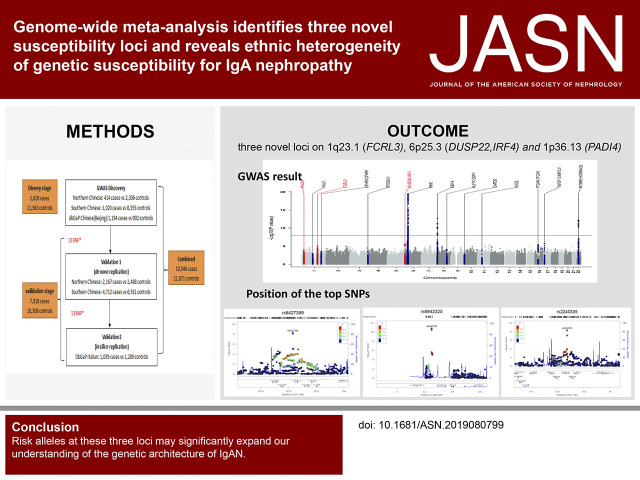
Significance Statement
Eighteen genetic risk loci for IgA nephropathy (IgAN) have been identified by genome-wide association studies (GWAS), but they only explain a small proportion of overall risk. By performing a three-stage meta-GWAS analysis in 10,546 patients and 21,871 healthy controls, the authors discovered three novel genetic risk loci on 1p36.13 (rs2240335), 1q23.1 (rs6427389), and 6p25.3 (rs6942325), implicating the roles of FCRL3, DUSP22.IRF4, and PADI4 in IgAN development. Through HLA imputation analyses, we revealed multiple independent associations within the MHC region. Besides the DEFA locus, they also discovered the genetic heterogeneity of six additional loci between Chinese and European populations. These findings have advanced the biologic understanding of IgAN and provided novel insight into the ethnic differences of genetic susceptibility.
Keywords: IgA nephropathy, meta-analysis, genome-wide association study, common variants
Visual Abstract
Abstract
Background
Eighteen known susceptibility loci for IgAN account for only a small proportion of IgAN risk.
Methods
Genome-wide meta-analysis was performed in 2628 patients and 11,563 controls of Chinese ancestry, and a replication analysis was conducted in 6879 patients and 9019 controls of Chinese descent and 1039 patients and 1289 controls of European ancestry. The data were used to assess the association of susceptibility loci with clinical phenotypes for IgAN, and to investigate genetic heterogeneity of IgAN susceptibility between the two populations. Imputation-based analysis of the MHC/HLA region extended the scrutiny.
Results
Identification of three novel loci (rs6427389 on 1q23.1 [P=8.18×10−9, OR=1.132], rs6942325 on 6p25.3 [P=1.62×10−11, OR=1.165], and rs2240335 on 1p36.13 [P=5.10×10−9, OR=1.114]), implicates FCRL3, DUSP22.IRF4, and PADI4 as susceptibility genes for IgAN. Rs2240335 is associated with the expression level of PADI4, and rs6427389 is in high linkage disequilibrium with rs11264799, which showed a strong expression quantitative trail loci effect on FCRL3. Of the 24 confirmed risk SNPs, six showed significant heterogeneity of genetic effects and DEFA showed clear evidence of allelic heterogeneity between the populations. Imputation-based analysis of the MHC region revealed significant associations at three HLA polymorphisms (HLA allele DPB1*02, AA_DRB1_140_32657458_T, and AA_DQA1_34_32717152) and two SNPs (rs9275464 and rs2295119).
Conclusions
A meta-analysis of GWAS data revealed three novel genetic risk loci for IgAN, and three HLA polymorphisms and two SNPs within the MHC region, and demonstrated the genetic heterogeneity of seven loci out of 24 confirmed risk SNPs. These variants may explain susceptibility differences between Chinese and European populations.
IgA nephropathy (IgAN) is a common form of GN around the world. First described by Berger in 1968,1 IgAN is diagnosed based on the renal biopsy specimen showing the deposition of IgA-containing immune complexes in the mesangial area of glomeruli, and the histopathologic lesions of mesangial cell proliferation and accumulation of extracellular matrix.2 About 20%–40% of IgAN cases progress to ESKD within 20 years after the disease was diagnosed.3–6
Prevalence of IgAN shows substantial variation globally; among patients undergoing renal biopsies, Asian populations have the highest frequency (40%–50%) of IgAN, with moderate frequency (20%–30%) in the European population, and low prevalence in the African population (<5%).7–10 The large difference in prevalence across world populations, together with the evidence of familial clustering and renal abnormalities among relatives of patients with IgAN, strongly suggest that genetic factors have significant contribution to IgAN risk.11–13
The genetic studies of IgAN, particularly genome-wide association studies (GWAS), have made great progress in recent years. Until now, there were five large GWAS performed in IgAN, which identified 18 susceptibility loci for IgAN and provided valuable insights into the genetic architecture of IgAN.14–18 However, these loci could only explain a small proportion of the disease risk and heritability, indicating that additional genetic risk factors remain to be discovered. Recently, meta-GWAS analysis of independent datasets across diverse populations has emerged as an effective and powerful approach for discovering additional genetic risk variants,19 which have identified several novel susceptibility loci in some complex diseases. However, no such analysis had yet been performed in IgAN.
Here, we performed a large-scale genome-wide meta-analysis (GWMA) of several published GWAS datasets, where 2628 patients with IgAN and 11,563 controls of Chinese ancestry were analyzed genome wide in the discovery stage, followed by a replication analysis of 6879 patients and 9019 controls of Chinese descent, and 1039 patients and 1289 controls of European descent (Supplemental Figure 1).15–18 We confirmed all of the known IgAN-associated loci and identified three novel loci on 1q23.1 (FCRL3), 6p25.3 (DUSP22.IRF4), and 1p36.13 (PADI4). These findings significantly expand our understanding of the disease susceptibility factor and provide novel biologic insights related to IgAN.
Methods
Study Subjects
We conducted a three-stage, case-control analysis for this study. The samples were of Chinese and European descent from four previously published GWAS (the studies of Gharavi et al.15,16 and ours17,18). The detailed sample information for each stage is shown in Table 1.
Table 1.
Summary of the sample information
| GWAS Cohorts | Population | Cases (N) | Controls (N) | Total | λGCa | λGC,1000 |
|---|---|---|---|---|---|---|
| Discovery cohorts | ||||||
| GWAS Northern18,b | Chinese | 414 | 2306 | 2720 | 1.052 | 1.046 |
| GWAS Southern18,b | Chinese | 1020 | 8355 | 9375 | 1.083 | 1.07 |
| Gharavi et al. study15 | Chinese | 1194 | 902 | 2096 | 1.035 | 1.034 |
| Total | 2,628 | 11,563 | 14,191 | 1.078 | 1.018 | |
| Validation cohorts | ||||||
| Validation Northern | Chinese | 2167 | 2488 | 4655 | — | — |
| Validation Southern | Chinese | 4712 | 6531 | 11,243 | — | — |
| Gharavi et al. study Europeans16,20 | European | 1039 | 1289 | 2328 | 1.039 | 1.034 |
| Total | 7,918 | 10,308 | 18,226 | — | — |
Adjusted for PCs except for Gharavi et al.’s GWAS because its patients were well matched with controls.
Only summary statistics were applied in this study.
The discovery study (stage 1) included three, independent, Chinese GWAS datasets from two published studies. The dataset of the Gharavi et al. study15,16 involved 2096 Chinese individuals (1194 patients with IgAN and 902 healthy controls). The two datasets of the Yu et al. study17,18 comprised 9375 individuals (1020 patients with IgAN and 8355 healthy controls) from Southern China, and 2720 (414 patients with IgAN and 2306 healthy controls) individuals from Northern China.
Replication analyses (stage 2) were performed de novo in two independent samples of individuals of Chinese Han descent, including 11,243 individuals (4712 patients with IgAN and 6531 healthy controls) from Southern China, and 4655 individuals (2167 patients with IgAN and 2488 healthy controls) from Northern China. We also used 2328 individuals of European ancestry for the in silico replication analysis (stage 3), where the data of 1039 Italian patients with IgAN were from the Gharavi et al. study16, and the data of 1289 Italian healthy controls were from the HYPERGENES study.20,21 In this study, the Chinese samples from the Gharavi et al. study15,16 were included in the GWMA discovery analysis, whereas the Italian samples from the Gharavi et al. study16 were included in the validation analysis.
As previously described,17,18 4137 patients were diagnosed by biopsy samples, according to histopathologic detection of immunofluorescence showing at least 2+ (scale 0–3+) mesangial deposition of IgA, with IgA comprising the dominant Ig deposited in the glomeruli, and excluding individuals with cirrhosis, Henoch–Schönlein purpura nephritis, hepatitis B–associated GN, HIV infection, and SLE.
