Abstract
Background
Enzyme assays have widespread applications in drug discovery from plants to natural products. The appropriate use of blanks in enzyme assays is important for assay baseline-correction, and the correction of false signals associated with background matrix interferences. However, the blank-correction procedures reported in published literature are highly inconsistent. We investigated the influence of using different types of blanks on the final calculated activity/inhibition results for three enzymes of significance in diabetes and obesity; α-glucosidase, α-amylase, and lipase. This is the first study to examine how different blank-correcting methods affect enzyme assay results. Although assays targeting the above enzymes are common in the literature, there is a scarcity of detailed published protocols. Therefore, we have provided comprehensive, step-by-step protocols for α-glucosidase-, α-amylase- and lipase-inhibition assays that can be performed in 96-well format in a simple, fast, and resource-efficient manner with clear instructions for blank-correction and calculation of results.
Results
In the three assays analysed here, using only a buffer blank underestimated the enzyme inhibitory potential of the test sample. In the absorbance-based α-glucosidase assay, enzyme inhibition was underestimated when a sample blank was omitted for the coloured plant extracts. Similarly, in the fluorescence-based α-amylase and lipase assays, enzyme inhibition was underestimated when a substrate blank was omitted. For all three assays, method six [Raw Data - (Substrate + Sample Blank)] enabled the correction of interferences due to the buffer, sample, and substrate without double-blanking, and eliminated the need to add substrate to each sample blank.
Conclusion
The choice of blanks and blank-correction methods contribute to the variability of assay results and the likelihood of underestimating the enzyme inhibitory potential of a test sample. This highlights the importance of standardising the use of blanks and the reporting of blank-correction procedures in published studies in order to ensure the accuracy and reproducibility of results, and avoid overlooked opportunities in drug discovery research due to inadvertent underestimation of enzyme inhibitory potential of test samples resulting from unsuitable blank-correction. Based on our assessments, we recommend method six [RD − (Su + SaB)] as a suitable method for blank-correction of raw data in enzyme assays.
Keywords: α-amylase, α-glucosidase, Blanks, Enzyme inhibition, Lipase, Methodology, Natural products, Plant extracts, Protocol, Standardisation
Background
Enzymes are the molecular targets of almost half of all marketed small-molecule drugs [1, 2]. Their protein structure affords a high level of druggability and target validation which makes enzymes an attractive target for novel drug discovery efforts [3]. Enzyme inhibitors form an important class of clinical drugs ranging in use from cancer [4, 5], cardiovascular disease [6, 7], diabetes [8, 9], neurological disorders [10–13] and obesity [14, 15]. Inhibitors of the endogenous carbohydrases α-glucosidase and α-amylase, reduces postprandial hyperglycaemia by delaying the digestion of dietary carbohydrates and are valuable therapeutics in the management of diabetes [8, 16]. Similarly, calorie restriction imposed by inhibition of carbohydrases and pancreatic lipase is useful for the prevention of weight gain and the treatment of obesity [14, 17]. Therefore, enzyme inhibition assays targeting α-glucosidase, α-amylase and lipase are widespread in research, and screening plant extracts and natural products for inhibitory activity against these enzymes is a common approach for the discovery of potential antidiabetic and antiobesity drugs to treat and manages these metabolic diseases [18–20].
Natural products are secondary metabolites produced by living organisms such as plants and microorganisms [21]. An abundant and easily accessible source of natural products is the kingdom of plants, a large proportion of which remains to be explored for potential bioactive metabolites [21]. These molecules possess high chemical and structural diversity unrivalled by synthetic compound libraries [22–24], have evolved intrinsic bioactive properties due to their evolutionary biological roles in living organisms [23–26], and are excluded from Lipinski’s rules of five [23, 24]. Therefore, natural products are an attractive source of therapeutic molecules and a significant body of research is devoted to the discovery of drugs from natural product extracts such as plant extracts [23, 24].
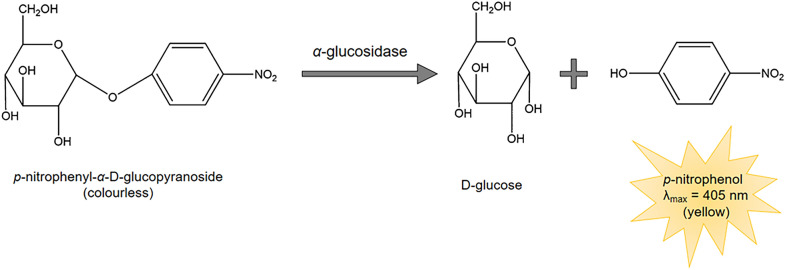
In-lab approaches for screening enzyme inhibitors from natural product extracts are based on spectroscopy and often determines enzyme activity using specially designed, labelled substrates which, upon enzymatic cleavage, produce a spectrometrically measurable signal either as colour (absorbance) or fluorescence of a defined wavelength [27–31]. For example, the chromogenic substrate p-nitrophenyl-α-d-glucopyranoside (pNPG) is widely used for the determination of α-glucosidase activity (Fig. 1). pNPG is a colourless molecule which contains a d-glucose residue linked to a p-nitrophenol moiety via a glycosidic bond. Hydrolysis of this glycosidic bond by α-glucosidase releases p-nitrophenol (coloured product), enabling the spectrophotometric determination of α-glucosidase activity at the absorbance maxima of pNPG; λ = 405 nm [27, 32].
Fig. 1.
Hydrolysis of colourless p-nitrophenyl-α-d-glucopyranoside to coloured p-nitrophenol by α-glucosidase
Adapted from [27]
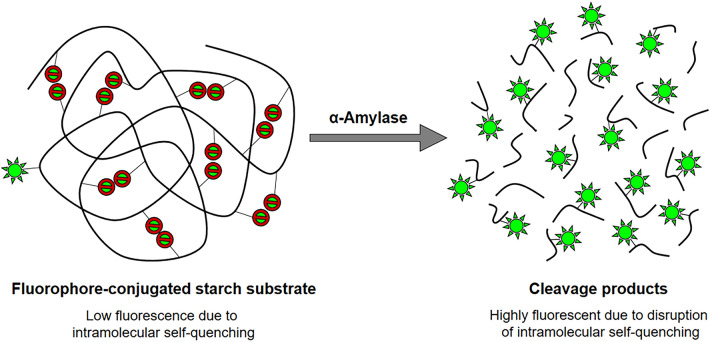
Alternatively, there are also fluorescence-based enzyme assays which make use of substrates linked to a fluorophore. Enzymatic cleavage releases the fluorophore which re-emits light (fluoresces) upon excitation at a specific wavelength [33–35]. A routinely used substrate for fluorescence-based α-amylase assays is the BODIPY® FL-DQ™ (boron-dipyrromethene fluorescent dye quenching) starch which consists of a starch derivative (DQ™ starch) conjugated to the green fluorescent BODIPY® FL fluorophore. The DQ™ starch is heavily labelled with BODIPY® FL to such an extent that the close proximity of the fluorophores to each other results in intramolecular self-quenching and the substrate fluorescence is almost completely quenched (Fig. 2). As the substrate is hydrolysed by α-amylase, the intramolecular self-quenching is disrupted causing an intense increase in green fluorescence [28, 36].
Fig. 2.
Schematic illustration of the BODIPY®FL-DQ™ starch assay concept
Adapted from [36]
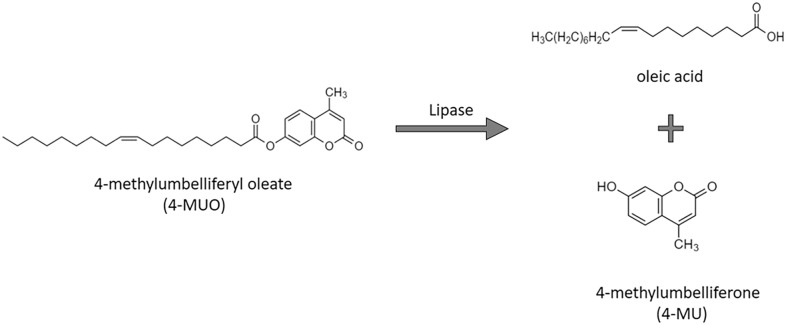
A commonly used fluorogenic substrate for lipase assays is 4-methylumbelliferyl oleate (4-MUO) [37]. Lipase-catalysed cleavage of 4-MUO liberates the fluorescent product, 4-methylumbelliferone (4-MU) (Fig. 3), in proportion to lipase activity [37–39].
Fig. 3.
Hydrolysis of 4-MUO by lipase
Modified from [39]
These enzyme assays can be performed in 96-well microplate readers [40–42], or in traditional cuvette-based spectrophotometers or spectrofluorometers [43–45]. However, miniaturisation of assays by adapting them to a 96-well format is beneficial as it reduces the volumes of reagents required, is cost-efficient and enables faster testing suitable for screening large libraries of plants, extracts or natural products [46].
Spectrometry-based assays are subject to spectral interference from sample colour, autofluorescence, and turbidity arising from poor solubility [47, 48]. Therefore, these factors must be taken into consideration when designing and carrying out enzyme-based assays with natural product extracts.
Plant extracts are often highly coloured as they contain natural, highly conjugated pigments. The colouration of plant extracts and natural products can cause interference with absorbance measurements in spectrophotometric assays [47]. Chlorophylls and carotenoids are the two major classes of photosynthetic plant pigments. Chlorophylls absorb light strongly in the 430–480 nm and 640–660 nm range [49]. However, chlorophylls are also capable of absorbing light at other wavelengths. Carotenoids encompass a wide range of compounds such as β-carotene, phytoene and lycopene, and have a wide visible light absorbance spectrum ranging from 400–530 nm [50]. Betalains, which includes betacyanins and betaxanthins [51]; and flavonoids such as anthocyanins and flavanols [52, 53] are the other classes of pigments contributing to colour in plants. All these phytochemical classes can interfere with assays which use wavelengths that overlap their absorption spectra.
In addition to the pigments intrinsic to plants, post-harvest changes can lead to the development of additional coloured products. For example, the action of polyphenol oxidases generates melanins and other brown pigments in harvested plant tissue exposed to oxygen. This process, referred to as enzymatic browning, is particularly common in fruits [54–56]. Samples with high sugar content, such as sugarcane (Saccharum spp.) extracts, are susceptible to intense browning caused by the Maillard reaction and caramelisation reactions [57]. The coloured products of such post-harvest reactions can be a significant source of interference in absorbance-based assays.
Autofluorescence is observed in some plants (e.g. Berberis vulgaris L., Humulus lupulus L., Matricaria chamomilla L., and Salvia officinalis L. [28]) and endogenous natural products [58–61] in a range of wavelengths which can interfere with fluorescence-based assays. For instance, anthranilates, alkaloids, coumarins, and stilbenes fluoresce in the blue-violet range (~ 400–520 nm), flavones and flavonoids in the green-yellow range (~ 520–590 nm), polycyclic aromatic quinones, tannins and some alkaloids in the orange range (~ 635–590 nm), and chlorophyll, porphyrins and certain quinones fluoresce in the red-far red range (~ 590–700 nm) [59, 60].
