Abstract
BACKGROUND
Accumulating evidence has revealed that several long non-coding ribonucleic acids (lncRNAs) are crucial in the progress of hepatocellular carcinoma (HCC).
AIM
To classify a long non-coding RNA, i.e., lncRNA W5, and to determine the clinical significance and potential roles of lncRNA W5 in HCC.
METHODS
The results showed that lncRNA W5 expression was significantly downregulated in HCC cell lines and tissues. Analysis of the association between lncRNA W5 expression levels and clinicopathological features suggested that low lncRNA W5 expression was related to large tumor size (P < 0.01), poor histological grade (P < 0.05) and serious portal vein tumor thrombosis (P < 0.05). Furthermore, Kaplan-Meier survival analysis showed that low expression of lncRNA W5 predicts poor overall survival (P = 0.016).
RESULTS
Gain-of-loss function experiments, including cell counting kit8 assays, colony formation assays, and transwell assays, were performed in vitro to investigate the biological roles of lncRNA W5. In vitro experiments showed that ectopic overexpression of lncRNA W5 suppressed HCC cell proliferation, migration and invasion; conversely, silencing of lncRNA W5 promoted cell proliferation, migration and invasion. In addition, acting as a tumor suppressor gene in HCC, lncRNA W5 inhibited the growth of HCC xenograft tumors in vivo.
CONCLUSION
These results showed that lncRNA W5 is down-regulated in HCC, and it may suppress HCC progression and predict poor clinical outcomes in patients with HCC. LncRNA W5 may serve as a potential HCC prognostic biomarker in addition to a therapeutic target.
Keywords: Hepatocellular carcinoma, Long non-coding ribonucleic acid, Long non-coding ribonucleic acid W5
Core Tip: Our results showed that the expression of long non-coding ribonucleic acid (lncRNA) W5 was considerably reduced in hepatocellular carcinoma (HCC) tissues, which suppressed proliferation, migration and invasion of tumor cells in vitro. It was also shown that low expression of lncRNA W5 correlated with tumor progression and poor prognosis. Furthermore, manipulation of lncRNA W5 expression affected the biological behavior of HCC. These results suggest that lncRNA W5 may serve as a tumor suppressor in the development and progression of HCC, and has the potential to be a diagnostic and therapeutic target in the clinical management of HCC.
INTRODUCTION
Around 90% of primary liver cancer cases are hepatocellular carcinoma (HCC)[1], approximately 600000 deaths occur globally each year, and half of all deaths occur in China[2,3]. In addition to hepatic resection, liver transplantation, chemotherapy and molecular targeted therapy are often carried out to improve outcomes in patients with HCC; however, the 5-year survival rate of patients with HCC still remains poor[4,5]. The specific signaling mechanisms underlying the development and progression of HCC remain to be defined.
The long non-coding ribonucleic acids (lncRNAs) are a class of noncoding RNAs with more than 200 nucleotides which have no protein-coding potential. Several reports have shown that lncRNAs are critical in numerous biological processes, including tumor development, differentiation, and tumorigenesis[6-10]. The expression of lncRNAs is dysregulated in cancer. Of note, specific lncRNAs are related to cancer recurrence, metastasis and prognosis in various cancers, including HCC[11-14]. To date, several lncRNA have been reported to be associated with the growth and advance of HCC, such as HULC[15], H19[16], MEG3[17], ZFAS1[18], P7[19], ATB[20], GAS8-AS1[21], and so on. In our previous study, we profiled the the expression of lncRNAs in influenza virus infected patients and identified panels of uncharacterized lncRNAs. In the current study, we classified the lncRNA W5 (mitochondrial translation optimization 1homologue; hsa_lncRNA_0007874/hsa_lncRNA_104135) which is notably down-regulated in HCC tissues and strictly associated with the prognosis of HCC patients. Furthermore, we investigated its roles, underlying mechanisms and clinical significance in HCC progression.
MATERIALS AND METHODS
Clinical samples
A total of 86 resected HCC tissues and matched tumor-adjacent tissues were kindly provided by the Department of Hepatobiliary Surgery of the Fifth Medical Center, Chinese PLA General Hospital between October 2013 and June 2018. Tumor tissues and adjacent non-tumor tissue specimens were obtained from the patients after informed consent in accordance with the institutional guidelines of the hospital’s Ethics Committee. Table 1 indicates the clinical and pathological characteristics of HCC patients obtained from clinical records.
Table 1.
Association of long non-coding ribonucleic acid W5 expression with clinicopathologic features in hepatocellular carcinoma patients
| Parameters | Group | Total |
W5 expression |
P value | |
| Low | High | ||||
| Gender | Male | 70 | 34 | 36 | 0.791 |
| Female | 16 | 10 | 6 | ||
| Age (yr) | ≤ 60 | 30 | 15 | 15 | 0.579 |
| > 60 | 56 | 28 | 28 | ||
| Tumor size (cm) | < 3 cm | 21 | 6 | 15 | 0.007b |
| ≥ 3 cm | 65 | 38 | 27 | ||
| AFP | < 20 | 39 | 21 | 18 | 0.328 |
| ≥ 20 | 47 | 22 | 25 | ||
| Histological grade | Well/moderate-poor | 52/34 | 5/28 | 47/6 | 0.046a |
| Clinical stage | I/II | 50 | 23 | 27 | 0.501 |
| Tumor number | III | 36 | 20 | 16 | |
| Solitary | 76 | 36 | 40 | 0.312 | |
| Multiple | 10 | 7 | 3 | ||
| Drinking status | Yes | 42 | 24 | 18 | 0.276 |
| No | 44 | 19 | 25 | ||
| Smoking status | Yes | 36 | 18 | 18 | 0.584 |
| No | 50 | 24 | 26 | ||
| PVTT | Yes | 32 | 22 | 10 | 0.024a |
| No | 54 | 21 | 33 | ||
| Microvascular invasion | Yes | 70 | 36 | 34 | 0.568 |
| No | 16 | 6 | 10 | ||
| Liver cirrhosis | Absence | 59 | 30 | 29 | 0.489 |
| HBV | Yes | 55 | 25 | 30 | 0.365 |
| No | 31 | 18 | 13 | ||
P < 0.05.
