Abstract
Here we present a platform for discovery of protease-activated prodrugs and apply it to antibiotics that target Gram-negative bacteria. Because cleavable linkers for prodrugs had not been developed for bacterial proteases, we used substrate phage to discover substrates for proteases found in the bacterial periplasm. Rather than focusing on a single protease, we used a periplasmic extract of E. coli to find sequences with the greatest susceptibility to the endogenous mixture of periplasmic proteases. Using a fluorescence assay, candidate sequences were evaluated to identify substrates that release native amine-containing payloads. We next designed conjugates consisting of (1) an N-terminal siderophore to facilitate uptake, (2) a protease-cleavable linker, and (3) an amine-containing antibiotic. Using this strategy, we converted daptomycin—which by itself is active only against Gram-positive bacteria—into an antibiotic capable of targeting Gram-negative Acinetobacter species. We similarly demonstrated siderophore-facilitated delivery of oxazolidinone and macrolide antibiotics into a number of Gram-negative species. These results illustrate this platform’s utility for development of protease-activated prodrugs, including Trojan horse antibiotics.
Graphical Abstract
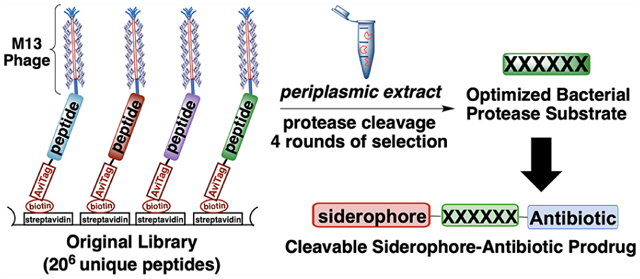
INTRODUCTION
The well-recognized term, “ESKAPEE” (previously ESKAPE),1 encompasses the names of seven species of clinically relevant pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., and Escherichia coli) that are associated with resistance to commonly prescribed antibiotics and are largely responsible for the world’s nosocomial infections.2 Five of these pathogens are Gram-negative species, whose outer membrane and associated resistance–nodulation–cell division (RND) efflux pumps render them resistant to many classes of antibiotics.3 Indeed, the outer membrane shields the bacteria from molecules that are unable to pass through porins,4 providing an effective barrier to many molecules that would otherwise be effective antibiotics against these pathogens.5 Gram-negative therapies can be delivered by siderophore-mediated antibiotic delivery using Nature’s Trojan-horse approach.6–8 Siderophores are small-molecule chelators that are produced by bacteria to sequester Fe(III),9 which is an essential nutrient required for bacterial growth and virulence.10 In the case of Gram-negative pathogens, outer membrane proteins (e.g., TonB-dependent transporters)11 bind ferric siderophores and provide opportunities for their active transport into the periplasm,12 where they may undergo further translocation into the cytoplasm by alternate transport mechanisms depending on the siderophore and the species of bacteria.13 Owing to the promiscuity of their transport systems, bacteria also use siderophores in warfare against other microbes.12,14 For example, streptomycetes produce albomycins, which are natural siderophore–antibiotic conjugates (SACs) and highly effective antibiotics against Gram-negative Enterobacteriaceae.15 Albomycins are recognized by siderophore uptake machinery, transported into the cytoplasm, and activated by peptidase N, which cleaves the N-terminal serine-amide bond and releases the serine-bound t-RNA synthetase inhibitor to bind to its target.16–19 Inspired by Nature’s strategy, we develop an unbiased platform for the discovery of linkers that are cleaved by periplasmic proteases,20 which demonstrates that this platform can produce SACs with both broad and narrow spectra of activity. We target proteases in the periplasm, a compartment that contains >20 known proteases, because all SACs pass through the periplasm.
There are two categories of SACs, depending on the type of linker they possess: non-cleavable and cleavable. There has been significant progress in the development of non-cleavable SACs,21,22b,23 with the first siderophore–β-lactam conjugate Fetroja (cefiderocol) recently approved by the FDA.24 However, their use is often limited to periplasmic-targeting antibiotics (e.g., daptomycin, vancomycin, and β-lactams). The few examples of cytoplasmic-targeting, non-cleavable SACs may be less effective than the parent antibiotic for two reasons:25,26b–d,27–34 (1) the conjugate may not pass through the inner membrane to reach the cytoplasm, or (2) the bulky siderophore component may interfere with binding to the target.12,25b,35 Therefore, cleavable linkers are traditionally thought to be required for SAC compatibility with cytoplasmic-targeting antibiotics.21 The majority of Gram-positive antibiotics are cytoplasmic-targeting and may require a cleavable linker to be converted into SACs for Gram-negative pathogens.12,21,36 We show that protease-cleavable linkers improve the activity of SACs containing periplasmic- and cytoplasmic-targeting antibiotics.
Over the past 30 years, only a few cleavable linker strategies have been developed for SACs and a number of challenges remain.22,25–27,35,21,37–39 Despite optimization for hydrolytic stability,35 ester linkers for SACs (e.g., A, Figure 1A) are susceptible to premature cleavage prior to bacterial-cell entry.25,26c,d,27,35,39 SACs with disulfide (e.g., B) and trimethyl-lock linkers based on reduction- (e.g., C), phosphatase (e.g., D), and esterase-triggered cleavage mechanisms were less active than the parent antibiotic.26,38 Recent work by Nolan demonstrated that cytoplasmic siderophore degradation by the siderophore-hydrolase IroD confers high activity to a conjugate with a non-cleavable linker.37 Miller and co-workers developed a dual drug conjugate with the cleavable β-lactam linker E.22a However, more alternatives are needed as the physiological instability of β-lactams can lead to hyper-sensitivity and allergic reactions in patients.40a Protease-cleavable linkers have the potential to overcome the limitations of cleavable SACs reported to date.
Figure 1.
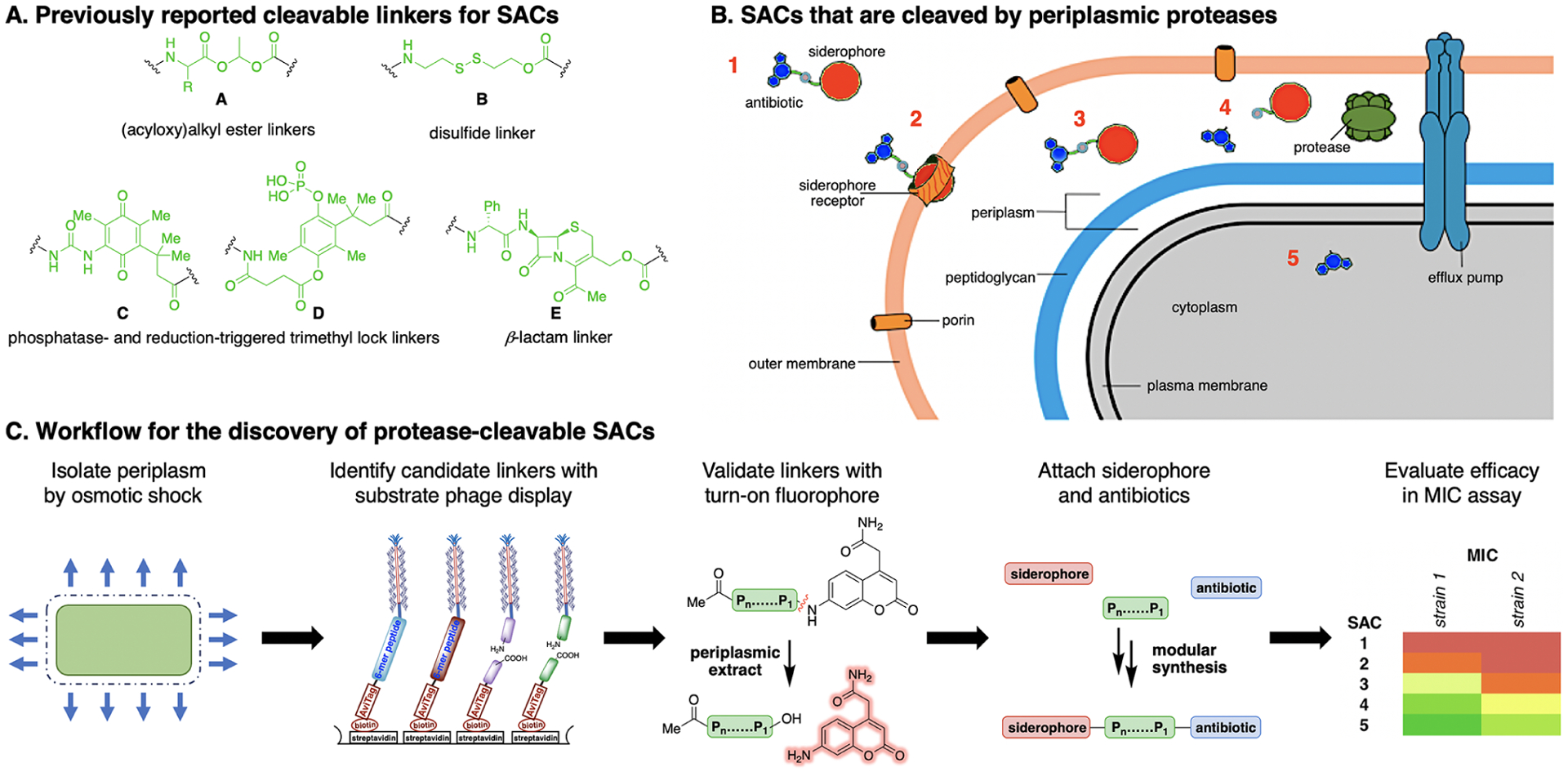
(A) Selected cleavable linkers that have previously been used for SACs. (B) Concept for SACs that contain a linker that can be cleaved by periplasmic proteases. (C) Workflow for the development of protease-cleavable SACs.
With few exceptions,22a,27,39 cleavable SACs have incorporated a DNA-gyrase-inhibiting fluoroquinolone antibiotic, ciprofloxacin or norfloxacin,40b,41 which are already active against Gram-negative pathogens. These SACs provide a method to study siderophore-mediated antibiotic delivery because release of the antibiotic from the conjugate is often required to observe significant growth inhibition.21,25,26,35,37,38 In line with this trend, we investigate a macrolide that is already active in Gram-negative bacteria as one strategy to interrogate SAC cleavage.
Protease-cleavable SACs are classified as prodrugs.42,43 Protease-activated prodrugs that are cleaved by mammalian proteases include antibody–drug conjugates,44–50 antibody–antibiotic conjugates,51,52 peptide–drug conjugates,53 macromolecular prodrugs,42,54 and protease-activatable photosensitizers,55,56 with the cathepsin B-sensitive valyl-citrulline (Val-Cit) linker being the most successful and widely known.47,49 Although most protease-activated prodrugs target cancer, antibody–antibiotic conjugates are undergoing clinical trials for intracellular bacterial infections associated with difficult-to-treat persisters.51,52 However, antibody–antibiotic conjugates have been limited to the treatment of intracellular S. aureus, using a linker that was optimized for cleavage by mammalian proteases. Protease-activated prodrugs that are activated by bacterial proteases would clearly be of value for treatment of a variety of Gram-negative bacterial infections.