Poor solubility of some extracts and compounds in the assay buffers results in turbidity due to the presence of undissolved, suspended particles and may lead to inaccurate results. Light passing through a turbid medium is subject to multiple scattering and absorption events [62]. Therefore, turbidity interferes with spectrophotometric measurements by increasing absorbance and can result in misleadingly high readings [63]. Similarly, the absorbance and scattering of photons in a turbid medium can also distort fluorescence measurements [62].
The substrate can also be a source of error in enzyme assays. For example, unstable substrates may gradually decay to form their product. Contamination of the substrate with the chromogenic or fluorogenic product introduces a false signal and can cause a misleading increase in absorbance or fluorescence which is unrelated to enzyme activity [64].
In summary, assay interference due to sample colour, autofluorescence and turbidity can contribute to errors in measurements and hence affect the accuracy and reproducibility of results [47, 63]. Therefore, it is essential to minimise the effects of these interferences by blank-correcting raw data (RD) using appropriate sample and reagent blanks.
A sample blank contains an equal concentration of the test sample—whether it be an extract, an isolated compound, or a drug used as a control—without the enzyme or substrate. The absorbance (or fluorescence) of the sample blank quantifies the absorbance (or fluorescence) contributed by the colour, autofluorescence and/or turbidity of the sample. Subtracting the sample blank reading from the test well (which contains the enzyme + substrate + test sample) reading provides the value of the absorbance or fluorescence which is due to the enzymatic reaction; i.e. the ‘true’ value contributed by the reaction product.
The optical properties of different test samples vary widely. Therefore, to ensure the accuracy of results, it is necessary to include a sample blank for each sample in multi-sample assays, a “positive control blank” (equivalent to a sample blank for the positive control), and a “negative control blank” (equivalent to a sample blank for the negative control), and to correct each measurement using their respective blanks. Sample blanks have been included in some published studies [45, 65, 66], however, many appear to overlook the use of a sample blank in their experiments.
A practical approach to minimise assay interference from substrate contamination and degradation is to use a reagent (substrate) blank [63]. Some published studies [65, 67] have included a substrate blank to eliminate any false signals due to colour (absorbance) or fluorescence of the substrate. However, as with the sample blanks, many studies appear to omit the use of a substrate blank. Instead, many report using either a buffer blank or a blank that is not defined in the publications.
Although the use of blanks in experiments is common practice, there is currently no consensus on which blanks should be used. Published studies vary widely with regards to which blanks are included for the calculation of blank-corrected data. Researchers have previously reported assays using a buffer blank [68–70], sample blank [66, 71–74], a substrate blank [75, 76], or even an enzyme blank [77, 78]. Some researchers have combined substrate and sample in a single blank to account for interference from both [65, 79, 80]. Therefore, there appears to be many ways of blank-correction, with no standardisation, which not only adds to the confusion of researchers attempting these enzyme inhibition assays, it also adds bias and makes it difficult to compare results between studies. There are no studies that have investigated whether the type of blank used influences the calculated enzyme activity or enzyme inhibition results, which makes the selection of appropriate blanks problematic. In addition, despite the availability of a range of assay methods and countless publications involving α-glucosidase, α-amylase, and lipase inhibition assays, there is a lack of comprehensive published protocols that detail the assays step-by-step.
The aims of this study were to (1) provide comprehensive protocols for carrying out a colorimetric (absorbance) α-glucosidase inhibition assay, and fluorometric α-amylase and lipase inhibition assays in 96-well microplates; and (2) investigate whether using a buffer blank, substrate blank, sample blank, or a combination of blanks for blank-correction will affect the final, calculated enzyme inhibition results.
Materials and methods
Plant samples
Six medicinal plants with evidence of enzyme inhibitory activity against α-glucosidase (Aegle marmelos (L.) Corrêa [81, 82] and Phyllanthus niruri L. [83–86]), α-amylase (Gardenia jasminoides Ellis. [87] and Nelumbo nucifera Gaertn [87]), and lipase (Camellia sinensis L. [14, 88–90] and Sophora japonica L. [14]) were selected for the enzyme assays, based on the literature, to determine how their pigmentation and the use of different blanks and blanking methods affect the final assessment of their bioactivity. These medicinal plants have previously been investigated by our research group for their antidiabetic and antiobesity properties. They were chosen for initial testing based on a systematic review conducted on promising bioactive plants which highlighted the above species. These plants were selected for inclusion in this study because their ability to inhibit the respective enzymes have already been established in studies published by others [81–86, 88–90] and in previous studies carried out by our research group [14, 87].
Aegle marmelos was identified by local experts and dry leaves were collected in Kandy, Sri Lanka on 30/05/2017 by Dr Tien Huynh. Dried P. niruri leaf samples were obtained from a commercial herbal products supplier in Malaysia (identified and collected by the supplier in Jalan Jelebu, Malaysia on 20/02/2017). Herbarium samples can be found at the Janaki Ammal Herbarium, Indian Institute of Integrative Medicine (Accession No. 18696) for A. marmelos and the University of South Florida Herbarium, Institute for Systematic Botany (Accession No. 66964) for P. niruri.
Commercially prepared herbal extract granules (Nong’s, HK) of S. japonica flowers (Batch No. A1601428; 1203A1601428071119), N. nucifera leaves (Batch No. A1601450; 1356A1601450060919), G. jasminoides fruit (Batch No. A1600810; 1033A1600810050719) and C. sinensis (matcha) leaves were provided by GL Natural Healthcare Clinic, Strathmore, Victoria, Australia.
Positive controls
Acarbose, a known pharmacological inhibitor of α-glucosidase and pancreatic α-amylase was used as the positive control in the α-glucosidase and α-amylase assays [45, 67, 91]. Orlistat, a known inhibitor of pancreatic lipase was used as the positive control in the lipase assay [92, 93]. Acarbose (Cat. No. ACR459080010), and orlistat (Cat. No. O4139) were purchased from Thermo Fisher Scientific Australia and Sigma-Aldrich, respectively.
Chemicals and reagents
Analytical grade ethanol (Cat. No. 111727), and dimethyl sulfoxide (DMSO) (Cat. No. 102952) were purchased from Merck Millipore Australia. The EnzChek™ Ultra Amylase assay kit (Cat. No. E33651) was purchased from Life Technologies Australia, α-amylase from porcine pancreas (EC Number 3.2.1.1, Cat. No. 10102814001), lipase from porcine pancreas (Type VI-S, EC Number 3.1.1.3, Cat. No. L0382), 4-methylumbelliferyl oleate (4-MUO) (Cat. No.75164), Tris base (Cat. No. 10708976001), intestinal acetone powders from rat (Cat. No. I1630), calcium chloride (CaCl2), and sodium chloride (NaCl) were purchased from Sigma-Aldrich Australia. p-nitrophenyl α-d-glucopyranoside (pNPG) (Cat. No. ACR337150050), and phosphate-buffered saline (PBS) tablets (Gibco) (Cat. No. 18912014), were purchased from Thermo Fisher Scientific Australia. Anhydrous disodium hydrogen phosphate (Na2HPO4, AnalaR®), and sodium dihydrogen phosphate dihydrate monobasic (NaH2PO4.H2O, UNIVAR®) were obtained from AJAX Chemicals Australia and BDH Chemicals Australia, respectively. Milli-Q® water was obtained from a Millipore Milli-Q® water purification system. Distilled water was used in the preparation of buffers and reagents unless otherwise stated.
Corning® Costar® tissue culture-treated clear, flat bottom 96-well plates (Cat. No. CLS3516) were purchased from Sigma-Aldrich Australia. Nunc™ Nunclon™ delta treated, black polystyrene, flat bottom 96-well plates (Cat. No. 137101) were purchased from Thermo Fisher Scientific Australia.
Methods
An absorbance assay with pNPG as the substrate was used for the determination of α-glucosidase activity, and fluorescence assays using the substrates BODIPY®FL-DQ™ starch, and 4-MUO were used for the determination of α-amylase and lipase activity, respectively. Acarbose was the positive control in the α-glucosidase and α-amylase assays while orlistat was the positive control in the lipase assay. The corresponding assay buffer was used as the negative control in each of the assays.
Absorbance assay: α-glucosidase
The α-glucosidase assay was adapted from [91, 93] with modifications. The method is described in detail in the proceeding section “α-glucosidase assay step-by-step.”
Extraction of plant samples
The leaves of Aegle marmelos and Phyllanthus niruri were separated from the dried plant samples, homogenized to a fine powder using a mortar and pestle or an electric blender (KitchenAid®), and the homogenized samples were stored in the dark at room temperature (RT = 25 °C) with silica gel desiccant.
Approximately 50 mg of the powdered leaf samples were further homogenized with 500 µL of 100% ethanol in lysing tubes using an MP Biomedicals™ FastPrep-24™ instrument. Each sample was then extracted for 15 min at 37 °C and 900 rpm using an Eppendorf ThermoMixer®, and then centrifuged for 15 min at 12,700 rpm at RT. The supernatant (ethanol extract) was subsequently transferred into a new Eppendorf tube. The pellet was re-suspended in 500 µL of Milli-Q water and allowed to extract for 15 min at 37 °C and 900 rpm using an Eppendorf ThermoMixer®. The supernatant (water extract) was then combined with the ethanol extract that was previously generated. The combined ethanol/water extract (50:50) was centrifuged for 10 min at 12,700 rpm at RT until a clear supernatant was obtained. The extracts were concentrated by evaporating solvents under reduced pressure using a rotational vacuum concentrator (Martin Christ RVC 2-33 CDplus) operating at 1500 rpm at 30 °C for 6–8 h. The average yields of the concentrated extracts were 0.3 mg for A. marmelos and 0.8 mg for P. niruri. The concentrated extracts were stored in darkness at RT with silica gel desiccant until required.
Preparation of buffers, reagent solutions, test samples and controls
α-glucosidase assay buffer I (for preparing extracts and acarbose) consisted of freshly prepared 100 mM sodium phosphate buffer with 2% DMSO (pH 6.9). All test extracts and acarbose (positive control) were prepared by dissolving in assay buffer I.
Comments: DMSO was used to aid in the solubility of the extracts in the buffer [94, 95]. Preliminary testing confirmed that 2% DMSO (final concentration of 0.001% DMSO) had no significant effect on rat intestinal α-glucosidase activity. It is also possible to use PBS (pH adjusted to 6.9) as the incubation buffer in this assay without affecting rat intestinal α-glucosidase activity.
α-glucosidase assay buffer II (for preparing assay reagents) consisted of a freshly prepared 100 mM sodium phosphate buffer without DMSO (pH 6.9). The enzyme and substrate solutions were prepared by dissolution in assay buffer II.
Test samples were prepared from the concentrated plant extracts by dissolving and diluting the extracts in α-glucosidase assay buffer I to obtain working test sample solutions of 1 mg/mL. These were freshly prepared immediately before experiments and used on the same day.
