Abstract
Bacterial chemoreceptors, the histidine kinase CheA, and the coupling protein CheW form transmembrane molecular arrays with remarkable sensing properties. The receptors inhibit or stimulate CheA kinase activity depending on the presence of attractants or repellants, respectively. We engineered chemoreceptor cytoplasmic regions to assume a trimer-of-receptor-dimers configuration that formed well-defined complexes with CheA and CheW and promoted a CheA kinase-off state. These mimics of core signaling units were assembled to homogeneity and investigated by site-directed spin-labeling with pulse-dipolar electron-spin resonance spectroscopy (PDS), small-angle x-ray scattering, targeted protein cross-linking, and cryo-electron microscopy. The kinase-off state was especially stable, had relatively low domain mobility, and associated the histidine substrate and docking domains with the kinase core, thus preventing catalytic activity. Together, the data provide an experimentally restrained model for the inhibited state of the core signaling unit and suggest that chemoreceptors indirectly sequester the kinase and substrate domains to limit histidine autophosphorylation.
INTRODUCTION
Many bacteria employ a complex transmembrane sensory apparatus to modulate their motility in response to the chemical environment (reviewed in (1-4)). Extensively studied in Escherichia coli (Ec), bacterial chemotaxis relies on transmembrane chemoreceptors to link extracellular signals to intracellular phosphorylation events (1-4). The receptors, also known as methyl-accepting chemotaxis proteins (MCPs), organize into a trimer-of-dimers (TOD) that further assemble with the cytoplasmic dimeric histidine kinase CheA and coupling protein CheW through their membrane-distal tips to produce an extended molecular lattice (Fig. 1A) capable of highly sensitive, cooperative responses (5-7). MCPs regulate CheA autophoshorylation rates in response to binding signals from external ligands. Phosphoryl transfer from autophosphorylated (activated) CheA to CheY activates CheY to switch the direction of flagellar rotation. MCPs also undergo methylation and demethylation on specific glutamate residues by the methyl transferase CheR and the methyl esterase CheB, respectively. In E. coli, MCP methylation activates CheA and thereby opposes the action of attractant binding, which inhibits the kinase. Despite considerable advances, it remains unclear how the ligand and methylation status of receptors regulates CheA autophosphorylation activity (1-4).
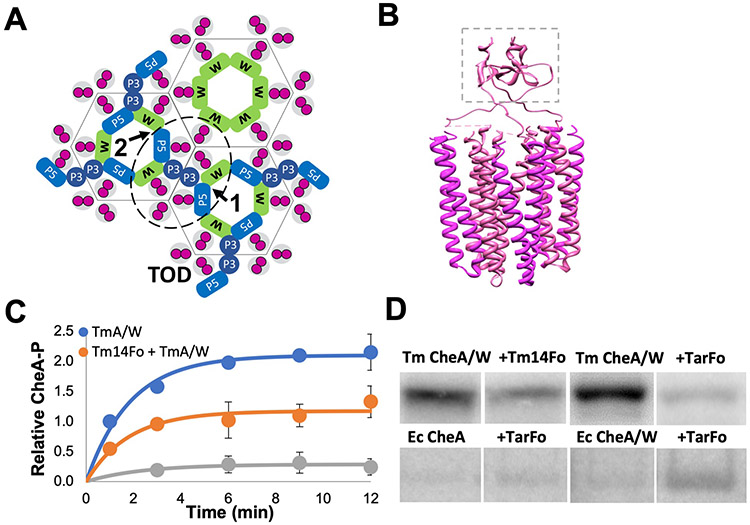
Fig. 1. Tar and Tm14 foldons mimic receptor trimer-of-dimers.
(A) Topography of a chemoreceptor array with a core signaling unit outlined (dotted line) as viewed from the intracellular side of the membrane. CheA (blue) and CheW (green) associate with chemoreceptor dimers (magenta) and form six-membered rings that are linked together in the repeating units of the array (grey outlines around receptor domains). CheA domain P5 (P5) and CheW (W) associate through interface 1 and interface 2 (arrows). Array symmetry is denoted by hexagons. CheW-only rings reside on the 6-fold symmetry axis of the arrays. CheA domain P3 (P3) dimerizes between two receptor trimers-of-dimers (TODs, magenta). (B) Side view of a model of a foldon TOD mimic trimerized by a fibritin motif (dashed box). (C) Quantification of T. maritima (Tm) CheA autokinase activity in the presence of CheW only (TmA/W) or CheW plus either the Tm14 foldon (Tm14Fo) or the E. coli Tar foldon (TarFo). Curves were fit to a = a0(1-exp(kt)), with a0*k representing the relative initial rates. Brackets represent 95% confidence values from global fits to three independent measurements at each time point. TmA/W: a0 = 2.1 [A-P] (1.9, 2.3), k = 0.5 sec−1 (0.3, 0.8), a0*k = 1.1 [A-P]sec−1; TmA/W +Tm14Fo: a0 = 1.1 [A-P] (0.9,1.4), k = 0.5 sec−1 (0.3,1.0), a0*k = 0.6 [A-P]sec−1; TmA/W + TarFo: a0 = 0.2 [A-P] (0.1,0.4), k = 0.4 sec−1 (0.0,1.3), a0*k = 0.1 [A-P]sec−1. (D) Autoradiography of T. maritima (Tm) CheA and E. coli CheA autophosphorylation in the presence of CheW and Tm14Fo or TarFo. 12 min assay for Tm CheA, 15 sec. for E. coli CheA, N=3 independent experiments. Samples were run in non-adjacent lanes on the same or parallel-processed gels that were imaged together. Complete autoradiograms are shown in fig. S4.
CheA is a complex dimeric kinase composed of five domains (P1 - P5), each with a distinct function. P1 contains the phosphate-accepting histidine residue, P2 docks CheY, P3 mediates CheA dimerization, P4 is an ATPase domain, and P5 interacts with CheW to form the ring structure that is integral to the array architecture. The CheA P1 domain contains the substrate histidine residue [His48 in Ec CheA and His45 in Thermotoga maritima (Tm) CheA] and consists of 5 αhelices (A-E), with A-D composing a four-helix bundle connected to helix E by a flexible linker (8, 9). The CheA domains are quite flexible in the free kinase, with the P1 and P2 modules sampling considerable conformational space (10-12). However, incorporation of CheA into the receptor arrays constrains and controls its conformation in ways that are not well understood (13-16).
Biophysical, biochemical, and genetic approaches have combined to define the architecture and sensing behavior of the arrays (reviewed in (1-3)). CheA sits at the interface of two TODs, with the helical P3 domain projecting between them toward the membrane (Fig. 1A). Paralogous P5 and CheW interact through conserved surfaces at the ends of their β-barrels to form rings composed from two types of interfaces: interface 1, which is proximal to P3 and involves P5 subdomain 1 and CheW subdomain 2; and interface 2, which is distal from P3 and involves P5 subdomain 2 and CheW subdomain 1 (Fig. 1A). Dimeric MCPs interact in grooves at the center of CheW and P5 on the external surfaces of the rings (Fig. 1A). CheW-only rings may form on the 6-fold symmetry axis of the hexagonal lattice (16). The P4 kinase domains suspend below the receptor tips and align underneath the P5 domains. Long, flexible linkers connect P1 and P2 to each other (L1) and P2 to P3 (L2), whereas linkers between P3 and P4 (L3), and P4 and P5 (L4), play an important role in kinase activity and regulation (17-19). Positions of the P1 and P2 domains are not formally known, although cryo-electron tomography (cryo-ET) suggests that they reside below the P4 domains of deactivated kinase (15, 20). However, the P1 and P2 positions are uncertain owing to limiting resolution in the cryo-ET reconstructions and/or domain mobility. The P4 domains are also not fully discernible in the tomograms of either in vivo or reconstituted arrays, but appear more ordered when the kinase is inactive (13, 15, 16, 20, 21). MD-assisted modeling against 12 Å resolution cryo-ET data has better defined two predominant conformations of the P4 domain below the P5-CheW layer and suggests that kinase regulation may involve transitions between these conformations (14).
The ability of receptors to alter Ec CheA phosphotransfer rates by several hundred fold suggests that the kinase assumes substantially different structural and dynamical states associated with these activity changes (1-4). Activity assays with P1 supplied as a separated substrate to the core kinase (domains P3, P4, and P5, referred to here as P3P4P5) in complex with MCPs indicate that activation primarily derives from increases in kcat and not changes to the P1 Michaelis constant (KM) (22, 23). Furthermore, both ATP-binding and autophosphorylation (which includes both P1 binding and phosphate transfer) are steps potentially regulated by the receptors (23). Removal of one of the two P1 domains from a CheA dimer increases autophosphorylation activity, suggesting that the P1 domains may interfere with each other (24, 25). Molecular dynamic simulations of array models based on cryo-ET data indicate that the P4 domains “dip” toward each other from a position where the P4 ATP-binding pocket and flexible ATP “lid” reside near the P5-CheW layer (14, 16). It follows that changes in kinase activity relate to P4 location or motion.
Despite the extensive coupling of components within the natural arrays, a core signaling unit comprised of two TODs, one dimeric CheA, and two CheW proteins shows considerable functionality in both nanodisc reconstitutions and cells (7, 26-28). In efforts to reconstitute a homogenous signaling particle of these components we previously developed a receptor engineering approach, wherein single chain versions of cytoplasmic receptor signaling domain “dimers” were trimerized by the foldon domain from bacteriophage T4 fibritin (12). These so-called receptor foldons produce trimeric species that are capable of modulating CheA activity in vitro (12). Here, we extended the foldon approach to reconstitute core signaling units in a highly inhibited state and characterized this species by a variety of biophysical methods, including small-angle X-ray scattering (SAXS), interface analysis by cross-linking and mass-spectrometry, and pulse-dipolar ESR spectroscopy (PDS). The results showed that inhibited CheA exhibits increased interactions among the P1, P2, and P4 domains and relatively well-ordered L1 and L2 connecting loops. Furthermore, we provide direct evidence for the P4 “dipped conformation” observed in MD simulations (16) and compose models of the foldon-associated signaling particles that capture this property.
RESULTS
TOD receptor foldons produce functional ternary complexes
Single-chain receptors that fuse the dimer subunits reduce the complexity and symmetry of the ternary complex (11, 12, 29). Soluble receptor fragments containing a non-native trimerization motif pre-form a stable TOD arrangement in the absence of the membrane and associate with CheA and CheW (12). The chimeras consist of a fused dyad of receptor cytoplasmic domains (each 71 residues) from either the Tm MCP Tm14 or Ec MCP Tar linked at the C-terminus to a “fold-on” trimerization motif from the T4 phage protein fibritin (Fig. 1B, fig. S1A). The receptor segment includes the ~36 residue highly conserved protein interaction region (PIR) that interacts with CheW and P5 of CheA and mediates the TOD trimer contact. Despite deriving from divergent bacteria, the Tm14 and Tar foldons share 44% identity and 67% sequence similarity over their 71 residue sequences, including 72% identity and 86% similarity over the 36 residue PIR that binds to CheA and CheW. Previous SAXS data and spin-labeling studies with the Tar foldon revealed a compact, globular shape with associated tips (12). Multi-angle light scattering (MALS) experiments with the purified foldons confirmed trimerization in solution (fig. S1B and D) and stable interactions with CheA and CheW (Fig. S1C and E).