P < 0.01. PVTT: Portal vein tumor thrombosis; HBV: Hepatitis B virus.
Cell lines
All human cell lines were provided by the Experimental Center of the Fifth Medical Center, Chinese PLA General Hospital (Beijing, China), and included the normal non-malignant liver cell line LO2 and HCC cell lines Huh7, MHCC-97L, MHCC-97H, PLC, Hep3B and HCCLM3. All cell lines were maintained in DMEM (Gibco, Beijing, China) and were supplemented with 10% fetal bovine serum (Gibco, Beijing, China) in an incubator at 37°C with 5% CO2.
RNA extraction and reverse transcription-polymerase chain reaction
TRIzol reagent (Invitrogen, United States) was used to isolate total RNA from HCC cells and tissues, and first strand complementary deoxyribonucleic acid (cDNA) was synthesized by the use of reverse transcriptase. Quantitative real-time polymerase chain reaction (PCR) was conducted using the SYBR Green PCR kit (Thermo Fisher Scientific, United States). All reactions were performed on the ABI 7500 system (Applied Biosystems). The lncRNA W5-specific reverse transcription-polymerase chain reaction (qRT-PCR) primers used were as follows: forward: 5'-AAGGAGAACACAAAACAGGCAT-3', reverse: 5'-TGTGAAGCCCTAG ATTTCCCAT-3'; GAPDH forward: 5'-AAG GAGAACACAAAACAG GCAT-3' reverse: 5'-TGTGAAGCCCTAGATTTCCCAT-3'. Human GAPDH gene was amplified as an internal control.
Vector construction
The lncRNA W5 vector was constructed and sub-cloned into the pcDNA3.1 (+) vector at the BamHI and EcoRI sites, which produced pcDNA3.1-lncRNA W5. The primers used were as follows: forward: 5’-GCGCGGATCCACTGACTCTTTTCGTTAAGC-3’, reverse: 5’-CGCGCGGAATTCATGTTGACTTAAGTTCAGG-3’. Empty vector pcDNA3.1 (+) was used as a negative control. The lncRNA W5 and control were transfected into HCC cells using Polyplus (Invitrogen) according to the manufacturer’s instructions and cultured on six-well plates, respectively.
Cell proliferation assay
Cell proliferation experiments were performed using the CCK-8 kit (Dojindo Laboratories) according to the manufacturer’s protocol. Briefly, a Huh7 or LM3 cell suspension was adjusted to a final cell concentration of 5 × 103/mL and then added to a 96-well plate. HCC cells were cultured for the indicated time points, and then 10 μL of CCK-8 (5 mg/mL) was added to each well. The cell culture plate was placed in the incubator for 1 h, and the absorbance was measured at 450 nm per well using a Thermomax plate reader (Thermo Fisher, China).
Colony formation assay
Huh7 or LM3 cells were completely dispersed into individual cells in 6-well plates and incubated at 37°C in DMEM with 10% fetal bovine serum, respectively. After 14 d, the cell colonies were washed with PBS, fixed in 4% paraformaldehyde for 20 min, and stained with crystal violet for 20 min. Photographs were subsequently taken and only colonies containing more than 50 cells were recorded.
Cell migration and invasion assays
A chamber assay with Matrigel (invasion) or without Matrigel (migration) was performed at least in triplicate. Twenty-four-well chambers with 8 μm pore size were used in this experiment. Briefly, cells were added to the top chamber without Matrigel (migration) or with Matrigel (invasion) in the 24-well plate (Corning). The medium with 15% forward-based system was added to the lower chambers. After incubator for 48-72 h, the DMEM medium was removed and the cells were washed with PBS, and carefully removed from the top chamber with a cotton swab. The cells were fixed with 4% paraformaldehyde, stained with crystal violet, and then photographed in five randomly selected microscope fields.
In vivo tumor growth
Athymic BALB/C mice (4-6 wk old) were purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China) and maintained in a SPF facility. Huh7 cells (5 × 106) over-expressing lncRNA W5 were subcutaneously injected into the flanks of nude mice. Tumor length (L) and width (W) were measured using calipers every 3 d up to 6 wk. Tumor volume was estimated using the formula: π × length × width 2/6. After 6 wk, the mice were sacrificed, and tumor volumes and weights were examined. Proliferation progression was examined and quantified using a noninvasive bioluminescence in vivo Imaging System (Xenogen Corporation, Alameda, CA, United States). All animal experiments were conducted with the approval of the Fifth Medical Center of Chinese PLA General Hospital’s Animal Care and Use Committee.
Statistical analysis
All statistical analyses were performed using the SPSS 20.0 software package (Chicago, IL, United States). Data were expressed as the mean ± SD. Kaplan-Meier analysis was used to determine whether there was a correlation between the expression of lncRNA W5 and overall survival rate of HCC patients. P < 0.05 was considered significant.
