Abstract
Background and Aims
Floral diversity as a result of plant–pollinator interactions can evolve by two distinct processes: shifts between pollination systems or divergent use of the same pollinator. Although both are pollinator driven, the mode, relative importance and interdependence of these different processes are rarely studied simultaneously. Here we apply a phylogenetic approach using the Balsaminaceae (including the species-rich genus Impatiens) to simultaneously quantify shifts in pollination syndromes (as inferred from the shape and colour of the perianth), as well as divergent use of the same pollinator (inferred from corolla symmetry).
Methods
For 282 species we coded pollination syndromes based on associations between floral traits and known pollination systems, and assessed corolla symmetry. The evolution of these traits was reconstructed using parsimony- and model-based approaches, using phylogenetic trees derived from phylogenetic analyses of nuclear ribosomal and plastid DNA sequence data.
Key Results
A total of 71 % of studied species have a bee pollination syndrome, 22 % a bimodal syndrome (Lepidoptera and bees), 3 % a bird pollination syndrome and 5 % a syndrome of autogamy, while 19 % of species have an asymmetrical corolla. Although floral symmetry and pollination syndromes are both evolutionarily labile, the latter shifts more frequently. Shifts in floral symmetry occurred mainly in the direction towards asymmetry, but there was considerable uncertainty in the pattern of shift direction for pollination syndrome. Shifts towards asymmetrical flowers were associated with a bee pollination syndrome.
Conclusion
Floral evolution in Impatiens has occurred through both pollination syndrome shifts and divergent use of the same pollinator. Although the former appears more frequent, the latter is likely to be underestimated. Shifts in floral symmetry and pollination syndromes depend on each other but also partly on the region in which these shifts take place, suggesting that the occurrence of pollinator-driven evolution may be determined by the availability of pollinator species at large geographical scales.
Keywords: Asymmetry, floral morphology, floral symmetry, Impatiens, Hydrocera, molecular phylogenetics, pollinator
Introduction
The evolution of floral diversity is traditionally thought to be driven by pollinators (Darwin, 1862; Grant, 1949, 1994; Grant and Grant, 1965; Stebbins, 1970). Two distinct processes of pollinator-driven evolution have been identified. First, populations may adapt to different pollination systems, resulting in the formation of pollination ecotypes (Armbruster, 1985; Johnson, 2006; Whitall and Hodges, 2007; Valente et al., 2012; Van der Niet et al., 2014a). Secondly, populations may diverge in the way in which a particular pollinator is utilized (divergent use of the same pollinators) (Armbruster et al., 1994; Waterman et al., 2011; Eaton et al., 2012). These processes may eventually result in pollinator-driven speciation either through ethological isolation via pollination system shifts, or mechanical isolation through divergent use of the same pollinator (Grant, 1994; Van der Niet et al., 2014a). Although the two processes are both driven by pollinators, they are fundamentally different in their underlying drivers and their effect on floral evolution (Grant, 1994). During pollination system shifts, populations diverge in floral traits which reflect the sensory bias and morphological differences between different most effective functional pollinator groups, or lack of pollinators in the case of a shift to autonomous self-pollination (Robertson and Wyatt, 1990; Johnson, 1997; Johnson et al., 1998; Moeller, 2006). This process is generally driven by geographical turnover in pollinator species (Johnson, 1997; Johnson and Steiner, 1997; Moeller, 2006; Van der Niet et al., 2014b; Duffy and Johnson, 2017). In contrast, divergent use of the same pollinator is mainly associated with divergence in floral traits which mediate the position on the pollinator where the pollen is deposited (character displacement), and is thought to be generally driven by local competition for pollinator services (Armbruster, 1985; Armbruster et al., 1994).
Pollinator-driven evolutionary processes are best studied at the population level (Kay et al., 2005; Boberg et al., 2014; Cosacov et al., 2014; Forest et al., 2014; Gómez et al., 2014; Peter and Johnson, 2014; Van der Niet et al., 2014b), but this is not the most suitable way to quantify their importance in macroevolution. This requires a comparative perspective in a phylogenetic context (e.g. Smith and Kriebel, 2018; Dellinger et al., 2019; Kriebel et al., 2019, 2020; Xiang et al., 2020). Inferences from such comparative studies have revealed that shifts in pollination systems are relatively frequent (reviewed in Van der Niet and Johnson, 2012). Fewer studies have considered the frequency of shifts in use of the same pollinator (e.g. Eaton et al., 2012), and no macroevolutionary analysis has been carried out so far that quantified the overall frequency of pollinator-driven evolution, and the mode of evolution and relative importance of both pollination system shifts and divergent use of the same pollinator.
Perhaps one of the greatest challenges associated with comparitive phylogenetic studies of pollinator-driven evolution is the need for information on pollination systems (Van der Niet, 2020), and an understanding of which floral traits may be involved in divergent use of the same pollinator, for a relatively large number of species. One approach that has been used in this context is to infer pollination systems based on species’ pollination syndromes. Although the use of pollination syndromes has been criticized (Ollerton, 1996; Waser et al., 1996; Ollerton et al., 2009; Van der Niet, 2020), there is evidence that in groups of species that are characterized by relatively high levels of specialization in pollination systems, floral syndrome traits may be indicative of pollination systems, at least at the level of functional pollinator groups (Johnson and Steiner, 2000; Fenster, 2004; Rosas-Guerrero et al., 2014; Johnson and Wester, 2017). Similarly, divergent use of the same pollinator may be inferred from distinct differences in floral morphology which are known to be associated with different pollen placement sites on pollinators (Armbruster, 1985; Armbruster et al., 1994; Waterman et al., 2011; Eaton et al., 2012).
Balsaminaceae consists of the monotypic Hydrocera, and its large sister genus Impatiens (Yuan et al., 2004). The >1000 Impatiens species are characterized by a tremendous floral diversity (Grey-Wilson, 1980). The general architecture of Balsaminaceae flowers comprises various zygomorphic perianth parts, which are partially fused in Impatiens and unfused in Hydrocera, including a nectar-producing spur that is part of the lower sepal (Grey-Wilson, 1980). Grey-Wilson (1980) classified Impatiens species into several pollination syndromes. He suggested that flat-type flowers with a narrow entrance, shallow lower sepals, long spur and pale to deep pink colour are pollinated by butterflies; funnel-type flowers with a large entrance, deep lower sepals, hood-like dorsal petals, short spurs and yellow, white or pale pink colour are pollinated by bees; red or orange flowers with a large entrance are pollinated by birds; and white funnel-type flowers with a very long spur are pollinated by moths. Comparative studies have confirmed that variation in size and shape of the perianth parts is associated with predictable differences in pollination systems (Grey-Wilson, 1980; Kato et al., 1991; Ruchisansakun et al., 2016; Abrahamczyk et al., 2017). Besides the presence of distinct pollination syndromes, species which share the same pollinator also vary in floral architecture. In particular, floral variation among these species is associated with precise placement of pollen on the pollinator bodies, as was confirmed independently among co-flowering bee-pollinated species in Asia and bird-pollinated species in Africa (Janeček et al., 2015; Ruchisansakun et al., 2016). In several cases, differential pollen placement on bees is achieved by a highly unusual mechanism of floral asymmetry in which the lower lateral petals are asymmetrical (Kato et al., 1991; Ruchisansakun et al., 2016). Floral asymmetry therefore mediates divergent use of the same pollinator. Based on the two distinct types of floral variation in association with different pollination systems and divergent use of the same pollinator, Balsaminaceae are an ideal family to evaluate the overall frequency and relative importance of two distinct processes of pollinator-driven evolution.
The aim of the current study is therefore to reconstruct the evolution of pollination syndromes and divergent use of the same pollinator in Balsaminaceae. For this study, we compiled all pollination studies of Balsaminaceae species to set up a predictive framework for assigning species sampled in the largest phylogeny of the genus to date to pollination syndromes. Together with data on floral symmetry, we used the phylogeny for evolutionary analyses of pollination syndromes and floral symmetry to assess their mode of evolution, the overall frequency and relative importance of shifts in these two features, and whether or not they evolved independently.
MATERIALS AND METHODS
Phylogenetic analyses
Taxon sampling.
To reconstruct the evolution of pollination systems and corolla symmetry in Balsaminaceae, we included 282 accessions of Balsaminaceae species. Accessions comprised 281 Impatiens (approx. 25 % of all species in the genus) and Hydrocera triflora. Among the Impatiens specimens included in this study, 251 are from Asia (representing approx. 29 % of all Asian species), while 30 species are from Africa, Europe, Madagascar and North America (representing approx. 8 % of all species from these regions). The analysis is therefore biased towards species from Asia, which comprises three out of the five informal hotspots of Impatiens diversity. Furthermore, the pollination ecology of these species is arguably the most well understood. Marcgravia umbellata is used as outgroup (see Supplementary data Table S1).
DNA sequencing and phylogeny reconstruction.
