Abstract
Background
Carex L. is one of the largest genera in the Cyperaceae family and an important vascular plant in the ecosystem. However, the genetic background of Carex is complex and the classification is not clear. In order to investigate the gene function annotation of Carex, RNA-sequencing analysis was performed. Simple sequence repeats (SSRs) were generated based on the Illumina data and then were utilized to investigate the genetic characteristics of the 79 Carex germplasms.
Results
In this study, 36,403 unigenes with a total length of 41,724,615 bp were obtained and annotated based on GO, KOG, KEGG, NR databases. The results provide a theoretical basis for gene function exploration. Out of 8776 SSRs, 96 pairs of primers were randomly selected. One hundred eighty polymorphic bands were amplified with a polymorphism rate of 100% based on 42 pairs of primers with higher polymorphism levels. The average band number was 4.3 per primer, the average distance value was 0.548, and the polymorphic information content was ranged from 0.133 to 0.494. The number of observed alleles (Na), effective alleles (Ne), Nei’s (1973) gene diversity (H), and the Shannon information index (I) were 2.000, 1.376, 0.243, and 0.391, respectively. NJ clustering divided into three groups and the accessions from New Zealand showed a similar genetic attribute and clustered into one group. UPGMA and PCoA analysis also revealed the same result. The analysis of molecular variance (AMOVA) revealed a superior genetic diversity within accessions than between accessions based on geographic origin cluster and NJ cluster. What’s more, the fingerprints of 79 Carex species are established in this study. Different combinations of primer pairs can be used to identify multiple Carex at one time, which overcomes the difficulties of traditional identification methods.
Conclusions
The transcriptomic analysis shed new light on the function categories from the annotated genes and will facilitate future gene functional studies. The genetic characteristics analysis indicated that gene flow was extensive among 79 Carex species. These markers can be used to investigate the evolutionary history of Carex and related species, as well as to serve as a guide in future breeding projects.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-020-02792-8.
Keywords: Carex L., Illumina RNA-sequencing, Gene function annotation, SSR marker, Marker polymorphism, Genetic cluster
Background
The genus Carex L. belongs to the Cyperaceae family and is an enormous genus. It is one of the most vital genera of vascular plants in the environment [1], with more than 2000 species widespread all over the world [2] and nearly 500 species in China [3]. Carex species are widely used as a ground cover for home lawns as well as for slope stabilization in many parts of the world because of cold and drought tolerance [4], trample resistance, and high ornamental value [5, 6].
Previous studies focused on geographical distribution, phylogeography, and origin area of Carex. Benítez-Benítez et al. [7] found obvious genetic differentiation between two Carex sister species in the western Mediterranean, and pointed out that geographic barriers played dominant role in restricting gene flow. Míguez et al. [8] revealed late Miocene-Pliocene aridification of the Mediterranean shaped the phylogeography of Carex sect. Rhynchocystis. Martín-Bravo et al. [9] proved that Carex originated in the late Eocene in East Asia which has a productive diversity of Carex. Previous studies clarified the phylogenetic structure of Carex into at least four major clades, including Siderostictae clade, core Carex, Vignea, and Caricoid clade [10]. However, a supermatrix analysis combining ETS, ITS and matK DNA regions indicated that over-reliance on morphological characters was inappropriate for the delimitation of natural groups [11].
Molecular markers are powerful tools for genetic diversity analysis which is the basis of accelerating plant breeding process. Currently, commonly used molecular markers mainly include ISSR, RAPD, RFLP, AFLP, SSR, and SRAP [12]. Due to the advantages of abundant, multi-allelic, highly polymorphic and codominant, simple repeat sequence (SSRs) for genetic research are a good choice to reveal the mechanism of genetic genes in plants [13, 14]. RNA-sequencing is an effective tool to obtain SSRs with higher rate of transferability for non-sequenced genomes and non-model organisms [15, 16]. It has been demonstrated that SSRs obtained from one species could be used to detect diversity in related species and even in other genera of the same family [17, 18]. SSR has been widely used in genetic mapping, relationship studies [19], cultivar identification [20, 21] and analysis of plant genetic diversity [22]. Hitherto SSR markers have been widely applied to plant research, such as Zea mays [23], Citrullus lanatus [24], Triticum aestivum and the genus Cerasus species [25].
In previous studies, there have been many molecular marker studies on Carex, which also included SSR method. M’Baya et al. [26] utilized 14 SSRs isolated in Carex kobomugi to test genetic structure of Carex hebes and Carex breviculmis and possessed a high level of genetic variation. Meanwhile, other useful molecular markers also used to investigate the diversity between Carex species. Liu et al. [3] reported 30 SSR markers from C. moorcroftii, which provide an available tool to explore the genetic structure and phylogenetic evolution. Starr et al. [27] found that matK barcoding could distinguish between 47% of Carex materials and clearly distinguish phylogenetic diversity relationships in NJ evolutionary trees. Ning et al. [28] proved that ISSR molecular markers are a powerful tool for studying the genetic diversity of Carex. They all found a result that the diversity between Carex species were complex. Nagasawa et al. [29] found that the evolutionary relationships of Carex populations could result in a low level of polymorphism in the populations. Man et al. [30] compared genetic variation and population structure of 15 C.breviculmis populations in Korea and indicated that gene flow was extensive. Although certain progress has been made in studying Carex genetics using molecular markers, there are few studies on SSR molecular markers based on Illumina RNA-sequencing. And the number of materials used are less in the previous research. Moreover, compared to the studies of crops and model plants, molecular studies of Carex are still lacking. It is of great economic value to research the relationships and diversity among Carex species at present.
In our previous study, we used the single-molecule long read sequencing method to investigate the transcriptional regulating network of Carex breviculmis in response to shade tolerance [31]. In the present study, we further explored the SSRs based on the previous Illumina sequencing dataset. The aims of this study were: (1) to enrich Carex transcriptome information and get a better understanding of the function categories from the annotated genes, (2) to develop SSR markers and validate their polymorphism levels, (3) to investigate the genetic background between Carex germplasms.
Results
Illumina sequencing and de novo assembly of transcriptome
The transcriptome of Carex breviculmis was sequenced using the HiSeqTM 2000 platform. A total of 43.67Gb clean data was obtained. The clean data of each sample reached 6.32Gb, and the Q30 base percentage was above 94.03%. A total of 36,403 unigenes were assembled, of which there were 12,657 unigenes with a length of more than 1 kb. The N50 of the unigene was 2016, indicating a high assembly integrity.
Gene annotation based on different databases
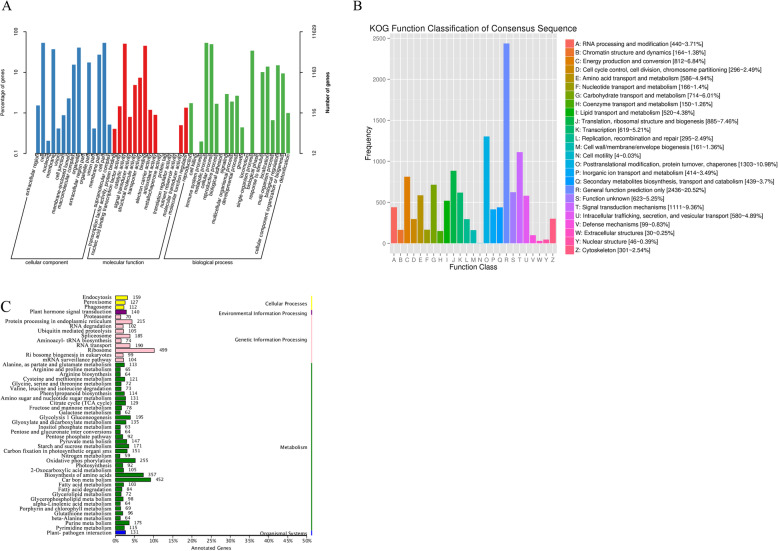
Based on homology analysis of the sequence, 11,629 unigenes (31.95%) were divided into three main GO categories and 50 sub-categories. The GO classification includes ‘Cellular process’, ‘Metabolic process’, and ‘Single-organism process’. (Fig. 1a). The ‘Cell’ was the largest subgroup of cellular components group. The next largest group was ‘Cell part,’ followed by ‘Organelle’, ‘Nucleoid’ and ‘Macromolecular complex’. The categories ‘Catalytic activity’ and ‘Binding’ among ten different molecular function categories for the Carex unigenes were also abundant. According to the KOG database, 11,871 unigenes were categorized into 25 functional groups and 20.52% of unigenes were annotated to ‘General function’ cluster. ‘Post-translational modification’, ‘Protein turnover’ and ‘Chaperones’ (1303 unigenes, 10.98%) was the next largest group and followed by ‘Signal transduction mechanisms’ (1111 unigenes, 9.36%). Alternatively, ‘Nuclear structure’ (46 unigenes, 0.39%), ‘Extracellular structures’ (30 unigenes, 0.25%). Unigenes in ‘Cell motility’ (4 unigenes, 0.03%) groups were significantly less than the above three groups (Fig. 1b). Based on the KEGG database, a total of 8440 unigenes were found, including 40 biological pathways belonging to five large groups (Cellular Processes, Genetic Information Processing, Environmental Information Processing, Metabolism, and Organismal Systems). Three main pathways included Ribosome (499, 5.91%), Carbohydrate metabolism (452, 5.36%), and Biosynthesis of amino acids (357, 4.23%) were in these 50 pathways (Fig. 1c).
