ABSTRACT
Under-recognition of dengue infection may lead to increased morbidity and mortality, whereas early detection is shown to help improve patient outcomes. Recent incidence and outbreak reports of dengue virus in the United States and other temperate regions where dengue was not typically seen have raised concerns regarding appropriate diagnosis and management by healthcare providers unfamiliar with the disease. This study aimed to describe self-reported clinical symptoms of dengue fever in a non-endemic cohort and to establish a clinically useful predictive algorithm based on presenting features that can assist in the early evaluation of potential dengue infection. Volunteers who experienced febrile illness while traveling in dengue-endemic countries were recruited for this study. History of illness and blood samples were collected at enrollment. Participants were classified as dengue naive or dengue exposed based on neutralizing antibody titers. Statistical analysis was performed to compare characteristics between the two groups. A regression model including joint/muscle/bone pain, rash, dyspnea, and rhinorrhea predicts dengue infection with 78% sensitivity, 63% specificity, 80% positive predictive value, and 61% negative predictive value. A decision tree model including joint/muscle/bone pain, dyspnea, and rash yields 77% sensitivity and 67% specificity. Diagnosis of dengue fever is challenging because of the nonspecific nature of clinical presentation. A sensitive predicting model can be helpful to triage suspected dengue infection in the non-endemic setting, but specificity requires additional testing including laboratory evaluation.
INTRODUCTION
Dengue, a viral disease with four serotypes (dengue virus [DENV] 1–4), is transmitted by Aedes mosquitoes (Aedes aegypti and Aedes albopictus). The febrile illness is endemic in tropical regions worldwide, including Southeast Asia, South and Central America, Oceana, Africa, and eastern Mediterranean regions. 1 Although less common, cases of dengue have also been described in North America and Europe, mostly imported by travelers from endemic areas. 2–4 Dengue-like cases have been documented in the United States, dating back to as early as 1,780. More recent outbreaks of autochthonous cases have been confirmed in Hawaii in 2001–2002, Texas in 2005, and Florida in 2009–2010.
Dengue is responsible for substantial economic, social, and political burdens worldwide. Millions of people are affected annually; incidence has increased over the past 50 years, and dengue remains on the WHO’s list of neglected tropical diseases. 1 The incidence of this mosquito-borne infection will likely continue to increase and expand geographically in parallel with increasing trends in population growth, urbanization, globalization, climate change, travel, and migration. 5
There have been several indicators that physicians in the United States will encounter dengue with increasing frequency in the future. After malaria, dengue is the most common cause of fever in returned travelers from dengue-endemic regions. 6 Aside from imported cases, climate and ecological change have created more suitable conditions for vector survival that may also contribute to an increase in local transmission. 7 Aedes albopictus was found to be mainly responsible for viral transmission during the dengue outbreak in Hawaii in 2001. 8 Aedes albopictus has a wider geographic range than Ae. aegypti, extending to temperate zones, and is highly prevalent in North America, raising the possibility of more widespread autochthonous dengue in previously non-endemic areas. 2,9,10
Infection with dengue virus results in a spectrum of illness, ranging from asymptomatic or mild and self-limiting fever to severe illness that can lead to death. Current diagnosis of dengue mostly relies on various laboratory methods, which may involve virus isolation and nucleic acid detection, or antibody analysis. 11 The different techniques require different levels of equipment and technician expertise, as well as time. In clinical settings where diagnosis confirmation is promptly needed to guide management, rapid tests might be chosen for their speed and simplicity, in exchange for sensitivity and specificity. For instance, a rapid antigen test that detects a dengue-specific marker such as DENV nonstructural protein 1, a protein secreted from infected vertebrate cells, is now commercially available and is being used clinically. 12 However, despite the accuracy and availability of these laboratory tests, diagnosis of dengue would still first require recognition by healthcare providers who will be ordering these tests. At this time, a predictive model based on clinical symptoms alone is not readily available. The 2009 WHO guidelines also stated that diagnosis based on clinical symptoms is unreliable because of the nonspecific symptoms. 11
Symptomatic dengue is described as a febrile illness with one or more of the following symptoms: high fever > 40°C, headache, retro-orbital pain, nausea/vomiting, myalgia, arthralgia, and rashes. 13 Additional objective findings listed on WHO guidelines that suggest probable dengue include signs of fluid accumulation, liver enlargement, positive tourniquet test, leukopenia, and thrombocytopenia. 11 These findings may not be most applicable in the triage assessment and initial formation of differential diagnoses when laboratory results are not yet available or where dengue is infrequently seen such that the tourniquet test is unlikely to be applied. Furthermore, a study has found that the sensitivity of the WHO classification scheme decreases with increasing patient age at the time of infection, making detection of dengue fever even more complicated in older adults. 14 This becomes even more relevant considering that the non-endemic population that develops dengue fever is mostly adults, as compared with the typical pediatric infection seen in endemic countries.
There have been examples of case reports describing dengue being misdiagnosed as acute appendicitis or acute cholecystitis, resulting in unnecessary surgery and even death from surgical wound bleeding complication. 4,15 A retrospective study in Texas on specimens collected for West Nile virus testing between 2003 and 2005 revealed DENV infections in 47 cases. 9 Further review revealed that dengue was not considered in the differential diagnosis in any of these cases. These instances highlight the concern regarding inaccurate clinical assessment and management, and under-recognition of the disease by clinicians working in non-endemic areas, which could increase the potential for dengue-related complications. 13 Although management of dengue typically involves only supportive therapy, early recognition of disease and appropriate fluid management can reduce the chance of developing dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS); thus, early detection helps decrease associated morbidity and mortality. 5,16
In the non-endemic setting, travel history can be the most informative aspect of history taking for clinicians. To aid this diagnosis process, a set of symptoms that patients experienced during dengue is evaluated in this study. Although symptoms are usually nonspecific, an algorithm based on a combination of symptoms that are collectively reasonably sensitive and specific for dengue may be helpful earlier in the evaluation, triggering suspicion of dengue and suggesting follow-up investigation with routine laboratory.