To reconstruct the phylogenetic tree of Balsaminaceae, DNA sequences from plastid (atpB–rbcL intergenic spacer, ‘atpB–rbcL’ hereafter) and nuclear (ribosomal internal transcribed spacer, ‘ITS’ hereafter) genomes were used as characters. Most DNA sequences used for phylogeny reconstruction were obtained from GenBank (Yuan et al., 2004; Ruchisansakun et al., 2015; Utami and Ardiyani, 2015; Yu et al., 2015; Shajitha, 2016a, b), but for 27 species DNA sequences were newly generated. For these species, genomic DNA was extracted from fresh or silica-dried leaf material and herbarium specimens using the cetyltrimethylammonium bromide (CTAB) protocol (Doyle and Doyle, 1987). The ITS and atpB–rbcL regions were amplified following the protocols of Yuan et al. (2004) and Janssens et al. (2006), respectively. Standard DNA sequencing using the original amplification primers was done by Macrogen (Amsterdam, the Netherlands). The chromatograms of forward and reverse sequences were combined using the De Novo Assembly tool to create contigs in Geneious 10.2.2 (Biomatters Ltd, New Zealand) with default settings. Sequences were aligned using default settings in MUSCLE 3.8 (Edgar, 2004) and manually edited in Geneious 10.2.2 (Biomatters Ltd, New Zealand).
To obtain a set of ultrametric trees for analyses of character evolution, two sets of Bayesian phylogenetic analyses were implemented. The methods for analyses of separate datasets comprising nuclear and plastid DNA sequences are provided in Supplementary data Methods S1. A total evidence dataset combining plastid and nuclear DNA sequences was analysed using BEAST v.1.8.4 (Drummond and Rambaut, 2007), which was run on the CIPRES Science Gateway v. 3.3 (www.phylo.org). All parameters for the BEAST analysis were set in BEAUTI v.1.8.0 (Drummond and Rambaut, 2007) as described below. Based on the Akaike information criterion (AIC) as implemented in jModelTest2 v0.1.1 (Darriba et al., 2012), the GTR+I+G model of sequence evolution was selected for ITS whereas the GTR+G model was specified for atpB–rbcL. We chose to analyse the combined dataset with a nuclear and plastid DNA sequence partition under the most complex model GTR+I+G with estimated base frequencies as suggested by Abadi et al. (2019). As input parameter, a log-normal relaxed clock model was implemented using the ‘estimate’ option in the clock model. In addition, a Yule speciation tree prior (Gernhard, 2008) was applied with a random starting tree. The length of the Markov chain Monte Carlo (MCMC) was set to 30 million generations. Parameters were sampled each 3000th generation, whereas trees were sampled each 30 000th generation with a 10 % burn-in, resulting in a set of 900 trees. The tree prior was set uniformly at 100 in order to create ultrametric trees with a similar root basis. A maximum clade credibility (MCC) tree was constructed in TreeAnnotator v1.8.0. Examination of chain convergence was carried out with Tracer v1.71 (Rambaut et al., 2018). This resulted in effective sampling size (ESS) parameters exceeding 100 for all parameters, and 200 for most parameters apart from parameters related to the Yule process and substitution rates, indicating an adequate sampling and subsequent mixing for the topology estimation.
Character coding
Pollination syndromes.
For 40 (14.2 %) of the Balsaminaceae species included in our analysis, the pollination systems are known, while all the other species included in the phylogenetic analysis had unknown pollination systems. We assigned species to pollination syndromes [suites of floral traits associated with attraction of, and pollination by, particular functional pollinator groups (Fenster et al., 2004)]. To assign the species that were not studied in the field to pollination syndromes, we reviewed all literature related to Balsaminaceae pollination to assess which floral characters were associated with particular pollination systems. We specifically focused on the following floral characters that were identified in previous comparative studies (Grey-Wilson, 1980; Ruchisansakun et al., 2016; Abrahamczyk et al., 2017): floral entrance width (<2 mm or >6 mm), spur length (shorter or longer than 10 mm, or spur absent) and petal colour. Pollination syndromes were assigned at the level of functional pollinator groups using a framework that integrates information from several comparative studies by Grey-Wilson (1980), Ruchisansakun et al., (2016) and Abrahamczyk et al. (2017), and our own pollinator observations. As functional groups, we used bee pollination, bimodal pollination by bees and Lepidoptera, bird pollination and autonomous self-pollination, as these groups are represented by more than one empirical study. Although Abrahamczyk et al. (2017) assigned species with small flowers to a fly pollination syndrome, we do not recognize this syndrome for the following reason. In a study by Lozada-Gobilard et al. (2019), several species that conformed to the fly pollination syndrome were autogamous and sometimes cleistogamous. Furthermore, Abrahamczyk et al. (2017) showed that species with the fly pollination syndrome produce hardly any nectar. These traits are similar to those of the autogamous species studied by Ruchisansakun et al. (2016). Given that there are no studies available in the public domain which provide empirical evidence for the existence of specialized fly pollination in small-flowered Impatiens species, these are rather inferred to be autonomously selfing. Based on characters that were associated with particular pollination systems for species for which pollinator data were available, we assigned species to the bee pollination syndrome if they had a large floral entrance and short spur; to a bimodal pollination syndrome if species had a small floral entrance and long spur; to the bird pollination syndrome if species had a large floral entrance, short spur, red flowers and were native to Africa; and to the autogamy syndrome if they were spurless (Table 1). We then scored each of these floral characters for species in our analysis without pollinator data, and used these characters to characterize their pollination syndromes.
Table 1.
Summary of associations between floral traits and pollination systems based on empirical studies of Impatiens
| Entrance | Spur | Colour | Bee | Bird | Lepidoptera | Fly | Bee and Lepidoptera | Bee and hoverfly | Lepidoptera and long proboscid fly | Autogamy | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Large | Short | Yellow to orange | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 |
| Large | Short | Pink | 10 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 11 |
| Large | Short | Red | 0 | 7 | 0 | 0 | 2 | 0 | 0 | 0 | 9 |
| Large | Short | White | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Large | Long | Yellow to orange | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 |
| Large | Long | Pink | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Large | Long | Red | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Large | Spurless | Yellow to orange | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Large | Spurless | White | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Small | Short | Yellow to orange | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Small | Short | White | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Small | Long | Pink | 0 | 0 | 1 | 0 | 11 | 0 | 1 | 0 | 13 |
| Small | Long | Red | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Small | Long | White | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 3 |
| Total | 24 | 7 | 1 | 2 | 18 | 1 | 1 | 1 | 55 |
Corolla symmetry.
To assess corolla symmetry (i.e. zygomorphic vs. asymmetrical flowers according to a different size or shape of lateral petals on either side of a vertical symmetry axis), we analysed fresh material, investigated photographs taken by the authors, or photographs of Balsaminaceae in books or on websites which were identified to species level by Balsaminaceae specialists, studied illustrations and descriptions in the literature and examined dried specimens or illustrations of herbarium specimens. Each species was classified as either ‘zygomorphic’ or ‘asymmetrical’. Based on our personal observations, Hydrocera is polymorphic for corolla symmetry in which the zygomorphic form is much more common than the asymmetrical form. We therefore scored Hydrocera as ‘zygomorphic’.
Character evolution analyses
Our character evolution analyses aimed at reconstructing ancestral character states, assessing patterns of transitions among both pollination syndromes and corolla symmetry, and testing for correlated evolution between these two characters.
A diverse array of methods exists for analysing character evolution, each with their own strengths and weaknesses. It is beyond the scope of this study to review these in detail, but we provide a brief justification for the choice of methods used here. Parsimony-based methods are known to perform well in datasets where character change is rare (e.g. Pirie et al., 2012), but have been criticized due to implicit assumptions of character evolution such as that only one shift per branch is permitted (e.g. Schluter et al., 1997). Approaches which explicitly implement models of character transitions are not constrained by the assumption of a single shift per branch, but have been shown to return model parameter values which are counter-intuitive, especially in cases where certain character states are rare (Pagel, 1999). Given the fact that two of the four pollination syndromes in our dataset are rare, we implemented both parsimony- and model-based methods. Although some authors have argued that accurate reconstruction of character evolution requires explicit modelling of whether and how character states affect diversification (through a family of methods referred to as SSE) (e.g. Maddison et al., 2007), these methods have also been criticized for their performance (Raboski and Goldberg, 2015). Our own preliminary analyses suggested little impact of character states on speciation and extinction rates in our dataset (results not shown). Given the problems associated with SSE models, we therefore decided to implement Fitch parsimony (Fitch, 1971) and SIMMAP (Bollback, 2006) for ancestral character state reconstruction, and BayesTraits (Pagel et al., 2004) for model selection and analyses of correlated evolution. These analyses were performed on the set of 900 BEAST trees from the analysis of combined nuclear and plastid DNA sequence data, as well as on similar sets of trees resulting from separate analyses of each genomic data partition (see Supplementary data Methods S1 for a description of the methods for the separate analyses and results).
