Abstract
RNA isolated from formalin-fixed, paraffin-embedded (FFPE) tissue is commonly evaluated in both investigative and diagnostic pathology. However, the quality of the data is directly impacted by RNA quality. The RNA integrity number (RIN), an algorithm based on a combination of electrophoretic features, is widely applied to RNA isolated from paraffin-embedded tissue, but it is a poor indicator of the quality of that RNA. Here we describe the novel paraffin-embedded RNA metric (PERM) for quantifying the quality of RNA from FFPE tissue. The PERM is based on a formula that approximates a weighted area-under-the-curve analysis of an electropherogram of the extracted RNA. Using biochemically degraded RNAs prepared from experimentally fixed mouse kidney specimens, we demonstrate that PERM values correlate with mRNA transcript measurements determined using the QuantiGene system. Furthermore, PERM values correlate with real-time PCR data. Our results demonstrate that the PERM can be used to qualify RNA for different end-point studies and may be a valuable tool for molecular studies using RNA extracted from FFPE tissue.
Keywords: formalin-fixed, paraffin-embedded, RNA integrity, molecular pathology
METHOD SUMMARY
Using an electropherogram created by the Agilent 2100 Bioanalyzer, the paraffin-embedded RNA metric (PERM) can be determined by a simple calculation based on the number of fluorescent units at specific time points. The PERM can be used to assess the integrity of RNA obtained from FFPE tissue in an application-specific manner.
The need to determine RNA quality is a result of advances in RNA-based assays, as well as the diversity of biospecimens from which RNA can be isolated. The advent of RNA-based biomarker assays of disease that interrogate the expression and sequences of RNAs in tissue has increased the demand for accurate measures of RNA quality. It is critical that the metric for RNA quality be defined in terms of its suitability for specific assays. Efforts to define RNA quality have thus accelerated with the development of microarrays (1). The RNA integrity number (RIN) has been widely adopted as a measure of RNA quality for RNA isolated from fresh and frozen tissue (2,3). However, the RIN remains an imperfect measure of quality; it lacks strong correlation to gene-specific measurements and cannot be accurately applied to RNA isolated from formalin-fixed, paraffin-embedded (FFPE) tissue (2).
The design of a robust metric of RNA quality requires the development of a model to explain the differences observed in RNA isolated from frozen and FFPE tissues (4). The nature of RNA degradation within FFPE tissue was unclear until our previous work demonstrated that the quality of the RNA obtained from FFPE tissue was affected by (i) tissue hypoxia during the fixation phase (5) and (ii) challenges in RNA isolation related to the impregnation protocols (6). These studies resulted in a new model of tissue preservation in which chemical fixation protocols result in prolonged tissue hypoxia/anoxia from the time the tissue is devitalized until the penetration and reaction of the fixative halts cellular processes. During this interval, cellular programming in response to hypoxia initiates mRNA degradation via the activation of RNases that randomly degrade mRNAs and also target for degradation their poly-A tails and 5′-caps. These processes seek to conserve energy that would otherwise be diverted to protein translation. Concurrent with mRNA degradation, all other classes of RNA are subject to degradation by RNases. On electropherograms, frozen tissues show a broad distribution of RNA lengths; however, this signal is overwhelmed by the 18S and 28S peaks in this frequency distribution representation. With formalin fixation and paraffin embedding, the 18S and 28S peaks are generally not observed, largely due to hypoxia related to immersion fixation, and the distribution of RNA fragments is left-biased, with a tail representing the longer RNA fragments extending to the right (6,7). This distribution has been described previously (4), but methods to define a metric of RNA quality that correlates with gene-specific measures have not been previously reported.
In the present study, we prepared specimens with varying RNA quality based on various fixation and tissue processing conditions, as previously described (5). In addition, we measured mRNA expression levels using the QuantiGene assay, which is based on branched-chain DNA technology. Furthermore, we created the paraffin-embedded RNA metric (PERM) formula, which is a novel quality assessment tool for RNA extracted from FFPE tissue. We also evaluated the value of PERM as an RNA quality tool and demonstrated the reliability of the PERM for RNA quality assessment in FFPE tissue specimens.