MALS was also used to find the optimal buffer conditions for producing soluble, homogenous ternary complexes. Complexes composed of either the Tm14 or Tar foldons were isolated by size-exclusion chromatography (SEC) after incubating equimolar amounts of the receptor mimetics with Tm CheA and CheW in various buffer conditions. Ternary complex yield increased with buffers of high KCl content (250 mM) and low pH (6.5) (fig. S2). In the case of the Tm14 foldon, subsequent MALS analyses of two resulting high molecular-weight SEC peaks revealed particles of masses 220 kDa and 440 kDa, respectively (fig. S3, A and B). Furthermore, these two species differed in their CheA activity, with the larger complex showing more inhibition (fig. S3C). Given the extended nature of the native of arrays, it is unsurprising that the Tar foldon complexes produced several different assembly states; nonetheless, they could be separated based on hydrodynamic properties (fig. S2, A and B).
Isolated ternary complexes were analyzed by size-exclusion chromatography coupled to SAXS (SEC-SAXS) to determine sample size, homogeneity, particle flexibility, and spatial extent. Complexes formed from receptor foldons, CheW, and full-length CheA or CheA P3P4P5 represented well-defined particles of expected sizes (table S1, fig. S3D). SAXS data collected across each respective SEC peak indicated separation of the reconstituted species into relatively homogenous complexes of different sizes (table S1). In general, the complex size decreased with P3P4P5 compared to full-length CheA, and particle size increased when CheW and the receptor foldons were included in the complexes.
Both the Tar Foldon and Tm14 Foldon were tested for their ability to modulate Tm CheA autophosphorylation activity by radioisotope incorporation from γ−32P-ATP (Fig. 1, C and D, fig. S4). Previous work indicates that Tar receptor foldons activate Ec CheA autophosphorylation (24); such behavior was recapitulated here and shown to be CheW-dependent (Fig. 1D, fig. S4A). In contrast, complexes formed by the Tar and Tm14 foldons with Tm CheA and CheW deactivated the kinase 20-fold and 2.5-fold, respectively (Fig. 1C, fig. S4B). At the level of assay detection, the Tar foldon turned off Tm CheA activity nearly completely. Given this ability of the Tar foldon to strongly inhibit the Tm kinase, Tar foldon complexes with Tm CheA and Tm CheW (produced at pH 6.5 and 150 mM KCl, fig. S2) were isolated for further analysis. To better evaluate the degree to which these particles mimicked native assembly states of core signaling particles, we evaluated the consequences of residue substitutions in the PIRs that are known to alter the functional properties of the native complex.
Single residue changes in receptor PIRs affected trimer stability and CheA responses. For example, Ec Tsr R388F and R388W (Tar R386F and R386W) and Tsr E391A (Tar E389A) alter cell tumbling bias and trimer stability (30). Tar foldon R386F was significantly more stable than the parental Tar foldon (fig. S5). This variant could be purified from cell extracts in much higher amounts than the wild-type Tar foldon (20 mg/L of cell culture vs. 6 mg/L), and the purified protein reached higher concentrations (~7 mg/ml compared to ~2 mg/ml; fig. S5). However, MALS and chemical crosslinking with the lysine-specific chemical crosslinker disuccinimidyl sulfoxide (DSSO) showed that the R386F variant neither trimerized nor produced stable complexes with CheA and CheW (fig. S5). Furthermore, autophosphorylation assays demonstrated that the R386F variant only exhibited ~1.5-fold deactivation of Tm CheA. Similarly, the R386W variant did not trimerize, failed to complex with CheA and CheW, and did not affect CheA autophosphorylation (fig. S5). The Tar E389A foldon was unstable and only present in high-molecular weight aggregates after purification on SEC. Mutagenesis studies and molecular dynamics (MD) simulations implicate the equivalent of Tm14 residue F395 in receptor on-off switching (31). The Tm14 F395W foldon produced similar complexes with Tm CheA as the wild-type Tm14 foldon, but did not alter CheA kinase activity (fig. S5). Thus, single residue substitutions in the receptor tips known to affect CheA activity in vivo also altered the integrity of the foldon complexes and showed perturbed effects on kinase activity, ruling against a largely non-native assembly of the particles.
Structural characterization by SAXS, cryo-EM, and interface mapping indicate a compact inhibited ternary complex
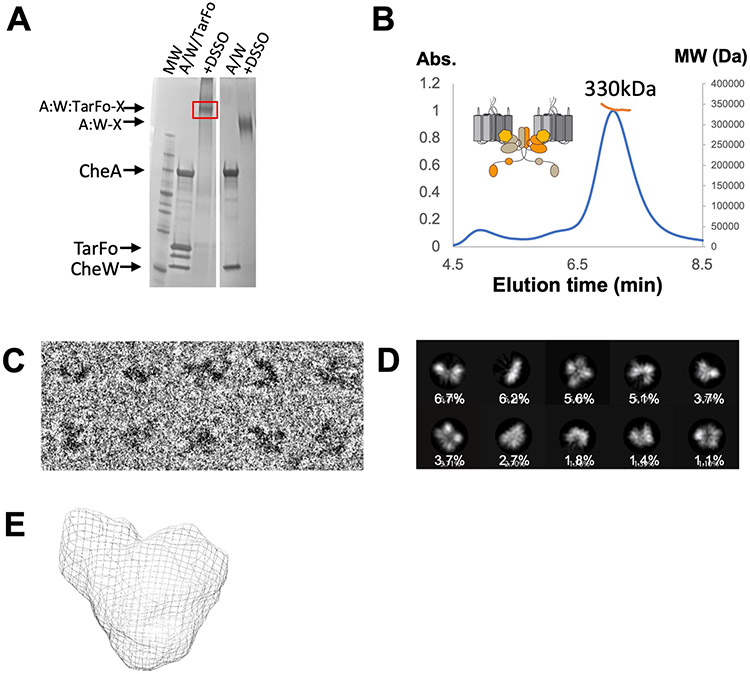
We further stabilized the deactivated ternary complex of the Tar Foldon, Tm CheA, and Tm CheW by DSSO cross-linking prior to SEC purification (Fig. 2A). The ~320 kDa MW of the complex (by MALS) agreed with the composition of a core signaling unit (2 Tar foldons, 2 CheA subunits, and 2 CheW subunits; Fig. 2B). SEC-SAXS of the cross-linked particles produced a MW similar to that found by MALS (MW (VC) = 305 kDa; MW (VP) = 328 kDa; table S1, fig. S6A). Some contribution from a higher MW species (~ 440 kDa) was evident in the SEC-SAXS data when analyzed by singular value decomposition with evolving factor analysis (32); however, the larger species component could be separated in the analysis. The dimensionless Kratky plot (fig. S6B) reflected a spherical particle with little indication of the flexibility found in the free CheA kinase (24).
Fig. 2. Isolation and analyses of foldon ternary complexes cross-linked with DSSO.
(A) SDS-PAGE Tm CheA (A) and Tm CheW (W) before and after addition of DSSO in the presence and absence of the Tar foldon (TarFo). Data are representative of N > 3 independent experiments. (B) SEC-MALS of the cross-linked TarFo-CheA-CheW complex, MW ~330 kDa +/− 15 kDa, which corresponds to a core signaling unit (schematic) containing two receptor foldons, one CheA dimer, and two CheW subunits (320 kDa). Data are representative of N > 3 indpendent experiments. (C) Cryo-EM of the cross-linked TarFo-CheA-CheW complexes. Enlarged views of single complexes were cropped from the micrograph in different orientations corresponding to the class averages. N=2 independent experiments. (D) Selected 2D class averages and their percentages over the total extracted particles used for alignment and classification (Box size = 268 Å, mask diameter = 140 Å). (E) The molecular envelope of the isolated cross-linked ternary complex generated by SEC-SAXS analyses. Data are representative of N=2 indepenent experiments.
Compositional and conformational homogeneity of the isolated cross-linked complexes was assessed by transmission cryo-electron microscopy (cryo-EM; Fig. 2C). When unlinked ternary complexes of the Tar foldon with Tm CheA and CheW were analyzed, attempts at two-dimensional (2D) classification of the particles were unsuccessful because the particles disassociated on grids and were conformationally heterogeneous (fig. S3E). However, micrographs of the species purified after DSSO cross-linking revealed monodispersed particles, and 2D classification generated averages that fit the expected size and shape of the ternary complexes (Fig. 2C). Indeed, the 2D classes (Fig. 2D) resembled the molecular envelopes constructed from SEC-SAXS experiments in size and general shape (Fig. 2E). An insufficient number of suitable particles for analyses currently limits 3D reconstruction of the 2D classes.
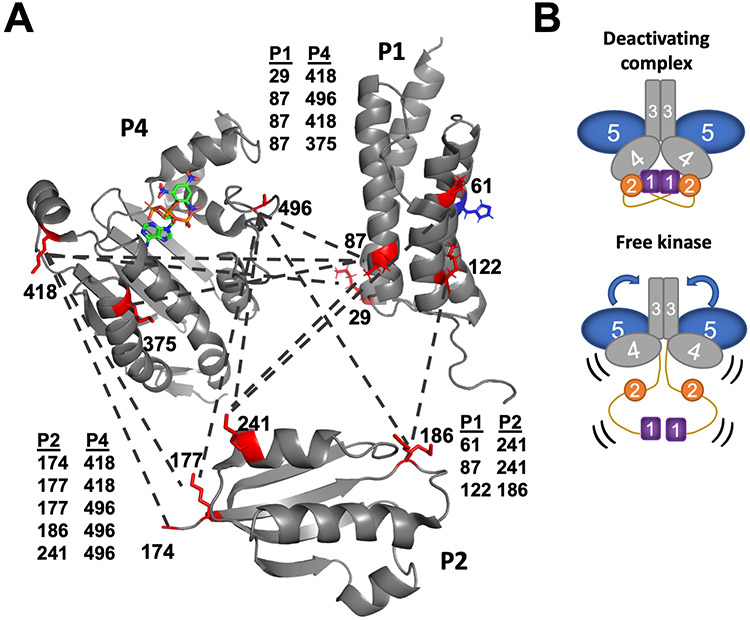
Domain proximity and component interfaces in free Tm CheA and in the foldon complexes were analyzed by DSSO crosslinking followed by mass spectrometry. DSSO is a cleavable chemical cross-linker that covalently binds two lysine residues within roughly 20 Å of each other (33). Tandem proteolysis of the cross-linked samples with trypsin and chymotrypsin followed by sequential fragmentation with mass spectrometry (MS) identified the peptides linked by DSSO. MS analysis of DSSO-treated Tm CheA in the absence and presence of CheW and the Tar foldon revealed key structural changes upon kinase inhibition by the Tar foldon. Among the many cross-links identified (fig. S7 and fig. S8), the most striking difference between the two samples was that inter-domain cross-links between P1 and P2, P1 and P4, and P2 and P4 were only detected when CheA associated with the deactivating receptor (fig. S7 and fig. S8). Indeed, the free kinase only produced cross-links within P1 or P2 (fig. S7), indicating that these domains are isolated from the rest of the protein. In contrast, the binding of receptor and CheW primarily to P5 associates P1, P2, and P4 to enhance their encounter frequency (Fig. 3A and fig. S8). The P4 sites that cross-linked to P1 and P2 mostly involved residues near the ATP-lid and ATP binding pocket. Furthermore, the cross-linking sites positioned the His45 substrate residue on P1 away from the P4 ATP-binding site. Instead, the lysine residue nearest His45 preferentially cross-linked to the P2 domain rather than the P4 domain (Fig. 3A).
Fig. 3. Component interactions in the foldon complexes revealed by DSSO cross-linking followed by mass spectrometry.
