Abstract
In this study, we constructed recombinant luminescent Escherichia coli with T7, T3, and SP6 promoters inserted between tol and lux genes as toluene biosensors and evaluated their sensitivity, selectivity, and specificity for measuring bioavailable toluene in groundwater and river water. The luminescence intensity of each biosensor depended on temperature, incubation time, ionic strength, and concentrations of toluene and coexisting organic compounds. Toluene induced the highest luminescence intensity in recombinant lux-expressing E. coli with the T7 promoter [T7-lux-E. coli, limit of detection (LOD) = 0.05 μM], followed by that in E. coli with the T3 promoter (T3-lux-E. coli, LOD = 0.2 μM) and SP6 promoter (SP6-lux-E. coli, LOD = 0.5 μM). Luminescence may have been synergistically or antagonistically affected by coexisting organic compounds other than toluene; nevertheless, low concentrations of benzoate and toluene analogs had no such effect. In reproducibility experiments, the biosensors had low relative standard deviation (4.3–5.8%). SP6-lux-E. coli demonstrated high adaptability to environmental interference. T7-lux-E. coli biosensor—with low LOD, wide measurement range (0.05–500 μM), and acceptable deviation (− 14.3 to 9.1%)—is an efficient toluene biosensor. This is the first study evaluating recombinant lux E. coli with different promoters for their potential application in toluene measurement in actual water bodies.
Keywords: Biosensor, Groundwater, Promoter, Toluene
Introduction
The large-scale consumption of petroleum-derived fuels has led to groundwater and soil contamination through their leakage from fuel tanks and pipelines. Because of its moderate solubility in water and toxicity, toluene is a petrochemical contaminant of particular concern [1]. Even at low concentrations, toluene can be carcinogenic, can exhibit mutagenic properties, and can damage the kidney, liver, and central nervous system [2]. In Taiwan, environmental agencies have set acceptable limits for toluene in drinking water and groundwater at considerably low levels (7.6–10.9 μM) [3, 4]. In addition, toluene measurement is paramount for the monitoring and clean-up of contaminated groundwater and surface water. Thus, the need for sensitive toluene detection is high, but its design is challenging. In particular, toluene is found in various water bodies, including rivers, as well as coastal water and groundwater; even drinking water contains toluene at trace concentrations (μM) [1].
Conventional analytical techniques, such as gas chromatography (GC) and high-performance liquid chromatography, are sensitive and reliable for toluene detection but are time-consuming, expensive, and laboratory-bound, and they require large equipment and specialized training [5, 6]. By contrast, biological methods can be useful alternatives for organics detection because they are low cost, easy to use, portable, small, and highly specific and can detect bioavailability [7–9]. Of the biological methods, biosensors are suitable for application as environmental sensors, even for on-field measurements.
Over the last 20 years, biosensors have been developed and are widely used as simple and practical approaches for the sensitive and specific detection of various compounds, including organic compounds (pesticides and chlorophenol), heavy metals (mercury, zinc, and cadmium), and some inorganic compounds [9–11]. Whole-cell biosensors rely on gene expression analysis: transcriptional fusions between a promoter and a reporter gene are created, and the extent of reporter gene expression is used to indicate the pollutant concentration [12]. Several engineered biosensors have specifically discriminated between alkyl-substituted benzene derivatives in water samples [13].
A biosensor of this type can be genetically engineered by placing a reporter gene, such as lacZ, gfp, luc, or lux, under the control of a transcriptional activator [11, 14]. Under appropriate conditions (e.g., in the presence of specific pollutant), the biosensor can produce a detectable signal (color or luminescence) that is directly correlated to the pollutant concentration [12, 15]. This property can aid in directly correlating the toluene concentration with the reporter enzyme activity. Various biosensors for benzene, toluene, ethylbenzene, and xylene detection have been developed on the basis of the tol plasmid of Pseudomonas putida mt-2 [16, 17]. In particular, bioluminescence is highly applicable as a reporter for pollutant detection because its instrumentation is sensitive for detecting light production [18]. However, Escherichia coli cells harboring this plasmid often express various response levels when constructed with different reporter genes or promoters that can lead to a range of linear measurement ranges and limits of detection (LODs) [19]. For instance, among induction-based biosensors, luc-, lux-, and aequorin-based biosensors have the LODs of 11, 7.5, and 1 μM, respectively [20–22]. Of the reporter genes, lux has acceptable sensitivity for signal production [18]. Measurement of toluene at very low concentration levels is a main goal of current environmental research; therefore, for the practical application of these biosensors, efforts toward overcoming the aforementioned limitations are warranted [23]. Rational selection of a suitable promoter or reporter gene is essential for increasing the sensitivity, signal intensity, and response speed of whole-cell biosensors.
SP6, T3, and T7 promoters, which are widely used for in vitro transcription, have similar but distinct promoter specificities [24]. They are classified as strong or weak promoters according to their RNA polymerase affinities. T7 is a strong promoter that maintains gene expression tuned to the highest level, thus potentially producing high signal intensity [25]. By contrast, weaker promoter (T3 and SP6) may adapt environmental variation, which produces different signal characteristics [26]. Thus, the linear measurement ranges and LODs of whole-cell biosensors would be expanded or improved if recombinant luminescent bacteria with suitable promoters are constructed.
In this study, we applied this strategy to construct recombinant E. coli strains carrying the tol plasmid from P. putida and including various promoters (T7, T3, or SP6) controlling lux expression. By optimizing the promoter and regulating the lux expression level in E. coli, the recombinant luminescent biosensors could detect bioavailable toluene under different environmental conditions.
