Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for an unprecedented global pandemic of COVID-19. Animal models are urgently needed to study the pathogenesis of COVID-19 and to screen vaccines and treatments. We show African green monkeys (AGMs) support robust SARS-CoV-2 replication and develop pronounced respiratory disease, which may more accurately reflect human COVID-19 cases than other nonhuman primate species. SARS-CoV-2 was detected in mucosal samples, including rectal swabs, as late as 15 days post-exposure. Marked inflammation and coagulopathy in blood and tissues were prominent features. Transcriptome analysis demonstrated stimulation of interferon and interleukin 6 pathways in bronchoalveolar lavage samples and repression of natural killer cell- and T cell-associated transcripts in peripheral blood. Despite a slight waning in antibody titers after primary challenge, enhanced antibody and cellular responses contributed to rapid clearance after re-challenge with an identical strain. These data support the utility of AGM for studying COVID-19 pathogenesis and testing medical countermeasures.
One Sentence Summary:
African Green monkeys infected with SARS-CoV-2 recapitulate pathological features of human COVID-19.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of coronavirus disease 2019 (COVID-19), emerged in Wuhan, China in late 2019 and rapidly swept the globe. As of October 12, 2020, over 37 million confirmed cases and 1 million deaths have been reported worldwide1. No licensed vaccines or therapeutics currently exist, although many clinical trials are underway. While clinical testing is ultimately needed to assess drug safety and efficacy, bypassing preclinical evaluation warrants caution due to the potential for disease enhancement2–4. A careful assessment in animal models could reveal possible immune complications elicited by vaccines and therapies before their release to the public. Moreover, animal models are critical to understanding aspects of pathogenesis and immunity that are not easily addressed or feasible in humans.
Several animal species including rodents, ferrets, and nonhuman primates (NHPs) were found to support SARS-CoV-2 replication and displayed mostly subclinical to mild illness post-challenge5–11. Syrian hamsters developed mild to moderate disease and pulmonary lesions that resolved within 2 weeks6,12. While each of these models has utility in the study of COVID-19, NHPs have the closest physiological resemblance to humans. This feature allows an accurate comparison of host responses to infection and enhances the predictive efficacy of medical countermeasures. Recently, studies evaluating the pathogenic potential of SARS-CoV-2 in cynomolgus and rhesus macaques were reported. Limited viral replication was observed in both models; rhesus macaques developed mild pneumonia and few clinical signs whereas disease in cynomolgus macaques was less pronounced8,10,11,13. These results suggest certain NHP species may serve as better models than others for coronavirus infections. For SARS-CoV, African green monkeys (AGMs) were found to support the highest level of viral replication followed by cynomolgus macaques and rhesus macaques when all three species were challenged in parallel14. Only AGMs challenged with SARS-CoV had marked replication in the lower respiratory tract in association with viral pneumonia consistent with human SARS. As SARS-CoV and SARS-CoV-2 share high genomic similarity and the same putative host receptor, angiotensin-converting enzyme 2 (ACE2)15,16, we reasoned AGMs might serve as a useful model for COVID-19.
Here, we exposed AGMs to low passage SARS-CoV-2 and evaluated their potential as a model for COVID-19. We demonstrate AGMs mimic several aspects of human disease including pronounced viral replication and pulmonary lesions, using a >2-fold lower dose of SARS-CoV-2 than has been employed in many rhesus and cynomolgus macaque studies. Transcriptomic analyses of bronchoalveolar lavage (BAL) and peripheral blood samples revealed AGMs exhibit similar immune profiles as human cases17,18. Moreover, our data show AGMs are protected from reinfection after re-challenge at 35 days post-exposure. Thus, the AGM model can be used to analyze the host immune response, conduct pathogenesis studies, and screen potential vaccines and therapeutics against COVID-19.
Results
Clinical disease in AGMs
In a narrowly focused study to examine acute pathogenesis and protection from back-challenge, we exposed six adult AGMs to 4.6 × 105 PFU of SARS-CoV-2 by combined intratracheal (i.t.) and intranasal (i.n.) routes. A cohort of three animals was euthanized at 5 dpi, while the remaining three animals were re-challenged via the same routes at 35 days post infection (dpi) (identical virus strain and dose). These group numbers are in line with previous SARS-CoV-2 NHP studies8–11,13,19,20. Back-challenged subjects were monitored for an additional 22 days. For each cohort, longitudinal blood and bronchoalveolar lavage (BAL) samples were collected throughout the study up until the respective study endpoint.
After primary challenge, AGMs experienced mild and varied clinical signs of disease. In 5/6 animals, monkeys exhibited decreased appetite compared to baseline (0 dpi), and a brief period of elevated body temperature suggestive of fever in 3/6 animals at 3-4 dpi (Table 1; Extended Data Fig. 1). A biphasic increase in partial CO2 pressures, indicative of hypercapnia, was noted in 4/6 animals following primary and back-challenge, but no overt changes in partial O2 pressures were noted. Transient lymphocytopenia and thrombocytopenia were observed in all AGMs, most prominently at 2-7 dpi. Markers for renal (BUN, CRE) and hepatic function (ALT, AST, ALP, GGT) remained mostly unchanged (< 2-fold increases); however, CRP, a marker of acute systemic inflammation, increased two to seven-fold in all animals 2-5 dpi (Table 1). No systemic increases in CRP were observed following back-challenge.
Table 1: Clinical findings of AGMs infected with SARS-CoV-2.
Days after SARS-CoV-2 challenge are in parentheses. Clinical findings following back challenge (d35/0) are in bold, with the day following primary challenge listed first and the day following back challenge listed second. All reported findings are in comparison to baseline (d0 for primary challenge, d35/0 for back challenge) values. Decreased appetite is defined as some food but not all food consumed from the previous day. Anorexia is defined as no food consumed from the previous day. Lymphocytopenia, monocytopenia, neutropenia, eosinopenia, basopenia, and thrombocytopenia are defined by a ≥ 35% drop in numbers of lymphocytes, monocytes, neutrophils, eosinophils, basophils, and platelets, respectively. Monocytosis, neutrophilia, eosinophilia, and basophilia are defined by a two-fold or greater increase in numbers of monocytes, neutrophils, eosinophils, or basophils, respectively. Hypoglycemia is defined by a ≥ 25% decrease in concentrations of glucose. Hypercapnia was defined as having a partial CO2 > 4 mmHg over d0 baseline values. (ALT) alanine aminotransferase, (GGT) gamma-glutamyl transferase, (CRE) creatinine, (CRP) C-reactive protein.
| Subject No. | Sex | Clinical illness | Clinical pathology |
|---|---|---|---|
| AGM-1 | F | Fever (d3), decreased appetite (d4,5). Subject survived to study endpoint (d5). | Monocytosis (d5); neutrophilia (d4); eosinophilia (d4); basophilia (d4); > 6-fold ↑ CRP (d4); > 1-fold ↑ CRP (d5) |
| AGM-2 | F | Decreased appetite (d1-5). Subject survived to study endpoint (d5). | Lymphocytopenia (d2-5); monocytopenia (d2, 3, 5); eosinopenia (d4); basopenia (d3, 4) thrombocytopenia (d4); > 1-fold ↑ in ALT (d3, 4); > 1-fold ↑ in GGT (d3-5); 1-fold ↑ in CRP (d3, 4) |
| AGM-3 | M | Fever (d3). Subject survived to study endpoint (d5). | Lymphocytopenia (d5); thrombocytopenia (d4);
monocytosis (d4); neutrophilia (d4); eosinophilia (d4); 1-fold ↑
in CRE (d4); > 4-fold ↑ in CRP (d4), hypercapnia (d3, 5) |
| AGM-4 | F | Fever (d4); decreased appetite (d1-10, 12, 13, 15-17, 20-24, 27, 29, 30, 33, 34, 36/1-39/4, 42/7-44/9), anorexia (d41/6). Subject survived to study endpoint (d57/22). | Lymphocytopenia (d2, 3); neutropenia (d2, 3, 5), thrombocytopenia (d2); neutrophilia (d4, 15); eosinophilia (d4, 7, 12, 15); monocytopenia (d21); eosinopenia (d2, 3, 38/3, 42/7); basopenia (d2, 3, 38/3, 40/5, 42/7); hypoglycemia (d7, 9, 12); > 6-fold ↑ CRP (d4); > 2-fold ↑ CRP (d5), hypercapnia (d7, 40/5, 42/7) |
| AGM-5 | M | Decreased appetite (d8, 10, 17, 22-24, 34, 36/1-39/4, ). Subject survived to study endpoint (d57/22). | Monocytopenia (2, 4, 9, 21, 28, 38/3, 49/14); neutropenia (d2, 28, 57/22); eosinopenia (d2, 3, 21, 28, 33, 40/5, 49/14); basopenia (d2-5, 7, 9, 21, 28, 33, 49/14); thrombocytopenia (d33); > 2-fold ↑ CRP (d4), hypoglycemia (d57/22), hypercapnia (d7, 15, 28, 38/3) |
| AGM-6 | F | Decreased appetite (d1-3, 5-10, 13-17, 22-24, 36/1); nasal exudate (d40), Subject survived to study endpoint (d57/22). | Lymphocytopenia (d7, 57/22); monocytosis (d12); eosinopenia (d2-5, 12, 21, 33); basopenia (2-5, 7, 9, 12, 15, 21, 28, 33); basophilia (d49/14); > 1-fold ↑ in CRP (d4), hypercapnia (d7, 12, 35/0, 40/5, 42/7, 57/22) |
Viral loads
Viral loads in blood, mucosal swabs, and BAL fluid were quantified following primary and secondary exposure by RT-qPCR (Fig. 1a,c,e,g) and plaque titration (Fig. 1b,d,f,h). Following primary challenge, all subjects had detectable quantities of viral RNA (vRNA) and infectious virus in nasal secretions at 2 dpi, with infectious viral titers ranging from ~2-4 log PFU/mL (Fig. 1a,b). Nasal shedding of vRNA occurred up to 9 dpi in AGM-4 and 12 dpi in AGM-5. Following secondary exposure to SARS-CoV-2, vRNA was detected in nasal swabs of AGM-4 at 38 dpi and AGM-5 at 38 and 40 dpi (3 and 5 days after re-challenge). Both vRNA and infectious virus were detected in oral swabs of three animals (AGM-2, AGM-5, AGM-6) 3 dpi, and persisted in AGM-5 up to 7 dpi (Fig. 1c,d). Infectious virus was only detected in the rectal swab of a single animal (AGM-3) 3-5 dpi; however, vRNA was detected in AGM-4 and AGM-5 rectal samples 12 and 15 dpi, respectively (Fig. 1e,f). Both vRNA and infectious virus was present in BAL fluid of all subjects 3-7 dpi (Fig. 1g,h). Neither vRNA nor infectious virus was detected in whole blood or plasma, respectively, indicating a lack of circulating cell-associated or free virus in the peripheral blood (data not shown). No infectious virus was evident in nasal samples after secondary exposure, nor was vRNA or infectious virus found in mucosal swabs or BAL fluid, (Fig. 1b–h), suggesting AGMs were protected from reinfection following SARS-CoV-2 back-challenge.
Fig. 1: Detection of SARS-CoV-2 vRNA and infectious virus in mucosal swabs and BAL fluid.
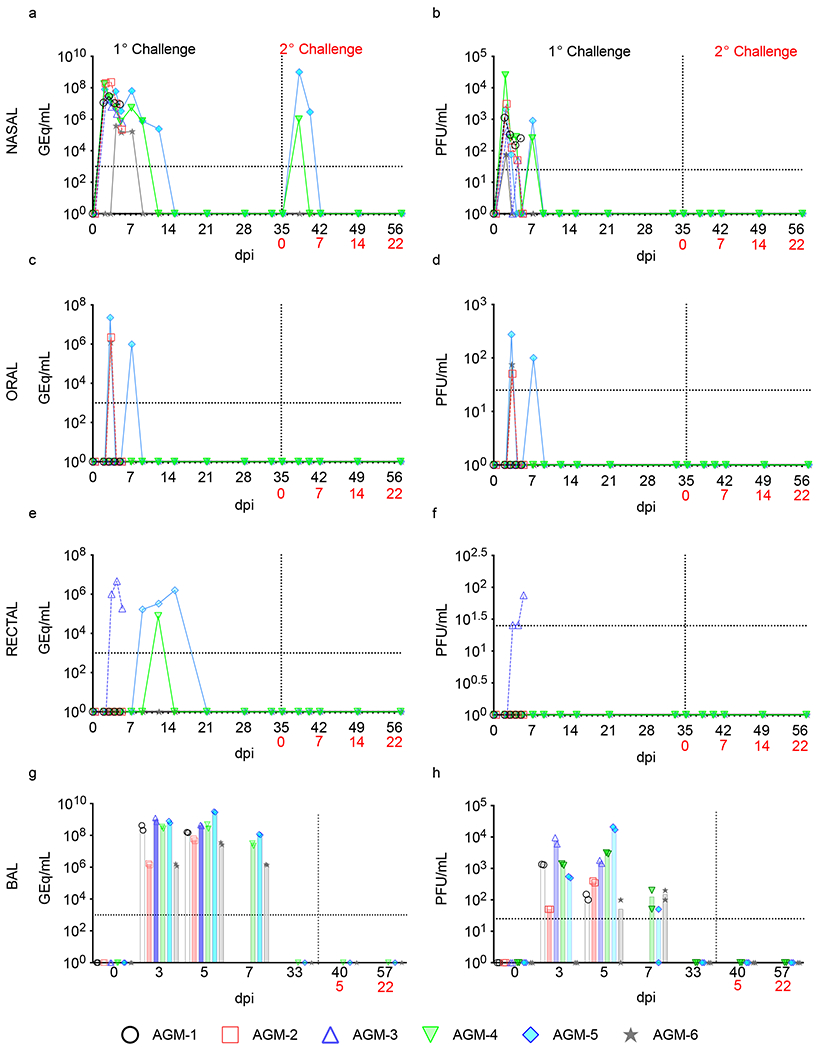
a,b) nasal swabs, c,d) oral swabs, e,f) rectal swabs, g,h) and BAL fluid from AGMs infected with SARS-CoV-2 were subjected to RT-qPCR (a,c,e,g) or plaque titration (b,d,f,h). For all graphs, dashed lines and open symbols (AGM-1, AGM-2, AGM-3) indicate AGMs sacrificed 5 dpi (n=3); solid lines (AGM-4, AGM-5, AGM-6) indicate AGMs held to 57 dpi or 22 days after re-challenge (n=3). Data plotted is each duplicate RT-qPCR reaction or well per subject/sample in a single experiment. Red text indicates day of back-challenge.
