SUMMARY
A certain number of epithelial cells in intestinal crypts are DNA damage resistant and contribute to regeneration. However, the cellular mechanism underlying intestinal regeneration remains unclear. Using lineage tracing, we show that cells marked by an Msi1 reporter (Msi1+) are right above Lgr5high cells in intestinal crypts and exhibit DNA damage resistance. Single-cell RNA sequencing reveals that the Msi1+ cells are heterogeneous with the majority being intestinal stem cells (ISCs). The DNA damage-resistant subpopulation of Msi1+ cells is characterized by low-to-negative Lgr5 expression and is more rapidly cycling than Lgr5high radiosensitive crypt base columnar stem cells (CBCs). This enables an efficient repopulation of the intestinal epithelium at early stage when Lgr5high cells are not emerging. Furthermore, relative to CBCs, Msi1+ cells preferentially produce Paneth cells during homeostasis and upon radiation repair. Together, we demonstrate that the DNA damage-resistant Msi1+ cells are cycling ISCs that maintain and regenerate the intestinal epithelium.
Graphical Abstract
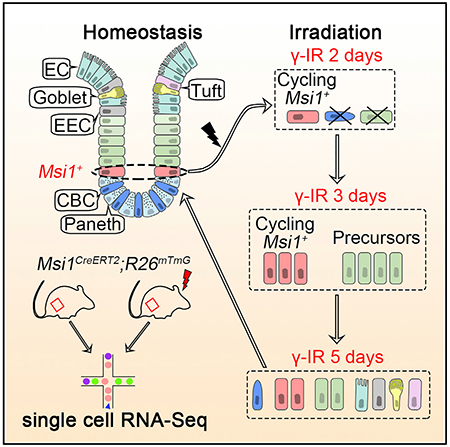
In Brief
Quiescent reserve stem cells in the intestine are thought to activate following irradiation to restore the depleted Lgr5high CBCs. Now, Sheng et al. demonstrate that cycling Msi1+ cells represent DNA damage-resistant ISCs that support efficient repopulation of the intestinal epithelium at the early stage of post-radiation repair, ahead of Lgr5high CBCs.
INTRODUCTION
The intestinal epithelium is a single-layer tissue organized into repetitive crypt-villus units. The cells that drive homeostatic intestinal renewal reside at the bottom of the crypt and move upward toward the villus tip, where they eventually die—a process referred to as the conveyer-belt model (Heath, 1996). The intestinal epithelium undergoes rapid turnover, with the majority of epithelial cells replaced in 3 to 5 days in mice (Heath, 1996). The rapid turnover of intestinal epithelial cells renders them sensitive to irradiation. Consequently, patients undergoing radiation therapy to the abdomen, pelvis, or rectum develop acute enteritis, displaying symptoms that include pain, bloating, nausea, fecal urgency, diarrhea, and rectal bleeding (Stacey and Green, 2014). Mucosal healing is critical for the remission of DNA damage-induced enteritis. Therefore, elucidating the cellular mechanisms of mucosal healing is necessary to develop new therapies for post-radiation enteritis.
Intestinal stem cells (ISCs), which reside within the proliferative compartment of crypts, are responsible for both intestinal homeostasis and epithelial regeneration after radiation exposure (Barker, 2014; Li and Clevers, 2010). Multiple studies have shown the existence of two functionally distinct ISC populations (Barker, 2014; Gehart and Clevers, 2015; Li and Clevers, 2010): mitotically active Lgr5high ISCs, commonly known as crypt base columnar stem cells, or CBCs (Barker et al., 2007; Sato et al., 2009), and more dormant, reserve ISCs, defined as +4 cells due to their location within crypts (Li et al., 2014; Montgomery et al., 2011; Powell et al., 2012; Sangiorgi and Capecchi, 2008; Takeda et al., 2011; Tian et al., 2011). Although CBCs mainly function to maintain physiological homeostasis of intestinal epithelium (Barker et al., 2007; Sato et al., 2009), they are also thought to be indispensable for epithelial regeneration (Metcalfe et al., 2014). In vitro, a single Lgr5high CBC can form a mini-gut structure that contains all mature intestinal cell types (Sato et al., 2009). Therefore, CBCs have been proposed to be bona fide ISCs. In contrast, considerable controversy exists regarding the precise identity of +4 cells and their lineage relationship to CBCs. It remains unclear whether +4 cells are bona fide ISCs. Several markers of +4 cells, including Bmi1, mTert, and Hopx, have been identified by in vivo lineage tracing, either by knockin of CreER into the gene or by randomly integrated transgenesis (Barker, 2014; Montgomery et al., 2011; Sangiorgi and Capecchi, 2008; Takeda et al., 2011; Tian et al., 2011). In contrast to CBCs, +4 cells are resistant to DNA damage and are activated to promote epithelial regeneration upon radiation-induced CBC depletion. In addition to +4 cells, progenitors of secretory and absorptive cells also contribute to the regeneration of damaged intestinal epithelium at relatively low efficiency (Buczacki et al., 2013; Tetteh et al., 2016; van Es et al., 2012; Yu et al., 2018). Importantly, +4 cells are thought to be reserve ISCs, and their cell-cycle quiescence has been proposed to be the main source of their radioresistance. The primary evidence for +4 cells’ quiescence is that the +4 position corresponds to label retaining cells (Potten et al., 1974, 2002), and +4 cells expressing Bmi1, Hopx, and mTert undergo slower kinetics of lineage tracing in comparison to Lgr5-expressing ISCs (Montgomery et al., 2011; Powell et al., 2012; Takeda et al., 2011; Yan et al., 2012). Further evidence is that HopxCreER cells were shown to reside in G0 (Li et al., 2016). However, three independent studies have demonstrated that label-retaining cells are, in fact, terminally differentiated Paneth cells or secretory progenitors (Buczacki et al., 2013; Li et al., 2016; Steinhauser et al., 2012). Additionally, it is worth mentioning that the primary DNA damage repair pathway in quiescent stem cells is non-homologous end joining (NHEJ), which is nonspecifically activated at all cell-cycle stages, and that it is error prone and unfavorable for tissue repair (Mohrin et al., 2010). In comparison, homologous recombination (HR)-mediated acute DNA repair can only occur in cycling cells during late Sand G2 phases (Maity et al., 1994; Moynahan and Jasin, 2010; Shaltiel et al., 2015). Therefore, the identity of +4 cells and the mechanisms underlying +4 cell-mediated epithelial regeneration remain uncertain.
Here, we generated an Msi1CreERT2 allele for lineage tracing and observe that Msi1CreERT2-marked cells are enriched at the +4 position in intestinal crypt, referred to as Msi1+ cells, and are resistant to DNA damage. Single-cell RNA sequencing (scRNA-seq) of Msi1+ cells further revealed that a subset of S/G2-phase stem cells, characterized by low-to-negative Lgr5 expression, exhibit DNA-damage resistance and repopulate radiation-damaged epithelium at early stage when Lgr5high cells are not emerging, which substantially differs from the classic theory that such +4 cells function as reserve stem cells, activate following irradiation to restore the depleted Lgr5high CBCs first, and then nascent CBCs rapidly divide to repair damaged intestinal epithelium. Furthermore, we observed that Msi1+ cells preferentially produce the Paneth lineage, relative to CBCs.
RESULTS
An Msi1 Reporter Is Enriched for DNA Damage-Resistant ISCs
Msi1 has been identified as a marker for ISCs, including both CBCs and +4 cells (Kayahara et al., 2003; Li et al., 2015). We first validated Msi1 expression pattern in CBCs and +4 cells at the protein level using immunohistochemistry (Figure S1A). At the RNA level, Msi1 expression was the strongest in +4 cells (Figure S1B). To track the fate of Msi1-expressing cells within intestinal epithelium, we generated a tamoxifen (TAM)-inducible Cre (CreERT2) knockin targeted just before the stop codon of the endogenous Msi1 locus (Figure S1C). We then crossed Msi1CreERT2 mice with R26Lax-Stop-Lox-LacZ (R26RLacZ) reporter mice. Fifteen hours after one pulse of TAM, X-gal staining showed that 15.7% of intestinal crypts were labeled, and Msi1 reporter-marked cells were mainly located at the +4 position of intestinal crypts (Figures 1A and 1B), which were further corroborated in Msi1CreERT2;R26mTmG mice (Figure 1C). Thus, Msi1CreERT2-marked cells are largely positionally distinct from Lgr5high CBCs (Figures 1A and 1B).
Figure 1. Msi1 Reporter-Marked Cells Are Enriched at +4 Position in Intestinal Crypts.
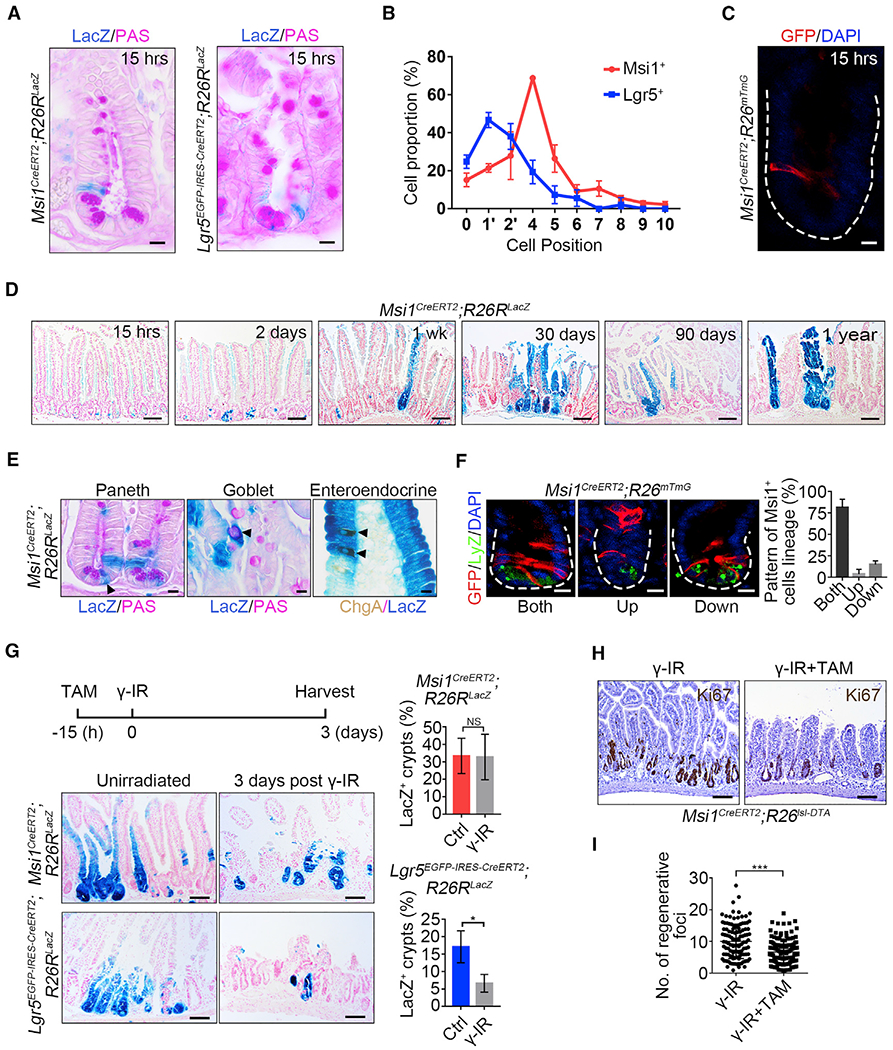
(A) Representative images of LacZ+ cells in Msi1CreERT2;R26RLacZ (302 crypts; n = 3 mice) and Lgr5EGFp-IRES-CreERT2;R26RLacZ (175 crypts; n = 3 mice) lineage-labeled small intestinal crypts 15 h after TAM induction. Scale bar, 10 μm.
(B) Quantification of the position of LacZ+ cells in intestinal crypts in (A). Data represent the mean value ± SD.
