Abstract.
Leishmania RNA virus (LRV) is a double-stranded RNA virus belonging to the Totiviridae family detected as cytoplasmic inclusions in some strains of the human parasite Leishmania spp. Experimental evidence supports the hypothesis that human coinfection with Leishmania spp.–LRV triggers an exacerbated immune response in the host that can be responsible for the observed complicated outcomes in cutaneous leishmaniasis (CL), such as mucosal leishmaniasis (ML) and treatment failure of CL. However, the reported frequencies of LRV associated with complicated outcomes in patient’s series are highly variable, diminishing the relevance on the virus presence in the pathogenesis of the disease. To assess whether or not the inconsistent information about the frequency of LRV associated with CL complicated outcomes could be related to the virus detection approach, the present study evaluated the LRV presence in clinical samples using a diagnostic algorithm according to the type of the sample. In 36 samples with diagnosis of complicated forms of CL (15 of ML and 21 of CL antimony treatment failure) and six samples with non–Leishmania spp. infection, the LRV presence was assessed by RT-PCR, RT-qPCR, and nested RT-PCR. Viral load was estimated in parasite clinical isolates. By combining the methods, LRV1 presence was confirmed in 45% (9/20) of isolates and 37.5% (6/16) of the incisional biopsies. Remarkably, in some cases (4/8), LRV1 was undetectable in the isolates but present in their respective biopsies, and less frequently, the opposite was observed (1/8), suggesting the possibility of loss of parasites harboring LRV1 during the in vitro growth.
INTRODUCTION
Leishmania species belonging to the Viannia subgenus are predominant in South America where they generate complicated forms of the disease such as mucosal leishmaniasis (ML)1,2 and treatment failure of cutaneous leishmaniasis (CL).3,4
The occurrence of the aforementioned complications has been associated with the presence of a cytoplasmic virus in the infecting parasite, known as Leishmania RNA virus (LRV). This virus belongs to the Totiviridae family, a group of RNA viruses present in other protozoa and fungi.5–9 Because of differences in sequence and genome organization between LRVs associated with Old World and New World Leishmania species, they are classified as LRV2 and LRV1, respectively.10–13
It has been demonstrated that metastasizing (but not non-metastasizing) strains of Leishmania (Viannia) guyanensis have high LRV1 burdens. In addition, there is experimental evidence that the LRV presence induces hyperimmune responses to Leishmania infection promoting pro-inflammatory responses with high levels of tumor necrosis factor α (TNFα, IL-6, chemokine (C-C motif) ligand 5 (CCL5), and chemokine (C-X-C motif) ligand (CXCL10), similar to the immunological profile observed in patients suffering from ML.14–16 Converging with this line of evidence is the observation that pro-inflammatory IL-17 levels are high in patients with chronic CL produced by L. (V.) guyanensis–LRV1+.17
Supporting the hypothesis that Leishmania spp.–LRV coinfection worsens disease prognosis through a Type 1 interferon response, murine model studies have shown that host coinfection with L. (V.) guyanensis–LRV1+ or L. (V.) guyanensis+ exogenous interferon-inducing viruses (e.g., lymphocytic choriomeningitis virus or Toscana virus) produces similar clinical disease, in which relapse risk is increased secondary to parasite reactivation.18
One clinical study has shown a strong association between LRV1 coinfection of Leishmania (Viannia) braziliensis and pentavalent antimony treatment failure of CL and ML.19 Furthermore, the risk of CL, associated with L. (V.) guyanensis, relapse appears to increase during pentamidine treatment when the parasite is infected with LRV1.20
At the epidemiologic level, LRV1-2 has been found in clinical samples from patients with leishmaniasis in variable frequencies, ranging from total absence21,22 to frequencies that can reach 87%.15,17,19,20,23–33 The strength of the association between viral presence and complication development varies according to the study24,28 and among different South American regions.22 The wide range in LRV1 frequency of detection in clinical samples may reflect diverse experimental approaches that do not account for differences in clinical specimens.
To overcome detection biases in identifying LRV1 in clinical samples, the present study was designed to determine LRV1 presence in samples from complicated Colombian patients, suffering from CL and presenting therapeutic failure to antimony treatment or ML, through the use of complementary approaches taking into account the sample source, the parasite species, and the viral load.
MATERIALS AND METHODS
Ethics statement.
Research in this study was subject to ethical review and approved by the ethics committees from the participant institutions, in accordance with national (resolution 008430 of the Colombian Health Ministry) and international (Declaration of Helsinki and amendments, World Medical Association, Korea 2008) guidelines. All clinical samples had been taken from patients as part of normal diagnosis and treatment, with no unnecessary invasive procedures and with written informed consent. Guiding Principles for Biomedical Research Involving Animals (Council for International Organizations of Medical Sciences) were followed regarding animal experimentation.
Type of study and samples.
The present study corresponds to a descriptive study with an experimental component using 36 samples from patients with diagnosis of complicated forms of CL, all previously collected for diagnostic purpose. Fifteen samples from 13 patients were diagnosed as ML, of which nine were frozen incisional biopsies and six correspond to parasite isolates, and 21 samples from 14 patients were diagnosed as CL with therapeutic failure to antimony, of which seven correspond to frozen incisional biopsies and 14 to parasite isolates.34 In addition, six samples were included as negative controls and correspond to frozen nasal incisional biopsies from patients with confirmed diagnosis different to ML such as lepromatous leprosy, traumatic piercing, sporotrichosis, chronic hyperplastic eosinophilic sinusitis, deep mycosis, and squamous cell carcinoma.
Parasite isolates.
Stocks of cryopreserved parasite isolates were thawed and seeded in Senekjie medium.35 Once adequate growth of the parasites was achieved, they were amplified until reaching the stationary phase in Schneider medium 10% fetal bovine serum (FBS), at a temperature of 26°C. Reference strains of L. (V.) guyanensis (MHOM/BR/75/M4147) and Leishmania (Viannia) panamensis (MHOM/PA/71/LS94) grown on Schneider medium 10% FBS were used as positive and negative controls for LRV1 infection, respectively.