Clinical information was collected from patients with IgAN at diagnosis, including serum IgA levels, history of gross hematuria, degree of microhematuria, proteinuria, eGFR (eGFR using the Modification of Diet in Renal Disease formula), and stage of CKD. On the basis of the collected clinical information, 3366 patients with IgAN were divided into three clinical subtypes: those with (1) asymptomatic hematuria or proteinuria, with urine protein <3.5 g in 24 hours, serum creatinine (Scr) <1.3 mg dl−1, and without edema or hypertension; (2) nephritic syndrome, with urine protein <3.5 g in 24 hours, with or without Scr >1.3 mg dl−1, edema, or hypertension; and (3) nephrotic syndrome, with urine protein ≥3.5 g in 24 hours. Gender and age information were collected from both patients and controls through questionnaires, and the patients and controls were matched for geographic origin and ethnicity.
The study was approved by the Institutional Review Board at the First Affiliated Hospital of Sun Yat-sen University. Written informed consent was obtained from all of the participants.
Sample Quality Control
In this study, we only performed the sample quality control (QC) in the Chinese and European datasets from the Gharavi et al. study15,16 separately, following the methods described in the previous publication.18 We first investigated autosomal single nucleotide polymorphisms (SNPs) by excluding SNPs from the X, Y, and mitochondrial chromosomes. Samples were removed if call rate <95% or heterozygosity rate was greater than the mean+3 SD. We performed identity-by-descent analysis using PLINK (http://www.cog-genomics.org/plink/1.9/) to identify the first- and second-degree relative pairs, and the relative with the low call rate was removed. The principal component (PC) analysis was done to identity outliers based on first five PCs. After sample QC, we were left with 1194 patients and 902 controls from Gharavi et al.’s Chinese dataset15,16 and with 1039 patients and 1289 controls from the Italian dataset16.
Whole-Genome Imputation and SNP QC
The Chinese and European datasets from the Gharavi et al. study15,16 were imputed separately. The software IMPUTE 2 (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html) was used for imputing untyped SNPs in each dataset after prephasing using SHAPEIT2 (https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html). We imputed only those genotyped SNPs that passed QC thresholds (call rate, >95%; minor allele frequency [MAF], >1%; Hardy–Weinberg equilibrium [HWE] P>1×10−4 in controls) in each dataset. The imputation was performed by using the multiethnic 1000 Genomes reference panel (dated October 2014) consisting of 2504 individuals from Africa, Southern Asia, East Asia, Europe, and the Americas. We performed stringent SNP QC after imputation: MAF >1%, impute information >0.5 if MAF >5%, impute information >0.8 if MAF <5%, HWE P in controls >1×10−8, and average maximum posterior probabilities >0.9. The final datasets for genotype dosage analyses were 5,092,149 SNPs and 6,067,310 SNPs for Gharavi et al.’s Chinese and Europeans, respectively.
HLA Imputation and Analysis
The imputation of HLA alleles and amino acid (AA) polymorphisms was performed in the Chinese and European datasets from the Gharavi et al. study15,16 using the SNP2HLA tool (http://software.broadinstitute.org/mpg/snp2hla/), based on the same set of directly genotyped SNPs within chromosome 6: 20–40 Mb that passed the QC thresholds. The Pan-Asian reference panel was used for the Chinese dataset,22,23 and the Type 1 Diabetes Genetic Consortium was used for the European dataset.24 For the Northern and Southern Chinese datasets of the Yu et al. study17,18, we obtained the imputed HLA data from the previously published study,18 which was performed using the same method and reference. We analyzed all imputed HLA alleles with high imputation confidence scores (r2>0.8). The tagged classic HLA alleles of the AA substitutions were defined by using the Sequence Alignment Tool (https://www.ebi.ac.uk/ipd/imgt/hla/align.html).
Genotyping and QC in the Validation Study
The genotyping analysis of selected SNPs for validation was performed by using Sequenom MassArray. The details of the assay were described in a previously published paper.18 All SNPs were confirmed to be of good quality by examining the clustering patterns of genotypes and selected mass peaks. All SNPs with call rates <95% and/or HWE <1×10−6 in controls and all samples with call rates <90% were removed from the Northern or Southern Chinese samples.
Association Analyses
The GWMA of the discovery stage was performed using the association statistics data from three GWAS datasets by conducting a fixed-effects, inverse variance meta-analysis using META (http://mathgen.stats.ox.ac.uk/genetics_software/meta/meta.html#), with or without correction for λ genomic control (GC) value. The association statistics data of the Northern and Southern Chinese datasets were from the previously published study,18 whereas the association statistics data of the Chinese dataset from the Gharavi et al. study15,16 was obtained using the SNPTEST implemented frequentist tests under a missing data logistic regression model. Because the patients and controls were shown to be genetically matched in the previous publication, the association analysis was performed without using PCs as covariates. λ1000 was calculated for individual and combined GWAS as a standardized genomic inflation, regardless of study sample size, using the following formula25: λ1000=1+(λobs−1)× (1/npatients+1/ncontrols)/(1/npatients,1000+1/ncontrols,1000).
For validation studies, we performed a trend test under a logistic regression model on samples collected from the northern or southern region of China to control for potential confounding of population stratification. We conducted inverse variance meta-analysis under a fixed-effects model to combine association statistics from GWAS, de novo replication samples, and in silico samples. The Cochrane Q test was used to test for heterogeneity in effect sizes between Chinese and European samples.
X Chromosome Analyses
We performed the chromosome X data analysis using software XWAS 3.0, and conducted SNP QC and imputation of chromosome X by following pipeline provided by the software. In Yu et al.’s Northern and Southern datasets17,18, we only obtained the genotypes of chromosome-X SNPs for 1343 patients and 4261 controls. These patients and controls were analyzed together as a single sample to maximize the power for association analysis. We also obtained the genotype data for Gharavi et al.’s Chinese samples (including 1194 patients and 902 controls)15,16. The general QC performed in either male or female was as follows: exclude SNPs if SNPs in pseudoautosomal regions, call rate <95%, MAF <0.005, HWE P<1×10−6 in controls, missingness P<1×10−6 in patients/controls. The sex-specific QC was as follows: exclude SNPs if the MAF difference has P<1×10−6 between male and female controls. We performed the association analysis in the two datasets independently using the logistic regression model in PLINK. For Gharavi et al.’s data15,16, only sex was included as covariates. For the Chinese dataset, both sex and five PCs (obtained from the previously published data) were included as covariates. We combined the association statistics of the two datasets through a fixed-effects, inverse variance meta-analysis.
Expression Quantitative Trait Loci and Functional Annotation Analyses
The expression quantitative trait loci (eQTL) analysis was done on the websites https://www.gtexportal.org/home/ and https://molgenis58.target.rug.nl/bloodeqtlbrowser/. The new tool GENE2FUNC (http://fuma.ctglab.nl/) was used for biologic annotation, and HaploReg (https://pubs.broadinstitute.org/mammals/haploreg), RegulomeDB (http://www.regulomedb.org/), and rVarBase (http://rv.psych.ac.cn/index.do) were used for annotation.
Fraction of Variance Explained by Loci
The percentage of the total variance explained by loci was estimated by the Nagelkerke pseudo-R2 using the fmsb package (https://cran.r-project.org/web/packages/fmsb/fmsb.pdf) from the logistic regression model with SNPs’ genotypes as predictors and disease state as an outcome in R (version 3.4.3).
Power Estimation and Heterogeneity of Effects
The statistical power of the SNP was estimated in the European dataset using software QUANTO (https://omictools.com/quanto-tool) under the following assumption: IgAN prevalence of 0.24%,26 a log-additive risk model, an effect size estimate observed in the combined Chinese population, and observed allele frequencies in European controls at 0.05 significance level.
Geospatial Risk Score of Ten Shared Gene Loci (13 SNPs) on the Human Genome Diversity Project
To investigate the geospatial distribution of genetic risk, we analyzed the publicly available genotype data from the Human Genome Diversity Project (HGDP), including 1043 samples among 52 worldwide populations and 660,918 markers typed on an Illumina 650Y array. Because three out of 13 shared SNPs were not available in the genotype data, they were imputed using the HGDP dataset. The QC was performed on the dataset before imputation. Briefly, 15 samples with an extreme high heterozygosity rate were removed, and 73 samples were removed because of first-degree cryptic relatedness. Markers were excluded with low call rate (<95%) and MAF <1%. The whole-genome imputation (software IMPUTE 2) was performed using the 1000 Genome Project reference panel (phase I, released in December 2013), using the remaining dataset including 955 individuals from 52 populations and 630, 559 markers. The three SNPs were imputed with high confidence (impute information >0.98). All 13 SNPs were extracted and converted to genotype data with probability 0.9. The call rate of all SNPs was higher than 95% and the MAF >0.2.
The genetic risk score (GRS) for each individual was calculated as the weighted sum of the number of risk alleles at each locus multiplied by the log of the corresponding meta odds ratio. The standardized GRS for each individual was estimated as follows:
The standardized risk score was compared among 52 worldwide populations by Kruskal–Wallis rank test. The correlation coefficients were measured between the standardized GRS and longitude or latitude of the population.
Analysis of Clinical Phenotypes
The three validated susceptibility SNPs were studied for association with clinical phenotypes by using different regression methods. We checked the normality of all quantitative phenotypes and the natural log transformation was performed for some continuous variables, including IgA, IgM, C3, C4, GFR, albumin, BUN, and Scr measurements. Samples with the extreme values (less than mean−3×SD or greater than mean+3×SD) were excluded from further association analyses. We used the linear regression test for association of these quantitative traits with different genotypes. Ordinal logistic regression was performed for the correlation analysis of clinical subtype and CKD stage. Regions were controlled for in all association tests. The means and SDs of serum IgA and proteinuria were estimated for each patient group stratified by genotype. All analysis was performed using R version 3.4.3.