Positive control, acarbose powder was dissolved in α-glucosidase assay buffer I to prepare a 100 mg/mL stock solution which was stored short-term at 4 °C. A working solution of 1 mg/mL acarbose was prepared by diluting the stock solution in α-glucosidase assay buffer I immediately before experiments.
Negative control (vehicle) was α-glucosidase assay buffer I.
-
Rat intestinal α-glucosidase enzyme solution was prepared by sonicating 0.625 g of rat intestinal acetone powder in 50 mL of assay buffer II at RT, followed by vigorous mixing at 800 rpm for 20 min at RT. The mixture was centrifuged at 14,000 rpm for 30 min at 4 °C (Beckman Coulter Avanti J-25i high-speed centrifuge), and the supernatant (working enzyme solution) was stored in aliquots in the dark at 4 °C until use. The working α-glucosidase solution can be refrigerated for storage for up to 3 months without any appreciable decline in enzyme activity. The working enzyme solution cannot be frozen as freezing inactivates the enzyme.
Comments: Note that the concentration of α-glucosidase in the enzyme solution is unknown. However, different volumes of the enzyme solution (10, 20 and 30 µL/well) were tested as part of the assay optimisation and 30 µL/well was optimal for the assay method described herein.
p-NPG substrate solution was prepared by dissolving and diluting p-NPG powder in assay buffer II to obtain a working substrate solution of 5 mM.
α-glucosidase assay step-by-step
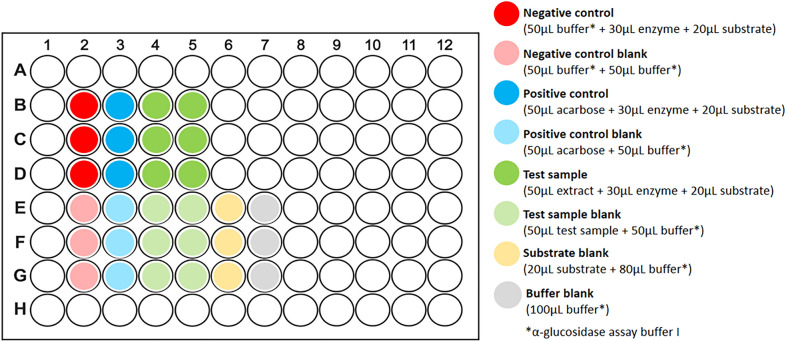
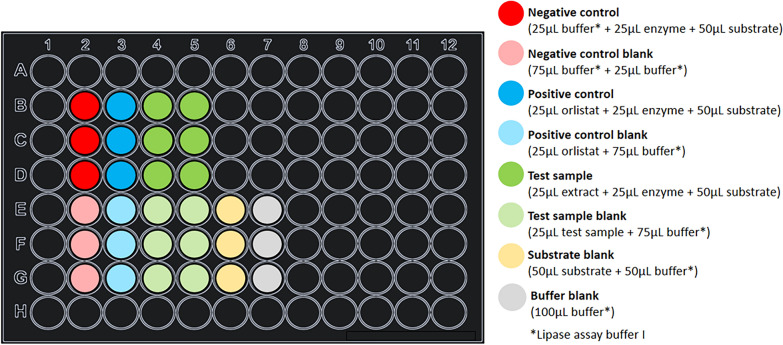
The absorbance-based α-glucosidase assay was performed in Corning® Costar® tissue culture-treated clear, flat-bottom 96-well plates. The assay method is described below including volumes of all test samples, controls, and assay reagents and a suggested plate layout is given in Fig. 4.
Fig. 4.
Suggested plate layout for the α-glucosidase assay
Step 1: Set up 96-well plate with test samples, controls, and blanks.
Firstly, assay buffer I was added to the wells for the negative control (50 µL/well) and the various blanks. Then the test samples and acarbose (50 µL/well of 1 mg/mL, final concentration 0.5 mg/mL) were added to their respective test wells and corresponding blank wells.
Comments: Assay buffer I was added to the sample blank wells and control blank wells to make their volumes and concentrations equal to the volumes and concentrations in the test wells. It was not necessary to add the substrate to the substrate blanks in Step 1 because, unlike the samples and controls which were all added at the start of the assay, the substrate was only added in Step 3 after pre-incubation. Temperature variation between outer and inner wells and increased evaporation in the outermost wells can give rise to edge effects which contribute to the degradation of assay results [96, 97]. To avoid this, only the central wells were used in the assay and the peripheral wells were excluded.
Step 2: Add enzyme and pre-incubate with test samples.
Using a multichannel pipette, the enzyme solution (30 µL/well) was added to all test wells and mixed by gentle pipetting. The plate was incubated in the dark at 37 °C for 10 min.
Comments: ‘Test wells’ refers to the wells containing enzyme + substrate and a test sample or control. Care must be taken not to create air bubbles and avoid foaming during mixing as aeration can denature the enzyme [98, 99]. In addition, entrapped air bubbles refract light and can therefore cause interference with absorbance measurements [47]. A temperature of 37 °C was chosen for all incubation steps in the α-glucosidase assay, as being close to body temperature, it is physiologically relevant and is optimal for mammalian enzyme activity [100].
Step 3: Add substrate to start reaction and incubate plate.
Using a multi-channel pipette, the substrate solution (20 µL/well of 5 mM, final concentration 1 mM) was added to all test wells and the substrate blank wells and mixed by gentle pipetting. The plate was incubated in the dark at 37 °C for 20 min.
Comments: Substrate was not added to the sample blanks, control blanks, or the buffer blanks. The plate was incubated in the microplate reader CLARIOstar® microplate reader (BMG LABTECH) set at 37 °C.
Step 4: Measure absorbance using plate reader.
Absorbance was measured at 405 nm using the microplate reader set to 37 °C. The plate was read from the top.
Fluorescence assays: α-amylase and lipase
Preparation of herbal extract granules for testing
To prepare the samples for testing, the herbal extract granules of Gardenia jasminoides and Nelumbo nucifera (for the α-amylase assay), and the granules of Camellia sinensis and Sophora japonica (for the lipase assay) were ground to a fine powder using a mortar and pestle, and 20 mg of the ground granules were dissolved in 4 mL of distilled water containing 2% DMSO to obtain 5 mg/mL stock solutions. These were vortexed for 5 min and sonicated for 10 min to aid solubilisation, and then filtered through a Millex-HP 0.45 µm filter (Millipore). The test sample stock solutions thus prepared were diluted in assay buffer I to prepare working solutions as described below.
α-amylase assay
Preparation of buffers, reagent solutions, test samples and controls
-
α-amylase assay buffer I (for diluting test samples and preparing the positive control acarbose) consisted of 10 mM sodium phosphate, 2.68 mM KCl, 140 mM NaCl and 1 mM CaCl2 (pH 6.9) and 2% DMSO.
Comments: DMSO was used to aid the solubility of the extracts in the buffer [94, 95]. Preliminary testing confirmed that 2% DMSO (final concentration of 0.001% DMSO) had no significant effect on α-amylase activity in this assay.
α-amylase assay buffer II (for diluting substrate and enzyme) consisted of 10 mM sodium phosphate, 2.68 mM KCl, 140 mM NaCl and 1 mM CaCl2 (pH 6.9).
Substrate buffer (solvent to dissolve substrate DQ ™ starch) provided with the EnzChek™ Ultra Amylase assay kit, consisted of 50 mM sodium acetate buffer, pH 4.0
Test samples (20 mg of herbal extract granules) were dissolved in distilled water with 2% DMSO (4 mL) to prepare a stock solution 5 mg/mL and diluted in α-amylase assay buffer I to obtain a working solution of 1.2 mg/mL (final concentration in well = 0.3 mg/mL). Samples were freshly prepared before experiments and used immediately.
Positive control acarbose (20 mg) was dissolved in α-amylase assay buffer I (4 mL) to obtain a 5 mg/mL stock solution and diluted to obtain a solution of 1.2 mg/mL (final concentration of 0.3 mg/mL) immediately before experiments.
Negative control (vehicle) was α-amylase assay buffer I.
Porcine pancreatic α-amylase (107 mU/mL) was serially diluted in α-amylase assay buffer II to obtain a working enzyme solution of 48 mU/mL.
BODIPY®FL-DQ™ starch substrate was prepared by dissolving first in substrate buffer, then in α-amylase assay buffer II to obtain a stock solution of 1 mg/mL as per the manufacturer’s instructions. A working substrate solution of 200 µg/mL was prepared by serial dilution.
α-amylase assay step-by-step
A fluorescence assay for determining porcine pancreatic α-amylase activity using DQ™ starch as a substrate was performed in Nunc™ Nunclon™ delta treated, black polystyrene, flat-bottom 96-well plates using a method adapted from [45].
Step 1: Set up 96-well plate with test samples, controls, and blanks.
Firstly, α-amylase assay buffer I was added to the negative control wells (25 µL/well) and the various blanks. Then the test samples and acarbose (25 µL/well of 1.2 mg/mL, final concentration 0.3 mg/mL) were added to their respective test wells and their corresponding sample blank or positive control blank wells.
Comments: Assay buffer I was added to the sample blank wells and control blank wells to make their volumes and concentrations equal to the volumes and concentrations of the test wells. Assay buffer I was also the negative control (vehicle). It was not necessary to add substrate to the substrate blanks in Step 1 because, unlike the samples and controls which were all added at the start of the assay, the substrate was only introduced in Step 3 after pre-incubation. Temperature variation between outer and inner wells and increased evaporation in the outermost wells can give rise to edge effects which contribute to the degradation of assay results [96, 97]. To avoid this, only the central wells were used in the assay and the peripheral wells were excluded.
Step 2: Add enzyme and pre-incubate with test samples.
Using a multichannel pipette, the amylase solution (25 µL/well of 48 mU/mL, final concentration 12 mU/mL) was added to all test wells and mixed by gentle pipetting. Assay buffer I (25 µL/well) was added to all test wells to bring up the volume to 100 µL. The plate was incubated in darkness at RT for 20 min.
Comment: ‘Test wells’ refer to the wells containing enzyme + substrate and a test sample or control. Note that the well volumes were adjusted to 100 µL with the addition of buffer to maintain the total reaction mixture volumes consistent across all three assays, for convenience and calculations, and to minimise the effects of evaporation during incubation. This is not strictly necessary and adjusting the volumes (and concentrations) of enzyme, substrate, and test substances to a total of 100 µL is suggested.
Step 3: Add substrate to start the reaction and incubate plate.
Using a multichannel pipette, the substrate solution (25 µL of 200 µg/mL, final concentration 50 µg/mL) was added to all test wells and the substrate blank wells and mixed thoroughly by gentle pipetting. Next, the plate was incubated in the dark at RT for 20 min.
Comments: Substrate was not added to the sample blanks, control blanks, or the buffer blanks.
Step 4: Measure fluorescence using a plate reader.
Fluorescence was measured with the CLARIOstar® microplate reader (BMG LABTECH) at excitation and emission wavelengths of 485 nm and 530 nm, respectively, over 30 min.