Tissues collected at necropsy from AGMs euthanized at day 5 dpi or 57/22 dpi were also examined for viral loads. For the cohort euthanized at 5 dpi, vRNA and infectious virus abundance was highest in tissues from the upper and lower respiratory tracts (Extended Data Fig. 2a,b). Substantial quantities of vRNA in some or all animals were detected in major organs, including lymphoid tissue (~104-106 GEq/g tissue), the heart (~105 GEq/g tissue), the digestive tract (~103-107 GEq/g tissue), and immunologically privileged sites such as the CNS, eyes, and urogenital tract (~104-106 GEq/g tissue). For the re-challenged cohort, AGM-4 had detectable vRNA (~103-107 GEq/g tissue) in inguinal and mesenteric lymph node, spleen, heart, and ileocecal junction tissues; AGM-5 had detectable vRNA (~105-106 GEq/g tissue) in liver, brain stem, ileum, ileocecal junction, eye, conjunctiva, and reproductive tissue; and AGM-6 had detectable vRNA (~106 GEq/g tissue) in duodenum tissue. However, considerable reductions of vRNA in specific tissues of the back-challenged cohort were noted, particularly in the upper and lower respiratory tracts. While infectious titers were prominent in the lungs of subjects euthanized at 5 dpi, no infectious virus was found in the lungs, or any other collected tissues, of re-challenged animals. Our data indicate SARS-CoV-2 replicates abundantly in the respiratory tract of AGMs, but viral loads are reduced in re-challenged subjects.
Gross and histological findings
Necropsy revealed varying degrees of pulmonary consolidation with hyperemia and hemorrhage in the lungs. (Fig. 2a–f). The lung from a historical SARS-CoV-2-negative AGM was included for comparison that was not subjected to BAL procedures (Fig. 2g). AGMs at 5 dpi had marked to severe locally extensive pulmonary lesions (Fig. 2a–c), whereas AGMs at 22 days post back-challenge had mild to moderate locally extensive pulmonary lesions (Fig. 2d–f). In all AGMs, the most severe lesions were located in dorsal aspects of the lower lung lobes. A board-certified veterinary pathologist approximated lesion severity for each lung lobe. Average lung severity scores were reduced in back-challenged AGMs versus animals sacrificed at 5 dpi (two-tailed t-test; p = 0.02; Supplementary Table 1). Thoracic radiographs taken on −1, 2, 3, 4, 5 dpi were inconclusive (Extended Data Fig. 3).
Fig. 2: Pulmonary gross changes in AGMs infected with SARS-CoV-2.

Dorsal view of lungs from a) AGM-1, b) AGM-2, c) AGM-3. a-c) subjects euthanized at 5 dpi exhibited marked locally extensive pulmonary consolidation with hyperemia and hemorrhage (circled regions). Dorsal view of lungs from d) AGM-4, e) AGM-5 & f) AGM-6. d-f) subjects euthanized at 57 dpi (22 days post re-challenge) exhibited mild locally extensive pulmonary consolidation with hyperemia and hemorrhage (circled regions). Dorsal view of a normal lung with no significant lesions from a g) SARS-CoV-2-negative AGM.
Histologically, all three AGMs euthanized at 5 dpi developed varying degrees of multifocal pulmonary lesions suggestive of inflammatory processes directly associated with the congestion and hemorrhage noted on gross examination. In the most severely affected animal (AGM-3), histologic features of acute bronchointerstitial pneumonia (Fig. 3a) included neutrophilic inflammation concentrated at terminal bronchioles in association with macrophages, numerous multinucleated giant cells (MNGCs) and syncytial cells that were positive by immunohistochemistry (IHC) for cytokeratin (an epithelial cell marker) (Fig. 4b and 4b inset). Continuous with the terminal bronchiolitis was evidence of focal alveolar damage with scant hyaline membrane formation (Fig 4e and 4e inset), type II pneumocyte hyperplasia, and flooding of alveolar spaces with pulmonary edema, hemorrhage and fibrin (Fig. 3c and 3c inset). Rarely, exuberant reparative lesions of collagenous tissue proliferations were noted protruding within the terminal airways reminiscent of early bronchiolitis obliterans organizing pneumonia (BOOP)-like lesions (Fig. 4d, 4d inset)21. Interstitial pneumonia was prominent in the lesser-affected regions of the lung along with congestion, increased numbers of alveolar macrophages, and microthrombi within alveolar capillaries (Fig. 3d, 3f). Modest amounts of immature loose collagen also diffusely expanded alveolar septae were observed and are highlighted with trichrome stain (Fig. 4h). Ulcerative tracheobronchitis was present and characterized by multifocal extensive epithelial erosion associated with hemorrhage, fibrin accumulation, and infiltrating mixed inflammatory cells.
Fig. 3: Pulmonary histologic changes in AGMs infected with SARS-CoV-2.
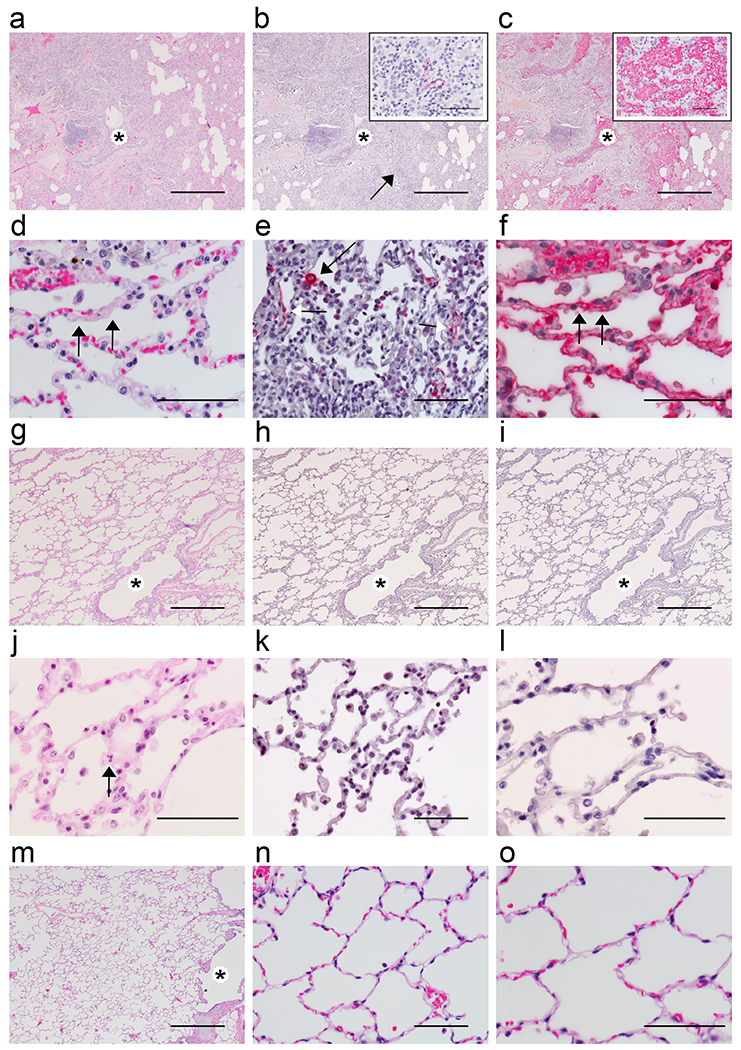
a-c) Low magnification of serial sections of left lower lung lobe of AGM-3. a) Locally extensive, marked, acute bronchiolitis (*) and interstitial pneumonia. b) SARS-CoV-2 IHC positive pneumocytes (arrow indicating the 60x inset) colocalized with bronchiolitis (*). c) Flooded alveolar sacs with fibrin (inset at 40x) that colocalized with acute bronchiolitis (*). Higher magnification of lung alveoli images of representative AGMs 5 dpi with SARS-CoV-2. d) interstitial pneumonia with fibrin microthrombi within alveolar capillaries (arrows). e) SARS-CoV-2 positive labeling of type I (white arrow) and type II (black arrow) pneumocytes. f) Serial section of tissue with positive fibrin labeling within alveolar capillaries (arrows). Low magnification of serial sections of right lower lung lobe of AGM-6 with g) mild interstitial pneumonia and patent bronchioles (*). h) no immunolabeling for anti-SARS-CoV-2 antigen. i) no immunolabeling for anti-fibrin antigen. Higher magnification of lung alveoli images of AGM-4. j) mild interstitial pneumonia, neutrophil indicated (arrow). k) no immunolabeling for anti-SARS-CoV-2 antigen. l) no immunolabeling for anti-fibrin antigen. SARS-CoV-2 negative control AGM lung at low magnification m) with patent bronchioles (*) and high magnification. n,o) with no significant lesions. H&E staining (a, d, g, j, m, n, & o), IHC labeling for anti-SARS-CoV-2 antigen (red) (b, e, h & k), IHC labeling for anti-fibrin antigen (red) (c, f, i & l). Images captured at 4x (a, b, c, g, h, i, & m), 40x (e, k, & n), 60x (d, f, j, l, & o). Scale bar 500um (a, b, c, g, h, i, & m) and 50um (b inset, c inset, d, e, f, j, k, l, n, & o).
Fig. 4: Additional histologic changes in AGMs infected with SARS-CoV-2.
