhPHD58 | 1031 | 114.246 | 33.5 | 8.459 | −0.299 | Nuclear |
| GhPHD59 | 1031 | 113.878 | 36.5 | 8.592 | −0.306 | Nuclear |
| GhPHD60 | 1299 | 144.45 | −11.5 | 5.886 | −0.711 | Nuclear |
| GhPHD61 | 290 | 32.821 | −8.5 | 4.832 | −0.419 | Nuclear |
| GhPHD62 | 216 | 24.835 | 8.5 | 8.262 | −0.789 | Nuclear |
| GhPHD63 | 699 | 79.319 | −4.5 | 6.21 | −0.283 | Nuclear |
| GhPHD64 | 345 | 39.311 | 13 | 8.745 | −0.544 | Nuclear |
| GhPHD65 | 1084 | 123.101 | 34 | 8.275 | −0.579 | Nuclear |
| GhPHD66 | 237 | 26.615 | −2 | 5.973 | −0.561 | Nuclear |
| GhPHD67 | 945 | 103.676 | −2 | 6.329 | −1.136 | Nuclear |
| GhPHD68 | 684 | 77.195 | 23 | 9.073 | −0.859 | Nuclear |
| GhPHD69 | 733 | 83.057 | 34.5 | 9.713 | −0.91 | Nuclear |
| GhPHD70 | 252 | 28.414 | −8.5 | 4.84 | −0.697 | Nuclear |
| GhPHD71 | 1731 | 194.29 | 53 | 8.282 | −0.675 | Nuclear |
| GhPHD72 | 252 | 28.35 | −7.5 | 4.894 | −0.661 | Nuclear |
| GhPHD73 | 224 | 25.664 | 7.5 | 8.248 | −0.774 | Cytoplasmic |
| GhPHD74 | 367 | 41.019 | 5 | 7.341 | −0.49 | Nuclear |
| GhPHD75 | 601 | 67.379 | 2 | 6.696 | −0.611 | Nuclear |
| GhPHD76 | 241 | 27.506 | −0.5 | 6.269 | −0.502 | Extracellular |
| GhPHD77 | 253 | 28.677 | −5.5 | 5.139 | −0.752 | Nuclear |
| GhPHD78 | 252 | 28.407 | −7.5 | 4.889 | −0.682 | Nuclear |
| GhPHD79 | 186 | 21.734 | 1.5 | 6.851 | −0.752 | Cytoplasmic |
| GhPHD80 | 237 | 27.077 | −4 | 5.221 | −0.689 | Extracellular |
| GhPHD81 | 1356 | 154.398 | 31 | 7.737 | −0.674 | Nuclear |
| GhPHD82 | 236 | 26.789 | −12 | 4.605 | −0.598 | Cytoplasmic |
| GhPHD83 | 676 | 74.819 | 22.5 | 8.253 | −0.665 | Nuclear |
| GhPHD84 | 1382 | 156.796 | 34 | 7.925 | −0.467 | Nuclear |
| GhPHD85 | 949 | 104.967 | 2.5 | 6.779 | −0.426 | Nuclear |
| GhPHD86 | 1653 | 183.656 | 37.5 | 8.15 | −0.449 | Nuclear |
| GhPHD87 | 1618 | 180.589 | 39 | 8.234 | −0.447 | Nuclear |
| GhPHD88 | 216 | 24.836 | 5.5 | 7.902 | −0.775 | Cytoplasmic |
| GhPHD89 | 822 | 88.688 | 0 | 6.506 | −0.531 | Nuclear |
| GhPHD90 | 1301 | 142.837 | −35.5 | 4.873 | −0.619 | Nuclear |
| GhPHD91 | 705 | 78.888 | 23.5 | 8.373 | −0.305 | Nuclear |
| GhPHD92 | 2182 | 241.654 | −34 | 5.362 | −0.441 | Nuclear |
| GhPHD93 | 226 | 25.984 | 6 | 8.086 | −0.767 | Nuclear |
| GhPHD94 | 1685 | 187.566 | −4 | 6.345 | −0.397 | Nuclear |
| GhPHD95 | 1237 | 137.855 | −0.5 | 6.486 | −0.718 | Nuclear |
| GhPHD96 | 253 | 28.613 | −6 | 5.13 | −0.75 | Nuclear |
| GhPHD97 | 696 | 78.217 | 18.5 | 8.023 | −0.17 | Nuclear |
| GhPHD98 | 503 | 55.441 | −5.5 | 5.455 | −0.597 | Nuclear |
| GhPHD99 | 385 | 44.394 | 31 | 10.216 | −0.428 | Nuclear |
| GhPHD100 | 812 | 91.657 | −29 | 4.818 | −0.838 | Nuclear |
| GhPHD101 | 859 | 95.86 | −32.5 | 4.789 | −1.023 | Nuclear |
| GhPHD102 | 1019 | 116.3 | 19 | 7.67 | −0.593 | Nuclear |
| GhPHD103 | 655 | 74.268 | 11.5 | 7.443 | −0.219 | Cytoplasmic |
| GhPHD104 | 1091 | 124.625 | 43 | 8.46 | −0.581 | Nuclear |
| GhPHD105 | 889 | 101.493 | −32 | 4.811 | −0.882 | Nuclear |
| GhPHD106 | 1305 | 145.225 | 9.5 | 7.121 | −0.424 | Nuclear |
| GhPHD107 | 801 | 90.038 | −12.5 | 5.251 | −0.805 | Nuclear |
| GhPHD108 | 252 | 28.259 | −6.5 | 5.136 | −0.619 | Cytoplasmic |
Phylogenetic analysis, chromosomal location, and gene duplication
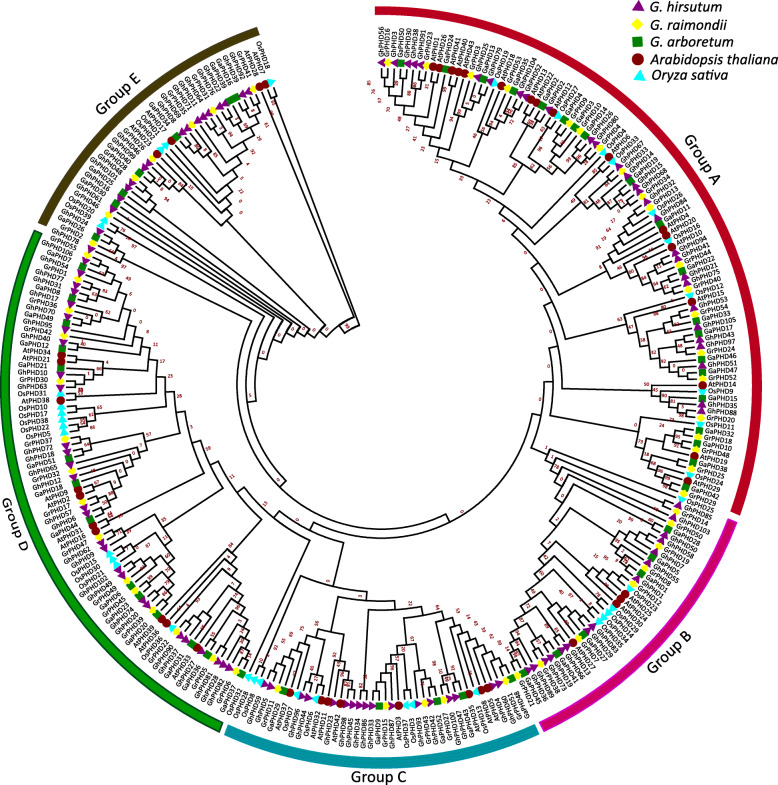
In order to understand the phylogenetic relationship of PHD proteins in rice, Arabidopsis, and cotton, we constructed a NJ phylogenetic tree and classified PHD proteins into five groups (A-E) (Fig. 1). Among them, most of the orthologous PHD proteins between the diploid and allotetraploid cotton are grouped in same clade exhibiting maximum homology in phylogenetic relationship. Each group contains PHD proteins of these five species, of which group A and D are the first and second largest groups, containing 97 and 79 members, respectively. While, there are relatively few PHD members in groups B, C, and E. Chromosome location analysis showed that 108 GhPHD genes are positioned on 26 chromosomes, including 13 chromosomes from the At subgenome and 13 chromosomes from the Dt subgenome (Fig. S1 and Table S2). Deeper insights indicated that At_05, At_07, and Dt_05 chromosomes contain more number of genes (eight GhPHD genes on each) and display a dense distribution at the top. However, some chromosomes contain only two GhPHD genes, such as At_10, At_11, Dt_03, and Dt_11.
