Abstract
Urinary Extracellular Vesicles (uEV) have emerged as a source for biomarkers of kidney damage, holding potential to replace the conventional invasive techniques including kidney biopsy. However, comprehensive studies characterizing uEV isolation methods with patient samples are rare. Here we compared performance of three established uEV isolation workflows for their subsequent use in transcriptomics analysis for biomarker discovery in diabetic kidney disease. We collected urine samples from individuals with type 1 diabetes with macroalbuminuria and healthy controls. We isolated uEV by Hydrostatic Filtration Dialysis (HFD), ultracentrifugation (UC), and a commercial kit‐ based isolation method (NG), each with different established urine clearing steps. Purified EVs were analysed by electron microscopy, nanoparticle tracking analysis, and Western blotting. Isolated RNAs were subjected to miRNA and RNA sequencing. HFD and UC samples showed close similarities based on mRNA sequencing data. NG samples had a lower number of reads and different mRNA content compared to HFD or UC. For miRNA sequencing data, satisfactory miRNA counts were obtained by all methods, but miRNA contents differed slightly. This suggests that the isolation workflows enrich specific subpopulations of miRNA‐rich uEV preparation components. Our data shows that HFD,UC and the kit‐based method are suitable methods to isolate uEV for miRNA‐seq. However, only HFD and UC were suitable for mRNA‐seq in our settings.
Keywords: diabetic kidney disease, exosomes, extracellular vesicles, isolation, miRNA sequencing, mRNA sequencing, urinary extracellular vesicles
1. INTRODUCTION
Diabetic kidney disease (DKD) is a progressing microvascular damage in diabetes patients, which leads to increasing functional abnormalities (Thomas et al., 2015). DKD diagnosis is based on albuminuria (measured by e.g., urinary albumin/creatinine ratio) and determining serum creatinine level to obtain a measure of reduced kidney function (decreased glomerular filtration rate) (Campion et al., 2017). However, these markers are typically altered only when there already is significant damage to the precious kidney filtration barrier (Gonzalez Suarez et al., 2013). Thus, earlier biomarkers of kidney damage in combination with easier, non‐invasive sample collection methods (to avoid kidney biopsies) are needed to manage and potentially reduce the risk of kidney disease in a timely manner.
Extracellular vesicles (EV) are lipid bilayer enclosed particles that are likely constitutively secreted by all cells of the body (Yanez‐Mo et al., 2015). Based on their biogenesis pathways, EV can be roughly classified into exosomes, microvesicles and apoptotic bodies (van Niel et al., 2018). Previous studies have shown that EV contain distinct proteins, lipids, various RNA species, DNA, and metabolites that may provide a snapshot of pathological and physiological changes of their parental cells in disease states (Puhka et al., 2017; Yanez‐Mo et al., 2015; Yuana et al., 2013). EV have been found in all bodily fluids studied to date, including urine (Campion et al., 2017). As urine forms in the kidney and can be collected non‐invasively, urinary extracellular vesicles (uEV) deriving from different parts of the kidney represent a promising source for DKD biomarker discovery (Erdbrugger & Le, 2016; Ranghino et al., 2015). EV can be isolated using methods such as ultracentrifugation, filtration, precipitation, and hydrostatic filtration dialysis and their combinations (Barreiro & Holthofer, 2017; Konoshenko et al., 2018). However, different isolation methods, including all the steps before the actual EV collection, tend to enrich EV into different subpopulations with a various degree of common urinary contaminants, which may all impact on sensitive downstream applications, such as transcriptomics and proteomics (Merchant et al. 2017; Svenningsen et al., 2020; Wachalska et al., 2016). Currently, there is no widely accepted standardization of pre‐analytical variables or isolation protocols and, in many cases, characterization of EV and methods are reported all too superficially. Thus, the interpretation, repeatability and comparison of reported results is challenging. However, thanks to the continuous efforts of the International Society for Extracellular Vesicles (ISEV) on reporting transparency and standardization (Thery et al., 2018; Van Deun et al., 2017), practices are improving in the rapidly growing field of EV studies.
Few systematic studies have compared uEV isolation methods for downstream effects on RNA transcriptomics results (Cheng et al., 2014; Mussack et al., 2019; Park et al., 2020; Srinivasan et al., 2019). However, these published studies have focused mainly in small/miRNA sequencing and most of them have used only urine from healthy donors, which may lead to misinterpretations of performance. Here we compared three established uEV isolation workflows including urines of both healthy donors as well as individuals with type 1 diabetes and existing kidney damage (macroalbuminuria). The uEV isolates were then compared with focus especially on their suitability for miRNA (miRNA‐seq) and mRNA sequencing (mRNA‐seq) for biomarker discovery studies. The methods we used included i) hydrostatic filtration dialysis (HFD) (Barreiro et al., 2020; Musante et al., 2014), a simple and inexpensive method to work efficiently with large volumes of sample, ii) ultracentrifugation (UC), the most used and characterized method to isolate EV (Puhka et al., 2017), and iii) a commercial kit‐based isolation method (NG), designed to be a fast and scalable method to work with larger number of samples (Cheng et al., 2014). Each method workflow was applied maintaining the recommended urine volumes and pre‐ isolation processing steps, while results were subsequently normalized to urine volumes or by using equal RNA quantities for sequencing.
2. MATERIALS AND METHODS
2.1. Urine samples
24 h urine was collected from five healthy controls and five participants of the Finnish Diabetic Nephropathy Study with type 1 diabetes and presenting constant macroalbuminuria. Calbiochem® EDTA‐free Protease Inhibitor Cocktail Set III (Merck Millipore, Burlington, MA) was added to voided urine at a dilution of 1/2000 before storage at −80°C. Donors information is given in Table 1.
TABLE 1.
Clinical information from individuals with type 1 diabetes and controls
| Sample ID | Group | Age | eGFR | UACR |
|---|---|---|---|---|
| 1 | Control | 59.80 | 80.30 | 0.52 |
| 2 | Control | 61.00 | 100.70 | 0.66 |
| 3 | Control | 61.00 | 95.90 | 5.00 |
| 4 | Control | 61.80 | 86.10 | 0.79 |
| 5 | Control | 51.70 | 101.30 | 0.68 |
| 6 | Macroalbuminuria | 41.90 | 106.70 | 57.00 |
| 7 | Macroalbuminuria | 67.90 | 45.70 | 92.60 |
| 8 | Macroalbuminuria | 54.00 | 98.50 | 47.00 |
| 9 | Macroalbuminuria | 43.50 | 58.30 | 102.10 |
| 10 | Macroalbuminuria | 47.00 | 107.70 | 13.50 |
eGFR, estimated glomerular filtration rate; UACR, urine albumin‐to‐creatinine ratio.
All experiments were performed in accordance with the Declaration of Helsinki and the study protocol was approved by the Ethical Committee of Helsinki and Uusimaa Hospital District. Participants signed a written informed consent prior to enrollment in the study.
2.2. Extracellular vesicle isolation
2.2.1. Hydrostatic filtration dialysis
For EV isolation with HFD, urine samples were melted at +37°C in a water bath and vortexed for 90 s. Urine was then centrifuged at 2000 × g at +4°C (no breaking) for 30 min using a fixed angle AG‐6512C rotor (Kubota Corp. Tokyo, Japan). uEV were isolated as previously described (Barreiro et al., 2020). Briefly, 50 ml of supernatant was poured into a dialysis membrane made of cellulose ester (CE) with molecular weight cut‐off (MWCO) 1,000 kDa (Repligen Corp., Waltham, MA, USA). The sample was first concentrated to 5–6 ml. After the first step, the membrane was refilled with 200 ml of deionized water to wash remaining analytes below the MWCO. When the sample inside the membrane reached about 1 ml, it was collected in protein or DNA LowBind tubes (Eppendorf, Hamburg, Germany) and stored at −80°C. HFD was run under vacuum (−20 kPa).
2.2.2. Ultracentrifugation
For the UC samples EV were isolated using differential centrifugation as previously described by Puhka et al, 2017 (Puhka et al., 2017); without sample dilution. Urine samples were melted at +37°C in a water bath and vortexed for 90 s. Then, centrifuged at 8000 × g at +4°C (breaking) for 15 min using a fixed angle AG‐6512C rotor (Kubota Corp.). Supernatants were filtered using Whatman 1.2 μm cellulose acetate syringe filters (GE Healthcare, Buckinghamshire, UK). 30 ml of processed urine was centrifuged at 4°C for 90 min at 100,000 × g (maximum breaking) in a SW28 rotor, k‐factor 254.5 (Beckman Coulter, Inc., Brea, CA, USA). Supernatants were discarded and the pellets were washed in 20 ml of PBS repeating the same ultracentrifugation. After the washing step, supernatants were removed and EV samples were suspended in filtered PBS (0,22 μm PES filter; Jet Bio‐Filtration, Guangzhou, China) and stored in protein or DNA LowBind tubes (Eppendorf) at −80°C.
2.2.3. Kit‐based isolation
Urine samples were melted at +37°C in a water bath. Clarified urine was prepared following Urine Exosome Purification and RNA Isolation Midi Kit (Norgen Biotech Corp., Thorold, ON, Canada) manufacturer's instructions. Briefly, urine was centrifuged at 200 × g at RT for 10 min. Then, supernatants were centrifuged at 1800 × g at RT for 10 min to remove any residual debris or bacterial cells. Both centrifugation steps were done using a fixed angle AG‐6512C rotor (Kubota Corp.). Supernatants were used to isolate extracellular vesicles employing the kit and following manufacturer's protocol. More details of uEV isolation protocols, respectively, are shown in Figure 1.