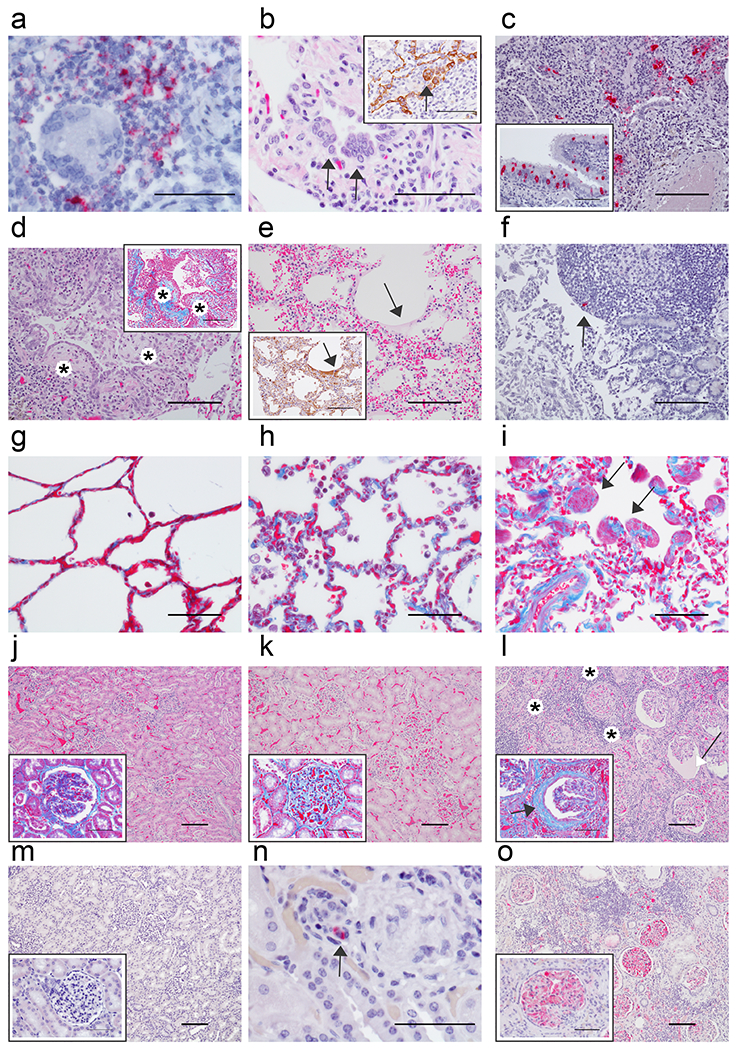
Pulmonary lesions in representative AGMs 5 dpi (a-e) and immunohistochemistry (IHC) of the (f) duodenum. a) Genomic SARS-CoV-2 RNA (red) detected by in situ hybridization in mononuclear cells near a multinucleated giant cell associated with acute bronchiolitis, 60x. b) Syncytial cells (arrows) within a terminal bronchiole, 60x and inset with pan-cytokeratin IHC positive syncytial cell indicating epithelial origin (arrow) within terminal bronchiole, 60x. c) SARS-CoV-2 positive IHC labeling (red) associated with acute bronchiolitis, 20x and in the inset SARS-CoV-2 IHC positive respiratory epithelium of the bronchus, 40x. d) Terminal bronchioles with multiple luminal protrusions of loose collagen covered by respiratory epithelium that is reminiscent of early formation of bronchiolitis obliterans organizing pneumonia (BOOP)-like lesions (*), 20x and inset serial section of tissue stained with trichrome highlighting immature collagen (*) (blue) 20x. e) Loss of alveolar architecture, marked expansion of septa and formation of faint hyaline membranes (arrow), 20x and inset IHC positive pan cytokeratin (brown) of hyaline membranes from serial section of tissue (arrow), 20x. f) IHC SARS-CoV-2 positive (red) mononuclear cell within the peyer’s patches of the duodenum (arrow), 20x. g) Trichrome stain of AGM alveolar septate basement membrane (blue) from a SARS-CoV-2 naïve AGM, 40x. h) Trichrome stain of alveolar septate with collagenous expansion (blue) from a SARS-CoV-2 AGM 5 dpi, 40x. i) and AGM 57 dpi and focal smooth muscle hyperplasia (arrows), 40x. j-o) Comparison of kidneys from naive AGMs, AGMs 5 dpi, and 57 dpi SARS-CoV-2. j) Renal congestion with NSL in naïve SARS-CoV-2 AGM, 10x and higher magnification in the inset of glomerulus with trichrome stain with no significant findings (NSF), 40x. k) Renal congestion with NSF in AGM 5dpi, 10x and higher magnification in the inset of glomerulus with trichrome stain with mild glomerular fibrosis, 40x. l) Marked renal interstitial lymphocytic inflammation (*) with glomerulopathy and expanded bowman’s space (white arrow) in AGM 57 dpi, 10x and higher magnification in the inset of glomerulus with trichrome stain with interstitial inflammation (*), glomerular fibrosis (blue), and marked periglomerular fibrosis (black arrow), 40x. m) Fibrin IHC negative AGM kidney 10x and higher magnification of fibrin negative glomerulus, 40x. n) IHC SARS-CoV-2 positive (red) mononuclear cell (arrow) within renal interstium of AGM 5 dpi, 60x. o) Fibrin IHC positive (red) multifocal within glomerular capillaries and renal interstitium of AGM 57 dpi, 10x and high magnification of fibrin positive glomerulus, 40x. H&E staining (b, d, e, j, k, & l), IHC labeling for anti-SARS-CoV-2 antigen (red) (c, c inset, f, & n), IHC labeling for anti-fibrin antigen (red) (m, m inset, o & o inset), IHC labeling for anti-pan cytokeratin (brown) (b inset and e inset), SARS-CoV-2 in situ hybridization (a), Trichrome (d inset, g, h, i, j inset, k inset, & l inset). Images captured at 10x (j, k, l, m & o), 20x (c, d, d inset, e, e inset & f), 40x (c inset, g, h, i, j inset, k inset, l inset, m inset & o inset), 60x (a, b, b inset, & n). Scale bar 100um (c, d, d inset, e, e inset, f, j, k, l, m & o) 50um (a, b, b inset, c inset, g, h, i, j inset, k inset, l inset, m inset, n, & o inset).
In the lesser-affected monkeys (AGM-1 AGM-2), pulmonary lesions lacked acute erosive inflammation within the trachea and bronchi; however, interstitial pneumonia with rare MNGCs, lymphocytic perivascular cuffs, and mild lymphocytic tracheobronchitis were present. Colocalization of SARS-CoV-2 antigen with pulmonary lesions presented as positive immunohistochemical labeling within the cytoplasm of respiratory epithelium of the bronchus, alveolar macrophages, and type I and type II pneumocytes (Fig. 3b, 3e, 4c, 4c inset). Genomic SARS-CoV-2 RNA was detected by in situ hybridization in pneumocytes and was associated with alveolar macrophages within acute inflammation centered on terminal bronchioles (Fig. 4a). Although, we did not observe overwhelming damage to pneumocytes from SARS-CoV-2 infection lacking inflammatory infiltrates in the primary tissues at the time of necropsy, it would be premature for us to wholly rule out that the virus as benign. There is likely a combination of events in which the virus, host inflammatory response, and host reparative/healing response are responsible for lesions noted early and later in the disease course.
Accordingly, polymerized fibrin was abundant and present within bronchial lumen associated with the acute inflammation at sites of epithelial erosion, flooding alveolar spaces associated with alveolar damage, within inflamed alveolar walls associated with microthrombi (Fig. 3c, 3f), and rarely along the pleural surface. Fibrin was prominent within large and small caliber vessels throughout the representative section of lung, but was not associated with any obvious adherent thrombus. Additional findings included positive IHC for SARS-CoV-2 antigen in rare mononuclear cells within the Peyer’s patches of the duodenum (Fig 4f) and associated with granulomatous foci within the renal interstitium (Fig. 4n). All other major organs were unremarkable.
To ascertain whether re-challenged AGMs were protected from pulmonary damage, we next performed histological staining and immunohistochemistry on tissues from this cohort. Twenty-two days after back-challenge, lesions indicative of previous infection with SARS-CoV-2 were evidenced as moderate interstitial pneumonia with lymphohistiocytic inflammation, congestion, increased numbers of alveolar macrophages, and rarely focal alveolar smooth muscle hypertrophy (Fig. 3g, 3j, 4i). Chronic ulcerative tracheobronchitis was rarely present and characterized by focal ulcerated respiratory epithelium with associated fibrin and inflammation. Multifocally, immature loose collagen diffusely expanded alveolar septae were present and are highlighted with trichrome stain (Fig. 4i). No significant immunolabeling for fibrin was detected in the alveoli of the examined lung sections (Fig 3i, 3l) and no immunolabeling for SARS-CoV-2 antigen was observed in the lung (Fig 3h, 3k) or any other examined organ.
In the liver, mild chronic inflammation and intravascular organizing fibrin was noted and characterized by mild sinusoidal leukocytosis, random lymphocytic infiltrates, congestion and organizing intravascular fibrin. In the kidney, all three subjects had lymphocytic interstitial nephritis and two of three monkeys (AGM-5, AGM-6) had evidence of chronic bilateral glomerulonephritis with hyalinzation of some glomeruli, fibrin accumulation within glomerular tufts, glomerular tuft atrophy with dilation of Bowman’s capsule, and periglomerular fibrosis (Fig 4l). Marked regions of interstitial lymphohistiocytic inflammation associated with fibrosis and contracture of the renal cortex were prominent. Thus, lung, liver and kidney tissue of re-challenged AGM seem to exhibit chronic reparative changes.
Humoral and cellular responses
Assessment of humoral responses in AGMs revealed five of six animals seroconverted, including all three animals held past 5 dpi, with the earliest detection of anti-SARS-CoV-2 IgG or neutralizing titers (50% plaque reduction values) occurring at 5 dpi (Fig. 5a). Total virus-specific IgG titers peaked at 15-21 dpi (1:200-1:6,400) in the back-challenged group and were followed by a period of slightly waning antibody titers in 2 of 3 animals that were amplified 1-3 weeks after re-challenge (1:12,800-1:25,600) (Fig. 5a). Notably, IgG titers directed at the nucleoprotein, full-length spike (S1+S2 ectodomain), and receptor-binding domain (RBD) appeared later than total IgG and followed a similar trend of waxing and waning (Fig. 5b–d). RBD-specific IgA in all three remaining monkeys was not detected until 12 dpi and remained relatively low (1:200-1:800) until 1-2 weeks after back-challenge, wherein titers increased to 1:1,1600-1:6400 (Fig. 5e). Interestingly, the appearance of RBD-specific IgG corresponded with neutralizing titers at 5 (AGM-1, AGM-3) and 7 (AGM-4) dpi, emphasizing the importance of the RBD domain in antibody-mediated virus neutralization (Fig. 5c,f). Neutralizing titers ranged from 1:8-1:32 after primary challenge and peaked at 1:512-1:1,024 post back-challenge (Fig. 5f). Strong anamnestic immune responses, coupled with the presence of vRNA in tissues and nasal swabs after secondary exposure to SARS-CoV-2, indicated a lack of sterilizing immunity. Similarly, cellular responses detected by ELISPOT (interferon-γ+ interleukin-2− (IFN-γ+ IL-2−) and IFN-γ+ IL-2+ cells) of peripheral blood mononuclear cells (PBMCs) to SARS-CoV-2 peptide pools increased over the course of the study relative to pre-challenge baselines (0 dpi) and were augmented upon re-challenge (Fig. 5g). By 21 dpi, mean spike-specific cellular responses were higher than nucleoprotein-specific responses, similar to human cases22. Unsurprisingly, IFN-γ− IL-2+ cells were not detected as they are understood as rare in circulation23. Together, these results suggest antibody and cellular responses slightly wane after primary challenge, but are enhanced after rechallenge to potentially protect animals from reinfection.
Fig. 5: Humoral and cellular responses in SARS-CoV-2-infected AGMs.
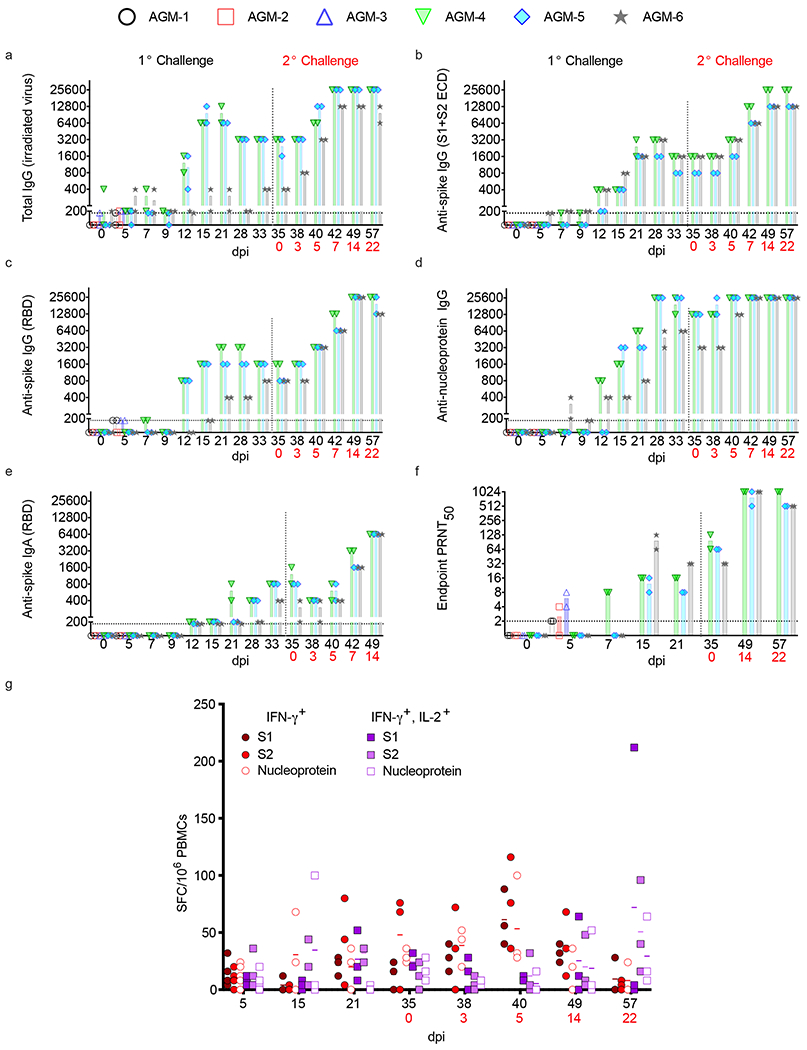
a-e) Anti-SARS-CoV-2 IgG binding titers. f) Neutralizing titers in AGM sera. PRNT50 values indicate 50% neutralization compared to virus control plates. All plaque counts were calculated from duplicate wells at each dilution. g) Peripheral blood mononuclear cells (PBMCs) were stimulated with 1 μg/ml of peptide pools spanning the SARS-CoV-2 nucleoprotein or spike (split into 2 pools). Unstimulated cells and PBMCs stimulated with pokeweed mitogen (PWM) served as negative and positive controls, respectively (data not shown). The spike pools contained 158 or 157 15mer peptides with 11 amino acid overlaps and the N pool contained 13mer peptides with 10 amino acid overlaps. Reported values were calculated by subtracting the number of spot-forming cells (SFCs) in a given unstimulated sample from its respective stimulated counterpart at the corresponding dpi. Values were subtracted from a pre-challenge baseline (0 dpi). Data plotted is each duplicate reciprocal dilution titer per subject in a single experiment. Red text indicates day of back-challenge.