Materials and methods
Tissue specimens
To create different levels of RNA degradation, we prepared FFPE tissue blocks as previously reported (5). Briefly, 6-week-old female Balb/c mouse kidneys were fixed and processed with varying fixation times, fixation buffers, and tissue processing times. A whole kidney was used as the standard tissue specimen for the experiment, and independent experiments were carried out in triplicate. Mice were acquired from the Small Animals Section, Veterinary Resources Branch, National Institutes of Health (NIH). The animals were housed and euthanized in accordance with NIH guidelines for care and use of laboratory animals (6).
RNA extraction
Total RNA was extracted as previously reported (5–7). Two 10 m-thick tissue sections were trimmed of excess wax and deparaffinized by three rounds of incubation in PROTOCOL Tissue Clearing Agent (Fisher Scientific, Kalamazoo, MI) for 15 m at 95°C with shaking followed by centrifugation at room temperature for 2 m at 10,000 × g. After deparaffinization, the sections were resuspended and ground in a solution of 4 M guanidine isothiocyanate, 20 mM sodium acetate, and 25 mM -mercaptoethanol (pH 5.5), followed by incubation for 72 h at 65°C with mild shaking. Subsequently, total RNA was isolated by phenol–chloroform extraction. In order to eliminate possible genomic DNA contamination, the isolated RNA was treated with TURBO DNase (Invitrogen, Carlsbad, CA).
The quantity of RNA extracted from FFPE tissue specimens was measured using a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). In addition, RNA was run on the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA), using the RNA 6000 LabChip kit (Agilent Technologies) to assess RNA integrity and the ratio of the ribosomal RNAs. Using the Agilent 2100 Expert Software, we measured the RIN.
Paraffin-embedded RNA metric (PERM)
To assess the RNA integrity of the isolated FFPE tissue specimen, we created a simple formula called the PERM. This novel RNA integrity assessment tool involved the use of an electropherogram created by the Agilent 2100 Bioanalyzer. The PERM is based on a formula that approximates a weighted area-under-the-curve approach (Figure 1A):
Figure 1. Paraffin-embedded RNA metric (PERM) for RNA extracted from formalin-fixed, paraffin-embedded (FFPE) tissues.
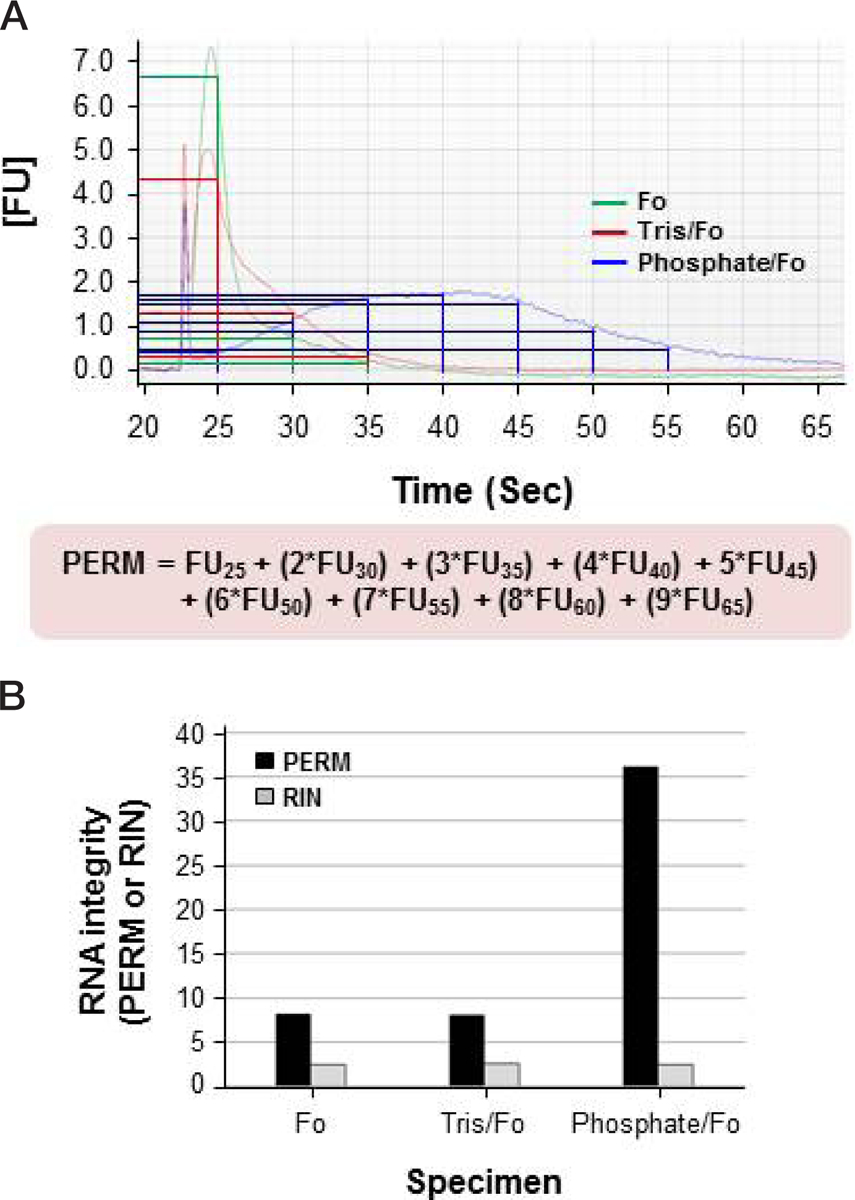
Total RNA was extracted from three different mouse kidney FFPE tissue specimens treated with different fixatives (not buffered, Tris-buffered, and phosphate-buffered formalin). (A) Calculation of the PERM. RNA quality was assessed using the Agilent 2100 Bioanalyzer combined with the RNA 6000 LabChip kit (Agilent Technologies), and the PERM was calculated using the formula shown underneath the electropherogram. FUn is the number of fluorescent units at time n (in seconds) in the electropherogram. (B) Comparison of the PERM and RNA integrity number (RIN). The RIN was calculated using the Agilent 2100 Expert Software. Black and gray bars represent the PERM and RIN, respectively. Fo, formalin; Tris/Fo, Tris-buffered formalin; Phosphate/Fo, phosphate-buffered formalin.