RESULTS
HCC cancer tissues and cell lines have low expression of lncRNA W5
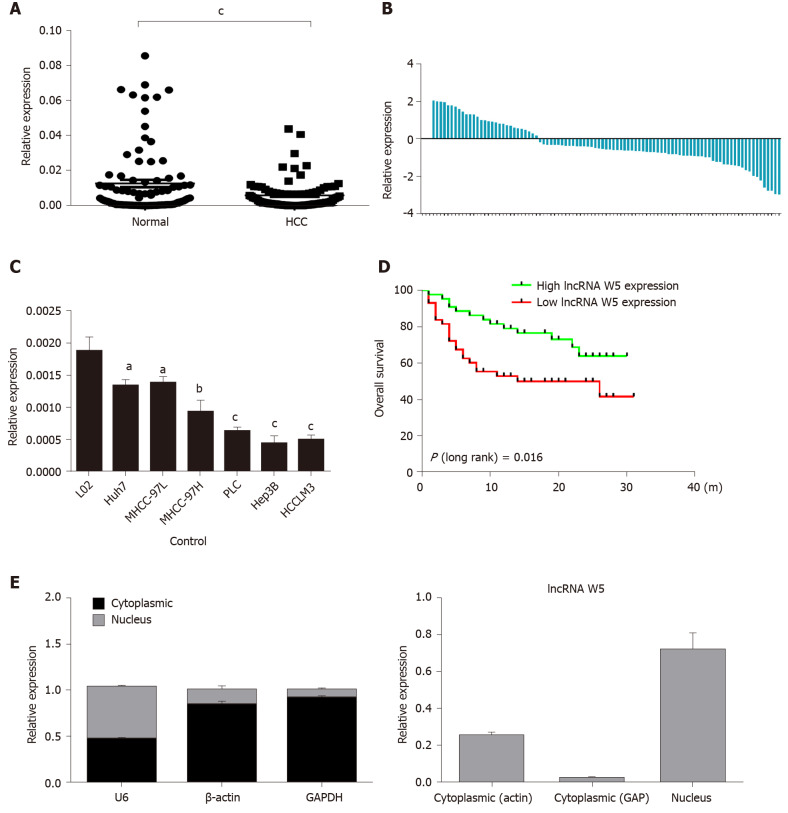
Initially, to investigate the potential role of lncRNA W5 in HCC tumorigenesis, we determined the expression of lncRNA W5 in 86 sets of HCC tissues and non-tumor tissues by qRT-PCR. As shown in Figure 1, the expression of lncRNA W5 was significantly reduced in HCC tissues compared with adjacent non-tumor tissues (P < 0.001, Figure 1A and B). In addition, we determined the expression of lncRNA W5 in regular liver cells (LO2) and six HCC cancer cell lines (Huh7, MHCC-97L, MHCC-97H, PLC, Hep3B and HCCLM3). The results revealed that lncRNA W5 expression was significantly downregulated in the six HCC cancer cells compared with the regular liver cell line LO2 (Figure 1C). Levels of lncRNA W5 expression were relatively lower in the Huh7 and LM3 HCC cell lines, and were used in subsequent studies. More importantly, a Kaplan-Meier survival analysis indicated that HCC patients with low expression of lncRNA W5 had shorter overall survival than those patients with high expression of lncRNA W5 (P = 0.016) (Figure 1D). Cox survival analysis was then used to further confirm the prognostic value of lncRNA W5 in HCC. Univariate analysis showed that the analyzed variables (lncRNA W5 expression, Pathologic-stage and Pathologic-TMN) were markedly associated with the overall survival time of HCC patients. Furthermore, multivariate analysis revealed that lncRNA W5 expression (P = 0.027), Pathologic-T (P = 0.014) and Pathologic-M (P = 0.005) were promising independent prognostic factors of HCC (Table 2). Thus, lncRNA W5 could be used as an independent prognostic factor. Finally, we also measured the expression of lncRNA W5 in nuclear and cytosolic fractions of Huh7 cells by qRT-PCR. The differential enrichments of GAPDH, β-actin and U1 RNA were used as fractionation indicators. Subcellular fractionation location results showed that lncRNA W5 was mainly located in the nucleus (Figure 1E), thus suggesting that lncRNA W5 might play an essential regulatory function at the transcriptional level.
Figure 1.
Expression of long non-coding ribonucleic acid W5 is downregulated in hepatocellular carcinoma tissues and cells. A: The expression of long non-coding ribonucleic acid (lncRNA) W5 was detected by reverse transcription-polymerase chain reaction (qRT-PCR) in tumor tissues and non-adjacent normal tissues of hepatocellular carcinoma (HCC) patients (n = 86). LncRNA W5 expression was normalized to GAPDH expression; B: The expression of lncRNA W5 was detected by qRT-PCR in tumor tissues and non-adjacent normal tissues of 86 HCC patients; C: The expression levels of lncRNA W5 in a series of HCC cell lines were reduced compared to that in LO2 cells; D: Analysis of overall survival based on lncRNA W5 expression levels is shown in 86 HCC patients; and E: Subcellular localization of lncRNA W5 in Huh7 cells was examined by qRT-PCR. GAPDH, β-actin and U1 were considered as the control markers, respectively. aP < 0.05; bP < 0.01; cP < 0.001. HCC: Hepatocellular carcinoma; lncRNA: Long non-coding ribonucleic acid; qRT-PCR: Reverse transcription-polymerase chain reaction.
Table 2.
Cox proportional hazards model analysis of clinicopathologic features related to overall survival in terms of long non-coding ribonucleic acid W5 expression in hepatocellular carcinoma patients
| Variables |
Univariate analysis |
Multivariate analysis |
||||
| P value | HR | 95%CI | P value | HR | 95%CI | |
| Expression (high/low) | 0.041a | 1.672 | 1.032-2.159 | 0.027a | 1.285 | 0.867-2.155 |
| Pathologic-Stage (I + II/III + IV) | 0.001a | 2.584 | 1.934-4.162 | 0.159 | 2.436 | 0.715-7.667 |
| Pathologic-T (T1 + T2/T3 + T4) | 0.004a | 4.068 | 1.573-8.869 | 0.014a | 10.638 | 2.314-57.649 |
| Pathologic-M (M0/M1) | 0.007a | 5.294 | 3.195-7.657 | 0.005a | 3.082 | 1.726-5.342 |
| Pathologic-N (N0/N1 + N2 + N3) | 0.002a | 2.413 | 1.519-3.969 | 0.246 | 0.719 | 0.312-1.911 |
| Age (< 60/ ≥ 60 yrs) | 0.342 | 1.402 | 0.914-2.357 | |||
| Gender (female/male) | 0.258 | 1.324 | 0.849-1.905 | |||
P < 0.05. HR: Hazard ratio; CI: Confidence interval.