(A) Reconstitution of CheA with the deactivating receptor and CheW promotes interactions among P1 (PDB code: 2LD6), P2 (1U0S), and P4 (1i5D) domains as revealed by cross-linked (dashed lines) lysine residues (red, numbered). Residues near helix D on P1, which is distal from the His45 substrate (blue), cross-link to sites near the P4 ATP-binding site. P1 sites near His45 cross-link to residues on the P2 CheY-binding interface. (B) In the free kinase, no inter-domain cross-links were identified for P1 or P2. However, additional cross-links were found between P3 and P5 and between P4 and P5 in the absence of the deactivating foldon. In the free kinase, the P4 domains underwent increased inter-domain cross-linking compared to the complexes.
Whereas many P1:P2:P4 cross-links were unique to the ternary complex, the CheA-only sample produced contacts between the P3 and P5 domains and the P4 and P5 domains not found in the foldon complex. Specifically, two lysine residues at the tip of the P3 domain cross-linked to P5 lysine residues that were more than 30 Å away in the crystal structure of CheA P3P4P5 (PDB code: 4XIV). In the ternary complex, the same P3 residues instead cross-linked to receptor residues predicted from array structures to be reasonably close (13, 34). The only P5 residue directed at P4 (Lys648), cross-linked to P4 in both species, but P5 Lys621, which is oriented away from the interface with P4, cross-linked with P4 Lys473 only in free CheA. Hence, the P5 domain is more flexible in the free kinase than in the foldon complex (Fig. 3B).
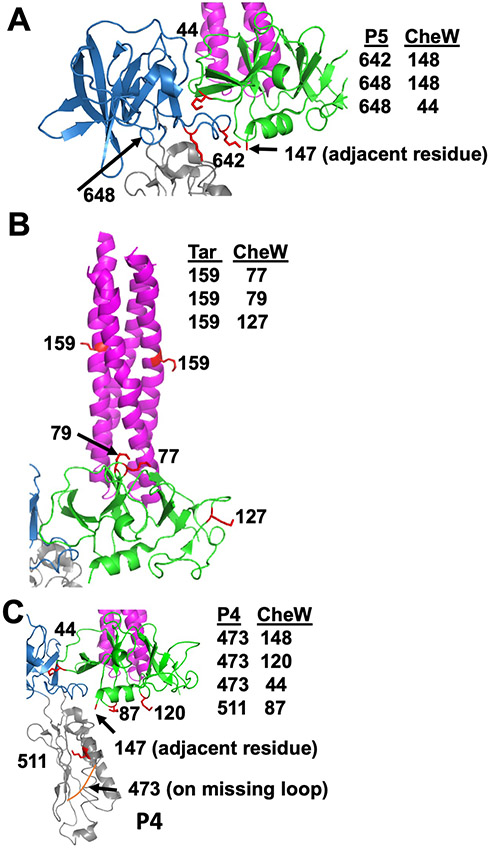
Inter-protein cross-links identified in the ternary complex were supported by crystal structures of complexes containing CheA P4P5, receptor fragments, and CheW (PDB ID: 3UR1) (Fig. 4, A to C). CheA P5 and CheW strictly cross-linked at binding interface 1, and residues on the upper portion of CheW cross-linked to the nearest lysine residue on the Tar foldon (Fig. 4, A and B). Only lysine residues on the bottom-directed face of CheW cross-linked to P4 residues as expected, but these sites were predicted to be more than 30 Å away based on array models (Fig. 4C) (13, 16). Thus, although the P4 domains juxtaposed the predicted face of CheW, they retained substantial flexibility in the foldon complexes. Nevertheless, taken together, the cross-linking data supports the conclusion that the foldon complexes have structural characteristics expected of core signaling units.
Fig. 4. Cross-links identified in the ternary complex among Tm CheA, CheW, and then Tar foldon.
Lysine residues involved in cross-links are shown with respect to the crystal structure (PDB code: 3UR1), which consists of CheA P4P5, CheW, and the receptor protein interaction region. (A) The CheA P5 domain (blue) interacts with CheW (green) at lysine residues adjacent to interface 1. (B) Cross-links identified between CheW and the Tar foldon (magenta) indicate that the ‘top’ portion of CheW orients toward the Tar lysine residue that is nearest the receptor tip. (C) Cross-links between the CheA P4 residues near the ATP binding pocket and at the ‘bottom’ portion of CheW indicate that P4 encounters CheW.
PDS provides evidence for P1 dimerization and P4 dipping within the foldon complexes
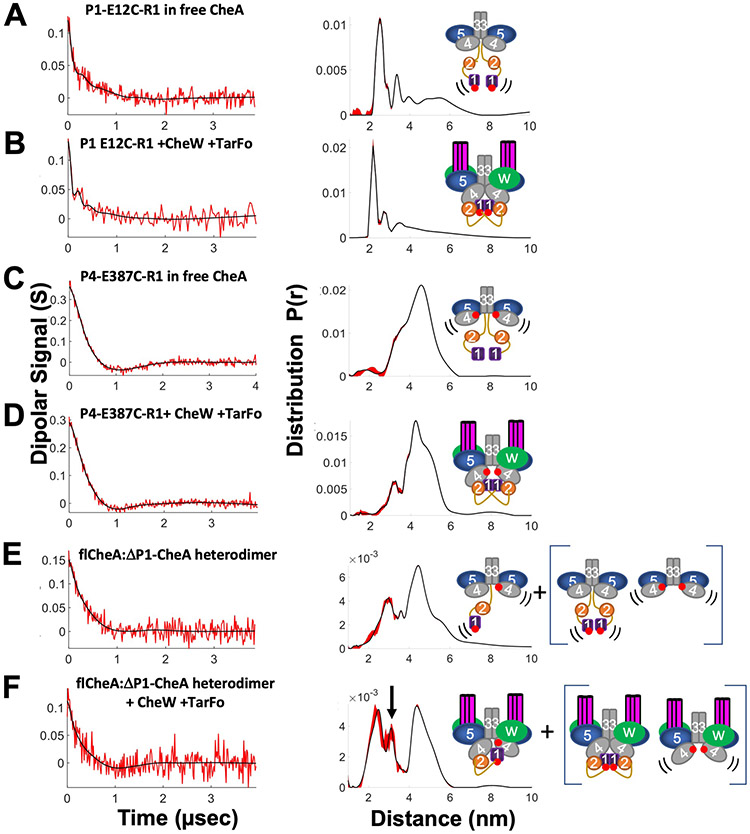
Double electron-electron resonance (DEER) spectroscopy of nitroxide-labeled CheA and nitroxide-labeled ADP was conducted to characterize CheA domain positioning within the Tar foldon complexes. Nitroxide spin-labels (also known as “R1”) were introduced at residues 12 (P1), 387 (P4), 588 (P5) by cysteine targeting of 1-oxyl-2,2,5,5-tetramethylpyrroline-3- methyl) methanethiosulfonate spin-label (MTSSL) and at the ADP-binding site by use of a previously reported spin-labeled ADP derivative, ADP-NO (35). Each labeling site per subunit generated two R1 moieties in the CheA dimer, between which distance distributions were determined.
In the free kinase, E12C-R1 CheA produced a wide range of distances including a short (~28 Å) distance not unlike those observed for previous P1 sites when CheA binds to single-chain “dimeric” receptor fragments (Fig. 5A) (11). The short distance became somewhat more prominent upon incorporation of E12C-CheA into the foldon complex with CheW and the larger, more distributed separations became less evident (Fig. 5B). PDS of other P1 sites (E76C-R1, E92C-R1 (11)) also produced distance distributions with maxima in the range of 30–50 Å and, taken together, these close separations indicate that the P1 domains associated with one another.
Fig. 5. PDS reveals conformational changes in CheA when reconstituted into foldon complexes.
All samples were reconstituted with ADP. Schematics (right) denote label positions (red dots represent the R1 nitroxide) and where PDS measurements reflect heightened domain mobility (black lines). Base-line corrected time domain data (left) before (red) and after (black) wavelet denoising and resulting distance distributions (right, black) with error bounds (red) for (A) P1-E12C-R1 in free CheA; (B) P1 E12C-R1 +CheW +Tar foldon (TarFo); (C) P4-E387C-R1 in free CheA; (D) P4-E387C-R1+ CheW +TarFo; (E) flCheA:ΔP1P2-CheA heterodimer; (F) flCheA:ΔP1P2-CheA heterodimer + CheW +TarFo. Arrow indicates new separation arising from presumed P4-to-P1 interaction. Heterodimer samples in (E) and (F) also contained contributions from the homodimeric species shown in brackets.
The P4 site, CheA E387C-R1, placed a spin label at the end of the first P4 β-strand, near the L3 linker and projecting toward the CheA dimer interface. As with previous PDS studies of CheA, the broad inter-subunit distance distribution centered at ~47 Å (Fig. 5C) (11, 36). In the foldon complex, the distance sharpened, and a component with a mean separation ~ 5 Å shorter than that of the free kinase predominated (Fig. 5D). To remove contributions from the 387-R1-to-387-R1 interaction and accentuate P1-to-P4 inter-subunit contacts, heterodimers were produced between one subunit of full-length E12C-R1 CheA and one subunit of CheA containing only P3P4P5 E387C-R1 (36). Heated incubation of the (flCheA-E12C-R1)2 and (P3P4P5-E387C-R1)2 dimers encourages subunit swapping; subsquent low-temperature trapping followed by SEC purification enriches for flCheA-E12C-R1:P3P4P5-E387C-R1 heterodimers (36). Removal of one P1 domain from the dimer increases CheA autophosphorylation, and thus may encourage more P1-P4 contacts (24). In the free kinase (Fig. 5E), the distance distribution primary showed the ~47 Å separation typical of the P4-P4 signal from remaining P3P4P5 homodimer and a shorter ~25 Å separation from full-length CheA dimers also not fully excluded by the SEC purification. There was also some hint of an intermediate distance at ~36 Å that intensified in the foldon complex (Fig. 5F). This 36 Å signal presumably derived from the interaction between 12-R1 and 387-R1. A 36 Å separation between P1 and P4 agrees well with previous interaction studies (10, 37). Previous models of P1 docked in a productive conformation with the P4 active site (24) (38) or in a non-productive inhibitory complex with P4 (39) could both produce such a distance.
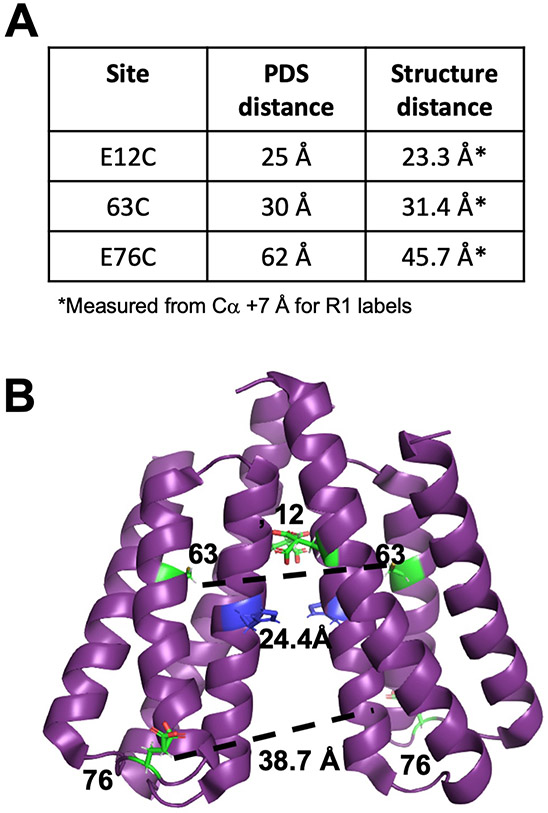
The short distances observed between symmetric spin labels on P1 in a full-length CheA dimer suggested that the P1 domains themselves may dimerize, especially in the inhibited state (Fig. 5B). Consistent with such an interaction, the crystal structure of the isolated P1 domain (1TQG) from T. maritima formed a parallel dimer that is generally compatible with these distance constraints (Fig. 6, A and B).