Lipase assay
Preparation of buffers, reagent solutions, test samples and controls
-
Lipase assay buffer I (for diluting test samples and preparing the positive control orlistat, and the substrate) consisted of 13 mM Tris-HCl, 75 mM NaCl, 1.3 mM CaCl2 (pH 8.0) and 2% DMSO.
Comments: DMSO was used to aid with the solubility of the extracts in the buffer [94, 95]. Preliminary testing confirmed that 2% DMSO (final concentration of 0.001% DMSO) had no significant effect on lipase activity in this assay.
Lipase assay buffer II (for preparing porcine pancreatic lipase enzyme) consisted of 13 mM Tris-HCl, 75 mM NaCl and 1.3 mM CaCl2 (pH 8.0).
Test samples (20 mg herbal extract granules) were dissolved in 4 mL distilled water containing 2% DMSO to prepare stock solutions of 5 mg/mL and diluted in lipase assay buffer I to 0.4 mg/mL (final concentration of 0.1 mg/mL). Samples were freshly prepared before experiments and used immediately.
Positive control orlistat (25 mg) was dissolved in lipase assay buffer I (5 mL) to obtain a 5 mg/mL stock solution and diluted in lipase assay buffer II to 0.4 mg/mL (final concentration of 0.1 mg/mL) immediately before experiments.
Negative control (vehicle) was the lipase assay buffer I.
Porcine pancreatic lipase enzyme powder was dissolved in lipase assay buffer II to obtain a stock solution 3.5 × 104 U/mL. Subsequently, the enzyme solution was serially diluted to obtain a working solution of 50 U/mL.
4-MUO substrate solution was prepared by dissolving in lipase assay buffer I to obtain a working substrate solution of 0.1 mM.
Lipase assay step-by-step
The assay was adapted from methods previously described [71, 101]. The volumes of test samples, controls, and assay buffer are illustrated below using an example plate layout (Fig. 5).
Fig. 5.
Suggested plate layout for lipase inhibition assay
Step 1: Set up 96-well plate with test samples, controls, and blanks.
Firstly, lipase assay buffer I was added to the negative control wells (25 µL/well) and the various blanks. Then the test samples and orlistat (25 µL/well of 0.4 mg/mL, final concentration 0.1 mg/mL) were added to their respective test wells and their corresponding sample blanks or positive control blanks.
Comments: Lipase assay buffer I was added to the sample blank wells and control blank wells to make their volumes and concentrations equal to the volumes and concentrations of the test wells. It was not necessary to add the substrate to the substrate blanks in Step 1 because, unlike the samples and controls which were all added at the start of the assay, the substrate was only introduced in Step 3 following pre-incubation. Temperature variation between outer and inner wells and increased evaporation in the outermost wells can give rise to edge effects which contribute to the degradation of assay results [96, 97]. To avoid this, only the central wells were used in the assay and the peripheral wells were excluded.
Step 2: Add enzyme and pre-incubate with test samples.
Using a multi-channel pipette, lipase solution (25 µL/well of 50 mU/mL, final concentration 12.5 mU/mL) was added to all test wells and mixed by gentle pipetting. Assay buffer I (25 µL/well) was added to all test wells to bring the volume up to 100 µL. The plate was incubated in darkness at RT for 5 min.
Comment: ‘Test wells’ refer to the wells containing enzyme + substrate and a test sample or control.
Step 3: Add substrate to start the reaction and incubate plate.
Using a multi-channel pipette, the substrate solution (50 µL of 200 µg/mL, final concentration 50 µg/mL) was added to all test wells and the substrate blanks and mixed thoroughly by gentle pipetting. Next, the plate was incubated in the dark at RT for 30 min.
Comments: Substrate was not added to the sample blanks, control blanks, or the buffer blanks.
Step 4: Measure fluorescence using plate reader.
Fluorescence was measured with the CLARIOstar® microplate reader (BMG LABTECH) at excitation and emission wavelengths of 355 nm and 460 nm, respectively.
Comments: The lipase substrate 4-MUO had poor solubility in aqueous-based buffer. DMSO increased solubility, however, there was still some turbidity which can interfere with fluorescence measurements [62]. A possible solution would be to use a stopping reagent, centrifuge the plate, and transfer the supernatant to a new plate before fluorescence measurement.
Calculation of results
Blank-correction and calculation of enzyme activity
For all three assays, enzyme activity was calculated using blank-corrected raw data. Enzyme activity was expressed as a percentage of the negative control which was normalised to 100% activity.
For comparison, raw data was blank-corrected in six different ways (Table 1) using a combination of buffer-only, substrate, and sample blanks:
Raw data corrected by subtracting only the buffer-only blank: RD–BB.
Raw data corrected using only the substrate blank: RD − SuB.
Raw data corrected using dedicated sample blanks for each sample, a positive control blank, and a negative control blank (“test substance blanks” for the samples and controls): RD − SaB.
Raw data corrected using both the buffer-only blank, and individual sample or control blanks: RD − (BB + SaB).
Raw data corrected using both the substrate blank, and individual sample or control blanks: RD − (SuB + SaB).
Raw data corrected with the difference in absorbance or fluorescence between the substrate blank and the buffer-only blank, and the individual sample and control blanks: RD − [(SuB − BB) + SaB].
Table 1.
Formulas for blank-correction of raw absorbance data
| Method | Formula for blank-correction | |
|---|---|---|
| α-glucosidase assay | α-amylase and lipase assays | |
| 1 | ||
| 2 | ||
| 3 | ||
| 4 | ||
| 5 | ||
| 6 | ||
A and F refers to absorbance and fluorescence respectively
Atest = the absorbance of treatment wells (enzyme + substrate + plant extract), positive control wells (enzyme + substrate + positive control acarbose) and negative control wells (enzyme + substrate + vehicle)
Abuffer blank = the absorbance of the buffer-only blank used to determine the absorbance due only to the buffer
Asubstrate blank = the absorbance of the substrate blank (substrate + buffer)
Asample blank = the absorbance of the sample blank, positive control blank, or negative control blank
Ftest = the fluorescence of treatment wells (enzyme + substrate + plant extract), positive control wells (enzyme + substrate + positive control acarbose) and negative control wells (enzyme + substrate + vehicle)
Fbuffer blank = the fluorescence of the buffer-only blank used to determine the absorbance due only to the buffer
Fsubstrate blank = the fluorescence of the substrate blank (substrate + buffer)
Fsample blank = the fluorescence of the sample blank, positive control blank, or negative control blank (Please see note below)
Note on sample blanks: In this study, each “test sample” (i.e. extracts, positive control (acarbose or orlistat), and negative control (vehicle) were given their own sample blank. The sample blank for the positive control, which may also be referred to as the “positive control blank” contained buffer + positive control. The sample blank for the negative control, which may also be referred to as the “negative control blank” contained buffer + negative control (vehicle). In methods 3–6, the positive and negative controls were blank-corrected using their respective positive control “sample” blank and negative control “sample” blank
The α-glucosidase activity and inhibition were calculated from the blank-corrected absorbance data as a percentage of the negative (uninhibited) control using the following formula:
The α-amylase and lipase activity and inhibition were calculated from the blank-corrected fluorescence data as a percentage of the negative (uninhibited) control using the following formula:
Statistical analyses
One-way ANOVA and Tukey’s multiple comparison test were applied to the data to establish statistical significance of differences in results between different treatment groups (i.e. negative control, positive control, and the different extracts) as well as between the different calculation methods within the same group.
Results and discussion
In all three assays, the positive controls—acarbose for α-glucosidase (Fig. 6) and α-amylase (Fig. 7), and orlistat for lipase (Fig. 8)—caused a significant reduction in enzyme activity when compared with the uninhibited control. This is as expected and provided validation of the assay methods and confirmed the suitability of the assays for their intended purpose [102].
Fig. 6.
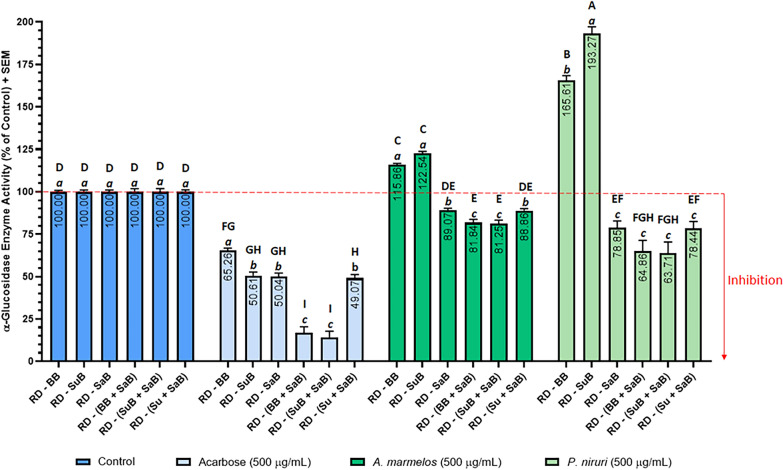
Effect of acarbose and plant extracts on rat intestinal α-glucosidase activity. Acarbose (positive control), and the Aegle marmelos and Phyllanthus niruri leaf extracts were tested at 0.5 mg/mL. The negative control (uninhibited control) was normalised to 100% activity. Data represent the mean + SEM of triplicate readings (n =3). Bars that do not share a letter were significantly different from each other as determined by one-way ANOVA and Tukey’s multiple comparison test (p < 0.05). Uppercase letters denote significance between treatment groups and lowercase letters denote significance within groups. RD: raw data, BB: buffer blank (contained buffer only), SuB: substrate blank (contained substrate + buffer), and SaB: sample blank. Sample blank composition: A. marmelos sample blank = A. marmelos extract + buffer, P. niruri sample blank = P. niruri extract + buffer, acarbose sample blank (positive control blank) = acarbose + buffer, and negative control sample blank (negative control blank) = vehicle + buffer (as the vehicle was buffer, this contained buffer only)
Fig. 7.
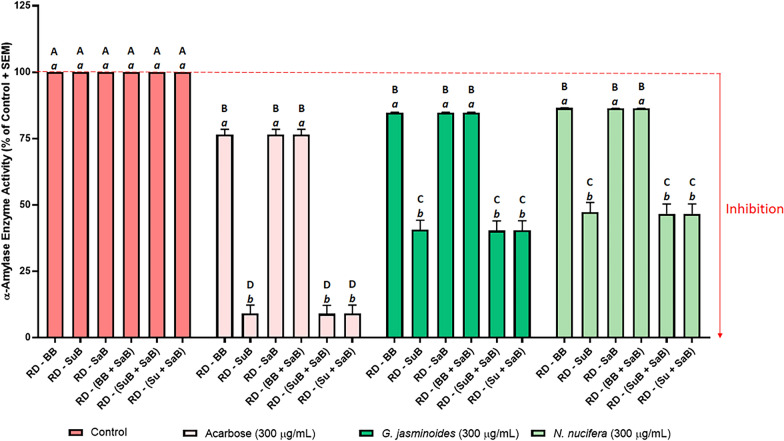
Effect of acarbose and plant extracts on porcine pancreatic α-amylase activity. Acarbose (positive control), and the Gardenia jasminoides and Nelumbo nucifera extracts were tested at 0.3 mg/mL. The negative control (uninhibited control) was normalised to 100% activity. Data represent the mean + SEM of readings from three independent experiments. Bars that do not share a letter were significantly different from each other as determined by one-way ANOVA and Tukey’s multiple comparison test (p < 0.05). Uppercase letters denotes significance between treatment groups and lowercase letters denote significance within groups. RD: raw data, BB: buffer blank (contained buffer only), SuB: substrate blank (contained substrate + buffer), and SaB: sample blank. Sample blank composition: G. jasminoides sample blank = G. jasminoides extract + buffer, N. nucifera sample blank = N. nucifera extract + buffer, acarbose sample blank (positive control blank) = acarbose + buffer, and control sample blank (negative control blank) = vehicle + buffer (as the vehicle was buffer, this contained buffer only)
Fig. 8.