To investigate the relationship between the expression of lncRNA W5 and clinicopathological characteristics, lncRNA W5 expression was detected in 86 HCC patients and distributed into two groups (high–high expression of lncRNA W5 and low–low expression of lncRNA W5) based on the median lncRNA W5 expression. The correlations between lncRNA W5 expression and clinical parameters were analyzed and it was found that low expression of lncRNA W5 was linked to large tumor size (P < 0.01), poor histological grade (P < 0.05) and serious portal vein tumor thrombosis (P < 0.05). Nevertheless, no significant correlation was observed between the expression of lncRNA W5 and other clinicopathological features, such as age, gender, AFP levels, tumor number and with/without HBV infection.
In vitro effects of lncRNA W5 on HCC cell proliferation
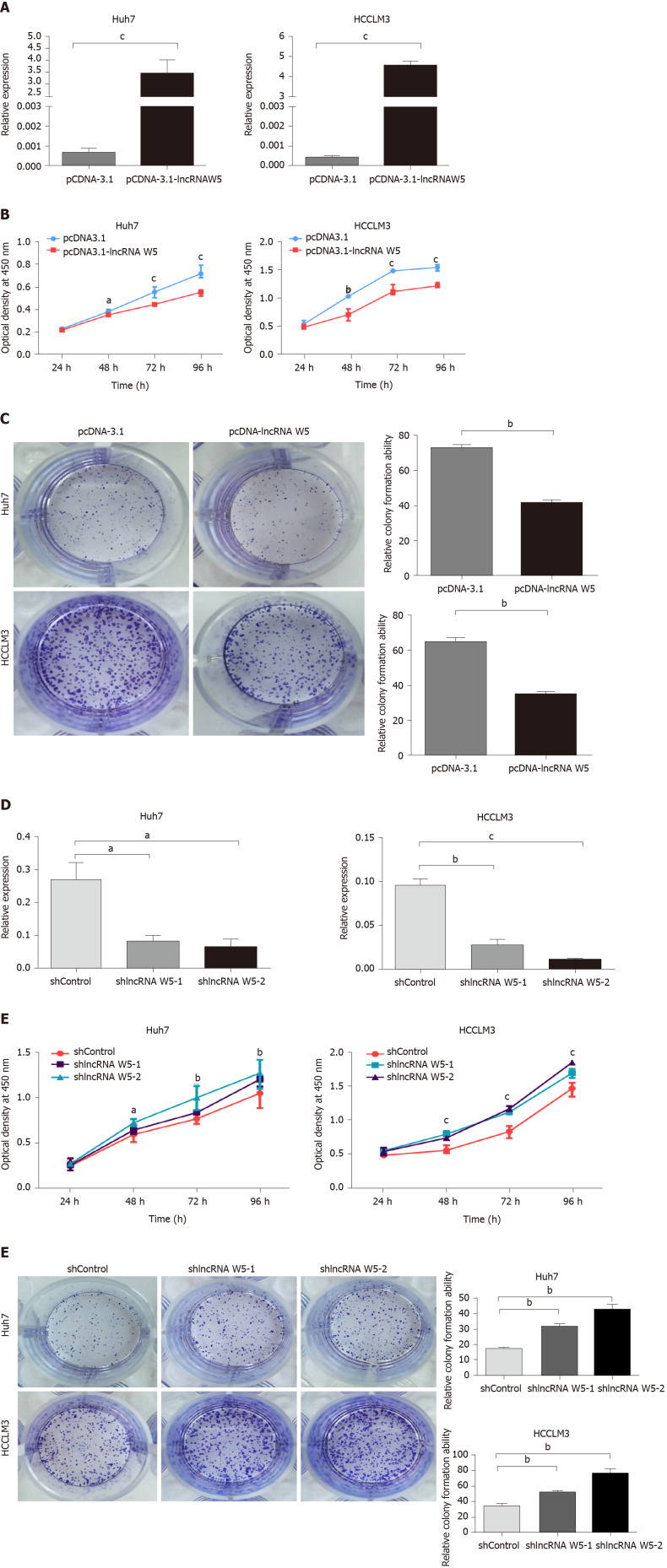
To assess the role of lncRNA W5 in regulating the biological behavior of HCC cells, we used the lncRNA W5 expression vector pcDNA3.1-lncRNA W5 and overexpressed lncRNA W5 in the HCC cell lines Huh7 and LM3. LncRNA W5 overexpression in the two cell lines was verified by RT-qPCR (Figure 2A). CCK-8 assays, which were used to show overexpression of lncRNA W5 in the HCC cell lines Huh7 and LM3, demonstrated a significant reduction in cell proliferation from 48 h to 96 h (Figure 2B). Accordingly, colony formation assays, which showed that Huh7 and LM3 cells transfected with lncRNA W5, resulted in significantly decreased clonogenic survival than empty vector control Huh7 and LM3 cell lines (Figure 2C). In addition, we constructed shRNA-1 and shRNA-2 containing the back-splicing region of lncRNA W5 for silencing. The efficiency of lncRNA W5 silencing was confirmed by qPCR following transfection with lncRNA W5 shRNA-1 or-2 in Huh7 and LM3 cells (Figure 2D). As expected, we found that lncRNA W5 silencing significantly promoted cell proliferation of Huh7 and LM3 cells as indicated by MTS (Figure 2E). Furthermore, a clone formation assay verified that following lncRNA W5 knockdown, the HCC population dependence and proliferation ability were considerably increased (Figure 2F). Overall, these data demonstrated that lncRNA W5 may inhibit HCC cell proliferation in vitro.
Figure 2.