Clinical samples and hamster biopsies.
Fragments of about 3 mm from patient’s biopsies were kept in a dry sterile tube at −80°C until RNA and DNA extraction. To standardize a protocol for LRV1 detection in tissue from biopsies, two young male golden hamsters (Mesocricetus auratus) were inoculated subcutaneously in their snouts and footpads with 1.8 × 106 (LRV1+)-MHOM/BR/75/M4147 metacyclic promastigotes. Three weeks after inoculation, the animals were sacrificed, and excisional biopsy was collected for proceeding to acid nucleic extraction.
RNA extraction and retrotranscription.
A Direct-zol™ RNA MiniPrep kit (Zymo Research, Irvine, CA) was used for the extraction of RNA from 3.5 × 107 to 3.4 × 108 stationary-phase promastigotes. For the biopsies, extraction was made using the Quick-RNA™ MiniPrep Plus kit (Zymo Research) following the manufacturer’s instructions. cDNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s recommendations in a final volume of 20 μL, using 2 μg and 1 μg of RNA from the isolates and the biopsies, respectively.
Primers and 18S-Leishmania spp. PCR amplification.
Real-time PCR and RT-qPCR for Leishmania spp.-18S were used for confirming the parasite presence and the cDNA quality in biopsies by following modified versions of the protocol originally described by van den Bogaart et al.36 In brief, RT-PCR was performed in 50 µL of mixture containing Taq buffer KCl; 1.65 mM MgCl2; 200 µM deoxyadenosine triphosphate (dATP), deoxycytidine triphosphate (dCTP), deoxythymidine triphosphate (dTTP), and deoxyguanosine triphosphate (dGTP); 0.4 µM of each primer; 2.5 units Taq DNA polymerase; and 1 µL of isolate cDNA. The amplification was performed in a Bio-Rad C1000 Touch thermocycler (BioRad, Hercules, CA) with one initial denaturalization step of 94°C for 5 minutes, followed by 32 cycles at 94°C for 30 seconds, 65°C for 30 seconds, and 72°C for 30 seconds. The amplified fragments were visualized on a 2% agarose gel stained with 0.5 µg/mL of ethidium bromide. Real-time qPCR was performed with SYBR Green Select Master mix (Thermo Fisher Scientific). Primer concentration was 0.6 µM, and the amplification was carried out in an Applied Biosystems 7500 Real-Time PCR System (Foster City, CA) with one initial uracil-DNA glycosylase (UDG) activation of 50°C for 2 minutes, one denaturalization step of 95°C for 2 minutes, followed by 45 cycles at 94°C for 30 seconds, and 60°C for 45 seconds. cDNAs from promastigotes of reference strain (LRV1+)-MHOM/BR/75/M4147 and from hamster snout biopsy infected with this were used as positive control for the RT-PCR and RT-qPCR, respectively. The mixture without cDNA corresponding to non-template control (NTC) was used as negative control. This RT-qPCR was used for generating a parasite standard curve with 10-fold serial dilutions of quantified promastigotes from 5 × 105 to five parasites/reaction, and then the load was calculated using Applied Biosystems 7500 Real-Time PCR software tools.
Leishmania RNA virus 1 detection in cDNA from Leishmania spp. isolates.
Real-time PCR with the set of primers described by Zangger et al.26 was performed. Those primers amplified a ∼485-bp capsid fragment. Reaction was set in 50 µL of mixture with KCl Taq buffer; 2.5 mM MgCl2; 200 µM dATP, dCTP, dTTP, and dGTP; 0.5 µM of each primer; 2.5 units Taq DNA polymerase; and 2 µL of the isolate complementary DNA (cDNA). The amplification was carried out in a Bio-Rad C1000 Touch thermocycler with one initial denaturalization step of 94°C for 3 minutes, followed by 30 cycles at 94°C for 1 minute, 50°C for 1 minute, and 72°C for 1 minute, and a final extension at 72°C for 5 minutes. The amplified fragments were visualized on 1% agarose gels stained with 0.5 µg/mL of ethidium bromide.
Also, RT-qPCRs were performed for the same set of samples using the two different sets of primers designed by Ito et al.27 and by Ramos Pereira et al.24 (Supplemental Table 2). Itos’ primers were used at 0.2 μM, and Ramos’ primers were used at 0.3 μM; they amplified fragments of ∼245 bp and 90 bp on open reading frame 1 (ORF1) respectively. Reactions were set in 10 µL of mixture contained SYBR Green Select Master mix™ 1× (Applied Biosystems) and the equivalent to 100 ng of isolate cDNA. The amplification was performed in an Applied Biosystems 7500 Real-Time PCR System with one initial UDG activation step of 50°C for 2 minutes and one denaturalization step of 95°C for 2 minutes, followed by 40 cycles at 94°C for 15 seconds and 60°C for 1 minute.
To rule out an isolate as negative for LRV1 presence, the published nested RT-PCR protocol24 with modifications as described in next section was carried out. For RNA derived from isolates, 2 µg of RNA was used for retrotranscription and conventional 18S gene PCR performed to verify the quality of the cDNA. Once the expected result was obtained, the nested PCR for viral detection was conducted using primers described in Supplemental Table 2, and the PCR products were evaluated on a 2% agarose gel stained with 0.5 µg/mL of ethidium bromide.
As positive control in RT-PCR, RT-qPCRs, and nested RT-PCR, cDNA from reference strain (LRV1+)-MHOM/BR/75/M4147 was used, and as negative control, NTC was used.
Leishmania RNA virus 1 detection in cDNA from biopsies of nested RT-PCR products.