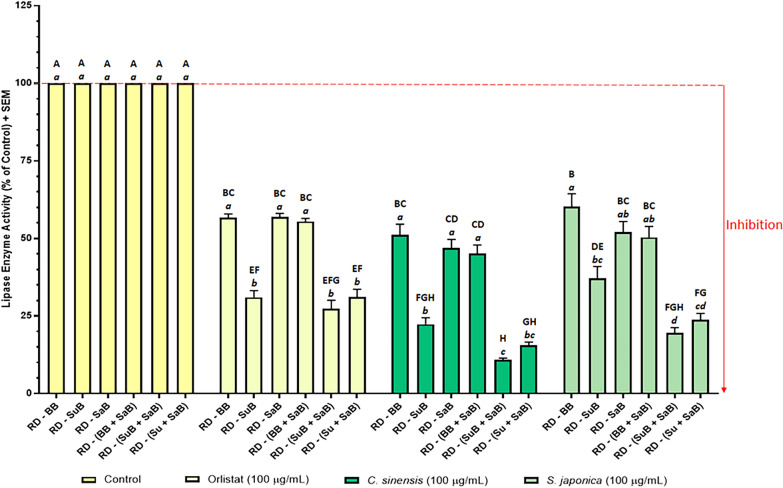
Effect of orlistat and plant extracts on porcine pancreatic lipase activity. Orlistat (positive control), and the Camellia sinensis and Sophora japonica extracts were tested at 0.1 mg/mL. The negative control (uninhibited control) was normalised to 100% activity. Data represent the mean + SEM of readings from three independent experiments. Bars that do not share a letter were significantly different from each other as determined by one-way ANOVA and Tukey’s multiple comparison test (p < 0.05). Uppercase letters denote significance between treatment groups and lowercase letters denotes significance within groups. RD: raw data, BB: buffer blank (contained buffer only), SuB: substrate blank (contained substrate + buffer), and SaB: sample blank. Sample blank composition: C. sinensis sample blank = C. sinensis extract + buffer, S. japonica sample blank = S. japonica extract + buffer, orlistat sample blank (positive control blank) = orlistat + buffer, and control sample blank (negative control blank) = vehicle + buffer (as the vehicle was buffer, this contained buffer only)
Blank-correction using just the buffer blank (RD − BB) yielded high enzyme activity, and therefore low enzyme inhibition results in all three assays (Figs. 6, 7 and 8). In the α-glucosidase assay, RD − BB exhibited the highest enzyme activity value for acarbose, and the second highest enzyme activity values for A. marmelos and P. niruri. Similarly, in the lipase assay, RD − BB yielded the highest enzyme activity for C. sinensis and S. japonica, and the second highest for orlistat. In the α-amylase assay, RD − BB gave very high enzyme activity, however, these enzyme activity values were almost identical to RD − SaB and RD − (BB + SaB).
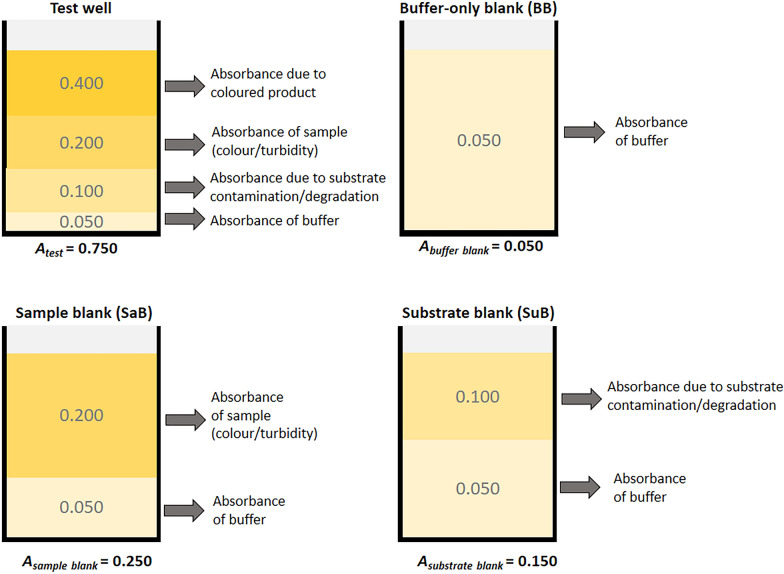
Buffer blanks are routinely used for baseline correction and account for the background absorbance or fluorescence of the assay buffer. However, if no substrate blank or sample blanks are included, this method (RD − BB) does not account for interference from the substrate or samples. Therefore, in RD − BB, the background absorbance (or fluorescence) of the substrate and the samples, which caused an additive effect on the test well measurements were uncorrected and resulted in misleadingly high enzyme activity results. This is also illustrated in Fig. 9 which shows the components contributing to the total absorbance (or fluorescence) of a test well, buffer blank, sample blank, and substrate blank. Table 2 provides an example of how the different blanking methods used in this study changed the blank-corrected absorbance (or fluorescence) of the test well.
Fig. 9.
Components contributing to absorbance. This figure illustrates the various components contributing to the absorbance in the test wells, buffer-only blank, sample blank, and substrate blank wells. This also applies to fluorescence assays. Absorbance values in this figure are for illustrative purposes only (not based on real values)
Table 2.
Effect of blank-correction methods on the test well absorbance values in Fig. 9
| Method | Formula | Calculation | Blank-corrected absorbance |
|---|---|---|---|
| 1 | RD − BB | 0.750 − 0.050 | 0.700 |
| 2 | RD − SuB | 0.750 − 0.150 | 0.600 |
| 3 | RD − SaB | 0.750 − 0.250 | 0.500 |
| 4 | RD − (BB + SaB) | 0.750 − (0.050 + 0.250) | 0.450 |
| 5 | RD − (SuB + SaB) | 0.750 − (0.150 + 0.250) | 0.350 |
| 6 | RD − [(SuB − BB) + SaB)] | 0.750 − [(0.150 − 0.050) + 0.250] | 0.400 |
RD: raw data (Atest); BB: buffer blank; SuB: substrate blank, SaB: sample blank
In the α-glucosidase assay (Fig. 6), enzyme inhibition by the two plant extracts A. marmelos and P. niruri was only observed when a sample blank was included [RD − SaB, RD − (BB + SaB), RD − (SuB + SaB), and RD − (Su + SaB)] to offset the interference due to the colour of the extracts (Fig. 10). When results were calculated without a sample blank (RD − BB and RD − SuB), A. marmelos and P. niruri appeared to promote enzyme activity. This apparent increase in enzyme activity can be attributed to the colour of the two extracts contributing an additive effect to the absorbance of the test wells resulting in an over-estimation of enzyme activity. Note that the interferences due to sample colour may be minimised by diluting the samples. However, if the sample is too dilute for inhibitory activity to be detected, screening samples at low concentrations increases the risk of missing potential hits.
Fig. 10.
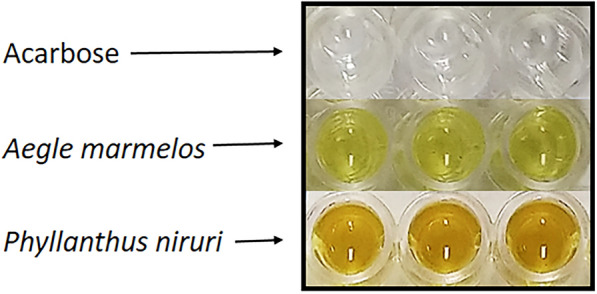
Acarbose, A. marmelos and P. niruri extracts at 0.5 mg/mL in a clear 96-well plate
In contrast to the plant extracts, acarbose inhibited α-glucosidase significantly regardless of whether or not a sample blank was included in the blank-correction. Acarbose was colourless and dissolved completely in α-glucosidase assay buffer I to give a clear, colourless solution (Fig. 10). Hence, any spectral interference due to the background absorbance of acarbose was negligible and not correcting this background absorbance did not affect the final results as much as the highly coloured extracts which had high background absorbances (sample blank absorbance).
Figures 7 and 8 illustrate the effects of the six blanking methods on the inhibition of α-amylase and lipase. Regardless of the blanking method used, all test samples and positive controls (acarbose for α-amylase, and orlistat for lipase) showed significant inhibition of their respective enzymes. This contrasted with the absorbance-based α-glucosidase assay, where not including a sample blank gave enzyme activity > 100% for the two plant extracts. The reason for this was that the colours of the extracts used in the α-glucosidase assay had high background absorbance values relative to the absorbance of the test wells and therefore contributed a larger error to the final results. Correcting this large error by subtracting the sample blanks caused a larger reduction in the final results. On the other hand, the background fluorescence of the samples in the amylase and lipase assays were much smaller relative to the fluorescence of the test wells, and therefore only contributed a smaller error to the final results.
Although the sample blank did not influence the enzyme activity values in the fluorescence assays, the inclusion or omission of a substrate blank significantly impacted the final enzyme activity values in both the α-amylase (Fig. 7) and lipases assays (Fig. 8). The enzyme activity results within each group (each of the positive controls and plant extracts) separated into two tiers depending on whether a substrate blank had been included in the blank-correction: the enzyme activity of all groups were significantly higher when a substrate blank was not included [RD − BB, RD − SaB, and RD − (BB + SaB)] than when a substrate blank was included (RD − SuB, RD − (SuB + SaB), and RD − (Su + SaB). This was attributed to the high background autofluorescence of the substrate (157,700 for α-amylase substrate, and 35,632 for lipase substrate) which contributed a proportionally larger error to the final results (e.g. for G. jasminoides, Ftest = 193222, Fsample blank = 179 vs Fsubstrate blank = 171765). Subtracting the high background autofluorescence of the substrate (substrate blank) proportionally reduced the blank-corrected fluorescence of the test wells (193,222 − 171,765 = 21,457 when substrate blank was included versus 193,222 − 179 = 193,044 when the sample blank was included) and therefore reduced the final calculated enzyme activity values. In the α-glucosidase assay, the substrate had a comparatively smaller effect on the enzyme activity results because the background absorbance due to the substrate was lower when compared to the raw absorbance data of the test wells. These results indicated that it is crucial to include a substrate blank to avoid the risk of missing potential enzyme inhibitory activity in drug candidates due to the overestimation of enzyme activity.
