Abstract
Deschampsia antarctica Desv. can be found in diverse Antarctic habitats which may vary considerably in terms of environmental conditions and soil properties. As a result, the species is characterized by wide ecotypic variation in terms of both morphological and anatomical traits. The species is a unique example of an organism that can successfully colonize inhospitable regions due to its phenomenal ability to adapt to both the local mosaic of microhabitats and to general climatic fluctuations. For this reason, D. antarctica has been widely investigated in studies analyzing morphophysiological and biochemical responses to various abiotic stresses (frost, drought, salinity, increased UV radiation). However, there is little evidence to indicate whether the observed polymorphism is accompanied by the corresponding genetic variation. In the present study, retrotransposon‐based iPBS markers were used to trace the genetic variation of D. antarctica collected in nine sites of the Arctowski oasis on King George Island (Western Antarctic). The genotyping of 165 individuals from nine populations with seven iPBS primers revealed 125 amplification products, 15 of which (12%) were polymorphic, with an average of 5.6% polymorphic fragments per population. Only one of the polymorphic fragments, observed in population 6, was represented as a private band. The analyzed specimens were characterized by low genetic diversity (uHe = 0.021, I = 0.030) and high population differentiation (F ST = 0.4874). An analysis of Fu's F S statistics and mismatch distribution in most populations (excluding population 2, 6 and 9) revealed demographic/spatial expansion, whereas significant traces of reduction in effective population size were found in three populations (1, 3 and 5). The iPBS markers revealed genetic polymorphism of D. antarctica, which could be attributed to the mobilization of random transposable elements, unique features of reproductive biology, and/or geographic location of the examined populations.
Keywords: Antarctic hairgrass, Antarctica, genetic diversity, iPBS, King George Island
Deschampsia antarctica is among the two angiosperms considered as a native for the Antarctic. The retrotransposon‐based iPBS markers applied in the study revealed low genetic variation of the species accompanied by surprisingly high population differentiation.

1. INTRODUCTION
Antarctic terrestrial biota occurs mainly in small, isolated ice‐free areas in the coastal zones of the Maritime Antarctic (Lee et al., 2017). The development of terrestrial ecosystems is generally limited by environmental factors such as low temperature, intermittent water supply, highly seasonal light regime, ground‐level wind speed, uneven distribution of nutrients, high salinity in locations with strong marine influence, and seasonal environmental stresses (Beyer et al., 2000; Convey & Peck, 2019). Soil properties are an important abiotic factor in the Antarctic ecosystem (Lachacz et al., 2018; Sierakowski et al., 2017; Znój et al., 2017). The Antarctic terrestrial ecosystem is a mosaic of microhabitats that differ in the availability of nutrients, water conditions, the influence of salty aerosols, and differences in exposition. As a result, ice‐free areas became colonized by highly heterogeneous and discontinuous plant communities that are interspersed with bare ground and are dominated by cryptogams. The only two native angiosperms are Deschampsia antarctica Desv. (Poaceae) (Antarctic hairgrass) (Figure 1) and Colobanthus quitensis (Kunth) Bartl. (Caryophyllaceae) (Ochyra et al., 2008; Olech, 2004). The distribution of D. antarctica is restricted to the Maritime Antarctic, including the west coast of the Antarctic Peninsula, its offshore islands and the South Sandwich, South Orkney, and South Shetland Islands. The species is also commonly encountered on sub‐Antarctic islands such as South Georgia Island, Heard Island, Crozet Islands, and the Kerguelen Archipelago in the Indian Ocean. Outside the sub‐Antarctic region, D. antarctica is found on the Falkland Islands and in South America in Tierra del Fuego and the Andes up to a latitude of around 34 degrees south (Convey, 1996).
FIGURE 1.

Deschampsia antarctica on the King George Island, South Shetland Islands. (fot. by I. Giełwanowska)
Genetic studies demonstrated very low levels of genetic variability in D. antarctica, even in populations separated by a considerable distance (Holderegger et al., 2003; Chwedorzewska & Bednarek, 2008, 2011; van de Wouw et al., 2008). Despite the above, D. antarctica is characterized by remarkable ecotypic variation and colonizes a wide range of habitats, from mineral soils to organic soils and eroded pits, from extremely dry windswept fellfields to waterlogged seepage areas that are occasionally inundated by sea water (Lewis‐Smith, 2003), as well as nutrient‐deficient sites or habitats that are enriched mainly with ammonium and nitrates by animals and sea spray (Lewis‐Smith, 2003; Nędzarek & Chwedorzewska, 2004). Even a small shift in environmental conditions such as exposition or distance from the source of nutrients (bird colonies) and sea can lead to the development of different terrestrial communities. Deschampsia antarctica populations originating from such sites differ significantly in morphological and anatomical traits (Chwedorzewska et al., 2008; Corner, 1971; Giełwanowska, 2003a,b).
Resistance to severe and diverse physiological stresses is essential for survival in the harsh Antarctic climate (Clemente‐Moreno et al., 2020). Stressors can induce a flexible response from an organism and lead to the development of a new phenotype. Environmentally induced genetic changes in transposable elements (TEs) are one of such mechanisms (Kalendar et al., 2000; Piacentini et al., 2014). Transposable elements are capable of changing their location and/or copy numbers, and they play an important role in the evolution of the plant genome (Finnegan, 1989). Transcriptional activation of TEs has been observed in many plant species that are exposed to various abiotic and biotic stressors (Moreau‐Mhiri et al., 1996; Takeda et al., 1998; Voronova et al., 2011), and it is regarded as a key mechanism that is responsible for genome plasticity under changing environmental conditions (Schrader et al., 2014). Intense stress may facilitate rapid changes in the structure, organization, and function of the genome through interactions with TEs, especially in populations with low genetic diversity (Stapley et al., 2015). The evolution of environmentally induced advantageous phenotypes through epigenetic mechanisms could be an immediate adaptive process, followed by TE‐induced genotypic changes that make these phenotypic variants heritable through the germline. Transposable elements could play different roles on the timescale of ecological variation (Pimpinelli & Piacentini, 2020), such as those related to diverse stress conditions in Antarctic habitats. The activation of TEs can induce genetic variability in response to environmental changes. Transposable elements also exert selective pressure on another genetic elements, thus contributing to rapid evolutionary processes and adaptation to local conditions.
Transposable elements are highly abundant and diverse mobile genetic elements that constitute up to 90% of eukaryotic genomes (San Miguel et al., 1996). Many features of TEs, such as their ubiquity, abundance, and dispersion in the eukaryotic genome, make them an attractive target for molecular marker systems (Kalendar et al., 2019). Various approaches have been proposed to explore polymorphisms in TE insertion patterns, including conventional or anchored PCR, and quantitative or digital PCR with primers designed for the 5' or 3' junction (Kalendar et al., 2019). The main drawback of TE‐based molecular markers techniques is the need for sequence information in designing element‐specific primers. In species where genomic data are scarce or absent, genetic polymorphism resulting from TE mobility can be detected based on their conserved sequences. The Inter Primer Binding Sequence (iPBS) technique developed by Kalendar et al. (2010) relies on the highly conserved domain of LTR retrotransposons for primer binding. This method proved to be a valuable tool for assessing retrotransposon‐based genetic variation in guava (Mehmood et al., 2016), beech, chestnut, and oak (Coutinho et al., 2018), barley (Bonchev et al., 2019) and Colobanthus quitensis populations from a wide geographic range (Koc et al., 2018), or sites exposed to diverse abiotic conditions such as the Maritime Antarctic (Androsiuk et al., 2015).
In the present study, iPBS markers were used to assess DNA polymorphism and genetic relationships between D. antarctica populations from patchy habitats of the Arctowski oasis (King George Island, South Shetlands, Antarctic).
2. MATERIALS AND METHODS
2.1. Material
A total of 165 D. antarctica plants sampled in nine sites with different microclimatic conditions and soil properties in the Arctowski oasis were subjected to molecular analyses (Figure 2, Table 1, Table S1). D. antarctica fresh, healthy leaves were stored at −20°C immediately after sampling in 2010. The population from each sampling site was represented by 10 to 33 specimens. The samples were not equal in size because the number of individuals was very small in some sites. In addition, many dry and exposed sites featured small individuals with high necromass content; therefore, sufficient quantities of fresh material for DNA extraction were difficult to collect.
FIGURE 2.
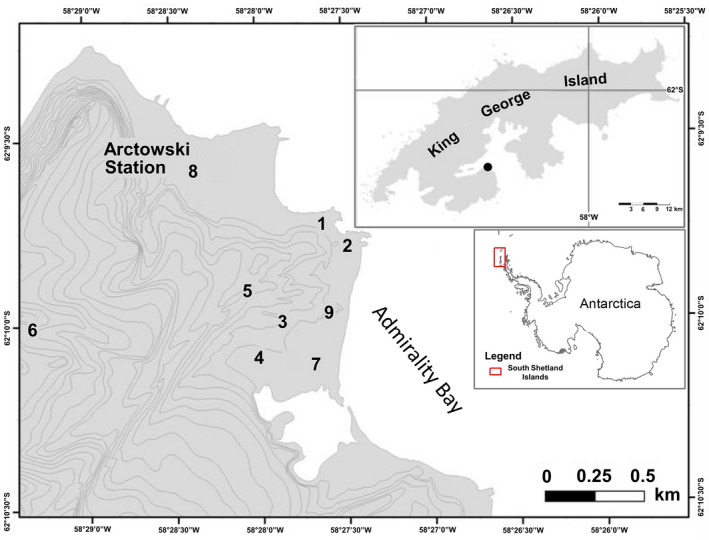
Study area showing sampling sites of Deschampsia antarctica on King George Island and contour map of Antarctica with marked location of South Shetland Islands. Numbers of populations according to Table 1
TABLE 1.
