Abstract
Background
The Gretchen Hagen 3 (GH3) genes encode acyl acid amido synthetases, many of which have been shown to modulate the amount of active plant hormones or their precursors. GH3 genes, especially Group III subgroup 6 GH3 genes, and their expression patterns in economically important B. oleracea var. oleracea have not been systematically identified.
Results
As a first step to understand regulation and molecular functions of Group III subgroup 6 GH3 genes, 34 GH3 genes including four subgroup 6 genes were identified in B. oleracea var. oleracea. Synteny found around subgroup 6 GH3 genes in B. oleracea var. oleracea and Arabidopsis thaliana indicated that these genes are evolutionarily related. Although expression of four subgroup 6 GH3 genes in B. oleracea var. oleracea is not induced by auxin, gibberellic acid, or jasmonic acid, the genes show different organ-dependent expression patterns. Among subgroup 6 GH3 genes in B. oleracea var. oleracea, only BoGH3.13–1 is expressed in anthers when microspores, polarized microspores, and bicellular pollens are present, similar to two out of four syntenic A. thaliana subgroup 6 GH3 genes. Detailed analyses of promoter activities further showed that BoGH3.13–1 is expressed in tapetal cells and pollens in anther, and also expressed in leaf primordia and floral abscission zones.
Conclusions
Sixty-two base pairs (bp) region (− 340 ~ − 279 bp upstream from start codon) and about 450 bp region (− 1489 to − 1017 bp) in BoGH3.13–1 promoter are important for expressions in anther and expressions in leaf primordia and floral abscission zones, respectively. The identified anther-specific promoter region can be used to develop male sterile transgenic Brassica plants.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-020-07345-9.
Keywords: Brassica oleraceea var. oleracea, TO1000, Gretchen Hagen 3, GH3, Anther, Promoter
Background
The Gretchen Hagen 3 (GH3) gene was first identified in Glycine max (soybean) as an early response gene, which is transcriptionally induced in less than 30 min by treatment of auxin plant hormone [1]. Later studies have found that GH3 genes are found in diverse plant species including mosses and fern, but not in two model algae, Chlamydomonas reinhardtii or Volvox carteri [2–7]. Like acyl CoA synthetases, non-ribosomal peptide synthetases, and luciferases in ANL superfamily proteins, GH3 proteins conjugate combinations of amino acids and acyl acids in two-step reactions [8, 9]. In the first half-reaction involving ATP and acyl acid, adenylated acyl acid is produced and pyrophosphate is released. In the second half-reaction, adenylated acyl acid intermediate reacts with amino acids, resulting in the release of acyl acid-amino acid amido conjugate and adenosine monophosphate. For example, Arabidopsis thaliana (Arabidopsis) GH3.11, jasmonate (JA) resistant 1 (JAR1), and Arabidopsis GH3.17, reversal of sav 2 (VAS2), catalyze the production of JA-isoleucine and indole acetic acid (IAA)–glutamate, respectively [10, 11].
GH3 proteins are involved in various developmental processes and environmental responses in plants, by modulating the activities or availabilities of plant hormones and related compounds, including precursors of plant hormones [12]. Abnormal expressions caused by null mutation or hyper- and mis-expression lead to various phenotypic defects. In Arabidopsis, atgh3.11 (jar1) mutant does not produce bioactive JA-Isoleucine and defective in JA signaling, while atgh3.17 (vas2) mutant over-accumulates free IAA at the expense of IAA-glutamate [11, 13]. In addition, atgh3.12 (avrPphB susceptible 3 (pbs3)) mutant was found to be more susceptible to bacterial pathogens because production of isochorismoyl glutamate, the precursor of salicylic acid (SA), catalyzed by PBS3, is compromised [14]. Over-expression of AtGH3.6 (Dwarf in Light 1 (DFL1)) or AtGH3.2 (Yadokari 1 (YDK1)), which are induced by auxin, causes hyper-sensitivity to light treatment leading to dwarfism [15, 16]. Over-expression of AtGH3.5 (WES1), which is induced by treatment of abscisic acid and SA, as well as auxin, leads to auxin resistant phenotypes [17]. In various plants, important roles played by plant GH3 enzymes have also been demonstrated: nodule numbers and sizes in soybean [18], resistance to Xanthomonas bacteria in citrus [19], drought and salt tolerance in cotton [20], and fruit softening in kiwi [21], were shown to be affected by GH3 gene expressions.
Phylogenetic analyses show that plant GH3 genes can be clustered into 3 groups (GroupI~ III) based on overall amino acid sequences or 8 subgroups (subgroup 1 ~ 8) based on acyl acid-binding site sequences of Arabidopsis, rice, soybean, maize, Selaginella, and moss GH3 proteins [7, 10, 12, 22]. However, only Group I and II GH3 genes have been identified in Gramineae genomes [23–25]. Using GH3 enzymes in various plant species, preferential substrates of GH3 enzymes in terms of acyl acids and amino acids have been determined [8, 14, 18, 22, 26–30]. In addition, a systematic evaluation of sixty GH3 enzymes from Arabidopsis, grape, rice, Physcomitrella, and Selaginella also revealed that not all the enzymes encoded by Group I GH3 genes are involved in JA signaling and 12 out of 16 enzymes encoded by Group II GH3 genes display clear substrate preferences for IAA among three acyl acid substrates - jasmonate, IAA, and 4 hydroxybenzoate (4-HBA) [31]. In case of Group III GH3 enzymes, which are encoded by the largest GH3 group in the plant genomes, no clear substrate preferences were established, except AtGH3.9 or OsGH3.13 for IAA and Arabidopsis PBS3 for 4-HBA. In case of Group III subgroup 6 GH3s, only AtGH3.15 in Arabidopsis was shown to have substrate preference for indole butyric acid (IBA), the auxin precursor [28]. Although decrease in IBA-mediated root elongation inhibition and lateral root formation were observed in transgenic plants constitutively expressing AtGH3.15, in vivo function(s) of other subgroup 6 GH3 genes have yet to be determined. In rapeseed (Brassica napus) and its diploid ancestors, Chinese cabbage (Brassica rapa) and cabbage (Brassica oleracea var. capitata), up to sixty-six GH3-coding genes have been identified [32, 33]. However, detailed study of GH3-coding genes in kale-type Brassica species (Brassica oleracea var. oleracea), TO1000, which serves as an excellent model for important vegetable crops in Brassica oleracea with various morphological and phytochemical traits [34], have not been performed yet.
The anther is a part of the stamen, the male reproductive organ in plants, and is connected to the flower receptacle by a filament, which is the other part of the stamen [35, 36]. Anther development is divided into two phases, culminating in the release of pollen grains, the male gametophytes in plants. Microsporogenesis, the first phase, includes establishment of anther morphology, cell and tissue differentiation, and meiosis of microspore mother cells. Tetrads of haploid microspores produced by meiotic divisions of diploid pollen mother cells are released as distinct unicellular microspores into locules by a mixture of enzymes produced from tapetum cells, which also provide nutrients and pollen wall materials for developing pollens [37, 38]. During microgametogenesis, the second phase, differentiation of microspores into pollen grains and tissue degeneration occur for the release of pollens. Microgametogenesis starts with the expansion of the microspore, which is often found with the formation of one large vacuole [39]. This involves movement of the microspore nucleus from the center of the cell to a position close to the cell wall, where the microspore produces two unequal cells, a large vegetative cell and a small generative cell, in a process called pollen mitosis (PM) I. Then, the generative cell, which is spatially separated from the pollen grain wall and engulfed by the vegetative cell, undergoes another round of cell division, called PM II [37]. Depending on whether PM II happens before or after pollen dispersal from the anther, the pollens are called tricellular or bicellular pollen [40]. Plant hormones - JA, auxin, gibberellic acid (GA), and ethylene – are known to play important roles in stamen maturation, locule opening, anther dehiscence, and pollen viability during stamen and pollen development [35, 41–43].
To expand our knowledge on the regulation and molecular functions of Group III GH3 genes in plants – especially those in subgroup 6 whose functions are still elusive – GH3 genes in kale-type B. oleracea var. oleracea were identified genome-wide, and expression patterns of subgroup 6 GH3 genes were investigated. It was found that subgroup 6 GH3 genes in B. oleracea var. oleracea, composed of four genes showing synteny with closely related Arabidopsis subgroup 6 GH3 genes, are not induced by auxin, GA, and JA treatment, but have different organ expression patterns. BoGH3.13–1, a subgroup 6 GH3 gene, is specifically expressed in tapetal cells in anther and pollens when microspores, polarized microspores, and bicellular pollens are produced, as well as in leaf primordia and floral abscission zones. Promoter bash experiments revealed that a 62 base pairs (bp) DNA sequence, − 340 to − 279 bp upstream of BoGH3.13–1 start codon, is required for anther-specific expression, while a ~ 450 bp region (− 1489 to − 1017) is necessary for expression in leaf primordia and floral abscission zones.
