Abstract
AIM
To present the multi-omics landscape of cutaneous melanoma (CM) and uveal melanoma (UM) from The Cancer Genome Atlas (TCGA).
METHODS
The differentially expressed genes (DEGs) between CM and UM were found and integrated into a gene ontology enrichment analysis. Besides, the differentially expressed miRNAs were also identified. We also compared the methylation level of CM with UM and identified the differentially methylated regions to integrate with the DEGs to display the relationship between the gene expression and DNA methylation. The differentially expressed transcription factors (TFs) were identified.
RESULTS
Though CM had more mutational burden than UM, they shared several similarities such as the same rankings in diverse variant types. Except GNAQ and GNA11, the other top 18 mutated genes of the combined group were mostly detected in CM instead of UM. On the transcriptomic level, 4610 DEGs were found and integrated into a gene ontology enrichment analysis. We also identified 485 differentially expressed miRNAs. The methylation analysis showed that UM had a significantly higher methylation level than CM. The integration of differentially methylated regions and DEGs demonstrated that most DEGs were downregulated in UM and the hypo- and hypermethylation presented no obvious difference within these DEGs. Finally, 116 hypermethylated TFs and 114 hypomethylated TFs were identified as differentially expressed TFs in CM when compared with UM.
CONCLUSION
This multi-omics study on comparing CM with UM confirms that they differ in all analyzed levels. Of notice, the results also offer new insights with implications for elucidating certain unclear problems such as the distinct role of epithelial mesenchymal transition in two melanomas, the different metastatic routes of CM and UM and the liver tropism of metastatic UM.
Keywords: cutaneous melanoma, uveal melanoma, genomics, transcriptomics, epigenetics
INTRODUCTION
Cutaneous melanoma (CM) and uveal melanoma (UM) are two fatal malignancies. They account for more than 90% and 5% of all melanomas respectively. UM is the most common eye tumor and constitutes about 85% of the intraocular malignancies. The incidences of CM and UM were reported to be 100-300 per 1 million and 7 per 1 million per year, respectively[1]. Though CM and UM arise in different tissues, both of them derive from melanocytes. Despite the common origin, the etiopathogenesis, biological processes, metastatic routes and clinical prognosis of these two melanomas differ greatly from each other[2]. Furthermore, some effective therapeutic methods applied in the treatment of CM have little effects on the UM patients. Thus, it's meaningful to elucidate the detailed distinctive mechanisms underlying these different behaviors of the two tumors[3]. However, cross-cancer studies are rarely performed because of many objective factors such as the limitation of collecting enough samples of varied cancers in a single institution and the huge expenses of large-scale cancer researches.
The Cancer Genome Atlas (TCGA) is a dataset that provides comprehensive, multi-level (gene mutation, methylation, mRNA, miRNA, proteins, clinical data) data of 43 selected cancer projects (until Oct. 2018). Because much data on it is publicly available, a lot of TCGA-based studies were already conducted to compare different tumors at a pan-cancer level[4]–[5]. To the best of our knowledge, this is the first TCGA-based study focusing on the comparison of CM with UM on multiple levels.
MATERIALS AND METHODS
In this study, we used R language (version 3.5.2) and its variety of packages (TCGAbiolinks, TCGAbiolinksGUI and ELMER etc.) to analyze the data from TCGA and illustrate figures[6]–[8]. In order to eliminate the effect of metastatic process, only the data of primary tumors were included in the study.
Multiple Genomic Alteration Analysis
In order to ensure the data quality, mutation annotation format (MAF) files were downloaded from the TCGA data portal via R. First, the package “TCGAbiolinks” was used to analyze the MAF files of CM (104 files) and UM (80 files) separately. Second, we pooled the MAF files together to identify the top 20 most frequently mutated genes of the combined group which were presented with additional information such as “disease”, “gender” and “race”. All variant coordinates were transferred to genome reference hg38.
Differentially Expressed Genes
First, we downloaded the combined mRNA data which contained 103 CM samples and 80 UM samples. The combined data was then analyzed with an Array Array Intensity correlation which defined a square symmetric matrix of Pearson correlation among all 183 samples. Second, the combined mRNA data was processed with within-lane normalization and between-lane normalization. Ultimately, with a threshold of 0.25, differentially expressed genes (DEGs) were identified through edgeR function of the package TCGAbiolinks.
Gene Ontology Enrichment Analysis
With the identified DEGs, we performed the gene ontology (GO) enrichment analysis in three perspectives, biological process, cellular component and molecular function. Besides, 10 most significantly enriched pathways were also identified. The results were plotted by the -log(P-value corrected FDR).