The origin of Deschampsia antarctica populations used in the study
| No. | Vegetation | Edaphic characteristics | Distance from the sea [m] | Altitude [m a.s.l.] | Location | Number of individuals |
|---|---|---|---|---|---|---|
| 1. | The area has abundant plant cover, mainly D. antarctica, C. quitensis, and Prasiola crispa, a green algae that develops rapidly during the growing season; ornithocoprophilous lichen species grow on rocks | Basalt rock outcrops where soils developed only in rock crevices; soil has high organic matter content originating mainly from fresh penguin guano; the habitat is humid and sheltered from the wind; area trampled by birds. | 100–120 | 10 | 62.1621S, 58.4606W | 14 |
| 2. | Many lichen species, including halophilous species Verrucaria tesselatula. D. antarctica and C. quitensis (less frequent than in site 1), as well as Prasiola crispa | Within the range of ocean waves during stormy weather; soils were classified as Eutric Skeletic Nudilithic Leptosols (Arenic, Humic, Ornithic, Protic); soil has high organic matter content originating mainly from fresh penguin guano; humid and exposed habitat | 1–2 | 0.5–1.0 | 62.1629S, 58.4567W | 21 |
| 3. | The site is covered by the Antarctic tundra; mosses, lichens and two species of flowering plants, D. antarctica and C. quitensis, were identified; plants are short (2.5–6.5 cm) | The oldest moraine of the Ecology Glacier; sloping edge of a relatively old fluted moraine; strongly graveled soils; habitat dry and exposed; the site occupies a former penguin rookery, with relict bird influences; within the range of sea water aerosols during stormy weather; soil was classified as Skeletic Protic Turbic Cryosol (Arenic, Eutric, Humic, Ornithic). | 400 | 40 | 62.1639S, 58.4599W | 21 |
| 4. | Lichens, D. antarctica and C. quitensis were identified, but plant cover was less extensive; flowering plants have a height of 3.5–7.5 cm; plant growth is relatively rapid | A moraine of the Ecology Glacier; dry and exposed habitat with a minor influence of penguin and seal rookeries; site 4 is similar to site 3, but younger; soil was classified as Eutric Protic Skeletic Regosol (Loamic, Turbic). | 400 | 35 | 62.1645S, 58.4603W | 11 |
| 5. | Typical Antarctic tundra; mosses, lichens and flowering plants form a dense carpet; flowering plants grow slowly and reach 1.5–2.5 cm in height | Near the grave of W. Puchalski; gravelly material is water permeable and well drained; old penguin rookery with relict ornithogenic soil; habitat dry and exposed; soil was classified as Skeletic Protic Turbic Cryosol (Arenic, Dystric, Humic, Ornithic). | 500 | 110 | 62.1635S, 58.4626W | 33 |
| 6. | The proportion of flowering plants is small, with a predominance of lichens | Near the Jersak Hills glacier; far from the coast (700 m); basalt rock outcrops with scree debris below; soil was classified as Eutric Protic Skeletic Leptic Regosol (Arenic, Humic, Turbic); habitat dry and exposed; soil has low nutrient and organic matter content | 700 | 200 | 62.1649S, 58.4874W | 10 |
| 7. | Numerous rocks with the smallest plant cover in all studied sites; plant age was determined based on flowering tussocks: Colobanthus quitensis plants appeared in this location approx. 10 years ago, and Deschampsia antarctica plants appeared in this location approx. 3–5 years ago | The youngest fluted moraine of the Ecology Glacier with rich petrographic composition; close to a fresh water lagoon from a melting glacier; very high moisture content due to the direct influence of sea water; minor influence of penguin and seal rookeries; soil was classified as Eutric Protic Skeletic Protic Regosol (Loamic). | 20 | 0.5 | 62.1682S, 58.4622W | 16 |
| 8. | Two native flowering plants, an invasive grass species (Poa annua) and chlorophyte algae (Prasiola crispa) | The area of Henryk Arctowski Station; the ground was transformed due to continuous human influence (road, water tank); humid habitat sheltered from the wind; sea sands and fluvioglacial sands were mechanically altered; vegetation cover is limited due to mechanical impact exerted by people and vehicles; soil was classified as Skeletic Eutric Fluvisol (Arenic). | 50 | 0.5 | 62.1598S, 58.4759W | 27 |
| 9. | Mostly lichens, mosses and scattered specimens of D. antarctica and C. quitensis, excluding the area surrounding Arctowski Station (site 8) | Mouth of the Ornithologists Creek; fertile and moist soil supplied with both fresh water from the Ornithologists Creek and sea water which is accumulated for several hours after a storm; influence of the penguin colony (guano is the main source of organic matter); human impact on soil and vegetation is minimal, limited to occasional trampling on routes to study sites | 30–40 | 1.0 | 62.1658S, 58.4589W | 12 |
2.2. Molecular analyses
DNA was extracted from individuals representing each population with the Syngen Plant DNA Mini Kit. The quality of DNA was verified on 1% agarose, and the purity of DNA samples was assessed spectrophotometrically.
The entire D. antarctica collection was genotyped with iPBS primers. According to the procedure described by Kalendar et al. (2010), 20 iPBS primers were initially screened. Seven iPBS primers which produced clearly identifiable and repeatable polymorphic bands were selected for further analyses (Table 2). The reproducibility of primer band profiles was verified based on comparison of the electrophoretic profiles of randomly selected D. antarctica samples. Data were generated and compared in two replicates. Gels were then checked to identify iPBS amplicons (bands) in one or both replicates. Seven iPBS primers were used individually in a polymerase chain reaction (PCR) according to the protocol described by Kalendar et al. (2010) with some modifications (Androsiuk et al., 2015; Koc et al., 2018). The reaction conditions for each primer (temperature of primer hybridization) were determined empirically. The amplification products were analyzed by electrophoresis on 1.5% (w/v) agarose gels with 1x TBE electrophoresis buffer at 100 V for 2 hr and were visualized by staining with 0.5 μg/ml ethidium bromide.
TABLE 2.
iPBS primers applied in the study and their specification
| Primer | Sequence (5’→3’) | Annealing temp. (°C) | No of scored bands | No of polymorphic bands |
|---|---|---|---|---|
| 2074 | GCTCTGATACCA | 52 | 18 | 1 |
| 2085 | ATGCCGATACCA | 51 | 17 | 3 |
| 2,224 | ATCCTGGCAATGGAACCA` | 52 | 16 | 2 |
| 2,251 | GAACAGGCGATGATACCA3` | 55 | 22 | 2 |
| 2,253 | TCGAGGCTCTAGATACCA3` | 53 | 24 | 2 |
| 2,374 | CCCAGCAAACCA | 55 | 14 | 1 |
| 2,376 | TAGATGGCACCA | 51 | 14 | 4 |
| Total | 125 | 15 |
2.3. iPBS data processing
All bands that were reliably identified across the studied individuals were scored as either present (1) or absent (0) across genotypes and treated as single dominant loci. Based on the obtained binary matrix of amplification products (Table S2), the following genetic parameters were estimated with the use of GenAlEx 6.5 software (Peakall & Smouse, 2006, 2012): total number of bands per population (NB), percentage of polymorphic bands (P), Shannon's Information Index (I), unbiased expected heterozygosity (uHe), and Nei's genetic distance (DN) (Nei, 1972). The genetic subdivision patterns of the analyzed D. antarctica populations were investigated by principal coordinate analysis (PCoA) based on DN values in GenAlEx 6.5. The genetic structure of the studied populations was inferred based on Bayesian model‐based clustering method implemented in STRUCTURE ver. 2.3.4. (Pritchard et al., 2000). The model assigns individual multilocus genotypes probabilistically to a user‐defined number of clusters (K), achieving linkage equilibrium within clusters. We performed 10 replicate runs for each K, ranging from 1 to 9, 500, 000 Markov Chain Monte Carlo repetitions and a burn‐in period of 500, 000. During the analysis, admixture model was used without any prior information on the original populations. To determine the optimal number of clusters, an ad hoc statistic ΔK was used (Evanno et al., 2005), estimated in Structure Harvester ver.0.6.94 (Earl & Vonholdt, 2012). Analysis of molecular variance (AMOVA) was also performed. In the analysis, the iPBS data were treated as haplotypic and composed of a combination of alleles at one or several loci (Excoffier et al., 2005). The significance of fixation indices was tested using a nonparametric permutation approach (Excoffier et al., 1992). The estimation of F ST and AMOVA were performed using Arlequin 3.5 software (Excoffier et al., 2005). Additionally, with the use of Hickory v.1.1 package (Holsinger & Lewis, 2003), two alternative F ST estimates were calculated: Gst‐B (Bayesian analog of Nei's G ST; Holsinger, 1999) and θ ( III ) which corresponds to a scaled allele frequency variance, where the variance is measured among contemporaneous populations (Song et al., 2006).
The possible effects of increased spatial distance and environmental heterogeneity on gene flow and genetic structure of the studied D. antarctica populations were also estimated. Spatial genetic structure was investigated by testing the significance of isolation by distance (IBD) in the Mantel test with 9,999 permutations of the relationship between the matrix of pairwise F ST/(1−F ST) and the matrix of log‐transformed geographic distances between populations (Rousset, 1997). The Mantel test with 9,999 permutations was also performed to compare the matrix of log‐transformed geographic distances between populations and the matrix of environmental distances to determine the degree of isolation by environment (IBE) of D. antarctica populations. Pairwise environmental distances (Euclidean distances) were calculated between the studied sampling sites based on the previously described data concerning general soil properties and nutrient content (Koc et al., 2018). The matrix containing pairwise environmental distances was standardized before the Mantel test based on the approach described by Nanninga et al. (2014). The pairwise environmental distance matrix was calculated and standardized in Statistica 12 software (StatSoft, Inc.). Sampling site 9 was not analyzed in the cited paper; therefore, only sites 1–8 were included in the IBE analysis. The Mantel test was performed in GenAlEx 6.5.
Tajima's D, Fu's F S neutrality test, mismatch distribution, and the demographic processes affecting populations were estimated in Arlequin 3.5. Recent population history was inferred by examining the departure from the drift–mutation equilibrium based on allele frequencies in the BOTTLENECK v. 1.2.02 program (Cornuet & Luikart, 1996; Piry et al., 1999) for each population. In populations that have experienced a recent reduction in effective size, the value of H e exceeds the heterozygosity expected at mutation–drift equilibrium. This effect was studied with the use of dominant markers in the infinite allele model (IAM) to test the mutation–drift hypothesis against the bottleneck hypothesis (Tero et al., 2003). The significance of potential bottleneck was estimated in the Sign test, the Standardized Differences test, and the one‐tailed Wilcoxon sign‐rank test for heterozygosity excess.
3. RESULTS
3.1. Efficiency of iPBS primers
The genotyping of D. antarctica populations with 7 iPBS primers supported the identification of 125 amplification products (bands). The highest number of 24 bands was revealed by primer 2,253, and the lowest number of amplification products (14) was obtained for primers 2,374 and 2,376. An average of 17.86 bands was obtained per primer. Fifteen of the identified loci (12%) were polymorphic (Table 2). A detailed analysis of genotypic data revealed one amplification product which could be represented as a potential private band for population 6. The band was amplified with primer 2,253, but it was identified in only one of 10 individuals.