Results
Discovery Study by GWMA
We first carried out a GWMA of autosomal SNPs using two independent Chinese GWAS datasets, including a total number of 2628 patients with IgAN and 11,563 controls. The first dataset (dataset 1) included 1434 patients and 10,661 controls, which was published in our previous study,18 and the second dataset (dataset 2) consisted of 1194 patients and 902 controls of Chinese ancestry, which were part of the GWAS dataset published by Gharavi et al.16 Dataset 1 consists of subjects recruited from Northern and Southern China, whereas the subjects of dataset 2 were all from Northern China. To combine the two independent datasets (generated using different SNP array platforms) and further increase the coverage of genetic variants for association testing, genome-wide imputation was performed in each dataset separately using the 1000 Genomes Project dataset as the reference panel. After applying stringent QCs, we tested the associations of 3,792,912 autosomal SNPs in dataset 1, 5,092,149 autosomal SNPs in dataset 2, and 3,611,808 shared autosomal SNPs in the GWMA of the combined dataset 1/2. To minimize the effect of population stratification caused by the known subpopulation structure of the Chinese population, we divided all of the samples of dataset 1 into northern (dataset 1_north: 414 patients and 2306 controls) and southern (dataset 1_south: 1020 patients and 8355 controls) clusters, and then performed a meta-analysis of all three independent datasets (dataset 1_north, dataset 1_south, and dataset 2) to test genetic association, where PC analysis was repeated within each cluster, and the significant PCs were used as covariates in the analysis. The information regarding the samples used in the GWMA was summarized in Table 1.
The genomic inflation factor for GWMA was 1.078. We used GC to derive more conservative statistics, bringing the genomic inflation down to <5% (Table 1). λ1000 was calculated for individual and combined GWAS as a standardized genomic inflation, regardless of study sample size. The inflation factor then reduced to 1.018, suggesting minimal effects of population stratification in our GWMA.
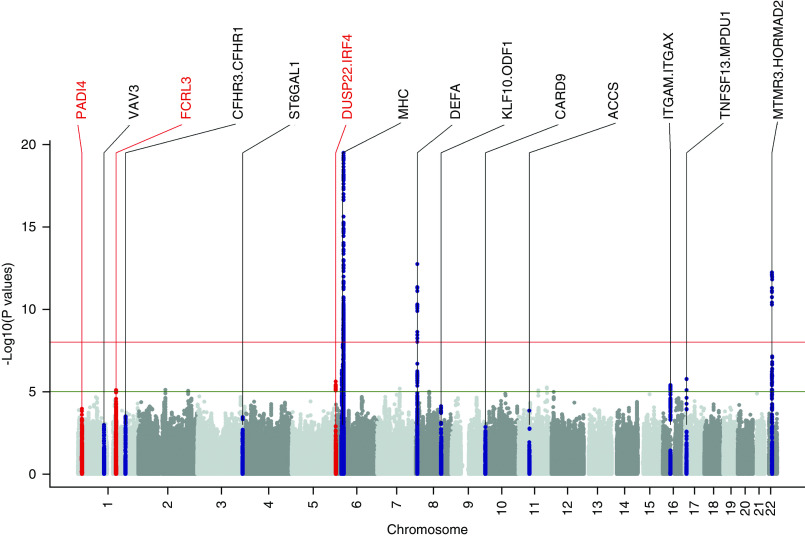
The quantile-quantile plots showed there is no strong evidence, beyond what is expected due to chance, for the excess of SNPs with small P values (Supplemental Figure 2). Supporting evidence of association were observed for all of the known loci (P<0.05) in the combined Chinese samples (Supplemental Table 1), and, in particular, the GWMA provided consistent evidence at genome-wide significance for the previously published associations within the MHC region, 8p23, and 22q12 (P<5×10−8) (Figure 1). We also performed the association analysis of chromosome X SNPs in the subset data of Yu et al.’s Northern and Southern Chinese samples17,18 (including 1343 patients and 4261 controls) and Gharavi et al.’s Chinese samples (including 1194 patients and 902 controls)15,16. However, our analysis did not reveal any X-linked associations with P<10−5 (Supplemental Figure 3).
Figure 1.
The three novel loci (red) and known loci (black) in the Manhattan plot of GWMA results at the discovery stage. Manhattan plot of the P values of association obtained by GWMA for IgAN based on 2628 patients and 11,563 controls. Plot obtained after dividing samples into three parts (Northern Chinese, Southern Chinese of our GWAS data17,18, and Northern Chinese in Gharavi et al.’s GWAS data15,16) in 3,611,808 SNPs. Negative log10-transformed P values for each SNP (y axis) are plotted by chromosomal position (x axis). The red and green lines represent the thresholds for genome-wide, statistically significant associations (P=5×10−8) and suggestive associations (P=1×10−5), respectively. A total of 11 known loci defined as loci that were previously published with genome-wide significance (P<5×10−8) were labeled black. Significant loci identified in the discovery stage of our study at genome-wide significance (P<5×10−8) were labeled red.
Validation Study of Novel Associations
After removing known loci, we extracted out the 34 top SNPs with a P value <1×10−5. These SNPs were clustered into 12 loci on the basis of their regional linkage disequilibrium (LD) patterns and physical location. We excluded five loci where association was supported by only a single SNP (“singleton signals,” defined by the absence of supporting signals in high LD [r2>0.8] with P<5×10−5 within each locus) (Supplemental Figure 4). For each of the remaining seven loci, we selected the top SNPs with lowest P value for further validation analysis.
In addition, we also selected six SNPs reported in the Gharavi et al.16 study, including three suggestive (rs12568771, rs2240335, and rs633059) (supplementary table 5 in the Gharavi et al. study16) and three confirmed SNPs (rs17019602, rs10086568, and rs4077515).
Therefore, a total of 13 SNPs were selected for the phase 1 validation analysis in independent Chinese samples. Thirteen SNPs were genotyped in two independent cohorts, totaling 6879 patients with IgAN and 9019 controls, with 4712 patients and 6531 controls from Southern Chinese populations, and 2167 patients and 2488 controls from Northern Chinese populations. In addition, we also included the Italian cohort of 1039 patients in the Gharavi et al.16 study and 1289 controls in HYPERGENE study20,21 as the third in silico replication dataset, which included 6,067,310 SNPs after imputation and SNP QC (λGC=1.037 with PC1 corrected).