Fig. 6. PDS identifies a P1 homodimer in the foldon complex.
(A) Summary of distance restraints from DEER experiments on CheA spin-labeled on P1 (this study and (11)). Distances were measured from Cα positions + 7 Å for R1 labels. (B) Structure of the parallel P1 crystallographic dimer (1TQG). Distances are shown between Cα positions. E76 is located on a flexible loop furthest from the dimer interface. The substrate His45 (blue) resides at the edge of the dimer interface.
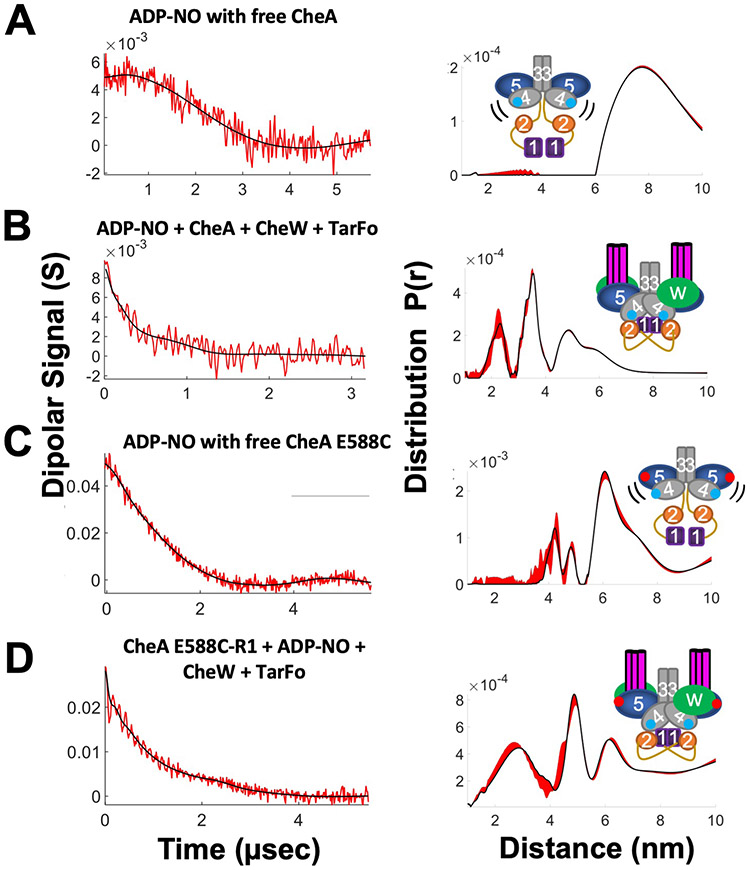
A spin-labeled ADP derivative (ADP-NO) that binds to CheA (35) was used to track the P4 domain by DEER. Treated with ADP-NO, the free kinase gave only very weak dipolar signals, indicative of long, broad distributions (Fig. 7A). In contrast, the foldon complex produced distances at ~22 Å and ~35 Å, in addition to some larger (~50 Å) separations (Fig. 7B). The low modulation depth of the time-domain data may derive from the fact that ATP binds Tm CheA with substantial negative cooperativity (40). Thus, even at mM concentrations, the ADP-NO is likely not bound to both subunits for a substantial fraction of the sample. Nonetheless, the short distances indicated that the Tar foldon caused the P4 domains to move together from their positions in free CheA. The bimodal distribution indicated that either the P4 domains or the nitroxide moieties themselves exhibited several related conformations.
Fig. 7. Spin-labeled ADP reports on the CheA ATP-binding pocket.
Schematics (right) denote label positions in free CheA or the foldon complexes (red dots represent R1 nitroxide, blue dots represent ADP-NO). Base-line corrected time domain data (left) before (red) and after (black) wavelet denoising and resulting distance distributions (right, black) with error bounds (red) for (A) ADP-NO with free CheA.; (B) ADP-NO, Tm CheA, CheW, and Tar foldon (TarFo); (C) ADP-NO with free CheA E588C; (D) CheA E588C-R1 with ADP-NO, CheW, and TarFo.
To further bound the positions of the ATP-binding site, CheA was labeled on the P5 domain at position 588 of subdomain 2. Free CheA E588C-R1 gave nearly no dipolar signals, in keeping with the predicted long (> 80 Å) separation between these sites. However, in the foldon complex, a very short distance component arose from E588C-R1 (fig. S9). Similar short distance components on the spin-labeled P5 domain are known to derive from CheA dimer association through symmetric subdomain 2 contacts (41). Such contacts mimic those of interface 2 in crystal structures and arrays but, in this case, were likely favored by the high-glycerol and low-temperature conditions of the DEER experiments. Binding the Tar foldon and CheW further increased these inter-dimer contacts relative to the free kinase (fig. S9), probably because the receptors aligned the P5 domains in planes compatible with symmetric binding at the P5 domain ends. The addition of ADP-NO to free CheA E588C-R1 produced some shorter distances at ~45 Å, but mainly large distances (> 60 Å) that represent the P5-to-P4 separation were evident (Fig. 7C). In contrast, the Tar foldon complex with E588C-R1 and ADP-NO (Fig. 7D) produced a mid-range distance at ~50 Å and a short distance at ~20–25 Å that matched overlapping contributions from the inter-dimer contact (fig. S9) and the ADP-NO contributions seen in absence of E588C-R1 (Fig. 7B). The 50 Å distance, which intensified relative to the long-distance component seen with the free kinase (Fig. 7C), likely reflects an ordering and contraction between P5 subdomain 2 and the ATP-binding pocket of P4 (Fig. 7D). Longer distances (> 60 Å) may represent greater separations of these units, as observed in the free kinase. In summary, the Tar foldon complexes enforce a kinase conformation that is largely consistent with array models. Importantly, the PDS data also provided direct evidence for the close interaction between the P4 domains previously predicted by MD simulations (14, 16).
Modeling of the foldon complexes provides insight into kinase inhibition
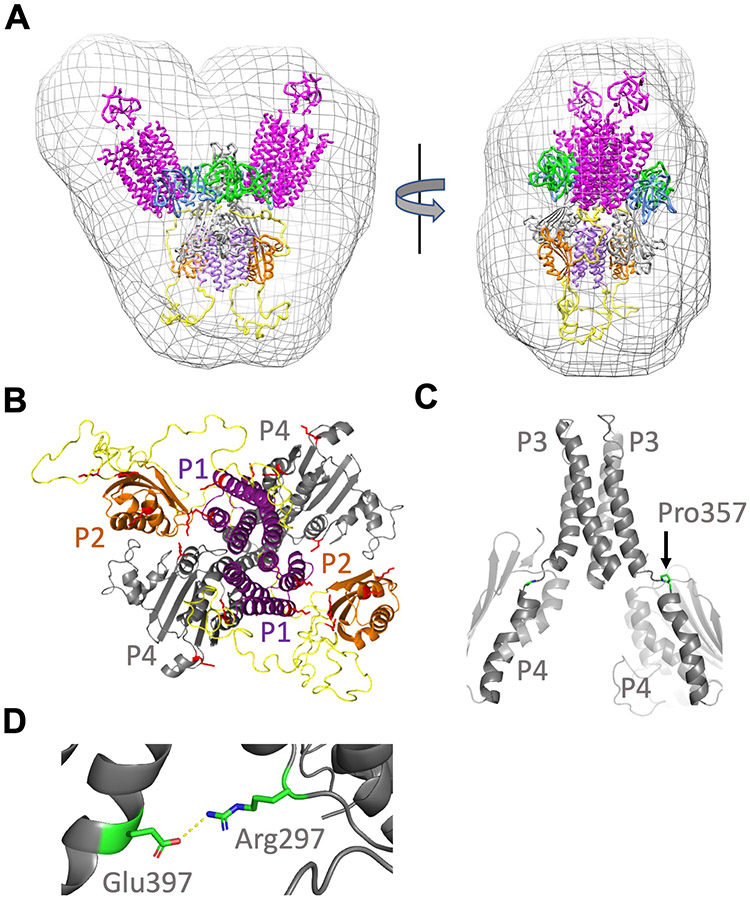
The foldon core complexes were computationally modeled in a step-wise manner that took into account their determined component stoichiometries, data from SAXS, cross-linking constraints, PDS restraints, and molecular structures determined from crystals and arrays in native and reconstituted forms (Fig. 8, A and B). Interactions among the receptor dimers and between CheA and CheW have been well defined by crystallographic structures, PDS, disulfide cross-linking studies, and modeling to cryo-ET images (1, 3, 14). However, the positioning of P1, P2, and P4, as well as the conformations of the interdomain linkers, are not well defined, especially for specific activity states. To place the P4 domains relative to P3 in the model of the inhibited foldon complex, two crystal structures that contain the CheA P3-P4 unit were considered: the dimer of P3P4 (24) and the dimer of P3P4P5 (42). Cryo-ET of arrays indicate that the P4 domains assume a conformation relative to the P3 domains that is similar to that observed in the crystal structure of CheA P3P4 (13, 15, 16). Thus, P4 was suspended below the P5-CheW layer based on the P3P4 structure (PDB code: 4XIV). Anchored by the P3-P4 domain positions, the L3 and L4 linkers were removed and rebuilt using the Rosetta KIC (Kinematic Closer) loop modeling algorithm. The L3 and L4 linker connections were constructed according to allowable sterochemical, spatial, and sequence constraints. The CheA model with the lowest energy loop conformations in Rosetta (see Methods and Materials) was then associated with CheW and receptor trimers based on known crystallographic interfaces and was then minimized against stereochemical and packing functions of the Rosetta Relax program with additional distance restraints from the PDS data. The output models were then ranked based on their Rosetta all-atom energy scores.
Fig. 8. Model of the ternary complex derived from crystal structures, PDS, SEC-SAXS, and cross-linking.
(A) The working model fits the size and shape of the molecular envelope generated by SEC-SAXS of the cross-linked ternary complex (purple, P1; orange, P2; grey, P3-P4; blue, P5; green, CheW; yellow, L1-L2). (B) The lysine residues identified in DSSO cross-links among the P1, P2, and P4 domains (red) localize to internal interfaces in the ternary complex model. (C) The L3 linker between the CheA P3 and P4 domains in both subunits forms a nearly continuous helix with a break at Pro357 (sidechains highlighted) in the highest scoring Rosetta model. (D) Salt bridge between L3 residue Arg297 and P4 residue Glu397 (shown in Chain A) has been implicated in receptor-coupled CheA activity and is maintained in the model.