Here, we take the unique approach of evaluating symptoms reported by a cohort of subjects living in a non-endemic locale who report a presumed history of dengue but have not had previous laboratory confirmation. Using standard virus neutralization assays, we tested subject sera against all four DENV serotypes and classified them as having serology consistent with primary dengue, secondary dengue, or naive (no prior dengue). We then used multiple logistic regression and decision tree analyses to determine whether reported symptoms could reliably predict serologic status for each subject. The results of our work offer unique insight into the capacity of reported symptoms to differentiate dengue from non-dengue disease when subjects suspect a dengue infection.
METHODS
Study population.
Data for this analysis were obtained from a cross-sectional observational cohort study conducted between 2014 and 2018 at the Oregon Health & Science University. The eligibility criteria included having a history of febrile illness consistent with dengue that was associated with travel to a dengue-endemic region. At the time of initial recruitment visit, informed consent was obtained. Participants were then provided with a questionnaire which covered demographic data, travel history, pertinent medical history, and a checklist of symptoms to describe each episode of illness with the presumed diagnosis if available. All information obtained was based on participants’ recollection of the illness event, and no official diagnosis or laboratory workup was required. Volunteers then proceeded to donate blood sample for serological studies. Subjects were recruited and samples collected under an Institutional Review Board-approved protocol (OHSU Institutional Review Board study #10212).
The symptoms’ checklist consisted of 18 “yes/no” questions, which included the following symptoms: fever, severe joint/bone/muscle pain, headache, eye pain, nausea, loss of appetite, sensitivity to light, rash, malaise/tiredness, bleeding from gums, vomiting blood or coffee grounds, bloody or tar-like diarrhea, cough, cough with green/yellow phlegm, shortness of breath, runny nose, vomiting (without blood or coffee grounds), and diarrhea (without blood or tar-like stools). Collected data were coded before analysis. Because each subject might have recalled more than one instance of illness, reported symptoms used for final analysis were derived from each reported symptomatic episode.
Laboratory methods.
The viruses used in this study were the reference strains DENV1 (WestPac′74), DENV2 (16803), DENV3 (rDENV3), and DENV4 (rDENV4). 17,18 Viruses were propagated in T-75 tissue culture flasks seeded with C6/36 mosquito cells overlaid with Minimal Essential Media supplemented with 5% fetal calf serum, nonessential amino acids, penicillin, streptomycin, and amphotericin B. Flasks were incubated at 32°C with 5% CO2 for 5–7 days and supernatants harvested, clarified by centrifugation, and then stored at −80°C in 20% SPG buffer.
Serum samples were initially screened by ELISA and confirmed by plaque reduction neutralization test (PRNT). Fifty percent PRNT50 titers were used to characterize sera. 19 With PRNT, participants’ immune sera were initially heat-inactivated for 30 minutes at 56°C, serially diluted 4-fold from a starting dilution of 1:10, and mixed with an equal volume of ∼25 plaque-forming units of DENV serotypes 1–4, giving a final starting serum dilution of 1:20. Each serum–virus mix was quadruplicated. Virus dilution mixes without sera were prepared as controls. After 1 hour of incubation at 37°C, virus mixes were inoculated into individual wells of 24-well plates seeded with Vero E6 cells, incubated for another hour at 37°C, and then overlaid with 1% methylcellulose. After inoculation, plates were incubated for 5 days at 37°C and 5% CO2. The overlay was then removed, monolayers were fixed with 80% methanol and stained with 2% crystal violet, and plaques were enumerated by visual review of each well. The proportion of virus neutralized per well was calculated, and the serum dilution that neutralized 50% of control input virus (PRNT50) was determined by sigmoidal dose-response curve fitting in GraphPad Prism® (version 7.0; GraphPad software, San Diego, CA). 20 Sera were classified as primary DENV immune if they only neutralized a single DENV serotype at a dilution ≥ 1:20 or if the titer against a single DENV serotype titer was ≥ 4 times higher than the titers against the three remaining serotypes. Sera were classified as secondary DENV immune if the sera neutralized more than one serotype and the two highest titers against individual serotypes were within 4-fold of each other. Sera were classified as naive if the sera did not neutralize any DENV at a 1:20 serum dilution. Plaque reduction neutralization test results were also evaluated in the context of travel and exposure history, vaccine history, and laboratory results at the time of illness (provided by participants) if available. In addition, using a similar neutralization technique, serum samples were tested against chikungunya, Zika, yellow fever, and Japanese encephalitis virus. The sample was considered positive for previous exposure if the PRNT50 was greater than 1:20 for a specific virus. In the case that more than one result was above 1:20, the one with the highest neutralizing was considered to be the main exposure, with others being the result of cross-neutralization.
Data analysis.
All subjects and cases that reported possible previous exposure to dengue and/or listed dengue virus as the suspected cause of certain illness episodes were selected for this analysis. Exclusion criteria included the following:
1. Subjects who did not report any symptomatic episodes, or were unable to provide answers to the symptom checklist
2. Symptomatic episodes without fever
3. Missing sample or incomplete serological results at the time of analysis
Of the total of 217 available cases, 140 cases from 126 subjects met the inclusion criteria.