Results
Thirty-four GH3-encoding genes (BoGH3s) are present in B. oleracea var. oleracea
In the Ensembl Plants database (http://plants.ensembl.org/index.html), protein sequences of 55 GH3 candidate genes in kale-type B. oleracea showed similarities to the 19 Arabidopsis GH3 proteins [10]. Among these, 34 GH3 proteins were found to have intact GH3 domains (pfam03321) and considered as GH3 proteins (Table S1; Figure S1). Although identical genomic sequences were used for annotation, only 30 B. oleracea GH3 candidate proteins, including two with truncations in GH3 domains, were found to have significant similarities to Arabidopsis GH3s in NCBI database (NCBI, http://ncbi.nlm.nih.gov) [34]. The 34 BoGH3 proteins with the intact GH3 domains in Ensembl Plants database include all 28 putative GH3 proteins with the intact GH3 domains identified in NCBI database (Table S1). For proteins showing different protein sequences between two databases, such as BoGH3.12–2 and BoGH3.17–1, NCBI protein models were adopted in our study because they are supported by RNA-seq data in NCBI. While 34 GH3 protein-coding genes were identified from B. oleracea var. oleracea in our study, 25 and 29 GH3 protein-coding genes were previously reported for cabbage-type B. oleracea var. capitata in the comparison with B. napus genes by two independent studies, respectively [32, 33].
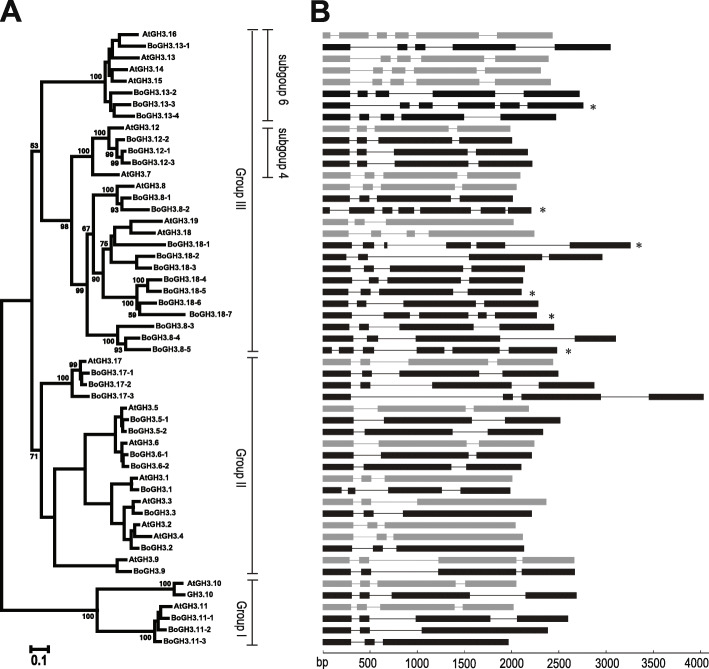
Similar to previous phylogenetic analyses of GH3 proteins including cabbage-type B. oleracea var. capitata, phylogenetic clustering of Arabidopsis and BoGH3 proteins demonstrated that BoGH3 proteins can be divided into three groups (Group I, II, and III) (Fig. 1a) [6, 10, 32, 33]. It was found that Group I consists of two Arabidopsis and four BoGH3 proteins, while Group II consists of eight Arabidopsis and 11 BoGH3 proteins. In the case of Group III, nine Arabidopsis GH3s and 19 BoGH3 proteins were clustered together. In general, exon/intron structures of BoGH3 genes were same to closely related counterparts in Arabidopsis with some exceptions (Fig. 1b). For example, four protein-coding exons were detected for BoGH3.1 in Group II, based on the distribution of RNA-seq reads in NCBI database, while three protein-coding exons of AtGH3.1 is reported in TAIR JBrowse (https://jbrowse.arabidopsis.org/). In case of BoGH3.11–2 and BoGH3.11–3, which are closely related to AtGH3.11 (JAR1) with four protein-coding exons, only three exons supported by RNA-seq reads were observed. Structural differences were also observed for five BoGH3 genes (BoGH3.8–2, BoGH3.8–5, BoGH3.13–3, BoGH3.18–1, and BoGH3.18–7) that were identified only in Ensembl Plants.
Fig. 1.
Phylogenetic relationships and exon/intron structures of GH3 proteins in Arabidopsis and B. oleracea var. oleracea. a Phylogenetic analysis of GH3 family members in Arabidopsis and B. oleracea var. oleracea. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The phylogenetic tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Bootstrap test percentages of 1000 replicates are shown next to the branches. b Gene structures for Arabidopsis and BoGH3 proteins were generated by gene structure display servers (http://gsds.cbi.pku.edu.cn/Gsds_about.php). Note that exons indicated here do not contain untranslated regions. Astertisks indicate exon/intron structures of genes, which were annotated only in Ensembl Plants. The black boxes and lines represent exons and introns in B. oleracea var. oleracea GH3 genes, while the gray boxes and lines represent exons and introns in Arabidopsis GH3 genes. Groups (I~ III) and subgroups (4 & 6) of GH3 proteins were designated based on the Staswick et al. (2002) and Westfall et al. (2012), respectively [10, 12]
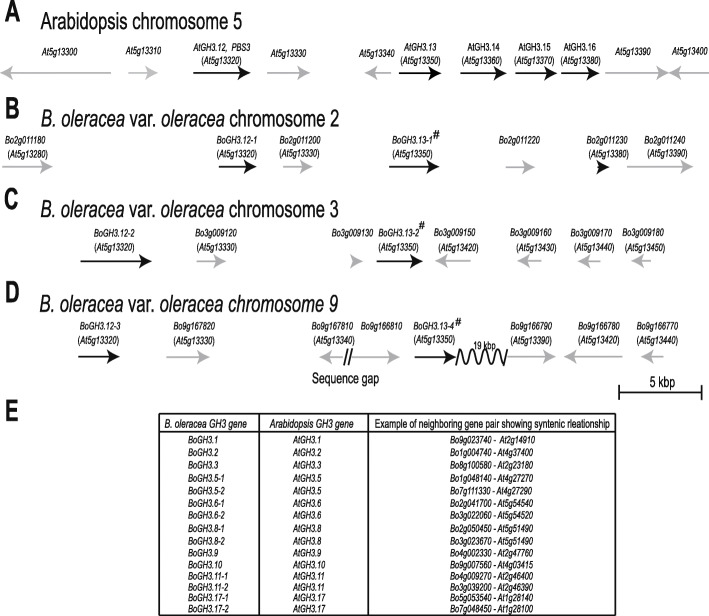
Synteny is observed for group III subgroup 6 GH3 genes between Arabidopsis and B. oleracea var. oleracea
In B. oleracea var. oleracea, 4 out of 34 Group III BoGH3 proteins (BoGH3.13–1, BoGH3.13–2, BoGH3.13–3, and BoGH3.13–4) show a close relationship with Arabidopsis subgroup 6 GH3 proteins (Fig. 1a). While the four BoGH3 genes are found on different chromosomes, four Arabidopsis GH3 genes (AtGH3.13, AtGH3.14, AtGH3.15, and AtGH3.16) in the same subgroup are located within 15,000 bp genomic region on Arabidopsis chromosome 5 (Fig. 2a). When genes located around Arabidopsis and B. oleracea var. oleracea subgroup 6 GH3 genes were compared, syntenies were detected around the AtGH3.13 ~ AtGH3.16 cluster and three BoGH3 genes (BoGH3.13–1, BoGH3.13–2, and BoGH3.13–4) (Fig. 2 b-d). In the upstream of three BoGH3 genes, Bo2g011200 (Fig. 2b), Bo3g009120 (Fig. 2c), and Bo9g167820 (Fig. 2d) showing sequence similarities to At5g13330, an RAP2.6 L transcription factor found upstream of the AtGH3.13 ~ AtGH3.16 cluster, were identified (Fig. 2a). Moreover, BoGH3.12–1, BoGH3.12–2, and BoGH3.12–3, which are clustered with AtGH3.12 (PBS3) in the phyologenetic tree as Group III subgroup 4 GH3 genes, were also found further upstream, same to AtGH3.12 (PBS3) located upstream of the AtGH3.13 ~ AtGH3.16 cluster. Consistent with the syntenic relationships in these genomic regions, sequence similarities were also observed downstream of the Arabidopsis GH3 cluster and the three BoGH3 genes on different chromosomes (Fig. 2b–d): Bo2g011240 and Bo9g166790 show sequence similarity to At5g13390, No Exine Formation 1. In addition to six subgroup 4 and subgroup 6 BoGH3 genes showing synteny (Fig. 2b – 2D), analyses for remaining 28 BoGH3 genes revealed that 15 more BoGH3 genes have syntenic relationships with AtGH3 genes (Fig. 2e).
Fig. 2.