(C) Representative images of GFP+ cells in Msi1CreERT2;R26mTmG lineage-labeled small intestinal crypts 15 h after TAM induction. n = 3 mice. Scale bar, 10 μm.
(D) Low-magnification images of the LacZ+ ribbon in Msi1CreERT2;R26RLacZ lineage-labeled small intestine at different time points following TAM induction. n ≥ 3 mice at each time point. Scale bar, 40 μm.
(E) Periodic acid-Schiff (PAS) staining and immunohistochemistry for ChgA in Msi1CreERT2;R26RLacZ lineage-labeled small intestine 1 week after TAM induction. n = 3 mice. Scale bar, 10 μm.
(F) Double immunofluorescence for GFP and lysozyme in Msi1CreERT2;R26mTmG lineage-labeled small intestinal crypts one day after TAM induction. The position of GFP+ cells is above the +4 position, referred to as “Up”; below the +4 position, referred to as “Down”; above and below the +4 position, referred to as “Both.” Quantification of the lineage pattern of Msi1-reporter+ cells (n = 91 crypts; n = 3 mice). Data represent the mean value ± SD. Scale bar, 10 μm.
(G) Representative images of LacZ+ ribbons in Msi1CreERT2;R26RLacZ (Ctrl-720 crypts, n = 3 mice; γ-IR-540 crypts, n = 4 mice) and Lgr5EGFP-IRES-CreERT2;R26RLacZ (Ctrl-808 crypts, n = 3 mice; γ-IR-852 crypts, n = 4 mice) lineage-labeled small intestines 4 days after TAM induction, or the mice were irradiated after 15h of TAM exposure, and harvested 3 days after 12 Gy γ-IR. Scale bar, 10 μm. Quantification of LacZ+ ribbons under the indicated conditions. Data represent the mean value ± SD. NS, not significant; *p < 0.05 (Student’s t test).
(H) Msi1CreERT2;R26Isl-DTA mice irradiated 15 h after TAM induction and then harvested 3 days after γ-IR. Immunohistochemistry for Ki67 under the indicated conditions. Scale bar, 100 μm.
(I) Quantification of regenerative foci in (H) (Ctrl, n = 117 crypts, n = 3 mice; DTA, n = 139 crypts, n = 3 mice). Data represent the mean value ± SD. ***p < 0.001 (Student’s t test).
See also Figures S1 and S2.
2 days after TAM induction, most labeled crypts contained 3–7 cells exhibiting β-galactosidase activity (Figure 1D). One week after induction, X-gal staining became more widespread (Figures 1D and S1D), and the labeling cells included differentiated cell lineages—Paneth, goblet, and enteroendocrine cells (EECs) (Figure 1E). The number of fully labeled crypt-villus ribbons and the percentage of LacZ+ crypts sustained over time (Figures 1D and S1E), and Msi1 reporter marked progeny existed for at least 1 year (Figure 1D). Next, we sought to examine how Msi1 reporter-marked cells give rise to distinct cell lineages. We quantified the positions of labeled cells 1 day after TAM induction, a time point when newly generated cells are emerging, and found that the majority of labeled cells move both upward and downward relative to +4 positions (Figure 1F). This distribution suggests that Msi1 reporter-marked cells concomitantly give rise to distinct lineages, including CBCs, Paneth cells, and villus cells. The expression of CBC markers and Wnt target genes (Lgr5, Axin2, Sox9, and Olfm4) in Msi1CreERT2-marked cells was similar to that of cells marked with HopxCreERT2, a well-established marker of +4 ISCs (Takeda et al., 2011), and distinct from that of Lgr5high CBCs (Figures S1F and S1G). Collectively, these data demonstrate that Msi1 reporter-marked cells are primarily located above the crypt base and the CBC compartment and exhibit multipotent stem cell properties.
To examine the DNA damage response by Msi1+ cells, we exposed Msi1CreERT2;R26RLacZ and Lgr5EGFP-IRES-CreERT2;R26R-LacZ mice to 12 Gy of ionizing radiation (γ-IR), 15 h after a single pulse of TAM. After γ-IR exposure, the number of LacZ+ ribbons produced by Msi1+ cells was similar to what we observed during homeostasis, whereas the number of LacZ+ ribbons from Lgr5 reporter-marked cells was strongly reduced (Figure 1G). Lineage-tracing analysis in Msi1CreERT2;R26mTmG mice also demonstrated a robust repopulating capacity of Msi1+ cells after exposure to γ-IR (Figure S1H). In order to rule out the interference of TAM remains, we isolated intestinal organoids of Msi1CreERT2;R26mTmG mice. The organoids were incubated with 4-hydroxytamoxifen (4-OH) for 9.5 h then replaced with 4-OH-free medium to make sure only the initial Msi1+ cells were labeled. Quantification of the position showed that the initial labeled Msi1+ cells were also enriched at +4 position (Figure S1I), similar to its pattern in vivo. Then, we exposed the labeled organoids from Msi1CreERT2;R26RmTmG and Lgr5EGFp-IRES-CreERT2;R26Isl-tdT mice to 6 Gy γ-IR to examine the regeneration ability of Msi1+ versus Lgr5+ cells. Six days after γ-IR, the number of traced organoids produced by Msi1+ cells was similar to that under control conditions, while the number of traced organoids from Lgr5+ cells was significantly reduced (Figures S2A–S2D). Those findings indicate that Msi1+ cells are radioresistant, able to survive γ-IR, and repopulate the damaged epithelium.
Furthermore, we used Msi1CreERT2;R26RIsl-DTA mice to examine the importance of Msi1+ cells during intestine damage regeneration. Twenty-four hours after a single pulse of TAM injection, apparent apoptosis was detected at the base of crypts (Figure S2E). We found that the depletion of Msi1+ cells significantly impaired intestinal epithelial regeneration following γ-IR (Figures 1H and 1I). Similarly, using Msi1CreERT2;R26mTmG;R26Isl-DTR mouse model, the depletion of Msi1 reporter+ cells is more efficient, and the impairment of intestinal regeneration becomes more obvious (Figures S2F–S2H). Taken together, these findings suggest that Msi1+ cells are DNA damage-resistant ISCs with the capacity to repopulate γ-IR-damaged epithelium.
Msi1+ Cells Are a Heterogeneous Population
Next, we sought to better characterize the identity of Msi1+ cells using scRNA-seq analysis. GFP-labeled cells from Msi1CreERT2; R26mTmG mice were sorted 15 h after TAM induction, and subjected to scRNA-seq (Figure S3A; Table S1). Unsupervised clustering (Duò et al., 2018) identified nine distinct cell clusters (Figure 2A). We utilized the differentially expressed gene signatures to assign putative cell type identities to these clusters (Figures 2B–2D, S3B, and S3C). Cluster H15h-C1 (at homeostasis, traced for 15 h) is enriched in cells expressing the highest levels of ISC marker gene Lgr5, as well as several other ISC marker genes, namely, Gkn3, Ascl2, Olfm4, Jun, Pdgfa, and 2210407c18Rik (Figures 2C and S3C). Thus, H15h-C1 cells were defined as Lgr5high ISCs. Clusters of H15h-C2 and H15h-C3 cells have low or negative Lgr5 status, but concomitantly express the ISC marker genes Igfbp4 and Ascl2 (Figure 2C), on which basis they are classified as Lgr5low/neg ISCs. In comparison to H15h-C2 cells, H15h-C3 cells highly express G2/M-phase marker genes (Figure 2D). Consistently, single-cell consensus clustering (SC3) analysis (Kiselev et al., 2017) of clusters H15h-C1, H15h-C2, and H15h-C3 revealed higher similarity between H15h-C2 and H15h-C3 cells, relative to H15h-C1 ISCs, and further divided H15h-C1 cells into two sub-clusters (Figure 2E). Cluster H15h-C4 cells are also enriched for G2/M-phase marker genes (Figure 2D), and principal component analysis (PCA) analysis shows that these cells are intermediate between ISCs and enterocytes (ECs) (Figure 2F). Thus, H15h-C4 cells were identified as EC precursor cells (EPs). The smaller clusters H15h-C5 through H15h-C9 were characterized as differentiated cells (Figure S3B). These differentiated cells are likely the early differentiated progeny of initially labeled Msi1+ cells produced over the 15-h period, suggesting that Msi1+ cells have started the differentiation program.
Figure 2. Msi1+ Cells Are a Heterogeneous Population.
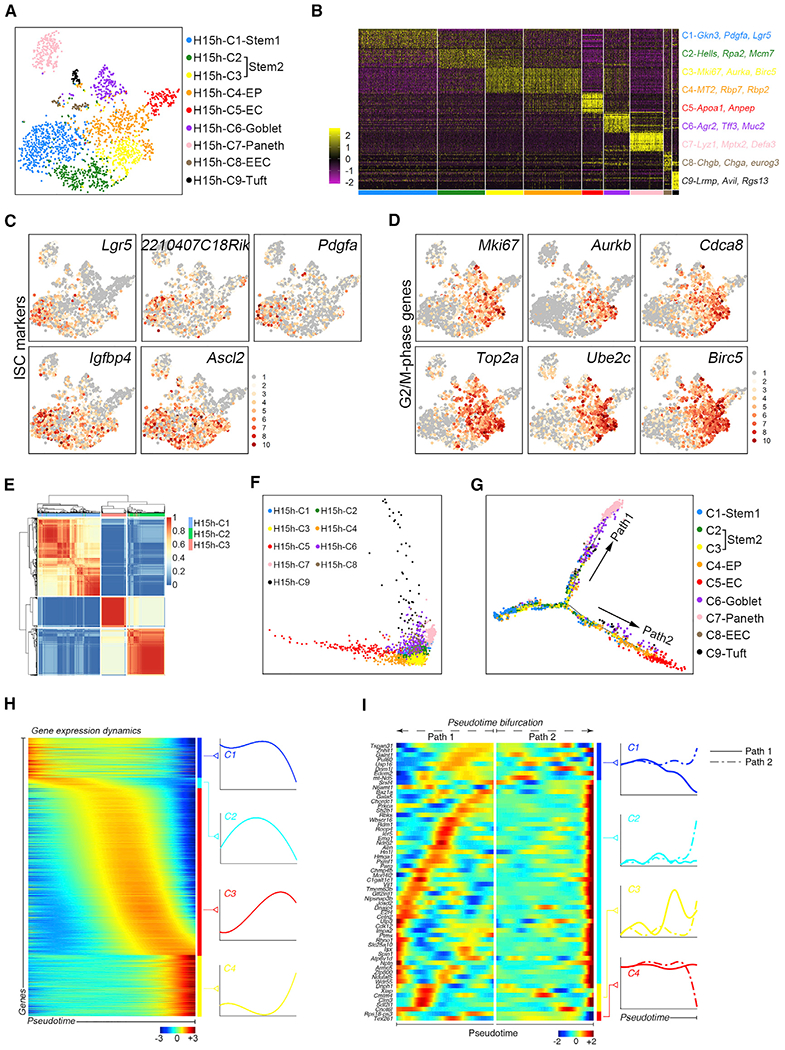
(A) A t-distributed stochastic neighbor embedding (t-SNE) plot revealed cellular heterogeneity of 2329 Msi1+ cells sorted from Msi1CreERT2;R26mTmG mice 15 h after TAM induction.
(B) Heatmap of differentially expressed genes in each cluster.
(C and D) Feature plots of expression distribution for ISC (C) and G2/M phase (D) marker genes.
(E) SC3 analysis showing the correlation of H15h-C1 to H15h-C3.
(F) PCA showing the association of distinct cell clusters.
(G) Pseudotime ordering on Msi1+ cells.
(H) scEpath analysis performed on pseudotime along the trajectory from stem cells to differentiated cells, identifying four gene clusters (C1–C4) of pseudotime-dependent genes.
(I) scEpath analysis identifying four gene clusters (C1–C4) of branching genes.