The published nested PCR protocol24 was optimized by the use of cDNA from the L. (V.) guyanensis–LRV+ infected tissue from hamsters. The thermal profile included a cycle of 95°C for 3 minutes, 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. The mixture was made to a final volume of 50 μL containing Taq buffer with KCl; 1.5 mM MgCl2; 200 μM dATP, dCTP, dTTP, and dGTP; 2.5 units of recombinant Taq polymerase; and 0.3 μM of each primer24 (Supplemental Table 2). The amplified fragments were 125 bp and 90 bp for the first and second round, respectively, and they were visualized in agarose gels. As a positive control, cDNA from (LRV1+)-MHOM/BR/75/M4147 promastigotes was used. Negative controls correspond to cDNA from the six samples of frozen biopsies from patients with confirmed diagnosis different to ML or CL. For this nested RT-PCR, negative controls given by NTC were needed every two to three samples to detect cross-contamination.
Sequencing nested RT-PCR products.
The 90-bp nested-PCR products were cloned into pGEM®-T easy Vector (Promega A137A, Madison, WI) in Escherichia coli JM109 cells. Plasmid DNA was obtained using a ZR Plasmid Miniprep™-Classic Kit (Zymo Research D4015). The sequencing was performed by Macrogen sequencing service using universal primers T7-SP6. Nucleotide sequences were deposited in the GenBank with accession numbers MK430135 (biopsy B8), MK430136 (biopsy B9), MK430137 (biopsy B11), MK430138 (biopsy B44), MK430139 (biopsy B312), and MK430140 (biopsy B333).
Viral load quantification.
A plasmid was constructed (TOP-LRV1-16-435) containing a fragment of 420 bp corresponding to positions 16–435 of LRV1 ORF1. The fragment was amplified from (LRV1+)-MHOM/BR/75/M4147 cDNA using the previously described forward primer27 and a new designed reverse primer (Supplemental Table 2). The PCR product was inserted into the PCR-4-TOPO® TA Vector (K457502, Invitrogen, Carlsbad, CA), following the manufacturer’s instructions and the construct amplified in E. coli strain One Shot™ TOP10 (Invitrogen). Standard curves based on nanograms of plasmid TOP-LRV1-16-435 were created using 10-fold serial dilutions, and the plasmid copy number was estimated according to the formula described from 6.6 × 107 to 66 plasmid copies/reaction (Supplemental Table 4).
To quantify the viral load in LRV1-positive isolates, Ito’s 27 and Ramos Pereira’s 24 primers were used to perform two independent modified RT-qPCRs. The viral load was calculated by Applied Biosystems 7500 Real-Time PCR software, using a linear regression equation for interpolating from the standard curves. Then the viral load was adjusted according to the parasite load estimated in each sample throughout the parasite standard curve.
Leishmania species identification.
Leishmania species identification was made using hsp70 PCR-restriction fragment length polymorphism (RFLP); species identification from isolates was performed according to Garcia et al.37 and Montalvo et al.38 and for biopsies according to Cruz-Barrera et al.39
RESULTS AND DISCUSSION
Colombia has one of the world’s highest CL incidences.40,41 Approximately 10,000 new cases are reported each year, with L. (V.) panamensis and L. (V.) braziliensis being the most common agents associated with human disease.42–44 The complication rates for ML and treatment failure are not well-known, but estimations made in other South American regions report that 10% of CL cases associated with L. (V.) braziliensis result in ML42,43 and 25% of CL cases treated with antimony salts result in therapeutic failure.45,46 Hence, a more detailed epidemiologic picture is urgently needed to better understand CL prognosis.
Understanding the role of LRV1 in human CL outcomes requires an ambitious epidemiologic design and robust detection methods using a multicenter study. Therefore, this study was aimed at evaluating the possible biases in LRV1 detection when using the simplest and cheapest available approaches.24–27
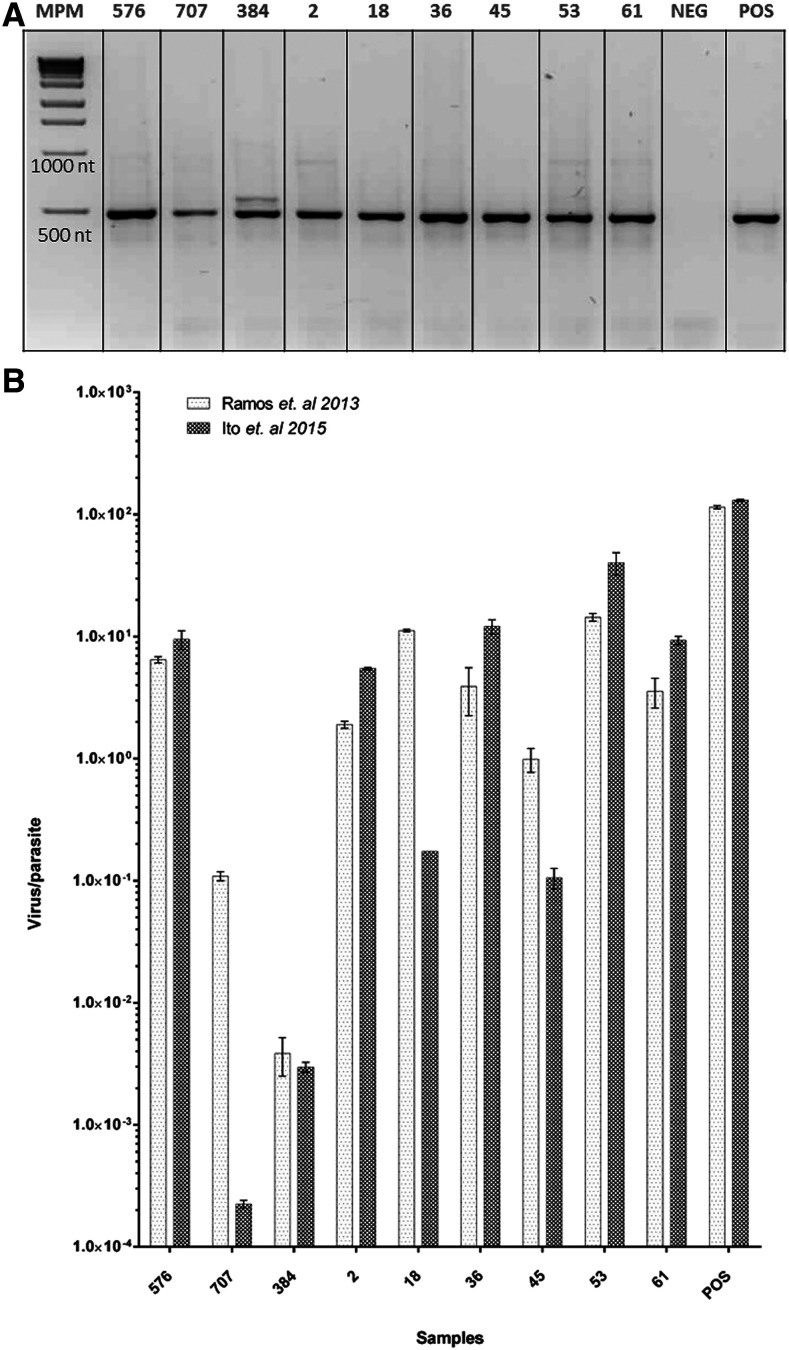
A multistep process was used to create an LRV1 detection algorithm based on the sample type. First, samples were classified as isolates (n = 20) or biopsies (n = 16). In case of isolates, RT-PCR26 was performed on cDNA from 19 L. (V.) braziliensis and one L. (V.) panamensis, isolates from lesions of patients with ML (n = 6) or CL with antimony therapeutic failure (n = 14). The expected product of ∼485 bp corresponding to a viral genome region located at positions 1089–1574 (ORF2) was observed in nine of 20 isolates (45%) (Figure 1A).