In the next step, the P1 and P2 domains were associated with the core complex. The P1 domains were dimerized based on the crystallographic structure and PDS restraints, and situated below the P4 domains to align the molecular symmetry axes. This configuration is generally consistent with cryo-ET density tentatively assigned to the P1 and P2 domains (20), as well as the spatial extent of the SAXS envelopes generated from the cross-linked complex (Fig. 8A). The P1 dimer was positioned to interact with P4 based on interdomain cross-links and a previously characterized inhibitory contact (39). The P2 domains where then situated beside the P1 and P4 domains to fill unmatched density in the SAXS envelopes and align the molecular faces that undergo interdomain cross-linking. Domain orientations were chosen to minimize distances given by DSSO cross-linking (20 ± 7 Å). The ADP-NO inter-subunit and P5-to-ADP-NO PDS distances provided strong evidence for dipped P4, and the Rosetta modeling confirmed that such conformations can be obtained with reasonable L3 and L4 geometries. Although placement of the P1 domains and especially the P2 domains are not precisely constrained by the cross-linking data, these domains clearly co-localized with P4 in the inhibited complex, and Lys residues on specific faces showed preferred cross-linking patterns (Fig. 8B). The L1 and L2 linker conformations were first built automatically as Cα traces, elaborated as full polypeptide chains, and then evaluated for consistency with the SAXS scattering curves. Configurations that placed the large L1 linker below the P1 domains and the L2 linkers to the side of P4 produced the best agreement; conformations related to these were not substantially distinguished by the SAXS data. The allowable linker conformations did not distinguish between an intra- or intersubunit orientation for the P1 dimer on the symmetry axis of the foldon complex, although the PDS data on the heterodimer (Fig. 5F) agreed best with an intersubunit contact. In support of a trans interaction, P1 only made intersubunit cross-links in an engineered disulfide study of the free E. coli kinase (10). The entire structure, including CheW and the foldon receptors, was then relaxed in Rosetta under constraints of the ESR data and the cross-linking distances. One hundred new models were generated, and each was evaluated for agreement against the SAXS data (fig. S10A).
In the final modeling step, the complex was further optimized against the SAXS data by configurational sampling about the P4-P5 linkage (P5-CheW and receptor foldons were treated as rigid bodies) to allow for flexibility in the absence of the constraints provided by native membrane insertion (fig. S10B). Neither multi-state modeling nor treating the L1 and L2 loops as highly disordered segments improved fits to the SAXS data. The current working model (Fig. 8, A to D) agrees reasonably well with the SAXS scattering data (χ2 = 4.5; fig. S10C), matches the size and shape of the SAXS-derived molecular envelope, and is consistent with current distance restraint data (fig. S11). Overall, the model encapsulates well-established structural features of the particle that include interfaces among P5, CheW and the PIRs defined by previous crystal structures; movement of the P4 domains toward one another relative to the free kinase; association of the P1, P2, and P4 domains; dimerization of P1 between the P4 domains; and localization of the L1 linker below the P1,P2, and P4 cluster.
DISCUSSION
The MCP foldons that we generated interacted with CheA and CheW to form complexes that could be isolated in milligram quantities as homogenous ternary complexes. Stability and uniformity of the complexes was improved through chemical cross-linking. The resulting cross-linked particles mimicked a core signaling unit (2 TODs, 2 CheA subunits, and 2 CheW subunits), which is the minimum component of the chemosensory array that maintains modulation of CheA kinase activity and some cooperative responses (27, 28). Thus, these particles are suitable for examining the domain arrangements of CheA in a receptor-regulated state. In particular, complexes of Tm CheA with foldons derived from the Ec Tar receptor produced an especially stable kinase-off configuration. Expressed in cells or reconstituted in vitro with CheA and CheW, MCP cytoplasmic domains are well known to affect CheA activity (43-49). Some in vitro complexes highly activate CheA (43, 44) and produce relatively homogeneous particles that are larger than a core unit (50). Complexes of CheA and CheW with MCP cytoplasmic domains characterized by pulsed ESR spectroscopy (11), hydrogen-deuterium exchange mass-spectrometry (HDX) (51), and solid-state NMR (52) reveal aspects of component interactions but have not provided holistic views of the core signaling unit. His-tagged mediated dimerization (53) and templating of cytoplasmic domains on lipids (16, 48) have been useful strategies for analysis by cryo-ET (16). The foldon approach leverages trimerization of single-chain cytoplasmic domain fused subunits to produce particles of well-defined size that contain the components of a single core module.
Cross-linking MS experiments on the inhibited CheA foldon complex indicate that the CheA P1, P2, and P4 domains co-localize in the deactivated kinase. This result is consistent with cryo-ET studies of receptor variants that deactivate CheA and show enhanced “keel” density below the P4 domains that depend on the presence of P1 (15, 20). Importantly, inter-domain cross-links between P5 and CheW, as well as between CheW and the receptor, matched expectations from crystal structures and array models, thereby indicating that the foldon complexes recapitulate functionally relevant contacts among the components. Although cross-linking revealed interactions among P1, P2, and P4 in the deactivated state, no such interactions were found without receptor foldons and CheW. The association of Tm CheA with TODs indirectly co-localized these domains because the receptors primarily interact with CheW and P5 and do not likely contact P1, P2, or P4. The inter-domain contacts between P1 and P4 orient P1 helix D toward the ATP-binding site of P4. Anchoring P1 here may contribute to kinase inhibition by shielding His45 from P4. Indeed, previous NMR-shift assays of P1 binding to P3P4 identified interaction sites between helix D of P1 and a cluster of residues close to the L3 linker on the C-termini of both α1 and β1 of P4 (39). The cross-linking pattern of the inhibited foldon complex was consistent with this interaction mode, and the resulting model places the P4 β1 site directly in the interface with P1 helix D and the α1 site in contact with the L3 linker that changes conformation upon P4 dipping. The PDS measurements surprisingly reveal P1 dimerization as found in the Tm P1 crystal structure (Fig. 6). The P1 dimer occludes access of the P4 ATP pocket to His45, but leaves helix D available for interaction with P4. Whereas interactions between P1 and P4 in the inhibitory complex were reasonably anticipated, interactions among P1, P2, and P4 were not. For phosphotransfer to CheY, P2 need not interact with P1 or P4, yet P2 preferentially cross-links to these domains and not to P5, CheW or the receptor foldons. P2 sites on the CheY interaction surface (54) cross-linked to a Lys residue near the P1 substrate His45. Thus, P1-to-P2 contacts could potentially interfere with both His45 phosphorylation and CheY binding. P1-P1 interactions are also supported by the observations that the P1 domains cross-link to each other (24) and increase the subunit affinity of the CheA dimer (55). P1 dimerization that reduces His45 accessibility may also explain why both Ec and Tm CheA activity increases upon removal of one P1 domain (24, 25).
Compared to their mobility in the free kinase, the P4 domains order and move toward each other in the foldon complexes. MD simulations of the core signaling particle composed of CheA P3P4P5 captured transitions between a CheA conformation in which the P4 domain interacts with the P5-CheW layer (undipped) and a conformation wherein P4 moves down toward the adjacent P4 domain (dipped) (14, 16). Positioning of the P4 domains based on the ADP-NO interaction, the E387C-R1 contact, and Rosetta loop modeling of the L3 and L4 linkers provide strong support for the dipped conformation of P4 in the inhibitory foldon complex. These configurations are consistent with high-resolution cryo-ET density of the P4 region in reconstituted arrays on lipid supports that contain CheA P3P4P5; however, disorder in the cryo-ET density limits precise positioning of P4 (14). The MD simulations further suggest that key salt bridges between the P3 domain (Ec Arg265) and P4 residues (Ec E368,D372) stabilize the dipped conformation (14, 16). Residue substitutions at these positions affect CheA activity and chemotaxis (56). In particular, charge reversals at several key sites substantially reduce receptor-coupled CheA activity and regulation by attractant (56). Thus, it has been suggested that the active state of CheA involves a dipped conformation (56). The MD simulations also indicate that the formation of a nearly continuous helix between P3 α2 and P4 α1 stabilizes the dipped conformation (Fig. 8C). The Rosetta data-restrained models also recapitulated the salt bridge between Ec Arg265 (Tm R297) and Ec E368 (Tm E397; Fig. 8D). However, continuous helicity breaks at Ec Val344-Pro345 (Tm V356-P357) and the P3 and P4 helices were not fully aligned (Fig. 8C).
Despite harboring a “dipped” P4 conformation, CheA autophosphorylation activity is low in the foldon complexes, at least in part because both P1 and P4 are sequestered. Could then a dipped conformation be associated with both an inhibited and activated state of CheA? Several related conformations that all suspend P4 away from the P5-CheW layer could be collectively important for both the inhibited and activated states. Both P4-P4 cross-linking, which decreased in the foldon complexes compared to free CheA, and the PDS distances rule against direct P4-P4 contacts in this particular inhibited state. However, negative allostery in nucleotide binding and the low activity of P4 monomers for P1 phosphorylation (24, 39) suggest that the P4 domains do influence each other, either directly or indirectly. When the P1 domains release to become substrates, the P4 domains may approach one another more closely. As has been suggested (56), transitions of P4 among conformational states may be key for the catalytic cycle. Under this assumption, restricting such motion, through interaction with P1, or possibly with the P5-CheW layer, would provide the means to curtail autophosphorylation. Whether the key regulatory conformations also involve a fully undipped conformation is unclear. Undipped P4 may allow for interaction of the ATP-binding regions with the P5-CheW layer, but it may also be favored by the absence of the P1 and P2 domains in the reconstituted arrays and simulations.
Thus, in the Tm inhibited foldon complex, the P4 domains are likely juxtaposed, and the P1, P2, and P4 domains are closely associated in a region distal from the P3, P5, CheW, and receptor components. Receptor binding to remote sites on P5 and CheW promoted these interactions. Consistent with structural signals transferring from the P5-CheW layer to the linkers and domains beneath, engineered disulfide cross-linking between P5 and CheW increase array stability by protecting the L1 and L2 linkers from proteolysis (57). It is important to note that core complexes composed with CheA P3P4P5 display normal receptor-mediated inhibition of CheA activity, even with free P1 as the substrate (22, 23). Thus, neither covalent attachment of P1 nor the presence of P2 is required for receptor-regulated kinase inhibition. Furthermore, saturating levels of free P1 do not curtail kinase activity (22, 23). Nonetheless, a P1 inhibitory site could be operative in the kinase-off state, if kinase activation then renders this site unavailable.
Changes in free P1 phosphorylation manifest from changes in the catalytic rate constant (kcat), not the Michaelis constant (KM) (22, 23). Thus, kinase inhibition may derive from a depopulation of CheA states capable of productive interactions with P1. A bound P1 dimer may contribute to stabilization of an inhibited state, provided such interactions do not depend on covalent attachment to the core complex and that they are relieved in the kinase-on state. Because an ultrastable fraction of purified arrays does not undergo L1 or L2 proteolysis for > 20 days yet remains active (58), it has been suggested that the linkers are ordered and protected in both the inhibited and activated states (3). The linkers may only become dynamic in the transition between these states, which may be distinguished by where P1 binds: either at the P4 active site or the P4 inhibitory site (3). Our model is consistent with such a view, but elaborates the P1 inhibitory interaction in the form of a symmetric dimer on the molecular symmetry axis and furthermore indicates that P2 interactions with the core kinase are also coupled to linker ordering. The constrained architecture of the foldon receptor complexes prevents access of the P4 ATP-binding pocket to the P1 substrate His. Reduced domain dynamics in the foldon complexes, communicated from the receptors to P4 through its linkers to P3, or through interactions with the P5-CheW layer, may favor P1 binding in this non-productive configuration. Indeed, increased order in the arrays, especially in those CheA regions suspended below the P5-CheW layer, appears to be a hallmark of the inactive state (15). Initial studies of CheA activity in membranes described a receptor-inhibited state that was designated as “sequestered” because it could not autophosphorylate P1 nor exchange phosphate with ADP once phosphorylated (59). The data presented here indicate that an inhibited core signaling unit sequesters both the P1 and P4 domains through their non-productive interaction and that alleviation of these restraints would be required for kinase activation.