To understand the hierarchy among distinct cell clusters, we performed pseudo-temporal ordering of scRNA-seq data using Monocle 2, which places cells along putative differentiation trajectories (Qiu et al., 2017). This analysis arranged most ISCs from the H15h-C1, H15h-C2, and H15h-C3 clusters into a major pseudotime trajectory that bifurcates toward ECs and differentiated secretory cells (Figures 2G and S3D). Consistent with cluster identity attribution, ECs are preceded by EPs (H15h-C4 cells) in Path2 of the pseudotime (Figure 2G). A large number of genes were differentially expressed in cells along the pseudotime trajectory (Figure 2H). Among them, a number of “branching” genes were identified, which are potentially important for EC versus secretory cell differentiation (Figure 2I). The scRNA-seq data and its computational analysis suggest that the Msi1CreERT2 allele might marks a heterogenous population of cells, consisting primarily of ISCs and a small number of differentiated cells and residing along the two major differentiation trajectories.
Cycling ISCs Initiate Epithelial Regeneration
Next, we sought to define the initial cells that repopulate the epithelium after γ-IR exposure. A minimal number (1–2) of proliferating cells exist in each crypt at 2 days after γ-IR, followed by rapid proliferative expansion between 72–96 h (Figures S4A and S4B). We performed scRNA-seq on the progeny of Msi1+ cells from Msi1CreERT2;R26mTmG mice 2 days after γ-IR (refer to IR2), a time point marking the initiation of epithelial regeneration (Kim et al., 2017). Ten distinct cell clusters were identified (Figures 3A, 3B, and S4C–S4E; Table S1). Importantly, the distribution of known ISC marker genes changed dramatically (Figure 3C). Compared to the distribution of Msi1+ cells during homeostasis, the Lgr5high cell cluster was depleted 2 days after γ-IR (Figure 3C). Consistently, the number of Lgr5high cells becomes markedly reduced 2 days after γ-IR or treatment with the DNA replication inhibitor and chemotherapeutic agent 5-fluorouracil (5-FU) (De Angelis et al., 2006). In contrast, the number of Msi1+ cells showed an increasing trend, albeit not significant, upon these treatments (Figures 3D and S4F). Cluster IR2-C1 cells are identified as ISCs, because they strongly expressed ISC marker genes Igfbp4 and Ascl2 (Figure 3C). IR2-C2 cells were identified as a transition cluster due to their intermediate position between ISCs and differentiated cells (Figure 3E). IR2-C1 and -C2 cells are enriched for genes functioning on DNA damage response (DDR) and cell survival (Figures 3F and S4G), suggesting a strong DDR. In the pseudotime trajectory, IR2-C1 and IR1-C2 cells are enriched at the starting point of the major branch, whereas IR2-C3 and IR2-C4 cells are enriched at the end of EC branch, with the remaining cells enriched at the end of the secretory/differentiated branch (Figure 3G). Consequently, few cells localize around the pseudotime bifurcation as compared with normal physiological conditions (Figure 3G). Given that WNT pathway activation is critical for regeneration of damaged intestinal epithelium after γ-IR. The distribution of WNT downstream target genes Axin2 and Ascl2, as well as surface receptor genes for Wnt ligand, Lrp5 and Lrp6 showed that WNT pathway is activated in IR2-C1/C2 stem cells (Figures 3C and 3H). Importantly, IR2-C1 and IR2-C2 cells are cycling, whereas the other cells primarily reside in G0/G1 phase (Figures 3I and S4H; Tables S2 and S3). These data suggest that IR2-C1 and IR2-C2 cells are cycling ISCs that initiate epithelial regeneration.
Figure 3. Cycling ISCs Initiate Intestinal Epithelial Regeneration.
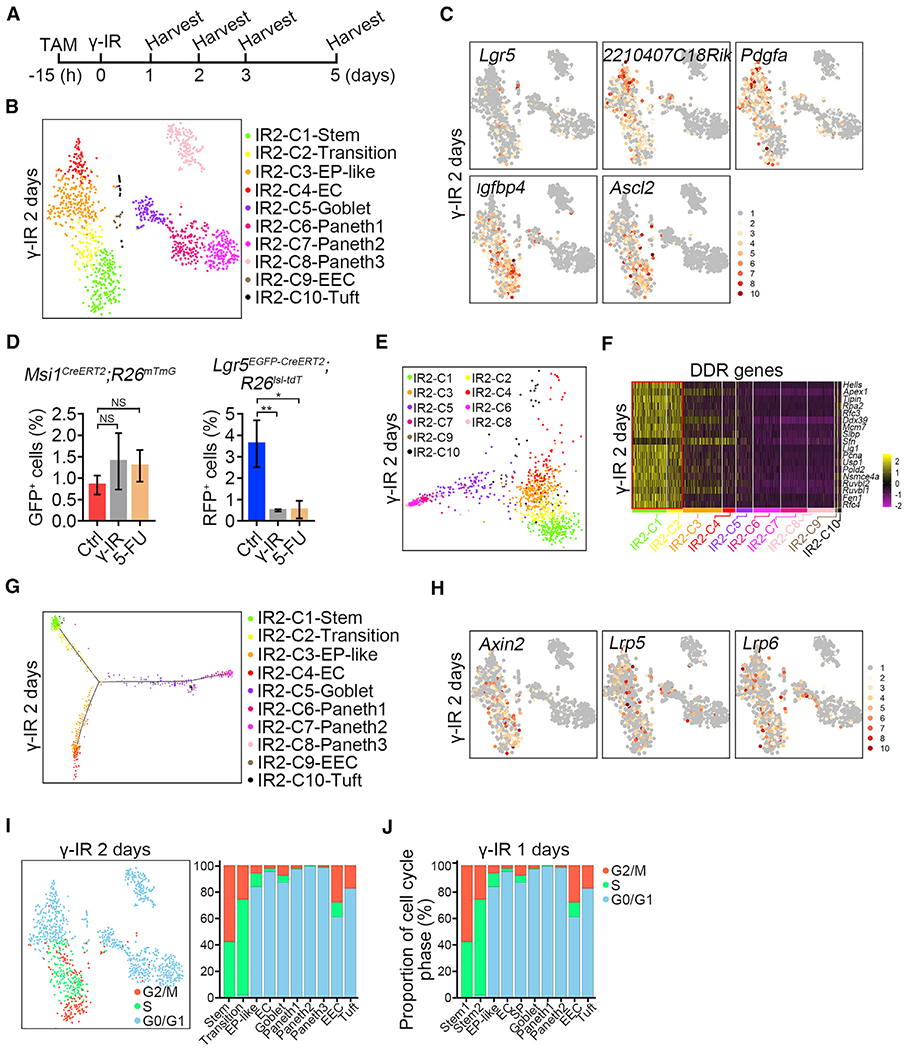
(A) Strategy of sample collection for scRNA-seq after γ-IR.
(B) A t-SNE plot revealed cellular heterogeneity of 1,335 Msi1+ cell progeny from Msi1CreERT2;R26mTmG mice 2 days after γ-IR. The mice were pretreated with TAM 15 h before γ-IR.
(C) Feature plots of expression distribution for ISC marker genes 2 days after γ-IR.
(D) Quantification of Msi1+ (n = 3 mice) and Lgr5+ (n = 3 mice) populations 2 days after treatment with γ-IR or 5-FU. Mice were treated with γ-IR or two consecutive doses of 5-FU and then induced by TAM 15 h before sacrifice, as shown in Figure S4F. Data represent the mean value ± SD. NS, not significant; **p < 0.01 (Student’s t test).
(E) PCA showing the association of distinct cell clusters 2 days after γ-IR.
(F) Heatmap of DDR genes in distinct clusters 2 days after γ-IR.
(G) Pseudotime ordering of Msi1+ cell progeny 2 days after γ-IR.
(H) Feature plots of expression distribution for WNT pathway-related genes 2 days after γ-IR.
(I) Cell-cycle metrics of Msi1+ cell progeny 2 days after γ-IR.
(J) Proportions of cell-cycle stages in each cluster 1 day after γ-IR.
Previous reports indicated that radioresistant +4 cells are dormant (Montgomery et al., 2011; Powell et al., 2012; Yan et al., 2012). Therefore, on scRNA-seq, we expected to see a certain number of quiescent ISCs (in G0/G1 phase), along with proliferatively active ISCs. In contrast, we found that ~42% of IR2-C1 cells were in S phase, and 58% were in G2/M phase. There were no G0/G1-phase IR2-C1 cells (Figure 3I; Table S2). IR2-C1 cluster also strongly expressed proliferating marker genes (Figure S4H; Table S3). These data suggest that quiescent ISCs are lacking at this stage. Next, we considered that 2 days after γ-IR might be too late to detect surviving quiescent ISCs. Thus, we performed scRNA-seq on the progeny of Msi1+ cells from Msi1CreERT2;R26mTme mice 1 day after γ-IR, a time point when the majority of intestinal cells are undergoing cell death. Two clusters of ISCs were identified (Figures S4I and S4J; Table S1), and surprisingly, they also exhibited a highly proliferative state, with no cells in G0/G1 phase (Figures 3J and S4K; Table S1). Together, these data demonstrate that cycling, rather than quiescent ISCs, initiate epithelial regeneration. It also raises the possibility that a population of cycling ISCs is resistant to and can survive γ-IR exposure.
Cycling Msi1+ ISCs Survive from Exposure to High Dose of γ-IR
To test whether cycling ISCs survive from exposure to high dose of γ-IR, we labeled S-phase Msi1+ cells using a 90 min pulse of 5-ethynyl-2′-deoxyuridine (EdU) at a very low dose of 0.017 mg/25 g body weight, which is insufficient to label all S-phase cells (Figure S5A), and then irradiated the mice. Indeed, we found that the labeled S-phase Msi1+ cells survived γ-IR and divided, and the EdU signals diluted over time (Figure 4A). We then went back to the homeostatic condition and analyzed cell-cycle phases of cells in intestinal crypts by quantifying the positions of PCNA+, EdU+, and pH3+ cells. Most cells in the +1 and +2 positions, which are usually considered to be Lgr5high CBCs (Barker et al., 2007), were in G1 phase (Figure 4B), whereas the cells from positions 4 to 6, referred to as Lgr5low/neg +4 cells with DNA damage resistance (Powell et al., 2012; Takeda et al., 2011), were in Sor G2/M phases (Figure 4B). We then analyzed the division kinetics using dual bromodeoxyuridine (BrdU)/EdU labeling and revealed that the average length of the cell cycle for +4 cells is 13.28 h (n = 177 crypts from 3 mice), whereas that of CBCs is 18.12 h (n = 249 crypts from 3 mice) (Figure 4C). Furthermore, EdU labeling assay revealed that more Msi1+ cells were in S phase compared to Lgr5high CBCs (Figures 4D and 4E). Similar findings were also observed in Hopx reporter marked cells (Figures 4D and 4E). In agreement with those results, the majority of Lgr5low/neg ISCs from H15h-C2 and H15h-C3 scRNA-seq clusters reside in S and G2/M phases during homeostasis, whereas the majority of Lgr5high ISCs reside in G1 phase (Figure 4F; Table S2). It has been shown that the signaling pathways regulating the DDR also activate during normal S phase for genome integrity maintenance (Ben-Yehoyada et al., 2007), and this property can increase cellular resistance to DNA damage. Accordingly, DDR genes are specifically enriched in cycling H15h-C2 cells (Figures 4G and S5B). H15h-C2 and H15h-C3 cells are enriched for genes functioning in cell survival and stress, which might facilitate cell survival after exposure to γ-IR (Figure S5C). The HR-mediated DNA repair only occurs in cycling cells at S and G2 phases, enabling an accurate repair of DNA damage (Maity et al., 1994; Moynahan and Jasin, 2010; Shaltiel et al., 2015). We found that the key components of HR-type repair such as Rad51, Rad51ap1, Brca1, Brca2, and Smc6 are highly expressed in the ISCs populations 1 and 2 days after γ-IR (Figures 4H and S5D), suggesting a strong HR-type repair response in S- and G2-phase ISCs. Taken together, these findings strongly suggest that the DNA damage-resistant Msi1+ cells are more rapidly cycling than Lgr5high CBCs.