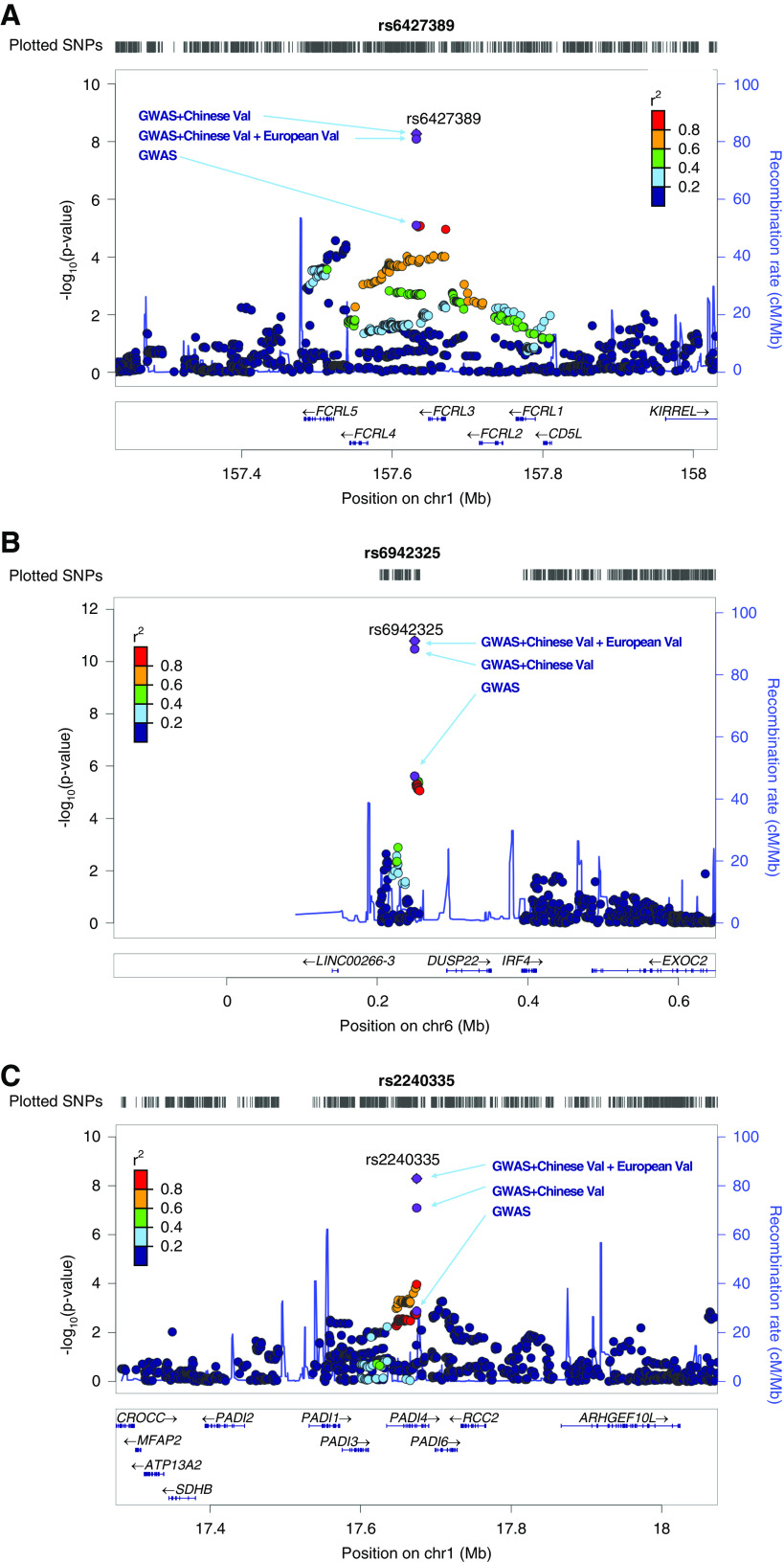
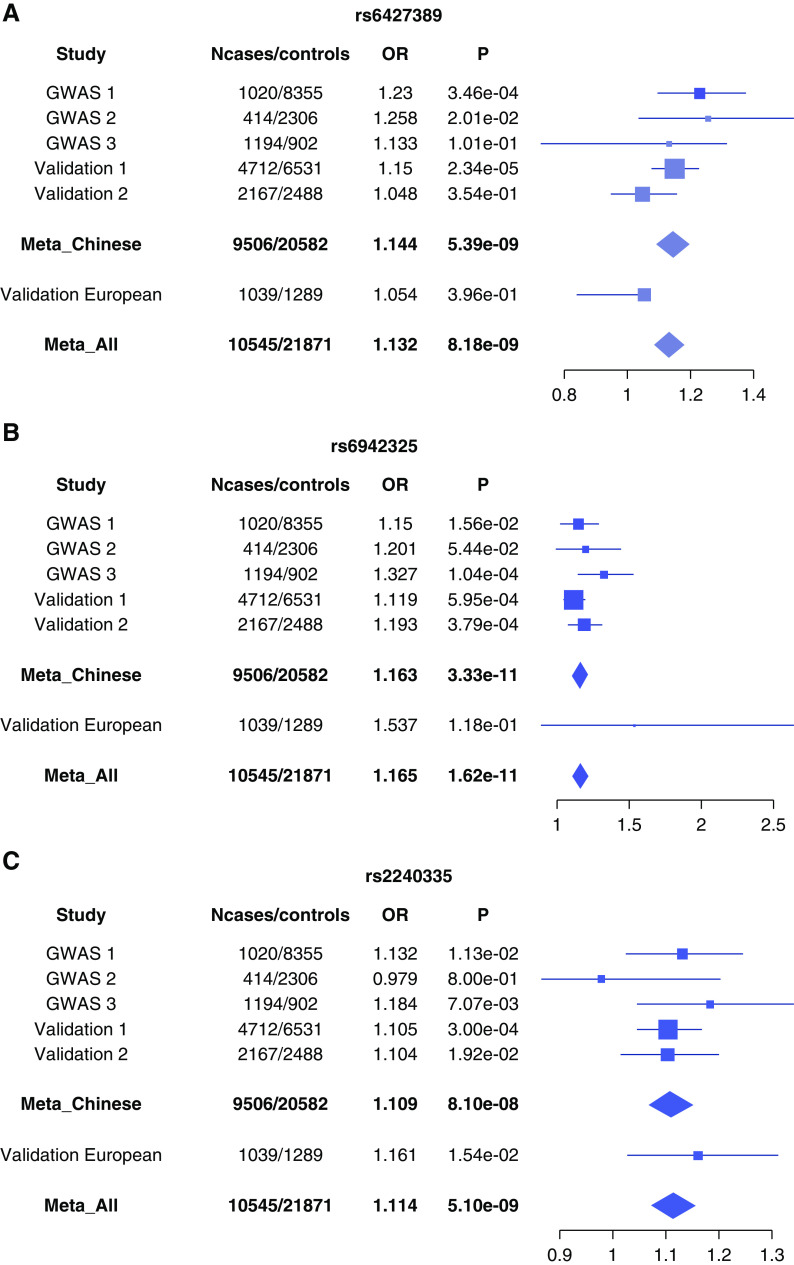
In the validation analysis, 7918 patients with IgAN and 10,308 healthy controls were involved in the joint association analysis using meta-analysis under a fixed-effects model. The association results of these 13 SNPs in discovery and/or combined samples were summarized in Supplemental Table 2. Of the 13 SNPs, only three were confirmed by the validation analysis, and the other ten SNPs failed to be validated. Three novel associations were identified to have genome-wide significance (P<5×10−8) with consistent effects across the discovery and validation samples (PQ>0.05), including rs2240335 on 1p36.13 (P=5.10×10−9, odds ratio [OR]=1.114, PADI4), rs6427389 on 1q23.1 (P=8.18×10−9, OR=1.132, FCRL3), and rs6942325 on 6p25.3 (P=1.62×10−11, OR=1.165, IRF4, DUSP22) (Figures 2 and 3, Table.2). The associations at the three novel loci remained the same level of significance after correction by λGC values (PGC in Table 2).
Figure 2.
Three novel genetic risk loci reaching genome-wide significance. (A) rs6427389 at 1q23.1 (FCRL3); (B) rs6942325 at 6p25.3 (DUSP22, IRF4); and (C) rs2240335 at 1p36.13 (PADI4). Shown are P values obtained in GWAS discovery, in the combined analysis of GWAS and Chinese validation samples, and/or in silico European samples (fixed-effects meta-analysis). Chr1, chromosome 1; chr6, chromosome 6; val, validation.
Figure 3.
The forest plots of three novel loci across the three discovery and two validation samples. (A) rs6427389 at 1q23.1 (FCRL3); (B) rs6942325 at 6p25.3 (DUSP22, IRF4); and (C) rs2240335 at 1p36.13 (PADI4). Shown are the associations in the discovery stage (across Northern Chinese samples, Southern Chinese samples, and the Chinese samples in Gharavi et al.’s study15,16), and the validation stage (Northern Chinese, Southern Chinese, and Gharavi et al.’s Italian population [Europeans]16). On the left, each study is indicated. The plots show the study-specific association estimates (ORs) and 95% confidence intervals for the discovery and validation stage studies, presented as bars. GWAS 1, Northern Chinese samples; GWAS 2, Southern Chinese samples; GWAS 3, Chinese samples in Gharavi et al.’s study; validation 1, Northern Chinese; validation 2, Southern Chinese; validation European, Gharavi et al.’s Italian population.
Table 2.
Meta-analysis results of the three novel IgAN susceptibility loci
| Information | TA | NTA | Study | TAF_A | TAF_U | P | PGC | OR (95% CI) | I2 | PQ |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2240335Chr1:17674537PADI4 | A | C | Chinese Southern | 0.606 | 0.586 | 1.13e−02 | 1.132 (1.028 to 1.245) | |||
| A | C | Chinese Northern | 0.401 | 0.408 | 8.00e−01 | 0.979 (0.867 to 1.203) | ||||
| A | C | Chinese dbGaP | 0.597 | 0.555 | 7.07e−03 | 1.184 (1.047 to 1.340) | ||||
| A | C | Validation Southern | 0.605 | 0.581 | 3.00e−04 | 1.105 (1.047 to 1.167) | ||||
| A | C | Validation Northern | 0.601 | 0.577 | 1.92e−02 | 1.104 (1.016 to 1.199) | ||||
| A | C | Meta_Chinese | 8.10e−08 | 1.09e−07 | 1.109 (1.068 to 1.152) | 0.00 | 4.76e−01 | |||
| A | C | Italian dbGaP | 0.405 | 0.374 | 1.54e−02 | 1.161 (1.029 to 1.311) | ||||
| A | C | Meta-analysis | 5.10e−09 | 7.64e−09 | 1.114 (1.074 to 1.155) | 0.00 | 4.78e−01 | |||
| rs6427389Chr1:157632011FCRL3 | C | A | Chinese Southern | 0.806 | 0.771 | 3.46e−04 | 1.230 (1.098 to 1.377) | |||
| C | A | Chinese Northern | 0.792 | 0.758 | 2.01e−02 | 1.258 (1.037 to 1.527) | ||||
| C | A | Chinese dbGaP | 0.790 | 0.769 | 1.01e−01 | 1.133 (0.727 to 1.315) | ||||
| C | A | Validation Southern | 0.793 | 0.769 | 2.34e−05 | 1.150 (1.078 to 1.227) | ||||
| C | A | Validation Northern | 0.783 | 0.775 | 3.54e−01 | 1.048 (0.950 to 1.156) | ||||
| C | A | Meta_Chinese | 5.39e−09 | 9.83e−09 | 1.144 (1.093 to 1.197) | 28.71 | 2.30e−01 | |||
| C | A | Italian dbGaP | 0.658 | 0.644 | 3.96e−01 | 1.054 (0.840 to 1.071) | ||||
| C | A | Meta-analysis | 8.18e−09 | 1.44e−08 | 1.132 (1.086 to 1.181) | 34.78 | 2.16e−01 | |||
| rs6942325Chr6:249714DUSP22, IRF4 | G | A | Chinese Southern | 0.792 | 0.769 | 1.56e−02 | 1.150 (1.027 to 1.288) | |||
| G | A | Chinese Northern | 0.777 | 0.761 | 5.44e−02 | 1.201 (0.997 to 1.446) | ||||
| G | A | Chinese dbGaP | 0.788 | 0.737 | 1.04e−04 | 1.327 (1.150 to 1.531) | ||||
| G | A | Validation Southern | 0.790 | 0.770 | 5.95e−04 | 1.119 (1.050 to 1.194) | ||||
| G | A | Validation Northern | 0.792 | 0.761 | 3.79e−04 | 1.193 (1.082 to 1.315) | ||||
| G | A | Meta_Chinese | 3.33e−11 | 5.62e−11 | 1.163 (1.112 to 1.216) | 20.83 | 2.82e−01 | |||
| G | A | Italian dbGaP | 0.988 | 0.983 | 1.18e−01 | 1.537 (0.896 to 2.637) | ||||
| G | A | Meta-analysis | 1.62e−11 | 2.80e−11 | 1.165 (1.114 to 1.218) | 2.12 | 3.12e−01 |
TA, test allele; NTA, non–test allele; TAF_A, frequency of TA in patients; TAF_U, frequency of TA in controls; PGC, association P value after adjusting λGC by META software; PQ, P value of heterogeneity test; Chr1, chromosome 1; Chr6, chromosome 6.