Subtracting two blanks [RD − (BB + SaB) and RD − (SuB + SaB)] generally resulted in lower enzyme activity than for the methods that only subtracted one blank. Note that both the substrate blank and the sample blank contains a certain volume of buffer (Fig. 9). Therefore, the absorbance or fluorescence of these blanks was in part, due to the buffer, and only a proportion of the absorbance was due to the substrate or sample. If two blanks were subtracted during blank-correction, as in the case with RD − (BB + SaB) and RD − (SuB + SaB), this “hidden” background due to the buffer would be subtracted twice, resulting in “double-blanking.” In the literature, some studies have combined sample and substrate in a single blank [65, 79, 80]. This approach prevents double-blanking the buffer as only one blank (containing buffer + sample + substrate) is subtracted from the raw data. However, this must be carried out with caution as unexpected results may occur when the substrate interacts with unknown constituents in the sample; especially in crude natural product extracts which contain complex mixtures of compounds and are a notorious, complicated biological matrix. Another drawback to this approach is that the substrate must be added to each sample blank and this would therefore require larger amounts of substrate per assay which adds to the cost of the assay. In method six [RD − (Su + SaB)], the buffer blank was subtracted from the substrate blank to obtain the absorbance or fluorescence contributed by the substrate only (Su = SuB − BB) and this value was subtracted from the raw data along with the sample blank [RD − (Su + SaB)]. This allowed for the correction of interference due to the sample and the substrate, without double-blanking the buffer, and without adding the substrate to each sample blank and any unexpected artefacts which may arise from doing so. Therefore, [RD − (Su + SaB)] was the most appropriate blanking method for all three assays.
Conclusion
This is the first study to investigate the effects of using different blanks and blanking methods on the results of enzyme assays, and to provide comprehensive, step-by-step protocols for α-amylase, α-glucosidase, and lipase-inhibition assays that can be performed in 96-well format, in a simple, fast, and resource-efficient manner, with clear instructions for blank-correction and calculation of results.
The type of blanks and blank-correction methods employed in the assays had a significant effect on the final, calculated enzyme activity (and inhibition) values in all three assays. The importance of including a sample blank when testing highly coloured samples, as well as the relevance of a substrate blank, and avoiding errors due to unintended double-blanking were highlighted in the results. Depending on the blank(s) used in the blank-correction of raw data, variation in the final calculated results can lead to either an over-estimation or under-estimation of the calculated enzyme activity. Not accounting for interferences due to the colour of the natural product extracts can result in misleadingly high enzyme activity values which underestimates the bioactivity of the target sample. Therefore, potential enzyme inhibitors can be inadvertently overlooked resulting in missed opportunities in the drug discovery process.
The variation in the final calculated results demonstrated that the same set of raw data can produce different results depending on the blank-correction method. This emphasises the importance of standardising the use of blanks and the reporting of blank-correction procedures in published literature in order to enhance the reproducibility of results and prevent misleading results in not only enzyme assays, but also in other spectrometry-based assays involving natural product extracts.
Out of the methods tested, the sixth blanking method [RD − (Su + SaB)] adequately accounted for interferences due to the background absorbance/fluorescence of both the substrate and sample without double blanking, and using lower volumes of substrate, and is therefore recommended for the blank-correction of raw data in enzyme assays.
Acknowledgements
Dr Daniel Anthony Dias gratefully acknowledges The American Society of Pharmacognosy for being awarded a Research Starter Grant and support from The Product Makers Pty Ltd (TPM Bioactives Division), Melbourne, Australia.
Abbreviations
- 4-MU
4-Methylumbelliferone
- 4-MUO
4-Methylumbelliferyl oleate
- Asubscript
Absorbance
- BB
Buffer blank
- BODIPY®
Boron-dipyrromethene
- BODIPY®FL-DQ™
Boron-dipyrromethene fluorescent dye quenching
- Cat. No.
Catalogue number
- DMSO
Dimethyl sulfoxide
- DQ™
Dye quenching
- DQ™ starch
Dye quenching starch
- EC
Enzyme commission number
- Fsubscript
Fluorescence
- PBS
Phosphate buffered saline
- pNPG
p-Nitrophenyl-α-d-glucopyranoside
- RD
Raw data
- RT
Room temperature (25 °C)
- SaB
Sample blank
- SuB
Substrate blank
Authors’ contributions
CL and SL designed and carried out the experiments, CL conducted data processing, statistical analyses, and created all graphs and figures, MF, GL, HG, TH, and DD provided materials for the experiments and assisted with experimental design, CL and SL drafted and wrote the manuscript, MF, GL, HG, TH and DD commented and provided critical feedback on the manuscript, TH and DD revised the text and structure and outlined it several times together with CL and SL. All authors read and approved the final manuscript.
Funding
The American Society of Pharmacognosy and The Product Makers Pty Ltd (TPM Bioactives Division), Melbourne, Australia.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1(9):727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Copeland RA. Why enzymes as drug targets? Evaluation of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists. 2. Hoboken: Wiley; 2013. pp. 1–23. [PubMed] [Google Scholar]
- 3.Copeland RA, Harpel MR, Tummino PJ. Targeting enzyme inhibitors in drug discovery. Expert Opin Ther Targets. 2007;11(7):967–978. doi: 10.1517/14728222.11.7.967. [DOI] [PubMed] [Google Scholar]
- 4.Seiler N. Thirty years of polyamine-related approaches to cancer therapy. Retrospect and prospect. Part 1. Selective enzyme inhibitors. Curr Drug Targets. 2003;4(7):537–564. doi: 10.2174/1389450033490885. [DOI] [PubMed] [Google Scholar]
- 5.Scatena R, Bottoni P, Pontoglio A, Mastrototaro L, Giardina B. Glycolytic enzyme inhibitors in cancer treatment. Expert Opin Investig Drugs. 2008;17(10):1533–1545. doi: 10.1517/13543784.17.10.1533. [DOI] [PubMed] [Google Scholar]
- 6.McFarlane SI, Kumar A, Sowers JR. Mechanisms by which angiotensin-converting enzyme inhibitors prevent diabetes and cardiovascular disease. Am J Cardiol. 2003;91(12):30–37. doi: 10.1016/S0002-9149(03)00432-6. [DOI] [PubMed] [Google Scholar]
- 7.Messerli FH, Bangalore S, Bavishi C, Rimoldi SF. Angiotensin-converting enzyme inhibitors in hypertension. J Am Coll Cardiol. 2018;71(13):1474–1482. doi: 10.1016/j.jacc.2018.01.058. [DOI] [PubMed] [Google Scholar]
- 8.Chiasson J-L, Josse RG, Gomis R, Hanefeld M, Karasik A, Laakso M. Acarbose for prevention of type 2 diabetes mellitus: the STOP-NIDDM randomised trial. Lancet. 2002;359(9323):2072–2077. doi: 10.1016/S0140-6736(02)08905-5. [DOI] [PubMed] [Google Scholar]
- 9.Van de Laar FA, Lucassen PLBJ, Akkermans RP, Van de Lisdonk EH, Rutten GEHM, Van Weel C. Alpha glucosidase inhibitors for type 2 diabetes mellitus. Cochrane Database Syst Rev. 2005;2:CD003639. doi: 10.1002/14651858.CD003639.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Youdim MBH, Edmondson D, Tipton KF. The therapeutic potential of monoamine oxidase inhibitors. Nat Rev Neurosci. 2006;7(4):295. doi: 10.1038/nrn1883. [DOI] [PubMed] [Google Scholar]
- 11.Finberg JPM, Rabey JM. Inhibitors of MAO-A and MAO-B in Psychiatry and Neurology. Front Pharmacol. 2016;7:340. doi: 10.3389/fphar.2016.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehta M, Adem A, Sabbagh M. New acetylcholinesterase inhibitors for Alzheimer’s disease. Int J Alzheimers Dis. 2012;2012:728983. doi: 10.1155/2012/728983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rolinski M, Fox C, Maidment I, McShane R. Cholinesterase inhibitors for dementia with Lewy bodies, Parkinson’s disease dementia and cognitive impairment in Parkinson’s disease. Cochrane Database Syst Rev. 2012;2012(3):CD006504. doi: 10.1002/14651858.CD006504.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo S, Gill H, Dias DA, Li M, Hung A, Nguyen LT, et al. The inhibitory effects of an eight-herb formula (RCM-107) on pancreatic lipase: enzymatic, HPTLC profiling and in silico approaches. Heliyon. 2019;5(9):e02453. doi: 10.1016/j.heliyon.2019.e02453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tucci SA, Boyand EJ, Halford JCG. The role of lipid and carbohydrate digestive enzyme inhibitors in the management of obesity. Diabetes Metab Syndr Obes. 2010;3:125–143. doi: 10.2147/DMSO.S7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal P, Gupta R. Alpha-amylase inhibition can treat diabetes mellitus. Res Rev J Med Health Sci. 2016;54:1–8. [Google Scholar]
- 17.Sellami M, Louati H, Kamoun J, Kchaou A, Damak M, Gargouri Y. Inhibition of pancreatic lipase and amylase by extracts of different spices and plants. Int J Food Sci Nutr. 2017;68(3):313–320. doi: 10.1080/09637486.2016.1237479. [DOI] [PubMed] [Google Scholar]
- 18.Buchholz T, Melzig MF. Medicinal plants traditionally used for treatment of obesity and diabetes mellitus—screening for pancreatic lipase and α-amylase inhibition. Phytother Res. 2016;30(2):260–266. doi: 10.1002/ptr.5525. [DOI] [PubMed] [Google Scholar]
- 19.Rajan L, Palaniswamy D, Mohankumar SK. Targeting obesity with plant-derived pancreatic lipase inhibitors: a comprehensive review. Pharmacol Res. 2020;155:104681. doi: 10.1016/j.phrs.2020.104681. [DOI] [PubMed] [Google Scholar]
- 20.Tundis R, Loizzo M. Natural products as alpha-amylase and alpha-glucosidase inhibitors and their hypoglycaemic potential in the treatment of diabetes: an update. Mini Rev Med Chem. 2010;10:315–331. doi: 10.2174/138955710791331007. [DOI] [PubMed] [Google Scholar]
- 21.Dias DA, Urban S, Roessner U. A historical overview of natural products in drug discovery. Metabolites. 2012;2(2):303–336. doi: 10.3390/metabo2020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cragg GM, Grothaus PG, Newman DJ. New horizons for old drugs and drug leads. J Nat Prod. 2014;77(3):703–723. doi: 10.1021/np5000796. [DOI] [PubMed] [Google Scholar]
- 23.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79(3):629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 24.Beutler JA. Natural Products as a Foundation for Drug Discovery. Curr Protoc Pharmacol. 2009;46(1):9–11. doi: 10.1002/0471141755.ph0911s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lahlou M. The Success of natural products in drug discovery. Pharmacol Pharm. 2013;4(3A):17–31. doi: 10.4236/pp.2013.43A003. [DOI] [Google Scholar]
- 26.Shen B. A New Golden Age of Natural Products Drug Discovery. Cell. 2015;163(6):1297–1300. doi: 10.1016/j.cell.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qurratul A, Ashiq U, Jamal RA, Saleem M, Mahroof-Tahir M. Alpha-glucosidase and carbonic anhydrase inhibition studies of Pd(II)-hydrazide complexes. Arabian J Chem. 2017;10(4):488–499. doi: 10.1016/j.arabjc.2015.02.024. [DOI] [Google Scholar]
- 28.Molecular Probes Inc. EnzChek Ultra Amylase Assay Kit (E33651). 2006. https://www.thermofisher.com/order/catalog/product/E33651. Accessed 10 Feb 2018.