Several technologies have been developed to screen libraries of protease substrates,57 including indexed arrays of fluorogenic substrates,58,59 positional scanning of synthetic combinatorial libraries,60,61 substrate phage,62,63 multiplex substrate profiling by mass spectrometry,57,64–66 and others.58,67–71 Given the large size of achievable libraries, substrate phage display62 provides an unbiased selection tool to discover cleavable linkers for SACs. Conventional phage display has been used in vitro and in vivo to design targeted-peptide conjugates.72 For example, Wilfred van der Donk used phage display to select for ribosomally synthesized and post-translationally modified peptides (RiPPs) that bind to lipid II.72g However, “substrate phage display” has not been applied to prodrug development,63 and its use for profiling complex biological mixtures is limited.73 Here, we extend substrate phage display to identify cleavable peptide linkers for SACs that are activated by periplasmic proteases.
In designing protease-activated prodrugs, there are advantages to targeting multiple proteases over an individual protease.74–76 For example, in vivo deletion mutants of cathepsin B retain the ability to release prodrugs from the combined activity of several proteases.77,78 Indeed, targeting multiple proteases might minimize resistance when designing antibiotic prodrugs. Thus, we screened broadly for peptides that are cleaved by the proteases present in an unfractionated periplasmic extract.
RESULTS
Substrate Phage Display Leads to WSPKYM-RFG and WSWC-KWASG as Substrates for Periplasmic Cleavage.
To discover efficient peptide substrates using the method of substrate phage,62 we built a random hexapeptide library genetically fused to the pIII gene of M13 bacteriophage. A phagemid vector allows monovalent display of the corresponding protein on the tip of the phage. A GGS spacer was incorporated at each end of the randomized peptide to enhance flexibility. An AviTag sequence was also incorporated at the N-terminus for biotinylation of the displayed peptides. The biotin is used to immobilize the phage library on a streptavidin-coated surface, and a protease can then cleave at favorable peptide sequences. Proteolysis releases the phage, which are then amplified and sequenced to determine the favorable substrates for the protease of interest.
The process of “biopanning” entails the following steps: (1) enzymatic biotinylation of the AviTag sequence,79,80 (2) immobilization of the biotinylated library on streptavidin 96-well plates, (3) cleavage of the immobilized library by incubation with the periplasmic extract of E. coli K12 MG1655 at 37 °C, (4) amplification of the eluted phage using E. coli TG-1 cells, and (5) isolation and purification of phage for the next round of selection. The periplasmic extract used in panning was obtained by osmotic shock of E. coli K12 MG1655.81 We carried out four rounds of selection (see Supporting Information, section VB, page S79), with the stringency being increased with each succeeding round by reducing extract concentration and decreasing the incubation time. The phagemids from the input library and final round of biopanning were isolated, barcoded, and submitted for next-generation sequencing.82 Sequences for further characterization were ranked based on the extent of enrichment relative to the original library (Table 1).
Table 1.
Highly Enriched Sequences Found through Substrate Phage Display
| sequence | reads | initial reads | enrichment factor |
|---|---|---|---|
| KNQSLG | 10652 | 0.5 | 21304 |
| GSDSSV | 9239 | 0.5 | 18478 |
| NHADVH | 8138 | 0.5 | 16276 |
| KSEMLS | 7742 | 0.5 | 15484 |
| WCKWAS | 15307 | 1 | 15307 |
| PKYMRF | 13192 | 1 | 13192 |
Six highly enriched sequences (SKNQSLGG, SGSDSSVG, SNHADVHG, SKSEMLSG, SWCKWASG, and SPKYMRFG) were synthesized with a Ser at the N-terminus and a Gly at the C-terminus to mimic the GGS spacers in the phage library. A tryptophan was added to the N-terminus to facilitate detection by HPLC. Each peptide was found to be cleaved to varying extents following treatment with periplasmic extract for 18 h at 37 °C. The cleavage sites and extent of proteolysis were evaluated by LC/MS (Table S5), which revealed that the sequences WSWC-KWASG (dash indicates site of cleavage) and WSPKYM-RFG may be optimal linkers for cleavable SACs.82 In addition to their promising cleavage profiles described in Table S5, the presence of these sequences in the original library contributed to their selection as potential linker candidates. In protease substrate nomenclature, the Cys and Met residues at the N-terminal side of the scissile bond for WSWC-KWASG and WSPKYM-RFG are designated as the P1 positions, while the Lys and Arg residues at the C-terminal side are designated as the P1′ positions. Although the residues on the P′ side are sometimes important for efficient cleavage,83 this is not always the case.84
WSPKYM Conjugates Are Efficiently Cleaved without a P′ Peptide.
With the candidate sequences WSWC-KWASG and WSPKYM-RFG in-hand, we asked whether the residues on the C-terminal P′ sequence were required for proteolysis (Figure 2A). To probe this question, we used a solid-phase method to synthesize fluorescent substrates. A 7-amino-4-carbamoylmethylcoumarin (acc)85,86 was coupled directly to the P1 Cys and Met residues as an antibiotic surrogate (see Supporting Information, section IIIF, pp S67–S72). We were indeed pleased to find that the treatment of peptide 1 (25 μM) with periplasmic extract led to rapid release of the fluorescent amino coumarin. HPLC analysis also showed the substrate was fully consumed at the end of the reaction (Figure S9). As expected, the rate of cleavage was dependent on the substrate concentration, showing partial saturation at concentrations over 12.5 μM (Figures S4 and S5). However, we did not attempt to fit a value of KM, given the heterogeneous nature of the proteolytic composition of the extract. On the other hand, the amount of acc released from peptide 2 (WSWC-coumarin) was comparably insignificant under these conditions (Figure 2B). Thus, WSPKYM was determined to be more suitable than WSWC for the development of cleavable SACs.
Figure 2.
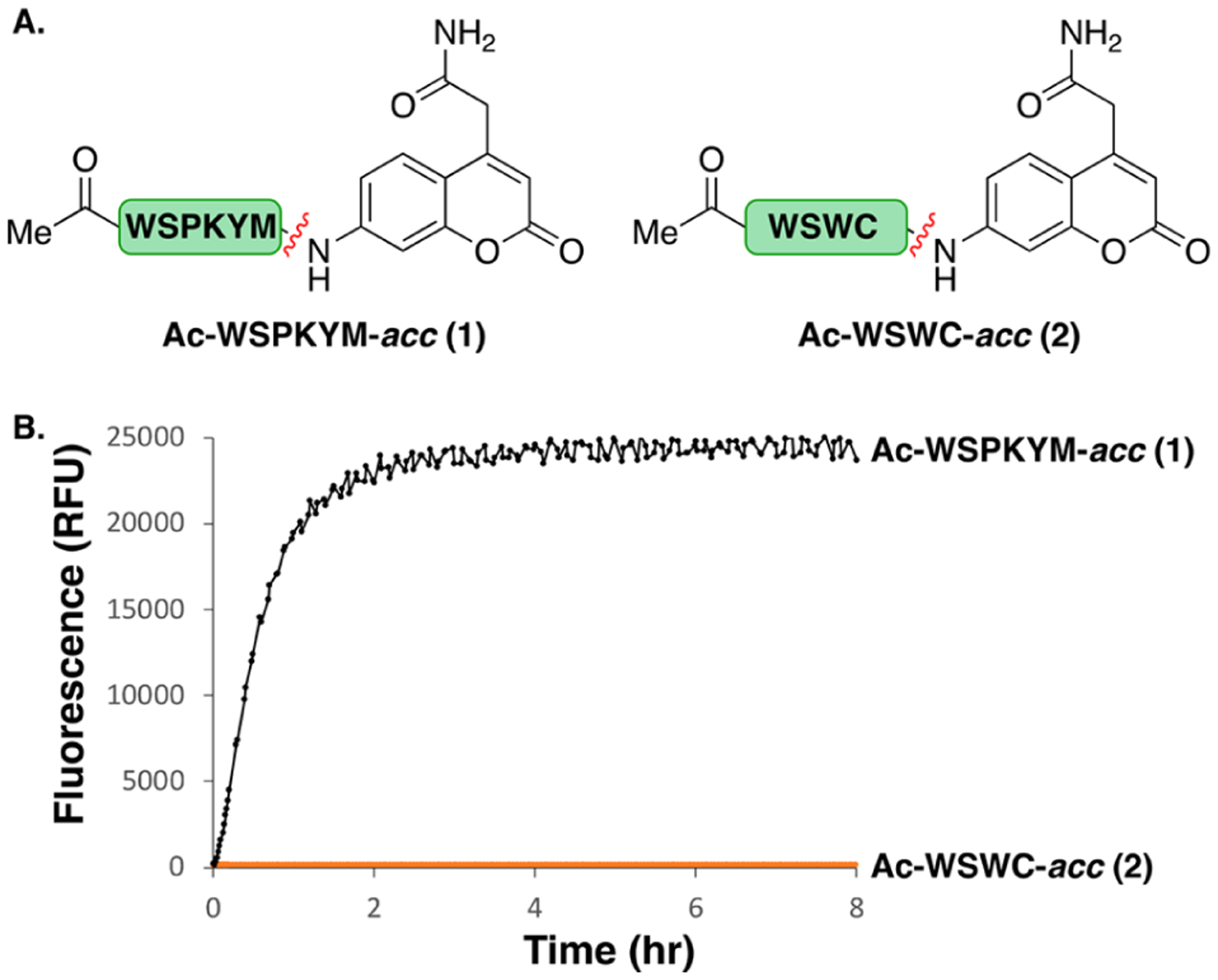
(A) Peptides with turn-on fluorophore, acc, as an antibiotic surrogate. (B) Evaluation of acc cleavage from peptides 1 and 2 in a periplasmic extract of E. coli K12 MG1655 (400 μg/mL) at 37 °C.
To determine the type of protease responsible for acc release in a periplasmic extract of E. coli, we evaluated a variety of protease inhibitors (Figure S6), which revealed that the enzymes of interest include a metalloprotease or a calcium-dependent protease. We also used alanine positional scanning to evaluate the sequence dependence for efficient cleavage. The starting peptide Ac-WSPKYM-acc (1) was the most efficient substrate (Figure S7). Individual substitutions of Ala at Tyr (P2), Trp (P6), and Lys (P3) resulted in large decreases in rate, while substitutions at other positions resulted in an approximately 2-fold decrease in rate. These findings indicate that substrate phage was effective in discovering an optimized substrate for cleavage.
We also examined the ability of human and mouse serum to cleave 1. We were pleased to find that under conditions where 1 is rapidly cleaved by periplasmic extract, human serum released acc with a half-life of approximately 3–4 h (Figure S8). However, the compound is cleaved more rapidly by mouse serum, indicating that some optimization would be necessary for applications using mouse models.
Design and Synthesis of SACs That Incorporate Solithromycin, Daptomycin, and Eperezolid-NH2.
To explore the versatility of SACs, we selected three structurally and mechanistically diverse antibiotics that act on targets in either the periplasm or the cytoplasm. Each antibiotic has an amine, which can be unmasked upon proteolysis of the WSPKYM linker. The lipopeptide daptomycin (4) interacts with the cytoplasmic membrane in Gram-positive bacteria, leading to increased membrane permeability and membrane depolarization.87a–c However, it is ineffective against Gram-negative bacteria and challenging to functionalize without loss of potency.87d Nevertheless, the Miller group has shown that daptomycin can gain activity in Gram-negative species if conjugated to a siderophore with a non-cleavable linker.22b,23 Here, we examine the use of a protease-cleavable linker for this system.