In vitro suppression of long non-coding ribonucleic acid W5 in hepatocellular carcinoma proliferation. A: Increased long non-coding ribonucleic acid (lncRNA) W5 expression in Huh7 and LM3 cells was confirmed after over-expressed lncRNA W5 transfection by reverse transcription-polymerase chain reaction. LncRNA W5 expression was normalized to GAPDH. cP < 0.001; B: Cell viability of pCDNA-3.1 LncRNAW5-transfected Huh7 and LM3 cells were detected by CCK-8 assays. Cell number was determined every 24 h up to 96 h. The results are shown as the mean ± SE from three independent experiments. aP < 0.05; bP < 0.01. cP < 0.001, compared with the control by two-sided t-test; C: Colony-forming assay was used to determine the effect of lncRNA W5 on the proliferation in Huh7 and LM3 cells; D: Decreased lncRNA W5 expression in Huh7 and LM3 cells was confirmed after sh-1 or sh-2 LncRNAW5 transfection by reverse transcription-polymerase chain reaction. LncRNA W5 expression was normalized to GAPDH. aP < 0.05; bP < 0.01. cP < 0.001; E: Cell viability of sh-1 or sh-2 LncRNAW5-transfected Huh7 and LM3 cells were detected by CCK-8 assays. Cell number was determined every 24 h up to 96 h. The results are shown as the mean ± SE from at least three independent experiments. aP < 0.05; bP < 0.01. cP < 0.001, compared with the control by two-sided t-test; and F: Colony-forming assay was used to determine the effect of sh-1 or sh-2 LncRNA W5 on the proliferation of Huh7 and LM3 cells.
In vitro effects of lncRNA W5 on HCC cell migration and invasion
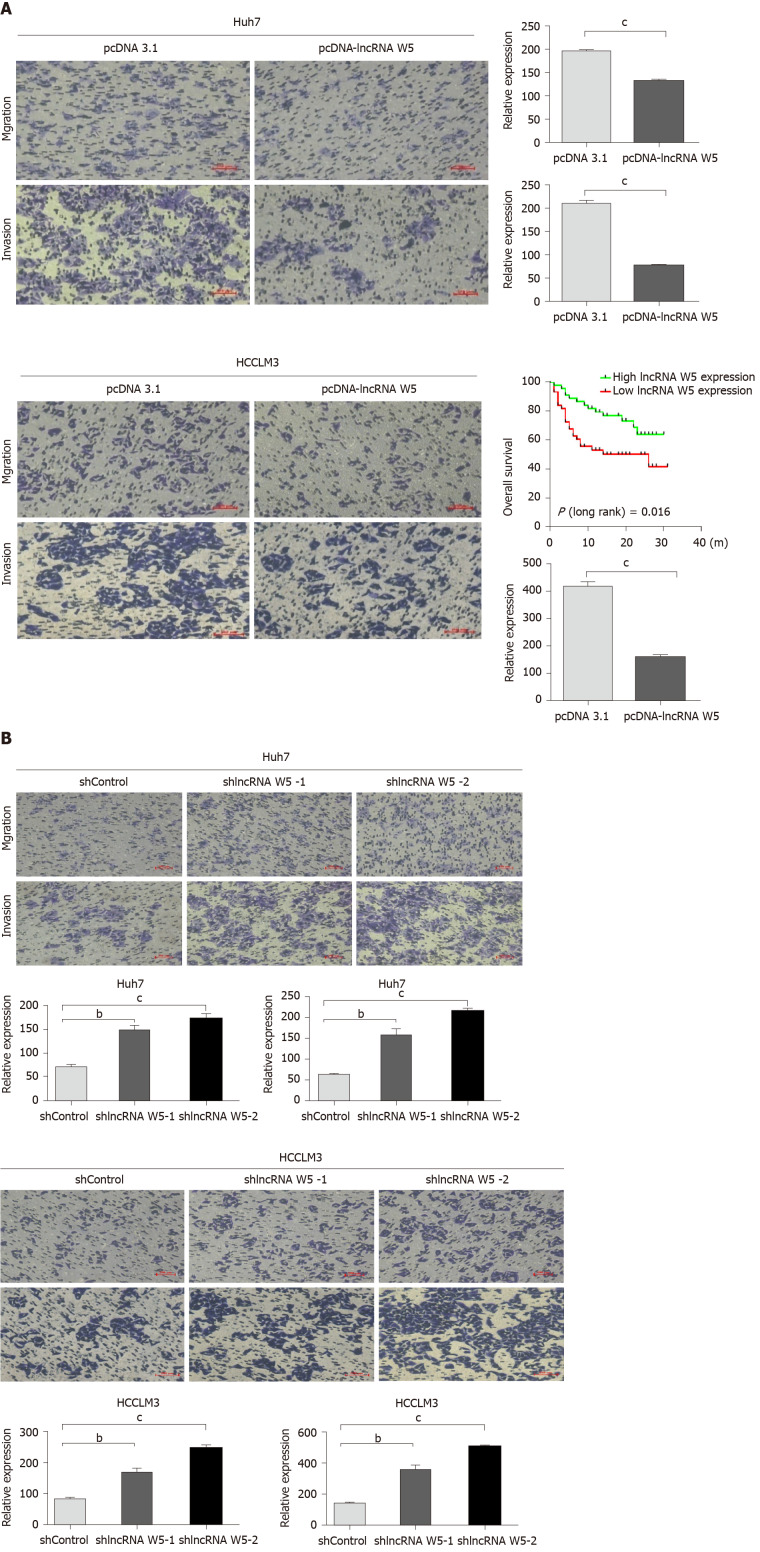
To evaluate the potential role of lncRNA W5 in HCC metastasis, and investigate the effect of lncRNA W5 on cell migration and invasion capacity, we performed transwell assays using Huh7 and LM3 cells. The results revealed that the migration and invasion ability of HCC cells that over-expressed lncRNA W5 was significantly decreased compared with the empty vector group (Figure 3A). On the contrary, lncRNA W5 knockdown with shRNA-1 or-2 significantly promoted cell migration and enhanced cell invasion of Huh7 and LM3 cells, respectively (Figure 3B). These results strongly suggest that lncRNA W5 has a critical effect on the inhibition of HCC cell migration and invasion.
Figure 3.