Fig. 1.
Phylogenetic tree displaying relationships between 108 G. hirsutum, 52 G. arboreum, 55 G. raimondii, 39 O. sativa and 43 A. thaliana PHD proteins. The phylogenetic tree was constructed in MEGA 6.0 using the neighbor-joining method. The bootstrap test was performed with 1000 iterations. The five subgroups are shown with different colours. At, Arabidopsis thaliana; Ga, Gossypium arboreum; Gr, Gossypium raimondii; Gh, Gossypium hirsutum; Os, Oryza sativa
We further investigated the whole genome duplication (WGD) event experienced by GhPHD genes. As a result, 73 GhPHD gene pairs depict segmental duplication and four gene pairs show tandem duplication events (Table 2), indicating that WGD is the main contributor of GhPHD gene family expansion. Duplication gene pairs may have undergone three alternative fates during the evolution process, namely non-functionalization, neo-functionalization, and sub-functionalization [44]. In order to study the evolutionary history of GhPHD genes, the Ka/Ks calculator 2.0 is used to calculate the synonymous and non-synonymous substitution rates. The Ka/Ks ratio of 76 duplicated gene pairs is less than 1, indicating that GhPHD genes underwent purification selection pressure with limited functional divergence. However, there is only one gene pair with the Ka/Ks greater than 1, indicating the occurrence of positive selection pressure. Collectively, these results indicated that the great contribution of purification selection pressure in the functional maintenance of GhPHD genes in upland cotton.
Table 2.
Ka/Ks analysis for the duplicated PHD gene pairs from G. hirsutum
| Duplicated gene 1 | Duplicated gene 2 | Ka | Ks | Ka/Ks | Purifying selection | Duplicate type |
|---|---|---|---|---|---|---|
| GhPHD1 | GhPHD11 | 0.064 | 0.533 | 0.119 | Yes | Segmental |
| GhPHD1 | GhPHD62 | 0.064 | 0.533 | 0.119 | Yes | Segmental |
| GhPHD2 | GhPHD58 | 0.011 | 0.043 | 0.248 | Yes | Segmental |
| GhPHD5 | GhPHD60 | 0.016 | 0.039 | 0.411 | Yes | Segmental |
| GhPHD5 | GhPHD95 | 0.126 | 0.389 | 0.324 | Yes | Segmental |
| GhPHD6 | GhPHD63 | 0.004 | 0.048 | 0.089 | Yes | Segmental |
| GhPHD9 | GhPHD61 | 0.007 | 0.032 | 0.212 | Yes | Segmental |
| GhPHD10 | GhPHD14 | 0.232 | 0.441 | 0.526 | Yes | Segmental |
| GhPHD10 | GhPHD47 | 0.245 | 0.488 | 0.502 | Yes | Segmental |
| GhPHD10 | GhPHD67 | 0.241 | 0.471 | 0.512 | Yes | Segmental |
| GhPHD10 | GhPHD101 | 0.235 | 0.446 | 0.527 | Yes | Segmental |
| GhPHD11 | GhPHD53 | 0.039 | 0.606 | 0.064 | Yes | Segmental |
| GhPHD11 | GhPHD62 | 0.004 | 0.014 | 0.282 | Yes | Segmental |
| GhPHD13 | GhPHD66 | 0.009 | 0.053 | 0.169 | Yes | Segmental |
| GhPHD14 | GhPHD47 | 0.376 | 0.666 | 0.564 | Yes | Segmental |
| GhPHD14 | GhPHD67 | 0.018 | 0.046 | 0.387 | Yes | Segmental |
| GhPHD15 | GhPHD68 | 0.030 | 0.063 | 0.468 | Yes | Segmental |
| GhPHD16 | GhPHD28 | 0.089 | 0.311 | 0.285 | Yes | Segmental |
| GhPHD16 | GhPHD77 | 0.045 | 0.352 | 0.129 | Yes | Segmental |
| GhPHD16 | GhPHD82 | 0.053 | 0.405 | 0.130 | Yes | Segmental |
| GhPHD17 | GhPHD71 | 0.008 | 0.034 | 0.235 | Yes | Segmental |
| GhPHD17 | GhPHD81 | 0.075 | 0.381 | 0.196 | Yes | Segmental |
| GhPHD19 | GhPHD25 | 0.081 | 0.447 | 0.180 | Yes | Segmental |
| GhPHD19 | GhPHD40 | 0.083 | 0.451 | 0.184 | Yes | Segmental |
| GhPHD19 | GhPHD73 | 0.026 | 0.053 | 0.492 | Yes | Segmental |
| GhPHD19 | GhPHD79 | 0.083 | 0.496 | 0.167 | Yes | Segmental |
| GhPHD19 | GhPHD93 | 0.085 | 0.451 | 0.188 | Yes | Segmental |
| GhPHD20 | GhPHD74 | 0.024 | 0.032 | 0.753 | Yes | Segmental |
| GhPHD22 | GhPHD51 | 0.080 | 0.398 | 0.200 | Yes | Segmental |
| GhPHD22 | GhPHD65 | 0.007 | 0.028 | 0.240 | Yes | Segmental |
| GhPHD22 | GhPHD104 | 0.079 | 0.391 | 0.201 | Yes | Segmental |
| GhPHD28 | GhPHD77 | 0.085 | 0.284 | 0.298 | Yes | Segmental |
| GhPHD28 | GhPHD82 | 0.042 | 0.037 | 1.121 | No | Segmental |
| GhPHD24 | GhPHD78 | 0.003 | 0.039 | 0.086 | Yes | Segmental |
| GhPHD25 | GhPHD73 | 0.057 | 0.434 | 0.131 | Yes | Segmental |
| GhPHD25 | GhPHD79 | 0.016 | 0.017 | 0.982 | Yes | Segmental |
| GhPHD26 | GhPHD44 | 0.204 | 0.368 | 0.553 | Yes | Segmental |
| GhPHD26 | GhPHD98 | 0.186 | 0.344 | 0.541 | Yes | Segmental |
| GhPHD29 | GhPHD83 | 0.019 | 0.041 | 0.473 | Yes | Segmental |
| GhPHD31 | GhPHD85 | 0.011 | 0.031 | 0.356 | Yes | Segmental |
| GhPHD32 | GhPHD86 | 0.015 | 0.027 | 0.554 | Yes | Segmental |
| GhPHD34 | GhPHD88 | 0.006 | 0.021 | 0.288 | Yes | Segmental |
| GhPHD36 | GhPHD89 | 0.015 | 0.031 | 0.499 | Yes | Segmental |
| GhPHD39 | GhPHD92 | 0.014 | 0.040 | 0.339 | Yes | Segmental |
| GhPHD40 | GhPHD73 | 0.051 | 0.442 | 0.116 | Yes | Segmental |
| GhPHD40 | GhPHD93 | 0.002 | 0.054 | 0.035 | Yes | Segmental |
| GhPHD41 | GhPHD94 | 0.013 | 0.031 | 0.416 | Yes | Segmental |
| GhPHD44 | GhPHD98 | 0.022 | 0.052 | 0.413 | Yes | Segmental |
| GhPHD46 | GhPHD54 | 0.127 | 0.418 | 0.304 | Yes | Segmental |
| GhPHD46 | GhPHD100 | 0.014 | 0.055 | 0.256 | Yes | Segmental |
| GhPHD46 | GhPHD107 | 0.101 | 0.395 | 0.256 | Yes | Segmental |
| GhPHD47 | GhPHD67 | 0.231 | 0.450 | 0.514 | Yes | Segmental |
| GhPHD47 | GhPHD101 | 0.016 | 0.030 | 0.530 | Yes | Segmental |
| GhPHD49 | GhPHD102 | 0.006 | 0.035 | 0.163 | Yes | Segmental |
| GhPHD50 | GhPHD103 | 0.011 | 0.048 | 0.233 | Yes | Segmental |
| GhPHD51 | GhPHD65 | 0.079 | 0.389 | 0.202 | Yes | Segmental |
| GhPHD51 | GhPHD104 | 0.010 | 0.036 | 0.270 | Yes | Segmental |
| GhPHD52 | GhPHD80 | 0.038 | 0.528 | 0.072 | Yes | Segmental |
| GhPHD53 | GhPHD62 | 0.039 | 0.606 | 0.064 | Yes | Segmental |
| GhPHD54 | GhPHD100 | 0.140 | 0.416 | 0.336 | Yes | Segmental |
| GhPHD54 | GhPHD107 | 0.136 | 0.405 | 0.335 | Yes | Segmental |
| GhPHD55 | GhPHD106 | 0.023 | 0.045 | 0.497 | Yes | Segmental |
| GhPHD56 | GhPHD76 | 0.236 | 0.639 | 0.369 | Yes | Segmental |
| GhPHD56 | GhPHD108 | 0.007 | 0.051 | 0.132 | Yes | Segmental |