Results and discussion
Comparison of time-dependent induction of our three recombinant luminescent E. coli strains with toluene
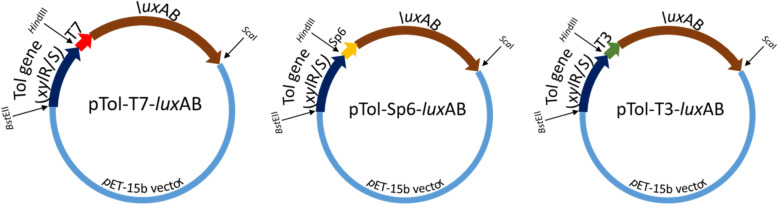
Figure 1 illustrates the construction of the three recombinant plasmids. According to the preliminary experiment, the logarithmic growth phases of the three recombinant E. coli strains occurred from 6 to 15 h of incubation, and the relationship between OD of bacterial growth and RLU (Relative Light Unit) emitted from the three recombinant E. coli strains was linear from 8 to 14 h of incubation. Accordingly, the inoculation time of the three recombinant E. coli strains for the subsequent experiment was set as 12 h after incubation. Figure 2 presents a comparison of the time-dependent induction of luminescence emitted from T3-lux-E. coli, SP6-lux-E. coli, and T7-lux-E. coli caused by different toluene concentrations. As shown in Fig. 2, the induction of luminescence caused by different toluene concentrations occurred time-dependently, regardless of the promoter type. The luminescence intensity continuously increased, leveled off, and then began to decrease considerably during incubation, all potentially due to the biochemical nature of the reporter gene lux [27].
Fig. 1.
Construction of pTOL-T3-lux, pTOL-SP6-lux and pTOL-T7-lux
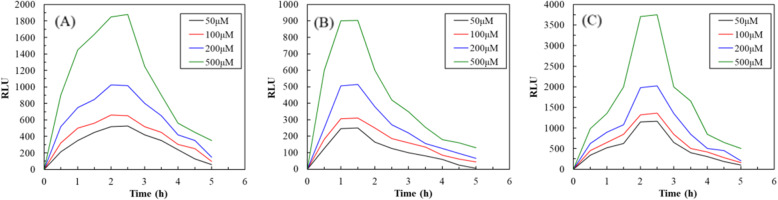
Fig. 2.
Comparison of time-dependent induction of luminescence from (a) T3-lux-E. coli, (b) SP6-lux-E. coli, and (c) T7-lux-E. coli; initial cell concentration: 5 × 107 cfu/mL, culture media: TMM with different toluene concentration, operational conditions: 37 °C and 200 rpm
The results demonstrated that luminescence was stable and the greatest at 2–2.5 h after incubation for T3-lux-E. coli and T7-lux-E. coli or 1–1.5 h after incubation for SP6-lux-E. coli. The time was equal to or shorter than that previously reported for the lux-based bioluminescent bioreporter P. putida TVA8 (2 h) and luminescence bacterial biosensors without the T7 promoter (3 h) for toluene measurement [6, 21]. Therefore, on average, 20-min consecutive measurements were recorded when T3-lux-E. coli and T7-lux-E. coli were cultured for 2 h and when SP6-lux-E. coli was cultured for 1 h. The maximum average luminescence induced by 200 μM toluene for T3-lux-E. coli, SP6-lux-E. coli, and T7-lux-E. coli was 1020 ± 20, 510 ± 10, and 2120 ± 60 RLU, respectively. Moreover, at the same toluene concentration, the signal intensity of luminescence decreased as follows: T7-lux-E. coli > T3-lux-E. coli > SP6-lux-E. coli. However, SP6-lux-E. coli had the shortest stable period for luminescence induction. Previous research demonstrated an increase in bioluminescence emission by fusing the T7 promoter to control expression of the lux operon [28].
Effects of culture conditions on luminescence
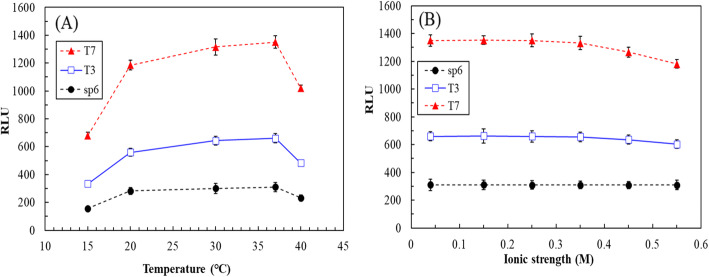
The effects of incubation temperature and ionic strength on the induction of luminescence biosensors for toluene were evaluated according to practical considerations. Figure 3a illustrates the effects of incubation temperature on the luminescence induced by 100 μM toluene for T7-lux-E. coli. The experimental results demonstrated the optimal temperature range of luminescence for T7-lux-E. coli to be 30–37 °C, with nonsignificant differences (p > 0.05). Similar results were observed for T3-lux-E. coli and SP6-lux-E. coli. Moreover, luminescences of the three recombinant E. coli strains at 20 and 40 °C were 12.1–15.3% and 24.4–26.8% lower than those at 37 °C, respectively. The effect of high temperature on the luminescence of the recombinant E. coli strain was more noticeable, a result attributable to the physiological characteristics of the E. coli [29]. Thus, subsequent experiments were performed at 37 °C for all three recombinant E. coli strains.
Fig. 3.
a Effects of incubation temperature on luminescence of T7-lux-E. coli induced by 100 μM toluene for 2 h. b Effects of ionic strength on the luminescence of T3-lux-E. coli, SP6-lux-E. coli and T7-lux-E. coli induced by 100 μM toluene for 2 h (T3-lux-E. coli and T7-lux-E. coli) or 1 h (SP6-lux-E. coli)
Figure 3b shows the effects of ionic strength on the luminescence of the recombinant E. coli with the T3, SP6, or T7 promoter that were induced by 100 μM toluene. The results demonstrated almost no effect of different ionic strengths on the luminescence for SP6-lux-E. coli, but the ionic strength had greater effects on that of T7-lux-E. coli. When the ionic strength was 0.55 M, the luminescence of T7-lux-E. coli decreased by 12.5% ± 0.6%. This inconsistency among the recombinant E. coli with different promoters was presumed to be related to promoter structure and composition, which determine the strength of various types of promoter–target DNA bonds [30]. Additional experiments to investigate these differences are planned. In general, the ranges of ionic strengths of groundwater, river water, seawater, and polluted water are 0.01–0.02 M, 10− 3–10− 2 M, 0.45–0.55 M, and > 10− 2 M, respectively. Thus, SP6-lux-E. coli is suitable for application in various water environments (groundwater, river water, and seawater), whereas T7-lux-E. coli is suitable for use in low ionic strength environments.