Figure 4. Msi1+ Cells Are More Rapidly Cycling Compared to CBCs.
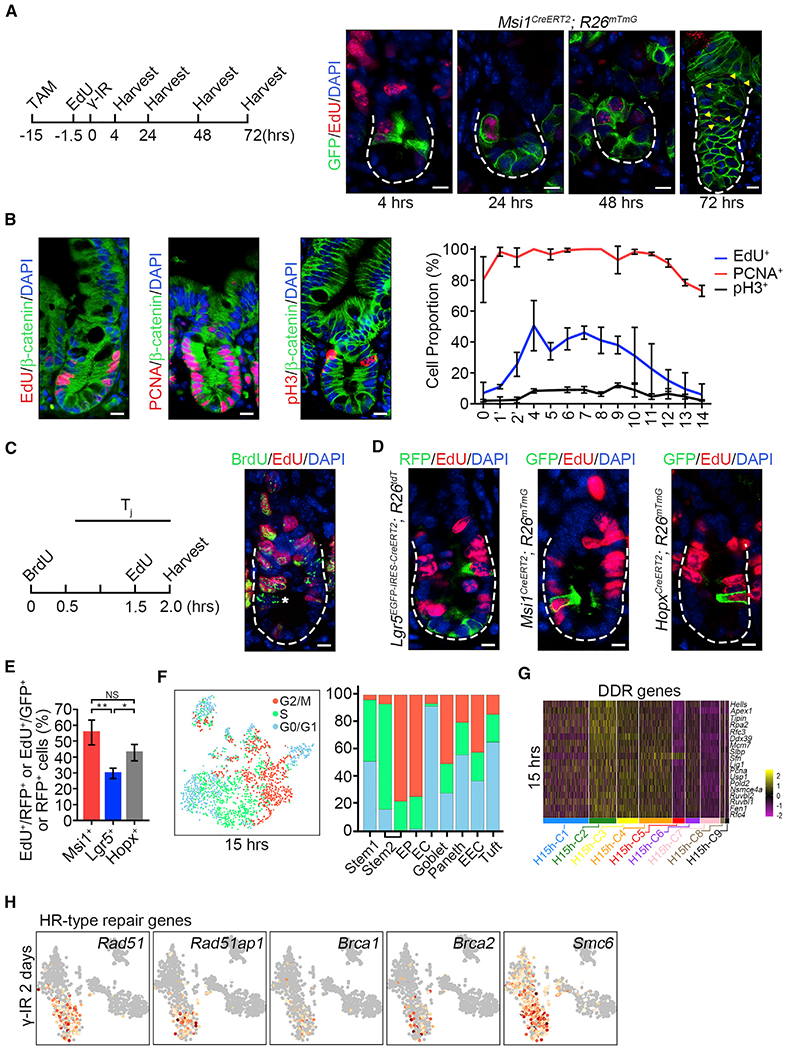
(A) Strategy of testing whether EdU-labeled S-phase Msi1+ cells survive from γ-IR exposure. Msi1CreERT2;R26mTmG mice were treated by TAM and labeled using a 90-min pulse of EdU at 0.017 mg/25 g bodyweight 13.5 h after TAM induction and then irradiated 15h after TAM induction. Immunofluorescence for EdU and GFP in intestinal crypts at the indicated time points. n = 3 mice at each time point. Scale bar, 10 μm.
(B) Immunofluorescence for EdU/β-catenin, PCNA/β-catenin, and phosphor-histone 3/β-catenin in the intestinal crypts of WT mice. Scale bar, 10 μm. Quantification of EdU+ (n = 100 crypts; n = 3 mice), PCNA+ (n = 59 crypts; n = 3 mice) and pH3+ (n = 202 crypts; n = 3 mice) cells at the indicated position of intestinal crypts. Data represent the mean value ± SD.
(C) Schematics of the EdU and BrdU temporary space pulse method to calculate the average length of cell-cycle times (left). Cells still in S phase during the labeled time were EdU+BrdU+, whereas cells that exited S phase were EdU−BrdU+, indicated by asterisks (right).
(D) Immunofluorescence for RFP and EdU in intestines from Lgr5EGFP-CreERT2;R26Isl-tdT mice 15 h after TAM induction, and immunofluorescence for GFP and EdU in intestines from Msi1CreERT2;R26mTmG and HopxCreERT2;R26mTmG mice 15 h after TAM induction. Scale bar, 10 μm.
(E) Quantification of RFP+/EdU+ cells in intestinal crypts (n = 268 cells; n = 3 mice) and GFP+/EdU+ cells in Msi1CreERT2;R26mTmG (n = 162 cells; n = 3 mice) and HopxCreERT2;R26mTmG (n = 200 cells; n = 3 mice) intestinal crypts in (D). Data represent the mean value ± SD. NS, not significant; *p < 0.05; **p < 0.01 (Student’s t test).
(F) Cell-cycle metrics of Msi1+ cells at homeostasis.
(G) Heatmap of DDR genes in distinct clusters of Msi1+ cells.
(F) Feature plots of expression distribution for the key genes functioning in HR-type DNA damage repair 2 days after γ-IR.
See also Figure S5.
Msi1+ Cells Repopulate the Intestinal Epithelium at Early Stage When Lgr5high Cells Are Not Emerging
To define the mechanism underlying Msi1+ ISC-mediated epithelial regeneration, we also performed scRNA-seq on Msi1+ cell progeny 3 and 5 days after γ-IR (refer to IR3 and IR5). Three days after γ-IR, considered as proliferative phase (Kim et al., 2017), ten distinct cell clusters were identified (Figures 5A, 5B, and S6A; Table S1). ISCs are subdivided into two clusters, IR3-C1 and IR3-C2. The first cluster is highly enriched for DDR genes (Figure 5C) and DNA helicases (Figure S6B), and most cells are in S phase (Figure S6C; Table S2). Compared to IR3-C1, IR3-C2 cells are characterized by reduced levels of DDR and DNA helicase genes (Figures 5C and S6B) and exhibit increased proliferative capacity, as evidenced by the enrichment for proliferating marker genes (Figure S6D; Table S3). Indeed, over 90% of IR3-C2 cells were in G2/M phase (Figure S6C). IR3-C3 cells localize in the EC branch before EP-like cells (Figure 5D), and most of them were in S and G2/M phases. Thus, IR3-C3 cells were identified as proliferating EPs. In comparison, IR3-C4 cells are dormant EP-like cells and are in G0/G1 phase (Figure S6C). Another important finding is that secretory precursors (SPs, IR3-C6) start to emerge at this stage. The cells are defined by DII1 expression (van Es et al., 2012) (Figure S6E), rapid proliferation (Figure S6C), and close relatedness to secretory differentiated cells in the pseudotime trajectory (Figure 5D) and on PCA analysis (Figure S6F). Many proliferating goblet cells were identified 3 days after γ-IR, compared to 2 days (Figures 5B and S6C). Interestingly, Lgr5high CBCs are not emerging at this stage (Figures 5B and S6G). In agreement, immunohistochemical assay showed that the proportion of Lgr5+ cells is the lowest 3 days after γ-IR (Figures 5E and S6H). Thus, it appears that surviving ISCs directly give rise to proliferating EPs and proliferating SPs at early stage when Lgr5high cells are not emerging.
Figure 5. Msi1+ Cells Repopulate the Intestinal Epithelium at Early Stage When Lgr5high Cells Are Not Emerging.
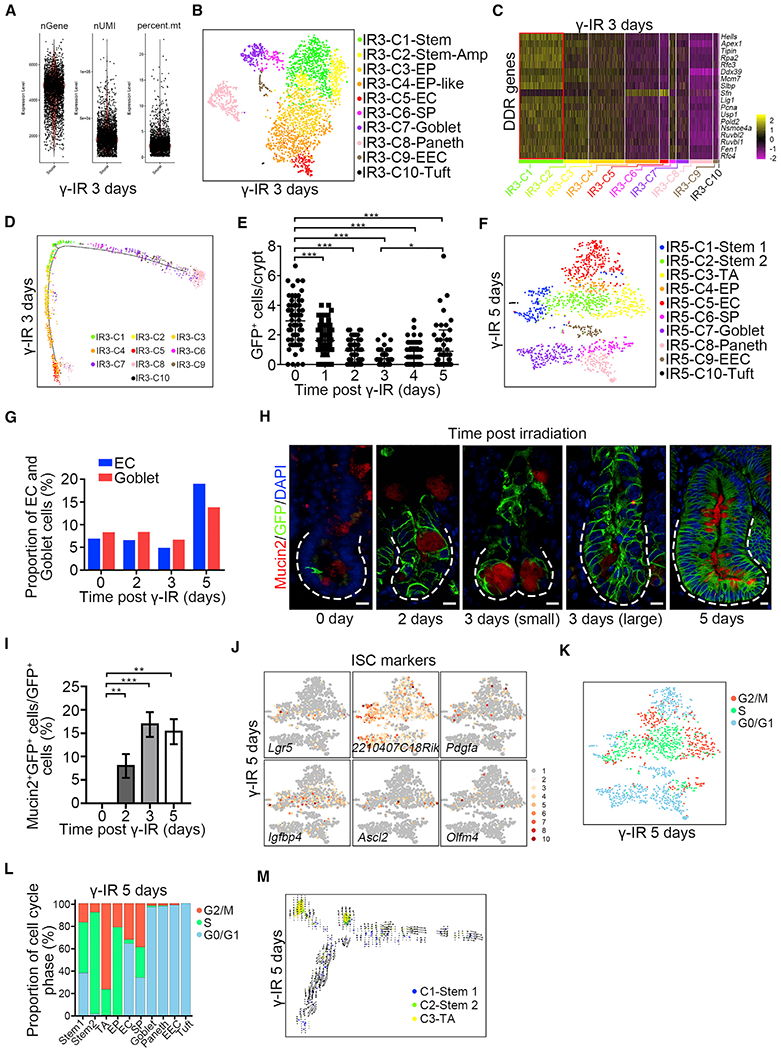
(A) scRNA-seq data quality control of Msi1+ cell progeny from Msi1CreERT2;R26mTmG mice 3 days after γ-IR. The mice were pretreated with tamoxifen 15 h before γ-IR.
(B) A t-SNE plot revealed cellular heterogeneity of 3124 Msi1+ cell progeny from Msi1CreERT2;R26mTmG mice 3 days after γ-IR.
(C) Heatmap of DDR genes in distinct clusters 3 days after γ-IR.
(D) Pseudotime ordering on Msi1+ cell progeny 3 days after γ-IR.
(E) Quantification of GFP+ cells in the intestinal crypts of Lgr5EGFP-IRES-CreERT2 mice at the indicated time points after γ-IR. 180 intestinal crypts (60 crypts/mouse, n = 3 mice) were quantified at each time point. Representative images were shown in Figure S6H. Data represent the mean value ± SD. *p < 0.05; ***p < 0.001 (Student’s t test).
(F) A t-SNE plot revealed cellular heterogeneity of 1,556 Msi1+ cell progeny from Msi1CreERT2;R26mTmG mice 5 days after γ-IR.
(G) The proportion of EC and goblet populations of scRNA-seq results at the indicated time points after γ-IR.
(H) Immunofluorescence for Mucin2 and GFP in Msi1CreERT2;R26mTmG normal intestinal crypts and regenerative foci at the indicated time points after γ-IR. “small” indicates small regenerative foci; “large” indicates large regenerative foci. Scale bar, 10 μm.
(I) Quantification of the percentage of Mucin2+GFP+ cells versus GFP+ cells (n = 365 crypts, n = 3 mice per chase time point) in (H). Data represent the mean value ± SD. **p < 0.01, ***p < 0.001 (Student’s t test).
(J) Feature plots of expression distribution for ISC marker genes 5 days after γ-IR.
(K) Cell-cycle metrics on Msi1+ cell progeny 5 days after γ-IR.
(L) Proportions of cell-cycle stages in each cluster 5 days after γ-IR.
(M) RNA velocity analysis of IR5-C1 to IR5-C3 across the pseudotime trajectory 5 days after γ-IR.
