ABSTRACT
Vector-borne diseases, such as dengue, Zika and malaria, are a major cause of morbidity and mortality worldwide. These diseases have proven difficult to control and currently available management tools are insufficient to eliminate them in many regions. Gene drives have the potential to revolutionize vector-borne disease control. This suite of technologies has advanced rapidly in recent years as a result of the availability of new, more efficient gene editing technologies. Gene drives can favorably bias the inheritance of a linked disease-refractory gene, which could possibly be exploited (i) to generate a vector population incapable of transmitting disease or (ii) to disrupt an essential gene for viability or fertility, which could eventually eliminate a population. Importantly, gene drives vary in characteristics such as their transmission efficiency, confinability and reversibility, and their potential to develop resistance to the drive mechanism. Here, we discuss recent advancements in the gene drive field, and contrast the benefits and limitations of a variety of technologies, as well as approaches to overcome these limitations. We also discuss the current state of each gene drive technology and the technical considerations that need to be addressed on the pathway to field implementation. While there are still many obstacles to overcome, recent progress has brought us closer than ever before to genetic-based vector modification as a tool to support vector-borne disease elimination efforts worldwide.
KEY WORDS: CRISPR, Cas9, Split drive, Medea, ClvR, Homing drives
Summary: Gene drives are an important emerging technology. This review discusses the current state of gene drive technologies for vector-borne disease control and their performance and safety features.
Introduction
Half of the global population is currently at risk of mosquito-borne diseases such as dengue (Brady et al., 2012; Kraemer et al., 2015) or malaria (World Health Organization, 2019). There have been outstanding reductions in global malaria transmission over the last two decades as a result of integrated mosquito management programs that have focused on reducing human–mosquito contact and suppressing mosquito populations (Bhatt et al., 2015; Cibulskis et al., 2016; World Health Organization, 2019), but other mosquito-borne diseases, such as dengue, are on the rise (Huang et al., 2019; Stanaway et al., 2016). For the majority of arboviruses, there are no available drugs or vaccines, and while there are drugs and vaccines available for malaria, their efficacy at the population level is limited. Mosquito-targeted interventions are the primary methods to prevent disease transmission; however, given the widespread resistance to insecticides, escalating burden of transmission and deaths worldwide related to mosquito-borne disease, new technologies are in high demand. A tool that has sparked significant enthusiasm is gene drive-modified mosquitoes (Champer et al., 2016; Esvelt et al., 2014). Gene drives have promise to be a transformative technology that leads to the elimination of burdensome vector-borne diseases (Feachem et al., 2019; Raban and Akbari, 2017).
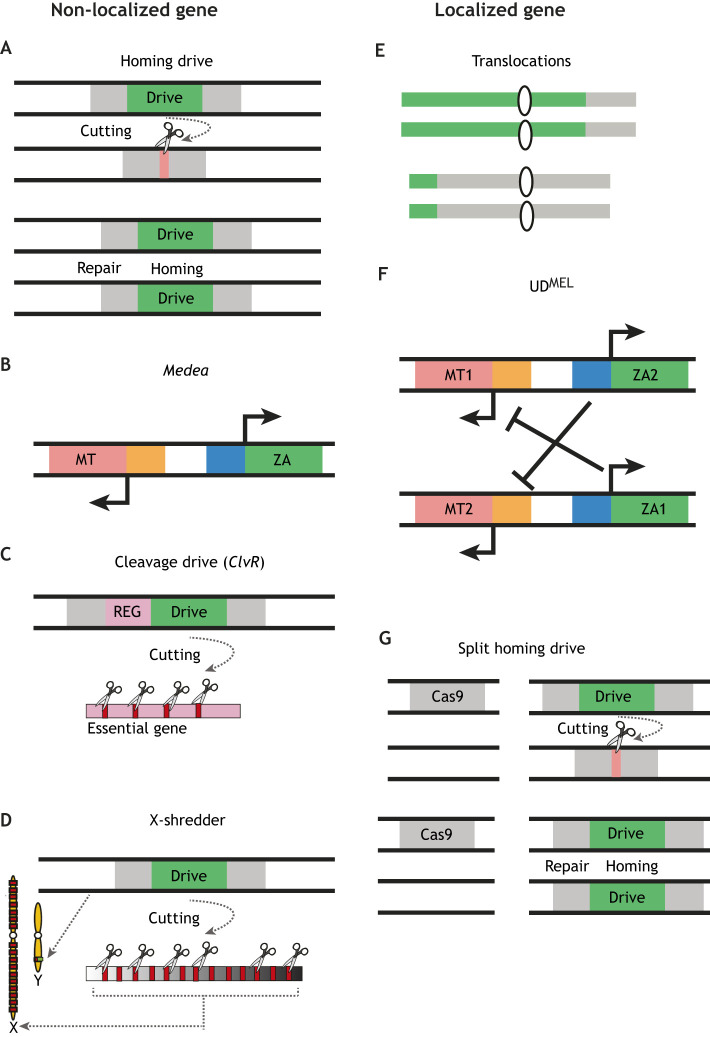
Many varieties of engineered gene drives have been developed to date (Fig. 1 and Table 1). Each of these drives are designed to bias the inheritance of a gene or gene modification, known as an effector, so it is inherited more frequently than would be expected by Mendelian segregation. Over time, the gene drive can therefore spread a linked effector into a wild population. In the context of vector–pathogen systems, gene drives are engineered to facilitate biased inheritance of an effector that either confers resistance to a pathogen or suppresses the vector population by generating a non-functional essential sterility or lethality gene. These approaches, known as population replacement and population suppression, respectively, aim to reduce, or possibly eliminate, the targeted mosquito-borne pathogens. While these gene drive systems were initially proposed for population replacement or suppression many years ago (Burt, 2003; Curtis, 1968; Marshall and Akbari, 2016), recent advances in gene editing technologies, principally the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9)-based genome engineering technologies (Jinek et al., 2012), have greatly accelerated the development of proof-of-concept gene drive systems (Esvelt et al., 2014) with demonstrations in mosquitoes (Gantz et al., 2015; Hammond et al., 2016; Kyrou et al., 2018; Li et al., 2019 preprint).
Fig. 1.