FIGURE 1.
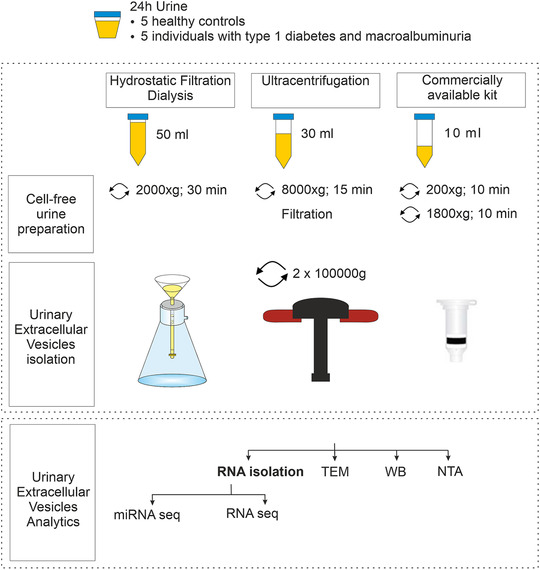
Experimental scheme used to compare urinary extracellular vesicle isolation workflows with focus on RNA sequencing. TEM, transmission electron microscopy; WB, Western blot; NTA, nanoparticle tracking analysis
2.3. Sample concentration
EV samples isolated by HFD and Norgen kit to be used for negative staining and Western blotting were further concentrated (HFD: 4X and Norgen Kit 8X). Samples were concentrated using Amicon Ultra‐0.5 ml Centrifugal Filters 10k (Merck Millipore) following manufacturer's instructions.
2.4. RNA isolation
Total RNA from HFD and UC EV samples was isolated using TRIzol LS (Life Technologies Corp., Carlsbad, CA, USA) following manufacturer's instructions up to phase separation and then combined to miRNeasy mini kit (Qiagen, Hilden, Germany) procedure following manufacturer's protocol for Purification of Total RNA, Including Small RNAs, from Animal Cells. RNA was eluted in 30 μl of RNase free water. Total RNA from NG samples was isolated following manufacturer's instructions. RNA obtained was analysed using Agilent RNA 6000 Pico kit (Agilent Technologies, Santa Clara, CA).
2.5. Protein quantification
Protein concentration was measured using Micro BCA™ Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Samples and standards were diluted in 2% SDS in PBS (to avoid the interference of lipids in the determination of protein using bicinchoninic acid). Samples were measured following microplate procedure according to manufacturer's instructions.
2.6. Western blotting
EV samples derived from 2 ml of urine were mixed with 5X Laemmli buffer, denatured for 5 min at 95°C, and loaded to 4–20% Mini‐PROTEAN® TGX Stain‐Free Protein Gels (Bio‐Rad laboratories Inc., Hercules, CA, USA). Gels were run 1 h at 150 V after which they were transferred to Immobilon®‐FL PVDF membrane pore size 0.45 μm (Merck Millipore) at 100 V, 90 min. Membranes were blocked with 3% Bovine Serum Albumin (BSA) (Amresco, Solon, OH) in TBS, 1 h at RT. After blocking, membranes were Incubated with primary antibody diluted in 3% BSA in TTBS (tween 20 0,1%, TBS) overnight at +4°C. Membranes were washed in TTBS and incubated with secondary antibody diluted in 3% BSA in TTBS. Secondary antibody was detected using Pierce™ ECL Plus Western Blotting Substrate (Thermo Fisher Scientific) and ChemiDoc Imager system (Bio‐Rad laboratories Inc.). Details on primary and secondary antibodies used in this study are provided in Table 2.
TABLE 2.
Primary and secondary antibodies used for Western blotting
| Primary antibody | Company | Code | Dilution |
|---|---|---|---|
| Tamm‐Horsfall protein | Santa Cruz Biotechnology | SC‐20631 | 1:3000 |
| Albumin | Santa Cruz Biotechnology | SC‐58688 | 1:1000 |
| Podocalyxin; clone 3D3 | Novus Biologicals | NBP2‐25219 | 1:500 |
| TSG‐101 | Santa Cruz Biotechnology | SC‐7964 | 1:400 |
| CD9 | Santa Cruz Biotechnology | SC‐13118 | 1:400 |
| Secondary antibody | Company | Code | Dilution |
|---|---|---|---|
| Rabbit anti mouse | Jackson Immuno Research | 315‐035‐045 | 1:6000 |
| Goat anti rabbit | Jackson Immuno Research | 111‐035‐045 | 1:6000 |
2.7. Transmission electron microscopy
Negative staining was performed as previously described (Barreiro et al., 2020; Puhka et al., 2017). Briefly, 5 μl of sample (equivalent to 600 μl of urine) was loaded on Formvar/Carbon 200 mesh TH, Copper grids (Ted Pella Inc., Redding, CA, USA) and fixed with 2% PFA (Electron Microscopy Sciences, Hatfield, PA, USA). Grids were negatively stained with 2% neutral uranyl acetate and embedded in methyl cellulose uranyl acetate mixture (1,8/0,4%). Images were acquired using Jeol JEM‐1400 (Jeol Ltd, Tokyo, Japan) operating at 80 kV, equipped with Gatan Orius SC 1000B CCD‐camera (Gatan Inc., Pleasanton, CA) and using image size of 4008 × 2672 pixels. Maximum diameter of EV was measured using imageJ (Schneider et al., 2012). 150 EV were measured in 4 to 15 randomly sampled images.
2.8. Cryo‐transmission electron microscopy
The vitrified samples were prepared from 3 μl aliquots with Leica EMGP vitrification device on freshly glow‐discharged Quantifoil R1.2/1.3 grids. The samples were observed in a FEI Talos Arctica microscope operated at 200 kV. Images were recorded at a nominal magnification of 57,000 x with a FEI Falcon 3 camera operated in a linear mode.
2.9. Nanoparticle tracking analysis (NTA)
Urinary EV samples were analysed using Nanosight model LM14 (Malvern Instruments Ltd, Malvern, UK) equipped with blue (404 nm, 70 mW) laser and SCMOS camera (Hamamatsu photonics K.K., Hamamatsu, Japan). Samples were diluted in 0.1 μm filtered (Millex VV, Millipore) DPBS to obtain 40–100 particles/view. Five 30 s videos were recorded using camera level 13. Videos were analysed using NTA software 3.0 (Malvern Instruments Ltd) with the detection threshold 5 and screen gain at 10.
2.10. mRNA sequencing
The cDNA libraries were prepared from 1 ng of total RNA using SMART‐Seq® v4 Ultra Low Input RNA Kit for Sequencing (Takara BIo Inc., Mountain View, CA) and Nextera XT kit (Ilumina Inc., San Diego, CA, USA). RNA was sequenced on the Illumina NextSeq 500 platform (Illumina Inc.).
2.11. miRNA sequencing
miRNA libraries were prepared using the QIAseq miRNA Library Kit (Qiagen) following the manufacturer's protocol, starting with ∼1 ng of input RNA for each sample.
Quality control and concentrations of individual libraries were assessed with Bioanalyzer 2100 Instrument and High Sensitivity DNA Kit (Agilent Technologies). miRNA‐sequencing was performed on the Illumina HiSeq 4000 platform (Illumina Inc.).
2.12. RNAseq data analysis
mRNA sequencing reads were mapped to the GRCh38.99 reference genome using STAR v2.7.1a (Dobin et al., 2013) . Gene quantification was done using STAR, selecting the parameter –quantMode GeneCounts. miRNA reads were quantified using featureCounts (Liao et al., 2014). Data from STAR log files was accessed using MultiQC (Ewels et al., 2016). Differential expression analysis was performed using DESeq2 (Anders & Huber, 2010) in R v3.6.3 (Team, 2020). Genes/miRNAs with an adjusted P‐value lower than 0.05 and a Log2 Fold change ≥ 0.75 or ≤ −0.75 were considered differentially expressed. Scatterplots, FPKM distributions and correlation values were obtained using ViDGER (McDermaid et al., 2019).We used miRWalk (Dweep et al., 2014) for miRNA target and enrichment analysis. miRNA validated targets were filtered by selecting miRTarBase option in miRWalk target mining page. Vesiclepedia (accession date 10.07.2019) was used to assess the presence or absence of miRNAs in EVs as reported in other studies (Pathan et al., 2019).
2.13. Statistics
IBM SPSS statistics V24 was used for statistical analysis. RNA yield, protein yield, particle to protein ratio (Log10 transformed), and size distribution data (arcsin transformed) mean differences, were tested using ANOVA and Bonferroni post hoc tests. Particle yield mean differences were tested using Kruskal‐Wallis test. Values are represented as mean ± SEM. P‐values < 0.05 were considered statistically significant. ggplot2 (Wickham, 2016) was used for data visualization.
2.14. Data availability
The raw sequencing data analysed in this study is not publicly available due to local regulations. We provide raw sequencing count data in Tables S5 and S6. Raw sequencing data is available to qualified investigators upon reasonable request.
3. RESULTS
We enriched uEV from healthy controls and individuals with type 1 diabetes (with macroalbuminuria) by HFD, UC and NG methods. uEV samples were then analysed by Western blot, nanoparticle tracking analysis, electron microscopy, and RNA sequencing. An overview of the experimental workflow used in this study is presented in Figure 1.
Data was first analysed considering isolation methods versus sample donor disease status that impacts the urine sample characteristics, for example, the protein content. However, no gross differences were observed between the healthy controls and type 1 diabetes samples for RNA yield, NTA, and RNA sequencing. Thus, samples were analysed comparing solely isolation methods.