Effects of coexisting carbon sources, intermediates, and toluene analogs on luminescence
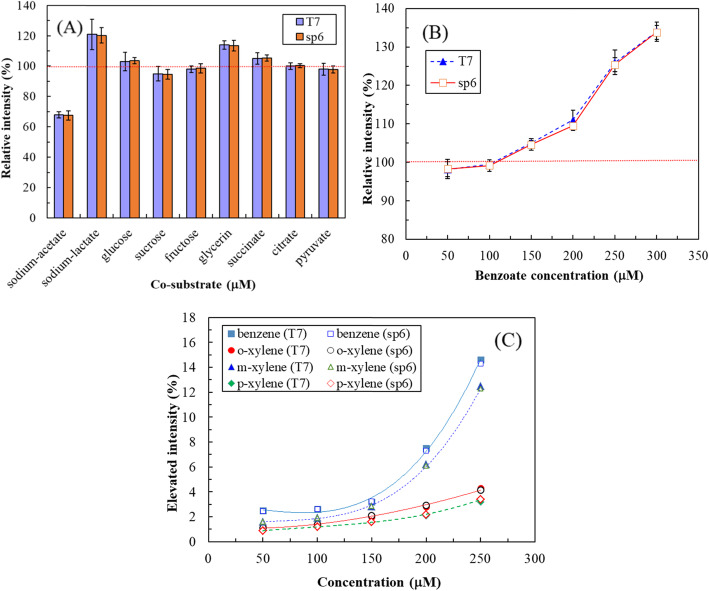
The xyl genes of the Pseudomonas putida TOL plasmid encode the genetic information required for the degradation of toluene and related aromatic compounds. The xylR and xylS genes of the xyl structural genes encode the regulatory proteins of the catabolic operons, whereas the XylR protein is the master regulator of TOL plasmid catabolic operons for the metabolism of toluene [31]. Transcription of the operon is positively regulated by the XylR/XylS protein activated by toluene, xylenes, or their alcohol catabolic products [32]. Figure 4a illustrates the effects of coexisting carbon sources at 100 μM on the luminescence of T7-lux-E. coli and SP6-lux-E. coli. The tested chemicals are considered potential inhibitors or activators (indirect or direct inducers) of xylS and xylR and may deviate significantly or have an additive effect in relation to theoretically expected effects, calculated on the basis of individual chemicals [12, 33, 34]. The current results demonstrated that the coexistence of lactate or glycerin with toluene induced greater luminescence than did toluene alone. Lactate at 100 μM increased luminescence by 21% ± 8.6% for T7-lux-E. coli and 20.3% ± 5.1% for SP6-lux-E. coli, while glycerin increased by 14% ± 1.8% for T7-lux-E. coli and 13.5% ± 3.5% for SP6-lux-E. coli, respectively. The increased luminescence disappeared when the concentrations were below 70 μM (lactate) or 85 μM (glycerin). By contrast, the coexistence of acetate with toluene induced lower luminescence than did toluene alone; luminescence decreased by 32% ± 1.5% for T7-lux-E. coli and 32.5% ± 2.9% for SP6-lux-E. coli. However, for other chemicals, the coexistence had negligible effect on the detection of toluene by T7-lux-E. coli and SP6-lux-E. coli.
Fig. 4.
Effects of (a) coexisting carbon sources (100 μM), (b) benzoate, and (c) toluene analogs and their concentrations on luminescence of T7-lux-E. coli and SP6-lux-E. coli induced by 100 μM toluene for 2 h or 1 h
Figure 4b illustrates the effects of the benzoate concentration on the luminescence of T7-lux-E. coli and SP6-lux-E. coli. Benzoate is the most important metabolite produced during toluene biodegradation [35], which may affect XylS expression [33]; thus, we evaluated the effect of the benzoate concentration on the luminescence of T7-lux-E. coli and SP6-lux-E. coli. The results demonstrated that a high benzoate concentration could induce higher luminescence than did toluene alone, as detected using T7-lux-E. coli and SP6-lux-E. coli. Although 50–150 μM benzoate did not affect luminescence, 250–300 μM benzoate increased luminescence by 26% ± 3.5% for T7-lux-E. coli (25.4% ± 1.8% for SP6-lux-E. coli) and 34% ± 2.8% for T7-lux-E. coli (33.8% ± 1.8% for SP6-lux-E. coli), respectively. In other words, the effect of low concentrations of benzoate on luminescence was limited when toluene was detected by T7-lux-E. coli or SP6-lux-E. coli.
Figure 4c illustrates the effects of toluene analogs and their concentrations on the luminescence of T7-lux-E. coli and SP6-lux-E. coli. The results demonstrated that the various concentrations of o-xylene and p-xylene had negligible effects on toluene detection by the recombinant E. coli biosensor; moreover, even when 250 μM o-xylene was used, only 4.15–4.30% increase in luminescence was observed. However, 250 μM m-xylene and 250 μM benzene induced T7-lux-E. coli or SP6-lux-E. coli to produce relatively high luminescence (12.3–12.5% and 14.3–14.6%, respectively). By contrast, the effect of the toluene analog concentration of ≤200 μM on toluene detection was limited (< 8%). The effect of the synergistic mode was far lower than that observed in the P. putida mt-2 KG1206 biosensor [12].
Taken together, these results illustrate that our recombinant luminescent biosensor possesses high selectivity and specificity when detecting a group of analytes with similar chemical structures. Because the included chemicals mainly affect the regulatory genes xylS or xylR, but not the T3, SP6, or T7 promoter, their effects on the magnitude of luminescence among all three recombinant E. coli biosensors were similar [12]. Figure 4 exemplifies the cases of T7-lux-E. coli and SP6-lux-E. coli.