Fig. 1.
The gene annotation results based on GO, KOG and KEGG databases. a Gene Ontology (GO) annotated graph of Carex. The results showed that 11,629 unigenes were divided into three main GO categories: ‘Cellular process’, ‘Metabolic process’, and ‘Single-organism process’. b EuKaryotic Ortholog Groups (KOG) classification of Carex. Eleven thousand eight hundred seventy-one unigenes were classified into 25 functional categories. c Kyoto Encyclopedia of Gene and Genomes (KEGG) classification of Carex. The X-axis and Y-axis represent the number of genes and the number of subgroups in each metabolic pathway respectively
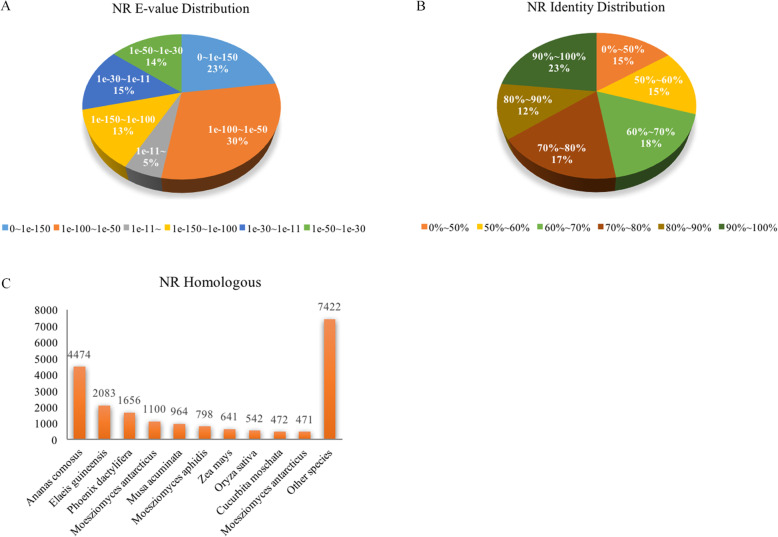
Based on the database of NCBI non-redundant nucleotide, the E-value distribution revealed that 23.00% of unigenes yielded significant hits (Fig. 2a), and approximately 35.00% of unigenes exhibited greater than 80% identity (Fig. 2b). NR protein sequences alignment results revealed that 21.69% could be aligned with Ananas comsous, 10.10% could be aligned with Elaeis guineensis, and 8.03% could be aligned with Phoenix dactylifera (Fig. 2c).
Fig. 2.
Homology searches of Carex breviculmis unigene and characteristics of non-redundant protein databases (Nr). a The E-value distribution of unigene BLASTx hits for every assembly. b BLASTx hit profiles for every assembly of unigene. c Distribution of acessions hit by BLASTx at the top of each assembly of unigene
Frequency and distribution of SSRs in the transcriptome
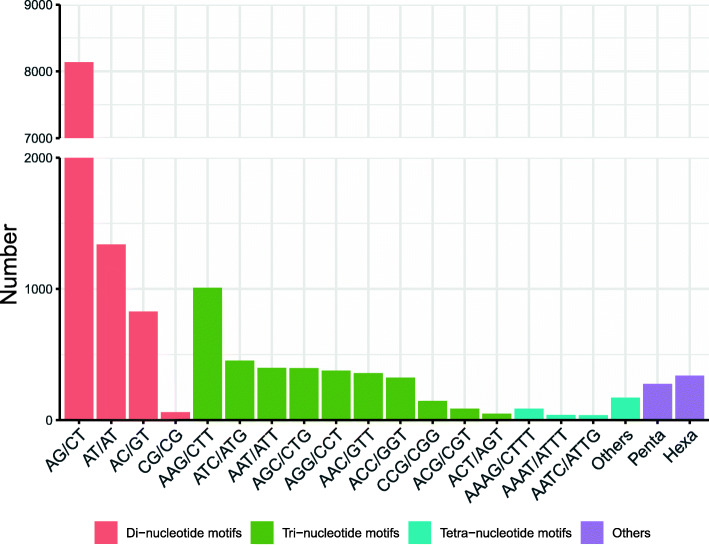
A total of 36,403 unigenes were scanned using the MISA software and 8776 SSR loci were detected (Table 1). The SSR locus in the transcriptome has six types and the number of each repeat type varies greatly. The single repeat motif accounting for 64.93% ranked the most abundant type, whereas the hexa-nucleotides accounting for 1.11% was the least abundant type. The most abundant Di-nucleotide repeats were AC/GT (8138; 18.02%) and followed by AT/AT (1339; 3.10%), AC/GT (827; 1.83%). The most plenty Tri-nucleotide repeats were AAG/CTT (1008; 2.23%) and followed by ATC/ATG (454; 1.01%). Meanwhile, the most affluent tetra-repeat motif types were AAAG/CTTT (87; 0.21%). The number of hexa- and penta-nucleotide motifs were 399 (0.88%) and 377 (0.83%), respectively (Fig. 3).
Table 1.
Prediction of SSRs out of our transcript datasets of Carex breviculmis
| Item | Number |
|---|---|
| Total number of sequences examined | 36,403 |
| Total size of examined sequences (bp) | 41,724,615 |
| Total number of identified SSRs | 8776 |
| Number of SSR containing sequences | 6018 |
| Number of sequences containing more than one SSR | 504 |
| Number of SSRs present in compound formation | 20 |
| Mono nucleotide | 5699 |
| Di nucleotide | 1873 |
| Tri nucleotide | 582 |
| Tetra nucleotide | 63 |
| Penta nucleotide | 15 |
| Hexa nucleotide | 20 |
Fig. 3.
Type distribution of SSRs identifed in Carex breviculmis unigenes
Development and transferability assessment of novel SSRs
We designed and synthesized 96 pairs to amplify 11 phenotypic difference Carex materials (Table S1). Among them, 42 (43.75%) pairs can amplify several bands and have high polymorphism. The primers indicated good transferability between different Carex species. The number of not amplified bands accounts for 15.6% and others showed low polymorphism or no polymorphism.
Genetic diversity statistics
In the set of 42 SSRs, we recognized 180 marker alleles across the 79 accessions. Among the 42 SSRs, PIC value ranged from 0.133 and 0.494, with an average of 0.259. SSRs displayed wide genetic variation among accessions. The genetic diversity between Carex materials was investigated by cluster analysis, principal component analysis. The polymorphic ratio was 100% and an average of 4.3 primers was amplified per primer. The number of observed alleles (Na), the number of effective alleles (Ne), Nei’s (1973) gene diversity (H), and the Shannon information index (I) were 2.000, 1.376, 0.243, and 0.391, respectively, indicating that the genetic diversity between the Carex accessions was high. We also calculated the genetic distance between accessions (Table S2), which ranged from 0.222 to 1.000. The genetic distance average value was 0.548.
Cluster analysis of Carex based on SSR markers
In order to reveal the classification information of Carex species, we obtained the allele frequency according to our original data. Instead of using a priori classification such as provenance or taxonomy, we used NJ, PCoA and UPGMA cluster analysis and combined the results to explore the genetic information and classification of all accessions.