BAL and blood transcriptome analyses
To unravel the AGM host immune response to SARS-CoV-2 infection, we temporally tracked transcriptional changes in blood and BAL samples during the acute phase of disease. For this analysis, blood and BAL RNA samples were assayed in parallel at 3 (n=6), 5 (n=6), and 7 (n=3) dpi to discriminate between localized and systemic immune responses to infection; additionally, we analyzed blood samples at an early (2 dpi; n=6) and convalescent time point (21 dpi; n=3).
Examination of normalized sample populations by principal component analysis (PCA) revealed time-dependent expression changes in BAL and blood samples with minimal dimensional separation between 3 and 5 dpi samples (Extended Data Fig. 4a,b). Blood 2 dpi samples mostly clustered with 3 and 5 dpi samples, whereas blood 7 dpi samples mostly clustered with pre-challenge and convalescent (21 dpi) samples.
SARS-CoV-2-specific gene products (envelope, membrane, nucleocapsid, orf1ab, orf3a, orf7a, orf8, spike) were also quantified in BAL samples. Copious reads of SARS-CoV-2-specific probes were detected in BAL samples at 3 and 5 dpi, but no significant abundance in any particular gene product was recognized at any time point (Extended Data Fig 4c).
Analysis of global changes revealed upregulated differentially expressed (DE) RNAs shared between BAL and blood samples were involved in interferon signaling (MX1, MX2, IFIT1, IFIT3, IFI44), RIG-I/MDA-5 pattern recognition (DDX58, IFIH1), and RNAseL signaling (OAS1, OAS2, OAS3, OASL) (Fig. 6a–f). Robust interferon signaling was observed in the blood as early as 2 dpi (Extended Data Fig. 5a,b). In contrast, shared repressed DE transcripts were implicated in major histocompatibility complex class II (MHC class II)-based antigen presentation (HLA-DQA1, HLA-DQB1) (Fig. 6a–c,f).
Fig. 6: Volcano plots indicating transcriptional changes in BAL and blood samples at selected time points.
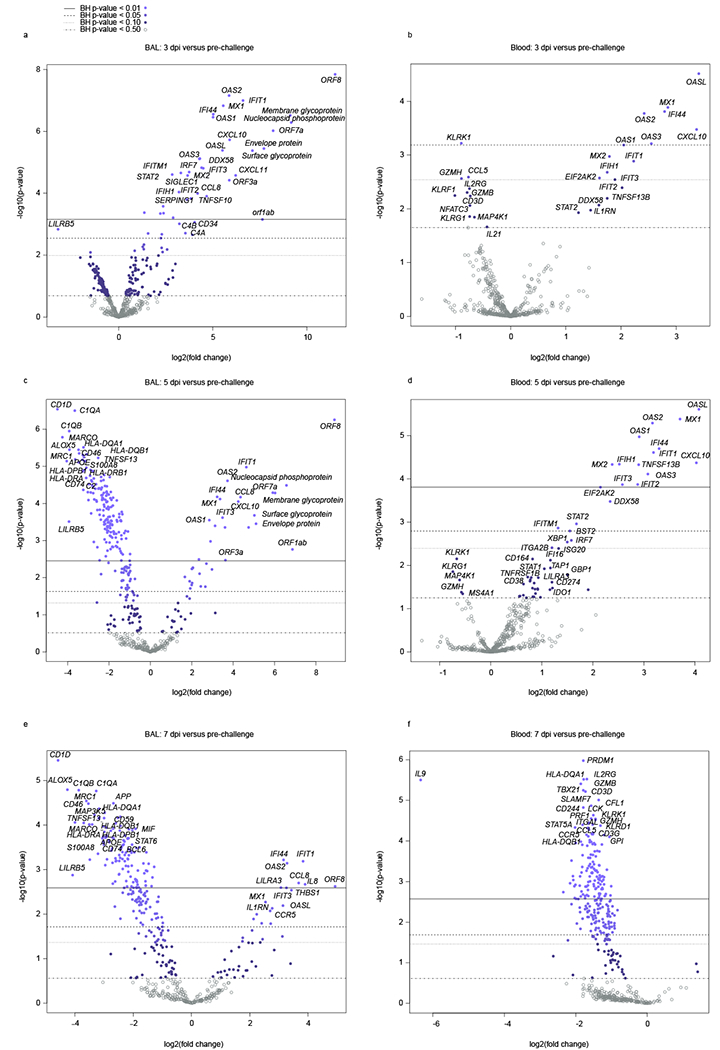
RNA expression changes were evaluated in a,c,e) BAL and b,d,f) blood samples from SARS-CoV-2-infected AGMs at 3, 5, and 7 dpi. Displayed are the mean −log10 (p-values) and log2 fold changes for each mRNA target from n=6 biological replicates in a single experiment for a-d) 3 and 5 dpi time points and e,f) n=3 biological replicates for the 7 dpi time point relative to a pre-challenge baseline (−8 dpi). Horizontal lines within each plot indicate adjusted p-value thresholds. Targets highlighted in blue indicate FDR adjusted p-values < 0.10. A Benjamini-Hochberg test was employed to derive FDR-adjusted p-values.
In BAL samples, expression of leukocyte immunoglobulin like receptor B5 (LILRB5), encoding a member of the leukocyte immunoglobulin-like receptor (LIR) family, was consistently decreased at 3, 5, and 7 dpi (Fig. 6a,c,e). Additional downregulated BAL DE mRNAs at 5 and 7 dpi are involved in lipid antigen presentation (CD1D), the complement system (C1QA, C1QB), calgranulin-mediated calcium sensing (S100A8, S100A9), or MHC class II antigen presentation (HLA-DPBl, HLA-DRB1, HLA-DRA). Increased expression of IL8 and CCR5 transcripts were measured in BAL samples at 7 dpi pointing to inflammation in the lung mediated by macrophages and/or neutrophil populations.
In the blood, upregulated immune signatures at 5 dpi suggested plasma cell differentiation (XBP1), lymphocyte activation (CD38), as well as immune tolerance/T cell exhaustion (CD274 (PD-1), IDO1). Repressed transcripts at 3 or 5 dpi pointed to a reduction in mitogen-activated protein kinase signaling (MAP4K1), NK cells (KLRK1, KLRF1, KLRG1), T cells (CD3D, IL2RG), NK and T cell cytotoxicity (GZMH, GZMB, GZMK, IL21), and T helper 1 (Th1) cells (TBX21) (Fig. 6b,d). The majority of DE mRNAs were downregulated in BAL and blood samples at 7 dpi (Fig. 6e,f), as well as the convalescent stage (Extended Data Fig. 5a–c). These data potentially reflect the onset of disease resolution in the lung and peripheral blood compartments at 7 dpi. This observation is consistent with a reduction in viral titers and SARS-CoV-2-specific gene transcripts in BAL samples at this time point.
Functional enrichment of DE transcripts was next used to decipher signaling networks associated with infection in the lungs and blood. For this analysis, each BAL or blood dataset was filtered by dpi (3, 5, 7). At all-time points other than the 7 dpi blood group, positive z-scores correlated with induction of canonical pathways related to stimulation of immunity and clearance of viral infections (Fig. 7a). Although a slight reduction in positive z-scores in BAL samples at 7 dpi was detected, these pathways were significantly downregulated (negative z-scores) in the blood at 7 dpi, again indicating rapid resolution of the acute phase of disease in the blood compartment. At 3 and 5 dpi, upstream regulators and causal networks predicted induction of interferon, tumor necrosis factor (TNF) and Toll-like receptor (TLR) signaling, which was resolved at 7 dpi in the blood. Causal networks associated with negative regulation of innate immunity (CACTIN) and ubiquitination (UBE3C, RNF216) were consistently downregulated in both BAL and blood samples at 3 and 5 dpi. To compare BAL and blood responses early in infection, we mapped JAK/STAT signaling pathways for each dataset. Unlike BAL samples, evidence of negative regulation of type I IFN mediated by downregulation of IFNAR2 signaling was evident in blood as early as 3 dpi (Fig. 7b). At 5 dpi, network maps depicting the relatedness of prominent gene clusters indicated sustained activation of innate immunity in BAL samples with evidence of regulation of cytokine production. In contrast, blood networks were predominantly involved in adaptive immunity or regulation of innate immunity (Fig. 7c). At 7 dpi, predicted activation of interferon alpha was noted in the BAL dataset, whereas predicted inhibition of interferon alpha was strongly apparent in the blood dataset (Fig. 7d). Collectively, these results suggest clearance of virus in the blood and lungs at 7 dpi with rapid resolution of immune responses in the blood.
Fig. 7: Functional enrichment of DE mRNAs in SARS-CoV-2-infected AGM BAL and blood samples.
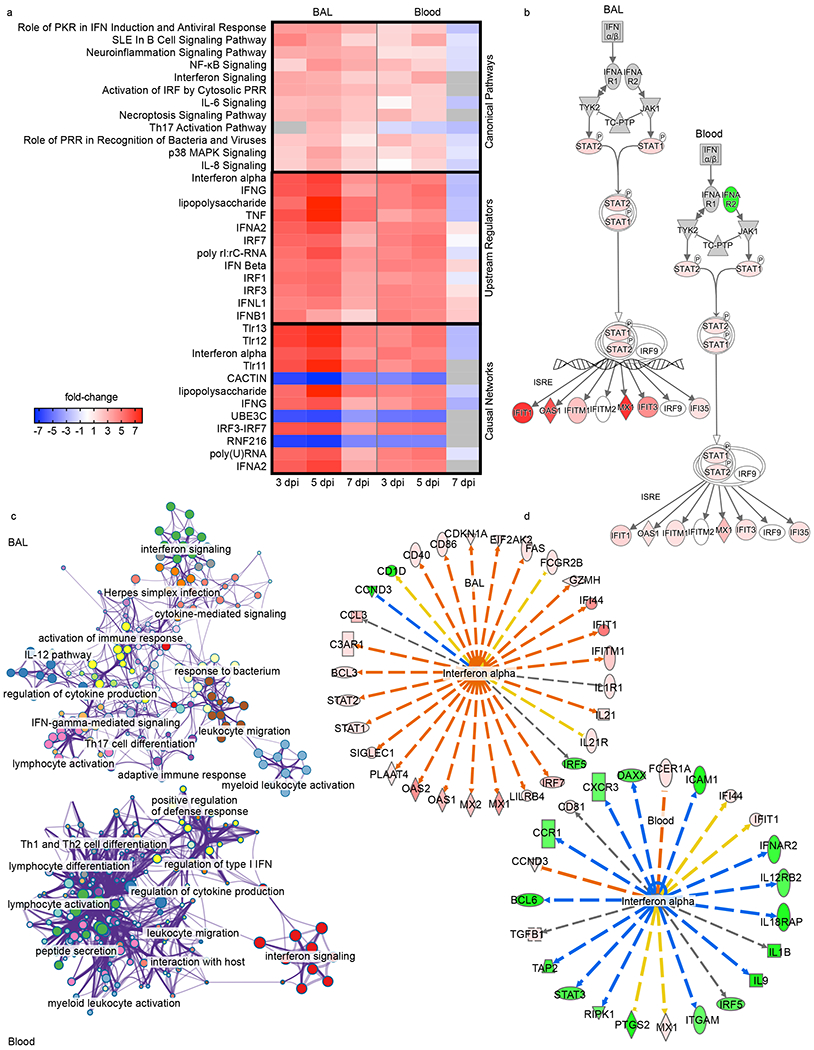
All data were normalized against a pre-challenge baseline (−8 dpi) for each time point and sample type. A higher degree of transparency denotes values that are less extreme in the dataset. Unless otherwise noted, red indicates an increased measurement, green indicates a decreased measurement. a) Heatmap depicting the most highly upregulated and downregulated canonical pathways, upstream regulators, and causal networks in BAL and blood samples at each time point; red indicates high expression (z-scores); blue indicates low expression; white indicates similar expression; gray indicates insufficient transcripts mapping to the indicated pathway. b) Jak/STAT signaling pathways illustrating differential responses in BAL and blood datasets at 3 dpi. c) Network maps depicting gene networks associated with SARS-CoV-2 infection in each dataset. d) Differential regulation of interferon alpha in BAL and blood samples at 7 dpi; orange lines indicate predicted activation, blue lines indicate predicted inhibition, yellow lines indicate findings inconsistent with state of downstream molecule, gray lines indicate effect not predicted.
Systemic cytokine and fibrinogen concentrations
Serum concentrations of pro-inflammatory IL-6, IL-8, monocyte chemoattractant protein (MCP-1), IP-10, IL-12, and immunoregulatory IL-10 largely peaked at 2 dpi for most animals corresponding to subsequent recruitment of monocytes and neutrophils in the blood (Extended Data Fig. 6a–f; Table 1). Although IFN-related transcripts were highly expressed in blood and BAL samples (Fig. 6a–f), animals only secreted modest amounts of IFN-beta (Extended Data Fig. 6g). As IL-6 is a main regulator of acute phase fibrinogen synthesis, and elevated fibrinogen and other coagulation abnormalities are thought to correlate with disease severity in hospitalized patients24,25, we also measured the abundance of this clotting factor. Circulating fibrinogen concentrations surged in 4 of 6 monkeys at 4 dpi indicating potential coagulation abnormalities in these animals (Extended Data Fig. 6h). This observed coagulopathy aligns with our gross pathology findings of substantial hemorrhage in the lung of monkeys euthanized at 5 dpi.