The PERM is calculated as the fluorescence units at 25 s, plus 2× the fluorescence units at 30 s, plus 3× the fluorescence units at 35 s, continuing in a multiplicative progression until the signal returns to base line. This approach provides a metric that places progressively greater value on the length of RNA. The PERM value is only applicable to RNA extracted from tissue that is subjected to chemical fixation via immersion and impregnation. We have validated the PERM against a variety of chemical fixatives paired with paraffin impregnation.
mRNA expression
Transcriptional expression levels in FFPE tissue specimens were assessed using the QuantiGene 2.0 Reagent System (Panomics, Fremont, CA). The QuantiGene assay was performed as previously described (5,8–10). To measure mRNA expression signals in FFPE tissue specimens, we used probes against glyceraldehye 3-phosphate dehydrogenase (GAPDH), cyclin-dependent kinase 4 (CDK4), and actin (ACTB), all specific for mouse RNA. Briefly, we used 200 ng of total RNA extracted from FFPE tissue samples for both the GAPDH and ACTB genes, whereas we used 500 ng total RNA for CDK4. The total RNA was resuspended in 10 μL RNA and then mixed with 40 μL capture buffer, 40 μL lysis mixture, and 10 μL target gene probe set (capture extender, label extender, and blocker). These mixtures were incubated at 53°C for 16–20 h within a 96-well capture plate. Subsequently, the plates were incubated with a branched DNA amplifier at 46°C for 1 h and then the label probe working reagents were added. After incubation with the substrate dioxetane solution at 46°C for 30 m, luminescence was measured using a GloRunnerTM Microplate Luminometer (TunerBiosystems, Sunnyvale, CA). Data represent the mean of three independent experiments.
We have additionally evaluated a series of alcohol-based alternative fixatives (11). The effect of fixatives on RNA integrity was evaluated by real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) amplification of the hypoxanthine-guanine phosphoribosyl transferase (HPRT) gene. To further evaluate the PERM as an FFPE RNA integrity tool, we examined the correlation between PERM values and Cq values of HPRT using a retrospective data set.
Statistical analysis
Statistical analyses were performed using IBM SPSS version 21.0 (IBM Corp., Armonk, NY). Pearson’s correlation coefficient analysis was used to evaluate the association not only between RNA measurement and the PERM or RIN but also the correlation between the PERM and RT-qPCR data. A P value <0.05 was considered statistically significant.