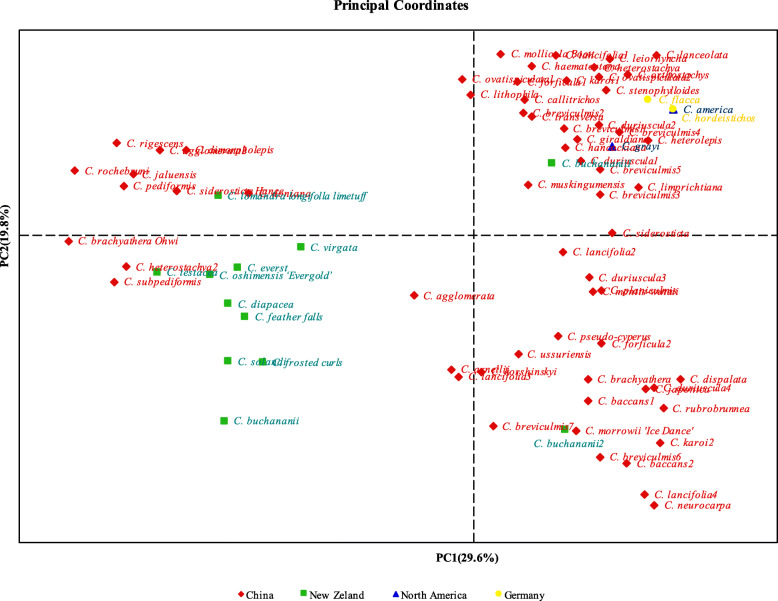
Principal coordinate (PCoA) results showed that Axis 2 separated and generated two genetically differentiated groups of Carex accessions (Fig. 4). The first principal component accounts for 29.6% and the second principal component accounts for 19.8%.
Fig. 4.
PCoA analysis of 79 materials of Carex. The first principal component accounts for 29.6% and the second principal component accounts for 19.8%. The red diamond represents the germplasm resources from China, the green square represents New Zealand, the blue triangle represents North America, and the yellow circle represents Germany
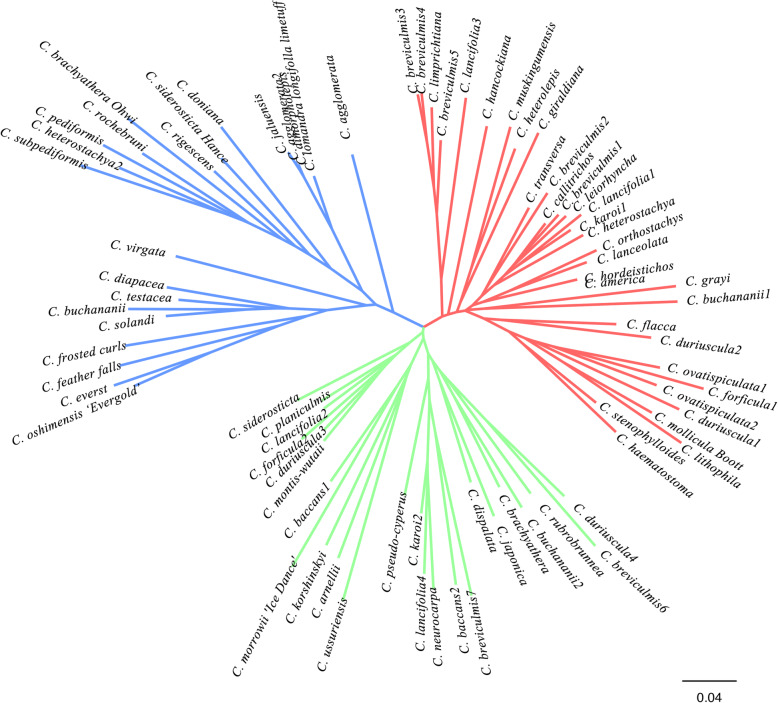
Based on the distance calculation method of Shered Allele, the Neibor-Joining phylogenetic analysis divided 79 Carex accessions into three groups (Fig. 5). The Group I has a total of 22 materials and C. jaluensis, C. dimorpholepis, C. agglomerata2 are grouped into very similar categories. Also the commercial plant materials from New Zealand are grouped into one category in Group I, including C. virgata, C. frosted curls, C. solandi, C. oshimensis’Evergold’, C. feather falls, C. buchananii, C. everst, C. diapacea, C. testacea, C. lomandra longifolla limetuff. The Group II includes 24 materials, C. subpediformis and C. jaluensis are the most similar accessions. The Group III includes 33 materials and most of which are from all over China, but also four materials are from Germany and North America. It is worth mentioning that the C. breviculmis collected from various provinces in China are all classified into this group.
Fig. 5.
N-J phylogenetic tree of 79 Carex accessions. N-J tree included three major clusters, including Group I (22 accessions) which is blue, II (24 accessions) green and III (33 accessions) red
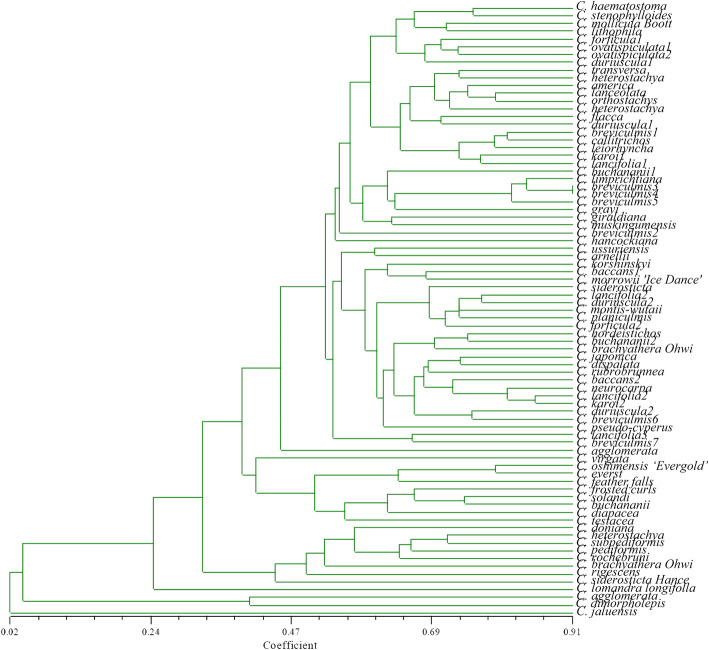
Based on the SSR original data, Dice coefficient method was used to calculate the similarity. A total of 79 materials were clustered using the UPGMA method (Fig. 6). The similarity between genotypes was 0.070 ~ 0.786. Through UPGMA clustering, 79 accessions of germplasm resources were divided into two major groups. C. jaluensis was assigned to a separate group. The remaining 78 domains of Carex accessions were divided into two subgroups, and C. agglomerata, C. dimorpholepis and C. lomandra longifolla limetuff were classified into one group. While nine accessions from New Zealand were clustered into one group when the genetic distance was 0.42, but C. lomandra longifolla limetuff was not clustered into this group. Although it was not clustered with the accessions from New Zealand, the genetic similarity was relatively high. Regarding the collection sources of Carex, there is a certain degree of gene exchange between Carex germplasm from China and New Zealand, Germany and North America. However, the resources cannot be divided completely by region in general. The Sorensen-Dice correlation coefficient was r = 0.941, indicating that the clustering results were reliable.
Fig. 6.
UPGMA clustering analysis of 79 Carex
AMOVA analysis of Carex accessions based on different methods
In order to evaluate the genetic differences between these germplasms, we calculated the FST values between all pairs of accessions for the two levels studied (origin and genetic classification). Through different analysis of source, the AMOVA indicated that 88% of the total genetic variation of 79 accessions was within populations, and 12% was among populations. The results of AMOVA analysis showed that some subspecies have large variations within countries, which was consistent with the above results of accessions from different countries. While the results of AMOVA analysis based on NJ cluster analysis showed that 89% within populations (Table 2). FST values and probability P (rand ≥ data) were 0.117 and 0.052 respectively.
Table 2.