DNA Methylation
DNA methylation analysis of both CM and UM were conducted on the platform “Illumina Human Methylation 450”. We searched for differentially methylated CpG sites, which were regarded as possible functional regions involving in gene transcriptional regulation. In order to find these regions, the beta-values of methylation were used for the comparison. First, the calculation of the mean methylation of each tumor was performed. Second, we calculated the P-value by using a Benjamini-Hochberg adjusted Wilcoxon test. Finally, a heatmap of the methylation of CM and UM was generated with R.
Integration of DNA Methylation and Gene Expression Data
We conducted the integration of DNA methylation and gene expression data with the results from step 2 and step 4 by using the “TCGAvisualize_starburst” function in the “TCGAbiolinks” package.
Differentially Expressed miRNAs
We downloaded the combined miRNA data which contained 103 CM samples and 80 UM samples. By using the function “TCGAanalyze_DEA”, we identified the differentially expressed miRNAs between CM and UM patients. The “fdr.cut” and “logFC.cut” were set to be 0.01 and 1, respectively.
ELMER Analysis on Identifying the Differentially Expressed Master Transcription Factors
With the ELMER package, a Multi Assay Experiment (MAE) object including a DNA methylation matrix and a gene expression matrix was generated. For instance, in the hypomethylated direction, the samples of CM and UM were ranked by the beta values of the DNA methylation and 20% of the samples with the lowest methylation levels were processed with an unpaired one-tailed t-test to find out the comparatively hypomethylated probes in CM. Afterwards, the methylation of the probes and the expression of their 10 nearest upstream and downstream genes were tested for an inverse correlation. The differentially methylated probes were identified via the Mann-Whitney U test and the non-parametric test. Then, we integrated the significant probe-gene pairs to perform the motif enrichment analysis which finally helped to identify the candidate differentially expressed master regulatory transcription factors (TFs). The same method was also applied in the analysis of the hypermethylated direction.
RESULTS
Multiple Genomic Alteration Analysis
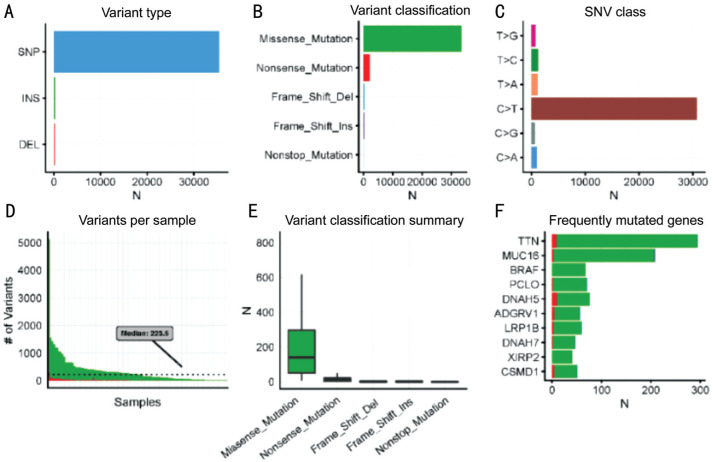
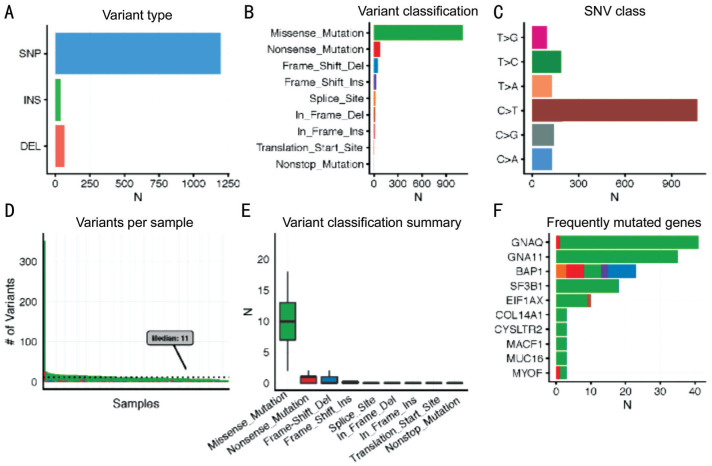
In Figure 1A and 2A, we noticed that single nucleotide polymorphism (SNP) was the most common variant type in both tumors when compared with insertion and deletion. Furthermore, in both tumors, “C>T” was the most frequent one in the six classes of single nucleotide variant (SNV) types. With regard to the variant classification, a same ranking order was observed in both groups, with missense mutation ranking first, followed by nonsense mutation, frameshift deletion and frameshift insertion. Besides, CM samples had a median variants' number of 223.5, with the maximum reaching more than 5000, whereas the median variants' number of UM was only 11 and the maximum was less than 400. This is concordant with a previous study which reported that CM had more mutational burden than UM[9].