Results and discussion
The reliability of the PERM for assessing RNA integrity was evaluated in experimental mouse kidney FFPE tissue blocks. Total RNA was isolated from 20 mouse kidney FFPE tissue specimens. Optimal A260/A280 ratios for RNA were obtained from all specimens (1.81–1.93, data not shown). All samples were run on the Agilent 2100 Bioanalyzer. For the initial assessment of the PERM, three RNA specimens were analyzed using the Agilent 2100 Expert Software, and the PERM was then calculated using electropherograms (Figure 1). As shown in Figure 1B, the PERM had a greater dynamic range (8.05–51.65) than the RIN (2.4–2.5). These data demonstrate that the RIN is not appreciably correlated with the quality of RNA extracted from FFPE tissue.
To evaluate the reliability of the PERM as an RNA integrity tool for FFPE tissue, we performed the QuantiGene assay using mouse GAPDH, CDK4, and ACTB probe sets on all of the specimens. There was an excellent correlation between mRNA expression measured by the QuantiGene assay and the PERM values for GAPDH (r = 0.963, P < 0.001) (Figure 2A), CDK4 (r = 0.869, P < 0.001) (Figure 2B), ACTB (r = 0.974, P < 0.001) (Figure 2C), and the average of the 3 genes (r = 0.974, P < 0.001) (Figure 2D). However, there was no correlation between the RIN and mRNA using the QuantiGene assay (r = 0.060, P = 0.801) (Figure 2E).
Figure 2. Correlation of the paraffin-embedded RNA metric (PERM) with mRNA expression levels in formalin-fixed, paraffin-embedded (FFPE) tissues.
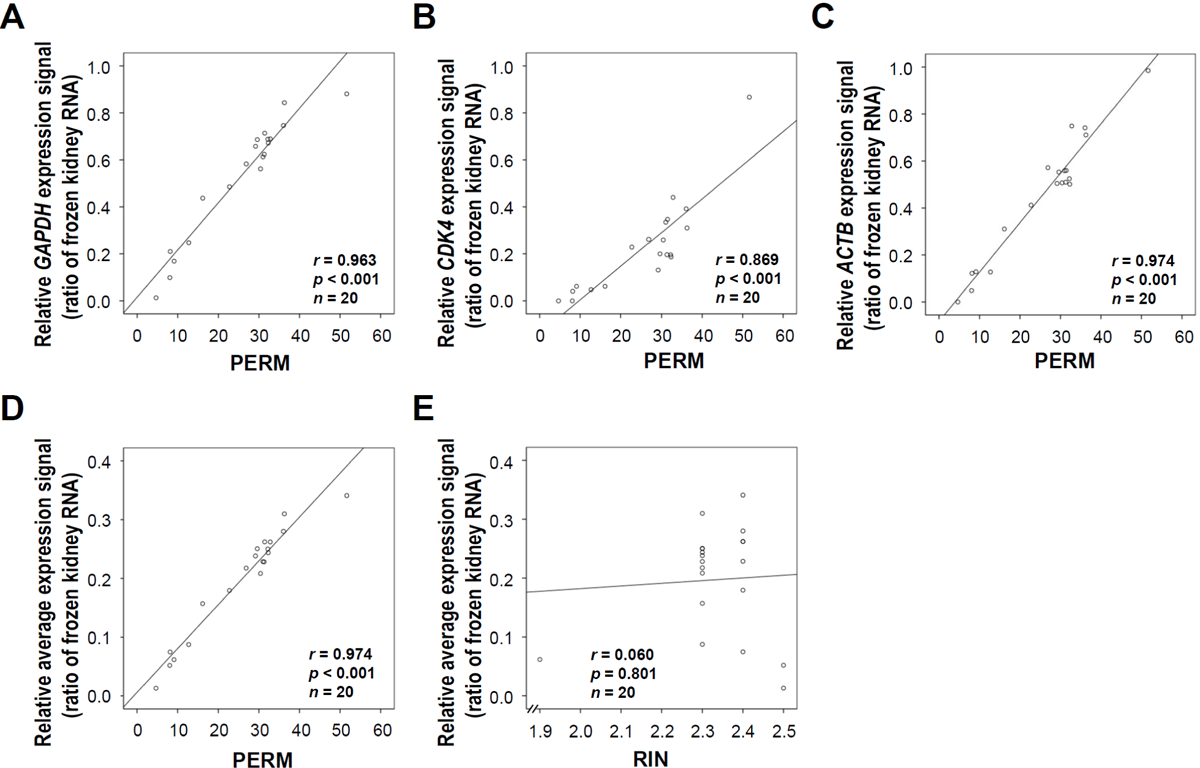
We extracted total RNA from 20 different mouse kidney FFPE tissue specimens. mRNA expression was measured using the QuantiGene assay with mouse GAPDH, CDK4, and ACTB gene-specific probe sets using the QuantiGene reagent system (Panomics). The relative expression level of each gene was normalized to that of frozen kidney tissue. (A) Scatter plot of the PERM versus GAPDH gene expression level (r = 0.963, P < 0.001). (B) Scatter plot of the PERM versus CDK4 gene expression level (r = 0.869, P < 0.001). (C) Scatter plot of the PERM versus ACTB gene expression level (r = 0.974, P < 0.001). (D) Scatter diagram of the PERM versus the average expression levels of the three genes (r = 0.974, P < 0.001). (E) Scatter plot of the RIN versus the average expression levels of the three genes (r = 0.060, P = 0.801). Data represent the mean of three independent experiments.