To assess the optimal model of character evolution in terms of transition rates among character states, we implemented Bayesian analyses using the BayesTraits V3.0 software (Pagel et al., 2004; Pagel and Meade, 2006) on the set of 900 BEAST trees. For all analyses in BayesTraits, we used the ‘ScaleTrees’ command (Pagel and Meade, 2006). For the binary character ‘floral symmetry’, we ran a test to assess the fit of two discrete models. The first included a discrete, unconstrained model, in which transition rates between the two states were allowed to vary independently. The second included a model in which only a single transition rate (equal forward and reverse rates) was modelled. Each model was run for 2 000 000 generations with a burn-in of 20 000 (ESSs of estimated parameters >200). For this analysis, we used an exponential hyperprior with a mean seeded from a uniform distribution on an interval ranging from 0 to 10. Marginal likelihoods were estimated by heating 100 stones for 10 000 iterations each. The Bayes factor (BF) between the independent and restricted model was subsequently calculated according to the formula: log BF = 2(log marginal likelihood complex model – log marginal likelihood restricted model). Log BF values of <2 indicate that the restricted (less complex) model should be preferred (and hence that transition rates between symmetrical and asymmetrical flowers are not significantly different).
Pollination syndrome is a multistate character with four states. Consequently, 12 transition rates and many model restrictions are possible, resulting in high model complexity. To accommodate this complexity, we used reverse jump (RJ) MCMC to select optimal models and parameters (Pagel and Meade, 2006). We ran 100 000 000 iterations (resulting in an ESS >200) with a burn-in of 10 000 000. The ten models that were most frequently sampled were considered for further interpretation. Both analyses also calculate the posterior probability of each state at the root node of Balsaminaceae, which is also reported.
The number and position of shifts under the parsimony criterion was reconstructed based on the set of 900 sampled BEAST trees. To summarize the frequency and direction of evolution, we used the function ‘summarize state changes over trees’ in Mesquite Version 3.6 (Maddison and Maddison, 2018) with 50 mappings sampled per tree.
To implement a model-based approach to reconstructing the frequency and position of shifts, we used the make.simmap function in the ‘phytools’ R package (Revell, 2012). Shifts were reconstructed onto a set of 100 randomly sampled post-burn-in BEAST trees using stochastic mapping. Prior to reconstruction, three different models were compared: an all rates different (ARD) model, an equal rates (ER) model and a symmetrical rate (SR) model. Model comparison was based on the AIC, and the most fitting model (ARD) was used in subsequent analyses. Transition rates were estimated with the ARD model. For all analyses, we estimated the prior distribution of state probabilities at the root node. The transition rate matrix and mean and median total number of estimated shifts between states were calculated. Based on ten simulations, average probabilities of each state at each node were estimated and plotted on the MCC tree using ‘ggtree’ in R (Yu et al., 2017).
To evaluate whether floral symmetry and pollination syndromes evolve in a correlated fashion or independently, we used Bayesian character evolution analyses implemented in BayesTraits V3.0 (Pagel and Meade, 2006). Analyses of correlated evolution for discrete traits can only be performed between two binary characters. We therefore first reduced the multistate character ‘pollination syndrome’ to multiple binary characters (e.g. bee pollination syndrome present/absent). Each of the four resulting binary character datasets (representing the four pollination syndromes present in our dataset) was then analysed separately for correlated evolution with the binary character ‘floral symmetry’. We used the discrete independent and dependent option in the software and ran 10 000 000 iterations with a burn-in length of 1 000 000. Due to low ESS for the dependent model of the Autogamy data set, we ran this for 50 000 000 iterations with a burn-in of 5 000 000 (ESS all >100). We used an RJ MCMC with an exponential hyperprior with a mean seeded from a uniform distribution on an interval ranging from 0 to 10. A stepping-stone sampling approach was used to calculate the marginal log-likelihood of both dependent and independent analyses. For this, a set of 100 stones were run for 10 000 iterations each. Two replicate runs were performed for each individual analysis to confirm consistency (only results from one run shown). The BF between the dependent and the independent model was then calculated. Log BFs <2 indicate that there is no evidence for correlated evolution between pollination syndrome and floral symmetry. The average and 95 % highest posterior density (HPD) transition rates among character states were subsequently calculated from the combined runs in the RJ MCMC analysis using Tracer v 1.71 and graphed.
RESULTS
Assignment of pollination syndromes and corolla symmetry
Data on pollination systems were available for 58 Impatiens species, 40 species of which were included in the phylogenetic analysis (Supplementary data Table S2). The majority of pollination studies were done on Asian species (n = 31), followed by African species (n = 10), species occurring in Europe (n = 8) and American species (n = 3). Most Impatiens species are pollinated by more than one pollinator species. Apart from African Impatiens species, the majority of species are predominantly bee pollinated. Furthermore, most species pollinated by Lepidoptera are also pollinated by bees (Fig. 1; Table 1; Supplementary data Table S2).
Fig. 1.
Balsaminaceae flower diversity and pollination syndromes. (A) Hydrocera triflora flower. (B–E) Bee-pollinated species: (B). I. jiewhoei, (C) I. psittacina, (D) I. daraneenae, (E) I. kerriae. (F–H) Species pollinated by Lepidoptera and bees (bimodal): (F) I. chiangdaoensis, (G and H) I. santisukii. (I and J) Bird-pollinated species: (I) I. hians, (J) I. niamniamensis. (K and L) Autogamous species: (K) I. decurva, (L) I. muscicola.
Variation in floral traits is associated with different pollination systems (Table 1). Asian, European and North American species with a short spur and large floral chamber are bee pollinated; species with a long spur and small floral chamber are mostly pollinated by both Lepidoptera and bees (bimodal pollination system) (Table 1); the species with small, spurless flowers are autogamous (Table 1). African species with a large floral entrance, short spur and red flowers are bird pollinated.
Based on these associations, among the Impatiens species sampled, 200 (70.9 %) are assigned to the bee pollination syndrome, 61 (21.6 %) species to the bimodal Lepidoptera and bee pollination syndrome, 14 (5.0 %) species to the autogamy syndrome and 7 (2.5 %) species to the bird pollination syndrome. Furthermore, 56 out of 282 (19.9 %) species have an asymmetrical corolla.
Phylogenetic analysis
The combined dataset consisted of 2703 bp: 1269 bp for the ITS region and 1434 bp for the atpB–rbcL region. Separate and combined BEAST MCC trees revealed some cases of topological incongruence (Supplementary data Figs S1 and S2), but the topologies are similar in terms of the placement of the main clades, and the MCC tree from the combined analysis is well resolved (Supplementary data Fig. S3). Impatiens can be divided into eight clades (Fig. 2). Species from outside Asia were distributed across several sections of Impatiens. In particular, African species were distributed among three clades: two of these are part of a big clade which comprises I. sect. Uniflorae, whereas some African species are members of the smaller clade which comprises I. sect. Tuberosae. Madagascan species form a monophyletic clade and are nested inside the largest African clade. North American species form a monophyletic clade within the clade of I. sect. Impatiens, whereas the European I. parviflora is part of I. sect. Racemosae. The remaining Asian species are scattered across all eight clades.
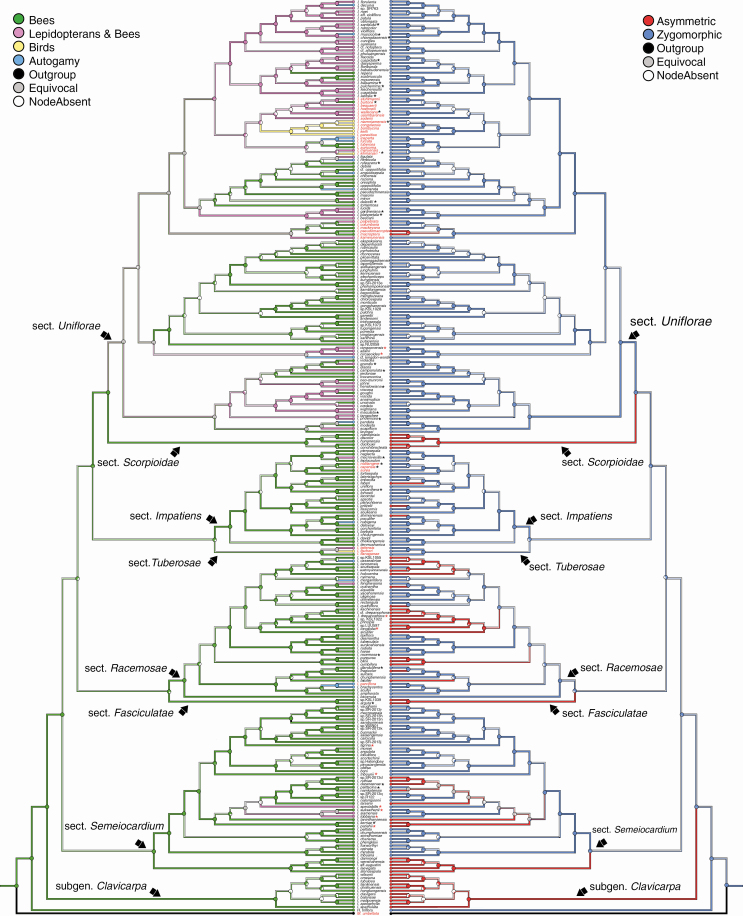
Fig. 2.