Data were analyzed using both descriptive and inferential statistics. Each case was classified as either positive or negative for dengue based on serology result, as described in the Laboratory Methods section, in conjunction with circumstantial information provided by the questionnaire. Clinical symptoms were compared between the positive and negative groups. Initially, each symptom was summarized into a 2 × 2 contingency table. These data were then used to calculate sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for each symptom.
Statistical analysis was performed using chi-square and Fisher’s exact test, univariate and multiple logistic regression analyses, and decision tree algorithm. Variables used were the 17 symptoms from the questionnaire checklist as mentioned earlier (excluding fever). First, chi-square and Fisher’s exact test were used to assess relationship between dengue and each individual symptom. Considering each symptom as an independent variable and dengue status as a categorical dependent variable, a multiple logistic regression model was developed from the data. Only variables with a P-value < 0.25 were initially included in the multiple logistic regression model via the forward selection approach. The preliminary model was composed of variables that when added yielded the lowest P-value, that is, < 0.05. Interaction effects between paired symptoms were then assessed, with every possible pair of the 17 symptoms assessed separately (see Table 1). If a two-symptom interaction yielded a P-value of < 0.25, then it was added to the preliminary model. The final algorithm was chosen from the simplest model with the least number of variables included, yet provided the best fit and was most inclusive, based on the area under the curve (AUC), sensitivity, specificity, and P-value, considered in that order. As a further analysis, we also constructed a model that statistic run based on new variables that combined all aches/pain and bleeding symptoms into one category. Finally, a decision tree algorithm was performed using the R package “tree” (Ripley B., Oxford, United Kingdom; CRAN repository, R Foundation for Statistical Computing, Vienna, Austria) based on available variables to visualize a simple flowchart of symptoms that suggest dengue infection. All statistical analyses were performed using R Studio Version 1.1.453 (R Foundation for Statistical Computing, Vienna, Austria).
Table 1.
Cohort characteristics categorized by serologic response to dengue virus
| DENV seropositive (n = 82) | DENV seronegative (n = 44) | P-value | |
|---|---|---|---|
| Gender, % women | 59.8 | 54.5 | 0.71 |
| Age at enrollment (years) | 0.72 | ||
| Median (range) | 39 (19–85) | 40 (19–79) | |
| Mean (SD), n (%) | 45.3 (18.5) | 44.1 (16.0) | |
| Age at first infection (years) | 0.46 | ||
| Median (range) | 26 (6–69) | 29 (7–89) | |
| Mean (SD), n (%) | 31.7 (17.3) | 34.1 (17.6) | |
| Ethnicity | |||
| Hispanic | 5 | 2 | |
| Non-Hispanic | 74 | 41 | |
| Decline | 3 | 1 | |
| Race | |||
| Asian | 11 | 2 | |
| Caucasian | 60 | 35 | |
| Multiracial/other | 6 | 3 | |
| Decline | 5 | 4 | |
| Yellow Fever Virus vaccinated (%) | 45.1 | 56.8 | 0.29 |
| Japanese Encephalitis Virus vaccinated (%) | 12.2 | 13.6 | > 0.99 |
RESULTS
Characteristics of the study population.
Overall, DENV seroprevalence in this cohort was 65.1% (82 of 126 participants). Among the 126 participants, 14 participants reported two symptomatic episodes fitting with dengue, and thus a total of 140 cases were included in the analysis. Ninety-one cases were classified as positive, meaning that the symptoms reported were considered to be caused by dengue virus, and 49 cases were classified as negative for dengue virus infection. Among the 14 participants who reported two symptomatic episodes, three participants were considered to have both negative cases, nine participants had both positive cases, and two participants had one positive and one negative cases, based on circumstantial information provided at the time of questionnaire.
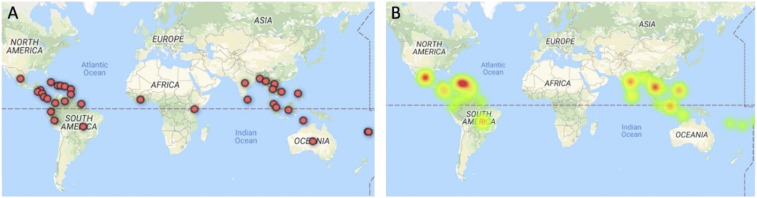
Demographics of participants are presented in Table 1, showing the comparison between the two groups categorized by serologic result. Dengue seronegative and dengue seropositive groups were not significantly different with respect to age, gender, or vaccination status. Countries visited by participants and reported as the suspected location of dengue acquisition are shown (Figure 1A), and countries associated with confirmed cases are represented with a heat map (Figure 1B). Of the 49 DENV-negative cases, three were found to be seropositive for chikungunya virus, and two were positive for having antibody against yellow fever virus without previous vaccination.
Figure 1.
Map of countries related to dengue cases. (A) Markers representing the countries where research participants visited and suspected to have contracted dengue. (B) Heat map showing countries associated with cases classified as dengue positive. This figure appears in color at www.ajtmh.org.
Epidemiological study.
Table 2 shows the summary of clinical symptoms from 140 cases totally reported by participants, grouping by infection status as determined by combination of serologic and circumstantial information. Table 3 shows the comparison of sensitivity, specificity, PPV, and NPV.
Table 2.