We also chose two ribosomal protein synthesis inhibitors, amino-oxazolidinone 5 and solithromycin 6, as examples of antibiotics that must gain access to the cytoplasm to be active.22a Since both oxazolidinones and macrolides bind deep within the large ribosomal subunit in fairly occluded binding sites, siderophore conjugates without cleavable linkers have been met with limited success, potentially due to interference of the linkers with binding.12,21,36,88,89 Our strategy would avoid this complication by enabling release of the parent antibiotics.
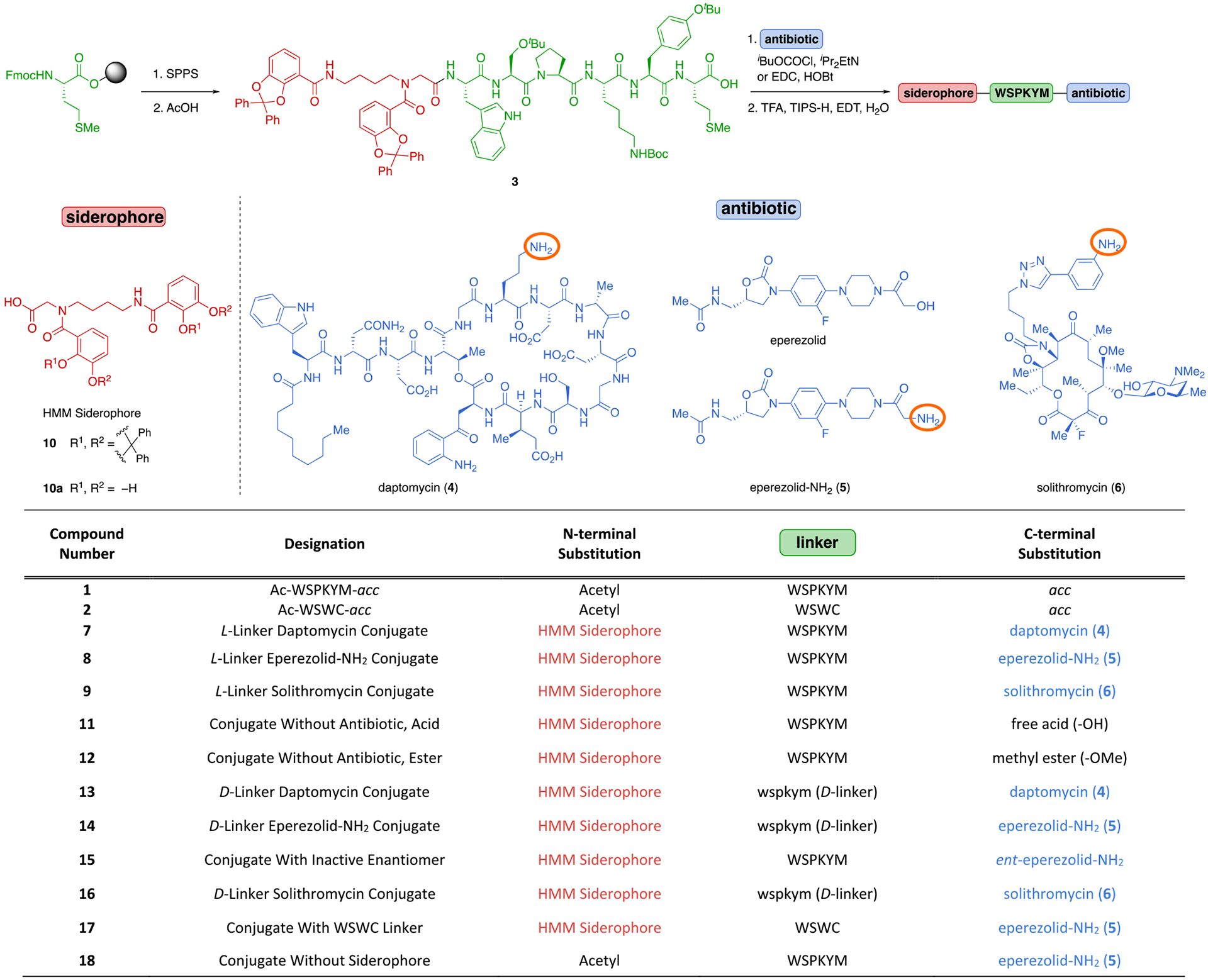
For attachment to the N-terminal side of the linker, we sought a siderophore that was synthetically accessible and compatible with a variety of bacterial siderophore uptake systems. The bis-catecholate, azotochelin-like90 siderophore (Heinisch–Möllmann–Miller (HMM) siderophore, Table 2)22,91a,b was selected due to its ease of synthesis and its ability to carry large cargo (e.g., daptomycin) into A. baumannii, E. coli, and P. aeruginosa.22,23 Furthermore, β-lactam conjugates with non-cleavable linkers that used tetra-acetate derivatives of siderophore 10a were active against MDR strains capable of efflux.91c We used a modified version of Miller’s protocol to access siderophore 10, which has acid-labile ketal protecting groups that can be removed concomitantly with tert-butyl and tert- butoxycarbonyl (Boc) protecting groups on the amino acid side chains.82
Table 2.
Modular Synthetic Platform for SAC Synthesis
|
For detailed synthetic methods, see Supporting Information.
We developed a modular synthetic route that enables the facile incorporation of a variety of linkers, antibiotics, and siderophores (Table 2). Gram-scale linker assembly and subsequent siderophore attachment were accomplished on solid-phase to provide the partially protected intermediate 3 in 50% overall yield, and the antibiotic was then coupled to the C-terminus in solution. Following acidolytic deprotection, the final SACs (7–9) were obtained in 12–53% yield over two to four steps.
Several aspects of our route merit further discussion. The majority of the synthesis proceeds on solid phase, simplifying purification and facilitating parallel synthesis of analogues. Antibiotics are directly attached in the penultimate step, enabling rapid access to the final antibiotic conjugates from intermediate 3.82 The synthesis requires only one HPLC purification, and the final products are purified by trituration. Daptomycin and solithromycin are commercially available and the oxazolidinones were synthesized following the protocols of Miller22a and Rafai Far.91d
We also synthesized several controls to probe the mechanism of action of 7, 8, and 9 using modifications of our existing protocol (11–18, Table 2). These included conjugates with d-amino acid linkers (e.g., 13, 14, and 16), which are not readily cleaved by proteases, enabling us to determine if proteolytic linker cleavage is responsible for the observed activity (see Supporting Information, section VIII, page S139).92 To confirm on-target activity of the antibiotic, we evaluated conjugates that lack an antibiotic or contain an inactive enantiomer of the antibiotic (e.g., 11, 12, and 15). To compare the effectiveness of conjugates containing a peptide linker that did not release acc in periplasmic extract (Figure 2B), we synthesized WSWC conjugate 17. We also synthesized a siderophore-free conjugate (18) to determine the dependence of activity on the siderophore.
Determination of the Antibacterial Activity of SACs 7–9 and Iron-Dependent Activity.
The minimum inhibitory concentrations (MICs) of conjugates 7–9 were evaluated according to the standard CLSI antimicrobial susceptibility testing guidelines in Meuller–Hinton-II (MH-II) broth with dipyridyl to sequester iron from the media and promote siderophore-mediated transport (Tables 3 and 4, Table S1A,B).82,93 Controls that lacked a siderophore did not show activity dependence on dipyridyl concentration, while the siderophore conjugate became increasingly active at higher concentrations of dipyridyl (Table S3A–C). This phenomenon can be explained by the enhanced expression of outer-membrane transport proteins for siderophore uptake in iron-deficient media.91e The absence of dipyridyl from the growth medium dramatically attenuated siderophore–conjugate activities without influencing the MIC of the free antibiotic.82 These results correlate well with expected growth-inhibitory activity of SACs.
Table 3.
Antibacterial Activity (MIC in μM) and In Vitro Cleavage of Eperezolid-NH2 Conjugate 8 and Derivatives Thereofa
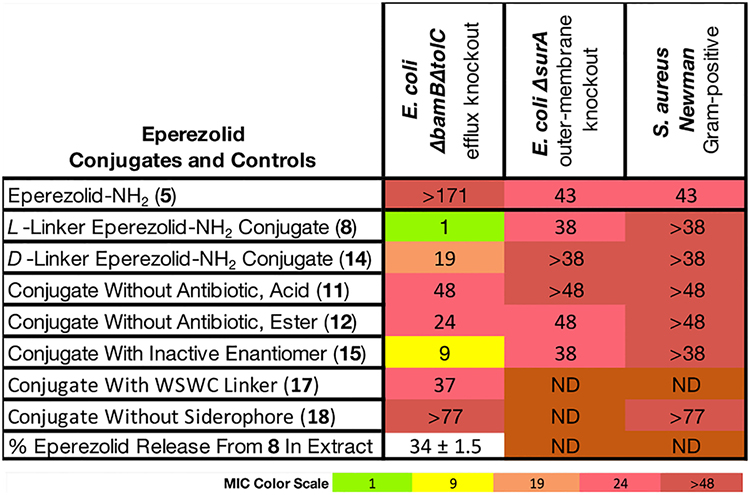
|
For strain descriptions and extract cleavage procedure, see Supporting Information, Table S1B. ND = not determined.
Table 4.
Antibacterial Activity (MIC in μM) of Daptomycin SAC 7 and Derivatives Thereofa
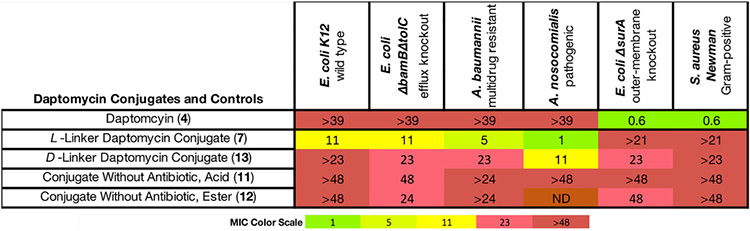
|
For strain descriptions, see Supporting Information. ND = not determined. Daptomycin is a calcium-dependent lipopeptide. MIC assays of daptomycin conjugates were conducted in the presence of 100 μg/mL CaCl2 in MH-II media.103
We included 15 bacterial strains in our assay (14 Gram-negative and one Gram-positive)94–98 and have highlighted selected activities below (for full-activity tables and strain details, see Table S1B). Two genetically modified strains of E. coli were included: a ΔsurA strain that is deficient in outer-membrane proteins and has increased permeability,95a and a ΔbamBΔtolC mutant, which has a deficient BamACDE outer-membrane-assembly complex and lacks the TolC-transport protein.95a,b This strain is widely used because it is defective in small-molecule efflux.