Figure 1.
Real-time PCR (RT-PCR) is useful for detection of low viral loads in Leishmania spp. isolates infected with Leishmania RNA virus 1 (LRV1). (A) Amplification of LRV RNA capsid fragment (∼485 bp) using modified RT-PCR.26 MM = molecular marker, Positive (POS): cDNA from (LRV1+)-MHOM/BR/75/M4147, Negative (NEG): non-template control (NTC). (B) Viral loads of the same samples presented in (A), estimated using two different modified RT-qPCRs, Ramos Pereira’s,24 and Itos.’27 Labels on the top of the gel in (A) and on x axis in (B) correspond to isolate ID according to Table 1.
Considering that different Leishmania strains can carry different LRV1 loads with slight sequence variation, an alternative detection method was used to determine whether a more conserved nucleotide sequence region, as it is LRV1-ORF1, could be used to improve detection sensitivity.10,11,13,33 This detection was accomplished by using a modified version of the RT-qPCR method described by Ito et al.27 and a modified RT-qPCR version from the one described by Ramos Pereira et al.24 Real-time qPCR results were consistent between them and with RT-PCR, detecting the LRV1 presence in the same nine isolates (Figure 1B, Table 1). When comparing the viral load estimated by each one of the two RT-qPCRs and the positive control ([LRV1+]-MHOM/BR/75/M4147), most of the isolates showed equivalent results (Figure 1B). However, the RT-qPCR using Ramos’ primers estimated a higher viral load per parasite in isolate numbers 18, 45, and 707, as compared with the estimates made of RT-qPCR using Ito’s primers (Figure 1B). This loss of accuracy could obey to a slight loss of specificity of Ito’s primers for the target sequence in those isolates. Efficiency, calculated using the TOP-LRV1-16-435 standard curve for both RT-qPCRs, is identical (Supplemental Table 3). Nevertheless, whereas the target sequence of both reverse primers and Ramos’ forward primer is conserved in all 36 available LRV1 sequences in GenBank, for the target region corresponding to Itos’ forward primer, the fragment is missing in 19 of the 36 sequences mentioned previously. Therefore, it is possible that LRV1 infecting isolates 18, 45, and 707 present some degree of polymorphism in the target region such that ito’s forward primers display a decrease in the qPCR efficiency, hammering the accuracy of the quantification.
Table 1.
Features of the clinical samples included in this study
| Clinical condition | Sample type | ORF1 RT-qPCR viral load (virus/parasite) | Nested RT-PCR | 18S RT-qPCR | Leishmania RNA virus 1 status | Lesion localization | Leishmania species | Region | |
|---|---|---|---|---|---|---|---|---|---|
| Ito et al.27 | Ramos et al.24 | CT | |||||||
| ML | Isolate | 9,50E+00 | 6,47E+00 | NA | NA | Positive | Nose | L. (V.) braziliensis | Amazon |
| ML | Isolate | 2,24E−04 | 1,09E−01 | NA | NA | Positive | Cheek | L. (V.) braziliensis | Andean |
| ML | Isolate | NA | NA | NA | NA | Negative | Nose | L. (V.) panamensis | Andean |
| ML | Isolate | NA | NA | NA | NA | Negative | Nose | L. (V.) braziliensis | Orinoquía |
| ML | Isolate | 2,97E−03 | 3,84E−03 | NA | NA | Positive | Nose | L. (V.) braziliensis | Andean |
| ML | Isolate | NA | NA | NA | NA | Negative | Nose | L. (V.) braziliensis | Andean |
| CLTF | Isolate | 5,47E+00 | 1,90E+00 | NA | NA | Positive | Face and neck | L. (V.) braziliensis | Amazon |
| CLTF | Isolate | NA | NA | NA | NA | Negative | Legs | L. (V.) braziliensis | Orinoquía |
| CLTF | Isolate | 1,74E−01 | 1,12E+01 | NA | NA | Positive | Upper limbs | L. (V.) braziliensis | Orinoquía |
| CLTF | Isolate | NA | NA | NA | NA | Negative | Upper limbs | L. (V.) braziliensis | Amazon |
| CLTF | Isolate | NA | NA | NA | NA | Negative | Multiple injuries | L. (V.) braziliensis | Orinoquía |
| CLTF | Isolate | NA | NA | NA | NA | Negative | Multiple injuries | L. (V.) braziliensis | Orinoquía |
| CLTF | Isolate | NA | NA | NA | NA | Negative | Upper limbs | L. (V.) braziliensis | Orinoquía |
| CLTF | Isolate | NA | NA | NA | NA | Negative | Upper limbs | L. (V.) braziliensis | Orinoquía |
| CLTF | Isolate | 1,21E+01 | 3,91E+00 | NA | NA | Positive | Upper limbs | L. (V.) braziliensis | Orinoquía |
| CLTF | Isolate | 1,06E−01 | 9,90E−01 | NA | NA | Positive | Multiple injuries | L. (V.) braziliensis | Orinoquía |
| CLTF | Isolate | NA | NA | NA | NA | Negative | Multiple injuries | L. (V.) braziliensis | Amazon |
| CLTF | Isolate | 4,02E+01 | 1,44E+01 | NA | NA | Positive | Upper limbs | L. (V.) braziliensis | Orinoquía |