Schematic illustration of the various non-localized and localized gene drives. (A) Homing endonuclease drives encode an endonuclease that cuts a target sequence in a homologous chromosome. Through a DNA repair mechanism called homology-directed repair (HDR), the drive is then used as a template and is copied into the cut location. (B) Maternal-effect dominant embryonic arrest (Medea) drives encode a maternal toxin (MT) that is deposited into eggs and a zygotic antidote (ZA) to this toxin that is encoded in the early-stage offspring. When mated to a wild-type mosquito, the drive kills all offspring that do not inherit the drive and over time this fitness advantage will drive it into the population. (C) Cleavage drive consists of a germline-specific Cas9 endonuclease and a guide RNAs (gRNAs) targeting a haplosufficient essential gene (toxin) located on another chromosome. The drive also has a cleavage-resistant recoded copy of the essential gene (REG; antidote), which protects offspring that inherit the drive from being killed. (D) X-shredders are engineered to express an endonuclease on the male Y chromosome during spermatogenesis that targets and destroys genes on the X chromosome. This generates male-only progeny, which can be used for population suppression. (E) Translocation drives generate a chromosomal rearrangement or inversion that gives a disadvantage to individuals heterozygous for the drive. This system creates a genetic condition called underdominance, whereby over time the fitness advantage of translocation homozygotes facilitates their spread into a population. (F) Maternal-effect lethal underdominance (UDMEL) drives encode two constructs each possessing a maternally expressed toxin gene (MT1 and MT2), which is active in the embryo, and a zygotic antidote gene (ZA1 and ZA2), which is capable of neutralizing the maternal toxin expressed by the other toxin–antidote construct. This drive is another underdominance system whereby heterozygous females generate mostly inviable offspring, while homozygous females are fully fertile and viable. (G) Split homing or daisy endonuclease drives contain the same components as the standard homing endonuclease drive, but the homing endonuclease and programmable guide components are separated into different lines. Gene conversion only occurs when these lines are crossed to each other and with many releases and over time can be used to drive effectors into a population.
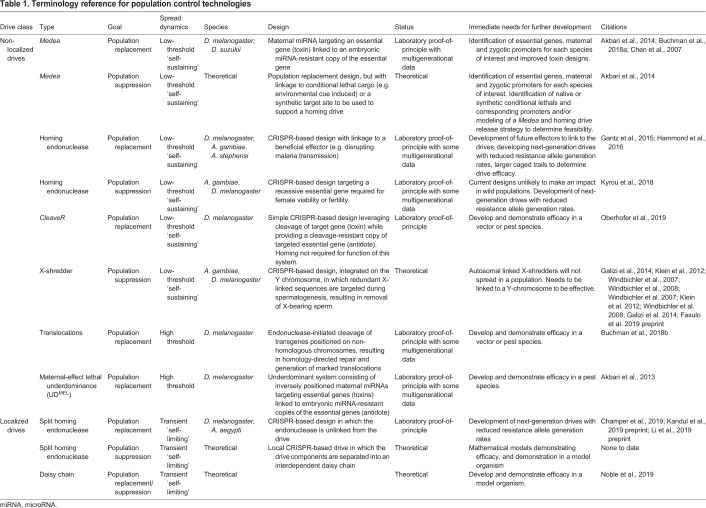
Table 1.
Terminology reference for population control technologies
Gene drives may be an essential tool to make genetic mosquito-borne disease control affordable at scale in the field. One of the principal advantages of gene drive systems is that they can be used to overcome fitness costs associated with the effectors and genetic elements required for the stable and heritable transmission of the desirable synthetic genes into a wild population (Burt, 2003; Unckless et al., 2015). In the absence of a gene drive, these fitness costs have caused the rapid elimination of effectors from cage populations (Catteruccia et al., 2003; Lambrechts et al., 2008; Marrelli et al., 2006). Without the biased inheritance directed by the gene drive, it is generally considered infeasible to generate enough individuals at a high enough release frequency to spread effectors to a 100% allele frequency, otherwise known as fixation, in a population (James, 2005).
In this review, we examine the different types of gene drive systems that have been developed to date and the current state of the technology. We focus on two broad categories of drives: ‘localized drives’ and ‘non-localized drives’. Non-localized drives are expected to spread beyond a release site and maintain themselves in the population for many generations. In contrast, localized drives are expected to spread only into local populations and, in some cases, eliminate themselves from the population over time. The ability to limit the spread of localized drives makes these technologies of interest during the trial phase of the technology, when localized control is otherwise desired, or when spread into other populations would be problematic. For instance, if gene drives spread across political borders, into ecologically sensitive areas or into areas with conflicting technologies, this may cause significant challenges for regulators and stakeholders. There are a number of varieties of both localized and non-localized drives that have unique attributes that affect their performance and safety and regulation implications.
Overview of the current state of gene drive development
Non-localized drives
Homing endonuclease drives
Homing endonuclease-based gene drives were first described over 15 years ago (Burt, 2003), but in recent years CRISPR-Cas9 genome editing has fostered a resurgence in the interest in these technologies because of their ease of use compared with other genome editing methods (e.g. zinc finger nucleases and transcription activator-like nucleases) and their adaptability to a wide range of organisms (Esvelt et al., 2014). Unsurprisingly, there are now Cas9 endonuclease-based gene drives in yeast (DiCarlo et al., 2015; Roggenkamp et al., 2018), flies (Gantz and Bier, 2015; Kandul et al., 2019 preprint; Oberhofer et al., 2018), mosquito disease vectors (Gantz et al., 2015; Hammond et al., 2016; Kyrou et al., 2018; Li et al., 2019 preprint), fungi (Shapiro et al., 2018) and mice (Grunwald et al., 2019). The fundamentals of these gene drive systems have been discussed extensively in other articles (Champer et al., 2016; Esvelt et al., 2014; Gantz and Akbari, 2018; Gantz and Bier, 2016), but in brief, Cas9 is a highly specific endonuclease that can be easily programmed to introduce double-stranded breaks in desired genome regions corresponding to a complementary guide RNA (gRNA) sequence (Fig. 1A). When these breaks are repaired, they can generate insertions or deletions (indels) through a process called non-homologous end joining (NHEJ), or they can be repaired by precise integration of a copy of a corresponding region in the homologous chromosome through a process called homology-directed repair (HDR). This HDR mechanism causes transformation of offspring that would be heterozygous for the drive, by normal Mendelian inheritance, to instead become homozygous for the drive. This process, also known as gene conversion, is key to the efficient, biased inheritance of these drive systems.