Solithromycin is active in many Gram-negative and Gram-positive species, and the strategy of using a Gram-negative antibiotic is frequently used to evaluate the efficiency of linker cleavage for cleavable SACs (see Introduction).21,25,26,35,37,38 The l-linker solithromycin conjugate 9 was comparably active to solithromycin in several pathogenic Gram-negative strains, and the d-linker conjugate 16 was inactive (Table S1A,B). In E. coli, however, conjugates 9 and 16 had similar activity, suggesting that the d-linker conjugate should not be used to evaluate linker cleavage in this species. We found that this may be due to the ability of the entire conjugate to inhibit the 70S E. coli ribosome (Figure S1A). However, the differences in activity between the d- and the l-solithromycin conjugates in pathogenic strains suggested that the linker may enable the release of Gram-positive antibiotics (Table S1A and Figure S1A). With this promising result in-hand, we then proceeded to investigate Gram-positive-only antibiotics (vide infra).
Oxazolidinone Conjugate 8 Was Active against E. coli ΔbamBΔtolC (Table 3).
The oxazolidinone class of antibiotics are active against Gram-positive bacteria, but members of this class lack activity against Gram-negative bacteria, due to the presence of endogenous efflux pumps (Figure S1B). Nevertheless, mutants of E. coli such as ΔbamBΔtolC are susceptible to oxazolidinones because these bacterial strains have disruptions in their efflux systems. This strain is susceptible to eperezolid (Table S1B), but the corresponding amine variant, eperezolid-NH2 (5), was inactive (MIC > 171 μM). This is likely a result of the inability of 5 to diffuse through the outer and inner membranes as indicated by the data in Table 3 (vide infra). We therefore asked whether conjugate 8 could deliver 5 into a ΔbamBΔtolC mutant of E. coli. We were pleased to discover an MIC of 1 μM for 8 in this mutant; the corresponding derivative 14 with an all-d linker showed strongly decreased activity with an MIC of 19 μM, which is consistent with low activities of previously reported oxazolidinone conjugates with non-cleavable linkers.39 In contrast to eperezolid-NH2 (5), eperezolid conjugate 8 displayed only 10% inhibition in a cell-free translation assay at a concentration of 38 μM (Figure S1A), indicating that the intact conjugate does not inhibit the ribosome. These findings suggest that potent inhibition of bacterial growth requires enzymatic cleavage of the linker. Supporting this suggestion, 34% cleavage of 8 to the parent antibiotic eperezolid-NH2 occurred after 11 h of incubation with bacterial periplasmic extract (Table 3). Finally, as expected, conjugate 8 was not active in wild-type strains with functional endogenous efflux pumps (Table S1B).
Eperezolid-NH2 (5) has only minimal activity (MIC = 43 μM) against both E. coli ΔsurA and S. aureus Newman, which lack outer membrane proteins, indicating that the cytoplasmic membrane of both Gram-positive and Gram-negative bacteria provide a barrier for the diffusion of 5 into the cytoplasm. There are two possible explanations for the potent activity of 8 given the lack of activity of 5 in E. coli ΔbamBΔtolC: (1) conjugate 8 may be actively transported to the cytoplasm and activated by a cytoplasmic protease99 or (2) cleavage in the periplasm may lead to large differences in the concentrations of molecules in the periplasm versus the cytoplasm, enhancing the effective concentration of 5 in the cytoplasm and hence potency.
We also synthesized a number of additional control molecules to probe the antibacterial mechanism of 8, including conjugate 17 with the WSWC linker (Table 3); this linker did not release an acc fluorophore from peptide 2 (Figure 2). Not surprisingly, this analogue had only weak (MIC = 37 μM) activity, as did compounds 11, 12, and 15 that lacked an active antibiotic payload. Similarly, the conjugate 18 without a siderophore was inactive. However, methyl ester 15 retained a modicum of activity (9 μM), possibly by a mechanism similar to many non-helical proline-containing cationic antimicrobial peptides.100
Miller and co-workers published a highly similar eperezolid conjugate using the same siderophore and eperezolid-NH2 derivative with a cleavable β-lactam linker.22a This conjugate exhibited activity against a number of strains of Gram-negative bacteria, including some with high-level β-lactam resistance. Against strains with high β-lactamase content for which the β-lactam linker did not measurably contribute to activity (up to 50 μM), the MIC of the conjugates was 6 μM. We did not observe activity of conjugate 8 in the same A. baumannii strain (ATCC BAA-1797), which could be due to inherent differences in the two linkers.
Daptomycin Conjugate 7 Exhibits High Activity against Acinetobacter Species (Table 4).
Daptomycin is used to treat Gram-positive infections, but it lacks activity against Gram-negative species. Therefore, we were gratified to find that the cleavable l-linker daptomycin conjugate 7 showed species-specific activity against A. nosocomialis, A. baumannii, and E. coli, with MIC values in the 1 to 10 μM range, while daptomycin itself was inactive against these species (MIC > 39 μM). The highest activities were observed in Acinetobacter species (MIC = 1–5 μM). Similarly, Miller and co-workers revealed that non-cleavable daptomycin conjugates displayed selective activity against A. baumannii.22b,23 These findings indicate that this approach has the potential to produce precision antibiotics with relatively narrow-spectrum activity. Moreover, as expected, 7 was inactive against S. aureus as it was not proteolytically activated due to the absence of a periplasm and periplasmic proteases. The activities of ornithine-functionalized daptomycin analogs in S. aureus are highly dependent on the side chain, and the WSPKYM linker reduces the activity of daptomycin in this species.87d The activity of 7 cannot be compared to previously reported non-cleavable linkers because the specific linker contributes to the activity of non-cleavable daptomycin conjugates (Figure S3).22b,23 Therefore, we limit our comparisons of the cleavable conjugate 7 to its closely related non-cleavable diastereomer 13.
The d-linker conjugate 13 was 2- to 11-fold less active than 7 against E. coli, A. baumannii, and A. nosocomialis and did not entirely lose activity as would be expected because daptomycin can still engage its target in the periplasm with a noncleavable linker.22,23 Nevertheless, the l-linker conjugate 7 improved the activity up to 11-fold, suggesting that cleavable linkers may benefit SACs containing periplasmic-targeting antibiotics. Finally, derivatives of 7 lacking the daptomycin payload (11 and 12) were essentially inactive. Taken together, the data in Table 4 indicates that we have successfully repurposed daptomycin for Gram-negative bacteria, and the enhanced activity of 7 relative to 13 is consistent with our guiding hypothesis of stereospecific proteolytic activation. It is also clear that proteolytic activation likely occurred in the periplasm, rather than by an extracellular protease in the medium given that daptomycin lacks activity against Gram-negative species.
To understand the lower activity observed for E. coli (MIC = 11 μM) relative to Acinetobacter species (MIC = 1–5 μM), we note that the release of daptomycin from conjugate 7 in periplasmic extract of E. coli was not observed by HPLC analysis (see Supporting Information, section VIII, pp S120–S124), which may be due to unfavorable steric interactions between the approaching protease(s)101 and the large daptomycin molecule. Following incubation of 7 in periplasmic extracts of A. baumannii and A. nosocomialis, however, the HPLC trace revealed cleavage products with retention times that overlapped with daptomycin (see Supporting Information, section VIII, pp S115–S119).
These results are also of interest with respect to the mechanism of action of daptomycin. It has been previously reported that the target for daptomycin (4) may be absent in Gram-negative species due to the differing membrane compositions between Gram-positive and Gram-negative bacteria.102 Given that 7 is active against E. coli, it would appear that daptomycin is able to act on the cytoplasmic membranes of these Gram-negative species once they gain access. Also, our finding that daptomycin itself is equipotent against S. aureus Newman and in the outer membrane-compromised E. coli ΔsurA (MIC = 0.6 μM) is consistent with this conclusion.
CONCLUSION
The strategy developed here should be broadly applicable for discovery of protease-activated peptide prodrugs for a variety of applications. Here, we focused on delivering antibiotics by designing protease-cleavable siderophore conjugates. By targeting E. coli periplasmic proteases, we were able to design conjugates that act against a broad (or narrow) spectrum of Gram-negative bacteria, illustrating the potential of this approach. Our results provide strong support for the overall mechanism of proteolytic release of the antibiotic from conjugates 7–9. Although we have not yet identified the proteases responsible for activity against our substrates, we purposefully avoided targeting a single protease to decrease the chances of resistance arising from mutants of a single protein. Moreover, the use of chemically stable amide linkers provides an advantage to targeting proteases over esterases and β-lactamases by avoiding the need for esters and β-lactams, which are chemically more labile. Importantly, the modular design and facile synthetic route provides opportunity for rapid synthesis of SACs with different siderophores, linkers, and antibiotics. This has led to the discovery of cleavable conjugates with activity against several clinically relevant Gram-negative pathogens.
Throughout the course of this work, we made a number of unexpected discoveries with impacts that extend beyond the scope of protease-cleavable prodrugs. The l-linker daptomycin conjugate 7 completely lacks the Gram-positive activity of daptomycin and has gained Gram-negative activity, effectively “flipping” the spectrum of activity of this potent antibiotic. We found that conjugates with d-linkers, which are unlikely to be cleaved proteolytically, have moderate activity against several strains of Gram-negative bacteria. Perhaps the most unexpected results are the activities of the solithromycin conjugates 9 and 16 in a cell-free translation assay, which indicate that these large (MW > 2000) conjugates may directly inhibit the ribosome in E. coli (see Supporting Information, Figure S1A). These results are extremely surprising in the context of solithromycin–ribosome structural data104 and may provide the basis for new macrolide–peptide hybrid antibiotics.
Additional studies will be required to optimize the current linkers for use as practical drugs. The current linker has reasonable stability in human serum, which could doubtlessly be enhanced by limited structure–activity relationships (Figure S8). The peptide WSPKYM also contains more than one cleavage site (Table S5), which may complicate the drug activation mechanism. Our conjugates do not show significant activity against P. aeruginosa (Table S1B and Figure S2), a pathogen prone to resistance, which could be due to the following: (1) insufficient linker cleavage may not lead to growth inhibition because the linker was optimized for E. coli, (2) the target for daptomycin may be present in minimal amounts, depending on the strain or species of Gram-negative bacteria,102 or (3) solithromycin and eperezolid might be efflux substrates in this organism.
In summary, this work provides a robust methodology for selection and screening of Trojan-horse prodrugs applied to the persistent and growing problem of antibacterial resistance. The resulting conjugates from this platform improve upon existing cleavable linkers for SACs. Using phage display, one can rapidly screen vast peptide libraries, and by varying the selection strategy one can screen for linkers with desired characteristics. For example, by using periplasmic extracts from different species of bacteria in succeeding selections, one can ensure broad activity over the desired range of bacteria. Alternatively, negative selection could be incorporated to select against cleavage of serum proteases or beneficial members of the microbiome. Thus, the potential for fine-tuning the protocol for future practical applications is substantial.