Syntenies are found between genomic regions around Arabidopsis AtGH3.13 and corresponding regions in B. oleracea var. oleracea. Each panel shows gene organization, in which GH3 and non-GH3 genes from start to stop codons are indicated by black and gray arrows, respectively. Direction of each arrow shows that of gene transcription. a The gene organization on Arabidopsis chromosome 5 around AtGH3.12. b-d The gene organizations of B. oleracea var. oleracea chromosome 2 near BoGH3.12–1, chromosome 3 near BoGH3.12–2, and chromosome 9 near BoGH3.12–4. Arabidopsis genes showing sequence similarities to BoGH3 genes are indicated in parenthesis below B. oleracea var. oleracea gene names. The BoGH3 genes similar to Arabidopsis AtGH3.13 ~ AtGH3.16 gene cluster are indicated with sharp (#) symbols. Bo2g011230 in (B) encodes a truncated protein with a sequence similarity to Arabidopsis GH3 genes in the cluster. e Syntenic relationships detected between other BoGH3 genes in B. oleracea var. oleracea and Arabidopsis
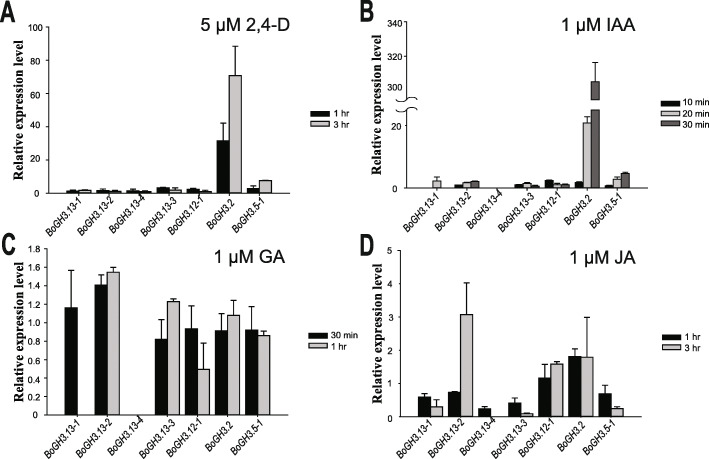
Subgroup 6 BoGH3 genes are not induced by auxin treatment in the seedling stage
In Arabidopsis, auxin treatment can induce transcription of some GH3 genes, such as AtGH3.2 (YDK1), AtGH3.5 (WES1), and AtGH3.6 (DFL1) [15–17]. However, expression conditions and functions of GH3 genes in other plants are largely unknown. To gain insights on the expression patterns and functions of four B. oleracea var. oleracea subgroup 6 GH3 identified in this study, we determined whether these genes can be induced by plant hormones and found that none of subgroup 6 BoGH3 genes were significantly induced by auxin (synthetic 2,4-Dichlorophenoxy acetic acid (2,4-D) or natural IAA), GA or JA treatment at the seedling stage, except BoGH3.13–2 that is weakly induced by JA (Fig. 3). One of subgroup 4 BoGH3 gene, BoGH3.12–1, also did not show expression changes responding to hormone treatments. In contrast, transcriptional inductions by auxin were evident for BoGH3 genes included as positive controls (BoGH3.2 and BoGH3.5–1), which are closely related auxin-inducible Arabidopsis GH3 genes [16, 17].
Fig. 3.
Subgroup 6 BoGH3 genes are not induced by auxin at the seedling stage. Relative expression levels of four subgroup 6 BoGH3 genes and three selected GH3 genes in other subgroups in response to treatments of 5 μM 2,4-D (a), 1 μM IAA (b), 1 μM GA (c), and 1 μM JA (d) were determined by qRT-PCR experiment with Actin control. The expression level of mock condition was set to value 1 and used as reference to compare expression level changes after hormone treatments. Bar graphs show average relative expression values with standard errors (SE). Averages values of two independent results for 2,4-D or JA treatments are shown, while representative results are shown for IAA or GA treatments. Bar graphs for genes without any significant amplification are not included
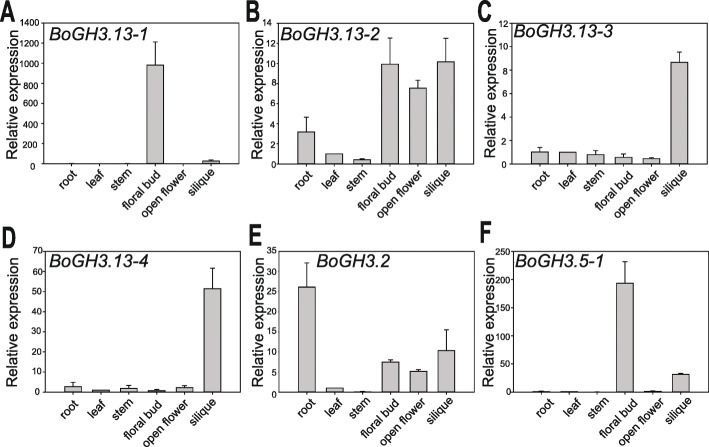
BoGH3.13–1 is strongly expressed in stamen at a specific stage during flower development
For four subgroup 6 and two auxin-inducible GH3 genes in B. oleracea var. oleracea, relative expression patterns in six different organs - root, leaf, stem, floral bud, opened flower, and silique - were determined. Among four subgroup 6 BoGH3 genes, BoGH3.13–1 was found to be most strongly expressed in floral bud, although significant expression was also observed in silique compared to that in leaf (Fig. 4a). Only negligible expressions of BoGH3.13–1 were detected in other organs, including open flowers. For the other three subgroup 6 BoGH3 genes, the strongest expression was commonly found in siliques (Fig. 4 b-d), while comparable expressions in floral bud and open flower were also observed for BoGH3.13–2 (Fig. 4b). For auxin-inducible BoGH3.2 and BoGH3.5–1, which were included as comparison, distinct relative expression patterns were detected: BoGH3.2 and BoGH3.5–1 were found to be most strongly expressed in root and floral bud, respectively (Fig. 4 e & f). For three subgroup 4 BoGH3 genes, stronger expressions were commonly observed in roots (Figure S2).
Fig. 4.
qRT-PCR results showing expression patterns of selected BoGH3 genes, including four subgroup 6 BoGH3 genes. Relative expression levels of BoGH3.13–1 (a), BoGH3.13–2 (b), BoGH3.13–3 (c), BoGH3.13–4 (d), BoGH3.2 (e), and BoGH3.5–1 (f) were determined by qRT-PCR experiment with Actin control in different organs and/or developmental stages. Bar graphs show average relative expression values with SEs. The expression level of leaf was set to value 1 and used as reference to compare expression levels in different organs
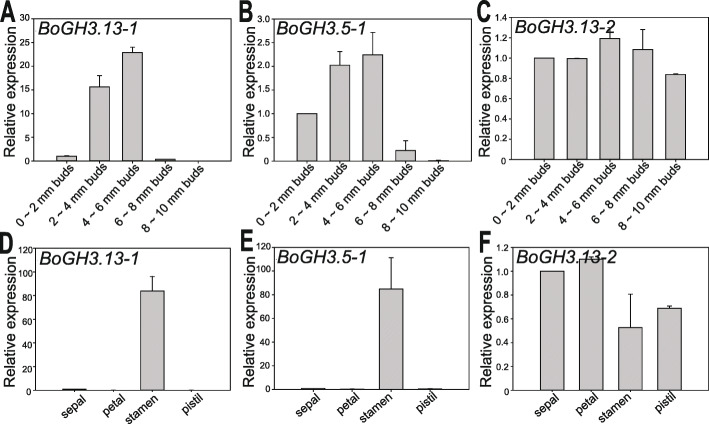
For BoGH3.13–1 and BoGH3.5–1, which show strong preferential expressions in floral bud (Fig. 4 a & f), it was also determined whether expressions of these genes are temporally regulated during floral bud development. When the expression levels were monitored for developing floral buds sorted by lengths (Figure S3), which reflect the progress of flower development [44], both genes showed stronger expression when bud lengths are about 2 to 6 mm, although BoGH3.13–1 in subgroup 6 GH3 showed more dramatic expression changes by developmental progress than BoGH3.5–1 (Fig. 5 a & b). In 4 ~ 6 mm-long floral buds, where the two genes are most strongly expressed, almost exclusive expression was detected in stamen among sepal, petal, stamen, and pistils (Fig. 5 d & e). In contrast, no significant developmental and organ-specific expression differences were observed for BoGH3.13–2, another subgroup 6 BoGH3 that are constitutively expressed in floral buds, open flowers, and siliques (Figs. 4b, 5c & f).
Fig. 5.
BoGH3.13–1 and BoGH3.5–1 are strongly expressed in anther. Steady-state expression levels of BoGH3.13–1 (a), BoGH3.5–1 (b), and BoGH3.13–2 (c) in developing floral buds and those of BoGH3.13–1 (d), BoGH3.5–1 (e), and BoGH3.13–2 (f) in sepal, petal, stamen, and pistil of 4 ~ 6 mm floral buds were determined with qRT-PCR. Bar graphs show average relative expression values with SEs. The expression level of 0 ~ 2 mm buds (a-c) and that of sepal (d-e), which were normalized to that of ACTIN, were set to value 1 and used as reference
BoGH3.13–1 and BoGH3.5–1 are expressed in tapetum and pollen grains
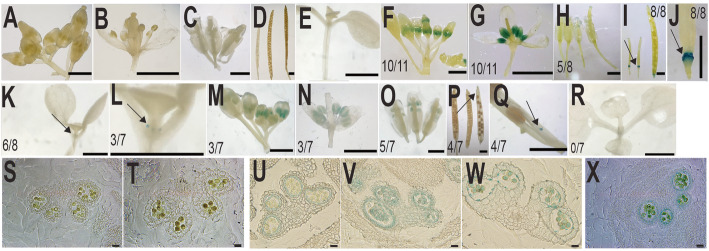
To narrow down spatial expression patterns of stamen-expressed BoGH3.13–1 and BoGH3.5–1, we generated transgenic plants, in which GUS (β-glucuronidase) reporter genes are expressed under the control of about 1500 bp putative promoter sequences of these BoGH3 genes. BoGH3.13–1 (− 1489 ~ − 1)::GUS and BoGH3.5–1 (− 1496 ~ − 1)::GUS are two transgenic plants, in which − 1489 ~ − 1 and − 1496 ~ − 1 bp DNA sequences upstream of BoGH3.13–1 and BoGH3.5–1 start codon, respectively, are fused to GUS reporter genes. In BoGH3.13–1 (− 1489 ~ − 1)::GUS, GUS expression was observed in anthers of developing floral buds (Fig. 6 f & g), consistent with the qRT-PCR (quantitative reverse transcription polymerase chain reaction) results (Figs. 4 & 5). Weak GUS stainings in some stigmas were found to be caused by stigma-attached pollens (Fig. 6h). GUS staining was also observed in siliques, but only in the floral organ abscission regions of petals, sepals, and stamens (Fig. 6 i & j). In addition, GUS expression was detected in leaf primordia of BoGH3.13–1 (− 1489 ~ − 1)::GUS seedlings (Fig. 6 k & l). In BoGH3.5–1 (− 1496 ~ − 1)::GUS, GUS expression was detected in developing anthers and unfertilized ovule or aborted seeds (Fig. 6m-q), but not in seedling leaf primordia (Fig. 6r). To further define the spatial expression patterns of BoGH3.13–1 and BoGH3.5–1 in anther, cross-sectioned floral buds were examined and specific expression in tapetum cells and pollen grains were detected for both genes (Fig. 6u-x). In BoGH3.13–1 (− 1489 ~ − 1)::GUS, GUS staining seems to appear in the tapetum first and pollens later (Fig. 6u & v).