Clinical symptoms reported by research participants with confirmed previous exposure to dengue compared with those in whom infection was excluded
| Positive* (n = 91), n (%) | Negative* (n = 49), n (%) | P-value† | |
|---|---|---|---|
| Aches and pain | 82 (90) | 46 (94) | 0.54 (F) |
| Severe joint/bone/muscle pain | 70 (77) | 45 (92) | 0.03 |
| Headache | 73 (80) | 43 (88) | 0.26 |
| Eye pain | 48 (53) | 22 (45) | 0.38 |
| Nausea | 45 (49) | 30 (61) | 0.18 |
| Loss of appetite | 80 (88) | 45 (92) | 0.47 |
| Sensitivity to light | 44 (48) | 21 (43) | 0.53 |
| Rash | 50 (55) | 14 (29) | < 0.01 |
| Malaise | 86 (95) | 48 (98) | 0.67 (F) |
| Bleeding symptoms | 3 (3) | 4 (8) | 0.24 (F) |
| Bleeding from gums | 2 (2) | 3 (6) | 0.34 (F) |
| Vomiting blood or coffee grounds | 0 | 1 (2) | 0.35 (F) |
| Bloody or tar-like diarrhea | 1 (1) | 0 | > 0.99 (F) |
| Cough | 14 (15) | 8 (16) | 0.88 |
| Cough with green/yellow phlegm | 4 (4) | 2 (4) | > 0.99 (F) |
| Shortness of breath | 26 (29) | 3 (6) | < 0.01 |
| Runny nose | 11 (12) | 10 (20) | 0.19 |
| Vomiting (without blood or coffee grounds) | 20 (22) | 10 (20) | 0.83 |
| Diarrhea (without blood or coffee grounds) | 29 (32) | 19 (39) | 0.41 |
Status of each case determined by combination of serologic and circumstantial information.
Chi-square test without Yates’ continuity correction or Fisher’s exact test (F) for categorical variables.
Table 3.
Sensitivity, specificity, and predictive values of each symptom
| Sensitivity | Specificity | Positive predictive value | Negative predictive value | |
|---|---|---|---|---|
| Aches and pain | 0.91 | 0.06 | 0.64 | 0.25 |
| Severe joint/bone/muscle pain | 0.77 | 0.08 | 0.61 | 0.16 |
| Headache | 0.80 | 0.12 | 0.63 | 0.25 |
| Eye pain | 0.53 | 0.55 | 0.69 | 0.39 |
| Nausea | 0.49 | 0.39 | 0.60 | 0.29 |
| Loss of appetite | 0.88 | 0.08 | 0.64 | 0.27 |
| Sensitivity to light | 0.48 | 0.57 | 0.68 | 0.37 |
| Rash | 0.55 | 0.71 | 0.78 | 0.46 |
| Malaise | 0.95 | 0.02 | 0.64 | 0.17 |
| Bleeding symptoms | 0.03 | 0.92 | 0.43 | 0.34 |
| Bleeding from gums | 0.02 | 0.94 | 0.40 | 0.34 |
| Vomiting blood or coffee grounds | 0.00 | 0.98 | 0.00 | 0.35 |
| Bloody or tar-like diarrhea | 0.01 | 1.00 | 1.00 | 0.35 |
| Cough | 0.15 | 0.84 | 0.64 | 0.35 |
| Cough with green/yellow phlegm | 0.04 | 0.96 | 0.67 | 0.35 |
| Shortness of breath | 0.29 | 0.94 | 0.90 | 0.41 |
| Runny nose | 0.12 | 0.80 | 0.52 | 0.33 |
| Vomiting (without blood or coffee grounds) | 0.22 | 0.80 | 0.67 | 0.35 |
| Diarrhea (without blood or coffee grounds) | 0.32 | 0.61 | 0.60 | 0.33 |
Symptoms that had a sensitivity > 0.75 included severe joint/bone/muscle pain, headache, loss of appetite, and malaise. Symptoms that had a specificity > 0.75 included gum bleeding, hematemesis, melena, cough, productive cough, shortness of breath, runny nose, and vomiting. Rash, melena, and shortness of breath represented the highest PPV among all symptoms. In addition, severe joint/bone/muscle pain, rash, and shortness of breath were the only symptoms that showed a statistically significant difference between the two groups based on chi-square analysis (see Supplemental Tables 2–4 for further details of infection status for these three symptoms). Headache, eye pain, nausea, anorexia, sensitivity to light, malaise, and gum bleeds had statistically similar rates of occurrence among the two groups in this adult non-endemic cohort.
Multiple logistic regression.
As described in the Methods, the final model acquired from regression analysis (Table 4) included only the symptoms and symptom interactions that resulted in an overall algorithm that remains significant and reasonably inclusive.
Table 4.
Variables in the multiple logistic regression model from data that included only febrile cases, with their estimated coefficients, standard error, P-value, unadjusted and adjusted ORs, and its 95% CIs
| Estimated coefficients | Standard error | P-value | Crude ORs | Estimated adjusted ORs | 95% CI | |
|---|---|---|---|---|---|---|
| Intercept | 1.273 | 0.563 | 0.024 | – | – | – |
| JT/bone/muscle pain | −1.520 | 0.612 | 0.013 | 0.296 | 0.219 | 0.066–0.725 |
| Rash | 1.087 | 0.416 | 0.009 | 3.049 | 2.965 | 1.312–6.700 |
| Sob | 2.557 | 0.796 | 0.001 | 6.133 | 12.90 | 2.71–61.47 |
| Rhinorrhea | −1.308 | 0.706 | 0.064 | 0.536 | 0.270 | 0.068–1.079 |
OR = odd ratio.