3.1. uEV characterization by electron microscopy and Western Blotting
By electron microscopy, uEV of typical morphology were observed with all three methods. However, in NG samples less uEV were observed despite that the loaded EVs were derived from equal urine starting volumes (EVs from 600 μl of urine in 5 μl of sample/EM grid) (Figure 2). Filament meshworks compatible with Tamm‐Horsfall protein (THP) complexes (Cheng et al., 2014; Fernández‐Llama et al., 2010; Musante et al., 2020; Pisitkun et al., 2006; Wachalska et al., 2016), highly present in normal urine, were likewise observed in samples isolated by all methods. By Cryo‐EM, vesicles of typical morphologies were also observed in all samples (Figure 2)
FIGURE 2.
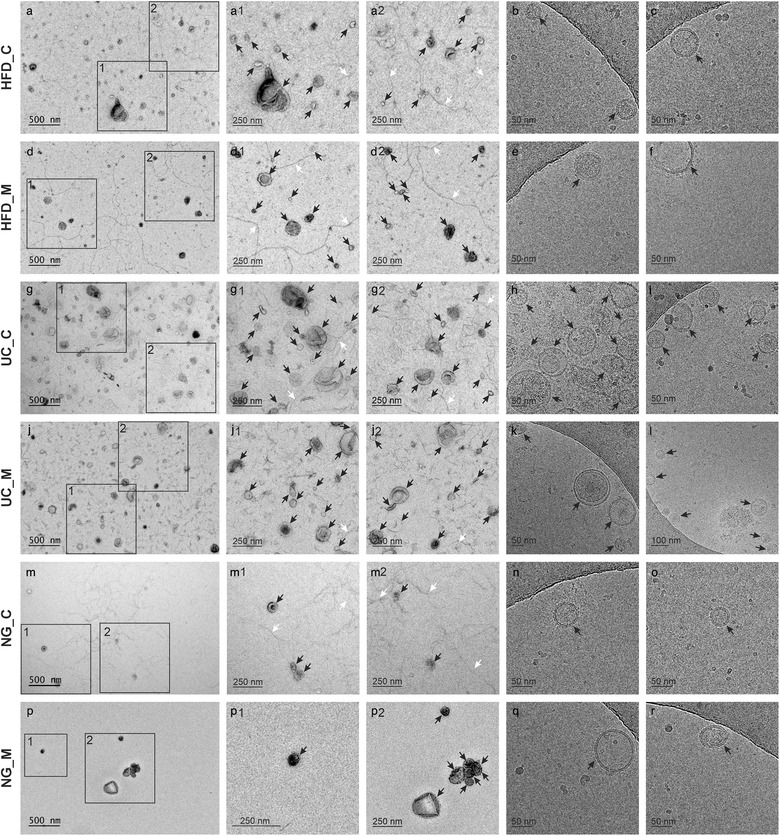
Transmission electron micrographs (a,d,g,j,m,p) and cryo‐electron micrographs (b,c,e,f,h,i,k,l,n,o,q,r) of EVs from the three methods. Black arrowheads: extracellular vesicles; white arrowheads: filamentous structures compatible with Tamm–Horsfall protein filaments. Hydrostatic filtration dialysis control (HFD_C), hydrostatic filtration dialysis macroalbuminuria (HFD_M), ultracentrifugation control (UC_C), ultracentrifugation macroalbuminuria (UC_M), norgen kit control (NG_C), norgen kit macroalbuminuria (NG_M)
In evidence of successful EV isolation including the exosome class, Western blotting showed EV markers TSG101, CD9 and, additionally, podocalyxin in HFD and UC processed samples, but not in NG samples (Figure 3). For comparability, gels were loaded with EVs derived from equal urine volumes (2 ml of urine). We also assessed the presence of common urine proteins that typically co‐isolate and contaminate with uEV (THP and Albumin). Thus, THP was detected in samples isolated with all three methods but it appeared to be somewhat less abundant in NG samples. Albumin was additionally detected in HFD and NG samples (Figure 3).
FIGURE 3.
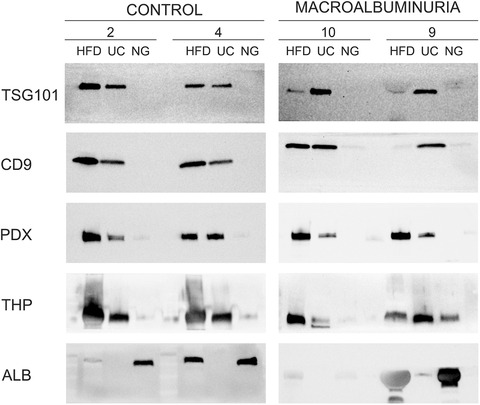
Western blotting of control and macroalbuminuria urinary extracellular vesicle samples isolated by Hydrostatic Filtration Dialysis (HD), ultracentrifugation (UC) and Norgen Kit (NG). EVs from 2 ml of urine were loaded from each method. Immunodetection of EV markers (TSG101, CD9 and podocalyxin, PDX) and two common proteins that co‐isolate with urinary extracellular vesicles (Tamm–Horsfall protein, THP and albumin, ALB) are shown
3.2. RNA yield per ml of urine did not differ between isolation methods
Total RNA from uEV isolated by HFD, UC and NG was profiled using Bioanalyzer pico Kit®. RNA electropherogram profiles were similar between methods, all the samples studied typically presented an RNA peak between 25–200 nt (Figure S1). As different starting volumes of urine were used to isolate uEV, respectively, we calculated the RNA yield per ml of original urine sample to compare the RNA yields between methods. RNA yield between HFD (767.9 ± 310.6 pg/ml, n = 10), UC (1308.6 ± 499.0 pg/ml, n = 10) and NG (1719.3 ± 572.0 pg/ml, n = 6) did not, however, differ significantly (Figure 4a).
FIGURE 4.
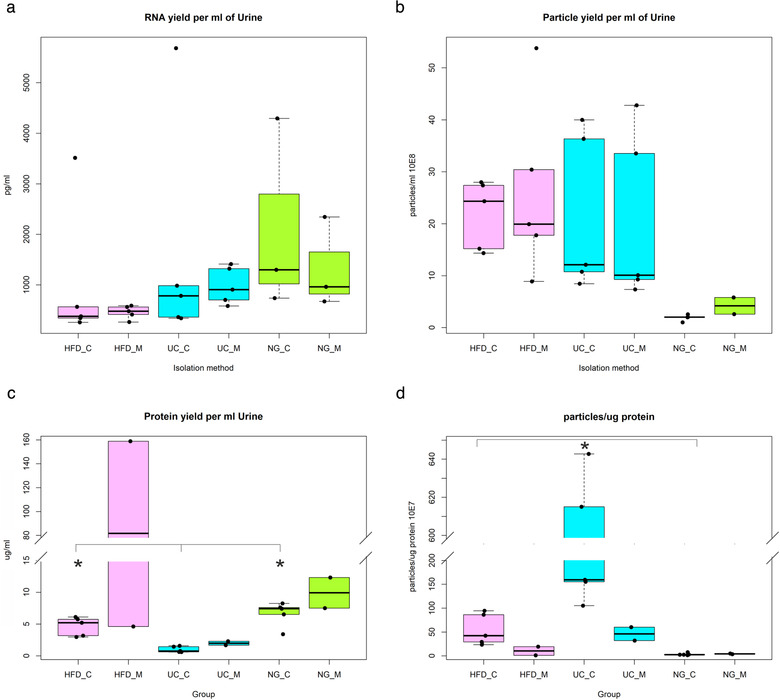
a. Total RNA yield per ml of urine quantified using pico 6000 RNA chip in Agilent 2100 Bioanalyzer b. Particle yield per ml of urine determined by Nanoparticle tracking analysis. c. Protein yield per ml of urine. d. Particle to protein ratio. Hydrostatic filtration dialysis control (HFD_C), hydrostatic filtration dialysis macroalbuminuria (HFD_M), ultracentrifugation control (UC_C), ultracentrifugation macroalbuminuria (UC_M), norgen kit control (NG_C), norgen kit macroalbuminuria (NG_M), P < 0.05 (*)
3.3. Particle and protein yield per ml of urine and size distribution of vesicles
The number and size of uEV isolated by HFD, UC and NG were quantified by NTA. To compare particle yields between methods, we calculated the particle yield relative to one ml of urine. The mean yields from HFD (2.4E+9 ± 4.0E+8; n = 10) and UC (2.1E+9 ± 4.7E+8; n = 10) were very close to each other, but significantly higher than the yields from NG (2.6E+8 ± 5.7E+7; n = 7) (Kruskal‐Wallis; pairwise comparisons; P < 0.05, Figure 4b).
Due to sample volume limitations, protein content could not be measured in three of the macroalbuminuria uEV samples and in consequence, statistical testing was performed only using control samples. Between isolation methods, we found that the quantity of protein per ml of urine in HFD (4.6 ± 0.66) and NG (6.7 ± 0.85) samples was significantly higher than in UC (0.1 ± 0.2, n = 5) (Bonferroni post hoc test, both P < 0.01) (Figure 4c). The amount of proteins showed a tendency to increase in macroalbuminuria samples when compared to control samples within an isolation method (Figure 4c). To assess uEV preparation purity, as reported by Webber and Clayton (Webber & Clayton, 2013), we calculated the particle to protein ratio. Comparing control samples, we found that particle to protein ratio from UC (3.4E+09 ± 1.2E+09, n = 5) was higher than from HFD (5.5E+08 ± 1.5E+08, n = 5) and NG (3.4E+07 ± 1.1E+07, n = 5) (Bonferroni post hoc test, both P < 0.01). Particle to protein ratio from HFD was also higher than from NG (Bonferroni post hoc test, both P < 0.01) (Figure 4d). Within an isolation method, the ratio appeared to decrease in macroalbuminuria samples with respect to control samples as shown in Figure 4d.