AMOVA analysis of Carex accessions
| Source | df | SS | MS | Est. Var. | % | |
|---|---|---|---|---|---|---|
| Regions | Among Pops | 3 | 181.719 | 60.573 | 3.667 | 12% |
| Within Pops | 75 | 2082.687 | 27.769 | 27.769 | 88% | |
| Total | 78 | 2264.405 | 31.436 | 100% | ||
| Taxa | Among Pops | 2 | 217.629 | 108.814 | 3.162 | 11% |
| Within Pops | 76 | 2046.777 | 26.931 | 26.931 | 89% | |
| Total | 78 | 2264.405 | 30.093 | 100% |
Note: AMOVA results for 4 regions of origin and 3 taxa. Fst values and probability P (ran≥data) were as follows: Region of Origin (0.117;0.052), Taxa (0.105;0.001)
Df Degrees of Freedom, SS Sum of squares, MS Mean squares, Est. Var. estimated variance, % proportion of genetic variability
Fingerprint
Results showed that 42 pairs of SSR markers can be efficient in differentiating between 79 accessions, and a fingerprint map, which can distinguish more varieties at a time, was established (Table S3).
Discussion
Our previous work reported the transcriptional regulation network of C. breviculmis in response to shade stress using single-molecule long-read sequencing [31]. In the present study, we utilized the Illumina RNA-seq data in order to generate SSR markers and thus to explore the genetic diversity of other 79 Carex accessions. We released a large quantity of expressed gene sequences (36,403 unigene sequences). In order to get a better understanding of the function categories of Carex genes, we searched the GO, KOG, KEGG pathway mapping databases. A total of 20,948 unigenes annotation results were obtained. The results showed that 57.54% of the genes were successfully annotated, reflecting the high transcriptome diversity of Carex. Gene Ontology is a global classification system of the gene function, classifies the characteristics of genes into groups of ‘biological processes’, ‘cellular components’ and ‘molecular functions’ [32]. KOG analysis was devoted to classify orthologous groups for eukaryotic complete genomes. KEGG analysis is one of the most frequently used method to analyze gene metabolic products and its putative functions [33]. As in the results of Li et al. [34], the GO database classifies annotation information into three categories: biological processes, cellular components, and molecular functions. The largest subcategory was ‘binding’, while followed by ‘catalytic activity’. We also found that the KEGG pathway had the largest group of genes belonging to the metabolism category, while the genetic information processing category was the second largest group. Based on NR alignment results, 21.69% of sequences could be aligned with those of Ananas comsous. It was indicated that C. breviculmis and Bromeliaceae family members has closest relationship with respect to protein alignment. Ananas comsous belongs to Bromeliaceae family, while C. breviculmis belongs to Cyperaceae. Considering the taxonomy of plants, A. comsous and C. breviculmis seem to be genetically distant species. The reason for the unexpected protein alignment could be the lack of data on Cyperaceae-related species in the current NR database. The results show that it is necessary and urgent to update the genetic database of this genus.
Gene structure analysis was performed based on the unigene library, in which SSR analysis obtained a total of 8776 SSR markers. Among the detected sites, 5699 single nucleotide repeats accounting for the total number of SSR sites of 64.93%. Followed by trinucleotides and dinucleotides (1873 and 582, respectively, 21.37 and 6.63%), which are different from the results that three Nucleotide repetitive sequences are the most abundant repetitive units in radish, with a frequency of 52% [35]. It indicated that the specificity of SSR sites is different in plants. Among all the repeat motifs of 303 functional primer pairs, AC/GT (18.02%) is the most abundant dinucleotide repeat in Carex breviculmis.
Although there are a lot of researches on Carex, more researches are on a certain kind or a restricted property of it. However, the genetic background and genetic relationship are unclear, especially between and among Carex species. It restricted the introduction of Carex resources, rational selection of hybrid parents, and genetic engineering breeding. In particular, studies on the genetic level of Carex have not revealed evolutionary issues such as genetic relationship.
In this research, we found that 42 SSR primer pairs amplified 178 alleles in 79 Carex accessions, with an average of 4.3 alleles per microsatellite. Compared with previous studies, the ratio of polymorphic 100% were higher than the study of C. sempervirens used RAPD markers [36]. It indicted that the development efficiency and polymorphism of SSR markers developed by transcriptome sequencing are more efficient than RAPD markers. We also found that PIC value in this research was lower than the value of 0.83 reported by Ning et al. [28] and similar to the value obtained by Nagasawa et al. [29]. PIC values are used to measure the level of population polymorphism in other plants and they depend on the accessions tested. Locus polymorphism can be divided into high level (PIC > 0.5), medium level (0.5 > PIC > 0.25) and low level (PIC < 0.25) according to their information content [37]. In case of the number of Carex and molecular markers used in Ning’s study were small, and all of the Carex accessions were from the same region in the Shandong Province. Secondly, ISSR used by Ning et al. [28] is probably more efficient than using specie specific SSR markers. The PIC value is similar to Nagasawa et al. [29] research results that 20 of EST-SSR markers developed with low polymorphism in C. angustisquama population and King et al. [38] results that identified 11 microsatellite loci from Carex macrocephala. Due to the species’ population dynamics, the low genetic polymorphism were obtained, rather than to null alleles at the developed markers. Carex germplasms are considered to have the lower PIC values which identified in this study appeared to reflect this low diversity and the low genetic variation in Carex resulted from the species history, and not from the characteristics of the used markers.
In cluster analysis, we used a combination of NJ, UPGMA and PCoA analysis methods. Firstly, by the reason of the growth environment is similar and the nine accessions are all commercial materials introduced and domesticated in New Zealand, the germplasm from New Zealand are consistent in the classification to one group. Same with the research of Ning et al. [28], the results of the species from the same area could be classified into one category, which is correlated with geographical distribution and environmental conditions. Secondly, because of similar genetic backgrounds and morphological characteristics or their common geographic origin, C. grayi and C. hordeisticho of NJ Group III had obvious characteristics. Also C. breviculmis and C. lancifolia from China were clustered into Group III of closely related species, which is consistent with the traditional classification method. The morphology of these two Carex species are very similar, and even hard to distinguish, but it can be achieved using the clustering method of molecular markers.
There was a gene exchange with the Chinese test materials during the cultivation process that C. buchananii1 and C. buchananii2 are mixed with Chinese materials to varying degrees. The materials from Germany and North America could not be classified into a single category due to the sample size, but we found that the collection place of the materials would have a certain impact on the classification of Carex. Furthermore, through the analysis of principal components, it can be seen that Carex plants are not clearly classified. Perhaps they still have large genetic differences among plants of the same genus and no definite result. Therefore, further studies involving more foreign germplasms are still needed in order to better interpret this phenomenon.
In addition, the PCoA based on these genotypic data clearly showed that there was an obvious genetic differentiation among Carex accessions. The clustering results of PCoA and UPGMA were partially consistent and showed significant differences among all the analyzed accessions. The result of fingerprint is valuable for Carex species with extremely similar appearance that don’t be easily distinguished. Previous studies have showed that SSR markers can be used for plant species diversity analysis and fingerprint development [39]. For example, they were used in establishing the fingerprint of 36 Chinese jujube cultivars with 12 pairs of newly developed SSR primers [40], and eight SSR loci were further recommended as a core marker set for fingerprinting of the tea plant [41]. In this study, we concluded that different combinations of primer pairs could be used to distinguish different species of Carex, which is of great significance in the selection of hybrids for breeding new varieties.
Conclusion
The transcriptomic analysis shed new light on the function categories from the annotated genes and will facilitate future gene functional studies. The genetic background analysis indicated that gene flow was extensive among 79 Carex species. These markers can be used to assess genetic diversity and to investigate the evolutionary history of Carex and related species, as well as to serve as a guide in future breeding projects.
Methods
Plant material and DNA extraction
A total of 79 Carex accessions were collected from different locations, including 64 from China, 11 from New Zealand, 2 from the North America, and 2 from Germany. The 64 accessions from China were sampled from the Beijing Botanical Garden, Mount Tai, Xi’an Botanical Garden, Hebei, Shanghai, and the base of the Beijing Academy of Agricultural and Forestry Sciences (Table 3). We have acquired a permission to purchase or collect all of the plant materials under the guidelines of local organizations. The formal identification of the samples was carried out by two plant taxonomists of our group, Dr. Chen and Dr. Xun, referring to the Flora Reipublicae Popularis Sinicae and based on the commercial introductions. Genomic DNA was extracted from healthy young shoots by mixing 10 individual plants of each culture separately using the CTAB method [42]. Using 1% agarose gel electrophoresis to examine the quality and integrity of genomic DNA. DNA concentration was determined through with the NanoDrop 2000 spectrophotometer (NanoDrop, Thermo Fisher Scientific, Wilmington, DE, USA). The extracted DNA was diluted with ddH2O to 20 ng/mL and used as the template and inventory for PCR amplification. The extracted DNA was stored at -20 °C [43].