| CLTF | Isolate | 9,31E+00 | 3,57E+00 | NA | NA | Positive | Upper limbs | L. (V.) braziliensis | Amazon |
| CLTF | Isolate | NA | NA | NA | NA | Negative | Face and neck | L. (V.) braziliensis | Pacific |
| ML | Biopsy | NA | NA | Positive | 32,22 | Positive | Nose | Leishmania spp. | Andean |
| ML | Biopsy | NA | NA | Negative | 34,73 | Negative | Nose | L. (V.) braziliensis | Orinoquía |
| ML | Biopsy | NA | NA | Negative | 34,47 | Negative | Nose | L. (V.) panamensis | Andean |
| ML | Biopsy | NA | NA | Positive | 35,11 | Positive | Nose | L. (V.) braziliensis | Andean |
| ML | Biopsy | NA | NA | Negative | 30,3 | Negative | Nose | Leishmania spp. | Caribbean |
| ML | Biopsy | NA | NA | Negative | 27,72 | Negative | Nose | L. (V.) panamensis | Andean |
| ML | Biopsy | NA | NA | Negative | 32,59 | Negative | Nose | L. (V.) braziliensis | Amazon |
| ML | Biopsy | NA | NA | Negative | 35,64 | Negative | Nose | L. (V.) braziliensis | Orinoquía |
| ML | Biopsy | NA | NA | Negative | 31,45 | Negative | Nose | L. (V.) braziliensis | Orinoquía |
| CLTF | Biopsy | NA | NA | Positive | 32,45 | Positive | Legs | L. (V.) braziliensis | Orinoquía |
| CLTF | Biopsy | NA | NA | Negative | 28,07 | Negative | Upper limbs | L. (V.) braziliensis | Amazon |
| CLTF | Biopsy | NA | NA | Positive | 24,85 | Positive | Multiple injuries | L. (V.) braziliensis | Orinoquía |
| CLTF | Biopsy | NA | NA | Positive | 21,98 | Positive | Multiple injuries | L. (V.) braziliensis | Orinoquía |
| CLTF | Biopsy | NA | NA | Positive | 27,08 | Positive | Upper limbs | L. (V.) braziliensis | Orinoquía |
| CLTF | Biopsy | NA | NA | Negative | 29,59 | Negative | Multiple injuries | L. (V.) braziliensis | Amazon |
| CLTF | Biopsy | NA | NA | Negative | 31,44 | Negative | Face and neck | L. (V.) braziliensis | Pacific |
| Lepromatous leprosy | Biopsy | NA | NA | Negative | Undetermined | Negative | Nose | NA | No data |
| Traumatic piercing | Biopsy | NA | NA | Negative | Undetermined | Negative | Nose | NA | No data |
| Sporotrichosis | Biopsy | NA | NA | Negative | Undetermined | Positive | Nose | NA | No data |
| Acute and chronic reactions secondary to insect bite | Biopsy | NA | NA | Negative | Undetermined | Negative | Nose | NA | No data |
| Deep mycosis | Biopsy | NA | NA | Negative | Undetermined | Negative | Nose | NA | No data |
| Squamous cell carcinoma | Biopsy | NA | NA | Negative | Undetermined | Negative | Nose | NA | No data |
CLTF = cutaneous leishmaniasis with therapeutic failure; L. (V.) braziliensis = Leishmania (Viannia) braziliensis; L. (V.) panamensis = Leishmania (Viannia) panamensis; ML = mucosal Leishamniasis; NA not applicable.
In the present study, before ruling out an isolate as LRV1-negative, multiple attempts for detection by RT-qPCR were carried out with variable amounts of RNA ranging from 6 to 200 ng, and conventional and nested RT-PCR were conducted (data not shown). For samples corresponding to biopsies, a different chain of procedures was performed. Previously, to process samples for LRV1 detection, RT-qPCR for Leishmania spp. 18S gene36
was carried out for all cDNA extracted from biopsy specimens (Table 1). This step allowed us to include in the analysis only cDNA from tissue where the parasite was detectable. Real-time qPCR for Leishmania spp. 18S was also useful to provide evidence of the absence of parasites in all of the six specimens used as true negative controls for detection of LRV1 in biopsies (Table 1).
To detect LRV1 in biopsies, it was necessary to implement a nested RT-PCR,24 given that the RT-PCR and RT-qPCR methods used for LRV1 detection in parasite isolates were not feasible for that purpose in the experimental positive control. The nested RT-PCR technique was applied to 22 biopsy specimens from patients with ML (n = 9), CL with antimony treatment failure (n = 7), and negative controls (n = 6). Leishmania RNA virus 1 was detected in six biopsies of 16 (37.5%): two from patients with ML and four from patients corresponding to CL with antimony treatment failure. The 90-bp fragments produced in each case were sequenced.
For the eight patients for whom both types of samples were available, incisional biopsy and parasite isolate (Supplemental Table 1), an analysis was conducted to evaluate simultaneously the LRV1 presence in biopsies and in their respective isolates. Leishmania RNA virus 1 presence was established in four biopsies from patients with CL, in which in their corresponding isolates, LRV1 was not detectable, as assessed by RT-PCR, nested RT-PCR, and RT-qPCR. The opposite was observed in one patient with diagnosis of ML (Supplemental Table 1).