We evaluated the contribution of the three novel SNPs to overall IgAN risk. Cumulatively, these three novel association signals explained about 0.5% of disease variance. About 5.0% of overall risk can be explained by these three novel and 20 previously reported SNPs.
Imputation-Based Analysis of the MHC/HLA Region
Our GWMA revealed extensive association within the MHC region (Figure 1). To fine map this extensive association, we did a stepwise conditional analysis of the MHC region in the GWMA discovery dataset. We identified four independent signals (rs9275464, rs2295119, rs9271611, and rs9275169) within the MHC region with a genome-wide significance (Supplemental Table 3). SNP rs2295119 has been reported previously.18 The top SNP, rs9275464, is in LD with other previously reported SNPs, rs660895 (D′=0.74, r2=0.31) and rs1794275 (D′=0.67, r2=0.39).
We also performed HLA imputation using the SNP2HLA tool and Pan-Asian reference panel, and tested the associations of HLA alleles and AA polymorphisms. We observed extensive associations of HLA alleles and AAs (data not shown), but stepwise conditional analysis discovered three independent associations at the HLA allele DPB1*02 (OR=1.31, P=7.93×10−12) and two AAs, AA_DRB1_140_32657458_T (OR=1.43, P=2.58×10−23) and AA_DQA1_34_32717152 (OR=1.25, P=2.80×10−10) (Supplemental Figure 5, Supplemental Table 4). The association of DPB1*02 has been reported before, and AA_DRB1_140_32657458_T and AA_DQA1_34_32717152 were novel. We also studied the two AA substitutions on three-dimensional protein structure of HLA-DR and DQ molecules, and found that AA_DRB1_140_32657458_T was in position 140, located in the β-turn of the HLA-DR protein, whereas AA_DQA1_34_32717152 was in position 34 in the β-sheet floor of the HLA-DQ protein (Supplemental Figure 6).
After conditioning the genetic effects of the three HLA allele/AAs, rs9271611 no longer showed association, and the evidence of association at rs9275169 was significantly weakened (from OR=1.43, P=3.65×10−13 to OR=1.23, P=1.85×10−5) (Supplemental Table 4). Associations at rs9275464 and rs2295119 remained at a level of genome-wide significance. We also observed significant associations with HLA_DRB1_04 and HLA_DRB1_1501, but their evidence for association was abolished or significantly weakened by conditioning on the association effect of AA_DRB1_140_32657458_T, indicating these HLA classic allele associations are due to the genetic effect of this AA polymorphism. Similarly, we also observed the significant associations with the classic alleles of DQA1 (*0101, *03, *0301), but these associations were due to the genetic effect of the AA polymorphism AA_DQA1_34_32717152 (Supplemental Table 5).
Together, our imputation-based analysis has revealed significant associations at three HLA polymorphisms and additional two SNPs within the MHC region.
Association with Clinical Phenotypes
The associations of the clinical phenotypes of IgAN with the three novel SNPs were investigated in 3747 patients where clinical information was collected (Supplemental Table 6). The results showed that the minor and protective allele A of rs6427389 was associated with lower serum albumin levels (P=0.019), and the minor and protective allele A of rs6942325 was associated with lower serum IgA levels (P=0.028). (Supplemental Tables 7 and 8). However, after the Bonferroni correction, neither of them was significant (P>1×10−3, 0.05/45). There are also no significant associations with other clinical symptoms, such as hypertension, microhematuria, GFR, and proteinuria, and the pathologic pattern in patients with IgAN.
Analysis of Ethnic Heterogeneity between Chinese and Europeans
To analyze the ethnic heterogeneity of genetic association, we compared the association evidence of 33 reported or proxy SNPs from 21 confirmed loci (18 previously reported and three newly discovered) between the Chinese and European datasets (Supplemental Table 1). We observed significant and consistent associations of all 33 SNPs in the combined Chinese samples (meta-analysis P values ranging from 9.36×10−14 to 9.36×10−3). We looked up the independence of these 33 SNPs in two populations. The LD pattern (r2) for all paired SNPs is consistent in both populations, except for two pairs of SNPs (rs12716641/rs10086568 and rs3803800/rs4227). Both rs12716641 and rs10086568 are independent in Chinese (r2<0.2), but are in moderate LD in European (r2=0.33) populations; rs3803800 and rs4227 are in moderate LD (r2=0.39) in Chinese, but independent in European (r2<0.2) populations. The heterogeneity analysis was performed for all 33 SNPs.
First, we compared the allele frequencies of 33 SNPs across the two populations. Although 31 SNPs showed similar allele frequency, rs11150612 at ITGAX-ITGAM locus and rs2240335 at the PADI4 locus had significant differences between the two populations (Supplemental Table 1). The minor allele G of rs11150612 had a much lower frequency in our Chinese samples (31.3%) than in the European samples (58.8%). Similarly, the minor allele C of rs2240335 had a much lower frequency in our Chinese samples (44.5%) than in the European samples (61.3%).
We then compared the genetic association effects of 33 SNPs detected in our Chinese and European samples. Of the 33 SNPs, 16 SNPs failed to show a significant association in the European samples. We estimated the statistical power of the European samples for detecting the association effects of these SNPs and found that the European samples of our study have sufficient statistical power (power >80%) for seven SNPs, but insufficient power (power <80%) for the remaining nine SNPs. We also compared three DEFA-reported SNPs (rs12716641, rs2738048, and rs10086568) between two populations. Only rs12716641 showed significance in Europeans, the genetic effect of which is consistent with the one detected in Chinese samples. SNP rs2738048 did not show significance in Europeans, although the European samples provided sufficient power (>80%) for detecting the genetic effect identified in the Chinese population. SNP rs10086568 did not show significance in Europeans, but the European samples did not have sufficient power (<80%) for detection. We did not observe other associations within the LD block of SNP rs2738048 in the European samples (Supplemental Figure 7A). Taking together, these results strongly suggest allelic heterogeneity at the DEFA locus between Chinese and European populations. For the remaining six SNPs where the European samples provided sufficient power (>80%) for detection, none of them showed significant association in the European samples, and significant heterogeneity of the OR estimate (PQ<0.05) was observed between the two populations. In addition, the regional plots of these six SNPs showed that only very moderate associations were detected in SNPs that were in moderate LD with the reported SNPs at nominal significance level (Supplemental Figure 7, B–G). Therefore, these six loci are either Chinese specific or have much weaker genetic effects in European populations.
A total of 17 SNPs within the loci of VAV3, CFH, DQB1/DRB1, DPA1/DPB2, XKR5, DEFA, CARD9, ITGAX/ITGAM, HORMAD2/MTMR3, and PADI4 showed significant association in European samples (P values ranging from 3.85×10−2 to 1.21×10−11). Of these 17 SNPs, 16 showed consistent association between the Chinese and European samples without evidence of heterogeneity (PQ>0.05). However, the OR estimates for rs114240894 near the DQB1/DRB1 gene were in the same direction, but significantly different between the Chinese (OR=1.305) and European samples (OR=1.644) (PQ<0.05). As expected, the meta-analysis of the combined Chinese and European samples revealed stronger associations at these SNPs than the individual ethnic samples under a fixed-effects model (Supplemental Table 1). We also compared the association evidence of classic HLA alleles and AAs between Chinese and European cohorts. There was no evidence for heterogeneity for AA_DRB1_140_32657458_T and AA_DQA1_34_32717152. Moderate heterogeneity was observed for HLA_DPB1_02 (OR=1.31 versus 1.14, I2=0.63), but the evidence was not statistically significant (Supplemental Table 9).