The receiver operating characteristic (ROC) curve was drawn from symptom scores by calculating from the regression equation as follows:
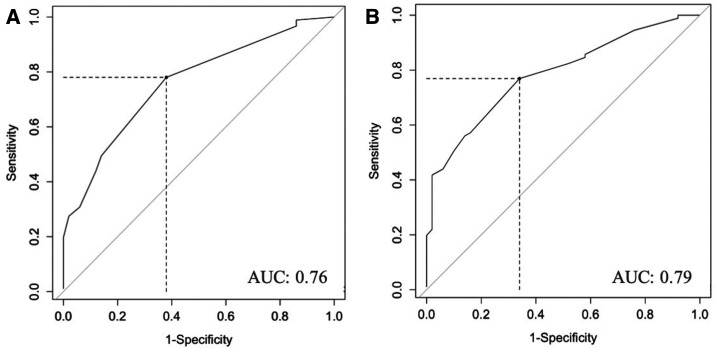
The ROC curve allows for evaluation of the model’s ability to classify the disease status via measurement of the AUC. The AUC for this regression model was estimated to be at 0.76 (Figure 2A). The cutoff point at the symptom score of 0.84 yielded sensitivity of 0.78 and specificity of 0.63, with a PPV of 0.80 and an NPV of 0.61.
Figure 2.
Receiver operating characteristic curves of logistic regression model. (A) Receiver operating characteristic curve based on the first model of multivariate regression, which included only single variables. (B) Receiver operating characteristic curve based on an alternative multivariate regression model, which included nausea/diarrhea interaction. AUC = area under the curve.
We evaluated an alternative regression model which included a nausea–diarrhea symptom interaction, in addition to the other individual variables already included in the model as earlier, with the result shown in the following text (Table 5). This new model yielded an AUC of 0.79 as shown in Figure 2B. At a cutoff point score of 0.51, the sensitivity, specificity, PPV, and NPV of this model are 0.77, 0.67, 0.81, and 0.61, respectively. Additional analysis with a model constructed from new combined aches/pain and bleeding symptoms as one category met the criteria for inclusion into a prelim model. The model that included joint pain, rash, shortness of breath, rhinorrhea, and bleeding yielded the AUC, sensitivity, and specificity that were comparable to the previous models.
Table 5.
Variables in the alternative regression model with their estimated coefficients, standard error, P-value, unadjusted and adjusted ORs, and its 95% CIs
| Estimated coefficients | Standard error | P-value | Crude ORs | Estimated adjusted ORs | 95% CI | |
|---|---|---|---|---|---|---|
| Intercept | 1.452 | 0.626 | 0.020 | – | – | – |
| JT/bone/muscle pain | −1.698 | 0.628 | 0.007 | 0.296 | 0.183 | 0.053–0.627 |
| Rash | 1.092 | 0.430 | 0.011 | 3.049 | 2.979 | 1.282–6.925 |
| Sob | 2.696 | 0.837 | 0.001 | 6.133 | 14.82 | 2.870–76.480 |
| Rhinorrhea | −1.111 | 0.751 | 0.139 | 0.536 | 0.329 | 0.076–1.436 |
| Nausea | 0.223 | 0.517 | 0.666 | 0.620 | 1.250 | 0.454–3.442 |
| Diarrhea | 1.042 | 0.925 | 0.260 | 0.739 | 2.835 | 0.463–17.366 |
| Nausea–diarrhea | −2.094 | 1.089 | 0.054 | 0.191 | 0.123 | 0.015–1.040 |
OR = odd ratio.
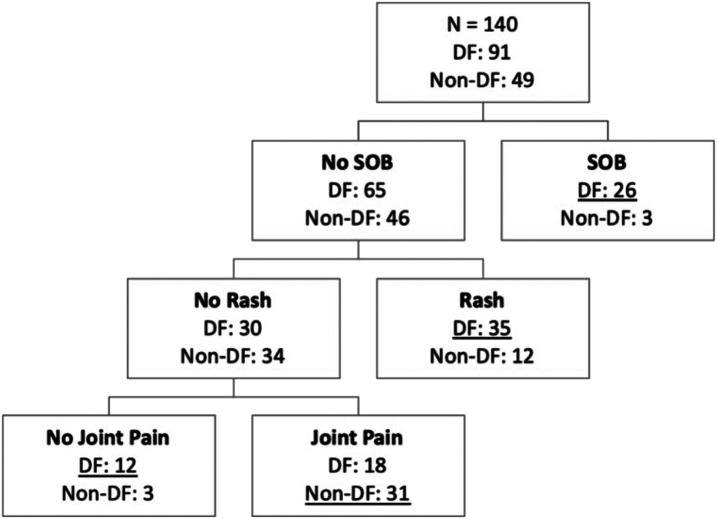
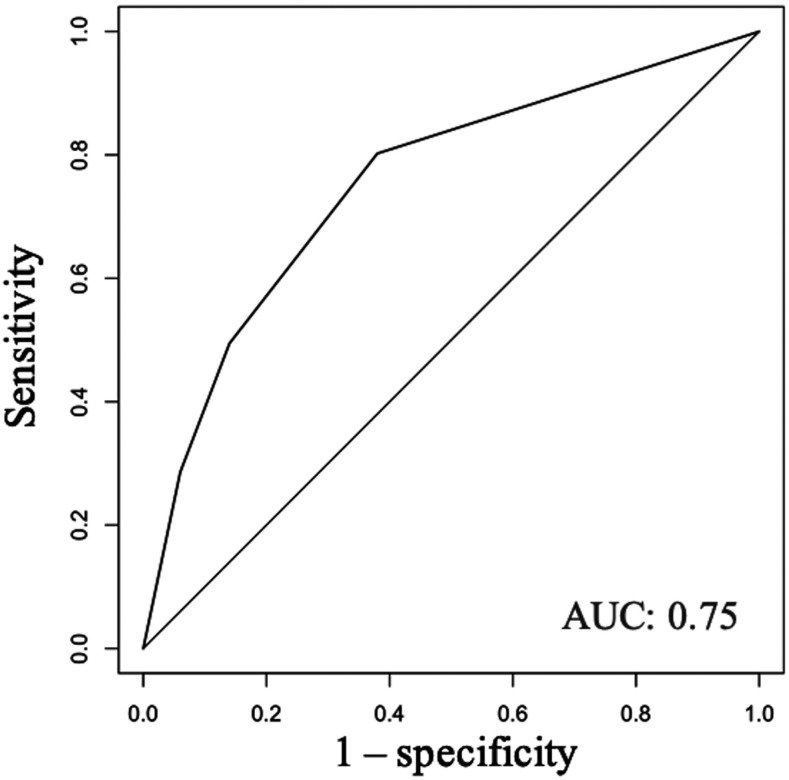
The clinical features included in the determination of the regression model were also used to develop a decision tree. The only symptoms included in the final decision tree were shortness of breath, rash, and joint/bone/muscle pain. Result is shown in Figure 3, which has an AUC of 0.75 (Figure 4). The sensitivity and specificity of the decision tree are 0.77 and 0.67, respectively.
Figure 3.
Decision tree for predicting dengue viral infection. The number of dengue fever (DF) and non-dengue fever (non-DF) cases in the study population is shown in each node.
Figure 4.
Receiver operating characteristic curve of decision tree model. Receiver operating characteristic curve based on decision tree model showing area under the curve (AUC) of 0.75.
DISCUSSION
The results presented here re-demonstrate the difficulties in diagnosing dengue based on clinical history alone because of the nonspecific signs and symptoms which are common to many acute febrile illnesses. Severe joint/bone/muscle pain, rash, and shortness of breath were the only symptoms that, by themselves, statistically differed in occurrence between the DENV-positive and DENV-negative groups. However, none of these symptoms individually had sufficient sensitivity or specificity to be useful. Symptoms that were more sensitive include severe joint/bone/muscle pain, headache, anorexia, and malaise. A more specific symptom included shortness of breath. Although it seemed that gum bleeding, hematemesis, melena, cough, rhinorrhea, and vomiting have high specificity and were reported with greater frequency in the positive dengue group, the differences between the two groups were statistically insignificant, and therefore these symptoms alone were not very helpful in predicting dengue.