Figure 1. Multiple genomic alteration analysis of CM.
A: The counts of 3 variant types; B: The counts of 5 gene variant classifications; C: The variants' counts of 6 SNP classes; D: The variants' counts per CM sample; E: The summary of 5 gene variant classification: box plots show median counts and the interquartile range; F: Top 10 most frequently mutated genes in the CM group.
Figure 2. Multiple genomic alteration analysis of uveal melanoma.
A: The counts of 3 variant types; B: The counts of 9 gene variant classifications; C: The variants' counts of 6 SNP classes; D: The variants' counts per UM sample; E: The summary of 9 gene variant classification: box plots show median counts and the interquartile range; F: Top 10 most frequently mutated genes in the UM group.
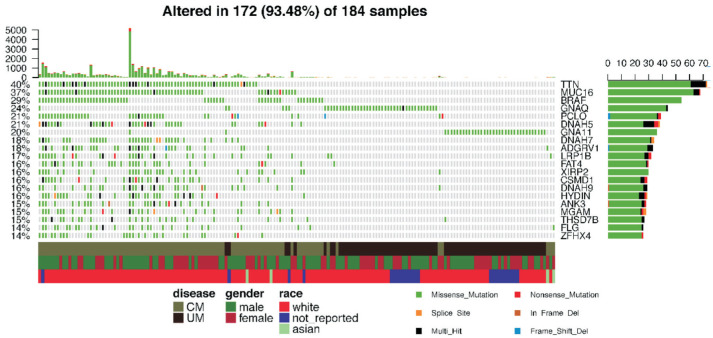
Furthermore, the CM and UM samples were combined together into a pooled analysis (Figure 3). We identified the 20 most frequently mutated genes in the combined group, with TTN (40%) ranking first, followed by MUC16 (37%), BRAF (29%), GNAQ (24%), PCLO (21%), DNAH5 (21%), GNA11 (20%), DNAH7 (18%), ADGRV1 (18%), LRP1B (17%), FAT4 (16%), XIRP2 (16%), CSMD1 (16%), DNAH9 (16%), HYDIN (16%), ANK3 (15%), MGAM (15%), THSD7B (15%), FLG (14%), ZFHX4 (14%). In the included 184 (104 CM and 80 UM) samples, 172 (93.48%) had at least one mutation in the abovementioned 20 genes.
Figure 3. Top 20 most frequently mutated genes in the combined group of CM and UM.
The upper histogram shows the variants' counts per sample. The percentiles at the left border of the main heatmap mean the occurrence ratio of each gene mutation. The right horizontal histogram presents the variant classifications of each gene. The covariate tracks show clusters for disease, gender, race and variant classification type.
Interestingly, mutation on titin, or TTN, occurred in 71 (68%) of 104 CM samples, whereas only 2 (2.5%) of 80 UM samples were identified to have TTN mutations. In our study, 68 (65 CM and 3 UM) samples were identified to have MUC16 mutations. We also noticed an obvious absence of BRAF mutation in the UM group. In contrast to these top three mutated genes, 40 of the 44 GNAQ-mutated samples originated from UM, with the left four deriving from CM. Thirty-nine samples with mutated PCLO were detected in our study. Among them, 38 were CM samples and only one UM sample. Within the 38 mutated CM samples, the number of male samples (n=26) was more than twice as large as the number of female samples (n=12). Three cytoskeletal dynein genes DNAH5, DNAH7 and DNAH9 were also presented in the top 20 mutated genes. We noticed that only one UM sample was detected with DNAH7 mutation while the others were all CM samples. Similarly, few UM samples were found in the XIRP2-, ADGRV1-, LRP1B-, CSMD1-, HYDIN-, MGAM-, FLG-, ZFHX4-, ANK3- and FAT4-mutated groups.
Transcriptomic Analysis
Totally 80 UM and 103 CM mRNA samples were included in the transcriptomic analysis. Finally, 4610 differentially expressed genes were identified. The top 20 differentially expressed genes (Table 1) were selected and ordered by the Delta value (the difference of gene expression between the two tumors multiplied logFC), i.e. KRT14, KRT6A, KRT16, KRT6B, KRT5, KRT17, KRT1, KRT6C, FN1, APOD, SERPINE2, KRT10, COL1A1, S100A9, SFN, TNC, S100A7, EEF2, S100A8, SPRR1B. Interestingly, 9 of the 20 identified genes belonged to the Keratin family which were abundant in the keratinocytes and were regarded as epithelial markers. And three members of S100 protein family (S100A7, S100A8 and S100A9) were also shown in Table 1.