Parsimony ancestral character state reconstruction of pollination syndromes (left) and corolla symmetry (right). Branch colours represent the most parsimonious ancestral character states reconstructed using the MCC tree of the combined analysis. Pie charts at nodes represent the proportion of BEAST trees for which a particular character state at each node was reconstructed. The names of species outside Asia are marked in red. Black asterisks (*) show species for which pollination systems are known (see Supplementary data Table S2). Red asterisks (*) show species for which pollinators were observed by Saroj Ruchisansakun.
Character evolution
Models of character evolution.
The analyses of floral symmetry evolution indicated that the simpler model with a single transition rate between zygomorphic and asymmetrical corollas was preferred over the model in which these rates vary independently for the combined (BF 0.43) and nuclear analyses (BF –0.51), but for the plastid analysis the unrestricted model was preferred (BF 8.99) with a mean rate bias in favour of transitions from asymmetrical to zygomorphic flowers of 6.3. The average probability of a zygomorphic ancestor in Balsaminaceae was 0.88 based on the combined dataset (nuclear, 0.95; plastid, 0.19). The RJ MCMC analysis of the combined dataset indicated that several transition rates among pollination syndromes were indistinguishable from 0 (Table 2), in particular rates involving shifts away from the bee pollination syndrome and shifts away from the Lepidoptera + bee pollination syndrome. Shifts towards the autogamy syndrome and from the autogamy syndrome to the bee and Lepidoptera + bee pollination syndromes were generally high and almost never 0. Results were similar for the separate analyses, although there were fewer cases where rates were 0 (Supplementary data Table S3). There was much uncertainty regarding the ancestral pollination syndrome in all three analyses, although for all of them the bee pollination syndrome had the highest probability (combined, 0.32; nuclear, 0.38; plastid, 0.29).
Table 2.
Top ten models of transition rates (q) among pollination syndromes, ranked according to their sampling frequency in the RJ MCMC analysis of the combined dataset
| Model | q 01 | q 02 | q 03 | q 10 | q 12 | q 13 | q 20 | q 21 | q 23 | q 30 | q 31 | q 32 | Proportion | Cumulative proportion |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Z | Z | 0 | Z | Z | 0 | Z | 0 | Z | 1 | 0 | 0 | 0.07 | 0.07 |
| 2 | Z | Z | 0 | Z | Z | 0 | Z | 0 | 0 | 1 | 0 | 0 | 0.06 | 0.12 |
| 3 | Z | Z | 0 | Z | Z | 0 | 0 | 0 | Z | 1 | 0 | 0 | 0.05 | 0.18 |
| 4 | Z | Z | 0 | Z | Z | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0.04 | 0.21 |
| 5 | Z | Z | 0 | 0 | 0 | 0 | Z | Z | 1 | 1 | 1 | Z | 0.02 | 0.24 |
| 6 | Z | Z | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Z | 0.02 | 0.26 |
| 7 | Z | Z | 0 | 0 | 0 | 0 | Z | 0 | 1 | 1 | 1 | Z | 0.02 | 0.28 |
| 8 | Z | Z | 0 | 0 | 0 | 0 | 0 | Z | 1 | 1 | 1 | Z | 0.02 | 0.30 |
| 9 | Z | Z | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | Z | 0.02 | 0.32 |
| 10 | Z | Z | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0.02 | 0.33 |
Subscript numbers indicate pollination syndromes (0 = bee, 1 = Lepidoptera + bee, 2 = bird, 3 = autogamy); numbers in sequence indicate shifts from one pollination syndrome to another. Symbols (Z, 0 or 1) in the table represent transition rates. A value of Z indicates a transition rate of 0, whereas values of 0 and 1 represent different positive transition rates. Within models, transitions sharing a symbol occur at the same rate, whereas transitions with different symbols occur at different rates
Shifts in floral symmetry and pollination syndromes.
Based on the parsimony analysis of the combined dataset, the ancestral state of corolla symmetry in Balsaminaceae was zygomorphic in 91 % of all trees and the remaining reconstructions were equivocal (nuclear, 92 % zygomorphic, remainder equivocal; plastid, 29 % zygomorphic, remainder equivocal). There was a higher average number of shifts from a zygomorphic to an asymmetrical corolla for the combined analysis (Fig. 2; Table 3) and both separate datasets (Supplementary data Table S4). The positions of corolla symmetry shifts are scattered across several clades (i.e. I. subgen. Clavicarpa, I. sect. Scorpioidae, I. sect. Impatiens, I. sect. Racemosae, I. sect. Fasciculatae and I. sect. Semeiocardium), comprising only bee-pollinated species (Fig. 2), and almost never occurred in I. sect. Uniflorae.
Table 3.
Summary of changes in corolla symmetry of Balsaminaceae
| Zygomorphic to asymmetrical | Asymmetrical to zygomorphic | Total number of shifts | |
|---|---|---|---|
| Parsimony | 17.93 (7–23) | 2.97 (0–15) | 20.90 |
| SIMMAP | 18.42 (20) ± 4.41 | 9.45 (5) ± 8.96 | 27.88 (26) ± 5.74 |
For parsimony, the numbers represent the average number of shifts (min–max) of each kind across 50 mappings on a set of 900 BEAST trees. For SIMMAP, the numbers represent the average number of shifts (median) ± s.d. resulting from ten stochastic mappings on a random sample of 100 post-burn-in BEAST trees.
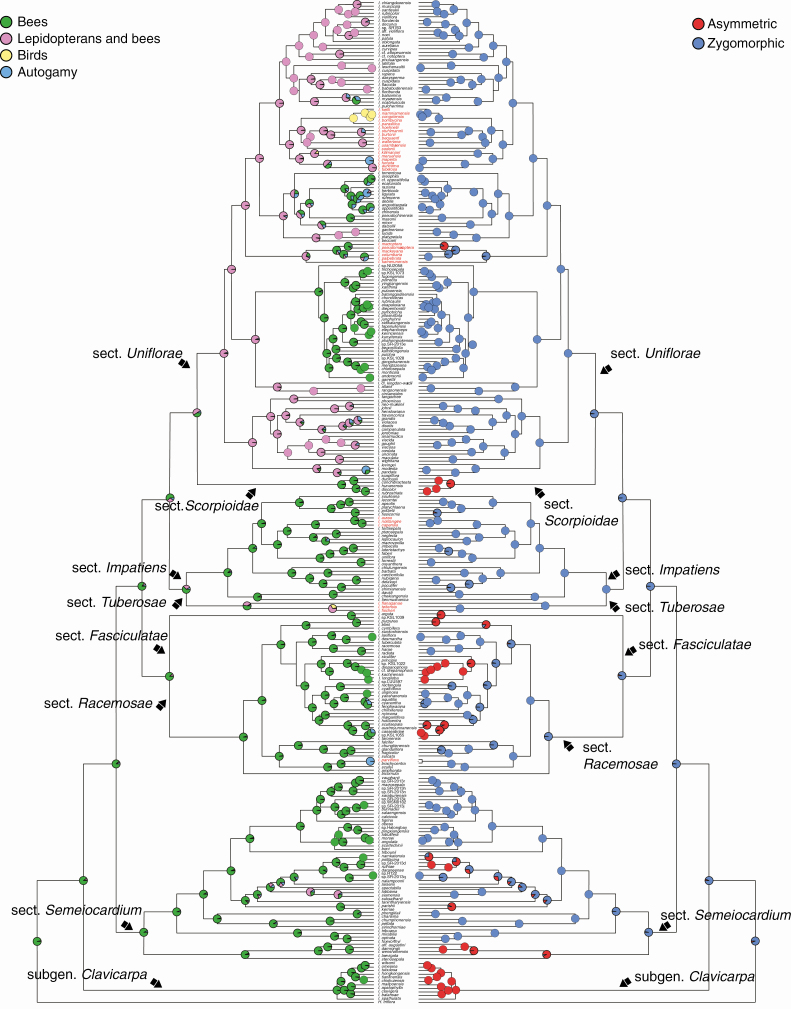
The results from the SIMMAP analysis of the combined dataset showed a much higher number of transitions from zygomorphic to asymmetrical flowers (Fig. 3; Table 3). The results from the nuclear dataset were consistent, but the number of shifts for the plastid dataset was higher in the opposite direction (Supplementary data Table S4).
Fig. 3.
SIMMAP ancestral character state reconstruction of pollination syndromes (left) and corolla symmetry (right) on the maximum clade credibility tree from the combined BEAST analysis. Pie charts at nodes represent the posterior probability of states based on ten stochastic mappings. The names of species outside Asia are marked in red.