Discussion
AGMs serve as disease models for several respiratory pathogens14,28–31. Guided by previous data suggesting that AGMs were a superior model for SARS-CoV in terms of exemplifying human viral replication kinetics and histopathological features14, we investigated their suitability as a model for SARS-CoV-2 infection. In our study, we show AGMs develop mild, moderate, or severe pulmonary lesions following SARS-CoV-2 infection. AGMs challenged with SARS-CoV-2 did not develop debilitating clinical illness; however, these animals exhibited an impressive array of disease features observed in humans. A low fraction of NHPs may develop severe disease similar to SARS-CoV-2-infected humans. The heterogeneous responses of AGMs are a chief attribute that makes them an attractive model compared to inbred, more genetically homogenous, rodent models. Furthermore, NHPs have the closest physiological resemblance to humans allowing an accurate comparison of host responses to infection and enhancing the predictive efficacy of medical countermeasures.
Other NHP species, including rhesus and cynomolgus macaques, were evaluated as animal models for COVID-19, with rhesus macaques more closely exhibiting human symptoms9–11,26. Data on these NHP models are difficult to interpret, as many early studies used high challenge doses. The limited viral replication reported in these studies may also represent artifact at the challenge and sampling site. Moreover, data from these studies do not reflect the high viral replication and/or shedding kinetics observed in humans10,11,19. A recent study evaluating the effect of various challenge doses in rhesus macaques down to 104 total PFU per animal was the first model to demonstrate clear evidence of replication in nasal swabs by viral sub-genomic mRNA (sgmRNA) content. However, no demonstration of infectious virus was reported in respiratory tissues of these animals. sgmRNA content did not appear challenge dose-dependent, nor was there evidence of clinical disease observed in humans (fever, weight loss, respiratory distress). However, immune responses to viral challenge were apparent13. Given the lack of infectious virus in the lower respiratory tract and only mild disease presentation, others have suggested the rhesus macaque model may be more appropriate for studying vaccination or therapeutic responses against SARS-CoV-2 rather than modeling human COVID-1927.
We demonstrate AGMs recapitulate human infection and recovery and rapidly cleared infection after re-challenge. Transient lymphopenia and thrombocytopenia, and elevated serum markers associated with systemic inflammation, were evident. Fever is estimated in ~78% of human patients presenting with COVID-1932. Using surgically implanted temperature data loggers, elevated body temperatures were detected in three animals at 3-4 dpi, indicative of mild transient fever. To date, only one study reported elevated core temperatures in SARS-CoV-2-infected NHPs immediately following challenge10. The rapid induction of fever in this study clouds interpretation to whether an acute response to inoculum or a bona fide response to viral replication was reported. Transient fever may be a feature of disease in other NHP models of COVID-19, but perhaps not consistently observed, as only infrequent periodic monitoring is common.
An important aspect of the AGM model for SARS-CoV-2 infection is the development of pronounced viral pneumonia. All AGMs in this study exhibited pulmonary consolidation with hemorrhage, varying in severity between animals and lung lobes. Histology and IHC of lung tissue revealed multifocal lesions of varying severity with co-localization of viral antigen, confirming that infection with SARS-CoV-2 resulted in marked viral pneumonia in these animals. Importantly, numerous histological features of the lungs from AGMs infected with SARS-CoV-2 mirrored those reported in human COVID-19 cases33–35. A caveat to this study is we did not directly compare vRNA, gross pathology, and histological changes at 57 days post primary challenge with a group that was not re-exposed. Therefore, it is unclear whether changes in the back-challenged AGMs were a result of primary challenge or back-challenge. Nevertheless, average lesion severity scores were lower in back-challenged AGMs and no significant immunolabeling for fibrin or SARS-CoV-2 antigen was observed in the lung, or any other examined organ, in these subjects.
We performed thoracic radiographs on all six SARS-CoV-2-infected AGMs. It is our opinion that radiographs from our study, while consistent with reports in SARS-CoV-2-infected macaques8,10,26, belie the degree of lesions and hemorrhage of the lungs seen at necropsy and do not convincingly demonstrate SARS-CoV-2-induced disease. Specifically, opacities and changes observed in radiographs are nonspecific and could be due to atelectasis, inadequate inspiratory effort (unable to control inspiratory effort in NHPs), or BAL collection procedures among other etiologies. Important to this discussion, the US Centers for Disease Control (CDC) does not recommend either radiography or CT as a primary screening tool nor does the American College of Radiology due to poor specificity and sensitivity36. A study of 636 ambulatory patients with COVID-19 did not detect chest radiograph abnormalities in 60% of confirmed cases37. Furthermore, a multidisciplinary panel comprised principally of radiologists and pulmonologists with experience managing COVID-19 patients recommended against chest radiography in cases with mild clinical features38.
Our data show that infection of AGMs with SARS-CoV-2 results in upregulation of IFN-stimulated genes and IL-6 and IL-8 signaling in the lungs and peripheral blood; in the blood, NK- and T cell-associated transcripts were repressed, which is consistent with human cases17,18,39,40. Functional enrichment of DE mRNAs pointed to rapid viral clearance with tightly controlled responses in the blood. Our results indicate evaluation of lung and blood samples is important to fully capture the state of disease, as some key differences were observed between these compartments particularly in terms of gene regulation. We observed increased serum concentrations of these interleukins as well as other pro-inflammatory cytokines and chemokines elevated in human cases. Additionally, we detected a rise in circulating fibrinogen in a majority of AGMs, which is implicated in thrombosis and vascular injury in human patients24,25.
The potential for re-infection in humans with SARS-CoV-2 is speculated, but the risk factors or incidence are unknown. This possibility is concerning as antibody titers to endemic human coronaviruses (e.g., HCoV-229E) are reported to gradually wane, and re-infection with homologous virus has been reported41,42. Others have demonstrated protection against back-challenge of rhesus macaques13,19. Similarly, we describe the capacity of AGM challenged with SARS-CoV-2 to achieve natural immunity. Importantly, all animals held past 5 dpi seroconverted and, despite relatively low neutralizing titers at back-challenge, rapidly cleared the secondary challenge inoculum. Immunity was not sterilizing as vRNA was detected in nasal swabs shortly after infection, yet infectious virus was not detected suggesting the risk for infectiousness post exposure may be present, albeit low. Nonetheless, anamnestic humoral and cellular responses were uniformly demonstrated in all back-challenged animals. Interestingly, the development of circulating anti-spike RBD binding IgG and IgA coincided with an increase in neutralizing sera potency. Future studies examining antibody type and potency from the mucosa may further inform compartmentalization of protective antibodies.
Our comprehensive evaluation indicates AGMs can be used to study pathogenesis and the host response to SARS-CoV-2 infection. Importantly, this model may prove valuable in streamlining the most promising medical countermeasures for human use.
Methods
Virus
The virus (SARS-CoV-2/INMIl-Isolate/2020/Italy) employed in this study was isolated on January 30, 2020 from the sputum of the first clinical case in Italy, a tourist visiting from the Hubei province of China that developed respiratory illness while traveling43. The virus was initially passaged twice (P2) on Vero E6 cells; the supernatant and cell lysate were collected and clarified following a freeze/thaw cycle. This isolate is certified mycoplasma-free. The complete sequence was submitted to GenBank (MT066156) and is available on the GISAID website (BetaCoV/Italy/INMI1-is1/2020: EPI_ISL_410545) upon registration. For in vivo challenge, the P2 virus was propagated on Vero E6 cells and the supernatant was collected and clarified by centrifugation making the virus used in this study a P3 stock.
Animal challenge
SARS-CoV-2 seronegative African green monkeys (Chlorocebus aethiops) (4 females, 2 males) (St Kitts origin, Worldwide Primates, Inc.) were randomized into two cohorts where one group (n = 3) was scheduled for euthanasia at 5 dpi and the other to be back-challenged (n = 3) with the same SARS-CoV-2 isolate and dose. Animals were anesthetized with ketamine and inoculated with a combined 4.6 × 105 PFU dose of SARS-CoV-2 with 2.3 × 105 PFU delivered by the intratracheal (i.t.) route in 5.0 ml and 2.3 × 105 PFU delivered by the intranasal (i.n.) route in 1.0 ml total (0.5 ml per nostril). All subjects were longitudinally monitored for clinical signs of illness including temperature (measured by surgically implanted DST micro-T small implantable thermo loggers (Star-Oddi), respiration quality, and clinical pathology. All measurements requiring physical manipulation of the animals were performed under sedation by ketamine or telazol. Animal protocols were approved by the University of Texas Medical Branch (UTMB) Institutional Animal Care and Use Committee and adhere to the NIH Guide for the Care and Use of Laboratory Animals. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Radiographic technique
All monkeys were imaged with a portable GE AMX-4+ computed radiography system using a DRTECH detector set at a 36-inch focal film distance. Images were captured and evaluated using the Maven Patient Image Voyance Software (version 2020) in ventral dorsal (VD) and right lateral (R LAT) positions at 50 kVp and 12.5mA as previously described44. Chest radiographs were captured and interpreted by a double board-certified clinical veterinarian and veterinary pathologist and reviewed by a MD board-certified radiologist.
RNA isolation from SARS-CoV-2-infected AGMs
On specified procedure days (days 0, 2, 3, 4, 5, 7, 12, 15, 21), 100 μl of blood or BAL fluid was added to 600 μl of AVL viral lysis buffer (Qiagen) for virus inactivation and RNA extraction. Following removal from the high containment laboratory, RNA was isolated from blood, BAL fluid, and mucosal swabs using the QIAamp viral RNA kit (Qiagen). Tissues were put into RNAlater, inactivated with Qiagen RLT buffer, and extracted using a Qiagen RNeasy Mini kit.
Detection of SARS-CoV-2 load
vRNA was quantified using CDC–designed SARS-CoV-2 N2 assay primers/probe for reverse transcriptase quantitative PCR (RT-qPCR)45. SARS-CoV-2 RNA was detected using One-step probe RT-qPCR kits (Qiagen) and a CFX96 detection system (Bio-Rad) with the following cycle conditions: 50 °C for 10 min, 95°C for 10 sec, and 45 cycles of 95 °C for 10 sec and 55 °C for 30 sec. Threshold cycle (CT) values representing SARS-CoV-2 genomes were analyzed with CFX Manager Software, and the data are presented as GEq. To generate the GEq standard curve, SARS-CoV-2 RNA from cell supernatants was serially diluted, and the number of genomes was calculated using Avogadro’s number and the molecular weight of the SARS-CoV-2 genome.
Virus titration was performed by plaque assay with Vero E6 cells (ATCC CRL-1586). Briefly, increasing 10-fold dilutions of samples were adsorbed to Vero E6 cell monolayers in duplicate wells (200 μl). Cells were overlaid with EMEM agar medium plus 1.25% Avicel, incubated for 2 days, and plaques were counted after staining with 1% crystal violet in formalin.
Hematology and serum biochemistry
Total white blood cell counts, white blood cell differentials, red blood cell counts, platelet counts, hematocrit values, total hemoglobin concentrations, mean cell volumes, mean corpuscular volumes, and mean corpuscular hemoglobin concentrations were analyzed from blood collected in tubes containing EDTA using a Vetscan HM5 hematologic analyzer (Abaxis). Serum samples were tested for concentrations of albumin, amylase, alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), blood urea nitrogen (BUN), calcium, creatinine (CRE), C-reactive protein (CRP), gamma-glutamyltransferase (GGT), glucose, total protein, and uric acid using a Piccolo point-of-care analyzer and Biochemistry Panel Plus analyzer discs (Abaxis). Partial pressures of CO2 and O2 were obtained using an iSTAT Alinity hematological analyzer (Abbott).
ELISA
SARS-CoV-2-specific IgG and IgA antibodies were measured in sera by ELISA at the indicated time points. Immunosorbent 96-well plates were coated overnight with each antigen. To detect spike IgA or IgG, plates were coated with 0.1 μg/ml spike S1 plus S2 ectodomain (Sino Biologicals; Cat: 40589-V08B1) or RBD (BEI; Cat: NR-52366). For total virus-specific IgG, coates were plated with a 1:1000 dilution of irradiated SARS-CoV-2 infected or normal Vero E6 lysate in PBS (pH 7.4) kindly provided by T.W. Ksiazek (UTMB). Nucleoprotein ELISA kits were kindly provided by Zalgen Labs, LLC. Sera were initially diluted 1:100 and then two-fold through 1:25,600 in 4% BSA in 1× PBS or in Zalgen-provided reagents. After a 1 h incubation, plates were washed six times with wash buffer (1× PBS with 0.2% Tween-20) and incubated for 1 h with a 1:5000 or 1:600 dilution of horseradish peroxidase (HRP)-conjugated anti-primate IgG antibody (Fitzgerald Industries International; Cat: 43R-IG020HRP) or anti-primate IgA antibody (Rockland Immunochemicals, Inc.; Cat: 617-103-006), respectively. RT SigmaFast O-phenylenediamine (OPD) substrate (P9187, Sigma) was added to the wells after six additional washes to develop the colorimetric reaction. The reaction was stopped with 3M sulfuric acid 5-10 min after OPD addition and absorbance values were measured at a wavelength of 492 nm on a spectrophotometer (Biotek Cytation5 system). For Zalgen kits, tetramethylbenzidine (TMB) was used to develop the reaction; the reaction was stopped with methanesulfonic acid and plates were read at a wavelength of 450 nm. Absorbance values were normalized by subtracting uncoated wells from antigen-coated wells at the corresponding serum dilution. End-point titers were defined as the reciprocal of the last adjusted serum dilution with a value ≥ 0.20.