Effects of long non-coding ribonucleic acid W5 on hepatocellular carcinoma migration and invasion. A: Cell migration and invasion abilities were determined after transfection with pcDNA-3.1 and pcDNA-3.1 long non-coding ribonucleic acid W5 in Huh7 and LM3 cell lines, respectively; B: Cell migration and invasion abilities were determined after transfection with sh-1 or sh-2 LncRNA W5 in Huh7 and LM3 cell lines, respectively. All experiments were performed in triplicate. aP < 0.05; bP < 0.01; cP < 0.001.
lncRNA W5 inhibits tumor growth in vivo
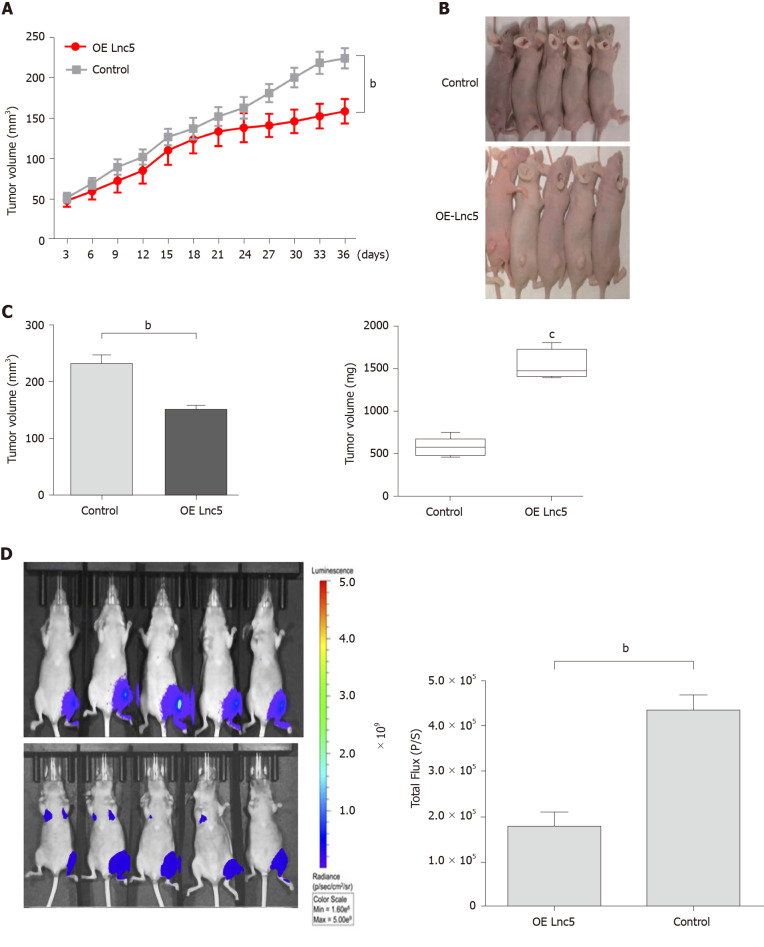
To elucidate the in vivo role of lncRNA W5 in tumorigenesis, we subcutaneously injected the flanks of nude mice with Huh7 cells over-expressing lncRNA W5 and stably expressing luciferase and monitored tumor growth every three days. As shown in Figure 4, mice injected with lncRNA W5 over-expressing cells had a significant decrease in tumor growth at 36 d post-injection as compared with mice injected with control cells. Six weeks after injection, the volumes and weights of tumors examined in mice injected with pcDNA3.1-lncRNA W5 were notably smaller than those in mice injected with the control. At 6 wk after injection, bioluminescent signals were weaker in mice with lncRNA W5 over-expression than in control mice, suggesting that lncRNA W5 may inhibit the growth of HCC xenograft tumors in vivo.
Figure 4.
Long non-coding ribonucleic acid W5 inhibits tumor growth in vivo. A: Huh7 cells (5 × 106) stably expressed with long non-coding ribonucleic acid W5 (lncRNA W5) were subcutaneously injected into the left flank of nude mice, and the effect of lncRNA W5 on hepatocellular carcinoma tumor growth was examined every 3 d during the course of the experiment (n = 5); B: A representative image of the xenograft-bearing mice; C: Tumors were isolated from the nude mice after sacrifice. The effects of lncRNA W5 on hepatocellular carcinoma growth were determined by tumor volume and tumor weight; D: LncRNA W5-overexpressing Huh7 cells which stably expressed luciferase were injected into nude mice (n = 5). The bioluminescence photographs of tumor were recorded with the in vivo 200 Imaging System. A representative luciferase signal was recorded from each group at 6 wk after injection. bP < 0.01.
DISCUSSION
Increasing evidence has shown that aberrant expression of numerous lncRNAs has been discovered in HCC. Previous studies showed that amplification of lncRNA ZFAS1 promotes metastasis in HCC[18]. Sun SH’s group observed in HCC that the lncRNA-activated by TGF-β (lncRNA-ATB) promoted the invasion-metastasis cascade[20]. Hur K’s group also reported that lncRNA-ATB could have potential as a biomarker for the prognosis of HCC and as a targeted therapy for HCC patients[22]. Another study showed that the MBNL3 splicing factor promoted HCC by increasing the expression of PXN by the alternative splicing of lncRNA-PXN-AS1[23]. The lncRNA lncHDAC2 may drive the self-renewal of liver cancer stem cells through the activation of Hedgehog signaling[24]. Super-enhancer associated lncRNA HCCL5 is activated by ZEB1 and promotes the malignancy of HCC[25]. Recently, Huang et al[26] identified oncofetal lncRNA Ptn-dt which might promote HCC proliferation by regulating the Ptn receptor[26]. These results indicate that lncRNAs may have critical roles in HCC progression and development and can be used in clinical applications.