Based on the parsimony analysis, the bee pollination syndrome is ancestral in Impatiens. Shifts in pollination syndrome appear phylogenetically concentrated in the clade that comprises sect. Uniflorae (Fig. 2). There were several shifts from a bee pollination syndrome to bimodal pollination syndrome, and towards the autogamy syndrome (Fig. 2; Table 4). Shifts from a bee to a bird pollination syndrome never occurred, but the shift from a bimodal to a bee pollination syndrome occurred relatively frequently. The bimodal pollination syndrome also shifted to the autogamy and bird pollination syndrome multiple times (Fig. 2; Table 4). Shifts between other pollination syndromes were almost absent (Fig. 2; Table 4). The average total number of shifts in pollination syndromes is 40.
Table 4.
Summary of changes in pollination syndromes of Balsaminaceae
| From: | To: | Parsimony | SIMMAP |
|---|---|---|---|
| Bees | Bimodal | 12.73 (5–20) | 3.51 (3) ± 2.91 |
| Bees | Birds | 0.69 (0–3) | 0 |
| Bees | Autogamy | 8.33 (5–10) | 36.87 (35) ± 14.57 |
| Bimodal | Bees | 11.03 (4–20) | 6.22 (6) ± 4.56 |
| Bimodal | Birds | 2.28 (0–3) | 3.71 (3) ± 1.09 |
| Bimodal | Autogamy | 3.65 (2–6) | 24.21 (24) ± 6.26 |
| Birds | Bees | 0.02 (0–1) | 0.14 (0) ± 0.82 |
| Birds | Bimodal | 0.37 (0–3) | 1.49 (1) ± 2.56 |
| Birds | Autogamy | 0.02 (0–1) | 0.46 (0) ± 1.64 |
| Autogamy | Bees | 0.43 (0–3) | 49.24 (46) ± 15.90 |
| Autogamy | Bimodal | 0.56 (0–3) | 13.86 (13) ± 5.23 |
| Autogamy | Birds | 0.00 (0–1) | 0 |
| Total shifts | 40.11 | 139.71 (135) ± 31.83 |
For parsimony, the numbers represent the average number of shifts (min–max) of each kind across all mappings on a set of 900 BEAST trees. For SIMMAP, the numbers represent the average number of shifts (median) ± s.d. resulting from ten stochastic mappings on 100 randomly selected post-burn-in trees from the BEAST analysis
The results from the SIMMAP analysis of the combined dataset differ in many ways. The most frequent shifts involve the autogamy syndrome, in particular shifts from a bee pollination syndrome towards an autogamy syndrome were common, as were shifts from a bimodal pollination syndrome to the autogamy syndrome, whereas shifts from the autogamy syndrome towards these two pollination syndromes were also frequent (Fig. 3; Table 4). All other types of shifts were rare or absent. The average total number of shifts was 140 (median 135) and thus much higher than those from the parsimony analysis. Results from the separate analyses are largely congruent with those of the combined analysis (Supplementary data Table S5).
Correlated evolution.
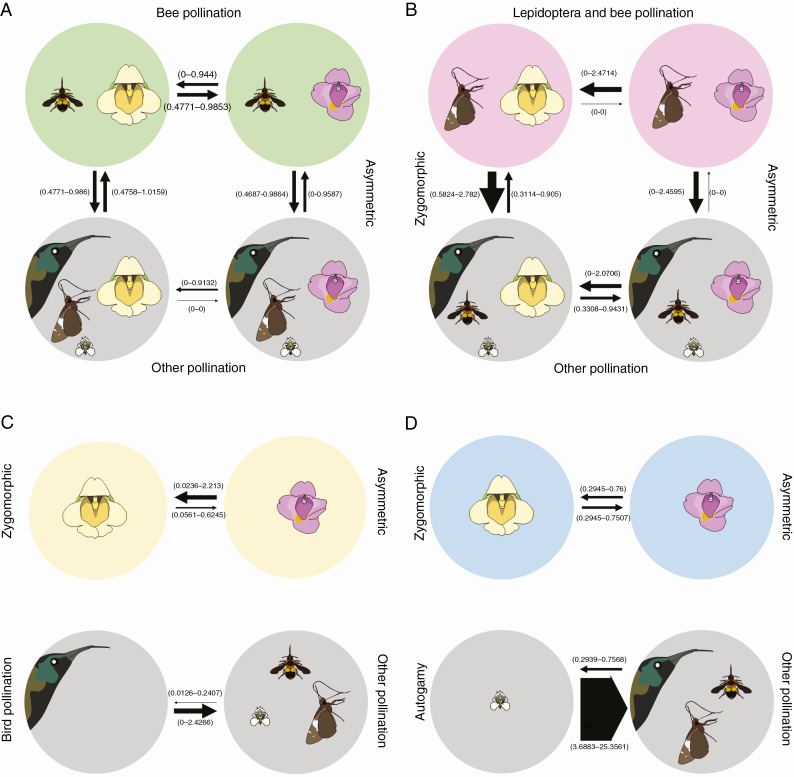
Results from analyses of correlated evolution between floral symmetry and pollination syndromes showed that when pollination syndrome was coded as bee vs. other and Lepidoptera + bee vs. other, BFs indicated that a model of dependent evolution was preferred over a model in which all shifts occur independently of background character states (Table 5; Supplementary data Table S6). This was not the case when pollination syndromes were coded as bird vs. other or autogamy vs. other (Table 5; Supplementary data Table S6).
Table 5.
RJ MCMC model scores for the combined dataset when pollination syndromes are scored as binary traits (present/absent)
| Pollinator type | Model | Log marginal likelihood | BF |
|---|---|---|---|
| Bee | Dependent | –192.153686 | 12.01 |
| Independent | –198.156587 | ||
| Butterfly | Dependent | –184.335912 | 27.64 |
| Independent | –198.156587 | ||
| Bird | Dependent | –106.385056 | –2.14 |
| Independent | –105.311592 | ||
| Autogamy | Dependent | –148.681805 | –6.53 |
| Independent | –145.415361 |
The dependent model indicates evolution of one trait dependent on the state in the other trait, whereas traits evolve independently in the independent model.
If pollination syndrome is coded as ‘bee vs other’, shifts towards asymmetrical flowers only happen against a background of a bee pollination syndrome (Fig. 4). This result was consistent across all three datasets. If pollination syndrome is coded ‘bimodal vs. other’, shifts towards asymmetrical flowers do not happen against a background of the bimodal syndrome, and neither do shifts towards this syndrome happen against a background of asymmetrical flowers (Fig. 4). This result was consistent across all three datasets.
Fig. 4.
Analyses of correlated evolution between pollination syndromes and floral symmetry. Pollination syndromes were coded as binary characters (present vs. other). (A and B) Cases where the evolution of pollination syndrome and floral symmetry are dependent on each other, whereas in (C) and (D) they evolve independently. Arrow thickness represents the magnitude of the average transition rates across the RJ MCMC analysis based on the set of 900 trees resulting from the combined BEAST analysis. Values in parentheses represent the 95 % HPD of transition rates. (A) Bee pollination syndrome vs. other pollination syndromes. (B) Bimodal (Lepidoptera + bee) pollination syndrome vs. other pollination syndromes. (C) Bird pollination syndrome vs. other pollination syndromes. (D) Autogamy syndrome vs. other pollination syndromes.
Discussion
Floral evolution in Impatiens is characterized by shifts in both pollination syndromes and floral symmetry (Figs 2 and 3). There are several uncertainties in terms of the frequency and direction of shifts, but some patterns were supported by almost all analyses. These included a predominance of shifts towards asymmetrical flowers with few reversals (Figs 2 and 3; Table 3; Supplementary data Table S4), as well as their occurrence in association with a bee pollination syndrome (Fig. 4).
The accuracy of our analyses of pollinator-driven evolution depends on the correct characterization of pollination syndromes, as the majority of sampled taxa have not been studied in the field. Although our inferences were based on pollination studies for >52 species (Table 1; Supplementary data Table S2), we acknowledge that our coding may still include some inaccuracies. In particular, pollination systems which are not represented among the studied species remain unknown and hence are likely to lead to underestimation of diversity in pollination systems and number of shifts in pollination syndromes (cf. Van der Niet, 2020). Given that most species in our analyses are from Asia (Figs 2 and 3), where most pollination studies were done (Supplementary data Table S2), we expect relatively few incorrect inferences. We also included pollination studies that were performed on non-native species (e.g. I. glandulifera, which is native in the Hymalayan mountains and invasive in many European countries, has been extensively studied in Europe) (Knuth, 1898; Erpenbach, 2006; Nienhuis et al., 2009; Ugoletti et al., 2013; Abrahamczyk et al., 2017). Given that pollinators of invasive species are unlikely to have selected for large changes in floral syndrome traits over the relatively short time since the invasion, we think that studies done on invasive species can reveal useful information on morphological fit and pollinator types (cf. Abrahamczyk et al., 2017). This is particularly the case if pollinators in the native and invaded range of an Impatiens species represent similar functional pollinator groups, as is the case, for instance, with bumble-bee species in Asia and Europe, which are both representatives of the genus Bombus [in fact, it is unlikely that a species with a relatively specialized floral morphology can invade a region without the presence of its functional pollinator niche, unless it is capable of autonomous self-pollination (see Duffy and Johnson, 2017)].