The indels caused by NHEJ repair, in contrast, are a primary cause of drive resistance in Cas9-mediated homing endonuclease drives. If the NHEJ-generated indels mutate or omit the genomic sequence targeted by the drive, then the drive may no longer cut at the site. Over time, with accumulation of NHEJ repair events that alter the drive cleavage sites, coupled with any fitness advantage for resistance to the drive, the population will become resistant to the drive. The particular importance of mitigating NHEJ repair in gene drives was recently exemplified in the first example of a CRISPR-Cas9-mediated super-Mendelian inheritance in mice (Grunwald et al., 2019). This was an important initial step towards demonstrating the feasibility of gene drive in mice, but the predilection of this system to strongly favor NHEJ repair in the offspring of all but a few female parental lines demonstrates the need for the identification and optimization of drive components. The HDR double-stranded DNA repair pathways are favored at specific time points in the cell cycle and in specific tissues (Branzei and Foiani, 2008; Heyer et al., 2010; van Gent et al., 2001; Zlotorynski, 2016). In the Grunwald et al. (2019) study, alterations in gene drive expression, timing and germline specificity are proposed as a way to promote increased HDR and drive efficiency in mice.
Another recent study in Drosophila melanogaster engineered a homing endonuclease drive to target transformer (tra), a gene that plays a key role in sex determination in many insects (KaramiNejadRanjbar et al., 2018). In this system, functional, drive-resistant alleles formed independently many times, and in 15 generations these resistant alleles accumulated to a point where the drive was not functional. Therefore, even though Cas9-based gene drive strategies are seemingly universally applicable, with the accelerated rate of resistance seen against Cas9-mediated homing-based drives in the laboratory, much work is still needed to optimize gene drive components and target selection before this technology can be used to produce reliable drive systems, especially in organisms with limited genetic engineering tools.
One recent advancement in homing endonuclease gene drives is a system that targets highly conserved regions of a sex determination gene, doublesex (dsx), to suppress the principal malaria vector in Africa, Anopheles gambiae (Kyrou et al., 2018). The dsx gene is sex-specifically spliced and encodes sex-specific transcription factors that regulate sexual development in insects (Burtis and Baker, 1989; Salvemini et al., 2011; Shukla and Nagaraju, 2010). In small-population cage studies targeting the female-specific alternatively spliced transcript in dsx, Kyrou et al. (2018) achieved >95% inheritance in males, which were fully fertile and viable, in addition to demonstrating 99% inheritance in females, which confers a sterile intersex female phenotype. Furthermore, these sterile females were unable to blood feed, which is required for egg production, thereby rendering them incapable of both reproducing and transmitting pathogens.
In the two small-population cage experiments, suppression of the wild-type population was achieved in 12 generations or fewer, but even in as little as 2–5 generations, indels at the target site developed that were resistant to the drive (Kyrou et al., 2018). In this case, these resistance mutants were found at a frequency not exceeding 1.16% and did not encode a functional dsx transcript required for normal female development, so females were still rendered intersex and sterile. The authors attribute targeting of highly conserved and functionally constrained target sites in dsx to the success of this drive despite resistance, and this should be an important consideration for target site selection in future designs, but as this experiment was very small scale (two cages, N=300 mosquitoes) and of limited time scale (12 generations), and therefore it is unknown whether resistance mutations that disrupt drive function and restore transcript function will develop in larger populations over a longer periods of time. Moreover, resistance mutation accumulation in other A. gambiae gene drive systems were predicted to be incapable of long-term population suppression (Hammond et al., 2016; Marshall et al., 2017).
Medea drives
Maternal-effect dominant embryonic arrest (Medea) population replacement drives were first demonstrated over a decade ago (Chen et al., 2007). The design and function of these drives is reviewed extensively elsewhere (Champer et al., 2016; Ward et al., 2011), but briefly, these systems bias inheritance through the tight linkage of a maternally expressed toxin, which to date has been microRNAs (miRNAs) targeting essential genes, and a linked zygotically or embryonically active antidote gene that results in the death of offspring that fail to inherit the Medea system (Fig. 1B). Essentially, females with a Medea drive deposit a toxin into all their eggs that must be counteracted by an antidote that is expressed early in development, or the offspring dies. In successive generations, this system results in a disadvantage for wild-type alleles and, therefore, if the Medea system is linked to an effector to make the population disease resistant, for example in mosquitoes (Buchman et al., 2019a preprint, 2019b; Marshall et al., 2019), or an effector targeting a conditional essential gene, or another conditional lethal effector such as one that confers temperature or small-molecule sensitivity to kill the population, it is predicted to favorably bias inheritance of the effector and therefore modify the population. The drive could also behave as a non-localized drive when released over a certain threshold, which is dependent on the fitness cost of the gene drive and its associated genetic elements as well as the rate of toxin resistance or natural genetic resistance in the population. The rate of drive resistance accumulation is expected to be lower with these drives compared with homing drives, as offspring that do not receive the full drive are removed from the population, but improvements in drive components are needed to move this technology forward in development. Specifically, there is a need to identify maternal and zygotic/embryonic promoters and appropriate toxin effectors/targets in pest species as the lack of these components has limited the development of these technologies (Akbari et al., 2014; Buchman et al., 2018a), but these needs are not insurmountable. The first Medea drive system in a crop pest species, Drosophila suzukii, was recently demonstrated in the laboratory (Buchman et al., 2018a). The Medea driven was successfully driven to high frequencies into laboratory populations for many generations although natural genetic variation in the laboratory population likely restricted the drive from going to fixation. CRISPR genome engineering technologies such as RNA targeting systems may provide the tools to move these technologies into other organisms (Terns, 2018).