| GhPHD60 | GhPHD95 | 0.125 | 0.397 | 0.314 | Yes | Segmental |
| GhPHD65 | GhPHD104 | 0.078 | 0.386 | 0.203 | Yes | Segmental |
| GhPHD67 | GhPHD101 | 0.225 | 0.456 | 0.492 | Yes | Segmental |
| GhPHD71 | GhPHD81 | 0.075 | 0.369 | 0.203 | Yes | Segmental |
| GhPHD73 | GhPHD79 | 0.054 | 0.486 | 0.111 | Yes | Segmental |
| GhPHD73 | GhPHD93 | 0.053 | 0.419 | 0.127 | Yes | Segmental |
| GhPHD76 | GhPHD108 | 0.237 | 0.630 | 0.376 | Yes | Segmental |
| GhPHD91 | GhPHD38 | 0.016 | 0.052 | 0.298 | Yes | Segmental |
| GhPHD100 | GhPHD107 | 0.104 | 0.379 | 0.276 | Yes | Segmental |
| GhPHD2 | GhPHD3 | 0.035 | 0.122 | 0.289 | Yes | Tandem |
| GhPHD32 | GhPHD33 | 0.031 | 0.076 | 0.407 | Yes | Tandem |
| GhPHD58 | GhPHD59 | 0.029 | 0.138 | 0.212 | Yes | Tandem |
| GhPHD86 | GhPHD87 | 0.027 | 0.062 | 0.428 | Yes | Tandem |
Gene structure and conserved motifs analysis
To better understand the similarity and diversity of GhPHD proteins in upland cotton, we analyzed the phylogenetic tree, exon-intron structure, and conserved motif. Phylogenetic tree grouped GhPHD proteins according to protein homology, conserved gene structure, and motif distribution (Fig. 2). GhPHD49 shows the longest genomic sequence with 26 exons, while GhPHD12 displays the shortest genomic sequence with only two exons (Fig. 2 and Table S3). Furthermore, a total of three motifs are identified in all GhPHD proteins, and all GhPHD proteins have a typical PHD domain (i.e., motif 1). Phylogenetic tree showed that 21 GhPHD proteins are clustered in a clade. Except for GhPHD28, all other GhPHD proteins contain three motifs with similar gene structure and motif distribution (Fig. 2).
Fig. 2.
Phylogenetic tree, gene structure, and conserved motif analysis of GhPHD proteins. a An unrooted phylogenetic tree was generated in MEGA 6.0 by neighbor-joining (NJ) method. b Exon-intron structure of GhPHD genes. The yellow boxes represent exons, black lines represent introns, and blue boxes represent the upstream/downstream UTRs. The sizes of exon and intron can be estimated using the scale bar at the bottom. c Motifs distribution of GhPHD proteins and different motif boxes are represented in different colors (motif 1 to 3). Motif 1 is the PHD domain
Protein sequence alignment shows that GhPHD proteins have a typical Cys4-His-Cys3 motif, which consists of about 60 amino acids and is accompanied by nine conserved amino acid residues (Fig. S2). The conserved histidine (H) is separated from the fourth conserved cysteine (C) by four amino acids and two amino acids from subsequent conserved cysteine (C) residue. The third and fourth conserved cysteine (C) before histidine (H) are separated by one or two amino acids, but the interval number between other conserved amino acids is uncertain. However, GhPHD17, GhPHD27, GhPHD71, and GhPHD81 exhibit maximum homology, but show less conserved PHD domain (Fig. 2 and Fig. S2).
Cis-acting element analysis
Many studies have showed that PHD genes are involved in various stress responses [30, 31, 37]. To elucidate the putative function of GhPHDs under different stresses, we first identified the cis-acting elements in the promoter region that respond to stresses and phytohormones. We identified many cis-acting elements that respond to ABA (ABRE), auxin (TGA and AuxRR-core), GA (TATC-box, P-box, CARE, and GARE), ethylene (ERE), SA (TCA), and MeJA (CGTCA). These results indicated that a total of 85 GhPHD genes are responsive to ethylene, followed by ABA, GA, and MeJA. 73 GhPHD genes have cis-acting elements that respond to three or more phytohormones. Interestingly, the promoters of GhPHD5, GhPHD47, GhPHD56, and GhPHD65 genes contain cis-elements that respond to the above six phytohormones. In addition, we found that many abiotic stresses response elements (TC-rich repeat, MBS, and LTR), circadian control elements, and light-responsive elements (G-box) are also present in the promoters of various GhPHD genes (Fig. 3 and Table S5). These results indicated that GhPHD genes may participate in various signal transduction pathways, such as phytohormones, light response, and abiotic stresses, and play important roles in regulating plant growth and development.
Fig. 3.
Distribution of stress-related and phytohormone-related cis-acting elements in the promoter regions of GhPHD genes. The locations of cis-acting elements were confirmed using PlantCARE database. Different cis-acting elements were represented by different color boxes
Tissue-specific expression pattern of GhPHD genes
To predict the physiological functions of GhPHD genes in cotton growth and development, we used the online transcriptome data to analyze the tissue-specific expression profile of GhPHD genes in different tissues such as root, stem, leaf, petal, stamen, pistil, ovule, and fiber. According to the expression features and hierarchical clustering (Fig. 4), GhPHD genes are mainly clustered into four groups (A-D). The nine GhPHD genes in group A are highly expressed in all tissues, indicating that they may play important roles in plant growth and development. In particular, GhPHD23 and GhPHD77 show maximum expression levels in ovule and fiber tissues, demonstrating that these two genes may be involved in the development of ovule and fiber. Further, 43 GhPHDs in group B show lower expression levels in all tissues, while six GhPHD genes (GhPHD56, GhPHD108, GhPHD40, GhPHD93, GhPHD19, and GhPHD73) are predominantly expressed in the early stage of ovule development, indicating that they may play important roles in ovule and seed development. Moreover, GhPHD genes in group C show higher expression levels in ovule. However, GhPHD genes in group D show poor expression in all observed tissues. These results indicated that GhPHDs may be involved in regulating cotton growth and development, especially in the development of ovule and fiber.