Supplementary Material
ACKNOWLEDGMENTS
We thank Adam Cotton, Peter Rowheder, Dr. Sam Ivry, and Dr. Matthew Ravalin for helpful discussions. We thank Bruk Mensa for helpful discussions and for providing P. aeruginosa ATCC 10145, S. typhi ATCC 700931, S. aureus Newman, E. coli K12 MG1655, E. coli BW25113 ΔtolC, and E. coli BW25113 ΔsurA. We thank Neha Prasad for helpful discussions and for providing K. pneumoniae MGH 78578, E. cloacae ATCC 13047, E. aerogenes ATCC 13048, P. aeruginosa PA01, P. aeruginosa PA14, and S. enterica 14028s. We thank Jenna Pellegrino for providing E. coli BW25113 ΔbamBΔtolC and for helpful discussions on in vitro translation assays. We thank Professor Joanne Engel for a generous gift of A. nosocomialis M2. J.H.B. was supported by the National Institutes of Health under the Ruth L. Kirschstein National Research Service Award 5T32HL007731-27 from the National Heart Lung and Blood Institute (NHLBI). Q.E. was supported by the National Science Foundation Graduate Research Fellowship Program under Grant no. 1650113. B.A. was supported by the National Institutes of Health under Grant no. T32 Al 0605357. This project was supported by the David and Lucile Packard Foundation (I.B.S.) and the National Institutes of Health under Grant no. R35 GM122603 (W.F.D.).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c06987.
Supplementary Tables S1–S5 and Figures S1–S9; general information; experimental procedures and compound charcterization; MIC, periplasmic cleavage assays, and cell-free translation; substrate phage display; references cited; select NMR spectra; and HPLC traces for cleavage in periplasmic extract (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.0c06987
The authors declare no competing financial interest.
Contributor Information
Jonathan H. Boyce, Department of Pharmaceutical Chemistry, University of California, San Francisco, California 94158, United States; Cardiovascular Research Institute, University of California, San Francisco, California 94158, United States;.
Bobo Dang, Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang 310024, China; Center for Infectious Disease Research, Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang 310024, China; Institute of Biology, Westlake Institute for Advanced Study, Hangzhou, Zhejiang 310024, China;.
Beatrice Ary, Department of Pharmaceutical Chemistry, University of California, San Francisco, California 94158, United States.
REFERENCES
- (1).Llaca-Díaz JM; Mendoza-Olazarán S; Camacho-Ortiz A; Flores S; Garza-Gonzalez E One-year surveillance of ESKAPE pathogens in an intensive care unit of Monterrey, Mexico. Chemotherapy 2012, 58, 475–481. [DOI] [PubMed] [Google Scholar]
- (2).Pendleton JN; Gorman SP; Gilmore BF Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther 2013, 11, 297–308. [DOI] [PubMed] [Google Scholar]
- (3).(a) Breijyeh Z; Jubeh B; Karaman R Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Li XZ; Plésiat P; Nikaido H The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev 2015, 28, 337–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).(a) Galdiero S; Falanga A; Cantisani M; Tarallo R; Della Pepa ME; D’Oriano V; Galdiero M Microbe-host interactions: structure and role of Gram-negative bacterial porins. Curr. Protein Pept. Sci 2012, 13, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nikaido H Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev 2003, 67, 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).(a) Choi U; Lee CR Distinct Roles of Outer Membrane Porins in Antibiotic Resistance and Membrane Integrity in Escherichia coli. Front. Microbiol 2019, 10, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Braun V; Braun M Active transport of iron and siderophore antibiotics. Curr. Opin. Microbiol 2002, 5, 194–201. [DOI] [PubMed] [Google Scholar]; (c) Livermore DM Antibiotic uptake and transport by bacteria. Scand. J. Infect Dis Suppl 1990, 74, 15–22. [PubMed] [Google Scholar]
- (6).(a) Perlman D The roles of the Journal of Antibiotics in determining the future of antibiotic research. Jpn. J. Antibiot 1977, 30, S201–S206. [PubMed] [Google Scholar]; (b) Diarra MS; Lavoie MC; Jacques M; Darwish I; Dolence EK; Dolence JA; Ghosh A; Ghosh M; Miller MJ; Malouin F Species selectivity of new siderophore-drug conjugates that use specific iron uptake for entry into bacteria. Antimicrob. Agents Chemother 1996, 40, 2610–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ji C; Juárez-Hernández RE; Miller MJ Exploiting bacterial iron acquisition: siderophore conjugates. Future Med. Chem 2012, 4, 297–313. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Page MG Siderophore conjugates. Ann. N. Y. Acad. Sci 2013, 1277, 115–126. [DOI] [PubMed] [Google Scholar]; (e) Tillotson GS Trojan Horse Antibiotics-A Novel Way to Circumvent Gram-Negative Bacterial Resistance? Infect. Dis.: Res. Treat 2016, 9, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zheng T; Nolan EM Enterobactin-mediated delivery of β-lactam antibiotics enhances antibacterial activity against pathogenic Escherichia coli. J. Am. Chem. Soc 2014, 136, 9677–9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).(a) Watanabe NA; Nagasu T; Katsu K; Kitoh K E-0702, a new cephalosporin, is incorporated into Escherichia coli cells via the tonB-dependent iron transport system. Antimicrob. Agents Chemother 1987, 31, 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Curtis NA; Eisenstadt RL; East SJ; Cornford RJ; Walker LA; White AJ Iron-regulated outer membrane proteins of Escherichia coli K-12 and mechanism of action of catechol-substituted cephalosporins. Antimicrob. Agents Chemother 1988, 32, 1879–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Silley P; Griffiths JW; Monsey D; Harris AM Mode of action of GR69153, a novel catechol-substituted cephalosporin, and its interaction with the tonB-dependent iron transport system. Antimicrob. Agents Chemother 1990, 34, 1806–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Hashizume T; Sanada M; Nakagawa S; Tanaka N Comparison of transport pathways of catechol-substituted cephalosporins, BO-1236 and BO-1341, through the outer membrane of Escherichia coli. J. Antibiot 1990, 43, 1617–1620. [DOI] [PubMed] [Google Scholar]; (e) Nikaido H; Rosenberg EY Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: study with beta-lactam antibiotics containing catechol and analogous groups. J. Bacteriol 1990, 172, 1361–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) McKee JA; Sharma SK; Miller MJ Iron transport mediated drug delivery systems: synthesis and antibacterial activity of spermidineand lysine-based siderophore-beta-lactam conjugates. Bioconjugate Chem 1991, 2, 281–291. [DOI] [PubMed] [Google Scholar]; (g) Dolence EK; Minnick AA; Lin CE; Miller MJ; Payne SM Synthesis and siderophore and antibacterial activity of N5-acetyl-N5-hydroxy-L-ornithine-derived siderophore-beta-lactam conjugates: iron-transport-mediated drug delivery. J. Med. Chem 1991, 34, 968–978. [DOI] [PubMed] [Google Scholar]; (h) Ji C; Miller PA; Miller MJ Iron transport-mediated drug delivery: practical syntheses and in vitro antibacterial studies of tris-catecholate siderophore-aminopenicillin conjugates reveals selectively potent antipseudomonal activity. J. Am. Chem. Soc 2012, 134, 9898–9901. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Kohira N; West J; Ito A; Ito-Horiyama T; Nakamura R; Sato T; Rittenhouse S; Tsuji M; Yamano Y In Vitro Antimicrobial Activity of a Siderophore Cephalosporin, S-649266, against Enterobacteriaceae Clinical Isolates, Including Carbapenem-Resistant Strains. Antimicrob. Agents Chemother 2016, 60, 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).(a) Miethke M; Marahiel MA Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev 2007, 71, 413–451. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hider RC; Kong X Chemistry and biology of siderophores. Nat. Prod. Rep 2010, 27, 637–657. [DOI] [PubMed] [Google Scholar]; (c) Chu BC; Garcia-Herrero A; Johanson TH; Krewulak KD; Lau CK; Peacock RS; Slavinskaya Z; Vogel HJ Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. BioMetals 2010, 23, 601–611. [DOI] [PubMed] [Google Scholar]
- (10).(a) Ribeiro M; Simões M Siderophores: A Novel Approach to Fight Antimicrobial Resistance In Pharmaceuticals from Microbes, Environmental Chemistry for a Sustainable World, Vol. 28; Arora D, Sharma C, Jaglan S, Lichtfouse E, Eds.; Springer: Cham, 2019. [Google Scholar]; (b) Ma L; Terwilliger A; Maresso AW Iron and zinc exploitation during bacterial pathogenesis. Metallomics 2015, 7, 1541–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Messenger AJM; Barclay R Bacteria, Iron and Pathogenicity. Biochem. Educ 1983, 11, 54–63. [Google Scholar]
- (11).Noinaj N; Guillier M; Barnard TJ; Buchanan SK TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol 2010, 64, 43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wencewicz TA; Miller MJ Sideromycins as Pathogen-Targeted Antibiotics In Antibacterials, Topics in Medicinal Chemistry, Vol. 26; Fisher J, Mobashery S, Miller M, Eds.; Springer: Cham, 2017. DOI: 10.1007/7355_2017_19 [DOI] [Google Scholar]
- (13). For references discussing the transport of siderophores or siderophore–antibiotic conjugates across the outer and inner membrane of Gram-negative bacteria, see:; (a) Page MGP The Role of Iron and Siderophores in Infection, and the Development of Siderophore Antibiotics. Clin. Infect. Dis 2019, 69, S529–S537. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schalk IJ Siderophore–antibiotic conjugates: exploiting iron uptake to deliver drugs into bacteria. Clin. Microbiol. Infect 2018, 24, 801–802. [DOI] [PubMed] [Google Scholar]; (c) Schalk IJ; Guillon L Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids 2013, 44, 1267–1277. [DOI] [PubMed] [Google Scholar]; (d) Schalk IJ; Mislin GL; Brillet K Structure, function and binding selectivity and stereoselectivity of siderophore-iron outer membrane transporters. Curr. Top. Membr 2012, 69, 37–66. [DOI] [PubMed] [Google Scholar]; (e) Faraldo-Gómez JD; Sansom MSP Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol 2003, 4, 105–116. [DOI] [PubMed] [Google Scholar]; (f) Tonziello G; Caraffa E; Pinchera B; Granata G; Petrosillo N Present and future of siderophore–based therapeutic and diagnostic approaches in infectious diseases. Infect. Dis. Rep 2019, 11, 8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).(a) Wilson BR; Bogdan AR; Miyazawa M; Hashimoto K; Tsuji Y Siderophores in Iron Metabolism: From Mechanism to Therapy Potential. Trends Mol. Med 2016, 22, 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Holden VI; Bachman MA Diverging roles of bacterial siderophores during infection. Metallomics 2015, 7, 986–995. [DOI] [PubMed] [Google Scholar]; (c) Braun V; Pramanik A; Gwinner T; Köberle M; Bohn E Sideromycins: tools and antibiotics. BioMetals 2009, 22, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Pramanik A; Stroeher UH; Krejci J; Standish AJ; Bohn E; Paton JC; Autenrieth IB; Braun B Albomycin is an effective antibiotic, as exemplified with Yersinia enterocolitica and Streptococcud pneumoniae. Int. J. Med. Microbiol 2007, 297, 459–469. [DOI] [PubMed] [Google Scholar]