Combinations of clinical symptoms were more informative than individual symptoms in predicting the diagnosis of dengue. When using multiple logistic regression analysis, the symptoms that were positively correlated with dengue included rash and shortness of breath, whereas joint/bone/muscle pain, rhinorrhea, and a combination of nausea and diarrhea demonstrated negative correlation. When adapting the variables into the decision tree model, variables similar to those of the regression model were included into the final result. The decision tree performed at a comparable level to the logistic regression model, with only minimally lower AUC, but with similar sensitivity and specificity values. However, a decision tree allowed for simpler interpretation and utilization of the model overall.
Shortness of breath had the highest correlation coefficients in the regression model and provided a strong positive prediction of dengue in this test cohort. Although this may differ from the objective findings of dyspnea or tachypnea, the results of our analysis suggested that subjective shortness of breath has value in helping with dengue diagnosis and should be explored during history intake. It is also important to note that shortness of breath was observed more with secondary infection, in which 44% of participants with secondary dengue developed the symptom, whereas only 18% of primary infection experienced shortness of breath. Based on the retrospective design of this study, the secondary infection cannot be proven as the cause of each symptomatic case, regardless this may suggest reasonable association with severe dengue infection, including DHF or DSS, which should be further explored. In addition, the negative correlation between rhinorrhea and dengue fever may be expected, and probably setting this mosquito-borne disease apart from other common cold and flu symptoms.
It is also an interesting finding that severe joint/bone/muscle pain was negatively correlated with dengue fever, considering that myalgia and arthralgia are considered one of the more traditional descriptive characteristics of dengue, also known as “breakbone” fever. 21 The study may be affected by type II error from the relatively small sample size, causing the true significance of the presence of each symptom to not be observed in this analysis. However, findings have also been inconsistent in other studies with larger sample sizes. A study by Low et al. 14 found the symptom of bone pain to be more common in patients suffering from dengue fever, whereas another study by Phuong et al. 22 showed that the symptom was similarly reported among the two groups of patients, more consistent with our own results. The difference in prevalence of muscle pain was ultimately not statistically significant in either study. This suggests that there may be an attribution bias for certain symptoms subjects believe should be associated with the presumptive diagnosis of dengue but, in our studies and others, empirically were not. It is also important to note that the study cohort specifically recruited subjects who had been presumptively diagnosed with dengue, which, rather than enriching for dengue positive subjects, may instead have enriched for subjects with bone and joint pain (an assumed common symptom for dengue as mentioned) but not necessarily for dengue. In addition, cultural differences in how subjects perceive and report pain may have contributed to the discrepancy being observed. 23,24 Last, severe joint/bone/muscle pain was a sensitive, but nonspecific, feature of dengue; thus, the association of myalgia and arthralgia with other mosquito-borne diseases, such as Zika and chikungunya, or even influenza and other tropical illnesses could also explain the negative correlation observed. 25 It was noted in this study that among the 49 cases considered negative for dengue infection, five of 45 cases which reported joint, bone, and muscle pain tested positive for previous exposure to other viruses (three for chikungunya and two for yellow fever). Similarly, this may be an explanation for poor specificity and negative correlation unexpectedly seen with bleeding symptoms, anorexia, and malaise.