To assess whether the isolation method affected enrichment of a particular size range of vesicles, we calculated the percentage of vesicles in six size categories (0‐100, 101–200, 201–300, 301–400, 401–500 and > 500 nm). By ANOVA test, we found significant differences in 0–100 nm and > 500 nm categories (both P < 0.05). For the smallest vesicle size category, the mean percentage of particles from HFD (0.5% ± 0.2%, n = 6) and UC (1.2% ± 1.0%, n = 6) were significantly lower than from NG (4.0% ± 0.8%, n = 4) (Bonferroni post hoc test, both P < 0.05). For the largest vesicle size category, the mean percentage of particles from HFD (4.3% ± 1.1%, n = 6) was significantly higher than from NG (0.5% ± 0.3% n = 4) (Bonferroni post hoc test, P < 0.05) (Figure 5). As some studies reported that particle size distribution differs depending on the method used to measure EV diameter (Bachurski et al., 2019; van der Pol et al., 2014), we also calculated the particle size distribution using EV size values measured in TEM images. We found that the results for > 400 nm EVs by TEM were consistent with the NTA data showing a higher proportion of large EVs from HFD and UC (Figure S2). In addition, HFD contained less EVs in the 60–140 nm size categories compared to UC and NG, again agreeing with NTA data. In the size range of very small EVs or alike particles (< 40 nm), which is not well detected by NTA, TEM data indicated that HFD contained more vesicles than UC and NG.
FIGURE 5.
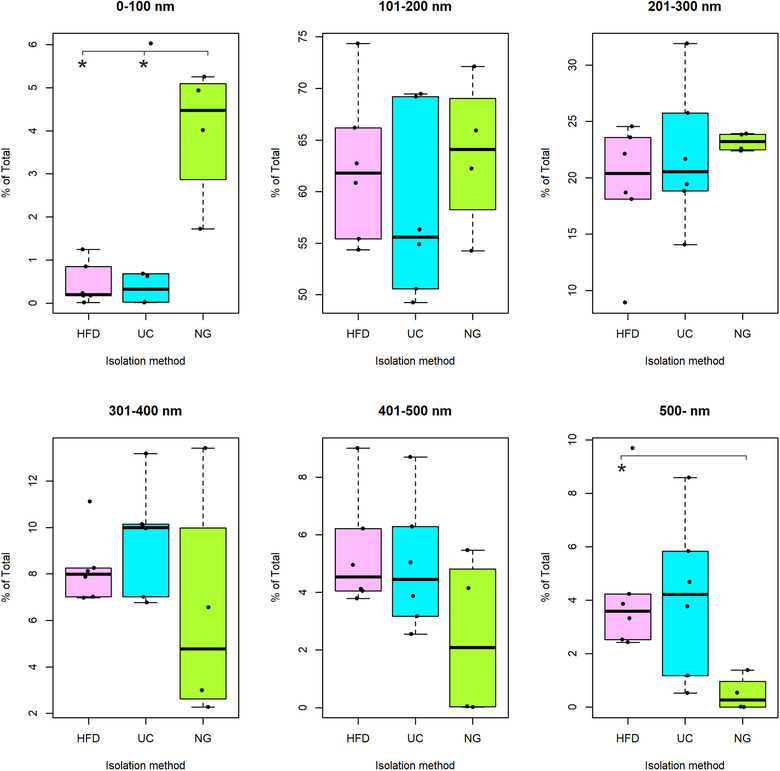
Size distribution of particles represented as percentage of the total. Size of particles determined by Nanoparticle tracking analysis. Hydrostatic Filtration Dialysis (HFD), ultracentrifugation (UC), Norgen Kit (NG), P < 0.05 (*)
3.4. RNA sequencing
The number of reads of mRNA sequencing from NG samples (2.1E+06 ± 0.3E+06) was lower than from HFD (7.8E+06 ± 1.2E+06) and UC (8.4E+06 ± 1.0E+06) samples (Figure 6). Exploratory analysis of mRNA sequencing data by principal component analysis (PCA) (12145 genes, which raw counts were > 5 in at least 50% of the samples) showed similarities between HFD and UC samples and a clear separation of NG samples. In agreement with PCA results, sample to sample correlation analysis showed two clusters of samples, a cluster containing HFD and UC samples and another cluster of NG samples only. By differential expression analysis, we found 16 differentially expressed genes (Table S1), all of them mitochondrial RNAs, between HFD and UC, and a high number of differentially expressed genes between HFD‐NG and UC‐NG (3344 and 3414, respectively) as shown in Figure 7. The correlation between HFD and UC (r = 0.97) was high. On the contrary, pairwise correlation analysis between NG and HFD or UC showed to be weak (0.45, 0.44 respectively) (Figure S3A).
FIGURE 6.
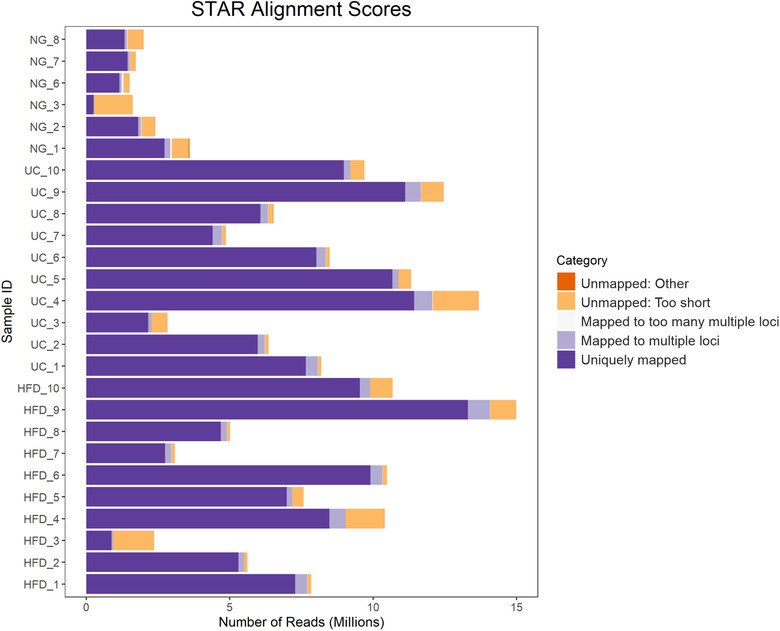
STAR alignment results for mRNA data
FIGURE 7.
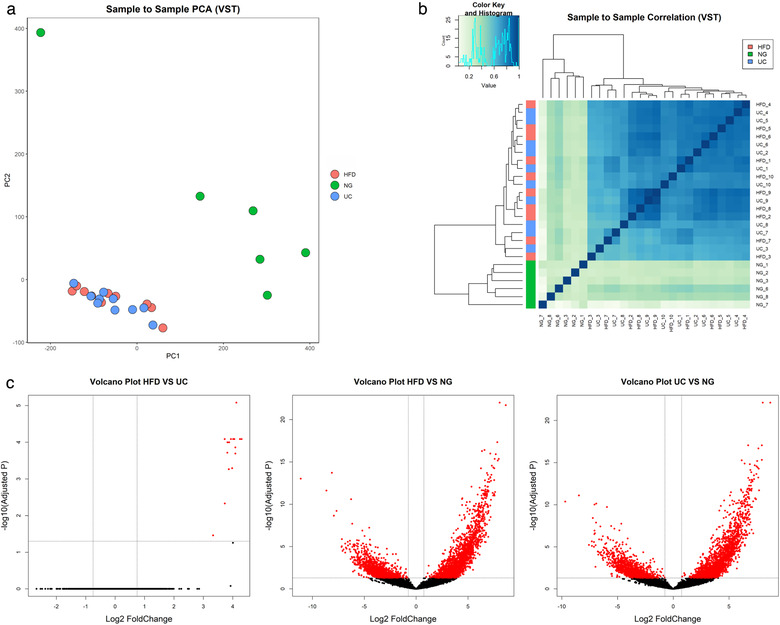
mRNA sequencing data analysis using DEseq. a. Principal component analysis (PCA), b. Correlation heatmap, c. Volcano plot of pairwise comparisons between isolation methods. Adjusted P‐values lower than 0.05 and a Log2 Fold change ≥ 0.75 or ≤ −0.75 are highlighted in red. Variance‐stabilizing transformation (VTS), Hydrostatic Filtration Dialysis (HFD), ultracentrifugation (UC), Norgen Kit (NG)
The total number of filter‐pass reads obtained by miRNA sequencing were higher for UC (6.5E+06 ± 1.2 E+06) than for HFD (3.5E+06 ± 0.5 E+06) and NG (4.4E+06 ± 0.8E+06) (Figure 8a). Similarly, the total raw miRNA counts showed to be higher for UC samples (1.1E+06 ± 1.9E+05, n = 10) than for HFD (3.1E+05 ± 2.7 E+04, n = 10) and NG (5.8E+05 ± 2.0E+05, n = 6) (Figure 8b). miRNA biotype distribution showed to be mostly similar within isolation methods (Figure S4). PCA analysis of miRNA sequencing data (1366 miRNAs, which raw counts were > 1 in at least one sample) did not show a clear separation of samples isolated by different methods (Figure 9a and Figure S5). In addition, no clear clusters were seen showing similarities between HFD, UC and NG by sample‐to‐sample correlation analysis (Figure 9b). For miRNA, the correlation values between methods was strong for all the pairwise analysis (HFD‐ UC: r = 0.99; HFD‐ NG: 0.97; and NG‐UC: 0.97) (Figure S3B). By differential expression analysis, we found differentially expressed miRNAs between HFD‐UC and HFD‐NG (19 and 9, respectively) (Table S2). No differentially expressed miRNAs were found between UC and NG samples (Figure 9c).