3.2. Genetic diversity and differentiation
The iPBS markers revealed the presence of genetic polymorphism between individuals within a population as well as genetic variation between populations (Table 3). The number of iPBS amplification products (bands) ranged from 120 in population 2 to 124 in populations 5 and 6. The polymorphic rate was highest in population 5 (7%) and lowest in populations 2, 6, and 9 (5%). Genetic variation was assessed based on the values of Shannon's Information Index and unbiased expected heterozygosity, and both values were highest in population 5 and lowest in population 9.
TABLE 3.
Population genetic characteristics for analyzed populations of Deschampsia antarctica
| Populations | NB | P | I | uHe ± SE |
|---|---|---|---|---|
| 1 | 122 | 6.40 | 0.041 | 0.030 ± 0.010 |
| 2 | 120 | 4.80 | 0.023 | 0.016 ± 0.010 |
| 3 | 122 | 5.60 | 0.032 | 0.022 ± 0.009 |
| 4 | 123 | 5.60 | 0.028 | 0.020 ± 0.008 |
| 5 | 124 | 7.20 | 0.044 | 0.031 ± 0.010 |
| 6 | 124 | 4.80 | 0.024 | 0.017 ± 0.007 |
| 7 | 123 | 5.60 | 0.026 | 0.017 ± 0.007 |
| 8 | 123 | 5.60 | 0.029 | 0.020 ± 0.008 |
| 9 | 122 | 4.80 | 0.021 | 0.014 ± 0.007 |
| Average over loci and populations | 122.55 | 5.60 | 0.030 | 0.021 |
Abbreviations: NB, number of bands; P, percentage of polymorphic bands; I, Shannon's Information Index; uHe, unbiased expected heterozygosity with standard error (SE).
The results of AMOVA revealed that 51% of the identified genetic variation occurred between individuals within populations, whereas the remaining 49% was attributed to variation among populations (Table 4). High genetic variation between population was consistent with the F ST value (0.4874), as well as Gst‐B (0.4349). The θ ( III ) estimate, though lower in value, has reasonably similar magnitude (0.3703).
TABLE 4.
Partitioning of diversity found in Deschampsia antarctica from all analyzed populations using AMOVA (F ST = 0.4874)
| Source of variation | Degrees of freedom | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|
| Among populations | 8 | 176.357 | 1.159 | 48.74 |
| Within populations | 156 | 190.225 | 1.219 | 51.26 |
| Total | 164 | 366.582 | 2.379 |
Significance tests (1,023 permutations); p < .001.
Nei's genetic distance was calculated to estimate genetic differentiation between D. antarctica populations (Table 5). This parameter ranged from 0.007 to 0.047 (0.021 on average). The results of PCoA based on DN values demonstrated that 81.75% of variation was explained by the first three components (48%, 23%, and 10%, respectively). The projection of the analyzed populations on the first two axes is shown in Figure 3. The groups identified in PCoA revealed a pattern of interpopulation genetic variation, where the analyzed populations were scattered along both axes. Two pairs of populations shared the highest degree of similarity: populations 4 and 5 which diverged along the first axis, and populations 1 and 3 which diverged along the second axis; populations 2, 8, and 9 had a peripheral position, population 7 was located somewhere in the middle of mentioned above groups of populations, whereas population 6 diverged from the other populations along the Coord.2.
TABLE 5.
Nei's genetic distance values between studied Deschampsia antarctica populations. Numbers of populations according to Table 1
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | *** | ||||||||
| 2 | 0.015 | *** | |||||||
| 3 | 0.009 | 0.016 | *** | ||||||
| 4 | 0.026 | 0.047 | 0.028 | *** | |||||
| 5 | 0.015 | 0.040 | 0.021 | 0.004 | *** | ||||
| 6 | 0.017 | 0.026 | 0.026 | 0.025 | 0.017 | *** | |||
| 7 | 0.011 | 0.018 | 0.013 | 0.036 | 0.022 | 0.018 | *** | ||
| 8 | 0.022 | 0.016 | 0.026 | 0.035 | 0.027 | 0.013 | 0.012 | *** | |
| 9 | 0.018 | 0.011 | 0.025 | 0.043 | 0.034 | 0.016 | 0.012 | 0.007 | *** |
refers to 0.0 Nei's genetic distance.
FIGURE 3.
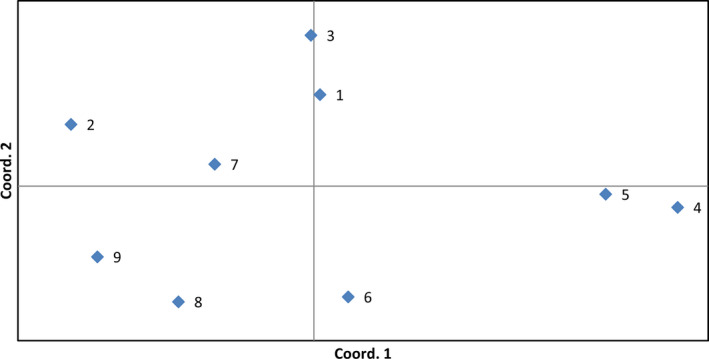
Projection of studied Deschampsia antarctica populations on the first two axes after principal coordinates analysis based on Nei's genetic distance values. Numbers of populations according to Table 1
In order to infer about the genetic structure of the studied D. antarctica populations Bayesian model‐based clustering method was also applied, and the optimal number of clusters was estimated with the ΔK method (Evanno et al., 2005) The ΔK produced the highest peak at K = 6 with a minor peak at K = 3 (Figure 4a). When K = 3, populations 2, 8, and 9 were assigned to one cluster, populations 4 and 5 to the second one, whereas the other populations appeared to be populations with a certain degree of substructure between the two. Additionally, population 6 (due to additional, unique admixture) was characterized by the highest proportion of membership of each predefined population in the third cluster (Figure 4b). When K = 6, populations 1 and 3 appeared to be the most admixed populations; moreover, most populations showed population substructure, with population 4 standing out as nearly homogenous population (Figure 4c). Although the highest peak of ΔK point at K = 6 as the most likely number of clusters, bar plot for each individual genotypes for K = 3, sorted by population, is somewhat similar in composition to the population grouping based on PCoA.
FIGURE 4.
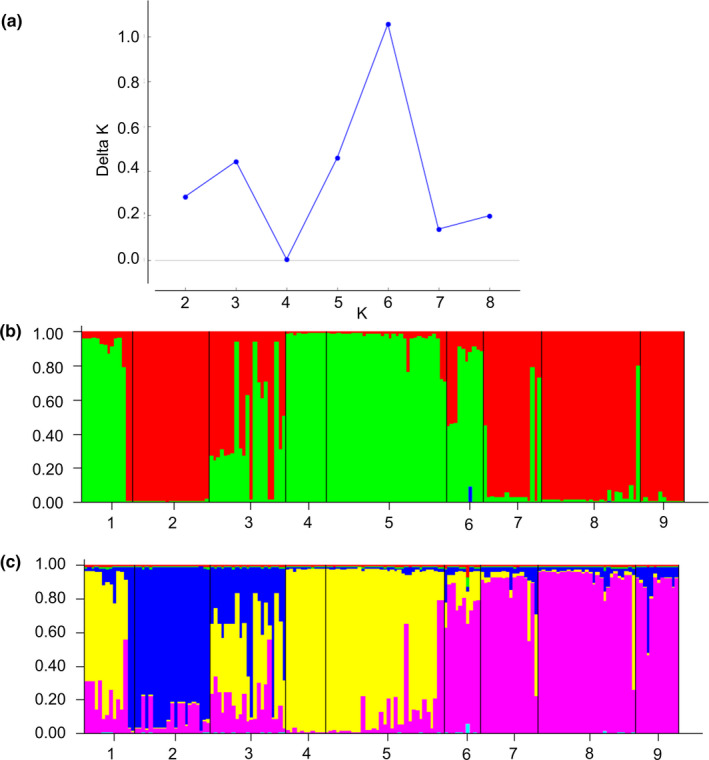
Structure of D. antarctica populations revealed by Bayesian analysis implemented in Structure. (a) Change (delta K) in likelihood for K = 1–9 (Evanno et al., 2005). (b) Individual probability assignment of each of the individuals sampled from 9 populations for K = 3. (c) Individual probability assignment of each of the individuals sampled from 9 populations for K = 6. Each individual is represented by a vertical bar broken into different colored genetic clusters, with length proportional to probability of assignment to each cluster
3.2.1. The effects of geographic and environmental isolation on population differentiation
Rousset's isolation by distance method did not reveal any correlations between genetic and geographic pairwise distances (R 2 = 0.044; p = .08), which suggests that geographic distance did not influence genetic structure. The IBE Mantel test comparing geographic distance with pairwise environmental distance (calculated based on combined data for 12 soil properties and 10 variables describing nutrient content) did not reveal significant correlations, either. However, when environmental variables were analyzed separately, a significant correlation was found between geographic distance versus. the content of P and N–NO3 (R 2 = 0.0616, p = .041) (Figure 5).
FIGURE 5.
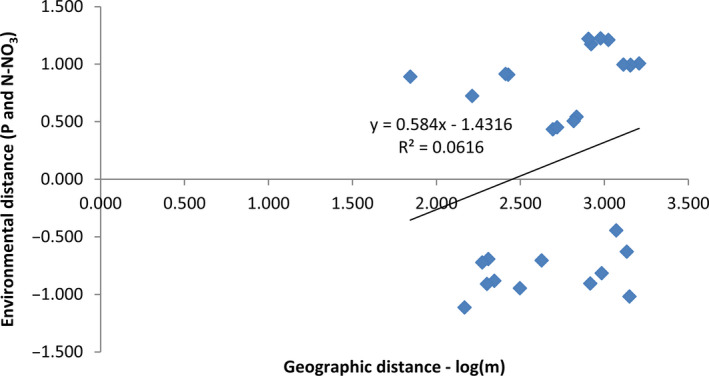
IBE analysis. The Mantel test scatterplot shows environmental distance (standardized to mean of zero and a standard deviation of one) as a function of logarithm of geographic distance
3.2.2. Neutrality tests and demography
Tajima's D did not reveal any deviations from 0, while Fu's F S was negative and significant for most populations excluding population 2, 6, and 9 (Table 6), for which p‐value was over 0.02. As it was described by Fu (1997), F S statistic should be considered as significant at the 5% level, if its p‐value is below .02, and not below .05. In the mismatch distribution test for demographic/spatial expansion, SSD values were not significant, and all samples had a very low raggedness index (Table 7).
TABLE 6.