In a retrospective study, PERM values were also correlated with RT-qPCR data (11). An alternative fixative showed the highest PERM values (mean = 121.23), while neutral buffered formalin showed the lowest PERM values (mean = 32.02) among tested fixatives (11). RNA isolated from tissue fixed in the alternative fixative resulted in a mean Cq value of 34.46 for HPRT, whereas the Cq value was higher in samples generated from formalin-fixed tissues (mean Cq = 41.27). Notably, the PERM values of RNA isolated from samples treated with different fixatives were negatively correlated with the Cq values for the HPRT gene (r = −0.900, P < 0.001) (Figure 3). Altogether, these data suggest that the PERM is a robust tool for RNA quality measurement in fixed tissues.
Figure 3. Correlation between the paraffin-embedded RNA metric (PERM) and RT-qPCR data.
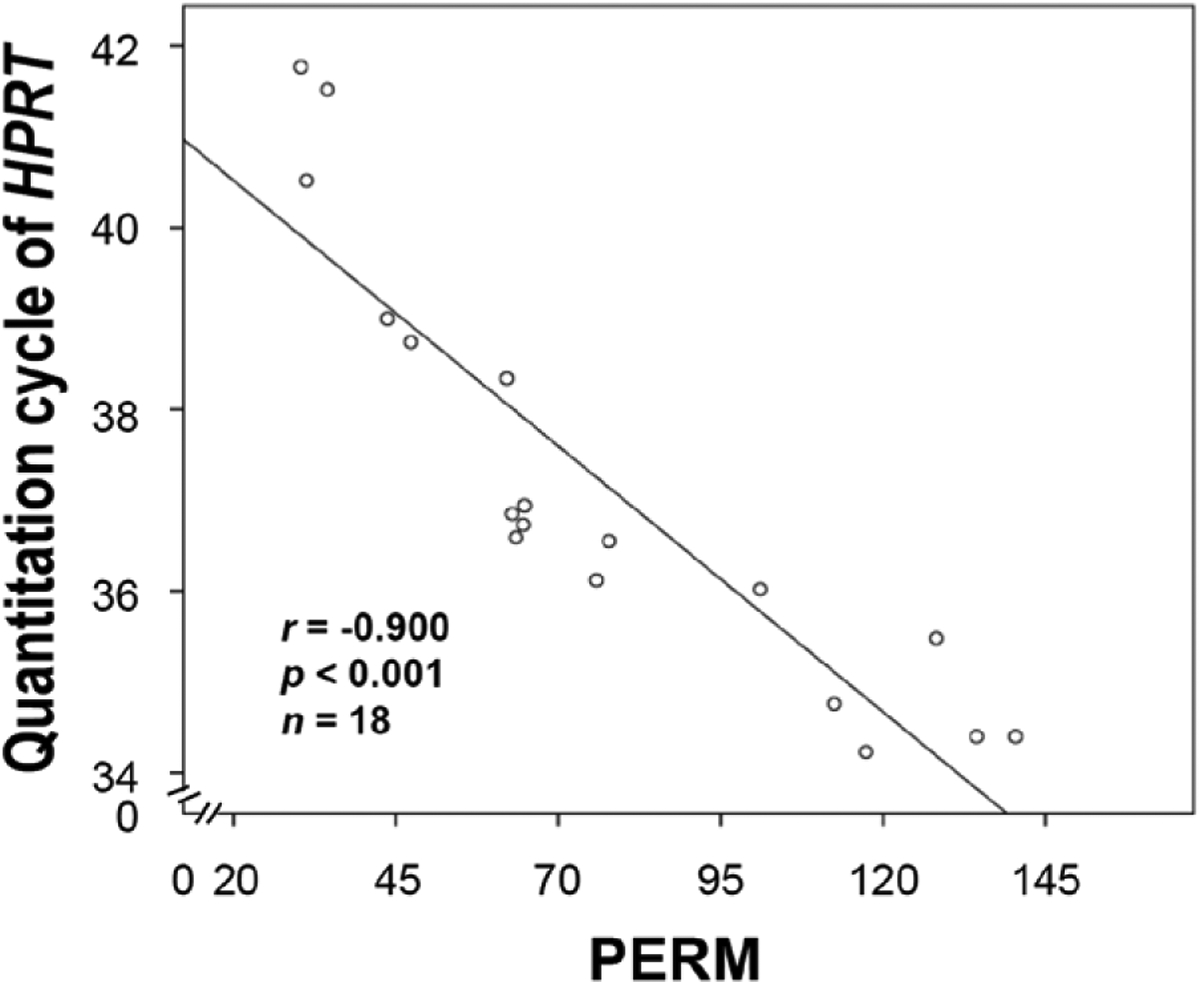
We applied the PERM to retrospective data for the evaluation of its value as an RNA quality control tool. The mean quantitation cycle (Cq) value of the housekeeping gene HPRT was determined in kidney tissue treated with different fixatives (n = 19). The scatter plot shows that the PERM number is negatively correlated with the Cq values for HPRT (r = −0.900, P < 0.001).
RNA obtained from FFPE tissue is subject to substantial degradation. Our previous studies suggest that degradation is a function of cellular hypoxia/anoxia during chemical fixation, as well as strand-breakage induced by the fixatives (5,6). We have utilized the PERM in as yet unpublished studies of alternative fixatives and demonstrated that it provides a simple and accurate measure of RNA quality when tested against PCR-based assays.
The quality of extracted RNA is a critical factor in downstream quantification assays such as microarray analysis and RT-qPCR (1,12). RNA integrity has conventionally been measured by the ratio of 28S to 18S rRNA, which is neither robust nor accurate. Furthermore, this methodology is unsuitable for highly degraded RNA from FFPE tissues. To overcome these