Table 3.
The 79 Carex materials information
| Code | Name | Origin |
|---|---|---|
| 1 | C. haematostoma | Yunnan, China |
| 2 | C. mollicula Boott | Beijing, China |
| 3 | C. transversa | Beijing, China |
| 4 | C. forficula1 | Beijing, China |
| 5 | C. breviculmis1 | Beijing, China |
| 6 | C. breviculmis2 | Beijing, China |
| 7 | C. ovatispiculata1 | Beijing, China |
| 8 | C. leiorhyncha | Beijing, China |
| 9 | C. lithophila | Beijing, China |
| 10 | C. ovatispiculata2 | Beijing, China |
| 11 | C. heterostachya | Beijing, China |
| 12 | C. karoi1 | Beijing, China |
| 13 | C. callitrichos | Beijing, China |
| 14 | C. lancifolia1 | Beijing, China |
| 15 | C. limprichtiana | Beijing, China |
| 16 | C. duriuscula1 | Beijing, China |
| 17 | C. breviculmis3 | Beijing, China |
| 18 | C. breviculmis4 | Beijing, China |
| 19 | C. stenophylloides | Beijing, China |
| 20 | C. breviculmis5 | Beijing, China |
| 21 | C. duriuscula2 | Beijing, China |
| 22 | C. giraldiana | Beijing, China |
| 23 | C. muskingumensis | Beijing, China |
| 24 | C. heterolepis | Beijing, China |
| 25 | C. lanceolata | Beijing, China |
| 26 | C. orthostachys | Beijing, China |
| 27 | C. hancockiana | Beijing, China |
| 28 | C. siderosticta | Beijing, China |
| 29 | C. lancifolia2 | Beijing, China |
| 30 | C. ussuriensis | Beijing, China |
| 31 | C. planiculmis | Beijing, China |
| 32 | C. arnellii | Beijing, China |
| 33 | C. lancifolia3 | Shandong, China |
| 34 | C. japonica | Beijing, China |
| 35 | C. montis-wutaii | Hebei, China |
| 36 | C. korshinskyi | Xi’an, China |
| 37 | C. forficula2 | Shandong, China |
| 38 | C. duriuscula3 | Beijing, China |
| 39 | C. baccans1 | Xi’an, China |
| 40 | C. dispalata | Xi’an, China |
| 41 | C. brachyathera | Xi’an, China |
| 42 | C. agglomerata | Xi’an, China |
| 43 | C. duriuscula4 | Xi’an, China |
| 44 | C. rubrobrunnea | Xi’an, China |
| 45 | C. morrowii ‘Ice Dance’ | Xi’an, China |
| 46 | C. breviculmis6 | Xi’an, China |
| 47 | C. baccans2 | Beijing, China |
| 48 | C. neurocarpa | Shaanxi, China |
| 49 | C. breviculmis7 | Shandong, China |
| 50 | C. lancifolia4 | Shandong, China |
| 51 | C. pseudo-cyperus | Shandong, China |
| 52 | C. karoi2 | Xi’an, China |
| 53 | C. agglomerata2 | Beijing, China |
| 54 | C. doniana | Xi’an, China |
| 55 | C. heterostachya2 | Xi’an, China |
| 56 | C. rochebruni | Xi’an, China |
| 57 | C. dimorpholepis | Xi’an, China |
| 58 | C. pediformis | Beijing, China |
| 59 | C. brachyathera Ohwi | Beijing, China |
| 60 | C. rigescens | Beijing, China |
| 61 | C. subpediformis | Shanghai, China |
| 62 | C. jaluensis | Shanghai, China |
| 63 | C. siderosticta Hance | Shanghai, China |
| 64 | C. buchananii1 | Shanghai, China |
| 65 | C. buchananii2 | ODERINGS, New Zealand |
| 66 | C. virgata | ZELANDIA, New Zealand |
| 67 | C. frosted curls | ODERINGS, New Zealand |
| 68 | C. solandi | ZELANDIA, New Zealand |
| 69 | C. oshimensis ‘Evergold’ | ZELANDIA, New Zealand |
| 70 | C. feather falls | ZELANDIA, New Zealand |
| 71 | C. buchananii | ODERINGS, New Zealand |
| 72 | C. everst | ODERINGS, New Zealand |
| 73 | C. diapacea | ODERINGS, New Zealand |
| 74 | C. testacea | ODERINGS, New Zealand |
| 75 | C. lomandra longifolla limetuff | ODERINGS, New Zealand |
| 76 | C. grayi | North America |
| 77 | C. america | North America |
| 78 | C. flacca | Germany |
| 79 | C. hordeistichos | Germany |
Transcriptome sequencing, de novo assembly and function annotation
Transcriptome sequencing of Carex breviculmis was performed by Biomarker Technologies for RNA extraction, and sequencing was performed on the HiSeq™ 2000 platform [31]. In this study, TRINITY Version 2.5.1 was used to detect contigs from the same transcript, to determine the distance between contigs and connect them together to obtain contigs with inextensible ends [44]. TGICL software was used to splice these single genes and remove their redundancy to obtain non-redundant single genes. Using the BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches (e-value <1E-5) of the assembled unigenes [45, 46]. The sequence is compared to the following databases: Gene Ontology (GO), Eukaryotic Orthologous Groups of proteins (KOG), the Kyoto Encyclopedia of Genes and Genomes (KEGG), and National Center for Biotechnology Information for non-redundant proteins (NR).
SSRs of the transcriptome were identified using MISA [47]. Using Primer3 (http://primer3.sourceforge.net/releases.php) for each SSR primer design. SSR loci contained motifs of two to six nucleotides in size were preferentially selected. The other principle of selection was that six minimum repeating units of di-nucleotide, five tri-nucleotides, and four of all higher order motifes, including tetra-nucleotide, penta-nucleotide, and hexa-nucleotide.
Identification of SSRs, and primer design
Amplification of SSR markers was carried out using DNA of 11 Carex species with large phenotypic differences. Ninety-six SSR primers, which were identified in Carex breviculmis using RNA-seq, were randomly selected for this research. All primers were synthesized by RuiBo Biotech. Among the ninty-six synthesized pairs of primers, 42 pairs had higher levels of polymorphisms and were used in further experimental research (Table 4). All reactions were conducted using BIO-RAD T100 Thermal Cycler™. The PCR reaction system were carried out in a total volume of 10 μL, including 5 μL of 2 × Taq Master Mix, 0.2 μL of primer, 2 μL of genomic DNA, and 2.6 μL of ddH2O. The thermal profile used for amplifications consisted of 10 min of initial denaturation at 94 °C, followed by 34 cycles of 30 s at 94 °C, 30 s at the optimized annealing temperature, 60 s of extension at 72 °C, and a final extension of 5 min at 72 °C. Used 8.0% non-denatured polyacrylamide gels with a 100-bp ladder marker (TRANSGEN BIOTECH, Beijing, China) to separate successful PCR products and visualized by silver staining [48]. DNA of 79 Carex individuals was amplified using SSR primers to analyze genetic diversity. Clear bands on the gel images under the light lamp were observed, with or without bands as (1) or (0).
Table 4.