FIGURE 8.
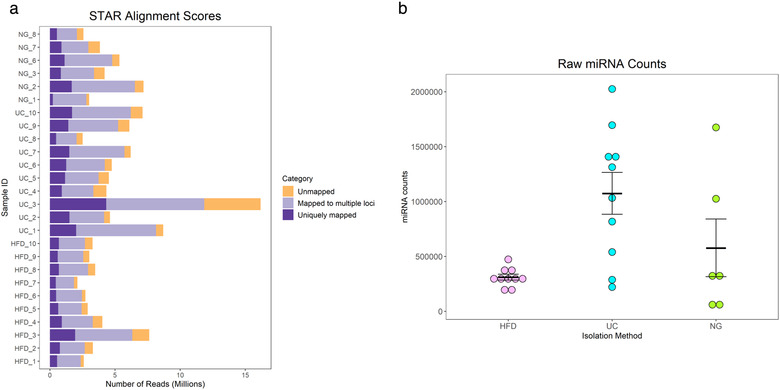
miRNA sequencing reads (a) and miRNA counts mean ± SEM (b). Hydrostatic Filtration Dialysis (HFD), ultracentrifugation (UC), Norgen Kit (NG)
FIGURE 9.
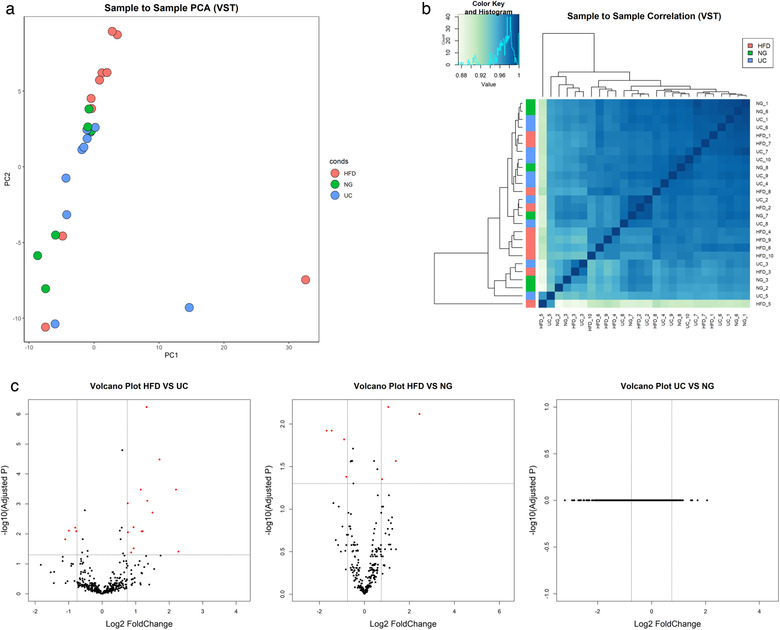
miRNA sequencing data analysis using DEseq. a. Principal component analysis (PCA), b. Correlation heatmap, c. Volcano plot of pairwise comparisons between isolation methods. Adjusted P‐values lower than 0.05 and a Log2 Fold change ≥ 0.75 or ≤ −0.75 are highlighted in red. Variance‐stabilizing transformation (VTS), Hydrostatic Filtration Dialysis (HFD), ultracentrifugation (UC), Norgen Kit (NG)
To compare the number of miRNAs detected by each method, we selected the miRNAs with normalized counts > 5 in all the samples. We found that under this condition, the three methods detected a similar number of miRNAs across all samples (HFD = 223, UC = 230 and NG = 234). Using this filtered data, we then determined which miRNAs were shared between methods and which were unique (Figure 10). We found that around 85–90% (201 miRNAs) of the miRNAs were shared between HFD, NG, and UC. GO Enrichment Analysis showed that miRNAs (validated target genes) not shared between methods contribute to diverse biological processes (Table 3). Additionally, we compared the top ten identified miRNAs between methods and found that most of the miRNAs were shared (90%) between methods (Figure S6A, Table S3). GO Enrichment Analysis showed that most of the biological process affected by the top ten miRNAs are shared between methods (Figure S6B and Table S4).
FIGURE 10.
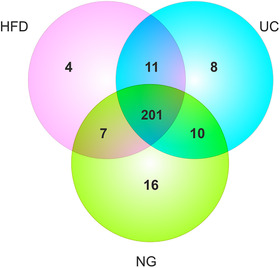
Venn diagram showing detected miRNAs (with > 5 normalized counts in all the samples within the isolation method group) in samples isolated by Hydrostatic Filtration Dialysis (HFD), ultracentrifugation (UC), and Norgen kit (NG)
TABLE 3.
Unique identified miRNAs per isolation method (when comparing miRNAs with normalized counts > 5 in all the samples within the isolation method group), entries of the respective miRNA in Vesiclepedia database and, GO biological process enrichment analysis of miRNA validated targets
| miRNA ID | Vesiclepedia | GO: biological process (P > 0.05) | |
|---|---|---|---|
| HFD | hsa‐miR‐92b‐5p | GO:0010468_regulation_of_gene_expression; GO:0043687_post‐translational_protein_modification; GO:0008285_negative_regulation_of_cell_proliferation; GO:0006468_protein_phosphorylation | |
| hsa‐miR‐99a‐3p | ✓ | ||
| hsa‐miR‐664b‐3p | ✓ | ||
| hsa‐miR‐5187‐5p | |||
| UC | hsa‐miR‐1270 | ✓ | GO:0030308_negative_regulation_of_cell_growth; GO:0061024_membrane_organization; GO:0071456_cellular_response_to_hypoxia; GO:0006629_lipid_metabolic_process; GO:0008283_cell_proliferation; GO:0010629_negative_regulation_of_gene_expression; GO:0030336_negative_regulation_of_cell_migration |
| hsa‐miR‐1843 | |||
| hsa‐miR‐455‐3p | ✓ | ||
| hsa‐miR‐28‐5p | ✓ | ||
| hsa‐miR‐26b‐3p | ✓ | ||
| hsa‐miR‐1287‐5p | ✓ | ||
| hsa‐miR‐628‐5p | ✓ | ||
| hsa‐miR‐200c‐5p | |||
| NG | hsa‐miR‐4662a‐5p | ✓ |
GO:0051781_positive_regulation_of_cell_division; GO:0000082_G1S_transition_of_mitotic_cell_cycle; GO:0001525_angiogenesis; GO:0006468_protein_phosphorylation; GO:0051897_positive_regulation_of_protein_kinase_B_signaling; GO:0035556_intracellular_signal_transduction; GO:0006366_transcription_by_RNA_polymerase_II; GO:0051301_cell_division |
| hsa‐miR‐4662b | |||
| hsa‐miR‐142‐3p | ✓ | ||
| hsa‐miR‐1269a | |||
| hsa‐miR‐577 | ✓ | ||
| hsa‐miR‐320d | ✓ | ||
| hsa‐miR‐1468‐5p | ✓ | ||
| hsa‐miR‐505‐3p | ✓ | ||
| hsa‐miR‐671‐3p | ✓ | ||
| hsa‐miR‐2116‐3p | ✓ | ||
| hsa‐miR‐424‐3p | ✓ | ||
| hsa‐miR‐576‐5p | ✓ | ||
| hsa‐miR‐4728‐3p | |||
| hsa‐miR‐653‐5p | |||
| hsa‐miR‐874‐5p | ✓ | ||
| hsa‐miR‐135a‐2‐3p |
HFD, Hydrostatic Filtration Dialysis; UC, ultracentrifugation; NG, Norgen kit.
GO Biological processes with P value < 0.05 are shown.
4. DISCUSSION
Interest in EV as source for disease biomarkers has increased remarkably. Accordingly, the number of publications analysing EV cargo through proteomics, transcriptomics, lipidomics, and metabolomics (Ghai et al., 2018; Harshman et al., 2016; Hough et al., 2018; Puhka et al., 2017) has increased exponentially in recent years. As urine can be collected non‐invasively, repeatedly and in large volumes, uEV hold a lucrative potential to replace conventional invasive procedures (e.g., kidney biopsies) by “liquid biopsies” from urine. Urine is rich in EV derived from cells lining the urinary tract, which highlights uEV as a promising source of biomarkers especially for kidney diseases (Erdbrugger & Le, 2016). Interestingly, there is also some evidence showing that uEV could be used to profile not only diseases of the genitourinary system, but also systemic diseases (Morrison et al., 2016).
Although uEV can be isolated using a broad selection of available methodologies (Barreiro & Holthofer, 2017), there is little published evidence of their respective key performance characteristics. Whether the proposed isolation method performs equally well in control and disease samples is a topic not explored to sufficient detail while this is a key aspect in evaluating their usefulness in general. This is especially important when working with diseases that greatly affect the composition of the biofluid under study. Diabetic kidney disease causes damage to the kidney filtration barrier and consequently proteins are leaking into the primary urine changing the surrounding milieu of uEV. Furthermore, the disease may cause major changes in the uEV secretion pattern and functional capacity of epithelial cells along the whole nephron and lower urinary tract (Ranghino et al., 2015). Therefore, the composition of urine in DKD, in terms of total proteins and other characteristics such as higher levels of oxidative DNA damage marker 8‐hydroxy‐deoxyguanosine (Van Deun et al., 2017), differs considerably between healthy controls and patients, offering the possibility for active disease monitoring. Thus, when comparing isolation methods, the inclusion of real patient samples in pilot studies is of key importance.