- (16).Stefanska AL; Fulston M; Houge-Frydrych CSV; Jones JJ; Warr SR A potent seryl tRNA synthetase inhibitor SB-217452 isolated from a Streptomyces species. J. Antibiot 2000, 53, 1346–1353. [DOI] [PubMed] [Google Scholar]
- (17). For select reports discussing the clinical use of albomycin, see:; (a) Gause GF Recent studies on albomycin, a new antibiotic. Br. J. Med 1955, 2, 1177–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kaliuzhnaia-Lukashova GM Clinical study of albomycin, colimycin and terramycin. Klinicheskaia meditsina 1958, 36, 30–5. [PubMed] [Google Scholar]; (c) Georgievskaia VS Clinical observations on the use of albomycin in purulent mastitis. Sovetskaia meditsina 1958, 22, 82–5. [PubMed] [Google Scholar]; (d) Danovoi ID Use of albomycin in an obstetrical–gynecological clinic. Akusherstvo i ginekologiia 1957, 33, 37–40. [PubMed] [Google Scholar]; (e) Uglova VM Comparative evaluation of the use of albomycin and furacillin in the treatment of infected wounds; experimental study. Vestnik khirurgii imeni I. I. Grekova 1956, 77, 73–80. [PubMed] [Google Scholar]; (f) Sigal AE Application of albomycin in the treatment of pulmonary suppurations. Klinicheskaia meditsina 1955, 33, 24–28. [PubMed] [Google Scholar]; (g) Berent IE; Gil’man KZ Experience in application of the new domestic antibiotic albomycin in dermatovenerology. Sovetskaia meditsina 1954, 18, 34–35. [PubMed] [Google Scholar]; (h) Raikher EA; El’man EF Application of albomycin in pneumonia in infants during their first months of life. Sovetskaia meditsina 1952, 16, 18–21. [PubMed] [Google Scholar]; (i) Gamburg RL Use of albomycin in pneumonia in children. Pediatriia 1951, 5, 37–44. [PubMed] [Google Scholar]; (j) Anonymous author. Antibiotic albomycin in the treatment of pneumonias and toxemias in infants during the first year of life. Fel’dsher i akusherka 1951, 12, 38. [PubMed] [Google Scholar]; (k) Krechmer BB; Val’ter EM; Baiandina SA Application of albomycin in pneumonia in infants. Sovetskaia meditsina 1951, 10, 10–13. [PubMed] [Google Scholar]
- (18).(a) Zeng Y; Kulkarni A; Yang Z; Patil PB; Zhou W; Chi X; Van Lanen S; Chen S Albomycin δ2 Provides a Template for Assembling Siderophore and Aminoacyl-tRNA Synthetase Inhibitor Conjugates. ACS Chem. Biol 2012, 7, 1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fiedler H-P; Walz F; Döhle A; Zähner H Albomycin: Studies on fermentation, isolation and quantitative determination. Appl. Microbiol. Biotechnol 1985, 21, 341–347. [Google Scholar]
- (19).Lin Z; Xu X; Zhao S; Yang X; Guo J; Zhang Q; Jing C; Chen S; He Y Total synthesis and antimicrobial evaluation of natural albomycins against clinical pathogens. Nat. Commun 2018, 9, 3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Merdanovic M; Clausen T; Kaiser M; Huber R; Ehrmann M Protein quality control in the bacterial periplasm. Annu. Rev. Microbiol 2011, 65, 149–168. [DOI] [PubMed] [Google Scholar]
- (21). For a recent, comprehensive overview of cleavable and non-cleavable SACs, see:; Negash KH; Norris JKS; Hodgkinson JT Siderophore – Antibiotic conjugate design: New drugs for bad bugs? Molecules 2019, 24, 3314–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22). For a list of references using the azotochelin-like Miller siderophore for SAC development, see:; (a) Liu R; Miller PA; Vakulenko SB; Stewart NK; Boggess WC; Miller MJ A synthetic dual drug sideromycin induces gram-negative bacteria to commit suicide with a gram-positive antibiotic. J. Med. Chem 2018, 61, 3845–3854. [DOI] [PubMed] [Google Scholar]; (b) Ghosh M; Lin Y-M; Miller PA; Mollmann U; Boggess WC; Miller M Siderophore conjugates of daptomycin are potent inhibitors of carbapenem resistant strains of Acinetobacter baumannii. ACS Infect. Dis 2018, 4, 1529–1535. [DOI] [PubMed] [Google Scholar]; (c) Miller MJ; Lin Y-M; Ghosh M; Patricia A; Moellmann U Antibacterial Sideromycins. Int. Patent Application PCT/IB2015/056915, February 25, 2016. [Google Scholar]; (d) Miller MJ; Cheng IJ Antibacterial Monobactams. Int. Patent Application PCT/US2018/053917, April 11, 2019. [Google Scholar]; (e) Carosso S; Liu R; Miller PA; Hecker SJ; Glinka T; Miller MJ Methodology for Monobactam Diversification: Syntheses and Studies of 4-Thiomethyl Substituted β-Lactams with Activity Against Gram–Negative Bacteria, Including Carbapenemase Producing Acinetobacter baumannii. J. Med. Chem 2017, 60, 8933–8944. [DOI] [PubMed] [Google Scholar]
- (23).Ghosh M; Miller PA; Möllmann U; Claypool WD; Schroeder VA; Wolter WR; Suckow M; Yu H; Li S; Huang W; Zajicek J; Miller MJ Targeted antibiotic delivery: selective siderophore conjugation with daptomycin confers potent activity against multidrug resistant Acinetobacter baumannii both in vitro and in vivo. J. Med. Chem 2017, 60, 4577–4583. [DOI] [PubMed] [Google Scholar]
- (24). https://www.fda.gov/news-events/press-announcements/fda-approves-new-antibacterial-drug-treat-complicated-urinary-tract-infections-part-ongoing-efforts.
- (25). For examples of (acyloxy)methyl ester linkers with low hydrolytic stability, see:; (a) Hennard C; Truong QC; Desnottes J-F; Paris J-M; Moreau NJ; Abdallah MA Synthesis and Activities of Pyoverdin-Quinolone Adducts: A Prospective Approach to a Specific Therapy Against Pseudomonas aeruginosa. J. Med. Chem 2001, 44, 2139–2151. [DOI] [PubMed] [Google Scholar]; (b) Rivault F; Liébert C; Burger A; Hoegy F; Abdallah MA; Schalk IJ; Mislin GLA Synthesis of pyochelin–norfloxacin conjugates. Bioorg. Med. Chem. Lett 2007, 17, 640–644. [DOI] [PubMed] [Google Scholar]; (c) Noël S; Gasser V; Pesset B; Hoegy F; Rognan D; Schalk IJ; Mislin GLA Synthesis and biological properties of conjugates between fluoroquinolones and a N3″-functionalized pyochelin. Org. Biomol. Chem 2011, 9, 8288–8300. [DOI] [PubMed] [Google Scholar]
- (26). For examples of esterase-, phosphatase-, and reduction-triggered linkers for fluoroquinolone release, see:; (a) Miller MJ; Cheng IJ Reduction-Triggered Antibacterial Sideromycins. U.S. Patent application US2016/0368878A1, April 23, 2016. [Google Scholar]; (b) Ji C; Miller MJ Siderophore-fluoroquinolone conjugates containing potential reduction-triggered linkers for drug release: synthesis and antibacterial activity. BioMetals 2015, 28, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Fardeau S; Dassonville-Klimpt A; Audic N; Sasaki A; Pillon M; Baudrin E; Mullié C; Sonnet P Synthesis and antibacterial activity of catecholate–ciprofloxacin conjugates. Bioorg. Med. Chem 2014, 22, 4049–4060. [DOI] [PubMed] [Google Scholar]; (d) Ji C; Miller MJ Chemical syntheses and in vitro antibacterial activity of two desferrioxamine B-ciprofloxacin conjugates with potential esterase and phosphatase triggered drug release linkers. Bioorg. Med. Chem 2012, 20, 3828–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27). For a desferridanoxamine-triclosan conjugate with a labile phenolic ester linker, see:; Wencewicz TA; Möllmann U; Long TE; Miller MJ Is drug release necessary for antimicrobial activity of siderophore-drug conjugates? Syntheses and biological studies of the naturally occurring salmycin “Trojan Horse” antibiotics and synthetic desferridanoxamine-antibiotic conjugates. BioMetals 2009, 22, 633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Md-Saleh SR; Chilvers EC; Kerr KG; Milner SJ; Snelling AM; Weber JP; Thomas GH; Duhme-Klair A-K; Routledge A Synthesis of citrate–ciprofloxacin conjugates. Bioorg. Med. Chem. Lett 2009, 19, 1496–1498. [DOI] [PubMed] [Google Scholar]
- (29).Juarez-Hernandez RE; Miller PA; Miller MJ Syntheses of Siderophore–Drug Conjugates Using a Convergent Thiol–Maleimide System. ACS Med. Chem. Lett 2012, 3, 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wencewicz TA; Miller MJ Biscatecholate–Monohydroxamate Mixed Ligand Siderophore–Carbacephalosporin Conjugates are Selective Sideromycin Antibiotics that Target Acinetobacter baumannii. J. Med. Chem 2013, 56, 4044–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wencewicz TA; Long TE; Möllmann U; Miller MJ Trihydroxamate Siderophore–Fluoroquinolone Conjugates Are Selective Sideromycin Antibiotics that Target Staphylococcus aureus. Bioconjugate Chem 2013, 24, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Milner SJ; Seve A; Snelling AM; Thomas GH; Kerr KG; Routledge A; Duhme-Klair A-K Staphyloferrin A as siderophore-component in fluoroquinolone-based Trojan horse antibiotics. Org. Biomol. Chem 2013, 11, 3461–3468. [DOI] [PubMed] [Google Scholar]
- (33).Souto A; Montaos MA; Balado M; Osorio CR; Rodriguez J; Lemos ML; Jimenez C Synthesis and antibacterial activity of conjugates between norfloxacin and analogues of the siderophore vanchrobactin. Bioorg. Med. Chem 2013, 21, 295–302. [DOI] [PubMed] [Google Scholar]
- (34).Fardeau S; Dassonville-Klimpt A; Audic N; Sasaki A; Pillon M; Baudrin E; Mullie C; Sonnet P Probing linker design in citric acid–ciprofloxacin conjugates. Bioorg. Med. Chem 2014, 22, 4049–4060. [DOI] [PubMed] [Google Scholar]
- (35).Zheng T; Nolan EM Evaluation of (acyloxy)alkyl ester linkers for antibiotic release from siderophore-antibiotic conjugates. Bioorg. Med. Chem. Lett 2015, 25, 4987–4991. [DOI] [PubMed] [Google Scholar]
- (36). For an oxazolidinone-conjugated SAC with a non-cleavable linker, see:; Paulen A; Hoegy F; Roche B; Schalk IJ; Mislin GLA Synthesis of conjugates between oxazolidinone antibiotics and a pyochelin analogue. Bioorg. Med. Chem. Lett 2017, 27, 4867–4870. [DOI] [PubMed] [Google Scholar]
- (37).Neumann W; Sassone-Corsi M; Raffatellu M; Nolan EM Esterase-catalyzed siderophore hydrolysis activates an enterobactin–ciprofloxacin conjugate and confers targeted antibacterial activity. J. Am. Chem. Soc 2018, 140, 5193–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Neumann W; Nolan EM JBIC, J. Biol. Inorg. Chem 2018, 23, 1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39). For a conjugate with a base-sensitive triazole-methylene carbamate linker, see:; Paulen A; Gasser V; Hoegy F; Perraud Q; Pesset B; Schalk IJ; Mislin GLA Synthesis and antibiotic activity of oxazolidinone–catechol conjugates against Pseudomonas aeruginosa. Org. Biomol. Chem 2015, 13, 11567–11579. [DOI] [PubMed] [Google Scholar]
- (40).(a) Khan DA; Banerji A; Bernstein JA; Bilgicer B; Blumenthal K; Castells M; Ein D; Lang DM; Phillips E Cephalosporin Allergy: Current Understanding and Future Challenges. J. Allergy Clin. Immunol. Pract 2019, 7, 2105–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gupta K; Hooton TM; Naber KG; Wullt B; Colgan R; Miller LG; Moran GJ; Nicolle LE; Raz R; Schaeffer AJ; Soper DE International Clinical Practice Guidelines for the Treatment of Acute Uncomplicated Cystitis and Pyelonephritis in Women: A 2010 Update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin. Infect. Dis 2011, 52, e103–e120. [DOI] [PubMed] [Google Scholar]
- (41).Castro W; Navarro M; Biot C Medicinal potential of ciprofloxacin and its derivatives. Future Med. Chem 2013, 5, 81–96. [DOI] [PubMed] [Google Scholar]