Altogether, we were able to analyze the genetic heterogeneity of 24 SNPs (seven plus 17) (Supplemental Table 1) and three HLA alleles and AAs. We found strong evidence for allelic heterogeneity at the DEFA locus between Chinese and Europeans. We also found significant evidence of genetic heterogeneity for seven additional SNPs within four loci (rs2523946, 6p21[MHC], HLA-A; rs2295119, 6p21[MHC], DPA1.DPB2; rs4227, 17p13.1, MPDU1.SOX15; rs114240894, 6p21[MHC], DQB1.DRB1; rs7190997, 16p11.3, ITGAX-ITGAM; rs2412971, 22q12, HORMAD2; and rs2412973, 22q12, HORMAD2), and all of the SNPs showed stronger genetic effect in Chinese populations than in the Europeans. The remaining 16 SNPs and HLA alleles and AAs showed similar genetic effects between the two populations.
We also studied the distribution of the mean of standardized GRS in HGDP populations. Standardized GRS is quite different among the 52 HGDP populations (P<2.2×10−16, Kruskal–Wallis test). The GRS is also positively correlated with both longitude (P=2.24×10−7) and latitude (P=1.26×10−5). In particular, the analysis confirmed that the GRS is higher in Chinese than in European populations (Supplemental Figure 8).
Functional Investigation and eQTL Analysis
The three novel SNPs (rs2240335, rs6427389, and rs6942325) identified in this study were located within LD blocks. To investigate the functional implication of these novel associations, we first determined the SNPs and indels that are in high LD (r2≥0.8) with three novel SNPs, using the LD information provided by HaploReg version 3 on the 1000 Genomes phase 1 Asian population, which identified 89 SNPs and small indels (Supplemental Table 10). The RegulomeDB annotation analysis of 92 variants suggested that rs2240335 likely affects transcription factor binding which may regulate the expression of PADI4 (Supplemental Table 11). rVarBase annotation analysis also suggested that rs2240335 may influence transcription factor binding, the gene target of which is PADI4 (Supplemental Table 12).
The mRNA expression level (eQTL) effects were evaluated through the GTEx portal (https://gtexportal.org/home/eqtls/tissue?tissueName=All). The eQTL analysis showed that the risk allele A of rs2240335 was strongly associated with increasing expression level of gene PADI4 in tissues such as brain, lung, spleen, muscle, adipose tissue, and thyroid. There was no direct eQTL evidence found for the other two SNPs. However, the C allele of rs11264799 (in high LD with the risk allele of rs6427389) showed strong association with the decreased expression level of gene FCRL3 in tissues of whole blood, esophagus mucosa, adipose tissue, lung, and thyroid (Supplemental Table 13).
We further investigated potential pleiotropic effects of three novel SNPs by documenting reported associations with other diseases and known biologic functions within the LD regions of the three loci (Table 3). The analysis found that these variants within the LD regions of the three novel SNPs are reported to be associated with immune response, chromatin organization, and transcription, and are involved in the regulatory functions of immune system.
Table 3.
Functional annotations of novel loci and associated SNPs
| SNP/locus | eQTL/ENCODE | Other disease | Known functions of genes |
|---|---|---|---|
| rs2240335 1p36.13 (chr1:17674537) SNP location: lies within the exon of PADI4 | Risk allele is strongly associated with increased expression level of PADI4 in tissues such as brain, lung, spleen, muscle, adipose tissue, and thyroid | Amyotrophic lateral sclerosis, rheumatoid arthritis | PADI4 encodes peptidylarginine deiminase 4 belonging to the peptidylarginine deiminase family. Responsible for the conversion of arginine residues to citrulline residues. Play key roles in various biologic processes, such as immune response, chromatin organization, transcription, and protein aggregation.27 |
| rs6427389 1q23.1 (chr1:157632011) SNP location: located in an intergenic region closest to the gene FCRL3 | Risk allele is in strong LD (r2>0.9 in our samples, HapMap Europeans, and Asia) with variant (rs11264799), associated with decreased expression level of FCRL3 in whole blood, esophagus, adipose tissue, lung, and thyroid | Not previously associated with any other trait | FCRL3 encodes Fc receptor like 3, a member of the Ig receptor superfamily. Promotes TLR9-mediated B cell activation, but inhibits antibody production and plasma cell differentiation by enhancing ERK-dependent suppression of BLIMP1. Enhances activation of NF-κB and MAPK signaling pathways in CpG/TLR9-stimulated B cells.28 |
| rs6942325 6p25.3 (chr6:249714) SNP location: located in an intergenic region closest to the genes DUSP22, IRF4 | No evidence for effects on gene expression levels | Not previously associated with any other trait | DUSP22 encodes dual specificity phosphatase 22. It is extensively expressed various mammalian cells such as T cells, B cells, and NK cells, indicating that JKAP may be involved in several important biologic processes, activates c-Jun N-terminal kinase pathway but not extracellular signal–related kinase pathway or p38 MAPK pathway in mammalian cells.29 IRF4 encodes IFN regulatory factor 4. Acts as transcription factors for IFNs, thus involved in regulatory functions in the immune system and in oncogenesis.30 Rearrangement of DUSP22/IRF4 gene was observed in some hematologic neoplasms, such as multiple myeloma, diffuse large B cell lymphoma, anaplastic large cell lymphoma, and transformed mycosis fungoides.31 |
Details on eQTLs and ENCODE annotations in Supplemental Tables 6–9. BLIMP1, B lymphocyte induced maturation protein 1; Chr1, chromosome 1; TLR9, toll-like receptor 9; ERK, extracellular signal–regulated kinase; MAPK, mitogen-activated protein kinase; chr6, chromosome 6; JKAP, JNK pathway associated phosphatase.
Discussion
By performing a powerful, three-stage GWMA of IgAN in a large sample consisting of 10,546 patients with IgAN and 21,871 healthy controls, we identified three novel associations on 1q23.1, 6p25.3, and 1p36.13. For each locus of association, the robustness of association is reflected by the genome-wide significance of evidence in the combined discovery and validation samples, significant evidence in the independent validation samples, and the consistent association effects across all of the independent samples. As a limitation, only 3.6M autosomal SNPs were investigated in our current study, because the GWMA discovery analysis was performed using published summary statistics. Further studies with more comprehensive coverage of genome-wide variants, particularly through deeper imputation using a larger reference panel of Asian population,32 can help to further advance the genetic study of IgAN in Asian populations. Although we have only modestly improved the portion of overall risk explained by genetic risk factors, the discovery of these novel loci has advanced the biologic insight of IgAN pathogenesis by revealing additional components of immune response, inflammation, and mucosal immunity that are related to IgAN.
The most significant locus was on 6p25.3, where the candidate genes DUSP22 and IRF4 were located within this LD region. IFN regulatory factor 4 (IRF4) was an inducible inhibitor of LPS signaling, which has multiple roles in innate and adaptive immunity. The deficiency of IRF4 could enhance systemic inflammation and the activation of antigen-presenting cells, and could prevent the maturation of plasma cells and effector T cells. It was reported that IRF4 may promote the development of lupus nephritis, despite suppressing the antigen-presenting cells.33 The gene of DUSP22 (dual specificity phosphatase 22), also named JKAP (JNK pathway-associated phosphatase), which specifically activates the kinase JNK34 and acts as a tyrosine phosphatase.35 DUSP22-knockout mice show enhanced T cell–mediated immune responses, leading to suppression of T cell–mediated immunity, and they spontaneously develop systemic inflammation and autoimmunity, including exacerbated renal damage.36 DUSP22 protein levels were significantly decreased in T cells of patients with active lupus nephritis, and the loss of DUSP22 in T cells may relate to dysregulation of the immune complement system and inflammatory responses in kidneys.37
For the locus on 1p36.13 (rs2240335), both functional and eQTL analyses have implicated PADI4 (peptidyl arginine deiminase 4) as a strong candidate for being a disease gene for IgAN. PADI4 is a member of the gene family which encodes post-translational modification enzymes responsible for the conversion of arginine residues to citrulline residues, mainly distributed in the cells of various hematopoietic lineages. PADI4 is a nuclear enzyme that is critically involved in the release of decondensed chromatin from neutrophils as neutrophil extracellular traps, which were implicated in host defense against pathogens.38 It was considered that PADI4 may play a role in granulocyte and macrophage development, leading to inflammation and immune response.39 In rheumatoid arthritis, PADI4 was found to be responsible for fibrin citrullination and involved in apoptosis.40
Within the locus on 1q23.1 (rs6427389), eQTL analysis has implicated FCRL3 (Fc receptor like 3) as the candidate gene for susceptibility to IgAN. FCRL3 is one of the Fc receptor–like glycoproteins. Mutations in FCRL3 have been associated with several autoimmune diseases, including rheumatoid arthritis, autoimmune thyroid disease, and SLE, which are considered to be involved in pathologic autoimmune reactions in these disease-specific ectopic lymphocyte aggregates.41 The gene expression level of FCRL3 was found to be decreased in esophagus mucosa, which was thought to be closely related to the mucosal immunity. These findings have expanded our understanding of the immune genetic pathogenesis of IgAN.