CleaveR drives
A newly developed gene drive system has taken a novel approach to reducing NHEJ-associated drive resistance by eliminating the need for the system to be copied via HDR (Fig. 1C). This CleaveR [Cleave and Rescue (ClvR)] drive has been engineered as proof of concept in D. melanogaster, where it is inserted at a separate location in the genome from the target gene and encodes both a germline-specific Cas9 and gRNA targeting a haplosufficient essential gene (toxin). The drive also encodes a cleavage-resistant, recoded version of the haplosufficient essential gene target (antidote) with limited homology to the wild-type allele to prevent recombination (Oberhofer et al., 2019). The resulting ClvR drive is lethal to offspring that do not inherit the antidote, thereby positively biasing inheritance of the ClvR drive and any linked components. This drive system is notable as its population introgression rate is predicted to be tunable by the introduction frequency. Furthermore, the efficiency of the drive is independent of the repair mechanism and the drive does not require HDR/homing for spread. As previously discussed, the NHEJ repair mechanism results in target disruption and drive resistance, but in this system, NHEJ repair causes lethality in haplosufficient targets and therefore removes these individuals from the population. These ClvR designs may be able to circumvent NHEJ-associated drive resistance, but further multigenerational long-term drive experiments in diverse populations need to be undertaken to determine the true robustness of this system. Additionally, given the requirement for defining key promoters to express the recoded rescue in this drive, the ease of developing this drive in pest species remains to be proven. Finally, it should be noted that while this drive is generally a population replacement type of drive (similar to Medea), it could possibly be used to spread conditional lethal effectors (e.g. temperature sensitivity or sensitivity to a small molecule) and therefore could also be used to replace then suppress populations.
X chromosome shredders
X chromosome shredders (X-shredders) are another potential approach to the development of population suppression drives (Papathanos et al., 2014). These sex-biasing systems can be used in organisms with two different sex chromosomes, known as heterogametic species, where females have two of the same sex chromosomes (XX) and males have two different sex chromosomes (XY). These drives are engineered by expressing an endonuclease from the Y chromosome to target and destroy genes on the X chromosome during spermatogenesis (Fig. 1D). Only the Y-bearing sperm survive to fertilize the egg, so this strategy generates male-only progeny and therefore could be used as a potential drive strategy. In theory, an X-shredder could be designed for any heterogametic species, which would preclude disease vectors such as Aedes aegypti, but be suitable for most species of interest. Although significant attempts to engineer an X-shredder drive in A. gambiae have been undertaken, a complete system has yet to be fully developed (Galizi et al., 2014; Klein et al., 2012; Windbichler et al., 2007, 2008). These efforts have demonstrated that, while targeting the X chromosome during spermatogenesis proved possible, efforts to position the nuclease on the Y chromosome while maintaining functionality is difficult. Future efforts to exploit randomly generated Y chromosome docking strains (Bernardini et al., 2014) or even generate new site-specific docking strains on the Y chromosome using CRISPR (Buchman and Akbari, 2019) may improve functionality of Y chromosome nucleases and thereby expedite the development of the first synthetic X chromosome shredders. Recent work in D. melanogaster unraveled some of the important considerations for designing X-shredder systems, including optimization considerations for Cas9 and gRNA timing and expression, target and target site selection and drive resistance mitigation (Fasulo et al., 2019 preprint). This work also demonstrated the adaptability of X-shredder systems to new organisms as they had only been previously used in one anopheline mosquito vector. As our understanding of X-shredder systems improves, we will certainly see the development of more of these drives.
Localized gene drives
Translocations
CRISPR-Cas9 technologies have also generated useful genome engineering tools that can advance the development of other drive systems, including translocation drives (Gantz and Akbari, 2018). Translocation drives are engineered to generate a reciprocal chromosome rearrangement or inversion that is unlike the chromosomal organization of the wild population (Fig. 1E). When mated to wild-type mosquitoes, translocation heterozygotes produce a large proportion of inviable offspring, as 50% of the offspring do not inherit a balanced set of chromosomes. Homozygotes for the translocation contain a balanced set of chromosomes, so their progeny are viable and consequently have higher fitness than the heterozygotes, a genetic condition known as underdominance. The fitness advantage of homozygotes allows the translocation to spread into the population along with any effector linked to translocation breakpoints (Curtis, 1968). This translocation drive is threshold dependent, however, which means it needs to be released at a high enough frequency to spread into a population. Below that threshold, which is determined by the fitness differentials between heterozygotes and homozygotes and the local population genetics (Marshall and Akbari, 2018), the drive will go extinct in the population.
Translocation drives are confineable to the target species as viable hybridizations between an individual harboring a translocation and another species are exceedingly unlikely. They are also confinable to a local population, as they need to be introduced into a new population at a frequency exceeding its threshold to be maintained in that population, otherwise they are actively driven out. Their introduction is also reversible, as releasing large numbers of wild-type individuals can push the drive below its threshold where over time it will become extinct in the population. This species specificity and ability to confine and reverse translocation drives make them desirable for field release. Moreover, once they spread into the target population at a high frequency, the drive can halt migration of non-drive alleles as long as these alleles remain below the threshold needed to maintain the drive. Despite these favorable attributes, previous attempts to generate translocations resulted in high fitness costs because of the low precision methods by which these translocations were generated (Asman et al., 1981; Curtis et al., 1972; Lorimer et al., 1972; Robinson, 1976). A recent study in D. melanogaster demonstrated the use of endonuclease engineering technologies to generate site-specific genome translocations that could be reliably integrated into known engineered docking sites in the genome (Buchman et al., 2018a). This is a substantial improvement on the essentially random translocation insertion methods used in the past and thus may be able to mitigate the insertional gene disruption that likely contributed to the fitness issues seen in past engineered translocations. Furthermore, unlike many of the other drives discussed in this review, this drive should be evolutionarily stable, as reversion to the wild-type chromosome arrangement should be exceedingly rare.
UDMEL
A synthetic maternal-effect lethal underdominance (UDMEL) drive is a dual toxin and dual antidote system. This system consists of two transgenic constructs each possessing a maternally expressed toxin gene, which is active in the embryo, and a zygotic antidote gene, which is capable of neutralizing the maternal toxin expressed by the other toxin–antidote construct (Fig. 1F). Similar to the translocation drive, the genetics of the system result in heterozygous females generating mostly inviable offspring, while homozygous females are fully fertile and viable. This disparity in fitness between drive heterozygotes and homozygotes results in bistable dynamics which are characteristic of underdominant systems and can be exploited to drive effectors into populations. To date, UDMEL drive has only been built in D. melanogaster as a proof of principle, but as designed, it successfully replaced laboratory populations in a threshold-dependent manner (Akbari et al., 2013). While this system was engineered using synthetic miRNA toxins, recently described CRISPR RNA targeting systems such as C2C2 and Cas13b could help expedite development of this system in other species (Terns, 2018).