- (42).Choi KY; Swierczewska M; Lee S; Chen X Protease-activated drug development. Theranostics 2012, 12, 156–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).(a) Baurain R; Masquelier M; Deprez-De Campeneere D; Trouet A Amino acid and dipeptide derivatives of daunorubicin. 2. Cellular pharmacology and antitumor activity on L1210 leukemic cells in vitro and in vivo. J. Med. Chem 1980, 23, 1171–1174. [DOI] [PubMed] [Google Scholar]; For a recent review of protease-activated prodrugs, see:; Poreba M Protease-activated prodrugs: strategies, challenges, and future directions. FEBS J 2020, 287, 1936–1969. [DOI] [PubMed] [Google Scholar]
- (44).Law CL; Cerveny CG; Gordon KA; Klussman K; Mixan BJ; Chace DF; Meyer DL; Doronina SO; Siegall CB; Francisco JA; Senter PD; Wahl AF Efficient elimination of B-lineage lymphomas by anti-CD20-auristatin conjugates. Clin. Cancer Res 2004, 10, 7842–7851. [DOI] [PubMed] [Google Scholar]
- (45).Diamantis N; Banerji U Antibody-drug conjugates–an emerging class of cancer treatment. Br. J. Cancer 2016, 114, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Weidle UH; Tiefenthaler G; Georges G Proteases as activators for cytotoxic prodrugs in antitumor therapy. Cancer Genomics Proteomics 2014, 11, 67–79. [PubMed] [Google Scholar]
- (47).Senter PD; Sievers EL The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat. Biotechnol 2012, 30, 631–637. [DOI] [PubMed] [Google Scholar]
- (48).Deeks ED Polatuzumab vedotin: first global approval. Drugs 2019, 79, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Beck A; Goetsch L; Dumontet C; Corvaia N Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discovery 2017, 16, 315–337. [DOI] [PubMed] [Google Scholar]
- (50).Jeffrey SC; Nguyen MT; Andreyka JB; Meyer DL; Doronina SO; Senter PD Dipeptide-based highly potent doxorubicin antibody conjugates. Bioorg. Med. Chem. Lett 2006, 16, 358–362. [DOI] [PubMed] [Google Scholar]
- (51).Lehar SM; Pillow T; Xu M; Staben L; Kajihara KK; Vandlen R; DePalatis L; Raab H; Hazenbos WL; Morisaki JH; Kim J; Park S; Darwish M; Lee BC; Hernandez H; Loyet KM; Lupardus P; Fong R; Yan D; Chalouni C; Luis E; Khalfin Y; Plise E; Cheong J; Lyssikatos JP; Strandh M; Koefoed K; Andersen PS; Flygare JA; Wah Tan M; Brown EJ; Mariathasan S Novel antibody–antibiotic conjugate eliminates intracellular S. aureus. Nature 2015, 527, 323–328. [DOI] [PubMed] [Google Scholar]
- (52).Mariathasan S; Tan MW Antibody-antibiotic conjugates: a novel therapeutic platform against bacterial infections. Trends Mol. Med 2017, 23, 135–149. [DOI] [PubMed] [Google Scholar]
- (53).de Groot FM; Broxterman HJ; Adams HP; van Vliet A; Tesser GI; Elderkamp YW; Schraa AJ; Kok RJ; Molema G; Pinedo HM; Scheeren HW Design, synthesis, and biological evaluation of a dual tumor-specific motive containing integrin-targeted plasmin-cleavable doxorubicin prodrug. Mol. Cancer Ther 2002, 1, 901–911. [PubMed] [Google Scholar]
- (54).Vhora I; Patil S; Bhatt P; Misra A Protein- and peptide-drug conjugates: an emerging drug delivery technology. Adv. Protein Chem. Struct. Biol 2015, 98, 1–55. [DOI] [PubMed] [Google Scholar]
- (55).Zheng G; Chen J; Stefflova K; Jarvi M; Li H; Wilson BC Photodynamic molecular beacon as an activatable photosensitizer based on protease-controlled singlet oxygen quenching and activation. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 8989–8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Lo PC; Chen J; Stefflova K; Warren MS; Navab R; Bandarchi B; Mullins S; Tsao M; Cheng JD; Zheng G Photodynamic molecular beacon triggered by fibroblast activation protein on cancer-associated fibroblasts for diagnosis and treatment of epithelial cancers. J. Med. Chem 2009, 52, 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Ivry SL; Meyer NO; Winter MB; Bohn MF; Knudsen GM; O’Donoghue AJ; Craik CS Global substrate specificity profiling of post-translational modifying enzymes. Protein Sci 2018, 27, 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Poreba M; Drag M Current strategies for probing substrate specificity of proteases. Curr. Med. Chem 2012, 17, 3968–3995. [DOI] [PubMed] [Google Scholar]
- (59).Janssen S; Jakobsen CM; Rosen DM; Ricklis RM; Reineke U; Christensen SB; Lilja H; Denmeade SR Screening a combinatorial peptide library to develop a human glandular kallikrein 2-activated prodrug as targeted therapy for prostate cancer. Mol. Cancer Ther 2004, 3, 1439–1450. [PubMed] [Google Scholar]
- (60).Thornberry NA; Rano TA; Peterson EP; Rasper DM; Timkey T; Garcia-Calvo M; Houtzager VM; Nordstrom PA; Roy S; Vaillancourt JP; Chapman KT; Nicholson DW A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem 1997, 272, 17907–17911. [DOI] [PubMed] [Google Scholar]
- (61).Choe Y; Leonetti F; Greenbaum DC; Lecaille F; Bogyo M; Bromme D; Ellman JA; Craik CS Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J. Biol. Chem 2006, 281, 12824–12832. [DOI] [PubMed] [Google Scholar]
- (62).Matthews DJ; Wells JA Substrate of protease substrates by monovalent phage display. Science 1993, 260, 1113–1117. [DOI] [PubMed] [Google Scholar]
- (63).Newman MR; Benoit DS In Vivo Translation of Peptide-Targeted Drug Delivery Systems Discovered by Phage Display. Bioconjugate Chem 2018, 29, 2161–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).O’Donoghue AJ; Eroy-reveles AA; Knudsen GM; Ingram J; Zhou M; Statnekov JB; Greninger AL; Hostetter DR; Qu G; Maltby DA; Anderson MO; DeRisi JL; McKerrow JH; Burlingame AL; Craik CS Global identification of peptidase specificity by multiplex substrate profiling. Nat. Methods 2012, 9, 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Li H; O’Donoghue AJ; van der Linden WA; Xie SC; Yoo E; Foe IT; Tilley L; Craik CS; da Fonseca PCA; Bogyo M Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature 2016, 530, 233–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Lapek JD Jr.; Jiang Z; Wozniak JM; Arutyunova E; Wang SC; Lemieux MJ; Gonzalez DJ; O’Donoghue AJ Quantitative Multiplex Substrate Profiling of Peptidases by Mass Spectrometry. Mol. Cell. Proteomics 2019, 18, 968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Vizovisek M; Vidmar R; Drag M; Fonovic M; Salvesen GS; Turk B Protease specificity: towards in vivo imaging applications and biomarker discovery. Trends Biochem. Sci 2018, 43, 829–844. [DOI] [PubMed] [Google Scholar]
- (68).Chen S; Yim JJ; Bogyo M Synthetic and biological approaches to map substrate specificities of proteases. Biol. Chem 2019, 401, 165–182. [DOI] [PubMed] [Google Scholar]
- (69).Vizovisek M; Vidmar R; Fonovic M; Turk B Current trends and challenges in proteomic identification of protease substrates. Biochimie 2016, 122, 77–87. [DOI] [PubMed] [Google Scholar]
- (70).Sobotic B; Vizovisek M; Vidmar R; Van Damme P; Gocheva V; Joyce JA; Gevaert K; Turk V; Turk B; Fonovic M Proteomic identification of cysteine cathepsin substrates shed from the surface of cancer cells. Mol. Cell. Proteomics 2015, 14, 2213–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Biniossek ML; Niemer M; Maksimchuk K; Mayer B; Fuchs J; Huesgen PF; McCafferty DG; Turk B; Fritz G; Mayer J; Haecker G; Mach L; Schilling O Identification of protease specificity by combining proteome-derived peptide libraries and quantitative proteomics. Mol. Cell. Proteomics 2016, 15, 2515–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72). For studies investigating phage display to design peptide conjugates for targets such as whole cell or protein mixtures, see:; (a) Laakkonen P; Akerman ME; Biliran H; Yang M; Ferrer F; Karpanen T; Hoffman RM; Ruoslahti E Antitumor activity of a homing peptide that targets tumor lymphatics and tumor cells. Proc. Natl. Acad. Sci. U. S. A 2004, 101, 9381–9386. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Laakkonen P; Porkka K; Hoffman JA; Ruoslahti E A tumor-homing peptide with a targeting specificity related to lymphatic vessels. Nat. Med 2002, 8, 751–755. [DOI] [PubMed] [Google Scholar]; (c) Jin W; Qin B; Chen Z; Liu H; Barve A; Cheng K Discovery of PSMA-specific peptide ligands for targeted drug delivery. Int. J. Pharm 2016, 513, 138–147. [DOI] [PubMed] [Google Scholar]; (d) Cieslewicz M; Tang J; Yu JL; Cao H; Zavaljevski M; Motoyama K; Lieber A; Raines EW; Pun SH Targeted delivery of proapoptotic peptides to tumor-associated macrophages improves survival. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 15919–15924. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Liu J; Liu J; Chu L; Wang Y; Duan Y; Feng L; Yang C; Wang L; Kong D Novel peptide-dendrimer conjugates as drug carriers for targeting nonsmall cell lung cancer. Int. J. Nanomed 2010, 6, 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Lempens EH; Merkx M; Tirrell M; Meijer EW Dendrimer display of tumor-homing peptides. Bioconjugate Chem 2011, 22, 397–405. [DOI] [PubMed] [Google Scholar]; (g) Hetrick KJ; Walker MC; van der Donk WA Development and Application of Yeast and Phage Display of Diverse Lanthipeptides. ACS Cent. Sci 2018, 4, 458–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).(a) Whitney M; Crisp JL; Olson ES; Aguilera TA; Gross LA; Ellies LG; Tsien RY Parallel in vivo and in vitro selection using phage display identifies protease-dependent tumor-targeting peptides. J. Biol. Chem 2010, 285, 22532–22541. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cloutier SM; Kündig C; Gygi CM; Jichlinski P; Leisinger HJ; Deperthes D Profiling of proteolytic activities secreted by cancer cells using phage display substrate technology. Tumor Biol 2004, 25, 24–30. [DOI] [PubMed] [Google Scholar]
- (74).Dubowchik GM; Firestone RA; Padilla L; Willner D; Hofstead SJ; Mosure K; Knipe JO; Lasch SJ; Trail PA Cathepsin B-labile dipeptide linkers for lysosomal release of doxorubicin from internalizing immunoconjugates: model studies of enzymatic drug release and antigen-specific in vitro anticancer activity. Bioconjugate Chem. 2002, 13, 855–869. [DOI] [PubMed] [Google Scholar]
- (75).Kasperkiewicz P; Poreba M; Groborz K; Drag M Emerging challenges in the design of selective substrates, inhibitors and activity-based probes for indistinguishable proteases. FEBS J. 2017, 284, 1518–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Kisselev AF; Goldberg AL Proteasome inhibitors: from research tools to drug candidates. Chem. Biol 2001, 8, 739–758. [DOI] [PubMed] [Google Scholar]
- (77).Caculitan NG; Dela Cruz Chuh J; Ma Y; Zhang D; Kozak KR; Liu Y; Pillow TH; Sadowsky J; Cheung TK; Phung Q; Haley B; Lee BC; Akita RW; Sliwkowski MX; Polson AG Cathepsin B is dispensable for cellular processing of cathepsin B-cleavable antibody-drug conjugates. Cancer Res. 2017, 77, 7027–7037. [DOI] [PubMed] [Google Scholar]
- (78).Akkari L; Gocheva V; Quick ML; Kester JC; Spencer AK; Garfall AL; Bowman RL; Joyce JA Combined deletion of cathepsin protease family members reveals compensatory mechanisms in cancer. Genes Dev. 2016, 30, 220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Kay BK, Thai S; Volgina VV High-throughput biotinylation of proteins In High Throughput Protein Expression and Purification; Doyle SA, Ed.; Methods in Molecular Biology, Vol. 498; Humana Press:: Totowa, NJ, 2009; pp 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Fairhead M; Howarth M Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol 2015, 1266, 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Neu HC; Heppel LA The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J. Chem. Biol 1965, 240, 3685–3692. [PubMed] [Google Scholar]
- (82). See Supporting Information for complete experimental details.