In this study, we reported an uncharacterized low expression of lncRNA W5 in HCC specimens and cell lines, suggesting that lncRNA W5 expression might be related to HCC carcinogenesis. Decreased expression of lncRNA W5 was associated with aggressive clinicopathological features of HCC tissues, including tumor size, histological grade and the presence of portal vein tumor thrombosis. Gain-and loss function experiments showed that over-expression of lncRNA W5 in Huh7 and LM3 cells decreased cell proliferation, and impaired cell migration and invasion. Moreover, lncRNA W5 over-expression inhibited tumor growth in HCC xenograft-bearing mice. These results demonstrated that lncRNA W5 is involved in the progression of HCC and may be a potential target for therapy. Furthermore, our studies showed that HCC patients with low expression of lncRNA W5 had shorter overall survival than patients with high expression of lncRNA W5, suggesting that lncRNA W5 might be a potential prognostic predictor. Of course, down-regulation of lncRNA W5 should be validated in more HCC cohorts. The interactions of this protein and downstream pathways also warrant further investigation in subsequent studies. In addition, whether lncRNA W5 exists in other solid tumors remains to be elucidated.
CONCLUSION
In conclusion, our results showed that the expression of lncRNA W5 was considerably reduced in HCC tissues, which suppressed proliferation, migration and invasion of tumor cells in vitro. The results also showed that low expression of lncRNA W5 correlated with tumor progression and poor prognosis. Furthermore, manipulation of lncRNA W5 expression impacted the biological behavior of HCC. These results suggest that lncRNA W5 may serve as a tumor suppressor in the development and progression of HCC, and has potential as a diagnostic and therapeutic target in the clinical management of HCC.
ARTICLE HIGHLIGHTS
Research background
Accumulating evidence has revealed that several long non-coding RNAs (lncRNAs) are crucial in the progress of hepatocellular carcinoma (HCC).
Research motivation
To determine the clinical significance and potential roles of lncRNA W5 in HCC.
Research objectives
We classified the long non-coding RNA, lncRNA W5, and examined its clinical significance and potential roles in HCC.
Research methods
Analysis of the association between lncRNA W5 expression levels and clinico-pathological features was performed. In addition, overall survival was determined using Kaplan-Meier survival analysis.
Research results
The results showed that lncRNA W5 was down-regulated in HCC, and it may suppress HCC progression and predict a poor clinical outcome in patients with HCC.
Research conclusions
lncRNA W5 may serve as a potential prognostic biomarker and therapeutic target in HCC.
Research perspectives
lncRNA W5 may serve as a tumor suppressor in the development and progression of HCC, and have potential as a diagnostic and therapeutic target in the clinical management of HCC.
Footnotes
Institutional animal care and use committee statement: All animal experiments were conducted with the approval of the Fifth Medical Center of Chinese PLA General Hospital’s Animal Care and Use Committee.
Conflict-of-interest statement: All authors declare no financial or commercial conflicts of interest.
Manuscript source: Unsolicited manuscript
Peer-review started: August 25, 2020
First decision: September 12, 2020
Article in press: November 13, 2020
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zhang L S-Editor: Zhang L L-Editor: Webster JR P-Editor: Ma YJ
Contributor Information
Guang-Lin Lei, Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China.
Hong-Xia Fan, Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China; College of Basic Medicine, Inner Mongolia Medical University, Hohhot 010110, Inner Mongolia, China.
Cheng Wang, First Medical Center of Chinese PLA General Hospital, Beijing 100853, China.
Yan Niu, College of Basic Medicine, Inner Mongolia Medical University, Hohhot 010110, Inner Mongolia, China.
Tie-Ling Li, First Medical Center of Chinese PLA General Hospital, Beijing 100853, China.
Ling-Xiang Yu, Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China.
Zhi-Xian Hong, Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China.
Jin Yan, Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China.
Xi-Liang Wang, State Key Laboratory of Pathogens and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing 100071, China.
Shao-Geng Zhang, Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China.
Ming-Ji Ren, College of Basic Medicine, Inner Mongolia Medical University, Hohhot 010110, Inner Mongolia, China.
Peng-Hui Yang, Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China; State Key Laboratory of Pathogens and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing 100071, China. yph3022017@163.com.
Data sharing statement
All data generated or analyzed during this study are included in this published article.
References
- 1.Hindupur SK, Colombi M, Fuhs SR, Matter MS, Guri Y, Adam K, Cornu M, Piscuoglio S, Ng CKY, Betz C, Liko D, Quagliata L, Moes S, Jenoe P, Terracciano LM, Heim MH, Hunter T, Hall MN. The protein histidine phosphatase LHPP is a tumour suppressor. Nature. 2018;555:678–682. doi: 10.1038/nature26140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J, Yin T, Xu Y, Lu XJ. Therapeutics for advanced hepatocellular carcinoma: Recent advances, current dilemma, and future directions. J Cell Physiol. 2019;234:12122–12132. doi: 10.1002/jcp.28048. [DOI] [PubMed] [Google Scholar]