Although nausea and diarrhea alone were not significant enough to be included in the model, the addition of their interaction to the model gave a negative prediction that contributed to a slightly more inclusive model (higher AUC), and thus, was shown here as the alternative regression model. This observation was likely due to the high incidence of gastrointestinal infections in travelers, with self-reported incidences being highest in Asia and Africa. 26 However, the impact of this interaction was quite small, and the first model remained the simplest model with the least symptoms included but with the highest inclusion values.
The models obtained from this analysis are not very specific to dengue but do have a relatively high sensitivity. According to the WHO guideline and other publications, hematocrit, complete blood count (especially neutrophils, platelets, and lymphocytes), as well as certain inflammatory markers can provide a more specific clue into definitive diagnosis and whether patient may be at risk of developing hemorrhagic symptoms and shock, and thus further guide the formation of management plan. 11,27,28 Future work on the algorithm may consider addition of laboratory results, such as white blood cells and platelets count, as independent variables that may improve specificity of the model. However, the objective in the development of these models was to provide help with initial evaluation, encouraging physicians to include dengue fever in the list of differential diagnoses and to pursue further examinations that can provide a more specific prediction value, as laboratory results are still required for confirmation of dengue diagnosis. A sensitive model at the initial step of evaluation will hopefully reduce the number of misdiagnosed dengue cases, and its related morbidity and mortality.
Limitations.
One limitation of this study involved the source of data being from volunteers who had suspected dengue infections during their previous oversea trips. This recruitment method may falsely elevate the prevalence of dengue among travelers, which would also be unquestionably higher than that found in the general non-endemic population. However, the model may still be applicable for travelers presenting with febrile symptoms, considering the higher pretest probability in patients with similar history. A previous study in missionaries returning from Jamaica found that 21% of survey respondents experienced acute febrile illness related to the trip, and 27% of available serum samples had seroconversion consistent with recent dengue. 29 Although the prevalence can affect the PPV and NPV of the model, the sensitivity and specificity should remain the same if the model should be applicable to other populations as anticipated. That said, our model would benefit from further prospective validation in the appropriate travel clinic settings and may prove most useful for excluding, rather than predicting, dengue disease.
This study may also be influenced by recall bias due to participant’s self-reported symptoms when answering the questionnaire. The effect of recall bias should have been minimized by using the same procedure of data collection in all participating volunteers. In addition, the questionnaire was delivered before the blood sample was collected, and subjects were blinded to serology results. However, because of the retrospective design of the study, the seropositive evidence could not completely prove that the symptomatic cases represent true dengue. For instance, participants who had a history of frequent travels may have been exposed to dengue before the reported illness episode. In this case, the seropositive result, of dengue or otherwise, would then be falsely associated with the reported symptoms. Additional potential problem related to the recollection of symptoms was that it was difficult to pinpoint which symptoms appeared earlier or later in the illness process. This can limit the actual application of the model in real clinical practice when patients are presenting to clinic earlier in symptoms manifestation, whereas the model may include symptoms that only appear later in the process of the disease. It would be essential for the model to be validated with different datasets for its application in different clinical settings.
CONCLUSION
Our study demonstrated that despite the seemingly nonspecific symptoms individually, a model including a combination of symptoms can help predict the diagnosis of dengue at a relatively high sensitivity, albeit with poor specificity. For clinicians in non-endemic areas evaluating patients for possible dengue infection with a positive travel history, we recommend an increased index of suspicion, especially in patients with fever, difficulty breathing, and absence of rhinorrhea; minor criteria may include rash, absence of paired nausea–diarrhea symptom, with or without joint, bone, and muscle pain. The purpose of this model was not to replace the existing criteria for dengue diagnosis, but rather to serve as a tool at triage or at the initial phase of history-taking and to help widen the differential diagnoses. Such a tool would prompt physicians to consider tropical infectious disease as a possibility in febrile patient with a recent travel history or in patients living in non-endemic areas with previous autochthonous cases and outbreaks. The model should aid in making the decision to perform additional examinations or order further laboratory tests. The goal was to provide the right management plan and close observation when necessary, as the consequence of missing the true diagnosis could be lethal. Under-recognition of dengue may lead to increased morbidity and mortality, whereas early detection is shown to help improve patient outcomes. With additional validation on other cohorts, the preliminary models developed in this study, including the regression-derived equation and the decision tree, have potential for clinical utilization.
Supplemental tables
ACKNOWLEDGMENTS
We would like to thank our colleagues from Siriraj Hospital, Mahidol University, for sharing their pearls of wisdom with us during the course of this research. We are grateful to Dumrong Mairiang, Panisadee Avirutnan, and Prida Malasit, who provided insight and expertise that greatly assisted the study, although they may not agree with all of the interpretations/conclusions of this article. We would like to thank Justin Denny and OHSU Global for making this research opportunity possible.
Note: Supplemental tables appear at www.ajtmh.org.