Fig. 4.
Tissue-specific expression patterns of GhPHD genes in upland cotton. A heatmap indicates the clustering of 108 GhPHD genes in eight tissues (shown at the bottom). DPA is days post anthesis. Gene names are shown on the right. Scale bars at the top show Log2 (FPKM+ 1) values of each gene
Identification of stress-related PHD genes in upland cotton
Analysis of the transcriptome data showed that 66 GhPHD genes have higher expression levels under heat, cold, salt, and drought treatments (Fig. S3). In order to further estimate the responses of GhPHDs under abiotic stresses, we treated four-week-old cotton seedlings with heat, cold, salt, and drought, and observed the relative expression level of 12 GhPHD genes (Fig. 5). The relative expression level of GhPHD18 is up-regulated under all stresses, indicating that GhPHD18 may be involved in multiple stresses response mechanisms. GhPHD23 is up-regulated only under heat treatment, indicating that GhPHD23 responds positively to heat stimuli. Further, GhPHD34, GhPHD40, and GhPHD43 are up-regulated after heat and salt treatment, while GhPHD80 and GhPHD88 are up-regulated after heat and drought tolerance at various time points. In addition, we found that GhPHD5 is up-regulated against salt and drought, while GhPHD72 and GhPHD107 are up-regulated against salt and heat, respectively. These results indicated that GhPHD genes may be involved in abiotic stress to improve plant tolerance in adverse environments.
Fig. 5.
The relative expression levels of 12 GhPHD genes under heat, cold, salt, and drought treatment. The relative expression levels were estimated by RT-qPCR. The error bars represent the standard deviations of three experiments
Identification of GhPHD genes in response to phytohormones
To further determine whether GhPHD genes respond to phytohormones, we treated four-week-old cotton seedlings with GA, MeJA, IAA, SA, and BL, and identified changes in the relative expression of GhPHD genes (Fig. 6). The relative expression level of GhPHD5 increases significantly after MeJA, IAA, and BL treatment. While GhPHD5 shows higher expression after 0.5 h after SA treatment indicating that GhPHD5 may respond to multiple phytohormones signal transduction pathway, which is consistent with the fact that GhPHD5 promoter contains cis-acting elements related to multiple phytohormones. GhPHD40 is significantly up-regulated under SA treatment, indicating that GhPHD40 responds positively to SA signal. Similarly, GhPHD43 is significantly up-regulated under all phytohormone treatments, especially under BL. The relative expression levels of GhPHD80 and GhPHD88 reach at peak after 0.5 h of GA treatment. The relative expression level of GhPHD88 increases gradually under SA treatment. Moreover, GhPHD107 expression significantly increases to the maximum level after 1 h of GA, IAA, and BL treatment. These results indicated that GhPHD genes are involved in regulating multiple phytohormone signal transduction pathways.
Fig. 6.
The relative expression levels of six GhPHD genes under GA, MeJA, IAA, SA, and BL treatment. The relative expression levels were estimated by RT-qPCR. The error bars show the standard deviation of three biological replicates
Co-expression network with functional modules for G. hirsutum and G. arboreum
Gene co-expression network analysis is a network diagram constructed on the basis of similarity of gene expression data, reflecting the relationship of expression regulation between genes [45]. We analyzed the co-expression network of GhPHD genes using ccNET software, and predicted many co-expressed genes and interaction proteins (Table S6). Among these, GhPHD5 is positively co-expressed with a plant-specific DNA ligase, which is related to seed germination and DNA repair. In addition, GhPHD5 is also positively co-expressed with SLOMO protein, which is a F-box protein required for auxin homeostasis and the normal timing of lateral organ initiation at the shoot meristem [46] illustrating that GhPHD5 may be involved in the regulation of auxin signal transduction pathway, and mediates seed germination and organ formation to regulate plant growth and development. Similarly, GhPHD18 interacts with highly hydrophilic proteins that regulate FLC (Flowering locus C) expression [47] and shows positively co-expressed with SHAGGY-related kinases involved in meristem organization, indicating that GhPHD18 may affect the flowering time of meristem. Further, GhPHD34 negatively co-expressed with ERF (Ethylene response factor) subfamily B-1, participating in ethylene signaling pathway and responding to abiotic stresses. GhPHD107 positively co-expressed with ARF-GAP and ERF genes, and may be involved in the signal pathways of auxin and ethylene. More interestingly, we predicted many proteins that interact with GhPHD88, such as leucine-rich repeat protein kinase (LRRK), late embryogenesis abundant (LEA) protein, AP2/B3 transcription factor, R2R3 factor, DREB subfamily A-2, cellulose synthase, gibberellin-regulated family protein (GRP), and ethylene response factor (ERF) (Fig. 7a and Table S6), suggesting that GhPHD88 may be involved in many physiological processes such as plant growth and development, phytohormone signal transduction, and stress response. Further, Gene Ontology (GO) analysis of GhPHDs indicated that protein binding and zinc ion binding are the most abundant functional terms (Fig. 7b), which is consistent with the existing results that the cysteine residues exhibit high affinity for zinc ions (Zn2+), and Zn2+-cysteine complexes are key medium for protein structure, catalysis, and regulation [48].
Fig. 7.
Co-expression networks analysis of GhPHD88 and GO enrichment analysis of 108 GhPHDs. a Co-expression network analysis of GhPHD88 with functional modules for G. hirsutum and G. arboreum. Yellow and green colour indicates that query protein and interaction proteins, respectively. There are four interaction lines, red lines indicated ortholog gene pairs in G. hirsutum and G. arboreum; pink lines and blue lines indicate proteins own interaction and positive/negative co-expression relationship with target protein; orange lines indicate proteins own interaction and protein-protein relationship with target protein. b GO enrichment analysis of all GhPHD genes
In summary, GhPHDs were involved in regulating cotton growth and development, especially ovule and fiber development. Further, GhPHDs not only respond to multiple phytohormones signal transduction pathways, but also improve cotton’s tolerance to adverse environments such as heat, salt, and drought. Particularly, GhPHD5, GhPHD80, GhPHD88 are prominent in their responses. Combining the predicted results of co-expressed genes and interacting proteins, we inferred that phytohormones could improve plant tolerance to abiotic stresses through GhPHD genes and their cofactors, but their regulatory mechanism and interaction network still need further research.
Discussion
Phylogenetic analysis and duplication
Phylogenetic tree was used to analyze the evolutionary relationship between PHD proteins in cotton, rice, and Arabidopsis. A total of 297 PHD proteins were divided into five groups (A-E). The relationship between cotton PHD proteins and AtPHD proteins was closer than that of OsPHD proteins, which is consistent with the evolutionary relationship between cotton, Arabidopsis, and rice. Although the G. arboreum genome is about twice that of the G. raimondii genome, however, more GrPHD proteins were identified than GaPHD proteins. Most PHD proteins from two diploids and one allotetraploid were closely distributed in phylogenetic tree, which is coherent with the fact that upland cotton evolved from the hybridization of A and D genomes [49].