- 3.Okusaka T, Ikeda M. Immunotherapy for hepatocellular carcinoma: current status and future perspectives. ESMO Open. 2018;3:e000455. doi: 10.1136/esmoopen-2018-000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog. 2017;16:1. doi: 10.4103/jcar.JCar_9_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Escudier B, Worden F, Kudo M. Sorafenib: key lessons from over 10 years of experience. Expert Rev Anticancer Ther. 2019;19:177–189. doi: 10.1080/14737140.2019.1559058. [DOI] [PubMed] [Google Scholar]
- 6.Maracaja-Coutinho V, Paschoal AR, Caris-Maldonado JC, Borges PV, Ferreira AJ, Durham AM. Noncoding RNAs Databases: Current Status and Trends. Methods Mol Biol. 2019;1912:251–285. doi: 10.1007/978-1-4939-8982-9_10. [DOI] [PubMed] [Google Scholar]
- 7.Lorenzi L, Avila Cobos F, Decock A, Everaert C, Helsmoortel H, Lefever S, Verboom K, Volders PJ, Speleman F, Vandesompele J, Mestdagh P. Long noncoding RNA expression profiling in cancer: Challenges and opportunities. Genes Chromosomes Cancer. 2019;58:191–199. doi: 10.1002/gcc.22709. [DOI] [PubMed] [Google Scholar]
- 8.Zeng Y, Ren K, Zhu X, Zheng Z, Yi G. Long Noncoding RNAs: Advances in Lipid Metabolism. Adv Clin Chem. 2018;87:1–36. doi: 10.1016/bs.acc.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Rafiee A, Riazi-Rad F, Havaskary M, Nuri F. Long noncoding RNAs: regulation, function and cancer. Biotechnol Genet Eng Rev. 2018;34:153–180. doi: 10.1080/02648725.2018.1471566. [DOI] [PubMed] [Google Scholar]
- 10.Idda ML, Munk R, Abdelmohsen K, Gorospe M. Noncoding RNAs in Alzheimer's disease. Wiley Interdiscip Rev RNA. 2018;9 doi: 10.1002/wrna.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu WD, Wang H, He QF, Xu Y, Wang XC. Long noncoding RNAs in cancer-immunity cycle. J Cell Physiol. 2018;233:6518–6523. doi: 10.1002/jcp.26568. [DOI] [PubMed] [Google Scholar]
- 12.Cheng D, Deng J, Zhang B, He X, Meng Z, Li G, Ye H, Zheng S, Wei L, Deng X, Chen R, Zhou J. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine. 2018;36:159–170. doi: 10.1016/j.ebiom.2018.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun QM, Hu B, Fu PY, Tang WG, Zhang X, Zhan H, Sun C, He YF, Song K, Xiao YS, Sun J, Xu Y, Zhou J, Fan J. Long non-coding RNA 00607 as a tumor suppressor by modulating NF-κB p65/p53 signaling axis in hepatocellular carcinoma. Carcinogenesis. 2018;39:1438–1446. doi: 10.1093/carcin/bgy113. [DOI] [PubMed] [Google Scholar]
- 14.Noh JH, Gorospe M. AKTions by Cytoplasmic lncRNA CASC9 Promote Hepatocellular Carcinoma Survival. Hepatology. 2018;68:1675–1677. doi: 10.1002/hep.30165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Chen F, Zhao M, Yang Z, Li J, Zhang S, Zhang W, Ye L, Zhang X. The long noncoding RNA HULC promotes liver cancer by increasing the expression of the HMGA2 oncogene via sequestration of the microRNA-186. J Biol Chem. 2017;292:15395–15407. doi: 10.1074/jbc.M117.783738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y, Fan RG, Qin CL, Jia J, Wu XD, Zha WZ. LncRNA-H19 activates CDC42/PAK1 pathway to promote cell proliferation, migration and invasion by targeting miR-15b in hepatocellular carcinoma. Genomics. 2019;111:1862–1872. doi: 10.1016/j.ygeno.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Q, Lin Z, Xu J, Lu Y, Meng Q, Wang C, Yang Y, Xin X, Li X, Pu H, Gui X, Li T, Xiong W, Lu D. Long noncoding RNA MEG3 suppresses liver cancer cells growth through inhibiting β-catenin by activating PKM2 and inactivating PTEN. Cell Death Dis. 2018;9:253. doi: 10.1038/s41419-018-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H, Zhan Q, Zhu Z. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015;75:3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 19.Cheng S, Li T, Wang C, Wang K, Lai C, Yan J, Fan H, Sun F, Wang Z, Zhang P, Yu L, Hong Z, Lei G, Sun B, Gao Y, Xiao Z, Ji X, Wang R, Wu J, Wang X, Zhang S, Yang P. Decreased long intergenic noncoding RNA P7 predicts unfavorable prognosis and promotes tumor proliferation via the modulation of the STAT1-MAPK pathway in hepatocellular carcinoma. Oncotarget. 2018;9:36057–36066. doi: 10.18632/oncotarget.23282. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Pan W, Zhang N, Liu W, Liu J, Zhou L, Liu Y, Yang M. The long noncoding RNA GAS8-AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8. J Biol Chem. 2018;293:17154–17165. doi: 10.1074/jbc.RA118.003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang SY, Kim G, Park SY, Lee YR, Kwon SH, Kim HS, Yoon JS, Lee JS, Kweon YO, Ha HT, Chun JM, Han YS, Lee WK, Chang JY, Park JG, Lee B, Tak WY, Hur K. Clinical significance of lncRNA-ATB expression in human hepatocellular carcinoma. Oncotarget. 2017;8:78588–78597. doi: 10.18632/oncotarget.21094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan JH, Liu XN, Wang TT, Pan W, Tao QF, Zhou WP, Wang F, Sun SH. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19:820–832. doi: 10.1038/ncb3538. [DOI] [PubMed] [Google Scholar]
- 24.Wu J, Zhu P, Lu T, Du Y, Wang Y, He L, Ye B, Liu B, Yang L, Wang J, Gu Y, Lan J, Hao Y, He L, Fan Z. The long non-coding RNA LncHDAC2 drives the self-renewal of liver cancer stem cells via activation of Hedgehog signaling. J Hepatol. 2019;70:918–929. doi: 10.1016/j.jhep.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Peng L, Jiang B, Yuan X, Qiu Y, Peng J, Huang Y, Zhang C, Zhang Y, Lin Z, Li J, Yao W, Deng W, Zhang Y, Meng M, Pan X, Li C, Yin D, Bi X, Li G, Lin DC. Super-Enhancer-Associated Long Noncoding RNA HCCL5 Is Activated by ZEB1 and Promotes the Malignancy of Hepatocellular Carcinoma. Cancer Res. 2019;79:572–584. doi: 10.1158/0008-5472.CAN-18-0367. [DOI] [PubMed] [Google Scholar]
- 26.Huang JF, Jiang HY, Cai H, Liu Y, Zhu YQ, Lin SS, Hu TT, Wang TT, Yang WJ, Xiao B, Sun SH, Ma LY, Yin HR, Wang F. Genome-wide screening identifies oncofetal lncRNA Ptn-dt promoting the proliferation of hepatocellular carcinoma cells by regulating the Ptn receptor. Oncogene. 2019;38:3428–3445. doi: 10.1038/s41388-018-0643-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.