Here we compared three commonly used uEV isolation workflows, in normal population and in individuals with type 1 diabetes and macroalbuminuria, with focus on applicability for downstream RNA sequencing and biomarker discovery.
Electron microscopy negative staining showed uEV of typical morphology in all samples regardless of the isolation method used. This testifies the general suitability of all three approaches to uEV isolation but with slightly different profiles. Accordingly, in NG micrographs we recorded less vesicles than in samples processed with HFD and UC. This finding was consistent with NTA data, where we observed higher yields of uEV per ml of urine in HFD and UC samples compared to NG samples. Cryo‐EM also showed uEV of typical morphology in all samples. However, the particle concentration was below the optimum for cryo‐EM in our uEV preparations. Thus, independently of the method used, a further concentration step may be needed to visualize more EV.
In control samples, protein concentration was higher in HFD and NG uEV preparations in comparison to UC samples. In addition, protein content appeared to increase in all the uEV preparations of patients with macroalbuminuria. This is not surprising considering that compared to healthy controls (albumin excretion rate (AER) < 30 mg per day), urine from patients with macroalbuminuria contain more proteins (AER > 300 mg per day) (Topham, 2009). Protein concentration in NG and HFD samples were quite similar. However, this observation did not match the protein profiles of stain free gels after electrophoresis (i.e., NG lanes were not stained as strong as HFD lanes) (Figure S7). A plausible explanation for these findings might be the presence of substances in the NG preparations not compatible with the Micro BCA protein assay. One of the approaches we used to assess purity of uEV preparations was particle to protein ratio (Webber & Clayton, 2013). In this approach, lower ratios are interpreted as being less pure samples. In control samples, we found that particle to protein ratio was higher for UC samples than for HFD and NG samples. This may suggest that UC produces somewhat better purity than HFD and NG
Immunodetection of EV markers by Western blotting showed the presence of exosome‐enriched markers (TSG101, CD9) (Tkach et al., 2018) and also podocalyxin, present in uEV and originated from podocytes (Hara et al., 2010). In NG samples, however, these markers were either faintly detected or not detected at all. While this aspect has not been widely shown in earlier studies, this finding is in agreement with the results published by Mussak et al. (Mussack et al., 2019). The absence of EV markers in NG samples can be partly explained by the low yield of vesicles in this protocol as measured by NTA. Tamm‐Horsfall protein (THP), the most abundant normal protein in urine of mammals, was clearly present in HFD and UC samples but in less degree in NG samples. As THP is recognized as the major protein leading to artifacts in many downstream urine analytics assays (Fernández‐Llama et al., 2010; Wachalska et al., 2016), it emphasizes the need to manage this “contaminant” efficiently in all urinary analytics as suggested (Xu et al., 2019). Albumin, also a common protein present in urine especially in samples from DKD patients (Kamińska et al., 2016), was detected in HFD and NG samples. However, as our focus was on RNA sequencing, we did not include any further purification step. As a reference, HFD in combination with protocols to reduce THP interference has been successfully used for uEV proteomics (Hu et al., 2018; Xu et al., 2019). The UC protocol presented in our study has not been used for proteomics, but UC in combination with reducing agents such as DTT (Rood et al., 2010; Wolley et al., 2017), or sucrose gradient centrifugation (Hogan et al., 2014), or size exclusion chromatography (Rood et al., 2010) have been used for uEV proteomics with good results.
Because the three isolation workflows had different preclearing steps and principles of EV isolation that all affect the resulting population of EVs, we compared the harvested EV sizes between methods. Analysis of the size distribution of vesicles by NTA showed that NG workflow appeared to enrich smaller vesicle classes, while the large vesicle fraction (> 500 nm) was underrepresented in comparison with HFD and UC. Results obtained using diameters measured in TEM images confirmed the presence of larger vesicles (> 400 nm) in HFD and UC samples and a lower proportion of small EVs (60‐140 nm) in HFD samples. In addition, based on TEM data, HFD captured more very small sized EV‐like particles (< 40 nm) than UC and NG. As this small size class is not reliably detected by NTA, our NTA and EM results agreed reasonably considering that several factors affect EV size distribution data obtained by NTA and EM. Factors interfering with NTA size measurement include: i) limit of detection of NTA (depending on the system used and the sample analysed) is about 50 nm (Bachurski et al., 2019; van der Pol et al., 2014). ii) Albumin (McNicholas et al., 2017) and THP (Musante et al., 2020) can also scatter light and could be mistakenly considered as vesicles by the system, and iii) Large aggregates in EV preparations could be detected as large EVs, or big EVs could mask smaller ones (van der Pol et al., 2014). On the other hand, TEM allows accurate quantification of smaller vesicles but it is also influenced by factors such as: i) The use of fixative and drying the samples for EM processing can cause a reduction of the EV size (van der Pol et al., 2014) and ii) adhesion of vesicles to TEM grids could not be uniform (van der Pol et al., 2014) thus affecting the particle size distribution. Considering both size distribution calculations, HFD workflow appears to capture a wider range of vesicle sizes.
We did not find significant differences in the RNA yield per ml of urine between isolation methods, which shows similar efficiencies for HFD, UC and NG. However, the number of vesicles per ml was considerably lower for NG. Thus, not observing differences in the RNA yield could be explained by factors such as: i) non‐EV RNA contamination, as previously mentioned by Cheng et al. (Cheng et al., 2014) and ii) the quantification method used. Bioanalyzer Pico chip quantitative accuracy is 30% (within a chip) and the qualitative range is from 50 to 5000 pg/ul. Considering that the starting volumes of urine in HFD and UC were higher than in NG, the total amount of RNA yielded for NG was lower than for HFD and UC. Therefore, the NG samples were measured near the lower end of the range and it is possible that NG yield values were overestimated. Lower RNA yields from NG compared to other isolation methods has been reported by previous studies (Mussack et al., 2019; Wachalska et al., 2016).
We compared the three uEV isolation methods for RNA transcriptomics using two modalities: mRNA‐seq and miRNA‐seq. For mRNA‐seq, libraries were prepared using the poly(A) enrichment approach which captures the polyadenylated RNA and thereby removes most of ribosomal RNA (rRNA, (Slomovic et al., 2006)). The mRNA‐seq results showed that the number of reads obtained from NG samples was remarkably low compared to HFD and UC. As poly(A) approach performance is poor for partially degraded mRNAs (Schuierer et al., 2017), a possible explanation for the low number of reads in NG samples could be partial RNA degradation. However, this should be further explored using another library preparation approach for example, one that includes random priming rather than poly(A) priming and an rRNA depletion step, which was not done here. Another explanation for the poor performance of NG in mRNA‐seq could be related to the low amount of RNA and possible overestimation of RNA concentration as discussed above.
Principal Component Analysis, correlation analysis, and differential gene expression analysis of mRNA‐seq data showed high level of similarity of transcript expression between HFD and UC. On the contrary, NG samples presented a high number of differentially expressed genes and grouped distinctly in PCA and correlation analysis. This difference between NG, HFD, and UC may be mostly explained by the low number of reads in NG samples. The 16 differentially expressed RNAs between HFD and UC samples were mitochondrial RNAs and in all cases upregulated in HFD samples relative to the UC samples. This could be reflecting the presence of higher amounts of apoptotic bodies (Jiang et al., 2017), vesicles carrying mitochondria (Zhang et al., 2020), vesicles rich in mitochondrial content (Jang et al., 2019) or secreted mitochondria (Al Amir Dache et al., 2020) in HFD samples.
PCA of miRNA‐seq data did not replicate the differences/similarities between methods from the mRNAseq analysis. Pair‐wise clustering of the samples in miRNA correlation heatmap showed no clear‐cut clustering. The total miRNA raw counts from UC samples were higher than for HFD and NG. However, miRNA counts from HFD and NG are in the range of counts reported in other uEV small RNA sequencing studies (Mussack et al., 2019). Identified miRNAs (> 5 normalized counts in all samples within a method) or top ten identified miRNAs showed to be around 90% similar between isolation methods. GO enrichment analysis of miRNA targets showed that miRNA that do not overlap between methods may modulate various biological processes. Taken together, this may indicate that each method enriches slightly different subpopulations of miRNA rich vesicles or other uEV preparation components.
We acknowledge that the use of different urine clarification protocols add a source of variation while a standard protocol for urine preclearing has not been established for the EV research field. Thus, currently it is relevant to compare the entire workflows, not just EV isolation steps. Therefore we compared protocols that were already established and published, without introducing additional modifications. Similarly, the initial volume of urine employed for each workflow was different. In particular, the volume of NG was the lowest. However, we wanted to include a method in the comparison that was designed for lower volumes of urine, fast, and suitable to process many samples in parallel. On the other hand, HFD and UC were included as methods suited to analytics for a variety of downstream purposes and, additionally, their capacity to process large urine volumes was a valuable asset. Larger sample volumes or higher EV yields may be of key importance to achieve parallel layers of results like proteomics, lipidomics, RNomics and simultaneously, for necessary quality control assays. To address the volume differences, we normalized results to the original urine volume or used equal RNA amounts in all our comparisons. Lastly, we used a relatively low number of samples. However, we also included disease samples, which is of key relevance when studying performance of different methods for the research of urological diseases, such as diabetic nephropathy.