We identified 108 GhPHD proteins in the G. hirsutum genome, which are more than previously identified PHD protein family members in Arabidopsis, maize, potato, and pear [30, 39, 40]. The main reason for the more number of GhPHDs is that upland cotton underwent polyploidization and promoted gene duplication. Upland cotton is an allotetraploid cotton produced by the hybridization between G. arboreum (A2 genome) and G. raimondii (D5 genome) [49]. The At and Dt subgenome donors of upland cotton are orthologous relatives and share the same number of ortholog genes, resulting in the duplication and doubling of GhPHD genes in upland cotton. Therefore, the sum total of GaPHD genes and GrPHD genes was approximately equal to the number of GhPHD genes. Previous studies have reported that gene duplication, including whole genome duplication, segment duplication, tandem duplication, and transposition events was the main reason for gene family expansion [50, 51]. In our study, a total of 77 duplicated gene pairs were identified in GhPHD family, including 73 segmental duplicated pairs and four tandem duplicated pairs (Table 2). The Ka/Ks values of most GhPHD duplication gene pairs was less than 1, which indicated that GhPHD family experienced strong purification selection pressure. Purification selection dominated the expansion of GhPHD genes, eliminated deleterious loss-of-function mutations at both duplicated loci, increased fixation, and retained the function of the new duplicated genes [52].
Conserved amino acid residues, protein motifs, and gene structure analysis
Conserved amino acid residues analysis showed that GhPHD domain was highly conserved during the process of evolution. The amino terminus of GhPHD domain contained the Cys4-His-Cys3 zinc finger motif composed of 50 to 80 amino acids with the regular arrangement of cysteine residues, an important medium for zinc ion binding and protein structure [48]. In addition, a total of three motifs were identified in GhPHD proteins and the motif distribution was relatively conservative, indicating that GhPHD proteins may play different physiological functions, and the subtle differences between GhPHD proteins in different clade may be related to cotton growth, development, and stress tolerance.
Gene structure may be determined by the insertion/deletion events and is an important parameter to predict gene evolution and new function generation [53]. Gene structure analysis indicated that the duplication genes showed similar gene structures with varied intron length indicating that the intron length may play major roles in the functional diversification of GhPHD genes. In this study, we found that the intron number varies from 1 to 25, but most GhPHD genes contained 2 to 11 introns which supported the previous research that cotton is a new evolution species that experience a decrease in the number of introns during the early stages of evolution [54].
GhPHD genes expression in tissues, abiotic and phytohormone stresses
Many studies demonstrated that PHD proteins are the main mediators of transcriptional regulation during plant developmental processes such as meiosis and postmeiotic events [55], germination [34], pollen maturation [56], flowering time [57], embryo meristem initiation, and root development [55, 58]. Gene’s expression profiles showed that GhPHDs may play important regulatory roles in cotton growth and development, especially during the development of ovule and fiber. In addition, we have also identified some GhPHD genes that respond to abiotic stress and phytohormones in upland cotton. The analysis of cis-acting elements and seedlings treatment experiments indicated that GhPHD genes may respond to abiotic stress and participate in the signal transduction of phytohormones. For example, GhPHD genes (GhPHD5, GhPHD40, GhPHD43, GhPHD80, and GhPHD88) respond positively to heat, salt, and drought and they may be important genetic materials for improving plant tolerance under adverse environments.
Research reports indicated that phytohormones may regulate the response to abiotic stress in plants. Auxin response factors (ARFs) are a type of transcription factors that regulate the expression of auxin-responsive genes [59, 60]. The significant up-regulation of the transcription level of ARFs under stress indicates that they are potential mediators for plants to respond to adverse environments [61, 62]. Ethylene response factors belong to the ERF subfamily of the AP2/ARF transcription factor family, and are widely involved in plant development, phytohormones response, disease resistance, and adversity response [63, 64]. In this study, co-expression network analysis indicated that GhPHD genes may improve plant tolerance to abiotic stresses by phytohormone signaling pathways. For instance, GhPHD5 may improve tolerance to heat, salt, and drought by regulating auxin homeostasis. Similarly, GhPHD34 and GhPHD107 may be involved in auxin and ethylene signal transduction pathways to improve heat tolerance and promote growth and development. GhPHD88 regulates the signal transduction of various phytohormones and abiotic stresses, and promotes growth and development. Although GhPHDs are indispensable in the course of life, the physiological functions of GhPHDs in crosstalk between abiotic stress and phytohormone need further study.
Conclusions
In this study, a total of 297 PHD proteins were identified in total five plant species including G. hirsutum, G. arboreum, G. raimondii, rice, and Arabidopsis. The PHD proteins were divided into five groups based on the phylogenetic analysis. Segmental duplication events were the main contributors toward the expansion of GhPHD gene family in upland cotton. Moreover, duplicated gene pairs of GhPHD gene family might have experienced functional divergence, since their expression patterns were different in different tissues. Tissues specific expression patterns indicated that GhPHDs are very important for growth and development, especially ovule and fiber development. The phytohormones and stresses treatment and co-expression network analysis showed that GhPHDs may improve the tolerance to adverse environments by phytohormones signal transduction pathway. Taken together, our study provides key basic knowledge to understand the functional mechanisms of cotton growth and development, as well as candidate genes for cotton breeding resistant to abiotic stresses and phytohormone stimulation.
Methods
Sequence retrieval, multiple sequence alignment, and phylogenetic analysis
The genome sequence and information of cotton (G. hirsutum, G. raimondii, and G. arboreum) were acquired from the CottonFGD (https://cottonfgd.org/) [65]. HMMER (https://www.ebi.ac.uk/Tools/hmmer/) software with default parameters was used to search for the corresponding protein sequences, and used the conserved PHD domain sequence as a query. We used BLAST program to further identify PHD sequences based on homology. The conserved domain of PHD proteins was predicted by Pfam [66] and SMART [67] software. Multiple sequence alignment of PHD proteins were performed using Clustal X [68]. MEGA 6.0 [69] was used to construct phylogenetic trees, using the neighbor-joining (NJ) algorithm with default parameters and 1000 bootstrap replicates. The molecular weight (MW), isoelectric point (pI), and GRAVY value of GhPHD proteins were predicted using ExPASy [70], and the subcellular localization of GhPHD proteins was predicted by the CELLO v2.5 server [71].
Chromosomal location, gene structure, and conserved motif
The positional information of GhPHD genes was obtained from the General Feature Format (GFF) file downloaded from the CottonFGD website [65]. GhPHDs were mapped on the chromosome using MapInspect (https://mapinspect.software.informer.com/). For the exon-intron structural analysis of GhPHD genes, the coding sequences were used to align their genomic DNA sequences and the structure diagram was drawn using the online Gene Structure Display Server (GSDS 2.0) program [72]. Conserved motifs of GhPHD proteins were investigated using the online toolkit Multiple Expectation maximization for Motif Elicitation (MEME 5.0.5) [73]. The optimized parameters of MEME are as follows: the number of repetitions, any; the maximum number of motifs, 50; and the optimum width of each motif, between 6 and 300 residues, and retaining only motifs associated with an E value < e− 5. The identified protein motifs were further annotated with TBtools [74].
Identification of cis-acting elements and gene expression pattern
The 1500 bp promoter sequence before the transcription start site of GhPHD genes were downloaded from the CottonFGD website [65]. The cis-acting elements in the GhPHD promoter regions were predicted using the Plant Cis-Acting Regulatory Element website [75]. The tissue expression patterns of GhPHD genes were analyzed using the online cotton transcriptome data, and heatmap was drawn by TBtools [74]. The transcriptome data of root, stem, leaf, petal, stamen, pistil, ovule (− 3, − 1, 0, 1, 3, 5, 10, 20, 25, 35 DPA) and fiber (5, 10, 20, 25 DPA) was used in this study. The ccNET software [76] was used to analyze the gene co-expression network relationship.