ELISPOT
To analyze cellular responses, AGM peripheral blood mononuclear cells (PBMCs) were rapidly thawed in a 37 °C water bath and resuspended in pre-warmed complete RPMI 1640 media with 10% fetal bovine serum (FBS), 1% GlutaMAX (ThermoFisher Scientific), and 1% penicillin-streptomycin (ThermoFisher Scientific). Cells were rested overnight at 37 °C and 5% CO2 and left unstimulated or stimulated for 24 h at 37 °C and 5% CO2 with either lectin (Sigma-Aldrich) from Phytolacca americana (pokeweed mitogen; PWM), one of two PepMix™ SARS-CoV-2 spike peptide pools (JPT) spanning the length of the protein, or a SARS-CoV-2 nucleoprotein peptide pool (BEI). The spike pools contained 158 or 157 15mer peptides with 11 amino acid overlaps and the N pool contained 13mer peptides with 10 amino acid overlaps. S and N pools were prepared in DMSO and used at a final concentration of 1 μg/ml. Unstimulated cells contained 0.2% DMSO by volume. As a positive control, PBMCs were stimulated with PWM at a final concentration of 2.5 μg/ml. For ELISPOT analysis, samples were stained using dual color primate IL-2 and IFN-γ kits (R&D Systems) according to the manufacturer’s recommendations. PBMCs were plated at 2.5 × 105 cells per well in 96-well plate coated with nonhuman primate IL-2 and IFN-γ capture antibodies. Following a 24 h incubation at 37 °C and 5% CO2, ELISpot plates were imaged using an Immunospot S6 UNIVERSAL Analyzer (Cellular Technology Limited). Reported values were calculated by subtracting the number of spot-forming cells (SFCs) in a given unstimulated sample from its respective stimulated counterpart at the corresponding dpi.
Serum neutralization assay
Neutralization titers were calculated by determining the dilution of serum that reduced 50% of plaques (PRNT50). We incubated a standard 100 PFU amount of SARS-CoV-2 with two-fold serial dilutions of serum samples for one hour. The virus-serum mixture was then used to inoculate Vero E6 cells for 60 minutes. Cells were overlaid with EMEM agar medium plus 1.25% Avicel, incubated for 2 days, and plaques were counted after staining with 1% crystal violet in formalin. End-point PRNT50 titers were defined as the reciprocal of the last serum dilution with an approximate PRNT50 value.
Bead-based cytokine and coagulation immunoassays
Concentrations of immune mediators and fibrinogen were determined by flow cytometry using LegendPlex multiplex technology (BioLegend). Serum concentrations of cytokines/chemokines and plasma concentrations of fibrinogen were quantified using Nonhuman Primate Inflammation 13-plex (1:4 dilution) and Human Thrombosis (1:100 dilution) kits or a Human Fibrinolysis (1:10,000 dilution) panel, respectively. Samples were processed in duplicate following the kit instructions and recommendations. Following bead staining and washing, 1500-4000 bead events were collected on a FACS Canto II cytometer (BD Biosciences) using BD FACS Diva software. The raw .fcs files were analyzed with BioLegend’s cloud-based LEGENDplex™ Data Analysis Software.
RNA sample preparation for transcriptomic analysis
NHPV2_Immunology reporter and capture probesets (Nanostring Technologies) were hybridized with ~5 μl of blood RNA or ~200 ng of BAL RNA at 65 °C for ~24 h. The RNA:probeset complexes were loaded into an nCounter microfluidics cartridge and assayed on a Nanostring nCounter® SPRINT Profiler. To estimate abundance of each of the 769 unique mRNA immune-related targets included in the NHPV2_Immunology panel, fluorescent reporter barcodes were imaged and counted in each sample lane. In conjunction with the predefined NHP targets, ACE2 expression and SARS-CoV-2-specific targets (envelope, membrane, nucleocapsid, orf1ab, orf3a, orf7a, orf8, spike) were analyzed in BAL samples with a Nanostring Covid-19 Plus panel plus kit.
Bioinformatics analysis
nCounter. RCC files were imported into Nanostring nSolver™ 4.0 software. All samples met the established QC criteria. To compensate for varying RNA inputs, housekeeping genes and spiked-in positive and negative controls were used to normalize raw counts. The data was analyzed using the Nanostring nSolver Advanced Analysis 2.0 package to generate the principal component analysis (PCA) figures and volcano plots, as well as to determine differential expression of transcripts compared to a pre-challenge baseline (a full list of probes detected for each sample group along with fold change values, standard error, confidence limits, and p-values is featured in Source Data File 1). Normalized data (log-fold change values and FDR-adjusted p-values) were exported as a .CSV file (Microsoft Excel Office for Mac version 14.1.0) for Ingenuity Pathway Analysis (IPA; Qiagen)-based functional enrichment of differentially expressed RNAs. Z-scores were imported into GraphPad Prism version 8 to produce canonical signaling, upstream regulator, and causal network heatmaps. Jak/STAT signaling pathways and interferon alpha regulation depictions were also generated with IPA. To generate the network maps, differentially expressed mRNAs with an FDR-adjusted p-value < 0.05 from each BAL or blood dataset were imported into Metascape and visualized using Cytoscape46.
Histopathology and immunohistochemistry
Necropsy was performed on all subjects euthanized at 5 dpi. Tissue samples of all major organs were collected for histopathologic and immunohistochemical (IHC) examination and immersion-fixed in 10% neutral buffered formalin for > 7 days. Specimens were processed and embedded in paraffin and sectioned at 5 μm thickness. For IHC, specific anti-SARS immunoreactivity was detected using an anti-SARS nucleocapsid protein rabbit primary antibody at a 1:800 dilution for 60 min (Novusbio). The tissue sections were processed for IHC using the ThermoFisher Scientific Lab Vision Autostainer 360 (ThermoFisher Scientific). Secondary biotinylated goat anti-rabbit IgG (Vector Laboratories) antibody was used at 1:200 for 30 min followed by Vector Streptavidin Alkaline Phosphatase at a dilution of 1:200 for 20 min (Vector Laboratories). Slides were developed with Bio-Red (Biopath) for 7 min and counterstained with hematoxylin for 1 min. For IHC, specific anti-fibrin was detected using an anti-fibrin monoclonal mouse primary antibody at a 1:3200 dilution for a 60 min incubation (Sekisui Diagnostics). The tissue sections were processed for IHC using the ThermoFisher Scientific Lab Vision Autostainer 360 (ThermoFisher Scientific). Secondary biotinylated goat anti-mouse IgG (Vector Laboratories) antibody was used at a concentration of 1:200 for 30 min followed by Vector Streptavidin Alkaline Phosphatase at a dilution of 1:200 for 20 min (Vector Laboratories). Slides were developed with Bio-Red (Biopath Laboratories) for 7 min and counterstained with hematoxylin for 1 min.
SARS-CoV-2 RNA in situ hybridization (ISH) in formalin-fixed paraffin embedded (FFPE) tissues was performed using an RNAscope 2.5 high definition (HD) RED kit (Advanced Cell Diagnostics according to the manufacturer’s instructions. 20 ZZ probe pairs targeting the genomic SARS-CoV-2 spike protein (S) gene were designed and synthesized by Advanced Cell Diagnostics (catalogue 848561). After sectioning, deparaffinization with xylene and graded ethanol washes and peroxidase blocking, the sections were heated in RNAscope target retrieval reagent buffer (Advanced Cell Diagnostics catalogue 322000) for 15 min and air-dried overnight. The sections were digested with Protease III (catalogue 322340) at 40 °C in a hydribidization oven (HybEZ, Advanced Cell Diagnostics catalogue 321711) for 15 min. Sections were exposed to ISH target probe and incubated at 40°C in a HybEZ oven for 2 h. After rinsing, the signal was amplified using the manufacturer provided pre-amplifier and amplifier conjugated to alkaline phosphatase and incubated with a red substrate-chromogen solution for 10 min, counterstained with hematoxylin, air-dried, and cover slipped. Tissues were stained following package instructions for collagen with the Trichrome One-Step Blue & Red Stain Kit (American MasterTech Scientific Laboratory Supplies).
Statistics and Reproducibility
Statistical analyses of data were implemented using GraphPad Prism v8.2.1. The data was fitted to a mixed model, which uses a compound symmetry covariance matrix, and is fit using Restricted Maximum Likelihood (REML), in the absence of missing values. This method gives the same P values and multiple comparisons tests as a repeated-measures ANOVA. Statistics were derived from n=6 individual animal subject samples for all time points prior to or on 5 dpi and n=3 individual animal subject samples for time points after 5 dpi in a single independent experiment (one cohort was sacrificed at 5 dpi (n=3); the other cohort was held to 57/22 dpi (n=3)). Statistics for all figures were calculated from individual animal data values, not technical, replicates. For experiments with technical replicates (e.g., duplicate RT-qPCR reactions/wells), only the mean was used to calculate statistical significance. Lung severity scores were analyzed using a two-tailed t-test (p = 0.0249; t = 3.500, df = 4). Tissue PCR and plaque assay titers were analyzed using multiple two-tailed t-tests with the Bonferroni-Dunn method. PCR: RLL (p = 0.0003 (adjusted); t = 4.593, df = 136); RML (p = 0.0017; t = 4.185, df = 136); RUL (p = 0.002; t = 4.146, df = 136); LML (p = 0.0021; t = 4.137, df = 136); LLL (p = 0.0021; t = 4.131, df = 136); Trac (p = 0.0025; t = 4.085, df = 136); NaMu (p = 0.007; t = 3.814, df = 136); Sto (p = 0.0162; t = 3.580, df = 136). Plaque: RLL (p = 0.0181 (adjusted); t = 3.548, df = 136); RML (p = 0.0102; t = 3.712, df = 136); RUL (p = 0.0007; t = 4.405, df = 136); LML (p = 0.0037; t = 3.983, df = 136); LLL (p = 0.0002; t = 4.690, df = 136); LUL (p < 0.0001; t = 4.908, df = 136). A mixed effects model with Geisser-Greenhouse correction and a Tukey’s multi-comparisons test was used to determine statistical significance between individual virus RNA probes revealing no significance between individual probes at each time point, but significant overall time-dependent effects (p = 0.0003; F (1.617, 73.32) = 10.56). A multiple hypothesis Benjamini-Hochberg false discovery rate (FDR) corrected p-value less than 0.05 was deemed significant for all RNA expression analyses, unless otherwise stated. No data points were excluded from our analyses.
Representative photomicrographs were qualitatively considered to display lesions that were nominally or ordinally measured by masking of the pathologist post-examination and ranking lesions to satiate the study objectives. Additionally, a thorough examination of multiple slides of the target tissues (e.g. 18 slides of lung) multiple times (up to 3 times per tissue) was performed in a timely manner to maintain interpretation consistency.
Data availability
RNA reads data and statistics are provided as an extended data file (Source Data File 1). Other data that support the findings of this study are available from the corresponding author, T.W.G., upon reasonable request.
Extended Data
Extended Data Fig. 1. Longitudinal temperature analysis of AGMs infected with SARS-CoV-2.
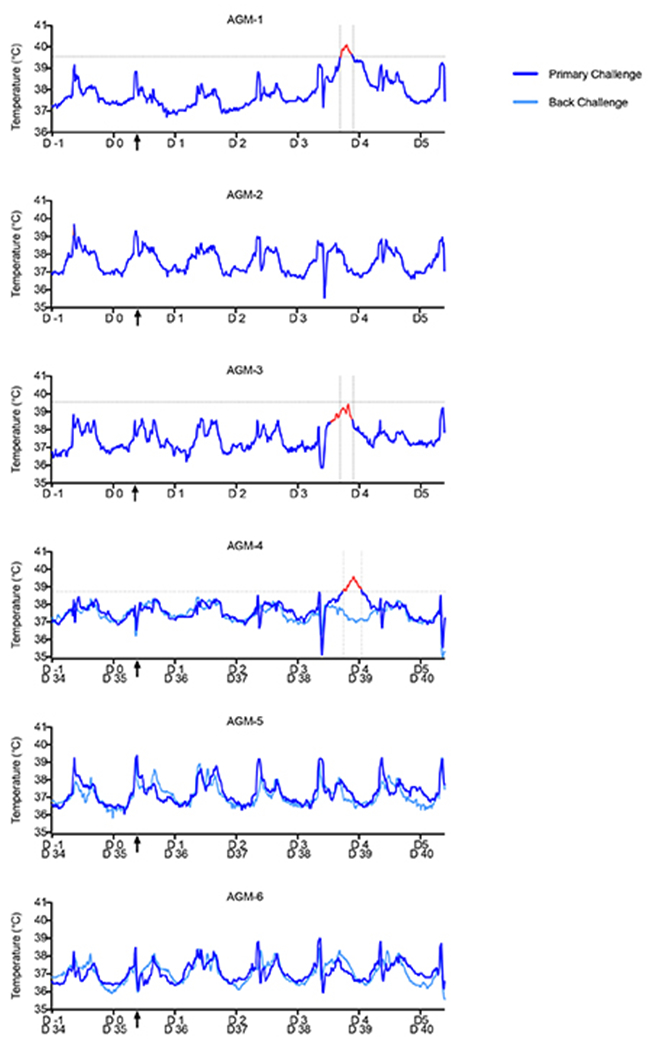
Prior to challenge, AGMs (n=6) were surgically implanted with a DST micro-T small implantable thermo logger (Star-Oddi), allowing body temperature measurements for each animal in 15-min increments (96 measurements/day) throughout the course of the study. AGM-1 and AGM-3 had elevated temperatures noticeably above baseline temperatures (1 day prior to challenge) at 3 dpi; AGM-4 exhibited increased temperature at 4 dpi. The “fever peak” for each subject is colored in red. Vertical dashed lines indicate the start and end of the fever peak. Horizontal dashed lines indicate the threshold temperature for classification as fever. Black arrows on the x-axis indicate time of challenge. Determination of the window of febrile temperatures was performed visually, with comparison of temperatures at all other points during the study duration (−1 dpi to 5 dpi).