Relationship of toluene concentration with luminescence
The function of these promoters (T7, T3, SP6) is to make the downstream reporter gene (lux) more strongly expressed. Therefore, xylR is first induced in the presence of toluene and activates gene expression, then the promoters and reporter gene (lux) follow. Under optimal operating conditions, we determined the relationships between the toluene concentration and the luminescence of the three recombinant E. coli strains. Two sets of linear relationships were observed between the toluene concentration and luminescence at different concentration ranges. Figure 5a presents a set of regression equations for the toluene concentration and the luminescence of T7-lux-E. coli, T3-lux-E. coli, and SP6-lux-E. coli when the toluene concentration was 10–500 μM: y = 6.140x + 724.9, y = 3.233x + 302.2, and y = 1.560x + 154.9, respectively. Figure 5b presents another set regression equations for T7-lux-E. coli, T3-lux-E. coli, and SP6-lux-E. coli when the toluene concentration was ≤10 μM: y = 40.515x + 46.9, y = 11.666x + 24.5, and y = 7.868x + 17.6, respectively. The coefficients of determination for these equations was high (> 0.99), indicating their reliability. The concentration-dependent differences in these linear relationships may have been due to differences in promoter characteristics [36]. Moreover, for T7-lux-E. coli, T3-lux-E. coli, and SP6-lux-E. coli, the LODs for toluene were 0.05, 0.2, and 0.5 μM, respectively. Therefore, T7-lux-E. coli was the most sensitive. Willardson et al. (1998) constructed a bacterial biosensor with the reporter gene luc, Casavant et al. (2003) constructed a site-specific recombination-based biosensor with tbuA1UBVA2C promoter, Li et al. (2008) constructed a lux-based bacterial biosensor, Zeinoddini et al. (2010) constructed a aequorin-based E. coli biosensor, Zhong et al. (2011) constructed a monooxygenase biosensor, and Ray et al. (2018) constructed a protein-based biosensor; their LODs for toluene were 10, 0.2, 7.5, 1, 3, and 3.3 μM, respectively [13, 20–22, 37, 38]. Compared with the aforementioned biosystems, T7-lux-E. coli has lower LOD (0.05 μM), indicating acceptable sensitivity. To develop a biosensor for detecting toluene, reporter genes such as luc, lux, and aequorin were often constructed downstream of the degradation gene. However, these biosensors could not measure trace levels of toluene contamination in wastewater. To improve the LOD, a promoter (T7, T3, or SP6) was inserted between the degradation gene and reporter gene. To our knowledge, little has been reported on applying this strategy to regulate the expression of the reporter gene and improve the LOD of a biosensor for toluene. In conclusion, the novel plasmid or biosensor with low LOD constructed here exhibited high potential for measuring bioavailable toluene.
Fig. 5.
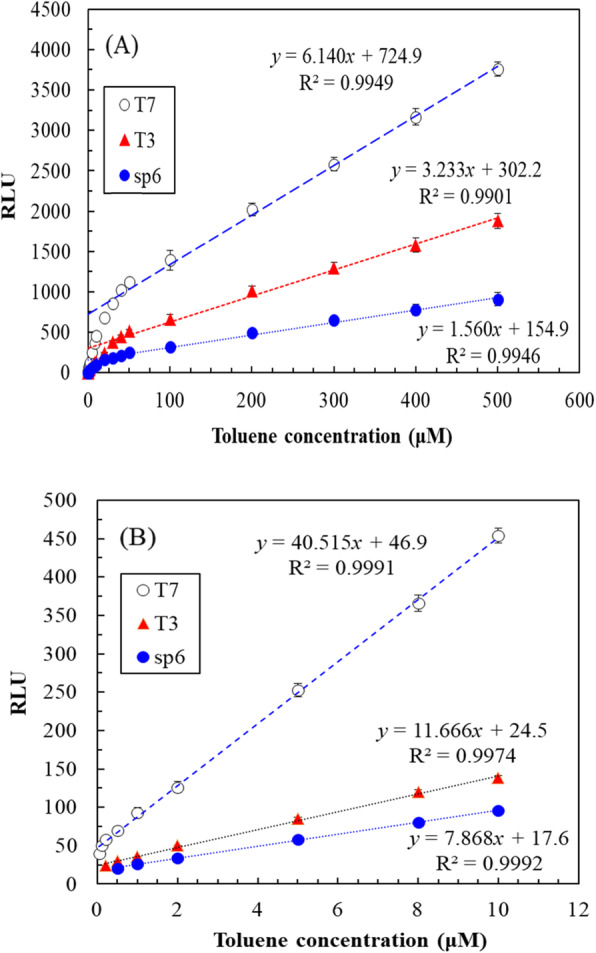
Relationship between toluene concentration [(a) 0.01–500 and (b) 0.05–10 μM] and luminescence of recombinant E. coli with different promoters (initial cell concentration: 5 × 107 cfu/mL, culture media: TMM, operational condition: 37 °C and 200 rpm, incubation time: 2 h for T3/T7-lux-E. coli and 1 h for SP6-lux-E. coli)
Hence, on the basis of the aforementioned reliable equations or the calibration curve for 0.05–10 or 10–500 μM toluene, the toluene concentration in the water samples can be rapidly determined. In addition, the broad detection ranges of T7-lux-E. coli indicate that it is a practical toluene measurement tool.