Plant material, abiotic stresses and phytohormones treatment
Upland cotton ZM24 is a short-season cotton variety selected by the Cotton Research Institute of Chinese Academy of Agricultural Sciences. Firstly, ZM24 seeds were pre-germinated in the conical flask filled with water at room temperature for 48 h. Pre-germinated seeds were then transferred to the liquid medium with a cultivation temperature of 30 °C, a photoperiod of 16 h light and 8 h dark. Four-week-old cotton seedlings were treated with brassinolide (BL, 10 μM), gibberellin (GA, 100 μM), indole-3-acetic acid (IAA, 100 μM), salicylic acid (SA, 10 μM), and methyl jasmonate (MeJA, 10 μM) for 0.5, 1, 3, and 6 h. Similarly, four-week-old cotton seedlings were treated with heat (38 °C), cold (4 °C), NaCl (200 mM), and polyethylene glycol (PEG) (20% mass fraction) for 1, 2, 4, and 6 h. In the experiment, the untreated sample was used as the control group. The collected leaves were immediately frozen in liquid nitrogen and stored at − 80 °C for RNA extraction and RT-qPCR analysis. For abiotic stresses and phytohormones treatment, a total of 20 cotton seedlings were used for each treatment and three biological replicates were performed for each experiment.
RNA extraction and RT-qPCR analysis
Total RNA of the collected cotton leaves was extracted using the RNAprep Pure Plant Kit (Polysaccharides & Polyphenolics-rich) (TianGen, Beijing, China). In order to synthesize the first-strand cDNA, the EasyScript All-in-One First-strand cDNA synthesis SuperMix for RT-qPCR kit (TransGen, Beijing, China) was used in accordance with the manufacturer’s protocol and the cDNA was used as template for subsequent RT-qPCR reaction. RT-qPCR was performed using TransStart Top Green qPCR SuperMix (TransGen, Beijing, China) in LightCycler 480 (Roche, Basel, Switzerland). Each PCR reaction was performed in triplicate, and three biological replicates were quantified. GhHistone 3 (GenBank accession no. AF024716) was used as an internal control [77]. The relative expression level was calculated as described previously [78]. The primers used for RT-qPCR analysis were listed in Table S7. For statistical analysis, the RT-qPCR data was considered as normal distribution and we conducted a two-tailed Student’s t-test in Microsoft Excel 2007.
Supplementary Information
Additional file 1: Fig. S1. Chromosomal location of GhPHD genes on 26 chromosomes in G. hirsutum. The chromosome numbers were shown on the top of each chromosome. The scale bar indicated the length in megabases (Mb)
Additional file 2: Fig. S2. Alignment results from the conserved domain of 108 GhPHD proteins and PHD motifs with a typical C4HC3 model
Additional file 3: Fig. S3. Expression profiles of GhPHD genes under cold, hot, salt, and drought. The expression characteristics of 108 GhPHD genes under four stress treatments were investigated using available transcriptomic data. 1 h, 3 h, 6 h, and 12 h indicate hours after different stress treatments. Gene names and the subfamilies are shown on the right. Blocks with colors represent the relative expression levels of GhPHDs
Additional file 4: Table S1. The PHD members from G. hirsutum, G. raimondii, G. arboreum, A. thaliana, and O. sativa
Additional file 5: Table S2. Chromosomal location and gene annotation of GhPHD genes in G. hirsutum
Additional file 6: Table S3. Transcript-features of 108 GhPHD genes
Additional file 7: Table S4. Distribution of major stress-related and phytohormone-related cis-acing elements in the promoter regions of GhPHD genes
Additional file 8: Table S5. Number of cis-acting elements in the promoters of GhPHD genes
Additional file 9: Table S6. Co-expression network analysis results
Additional file 10: Table S7. Primers for RT-qPCR in this study
Acknowledgements
None.
Abbreviations
- Gh
Gossypium hirsutum
- Ga
Gossypium arboreum;
- Gr
Gossypium raimondii
- Os
Oryza sativa
- At
Arabidopsis thaliana
- DPA
Day post-anthesis
- PHD
Plant homeodomain
- NJ
Neighbor-joining
- ARF
Auxin response factor
- MMD1
Male meiocyte death 1
- VIM1
Variant in methylation 1
- VIN3
Vernalization insensitive 3
- GSR1
Germostatin resistance locus 1
- BR
Brassinosteroide
- BL
Brassinolide
- GA
Gibberellin
- IAA
Indole-3-acetic acid
- SA
Salicylic acid
- MeJA
Methyl jasmonate
- ABA
Abscisic acid
- ET
Ethylene
- CK
Cytokinin
- PEG
Polyethylene glycol
- RT-qPCR
Quantitative real-time polymerase chain reaction
- GO
Gene Ontology
- WGD
Whole genome duplication
- FLC
Flowering locus C
- ERF
Ethylene response factor
- GRP
Gibberellin-regulated family protein
- LEA
Late embryogenesis abundant
- LRRK
Leucine-rich repeat protein kinase
Authors’ contributions
H.W. and G.Q. conceived and designed the experiments. H.W. and M.G. performed the experiment. H.W. and L.Z. analyzed the data. H.W. wrote the paper. Z.Y. and Z.W. revised the paper. All of the authors read and approved the final the manuscript.
Funding
This study was supported by the Major Research Plan of National Natural Science Foundation of China (grant number 31690093). The funder was not involved in the experimental design of the study, data collection and interpretation, and in writing the manuscript.
Availability of data and materials
The data used or analyzed during the current study has been included in this article and additional materials. The genome sequence and annotation datasets that supported the findings of this study are available in:
A. thaliana: https://www.arabidopsis.org
O. sativa: http://plants.ensembl.org/index.html
G. hirsutum, G. arboreum, and G. raimondii: https://cottonfgd.org/
Ethics approval and consent to participate
Not applicable. Our research did not involve in any human or animal subjects, material, or data. The plant materials used in this study were provided by the Institute of Cotton Research of Chinese Academy of Agricultural Sciences and are freely available for research purposes following institutional, national and international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Huanhuan Wu, Email: wuhuan621@163.com.
Lei Zheng, Email: zhengleiwangyi@126.com.
Ghulam Qanmber, Email: gqkhan12@gmail.com.
Mengzhen Guo, Email: gmz6600@126.com.
Zhi Wang, Email: wangzhi.12@163.com.
Zuoren Yang, Email: yangzuoren4012@163.com.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-020-02787-5.