Extended Data Fig. 2. Comparison of viral loads of tissues from primary and re-challenged AGMs.
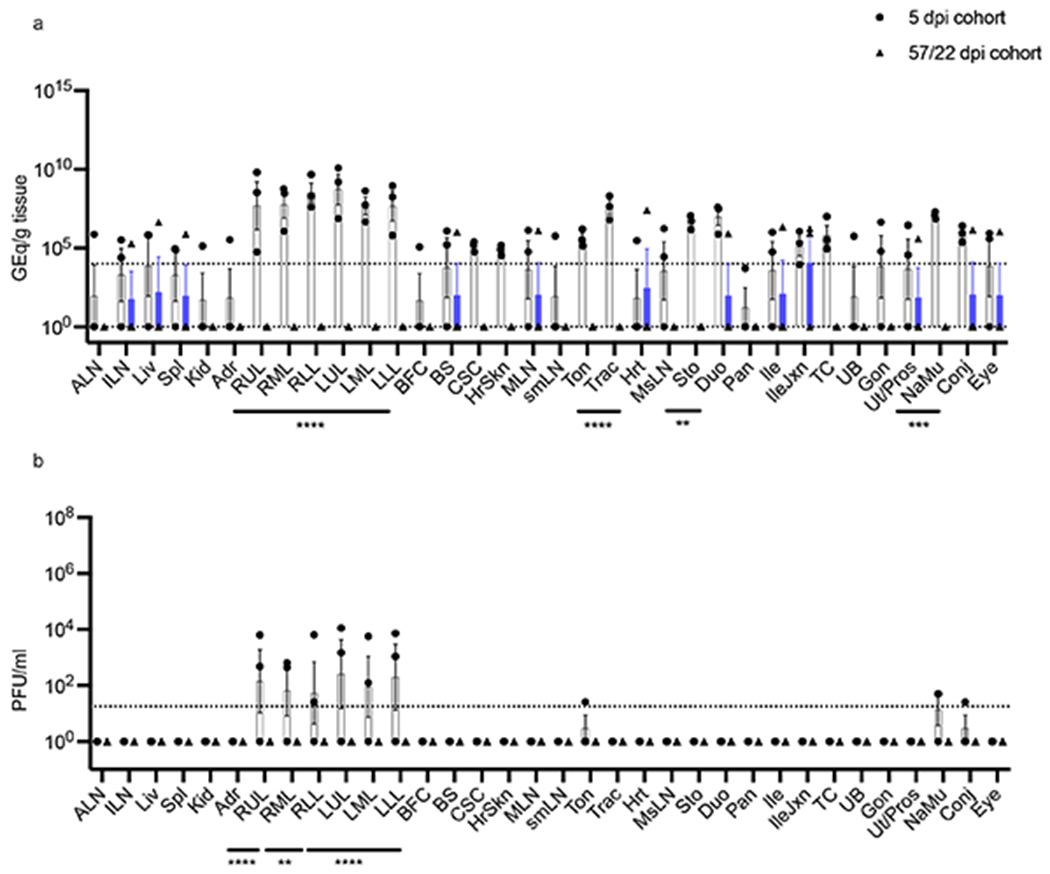
Tissues harvested at necropsy from SARS-CoV-2 infected-AGMs (n=6) were processed to determine viral loads by a) RT-qPCR and b) plaque titration. Tissues from animals sacrificed at 5 dpi (n=3) were compared to those re-challenged at 35 dpi and sacrificed at 57/22 dpi (n=3). Abbreviations for tissues: ALN: Axillary lymph node, ILN: inguinal lymph node, Liv: liver, Spl: spleen, Kid: kidney, Adr: adrenal gland, RUL: right upper lung, RML: right middle lung, RLL: right lower lung, LUL: left upper lung, LML: left middle lung, LLL: left lower lung, BFC: brain frontal cortex, BS: brain stem, CSC: cervical spinal cord, MLN: mandibular lymph node, smLN: submandibular lymph node, Ton: tonsil, Trac: trachea, Hrt: heart, MsLN: mesenteric lymph node, Sto: stomach, Duo: duodenum, Pan: pancreas, Ile: ileum, IleJxn: ileocecal junction, TC: transverse colon, UB: urinary bladder, Gon: gonad, Ut/Pros: uterus/prostate, NaMu: nasal mucosa, Conj: conjunctiva. The horizontal dashed line indicates the LOD for the assay. Multiple two-tailed t-tests using the Bonferroni-Dunn method: p= 0.0332 (*), 0.0021 (**), 0.0002 (***), <0.0001 (****). Data are presented as mean values +/− SEM. Statistics were derived from the mean of duplicate RT-qPCR reactions or wells of each tissue per animal (n=6 biologically independent animals/samples per tissue in a single experiment; n=3 animals per cohort (5 or 57/22 dpi).
Extended Data Fig. 3. Temporal radiographs of SARS-CoV-2-infected AGMs.
AGMs were imaged with a portable radiography system and detector. Images were captured and evaluated over the course of the study in ventral dorsal (VD) and right lateral (R LAT) positions. Chest radiographs were captured and interpreted by a double board-certified clinical veterinarian and veterinary pathologist and reviewed by a MD board-certified radiologist.
Extended Data Fig. 4. Analysis of temporal host RNA and virus-specific probe expression in SARS-CoV-2-infected AGM BAL and blood samples.
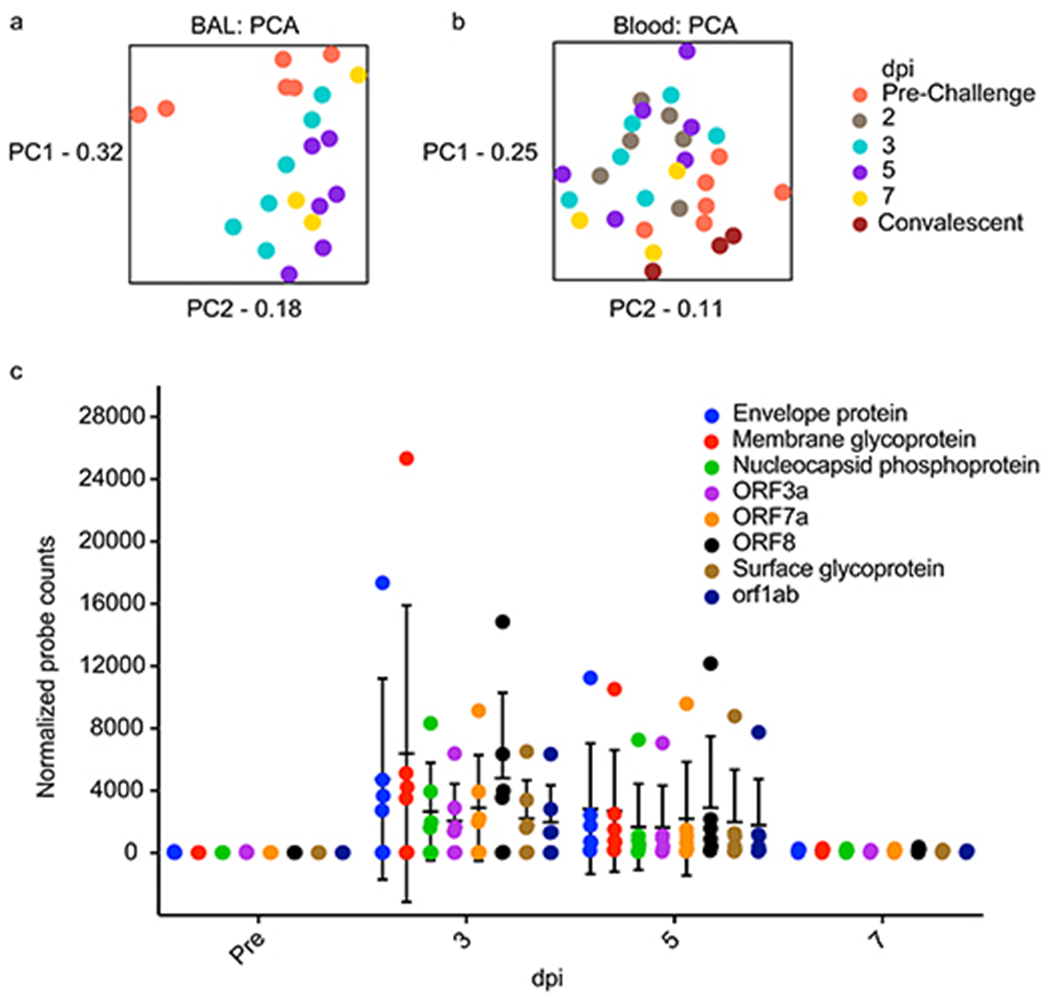
Principal Component Analyses (PCA) indicate overall sample variance in a) BAL and b) blood AGM transcriptomes when filtered by day post infection (dpi) and are shown to highlight time-dependent host transcriptional changes. PC1 (principal component 1), PC2 (principal component 2). c) At the indicated time point, the expression of individual virus-specific probes in BAL samples of each subject is plotted. Data are presented as mean values +/− SD. Statistics were derived from n=6 biologically independent animals/samples for pre-, 3, and 5 dpi time points and n=3 biologically independent animals/samples for the 7 dpi time point in a single experiment. A mixed effects model with Geisser-Greenhouse correction and a Tukey’s multi-comparisons test revealed no statistical significance between individual probes at any particular time point, but significant overall time-dependent effects (p=0.0003).
Extended Data Fig. 5. Early and convalescent stage transcriptional changes in SARS-CoV-2-infected AGMs.
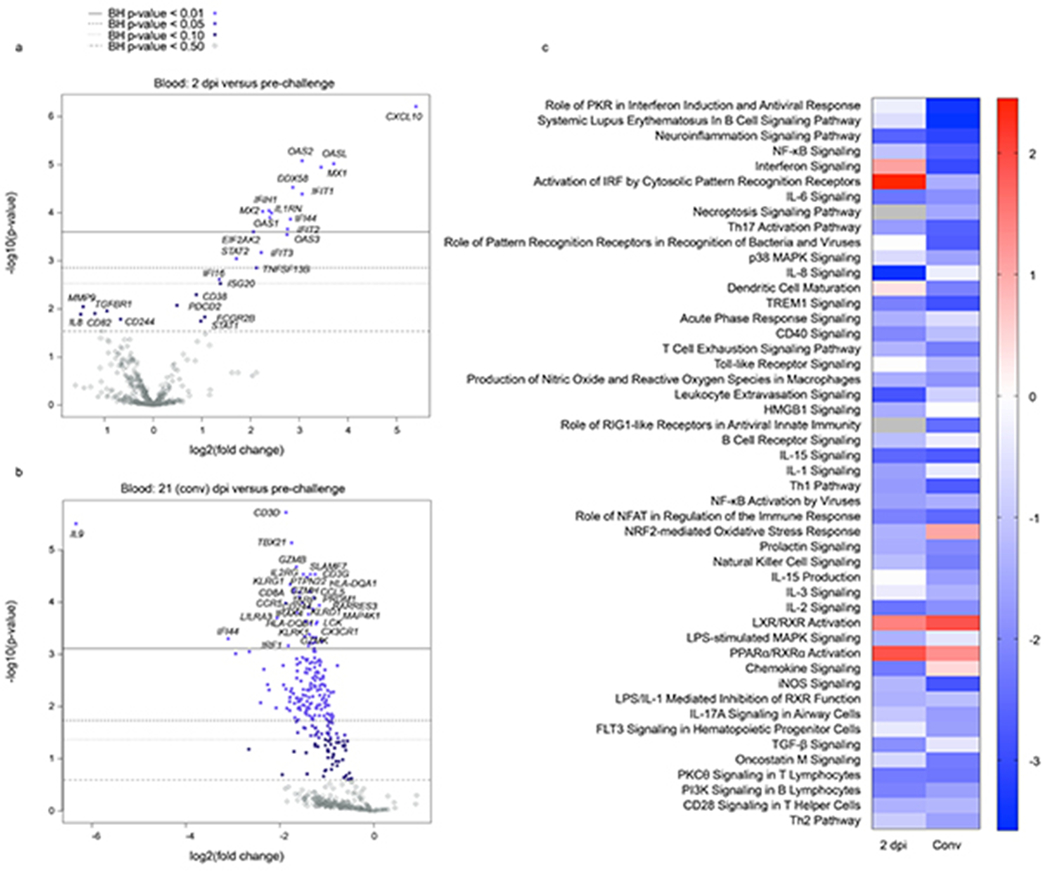
a, b) Volcano plots and a c) heatmap indicating early and convalescent stage transcriptional changes in SARS-CoV-2-infected AGMs. a,b) Displayed are −log10(p-values) and log2 fold changes for each mRNA target. Horizontal lines within the plot indicate FDR-adjusted p-value thresholds. Targets highlighted in blue indicate adjusted p-values < 0.10. A Benjamini-Hochberg test was employed to derive FDR-adjusted p-values. c) Heatmap demonstrating the most highly upregulated and downregulated canonical pathways at each time point. Only differentially expressed transcripts with an FDR-corrected p-value of less than 0.1 were enriched with Ingenuity Pathway Analysis (Qiagen). The data were normalized against a day 0 pre-challenge baseline for each NHP subject. Red indicates high expression (z-scores); blue indicates low expression; white indicate similar expression; gray indicates insufficient transcripts mapping to the indicated pathway.