Reproducibility
To evaluate the reproducibility of the biosensors for detecting toluene, T7-lux-E. coli, T3-lux-E. coli, and SP6-lux-E. coli were tested under identical conditions by using TMM containing 10 μM toluene. Relative standard deviation (RSD) for T7-lux-E. coli, T3-lux-E. coli, and SP6-lux-E. coli was 4.3, 5.1, and 5.8%, respectively (n = 10). Batch-to-batch variation was also tested by comparing the luminescence from the five sets, which was tested using TMM containing 10 μM toluene, and the RSD for T7-lux-E. coli, T3-lux-E. coli, and SP6-lux-E. coli was 6.2, 6.5, and 9.4%, respectively. These results are comparable to the reproducibility reported for two induction-based toluene biosensors: RSD = 9.5% for n = 3 (with 21.7 μM toluene) and RSD = 7.4% for n = 8 (with 92 μM toluene) [38, 39]. Thus, our recombinant luminescent E. coli biosensors demonstrated operational stability. Similar results were obtained when these biosensors were applied for measuring 10 μM toluene after a 3-month cryogenic storage period.
Toluene measurement in groundwater and river water by using our three recombinant luminescent E. coli biosensors
Most luminescent biosensors have been applied for measuring toluene availability in artificial wastewater, but few have been applied in actual wastewater. Table 1 summarizes the measured toluene concentrations in seven groundwater samples and three river water samples using our three recombinant luminescent E. coli biosensors and the standard GC–MS method. The results demonstrated that the toluene concentration determined using our biosensors and through GC–MS demonstrated excellent correlation (r2 > 0.998); moreover, the deviation between the toluene concentrations measured through GC–MS and those measured using T7-lux-E. coli, T3-lux-E. coli, and SP6-lux-E. coli was − 14.3 to 9.1%, − 10.7 to 26.7%, and − 3.6 to 4.2%, respectively. Considering the measurement ranges and accuracy, T7-lux-E. coli provided the accurate and reliable toluene measurement in these aqueous matrices. However, under appropriate toluene concentration ranges, SP6-lux-E. coli could be the best biosensor in terms of accuracy, and its genetic assembly is relatively less susceptible to environmental interference [26]. The measurement deviation of T7-lux-E. coli and SP6-lux-E. coli were comparable to that (− 16.7 to 7.5%) of electrochemical inhibition bacterial sensor array for toluene detection [40]. Taken together, these results indicate that the developed recombinant luminescent bacterial biosensors can determine toluene concentration in different water bodies.
Table 1.
Toluene measurement from groundwater and river water by using the GC–MS method and biosensors
| Groundwater | River water | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GC–MS | 0.15a | 0.56 | 1.20 | 9.5 | 5.6 | 15.6 | 0.082 | 0.12 | 20.6 | 31.5 |
| T7-biosensor | 0.16 (6.7%)b | 0.61 (8.9%) | 1.31 (9.1%) | 8.6 (−9.5%) | 4.8 (−14.3%) | 15.2 (−2.6%) | 0.078 (−4.9%) | 0.13 (8.3%) | 18.5 (−10.2%) | 30.6 (−2.9%) |
| T3-biosensor | NDc | 0.50 (−10.7%) | 1.52 (26.7%) | 10.1 (6.3%) | 6.1 (8.9%) | 16.5 (5.8%) | ND- | ND- | 21.8 (5.8%) | 32.6 (3.5%) |
| SP6-biosensor | ND- | 0.54 (−3.6%) | 1.25 (4.2%) | 9.8 (3.2%) | 5.8 (3.6%) | 15.9 (1.9%) | ND- | ND- | 20.9 (1.5%) | 32.1 (1.9%) |
aUnit: μM
bDeviation compared with GC–MS-measured value
cND meaning Not Detected, the value < LOD
Conclusions
In this study, recombinant luminescent E. coli biosensors containing different promoters (T3, T7, and SP6) positioned before the reporter gene lux were developed for the accurate measurement of toluene concentrations in groundwater and river water. Of these biosensors, T7-lux-E. coli was the most sensitive to toluene, with optimal LOD and widest measurement range for toluene concentrations. Moreover, SP6-lux-E. coli had the shortest reaction time and highest adaptability to environmental interference but the poorest LOD. T7-lux-E. coli exhibited competitive advantages over previously reported biosystems, particularly for optimal LOD and wide measurement range. According to the results of reproducibility experiments and the test on actual water samples, our lux-based biosensors exhibited the high operational stability (i.e., low RSD) and acceptable measurement deviation. In conclusion, our biosensors, particularly T7-lux-E. coli, are sensitive, reliable, specific, and stable systems for preliminary in-field detection of toluene in water samples.
Materials and methods
Bacterial strains, gene cloning, and biosensor plasmid construction
To clone the tol gene, partial tol in P. putida (ATCC 33015) was amplified using the primer set (forward 5′-GTTAACTGCATCCAGCCC-3′, reverse 5′-CCGGGCGATGCCAACCC-3′) through polymerase chain reaction (PCR). To clone T3-lux, SP6-lux, or T7-lux, lux in Vibrio vulnificus was amplified with the primer set for the corresponding genes (T7-lux, forward 5′- TAATACGACTCACTATAGGTCGACTTTATCGAGCCTGA-3′ and reverse 5′-CAGCTGTTTTTGCTCCT-3′; T3-lux, forward 5′- ATTAACCCTCACTAAAGGTCGACTTTATCGAGCCTGA-3′ and reverse 5′-CAGCTGTTTTTGCTCCT-3′; SP6-lux, forward 5′-ATTTAGGTGACACTATAGGTCGACTTTATCGAGCCTGA-3′ and reverse 5′-CAGCTGTTTTTGCTCCT-3′) through PCR. All the resultant DNA fragments were inserted into the pET-15b vector plasmid (Promega, Madison, WI, USA). The recombinant plasmids were named pTOL, pT3-lux, pSP6-lux, and pT7-lux. In brief, the plasmids were then transferred to the expression host E. coli DH5α and plated on Luria–Bertani (LB) agar plates. Then, isolated pTOL, pT3-lux, pSP6-lux, and pT7-lux plasmids were cut at cleavage sites using BstEII/HindIII and HindIII/SanI. Next, pTOL-T3-lux, pTOL-SP6-lux, and pTOL-T7-lux were constructed by ligating pTOL to pT3-lux, pSP6-lux, and pT7-lux fragments by using T4 DNA ligase (New England BioLabs, Beverly, MA, USA), respectively. The resulting plasmids were inserted into the pET-15b vector plasmid. Next, the plasmids were transformed into E. coli DH5α to create the corresponding whole-cell biosensors. All the restriction enzymes were purchased from New England BioLabs. Vector DNA was prepared using the QIAEX II gel extraction kit (Qiagen, Hilden, Germany).