References
- 1.Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014;19(6):371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hossain MA, Li Z-G, Hoque TS, Burritt DJ, Fujita M, Munné-Bosch S. Heat or cold priming-induced cross-tolerance to abiotic stresses in plants: key regulators and possible mechanisms. Protoplasma. 2018;255(1):399–412. doi: 10.1007/s00709-017-1150-8. [DOI] [PubMed] [Google Scholar]
- 3.Saeed M, Dahab A, Wangzhen G, Tianzhen Z. A cascade of recently discovered molecular mechanisms involved in abiotic stress tolerance of plants. Omics. 2012;16(4):188–199. doi: 10.1089/omi.2011.0109. [DOI] [PubMed] [Google Scholar]
- 4.Planas-Riverola A, Gupta A, Betegón-Putze I, Bosch N, Ibañes M, Caño-Delgado AI. Brassinosteroid signaling in plant development and adaptation to stress. Development. 2019;146(5):dev151894. doi: 10.1242/dev.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishna P, Prasad BD, Rahman T. Brassinosteroid Action in Plant Abiotic Stress Tolerance. Methods Mol Biol (Clifton, NJ) 2017;1564:193–202. doi: 10.1007/978-1-4939-6813-8_16. [DOI] [PubMed] [Google Scholar]
- 6.Farhangi-Abriz S, Ghassemi-Golezani K. Jasmonates: mechanisms and functions in abiotic stress tolerance of plants. Biocatalysis Agric Biotechnol. 2019;20:101210. doi: 10.1016/j.bcab.2019.101210. [DOI] [Google Scholar]
- 7.Colebrook EH, Thomas SG, Phillips AL, Hedden P. The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol. 2014;217(1):67–75. doi: 10.1242/jeb.089938. [DOI] [PubMed] [Google Scholar]
- 8.Khan MIR, Fatma M, Per TS, Anjum NA, Khan NA. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front Plant Sci. 2015;6:462. doi: 10.3389/fpls.2015.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey V, Bhatt ID, Nandi SK. Chapter 20 - Role and Regulation of Auxin Signaling in Abiotic Stress Tolerance. In: Khan MIR, Reddy PS, Ferrante A, Khan NA, editors. Plant Signaling Molecules. Cambridge: Woodhead Publishing; 2019. p. 319–31.
- 10.Leng P, Yuan B, Guo Y. The role of abscisic acid in fruit ripening and responses to abiotic stress. J Exp Bot. 2014;65(16):4577–4588. doi: 10.1093/jxb/eru204. [DOI] [PubMed] [Google Scholar]
- 11.Mehrotra R, Bhalothia P, Bansal P, Basantani MK, Bharti V, Mehrotra S. Abscisic acid and abiotic stress tolerance - different tiers of regulation. J Plant Physiol. 2014;171(7):486–496. doi: 10.1016/j.jplph.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Takatsuji H, Mori M, Benfey PN, Ren L, Chua NH. Characterization of a zinc finger DNA-binding protein expressed specifically in Petunia petals and seedlings. EMBO J. 1992;11(1):241–249. doi: 10.1002/j.1460-2075.1992.tb05047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai H, Medrano LJ, Meyerowitz EM. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature. 1995;378(6553):199–203. doi: 10.1038/378199a0. [DOI] [PubMed] [Google Scholar]
- 14.Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261(5120):438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- 15.Daniel-Vedele F, Caboche M. A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol Gen Genet. 1993;240(3):365–373. doi: 10.1007/BF00280388. [DOI] [PubMed] [Google Scholar]
- 16.Yanagisawa S. Dof DNA-binding proteins contain a novel zinc finger motif. Trends Plant Sci. 1996;1(7):213–214. doi: 10.1016/1360-1385(96)86897-X. [DOI] [Google Scholar]
- 17.Yanagisawa S, Izui K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J Biol Chem. 1993;268(21):16028–16036. doi: 10.1016/S0021-9258(18)82353-5. [DOI] [PubMed] [Google Scholar]
- 18.von Arnim AG, Deng XW. Ring finger motif of Arabidopsis thaliana COP1 defines a new class of zinc-binding domain. J Biol Chem. 1993;268(26):19626–19631. doi: 10.1016/S0021-9258(19)36562-7. [DOI] [PubMed] [Google Scholar]
- 19.Schindler U, Beckmann H, Cashmore AR. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 1993;4(1):137–150. doi: 10.1046/j.1365-313X.1993.04010137.x. [DOI] [PubMed] [Google Scholar]
- 20.Bellmann R, Werr W. Zmhox1a, the product of a novel maize homeobox gene, interacts with the shrunken 26 bp feedback control element. EMBO J. 1992;11(9):3367–3374. doi: 10.1002/j.1460-2075.1992.tb05415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Garcia I, Rabbitts TH. The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10(9):315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 22.Baltz R, Domon C, Pillay DT, Steinmetz A. Characterization of a pollen-specific cDNA from sunflower encoding a zinc finger protein. Plant J. 1992;2(5):713–721. [PubMed] [Google Scholar]
- 23.Kaadige MR, Ayer DE. The polybasic region that follows the plant homeodomain zinc finger 1 of Pf1 is necessary and sufficient for specific phosphoinositide binding. J Biol Chem. 2006;281(39):28831–28836. doi: 10.1074/jbc.M605624200. [DOI] [PubMed] [Google Scholar]
- 24.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31(1):35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol. 2006;26(21):7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy TV, Kaur J, Agashe B, Sundaresan V, Siddiqi I. The DUET gene is necessary for chromosome organization and progression during male meiosis in Arabidopsis and encodes a PHD finger protein. Development. 2003;130(24):5975–5987. doi: 10.1242/dev.00827. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Makaroff CA, Ma H. The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell. 2003;15(6):1281–1295. doi: 10.1105/tpc.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreuzza S, Nishal B, Singh A, Siddiqi I. The chromatin protein DUET/MMD1 controls expression of the meiotic gene TDM1 during male meiosis in Arabidopsis. PLoS Genet. 2015;11(9):e1005396. doi: 10.1371/journal.pgen.1005396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo HR, Pontes O, Pikaard CS, Richards EJ. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 2007;21(3):267–277. doi: 10.1101/gad.1512007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Liu J, Wang Y, Zhao Y, Jiang H, Cheng B. Systematic analysis of the maize PHD-finger gene family reveals a subfamily involved in abiotic stress response. Int J Mol Sci. 2015;16(10):23517–23544. doi: 10.3390/ijms161023517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alam I, Liu CC, Ge HL, Batool K, Yang YQ, Lu YH. Genome wide survey, evolution and expression analysis of PHD finger genes reveal their diverse roles during the development and abiotic stress responses in Brassica rapa L. BMC Genomics. 2019;20(1):773. doi: 10.1186/s12864-019-6080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei W, Huang J, Hao YJ, Zou HF, Wang HW, Zhao JY, Liu XY, Zhang WK, Ma B, Zhang JS, et al. Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS One. 2009;4(9):e7209. doi: 10.1371/journal.pone.0007209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim DH, Sung S. Accelerated vernalization response by an altered PHD-finger protein in Arabidopsis. Plant Signal Behav. 2017;12(5):e1308619. doi: 10.1080/15592324.2017.1308619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molitor AM, Bu Z, Yu Y, Shen WH. Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 2014;10(1):e1004091. doi: 10.1371/journal.pgen.1004091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye Y, Gong Z, Lu X, Miao D, Shi J, Lu J, Zhao Y. Germostatin resistance locus 1 encodes a PHD finger protein involved in auxin-mediated seed dormancy and germination. Plant J. 2016;85(1):3–15. doi: 10.1111/tpj.13086. [DOI] [PubMed] [Google Scholar]
- 36.Wu S, Wu M, Dong Q, Jiang H, Cai R, Xiang Y. Genome-wide identification, classification and expression analysis of the PHD-finger protein family in Populus trichocarpa. Gene. 2016;575(1):75–89. doi: 10.1016/j.gene.2015.08.042. [DOI] [PubMed] [Google Scholar]
- 37.Gao Y, Liu H, Wang Y, Li F, Xiang Y. Genome-wide identification of PHD-finger genes and expression pattern analysis under various treatments in moso bamboo (Phyllostachys edulis) Plant Physiol Biochem. 2018;123:378–391. doi: 10.1016/j.plaphy.2017.12.034. [DOI] [PubMed] [Google Scholar]
- 38.Wu X-J, Li M-Y, Que F, Wang F, Xu Z-S, Xiong A-S. Genome-wide analysis of PHD family transcription factors in carrot (Daucus carota L.) reveals evolution and response to abiotic stress. Acta Physiol Plant. 2016;38(3):67. doi: 10.1007/s11738-016-2086-x. [DOI] [Google Scholar]