Tajima's D test and Fu's F S neutrality tests of nine populations of Deschampsia antarctica
| Test | Description | Populations | Statistics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Mean | SD | ||
| Tajima's D test | S | 8 | 6 | 7 | 7 | 9 | 6 | 7 | 7 | 6 | 7 | 1 |
| Pi | 2.593 | 1.981 | 2.591 | 2.327 | 3.144 | 2.289 | 2.408 | 2.245 | 1.485 | 2.340 | 0.454 | |
| Tajima's D | 0.117 | 0.590 | 1.074 | −0.106 | 1.271 | 0.328 | 0.499 | 0.716 | −0.964 | 0.392 | 0.667 | |
| Tajima's D p‐value | 0.592 | 0.740 | 0.873 | 0.503 | 0.917 | 0.657 | 0.707 | 0.793 | 0.174 | 0.662 | 0.224 | |
| Fu's F S test | Theta pi | 2.593 | 1.981 | 2.590 | 2.327 | 3.144 | 2.289 | 2.408 | 2.245 | 1.485 | 2.340 | 0.454 |
| Exp. no. of alleles | 5.266 | 5.353 | 6.199 | 4.508 | 8.160 | 4.288 | 5.366 | 6.261 | 3.774 | 5.464 | 1.306 | |
| F S | −5.282 | −3.194 | −5.771 | −8.151 | −15.509 | −2.845 | −6.452 | −8.210 | −2.145 | −6.395 | 4.062 | |
| F S p‐value | 0.001 | 0.022 | 0.004 | 0.000 | 0.000 | 0.032 | 0.001 | 0.000 | 0.027 | 0.010 | 0.013 | |
S, number of segregating sites; Pi, mean number of pairwise differences; Theta pi, Watterson's theta based on S; F S, Fu's F S; SD, standard deviation.
TABLE 7.
Estimates of mismatch analysis of nine populations of Deschampsia antarctica
| Model | Statistic | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Mean | SD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Demographic expansion | SSD | 0.048 | 0.011 | 0.003 | 0.027 | 0.006 | 0.009 | 0.008 | 0.002 | 0.017 | 0.015 | 0.015 |
| Model (SSD) p‐value | 0.067 | 0.361 | 0.631 | 0.228 | 0.192 | 0.717 | 0.446 | 0.883 | 0.421 | 0.439 | 0.265 | |
| Raggedness index | 0.086 | 0.054 | 0.032 | 0.129 | 0.049 | 0.070 | 0.066 | 0.014 | 0.140 | 0.071 | 0.042 | |
| Raggedness p‐value | 0.430 | 0.560 | 0.761 | 0.190 | 0.136 | 0.575 | 0.373 | 0.995 | 0.284 | 0.478 | 0.277 | |
| Spatial expansion | SSD | 0.020 | 0.008 | 0.003 | 0.027 | 0.006 | 0.009 | 0.008 | 0.004 | 0.017 | 0.011 | 0.008 |
| Model (SSD) p‐value | 0.220 | 0.540 | 0.606 | 0.171 | 0.157 | 0.623 | 0.354 | 0.797 | 0.343 | 0.423 | 0.227 | |
| Raggedness index | 0.086 | 0.054 | 0.032 | 0.129 | 0.049 | 0.070 | 0.066 | 0.014 | 0.140 | 0.071 | 0.042 | |
| Raggedness p‐value | 0.298 | 0.630 | 0.730 | 0.190 | 0.126 | 0.533 | 0.345 | 0.998 | 0.275 | 0.458 | 0.286 |
Abbreviations: SD, standard deviation; SSD, sum of squared deviations; Raggedness index, Harpending's raggedness index.
3.2.3. The mutation−drift model versus the bottleneck hypothesis
All three tests used in this study analyze bottleneck effects in populations that develop transient heterozygosity excess. If the loci evolve in a strict one‐step mutation model, heterozygosity excess and deficiency can occur depending on locus variability and the time elapsed since the beginning of the bottleneck. In three of the analyzed populations (1, 3, and 5), significant traces of bottleneck effect were noted in populations 1 and 5 based on the results of all three tests (Sign test, Standardized Differences test, and Wilcoxon test), and in population 3 based on the results of the Standardized Differences test and the Wilcoxon test, but the p‐value in the Sign test was very close to the significance level (Table 8).
TABLE 8.
Testing bottleneck versus mutation–drift equilibrium hypotheses for all analyzed populations (IAM mutation model)
| Population | SIGN Test | Standardized test | Wilcoxon test |
|---|---|---|---|
| 1 |
Hee = 3.19 Hd = 0 He = 7 p = .00409 |
T2 = 3.058 p = .00112 |
One tail for heterozygosity deficiency: 1.00000 |
| One tail for heterozygosity excess: 0.00391 | |||
| Two tails for heterozygosity excess and deficiency: 0.00781 | |||
| 2 |
Hee = 2.09 Hd = 2 He = 3 p = .34933 |
T2 = 0.343 p = .36589 |
One tail for heterozygosity deficiency: 0.59375 |
| One tail for heterozygosity excess: 0.5000 | |||
| Two tails for heterozygosity excess and deficiency: 1.00000 | |||
| 3 |
Hee = 2.56 Hd = 1 He = 5 p = .05504 |
T2 = 1.741 p = .04081 |
One tail for heterozygosity deficiency: 0.97656 |
| One tail for heterozygosity excess: 0.03906 | |||
| Two tails for heterozygosity excess and deficiency: 0.07813 | |||
| 4 |
Hee = 2.39 Hd = 3 He = 3 p = .45257 |
T2=−0.216 p = .41430 |
One tail for heterozygosity deficiency: 0.50000 |
| One tail for heterozygosity excess: 0.57813 | |||
| Two tails for heterozygosity excess and deficiency: 1.0000 | |||
| 5 |
Hee = 3.45 Hd = 1 He = 7 p = .01388 |
T2 = 3.273 p = .00053 |
One tail for heterozygosity deficiency: 0.99805 |
| One tail for heterozygosity excess: 0.00391 | |||
| Two tails for heterozygosity excess and deficiency: 0.00781 | |||
| 6 |
Hee = 2.74 Hd = 1 He = 4 p = .25066 |
T2 = 0.939 p = .17374 |
One tail for heterozygosity deficiency: 0.92188 |
| One tail for heterozygosity excess: 0.10938 | |||
| Two tails for heterozygosity excess and deficiency: 0.21875 | |||
| 7 |
Hee = 2.92 Hd = 2 He = 4 p = .31898 |
T2 = 0.608 p = .27153 |
One tail for heterozygosity deficiency: 0.71875 |
| One tail for heterozygosity excess: 0.34375 | |||
| Two tails for heterozygosity excess and deficiency: 0.68750 | |||
| 8 |
Hee = 2.85 Hd = 2 He = 4 p = .29867 |
T2 = 1.107 p = .13412 |
One tail for heterozygosity deficiency: 0.94531 |
| One tail for heterozygosity excess: 0.07813 | |||
| Two tails for heterozygosity excess and deficiency: 0.15625 | |||
| 9 |
Hee = 2.08 Hd = 2 He = 3 p = .34541 |
T2 = 0.269 p = .39377 |
One tail for heterozygosity deficiency: 0.68750 |
| One tail for heterozygosity excess: 0.40625 | |||
| Two tails for heterozygosity excess and deficiency: 0.81250 |
Abbreviatins: Hee, expected heterozygosity excess; Hd, heterozygosity deficiency; He, heterozygosity excess.
4. DISCUSSION
Antarctic hairgrass is one of the most intriguing plant species in the world whose unique morphological and physiological features enable survival in extreme environments and colonization of the remote and inhospitable areas of the Maritime Antarctic. The species has large disjunctive distribution from north Patagonia in South America (around 38°S) to the Antarctic Peninsula, with the southernmost known locality in Lazer Bay (Alexander Island, 69°22.0’S, 71°50.7’W; Convey, 2011). Extensive geographic distribution has contributed to high morphological and anatomical variation of the species (Chwedorzewska et al., 2008; Giełwanowska & Szczuka, 2005; Giełwanowska et al., 2005). However, morphological plasticity is not accompanied by equally extensive genetic variability, which is generally low in the entire species range. A discrete decrease in genetic diversity is observed from the north to the south, with minimum values in the area of the Antarctic Peninsula (Holderegger et al., 2003; Chwedorzewska & Bednarek, 2008; van de Wouw et al., 2008).
According to the literature, D. antarctica colonized Antarctica during the Holocene (Chapman, 1996; Lewis‐Smith, 1984). In the paleobotanical research conducted by Birkenmajer et al. (1985), the fragments and pollen of D. antarctica isolated from peat cores were dated back at least five millennia. Reliable conclusions about the evolutionary history of Antarctic lichens, bryophytes, and flowering plants are difficult to draw due to insufficient data on contemporary species distribution. Recent molecular phylogeographic studies and classical biogeographic studies provided strong evidence that the persistence of Antarctica's extant terrestrial biota spans hundreds of thousands to millions of years (Convey, Bindschadler et al., 2009; De Wever et al., 2009; Domaschke et al., 2013; Fraser et al., 2014; McGaughran et al., 2010; Pisa et al., 2014; Romeike et al., 2002; Vyverman et al., 2010; Chong et al. 2015). The hypothesis postulating the presence of Antarctic glacial refugia during the Pleistocene was recently supported by glaciological evidence and population genetics data from various groups of organisms (Convey, Stevens et al., 2009; De Wever et al., 2009; McGaughran et al., 2010). For example, a recent population genetics study of the cosmopolitan moss Bryum argenteum Hedw. suggested its long‐term persistence in the Antarctic, which was reflected in its low genetic diversity (Clarke et al., 2009; Hills et al., 2010; Pisa et al., 2014).
To date, most molecular studies of D. antarctica have relied on the AFLP technique AFLP data (Chwedorzewska et al., 2004, 2008) demonstrated that D. antarctica populations which originated in close vicinity share considerable genetic similarity, but have evident morphological and anatomical differences. In the cited studies, the total percentage of polymorphic loci in the AFLP analysis did not exceed 39%. In studies that covered a wider geographic range of the species, polymorphism varied significantly from 13% (in ten populations from Signy Island, Anchorage Island, Lagoon Island, and Léonie Island in northern and southern Maritime Antarctic, respectively; Holderegger et al., 2003) to 92% within 38 D. antarctica populations from the sub‐Antarctic islands in the Indian Ocean, the Falklands, South Georgia Island, and the Antarctic Peninsula with the adjacent islands (van de Wouw et al., 2008). The high polymorphism demonstrated by AFLP was not confirmenoncoding regions of chloroplast genomesd by analyses of genetic variation within selected (van de Wouw et al., 2008).