We performed the imputation-based analysis of the extensive association observed within the MHC region. Through stepwise conditional analysis and HLA imputation, we revealed significant associations at three HLA polymorphisms and two additional SNPs. As the largest fine-mapping analysis of the MHC region, this study revealed the extensive involvement of HLA-mediated immunity in IgAN development by discovering three HLA variants. Further study in independent samples, particularly non-Chinese samples, will be needed to confirm these multiple associations within the MHC/HLA region. We also observe significant associations with HLA-DRB1*04 and HLA-DQA1 *0101, but their evidence for association was abolished by conditioning on the association effect of AA_DRB1_140_32657458_T and AA_DQA1_34_32717152 separately, indicating that these HLA classic allele associations are due to the genetic effect of the single AA polymorphism. These two classic HLA alleles were related to some autoimmune diseases, such as multiple sclerosis, autoimmune thyroid disease, and rheumatoid arthritis. To further investigate the HLA region, we will do fine mapping of HLA using next-generation sequencing in the future.
In our genotype to clinical phenotypes analysis, we altogether tested three SNPs and 15 phenotypes for association. Although two phenotypes showed association at P value of <0.05, none of them were significant after Bonferroni correction (P>1×10−3, 0.05/45). The findings need to be validated in additional patients with IgAN with detailed clinical phenotypes.
In this study, we performed a comprehensive genetic heterogeneity analysis of all of the confirmed IgAN risk SNPs, and revealed significant differences of genetic susceptibility between Chinese and European populations. Of the 24 confirmed risk SNPs for IgAN, where genetic heterogeneity analysis could be performed, only 17 showed consistent associations between Chinese and European populations. Six SNPs showed evidence of genetic heterogeneity between the two populations, and the DEFA locus seems to be Chinese specific. The evidence for the genetic heterogeneity of these six SNPs is solid. First, our analysis included all of the known loci, including the ones initially discovered in the European population. Second, the estimates of genetic effects from the current Chinese cohorts (used in the power calculation) should not be significantly overestimated, because (1) these six loci are the known loci discovered by previous studies, instead of this study, and (2) the Chinese samples of this study also included a significant amount of Chinese samples that are independent from the previous Chinese discovery samples. So, 30% of the confirmed risk SNPs (seven of 24 SNPs) showed significant differences in genetic association effects between the two populations. In addition, the GRS showed a positive correlation with both longitude and latitude, and the Chinese population has a higher GRS than Europeans. Taken together, these results clearly suggest significant differences in term of genetic susceptibility among ethnic populations, which may, at least partially, explain the global variation in the prevalence of IgAN.
Although the genetic heterogeneity of DEFA is known, this study performed a more comprehensive analysis of the genetic heterogeneity in all of the known loci. As a result, our study has revealed substantial genetic heterogeneity beyond the DEFA locus between Chinese and European populations. Given that the risk/protective alleles at the DEFA locus are thought to be conferred by patterns of copy number variations (CNVs) at this complex locus, we acknowledged that the observed allelic heterogeneity at the locus could either result from differences in LD between SNPs and CNVs (i.e., apparent allelic heterogeneity), or it could result from different effects of the same CNVs (i.e., functional allelic heterogeneity). We also acknowledge that, as a limitation, the Italian samples used in this study are smaller and less powerful than the Chinese samples. Evidence for genetic heterogeneity of IgAN across populations can be strengthened by analyzing additional and diverse European samples.
In summary, we have conducted the first GWMA in IgAN and discovered three novel susceptibility loci. The discovery of these new loci and the imputation-based analysis of the MHC region has advanced the understanding of the genetic architecture of IgAN and provided further biologic insight by implicating the involvement of mucosal immunity and inflammation in the development of IgAN. Our genetic heterogeneity analysis has also provided strong evidence for ethnic differences in terms of genetic susceptibility to IgAN. Such ethnic heterogeneity may help to explain the difference in prevalence of IgAN across ethnic populations, and also highlights the importance of conducting genetic studies of IgAN in diverse populations.
Disclosures
All authors have nothing to disclose.
Funding
This work was funded by the National Key Research and Development Program grant 2016YFC0906100; Guangdong Provincial Key Laboratory operational grant 2017B030314019; Guangdong-Hong Kong Joint Laboratory on Immunological and Genetic Kidney Diseases grant 2019B121205005; Science and Technology Planning Project of Guangdong Province grants 2017A050503003 and 2017B020227006; National Natural Science Foundation of China grants 81770661, 81570599, and 81920108008; Science and Technology Planning Project of Guangzhou City grant 2016201604030005; National Research Foundation Singapore fellowship grant NRF-NRFF2016-03; and the Agency for Science, Technology and Research (A*STAR) of Singapore (to J. Liu).
Supplementary Material
Acknowledgments
We are grateful to all of the subjects and healthy volunteers who participated in this work. We thank the staff in the First Affiliated Hospital of Sun Yat-sen University for help with sample collection, DNA extraction, and clinical data collection.
We thank the Susceptibility Loci for IgAN Study Investigators for providing the database of Genotypes and Phenotypes (dbGaP) data (dbGaP accession, phs000431.v2.p1).
The Susceptibility Loci for IgAN Study was conducted by the Susceptibility Loci for IgAN Study Investigators and supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
The data from the Susceptibility Loci for IgAN Study reported here were supplied by dbGAP. This manuscript was not prepared in collaboration with investigators of the Susceptibility Loci for IgAN Study and it does not necessarily reflect the opinions or views of the Susceptibility Loci for IgAN Study, dbGAP, or the NIDDK.
Dr. Xue-Qing Yu and Dr. Jian-Jun Liu conceived of and supervised the project; Dr. Jian-Jun Liu, Dr. Xue-Qing Yu, Dr. Ming Li, and Dr. Ling Wang prepared the manuscript; Dr. Ming Li, Dr. Ling Wang, Dr. Jia-Nee Foo, Dr. Chiea-Chuen Khor, and Dr. Jin-Xin Bei conducted and supervised the genotyping of samples; Dr. Jian-Jun Liu, Dr. Ling Wang, and Dr. Jia-Nee Foo contributed to the design and execution of statistical analyses; Dr. Li Wang, Dr. Yun-Hua Liao, Dr. Jian Chen, Dr. Qin-kai Chen, Dr. Gang Xu, Dr. Geng-Ru Jiang, Dr. Jian-Xin Wan, Dr. Meng-Hua Chen, Dr. Nan Chen, Dr. Hong Zhang, Dr. Yi-Xin Zeng, and Dr. Zhi-Hong Liu performed clinical characterization, were responsible for recruitment of subjects, and contributed samples; Dr. Chiara Lanzani, Dr. Lorena Citterio, and Dr. Erika Salvi contributed to providing the data of European samples in the validation study; Ms. Dian-Chun Shi, Mr. Zhong Zhong, and Ms. Pei-Ran Yin contributed to DNA extraction and clinical data collection.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2019080799/-/DCSupplemental.
Supplemental Table 1. Look-up of 33 reported SNPs within known and newly discovered loci of IgAN in current Chinese and European (Italian) genome-wide meta-analyses.
Supplemental Table 2. Thirteen SNPs being analyzed in de novo validation samples.
Supplemental Table 3. Meta results of conditional analyses within MHC region in discovery stage.
Supplemental Table 4. Meta results of conditional analyses on HLA alleles in discovery stage.
Supplemental Table 5. Stepwise conditional analysis of three independent HLA signals.
Supplemental Table 6. Clinical characteristics of the IgAN Patients of the Meta-GWAS and replication cohorts.
Supplemental Table 7. Clinical phenotypes-genotype association of IgAN cases with three identified SNPs.
Supplemental Table 8. Mean of three genotypes for two clinical phenotypes of IgAN.
Supplemental Table 9. Summary of three independent signals in HLA region between Chinese and European populations.
Supplemental Table 10. HaploRegv2 annotation of SNPs in novel and suggestive loci in LD (r2>0.8) with three identified SNPs in Asian population.
Supplemental Table 11. RegulomeDB annotation of SNPs in novel and suggestive loci in LD (r2>0.8) with three identified SNPs.
Supplemental Table 12. Look-up of three novel IgAN-associated SNPs in rVarBase.
Supplemental Table 13. Significant eQTLs effects for GWAS lead SNPs identified in the study based on GTEx (v7) data (48 different tissue types across 30 general tissue types).
Supplemental Figure 1. Flow chart of GWMA for IgAN.
Supplemental Figure 2. Quantile-quantile plot of the association.
Supplemental Figure 3. X chromosome data analysis in all Chinese samples or each gender group.
Supplemental Figure 4. Regional plots of the five loci with singleton signals.
Supplemental Figure 5. Regional plots of the stepwise conditional analysis of HLA signals in each conditioning step.
Supplemental Figure 6. The three-dimensional protein structure of HLA-DR, HLA-DQ moleculars with the two amino acid substitutions.
Supplemental Figure 7. Regional plots of seven reported SNPs associations in European (Italian samples).
Supplemental Figure 8. The distribution of mean of standardized genetic risk score (GRS) among Chinese and European in HGDP.