Split homing endonuclease drives
Split homing endonuclease drives (also referred to as daisy drives when arranged in a chain consisting of several components; Noble et al., 2019) separate the homing endonuclease and gRNA components of a homing endonuclease drive into separate lines, rendering the cleavage and homing inactive until the lines are genetically crossed. These drives are more geographically confinable than standard homing endonuclease drives, and therefore have been recommended as a safer alternative for initial field release (Akbari et al., 2015; Esvelt et al., 2014; Noble et al., 2019). While initially demonstrated in yeast (DiCarlo et al., 2015), split homing endonuclease drives in D. melanogaster were recently developed that had comparable gene conversion efficiencies to a standard homing endonuclease drive (Champer et al., 2019; Kandul et al., 2019 preprint). The rate of resistance allele formation was higher in the split drive, but this difference is likely due to differences in Cas9 expression between the drives. A split homing endonuclease drive engineered in A. aegypti also demonstrated high transmission rates (up to 94%), but resistance allele formation was also seen in this drive system. Mathematical modeling predicted that this drive could still be maintained at a high population frequency along with its associated cargo for up to 4 years, or 56 generations assuming 14 generations per year, and therefore may be suitable for field trials once effectors are linked (Li et al., 2019 preprint).
While this initial result focuses on split replacement drive, a split homing suppression drive has not been demonstrated yet in the lab but can be designed in multiple ways. Split suppression drives can be developed to target conserved sex-determination genes such as dsx, similar to the drive recently developed for A. gambiae (Kyrou et al., 2018). Alternatively, essential genes could be targeted in this approach (e.g. genes important for female viability, female flight, or fecundity). These results indicate that split drives are likely as adaptable as standard homing endonuclease drives but have the potential to be a safer alternative to these drives. Further research is needed in non-model organisms and in long-term, multi-generational laboratory cage trials to evaluate their efficiency and safety compared with other drives.
Open questions regarding field performance of drive systems
To date, gene drives have only been tested under laboratory conditions that are highly uniform and not at all reflective of field conditions. Scaled, confined field trials are desirable to generate the requisite data to better assess the efficacy and safety of these technologies prior to wider scale release. Gene drive technologies have not progressed past the initial proof-of-concept studies in the lab, but there are multiple research groups that plan to move this technology into the field, possibly in the not so distant future. These technologies have the potential to have an enormous impact on human health and livelihood, so their expedited development is essential, but not at the expense of prudent biosafety research and engagement with community members and other relevant stakeholders. From this review, we can see that there are many flavors of gene drives with differing mechanisms and spread dynamics. This suite of drive technologies may prove useful in different contexts. As these technologies advance, there are a number of open questions related to their regulation and implementation. Here, we discuss some of the technical ones.
How do drives and their components behave over long time scales?
Studies of gene drive systems thus far tend to be of the order of a few to a dozen generations. There are few if any long-term (>5 years) studies of any gene drives. These studies are essential to evaluate the expected duration of impact and stability of gene drive technologies. Resistant mutants have been identified in homing-based drives over short time scales (1–2 generations) (Champer et al., 2017; Gantz et al., 2015; Hammond et al., 2016; Kandul et al., 2019 preprint; KaramiNejadRanjbar et al., 2018; Kyrou et al., 2018; Li et al., 2019 preprint) and it is expected that these resistant alleles will proliferate over longer time periods and spatial scales. Presuming that resistance allele generation can be significantly reduced, perhaps through some combination of multiplexing of guide RNAs (Marshall et al., 2017) and targeting highly conserved genes (Kyrou et al., 2018), will this be sufficient to interrupt disease transmission in real populations? For homing-based population suppression, even very rare resistance allele generation is expected to interfere with vector population control (Marshall et al., 2017). Larger scale, longer-term studies will be required to determine the resistance allele generation rate and their long-term impact on population suppression strategies. For homing-based population replacement, higher resistance allele generation rates can be tolerated (Noble et al., 2017); but an additional concern is the stability of the drive system and effector gene cargo over time (Marshall et al., 2019). If some time elapses between the system reaching fixation in one population and an individual migrating from that population to another, then will the gene drive system and effector still be functional and seed a new wave of spread? What happens if the linked effector forces selection for pathogens that are more virulent (Marshall et al., 2019)? Given these issues and concerns, longer-term studies will be required to determine this as well.
How do drives perform in more genetically diverse populations?
Recent laboratory experiments have demonstrated that the genetic strain that a drive is present in can have a significant impact on drive efficacy (Champer et al., 2017; Hammond et al., 2016; Li et al., 2019 preprint). Populations with higher genetic diversity will have a higher probability of containing resistant drive alleles that could block the spread of the drive. Given that the genetic diversity in the wild cannot be entirely captured in a laboratory experiment, it is expected that genetic diversity will have an even more significant impact on the result of gene drive trials and interventions in wild populations. Substantial ecological work and laboratory studies will be required to refine our understanding of the behavior of drives in diverse populations and to ensure that any gene drive released in the wild will be efficacious over the desired treatment area.
The impact of genetic diversity is also a concern for other gene drive systems. A recent study of Medea drive in D. suzukii, a global crop pest, demonstrated that natural genetic variation in D. suzukii populations likely resulted in selection of toxin-resistant variants, which in multi-generational population cage studies resulted in limited drive spread (Buchman et al., 2018a). Similar genetic variation and toxin targeting issues will likely also arise when adapting these technologies to other species, especially as the genetics and population structure of most species are less defined than in D. melanogaster. Potential solutions to these issues include targeting of conserved gene regions, validation of target sites to optimize targeting efficiency and increased multiplexing, but substantial efforts are still needed to identify the minimal requisite components of these Medea systems in other species, including maternal and zygotic/embryonic promoters and appropriate toxin effectors and targets.
Next steps for gene drive technology
For many important vectors, we have developed the genetic tools needed to effectively produce proof-of-principle genetic effectors and gene drive systems for population replacement or suppression. At the current state of the technology, some gene drive technologies seem very adaptable to new species and populations (e.g. homing endonuclease-based drives) while others still need additional genetic innovations before they can be easily transferred to other species of interest (e.g. Medea and other toxin–antidote-based systems).