- 29.Goddard JP, Reymond JL. Enzyme assays for high-throughput screening. Curr Opin Biotechnol. 2004;15(4):314–322. doi: 10.1016/j.copbio.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Reymond JL, Fluxa VS, Maillard N. Enzyme assays. Chem Commun. 2009;1:34–46. doi: 10.1039/b813732c. [DOI] [PubMed] [Google Scholar]
- 31.Lankatillake C, Huynh T, Dias DA. Understanding glycaemic control and current approaches for screening antidiabetic natural products from evidence-based medicinal plants. Plant Methods. 2019;15(1):105. doi: 10.1186/s13007-019-0487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granados-Guzman G, Castro-Rios R, Waksman de Torres N, Salazar-Aranda R. Optimization and validation of a microscale in vitro method to assess alpha-glucosidase inhibition activity. Curr Anal Chem. 2018;14(5):458–464. doi: 10.2174/1573411013666170911154755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beneyton T, Wijaya IP, Postros P, Najah M, Leblond P, Couvent A, et al. High-throughput screening of filamentous fungi using nanoliter-range droplet-based microfluidics. Sci Rep. 2016;6:27223. doi: 10.1038/srep27223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Held P. Enzymatic digestion of polysaccharides. Part 1: monitoring polymer digestion and glucose production in microplates: BioTek Instruments, Inc. 2012. https://www.biotek.com/assets/tech_resources/Cellulosic_App_Note_Part_I.pdf. Accessed 15 Aug 2020.
- 35.Koyama K, Hirao T, Toriba A, Hayakawa K. An analytical method for measuring alpha-amylase activity in starch containing foods. Biomed Chromatogr. 2013;27(5):583–588. doi: 10.1002/bmc.2831. [DOI] [PubMed] [Google Scholar]
- 36.Thermo Fisher Scientific Inc. Detecting Peptidases and Proteases—Section 10.4. 2020. https://www.thermofisher.com/au/en/home/references/molecular-probes-the-handbook/enzymesubstrates/detecting-peptidases-and-proteases.html. Accessed 16 Aug 2020.
- 37.Buchholz T, Melzig MF. Polyphenolic compounds as pancreatic lipase inhibitors. Planta Med. 2015;81(10):771–783. doi: 10.1055/s-0035-1546173. [DOI] [PubMed] [Google Scholar]
- 38.Ortiz-Miranda S, Ji R, Jurczyk A, Aryee KE, Mo S, Fletcher T, et al. A novel transgenic mouse model of lysosomal storage disorder. Am J Physiol Gastrointest Liver Physiol. 2016;311(5):G903–G919. doi: 10.1152/ajpgi.00313.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jahn M, Zerr A, Fedorowicz FM, Brigger F, Koulov A, Mahler HC. Measuring lipolytic activity to support process improvements to manage lipase-mediated polysorbate degradation. Pharm Res. 2020;37(6):118. doi: 10.1007/s11095-020-02812-0. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Zhong X, Wang X, Li J, Liu J, Wang K, et al. Bioassay-guided isolation of triterpenoids as α-glucosidase inhibitors from Cirsium setosum. Molecules. 2019;24(10):1844. doi: 10.3390/molecules24101844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Sotto A, Locatelli M, Macone A, Toniolo C, Cesa S, Carradori S, et al. Hypoglycemic, antiglycation, and cytoprotective properties of a phenol-rich extract from waste peel of Punica granatum L. Var. Dente di cavallo DC2. Molecules. 2019;24(17):3103. doi: 10.3390/molecules24173103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ostberg-Potthoff JJ, Berger K, Richling E, Winterhalter P. Activity-guided fractionation of red fruit extracts for the identification of compounds influencing glucose metabolism. Nutrients. 2019;11(5):1166. doi: 10.3390/nu11051166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olaokun OO, Alaba AE, Ligege K, Mkolo NM. Phytochemical content, antidiabetes, anti-inflammatory antioxidant and cytotoxic activity of leaf extracts of Elephantorrhiza elephantina (Burch.) Skeels. S Afr J Bot. 2020;128:319–325. doi: 10.1016/j.sajb.2019.11.030. [DOI] [Google Scholar]
- 44.Popović BM, Blagojević B, Ždero Pavlović R, Mićić N, Bijelić S, Bogdanović B, et al. Comparison between polyphenol profile and bioactive response in blackthorn (Prunus spinosa L.) genotypes from north Serbia-from raw data to PCA analysis. Food Chem. 2020;302:125373. doi: 10.1016/j.foodchem.2019.125373. [DOI] [PubMed] [Google Scholar]
- 45.Yilmazer-Musa M, Griffith AM, Michels AJ, Schneider E, Frei B. Grape seed and tea extracts and catechin 3 gallates are potent inhibitors of alpha-amylase and alpha-glucosidase activity. J Agric Food Chem. 2012;60(36):8924–8929. doi: 10.1021/jf301147n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auld DS, Coassin PA, Coussens NP, Hensley P, Klumpp-Thomas C, Michael S, et al. Microplate Selection and Recommended Practices in High-throughput Screening and Quantitative Biology. In: Assay Guidance Manual. Bethesda: Eli Lilly & Company and the National Center for Advancing Translational Sciences. 2020. [PubMed]
- 47.Simeonov A, Davis MI. Interference with Fluorescence and Absorbance. In: Assay Guidance Manual. Bethesda: Eli Lilly & Company and the National Center for Advancing Translational Sciences. 2018 [PubMed]
- 48.Thorne N, Auld DS, Inglese J. Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr Opin Chem Biol. 2010;14(3):315–324. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chlorophylls Gross J. Pigments in Vegetables. Boston: Springer; 1991. pp. 3–74. [Google Scholar]
- 50.Carotenoids Gross J. Pigments in Vegetables. Boston: Springer; 1991. pp. 75–278. [Google Scholar]
- 51.Azeredo HMC. Betalains: properties, sources, applications, and stability—a review. Int J Food Sci Technol. 2009;44(12):2365–2376. doi: 10.1111/j.1365-2621.2007.01668.x. [DOI] [Google Scholar]
- 52.Mlodzinska E. Survey of plant pigments: molecular and environmental determinants of plant colors. Acta Biol Crac Ser Bot. 2009;51(1):7–16. [Google Scholar]
- 53.Glover BJ, Martin C. Anthocyanins. Curr Biol. 2012;22(5):R147–R150. doi: 10.1016/j.cub.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 54.Moon KM, Kwon EB, Lee B, Kim CY. Recent trends in controlling the enzymatic browning of fruit and vegetable products. Molecules. 2020;25(12):2754. doi: 10.3390/molecules25122754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicolas JJ, Richard-Forget FC, Goupy PM, Amiot MJ, Aubert SY. Enzymatic browning reactions in apple and apple products. Crit Rev Food Sci Nutr. 1994;34(2):109–157. doi: 10.1080/10408399409527653. [DOI] [PubMed] [Google Scholar]
- 56.Toledo L, Aguirre C. Enzymatic browning in avocado (Persea americana) revisited: history, advances, and future perspectives. Crit Rev Food Sci Nutr. 2017;57(18):3860–3872. doi: 10.1080/10408398.2016.1175416. [DOI] [PubMed] [Google Scholar]
- 57.Geremias-Andrade IM, Rocheto AC, Gallo FA, Petrus RR. The shelf life of standardized sugarcane juice stored under refrigeration. Food Sci Technol. 2020;40(1):95–101. doi: 10.1590/fst.33918. [DOI] [Google Scholar]
- 58.Roshchina VV, Kuchin AV, Yashin VA. Application of autofluorescence for analysis of medicinal plants. Int J Spectrosc. 2017;2017:1–8. doi: 10.1155/2017/7159609. [DOI] [Google Scholar]
- 59.Duval R, Duplais C. Fluorescent natural products as probes and tracers in biology. Nat Prod Rep. 2017;34(2):161–193. doi: 10.1039/C6NP00111D. [DOI] [PubMed] [Google Scholar]
- 60.Donaldson L. Autofluorescence in plants. Molecules. 2020;25(10):2393. doi: 10.3390/molecules25102393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muller SM, Galliardt H, Schneider J, Barisas BG, Seidel T. Quantification of Forster resonance energy transfer by monitoring sensitized emission in living plant cells. Front Plant Sci. 2013;4:413. doi: 10.3389/fpls.2013.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Müller MG, Georgakoudi I, Zhang Q, Wu J, Feld MS. Intrinsic fluorescence spectroscopy in turbid media: disentangling effects of scattering and absorption. Appl Opt. 2001;40(25):4633–4646. doi: 10.1364/AO.40.004633. [DOI] [PubMed] [Google Scholar]
- 63.Hach. What is the difference between a reagent blank and a sample blank? 2019. https://support.hach.com/app/answers/answer_view/a_id/1000699/loc/en_US#__highlight. Accessed 28 Apr 2020.
- 64.Földesi B. Guide to enzyme unit definitions and assay design. 2019. https://www.biomol.com/resources/biomol-blog/guide-to-enzyme-unit-definitions-and-assay-design. Accessed 16 Aug 2020.