References
- 1.Berger J, Hinglais N: [Intercapillary deposits of IgA-IgG]. J Urol Nephrol (Paris) 74: 694–695, 1968. [PubMed] [Google Scholar]
- 2.Wyatt RJ, Julian BA: IgA nephropathy. N Engl J Med 368: 2402–2414, 2013. [DOI] [PubMed] [Google Scholar]
- 3.Zhou FD, Zhao MH, Zou WZ, Liu G, Wang H: The changing spectrum of primary glomerular diseases within 15 years: A survey of 3331 patients in a single Chinese centre. Nephrol Dial Transplant 24: 870–876, 2009. [DOI] [PubMed] [Google Scholar]
- 4.Li LS, Liu ZH: Epidemiologic data of renal diseases from a single unit in China: Analysis based on 13,519 renal biopsies. Kidney Int 66: 920–923, 2004. [DOI] [PubMed] [Google Scholar]
- 5.Pan X, Xu J, Ren H, Zhang W, Xu Y, Shen P, et al. : Changing spectrum of biopsy-proven primary glomerular diseases over the past 15 years: A single-center study in China. Contrib Nephrol 181: 22–30, 2013. [DOI] [PubMed] [Google Scholar]
- 6.Rodrigues JC, Haas M, Reich HN: IgA nephropathy. Clin J Am Soc Nephrol 12: 677–686, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai KN, Tang SC, Schena FP, Novak J, Tomino Y, Fogo AB, et al. : IgA nephropathy. Nat Rev Dis Primers 2: 16001, 2016. [DOI] [PubMed] [Google Scholar]
- 8.Jarrick S, Lundberg S, Welander A, Carrero JJ, Höijer J, Bottai M, et al. : Mortality in IgA nephropathy: A nationwide population-based cohort study. J Am Soc Nephrol 30: 866–876, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magistroni R, D’Agati VD, Appel GB, Kiryluk K: New developments in the genetics, pathogenesis, and therapy of IgA nephropathy. Kidney Int 88: 974–989, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schena FP, Nistor I: Epidemiology of IgA nephropathy: A global perspective. Semin Nephrol 38: 435–442, 2018. [DOI] [PubMed] [Google Scholar]
- 11.Scolari F, Amoroso A, Savoldi S, Mazzola G, Prati E, Valzorio B, et al. : Familial clustering of IgA nephropathy: Further evidence in an Italian population. Am J Kidney Dis 33: 857–865, 1999. [DOI] [PubMed] [Google Scholar]
- 12.Paterson AD, Liu XQ, Wang K, Magistroni R, Song X, Kappel J, et al. : Genome-wide linkage scan of a large family with IgA nephropathy localizes a novel susceptibility locus to chromosome 2q36. J Am Soc Nephrol 18: 2408–2415, 2007. [DOI] [PubMed] [Google Scholar]
- 13.Karnib HH, Sanna-Cherchi S, Zalloua PA, Medawar W, D’Agati VD, Lifton RP, et al. : Characterization of a large Lebanese family segregating IgA nephropathy. Nephrol Dial Transplant 22: 772–777, 2007. [DOI] [PubMed] [Google Scholar]
- 14.Feehally J, Farrall M, Boland A, Gale DP, Gut I, Heath S, et al. : HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol 21: 1791–1797, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, et al. : Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 43: 321–327, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, et al. : Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 46: 1187–1196, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu XQ, Li M, Zhang H, Low HQ, Wei X, Wang JQ, et al. : A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet 44: 178–182, 2011. [DOI] [PubMed] [Google Scholar]
- 18.Li M, Foo JN, Wang JQ, Low HQ, Tang XQ, Toh KY, et al. : Identification of new susceptibility loci for IgA nephropathy in Han Chinese. Nat Commun 6: 7270, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evangelou E, Ioannidis JP: Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet 14: 379–389, 2013. [DOI] [PubMed] [Google Scholar]
- 20.Salvi E, Kutalik Z, Glorioso N, Benaglio P, Frau F, Kuznetsova T, et al. : Genomewide association study using a high-density single nucleotide polymorphism array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of endothelial NO synthase. Hypertension 59: 248–255, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aung T, Ozaki M, Lee MC, Schlötzer-Schrehardt U, Thorleifsson G, Mizoguchi T, et al. : Genetic association study of exfoliation syndrome identifies a protective rare variant at LOXL1 and five new susceptibility loci. Nat Genet 49: 993–1004, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pillai NE, Okada Y, Saw W-Y, Ong RT-H, Wang X, Tantoso E, et al. : Predicting HLA alleles from high-resolution SNP data in three Southeast Asian populations. Hum Mol Genet 23: 4443–4451, 2014. [DOI] [PubMed] [Google Scholar]
- 23.Okada Y, Kim K, Han B, Pillai NE, Ong RTH, Saw W-Y, et al. : Risk for ACPA-positive rheumatoid arthritis is driven by shared HLA amino acid polymorphisms in Asian and European populations. Hum Mol Genet 23: 6916–6926, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rich SS, Concannon P, Erlich H, Julier C, Morahan G, Nerup J, et al. : The Type 1 diabetes genetics consortium. Ann N Y Acad Sci 1079: 1–8, 2006 [DOI] [PubMed] [Google Scholar]
- 25.de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF: Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet 17: R122–R128, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon P, Ramee MP, Boulahrouz R, Stanescu C, Charasse C, Ang KS, et al. : Epidemiologic data of primary glomerular diseases in western France. Kidney Int 66: 905–908, 2004. [DOI] [PubMed] [Google Scholar]
- 27.Tanikawa C, Ueda K, Suzuki A, Iida A, Nakamura R, Atsuta N, et al. : Citrullination of RGG motifs in FET proteins by PAD4 regulates protein aggregation and ALS susceptibility. Cell Rep 22: 1473–1483, 2018. [DOI] [PubMed] [Google Scholar]
- 28.Li FJ, Schreeder DM, Li R, Wu J, Davis RS: FCRL3 promotes TLR9-induced B-cell activation and suppresses plasma cell differentiation. Eur J Immunol 43: 2980–2992, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou R, Chang Y, Liu J, Chen M, Wang H, Huang M, et al. : JNK pathway-associated phosphatase/DUSP22 suppresses CD4+ T-cell activation and Th1/Th17-cell differentiation and negatively correlates with clinical activity in inflammatory bowel disease. Front Immunol 8: 781, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam S, Lim JS: Essential role of interferon regulatory factor 4 (IRF4) in immune cell development. Arch Pharm Res 39: 1548–1555, 2016. [DOI] [PubMed] [Google Scholar]
- 31.Wang RC, Sakata S, Chen BJ, Chang ST, Hsieh PP, Yang CS, et al. : Mycosis fungoides in Taiwan shows a relatively high frequency of large cell transformation and CD56 expression. Pathology 50: 718–724, 2018. [DOI] [PubMed] [Google Scholar]
- 32.Wu D, Dou J, Chai X, Bellis C, Wilm A, Shih CC, et al. : Large-scale whole-genome sequencing of three diverse Asian populations in Singapore. Cell 179: 736–749.e715, 2019 [DOI] [PubMed] [Google Scholar]
- 33.Lech M, Weidenbusch M, Kulkarni OP, Ryu M, Darisipudi MN, Susanti HE, et al. : IRF4 deficiency abrogates lupus nephritis despite enhancing systemic cytokine production. J Am Soc Nephrol 22: 1443–1452, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen AJ, Zhou G, Juan T, Colicos SM, Cannon JP, Cabriera-Hansen M, et al. : The dual specificity JKAP specifically activates the c-Jun N-terminal kinase pathway. J Biol Chem 277: 36592–36601, 2002. [DOI] [PubMed] [Google Scholar]
- 35.Li JP, Fu YN, Chen YR, Tan TH: JNK pathway-associated phosphatase dephosphorylates focal adhesion kinase and suppresses cell migration. J Biol Chem 285: 5472–5478, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li JP, Yang CY, Chuang HC, Lan JL, Chen DY, Chen YM, et al. : The phosphatase JKAP/DUSP22 inhibits T-cell receptor signalling and autoimmunity by inactivating Lck. Nat Commun 5: 3618, 2014. [DOI] [PubMed] [Google Scholar]
- 37.Chuang HC, Chen YM, Hung WT, Li JP, Chen DY, Lan JL, et al. : Downregulation of the phosphatase JKAP/DUSP22 in T cells as a potential new biomarker of systemic lupus erythematosus nephritis. Oncotarget 7: 57593–57605, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honda M, Kubes P: Neutrophils and neutrophil extracellular traps in the liver and gastrointestinal system. Nat Rev Gastroenterol Hepatol 15: 206–221, 2018. [DOI] [PubMed] [Google Scholar]
- 39.Wong SL, Wagner DD: Peptidylarginine deiminase 4: A nuclear button triggering neutrophil extracellular traps in inflammatory diseases and aging [published online ahead of print June 20, 2018]. FASEB J doi:10.1096/fj.201800691R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang X, Yamada R, Suzuki A, Sawada T, Yoshino S, Tokuhiro S, et al. : Localization of peptidylarginine deiminase 4 (PADI4) and citrullinated protein in synovial tissue of rheumatoid arthritis. Rheumatology (Oxford) 44: 40–50, 2005. [DOI] [PubMed] [Google Scholar]
- 41.Kochi Y, Yamada R, Suzuki A, Harley JB, Shirasawa S, Sawada T, et al. : A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet 37: 478–485, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.