Impatiens is characterized by pollination by bees, Lepidoptera + bees, birds and autogamy (Grey-Wilson, 1980; Ruchisansakun et al., 2016). In our analysis with a focus on Asian species, bee pollination is the most common pollination system, followed by bimodal pollination by Lepidoptera + bees, and autogamy. Bird pollination is the least common and only found in Africa. The distribution of pollination systems among Asian species appears to be different from that in African species, in which pollination by Lepidoptera + bees and pollination by birds is more common (Grey-Wilson, 1980; Janssens, 2008). This difference may to some extent reflect different animal distributions between the areas. For example, bee genera such as Bombus and Apis, which are important pollinators of Asian Impatiens species, are uncommon in tropical Africa (Williams, 1998; Gupta, 2014). Sunbirds, on the other hand, are an important pollination niche for tropical African plants, including many African Impatiens species (Bartoš and Janeček, 2014; Janecek et al., 2015). However, pollinator distributions cannot be the only explanation for the differences in pollination systems as, despite the presence of sunbirds in Asia (Del Hoyo et al., 2018), these do not pollinate Impatiens (Ruchisansakun et al., 2016). For instance, based on floral traits, I. phoenicea, I. coelotropis and I. platyadena appear attractive to birds, but these species are pollinated by bees (Ramasubbu et al., 2009, 2011) or by bees and Lepidoptera (bimodal) (Sreekala et al., 2008a, b). We propose that the different frequencies of pollination systems can be understood in a context of historical biogeography. Phylogenetic evidence suggests that Balsaminaceae originated in Asia and dispersed from there to other regions (Janssens et al., 2009). This, combined with the distribution of bees, may explain why bee pollination is so common in Asia. Colonization of tropical Africa, where a limited number of large (forest) bee species currently occur, would then have to be associated with shifts to a new, locally available pollination niche (cf. Johnson, 1997; Van der Niet et al., 2014a). Indeed, pollination syndrome shifts occur mostly in the clade of I. sect. Uniflorae which comprises most of the African and Malagasy Impatiens species in our analysis (Yuan et al., 2004; Yu et al., 2015). However, since some African species have a bee pollination syndrome, and many Asian species have a bimodal syndrome, this scenario does not provide a full explanation for the pattern of shifts in pollination syndrome.
The relatively smaller number of pollination syndrome shifts among Asian Impatiens species does not necessarily signal a low frequency of pollinator-driven evolution. Indeed, shifts in floral symmetry between zygomorphic and asymmetrical corollas occurred several times in Asian Impatiens. These shifts appear more rarely in African Impatiens as only a few asymmetric-flowered Impatiens species are described from there (but see Janssens et al., 2015). Shifts in floral symmetry have occurred at least 20 times and appear to be reversible, although the number of shifts toward asymmetrical corollas is much higher than shifts toward zygomorphic corollas (Table 3) (apart from the model-based analysis of the plastid dataset). Furthermore, shifts in floral symmetry are biased to species with a bee pollination syndrome, and are almost entirely confined to Asian species (Figs 2–4) [interestingly, a mechanism of divergent use of the same pollinator appears to have evolved in African bird-pollinated Impatiens species, although the floral mechanism is not associated with asymmetrical corollas (Bartoš and Janeček, 2014)]. The specific evolutionary association between bee pollination and corolla asymmetry may be explained by the suitability of pollinator bodies for differential pollen placement. The relatively hairy bee bodies have many potential pollen placement sites (Armbruster et al., 2014), whereas pollen placement sites on butterflies are more limited to the central region and proboscis (but see Butler and Johnson, 2020). Based on the presumed function of asymmetrical corollas in terms of mediating precise and different pollen placement, our result suggests several independent shifts in the use of the same pollinator (cf. Stebbins, 1970; Johnson, 2010; Eaton et al., 2012; Armbruster, 2014). We probably underestimated the evolution of the divergent use of the same pollinator for two reasons. Firstly, divergent use of the same pollinator is not limited to asymmetrical flowers, as symmetrical bee-pollinated flowers also deposit pollen on different parts of the bee body (Ruchisansakun et al., 2016). Secondly, the binary coding of floral symmetry probably does not do justice to cases where divergent use of the same pollinator may be mediated by the degree of corolla asymmetry. More field-based studies are required to fully understand the mechanism of corolla asymmetry in mediating divergent use of the same pollinator.
The presence of species with asymmetrical corollas in Balsaminaceae is relatively uncommon in angiosperms (Etcheverry et al., 2008; Endress, 2012), and therefore provides a unique opportunity to investigate what drives its evolution. Ruchisansakun et al. (2016) showed that floral asymmetry results in different pollen placement sites on shared bee pollinators in species-rich Impatiens communities. It was hypothesized that asymmetrical flowers evolve in response to selection to avoid heterospecific pollen transfer. Three further hypotheses were proposed to explain the observation of multiple species with asymmetrical flowers. The phylogenetic analysis presented here can be used to distinguish among these hypotheses. The ‘community invasion’ hypothesis (cf. Sargent and Ackerly, 2008) postulates that asymmetrical flowers provide a key innovation which may facilitate successful invasion into Balsaminaceae communities, and hence allow clades with this trait to proliferate. Indeed, several relatively species-rich clades, such as sect. Clavicarpa, sect. Scorpioidae and several subclades of Sect. Racemosae, are characterized by the evolution of asymmetrical flowers in their respective common ancestors. However, it remains to be tested whether this trait promotes diversification compared with lineages with zygomorphic flowers by performing formal analyses of state-dependent diversification. Frequent, repeated evolution of asymmetrical flowers may also be a signature of reproductive character displacement, either among closely related species (reinforcement) or not. Our phylogenetic analysis provides unambiguous support for repeated evolution of asymmetrical flowers, confirming ideas proposed in Ruchisansakun et al. (2016) based on the non-homologous nature of corolla asymmetry. To distinguish reinforcement from general character displacement requires an analysis of whether species with asymmetrical flowers occur sympatrically with species with zygomorphic flowers, and whether they are cross-compatible. This is analysis was beyond the scope of this study, as it requires complete species-level sampling and data on the distribution of species and information on interspecific fertility. However, an informal analysis including five independently evolved asymmetrical species that are each sister to a (pair of) zygomorphic species suggests that only in one case (I. faberi vs. I. imbecilla/I. lateristachys) do the ranges of sister taxa overlap. On the other hand, in the remaining comparisons, the distribution of the asymmetrical species never overlaps with the (pair) of zygomorphic sister species. Our preliminary conclusion is therefore that the character displacement hypothesis (cf. Armbruster and Muchhala, 2009) would apply more frequently than the ‘reinforcement’ hypothesis (cf. Grant, 1994).
Shifts between pollination systems and floral symmetry can only partially explain the high diversity of Impatiens. In a meta-analysis of shifts in pollination systems, on average 25 % of cladogenic events were associated with pollination system shifts (Van der Niet and Johnson, 2012). In our study, we found that many branches in the phylogenetic tree are characterized by an absence of shifts. Although we argue that we may have underestimated the extent of pollinator-driven evolution, it seems likely that other drivers of speciation may be at play in Impatiens. Many Impatiens species are habitat specialists and consequently have a narrow and fragmented distribution. Such isolation may drive allopatric speciation (Janssens, 2008). Moreover, Yuan et al. (2004) showed that Impatiens species vary widely in their chromosome number, and that this trait is evolutionarily labile. This may be indicative of fast chromosomal evolution, leading to reproductive isolation and the evolution of species diversity without appreciable divergence in floral traits (White, 1968). More research into the relative importance of these different types of speciation is required to reconstruct and understand the evolution of species diversity in Balsaminaceae.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: species, localities, vouchers and GenBank accession numbers for the sequences used in this study. Table S2: list of studied Balsaminaceae species by country, floral traits and pollinators. Methods S1: methods and results associated with character evolution analyses based on separate nuclear and plastid datasets. Table S3: top five models ranked according to their sampling frequency. Table S4: summary of changes over trees in corolla symmetry of Balsaminaceae. Table S5: summary of changes over trees in pollination syndromes of Balsaminaceae. Table S6: RJ MCMC model scores for separate datasets when pollination syndromes are scored as binary traits. Figure S1: MCC nuclear dataset. Figure S2: MCC plastid dataset. Figure S3: MCC combined dataset.
ACKNOWLEDGEMENTS
We thank an anonymous reviewer for useful comments on an earlier draft of this manuscript.
Funding
This research was funded by a Development and Promotion of Science and Technology talents project (DPST) scholarship, provided by the Institute for the Promotion of Teaching Science and Technology (IPST) to S.R. The authors declare that they have no conflict of interest.