Extended Data Fig. 6. Soluble inflammatory mediators and coagulation markers detected in SARS-CoV-2 infected AGM sera following primary challenge.
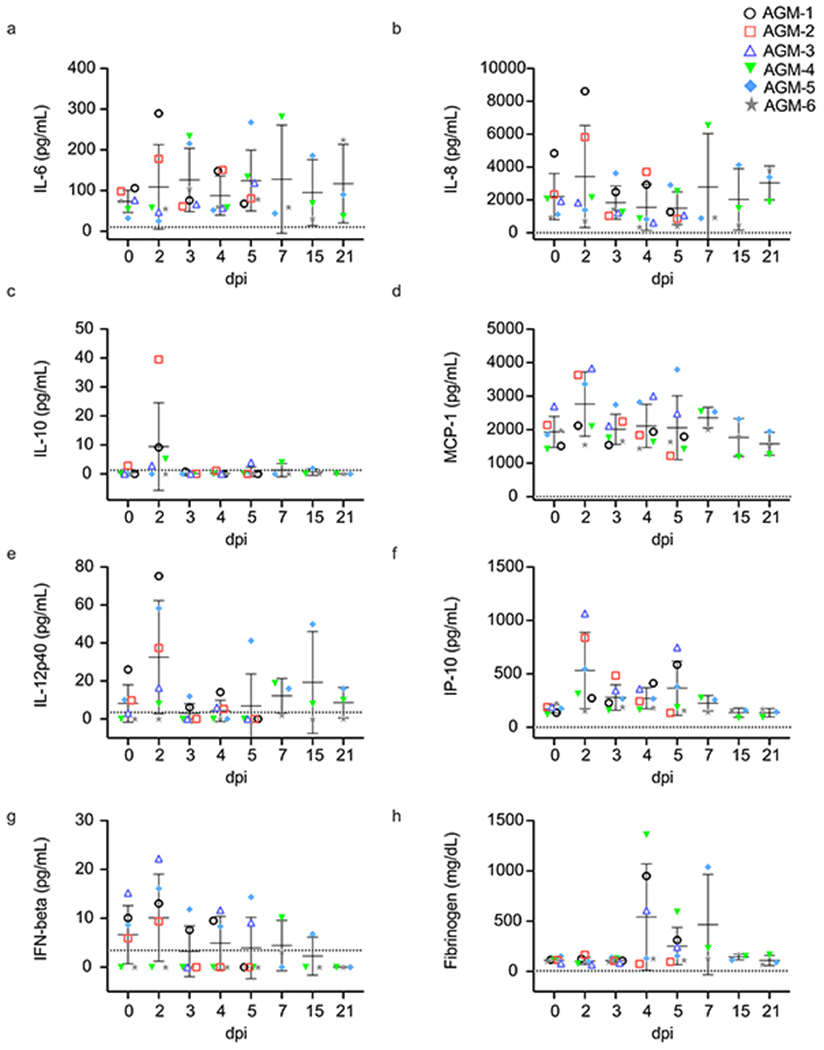
a-g) Cytokine and h) fibrinogen fold-changes relative to a pre-challenge (0 dpi) baseline in serum or plasma of AGMs infected with SARS-CoV-2 (n=6 biologically independent animals in a single experiment for 0, 2, 3, 4, and 5 dpi time points; n=3 for 7, 15, and 21 time points). Data are presented as mean values +/− SD of duplicate samples per subject per analyte in a single experiment.
Supplementary Material
Supplementary Table 1.
Normalized host and virus expression values and statistics in BAL and blood RNA samples from SARS-CoV-2-infected AGMs.
Acknowledgements
The authors wish to thank the UTMB Animal Resource Center for veterinary support and surgical implantation of temperature data loggers, as well as husbandry support. We thank V. Menachery, S. Makino, C.-T. Tseng, T. Ksiazek, and K. Narayanan for their valuable insight and technical assistance with coronavirus protocols. The virus used in this publication was kindly provided by the European Virus Archive goes Global (EVAg) project that has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 653316. We wish to thank L. Branco and M. Boisen (Zalgen Labs) for generously providing the SARS-CoV-2 anti-nucleoprotein ELISA assays. The following reagent was produced under HHSN272201400008C and obtained through BEI Resources, NIAID, NIH: Spike Glycoprotein Receptor Binding Domain (RBD) from SARS-Related Coronavirus 2, Wuhan-Hu-1 with C-Terminal Histidine Tag, Recombinant from HEK293F Cells, NR-52366. This study was supported by funds from the Department of Microbiology and Immunology, University of Texas Medical Branch at Galveston, Galveston, TX to T.W.G. Operations support of the Galveston National Laboratory was supported by NIAID/NIH grant UC7AI094660.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.World Health Organization. Coronavirus disease (COVID-19) Situation Report. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20201012-weekly-epi-update-9.pdf (2020). [Google Scholar]
- 2.Tseng CT et al. , Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One 7, e35421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolles M et al. , A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol 85, 12201–12215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weingartl H et al. , Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol 78, 12672–12676 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao L et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature (2020). doi: 10.1038/s41586-020-2312-y [DOI] [PubMed] [Google Scholar]
- 6.Chan JFW et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis (2020). doi: 10.1093/cid/ciaa325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim Y I1 et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 27(5), 704–709.e2 (2020). doi: 10.1016/j.chom.2020.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rockx B et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 368(6494), 1012–1015 (2020). doi: 10.1126/science.abb7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu P et al. Age-related rhesus macaque models of COVID-19. Anim. Model. Exp. Med 3, 93–97 (2020). doi: 10.1002/ame2.12108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munster VJ et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 585(7824), 268–272 (2020). doi: 10.1038/s41586-020-2324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shan C et al. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques. Cell Res. 30, 670–677 (2020). doi: 10.1038/s41422-020-0364-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai M et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci 117, 16587–16595 (2020). doi: 10.1073/pnas.2009799117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandrashekar A et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 369(6505), 812–817 (2020). doi: 10.1126/science.abc4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAuliffe J et al. , Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology 330, 8–15 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Letko M, Marzi A & Munster V Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol 5, 562–569 (2020). doi: 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang J et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci 117, (2020). doi: 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Z et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 27, 883–890.e2 (2020). doi: 10.1016/j.chom.2020.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen W et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 6, (2020). doi: 10.10388/s41421-020-0168-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng W et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science 369, 818–823 (2020). doi: 10.1126/science.abc5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Wit E et al. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci 117, 6771–6776 (2020). doi: 10.1073/pnas.1922083117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caswell JL & Williams K The respiratory system In: Jubb, Kennedy, and Palmer’s Pathology of Domestic Animals, 5th ed. Maxie MG et al. (eds). Pp. 523–655 (Elsevier, Edinburgh, 2007). [Google Scholar]
- 22.Grifoni A et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181, 1489–1501.e15 (2020). doi: 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letsch A & Scheibenbogen C Quantification and characterization of specific T-cells by antigen-specific cytokine production using ELISPOT assay or intracellular cytokine staining. Methods 31, 143–149 (2003). doi: 10.1016/S1046-2023(03)00124-5 [DOI] [PubMed] [Google Scholar]
- 24.Connors JM & Levy JH COVID-19 and its implications for thrombosis and anticoagulation. Blood 135, 2033–2040 (2020). doi: 10.1182/BLOOD.2020006000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ranucci M et al. The procoagulant pattern of patients with COVID-19 acute respiratory distress syndrome. J. Thromb. Haemost 18, 1747–1751 (2020). doi: 10.1111/jth.14854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williamson BN et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature 585, 273–276 (2020). doi: 10.1038/s41586-020-2423-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel A A prefusion SARS-CoV-2 spike RNA vaccine is highly immunogenic and prevents lung infection in non-human primates. Preprint at: https://www.biorxiv.org/content/10.1101/2020.09.08.280818v1 (2020).
- 28.Kakuk TJ et al. A human respiratory syncytial virus (RSV) Primate model of enhanced pulmonary pathology induced with a formalin-inactivated RSV vaccine but not a recombinant FG subunit vaccine. J. Infect. Dis 167, 553–561 (1993). doi: 10.1093/infdis/167.3.553 [DOI] [PubMed] [Google Scholar]
- 29.Nalca A, Totura A, Livingston V, Frick O & Dyer D African green monkey model of Middle East respiratory syndrome coronavirus (MERS-CoV) infection. Int. J. Infect. Dis 79, 99–100 (2019). doi: 10.1016/j.ijid.2018.11.249 [DOI] [Google Scholar]
- 30.Durbin AP, Elkins WR & Murphy BR African green monkeys provide a useful nonhuman primate model for the study of human parainfluenza virus types-1, -2, and -3 infection. Vaccine 18, 2462–2469 (2000). doi: 10.1016/S0264-410X(99)00575-7 [DOI] [PubMed] [Google Scholar]
- 31.Prasad AN et al. Resistance of Cynomolgus Monkeys to Nipah and Hendra Virus Disease Is Associated with Cell-Mediated and Humoral Immunity. J. Infect. Dis 221, S436–S447 (2020). doi: 10.1093/infdis/jiz613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant MC et al. The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS-CoV-2; COVID-19): A systematic review and meta-analysis of 148 studies from 9 countries. PLoS One 15, (2020). doi: 10.1371/journal.pone.0234765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian S et al. Pulmonary Pathology of Early-Phase 2019 Novel Coronavirus (COVID-19) Pneumonia in Two Patients With Lung Cancer. J. Thorac. Oncol 15, 700–704(2020). doi: 10.1016/j.jtho.2020.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ackermann M et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med 383, 120–128 (2020). doi: 10.1056/NEJMoa2015432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martines RB et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, united states. Emerg. Infect. Dis 26, 2005–2015 (2020). doi: 10.3201/eid2609.202095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.American College of Radiology, ACR Recommendations for the use of Chest Radiography and Computed Tomography (CT) for Suspected COVID-19 Infection. Accessed 20 May 2020 https://www.acr.org/Advocacv-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection (2020).
- 37.Weinstock ME et al. Chest X-Ray Findings in 636 Ambulatory Patients with COVID-19 Presenting to an Urgent Care Center: A Normal Chest X-Ray Is no Guarantee. J Urgent Care Med. 14, 13–18 (2020). [Google Scholar]
- 38.Rubin GD et al. The role of chest imaging in patient management during the covid-19 pandemic: A multinational consensus statement from the fleischner society. Radiology 296, 172–180 (2020). doi: 10.1148/radiol.2020201365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diao B et al. Human Kidney is a Target for Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection. Preprint at https://www.medrxiv.org/content/10.1101/2020.03.04.20031120v4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin. Infect. Dis ciaa449 (2020). doi: 10.1093/cid/ciaa449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Callow KA, Parry HF, Sergeant M & Tyrrell DAJ The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect 105, 435–446 (1990). doi: 10.1017/S0950268800048019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galanti M & Shaman J Direct Observation of Repeated Infections With Endemic Coronaviruses. J. Infect. Dis XX, 1–7 (2020). doi: 0.1093/infdis/jiaa392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 43.Capobianchi MR et al. Molecular characterization of SARS-CoV-2 from the first case of COVID-19 in Italy. Clinical Microbiology and Infection 26, 954–956 (2020). doi: 10.1016/j.cmi.2020.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geisbert TW et al. Development of an acute and highly pathogenic nonhuman primate model of nipah virus infection. PLoS One 5, 10690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.CDC CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel. Accessed 10 July 20 https://www.cdc.gov/coronavirus/2019-ncov/downloads/List-of-Acceptable-Commercial-Primers-Probes.pdf (2020).
- 46.Zhou Y et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10(1), (2019). doi: 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Normalized host and virus expression values and statistics in BAL and blood RNA samples from SARS-CoV-2-infected AGMs.
Data Availability Statement
RNA reads data and statistics are provided as an extended data file (Source Data File 1). Other data that support the findings of this study are available from the corresponding author, T.W.G., upon reasonable request.