Unfortunately, samples of the two types, parasite isolates and incisional biopsies, were available only for eight of 36 patients; interestingly, in five of them, the results of LRV1 presence were not concordant (Supplemental Table 1). The use of complementary approaches in this study led us to the important finding that LVR1 was not detectable in some parasite isolates growing in vitro, whereas their corresponding biopsies presented LRV1-detectable levels. The cause for the biological events responsible for this loss of LRV1 signal was not addressed in the present study. However, it is important to note whether loss of viral detection randomly occurs, for example, because of the media growth selection among Leishmania, thus allowing the expansion of LRV1-negative over LRV1-positive parasites; previous studies using LRV detection exclusively in isolates may have underestimated the rate of viral coinfection.17,19,21,22,29,31,47,48
The same can occur when LRV detection is performed in samples with low parasite load as biopsies, smears, or swaps, taken from patients with ML.23,24,27,28 In the present study, one biopsy turned out to be negative for LRV1 presence when assessed by nested RT-PCR, whereas its corresponding isolate was positive (Supplemental Table 1). We speculated that this may be due to the low parasite load in the sample according to the threshold cycle (CT) value observed when detecting parasites using 18S primers (Table 1, sample encoded as M354). Although the purpose of the present work was not to compare the parasite load between different types of samples, this brings our attention to the observation that the average CT value for 18S when detecting the parasite in CL biopsies was about 27.92 (±3.68), whereas in biopsies for ML lesions, it was 32.73 (±2.60), suggesting a lower parasite burden in theses samples as previously demonstrated.49 Further studies will be necessary to assess the tissue’s parasitic load threshold for LRV1 detection.
The predominant species analyzed in this study was L. (V.) braziliensis (31/36) followed by L. (V.) panamensis (3/36). In two cases, it was not possible to discriminate the Leishmania species, given the hsp70-RFLP mixed pattern obtained (Table 1). Although there is a geographic bias in this study and a reduced number of samples, our findings suggest that Leishmania strains harboring LRV1 may have spread in Colombia (Table 1) since 1996, when a report of 69 Leishmania strains showed low-LRV1 parasite infection rates found exclusively in the Amazon region.29
As mentioned, the predominant species in this set of samples was L. (V.) braziliensis. Available evidence suggests that the frequency of LRV1 infection in isolates from this species ranges from total absence to 33%.19,21,22,29,47 Nevertheless, LRV1 infecting L. (V.) braziliensis show regional differences in South America as reported in a study carried out in French Guiana where 80% of isolates belonging to this species was LRV1 infected,31 and in this same area, the virus has been found associated with isolates of L. (V.) guyanensis at a frequency ranging from 38.5% to 88%.7,20,31,48 Despite the absence of data available for the LRV1 rate of occurrence in L. (V.) guyanensis outside French Guiana, we have not found LRV1 in Colombian L. (V.) guyanensis isolates from CL patients (data not published). Therefore, our findings follow the line of evidence that outside of the Guiana Shield, the circulation of LRV1 infected parasites could be low.
Moreover, if Leishmania human infection is produced by a mixed population of LRV1 (+) and LVR1 (−) clones and dominant clones are in vitro selected, the probability for selecting LRV1 (+) isolates would be influenced by the geographical origin. Therefore, selection of LRV (−) clones can be favored, diminishing the clinical relevance of LRV1 infection.
Findings from the present work suggest that LRV1-infected Leishmania parasites can harbor viral loads as low as 0.1 virus per parasite (Figure 1B). In this scenario, it is unlikely that methods aimed at detecting the virus by immunological approaches could be more sensitive than methods detecting the viral RNA.
CONCLUSION
Findings in the present study suggest that detection of LRV1 in Leishmania parasite isolates can be performed using a simple RT-PCR.26 However, epidemiological studies aimed at determining an association between LRV1 presence and complicated outcomes of CL should consider the evaluation of LRV1 occurrence using samples coming directly from the patients and assessing the parasite load to rule out LRV false negatives. Thus, it would be necessary to develop methods that are simpler and more sensitive than the nested RT-PCR used in this study.
Supplemental tables
Acknowledgments:
We would like to thank Stephen Beverley for his help in improving the detection methods and data discussion, and Jussep Salgado for helping with Leishmania species identification. Colciencias grant number 110177758491, and Universidad Nacional de Colombia grants HERMES-32319 and HERMES-28542 supported this work.
Note: Supplemental tables appear at www.ajtmh.org.