In this study, iPBS markers supported the identification of 15 (12%) polymorphic loci, and average polymorphism was determined at 5.6% (across loci and populations). Previous studies relied on iPBS markers demonstrated up to 97% polymorphism in Myrica rubra (Chen & Liu, 2014). Lower polymorphism for iPBS markers (5%) was reported only by Baránek et al. (2012), but their study aimed to identify the clones of an apricot cultivar.
iPBS markers were previously applied to investigate the genetic diversity of Antarctic pearlwort (Colobanthus quitensis) (Androsiuk et al., 2015). The genotyping of individuals from eight C. quitensis populations in the vicinity of Henryk Arctowski Station revealed higher polymorphism than in D. antarctica, where 55 of 143 loci (39%) were polymorphic, with 14% of polymorphic loci per population on average. The average values of H e and I were determined at 0.040 and 0.061, respectively, which indicates that genetic diversity in C. quitensis was twice higher than that noted in D. antarctica in this study (average values of uHe = 0.021, I = 0.030). The results of AMOVA revealed 84% of the polymorphisms within the studied populations of C. quitensis and only 51% within populations of D. antarctica. As a result, the hierarchical analysis of population structure produced clearly higher values of the F‐statistic for D. antarctica (F ST = 0.4874) than C. quitensis (F ST = 0.164). Our results are highly consistent with the observations made by van de Wouw et al. (2008), in whose study, AMOVA revealed 46% of total genetic diversity among D. antarctica populations from Antarctic sites. Holderegger et al. (2003) also reported that 45% of genetic variation in D. antarctica from the Southern Maritime Antarctic was partitioned between populations. Unsurprisingly, genetic diversity among populations clearly decreased when a wider geographic range was analyzed, whereas the variation among regions increased to 37% (northern versus southern Maritime Antarctic; Holderegger et al., 2003) or even 75% when D. antarctica from the sub‐Antarctic islands in the Indian Ocean, the Falklands, South Georgia Island, and the Antarctic Peninsula with the adjacent islands were considered (van de Wouw et al., 2008).
Deschampsia antarctica is characterized by wide ecotypic variation (Lewis‐Smith, 2003; Nędzarek & Chwedorzewska, 2004); however, the genetic variation associated with the observed phenotypic dissimilarities has not been elucidated to date (van Fasanella et al., 2017; de Wouw et al., 2008). Polar plants have developed a number of response mechanisms to various biotic and abiotic stresses (Bruce et al., 2007; Cui et al., 2020; Giełwanowska et al., 2015). Some of these mechanisms can contribute to phenotypic variation caused by mutation (Rout et al., 2006) or modification of the DNA methylation pattern (Chinnusamy & Zhu, 2009). Chwedorzewska and Bednarek (2011), using methylation sensitive AFLP approach (metAFLP platform), found that methylation played a crucial role in the phenotypic variation of the D. antarctica specimens from different habitats of King George Island. Inconspicuous polymorphisms in the methylation pattern that have emerged in response to various stresses may be crucial in acclimatization to a range of environmental conditions, and they could be responsible for the differentiation of particular populations into local ecotypes (Cubas et al., 1999; Stajic & Bank, 2020).
A different approach was used in our previous study (Androsiuk et al., 2015), where the applicability of retrotransposon‐based molecular markers (iPBS technique) was verified in an analysis of genetic variation in C. quitensis. Similarly to other TEs, retrotransposons are mobilized in response to various stress factors (Capy et al., 2000; Schrader et al., 2014). Despite their random nature, genome rearrangements caused by TE activation could be beneficial because newly arisen genetic variation may be associated with adaptation to certain abiotic stressors (Finatto et al., 2015). A study of Hordeum spontaneum from the Evolution Canyon microsite in Lower Nahal Oren, Mount Carmel in Israel provided highly valuable insights (Kalendar et al., 2000). The authors reported an increase in the activity of BARE‐1 retrotransposons in H. spontaneum individuals growing on a slope exposed to high temperatures and drought. Retrotransposon‐based polymorphism allowed the identification of individuals that were and were not stressed by drought, even though the two sites were separated by a distance of only 300 m. The authors observed that local data were consistent with BARE‐1 trends in H. spontaneum throughout Israel and, therefore, could reflect adaptive selection for increasing genome size through retrotransposon activity (Kalendar et al., 2000). Similar observations were made in our previous study, where the genetic polymorphism analysis of C. quitensis revealed that TEs could be mobilized in response to various abiotic stressors (Androsiuk et al., 2015).
The results of the present study clearly indicate that despite low genetic polymorphism assessed with iPBS markers (6% on average) and low genetic diversity (H e = 0.020), the analyzed populations of D. antarctica are characterized by relatively high genetic differentiation (F ST = 0.4874, G st‐B = 0.4349, θ ( III ) = 0.3703, results of STRUCTURE analysis which assigned the studied individuals into 6 clusters, and high population dispersal revealed by PCoA), which could be attributed to limited gene transfer between these populations. This is a surprising observation in view of the fact that the analyzed populations were sampled in close proximity. However, local differences in landform and climatic conditions, such as strong winds blowing mainly from one direction (Wierzbicki, 2009), could be responsible for hampering seed dispersal and seedling establishment. These observations suggest that gene flow was obstructed by environmental heterogeneity or circulation boundaries in the studied area rather than the physical distance between sampling sites (no evidence for IBD was found).
The observed phenomena could also be attributed to the unique characteristics of the study area. The Maritime Antarctic is an extraordinary region not only due to extreme environmental conditions, but also their dynamics and diversity. Plants which colonized the studied area had to adapt to the local mosaic of microhabitats as well as general climatic fluctuations (Convey, 1996). Therefore, each site is characterized by dozens of factors describing microclimate conditions and soil properties that may vary considerably even between closely located sites. In extreme cases, even parts of the same population can experience different environmental conditions (Lachacz et al., 2018). For this reason, the genetic polymorphisms in the studied D. antarctica populations could have been shaped independently due to random TE mobilization and the spatial distribution of populations in an area with diverse abiotic stressors. The results of the IBE analysis revealed that environmental heterogeneity could have influenced the observed genetic differentiation. Most notably, the presence of a significant correlation between P and N–NO3 content versus. F ST/(1−F ST) indicates that differences in nutrient content could be associated with population divergence. However, further tests are needed to fully determine whether the differences in the composition and nutrient content of soils in the Antarctic oasis (Lachacz et al., 2018) are responsible for promoting certain genotypes.
Cytological analysis revealed that the common chromosome counts in D. antarctica is 2n = 26 (Amosova et al., 2015; Cardone et al., 2009; Volkov et al., 2010), although some counts reported also polyploid populations of 2n = 52 in Patagonia, which has not been reported in Antarctic populations (e.g., González et al., 2016). It has been suggested that polyploidization is one of the main factors responsible for shaping diversity in angiosperms (Leitch & Leitch, 2008). According to some studies, polyploidization could result in higher level of polymorphism revealed by molecular markers (e.g., Budak et al., 2005; Gulsen et al., 2009; Milla‐Lewis et al., 2013). However, there are also other studies which show lack of significant influence of polyploidization on genetic variation (e.g., Liu et al., 2001; Zeng et al., 2011; Zhang et al., 2017). Research conducted in marine Antarctica has shown that diploid individuals of D. antarctica predominate in the populations of this area. However, especially in the Southernmost populations the hypotriploid individuals and genotypes that had the B chromosome were also reported (Amosova et al., 2015; Navrotska et al., 2017). The presence of mixoploid plants in D. antarctica populations is maintained by its capacity of vegetative and apomictic propagation common in populations from the range limit, where environmental stress conditions prevail (e.g., Amosova et al., 2015). However, as it was shown by Navrotska et al. (2017), genetic differentiation between D. antarctica individuals with anomalous karyotype and diploids did not differ significantly from genetic differentiation between individuals representing typical diploid plants. Therefore, more detailed cytological and molecular studies with extensive sampling are needed to check whether polyploidization in D. antarctica is common or rather rare event, and to test how the change in ploidy level may affect the genetic diversity of the species.
The reproductive biology of D. antarctica is yet another important factor which should be considered in evaluations of the evolution and maintenance of genetic diversity and differentiation in the species. D. antarctica is self‐compatible plant species which can reproduce by self‐pollinating cleistogamous flowers or by vegetative reproduction (Giełwanowska & Kellmann‐Sopyła, 2015; Parnikoza et al., 2018) which does not contribute to genetic variation. In favorable circumstances, the species can produce viable seeds by outcrossing (Convey, 1996). However, outcrossing is highly unlikely in the Maritime Antarctic due to adverse climate conditions which inhibit generative reproduction or prolong the development of viable seeds even to two growing seasons (Parnikoza et al., 2018). Very strong winds may also impede pollen and seed dispersal. Therefore, new genotypes have limited dispersal opportunities because the seeds of D. antarctica do not have any structures that could promote dispersal across long distances. The patchy nature of ice‐free areas that are often separated by impassable barriers such as glaciers or open sea waters can also limit gene flow. Parnikoza et al. (2008) reported that some Antarctic bird species (Larus dominicanus, Catharacta maccormicki and C. lonnbergi) use plants, to build nests, which suggests that zoochory could play a role in the local dispersal of D. antarctica.
Regardless of its dispersal mechanism, a newly established population often has a limited number of individuals, which reproduce mainly by self‐fertilization and/or vegetative propagation (Holderegger et al., 2003). In most cases, these individuals are the only source of genes for the successive generations. In the current study, the negative values of Fu's F S statistic revealed a demographic expansion of most studied populations (except populations 2, 6 and 9), whereas significant traces of reduced effective population size were noted only in three populations (1, 3, and 5). The observed variations indicate that the studied D. antarctica populations had different demographic histories, which could be attributed to recurring adverse environmental conditions (that are different even for closely located, neighboring populations) or even recent local extinction–recolonization events. These processes could explain why bottlenecks were detected in only some of the analyzed populations. However, the difference between bottleneck and founder effects is difficult to identify with the use of dominant markers.
Our molecular data are consistent with the results of other studies on the genetic characteristic of D. antarctica. The present findings have confirmed the low genetic variation of D. antarctica and have demonstrated surprising differences between populations. The observed pattern of genetic differentiation probably reflects the local mosaic of microhabitats on King George Island. However, an analysis of the most southern range of the species indicates that genetic differentiation could also be attributed to specific landform which can isolate even close populations and limit generative propagation. Further research is needed to explore retrotransposon‐based polymorphism in greater detail and to confirm the association between abiotic stress factors and the polymorphisms revealed by iPBS markers.