REFERENCES
- 1. Guzman MG, Harris E, 2015. Dengue. Lancet 385: 453–465. [DOI] [PubMed] [Google Scholar]
- 2. Bouri N, Sell TK, Franco C, Adalja AA, Henderson DA, Hynes NA, 2012. Return of epidemic dengue in the United States: implications for the public health practitioner. Public Health Rep 127: 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Overbosch FW, Schinkel J, Stolte IG, Prins M, Sonder GJB, 2018. Dengue virus infection among long-term travelers from The Netherlands: a prospective study, 2008–2011. PLoS One 13: e0192193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmidt-Chanasit J, Tenner-Racz K, Poppert D, Emmerich P, Frank C, Dinges C, Penning R, Nerlich A, Racz P, Günther S, 2011. Fatal dengue hemorrhagic fever imported into Germany. Infection 40: 441–443. [DOI] [PubMed] [Google Scholar]
- 5. Gubler DJ, 1998. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 11: 480–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leder K, et al. 2013. GeoSentinel surveillance of illness in returned travelers, 2007–2011. Ann Intern Med 158: 456–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anez G, Rios M, 2013. Dengue in the United States of America: a worsening scenario? Biomed Res Int 2013: 678645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Effler PV, et al. Hawaii Dengue Outbreak Investigation Team , 2005. Dengue fever, Hawaii, 2001–2002. Emerg Infect Dis 11: 742–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beaumier C, Garcia MN, Murray KO, 2014. The history of dengue in the United States and its recent emergence. Curr Trop Med Rep 1: 32–35. [Google Scholar]
- 10. Kamal M, Kenawy MA, Rady MH, Khaled AS, Samy AM, 2018. Mapping the global potential distributions of two arboviral vectors Aedes aegypti and Ae. albopictus under changing climate. PLoS One 13: e0210122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. WHO , 2009. Dengue Guidelines for Diagnosis, Treatment, Prevention, and Control. Geneva, Switzerland: World Health Organization. [PubMed] [Google Scholar]
- 12. Nguyen MT, et al. 2017. An evidence-based algorithm for early prognosis of severe dengue in the outpatient setting. Clin Infect Dis 64: 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yacoub S, Wills B, 2015. Dengue: an update for clinicians working in non-endemic areas. Clin Med (Lond) 15: 82–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Low JG, et al. 2011. The early clinical features of dengue in adults: challenges for early clinical diagnosis. PLoS Negl Trop Dis 5: e1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McFarlane ME, Plummer JM, Leake PA, Powell L, Chand V, Chung S, Tulloch K, 2013. Dengue fever mimicking acute appendicitis: a case report. Int J Surg Case Rep 4: 1032–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Verma S, Kanga A, Singh D, Verma G, Mokta K, Ganju S, Sharma V, 2014. Emergence of travel: associated dengue fever in a non-endemic, Hilly state. Adv Biomed Res 3: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Widman DG, et al. 2017. Transplantation of a quaternary structure neutralizing antibody epitope from dengue virus serotype 3 into serotype 4. Sci Rep 7: 17169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Messer WB, Yount B, Hacker KE, Donaldson EF, Huynh JP, de Silva AM, Baric RS, 2012. Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization. PLoS Negl Trop Dis 6: e1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Russell PK, Nisalak A, Sukhavachana P, Vivona S, 1967. A plaque reduction test for neutralizing antibodies. J Immunol 9: 285–290. [PubMed] [Google Scholar]
- 20. Kraus AA, Messer W, Haymore LB, de Silva AM, 2007. Comparison of plaque- and flow cytometry-based methods for measuring dengue virus neutralization. J Clin Microbiol 45: 3777–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilder-Smith A, Schwartz E, 2005. Dengue in travelers. N Engl J Med 353: 924–932. [DOI] [PubMed] [Google Scholar]
- 22. Phuong CXNN, Kneen R, Thuy PT, van Thien C, Nga NT, Thuy TT, Solomon T, Stepniewska K, Wills B, Dong Nai Study Group , 2004. Clinical diagnosis and assessment of severity of confirmed dengue infections in Vietnamese children: is the World Health Organization classification system helpful? Am J Trop Med Hyg 70: 172–179. [PubMed] [Google Scholar]
- 23. Callister LC, 2016. Cultural influences on pain perceptions and behaviors. Home Health Care Manage Pract 15: 207–211. [Google Scholar]
- 24. Campbell CM, Edwards RR, 2012. Ethnic differences in pain and pain management. Pain Manag 2: 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carrillo-Hernandez MY, Ruiz-Saenz J, Villamizar LJ, Gomez-Rangel SY, Martinez-Gutierrez M, 2018. Co-circulation and simultaneous co-infection of dengue, chikungunya, and Zika viruses in patients with febrile syndrome at the Colombian-Venezuelan border. BMC Infect Dis 18: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dahl V, Wallensten A, 2017. Self-reported infections during international travel and notifiable infections among returning international travellers, Sweden, 2009–2013. PLoS One 12: e0181625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brasier AR, et al. 2012. A three-component biomarker panel for prediction of dengue hemorrhagic fever. Am J Trop Med Hyg 86: 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wangrangsimakul T, Althaus T, Mukaka M, Kantipong P, Wuthiekanun V, Chierakul W, Blacksell SD, Day NP, Laongnualpanich A, Paris DH, 2018. Causes of acute undifferentiated fever and the utility of biomarkers in Chiangrai, northern Thailand. PLoS Negl Trop Dis 12: e0006477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moncayo AC, et al. 2015. Dengue among American missionaries returning from Jamaica, 2012. Am J Trop Med Hyg 92: 69–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.