In many regions of the world, the benefits of current vector-borne disease interventions have plateaued while disease transmission persists. New technologies are needed to control these vectors and eliminate the diseases they transmit. Modeling has demonstrated that gene drive technologies, if they reach certain design requirements, are capable of generating long-term, sustainable vector-borne disease control (Eckhoff et al., 2017); however, drive resistance, genetic diversity in the target populations and other concerns pose challenges to achieving the level of control required to achieve disease elimination. Potential solutions for homing endonuclease-based drives include multiplexing of guide RNAs and targeting highly conserved genes. Work is also required to determine the target profile of each gene drive to meet specific performance and safety needs (Carballar-Lejarazú and James, 2017; James et al., 2018) and also to update guidance documents related to research, development and testing of genetically engineered insects and arthropods (Adelman et al., 2017). These profiles and guidance documents may differ between field trials and wide-scale disease control programs and will help to determine the goals that molecular technologies must meet.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by funding from a Defense Advanced Research Project Agency (DARPA) Safe Genes Program Grant (HR0011-17-2-0047) awarded to O.S.A. and J.M.M. and by National Institutes of Health award number R21AI123937 awarded to O.S.A.
References
- Adelman Z., Akbari O., Bauer J., Bier E., Bloss C., Carter S. R., Callender C., Denis A. C.-S., Cowhey P., Dass B. et al. (2017). Rules of the road for insect gene drive research and testing. Nat. Biotechnol. 35, 716-718. 10.1038/nbt.3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O. S., Matzen K. D., Marshall J. M., Huang H., Ward C. M. and Hay B. A. (2013). A synthetic gene drive system for local, reversible modification and suppression of insect populations. Curr. Biol. 23, 671-677. 10.1016/j.cub.2013.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O. S., Chen C.-H., Marshall J. M., Huang H., Antoshechkin I. and Hay B. A. (2014). Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of Medea-dependent population suppression. ACS Synth. Biol. 3, 915-928. 10.1021/sb300079h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O. S., Bellen H. J., Bier E., Bullock S. L., Burt A., Church G. M., Cook K. R., Duchek P., Edwards O. R., Esvelt K. M. et al. (2015). BIOSAFETY. Safeguarding gene drive experiments in the laboratory. Science 349, 927-929. 10.1126/science.aac7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asman S. M., McDonald P. T. and Prout T. (1981). Field studies of genetic control systems for mosquitoes. Annu. Rev. Entomol. 26, 289-318. 10.1146/annurev.en.26.010181.001445 [DOI] [PubMed] [Google Scholar]
- Bernardini F., Galizi R., Menichelli M., Papathanos P.-A., Dritsou V., Marois E., Crisanti A. and Windbichler N. (2014). Site-specific genetic engineering of the Anopheles gambiae Y chromosome. Proc. Natl. Acad. Sci. USA 111, 7600-7605. 10.1073/pnas.1404996111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S., Weiss D. J., Cameron E., Bisanzio D., Mappin B., Dalrymple U., Battle K., Moyes C. L., Henry A., Eckhoff P. A. et al. (2015). The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 526, 207-211. 10.1038/nature15535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady O. J., Gething P. W., Bhatt S., Messina J. P., Brownstein J. S., Hoen A. G., Moyes C. L., Farlow A. W., Scott T. W. and Hay S. I. (2012). Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl. Trop. Dis. 6, e1760 10.1371/journal.pntd.0001760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D. and Foiani M. (2008). Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 9, 297-308. 10.1038/nrm2351 [DOI] [PubMed] [Google Scholar]
- Buchman A. and Akbari O. S. (2019). Site-specific transgenesis of the Drosophila melanogaster Y-chromosome using CRISPR/Cas9. Insect Mol. Biol. 28, 65-73. 10.1111/imb.12528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A., Marshall J. M., Ostrovski D., Yang T. and Akbari O. S. (2018a). Synthetically engineered Medea gene drive system in the worldwide crop pest Drosophila suzukii. Proc. Natl. Acad. Sci. USA 115, 4725-4730. 10.1073/pnas.1713139115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. B., Ivy T., Marshall J. M., Akbari O. S. and Hay B. A. (2018b). Engineered reciprocal chromosome translocations drive high threshold, reversible population replacement in Drosophila. ACS Synth. Biol. 7, 1359-1370. 10.1021/acssynbio.7b00451 [DOI] [PubMed] [Google Scholar]
- Buchman A., Gamez S., Li M., Antoshechkin I., Lee S.-H., Wang S.-W., Chen C.-H., Klein M. J., Duchemin J.-B., Crowe J. E. et al. (2019a). Broad dengue neutralization in mosquitoes expressing an engineered antibody. BioRxiv. 10.1101/645481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A., Gamez S., Li M., Antoshechkin I., Li H.-H., Wang H.-W., Chen C.-H., Klein M. J., Duchemin J.-B., Paradkar P. N. et al. (2019b). Engineered resistance to Zika virus in transgenic expressing a polycistronic cluster of synthetic small RNAs. Proc. Natl. Acad. Sci. USA 116, 3656-3661. 10.1073/pnas.1810771116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A. (2003). Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 270, 921-928. 10.1098/rspb.2002.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis K. C. and Baker B. S. (1989). Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56, 997-1010. 10.1016/0092-8674(89)90633-8 [DOI] [PubMed] [Google Scholar]
- Carballar-Lejarazú R. and James A. A. (2017). Population modification of anopheline species to control malaria transmission. Pathog. Glob. Health 111, 424-435. 10.1080/20477724.2018.1427192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catteruccia F., Godfray H. C. J. and Crisanti A. (2003). Impact of genetic manipulation on the fitness of Anopheles stephensi mosquitoes. Science 299, 1225-1227. 10.1126/science.1081453 [DOI] [PubMed] [Google Scholar]
- Champer J., Buchman A. and Akbari O. S. (2016). Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 17, 146-159. 10.1038/nrg.2015.34 [DOI] [PubMed] [Google Scholar]