MATERIALS AND METHODS
Cloning and protein purification
Proteins were cloned into pet28a vector and transformed into E. coli (DE3) competent cells. After plating the transformation on agar containing kanamycin, a single colony was chosen for cell culture growth. 8 liters of cells were grown at 37°C until an O.D. at 600 nm reached 0.6. 1mM IPTG was added to each flask and the cells were grown for 16 hours at room temperature. After pelleting via centrifugation, the cells were resuspended in 50 mL of lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5mM Imidazole) and sonicated for 6 minutes. The lysed cells were then centrifuged to remove the insoluble fraction from the lysate. The lysate was then run over Nickel-NTA affinity resin, washed with 20 mM imidazole buffer to remove non-specific binding proteins, and eluted using buffer with 200 mM imidazole. The protein was further purified by size-exclusion chromatography (prep-grade Sephadex S200 or s75) in 50 mM Tris pH 7.5 150 mM NaCl. The fractions containing the protein were pooled and concentrated via high-speed centrifugation. Receptor foldons were kept at 4°C.
Protein spin-labeling
2 mg of 1-oxyl-2,2,5,5-tetramethylpyrroline-3- methyl) methanethiosulfonate spin-label (MTSSL) was added in 5 mL of wash buffer (50 mM Tris pH 7.5, 150 mM NaCl, 20 mM imidazole) to the protein bound on nickel resin and the column was rocked overnight at 4° C. 10 mL of wash buffer was flowed through the column to remove unattached spin label and the protein was eluted with 10 ml of wash buffer containing 200 mM imidazole. The sample was then purified by SEC on an S200 column as above. With a spin filter concentrator the sample was exchanged into 50 mM Tris pH 7.5, 150 mM NaCl, 20% deuterated glycerol, made from D2O and concentrated to 100 μM. 30 μL was placed in ESR tubes and flash-frozen in liquid N2.
Phosphorylation assays
25 μl samples containing 2 μM CheA (subunit) were incubated in the presence or absence of 2 μM CheW and 6 μM receptor foldon (subunit) in 50 mM MOPS pH 7.5, 150 mM KCl, 10 mM MgCl2. Phosphorylation of CheA was initiated by the addition of 1 mM ATP mixed with radiolabeled γ−32P ATP at a final radioactivity of 0.15 ± 0.05 mCi and quenched at 1–12 min for Tm CheA and 15 s for Ec CheA with 25 μl of 3X LDS buffer containing 100 mM EDTA. The samples were run on a native Tris-glycine gel for 2 hours at 120 volts. The gels were dried, placed in a radiocassette for 24 hours, and then imaged with a Typhoon phosphor-imager. Band intensities were quantified using Image J software. A monoexponential growth function was globally fit to all replicate data for each condition using non-linear least squares fitting in Matlab.
Multi-angle light scattering
Reverse-phase chromatography coupled to multi-angle light scattering experiments was used to determine the molecular weight of isolated ternary complexes. For each sample, 2–5mg/ml of protein was injected onto a Phenomenex reverse-phase column pre-equilibrated with 50mM Tris pH 7.5, 150 mM NaCl at room temperature. Bovine serum albumin (sigma) was used as a protein standard. Wyatt technologies ASTRA 6 program was used for data analysis and molecular weight calculations.
Preparation of isolated ternary complexes
Ternary complexes consisting of receptor foldon, CheA and CheW were produced in milligram quantities by mixing the composite purified proteins in a 1:1:1 stoichiometric subunit ratio in 50 mM Tris or HEPES pH 7.5, 250 mM KCl, 5mM MgCl2, 10% glycerol and allowing the mixture to incubate at 4°C for at least 1 hour. After incubation, the sample is injected onto two prep-grade Sephadex columns (s200 and s300) attached to run in tandem and the elutant was collected in 6 mL fractions. Protein components in each fraction were evaluated by SDS-PAGE. Fractions of interest were concentrated to ~ 8 mg/ml via high-speed centrifugation, flash frozen, and stored at −80°C.
Size-exclusion chromatography small-angle x-ray scattering (SEC-SAXS)
SAXS data were collected at CHESS G1 line on the Finger Lakes CCD detector. For each sample, ~2–3 mg/ml of protein was injected onto a Superdex 5/150 size-exclusion increase column pre-equilibrated with sample buffer (50 mM Tris pH 7.5, 250 mM KCl, 1 mM MgCl2, 4% glycerol) coupled to the G1 SAXS sample cell. The eluted sample was collected with 2 second exposure times to the x-ray beam at a flow rate of 0.15 ml/min. AMBIMETER (60) and GNOM (61) were used to initially assess the quality of the scattering data and particle homogeneity. RAW (62) and PRIMUS (63) were used for processing of the SEC-SAXS data and to generate Kratky plots. Further analysis including dimensionless Kratky were carried out with ScÅtter (64). Background scattering was determined from SEC fractions containing primarily buffer. Molecular weight estimates were made from the Porod volume (VP), volume of correlation (VC) (64) and from envelope reconstructions. Envelope reconstructions were calculated with DAMMIF (65) and DAMAVER (66). Ten models were independently generated and then averaged into a consensus envelope without assuming particle symmetry. For the cross-linked ternary complex singular value decomposition with evolving factor analysis (32) was applied to separate the minor contribution of a higher MW species from the leading edge of the SEC curve. Model agreement to SAXS scattering curves was evaluated with FoXS (67).
Cryo-EM and image analysis
For cryo-specimens, 3 μL aliquots of 0.1 mg/ml cross-linked ternary complexes were applied to freshly glow-discharged R2/2 Quantifoil 200 mesh copper grids (Quantifoil Micro Tools). Grids were blotted in a climate chamber set to 20 °C and at 95% humidity before plunge frozen in liquid ethane set at −183 °C using a Leica EM GP system (Leica Microsystems). Images containing complexes were collected manually with a 120 kV TALOS transmission electron microscope (Thermo Fisher Scientific) at a magnification corresponding to a pixel size of 1.4 Å in a defocus range between 2 to 4 microns. Image analysis was done with RELION −3.0.2 using ~ 34,000 particles from 65 images. 2D classification results were generated with a selection of ~ 22,000 particles.
Protein cross-linking
After incubation of the receptor foldons with CheA and CheW in a 1:1:1 stoichiometric ratio to a final concentration of 10 μM of each protein in 50 mM HEPES pH 7.5, 250 mM KCl, 5 mM MgCl2, 100 μM ADP, 10% glycerol for at least 30 minutes at 4°C, the chemical crosslinker DSSO is added to the protein mixture to a final concentration of 1 mM and crosslinking proceeds for 30 min to 1 hour at room temperature. The cross-linking reaction was quenched by the addition of Tris pH 8.0 to a final concentration of 20 mM. The reaction mixture is run through SEC on two prep-grade sephadex columns (s200 and s300) attached to run in tandem with a 6 ml fraction volume. Fractions of interest were selected to for further analysis by SDS-PAGE and MALS.
Mass spectrometry of cross-linked protein complexes
For cross-linking of free CheA with DSSO (33, 68) ~50 μg of of the protein was cross-linked in solution (50 mM HEPES pH 7.5, 250 mM KCl, 10% glycerol, 100 μM ADP, 5 mM MgCl2) and then dried. The sample was then denatured and reduced with the addition of 20 μl 6M guanidinium-HCL, 50 mM ammonium bicarbonate and 2 μl 0.110M DTT (final concentration = 10 mM DTT) and incubated at 60°C for 1 hour. The sample was then alkylated with the addition of 2.5 μl of 0.55 M iodoacetamide (IDA) followed by 45 minutes of incubation at room temperature in the dark. The alkylation reaction was quenched with 5.5 μl 0.2 M DTT. The sample was then diluted by the addition of 100 μl of 50 mM ammonium bicarbonate (Ambic). 12.5 μl of Trypsin at 0.2 μg/μl (2.5 μg) was added and incubated overnight at 37°C. The sample was heated to 90°C for 5 minutes to deactivate the trypsin. A second digestion with chymotrypsin was then carried out by the addition of 5 μl of 1 μg/μl chymotrypsin followed by an overnight incubation at 37°C. The digestion was stopped with the addition of 150 μl of 0.5% TFA. The pH after addition was < 2.5. The samples were then applied to a SepPak C18 1cc/ 50 mg (waters) cartridge for cleanup.
For the ternary complex, a protein sample containing a 1:1:1 ratio of the composite proteins was incubated and cross-linked in solution (50 mM HEPES pH 7.5, 250 mM KCl, 10% glycerol, 100 μM ADP, 5 mM MgCl2) and then run on an SDS page gel. A gel band containing ~50 μg the ternary complex was excised from the gel for MS/MS analysis. First, the gel band was washed with 400 μl ddH2O, then 400 μl of 50% acetonitrile (ACN) and 50 mM Ambic, then 400 μl 100% CAN. The gel was exposed to air in a chemical hood until dryness. The sample was then reduced with the addition of 200 μl of 10 mM DTT in 100 mM Ambic at 60°C for 1 hour. The sample was alkylated with the addition of 200 μl of 55 mM IDA in 100 mM Ambic and incubated for 45 minutes at room temperature in the dark. The washing steps were then repeated. 120 μl Trypsin at 10 ng/μl (1.2 μg) in 50 mM Ambic and 10% ACN was added and then incubated on ice for 20 minutes. The sample was then overlayed with another 150 μl of 50 mM Ambic with 10% ACN and incubated overnight at 37°C. After 18 hours of digestion, the reaction was stopped with the addition of 5 μl of 100% Formic acid. The sample was then extracted from the gel by first vortexing in a mixture of 200 μl of 50% ACN and 5% formic acid for 30 minutes at 1800 rpm followed by sonication for 10 minutes. This step was repeated once more and the sample was dried in a speed vacuum. The sample was then reconstituted with 45 μl of 50 mM Ambic and heated to 90°C for 5 minutes. The second digestion with chymotrypsin was then conducted by adding 5 μl chymotrypsin at 1 μg/μl (3 μg) and incubated overnight at 37°C. The digestion was stopped with the addition of 2 μl of 100% Formic acid. The resulting pH was <2.5. The sample was dried to dryness in a speed vacuum and then resuspended into 0.5% Formic acid.
Peptides and cross-linked peptides were analyzed using an UltiMate3000 RSLCnano (Dionex, Sunnyvale, CA) coupled to an Orbitrap Fusion (Thermo-Fisher Scientific, San Jose, CA) mass spectrometer equipped with a nanospray Flex Ion Source. Each sample was loaded onto an Acclaim PepMap 100 C18 trap column (5 μm, 100 μm × 20 mm, 100 Å, Thermo Fisher Scientific) at 20 μL/min of 0.5% formic acid (FA). After 3 minutes, the valve switched to allow peptides to be separated on an Acclaim PepMap C18 nano column (3 μm, 75μm x 25cm, Thermo Fisher Scientific), in a 120 min gradient of 5% to 40% B at 300 nL/min. The Orbitrap Fusion was operated in positive ion mode with nano spray voltage set at 1.7 kV and source temperature at 275 °C. External calibrations for Fourier transform, ion-trap and quadrupole mass analyzers were performed prior to the analysis. Samples were analyzed using the CID–MS2–MS3 workflow, in which peptides with charge states 4–10 were selected for CID MS2 acquisitions in Orbitrap analyzer with a resolution of 30,000 and an AGC target of 5 × 104. MS scan range was set to 375–1575 m/z and the resolution was set to 60,000. The precursor isolation width was 1.6 m/z and the maximum injection time was 100 ms. The CID MS2 normalized collision energy was set to 25%. Targeted mass-difference-dependent CID–MS3 spectra were triggered for acquisition in the ion trap with CID collision energy of 35%; AGC target of 2 × 104 when a unique mass difference (Δ=31.9721 Da) was observed in the CID–MS2 spectrum. MS2 isolation window of 3 m/z with the maximum injection time set to 100 ms. All data were acquired under Xcalibur 3.0 operation software and Orbitrap Fusion Tune Application v. 2.1 (Thermo-Fisher Scientific).