The SSRs primer information identified for validation in this study
| Primer | #Gene_ID | SSR | Forward primer(5′-3′) | Reverse primer(5′-3′) |
|---|---|---|---|---|
| CAREX001 | F01.1 | (AG)6 | CACTGGAGAACCTAGCGACC | TTGTACAAGGTCCAGGGAGAA |
| CAREX006 | F01.141 | (TC)7 | CGTTCCCCGTTTTCTTCTCT | GCCGTCTTCTTTGAAAACCA |
| CAREX008 | F01.166 | (TC)9 | CAGTATGGTGGTGAGAGCGA | CACAGACCGAACCTAACAACAA |
| CAREX010 | F01.180 | (TC)7 | GCTTCGTTGTCTACTAGCCCC | GACCAATCCAGCTGAGAAGC |
| CAREX012 | F01.240 | (CT)15 | CCACACAGCTTATTGCTTGC | TGATAGGTGGGTTTCTTGCC |
| CAREX015 | F01.339 | (AG)10 | TGATTTTTCCAATGCGTGAA | TGCCAGTTGAATCTCAGTGC |
| CAREX016 | F01.3449 | (AG)7 | CCCCTTCAATTCAATGCTGT | GCAATGAGAGGGAAAATCCA |
| CAREX017 | F01.3529 | (GA)10 | TGAATCATTGAAGGAGAGAGCA | GGTTGTTGCAAAGGAAGAGC |
| CAREX018 | F01.357 | (AG)7 | TTTCTAACCCTTTATCGCCG | AAAATTGCCTGGAGGAGGAG |
| CAREX019 | F01.364 | (AT)6 | ATCATGCGGCCAAGATAAAG | CAAGCAGGGGTGGAGAATAG |
| CAREX023 | F01.463 | (TC)6 | TAGTGCTGCCAGAAAGAGCA | AAACTCAACCCGAAAAGGCT |
| CAREX024 | F01.501 | (CT)8 | GTCCCCAAAACCTCTGTAGC | TGCTTGTTGTTCGCTTCATC |
| CAREX025 | F01.5079 | (AG)10 | TGCATGCAGCTGGAATAGAG | CTCCAAATCCGAACTATCCG |
| CAREX026 | F01.5099 | (CT)6 | AGCCTCTCTCTCTCTCCGCT | GCAAAAATGCCTGAGTGGAT |
| CAREX030 | F01.608 | (CT)14 | CGCCTCTTCATCGATCTTTC | GCCCCAATAATGGAGAGGAT |
| CAREX031 | F01.612 | (CT)9 | CCCAATCTCCAAAGAGCAAA | AGAACGAGACCTGGAGCTGA |
| CAREX037 | F01.735 | (AT)6 | GCAGGGTTGTTGAAGGTTTG | TTGTGGATGCAAAACAGCAT |
| CAREX040 | F01.789 | (TG)17 | TGCTGTTCTCATGGCTTCTG | CCTTTCATTTTGATGAGGCAA |
| CAREX041 | F01.792 | (AG)7 | CAAAAAGGAAGCGAAAGGAA | TGAGAGAGGAGATCGGAGGA |
| CAREX042 | F01.809 | (TA)8 | ACAAAAGAGCTCGCTGGAAA | TCTGATTGCTGCTCAACTATCTC |
| CAREX048 | F01.1070 | (GCT)5 | CTTCATTTCCGCCTCTCTTG | GCAATCATTATGCAATGCCA |
| CAREX044 | F01.854 | (CT)6 | ACACAGGGACAAGCCGATAG | CAACAAGCACAACAATAACCA |
| CAREX045 | F01.855 | (CT)6 | CCAAAAGATATCATCATCTCCGA | TGAGCAGCGATCTCTTTGAA |
| CAREX046 | F01.930 | (AT)6 | TCTTTTTGCCAAGATGGTGA | GTGCCAAGCATCAATCAGAA |
| CAREX050 | F01.151 | (CTC)6 | TCATTAGCTGGTCGCTTCCT | CATGCCCATTGTTCTTGATG |
| CAREX053 | F01.3450 | (CTG)5 | ACCCAGTGATCGTACTTCGG | AGATTCAATTTCCACCGTCG |
| CAREX054 | F01.3506 | (ACC)5 | TCAACCCGCTACACCTAACC | ACACGCTCCAGGTCAGAGAT |
| CAREX055 | F01.3514 | (AAC)5 | CCACACCTCCTACTCCTCCA | CTCCCCGTTGAAGTTGTTGT |
| CAREX060 | F01.5072 | (ATT)5 | TACGCCATTGTCAACGAAAA | GCACGAGACACCTGAACACA |
| CAREX062 | F01.5103 | (CAG)5 | AGGCACCACAAGATCCAAAC | GCTCCCATCCATACAGCTTC |
| CAREX064 | F01.511 | (CAC)9 | TCCTCAGGTAGCGAAAGCAG | TACCTTAATTGCGGAATCGG |
| CAREX069 | F01.52166 | (ATA)5 | GTACCTGCCCTGGATTCTGA | GGCGCTTTTACCAAATCAAA |
| CAREX070 | F01.592 | (ATT)6 | ATTCATCAGGCAATTCTGGC | GGCTAAACCAACTCCTGCAC |
| CAREX072 | F01.675 | (GAT)5 | CTGATCTCAAAAGGCGAAGG | CACGTAGGGATCACCCAATC |
| CAREX073 | F01.719 | (AGA)5 | TAAAATGAAGGGCGAGGATG | CACTCCGTGATGATGTGGTC |
| CAREX076 | F01.785 | (GAT)8 | TGAAGAAGGGCCTGGTACTG | AACCACCAGATCCCACAGTC |
| CAREX083 | F01.1429 | (AGA)5 | GGGAATCACAGACAAAGGGA | ATACTGGCAGAACCAATGGC |
| CAREX086 | F01.11991 | (CTT)6 | TACGGTACAGGCGTCTCTCA | CAGAAGCAACGCAACACATT |
| CAREX094 | F01.9172 | (GAAT)5 | ATGGTTTCTGATTTCCTGCG | GGGGTCAAATGTAGTTGCAGA |
| CAREX095 | F01.9272 | (TAAT)8 | CCTTCAAAAGAGAACCGAGC | TGTCGCCTTTGTGAGCATAG |
| CAREX096 | F01.17075 | (TCTCT)5 | GACTCCGACTCCAGTTGAGC | TCGAGGAGCTGTCCTTGAAT |
| CAREX097 | F01.21485 | (CCTCT)5 | CCCATCATCGATCAATCACA | GGAACAACGATCGGAAAGAA |
Genetic diversity analysis
A binary qualitative data matrix was constructed and analyzed using POPGENE Version 1.3.2. Genetic diversity of different materials was determined by calculating the percentage of polymorphic bands (PPB), the effective number of alleles (Ne), observed number of alleles (Na), Nei’s gene diversity (H), and Shannon’s information index (I). The polymorphic information (PIC) of a band was calculated by the following formula: (Pi and Pj are the frequencies of the ith and jth alleles at one locus). We used the software NTSYS Version 2.1 [49] to construct a cluster analysis description of selected group Q pattern based on Nei’s genetic distances [50]. Afterwards, the unweighted pairwise method of arithmetic mean is used to analyze the parameters.
Cluster analysis and AMOVA analysis of 79 accessions
The similarity of Carex was evaluated using NTSYS Version 2.1. According to the similarity matrix of SSR data set, the UPGMA clustering method was used to construct the dendrogram [51]. Using PowerMarker version 3.25 and MEGA 5, the unweighted phylogenetic tree was constructed based on the Dice dissimilarity matrix between 79 individuals [52, 53]. Based on the Bootstrap function of FreeTree program, the robustness of phylogenetic trees was evaluated through 1000 repeated bootstrap analyses [54]. Principal coordinate analysis (PCoA) was performed according to the anastomotic differences between binary genotypic profiles using the GenAlEx 6.5 program based on the pairwise distance matrix [55]. Both distance and covariance were standardized.
The tratified genetic variation between and within geographic groups was analyzed using Analysis of Molecular Variance (AMOVA) [56]. F statistic was used to analyze the genetic differentiation between populations. Both analyses were performed using the GenAlEx 6.5 software [55].
Supplementary Information
Additional file 1: Table S1. The information of 11 Carex materials to select primers.
Additional file 2: Table S2. The Nei’s genetic distance between Carex species.
Additional file 3: Table S3. The fingerprint of 79 Carex materials.
Acknowledgements
We acknowledge the Biomarker Corporation (Beijing, China) for the facilities and expertise of Illumina platform for libraries construction and sequencing. We also thank the Editage Company (www.editage.com) for language editing.
Abbreviations
- AFLP
Amplified fragment length polymorphism
- AMOVA
Analysis of molecular variance
- GO
Gene ontology
- H
The gene diversity index
- I
The Shannon information index
- ISSR
Inter-simple sequence repeat
- KEGG
Kyoto encyclopedia of genes and genomes
- KOG
EuKaryotic ortholog groups
- Na
The average number of observed alleles
- Ne
The effective alleles
- NJ
Neighbor-joining
- NR
NCBI non-redundant protein databases
- PCoA
Principal coordinate analysis
- PCR
Polymerase chain reaction
- PIC
The polymorphic information content
- RAPD
Random amplified polymorphic
- RFLP
Restriction fragment length polymorphism
- SRAP
Sequence related amplified polymorphism
- SSR
Simple sequence repeat
Authors’ contributions
ZC and XF conceived the study and designed the experiments. LL performed the experiment. KT, PT and LL analyzed the data with suggestions by JW, HZ, CH, CC, LX, and WG. LL and KT wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding
This research was supported by the Beijing Natural Science Foundation (No.6204039), National Natural Science Foundation of China (31901397), and Scientific Funds of Beijing Academy of Agriculture and Forestry Sciences to KT. Each of the funding bodies granted the funds based on a research proposal. They had no influence over the experimental design, data analysis or interpretation, or writing the manuscript.