Five days after γ-IR, tissue enters the normalization phase (Kim et al., 2017), and dramatic changes were observed at this time in Msi1+ progeny on scRNA-seq (Figures 5F, S6I, and S6J; Table S1). Compared to 3 days after γ-IR, the populations of EC and goblet cells expand dramatically (Figures 5F and 5G; Table S4), whereas EP-like cells almost entirely disappear (Figure 5F). The increase in goblet cells was further confirmed by immunofluorescence (Figures 5H and 5I). Another striking finding was the emergence of a new type of stem cell (IR5-C1), which is very similar to the Lgr5high ISC population in physiology and is characterized by the enrichment of Lgr5+, 2210407C18Ric+, and Pdgfa+ accompanied by the appearance of Igfbp4+, Ascl2+, and Olfm4+ (Figure 5J). Similar to homeostatic Lgr5high ISCs, a large number of IR5-C1 cells reside in G1 phase (Figures 5K and 5L; Table S2). Furthermore, the RNA velocity (La Manno et al., 2018) on IR5-C1, IR5-C2, and IR5-C3 clusters revealed that IR5-C1 cells are likely derived from IR5-C2 and IR5-C3 cells (Figures 5M and S6K). Together, our data indicate that new IR5-C1 cells are nascent Lgr5high ISCs. Overall, we posit that during epithelial regeneration, surviving ISCs directly give rise to proliferative precursors of differentiated lineages, and only later do they regenerate relatively slowly cycling Lgr5high ISCs. We conclude that the Msi1+ cells repopulate the intestinal epithelium at early stage when Lgr5high cells are not emerging and give rise to nascent Lgr5high cells only at later time.
Msi1+ ISCs Preferentially Produce Paneth Cells
Another striking finding that drew our attention was the dynamic change in Paneth cells during epithelial regeneration. Two days after γ-IR, Paneth cells are the most abundant cell type, accounting for 39% (Figures 6A and S4D; Table S4), with this proportion decreasing to ~10% 3–5 days after γ-IR (Figures 6A, S7A, and S7B; Table S4). Lineage-tracing analysis revealed a large number of Msi1+ cell-derived Paneth cells (over 25%) residing in the regenerative unit 2 days after γ-IR (Figures 6B and 6C). Three days after γ-IR, small and large regenerative units existed (Figure 6B). The proportion of Msi1+ cell-derived Paneth cells is much higher in the small regenerative units than in the large ones (Figure 6D). On scRNA-seq, 2 days after γ-IR, Paneth cells can be divided into three distinct clusters based on marker genes (Figure 6E). Compared with type 1 and type 2 Paneth cells, type 3 cells exhibit increased levels of Gm14851 and Defa22 and reduced level of Mptx2. Expression levels of AY761184 and Defa3 appear to gradually increase from Paneth cell type 1 to type 3 (Figure S7C). Type 1 Paneth cells, which were transcriptionally closest to goblet cells, gradually changed to type 2 and finally to type 3 (Figure S4D). This finding suggests a gradual maturation process in the direction of Paneth cell type 1 to type 2 to type 3. We also noticed that Paneth cell markers, such as Lyz1, Defa17, and Gm15284 were expanded in the goblet cell population (Figure 6E), whereas they are usually specific for Paneth cells during homeostasis (Figure S3B). At this stage, Paneth cells are preferentially generated relative to goblet cells. Paneth cells have been identified as a niche for ISCs under physiological conditions (Sato et al., 2011). Accordingly, the ISC ligand Wnt3 was highly enriched in these Paneth cells (Figure 6F). Together, our findings indicate that Msi1+ cells preferentially give rise to Paneth cells upon exposure to γ-IR.
Figure 6. Msi1+ ISCs Preferentially Generate Paneth Cells 2 Days after γ-IR.
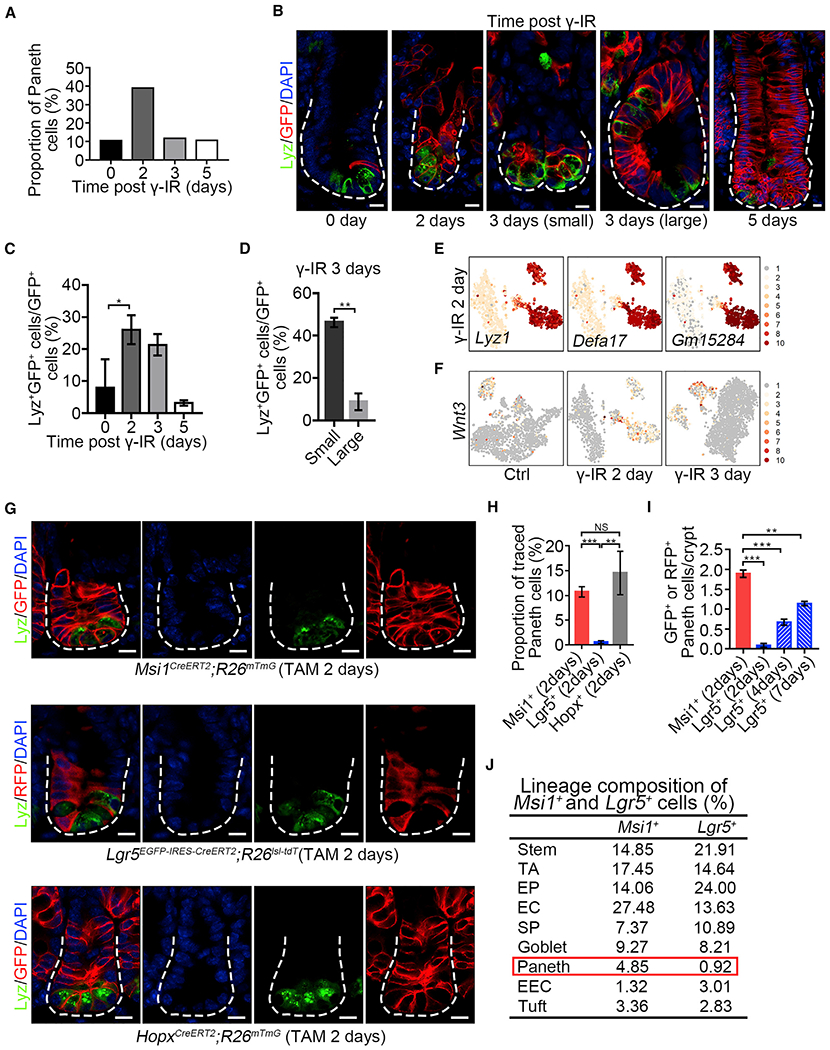
(A) The proportion of Paneth cells in scRNA-seq results at indicated time points after γ-IR.
(B) Immunofluorescence for lysozyme and GFP in normal intestinal crypts and regenerative foci from Msi1CreERT2;R26mTmG mice at indicated time points after γ-IR. “small” indicates small regenerative foci; “large” indicated large regenerative foci. Scale bar, 10 μm.
(C) Quantification of Lyz+/GFP+ versus GFP+ cells at indicated time points in (B) (n = 562 crypts, n = 3 mice per chase time point). Data represent the mean value ± SD. *p < 0.05 (Student’s t test).
(D) Quantification of Lyz+/GFP+ versus GFP+ cells in small (n = 133 crypts; n = 3 mice) and large (n = 66 crypts; n = 3 mice) regenerative foci 3 days after γ-IR. Data represent the mean value ± SD. **p < 0.01 (Student’s t test).
(E) Feature plots of expression distribution for Paneth cell marker genes 2 days after γ-IR.
(F) Feature plots of wnt3 distribution at indicated time points after γ-IR.
(G) Immunofluorescence for lysozyme and GFP/RFP in intestinal crypts from Msi1CreERT2;R26mTmG, Lgr5EGFP-CreERT2;R26Isl-tdT, and HopxCreERT2;R26mTmG mice 2 days after TAM induction. Scale bar, 10 μm.
(H) Quantification of Lyz+/GFP+ versus GFP+ or Lyz+/RFP+ versus RFP+ cells in (G) (n = 365 crypts, n = 3 mice per chase time point). Data represent the mean value ± SD. **p < 0.01; ***p < 0.001; NS, not significant (Student’s t test).
(I) Quantification of the number of GFP+ or RFP+ Paneth cells in each crypt from Msi1CreERT2;R26mTmG and Lgr5EGFP-CreERT2;R26Isl-tdT mice at indicated time points after TAM induction (n = 409 crypts, n = 3 mice per chase time point). Representative images are shown in Figure S7D. Data represent the mean value ± SD. **p < 0.01; ***p < 0.001 (Student’s t test).
(J) scRNA sequencing revealed lineage composition of Msi1CreERT2;R26mTmG and Lgr5EGFP-CreERT2;R26Isl-tdT mice 2 days after TAM induction. The t-SNE plots are shown in Figures S7G and S7H.
See also Figure S7.
Considering the increase in Msi1+ cell-derived Paneth cells 2 days after γ-IR, we sought to examine whether Msi1+ cells preferentially produce Paneth cells under normal physiological conditions. We quantified the number of Paneth cells after lineage tracing in Lgr5EGFP-IRES-CreERT2;R26Isl-tdT and Msi1CreERT2;R26mTmG mice 2 days after TAM induction. Strikingly, we found that the proportion of Msi1+ cell-derived Paneth cells is ~10.76%, whereas Lgr5+ cell-derived Paneth cells are just 0.58% (Figures 6G and 6H). We also observed that the proportion of Lgr5+ cell-derived Paneth cells increased with the lineage tracing time (Figures 6I and S7D), most likely due to the increase of Msi1+ cells derived from Lgr5+ cells with time. The finding of +4 cells preferentially generating Paneth cells was further confirmed by lineage tracing in HopxCreERT2;R26mTmG mice 2 days after TAM treatment (Figures 6G and 6H). These data suggest that Msi1+ cells preferentially generate Paneth cells as compared to Lgr5+ cells. To further confirm this idea, we performed scRNA-seq on labeled cells in intestinal crypt from Lgr5EGFP-IRES-CreERT2;R26Isl-tdT and Msi1CreERT2;R26mTmG mice 2 days after TAM induction. Consistently, we also found that the proportion of Paneth cells in Msi1+ cell progeny is much higher than that of the Lgr5+ progeny (Figures 6J and S7E–S7H; Table S4). In comparison, the proportions of goblet, tuft, and EC cells were similar between them (Figure 6J). Collectively, our findings strongly indicate that Msi1+ cells preferentially produce Paneth cells during homeostasis relative to Lgr5+ cells.
DISCUSSION
Our findings strongly indicate that the DNA damage-resistant subset of Msi1+ ISCs, most likely Lgr5low/neg ISCs, are more rapidly cycling than Lgr5high CBCs (Figure 7), rather than quiescent, which substantially differs from the current intestinal stem cell theory. Classically, +4 cells have been identified as quiescent ISCs, whereas Lgr5+ CBCs were thought to be rapidly cycling (Montgomery et al., 2011; Powell et al., 2012; Sangiorgi and Capecchi, 2008; Takeda et al., 2011; Yan et al., 2012). The notion of +4 ISCs dormancy was mainly supported by their colocalization with label-retaining cells in pulse-chase experiments. However, the +4 location of label-retaining cells (Potten et al., 1974, 2002) has been challenged by a number of subsequent studies. Three independent works demonstrated that the long-term label-retaining cells in intestinal crypts were Paneth cells, and short-term label-retaining cells were SPs undergoing commitment toward Paneth and EEC lineages (Buczacki et al., 2013; Li et al., 2016; Steinhauser et al., 2012). Likewise, Bmi1-expressing cells were recently identified as EEC lineage cells that possess ISC activity (Yan et al., 2017), although they were considered slow-cycling ISCs resistant to irradiation (Yan et al., 2012). Those conclusions contrast the notion that +4 cells are quiescent label-retaining ISCs. Our findings that +4 cells cycle faster than Lgr5high cells is further supported by recent work, showing that Lgr5high CBCs are in an unlicensed G1 phase, whereas most cells in the +4 to +8 positions are in S phase (Carroll et al., 2018).