REFERENCES
- 1.Strazzulla A, Cocuzza S, Pinzone MR, Postorino MC, Cosentino S, Serra A, Cacopardo B, Nunnari G, 2013. Mucosal leishmaniasis: an underestimated presentation of a neglected disease. Biomed Res Int 2013: 805108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Oliveira Guerra JA, Prestes SR, Silveira H, Coelho CLIdAR, Gama P, Moura A, Amato V, Barbosa MdGV, de Lima Ferreira LC, 2011. Mucosal leishmaniasis caused by Leishmania (Viannia) braziliensis and Leishmania (Viannia) guyanensis in the Brazilian Amazon. PLoS Negl Trop Dis 5: e980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haldar AK, Sen P, Roy S, 2011. Use of antimony in the treatment of leishmaniasis: current status and future directions. Mol Biol Int 2011: 571242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanaerschot M, Dumetz F, Roy S, Ponte-Sucre A, Arevalo J, Dujardin JC, 2014. Treatment failure in leishmaniasis: drug-resistance or another (epi-) phenotype? Expert Rev Anti Infect Ther 12: 937–946. [DOI] [PubMed] [Google Scholar]
- 5.Ghabrial SA, 2008. Totiviruses. van Regenmortel HV, Mahy BWJ, eds. Encyclopedia of Virology, 3rd edition Oxford, United Kingdom: Academic Press, 163–174. [Google Scholar]
- 6.Diamond LS, Mattern CFT, 1976. Protozoal Viruses. Lauffer MA, Kenneth MS, eds. Advances in Virus Research. New York, San Francisco, London: Academic Press, 87–112. [DOI] [PubMed] [Google Scholar]
- 7.Akopyants NS, Lye LF, Dobson DE, Lukeš J, Beverley SM, 2016. A novel bunyavirus-like virus of trypanosomatid protist parasites. Genome Announc 4: 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mertens P, 2004. The dsRNA viruses. Virus Res 101: 3–13. [DOI] [PubMed] [Google Scholar]
- 9.Wang A, Wang C, 1991. Viruses of the protozoa. Annu Rev Microbiol 45: 251–263. [DOI] [PubMed] [Google Scholar]
- 10.Stuart KD, Weeks R, Guilbride L, Myler PJ, 1992. Molecular organization of Leishmania RNA virus 1. Proc Natl Acad Sci USA 89: 8596–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guilbride L, Myler PJ, Stuart K, 1992. Distribution and sequence divergence of LRV1 viruses among different Leishmania species. Mol Biochem Parasitol 54: 101–104. [DOI] [PubMed] [Google Scholar]
- 12.Scheffter SM, Ro YT, Chung IK, Patterson J, 1995. The complete sequence of Leishmania RNA virus LRV2-1, a virus of an Old World parasite strain. Virology 212: 84–90. [DOI] [PubMed] [Google Scholar]
- 13.Scheffter S, Widmer G, Patterson JL, 1994. Complete sequence of Leishmania RNA virus 1-4 and identification of conserved sequences. Virology 199: 479–483. [DOI] [PubMed] [Google Scholar]
- 14.Ives A, et al. 2011. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science 331: 775–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kariyawasam R, Grewal J, Lau R, Purssell A, Valencia BM, Llanos-Cuentas A, Boggild AK, 2017. Influence of Leishmania RNA virus-1 on pro-inflammatory biomarker expression in a human macrophage model of American tegumentary leishmaniasis. J Infect Dis 216: 877–886. [DOI] [PubMed] [Google Scholar]
- 16.Soliman MF, Ibrahim MM, 2017. Do parasite viruses affect the relationship of parasites with their host?. Edorium J Infect Dis 3: 9–11. [Google Scholar]
- 17.Hartley M-A, Bourreau E, Rossi M, Castiglioni P, Eren RO, Prevel F, Couppié P, Hickerson SM, Launois P, Beverley SM, 2016. Leishmania virus-dependent metastatic leishmaniasis is prevented by blocking IL-17A. PLoS Pathog 12: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossi M, et al. 2017. Type I interferons induced by endogenous or exogenous viral infections promote metastasis and relapse of leishmaniasis. Proc Natl Acad Sci USA 114: 4987–4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adaui V, Lye LF, Akopyants NS, Zimic M, Llanos-Cuentas A, Garcia L, Maes I, De Doncker S, Dobson DE, Arevalo J, 2015. Association of the endobiont double-stranded RNA virus LRV1 with treatment failure for human leishmaniasis caused by Leishmania braziliensis in Peru and Bolivia. J Infect Dis 213: 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourreau E, Marine Ginouves M, Prévot G, 2015. Leishmania-RNA virus presence in L. guyanensis parasites increases the risk of first-line treatment failure and symptomatic relapse. J Infect Dis 213: 105–111. [DOI] [PubMed] [Google Scholar]
- 21.Alves-Ferreira EVC, et al. 2015. Differential gene expression and infection profiles of cutaneous and mucosal Leishmania braziliensis isolates from the same patient. PLoS Negl Trop Dis 9: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macedo DH, Menezes-Neto A, Rugani JM, Rocha AC, Silva SO, Melo MN, Lye L-F, Beverley SM, Gontijo CM, Soares RP, 2016. Low frequency of LRV1 in Leishmania braziliensis strains isolated from typical and atypical lesions in the State of Minas Gerais, Brazil. Mol Biochem Parasitol 210: 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogg MM, Carrion R, Botelho ACDC, Mayrink W, Correa-Oliveira R, Patterson JL, 2003. Short report: quantification of leishmaniavirus RNA in clinical samples and its possible role in pathogenesis. Am J Trop Med Hyg 69: 309–313. [PubMed] [Google Scholar]
- 24.Ramos Pereira LdO, Maretti-Mira AC, Rodrigues KM, Lima RB, de Oliveira-Neto MP, Cupolillo E, Pirmez C, de Oliveira MP, 2013. Severity of tegumentary leishmaniasis is not exclusively associated with Leishmania RNA virus 1 infection in Brazil. Mem Inst Oswaldo Cruz 108: 665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zangger H, et al. 2013. Detection of Leishmania RNA virus in Leishmania parasites. PLoS Negl Trop Dis 7: e2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zangger H, Hailu A, Desponds C, Lye LF, Akopyants NS, Dobson DE, Ronet C, Ghalib H, Beverley SM, Fasel N, 2014. Leishmania aethiopica field isolates bearing an endosymbiontic dsRNA virus induce pro-inflammatory cytokine response. PLoS Negl Trop Dis 8: e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito MM, Catanhêde ML, Katsuragawa TH, da Silva Junior CF, Camargo MLA, de Godoi Mattos R, Vilallobos-Salcedo JM, 2015. Correlation between presence of Leishmania RNA virus 1 and clinical characteristics of nasal mucosal leishmaniosis. Braz J Otorhinolaryngol 81: 533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cantanhêde ML, da Silva Júnior CF, Ito MM, Felipin KP, Nicolete R, Salcedo JMV, Porrozzi R, Cupolillo E, Ferreira RdGM, 2015. Further evidence of an association between the presence of Leishmania RNA virus 1 and the mucosal manifestations in tegumentary leishmaniasis patients. PLoS Negl Trop Dis 9: e0004079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salinas G, Zamora M, Stuart K, Saravia N, 1996. Leishmania RNA viruses in Leishmania of the Viannia subgenus. Am J Trop Med Hyg 54: 425–429. [DOI] [PubMed] [Google Scholar]