problems, several publications have discussed RNA quality control (QC) tools, for example the RIN (2,3), RNA integrity score (RIS) (13), DV200 (2014. Expression Analysis of FFPE Samples. Illumina Technical Note Pub. No. 470-2013-002), RT-qPCR assay (14,15), and multiplex RT-qPCR (16). Although the PCR-based assays are the most reliable and sensitive assays among these RNA QC tools, challenges remain, such as the appropriate choice of target genes, primer design, optimization of PCR conditions, and long processing times. On the other hand, microcapillary electrophoresis-based methods have been recently developed and introduced as RNA QC tools. The RIN is an algorithm based on a combination of electrophoretic features including the total rRNA ratio, the height of the 18S peak, the fast area ratio, and the height of the lower marker. The RIN is a reliable RNA QC method for RNA prepared from fresh and frozen fresh tissue and used in gene expression experiments. However, prior studies have suggested that the RIN is not good predictor of the success of gene expression experiments, especially in cases where a small number of RIN values are calculated. In addition, Unger et al. recently demonstrated that the precision of the RIN as an RNA QC test depends on the biochemical or biophysical RNA degradation method used, such as heat, enzymatic, or UV light. Recently, RIS (13) and DV200 (Illumina Technical Note, Pub. No 470-2013-002) metrics were described in the latest versions of analysis software using QIAxcel Screen Gel software (Ver. 1.2.0) and Agilent 2100 Expert software (Ver. B. 02.07. S1532), respectively, for automated capillary electrophoresis of extracted RNA. The RIS uses a similar scoring system (ranging from 1 to 10) as the RIN, whereas DV200 calculates the percentage of RNA fragments >200 nucleotides in electrophoretic measurements. Although these latest RNA QC tools potentially improve the assessment of the integrity of RNA extracted from archival FFPE tissue, both systems require a special kit and sophisticated instrumentation.