Figure 7. A Model of Msi1+ Cells in Maintaining and Regenerating Intestinal Epithelium.
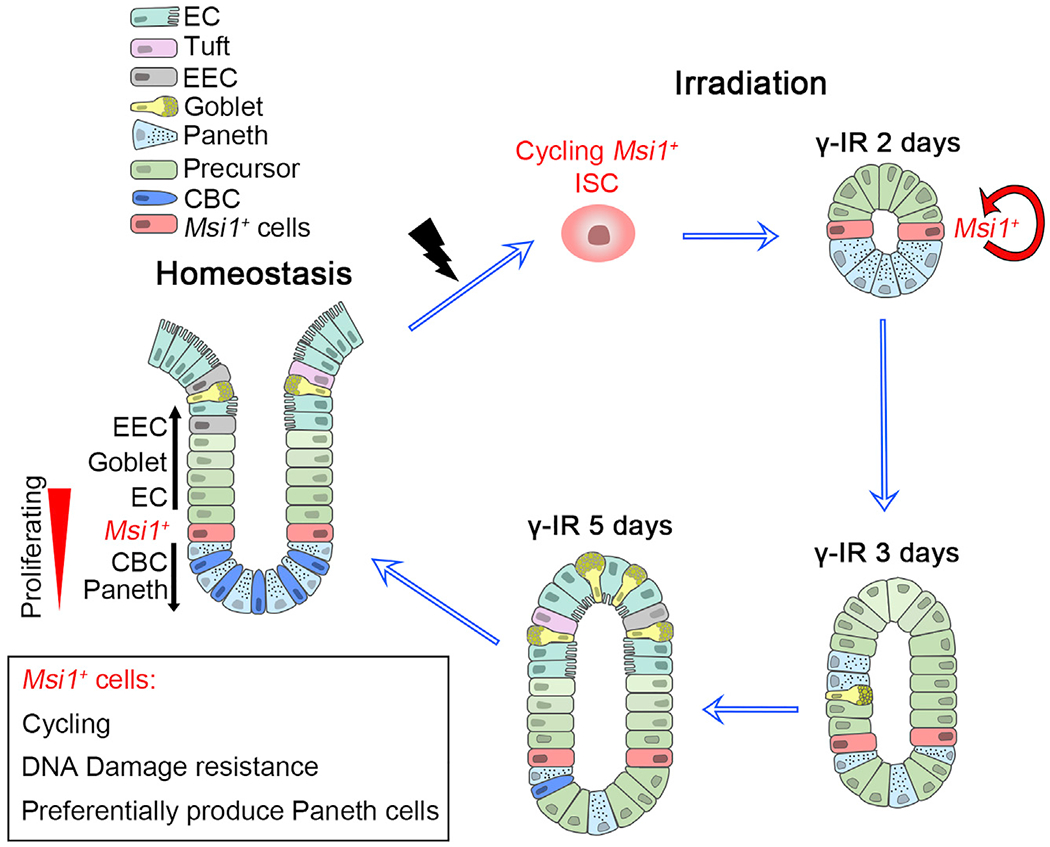
A subset of Msi1+ ISCs that exhibit DNA-damage resistance are cycling faster than Lgr5high CBCs, and fast repopulation of the intestinal epithelium at early stage when Lgr5high cells are not emerging. Msi1+ cells preferentially produce Paneth cells during homeostasis and upon radiation repair.
Classic +4 ISC theory states that quiescent +4 cells become activated in response to γ-IR. However, to the best of our knowledge, this idea lacks direct evidence. In contrast, it is well established that following γ-IR exposure, cells either transiently block cell-cycle progression to allow time for DNA repair, or exist cell cycle permanently (Shaltiel et al., 2015). G1 arrest, S-phase delay, or G2 arrest can all take place following γ-IR-induced damage. Importantly, G1 arrest typically occurs at lower doses of γ-IR, whereas S-phase delay and G2 arrest are common at higher doses to allow for cells to repair DNA damage (Maity et al., 1994). Accordingly, HR-mediated DNA repair, which enables an accurate repair using the sister chromatid as the template, can only occur in cycling cells during late S and G2 phases to repair DNA damage, making the cells survive from γ-IR exposure (Moynahan and Jasin, 2010). Another important factor in rendering S-phase cells resistant to DNA damage is that the signaling pathways regulating response to acute DNA damage also operate during normal S phase to maintain genome integrity in the presence of low levels of replication-associated damage (Ben-Yehoyada et al., 2007). Indeed, S-phase cells have been shown to be the least sensitive to γ-IR (Pawlik and Keyomarsi, 2004). Consistently, we found that DNA damage repair genes are enriched in a subset of Msi1+ cells during S and G2/M phases. Thus, we posit that the Msi1+ ISCs during S and G2/M phase possess the capacity to resist DNA damage.
Although our data indicate that it is the cycling Msi1+ ISCs that survive γ-IR exposure and repopulate damaged epithelium, we cannot formally rule out the previously proposed model that quiescent ISCs and/or precursors also contribute to epithelial regeneration (Ayyaz et al., 2019; Chaves-Pérez et al., 2019; Yan et al., 2017). It is noteworthy that, although many secretory progenitor cells, marked by DII1CreERT (van Es et al., 2012), Prox1CreERT (Yan et al., 2017), or H2B-label (Buczacki et al., 2013), have regenerative capacity, the contribution of these cells to epithelial regeneration are limited (Bankaitis et al., 2018). In comparison, the cycling Msi1+ ISCs might represent a primary source for regenerating intestinal epithelium. The quiescent LRCs can also generate Paneth cells and participate in the regeneration of damaged intestinal epithelium (Buczacki et al., 2013), but they are significantly different from Msi1+ ISCs. First, the majority of quiescent LRCs were secretory progenitors and committed to differentiated secretory cells mostly within a week (Buczacki et al., 2013). In comparison, the Msi1 reporter+ cells were cycling stem cells that can contribute the whole lineage of intestinal epithelium, and Msi1 reporter-marked progeny existed for at least 1 year. Second, only a few clones were traced by the initial labeled LRCs 2 weeks after damage (Buczacki et al., 2013), suggesting a low efficiency of regeneration. In comparison, Msi1 reporter+ cells enable a quick repopulation of the damaged intestinal epithelium with a high efficiency. Thus, the two types of cells are apparently different, both at homeostasis and damage regeneration. Furthermore, it is worth mentioning that the primary DNA damage repair pathway in quiescent stem cells in other tissues such as hematopoietic system — NHEJ — is error-prone, resulting in genome instability due to the accumulation of subtle mutations and chromosomal aberrations (Mohrin et al., 2010). If quiescent ISCs also use the same mechanism, many DNA mutations and chromosomal aberrations would exist in the surviving quiescent ISCs after γ-IR exposure. This would be detrimental to normal epithelial regeneration and would contribute to tumorigenesis. Therefore, we believe that cycling ISCs survive γ-IR exposure due to the high-fidelity HR-type repair.
Our data also demonstrate that the surviving Msi1+ cells repopulate damaged intestinal epithelium at early stage when Lgr5high cells are not emerging and give rise to nascent Lgr5high cells only at later time. This observation substantially differs from the prevailing idea that dormant surviving +4 cells function as reserve stem cells that, upon activation, generate rapidly cycling Lgr5high cells that then go on to produce all differentiated lineages (Li and Clevers, 2010). Indeed, in our lineage studies, progeny of Msi1+ cells can initially move both up and down the crypt relative to +4 positions in normal physiology, suggesting that they generate their progeny independent of Lgr5high CBCs during homeostasis. In agreement with our observation, classic cell migration tracing studies also demonstrated that all crypt cells ultimately derive from cells located at approximately the +4 position (Kaur and Potten, 1986; Potten, 1998; Qiu et al., 1994). In other words, Lgr5high ISCs are not the only direct progeny of +4 ISCs. Thus, we posit that a subset of Msi1+ cells might be bona fide ISCs responsible for both normal homeostasis and epithelial regeneration (Figure 7).
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Zhengquan Yu (zyu@cau.edu.cn).
Materials Availability
Mouse lines generated in this study are available upon request to Lead Contact provided the requestor covers shipping costs.
Data and Code Availability
All scRNA-seq data from this study are available at NCBI Gene Expression Omnibus (GEO). The accession number for data reported in this paper is GEO: GSE145866.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All mouse experiment procedures and protocols were evaluated and authorized by the Regulations of Beijing Laboratory Animal Management and were strictly in accordance with the guidelines of the Institutional Animal Care and Use Committee of China Agricultural University (approval number: SKLAB-2015-04-03). Msi1CreERT2 mice were generated at the Model Animal Research Center of Nanjing University. Lgr5EGFP-IRES-CreERT2, R26mTmG, R26tdT and R26RLacZ mice were purchased from Jackson Laboratories (stock number: 008875, 007676, 007914, 009427). HopxCreERT2 mice were obtained from John Epstein’s laboratory at the University of Pennsylvania. R26Isl-DTA mice were obtained from Sen Wu’s laboratory at China Agricultural University. R26Isl-DTR mice were obtained from Hua Zhang’s laboratory at China Agricultural University. To evaluated the identity of Msi1+ cells, Msi1CreERT2 mice were crossed with the mouse models listed above and the detailed description were shown in the figure legends and method details. Male and female age-matched mice between 8-10 weeks were utilized for all experiments.
METHOD DETAILS
Lineage tracing
For lineage tracing, eight-week-old mice were injected with a single pulse of tamoxifen (4 mg/25 g body weight, Sigma-Aldrich, T5648). To label the Msi1+ cells at homeostasis, Msi1CreERT2;R26mTmG mice were administered with tamoxifen for fifteen hours before sacrifice. For the injury study, Msi1CreERT2;R26RLacZ and Lgr5EGFP-IRES-CreERT2;R26RLacZ mice were treated with 12 Gy γ-IR fifteen hours after a single pulse of tamoxifen, and sacrificed at indicated time points. In order to examine the survival after high doses of irradiation or cytotoxic damage, Msi1CreERT2;R26mTmG, and Lgr5EGFP-IRES-CreERT2;R26Isl-tdT mice were injected intraperitoneally with two doses of 5-FU (100 mg/Kg body weight, Sigma-Aldrich, F6627) within two days or 12 Gy γ-IR once and analyzed with FACS after two days. To exam the influence with the absence of Msi1+ cells during regeneration, Msi1CreERT2;R26mTmG;R26Isl-DTR mice model were treated with TAM every other day, and four consecutive DT induction (50 μg/Kg body weight, Sigma-Aldrich, D0564). Twenty four hours after the last DT injection, Msi1CreERT2;R26mTmG and Msi1CreERT2;R26mTmG;R26Isl-DTR mice were exposed to 12 Gy γ-IR and analyzed three days after irradiation. For cell proliferation assay, Msi1CreERT2;R26mTmG and Lgr5EGFP-IRES-CreERT2;R26Isl-tdT mice were intraperitoneally injected with EdU (0.2 mg/25 g body weight, Thermo Fisher, A10044) for 1.5 hours before sacrifice.
To test whether S-phase Msi1+ cells survived from exposure of 12Gy γ-IR, Msi1CreERT2;R26mTmG were pretreated with tamoxifen, intraperitoneally injected with EdU (0.017 mg/25 g body weight) 13.5 hours after tamoxifen induction, and then exposed to 12 Gy γ-IR 1.5 hours after EdU injection. The intestinal samples were harvested four hours, one day, two days and three days after exposure to γ-IR.
LacZ staining
Tissues were fixed in fixative solution (0.2% glutaraldehyde, 5 mM EGTA, 2 mM MgCl2 in PBS) for two hours on ice, rinsed for ten minutes with detergent rinsing solution (0.02% NP40, 0.01% sodium deoxycholate, 2 mM MgCl2 in PBS) for three times and immersed in X-gal staining solution (5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 0.02% NP40, 0.01% sodium deoxycholate, 2 mM MgCl2 1 mg/mL X-gal in PBS) overnight at 37°C. The stained tissues were fixed in 4% paraformaldehyde (PFA) and dehydrated for paraffin embedding.