Bacterial growth
E. coli with pTOL-T3-lux (T3-lux-E. coli), pTOL-SP6-lux (SP6-lux-E. coli), and pTOL-T7-lux (T7-lux-E. coli) (all initial concentration = 2 × 105 cfu/mL) were cultivated in LB broth containing 50 mg/L ampicillin at 37 °C at 200 rpm on an orbital shaker. Overnight cultures were then diluted 100-fold into toluene-mineral medium (TMM) containing 0.43 g/L K2HPO4, 0.23 g/L KH2PO4, 1 g/L NH4NO3, 0.2 g/L MgSO4.7H2O, 0.1 g/L CaCl2, 0.05 mg/L Fe2(SO4)3, 0.25 mg/L NaMoO4.2H2O, 50 mg/L ampicillin, and a specific concentration of toluene (in this case: 10 mg/L). The cultures were incubated at 37 °C at 200 rpm on an orbital shaker. The optical density (OD) measurements of the bacterial growth and the luminescence intensity released from recombinant E. coli were conducted at specific intervals. The OD of the cultures was measured at 600 nm on a UV-vis spectrophotometer (Shimadzu, Kyoto, Japan). The luminescence intensity [in relative light units (RLU)] was measured by adding 200 μL of the culture to a 96-well microplate and then placing it under a microplate luminometer (Titertek-Berthold, Pforzheim, Germany). All chemicals used in the experiment were of analytical grade (purity > 99%). Toluene was purchased from Sigma-Aldrich Corporation (St. Louis, MI, USA).
Determination of optimum conditions
After 12 h of cultivation in TMM, 1 mL of culture containing 5 × 107 cfu/mL T3-lux-E. coli, SP6-lux-E. coli, or T7-lux-E. coli was inoculated into 200 mL of TMM [with different final concentrations (50–500 μM) of toluene] and incubated at 37 °C on an orbital shaker (200 rpm) for 5 h. The luminescence intensity was continuously measured until the luminescence intensity approached zero. The effects of temperature and ionic strength on bioluminescence emissions of the three recombinant E. coli strains were evaluated separately, and 100 μM toluene was used as an inducer in TMM. During incubation, temperature (15–40 °C) was controlled using thermostat, and ionic strength (0.04–0.55 M) was adjusted using aqueous NaCl. After 2-h incubation for T3-lux-E. coli and T7-lux-E. coli and 1-h incubation for SP6-lux-E. coli, 200 μL of the cultures were sampled, and the luminescence intensity (in RLU) of these biosensors was measured immediately. On average, 20-min consecutive measurements were recorded (i.e., one measurement every 0.5 s).
Various carbon sources (i.e., acetate, lactate, glucose, sucrose, fructose, glycerin, succinate, citrate, and pyruvate) were added to TMM to evaluate their effects on bioluminescence emissions of the three recombinant E. coli strains. In medium, final concentrations of coexisting carbon sources and toluene were 100 μM. After 2-h incubation for T3-lux-E. coli and T7-lux-E. coli and 1-h incubation for SP6-lux-E. coli, the luminescence intensity of each biosensor was measured immediately. Toluene analogs (i.e., benzene, o-xylene, p-xylene, and m-xylene) and intermediates of toluene degradation (benzoate) were added to TMM to evaluate the effects on the bioluminescence emissions of the three recombinant E. coli strains. Based on their solubility, o-xylene, p-xylene, and m-xylene were predissolved in 95% ethanol and added to TMM. The final concentrations of the toluene analogs, benzoate, and toluene in medium were 50–250, 50–300, and 100 μM, respectively. The cells were incubated for 2 h (T3-lux-E. coli and T7-lux-E. coli) or 1 h (SP6-lux-E. coli) at 37 °C; the luminescence intensity (in RLU) of these biosensors was then measured, as described above. Measurements were obtained from at least three independent experiments, each performed at least in triplicate.
Establishment of calibration curve and measurement of real water sample
To establish the relationships between the toluene concentration and the luminescence intensity of the three recombinant E. coli biosensors, we mixed 100 μL of toluene (0.01–500 μM), 50 μL of 4× TMM (without toluene), and 50 μL of recombinant luminescent E. coli cells (final concentration after mixing: 5 × 107 cfu/mL). We then operated at the optimal incubation time and conditions determined in previous experiments. Standard curves (known as calibration curves) were plotted from the linear regression of average luminescence intensity at each toluene concentration. To obtain the LOD concentration, we calculated the SD from the average of the three blank measurements, multiplied the SD by 3, and then used the standard curve to determine the LOD concentration. To ensure that the established curves and methods were valid, we prepared similar solutions as mentioned above, but used groundwater (from Lin-Yuan Industrial Park, Kaohsiung City, Taiwan) and river water (from Tamsui River, New Taipei City, Taiwan) instead of pure toluene. The toluene concentration in the prepared solution was separately measured using the established GC–mass spectrometry (MS) method [41] as well as using our three recombinant E. coli biosensors. Considering practical application, the retention of illuminance of recombinant E. coli after its cryogenic storage is essential for biosensor usage; thus, similar experiments were conducted when the biosensors were cryogenically stored for 3 months. Data were obtained from at least three independent experiments, with each performed at least in triplicate.
Acknowledgments
The authors thank J.T. Kuo and Y.J. Liao for help with partially analytical measurements.