Fig. 6.
GUS staining patterns in Arabidopsis transgenic plants with two BoGH3 promoter::GUS transgene. a-e Samples from wild-type Arabidopsis plants (WT) - floral buds (a), a dissected floral bud (b), open flowers (c), siliques (d), and 8-day old seedling (e). f-l Samples from BoGH3.13–1 (− 1489 ~ − 1)::GUS transgenic plants - floral buds (f), a dissected floral bud (g), open flowers (h), siliques (i-j), and 8-day old seedling (k-l). m-r Samples from BoGH3.5–1 (− 1496 ~ − 1)::GUS transgenic - floral buds (m), a dissected floral bud (n), open flowers (o), siliques (p-q), and 8-day old seedling (r). Transverse sections of GUS-stained floral buds of WT plants (s-t). Transverse sections of GUS-stained floral buds of BoGH3.13–1 (− 1489 ~ − 1)::GUS (u-w). Transverse sections of GUS-stained floral buds of BoGH3.5–1 (− 1496 ~ − 1)::GUS (x). Dividends and denominators of fractions in the pictures are transgenic plants with the GUS staining and all the transgenic plants examined, respectively. Arrows indicate GUS stained parts. Scale bars in (a-r): 1 mm. Scale bars in (s-x): 20 μm
BoGH3.13–1 and BoGH3.5–1 are most strongly expressed around when polarized microspores are generated
To investigate which milestone events in microsprogenesis or microgametogenesis occur in pollens when BoGH3.13–1 and BoGH3.5–1 are expressed (Fig. 5), developing pollens were collected from floral buds and open flowers. Based on the numbers and organization of 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei, it was found that tetrads and microspores are observed in less than 2 mm floral buds (Fig. 7 a & d), in which the two anther-expressed GH3 genes, BoGH3.13–1 and BoGH3.5–1, are weakly expressed (Fig. 5). In 2 ~ 6 mm floral buds, in which the two anther-expressed GH3 genes are most strongly expressed, microspores, polarized microspores, and bicellular pollens were observed (Fig. 7b-c & e-f). While bicellular and tricellular pollens were observed in 6 ~ 8 mm buds, only tricellular pollens were observed in 8 ~ 10 mm buds and opened flowers (Fig. 7g- l). These data show that BoGH3.13–1 and BoGH3.5–1 are strongly induced when polarized microspores are mainly produced during early microgametogenesis [45, 46].
Fig. 7.
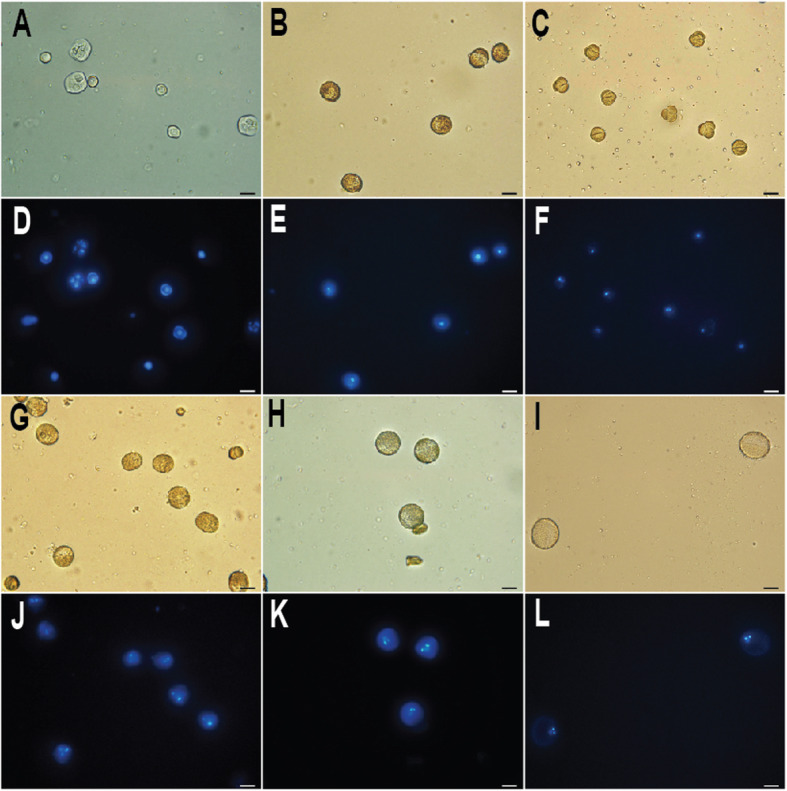
DAPI-stained developing pollen grains in B. oleracea var. oleracea floral buds. Bright-field images of pollens in less than 2 mm floral buds (a), 2 ~ 4 mm buds (b), 4 ~ 6 mm buds (c), 6 ~ 8 mm buds (g), 8 ~ 10 mm buds (h), and open flowers (i). Fluorescence images of DAPI-stained pollens in less than 2 mm floral buds (d), 2 ~ 4 mm buds (e), 4 ~ 6 mm buds (f), 6 ~ 8 mm buds (j), 8 ~ 10 mm buds (k), and open flowers (l). Scale bar: 20 μm
One hundred eighty-six bp region upstream of BoGH3.13–1 is sufficient for anther-specific expression
DNA sequences responsible for tissue-specific expression of BoGH3.13–1 was investigated with different DNA regions upstream of the start codon (Fig. 8a). When P1, in which − 1017 ~ − 1 bp region was fused upstream of GUS reporter gene, was used to generate P1 transgenic plants, GUS expressions in anthers and pollens were still detected (Fig. 8b-d), but those in floral abscission zones and leaf primordia were lost, except one case showing GUS staining in the floral abscission zone (Fig. 8 e & f). When P2 (− 418 ~ − 1) and P3 (− 340 ~ − 155), without − 155 ~ − 1 bp putative 5′ untranslated region based on RNA-seq reads in SRX209697 (NCBI), were used, anther-specific GUS expressions were found to be maintained (Fig. 8 g & h). While P4 (− 278~ − 155) did not show GUS expression in all twelve lines, five out of twelve P5 (− 418 ~ 279) showed GUS expression, suggesting sixty-two bp region (− 340 ~ − 279) in P3 sequence is important for anther-specific expression of BoGH3.13–1 (Fig. 8 i & j).
Fig. 8.

Representative GUS staining patterns to define a promoter region directing anther-specific expression of BoGH3.13–1. a Genomic DNA regions used in transgenic lines for promoter analysis. Names of transgenic plants used are written in italic and different regions upstream of BoGH3.13–1 start codon to direct GUS reporter expression are indicated in parentheses. b-f GUS staining of floral buds (b), a dissected floral bud (c), open flowers (d), siliques (e), and 8-day old seedling (f) of P1 transgenic plants, and floral buds from P2 (g), P3 (h), P4 (i), and P5 (j) are shown. Arrows indicate GUS-stained pollen (d) or floral abscission zones in a silique (e). Dividends and denominators of fractions in the pictures are numbers of transgenic plants with the GUS staining and all the transgenic plants examined, respectively. Scale bars: 1 mm
Discussion
Thirty-four GH3-coding genes of kale-type B. oleracea var. oleracea, which have intact GH3 domains, were identified from Ensembl plants database (Fig. 1a). Among these, 28 gene models were also found in NCBI database, which had used the identical genomic sequence for annotation [34]. The discrepancy in BoGH3 gene numbers between Ensembl plants and NCBI database may result from the use of different gene prediction algorithms or validations. Recently, twenty-nine GH3 protein-coding gene models related to cabbage-type B. oleracea var. capitata GH3 genes were identified from the investigation of genomic sequence of B. napus [32, 33], and twenty-eight genes were found to have intact GH3 domains and meet our criteria, while Bol042635 was found to encode a truncated GH3 domain with only 224 amino acids (Table S2) [33]. Among 34 BoGH3 coding-genes reported in this study, putative orthologs of cabbage-type B. oleracea were identified for 19 genes, but clear orthologous relationship could not be determined for the other 15 B. oleracea var. oleracea GH3 genes, based on amino acid sequence identities of over 95%. Considering 6 B. oleracea var. oleracea GH3 genes, whose expression could not be confirmed in NCBI database, are included in these 15 cabbage-type GH3 genes without putative orthologs, we speculate these 6 genes are pseudogenes and lost in B. oleracea var. capitata. In addition, orthologs of 9 cabbage-type B. oleracea GH3 genes could not be determined in B. oleracea var. oleracea (Table S2).