CONFLICT OF INTEREST
All authors declare no conflict of interest.
AUTHOR CONTRIBUTION
Piotr Androsiuk: Conceptualization (equal); Data curation (lead); Formal analysis (lead); Investigation (equal); Methodology (equal); Resources (supporting); Supervision (lead); Validation (equal); Visualization (lead); Writing‐original draft (lead); Writing‐review & editing (lead). Katarzyna J. Chwedorzewska: Conceptualization (equal); Data curation (supporting); Formal analysis (supporting); Investigation (supporting); Methodology (equal); Resources (equal); Validation (equal); Visualization (supporting); Writing‐original draft (equal); Writing‐review & editing (equal). Justyna Dulska: Data curation (supporting); Investigation (equal); Writing‐review & editing (supporting). Sylwia Milarska: Data curation (supporting); Investigation (equal); Writing‐review & editing (supporting). Irena Giełwanowska: Conceptualization (equal); Data curation (supporting); Formal analysis (supporting); Investigation (supporting); Methodology (equal); Resources (equal); Supervision (equal); Validation (equal); Visualization (supporting); Writing‐original draft (equal); Writing‐review & editing (equal).
Supporting information
Table S1
Table S2
Androsiuk P, Chwedorzewska KJ, Dulska J, Milarska S, Giełwanowska I. Retrotransposon‐based genetic diversity of Deschampsia antarctica Desv. from King George Island (Maritime Antarctic). Ecol Evol.2021;11:648–663. 10.1002/ece3.7095
DATA AVAILABILITY STATEMENT
Binary matrix of iPBS data is available among Supplementary Materials (iPBS_data.xlsx) attached to this paper. Raw data (photographs of horizontal agarose gel electrophoresis) are available from the Dryad Digital Repository (https://doi.org/10.5061/dryad.cfxpnvx47).
REFERENCES
- Amosova, A. V. , Bolsheva, N. L. , Samatadze, T. E. , Twardovska, M. O. , Zoshchuk, S. A. , Andreev, I. O. , Badaeva, E. D. , Kunakh, V. A. , & Muravenko, O. V. (2015). Molecular cytogenetic analysis of Deschampsia Antarctica Desv. (Poaceae), Maritime Antarctic. PLoS One, 10(e0138878). 10.1371/journal.pone.0138878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androsiuk, P. , Chwedorzewska, K. , Szandar, K. , & Giełwanowska, I. (2015). Genetic variability of Colobanthus quitensis from King George Island (Antarctica). Polish Polar Research, 36, 281–295. 10.1515/popore-2015-0017 [DOI] [Google Scholar]
- Baránek, M. , Mészáros, M. , Sochorová, J. , Čechová, J. , & Raddová, J. (2012). Utility of retrotransposon−derived marker systems for differentiation of presumed clones of the apricot cultivar Velkopavlovická. Scientia Horticulturae, 143, 1–6. 10.1016/j.scienta.2012.05.022 [DOI] [Google Scholar]
- Beyer, L. , Bölter, M. , & Seppelt, R. D. (2000). Nutrient and thermal regime, microbial biomass and vegetation of Antarctic soils in the Windmill Islands Region of east Antarctica (Wilkes Land). Arctic Antarctic and Alpine Research, 32, 30–39. 10.1080/15230430.2000.12003336 [DOI] [Google Scholar]
- Birkenmajer, K. , Ochyra, R. , Olsson, I. U. , & Stuchlik, L. (1985). Mid‐Holocene radiocarbon‐dated peat at Admiralty Bay, King George Island (South Shetland Islands, West Antarctic). Bulletin Polish Academy of Sciences: Earth Sciences, 33, 7–12. [Google Scholar]
- Bonchev, G. , Dušinský, R. , Hauptvogel, P. , Gaplovska‐Kysela, K. , & Švec, M. (2019). On the diversity and origin of the barley complex agriocrithon inferred by iPBS transposon markers. Genetic Resources and Crop Evolution, 66, 1573–1586. 10.1007/s10722-019-00814-5 [DOI] [Google Scholar]
- Bruce, T. A. , Matthes, M. C. , Napier, J. A. , & Pickett, J. A. (2007). Stressful “memories” of plants: Evidence and possible mechanisms. Plant Science, 173, 603–608. 10.1016/j.plantsci.2007.09.002 [DOI] [Google Scholar]
- Budak, H. , Shearman, R. C. , Gulsen, O. , & Dweikat, I. (2005). Understanding ploidy complex and geographic origin of the Buchloe dactyloides genome using cytoplasmic and nuclear marker systems. Theoretical and Applied Genetics, 111, 1545–1552. 10.1007/s00122-005-0083-3 [DOI] [PubMed] [Google Scholar]
- Capy, P. , Gasperi, G. , Biemont, C. , & Bazin, C. (2000). Stress and transposable elements: Co−evolution or useful parasites? Heredity, 85, 101–106. 10.1046/j.1365-2540.2000.00751.x [DOI] [PubMed] [Google Scholar]
- Cardone, S. , Sawatani, P. , Rush, P. , García, A. M. , Poggio, L. , & Schrauf, G. (2009). Karyological studies in Deschampsia antarctica Desv. (Poaceae). Polar Biology, 32, 427–433. 10.1007/s00300-008-0535-8 [DOI] [Google Scholar]
- Chapman, G. P. (1996). The biology of grasses (p. 272). CABI. [Google Scholar]
- Chen, F. Y. , & Liu, J. H. (2014). Germplasm genetic diversity of Myrica rubra in Zhejiang Province studied using inter−primer binding site and start codon−targeted polymorphism markers. Scientia Horticulturae, 170, 169–175. 10.1016/j.scienta.2014.03.010 [DOI] [Google Scholar]
- Chinnusamy, V. , & Zhu, J. K. (2009). Epigenetic regulation of stress response in plants. Current Opinion in Plant Biology, 12, 133–139. 10.1016/j.pbi.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, C.‐W. , Pearce, D. , & Convey, P. (2015). Emerging spatial patterns in Antarctic prokaryotes. Frontiers in Microbiology, 6, 1058 10.3389/fmicb.2015.01058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwedorzewska, K. J. , & Bednarek, P. T. (2008). Genetic variability in the Antarctic hairgrass Deschampsia antarctica Desv. from maritime Antarctic and sub‐Antarctic sites. Polish Journal of Ecology, 56, 209–216. [Google Scholar]
- Chwedorzewska, K. J. , & Bednarek, P. T. (2011). Genetic and epigenetic studies on populations of Deschampsia antarctica Desv. from contrasting environments at King George Island (Antarctic). Polish Polar Research, 32, 15–26. 10.2478/v10183"12011"120005"129 [DOI] [Google Scholar]
- Chwedorzewska, K. J. , Bednarek, P. T. , & Puchalski, J. (2004). Molecular variation of Antarctic grass Deschampsia antarctica Desv. from King George Island (Antarctica). Acta Societatis Botanicorum Poloniae, 73, 23–29. 10.5586/asbp.2004.004 [DOI] [Google Scholar]
- Chwedorzewska, K. J. , Giełwanowska, I. , Szczuka, E. , & Bochenek, A. (2008). Anatomical and genetic variation of Deschampsia antarctica Desv. from King George Island (the Antarctic). Polish Polar Research, 4, 377–386. [Google Scholar]
- Clarke, L. C. , Ayre, D. J. , & Robinson, S. A. (2009). Genetic structure of East Antarctic populations of the moss Ceratodon purpureus . Antarctic Science, 21(1), 51–58. 10.1017/S0954102008001466 [DOI] [Google Scholar]
- Clemente‐Moreno, M. J. , Omranian, N. , Sáez, P. L. , Figueroa, C. M. , Del‐Saz, N. , Elso, M. , Poblete, L. , Orf, I. , Cuadros‐Inostroza, A. , Cavieres, L. A. , Bravo, L. , Fernie, A. R. , Ribas‐Carbó, M. , Flexas, J. , Nikoloski, Z. , Brotman, Y. , & Gago, J. (2020). Low‐temperature tolerance of the Antarctic species Deschampsia antarctica: A complex metabolic response associated with nutrient remobilization. Plant Cell and Environment, 43, 1376–1393. 10.1111/pce.13737 [DOI] [PubMed] [Google Scholar]
- Convey, P. (1996). The influence of environmental characteristicos on life history attributes of Antarctic terrestrial biota. Biological Reviews, 71, 191–225. 10.1111/j.1469-185X.1996.tb00747.x [DOI] [Google Scholar]
- Convey, P. (2011). Antarctic terrestrial biodiversity in a changing world. Polar Biology, 34, 1629–1641. 10.1007/s00300-011-1068-0 [DOI] [Google Scholar]
- Convey, P. , Bindschadler, R. , Di Prisco, G. , Fahrbach, E. , Gu, J. , Hodgson, D. A. , Mayewski, P. A. , Summerhayes, C. P. , & Turner, J. (2009). Antarctic climate change and the environment. Antarctic Science, 21, 541–563. 10.1017/S0954102009990642 [DOI] [Google Scholar]
- Convey, P. , & Peck, L. S. (2019). Antarctic environmental change and biological responses. Science Advence, 11, eaaz0888 10.1126/sciadv.aaz0888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Convey, P. , Stevens, M. I. , Hodgson, D. A. , Smellie, J. L. , Hillenbrand, C.‐D. , Barnes, D. K. A. , Clarke, A. , Pugh, P. J. A. , Linse, K. , & Cary, S. C. (2009). Exploring biological constraints on the glacial history of Antarctica. Quaternary Science Reviews, 28, 3035–3048. 10.1016/j.quascirev.2009.08.015 [DOI] [Google Scholar]
- Corner, R. W. M. (1971). Studies in Colobanthus quitensis (Kunth) Bartl. and Deschampsia antarctica Desv.: IV. Distribution and reproductive performance in Argentine islands. British Antarctic Survey B, 26, 41–50. [Google Scholar]
- Cornuet, J. M. , & Luikart, G. (1996). Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics, 144, 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho, J. P. , Carvalho, A. , Martin, A. , & Lima‐Brito, J. (2018). Molecular characterization of Fagaceae species using inter‐primer binding site (iPBS) markers. Molecular Biology Reports, 45, 133–142. 10.1007/s11033-018-4146-3 [DOI] [PubMed] [Google Scholar]
- Cubas, P. , Vincent, C. , & Coen, E. (1999). An epigenetic mutation responsible for natural variation in floral symmetry. Nature, 401(6749), 157–161. 10.1038/43657 [DOI] [PubMed] [Google Scholar]