Dual-staining for EdU and BrdU
Five-micrometer tissue paraffin sections were dewaxed, hydrated, incubated in 1 M hydrochloric acid at 37°C for twenty minutes, washed with PBS for three times and antigen retrieval was performed in 10 mM citric acid. The sections were then stained according to the manufacturer’s instructions using the Click-iT EdU Alexa Flour 594 kit (Beyotime, C0078S). After staining, the sections were incubated with blocking solution (Beyotime, P0102) for one hour at room temperature and incubated with primary antibody against BrdU (Abcam, ab6326,1:100) overnight at 37°C. The sections were washed for three times, and incubated with 488-conjugated secondary antibodies (Thermo Fisher, A11006,1:400) for one hour at room temperature, stained with DAPI for eight minutes, and finally mounted with anti-fluorescence quenching sealing medium.
Histology, Immunohistochemistry (IHC) and Immunofluorescence (IF) assays
For histological staining, paraffin-embedded and 5-μm sections were stained with hematoxylin and eosin (H&E). Periodic acid-Schiff (PAS) staining was performed using standard methods. For immunohistochemistry staining, the sections were deparaffinized with xylene followed by treatment with serial dilutions of ethanol. Antigen-retrieval was performed by heating slides to 95°C for 10 min in 0.01 M citrate buffer (pH 6) in a microwave oven. After cooling to room temperature, sections were incubated with blocking solution for 1 hour after administration of 3% H2O2 to eliminate endogenous peroxidase activity. Then, the sections were incubated with primary antibody overnight at 4°C. The sections were then immunostained by the ABC peroxidase method (Vector Laboratories) with diaminobenzidine as the enzyme substrate and hematoxylin as a counterstain. For immunofluorescence staining, paraffin sections were microwave pretreated in 0.01 M citrate buffer (pH 6.0), and incubated with primary antibodies, then incubated with secondary antibodies (invitrogen) and counterstained with DAPI in mounting media. The primary antibodies included Ki67 (thermo fisher, RM-9106-S1,1:500), cleaved caspase-3 (CST, 9664s, 1:1000), lysozyme C (Santa Cruz, sc-27958, 1:500), ChgA (Abcam, ab15160, 1:400), Mucin2 (Santa Cruz, sc-15334, 1:500), pH3 (Abcam, ab14955, 1:200), BrdU (Abcam, ab6326, 1:100), PCNA (Abcam, ab92552, 1:200), Msi1 (MBL, D270-3, 1:1000), GFP (Abcam, ab13970, 1:800), GFP (Abcam, ab290, 1:800), RFP (ROCKLAND, 600-401-379,1:200).
Cell cycle calculation
Ten-week-old mice were intraperitoneally injected with EdU (0.2 mg/25 g body weight) 1.5 hours after a pulse of BrdU (1 mg/25 g body weight, Sigma-Aldrich, B5002) and sacrificed 0.5 hours later. The calculation was based on the assumption that EdU and BrdU could not be detected within thirty minutes after administration into mice. The number of cells still in S phase during the labeled time were EdU+BrdU+ (Scells) whereas cells that had exited S phase were BrdU+ (Lcells). The average cell cycle time (Tc) and S phase length (Ts) of +4 cells and CBCs were calculated according to the formulas below. The number of proliferating cells (Pcells) was calculated based on the percentage of PCNA+ cells in each stem cell in Figure 4B. Tj is the time during which cells can labeled with BrdU but not EdU (Jones et al., 2019; Shibui et al., 1989).
In situ hybridization
The small intestine of 10-week-old mice was harvested and fixed in neutral buffered formalin (NBF) at room temperature (RT) for twenty four hours before paraffin embedding. The tissues were chopped into 5 μm sections and handled using Advanced Cell Diagnostics RNAscope 2.5 HD detection Reagents-RED kit (ACD) with mouse Msi1 probe (ACD, 469801). The detailed operation steps of in situ hybridization were followed according to the manufacturer’s instructions (322360-USM).
Flow cytometry
The single-cell suspension of intestinal epithelium was collected as described previously (Sato et al., 2009). The fresh mouse intestine was cut open longitudinally and the villi were scraped off. The tissue was chopped into 5 mm pieces and incubated with 10 mM EDTA in PBS for thirty minutes at 4°C. The crypt fractions were collected by pipetting and filtered through a 70 μm cell strainer (BD biosciences). The gathered crypts were centrifuged at 1200 rpm for five minutes and digested with dispase (1 U/ml, Stem Cell Technologies). The single cell suspension was passed through a 40μm cell strainer (BD biosciences) and stained with Fixable Viability Dye (eBioscience, 65-0863-14) to remove dead cells. The flow cytometry analysis was performed on a BD FACS Arial 3.0. Msi1+ cells were quantified by cells separated from Msi1CreERT2; R26mTmG mice 15 hours after tamoxifen induction. Lgr5high cells were sorted by flow cytometry from Lgr5EGFP-IRES-CreERT2 mice.
Intestinal organoids culture
The isolation of intestinal crypts was described above. The gathered crypts were washed twice with PBS and collected by centrifuged at 800 rpm for five minutes. The supernatant was removed and the crypts were resuspended into Matrigel (Corning, 356231) and Medium (STEMCELL Technologies, 06005) (1:1 ratio) and plated into 48 well plates. The medium was replaced every other day. To label the initial Msi1+ cells in vitro, the intestinal organoids of Msi1CreERT2;R26mTmG and Lgr5EGFP-IRES-CreERT2; R26Isl-tdT mice were cultured and induced with 4-OH (1 μM, Sigma-Aldrich, H6278) for 9.5 hours and then replaced with 4-OH-free medium. To test the contribution of Msi1+ cells to regeneration, the organoids were exposed with 6 Gy γ-IR immediately after Msi1+ cells were labeled.
qRT-PCR analysis
All collected cells were sorted into TRIzol (Invitrogen, 10296010) immediately and total RNA was extracted using the RNeasy Plus Mini Kit (QIAGEN, 74134). Real-time PCR was performed on a LightCycler 480 real-time PCR system (Roche) combined with the LightCycler 480 SYBR Green I master mix (Roche, 04887352001). The primers used for the gene expression assessment were as follows:
Olfm4-forward, 5′-CAGCCACTTTCCAATTTCACTG-3′; Olfm4-reverse, 5′-GCTGGACATACTCCTTCACCTTA-3′;
Lgr5-forward, 5′-CCTACTCGAAGACTTACCCAGT-3′; Lgr5-reverse, 5′-GCATTGGGGTGAATGATAGCA-3′;
Axin2-forward, 5′-TGACTCTCCTTCCAGATCCCA-3′; Axin2-reverse, 5′-TGCCCACACTAGGCTGACA-3′;
Sox9-forward, 5′-GCAGACCAGTACCCGCATCT-3′; Sox9-reverse, 5′-CGCTTGTCCGTTCTTCACC-3′;
Single-cell mRNA sequencing
A single-cell suspension of intestinal epithelium was prepared as described above. The cells were stained with Fixable Viability Dye (eBioscience, 65-0863-14), CD45 (eBioscience, 17-0451-82), CD31 (eBioscience, 17-0311-82), TER119 (eBioscience, 17-5921-82), to remove dead and lin− cells, and GFP+ cells were sorted into EP tubes in single-cell mode by FACS. The collected cells were held on ice before loaded for GemCode single cell platform (10X). Chromium Single Cell 3′ v2 libraries were sequenced with a Novaseq 6000 sequencer, with the following sequencing parameters: read 1, 150 cycles; i7 index, 8 cycles and read 2,150 cycles.
Primary computational analysis
Raw Illumina data were demultiplexed and processed using Cell Ranger (10X Genomics version Cell Ranger 2.0.1). The MM10 reference transcriptome provided by 10X genomics was used for mapping. Seurat version 2.3.4 was used for filtering and subsequent clustering (Butler et al., 2018). In order to remove partial cells and doublets, cells with less than 1000 genes or more than 7000 genes were removed. Additionally, cells with more than 10% of mitochondrial unique molecular identifiers (UMIs) were removed, as a high proportion of mitochondrial expression in cells is indicative of cell stress/damage during isolation. In order to reduce gene expression noise, genes that are expressed in 6 cells or less are removed. Gene-cell matrices were normalized and scaled in Seurat using default parameters for UMIs. Highly variable genes were found using a lower x threshold of 0.0125 and ay threshold of 0.5. Principal Component Analysis (PCA) was performed using the highly variable genes identified. T-distributed stochastic neighbor embedding (t-SNE) was performed using the PCA reduction. PCA reduction was also used to clusters with standard modularity function. Because of their low numbers, tuft cells in each time point were manually identified based on expression of canonical markers. A likelihood-ratio test for single cell gene expression was used to identify marker genes for each population (McDavid et al., 2013). Single-cell consensus clustering (SC3) analysis was used to validate the robustness of some clusters (Kiselev et al., 2017). Cell cycle analysis was carried out in Seurat using a list of cell cycle genes from the Regev laboratory (Kowalczyk et al., 2015).
Pseudotime
Monocle version 2.10.1 was used on cells filtered from Seurat to infer differentiation trajectories (Qiu et al., 2017). An expression threshold of 0.1 was applied. The highly variable genes identified from Seurat were used as the ordering filter. DDRTree was used for dimension reduction. Initially, no root state was specified and the cells were ordered in an unsupervised manner. After the trajectory was obtained, a root state was specified based on where the stem cell populations are for subsequent systematic identification of pseudotime-dependent genes.