- 39.Qin M, Luo W, Zheng Y, Guan H, Xie X. Genome-wide identification and expression analysis of the PHD-finger gene family in Solanum tuberosum. PLoS One. 2019;14(12):e0226964. doi: 10.1371/journal.pone.0226964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao Y, Han Y, Meng D, Abdullah M, Li D, Jin Q, Lin Y, Cai Y. Systematic analysis and comparison of the PHD-finger gene family in Chinese pear (Pyrus bretschneideri) and its role in fruit development. Funct Integr Genomics. 2018;18(5):519–531. doi: 10.1007/s10142-018-0609-9. [DOI] [PubMed] [Google Scholar]
- 41.Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 42.Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46(6):567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 43.Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492(7429):423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 44.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 45.Rao X, Dixon RA. Co-expression networks for plant biology: why and how. Acta Biochim Biophys Sin. 2019;51(10):981–988. doi: 10.1093/abbs/gmz080. [DOI] [PubMed] [Google Scholar]
- 46.Lohmann D, Stacey N, Breuninger H, Jikumaru Y, Muller D, Sicard A, Leyser O, Yamaguchi S, Lenhard M. SLOW MOTION is required for within-plant auxin homeostasis and normal timing of lateral organ initiation at the shoot meristem in Arabidopsis. Plant Cell. 2010;22(2):335–348. doi: 10.1105/tpc.109.071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20(7):898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pace NJ, Weerapana E. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014;4(2):419–434. doi: 10.3390/biom4020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wendel J, Grover C. Taxonomy and evolution of the cotton genus, Gossypieum. 2015. [Google Scholar]
- 50.Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16(7):1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183(3):557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka KM, Takahasi KR, Takano-Shimizu T. Enhanced fixation and preservation of a newly arisen duplicate gene by masking deleterious loss-of-function mutations. Genet Res. 2009;91(4):267–280. doi: 10.1017/S0016672309000196. [DOI] [PubMed] [Google Scholar]
- 53.Xu G, Guo C, Shan H, Kong H. Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci U S A. 2012;109(4):1187–1192. doi: 10.1073/pnas.1109047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roy SW, Penny D. Patterns of intron loss and gain in plants: intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol Biol Evol. 2007;24(1):171–181. doi: 10.1093/molbev/msl159. [DOI] [PubMed] [Google Scholar]
- 55.Sebastian J, Ravi M, Andreuzza S, Panoli AP, Marimuthu MP, Siddiqi I. The plant adherin AtSCC2 is required for embryogenesis and sister-chromatid cohesion during meiosis in Arabidopsis. Plant J. 2009;59(1):1–13. doi: 10.1111/j.1365-313X.2009.03845.x. [DOI] [PubMed] [Google Scholar]
- 56.Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson ZA, Zhang D. PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal CELL death and pollen development in rice. Plant Physiol. 2011;156(2):615–630. doi: 10.1104/pp.111.175760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopez-Gonzalez L, Mouriz A, Narro-Diego L, Bustos R, Martinez-Zapater JM, Jarillo JA, Pineiro M. Chromatin-dependent repression of the Arabidopsis floral integrator genes involves plant specific PHD-containing proteins. Plant Cell. 2014;26(10):3922–3938. doi: 10.1105/tpc.114.130781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlereth A, Moller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jurgens G, Weijers D. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464(7290):913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- 59.Chandler JW. Auxin response factors. Plant Cell Environ. 2016;39(5):1014–1028. doi: 10.1111/pce.12662. [DOI] [PubMed] [Google Scholar]
- 60.Roosjen M, Paque S, Weijers D. Auxin response factors: output control in auxin biology. J Exp Bot. 2018;69(2):179–188. doi: 10.1093/jxb/erx237. [DOI] [PubMed] [Google Scholar]
- 61.Bouzroud S, Gouiaa S, Hu N, Bernadac A, Mila I, Bendaou N, Smouni A, Bouzayen M, Zouine M. Auxin response factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum) PLoS One. 2018;13(2):e0193517. doi: 10.1371/journal.pone.0193517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Z, Yuan Y, Fu D, Shen C, Yang Y. Identification and Expression Profiling of the Auxin Response Factors in Dendrobium officinale under Abiotic Stresses. Int J Mol Sci. 2017;18(5):927. doi: 10.3390/ijms18050927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Müller M, Munné-Bosch S. Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 2015;169(1):32–41. doi: 10.1104/pp.15.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang PY, Catinot J, Zimmerli L. Ethylene response factors in Arabidopsis immunity. J Exp Bot. 2016;67(5):1231–1241. doi: 10.1093/jxb/erv518. [DOI] [PubMed] [Google Scholar]
- 65.Zhu T, Liang C, Meng Z, Sun G, Meng Z, Guo S, Zhang R. CottonFGD: an integrated functional genomics database for cotton. BMC Plant Biol. 2017;17(1):101. doi: 10.1186/s12870-017-1039-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26(1):320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(D1):D493–d496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 69.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(Web Server issue):W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu C-S, Lin C-J, Hwang J-K. Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13(5):1402–1406. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen C, Xia R, Chen H, He Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv. 2018:289660.
- 75.Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.You Q, Xu W, Zhang K, Zhang L, Yi X, Yao D, Wang C, Zhang X, Zhao X, Provart NJ, et al. ccNET: Database of co-expression networks with functional modules for diploid and polyploid Gossypium. Nucleic Acids Res. 2017;45(D1):D1090–d1099. doi: 10.1093/nar/gkw910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wan Q, Guan X, Yang N, Wu H, Pan M, Liu B, Fang L, Yang S, Hu Y, Ye W, et al. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol. 2016;210(4):1298–1310. doi: 10.1111/nph.13860. [DOI] [PubMed] [Google Scholar]
- 78.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Chromosomal location of GhPHD genes on 26 chromosomes in G. hirsutum. The chromosome numbers were shown on the top of each chromosome. The scale bar indicated the length in megabases (Mb)
Additional file 2: Fig. S2. Alignment results from the conserved domain of 108 GhPHD proteins and PHD motifs with a typical C4HC3 model
Additional file 3: Fig. S3. Expression profiles of GhPHD genes under cold, hot, salt, and drought. The expression characteristics of 108 GhPHD genes under four stress treatments were investigated using available transcriptomic data. 1 h, 3 h, 6 h, and 12 h indicate hours after different stress treatments. Gene names and the subfamilies are shown on the right. Blocks with colors represent the relative expression levels of GhPHDs
Additional file 4: Table S1. The PHD members from G. hirsutum, G. raimondii, G. arboreum, A. thaliana, and O. sativa
Additional file 5: Table S2. Chromosomal location and gene annotation of GhPHD genes in G. hirsutum
Additional file 6: Table S3. Transcript-features of 108 GhPHD genes
Additional file 7: Table S4. Distribution of major stress-related and phytohormone-related cis-acing elements in the promoter regions of GhPHD genes
Additional file 8: Table S5. Number of cis-acting elements in the promoters of GhPHD genes
Additional file 9: Table S6. Co-expression network analysis results
Additional file 10: Table S7. Primers for RT-qPCR in this study
Data Availability Statement
The data used or analyzed during the current study has been included in this article and additional materials. The genome sequence and annotation datasets that supported the findings of this study are available in:
A. thaliana: https://www.arabidopsis.org
O. sativa: http://plants.ensembl.org/index.html
G. hirsutum, G. arboreum, and G. raimondii: https://cottonfgd.org/