- Cui, L. H. , Byun, M. Y. , Oh, H. G. , Kim, S. J. , Lee, J. , Park, H. , Lee, H. , & Kim, W. T. (2020). Poaceae Type II Galactinol Synthase 2 from Antarctic Flowering Plant Deschampsia antarctica and Rice Improves Cold and Drought Tolerance by Accumulation of Raffinose Family Oligosaccharides in Transgenic Rice Plants. Plant and Cell Physiology, 61(1), 88–104. 10.1093/pcp/pcz180 [DOI] [PubMed] [Google Scholar]
- De Wever, A. , Leliaert, F. , Verleyen, E. , Vanormelingen, P. , Van der Gucht, K. , Hodgson, D. A. , Sabbe, K. , & Vyverman, W. (2009). Hidden levels of phylodiversity in Antarctic green algae: further evidence for the existence of glacial refugia. Proceedings of the Royal Society B: Biological Sciences, 276(1673), 3591–3599. 10.1098/rspb.2009.0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domaschke, S. , Fernández‐Mendoza, F. , García, M. A. , Martín, M. P. , & Printzen, C. (2013). Low genetic diversity in Antarctic populations of the lichen‐forming ascomycete Cetraria aculeate and its photobiont. Polar Research, 31, polar.v31i0.17353. [Google Scholar]
- Earl, D. A. , & Vonholdt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizingSTRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Evanno, G. , Regnaut, S. , & Goudet, J. (2005). Detecting the number of clusters of individuals usingthe software STRUCTURE: A simulation study. Molecular Ecology, 14(8), 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Excoffier, L. , Laval, G. , & Schneider, S. (2005). Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics, 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- Excoffier, L. , Smouse, P. , & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction sites. Genetics, 131, 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanella, M. , Premoli, A. C. , Urdampilleta, J. D. , González, M. L. , & Chiapella, J. O. (2017). How did a grass reach Antarctica? The Patagonian connection of Deschampsia antarctica (Poaceae). Botanical Journal of the Linnean Society, 185, 511–524. 10.1093/botlinnean/box070 [DOI] [Google Scholar]
- Finatto, T. , de Oliveira, A. C. , Chaparro, C. , da Maia, L. C. , Farias, D. R. , Woyann, L. G. , Mistura, C. C. , Soares‐Bresolin, A. P. , Llauro, C. , Panaud, O. , & Picault, N. (2015). Abiotic stress and genome dynamics: Specific genes and transposable elements response to iron excess in rice. Rice (NY), 8, 13 10.1186/s12284-015-0045-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, D. J. (1989). Eukaryotic transposable elements and genome evolution. Trends Genetics, 5, 103–107. 10.1016/0168-9525(89)90039-5 [DOI] [PubMed] [Google Scholar]
- Fraser, C. I. , Terauds, A. , Smellie, J. , Convey, P. , & Chown, S. L. (2014). Geothermal activity helps life survive glacial cycles. Proceedings of the National Academy of Sciences, 111(15), 5634–5639. 10.1073/pnas.1321437111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. X. (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147, 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giełwanowska, I. (2003a). Responses of Deschampsia antarctica to abiotic stress factors. Polish Journal of Natural Sciences, 1, 108–111. [Google Scholar]
- Giełwanowska, I. (2003b). Ultrastructural investigations of Deschampsia antarctica mesophyll In Olech M. (Ed.), The functioning of polar ecosystems as viewed against global environmental changes (pp. 47–49). XXXIX International Polar Symposium. [Google Scholar]
- Giełwanowska, I. , & Kellmann‐Sopyła, W. (2015). Generative reproduction of Antarctic grasses, the native species Deschampsia antarctica Desv. and the alien species Poa annua L. Polish Polar Research, 36, 261–279. 10.1515/popore"122015"120016 [DOI] [Google Scholar]
- Giełwanowska, I. , Pastorczyk, M. , Kellmann‐Sopyła, W. , Gorniak, D. , & Gorecki, R. J. (2015). Morphological and ultrastructural changes of organelles in leaf mesophyll cells of the Arctic and Antarctic plants of Poaceae family under cold influence. Arctic Antarctic and Alpine Research, 47, 17–25. 10.1657/AAAR0014-019 [DOI] [Google Scholar]
- Giełwanowska, I. , & Szczuka, E. (2005). New ultrastructural features of organelles in leaf cell of Deschampsia antarctica Desv. Polar Biology, 28, 951–955. 10.1007/s00300-005-0024-2 [DOI] [Google Scholar]
- Giełwanowska, I. , Szczuka, E. , Bednara, J. , & Górecki, J. (2005). Anatomical Features and Ultrastructure of Deschampsia antarctica (Poaceae) Leaves from Different Growing Habitats. Annals of Botany‐London, 96, 1109–1119. 10.1093/aob/mci262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, M. L. , Urdampilleta, J. D. , Fasanella, M. , Premoli, A. C. , & Chiapella, J. C. (2016). Distribution of rDNA and polyploidy in Deschampsia antarctica E. Desv. in Antarctic and Patagonic populations. Polar Biology, 39, 1663–1677. 10.1007/s00300-016-1890-5 [DOI] [Google Scholar]
- Gulsen, O. , Sever‐Mutlu, S. , Mutlu, N. , Tuna, M. , Karaguzel, O. , Shearman, R. C. , Riordan, T. P. , & Heng‐Moss, T. M. (2009). Polyploidy creates higher diversity among Cynodon accessions as assessed by molecular markers. Theoretical and Applied Genetics, 118, 1309–1319. 10.1007/s00122-009-0982-9 [DOI] [PubMed] [Google Scholar]
- Hills, S. F. , Stevens, M. I. , & Gemmill, C. E. C. (2010). Molecular support for Pleistocene persistence of the continental Antarctic moss Bryum argenteum . Antarctic Science, 22, 721–726. 10.1017/S0954102010000453 [DOI] [Google Scholar]
- Holderegger, R. , Stehlik, I. , Lewis‐Smith, R. I. , & Abbott, R. J. (2003). Population of Antarctic hairgrass (Deschampsia antarctica) show low genetic diversity. Arctic Antarctic and Alpine Research, 35, 214–217. [Google Scholar]
- Holsinger, K. E. (1999). Analysis of genetic diversity in geographically structured populations: A Bayesian perspective. Hereditas, 130, 245–255. 10.1111/j.1601-5223.1999.00245.x [DOI] [Google Scholar]
- Holsinger, K. , & Lewis, P. (2003). HICKORY: A package for analysis of population genetic data, Vol. 1.0 University of Connecticut. [Google Scholar]
- Kalendar, R. , Amenov, A. , & Daniyarov, A. (2019). Use of retrotransposon‐derived genetic markers to analyse genomic variability in plants. Functional Plant Biology, 46(1), 15–29. 10.1071/FP18098 [DOI] [PubMed] [Google Scholar]
- Kalendar, R. , Antonius, K. , Smýkal, P. , & Schulman, A. H. (2010). iPBS: A universal method for DNA fingerprinting and retrotransposon isolation. Theoretical and Applied Genetics, 121(1419–1430), 10.1007/s00122-010-1398-2 [DOI] [PubMed] [Google Scholar]
- Kalendar, R. , Tanskanen, J. , Immonen, S. , Nevo, E. , & Schulman, A. H. (2000). Genome evolution of wild barley (Hordeum spontaneum) by BARE−1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences of the United States of America, 97, 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc, J. , Androsiuk, P. , Chwedorzewska, K. J. , Cuba‐Díaz, M. , Górecki, R. , & Giełwanowska, I. (2018). Range‐wide pattern of genetic variation in Colobanthus quitensis . Polar Biology, 41, 2467–2479. 10.1007/s00300-018-2383-5 [DOI] [Google Scholar]
- Lachacz, A. , Kalisz, B. , Giełwanowska, I. , Olech, M. , Chwedorzewska, K. J. , & Kellmann‐Sopyła, W. (2018). Nutrient abundance and variability from Antarctic soils in the coastal of King George Island. Journal of Soil Science and Plant Nutrition, 18, 294–311. 10.4067/S0718-95162018005001101 [DOI] [Google Scholar]
- Lee, J. R. , Raymond, B. , Bracegirdle, T. J. , Chadès, I. , Fuller, R. A. , Shaw, J. D. , & Terauds, A. (2017). Climate change drives expansion of Antarctic ice‐free habitat. Nature, 547, 49–54. 10.1038/nature229962 [DOI] [PubMed] [Google Scholar]
- Leitch, A. R. , & Leitch, I. J. (2008). Genomic plasticity and the diversity of polyploid plants. Science, 320, 481–483. 10.1126/science.1153585 [DOI] [PubMed] [Google Scholar]
- Lewis‐Smith, R. I. (1984). Plant biology of the Sub‐Antarctic and Antarctic In Laws R. M. (Ed.), Antarctic Ecology I (pp. 61–162). Academic Press. [Google Scholar]
- Lewis‐Smith, R. I. (2003). The enigma of Colobanthus quitensis and Deschampsia antartica in Antarctica In Huiskes A. H. L., Gieskes W. W. C., Rozema J., Schorno R. M. L., van der Vies S. M., & Wolff W. J. (Eds.), Antarctic biology in a global context (pp. 234–239). Backhuys Publishers. [Google Scholar]
- Liu, B. , Brubaker, C. L. , Mergeai, G. , Cronn, R. C. , & Wendel, J. F. (2001). Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome, 44, 321–330. 10.1139/g01-011 [DOI] [PubMed] [Google Scholar]
- McGaughran, A. , Stevens, M. I. , & Holland, B. (2010). Biogeography of circum‐Antarctic springtails. Molecular Phylogenetics and Evolution, 57, 48–58. 10.1016/j.ympev.2010.06.003 [DOI] [PubMed] [Google Scholar]
- Mehmood, A. , Luo, S. , Ahmad, N. M. , Dong, C. , Mahmood, T. , Sajjad, Y. , Jaskani, M. J. , & Sharp, P. (2016). Molecular variability and phylogenetic relationships of guava (Psidium guajava L.) cultivars using inter‐primer binding site (iPBS) and microsatellite (SSR) markers. Genetic Resources and Crop Evolution, 63, 1345–1361. 10.1007/s10722-015-0322-7 [DOI] [Google Scholar]
- Milla‐Lewis, S. R. , Carolina Zuleta, M. , Van Esbroeck, G. A. , Quesenberry, K. H. , & Kenworthy, K. E. (2013). Cytological and molecular characterization of genetic diversity in Stenotaphrum . Crop Science, 53, 296–308. 10.2135/cropsci2012.04.0234 [DOI] [Google Scholar]