Group III subgroup 6 GH3 genes in B. oleracea var. oleracea and Arabidopsis seem to have evolved by duplications. Four Arabidopsis GH3 genes in subgroup 6 are located within 15 kbp region on the same Arabidopsis chromosome, while 4 BoGH3 genes in the same subgroup are located on 4 different chromosomes of B. oleracea var. oleracea (Fig. 2). Syntenies found between genomic regions around the subgroup 6 AtGH3 and BoGH3 genes suggest that AtGH3.13 ~ AtGH3.16 cluster in Arabidopsis was generated by tandem duplication after Brassica lineage-specific whole genome triplication and/or other BoGH3 genes around BoGH3.13–1, BoGH3.13–2, and BoGH3.13–4 might have been lost after divergence of Arabidopsis and Brassica lineages [47]. Consistent with this idea, one intact and one truncated form of GH3 genes in subgroup 6, BoGH3.13–1 and B02g011230, were identified within 15 kb region on the chromosome 2 of B. oleracea var. oleracea (Fig. 2b). Members in gene family in plants are known to evolve through both tandem (local) duplication and whole genome duplication, which were followed by gene loss or gene retention leading to functional diversification [48]. Nonetheless, close genomic locations of subgroup 4 and subgroup 6 GH3 genes in Arabidopsis and B. oleracea var. oleracea indicate both AtGH3.12-like and AtGh3.13-like GH3 genes were present in proximity before the separation of Arabidopsis and Brassica lineages. For exon/intron structures of BoGH3 and AtGH3 genes, overall similarities were observed for the evolutionarily related genes. However, distributions of RNA-seq reads in NCBI database revealed that protein-coding exons of BoGH3.1, BoGH3.11–2, and BoGH3.11–3 are differently organized compared to those of related Arabidopsis GH3 genes (Fig. 1b). Differences in the structures observed for five BoGH3 genes (BoGH3.8–2, BoGH3.8–5, BoGH3.13–3, BoGH3.18–1, and BoGH3.18–7) and related Arabidopsis genes might result from deletions/insertions and incorrect annotations, considering that these five BoGH3 genes are identified only in Ensembl Plants, not supported by RNA-seq data in NCBI database, and encode predicted GH3 proteins with multiple deletions (Fig. 1b & S1).
Four subgroup 6 BoGH3 genes, which seem to be generated from same ancestor gene(s), show distinct expression patterns. At the organ level, BoGH3.13–1 is almost exclusively detected in floral buds by qRT-PCR, while the strongest expressions of BoGH3.13–3 and BoGH3.13–4 are observed in siliques (Fig. 4 a, c & d). In case of BoGH3.13–2, no significant expression preference is found among different organs and constitutively expressed in all parts of flowers (Figs. 4b, 5 c & f). In developing floral buds, BoGH3.13–1 is strongly expressed in stamen when floral buds are about 2 ~ 6 mm long (Fig. 5). However, investigation of BoGH3.13–1 promoter activity using GUS reporter revealed that BoGH3.13–1 is also expressed in abscission zones in siliques and leaf primordia, in addition to tapetal cells in stamen and pollen grains (Fig. 6f–l). Relatively weak detection of BoGH3.13–1 in siliques by qRT-PCR may be related to the facts that the gene is expressed only in a small portion of siliques cells, although we do not exclude the possibility that the expression level is also lower in siliques than in stamen. In 2 ~ 6 mm floral buds, in which BoGH3.13–1 is most strongly expressed, microspores, polarized microspores, and bicellular pollens are mainly observed in anthers (Fig. 7). Similar to BoGH3.13–1, two syntenic subgroup 6 Arabidopsis GH3 genes, AtGH3.13 and AtGH3.16, are expressed in flower stage 9 ~ 11 floral buds and flower stage 12, respectively [49]. More specifically, AtGH3.16 is expressed in polarized microspore and AtGH3.13 is expressed bicellular pollens. Based on the numbers of pollen nuclei and floral bud phenotypes [50], the flower stages, when BoGH3.13–1 is strongly expressed, roughly correspond to stages 8 ~ 12 of Arabidopsis flower and overlap with the periods when AtGH3.13 and AtGH3.16 are expressed (Fig. 7 & S2). Although BoGH3.5–1, a group II BoGH3 gene, is also specifically expressed in stamen like BoGH3.13–1 (Figs. 3a-b & 6u-x), BoGH3.5–1 seems to be expressed in a longer time period compared to BoGH3.13–1 (Figs. 5a - b, 6h & o). Different from BoGH3.13–1, neither in floral abscission zones nor in leaf primordia is expression of BoGH3.5–1 observed (Fig. 6 p & r). It needs to be determined which substrate(s) are preferentially used by BoGH3.13–1 and BoGH3.5–1.
BoGH3.13–1 is not induced by auxin (IAA or 2,4-D), JA, or GA, but expressed in a tissue-specific manner. Different from many GH3 genes in other plants, which have been found to be induced by various plant hormones [1, 17, 33, 51, 52], no expression changes for BoGH3.13–1 and 3 other subgroup 6 BoGH3 genes were detected in our experimental conditions (Fig. 3). In contrast, expression levels of BoGH3.2 was found to be elevated upon exposure to auxin in the same condition. Similar to our findings, all subgroup 6 GH3 genes in B. napus, an allotetraploid carrying chromosomes with B. oleracea origin, did not show any significant expression changes in response to IAA treatment in leaves [33]. Although BoGH3.13–1 expression is not induced by auxin in our experimental condition, tissues or cells, in which BoGH3.13–1 promoter activity is detected, largely overlap with the regions where auxin-responsive DR5 promoter is activated in Arabidopsis and rice (Figs. 3 & 6) [53–56]. We do not exclude the possibility that BoGH3.13–1 promoter is less sensitive to auxin treatment than BoGH3.2, but we prefer the idea that expression of BoGH3.13–1 is induced by a transcription factor that is activated in tissue-specific manners downstream of auxin signaling pathway. When expression patterns of BoGH3 genes were probed at the organ level using EMBL-EBI expression atlas (https://www.ebi.ac.uk/gxa/experiments/E-GEOD-42891/Results), BoGH3.13–1 was found to be specifically expressed in floral buds, similar to our qRT-PCR results (Fig. 4 & Table S3). However, expression levels of BoGH3.5–1 was found to be higher in silique than in floral bud, different from our results. Although differences in growth conditions and sampling times might have affected gene expressions, transcription profiling based on RNA-seq could have been confounded by sequence reads produced from highly homologous BoGH3 gene family members. Given that expressions of BoGH3.13–1 in leaf primordia and floral abscission zone could not be detected by transcription profiling, complete understanding of some BoGH3 expression patterns seem to require both qRT-PCR and investigation of promoter activity using promoter-reporter system.
Anther-specific expression of BoGH3.13–1 is directed by 62 bp DNA sequence, from − 340 to − 279 bp from the start codon. Determination of promoter regions important for tissue-specific expressions revealed that about 180 bp P3 region (− 340 ~ − 155) close to the transcription start site is sufficient for anther-specific expression (Fig. 8). The observation that P4 region (− 278 ~ − 155) does not supports anther-specific expression suggests that cis-acting element necessary for anther-specific expression is included by 62 bp DNA sequence from − 340 to − 279 bp. GUS expression detected in 5 out of 12 P5 transgenic lines containing − 418 to − 279 bp region further supported this idea. We suspect that deletion of promoter sequences (− 278 ~ − 155) close to the transcription start site makes anther-specific expression depend on the genomic positions where transgene is inserted. In Arabidopsis, Male Sterility 1, a plant homeodomain-finger, and MYB99 transcription factors functioning in anther and pollen development pathway are expressed in microspores, polarized microspores, and bicellular pollens [49, 57]. The findings (1) that BoGH3.13–1 is strongly expressed when microspores, polarized microspores, and bicellular pollens are produced and (2) that MYB core cis-acting element (CTGTTA) is located at − 293 ~ − 288 raises a possibility that Brassica oleracea var. oleracea ortholog of Arabidopsis MYB99 plays an important role for anther-specific expression of BoGH3.13–1 [58]. Because GUS expressions in leaf primordia and floral abscission zones are lost without any obvious effect on anther-specific expression, cis-acting element important for leaf primordia and floral abscission zone expressions must be located in the − 1489 to − 1017 region in BoGH3.13–1 promoter and independent of cis-acting element for anther-specific expression (Fig. 8a – f).
Conclusions
In this study, we identified 34 GH3 genes in Brassica oleracea var. oleracea, including four subgroup 6 GH3 genes, and a critical promoter region for anther-specific expression of a subgroup 6 BoGH3 gene, BoGH3.13–1. The information will broaden our understanding of transcriptional regulations during anther development and can be used to develop transgenic male sterile lines for economically important Brassica plants.
Methods
Plant growth
Brassica oleracea var. oleracea (TO1000 seeds, stock number CS29002) were obtained from the Arabidopsis Biological Resource Center. Brassica oleracea var. oleracea and Arabidopsis plants were grown on soil or a half-strength liquid Murashige and Skoog (MS) media (pH 5.7) with vitamins made with Duchefa Biochemie M0222 (Haarlem, Netherlands). Plants were grown under a 16 h (hr) light/8 h dark photoperiod at 22 C°. Organ samples of Brassica oleracea var. oleracea were collected from 50-day old soil-grown plants.
Transgenic Arabidopsis plants (ecotype Columbia) carrying β-glucuronidase (GUS)-coding sequences expressed by GH3 promoter sequences were selected on half-strength solid MS media containing 0.8% Duchefa Plant agar P1001 (Haarlem, Netherlands) and 20 μg/ml Kanamycin, and transferred to soil for flowering.