Availability of data and materials
The Illumina NGS reads generated in this study has been submitted to the BioProject database of National Center for Biotechnology Information (PRJNA488506).
Ethics approval and consent to participate
The materials used in this study were collected by ourselves. And we complied with all relevant institutional, national and international guidelines and specify the appropriate permissions obtained. We also have acquired a permission to collect all of the plant materials.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lingyun Liu, Email: liulingyun01@163.com.
Xifeng Fan, Email: fanxifengcau@163.com.
Penghui Tan, Email: penghuitan@bjfu.edu.cn.
Juying Wu, Email: wujuying@grass-env.com.
Hui Zhang, Email: peakzeal@163.com.
Chao Han, Email: han_chao1988@126.com.
Chao Chen, Email: xiaoyingwulu@163.com.
Lulu Xun, Email: xunlulu20032006@126.com.
Weier Guo, Email: weguo@ucdavis.edu.
Zhihui Chang, Email: changzh@bjfu.edu.cn.
Ke Teng, Email: tengke.123@163.com.
References
- 1.Pedersen MAT, Nowak MD, Brysting AK, Elven R, Bjorå CS. Hybrid origins of Carex rostrata var. borealis and C. stenolepis, two problematic taxa in carex section vesicariae (cyperaceae). PloS One. 2016;11(10):1. [DOI] [PMC free article] [PubMed]
- 2.Gillespie EL, Pauley AG, Haffner ML, Hay NM, Estep MC, Murrell ZE. Fourteen polymorphic microsatellite markers for a widespread limestone endemic, Carex eburnea (Cyperaceae: Carex sect. Albae). Appl Plant Sci. 2017;5(8):1700031. [DOI] [PMC free article] [PubMed]
- 3.Liu W, Zhou Y, Liao H, Zhao Y, Song Z. Microsatellite primers in Carex moorcroftii (Cyperaceae), a dominant species of the steppe on the Qinghai-Tibetan plateau. Am J Bot. 2011;98(12):e382–4. [DOI] [PubMed]
- 4.Ye YR, Wang WL, Zheng CS, Fu J, Liu HW. Evaluation of cold resistance of four wild Carex speices. Chin J Appl Ecol. 2017;28(1):89–95. [DOI] [PubMed]
- 5.Wang Y, Yang H, Li X, Wang W, Bai C, Liu G. Carex jianfengensis (Carex sect. Rhomboidales, Cyperaceae), a New Species from Hainan, China. Plos One. 2015;10(9):e0136373. [DOI] [PMC free article] [PubMed]
- 6.Léveillé-Bourret É, Starr JR, Ford BA. Why are there so many sedges? Sumatroscirpeae, a missing piece in the evolutionary puzzle of the giant genus Carex (Cyperaceae). Mol Phylogenet Evol. 2018;119:93–104. [DOI] [PubMed]
- 7.Benítez-Benítez C, Escudero M, Rodríguez-Sánchez F, Martín-Bravo S, Jiménez-Mejías P. Pliocene-Pleistocene ecological niche evolution shapes the phylogeography of a Mediterranean plant group. Mol Ecol. 2018;27(7):1696–1713. doi: 10.1111/mec.14567. [DOI] [PubMed] [Google Scholar]
- 8.Míguez M, Gehrke B, Maguilla E, Jiménez-Mejías P, Martín-Bravo S. Carex sect. Rhynchocystis (Cyperaceae): a Miocene subtropical relict in the Western Palaearctic showing a dispersal-derived Rand Flora pattern. J Biogeography. 2017.
- 9.Martín-Bravo S, Jiménez-Mejías P, Villaverde T, Escudero M, Hahn M, Spalink D, Roalson EH, Hipp AL, Benítez-Benítez C, Bruederle LP, et al. A tale of worldwide success: Behind the scenes of Carex (Cyperaceae) biogeography and diversification. J Syst Evol. 2019;57(6).
- 10.Group GC. Making Carex monophyletic (Cyperaceae, tribe Cariceae): a new broader circumscription. Bot J Linn Soc. 2015;179(1).
- 11.Group TGC, Jiménez-Mejías P, Hahn M, Lueders K, Starr JR, Brown BH, Chouinard BN, Chung KS, Escudero M, Ford BA. Megaphylogenetic specimen-level approaches to the Carex (Cyperaceae) phylogeny using ITS, ETS, and matK sequences: implications for classification. Syst Bot. 2016.
- 12.Jones CJ, Edwards KJ, Castaglione S, Winfield MO, Sala F, Wiel CVD, Bredemeijer G, Vosman B, Matthes M, Daly A. Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Mol Breed. 1997;3(5):381–390. [Google Scholar]
- 13.Hokanson SC, Szewc-McFadden AK, Lamboy WF, McFerson JR. Microsatellite (SSR) markers reveal genetic identities, genetic diversity and relationships in a Malus x domestica borkh. Core subset collection. Theor Appl Genet. 1998;97(5–6):671–83.
- 14.Escudero M, Vargas P, Arens P, Ouborg NJ, Luceno M. The east-west-north colonization history of the Mediterranean and Europe by the coastal plant Carex extensa (Cyperaceae). Mol Ecol. 2010;19(2):352–70. [DOI] [PubMed]
- 15.Zhang Z, Xie W, Zhao Y, Zhang J, Wang Y. EST-SSR marker development based on RNA-sequencing of E. sibiricus and its application for phylogenetic relationships analysis of seventeen Elymus species. BMC Plant Biol. 2019;19(1). [DOI] [PMC free article] [PubMed]
- 16.Pan L, Huang T, Yang Z, Tang L, Cheng Y, Wang J, Ma X, Zhang X. EST-SSR marker characterization based on RNA-sequencing of Lolium multiflorum and cross transferability to related species. Mol Breed. 2018;38(6).
- 17.Alisoltani A, Ebrahimi S, Azarian S, Hematyar M, Rafiei F. Parallel consideration of SSRs and differentially expressed genes under abiotic stress for targeted development of functional markers in almond and related Prunus species. Sci Hortic-Amsterdam. 2016;198:462–72.
- 18.Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK. Microsatellite markers: an overview of the recent progress in plants. Euphytica. 2011;177(3):309–334. [Google Scholar]
- 19.Senthilvel S, Shaik M, Anjani K, Shaw RK, Kumari P, Sarada C, Kiran BU. Genetic variability and population structure in a collection of inbred lines derived from a core germplasm of castor. J Plant Biochem Biot. 2017;26(1):27–34. [Google Scholar]
- 20.Chapman MA, Hvala J, Strever J, Matvienko M, Kozik A, Michelmore RW, Tang S, Knapp SJ, Burke JM. Development, polymorphism, and cross-taxon utility of EST–SSR markers from safflower (Carthamus tinctorius L.). Theor Appl Genet. 2009;120(1):85–91. [DOI] [PubMed]
- 21.Zhou S, Wang C, Frazier TP, Yan H, Chen P, Chen Z, Huang L, Zhang X, Peng Y, Ma X. The first Illumina-based de novo transcriptome analysis and molecular marker development in Napier grass (Pennisetum purpureum). Mol Breed. 2018;38(7):95.
- 22.Nie G, Tang L, Zhang Y, Huang L, Ma X, Cao X, Pan L, Zhang X, Zhang X. Development of SSR markers based on transcriptome sequencing and association analysis with drought tolerance in perennial grass Miscanthus from China. Front Plant Sci. 2017;8. [DOI] [PMC free article] [PubMed]
- 23.Choi JK, Sa KJ, Park DH, Lim SE, Ryu SH, Park JY, Park KJ, Rhee HI, Lee M, Lee JK. Construction of genetic linkage map and identification of QTLs related to agronomic traits in DH population of maize ( Zea mays L.) using SSR markers. Genes Genom. 2019;41(6):667–78. [DOI] [PubMed]
- 24.Che KP, Xu Y, Liang CY, Gong GY, Weng ML, Zhang HY, Jin DM, Wang B. AFLP fingerprint and SCAR marker of watermelon core collection. Acta Bot Sin. 2003;45(6):731–735. [Google Scholar]
- 25.Kato S, Matsumoto A, Yoshimura K, Katsuki T, Iwamoto K, Kawahara T, Mukai Y, Tsuda Y, Ishio S, Nakamura K, et al. Origins of Japanese flowering cherry (Prunus subgenus Cerasus) cultivars revealed using nuclear SSR markers. Tree Genet Genomes. 2014;10(3):477–87.