As all vesicle classes appear to contain at least partially different RNA contents (Lunavat et al., 2015), a non‐selective method to harvest majority of RNA contents without causing unacceptable sampling bias or unwanted losses may be preferred for evaluation of the total EVome. In our study, all the methods yielded representative vesicle repertoires and RNA, respectively, for sequencing and downstream sequence analysis. The workflows used here for HFD and UC were highly suitable to isolate uEVs for mRNA‐seq. Furthermore, the transcript expression data of both methods shared a high level of similarity. The miRNA‐seq results indicated that all three methods performed satisfactorily. However, lack of perfect correlation between the same individual samples is of concern. This suggests that the three different workflows and isolation principles may enrich different miRNA‐rich uEV preparation components. Variable levels of co‐isolating urinary proteins were also found depending on the albuminuria status and method used. Thus, for sensitive applications such as proteomics, further purification, especially management of Tamm‐Horsfall glycoprotein, is needed regardless of the isolation method. In conclusion, the different workflows and isolation methods enrich partially distinct EV preparation components – the lack of concordance may compromise attempts to cross‐compare studies, unless the basic performance of the methods is pre‐determined with studies such as ours.
CONFLICT OF INTEREST
KB, ODP, GL, MR, DD, CF, PG, LG, MP and HH, have no conflict of interest to declare.
Supporting information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGEMENTS
We thank Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki for providing laboratory facilities and the EV Core of University of Helsinki for performing NTA. We thank Benita Löflund and Pasi Laurinmäki (University of Helsinki) for technical assistance in cryoEM. The facilities and expertise of the HiLIFE CryoEM unit at the University of Helsinki, a member of Instruct‐ERIC Centre Finland, FINStruct, and Biocenter Finland are gratefully acknowledged. This study was supported by NovoNordisk Foundation grant no. NNF18OC0034200 (HH). This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement no. 115974 (BEAt‐DKD). This Joint Undertaking receives support from the European Union's Horizon 2020 research and innovation programme and EFPIA with JDRF. The research was also supported by the Academy of Finland (grants no. 263401, 267882, 312063 to L.G., and 317599 to O.P.D.)
Barreiro K, Dwivedi OP, Leparc G, et al. Comparison of urinary extracellular vesicle isolation methods for transcriptomic biomarker research in diabetic kidney disease. J Extracell Vesicles. 2020;10:e12038 10.1002/jev2.12038
REFERENCES
- Al Amir Dache, Z. , Otandault, A. , Tanos, R. , Pastor, B. , Meddeb, R. , Sanchez, C. , Arena, G. , Lasorsa, L. , Bennett, A. , Grange, T. , El Messaoudi, S. , Mazard, T. , Prevostel, C. , & Thierry, A. R. (2020). Blood contains circulating cell‐free respiratory competent mitochondria. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 34(3), 3616–3630. [DOI] [PubMed] [Google Scholar]
- Anders, S. , & Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biology, 11(10), R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachurski, D. , Schuldner, M. , Nguyen, P.‐H. , Malz, A. , Reiners, K. S , Grenzi, P. C , Babatz, F. , Schauss, A. C , Hansen, H. P , Hallek, M. , & Pogge Von Strandmann, E. (2019). Extracellular vesicle measurements with nanoparticle tracking analysis—An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. Journal of Extracellular Vesicles, 8(1), 1596016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro, K. , & Holthofer, H. (2017). Urinary extracellular vesicles. A promising shortcut to novel biomarker discoveries. Cell and Tissue Research, 369(1), 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro, K. , Huber, T. B. , & Holthofer, H. (2020). Isolating urinary extracellular vesicles as biomarkers for diabetic disease. Methods in Molecular Biology (Clifton, NJ), 2067, 175–188. [DOI] [PubMed] [Google Scholar]
- Campion, C. G. , Sanchez‐Ferras, O. , & Batchu, S. N. (2017). Potential role of serum and urinary biomarkers in diagnosis and prognosis of diabetic nephropathy. Canadian Journal of Kidney Health and Disease, 4, 205435811770537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L. , Sun, X. , Scicluna, B. J. , Coleman, B. M. , & Hill, A. F. (2014). Characterization and deep sequencing analysis of exosomal and non‐exosomal miRNA in human urine. Kidney International, 86(2), 433–444. [DOI] [PubMed] [Google Scholar]
- Dobin, A. , Davis, C. A. , Schlesinger, F. , Drenkow, J. , Zaleski, C. , Jha, S. , Batut, P. , Chaisson, M. , & Gingeras, T. R. (2013). STAR: ultrafast universal RNA‐seq aligner. Bioinformatics (Oxford, England), 29(1), 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep, H. , Gretz, N. , & Sticht, C. (2014). miRWalk database for miRNA‐target interactions. Methods in Molecular Biology (Clifton, NJ), 1182, 289–305. [DOI] [PubMed] [Google Scholar]
- Erdbrugger, U. , & Le, T. H. (2016). Extracellular vesicles in renal diseases: more than novel biomarkers? Journal of the American Society of Nephrology: JASN, 27(1), 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels, P. , Magnusson, M. , Lundin, S. , & Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics (Oxford, England), 32(19), 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Llama, P. , Khositseth, S. , Gonzales, P. A. , Star, R. A. , Pisitkun, T. , & Knepper, M. A. (2010). Tamm‐Horsfall protein and urinary exosome isolation. Kidney International, 77(8), 736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai, V. , Wu, X. , Bheda‐Malge, A. , Argyropoulos, C. P. , Bernardo, J. F. , Orchard, T. , Galas, D. , & Wang, K. (2018). Genome‐wide profiling of urinary extracellular vesicle microRNAs associated with diabetic nephropathy in type 1 diabetes. Kidney International Reports, 3(3), 555–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Suarez, M. L. , Thomas, D. B. , Barisoni, L. , & Fornoni, A. (2013). Diabetic nephropathy: is it time yet for routine kidney biopsy? World Journal of Diabetes, 4(6), 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, M. , Yanagihara, T. , Hirayama, Y. , Ogasawara, S. , Kurosawa, H. , Sekine, S. , & Kihara, I. (2010). Podocyte membrane vesicles in urine originate from tip vesiculation of podocyte microvilli. Human Pathology, 41(9), 1265–1275. [DOI] [PubMed] [Google Scholar]
- Harshman, S. W. , Canella, A. , Ciarlariello, P. D. , Agarwal, K. , Branson, O. E. , Rocci, A. , Cordero, H. , Phelps, M. A. , Hade, E. M. , Dubovsky, J. A. , Palumbo, A. , Rosko, A. , Byrd, J. C. , Hofmeister, C. C. , Benson, D. M. , Paulaitis, M. E. , Freitas, M. A. , & Pichiorri, F. (2016). Proteomic characterization of circulating extracellular vesicles identifies novel serum myeloma associated markers. Journal of Proteomics, 136, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, M. C. , Johnson, K. L. , Zenka, R. M. , Cristine Charlesworth, M. , Madden, B. J. , Mahoney, D. W. , Oberg, A. L. , Huang, B. Q. , Leontovich, A. A. , Nesbitt, L. L. , Bakeberg, J. L. , Mccormick, D. J. , Robert Bergen, H. , & Ward, C. J. (2014). Subfractionation, characterization, and in‐depth proteomic analysis of glomerular membrane vesicles in human urine. Kidney International, 85(5), 1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough, K. P. , Wilson, L. S. , Trevor, J. L. , Strenkowski, J. G. , Maina, N. , Kim, Y.‐I. , Spell, M. L. , Wang, Y. , Chanda, D. , Dager, J. R. , Sharma, N. S. , Curtiss, M. , Antony, V. B. , Dransfield, M. T. , Chaplin, D. D. , Steele, C. , Barnes, S. , Duncan, S. R. , Prasain, J. K. , … Deshane, J. S. (2018). Unique lipid signatures of extracellular vesicles from the airways of asthmatics. Scientific Reports, 8(1), 10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, S. , Musante, L. , Tataruch, D. , Xu, X. , Kretz, O. , Henry, M. , Meleady, P. , Luo, H. , Zou, H. , Jiang, Y. , & Holthofer, H. (2018). Purification and identification of membrane proteins from urinary extracellular vesicles using Triton X‐114 phase partitioning. Journal of Proteome Research, 17(1), 86–96. [DOI] [PubMed] [Google Scholar]
- Jang, S. C. , Crescitelli, R. , Cvjetkovic, A. , Belgrano, V. , Olofsson Bagge, R. , Sundfeldt, K. , Ochiya, T. , Kalluri, R. , & Lötvall, J. (2019). Mitochondrial protein enriched extracellular vesicles discovered in human melanoma tissues can be detected in patient plasma. Journal of Extracellular Vesicles, 8(1), 1635420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L. , Paone, S. , Caruso, S. , Atkin‐Smith, G. K. , Phan, T. K. , Hulett, M. D. , & Poon, I. K. H. (2017). Determining the contents and cell origins of apoptotic bodies by flow cytometry. Scientific Reports, 7(1), 14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamińska, A. , Platt, M. , Kasprzyk, J. , Kuśnierz‐Cabala, B. , Gala‐Błądzińska, A. , Woźnicka, O. , Jany, B. R. , Krok, F. , Piekoszewski, W. , Kuźniewski, M. , & Stępień, E. Ł. (2016). Urinary extracellular vesicles: potential biomarkers of renal function in diabetic patients. Journal of Diabetes Research, 2016, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konoshenko, M. Y. , Lekchnov, E. A. , Vlassov, A. V. , & Laktionov, P. P. (2018). Isolation of extracellular vesicles: general methodologies and latest trends. BioMed Research International, 2018, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G. K. , & Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England), 30(7), 923–930. [DOI] [PubMed] [Google Scholar]