Identification of genes encoding putative GH3 family proteins in Brassica oleracea var. oleracea
To identify putative GH3-coding genes in Brassica oleracea var. oleracea, 19 Arabidopsis GH3 protein sequences downloaded from The Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/) were used for BLAST search in the Ensembl Plants database (http://plants.ensembl.org/index.html) and the National Centre for Biotechnology Information (NCBI, http://ncbi.nlm.nih.gov). In the Ensembl Plants and NCBI database search, E-value thresholds for candidates were set on 1e− 1 and 0.1, respectively. BoGH3 proteins were further determined by the presence of the intact GH3 domains, and their exon/intron structures were determined based on RNA-seq exon coverage and RNA-seq intron spanning reads from NCBI B. oleracea annotation Release 100. Similarly, GH3 protein sequences in B. oleracea var. capitata were identified using the sequence in Bolbase (http://ocri-genomics.org/bolbase/blast/blast.html) [59].
Multiple sequence alignment and construction of phylogenetic tree
The multiple sequence alignment of GH3 proteins was performed using Clustal Omega and visualized using Jalview [60, 61]. Phylogenetic analysis was performed using the molecular evolutionary genetics analysis (MEGA) software [62]. The evolutionary history was inferred by using maximum likelihood method based on the JTT matrix-based model [63]. All positions with less than 90% site coverage were eliminated. There were a total of 549 positions in the final dataset. The bootstrap test was repeated 1000 times. An orthologous relationship for synteny between B. oleracea var. oleracea and Arabidopsis was determined using gene information in the Ensembl database (https://plants.ensembl.org/Brassica_oleracea/Info/Index).
Hormone treatment
For hormone treatment, surface sterilized Brassica oleracea var. oleracea seeds were germinated and grown in 24 well plates containing 1 ml half-strength liquid MS media for 5 days. 2,4-D (D0901), IAA (I0901), GA (G0907) and JA (J0936) from Duchefa (Haarlem, Netherlands) were treated to whole seedlings, after the seedlings were further grown in 2 ml fresh liquid media for 6 h.
Sample collection
Five-day-old seedlings were used to determine whether the BoGH3 gene of interest is induced by hormone treatment. For gene expression analysis by qRT-PCR, root, leaf, stem, floral bud, open flower, and silique were obtained from 3 individual plants: more specifically, 11th to 13th leaves, fifth to seventh node for stems, a mix of unopened floral buds without white petals exposed (bud length less than about 8 mm), a mix of open flowers (bud length larger than 8 mm) with white petals exposed, and siliques with various sizes were collected. Samples for floral buds were further divided into 5 categories by lengths: 0 ~ 2, 2 ~ 4, 4 ~ 6, 6 ~ 8, and 8 ~ 10 mm sizes (Figure S3). Sepals, petals, anthers, and pistils were collected from 4 ~ 6 mm -long unopened floral buds. After collection, samples were frozen in liquid nitrogen and stored at − 80 C° until RNA isolation. Samples for GUS staining were collected when transgenic Arabidopsis seedlings were 8 days old, or later when inflorescence and siliques were mature enough.
RNA isolation, reverse transcription, and qRT-PCR analysis
Total RNA was extracted using PhileKorea E-Zol RNA Reagent (Seoul, Korea) or Ambion TRIzol® Reagent (Austin, USA) following the manufacturer’s instructions. For silique samples, Invitrogen Plant RNA Purification Reagent (Carlsbad, USA) was used to. cDNA was synthesized from RNA with 260/280 ratios between 1.8 and 2.1. First stand cDNA was synthesized with Toyobo ReverTra Ace -α (Osaka, Japan) and 1.0 μg of total RNA, according to the manufacturer’s instructions. In case of hormone-treated seedlings, 0.5 μg of total RNA was used. As described in Nam et al. (2019), qRT-PCR was performed with a two-step reaction: 3 min (min) at 95 °C, followed by 50 cycles of 10 s at 95 °C and 30 s at 60 °C. Primer sequences used are listed in Table S4. For each analysis, three technical replicates of at least two independent biological replicates were used.
Construction of GH3 promoter-GUS reporter vector and plant transformation
DNA regions upstream of the start codon of GH3 genes used for promoter analyses are as follows: − 1489 ~ − 1, − 1017 ~ − 1, − 500 ~ − 1, − 418 ~ − 1, − 340 ~ − 155, − 278 ~ − 155 bp of BoGH3.13–1, and − 1496 ~ − 1 bp of BoGH3.5–1. Putative promoter regions were PCR-amplified with specific primers with SalI or BamHI recognition sequence for cloning (Table S5). After SalI and BamHI digestion, the PCR fragments were cloned into pBI101.1 vector between SalI and BamHI sites. The construct was transformed into Arabidopsis by the floral dip method [64].
Histochemical GUS staining and paraffin section of GUS-stained samples
Histochemical GUS staining was performed with 0.5 mg/ml MBcell 5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid-cyclohexylammonium salt (Seoul, Republic of Korea), as previously described [65]. The floral buds of T1 or T2 transgenic plants carrying a GH3 promoter::GUS fusion transgene were immersed in GUS reaction buffer in the dark condition for 1 day at 37 °C, after which samples were washed in 95% ethanol for 1 ~ 2 h. At least 7 individual transgenic lines were used to analyze GUS expression patterns.
To perform the paraffin section, GUS-stained samples were fixed in FAA solution (Formaline: ethanol: glacial acetic acid: water = 10: 50: 5: 35) for at least 24 h and washed in water for 24 ~ 48 h. Then the samples were dehydrated in 50, 60, 70, 80, 90% ethanol series for 20 min once, and 100% ethanol for 20 min twice. The samples were incubated in a series of ethanol:xylene mix (75:25, 50:50, and 25:75) for 30 min in each mix, and to a series of xylene:paraffin mix (2:1, 1:1, and 1:2) for 1 h twice in each mix. The samples were incubated in molten paraffin for 24 h and poured into blocks on a slide warmer at 70 °C and cooled down to 25 °C. Eight μm-thick transverse sections of paraffin-embedded samples were made with a microtome. Ribbons of serial sections floated on warm water (50 °C) were transferred to slide glasses on the slide warmer at 70 °C and cooled down to 25 °C. Paraffin in the sections was removed with xylene.
DAPI staining of pollen grains
For DAPI staining, pollen in 0 ~ 2 mm, 2 ~ 4 mm, 4 ~ 6 mm, 6 ~ 8 mm, and 8 ~ 10 mm TO1000 floral buds were put on microscope slides and stained with several drops of DAPI-staining solution, as described [66]. The pollen nuclei were inspected under an Olympus BX51 fluorescence microscope (Tokyo, Japan) with a DAPI filter.
Supplementary Information
Additional file 1: Supplementary Table 1 Protein identifiers and genomic locations of kale-like type B. oleracea var. oleracea GH3 proteins identified in Ensembl Plants and NCBI database.
Additional file 2: Supplementary Table 2 GH3 proteins in B. oleracea var. oleracea and putative orthologs in B. oleracea var. capitata.
Additional file 3: Supplementary Table 3 Transcription profiling calculated from high throughput sequencing results.
Additional file 4: Supplementary Table 4 Sequences of qRT-PCR primers.
Additional file 5: Supplementary Table 5 Sequences of primers used to clone putative promoter regions of BoGH3.13–1.
Additional file 6: Supplementary Figure 1. Multiple sequence alignment of thirty-four B. oleracea var. oleracea and nineteen Arabidopsis GH3 proteins.
Additional file 7: Supplementary Figure 2. qRT-PCR results showing expression patterns of three subgroup 4 BoGH3 genes. qRT-PCR results showing expression patterns in different organs. Relative steady-state expression levels of BoGH3 genes were determined by qRT-PCR experiment with Actin control. Bar graphs show average relative expression values with SEs. The expression level of leaf was set to value 1 and used as reference to compare expression levels in different organs.
Additional file 8: Supplementary Figure 3. Morphology of B. oleracea var. oleracea floral buds used in this study. Upper panels show representative intact floral buds. Lower panels show representative anthers and pistils after sepals and petals were removed. Scale bar shown with fully opened flower is 1 cm.
Acknowledgements
The authors appreciate Yoonkang Hur, Jeong-Won Nam, Yeon Lee, Byugwook Kang, Jinouk Yeon, and Jaebeom Lim for their helpful discussions.
Abbreviations
- GH3
Gretchen Hagen 3
- bp
Base pairs
- qRT-PCR
Quantitative reverse transcription polymerase chain reaction
- GUS
β-glucuronidase
- DAPI
4′,6-diamidino-2-phenylindole
- 2,4-D
2,4-Dichlorophenoxy acetic acid
- IAA
Indole-3-acetic acid
- GA
Gibberellic acid 3
- JA
Jasmonic acid
Authors’ contributions
JJ and HY designed experiments and wrote manuscript. JJ, SP, and JI conducted experiments. All authors read and approved the final manuscript.
Funding
This work was supported by a grant (2017–1902-01) from Chungnam National University in Republic of Korea.