Much like the RIN, the PERM was originally developed without comparison to gene-specific measures as an intermediate assay of RNA quality in investigations of tissue preservation. In our evaluation of the PERM, we demonstrated a correlation of PERM values with mRNA expression and RT-qPCR data. Notably, the PERM is applicable to FFPE tissue and indicates the suitability of isolated RNA for downstream applications. In practice, PERM values should be interpreted as PERM ± 2.5 units. We primarily applied the PERM to the Agilent 2100 Bioanalyzer; however, instruments from other vendors can be used, and simple densitometry of gel images would also be sufficient. Although initially used with the Bioanalyzer, the PERM can be used as an RNA quality metric for any method of quantifying RNA fragment length. One weakness of the PERM is that it requires laboratory validation to correlate the efficiency and performance of RNA extraction protocols with downstream assays. However, a laboratory can validate the PERM for its specific protocols and then use it as a robust method of qualifying the integrity of RNA prepared from FFPE tissues.
Acknowledgments
This study was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The authors would like to thank Kris Ylaya and Candice Perry for excellent technical assistance. This paper is subject to the NIH Public Access Policy.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, and Kornacker K. 2003. Chipping away at the chip bias: RNA degradation in microarray analysis. Nat. Genet 35:292–293. [DOI] [PubMed] [Google Scholar]
- 2.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, et al. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, and Auffray C. 2005. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 33:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hewitt SM, Lewis FA, Cao Y, Conrad RC, Cronin M, Danenberg KD, Goralski TJ, Langmore JP, et al. 2008. Tissue handling and specimen preparation in surgical pathology: issues concerning the recovery of nucleic acids from formalin-fixed, paraffin-embedded tissue. Arch. Pathol. Lab. Med 132:1929–1935. [DOI] [PubMed] [Google Scholar]
- 5.Chung JY, Braunschweig T, Williams R, Guerrero N, Hoffmann KM, Kwon M, Song YK, Libutti SK, and Hewitt SM. 2008. Factors in tissue handling and processing that impact RNA obtained from formalin-fixed, paraffin-embedded tissue. J Histochem Cytochem. 56:1033–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung JY, Braunschweig T, and Hewitt SM. 2006. Optimization of recovery of RNA from formalin-fixed, paraffin-embedded tissue. Diagn Mol Pathol. 15:229–236. [DOI] [PubMed] [Google Scholar]
- 7.Chung JY and Hewitt SM. 2010. An optimized RNA extraction method from archival formalin-fixed paraffin-embedded tissue. Methods Mol. Biol 611:19–27. [DOI] [PubMed] [Google Scholar]
- 8.Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin JJ, Sheridan P, Urdea M, et al. 1996. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol 34:3196–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, et al. 2006. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol 24:1115–1122. [DOI] [PubMed] [Google Scholar]
- 10.Knudsen BS, Allen AN, McLerran DF, Vessella RL, Karademos J, Davies JE, Maqsodi B, McMaster GK, and Kristal AR. 2008. Evaluation of the branched-chain DNA assay for measurement of RNA in formalin-fixed tissues. The Journal of molecular diagnostics. J Mol Diagn. 10:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perry C, Chung JY, Ylaya K, Choi CH, Simpson A, Matsumoto KT, Smith WA, and Hewitt SM. A buffered alcohol-based fixative for histomorphologic and molecular applications. J Histochem Cytochem. (In press.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleige S, Walf V, Huch S, Prgomet C, Sehm J, and Pfaffl MW. 2006. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol. Lett 28:1601–1613. [DOI] [PubMed] [Google Scholar]
- 13.Unger C, Kofanova O, Sokolowska K, Lehmann D, and Betsou F. 2015. Ultraviolet C radiation influences the robustness of RNA integrity measurement. Electrophoresis 36:2072–2081. [DOI] [PubMed] [Google Scholar]
- 14.Nolan T, Hands RE, and Bustin SA. 2006. Quantification of mRNA using real-time RT-PCR. Nat. Protoc 1:1559–1582. [DOI] [PubMed] [Google Scholar]
- 15.Penland SK, Keku TO, Torrice C, He X, Krishnamurthy J, Hoadley KA, Woosley JT, Thomas NE, et al. 2007. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab. Invest 87:383–391. [DOI] [PubMed] [Google Scholar]
- 16.Takano EA, Mikeska T, Dobrovic A, Byrne DJ, and Fox SB. 2010. A multiplex endpoint RT-PCR assay for quality assessment of RNA extracted from formalin-fixed paraffin-embedded tissues. BMC Biotechnol. 10:89. [DOI] [PMC free article] [PubMed] [Google Scholar]