- Lunavat, T. R , Cheng, L. , Kim, D.‐K. , Bhadury, J. , Jang, S. C. , Lässer, C. , Sharples, R. A , López, M. D. , Nilsson, J. , Gho, Y. S. , Hill, A. F , & Lötvall, J. (2015). Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells–Evidence of unique microRNA cargos. RNA Biology, 12(8), 810–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermaid, A. , Monier, B. , Zhao, J. , Liu, B. , & Ma, Q. (2019). Interpretation of differential gene expression results of RNA‐seq data: Review and integration. Briefings in Bioinformatics, 20(6), 2044–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNicholas, K. , Li, J. Y. , Michael, M. Z. , & Gleadle, J. M. (2017). Albuminuria is not associated with elevated urinary vesicle concentration but can confound nanoparticle tracking analysis. Nephrology (Carlton, Vic), 22(11), 854–863. [DOI] [PubMed] [Google Scholar]
- Merchant, M. L. , Rood, I. M. , Deegens, J. K. J. , & Klein, J. B. (2017). Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nature Reviews Nephrology, 13(12), 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, E. E. , Bailey, M. A. , & Dear, J. W. (2016). Renal extracellular vesicles: from physiology to clinical application. Journal of Physiology, 594(20), 5735–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante, L. , Bontha, S. V. , La Salvia, S. , Fernandez‐Piñeros, A. , Lannigan, J. , Le, T. H. , Mas, V. , & Erdbrügger, U. (2020). Rigorous characterization of urinary extracellular vesicles (uEVs) in the low centrifugation pellet–a neglected source for uEVs. Scientific Reports, 10(1), 3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante, L. , Tataruch, D. , Gu, D. , Benito‐Martin, A. , Calzaferri, G. , Aherne, S. , & Holthofer, H. (2014). A simplified method to recover urinary vesicles for clinical applications, and sample banking. Scientific Reports, 4, 7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussack, V. , Wittmann, G. , & Pfaffl, M. W. (2019). Comparing small urinary extracellular vesicle purification methods with a view to RNA sequencing‐Enabling robust and non‐invasive biomarker research. Biomolecular Detection and Quantification, 17, 100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S. , Lee, K. , Park, I. B. , Kim, N. H. , Cho, S. , Rhee, W. J. , Oh, Y. , Choi, J. , Nam, S. , & Lee, D. H. (2020). The profiles of microRNAs from urinary extracellular vesicles (EVs) prepared by various isolation methods and their correlation with serum EV microRNAs. Diabetes Research and Clinical Practice, 160, 108010. [DOI] [PubMed] [Google Scholar]
- Pathan M., Fonseka P., Chitti S. V., Kang T., Sanwlani R., Van Deun J., Hendrix A., Mathivanan S. (2019). Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Research, 47, (D1), D516–D519. 10.1093/nar/gky1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun, T. , Johnstone, R. , & Knepper, M. A. (2006). Discovery of urinary biomarkers. Molecular & Cellular Proteomics: MCP, 5(10), 1760–1771. [DOI] [PubMed] [Google Scholar]
- Puhka, M. , Nordberg, M‐E. , Valkonen, S. , Rannikko, A. , Kallioniemi, O. , Siljander, P. , & Af Hällström, T. M. (2017). KeepEX, a simple dilution protocol for improving extracellular vesicle yields from urine. European Journal of Pharmaceutical Sciences: Official Journal of the European Federation for Pharmaceutical Sciences, 98, 30–39. [DOI] [PubMed] [Google Scholar]
- Puhka, M. , Takatalo, M. , Nordberg, M.‐E. , Valkonen, S. , Nandania, J. , Aatonen, M. , Yliperttula, M. , Laitinen, S. , Velagapudi, V. , Mirtti, T. , Kallioniemi, O. , Rannikko, A. , Siljander, P. R‐M , & Af Hällström, T. M. (2017). Metabolomic profiling of extracellular vesicles and alternative normalization methods reveal enriched metabolites and strategies to study prostate cancer‐related changes. Theranostics, 7(16), 3824–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranghino, A. , Dimuccio, V. , Papadimitriou, E. , & Bussolati, B. (2015). Extracellular vesicles in the urine: markers and mediators of tissue damage and regeneration. Clinical Kidney Journal, 8(1), 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood, I. M. , Deegens, J. K.J. , Merchant, M. L. , Tamboer, W. P.M. , Wilkey, D. W. , Wetzels, J. F.M. , & Klein, J. B. (2010). Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney International, 78(8), 810–816. [DOI] [PubMed] [Google Scholar]
- Schneider, C. A. , Rasband, W. S. , & Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9(7), 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuierer, S. , Carbone, W. , Knehr, J. , Petitjean, V. , Fernandez, A. , Sultan, M. , & Roma, G. (2017). A comprehensive assessment of RNA‐seq protocols for degraded and low‐quantity samples. BMC Genomics, 18(1), 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic, S. , Laufer, D. , Geiger, D. , & Schuster, G. (2006). Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Research, 34(10), 2966–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, S. , Yeri, A. , Cheah, P. S. , Chung, A. , Danielson, K. , De Hoff, P. , Filant, J. , Laurent, C. D. , Laurent, L. D. , Magee, R. , Moeller, C. , Murthy, V. L. , Nejad, P. , Paul, A. , Rigoutsos, I. , Rodosthenous, R. , Shah, R. V. , Simonson, B. , To, C. , … Laurent, L. C. (2019). Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell, 177(2), 446–462.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen, P. , Sabaratnam, R. , & Jensen, B. L. (2020). Urinary extracellular vesicles: origin, role as intercellular messengers and biomarkers; efficient sorting and potential treatment options. Acta Physiologica (Oxford, England), 228(1), e13346. [DOI] [PubMed] [Google Scholar]
- Team, R. C. (2020). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; Retrieved from http://www.R-project.org/ [Google Scholar]
- Thery, C. , Witwer, K. W , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K , Ayre, D C. , Bach, J.‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N , Baxter, A. A , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, M. C. , Brownlee, M. , Susztak, K. , Sharma, K. , Jandeleit‐Dahm, K. A. M. , Zoungas, S. , Rossing, P. , Groop, P.‐H. , & Cooper, M. E. (2015). Diabetic kidney disease. Nature Reviews Disease Primers, 1, 15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach, M. , Kowal, J. , & Thery, C. (2018). Why the need and how to approach the functional diversity of extracellular vesicles. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences, 373(1737), 10.1098/rstb.2016.0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topham, P. (2009). Proteinuric renal disease. Clinical Medicine (London, England), 9(3), 284–287; quiz 8–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pol, E. , Coumans, F. A. W. , Grootemaat, A. E. , Gardiner, C. , Sargent, I. L. , Harrison, P. , Sturk, A. , Van Leeuwen, T. G. , & Nieuwland, R. (2014). Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. Journal of Thrombosis and Haemostasis: JTH, 12(7), 1182–1192. [DOI] [PubMed] [Google Scholar]
- Van Deun, J. , Mestdagh, P. , Agostinis, P. , Akay, Ö. , Anand, S. , Anckaert, J. , Martinez, Z. A. , Baetens, T. , Beghein, E. , Bertier, L. , Berx, G. , Boere, J. , Boukouris, S. , Bremer, M. , Buschmann, D. , Byrd, J. B , Casert, C. , Cheng, L. , Cmoch, A. , Daveloose, D. , … Hendrix, A. (2017). EV‐TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nature Methods, 14(3), 228–232. [DOI] [PubMed] [Google Scholar]
- van Niel, G. , D'Angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology, 19(4), 213–228. [DOI] [PubMed] [Google Scholar]
- Wachalska, M. , Koppers‐Lalic, D. , Van Eijndhoven, M. , Pegtel, M. , Geldof, A. A. , Lipinska, A. D. , Van Moorselaar, R. J. , & Bijnsdorp, I. V. (2016). Protein complexes in urine interfere with extracellular vesicle biomarker studies. Journal of Circulating Biomarkers, 5, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, J. , & Clayton, A. (2013). How pure are your vesicles? Journal of Extracellular Vesicles, 2, 19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis: New York: Springer‐Verlag. [Google Scholar]
- Wolley, M. J. , Wu, A. , Xu, S. , Gordon, R. D. , Fenton, R. A. , & Stowasser, M. (2017). In primary aldosteronism, mineralocorticoids influence exosomal sodium‐chloride cotransporter abundance. Journal of the American Society of Nephrology: JASN, 28(1), 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Barreiro, K. , Musante, L. , Kretz, O. , Lin, H. , Zou, H. , Huber, T. B. , & Holthofer, H. (2019). Management of tamm‐horsfall protein for reliable urinary analytics. Proteomics Clinical Applications, 13(6), 1900018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez‐Mo, M. , Siljander, P. R.‐M. , Andreu, Z. , Bedina Zavec, A. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Cordeiro‐Da Silva, A. , Fais, S. , Falcon‐Perez, J. M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. , Gursel, I. , … De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles, 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuana, Y. , Sturk, A. , & Nieuwland, R. (2013). Extracellular vesicles in physiological and pathological conditions. Blood Reviews, 27(1), 31–39. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Hubal, M. J. , & Kraus, V. B. (2020). Immune cell extracellular vesicles and their mitochondrial content decline with ageing. Immunity & Ageing: I & A, 17, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Data Availability Statement
The raw sequencing data analysed in this study is not publicly available due to local regulations. We provide raw sequencing count data in Tables S5 and S6. Raw sequencing data is available to qualified investigators upon reasonable request.