- (83).Lin K-H; Nalivaika EA; Prachanronarong KL; Yilmaz NK; Schiffer CA Dengue protease substrate recognition: binding of the prime side. ACS Infect. Dis 2016, 2, 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).ZHONG Y-J; SHAO L-H; LI Y Cathepsin B-cleavable doxorubicin prodrugs for targeted cancer therapy. Int. J. Oncol 2013, 42, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Harris JL; Backes BJ; Leonetti F; Mahrus S; Ellman JA; Craik CS Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc. Natl. Acad. Sci. U. S. A 2000, 97, 7754–7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Maly DJ; Leonetti F; Backes BJ; Dauber DS; Harris JL; Craik CS; Ellman JA Expedient solid-phase synthesis of fluorogenic protease substrates using the 7-amino-4-carbamoylmethylcoumarin (ACC) fluorophore. J. Org. Chem 2002, 67, 910–915. [DOI] [PubMed] [Google Scholar]
- (87).(a) Mensa B; Howell GL; Scott R; DeGrado WF Comparative Mechanistic Studies of Brilacidin, Daptomycin, and the Antimicrobial Peptide LL16. Antimicrob. Agents Chemother 2014, 58, 5136–514. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Beriashvili D; Taylor R; Kralt B; Abu Mazen N; Taylor SD; Palmer M Mechanistic Studies on the Effect of Membrane Lipid Acyl Chain Composition on Daptomycin Pore Formation. Chem. Phys. Lipids 2018, 216, 73–79. [DOI] [PubMed] [Google Scholar]; (c) Müller A; Wenzel M; Strahl H; Grein F; Saaki TNV; Kohl B; Siersma T; Bandow JE; Sahl H-G; Schneider T; Hamoen LW Daptomycin Inhibits Cell Envelope Synthesis by Interfering With Fluid Membrane Microdomains. Proc. Natl. Acad. Sci. U. S. A 2016, 113, E7077–E7086. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Hill J; Siedlecki J; Parr I; Morytko M; Yu X; Zhang Y; Silverman J; Controneo N; Laganas V; Li T; Lai JJ; Keith D; Shimer G; Finn J Synthesis and Biological Activity of N-Acylated Ornithine Analogues of Daptomycin. Bioorg. Med. Chem. Lett 2003, 13, 4187–4191. [DOI] [PubMed] [Google Scholar]
- (88).Ghosh M; Miller MJ Design, synthesis, and biological evaluation of isocyanurate-based antifungal and macrolide antibiotic conjugates: iron transport-mediated drug delivery. Bioorg. Med. Chem 1995, 3, 1519–1525. [DOI] [PubMed] [Google Scholar]
- (89).Daher SS; Jin X; Patel J; Freundlich JS; Buttaro B; Andrade RB Synthesis and biological evaluation of solithromycin analogs against multidrug resistant pathogens. Bioorg. Med. Chem. Lett 2019, 29, 1386–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Bellenger J-P; Arnaud-Neu F; Asfari Z; Myneni SCB; Stiefel EI; Kraepiel AML Complexation of oxoanions and cationic metals by the biscatecholate siderophore azotochelin. JBIC, J. Biol. Inorg. Chem 2007, 12, 367–376. [DOI] [PubMed] [Google Scholar]
- (91).(a) Heinisch L; Wittmann S; Stoiber T; Berg A; Ankel-Fuchs D; Möllmann U Highly antibacterial active aminoacyl penicillin conjugates with acylated bis-catecholate siderophores based on secondary diamino acids and related compounds. J. Med. Chem 2002, 45, 3032–3040. [DOI] [PubMed] [Google Scholar]; (b) Heinisch L; Moellmann U; Schnabelrachk M; Reisbrodt R Synthetic Catechol Derivatives, Method for Production and Use Thereof. United States Patent US6380181B1, April 30, 2002. [Google Scholar]; (c) Mollmann U; Heinisch L; Bauernfeind A; Kohler T; Ankel-Fuchs D Siderophores as drug delivery agents: application of the trojan horse strategy. BioMetals 2009, 22, 615–624. [DOI] [PubMed] [Google Scholar]; (d) Delorme D; Houghton T; Lafontaine Y; Tanaka K; Deitrick E; Kang T; Rafai Far A Phosphonated Oxazolidinones and Uses Thereof for the Prevention and Treatment of Bone and Joint Infections. Int. Patent Application PCT/IB2006/004233, December 6, 2006. [Google Scholar]; (e) Miller MJ; McKee JA; Minnick AA; Dolence EK The Design, Synthesis and Study of Siderophore-Antibiotic Conjugates. Siderophore Mediated Drug Transport. Biol. Met 1991, 4, 62. [DOI] [PubMed] [Google Scholar]
- (92).Carmona G; Rodriguez A; Juarez D; Corzo G; Villegas E Improved protease stability of the antimicrobial peptide Pin2 substituted with D-amino acids. Protein J 2013, 32, 456–466. [DOI] [PubMed] [Google Scholar]
- (93).Zheng T; Bullock JL; Nolan EM Siderophore-mediated cargo delivery to the cytoplasm of escherichia coli and pseudomonas aeruginosa: syntheses of monofunctionalized enterobactin scaffolds and evaluation of enterobactin–cargo conjugate uptake. J. Am. Chem. Soc 2012, 134, 18388–18400. [DOI] [PubMed] [Google Scholar]
- (94).Sklar JG; Wu T; Kahne D; Silhavy TJ Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007, 21, 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).(a) Hagan CL; Kim S; Kahne D Reconstitution of Outer Membrane Protein Assembly From Purified Components. Science 2010, 328, 890–892. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mahoney TF; Ricci DP; Silhavy TJ Classifying ß-barrel assembly substrates by manipulating essential Bam complex members. J. Bacteriol 2016, 198, 1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Mikkelsen H; McMullan R; Filloux A The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One 2011, 6, e29113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Knight DB; Rudin SD; Bonomo RA; Rather PN Acinetobacter nosocomialis: Defining the role of efflux pumps in resistance to antimicrobial therapy, surface motility, and biofilm formation. Front. Microbiol 2018, 9, 1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Chen TL; Lee YT; Kuo SC; Yang SP; Fung CP; Lee SD Rapid identification of Acinetobacter baumannii, Acinetobacter nosocomialis and Acinetobacter pittii with a multiplex PCR assay. J. Med. Microbiol 2014, 63, 1154–1159. [DOI] [PubMed] [Google Scholar]
- (99).(a) Miller CG Peptidases and Proteases of Escherichia Coli and Salmonella Typhimurium. Annu. Rev. Microbiol 1975, 29, 485–504. [DOI] [PubMed] [Google Scholar]; (b) Lazdunski AM Peptidases and proteases of escherichia coli and salmonella typhimurium. FEMS Microbiol. Rev 1989, 63, 265–276. [DOI] [PubMed] [Google Scholar]
- (100).(a) Koehbach J; Craik DJ The vast structural diversity of antimicrobial peptides. Trends Pharmacol. Sci 2019, 40, 517–528. [DOI] [PubMed] [Google Scholar]; (b) Lai PK; Tresnak DT; Hackel BJ Identification and elucidation of proline-rich antimicrobial peptides with enhanced potency and delivery. Biotechnol. Bioeng 2019, 116, 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Li WF; Ma GX; Zhou XX Apidaecin-type peptides: biodiversity, structure-function relationships and mode of action. Peptides 2006, 27, 2350–2359. [DOI] [PubMed] [Google Scholar]
- (101).Dubowchik GM; Firestone RA Cathepsin B-sensitive dipeptide prodrugs. 1. A model study of structural requirements for efficient release of doxorubicin. Bioorg. Med. Chem. Lett 1998, 8, 3341–3346. [DOI] [PubMed] [Google Scholar]
- (102).Randall CP; Mariner KR; Chopra I; O’Neill AJ The target of daptomycin is absent from Escherichia coli and other gram-negative pathogens. Antimicrob. Agents Chemother 2013, 57, 637–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Miller WR; Bayer AS; Arias CA Mechanism of Action and Resistance to Daptomycin in Staphylococcus aureus and Enterococci. Cold Spring Harbor Perspect. Med 2016, 6, a026997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Llano-Sotelo B; Dunkle J; Klepacki D; Zhang W; Fernandes P; Cate JH; Mankin AS Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis. Antimicrob. Agents Chemother 2010, 54, 4961–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