LITERATURE CITED
- Abadi S, Azouri D, Pupko T, Mayrose I. 2019. Model selection may not be a mandatory step for phylogeny reconstruction. Nature Communications 10: 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamczyk S, Lozada-Gobilard S, Ackermann M, et al. 2017. A question of data quality—testing pollination syndromes in Balsaminaceae. PLoS One 12: e0186125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster WS. 1985. Patterns of character divergence and the evolution of reproductive ecotypes of Dalechampia scandens (Euphorbiaceae). Evolution 39: 733–752. [DOI] [PubMed] [Google Scholar]
- Armbruster WS. 2014. Floral specialization and angiosperm diversity: phenotypic divergence, fitness trade-offs and realized pollination accuracy. AoB PLANTS 6: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster WS, Muchhala N. 2009. Associations between floral specialization and species diversity: cause, effect, or correlation? Evolution and Ecology 23: 159–179. [Google Scholar]
- Armbruster WS, Edwards ME, Debevec EM. 1994. Character displacement generates assemblage structure of Western Australian triggerplants (Stylidium). Ecology 75: 315–329. [Google Scholar]
- Armbruster WS, Shi XQ, Huang SQ. 2014. Do specialized flowers promote reproductive isolation? Realized pollination accuracy of three sympatric Pedicularis species. Annals of Botany 113: 331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoš M, Janeček S. 2014. Pollinator-induced twisting of flowers sidesteps floral architecture constraints. Current Biology 24: 793–795. [DOI] [PubMed] [Google Scholar]
- Boberg E, Alexandersson R, Jonsson M, Maad J, Ågren J, Nilsson LA. 2014. Pollinator shifts and the evolution of spur length in the moth-pollinated orchid Platanthera bifolia. Annals of Botany 113: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollback JP. 2006. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler HC, Johnson SD. 2020. Butterfly-wing pollination in Scadoxus and other South African Amaryllidaceae. Botanical Journal of the Linnean Society 193: 3363–3374. [Google Scholar]
- Cosacov A, Cocucci AA, Sérsic AN. 2014. Geographical differentiation in floral traits across the distribution range of the Patagonian oil-secreting Calceolaria polyrhiza: do pollinators matter? Annals of Botany 113: 251–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin CR. 1862. On the various contrivances by which British and foreign orchids are fertilized by insects. London: John Murray. [PMC free article] [PubMed] [Google Scholar]
- Del Hoyo J, Elliott A, Sargatal J, Christie DA, de Juana E. 2018. Handbook of the birds of the world alive. Barcelona: Lynx Edicions. [Google Scholar]
- Dellinger AS, Chartier M, Fernández-Fernández D, et al. 2019. Beyond buzz-pollination – departures from an adaptive plateau lead to new pollination syndromes. New Phytologist 221: 1136–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy KJ, Johnson SD. 2017. Specialized mutualisms may constrain the geographical distribution of flowering plants. Proceedings of the Royal Society B: Biological Sciences 284: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton DAR, Fenster CB, Hereford J, Huang SQ, Ree RH. 2012. Floral diversity and community structure in Pedicularis (Orobanchaceae). Ecology 93: S182–S194. [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. 2012. The immense diversity of floral monosymmetry and asymmetry across angiosperms. Botanical Review 78: 345–397. [Google Scholar]
- Erpenbach A. 2006. Blütenökologie madagassischer Springkräuter (Impatiens, Balsaminaceae). Diploma Thesis, University of Bonn. [Google Scholar]
- Etcheverry AV, Alemán MM, Fleming TF. 2008. Flower morphology, pollination biology and mating system of the complex flower of Vigna caracalla (Fabaceae: Papilionoideae). Annals of Botany 102: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. 2004. Pollination syndromes and floral specialization. Annual Review of Ecology, Evolution, and Systematics 35: 375–403. [Google Scholar]
- Fitch WM. 1971. Toward defining the course of evolution: minimum change for a specified tree topology. Systematic Zoology 20: 406–416. [Google Scholar]
- Forest F, Goldblatt P, Manning JC, et al. 2014. Pollinator shifts as triggers of speciation in painted petal irises (Lapeirousia: Iridaceae). Annals of Botany 113: 357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gernhard T. 2008. The conditioned reconstructed process. Journal of Theoretical Biology 253: 769–778. [DOI] [PubMed] [Google Scholar]
- Gómez JM, Muños-Pajares AJ, Abdelaziz M, Lorite J, Perfectti F. 2014. Evolution of pollination niches and floral divergence in the generalist plant Erysimum mediohispanicum. Annals of Botany 113: 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V. 1949. Pollination systems as isolating mechanisms in angiosperms. Evolution 3: 82–97. [DOI] [PubMed] [Google Scholar]
- Grant V. 1994. Modes and origins of mechanical and ethological isolation in angiosperms. Proceedings of the National Academy of Sciences,USA 91: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V, Grant KA. 1965. Flower pollination in the phlox family. New York: Columbia University Press. [Google Scholar]
- Grey-Wilson C. 1980. Impatiens of Africa. Rotterdam: Balkema. [Google Scholar]
- Gupta RK. 2014. Taxonomy and distribution of different honeybee species. In: Gupta R, Reybroeck W, van Veen J, Gupta A eds. Beekeeping for poverty alleviation and livelihood security. Dordrecht: Springer, 3–62. [Google Scholar]
- Janeček Š, Bartoš M, Njabo KY. 2015. Convergent evolution of sunbird pollination systems of Impatiens species in tropical Africa and hummingbird systems of the New World. Biological Journal of the Linnean Society 115: 127–133. [Google Scholar]
- Janssens SB. 2008. Evolutionary studies in Balsaminaceae: integration of evidence from molecular and morphological data. PhD Thesis, K.U. Leuven. [Google Scholar]
- Janssens SB, Geuten K, Yuan YM, Song Y, Kupfer P, Smets E. 2006. Phylogenetics of Impatiens and Hydrocera (Balsaminaceae) using chloroplast atpB-rbcL spacer sequences. Systematic Botany 31: 171–180. [Google Scholar]
- Janssens SB, Knox EB, Huysmans S, Smets EF, Merckx VS. 2009. Rapid radiation of Impatiens (Balsaminaceae) during Pliocene and Pleistocene: result of a global climate change. Molecular Phylogenetics and Evolution 52: 806–824. [DOI] [PubMed] [Google Scholar]
- Janssens SB, Sonké B, Lachenaud O, Lemaire B, Simo-Droissart M, Smets E. 2015. Morphology, molecular phylogenetics and biogeography of Impatiens akomensis (Balsaminaceae), a new species from Cameroon. Plant Ecology and Evolution 148: 397–408. [Google Scholar]
- Johnson SD. 1997. Pollination ecotypes of Satyrium hallackii (Orchidaceae) in South Africa. Botanical Journal of the Linnean Society 123: 225–235. [Google Scholar]
- Johnson SD. 2006. Pollinator-driven speciation in plants. In: Harder LD, Barrett SCH, eds. The ecology and evolution of flowers. Oxford: Oxford University Press, 295–310. [Google Scholar]
- Johnson SD. 2010. The pollination niche and its role in the diversification and maintenance of the southern African flora. Philosophical Transactions of the Royal Society B: Biological Sciences 365: 499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SD, Steiner KE. 1997. Long-tongued fly pollination and evolution of floral spur length in the Disa draconis complex (Orchidaceae). Evolution 51: 45–53. [DOI] [PubMed] [Google Scholar]
- Johnson SD, Steiner KE. 2000. Generalization versus specialization in plant pollination systems. Trends in Ecology & Evolution 15: 140–143. [DOI] [PubMed] [Google Scholar]
- Johnson SD, Wester P. 2017. Stefan Vogel’s analysis of floral syndromes in the South African flora: an appraisal based on 60 years of pollination studies. Flora 232: 200–206. [Google Scholar]
- Johnson SD, Linder HP, Steiner KE. 1998. Phylogeny and radiation of pollination systems in Disa (Orchidaceae). American Journal of Botany 85: 402–411. [PubMed] [Google Scholar]
- Kato M, Itino T, Hotta M, Inoue T. 1991. Pollination of four Sumatran Impatiens species by hawkmoths and bees. Tropics 1: 59–73. [Google Scholar]
- Kay KM, Reeves PA, Olmstead RG, Schemske DW. 2005. Rapid speciation and the evolution of hummingbird pollination in neotropical Costus subgenus Costus (Costaceae): evidence from nrDNA ITS and ETS sequences. American Journal of Botany 92: 1899–1910. [DOI] [PubMed] [Google Scholar]
- Knuth P. 1898. Die bisher in Europa und in arktischen Gebiet gemachten blutenbiologischen. In: Knuth P. ed. Handbuch der Blütenbiologie. Leipzig: Wilhelm Engelmann Verlag, 245–248. [Google Scholar]
- Kriebel R, Drew BT, Drummond CP, et al. 2019. Tracking temporal shifts in area, biomes, and pollinators in the radiation of Salvia (sages) across continents: leveraging anchored hybrid enrichment and targeted sequence data. American Journal of Botany 106: 573–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriebel R, Drew B, González-Gallegos JG, et al. 2020. Pollinator shifts, contingent evolution, and evolutionary constraint drive floral disparity in Salvia (Lamiaceae): evidence from morphometrics and phylogenetic comparative methods. Evolution 74: 1335–1355. [DOI] [PubMed] [Google Scholar]
- Lozada-Gobilard S, Weigend M, Fischer E, Janssens SB, Ackermann M, Abrahamczyk S. 2019. Breeding systems in Balsaminaceae in relation to pollen/ovule ratio, pollination syndromes, life history and climate zone. Plant Biology 21: 157–166. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. 2018. Mesquite: a modular system for evolutionary analysis. Version 3.40 http://mesquiteproject.org.
- Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character’s effect on speciation and extinction. Systematic Biology 56: 701–710. [DOI] [PubMed] [Google Scholar]
- Moeller DA. 2006. Geographic structure of pollinator communities, reproductive assurance, and the evolution of self-pollination. Ecology 87: 1510–1522. [DOI] [PubMed] [Google Scholar]
- Nienhuis CM, Dietzsch AC, Stout JC. 2009. The impacts of an invasive alien plant and its removal on native bees. Apidologie 40: 450–463. [Google Scholar]
- Ollerton J. 1996. Reconciling ecological processes with phylogenetic patterns: the apparent paradox of plant–pollinator systems. Journal of Ecology 84: 767–769. [Google Scholar]
- Ollerton J, Alarcón R, Waser NM, et al. 2009. A global test of the pollination syndrome hypothesis. Annals of Botany 103: 1471–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. 1999. The Maximum Likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Systematic Biology 48: 612–622. [Google Scholar]
- Pagel M, Meade A. 2006. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. The American Naturalist 167: 808–825. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A, Barker D. 2004. Bayesian estimation of ancestral character states on phylogenies. Systematic Biology 53: 673–684. [DOI] [PubMed] [Google Scholar]
- Peter CI, Johnson SD. 2014. A pollinator shift explains floral divergence in an orchid species complex in South Africa. Annals of Botany 113: 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirie MD, Humphreys AM, Antonelli A, Galley C, Linder HP. 2012. Model uncertainty in ancestral area reconstruction: a parsimonious solution? Taxon 61: 652–664. [Google Scholar]
- Rabosky DL, Goldberg EE. 2015. Model inadequacy and mistaken inferences of trait-dependent speciation. Systematic Biology 64: 340–355. [DOI] [PubMed] [Google Scholar]
- Ramasubbu R, Sreekala AK, Pandurangan AG, Kulloli SK. 2009. Floral phenology, pollination and pollen–pistil interactions of Impatiens phoenicea Bedd. from the Southern Western Ghats. Advances in Pollen Spore Research 27: 183–194. [Google Scholar]
- Ramasubbu R, Sreekala AK, Pandurangan AG, Kulloli SK. 2011. Reproductive ecology of Impatiens platyadena Fischer, a critically endangered balsam of Western Ghats. Current Science 100: 1550–1554. [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Systematic Biology 67: 901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Robertson JL, Wyatt R. 1990. Evidence for pollination ecotypes in the yellow-fringed orchid, Platanthera ciliaris. Evolution 44: 121–133. [DOI] [PubMed] [Google Scholar]
- Rosas-Guerrero V, Aguilar R, Martén-Rodríguez S, et al. 2014. A quantitative review of pollination syndromes: do floral traits predict effective pollinators? Ecology Letters 17: 388–400. [DOI] [PubMed] [Google Scholar]
- Ruchisansakun S, Van der Niet T, Janssens SB, et al. 2015. Phylogenetic analyses of molecular data and reconstruction of morphological character evolution in Asian Impatiens section Semeiocardium (Balsaminaceae). Systematic Botany 40: 1063–1074. [Google Scholar]
- Ruchisansakun S, Tangtorwongsakul P, Cozien RJ, Smets EF, Van Der Niet T. 2016. Floral specialization for different pollinators and divergent use of the same pollinator among co-occurring Impatiens species (Balsaminaceae) from Southeast Asia. Botanical Journal of the Linnean Society 181: 651–666. [Google Scholar]
- Sargent RD, Ackerly DD. 2008. Plant–pollinator interactions and the assembly of plant communities. Trends in Ecology & Evolution 23: 123–130. [DOI] [PubMed] [Google Scholar]
- Schluter D, Price T, Mooers AØ, Ludwig D. 1997. Likelihood of ancestor states in adaptive radiation. Evolution 51: 1699–1711. [DOI] [PubMed] [Google Scholar]
- Shajitha PP, Dhanesh NR, Ebin PJ, et al. 2016. a A combined chloroplast atpB–rbcL and trnL-F phylogeny unveils the ancestry of balsams (Impatiens spp.) in the Western Ghats of India. 3 Biotech 6: 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajitha PP, Dhanesh NR, Ebin PJ, et al. 2016. b Molecular phylogeny of balsams (genus Impatiens) based on ITS regions of nuclear ribosomal DNA implies two colonization events in South India. Journal of Applied Biology & Biotechnology 4: 1–9. [Google Scholar]
- Smith SD, Kriebel R. 2018. Convergent evolution of floral shape tied to pollinator shifts in Iochrominae (Solanaceae). Evolution 72: 688–697. [DOI] [PubMed] [Google Scholar]
- Sreekala AK, Ramasubbu R, Pandurangan AG, Kulloli SK. 2008a Pollination biology of Impatiens campanulata Wight. (Balsaminaceae). Advances in Pollen Spore Research 26: 9–19. [Google Scholar]
- Sreekala AK, Pandurangan AG, Ramasubbu R, Kulloli SK. 2008b Reproductive biology of Impatiens coelotropis – a critically endangered balsam from the Western Ghats. Current Science 95: 386–388. [Google Scholar]
- Stebbins GL. 1970. Adaptive radiation of reproductive characteristics in angiosperms. I. Pollination mechanisms. Annual Review of Ecology and Systematics 1: 307–326. [Google Scholar]
- Ugoletti P, Reidy D, Jones MB, Stout JC. 2013. Do native bees have the potential to promote interspecific pollination in introduced Impatiens species? Journal of Pollination Ecology 11: 1–8. [Google Scholar]
- Utami N, Ardiyani. 2015. Phylogenetic study of Sumatran Impatiens (Balsaminaceae) using nuclear and plastid DNA sequences. Acta Phytotaxonomica et Geobotanica 66: 81–90. [Google Scholar]
- Valente LM, Manning JC, Goldblatt P, Vargas P. 2012. Did pollination shifts drive diversification in southern African Gladiolus? Evaluating the model of pollinator-driven speciation. The American Naturalist 180: 83–98. [DOI] [PubMed] [Google Scholar]
- Van der Niet T. 2020. Paucity of natural history data impedes phylogenetic analyses of pollinator-driven evolution. New Phytologist doi: 10.1111/nph.16813. [DOI] [PubMed] [Google Scholar]
- Van der Niet T, Johnson SD. 2012. Phylogenetic evidence for pollinator-driven diversification of angiosperms. Trends in Ecology & Evolution 27: 353–361. [DOI] [PubMed] [Google Scholar]
- Van der Niet T, Peakall R, Johnson SD. 2014a Pollinator-driven ecological speciation in plants: new evidence and future perspectives. Annals of Botany 113: 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Niet T, Pirie MD, Shuttleworth A, Johnson SD, Midgley JJ. 2014b Do pollinator distributions underlie the evolution of pollination ecotypes in the Cape shrub Erica plukenetii? Annals of Botany 113: 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waser NM, Chittka L, Price MV, Williams NM, Ollerton J. 1996. Generalization in pollination systems, and why it matters. Ecology 77: 1043–1060. [Google Scholar]
- Waterman RJ, Bidartondo MI, Stofberg J, et al. 2011. The effects of above- and belowground mutualisms on orchid speciation and coexistence. The American Naturalist 177: E54–E68. [DOI] [PubMed] [Google Scholar]
- White MJD. 1968. Models of speciation. Science 159:1065–1070. [DOI] [PubMed] [Google Scholar]
- Whittall JB, Hodges SA. 2007. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature 447: 706–709. [DOI] [PubMed] [Google Scholar]
- Williams PH. 1998. An annotated checklist of bumblebees with an analysis of patterns of description (Hymenoptera: Apidae, Bombini). Bulletin of the Natural History Museum (Entomology) 67: 79–152. [Google Scholar]
- Xiang GJ, Guo YH, Yang CF. 2020. Diversification of floral orientation in Lonicera is associated with pollinator shift and flowering phenology. Journal of Systematics and Evolution. [Google Scholar]
- Yu G, Smith D, Zhu H, Guan Y, Lam TT. 2017. Ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 8: 28–36. [Google Scholar]
- Yu SX, Janssens SB, Zhu XY, Lidén M, Gao TG, Wang W. 2015. Phylogeny of Impatiens (Balsaminaceae): integrating molecular and morphological evidence into a new classification. Cladistics 32: 179–197. [DOI] [PubMed] [Google Scholar]
- Yuan YM, Song Y, Geuten K, et al. 2004. Phylogeny and biogeography of Balsaminaceae inferred from ITS sequence data. Taxon 53: 391–404. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.