Availability of data and materials
The accession numbers of BoGH3 genes, which were retrieved from Ensembl Plants repository (http://plants.ensembl.org/index.html) and analyzed during the current study, are indicated in parenthesis after gene names: BoGH3.2 (Bo1g004760), BoGH3.3 (Bo8g100590), BoGH3.5–1 (Bo1g048130), BoGH3.5–2 (Bo7g111320), BoGH3.6–1 (Bo2g041710), BoGH3.6–2 (Bo3g022080), BoGH3.8–2 (Bo3g023700), BoGH3.8–3 (Bo1g008000), BoGH3.8–5 (Bo7g116230), BoGH3.10 (Bo9g007560), BoGH3.11–1 (Bo4g009300), BoGH3.11–2 (Bo3g039200), BoGH3.12–1 (Bo2g011190), BoGH3.12–3 (Bo9g167830), BoGH3.13–1 (Bo2g011210), BoGH3.13–2 (Bo3g009140), BoGH3.13–3 (Bo7g011450), BoGH3.13–4 (Bo9g166800), BoGH3.17–3 (Bo8g039460), BoGH3.18–1 (Bo4g164910), BoGH3.18–2 (Bo9g052150), BoGH3.18–3 (Bo9g117680), BoGH3.18–5 (Bo8g109440), BoGH3.18–6 (Bo8g109480), and BoGH3.18–7 (Bo8g109490). The accession numbers of BoGH3 proteins, which were retrieved from NCBI repository (https://www.ncbi.nlm.nih.gov/) and analyzed during the current study, are indicated in parenthesis after protein names: BoGH3.1 (XP_013608568.1), BoGH3.8–1 (XP_013619802.1), BoGH3.8–4 (XP_013596331.1), BoGH3.9 (XP_013632208.1), BoGH3.11–3 (XP_013632135.1), BoGH3.12–2 (XP_013623633.1), BoGH3.17–1 (XP_013583597.1), BoGH3.17–2 (XP_013594064.1), and BoGH3.18–4 (XP_013603489.1). Accession numbers of BoGH3 proteins with additional information are also found in Table S1.
RNA-seq data for transcription profiling shown in Table S3 was retrieved from GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42891).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiseong Jeong, Email: jseong1268@gmail.com.
Sunhee Park, Email: zhdzhd000@naver.com.
Jeong Hui Im, Email: shfkstor97@naver.com.
Hankuil Yi, Email: hankuil.yi@cnu.ac.kr.
References
- 1.Hagen G, Kleinschmidt A, Guilfoyle T. Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta. 1984;162:147–153. doi: 10.1007/BF00410211. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW. FIN219, an auxin regulated gene, defines a link between phytochrome a and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 2000;14:1958–1970. [PMC free article] [PubMed] [Google Scholar]
- 3.Ludwig-Müller J, Jülke S, Bierfreund NM, Decker EL, Reski R. Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol. 2009;181:323–338. doi: 10.1111/j.1469-8137.2008.02677.x. [DOI] [PubMed] [Google Scholar]
- 4.Okrent RA, Wildermuth MC. Evolutionary history of the GH3 family of acyl adenylases in rosids. Plant Mol Biol. 2011;76:489–505. doi: 10.1007/s11103-011-9776-y. [DOI] [PubMed] [Google Scholar]
- 5.Roux C, Perrot-Rechenmann C. Isolation by differential display and characterization of a tobacco auxin-responsive cDNA Nt-gh3, related to GH3.FEBS Lett. 1997;419:131-36. [DOI] [PubMed]
- 6.Yuan H, Zhao K, Lei H, Shen X, Liu Y, Liao X, Li T. Genome-wide analysis of the GH3 family in apple (Malus × domestica) BMC Genomics. 2013;14:297. doi: 10.1186/1471-2164-14-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Zhang L, Wang D, Ma H, Liu B, Shi Z, Ma X, Chen Y, Chen Q. Evolutionary history of the glycoside hydrolase 3 (GH3) family based on the sequenced genomes of 48 plants and identification of jasmonic acid-related GH3 proteins in Solanum tuberosum. Int J Mol Sci. 2018;19:1850. doi: 10.3390/ijms19071850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Westfall CS, Hicks LM, Wang S, Jez JM. Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J Biol Chem. 2010;285:29780–29786. doi: 10.1074/jbc.M110.146431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulick AM. Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staswick PE, Tiryaki I, Rowe ML. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell. 2002;14:1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Z, Guo Y, Novák O, Chen W, Ljung K, Noel JP, Chory J. Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants. 2016;2:16025. doi: 10.1038/nplants.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westfall CS, Zubieta C, Herrmann J, Kapp U, Nanao MH, Jez JM. Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science. 2012;336:1708–1711. doi: 10.1126/science.1221863. [DOI] [PubMed] [Google Scholar]
- 13.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torrens-Spence MP, Bobokalonova A, Carballo V, Glinkerman CM, Pluskal T, Shen A, Weng JK. PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. Mol Plant. 2019;12:1577–1586. doi: 10.1016/j.molp.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001;25:213–221. doi: 10.1046/j.1365-313x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- 16.Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, Shimada H, Manabe K, Matsui M. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004;37(4):471–483. doi: 10.1046/j.1365-313X.2003.01973.x. [DOI] [PubMed] [Google Scholar]
- 17.Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem. 2007;282:10036–10046. doi: 10.1074/jbc.M610524200. [DOI] [PubMed] [Google Scholar]
- 18.Damodaran S, Westfall CS, Kisely BA, Jez JM, Subramanian S. Nodule-enriched GRETCHEN HAGEN 3 enzymes have distinct substrate specificities and are important for proper soybean nodule development. Int J Mol Sci. 2017;18:E2547. doi: 10.3390/ijms18122547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou X, Long J, Zhao K, Peng A, Chen M, Long Q, He Y, Chen S. Overexpressing GH3.1 and GH3.1L reduces susceptibility to Xanthomonas citri subsp. citri by repressing auxin signaling in citrus (Citrus sinensis Osbeck) PLoS One. 2019;14:e0220017. doi: 10.1371/journal.pone.0220017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirungu JN, Magwanga RO, Lu P, Cai X, Zhou Z, Wang X, Peng R, Wang K, Liu F. Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton. BMC Genet. 2019;20:62. doi: 10.1186/s12863-019-0756-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gan Z, Fei L, Shan N, Fu Y, Chen J. Identification and expression analysis of Gretchen Hagen 3 (GH3) in Kiwifruit (Actinidia chinensis) during postharvest process. Plants (Basel) 2019;8:473. doi: 10.3390/plants8110473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okrent RA, Brooks MD, Wildermuth MC. Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J Biol Chem. 2009;284:9742–54. [DOI] [PMC free article] [PubMed]
- 23.Feng S, Yue R, Tao S, Yang Y, Zhang L, Xu M, Wang H, Shen C. Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J Integr Plant Biol. 2015;57:783–795. doi: 10.1111/jipb.12327. [DOI] [PubMed] [Google Scholar]
- 24.Jain M, Kaur N, Tyagi AK, Khurana JP. The auxin-responsive GH3 gene family in rice (Oryza sativa) Funct Integr Genomics. 2006;6:36–46. doi: 10.1007/s10142-005-0142-5. [DOI] [PubMed] [Google Scholar]
- 25.Kong W, Zhang Y, Deng X, Li S, Zhang C, Li Y. Comparative genomic and transcriptomic analysis suggests the evolutionary dynamic of GH3 genes in Gramineae crops. Front Plant Sci. 2019;10:1297. [DOI] [PMC free article] [PubMed]
- 26.Holland CK, Westfall CS, Schaffer JE, De Santiago A, Zubieta C, Alvarez S, Jez JM. Brassicaceae-specific Gretchen Hagen 3 acyl acid amido synthetases conjugate amino acids to chorismate, a precursor of aromatic amino acids and salicylic acid. J Biol Chem. 2019;294:16855–16864. doi: 10.1074/jbc.RA119.009949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peat TS, Böttcher C, Newman J, Lucent D, Cowieson N, Davies C. Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant Cell. 2012;24:4525–4538. doi: 10.1105/tpc.112.102921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherp AM, Westfall CS, Alvarez S, Jez JM. Arabidopsis thaliana GH3.15 acyl acid amido synthetase has a highly specific substrate preference for the auxin precursor indole-3-butyric acid. J Biol Chem. 2018;293:4277–4288. doi: 10.1074/jbc.RA118.002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westfall CS, Sherp AM, Zubieta C, Alvarez S, Schraft E, Marcellin R, Ramirez L, Jez JM. Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc Natl Acad Sci U S A. 2016;113(48):13917–13922. doi: 10.1073/pnas.1612635113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiu LW, Heckert MJ, You Y, Albanese N, Fenwick T, Siehl DL, Castle LA, Tao Y. Members of the GH3 family of proteins conjugate 2,4-D and dicamba with aspartate and glutamate. Plant Cell Physiol. 2018;59:2366–2380. doi: 10.1093/pcp/pcy160. [DOI] [PubMed] [Google Scholar]
- 32.Wang R, Li M, Wu X, Wang J. The gene structure and expression level changes of the GH3 gene family in Brassica napus relative to its diploid ancestors. Genes (Basel) 2019;10:58. doi: 10.3390/genes10010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei L, Yang B, Jian H, Zhang A, Liu R, Zhu Y, Ma J, Shi X, Wang R, Li J, Xu X. Genome-wide identification and characterization of Gretchen Hagen3 (GH3) family genes in Brassica napus. Genome. 2019;62:597–608. doi: 10.1139/gen-2018-0161. [DOI] [PubMed] [Google Scholar]