- 26.Mbaya JBMJ, Hoffmann AA. Genetic structure of Carex species from the Australian Alpine region along elevation gradients: patterns of reproduction and gene flow. Int J Plant Sci. 2013;174(2):189–99.
- 27.Starr JR, Naczi RF, Chouinard BN. Plant DNA barcodes and species resolution in sedges (Carex, Cyperaceae) Mol Ecol Resour. 2010;9(s1):151–163. doi: 10.1111/j.1755-0998.2009.02640.x. [DOI] [PubMed] [Google Scholar]
- 28.Ning H, Wang W, Zheng C, Li Z, Zhu C, Zhang Q. Genetic diversity analysis of sedges ( Carex spp.) in Shandong, China based on inter-simple sequence repeat. Biochem Systematics Ecol. 2014;56:158–64.
- 29.Nagasawa K, Setoguchi H, Maki M, Goto H, Fukushima K, Isagi Y, Sakaguchi S. Development and characterization of EST-SSR markers for Carex angustisquama (Cyperaceae), an extremophyte in solfatara fields. Appl Plant Sci. 2018;6(10):e01185-n/a. doi: 10.1002/aps3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Man KH, Lee HY, Mishra SN, Hong WH. Genetic variation and population structure of Carex breviculmis (Cyperaceae) in Korea. J Plant Biol. 2000;43(3):136–42.
- 31.Teng K, Teng WJ, Wen HF, Yue YS, Guo WE, Wu JY, Fan XF. PacBio single-molecule long-read sequencing shed new light on the complexity of the Carex breviculmis transcriptome. BMC Genom. 2019;20(1):789. [DOI] [PMC free article] [PubMed]
- 32.Zhou F, Wang Z, Yu G, Shi B, Wang X, Qiang H, Gao H. Development and Characterization of Simple Sequence Repeat (SSR) Markers Based on RNA-Sequencing of Medicago sativa and In silico Mapping onto the M. truncatula Genome. PLoS One. 2014;9(3):e92029. [DOI] [PMC free article] [PubMed]
- 33.Liu Z-J, Torre S, Tattini M, Brunetti C, Fineschi S, Fini A, Ferrini F, Sebastiani F. RNA-Seq analysis of Quercus pubescens leaves: De novo transcriptome assembly, annotation and functional markers development. PLoS One. 2014;9(11):e112487. doi: 10.1371/journal.pone.0112487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li MA, Long RC, Feng ZR, Liu FQ, Sun Y, Zhang K, Kang JM, Wang Z, Cao SH. Transcriptome analysis of salt-responsive genes and SSR marker exploration in Carex rigescens using RNA-seq. J Integr Agr. 2018;17(1):184–96.
- 35.Wang S, Wang X, He Q, Liu X, Xu W, Li L, Gao J, Wang F. Transcriptome analysis of the roots at early and late seedling stages using Illumina paired-end sequencing and development of EST-SSR markers in radish. Plant Cell Rep. 2012;31(8):1437–1447. doi: 10.1007/s00299-012-1259-3. [DOI] [PubMed] [Google Scholar]
- 36.Yu F-H, Kruesi BO, Schneller JJ, Schuetz M, Tang M, Wildi O. Positive correlation between vegetation dissimilarity and genetic differentiation of Carex sempervirens. Flora. 2009;204(9):651–7.
- 37.Yan D, Zhao X, Cheng Y, Ma X, Huang L, Zhang X. Phylogenetic and diversity analysis of Dactylis glomerata subspecies using SSR and IT-ISJ markers. Molecules. 2016;21(12):1459. [DOI] [PMC free article] [PubMed]
- 38.King MG, Roalson EH. Isolation and characterization of 11 microsatellite loci from Carex macrocephala (Cyperaceae). Conserv Genet. 2009;10(3):531–3.
- 39.Bharti R, Kumar S, Parekh MJ. Development of genomic simple sequence repeat (gSSR) markers in cumin and their application in diversity analyses and cross-transferability. Ind Crop Prod. 2018;111:158–164. [Google Scholar]
- 40.Ma LY, Kong DC, Liu HB, Wang SQ, Li YY, Pang XM. Construction of SSR fingerprint on 36 Chinese jujube cultivars. Acta Horticulturae Sinica. 2012;39(04):647–654. [Google Scholar]
- 41.Tan LQ, PengM, Xu LY,Wang LY, Chen SX, Zou Y, Qi GN, Cheng H. Fingerprinting 128 Chinese clonal tea cultivars using SSR markers provides new insights into their pedigree relationships. Tree Genet Genomes. 2015;11(5):1–12.
- 42.Kim C, Jung J, Choi H-K. Molecular identification of Schoenoplectiella species (Cyperaceae) by use of microsatellite markers. Plant Syst Evol. 2012;298(4):811–7.
- 43.Baruah J, Gogoi B, Das K, Ahmed NM, Sarmah DK, Lal M, Bhau BS. Genetic diversity study amongst Cymbopogon species from NE-India using RAPD and ISSR markers. Ind Crop Prod. 2017;95:235–43.
- 44.Haynsen MS, Vatanparast M, Mahadwar G, Zhu D, Moger-Reischer RZ, Doyle JJ, Crandall KA, Egan AN. De novo transcriptome assembly of Pueraria montana var. lobata and Neustanthus phaseoloides for the development of eSSR and SNP markers: narrowing the US origin(s) of the invasive kudzu. BMC Genomics. 2018;19(1):439. doi: 10.1186/s12864-018-4798-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evangelistella C, Valentini A, Ludovisi R, Firrincieli A, Fabbrini F, Scalabrin S, Cattonaro F, Morgante M, Mugnozza GS, Keurentjes JJB, et al. De novo assembly, functional annotation, and analysis of the giant reed (Arundo donax L.) leaf transcriptome provide tools for the development of a biofuel feedstock. Biotechnol Biofuels. 2017;10:138. doi: 10.1186/s13068-017-0828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ndjiondjop MN, Semagn K, Gouda AC, Kpeki SB, Tia DD, Sow M, Goungoulou A, Sie M, Perrier X, Ghesquiere A, et al. Genetic variation and population structure of Oryzaglaberrima and development of a mini-core collection using DArTseq. Front Plant Sci. 2017;8. [DOI] [PMC free article] [PubMed]
- 49.Tohme J, Gonzalez DO, Beebe S, Duque MC. AFLP analysis of gene pools of a wild bean core collection. Crop Sci. 1996;36(5):1375–1384. [Google Scholar]
- 50.Nie M, Bolland HR. Spermatogenesis, chromosomes and sex determination of four Rhipicephalus species (Acari: Ixodidae) from East Africa. Genetica. 1978;48(3):233–238. [Google Scholar]
- 51.Dawyndt P, Meyer HD, Baets BD. UPGMA clustering revisited: a weight-driven approach to transitive approximation. 2006. [Google Scholar]
- 52.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput Appl Bio. 1994;10(2):189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 53.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;9:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 54.Hampl V, Pavlícek A, Flegr J. Construction and bootstrap analysis of DNA fingerprinting-based phylogenetic trees with the freeware program FreeTree: application to trichomonad parasites. Int J Syst Evol Microbiol. 2001;51(Pt 3):731. doi: 10.1099/00207713-51-3-731. [DOI] [PubMed] [Google Scholar]
- 55.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28(28):2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jia H, Yang H, Sun P, Li J, Zhang J, Guo Y, Han X, Zhang G, Lu M, Hu J. De novo transcriptome assembly, development of EST-SSR markers and population genetic analyses for the desert biomass willow, Salix psammophila. Sci Rep. 2016;6(1):39591. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The information of 11 Carex materials to select primers.
Additional file 2: Table S2. The Nei’s genetic distance between Carex species.
Additional file 3: Table S3. The fingerprint of 79 Carex materials.
Data Availability Statement
The Illumina NGS reads generated in this study has been submitted to the BioProject database of National Center for Biotechnology Information (PRJNA488506).