- 65.Yang X, Kong F. Evaluation of the in vitro α-glucosidase inhibitory activity of green tea polyphenols and different tea types. J Sci Food Agric. 2016;96(3):777–782. doi: 10.1002/jsfa.7147. [DOI] [PubMed] [Google Scholar]
- 66.Özek G, Özbek MU, Yur S, Göger F, Arslan M, Özek T. Assessment of endemic cota fulvida (Asteraceae) for phytochemical composition and inhibitory activities against oxidation, α-amylase, lipoxygenase, xanthine oxidase and tyrosinase enzymes. Rec Nat Prod. 2019;13(4):333–345. doi: 10.25135/rnp.109.18.09.875. [DOI] [Google Scholar]
- 67.Tan Y, Chang SKC, Zhang Y. Comparison of α-amylase, α-glucosidase and lipase inhibitory activity of the phenolic substances in two black legumes of different genera. Food Chem. 2017;214:259–268. doi: 10.1016/j.foodchem.2016.06.100. [DOI] [PubMed] [Google Scholar]
- 68.Tunsaringkarn T, Rungsiyothin A, Ruangrungsi N. α-Glucosidase inhibitory activity of Thai mimosaceous plant extracts. J Health Res. 2008;22(1):29–33. [Google Scholar]
- 69.Cardullo N, Muccilli V, Pulvirenti L, Cornu A, Pouysegu L, Deffieux D, et al. C-glucosidic ellagitannins and galloylated glucoses as potential functional food ingredients with anti-diabetic properties: a study of alpha glucosidase and alpha-amylase inhibition. Food Chem. 2020;313:126099. doi: 10.1016/j.foodchem.2019.126099. [DOI] [PubMed] [Google Scholar]
- 70.Rubilar M, Jara C, Poo Y, Acevedo F, Gutierrez C, Sineiro J, et al. Extracts of Maqui (Aristotelia chilensis) and Murta (Ugni molinae Turcz.): sources of antioxidant compounds and alpha-Glucosidase/alpha-Amylase inhibitors. J Agric Food Chem. 2011;59(5):1630–1637. doi: 10.1021/jf103461k. [DOI] [PubMed] [Google Scholar]
- 71.Podsedek A, Majewska I, Redzynia M, Sosnowska D, Koziolkiewicz M. In vitro inhibitory effect on digestive enzymes and antioxidant potential of commonly consumed fruits. J Agric Food Chem. 2014;62(20):4610–4617. doi: 10.1021/jf5008264. [DOI] [PubMed] [Google Scholar]
- 72.Sosnowska D, Podsędek A, Redzynia M, Kucharska AZ. Inhibitory effect of black chokeberry fruit polyphenols on pancreatic lipase – Searching for most active inhibitors. J Funct Foods. 2018;49:196–204. doi: 10.1016/j.jff.2018.08.029. [DOI] [Google Scholar]
- 73.Yuan Y, Zhang J, Fan J, Clark J, Shen P, Li Y, et al. Microwave assisted extraction of phenolic compounds from four economic brown macroalgae species and evaluation of their antioxidant activities and inhibitory effects on alpha-amylase, alpha-glucosidase, pancreatic lipase and tyrosinase. Food Res Int. 2018;113:288–297. doi: 10.1016/j.foodres.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 74.Hao W, Wang M, Lv M. The inhibitory effects of Yixing Black Tea extracts on A-Glucosidase. J Food Biochem. 2017;41(1):e12269. doi: 10.1111/jfbc.12269. [DOI] [Google Scholar]
- 75.Jacobson RL, Schlein Y. Phlebotomus papatasi and Leishmania major parasites express α-amylase and α-glucosidase. Acta Trop. 2001;78(1):41–49. doi: 10.1016/S0001-706X(00)00164-9. [DOI] [PubMed] [Google Scholar]
- 76.Khalil AT, Ovais M, Ullah I, Ali M, Jan SA, Shinwari ZK, et al. Bioinspired synthesis of pure massicot phase lead oxide nanoparticles and assessment of their biocompatibility, cytotoxicity and in vitro biological properties. Arabian J Chem. 2020;13(1):916–931. doi: 10.1016/j.arabjc.2017.08.009. [DOI] [Google Scholar]
- 77.Sun L, Warren FJ, Gidley MJ. Soluble polysaccharides reduce binding and inhibitory activity of tea polyphenols against porcine pancreatic α-amylase. Food Hydrocoll. 2018;79:63–70. doi: 10.1016/j.foodhyd.2017.12.011. [DOI] [Google Scholar]
- 78.Dou F, Xi M, Wang J, Tian X, Hong L, Tang H, et al. α-Glucosidase and-amylase inhibitory activities of saponins from traditional Chinese medicines in the treatment of diabetes mellitus. Die Pharmazie-Int J Pharm Sci. 2013;68(4):300–304. [PubMed] [Google Scholar]
- 79.Awosika TO, Aluko RE. Inhibition of the in vitro activities of α-amylase, α -glucosidase and pancreatic lipase by yellow field pea (Pisum sativum L.) protein hydrolysates. Int J Food Sci Technol. 2019;54(6):2021–2034. doi: 10.1111/ijfs.14087. [DOI] [Google Scholar]
- 80.Hamdani SS, Khan BA, Ahmed MN, Hameed S, Akhter K, Ayub K, et al. Synthesis, crystal structures, computational studies and α-amylase inhibition of three novel 1,3,4-oxadiazole derivatives. J Mol Struct. 2020;1200:127085. doi: 10.1016/j.molstruc.2019.127085. [DOI] [Google Scholar]
- 81.Phuwapraisirisan P, Puksasook T, Jong-Aramruang J, Kokpol U. Phenylethyl cinnamides: a new series of alpha glucosidase inhibitors from the leaves of Aegle marmelos. Bioorg Med Chem Lett. 2008;18(18):4956–4958. doi: 10.1016/j.bmcl.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 82.Ansari P, Afroz N, Jalil S, Azad SB, Mustakim MG, Anwar S, et al. Anti-hyperglycemic activity of Aegle marmelos (L.) corr. is partly mediated by increased insulin secretion, α-amylase inhibition, and retardation of glucose absorption. J Pediatr Endocrinol Metab. 2017;30(1):37–47. doi: 10.1515/jpem-2016-0160. [DOI] [PubMed] [Google Scholar]
- 83.Beidokhti MN, Andersen MV, Eid HM, Sanchez Villavicencio ML, Staerk D, Haddad PS, et al. Investigation of antidiabetic potential of Phyllanthus niruri L. using assays for α-glucosidase, muscle glucose transport, liver glucose production, and adipogenesis. Biochem Biophys Res Commun. 2017;493(1):869–874. doi: 10.1016/j.bbrc.2017.09.080. [DOI] [PubMed] [Google Scholar]
- 84.Guillen Quispe YN, Hwang SH, Wang Z, Zuo G, Lim SS. Screening in vitro targets related to diabetes in herbal extracts from Peru: identification of active compounds in hypericum laricifolium juss by offline high performance liquid chromatography. Int J Mol Sci. 2017;18(12):2512. doi: 10.3390/ijms18122512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Okoli CO, Obidike IC, Ezike AC, Akah PA, Salawu OA. Studies on the possible mechanisms of antidiabetic activity of extract of aerial parts of Phyllanthus niruri. Pharm Biol. 2011;49(3):248–255. doi: 10.3109/13880209.2010.501456. [DOI] [PubMed] [Google Scholar]
- 86.Ranilla LG, Kwon YI, Apostolidis E, Shetty K. Phenolic compounds, antioxidant activity and in vitro inhibitory potential against key enzymes relevant for hyperglycemia and hypertension of commonly used medicinal plants, herbs and spices in Latin America. Bioresour Technol. 2010;101(12):4676–4689. doi: 10.1016/j.biortech.2010.01.093. [DOI] [PubMed] [Google Scholar]
- 87.Luo S, Lenon GB, Gill H, Hung A, Dias DA, Li M, et al. Inhibitory effect of a weight-loss Chinese herbal formula RCM-107 on pancreatic alpha-amylase activity: enzymatic and in silico approaches. PLoS ONE. 2020;15(4):e0231815. doi: 10.1371/journal.pone.0231815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.He Q, Lv Y, Yao K. Effects of tea polyphenols on the activities of α-amylase, pepsin, trypsin and lipase. Food Chem. 2007;101(3):1178–1182. doi: 10.1016/j.foodchem.2006.03.020. [DOI] [Google Scholar]
- 89.Ramirez G, Zavala M, Perez J, Zamilpa A. In vitro screening of medicinal plants used in Mexico as antidiabetics with glucosidase and lipase inhibitory activities. Evid Based Complement Alternat Med. 2012;2012:701261. doi: 10.1155/2012/701261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gondoin A, Grussu D, Stewart D, McDougall GJ. White and green tea polyphenols inhibit pancreatic lipase in vitro. Food Res Int. 2010;43(5):1537–1544. doi: 10.1016/j.foodres.2010.04.029. [DOI] [Google Scholar]
- 91.Lordan S, Smyth TJ, Soler-Vila A, Stanton C, Ross RP. The alpha-amylase and alpha-glucosidase inhibitory effects of Irish seaweed extracts. Food Chem. 2013;141(3):2170–2176. doi: 10.1016/j.foodchem.2013.04.123. [DOI] [PubMed] [Google Scholar]
- 92.Herrera T, Navarro del Hierro J, Fornari T, Reglero G, Martin D. Inhibitory effect of quinoa and fenugreek extracts on pancreatic lipase and α-amylase under in vitro traditional conditions or intestinal simulated conditions. Food Chem. 2019;270:509–517. doi: 10.1016/j.foodchem.2018.07.145. [DOI] [PubMed] [Google Scholar]
- 93.Zhang B, Deng Z, Ramdath DD, Tang Y, Chen PX, Liu R, et al. Phenolic profiles of 20 Canadian lentil cultivars and their contribution to antioxidant activity and inhibitory effects on alpha-glucosidase and pancreatic lipase. Food Chem. 2015;172:862–872. doi: 10.1016/j.foodchem.2014.09.144. [DOI] [PubMed] [Google Scholar]
- 94.Di L, Kerns EH. Biological assay challenges from compound solubility: strategies for bioassay optimization. Drug Discov Today. 2006;11(9–10):446–451. doi: 10.1016/j.drudis.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 95.Johnston PA, Johnston PA. Cellular platforms for HTS: three case studies. Drug Discov Today. 2002;7(6):353–363. doi: 10.1016/S1359-6446(01)02140-7. [DOI] [PubMed] [Google Scholar]
- 96.Lundholt BK, Scudder KM, Pagliaro L. A simple technique for reducing edge effect in cell-based assays. J Biomol Screen. 2003;8(5):566–570. doi: 10.1177/1087057103256465. [DOI] [PubMed] [Google Scholar]
- 97.Lau CY, Zahidi AAA, Liew OW, Ng TW. A direct heating model to overcome the edge effect in microplates. J Pharm Biomed Anal. 2015;2015(102):199–202. doi: 10.1016/j.jpba.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 98.Clarkson JR, Cui ZF, Darton RC. Protein denaturation in Foam: II. Surface activity and conformational change. J Coll Interface Sci. 1999;1999(215):333–338. doi: 10.1006/jcis.1999.6256. [DOI] [PubMed] [Google Scholar]
- 99.Clarkson JR, Cui ZF, Darton RC. Effect of solution conditions on protein damage in foam. Biochem Eng J. 2000;2000(4):107–114. doi: 10.1016/S1369-703X(99)00038-8. [DOI] [Google Scholar]
- 100.Bisswanger H. Enzyme assays. Perspect Sci. 2014;1(1–6):41–55. doi: 10.1016/j.pisc.2014.02.005. [DOI] [Google Scholar]
- 101.Nakai M, Fukui Y, Asami S, Toyoda-Ono Y, Iwashita T, Shibata H, et al. Inhibitory effects of oolong tea polyphenols on pancreatic lipase in vitro. J Agric Food Chemi. 2005;53(11):4593–4598. doi: 10.1021/jf047814+. [DOI] [PubMed] [Google Scholar]
- 102.Gorovits B, Roldan MA, Baltrukonis D, Cai CH, Donley J, Jani D, et al. Anti-drug antibody assay validation: improved reporting of the assay selectivity via simpler positive control recovery data analysis. AAPS J. 2019;21(5):76. doi: 10.1208/s12248-019-0347-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request.