- 34.Parkin IA, Koh C, Tang H, Robinson SJ, Kagale S, Clarke WE, et al. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 2014;15:R77. doi: 10.1186/gb-2014-15-6-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldberg RB, Beals TP, Sanders PM. Anther development: basic principles and practical applications. Plant Cell. 1993;5:1217–1229. doi: 10.1105/tpc.5.10.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marciniak K, Przedniczek K. Comprehensive Insight into gibberellin- and jasmonate-mediated stamen development. Genes (Basel) 2019;10:811. doi: 10.3390/genes10100811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borg M, Brownfield L, Twell D. Male gametophyte development: a molecular perspective. J Exp Bot. 2009;60:1465–1478. doi: 10.1093/jxb/ern355. [DOI] [PubMed] [Google Scholar]
- 38.Scott RJ, Spielman M, Dickinson HG. Stamen structure and function. Plant Cell. 2004;16:S46–S60. doi: 10.1105/tpc.017012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto Y, Nishimura M, Hara-Nishimura I, Noguchi T. Behavior of vacuoles during microspore and pollen development in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:1192–1201. doi: 10.1093/pcp/pcg147. [DOI] [PubMed] [Google Scholar]
- 40.Williams JH, Taylor ML, O'Meara BC. Repeated evolution of tricellular (and bicellular) pollen. Am J Bot. 2014;101:559–571. doi: 10.3732/ajb.1300423. [DOI] [PubMed] [Google Scholar]
- 41.Acosta IF, Przybyl M. Jasmonate signaling during Arabidopsis stamen maturation. Plant Cell Physiol. 2019;60:2648–2659. doi: 10.1093/pcp/pcz201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubert R, Grunewald S, von Sivers L, Hause B. Effects of jasmonate on ethylene function during the development of tomato stamens. Plants (Basel) 2019;8:E277. doi: 10.3390/plants8080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao X, Tian L, Yang J, Zhao YN, Zhu YX, Dai X, Zhao Y, Yang ZN. Auxin production in diploid microsporocytes is necessary and sufficient for early stages of pollen development. PLoS Genet. 2018;14:e1007397. doi: 10.1371/journal.pgen.1007397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB. Different temporal and spatial gene expression patterns occur during anther development. Plant Cell. 1990;2:1201–1224. doi: 10.2307/3869340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol. 2005;56:393–434. doi: 10.1146/annurev.arplant.55.031903.141717. [DOI] [PubMed] [Google Scholar]
- 46.Twell D. Male gametogenesis and germline specification in flowering plants. Sex Plant Reprod. 2011;24:149–160. doi: 10.1007/s00497-010-0157-5. [DOI] [PubMed] [Google Scholar]
- 47.Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panchy N, Lehti-Shiu M, Shiu SH. Evolution of gene duplication in plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearce S, Ferguson A, King J, Wilson ZA. FlowerNet: a gene expression correlation network for anther and pollen development. Plant Physiol. 2015;167:1717–1730. doi: 10.1104/pp.114.253807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu Y, Lee PY, Truong MT, Beals TP, Goldberg RB. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod. 1999;11:297–322. doi: 10.1007/s004970050158. [DOI] [Google Scholar]
- 51.Yang Y, Yue R, Sun T, Zhang L, Chen W, Zeng H, Wang H, Shen C. Genome-wide identification, expression analysis of GH3 family genes in Medicago truncatula under stress-related hormones and Sinorhizobium meliloti infection. Appl Microbiol Biotechnol. 2015;99:841–854. doi: 10.1007/s00253-014-6311-5. [DOI] [PubMed] [Google Scholar]
- 52.Yu D, Qanmber G, Lu L, Wang L, Li J, Yang Z, Liu Z, Li Y, Chen Q, Mendu V, Li F, Yang Z. Genome-wide analysis of cotton GH3 subfamily II reveals functional divergence in fiber development, hormone response and plant architecture. BMC Plant Biol. 2018;18:350. doi: 10.1186/s12870-018-1545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Basu MM, González-Carranza ZH, Azam-Ali S, Tang S, Shahid AA, Roberts JA. The manipulation of auxin in the abscission zone cells of Arabidopsis flowers reveals that indoleacetic acid signaling is a prerequisite for organ shedding. Plant Physiol. 2013;162:96–106. doi: 10.1104/pp.113.216234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell. 2008;20:1760–1774. doi: 10.1105/tpc.107.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cecchetti V, Celebrin D, Napoli N, Ghelli R, Brunetti P, Costantino P, Cardarelli M. An auxin maximum in the middle layer controls stamen development and pollen maturation in Arabidopsis. New Phytol. 2017;213:1194–1207. doi: 10.1111/nph.14207. [DOI] [PubMed] [Google Scholar]
- 56.Yang J, Yuan Z, Meng Q, Huang G, Périn C, Bureau C, Meunier AC, Ingouff M, Bennett MJ, Liang W, Zhang D. Dynamic regulation of auxin response during rice development revealed by newly established hormone biosensor markers. Front Plant Sci. 2017;8:256. doi: 10.3389/fpls.2017.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gómez JF, Talle B, Wilson ZA. Anther and pollen development: a conserved developmental pathway. J Integr Plant Biol. 2015;57:876–891. doi: 10.1111/jipb.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chow C, Zheng H, Wu N, Chien C, Huang H, Lee T, Chiang-Hsieh Y, Hou P, Yang T, Chang W. PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2015;44:D1154–D1160. doi: 10.1093/nar/gkv1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu J, Zhao M, Wang X, Tong C, Huang S, Tehrim S, Liu Y, Hua W, Liu S. Bolbase: a comprehensive genomics database for Brassica oleracea. BMC Genomics. 2013;14:664. doi: 10.1186/1471-2164-14-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2-- a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X. Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–49. [DOI] [PMC free article] [PubMed]
- 63.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 64.Clough SJ, Bent AF. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 65.Hong J, Lee J, Jeong CW, Brooks JS, Choi Y, Lee JS. Characteristics and regulating role in thermotolerance of the heat shock transcription factor ZmHsf12 from Zea mays L. J Plant Biol. 209;62:329-41.
- 66.Dong X, Nou IS, Yi H, Hur Y. Suppression of ASKβ (AtSK32), a clade III Arabidopsis GSK3, leads to the pollen defect during late pollen development. Mol Cells. 2015;38:506–517. doi: 10.14348/molcells.2015.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1 Protein identifiers and genomic locations of kale-like type B. oleracea var. oleracea GH3 proteins identified in Ensembl Plants and NCBI database.
Additional file 2: Supplementary Table 2 GH3 proteins in B. oleracea var. oleracea and putative orthologs in B. oleracea var. capitata.
Additional file 3: Supplementary Table 3 Transcription profiling calculated from high throughput sequencing results.
Additional file 4: Supplementary Table 4 Sequences of qRT-PCR primers.
Additional file 5: Supplementary Table 5 Sequences of primers used to clone putative promoter regions of BoGH3.13–1.
Additional file 6: Supplementary Figure 1. Multiple sequence alignment of thirty-four B. oleracea var. oleracea and nineteen Arabidopsis GH3 proteins.
Additional file 7: Supplementary Figure 2. qRT-PCR results showing expression patterns of three subgroup 4 BoGH3 genes. qRT-PCR results showing expression patterns in different organs. Relative steady-state expression levels of BoGH3 genes were determined by qRT-PCR experiment with Actin control. Bar graphs show average relative expression values with SEs. The expression level of leaf was set to value 1 and used as reference to compare expression levels in different organs.
Additional file 8: Supplementary Figure 3. Morphology of B. oleracea var. oleracea floral buds used in this study. Upper panels show representative intact floral buds. Lower panels show representative anthers and pistils after sepals and petals were removed. Scale bar shown with fully opened flower is 1 cm.
Data Availability Statement
The accession numbers of BoGH3 genes, which were retrieved from Ensembl Plants repository (http://plants.ensembl.org/index.html) and analyzed during the current study, are indicated in parenthesis after gene names: BoGH3.2 (Bo1g004760), BoGH3.3 (Bo8g100590), BoGH3.5–1 (Bo1g048130), BoGH3.5–2 (Bo7g111320), BoGH3.6–1 (Bo2g041710), BoGH3.6–2 (Bo3g022080), BoGH3.8–2 (Bo3g023700), BoGH3.8–3 (Bo1g008000), BoGH3.8–5 (Bo7g116230), BoGH3.10 (Bo9g007560), BoGH3.11–1 (Bo4g009300), BoGH3.11–2 (Bo3g039200), BoGH3.12–1 (Bo2g011190), BoGH3.12–3 (Bo9g167830), BoGH3.13–1 (Bo2g011210), BoGH3.13–2 (Bo3g009140), BoGH3.13–3 (Bo7g011450), BoGH3.13–4 (Bo9g166800), BoGH3.17–3 (Bo8g039460), BoGH3.18–1 (Bo4g164910), BoGH3.18–2 (Bo9g052150), BoGH3.18–3 (Bo9g117680), BoGH3.18–5 (Bo8g109440), BoGH3.18–6 (Bo8g109480), and BoGH3.18–7 (Bo8g109490). The accession numbers of BoGH3 proteins, which were retrieved from NCBI repository (https://www.ncbi.nlm.nih.gov/) and analyzed during the current study, are indicated in parenthesis after protein names: BoGH3.1 (XP_013608568.1), BoGH3.8–1 (XP_013619802.1), BoGH3.8–4 (XP_013596331.1), BoGH3.9 (XP_013632208.1), BoGH3.11–3 (XP_013632135.1), BoGH3.12–2 (XP_013623633.1), BoGH3.17–1 (XP_013583597.1), BoGH3.17–2 (XP_013594064.1), and BoGH3.18–4 (XP_013603489.1). Accession numbers of BoGH3 proteins with additional information are also found in Table S1.
RNA-seq data for transcription profiling shown in Table S3 was retrieved from GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42891).