- 30.Saiz M, Llanos-Cuentas A, Echevarria J, Roncal N, Cruz M, Muniz MT, Lucas C, Wirth D, Scheffter S, Magill A, 1998. Short report: detection of Leishmania virus in human biopsy samples of leishmaniasis from Peru. Am J Trop Med Hyg 58: 192–194. [DOI] [PubMed] [Google Scholar]
- 31.Ginouvès M, Simon S, Bourreau E, Lacoste V, Ronet C, Couppié P, Nacher M, Demar M, Prévot G, 2015. Prevalence and distribution of Leishmania RNA virus 1 in Leishmania parasites from French Guiana. Am J Trop Med Hyg 94: 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hajjaran H, Mahdi M, Mohebali M, Samimi-Rad K, Ataei-Pirkooh A, Kazemi-Rad E, Naddaf SR, Raoofian R, 2016. Detection and molecular identification of leishmania RNA virus (LRV) in Iranian Leishmania species. Arch Virol 161: 3385–3390. [DOI] [PubMed] [Google Scholar]
- 33.Tirera S, Ginouves M, Donato D, Caballero IS, Bouchier C, Lavergne A, Bourreau E, Mosnier E, Vantilcke V, Couppié P, 2017. Unraveling the genetic diversity and phylogeny of Leishmania RNA virus 1 strains of infected Leishmania isolates circulating in French Guiana. PLoS Negl Trop Dis 11: e0005764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Franco JE, Cruz-Barrera ML, Robayo ML, Lopez MC, Daza CD, Bedoya A, Mariño ML, Saavedra CH, Echeverry MC, 2016. Clinical and parasitological features of patients with American cutaneous leishmaniasis that did not respond to treatment with meglumine antimoniate. PLoS Negl Trop Dis 10: e0004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walton BC, Shaw JJ, Lainson R, 1977. Observations on the in vitro cultivation of Leishmania braziliensis. J Parasitol 63: 1118–1119. [PubMed] [Google Scholar]
- 36.van den Bogaart E, Schoone GJ, Adams ER, Schallig HD, 2014. Duplex quantitative reverse-transcriptase PCR for simultaneous assessment of drug activity against Leishmania intracellular amastigotes and their host cells. Int J Parasitol Drugs Drug Resist 4: 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia L, Kindt A, Bermudez H, Llanos-Cuentas A, De Doncker S, Arevalo J, Tintaya KWQ, Dujardin J-C, 2004. Culture-independent species typing of neotropical Leishmania for clinical validation of a PCR-based assay targeting heat shock protein 70 genes. J Clin Microbiol 42: 2294–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montalvo AM, Fraga J, Monzote L, Montano I, De Doncker S, Dujardin JC, Van der Auwera G, 2010. Heat-shock protein 70 PCR-RFLP: a universal simple tool for Leishmania species discrimination in the New and Old World. Parasitology 137: 1159–1168. [DOI] [PubMed] [Google Scholar]
- 39.Cruz Barrera M, Ovalle Bracho C, Ortegón Vergara V, Pérez Franco JE, Echeverry MC, 2015. Improving Leishmania species identification in different types of samples from cutaneous lesions. J Clin Microbiol 53: 1339–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.WHO , 2012. Status of Endemicity of Cutaneous Leishmaniasis Worldwide, 2012. Geneva, Switzerland: World Health Organization; Available at: http://apps.who.int/neglected_diseases/ntddata/leishmaniasis/leishmaniasis.html. Accessed August 20, 2020. [Google Scholar]
- 41.WHO , 2009. Leishmaniasis: Burden of Disease. Geneva, Switzerland: World Health Organization; Available at: http://www.who.int/leishmaniasis/burden/en. Accessed August 20, 2020. [Google Scholar]
- 42.Ovalle CE, Porras L, Rey M, Ríos M, Camargo YC, 2006. Distribución geográfica de especies de Leishmania aisladas de pacientes consultantes al Instituto Nacional de Dermatología Federico Lleras Acosta, ESE, 1995–2005. Biomédica 26: 145–151. [PubMed] [Google Scholar]
- 43.Corredor A, Kreutzer RD, Tesh RB, Boshell J, Palau MT, Caceres E, Duque S, Pelaez D, Rodriguez G, Nichols S, 1990. Distribution and etiology of leishmaniasis in Colombia. Am J Trop Med Hyg 42: 206–214. [DOI] [PubMed] [Google Scholar]
- 44.Salgado-Almario J, Hernández CA, Ovalle CE, 2019. Distribución geográfica de las especies de Leishmania en Colombia, 1985–2017. Biomédica 39: 278–290. [DOI] [PubMed] [Google Scholar]
- 45.Llanos-Cuentas A, Tulliano G, Araujo-Castillo R, Miranda-Verastegui C, Santamaria-Castrellon G, Ramirez L, Lazo M, De Doncker S, Boelaert M, Robays J, 2008. Clinical and parasite species risk factors for pentavalent antimonial treatment failure in cutaneous leishmaniasis in Peru. Clin Infect Dis 46: 223–231. [DOI] [PubMed] [Google Scholar]
- 46.Tuon FF, Amato VS, Graf ME, Siqueira AM, Nicodemo AC, Neto VA, 2008. Treatment of New World cutaneous leishmaniasis–a systematic review with a meta‐analysis. Int J Dermatol 47: 109–124. [DOI] [PubMed] [Google Scholar]
- 47.Kariyawasam R, Lau R, Valencia B, Llanos-Cuentas A, Boggild A, 2020. Leishmania RNA virus 1 (LRV-1) in Leishmania (Viannia) braziliensis isolates from Peru: a description of demographic and clinical correlate. Am J Trop Med Hyg 102: 280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ginouvès M, et al. 2020. Leishmania virus genetic diversity is not related to leishmaniasis treatment failure. Clin Microbiol Infect 26: 18. [DOI] [PubMed] [Google Scholar]
- 49.Jara M, et al. 2013. Real-time PCR assay for detection and quantification of Leishmania (Viannia) organisms in skin and mucosal lesions: exploratory study of parasite load and clinical parameters. J Clin Microbiol 51: 1826–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.