- Moreau‐Mhiri, C. , Morel, J. B. , Audeon, C. , Ferault, M. , Grandbastien, M. A. , & Lucas, H. (1996). Regulation of expression of the tobacco Tnt1 retrotransposon in heterologous species following pathogen‐related stresses. Plant Journal, 9, 409–419. 10.1046/j.1365-313X.1996.09030409.x [DOI] [Google Scholar]
- Nanninga, G. B. , Saenz‐Agudelo, P. , Manica, A. , & Berumen, M. L. (2014). Environmental gradients predict the genetic population structure of a coral reef fish in the Red Sea. Molecular Ecology, 23(3), 591–602. 10.1111/mec.12623 [DOI] [PubMed] [Google Scholar]
- Navrotska, D. O. , Andreev, I. O. , Parnikoza, I. Y. , Spiridonova, K. V. , Poronnik, O. O. , Miryuta, N. Y. , Myryuta, G. Y. , Zahrychuk, O. M. , Drobyk, N. M. , & Kunakh, V. A. (2017). Comprehensive Characterization of Cultivated in vitro Deschampsia antarctica E. Desv. Plants with Different Chromosome Numbers. Cytology and Genetics, 51, 422–431. [Google Scholar]
- Nędzarek, A. , & Chwedorzewska, K. J. (2004). Nutrients content in water flash chosen sites of Deschampsia antarctica (King George Island, Antarctica). Folia Universitatis Agriculture Stetinensis, 234, 299–304. [Google Scholar]
- Nei, M. (1972). Genetic distance between populations. American Naturalist, 106, 282–283. 10.1086/282771 [DOI] [Google Scholar]
- Ochyra, R. , Lewis‐Smith, R. I. , & Bednarek‐Ochyra, H. (2008). The illustrated moss flora of Antarctica (p. 704). Cambridge University Press. [Google Scholar]
- Olech, M. (2004). Lichens of King George Island Antarctica (p. 391). The Institute of Botany of the Jagiellonian University. [Google Scholar]
- Parnikoza, I. , Kozeretska, O. , & Kozeretska, I. (2008). Is a Translocation of Indigenous Plant Material Successful in the Maritime Antarctic? Polarforschung, 78, 25–27. 10.2312/polarforschung.78.1-2.25 [DOI] [Google Scholar]
- Parnikoza, I. , Rozhok, A. , Convey, P. , Veselski, P. M. , Esefeld, J. , Ochyra, R. , Mustafa, O. , Braun, C. H. , Peter, H.‐U. , Smykla, J. , Kunakh, V. , & Kozeretska, I. (2018). Spread of Antarctic vegetation by the kelp gull: Comparison of two maritime Antarctic regions. Polar Biology, 41, 1143–1155. 10.1007/s00300-018-2274-9 [DOI] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2006). GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288–295. 10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2012). GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research‐an update. Bioinformatics, 28, 2537–2539. 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini, L. , Fanti, L. , Specchia, V. , Bozzetti, M. P. , Berloco, M. , Palumbo, G. , & Pimpinelli, S. (2014). Transposons, environmental changes, and heritable induced phenotypic variability. Chromosoma, 123, 345–354. 10.1007/s00412-014-0464-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli, S. , & Piacentini, L. (2020). Environmental change and the evolution of genomes: Transposable elements as translators of phenotypic plasticity into genotypic variability. Functional Ecology, 34, 428–441. 10.1111/1365-2435.13497 [DOI] [Google Scholar]
- Piry, S. , Luikart, G. , & Cornuet, J. M. (1999). BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity, 90, 502–503. 10.1093/jhered/90.4.502 [DOI] [Google Scholar]
- Pisa, S. , Biersma, E. M. , Convey, P. , Patiño, J. , Vanderpoorten, A. , Werner, O. , & Ros, R. M. (2014). The cosmopolitan moss Bryum argenteumin Antarctica: Recent colonisation or in situ survival? Polar Biology, 37, 1469–1477. 10.1007/s00300-014-1537-3 [DOI] [Google Scholar]
- Pritchard, J. K. , Stephens, P. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeike, J. , Friedl, T. , Helms, G. , & Ott, S. (2002). Genetic diversity of algal and fungal partners in four species of Umbilicaria (lichenized ascomycetes) along a transect of the Antarctic Peninsula. Molecular Biology and Evolution, 19, 1209–1217. 10.1093/oxfordjournals.molbev.a004181 [DOI] [PubMed] [Google Scholar]
- Rousset, F. (1997). Genetic differentiation and estimation of gene flow from F‐statistics under isolation by distance. Genetics, 145, 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, G. R. , Mohapatra, A. , & Jain, S. M. (2006). Tissue culture of ornamental pot plant: A critical review on present scenario and future prospects. Biotechnology Advances, 24, 531–560. 10.1016/j.biotechadv.2006.05.001 [DOI] [PubMed] [Google Scholar]
- San Miguel, P. , Tikhonov, A. , Jin, Y. K. , Motchoulskaia, N. , Zakharov, D. , Melake‐Berhan, A. , Springer, P. S. , Edwards, K. J. , Lee, M. , Avramova, Z. , & Bennetzent, J. L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science, 274, 765–768. 10.1126/science.274.5288.765 [DOI] [PubMed] [Google Scholar]
- Schrader, L. , Kim, J. W. , Ence, D. , Zimin, A. , Klein, A. , Wyschetzki, K. , Weichselgartner, T. , Kemena, C. , Stokl, J. , Schultner, E. , Wurm, Y. , Smith, C. D. , Yandell, M. , Heinze, J. , Gadau, J. , & Oettler, J. (2014). Transposable element islands facilitate adaptation to novel environments in an invasive species. Nature Communications, 5, 5495 10.1038/ncomms6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierakowski, K. , Korczak‐Abshire, M. , & Jadwiszczak, P. (2017). Changes in bird communities of Admiralty Bay, King George Island (West Antarctica): insights from monitoring data (1977–1996). Polish Polar Research, 38, 231–262. 10.1515/popore-2017-0010 [DOI] [Google Scholar]
- Song, S. , Dey, D. K. , & Holsinger, K. E. (2006). Hierarchical models with migration, mutation, and drift: Implications for genetic inference. Evolution, 60, 1–12. 10.1111/j.0014-3820.2006.tb01076.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajic, D. , & Bank, C. (2020). Evolutionary consequences of epigenetically induced phenotypic switching. EcoEvoRxiv Preprints. April 1. 10.32942/osf.io/6yf4u [DOI] [Google Scholar]
- Stapley, J. , Santure, A. W. , & Dennis, S. R. (2015). Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Molecular Ecology, 24, 2241–2252. 10.1111/mec.13089 [DOI] [PubMed] [Google Scholar]
- Takeda, S. , Sugimoto, K. , Otsuki, H. , & Hirochika, H. (1998). Transcriptional activation of the tobacco retrotransposon Tto1 by wounding and methyl jasmonate. Plant Molecular Biology, 36, 365–376. 10.1023/A:1005911413528 [DOI] [PubMed] [Google Scholar]
- Tero, N. , Aspi, J. , Siikamäki, P. , Jäkäläniemi, A. , & Tuomi, J. (2003). Genetic structure and gene flow in a metapopulation of an endangered plant species, Silene tatarica . Molecular Ecology, 12, 2073–2085. 10.1046/j.1365-294X.2003.01898.x [DOI] [PubMed] [Google Scholar]
- van de Wouw, M. , van Dijk, P. , & Huiskes, A. H. L. (2008). Regional genetic diversity in Antarctic hairgrass (Deschampsia antarctica Desv.). Journal of Biogeography, 35, 365–376. 10.1111/j.1365-2699.2007.01784 [DOI] [Google Scholar]
- Volkov, R. A. , Kozeretska, I. A. , Kyryachenko, S. S. , Andreev, I. O. , Maidanyuk, D. N. , Parnikoza, I. Y. , & Kunakh, V. A. (2010). Molecular evolution and variability of ITS1–ITS2 in populations of Deschampsia Antarctica from two regions of the maritime Antarctic. Polar Science, 4, 469–478. [Google Scholar]
- Voronova, A. , Jansons, Ā. , & Ruņģis, D. (2011). Expression of retrotransposon‐like sequences in Scots pine (Pinus sylvestris) in response to heat stress. Environmental and Experimental Biology, 9, 121–127. [Google Scholar]
- Vyverman, W. , Verleyen, E. , Wilmotte, A. , Hodgson, D. A. , Willems, A. , Peeters, K. , Van de Vijver, B. , De Wever, A. , Leliaert, F. , & Sabbe, K. (2010). Evidence for widespread endemism among Antarctic micro‐organisms. Polar Science, 4, 103–113. 10.1016/j.polar.2010.03.006 [DOI] [Google Scholar]
- Wierzbicki, G. (2009). Wiatry huraganowe w 2008 roku w Zatoce Admiralicji, Wyspa Króla Jerzego, Antarktyda Zachodnia [Hurricane winds of 2008 in Admiralty Bay, King George Island, west Antarctica]. Przegląd Naukowy Inżynieria I Kształtowanie Środowiska, 44, 47–55. [Google Scholar]
- Zeng, B. , Zuo, F. Y. , Zhang, X. Q. , Fan, Y. , & Yu, Y. X. (2011). SRAP analysis of genetic diversity and relationship in Orchardgrass. Journal of Plant Genetic Resources, 12(5), 709–715. 10.4141/CJPS07017 [DOI] [Google Scholar]
- Zhang, J. , Yan, J. , Shen, X. , Chang, D. , Bai, S. , Zhang, Y. , & Zhang, J. (2017). How genetic variation is affected by geographic environments and ploidy level in Erianthus arundinaceus? PLoS One, 12(5), e0178451 10.1371/journal.pone.0178451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Znój, A. , Chwedorzewska, K. J. , Androsiuk, P. , Cuba‐Diaz, M. , Giełwanowska, I. , Koc, J. , Korczak‐Abshire, M. , Grzesiak, J. , & Zmarz, A. (2017). Rapid environmental changes in the Western Antarctic peninsula region due to climate change and human activity. Applied Ecology and Environmental Research, 15, 525–539. 10.15666/aeer/1504_525539 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Data Availability Statement
Binary matrix of iPBS data is available among Supplementary Materials (iPBS_data.xlsx) attached to this paper. Raw data (photographs of horizontal agarose gel electrophoresis) are available from the Dryad Digital Repository (https://doi.org/10.5061/dryad.cfxpnvx47).