Authors’ contributions
All authors collaborated to carry out the work presented here. Conceptualization, Guey-Horng Wang and Ying-Chien Chung; Formal analysis, Chun-Chi Kui and Chiu-Yu Cheng; Funding acquisition, Ying-Chien Chung; Investigation, Teh-hua Tsai, Chun-Chi Kui and Tzu-Ling Huang; Writing – original draft, Ying-Chien Chung; Writing – review & editing, Guey-Horng Wang, Teh-hua Tsai and Ying-Chien Chung All authors read and approved the manuscript.
Funding
This research was supported by NSC 99–2815-C-157-005-E and MOST 109–2313-B-157-001.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guey-Horng Wang, wanggh@livemail.tw.
Teh-Hua Tsai, Email: thtsai@ntut.edu.tw.
Chun-Chi Kui, Email: cckui2020@gmail.com.
Chiu-Yu Cheng, Email: cycheng@cc.cust.edu.tw.
Tzu-Ling Huang, Email: tlhuang2020bb@gmail.com.
Ying-Chien Chung, Email: ycchung@cc.cust.edu.tw.
References
- 1.Barberes GA, dos Reis RP, Spigolon ALD, Fonseca PE, de Mello CB, Barata MT. Groundwater natural contamination by toluene in Beja and Faro districts. Portugal Geosciences. 2018;8:9. doi: 10.3390/geosciences8010009. [DOI] [Google Scholar]
- 2.Wang X, Bartha R. Effects of bioremediation on toxicity, mutagenesis, and microbiota in hydrocarbon-contaminated soils. In Remediation of Hazardous Waste Contaminated Soils, Wise, D.L.; Trantolo, D.J., Eds; Marcel Dekker; New York, USA, 1998, pp. 175–197.
- 3.EPA in Taiwan. Groundwater Pollution Control Standards. 2013. https://law.moj.gov.tw/ENG/LawClass/LawAll.aspx?pcode=O0110006. Accessed 15 Mar 2020.
- 4.EPA in Taiwan. Drinking Water Quality Standards. 2017. https://law.moj.gov.tw/ENG/LawClass/LawAll.aspx?pcode=O0040019. Accessed 20 Feb 2020.
- 5.Namieśnik J. Trends in environmental analytics and monitoring. Crit Rev Anal Chem. 2000;30:221–269. doi: 10.1080/10408340091164243. [DOI] [Google Scholar]
- 6.Kuncová G, Ishizaki T, Solovyev A, Trögl J, Ripp S. The repetitive detection of toluene with bioluminescence bioreporter Pseudomonas putida TVA8 encapsulated in silica hydrogel on an optical fiber. Materials. 2016;9:467. doi: 10.3390/ma9060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Mozaz S, de Alda MJL, Barceló D. Biosensors as useful tools for environmental analysis and monitoring. Anal Bioanal Chem. 2006;386:1025–1041. doi: 10.1007/s00216-006-0574-3. [DOI] [PubMed] [Google Scholar]
- 8.Xu TT, Close DM, Sayler GS, Ripp S. Genetically modified whole-cell bioreporters for environmental assessment. Ecol Indic. 2013;28:125–141. doi: 10.1016/j.ecolind.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashemi GN, Mirzaei H, Sahebkar A, Poursadeghiyan M, Malekshahi ZV MA, Negahdari B. Biosensors for the detection of environmental and urban pollutions. J Cell Biochem. 2018;119:207–212. doi: 10.1002/jcb.26030. [DOI] [PubMed] [Google Scholar]
- 10.Cheng CY, Kuo JT, Lin YC, Liao YR, Chung YC. Comparisons of Vibrio fischeri, Photobacterium phosphoreum, and recombinant luminescent using Escherichia coli as BOD measurement. J Environ Sci Health A. 2010;45:233–238. doi: 10.1080/10934520903430020. [DOI] [PubMed] [Google Scholar]
- 11.Gui Q, Lawson T, Shan S, Yan L, Liu Y. The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics. Sensors. 2017;17:1623. doi: 10.3390/s17071623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong IC. Effects of binary mixtures of inducers (toluene analogs) and of metals on bioluminescence induction of a recombinant bioreporter strain. Sensors. 2014;14:18993–19006. doi: 10.3390/s141018993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray S, Panjikar S, Anand R. Design of protein-based biosensors for selective detection of benzene groups of pollutants. ACS Sens. 2018;3:1632–1638. doi: 10.1021/acssensors.8b00190. [DOI] [PubMed] [Google Scholar]
- 14.Burlage RS. Emerging technologies: Bioreporters, biosensors, and microprobes. In: Hurst CJ, editor. Manual of Environmental Microbiology. 2. Washington, DC: ASM; 2002. pp. 147–157. [Google Scholar]
- 15.Zhou Y, Fang Y, Ramasamy RP. Non-covalent functionalization of carbon nanotubes for electrochemical biosensor development. Sensors. 2019;19:392. doi: 10.3390/s19020392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim MN, Park HH, Lim WK, Shin HJ. Construction and comparison of Escherichia coli whole-cell biosensors capable of detecting aromatic compounds. J Microbiol Methods. 2005;60:235–245. doi: 10.1016/j.mimet.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 17.Burlage RS, Tillmann J. Biosensors of bacterial cells. J Microbiol Methods. 2017;138:2–11. doi: 10.1016/j.mimet.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 18.Axelrod T, Eltzov E, Marks RS. Bioluminescent bioreporter pad biosensor for monitoring water toxicity. Talanta. 2016;149:290–297. doi: 10.1016/j.talanta.2015.11.067. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura H. Current status of water environment and their microbial biosensor techniques-part II: recent trends in microbial biosensor development. Anal Bioanal Chem. 2018;410:3967–3989. doi: 10.1007/s00216-018-1080-0. [DOI] [PubMed] [Google Scholar]