- Champer J., Reeves R., Oh S. Y., Liu C., Liu J., Clark A. G. and Messer P. W. (2017). Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 13, e1006796 10.1371/journal.pgen.1006796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champer J., Chung J., Lee Y. L., Liu C., Yang E., Wen Z., Clark A. G. and Messer P. W. (2019). Molecular safeguarding of CRISPR gene drive experiments. Elife 8, e41439 10.7554/eLife.41439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-H., Huang H., Ward C. M., Su J. T., Schaeffer L. V., Guo M. and Hay B. A. (2007). A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science 316, 597-600. 10.1126/science.1138595 [DOI] [PubMed] [Google Scholar]
- Cibulskis R. E., Alonso P., Aponte J., Aregawi M., Barrette A., Bergeron L., Fergus C. A., Knox T., Lynch M., Patouillard E. et al. (2016). Malaria: global progress 2000-2015 and future challenges. Infect. Dis. Poverty 5, 61 10.1186/s40249-016-0151-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C. F. (1968). Possible use of translocations to fix desirable genes in insect pest populations. Nature 218, 368-369. 10.1038/218368a0 [DOI] [PubMed] [Google Scholar]
- Curtis C. F., Southern D. I., Pell P. E. and Craig-Cameron T. A. (1972). Chromosome translocations in Glossina austeni. Genet. Res. 20, 101-113. 10.1017/S0016672300013616 [DOI] [PubMed] [Google Scholar]
- DiCarlo J. E., Chavez A., Dietz S. L., Esvelt K. M. and Church G. M. (2015). Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 33, 1250-1255. 10.1038/nbt.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhoff P. A., Wenger E. A., Godfray H. C. J. and Burt A. (2017). Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics. Proc. Natl. Acad. Sci. USA 114, E255-E264. 10.1073/pnas.1611064114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt K. M., Smidler A. L., Catteruccia F. and Church G. M. (2014). Concerning RNA-guided gene drives for the alteration of wild populations. Elife 3, e03401 10.7554/eLife.03401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasulo B., Meccariello A., Morgan M., Borufka C., Papathanos P. A. and Windbichler N. (2019). A fly model establishes distinct mechanisms for synthetic CRISPR/Cas9 sex distorters. BioRxiv. 10.1101/834630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feachem R. G. A., Chen I., Akbari O., Bertozzi-Villa A., Bhatt S., Binka F., Boni M. F., Buckee C., Dieleman J., Dondorp A. et al. (2019). Malaria eradication within a generation: ambitious, achievable, and necessary. Lancet 394, 1056-1112. 10.1016/S0140-6736(19)31139-0 [DOI] [PubMed] [Google Scholar]
- Galizi R., Doyle L. A., Menichelli M., Bernardini F., Deredec A., Burt A., Stoddard B. L., Windbichler N. and Crisanti A. (2014). A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat. Commun. 5, 3977 10.1038/ncomms4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz V. M. and Akbari O. S. (2018). Gene editing technologies and applications for insects. Curr. Opin. Insect Sci. 28, 66-72. 10.1016/j.cois.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz V. M. and Bier E. (2015). Genome editing. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442-444. 10.1126/science.aaa5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz V. M. and Bier E. (2016). The dawn of active genetics. BioEssays 38, 50-63. 10.1002/bies.201500102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz V. M., Jasinskiene N., Tatarenkova O., Fazekas A., Macias V. M., Bier E. and James A. A. (2015). Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 112, E6736-E6743. 10.1073/pnas.1521077112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald H. A., Gantz V. M., Poplawski G., Xu X.-R. S., Bier E. and Cooper K. L. (2019). Super-Mendelian inheritance mediated by CRISPR-Cas9 in the female mouse germline. Nature 566, 105-109. 10.1038/s41586-019-0875-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A., Galizi R., Kyrou K., Simoni A., Siniscalchi C., Katsanos D., Gribble M., Baker D., Marois E., Russell S. et al. (2016). A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78-83. 10.1038/nbt.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W.-D., Ehmsen K. T. and Liu J. (2010). Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113-139. 10.1146/annurev-genet-051710-150955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-J. S., Higgs S. and Vanlandingham D. L. (2019). Emergence and re-emergence of mosquito-borne arboviruses. Curr. Opin. Virol. 34, 104-109. 10.1016/j.coviro.2019.01.001 [DOI] [PubMed] [Google Scholar]
- James A. A. (2005). Gene drive systems in mosquitoes: rules of the road. Trends Parasitol. 21, 64-67. 10.1016/j.pt.2004.11.004 [DOI] [PubMed] [Google Scholar]
- James S., Collins F. H., Welkhoff P. A., Emerson C., Godfray H. C. J., Gottlieb M., Greenwood B., Lindsay S. W., Mbogo C. M., Okumu F. O. et al. (2018). Pathway to deployment of gene drive mosquitoes as a potential biocontrol tool for elimination of Malaria in sub-Saharan Africa: recommendations of a scientific working group†. Am. J. Trop. Med. Hyg. 98, 1-49. 10.4269/ajtmh.18-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A. and Charpentier E. (2012). A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandul N. P., Liu J., Buchman A., Gantz V. M., Bier E. and Akbari O. S. (2019). Assessment of a split homing based gene drive for efficient knockout of multiple genes. BioRxiv. 10.1101/706929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KaramiNejadRanjbar M., Eckermann K. N., Ahmed H. M. M., Sánchez C H. M., Dippel S., Marshall J. M. and Wimmer E. A. (2018). Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management. Proc. Natl. Acad. Sci. USA 115, 6189-6194. 10.1073/pnas.1713825115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T. A., Windbichler N., Deredec A., Burt A. and Benedict M. Q. (2012). Infertility resulting from transgenic I-PpoI male Anopheles gambiae in large cage trials. Pathog. Glob. Health 106, 20-31. 10.1179/2047773212Y.0000000003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer M. U. G., Sinka M. E., Duda K. A., Mylne A. Q. N., Shearer F. M., Barker C. M., Moore C. G., Carvalho R. G., Coelho G. E., Van Bortel W. et al. (2015). The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife 4, e08347 10.7554/eLife.08347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrou K., Hammond A. M., Galizi R., Kranjc N., Burt A., Beaghton A. K., Nolan T. and Crisanti A. (2018). A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062-1066. 10.1038/nbt.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts L., Koella J. C. and Boëte C. (2008). Can transgenic mosquitoes afford the fitness cost? Trends Parasitol. 24, 4-7. 10.1016/j.pt.2007.09.009 [DOI] [PubMed] [Google Scholar]