All MS, MS2 and MS3 raw spectra from each sample were searched using Proteome Discoverer 2.2 (Thermo-Fisher Scientific, San Jose, CA) with XlinkX v2.0 algorithm for identification of cross-linked peptides. The search parameters were as follow: three missed cleavage for full trypsin digestion or double digestion with fixed carbamidomethyl modification of cysteine, variable modifications of methionine oxidation. The peptide mass tolerance was 10 ppm, and MS2 and MS3 fragment mass tolerance was 20 ppm and 0.6 Da, respectively. The Ecoli database with added targeted protein sequences was used for PD 2.2 database search with 1% FDR for report of cross-link results. In addition, the search was also performed using the full sequences of the proteins including the recombinant tags. Identified cross-linked peptides were filtered for Max. XlinkX Score >20 containing at least two identified MS3 spectra for each pair of cross-linked peptides. Results of the search with were exported by the software as a spreadsheet.
Pulse Dipolar ESR Spectroscopy (PDS)
PDS measurements were conducted by four pulse double electron electron resonance (DEER) at 60 K based on a 17.3 GHz FT ESR spectrometer (69) modified for PDS ESR (70) and at 34 GHz in Q-band on on a Elexsys E580 spectrometer equipped with a 10W solid state amplifier (150W equivalent TWTA) and Arbitrary Waveform Generator (AWG) to generate MW pulses at the detection and pumping frequency offsets to suppress spurious echoes. Q-band DEER measurements were performed at 60 K (ER 4118HV-CF10-L FlexLine Cryogen-Free VT System) in an EN 5107D2 cavity using four pulses (π/2-τ1-π- τ1-πpump- τ2-π - τ2-echo) with 16-step phase cycling. Data were collected on 100 μM protein over 16 hours for each sample. The signal background was approximated by a polynomial function in the semi-log scale and subtracted out (71). Noise from the time domain data was then removed by the WavPDS method (72) (a wavelet denoising procedure for PDS) and distance distributions of spin separations calculated by the new singular-value decomposition (SVD) method (73, 74) developed to solve ill-posed problems such as for the PDS signal and estimate uncertainty in their measurement.
Model Building of the Ternary Complex
For the dipped CheA models, ten and seven residues were modeled for reconnecting the P3-P4 (L3) loop and the P4-P5 (L4) loop, respectively. The chain breaks were located one residue further away from P4 domain in both loops to allow more dynamic motion of the domain. In productive run, 250 KIC build attempts were made with 500 models generated. The best model was selected based on the Rosetta Energy Unit (REU) score and finalized after structural comparison with the other top models. The new P3-P4 and P4-P5 loop conformations of the dipped CheA models were reintroduced in the original models before Rosetta Relax.
The Rosetta Relax productive run of the dipped CheA-complex (including CheW and foldons) generated 100 models with five cycles of minimization including SDSL-PDS distance restraints (table S2) and sidechain repacking. A suitable function based on the “motion-on-a-cone” model (75) was implemented in RosettaEPR for the spin label distance. The function describes the relationship between the experimental measured spin label distance (dSL) and Cβ-Cβ distance (dCβ) in which an acceptable range of dCβ is determined from dSL (dCβ ϵ [dSL-12.5 Å, dSL +2.5Å]). Top scoring models based on the REU scores were selected and analyzed. The best model of the dipped CheA complex has an REU score of −4845.83, with the average score of 100 models at −4487.6. Top ten scoring models of dipped CheA were selected for analysis. In comparison to the P3, P4 and P5 domains of the best model, the rest of the models have a root mean square deviation (RMSD) of 1.90–2.75 Å (chain A) and 2.09–4.63 Å (chain B) for 373 Cα atoms.
The crystallographic P1 dimer was placed in the Rosetta model with CHIMERA in order to be consistent with the SAXS envelope and cross-linking data. The P2 domains were added symmetrically under similar considerations. The L1 and L2 loops were built with EOM (76) as Cα traces. Families of configurations were evaluated against the SAXS data with FoXS (67) after replacing the traces with the correctly sequenced polypeptide. Six general families of loop positions that extended into different regions of the SAXS envelopes were considered in this manner. Only those configurations that extended the L1 loops below the P1 domains sufficiently extended the radius of gyration to produce reasonable agreement with the SAXS data. With the L1 and L2 loops explicitly built and the receptor foldons incorporated, Rosetta Relax was used to produce 100 new models under RosettaEPR constraints of the PDS distances and bounded-function constraints of the cross-linking data. Protein cross-links were assumed to occur over a range of 20 ± 7 Å. Each of the 100 final models was evaluated against the SAXS data with FoXs. The model with the lowest Rosetta energy and the model with the closest agreement to the SAXS data have a REU score of −5863.93 and −5679.7 respectively. Their average scores out of 100 models is at −5333.27. Conformations with high (> 0) Rosetta energy scores were excluded. The best model that maintained the expected domain interfaces was then subjected configurational sampling about the L4 linker, while keeping P5, CheW and receptor foldons as rigid bodies. This sampling, implemented in MultiFoxS (67) maximized agreement with the SAXS data and the subsequent model was fit against the SAXS-derived molecular envelope in Chimera. Restraint distances in the final model are given in fig. S11.
Supplementary Material
fig. S1. Preliminary purification and characterization of receptor foldons and their complexes.
fig. S2. Buffer conditions that encourage interactions among the TarFo, CheA, and CheW.
fig. S3. Properties of Tm14Fo complexes with Tm CheA and Tm CheW.
fig. S4. Autoradiographs of CheA autophosphorylation experiments from Fig. 1.
fig. S5. Properties of single-residue foldon variants.
fig. S6. SEC-SAXS data of the cross-linked foldon complex.
fig. S7. Inter-domain cross-links in free CheA.
fig. S8. Inter-domain cross-links in the receptor foldon complex.
fig. S9. PDS of Tm CheA E588C-R1 with CheW and TarFo.
fig. S10. Modeling of the foldon complex.
fig. S11. Summary of model restraints in Rosetta refinements.
table S1. Real space SAXS parameters generated from SEC-SAXS for foldon complexes.
table S2. List of PDS distance restraints for Rosetta modeling.
Acknowledgements:
We thank Estella Yee for help with kinetics fits, the Cornell High Energy Synchrotron Source (CHESS) for access to data collection facilities.
Funding: This work was supported by grants from the National Institutes of Health: R35GM122535 to BRC and P41GM103521 for the National Biomedical Center for Advanced ESR Technologies (ACERT) to JHF, and was supported by the European Union under a Marie-Sklodowska-Curie COFUND LEaDing fellowship to ARM and a National Science Foundation Graduate Research Fellowship Program (NSF-GRFP) grant to ARM (2014155468) CHESS is supported by NSF award DMR-1332208 and NIH/NIGMS award P30GM103485:
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019599. Raw SAXS and PDS data is available upon request. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Muok AR, Briegel A, Crane BR, Regulation of the chemotaxis histidine kinase CheA: A structural perspective. Biochimica et Biophysica Acta Biomembranes 1862, 183030 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkinson JS, Hazelbauer GL, Falke JJ, Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends in Microbiology 23, 257–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falke JJ, Piasta KN, Architecture and signal transduction mechanism of the bacterial chemosensory array: Progress, controversies, and challenges. Current Opinion in Structural Biology 29, 85–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazelbauer GL, Lai WC, Bacterial chemoreceptors: providing enhanced features to two-component signaling. Current Opinion in Microbiology 13, 124–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mello BA, Tu YH, Quantitative modeling of sensitivity in bacterial chemotaxis: The role of coupling among different chemoreceptor species. Proceedings Of The National Academy Of Sciences Of The United States Of America 100, 8223–8228 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sourjik V, Berg HC, Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. U S A 99, 123–127 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinas GE, Frank V, Vaknin A, Parkinson JS, The source of high signal cooperativity in bacterial chemosensory arrays. Proceedings of the National Academy of Sciences of the United States of America 113, 3335–3340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou H, Lowry DF, Swanson RV, Simon MI, Dahlquist FW, NMR studies of the phosphotransfer domain of the histidine kinase CheA from Escherichia coli: assignments, secondary structure, general fold, and backbone dynamics. Biochemistry 34, 13858–13870 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Quezada CM, Gradinaru C, Simon MI, Bilwes AM, Crane BR, Helical shifts generate two distinct conformers in the atomic resolution structure of the CheA phosphotransferase domain from Thermotoga maritima. Journal of Molecular Biology 341, 1283–1294 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Gloor SL, Falke JJ, Thermal Domain Motions of CheA Kinase in Solution: Disulfide Trapping Reveals the Motional Constraints Leading to Trans-autophosphorylation. Biochemistry 48, 3631–3644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhatnagar J, Borbat P, Pollard AM, Bilwes AM, Freed JH, Crane BR, Structure of the ternary complex formed by a chemotaxis receptor signaling domain, the CheA histidine kinase and the coupling protein CheW as determined by pulsed dipolar ESR spectroscopy. Biochemistry 49, 3824–3841 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenswag AR, Li X, Borbat PP, Samanta D, Watts KJ, Freed JH, Crane BR, Preformed Soluble Chemoreceptor Trimers That Mimic Cellular Assembly States and Activate CheA Autophosphorylation. Biochemistry 54, 3454–3468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen G, C. B.R., Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. USA 109, 3766–3771 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cassidy CK, Himes BA, Sun D, Ma J, Zhao G, Parkinson JS, Stansfeld PJ, Luthey-Schulten Z, Zhan P, Structure and dynamics of the E. coli chemotaxis core signaling complex by cryo-electron tomography and molecular simulations. Communications Biology 3, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W, Cassidy K, Ames P, Diebolder CA, Schulten K, Luthey-Schulten Z, Parkinson JS, Briegel A, In situ conformational changes of the Escherichia coli serine chemoreceptor in different signaling states. mBio 10, e00973–00919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cassidy CK, Himes BA, Alvarez FJ, Ma J, Zhao G, Perilla JR, Schulten K, Zhang P, CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling. eLife 4, e08419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang XQ, Wu C, Anh Vu JES, Dahlquist FW, Computational and Experimental Analyses Reveal the Essential Roles of Interdomain Linkers in the Biological Function of Chemotaxis Histidine Kinase CheA. Journal of the American Chemical Society 134, 16107–16110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang XQ, Vallurupalli P, Vu A, Lee K, Sun S, Bai WJ, Wu C, Zhou HJ, Shea JE, Kay LE, Dahlquist FW, The Linker between the Dimerization and Catalytic Domains of the CheA Histidine Kinase Propagates Changes in Structure and Dynamics That Are Important for Enzymatic Activity. Biochemistry 53, 855–861 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding XY, He Q, Shen FL, Dahlquist FW, Wang XQ, Regulatory Role of an Interdomain Linker in the Bacterial Chemotaxis Histidine Kinase CheA. Journal of Bacteriology 200, e00052–00018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briegel A, Ames P, Gumbart JC, Oikonomou CM, Parkinson JS, Jensen GJ, The mobility of two kinase domains in the Escherichia coli chemoreceptor array varies with signalling state. Molecular Microbiology 89, 831–841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W, Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proceedings of the National Academy of Sciences of the United States of America 109, 1481–1488 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan W, Dahlquist FW, Hazelbauer GL, Signaling complexes control the chemotaxis kinase by altering its apparent rate constant of autophosphorylation. Protein Science 26, 1535–1546 