- 20.Willardson BM, Wilkins JF, Rand TA, Schupp JM, Hill KK, Keim P, Jackson PJ. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl Environ Microbiol. 1998;64:1006–1012. doi: 10.1128/AEM.64.3.1006-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li YF, Li FY, Ho CL, Liao VHC. Construction and comparison of fluorescence and bioluminescence bacterial biosensors for the detection of bioavailable toluene and related compounds. Environ Pollut. 2008;152:123–129. doi: 10.1016/j.envpol.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Zeinoddini M, Khajeh K, Behzadian F, Hosseinkhani S, Saeedinia AR, Barjesteh H. Design and characterization of an aequorin-based bacterial biosensor for detection of toluene and related compounds. Photochem Photobiol. 2010;86:1071–1075. doi: 10.1111/j.1751-1097.2010.00775.x. [DOI] [PubMed] [Google Scholar]
- 23.Akbar M, Narayanan S, Restaino M, Agah M. A purge and trap integrated microGC platform for chemical identification in aqueous samples. Analyst. 2014;139:3384–3392. doi: 10.1039/C4AN00254G. [DOI] [PubMed] [Google Scholar]
- 24.Eun HM. Enzymology Primer for Recombinant DNA Technology. San Diego: Elsevier Science Publishing Co Inc; 1996. RNA polymerases; pp. 491–566. [Google Scholar]
- 25.Martin CT, Esposito EA, Theis K, Gong P. Structure and function in promoter escape by T7 RNA polymerase. Prog Nucleic Acid Res Mol Biol. 2005;80:323–347. doi: 10.1016/S0079-6603(05)80008-X. [DOI] [PubMed] [Google Scholar]
- 26.Zhu LP, Li ZF, Sun X, Li SG, Li YZ. Characteristics and activity analysis of epothilone operon promoters from Sorangium cellulosum strains in Escherichia coli. Appl Microbiol Biotechnol. 2013;97:6857–6866. doi: 10.1007/s00253-013-4830-0. [DOI] [PubMed] [Google Scholar]
- 27.Dollard MA, Billard P. Whole-cell bacterial sensors for the monitoring of phosphate bioavailability. J Microbiol Methods. 2003;55:221–229. doi: 10.1016/S0167-7012(03)00164-7. [DOI] [PubMed] [Google Scholar]
- 28.de Las HA, de Lorenzo V. Engineering whole-cell biosensors with no antibiotic markers for monitoring aromatic compounds in the environment. Methods Mol Biol. 2012;834:261–281. doi: 10.1007/978-1-61779-483-4_17. [DOI] [PubMed] [Google Scholar]
- 29.Akkermans S, Logist F, Van Impe JF. An interaction model for the combined effect of temperature, pH and water activity on the growth rate of E. coli K12. Food Res Int. 2018;106:1123–1131. doi: 10.1016/j.foodres.2017.11.026. [DOI] [PubMed] [Google Scholar]
- 30.Blazeck J, Alper HS. Promoter engineering: recent advances in controlling transcription at the most fundamental level. Biotechnol J. 2013;8:46–58. doi: 10.1002/biot.201200120. [DOI] [PubMed] [Google Scholar]
- 31.Marqués S, Gallegos MT, Manzanera M, Holtel A, Timmis KN, Ramos JL. Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J Bacteriol. 1998;180:2889–2894. doi: 10.1128/JB.180.11.2889-2894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallegos MT, Marqués S, Ramos JL. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from σ70-dependent promoter or from alpha σ70- and σ54-dependent tandem promoters according to the compound used for growth. J Bacteriol. 1996;178:2356–2361. doi: 10.1128/JB.178.8.2356-2361.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong IC, Suh H, Yang Z, Burlage RS. A bioluminescent reporter strain utilizing the lower pathway promoter (pm) of the xyl operon of Pseudomonas: optimization of a bioassay for m-toluate. Adv Environ Res. 2004;8:647–654. doi: 10.1016/S1093-0191(03)00037-6. [DOI] [Google Scholar]
- 34.Ikuma K, Gunsch C. Effect of carbon source addition on toluene biodegradation by an Escherichia coli DH5α transconjugant harboring the TOL plasmid. Biotechnol Bioeng. 2010;107:269–277. doi: 10.1002/bit.22808. [DOI] [PubMed] [Google Scholar]
- 35.Martínez-Lavanchy PM, Müller C, Nijenhuis I, Kappelmeyer U, Buffing M, McPherson K, Heipieper HJ. High stability and fast recovery of expression of the TOL plasmid-carried toluene catabolism genes of Pseudomonas putida mt-2 under conditions of oxygen limitation and oscillation. Appl Environ Microbiol. 2010;76:6715–6723. doi: 10.1128/AEM.01039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Espinosa-Urgel M, Serrano L, Ramos JL, Ferna’ndez-Escamilla AM. Engineering biological approaches for detection of toxic compounds: a new microbial biosensor based on the Pseudomonas putida TtgR Repressor Mo Biotechnol 2015;57:558–564. [DOI] [PubMed]
- 37.Casavant NC, Thompson D, Beattie GA, Phillips GJ, Halverson LJ. Use of a site-specific recombination-based biosensor for detecting bioavailable toluene and related compounds on roots. Environ Microbiol. 2003;5:238–249. doi: 10.1046/j.1462-2920.2003.00420.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhong Z, Fritzsche M, Pieper SB, Wood TK, Lear KL, Dandy DS, Reardon KF. Fiber optic monooxygenase biosensor for toluene concentration measurement in aqueous samples. Biosens Bioelectron. 2011;26:2407–2412. doi: 10.1016/j.bios.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 39.Stiner L, Halverson LJ. Development and characterization of a green fluorescent protein-based bacterial biosensor for bioavailable toluene and related compounds. Appl Environ Microbiol. 2002;68:1962–1971. doi: 10.1128/AEM.68.4.1962-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abu-Ali H, Nabok A, Smith TJ. Electrochemical inhibition bacterial sensor array for detection of water pollutants: artificial neural network (ANN) approach. Anal Bioanal Chem. 2019;411:7659–7668. doi: 10.1007/s00216-019-01853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.USEPA . Method 624.1: Purgeables by GC/MS. 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.