- Li M., Yang T., Kandul N. P., Bui M., Gamez S., Raban R., Bennett J., Sánchez C. H. M., Lanzaro G. C., Schmidt H.. et al. (2019). Development of a confinable gene drive system in the human disease vector, Aedes aegypti. BioRxiv. 10.1101/645440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorimer N., Hallinan E. and Rai K. S. (1972). Translocation homozygotes in the yellow fever mosquito, Aedes aegypti. J. Hered. 63, 159-166. 10.1093/oxfordjournals.jhered.a108261 [DOI] [PubMed] [Google Scholar]
- Marrelli M. T., Moreira C. K., Kelly D., Alphey L. and Jacobs-Lorena M. (2006). Mosquito transgenesis: what is the fitness cost? Trends Parasitol. 22, 197-202. 10.1016/j.pt.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Marshall J. M. and Akbari O. S. (2016). Gene drive strategies for population replacement. Genetic Control of Malaria and Dengue 169-200. 10.1016/B978-0-12-800246-9.00009-0 [DOI] [Google Scholar]
- Marshall J. M. and Akbari O. S. (2018). Can CRISPR-based gene drive be confined in the wild? A question for molecular and population biology. ACS Chem. Biol. 13, 424-430. 10.1021/acschembio.7b00923 [DOI] [PubMed] [Google Scholar]
- Marshall J. M., Buchman A., Sánchez C H. M. and Akbari O. S. (2017). Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci. Rep. 7, 3776 10.1038/s41598-017-02744-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J. M., Raban R. R., Kandul N. P., Edula J. R., León T. M. and Akbari O. S. (2019). Winning the tug-of-war between effector gene design and pathogen evolution in vector population replacement strategies. Front. Genet. 10, 1072 10.3389/fgene.2019.01072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble C., Olejarz J., Esvelt K. M., Church G. M. and Nowak M. A. (2017). Evolutionary dynamics of CRISPR gene drives. Sci. Adv. 3, e1601964 10.1126/sciadv.1601964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble C., Min J., Olejarz J., Buchthal J., Chavez A., Smidler A. L., DeBenedictis E. A., Church G. M., Nowak M. A. and Esvelt K. M. (2019). Daisy-chain gene drives for the alteration of local populations. Proc. Natl. Acad. Sci. USA 116, 8275-8282. 10.1073/pnas.1716358116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhofer G., Ivy T. and Hay B. A. (2018). Behavior of homing endonuclease gene drives targeting genes required for viability or female fertility with multiplexed guide RNAs. Proc. Natl. Acad. Sci. USA 115, E9343-E9352. 10.1073/pnas.1805278115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhofer G., Ivy T. and Hay B. A. (2019). Cleave and Rescue, a novel selfish genetic element and general strategy for gene drive. Proc. Natl. Acad. Sci. USA 116, 6250-6259. 10.1073/pnas.1816928116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papathanos P. A., Windbichler N. and Akbari O. S. (2014). Sex ratio manipulation for insect population control. In Transgenic Insects: Techniques and Applications (ed. M. Q. Benedict), pp. 83-100. CIBA Publishing; 10.1079/9781780644516.0083 [DOI] [Google Scholar]
- Raban R. and Akbari O. S. (2017). Gene drives may be the next step towards sustainable control of malaria. Pathog. Glob. Health 111, 399-400. 10.1080/20477724.2017.1453587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. S. (1976). Progress in the use of chromosomal translocations for the control of insect pests. Biol. Rev. Camb. Philos. Soc. 51, 1-24. 10.1111/j.1469-185X.1976.tb01118.x [DOI] [PubMed] [Google Scholar]
- Roggenkamp E., Giersch R. M., Schrock M. N., Turnquist E., Halloran M. and Finnigan G. C. (2018). Tuning CRISPR-Cas9 gene drives in Saccharomyces cerevisiae. G3 8, 999-1018. 10.1534/g3.117.300557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvemini M., Mauro U., Lombardo F., Milano A., Zazzaro V., Arcà B., Polito L. C. and Saccone G. (2011). Genomic organization and splicing evolution of the doublesex gene, a Drosophila regulator of sexual differentiation, in the dengue and yellow fever mosquito Aedes aegypti. BMC Evol. Biol. 11, 41 10.1186/1471-2148-11-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R. S., Chavez A., Porter C. B. M., Hamblin M., Kaas C. S., DiCarlo J. E., Zeng G., Xu X., Revtovich A. V., Kirienko N. V. et al. (2018). A CRISPR–Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat. Microbiol. 3, 73-82. 10.1038/s41564-017-0043-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla J. N. and Nagaraju J. (2010). Doublesex: a conserved downstream gene controlled by diverse upstream regulators. J. Genet. 89, 341-356. 10.1007/s12041-010-0046-6 [DOI] [PubMed] [Google Scholar]
- Stanaway J. D., Shepard D. S., Undurraga E. A., Halasa Y. A., Coffeng L. E., Brady O. J., Hay S. I., Bedi N., Bensenor I. M., Castañeda-Orjuela C. A. et al. (2016). The global burden of dengue: an analysis from the global burden of disease study 2013. Lancet Infect. Dis. 16, 712-723. 10.1016/S1473-3099(16)00026-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M. P. (2018). CRISPR-Based technologies: impact of RNA-targeting systems. Mol. Cell 72, 404-412. 10.1016/j.molcel.2018.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless R. L., Messer P. W., Connallon T. and Clark A. G. (2015). Modeling the manipulation of natural populations by the mutagenic chain reaction. Genetics 201, 425-431. 10.1534/genetics.115.177592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C., Hoeijmakers J. H. and Kanaar R. (2001). Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2, 196-206. 10.1038/35056049 [DOI] [PubMed] [Google Scholar]
- Ward C. M., Su J. T., Huang Y., Lloyd A. L., Gould F. and Hay B. A. (2011). Medea selfish genetic elements as tools for altering traits of wild populations: a theoretical analysis. Evolution 65, 1149-1162. 10.1111/j.1558-5646.2010.01186.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbichler N., Papathanos P. A., Catteruccia F., Ranson H., Burt A. and Crisanti A. (2007). Homing endonuclease mediated gene targeting in Anopheles gambiae cells and embryos. Nucleic Acids Res. 35, 5922-5933. 10.1093/nar/gkm632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbichler N., Papathanos P. A. and Crisanti A. (2008). Targeting the X chromosome during spermatogenesis induces Y chromosome transmission ratio distortion and early dominant embryo lethality in Anopheles gambiae. PLoS Genet. 4, e1000291 10.1371/journal.pgen.1000291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2019). World Malaria Report 2018. World Health Organization. [Google Scholar]
- Zlotorynski E. (2016). DNA repair: the cell cycle flavours of repair. Nat. Rev. Genet. 17, 65 10.1038/nrg.2015.35 [DOI] [PubMed] [Google Scholar]