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mello BA, Pan WL, Hazelbauer GL, Tu YH, A dual regulation mechanism of histidine kinase CheA identified by combining network-dynamics modeling and system-level input-output data. Plos Computational Biology 14, e1006305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenswag AR, Muok A, Li X, Crane BR, Conformational Transitions that Enable Histidine Kinase Autophosphorylation and Receptor Array Integration. Journal of Molecular Biology 427, 3890–3907 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levit M, Liu Y, Surette M, Stock J, Active Site Interference and Asymmetric Activation in the Chemotaxis Protein Histidine Kinase CheA. J. Biol. Chem 271, 32057–32063 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Frank V, Pinas GE, Cohen H, Parkinson JS, Vaknin A, Networked Chemoreceptors Benefit Bacterial Chemotaxis Performance. Mbio 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Hazelbauer GL, Selective allosteric coupling in core chemotaxis signaling complexes. Proceedings of the National Academy of Sciences of the United States of America 111, 15940–15945 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li MS, Hazelbauer GL, Core unit of chemotaxis signaling complexes. Proceedings of the National Academy of Sciences of the United States of America 108, 9390–9395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mowery P, Ames P, Reiser RH, Parkinson JS, Chemotactic Signaling by Single-Chain Chemoreceptors. Plos One 10, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mowery P, Ostler JB, Parkinson JS, Different Signaling Roles of Two Conserved Residues in the Cytoplasmic Hairpin Tip of Tsr, the Escherichia coli Serine Chemoreceptor. Journal of Bacteriology 190, 8065–8074 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortega DR, Yang C, Ames P, Baudry J, Parkinson JS, Zhulin IB, A phenylalanine rotameric switch for signal-state control in bacterial chemoreceptors. Nature Communications 4, e2881 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meisburger SP, Taylor AB, Khan CA, Zhang SN, Fitzpatrick PF, Ando N, Domain Movements upon Activation of Phenylalanine Hydroxylase Characterized by Crystallography and Chromatography-Coupled Small-Angle X-ray Scattering. Journal of the American Chemical Society 138, 6506–6516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kao AH, Chiu CL, Vellucci D, Yang YY, Patel VR, Guan SH, Randall A, Baldi P, Rychnovsky SD, Huang L, Development of a Novel Cross-linking Strategy for Fast and Accurate Identification of Cross-linked Peptides of Protein Complexes. Molecular & Cellular Proteomics 10, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Fleetwood AD, Bayas C, Bilwes AM, Ortega DR, Falke JJ, Zhulin IB, Crane BR, The 3.2 angstrom Resolution Structure of a Receptor:CheA:CheW Signaling Complex Defines Overlapping Binding Sites and Key Residue Interactions within Bacterial Chemosensory Arrays. Biochemistry 52, 3852–3865 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muok AR, Chua TK, Le H, Crane BR, Nucleotide Spin Labeling for ESR Spectroscopy of ATP-Binding Proteins. Applied Magnetic Resonance 49, 1385–1395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Freed JH, Bilwes AM, Crane BR, Reconstruction of the chemotaxis receptor:kinase assembly. Nat. Struct. Mol. Biol 13, 400–407 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Nishiyama S, Garzon A, Parkinson JS, Mutational Analysis of the P1 Phosphorylation Domain in Escherichia coli CheA, the Signaling Kinase for Chemotaxis. Journal of Bacteriology 196, 257–264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Xu YC, Shen JH, Luo XM, Chen JG, Chen KX, Zhu WL, Jiang HL, Dynamic mechanism for the autophosphorylation of CheA histidine kinase: Molecular dynamics simulations. Journal of The American Chemical Society 127, 11709–11719 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Hamel DJ, Zhou H, Starich MR, Byrd A, Dahlquist FW, Chemical-shit-perturbation mapping of the phosphotransfer and catalytic domain interaction in the histidine autokinase CheA from Thermotoga maritima. Biochemistry 45, 9509–9517 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Eaton AK, Stewart RC, The Two Active Sites of Thermotoga maritima CheA Dimers Bind ATP with Dramatically Different Affinities. Biochemistry 48, 6412–6422 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhatnagar J, Sircar R, Borbat P, Freed JR, Crane BR, Self association of the histidine Kknase CheA as studied by pulsed dipolar ESR spectroscopy. Biophysical J. 102, 2192–2201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bilwes AM, Alex LA, Crane BR, Simon MI, Structure of CheA, a signal-transducing histidine kinase. Cell 96, 131–141 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Surette MG, Levit M, Liu Y, Lukat G, Ninfa EG, Ninfa A, S. J.B., Dimerization is required for the activity of the Protein Histidine Kinase CheA that mediates signal transduction in bacterial chemotaxis. J. Biol. Chem 271, 939–945 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Levit MN, Grebe TW, Stock JB, Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J. Biol. Chem 277, 36748–36754 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Asinas AE, Weis RM, Competitive and cooperative interactions in receptor signaling complexes. Journal Of Biological Chemistry 281, 30512–30523 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Ames P, Parkinson JS, Constitutively signaling fragments of Tsr, the E.coli serine chemoreceptor. J. Bacteriol 176, 6340–6348 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montefusco DJ, Shrout AL, Besschetnova TY, Weis RM, Formation and activity of template-assembled receptor signaling complexes. Langmuir 23, 3280–3289 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Shrout AL, Montefusco DJ, Weis RM, Template-directed assembly of receptor signaling complexes. Biochemistry 42, 13379–13385 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Airola MV, Sukomon N, Samanta D, Borbat PP, Freed JH, Watts KJ, Crane BR, HAMP domain conformers that propagate opposite signals in bacterial chemoreceptors. PLoS Biol 11, e1001479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolanin PM, Baker MD, Francis NR, Thomas DR, DeRosier DJ, Stock JB, Self-assembly of receptor/signaling complexes in bacterial chemotaxis. Proceedings Of The National Academy Of Sciences Of The United States Of America 103, 14313–14318 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koshy SS, Li XN, Eyles SJ, Weis RM, Thompson LK, Hydrogen Exchange Differences between Chemoreceptor Signaling Complexes Localize to Functionally Important Subdomains. Biochemistry 53, 7755–7764 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris MJ, Struppe JO, Wylie BJ, McDermott AE, Thompson LK, Multidimensional Solid-State Nuclear Magnetic Resonance of a Functional Multiprotein Chemoreceptor Array. Biochemistry 55, 3616–3624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haglin ER, Yang W, Briegel A, Thompson LK, His-Tag-Mediated Dimerization of Chemoreceptors Leads to Assembly of Functional Nanoarrays. Biochemistry 56, 5874–5885 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park SY, Beel BD, Simon MI, Bilwes AM, Crane BR, In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved. Proc. Natl. Acad. Sci. U S A 101, 11646–11651 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kott L, Braswell EH, Shrout AL, Weis RM, Distributed subunit interactions in CheA contribute to dimer stability: a sedimentation equilibrium study. Biochimica Et Biophysica Acta-Proteins and Proteomics 1696, 131–140 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Pinas GE, Parkinson JS, Identification of a Kinase-Active CheA Conformation in Escherichia coli Chemoreceptor Signaling Complexes. Journal of Bacteriology 201, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piasta KN, Falke JJ, Increasing and Decreasing the Ultrastability of Bacterial Chemotaxis Core Signaling Complexes by Modifying Protein-Protein Contacts. Biochemistry 53, 5592–5600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slivka PF, Falke JJ, Isolated Bacterial Chemosensory Array Possesses Quasi- and Ultrastable Components: Functional Links between Array Stability, Cooperativity, and Order. Biochemistry 51, 10218–10228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borkovich KA, Simon MI, The dynamics of protein phosphorylation in bacterial chemotaxis. Cell 63, 1339–1348 (1990). [DOI] [PubMed] [Google Scholar]
- 60.Svergun DI, Determination of the regularization parameter in indirect-transform methods using perceptual criteria. Journal of Applied Crystallography 25, 495–503 (1992). [Google Scholar]
- 61.Semenyuk AV, Svergun DI, GNOM - A program pacakge for small-angle scattering data processing. Journal of Applied Crystallography 24, 537–540 (1991). [Google Scholar]
- 62.Hopkins JB, Gillilan RE, Skou S, BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. Journal of Applied Crystallography 50, 1545–1553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI, PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography 36, 1277–1282 (2003). [Google Scholar]
- 64.Rambo RP, Tainer JA, Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477-+ (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Franke D, Svergun DI, DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. Journal of Applied Crystallography 42, 342–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Volkov VV, Svergun DI, Uniqueness of ab initio shape determination in small-angle scattering. Journal of Applied Crystallography 36, 860–864 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A, FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Research 44, W424–W429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mendes ML, Fischer L, Chen ZA, Barbon M, O’Reilly FJ, Giese SH, Bohlke-Schneider M, Belsom A, Dau T, Combe CW, Graham M, Eisele MR, Baumeister W, Speck C, Rappsilber J, An integrated workflow for crosslinking mass spectrometry. Molecular Systems Biology 15, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borbat PP, Crepeau RH, Freed JH, Multifrequency two-dimensional Fourier transform ESR: an X/Ku-band spectrometer. J Magn Reson 127, 155–167 (1997). [DOI] [PubMed] [Google Scholar]
- 70.Borbat PP, Georgieva ER, Freed JH, Improved Sensitivity for Long-Distance Measurements in Biomolecules: Five-Pulse Double Electron-Electron Resonance. Journal of Physical Chemistry Letters 4, 170–175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borbat PP, Mchaourab HS, Freed JH, Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme. J Am Chem Soc 124, 5304–5314 (2002). [DOI] [PubMed] [Google Scholar]
- 72.Srivastava M, Georgieva ER, Freed JH, A New Wavelet Denoising Method for Experimental Time-Domain Signals: Pulsed Dipolar Electron Spin Resonance. Journal of Physical Chemistry A 121, 2452–2465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Srivastava M, Freed JH, Singular Value Decomposition Method To Determine Distance Distributions in Pulsed Dipolar Electron Spin Resonance: II. Estimating Uncertainty. Journal of Physical Chemistry A 123, 359–370 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Srivastava M, Freed JH, Singular Value Decomposition Method to Determine Distance Distributions in Pulsed Dipolar Electron Spin Resonance. Journal of Physical Chemistry Letters 8, 5648–5655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alexander N, Bortolus M, Al-Mestarihi A, McHaourab H, Meilerl J, De novo high-resolution protein structure determination from sparse spin-labeling EPR data. Structure 16, 181–195 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI, Structural characterization of flexible proteins using small-angle X-ray scattering. Journal of the American Chemical Society 129, 5656–5664 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
fig. S1. Preliminary purification and characterization of receptor foldons and their complexes.
fig. S2. Buffer conditions that encourage interactions among the TarFo, CheA, and CheW.
fig. S3. Properties of Tm14Fo complexes with Tm CheA and Tm CheW.
fig. S4. Autoradiographs of CheA autophosphorylation experiments from Fig. 1.
fig. S5. Properties of single-residue foldon variants.
fig. S6. SEC-SAXS data of the cross-linked foldon complex.
fig. S7. Inter-domain cross-links in free CheA.
fig. S8. Inter-domain cross-links in the receptor foldon complex.
fig. S9. PDS of Tm CheA E588C-R1 with CheW and TarFo.
fig. S10. Modeling of the foldon complex.
fig. S11. Summary of model restraints in Rosetta refinements.
table S1. Real space SAXS parameters generated from SEC-SAXS for foldon complexes.
table S2. List of PDS distance restraints for Rosetta modeling.