Identification of pseudotime-dependent gene dynamics
We performed scEpath (Jin et al., 2018) on Monocle-ordered cells to identify pseudotime-dependent gene expression changes as before (Guerrero-Juarez et al., 2019). Briefly, we compared the standard deviation of the observed gene expressions by randomly permuting the cell order (nboot = 100 permutations). Genes with a standard deviation greater than 0.5 and a Bonferroni-corrected p-value below a significance level α = 0.01 were considered to be pseudotime-dependent. Pseudotime-dependent mouse transcription factors were annotated using the Animal Transcription Factor Database (AnimalTFDB 2.0).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using GraphPad Prism 7 software. All data are shown as mean value ± SD. Unpaired two-tailed Student’s t test and two-way ANOVA analysis were performed for statistical analyses (*p < 0.05, **p < 0.01, ***p < 0.001).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-BrdU | Abcam | Cat# ab6326; RRID: AB_305426 |
| Rabbit anti-Ki67 | Thermo Fisher | Cat# RM-9106-S1; RRID: AB_149792 |
| Rabbit anti-Cleaved Caspase-3 | Cell Signaling Technology | Cat# 9664S; RRID: AB_2070042 |
| Goat anti-Lysozyme C | Santa Cruz | Cat# sc-27958; RRID: AB_2138790 |
| Rabbit anti-Chromogranin A | Abcam | Cat# ab15160; RRID: AB_301704 |
| Rabbit anti-Mucin2 | Santa Cruz | Cat# sc-15334; RRID: AB_2146667 |
| Mouse anti-Histone H3, phospho (Ser10) | Abcam | Cat# ab14955; RRID: AB_443110 |
| Rabbit anti-PCNA | Abcam | Cat# ab92552; RRID: AB_10561973 |
| Rat anti-Msi1 | MBL | Cat# D270-3; RRID: AB_1953023 |
| Chicken anti-GFP | Abcam | Cat# ab13970; RRID: AB_300798 |
| Rabbit anti-GFP | Abcam | Cat# ab290; RRID: AB_303395 |
| Rabbit anti-RFP | ROCKLAND | Cat# 600-401-379; RRID: AB_2209751 |
| Rat anti-CD45 | eBioscience | Cat# 17-0451-82; RRID: AB_469392 |
| Rat anti-CD31 | eBioscience | Cat# 17-0311-82; RRID: AB_657735 |
| Rat anti-TER 119 | eBioscience | Cat# 17-5921-82; RRID: AB_469473 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen (TAM) | Sigma-Aldrich | Cat# T5648 |
| 5-fluorouracil (5-FU) | Sigma-Aldrich | Cat# F6627 |
| Diphtheria Toxin (DT) | Sigma-Aldrich | Cat# D0564 |
| 5-ethynyl-20-deoxyuridine (EdU) | Thermo Fisher | Cat# A10044 |
| Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA) | Sigma-Aldrich | Cat# E4378 |
| 5-bromo-20-deoxyuridine (BrdU) | Sigma-Aldrich | Cat# B5002 |
| Ethylenedinitrilotetraacetic acid (EDTA) | Sigma-Aldrich | Cat# E9884 |
| Dispase | Stem Cell Technologies | Cat# 07913 |
| Fixable Viability Dye | eBioscience | Cat# 65-0863-14 |
| IntestiCult™ Organoid Growth Medium (Mouse) | Stem Cell Technologies | Cat# 06005 |
| 4-hydroxytamoxifen (4-OH) | Sigma-Aldrich | Cat# H6278 |
| TRIzol | Invitrogen | Cat# 10296010 |
| LightCycler 480 SYBR Green I master mix | Roche | Cat# 04887352001 |
| Matrigel | Corning | Cat# 356231 |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Flour 594 Kit | Beyotime | Cat# C0078S |
| Advanced Cell Diagnostics RNAscope 2.5 HD detection Reagents-RED kit | ACD | Cat# 322360 |
| RNeasy Plus Mini Kit | QIAGEN | Cat# 74134 |
| Single cell 3 ‘Library and Gel Bead Kit V2 | 10x Genomics | Cat# 120237 |
| Chromium Single Cell A Chip Kit | 10x Genomics | Cat# 120236 |
| Deposited Data | ||
| Single-cell RNA sequencing data | This paper | GEO: GSE145866 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Msi1CreERT2 | This paper | N/A |
| Mouse: Lgr5EGFP-IRES-CreERT2 | The Jackson Laboratory | JAX# 008875 |
| Mouse: R26mTmG | The Jackson Laboratory | JAX# 007676 |
| Mouse: R26tdT | The Jackson Laboratory | JAX# 007914 |
| Mouse: R26RLacZ | The Jackson Laboratory | JAX# 009427 |
| Mouse: R26Isl-DTA | The Jackson Laboratory | JAX# 010527 |
| Mouse: R26Isl-DTR | The Jackson Laboratory | JAX# 007900 |
| Mouse: HopxCreERT2 | The Jackson Laboratory | JAX# 017606 |
| Oligonucleotides | ||
| ISH Msi1 probe | ACD | Cat# 469801 |
| Genotyping primer, Msi1CreERT2-forward: TGGTTTCGGCCACAGTCTTG | This paper | N/A |
| Genotyping primer, Msi1CreERT2-resersw: TCCAGCTCGACCAGGATGGG-3 | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism 7 | GraphPad | N/A |
| Cellranger 2.0.1 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Seurat 2.3.4 | Butler et al., 2018 | https://satijalab.org/seurat |
| Monocle 2.10.1 | Qiu et al., 2017 | http://cole-trapnell-lab.github.io/monocle-release/ |
| scEpath | Jin et al., 2018 | https://github.com/sqjin/scEpath |
| SC3 | Kiselev et al., 2017 | https://www.bioconductor.org/packages/release/bioc/html/SC3.html |
| Velocyto.R | La Manno et al., 2018 | https://github.com/velocyto-team/velocyto.R |
Highlights.
Cycling Msi1+ ISCs in the intestinal crypt are DNA damage resistant
Msi1+ ISCs enable fast intestinal repair ahead of Lgr5high CBCs
Paneth cells preferentially arise from Msi1+ ISCs during homeostasis and repair
ACKNOWLEDGMENTS
Z.Y. is funded by grants from the National Natural Science Foundation of China (81772984 and 81572614); the Major Project for Cultivation Technology (2016ZX08008001 and 2014ZX08008001); Basic Research Program (2015QC0104, 2015TC041, 2016SY001, 2016QC086, 2019TC227, and 2019TC088); and an SKLB Open Grant (2020SKLAB6-18). C.F.G.-J. is supported bythe University of California, Irvine Chancellor’s ADVANCE Postdoctoral Fellowship Program. Z. Lin. is supported by NIH T32-Training Program in Cancer Biology and Therapeutics. B.A. is supported by RO1AR44882.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107952.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ayyaz A, Kumar S, Sangiorgi B, Ghoshal B, Gosio J, Ouladan S, Fink M, Barutcu S, Trcka D, Shen J, et al. (2019). Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125. [DOI] [PubMed] [Google Scholar]
- Bankaitis ED, Ha A, Kuo CJ, and Magness ST (2018). Reserve Stem Cells in Intestinal Homeostasis and Injury. Gastroenterology 155, 1348–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N (2014). Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol 15, 19–33. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, and Clevers H (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. [DOI] [PubMed] [Google Scholar]
- Ben-Yehoyada M, Gautier J, and Dupre A (2007). The DNA damage response during an unperturbed S-phase. DNA Repair (Amst.) 6, 914–922. [DOI] [PubMed] [Google Scholar]
- Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, and Winton DJ (2013). Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll TD, Newton IP, Chen Y, Blow JJ, and Näthke I (2018). Lgr5+ intestinal stem cells reside in an unlicensed G-ι phase. J. Cell Biol 217, 1667–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves-Pérez A, Yilmaz M, Perna C, de la Rosa S, and Djouder N (2019). URI is required to maintain intestinal architecture during ionizing radiation. Science 364, eaaq1165. [DOI] [PubMed] [Google Scholar]
- De Angelis PM, Svendsrud DH, Kravik KL, and Stokke T (2006). Cellular response to 5-fluorouracil (5-FU) in 5-FU-resistant colon cancer cell lines during treatment and recovery. Mol. Cancer 5, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duò A, Robinson MD, and Soneson C (2018). A systematic performance evaluation of clustering methods for single-cell RNA-seq data. F1000Res. 7, 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehart H, and Clevers H (2015). Repairing organs: lessons from intestine and liver. Trends Genet. 31, 344–351. [DOI] [PubMed] [Google Scholar]
- Guerrero-Juarez CF, Dedhia PH, Jin S, Ruiz-Vega R, Ma D, Liu Y, Yamaga K, Shestova O, Gay DL, Yang Z, et al. (2019). Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun 10, 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath JP (1996). Epithelial cell migration in the intestine. Cell Biol. Int 20, 139–146. [DOI] [PubMed] [Google Scholar]
- Jin S, MacLean AL, Peng T, and Nie Q (2018). scEpath: energy landscape-based inference of transition probabilities and cellular trajectories from single-cell transcriptomic data. Bioinformatics 34, 2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KB, Furukawa S, Marangoni P, Ma H, Pinkard H, D’Urso R, Zilionis R, Klein AM, and Klein OD (2019). Quantitative Clonal Analysis and Single-Cell Transcriptomics Reveal Division Kinetics, Hierarchy, and Fate of Oral Epithelial Progenitor Cells. Cell Stem Cell 24, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P, and Potten CS (1986). Circadian variation in migration velocity in small intestinal epithelium. Cell Tissue Kinet. 19, 591–599. [DOI] [PubMed] [Google Scholar]
- Kayahara T, Sawada M, Takaishi S, Fukui H, Seno H, Fukuzawa H, Suzuki K, Hiai H, Kageyama R, Okano H, and Chiba T (2003). Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 535, 131–135. [DOI] [PubMed] [Google Scholar]
- Kim CK, Yang VW, and Bialkowska AB (2017). The Role of Intestinal Stem Cells in Epithelial Regeneration Following Radiation-Induced Gut Injury. Curr. Stem Cell Rep 3, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, and Hemberg M (2017). SC3: consensus clustering of single-cell RNA-seq data. Nat. Methods 14, 483–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, and Regev A (2015). Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, and Clevers H (2010). Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Yousefi M, Nakauka-Ddamba A, Jain R, Tobias J, Epstein JA, Jensen ST, and Lengner CJ (2014). Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Reports 3, 876–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Yousefi M, Nakauka-Ddamba A, Li F, Vandivier L, Parada K, Woo DH, Wang S, Naqvi AS, Rao S, et al. (2015). The Msi Family of RNA-Binding Proteins Function Redundantly as Intestinal Oncoproteins. Cell Rep. 13, 2440–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Nakauka-Ddamba A, Tobias J, Jensen ST, and Lengner CJ (2016). Mouse Label-Retaining CellsAre Molecularly and Functionally Distinct From Reserve Intestinal Stem Cells. Gastroenterology 151, 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity A, McKenna WG, and Muschel RJ (1994). The molecular basis for cell cycle delays following ionizing radiation: a review. Radiother. Oncol 31, 1–13. [DOI] [PubMed] [Google Scholar]
- McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, and Gottardo R (2013). Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29, 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe C, Kljavin NM,Ybarra R, and deSauvage FJ (2014). Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell 14, 149–159. [DOI] [PubMed] [Google Scholar]
- Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, Morrison CG, and Passegué E (2010). Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 7, 174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S, and Breault DT (2011). Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl. Acad. Sci. USA 108, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, and Jasin M (2010). Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol 11, 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlik TM, and Keyomarsi K (2004). Role of cell cycle in mediating sensitivity to radiotherapy. Int. J. Radiat. Oncol. Biol. Phys 59, 928–942. [DOI] [PubMed] [Google Scholar]
- Potten CS (1998). Stem cells in gastrointestinal epithelium: numbers, characteristics and death. Philos. Trans. R. Soc. Lond. B Biol. Sci 353, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Kovacs L, and Hamilton E (1974). Continuous labelling studies on mouse skin and intestine. Cell Tissue Kinet. 7, 271–283. [DOI] [PubMed] [Google Scholar]
- Potten CS, Owen G, and Booth D (2002). Intestinal stem cells protect their genome by selective segregation of template DNA strands. J. Cell Sci 115, 2381–2388. [DOI] [PubMed] [Google Scholar]
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, et al. (2012). The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149, 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JM, Roberts SA, and Potten CS (1994). Cell migration in the small and large bowel shows a strong circadian rhythm. Epithelial Cell Biol. 3, 137–148. [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C (2017). Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiorgi E, and Capecchi MR (2008). Bmi1 isexpressed in vivo in intestinal stem cells. Nat. Genet 40, 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, and Clevers H (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, and Clevers H (2011). Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaltiel IA, Krenning L, Bruinsma W, and Medema RH (2015). The same, only different - DNA damage checkpoints and their reversal throughout the cell cycle. J. Cell Sci 128, 607–620. [DOI] [PubMed] [Google Scholar]
- Shibui S, Hoshino T, Vanderlaan M, and Gray JW (1989). Double labeling with iodo- and bromodeoxyuridine for cell kinetics studies. J. Histochem. Cytochem 37, 1007–1011. [DOI] [PubMed] [Google Scholar]
- Stacey R, and Green JT (2014). Radiation-induced small bowel disease: latest developments and clinical guidance. Ther. Adv. Chronic Dis 5, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser ML, Bailey AP, Senyo SE, Guillermier C, Perlstein TS, Gould AP, Lee RT, and Lechene CP (2012). Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature 481,516–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, and Epstein JA (2011). Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, van den Born M, Korving J, de Sauvage F, van Es JH, et al. (2016). Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell 18, 203–213. [DOI] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, and de Sauvage FJ (2011). A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, Sato T, van de Wetering M, Lyubimova A, Yee Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, et al. (2012). Dll1 + secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol 14, 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, et al. (2012). The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA 109, 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, Davies PS, Cheng ZF, Kaddis JS, Han A, et al. (2017). Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell Stem Cell 21, 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Tong K, Zhao Y, Balasubramanian I, Yap GS, Ferraris RP, Bonder EM, Verzi MP, and Gao N (2018). Paneth Cell Multipotency Induced by Notch Activation following Injury. Cell Stem Cell 23, 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scRNA-seq data from this study are available at NCBI Gene Expression Omnibus (GEO). The accession number for data reported in this paper is GEO: GSE145866.