Table 1. Top 20 differentially expressed genes.
| mRNA | logFC | FDR | Expression level |
Deltaa | |
| CM gene | UM gene | ||||
| KRT14 | -11.346077 | 1.38E-55 | 180056.165 | 65.2375 | 2042931.18 |
| KRT6A | -12.533693 | 1.66E-31 | 128946.2816 | 14.25 | 1616173.15 |
| KRT16 | -13.59958 | 2.57E-40 | 100335.6893 | 5.65 | 1364523.22 |
| KRT6B | -13.709598 | 2.20E-44 | 98880.99029 | 5.4375 | 1355618.67 |
| KRT5 | -12.528266 | 2.18E-47 | 94258.47573 | 11.0625 | 1180895.26 |
| KRT17 | -11.839915 | 2.14E-57 | 83843.80583 | 15.95 | 992703.557 |
| KRT1 | -15.248924 | 8.06E-38 | 56077.99029 | 1.1375 | 855129.009 |
| KRT6C | -13.776406 | 2.61E-38 | 55547.1068 | 2.775 | 765239.475 |
| FN1 | -4.6306564 | 8.58E-77 | 154051.5728 | 5163.1125 | 713359.903 |
| APOD | -5.0761006 | 5.21E-63 | 102886.9612 | 2448.025 | 522264.567 |
| SERPINE2 | -2.893698 | 2.24E-42 | 126185.0874 | 15050.6375 | 365141.539 |
| KRT10 | -6.3170001 | 6.68E-53 | 55498.26214 | 656.85 | 350582.525 |
| COL1A1 | -4.0338005 | 6.78E-64 | 85640.96117 | 4216.8875 | 345458.553 |
| S100A9 | -8.4467583 | 8.08E-66 | 33439.74757 | 79.5375 | 282457.466 |
| SFN | -11.525871 | 4.37E-63 | 24372.58252 | 6.5375 | 280915.246 |
| TNC | -7.1195055 | 1.12E-105 | 37638.42718 | 234.8875 | 267966.988 |
| S100A7 | -14.921512 | 2.10E-46 | 15983.15534 | 0.3125 | 238492.842 |
| EEF2 | 1.41987278 | 2.07E-38 | 163253.2816 | 363191.325 | 231798.891 |
| S100A8 | -10.191145 | 8.08E-75 | 22144.60194 | 16.0625 | 225678.852 |
| SPRR1B | -13.252227 | 1.82E-35 | 13745.35922 | 0.8625 | 182156.624 |
FC: Fold change; FDR: False discovery rate. aDelta=|logFC×(CM gene expression level-UM gene expression level)|.
GO Enrichment Analysis
Basing on the results of DEGs, we performed a GO enrichment analysis to classify the DEGs. Three GO types (biological processes, cellular components, molecular functions) and pathways were investigated. The results were ordered by -log10(FDR).
In the 10 identified biological processes, we observed three processes with epidermal characteristics, i.e. the epidermal cell differentiation, epidermis development and keratinocyte differentiation. The DEGs also influence the cell and biological adhesion process. Additionally, two sensory organ-associated process, i.e. sensory perception and cognition, were also identified in the top 10 most significant biological processes.
From the perspective of the cellular component, it's noteworthy that eight of the 10 components were associated with cellular membrane, including plasma membrane, extracellular region part, extracellular region, intrinsic to plasma membrane, integral to plasma membrane, proteinaceous extracellular matrix, extracellular matrix and extracellular space.
With regard to the molecular function, the DEGs were enriched majorly in the binding functions. Despite the eight identified binding functions, the structural molecule activity and chemokine activity were also presented in the top 10 functions. Finally, the DEGs were integrated to identify the most significantly enriched pathways. We noticed that four of the 10 identified pathways were associated with the immune system, including the granulocyte adhesion and diapedesis, agranulocyte adhesion and diapedesis, T helper cell differentiation and the role of nuclear factor of activated T cells (NFAT) in regulation of the immune response. Other presented pathways like hepatic fibrosis/hepatic stellate cell activation and colorectal cancer metastasis signaling were also identified as DEGs-enriched pathways.
DNA Methylation
The methylation data of both tumors (104 CM and 80 UM samples) were acquired through the platform of Infinium Human Methylation 450. First, a comprehensive study was performed to check the mean DNA methylation level of the two tumors. A significant difference of DNA methylation level between the two tumors was confirmed by the t test with P<0.001. Second, a heatmap (Figure 4) was generated to visualize the DNA methylation level across all CM and UM samples. While notable methylation differences could be observed between CM and UM, no obvious disparities were detected in genders and races.
Figure 4. DNA methylation landscape across all CM and UM samples.
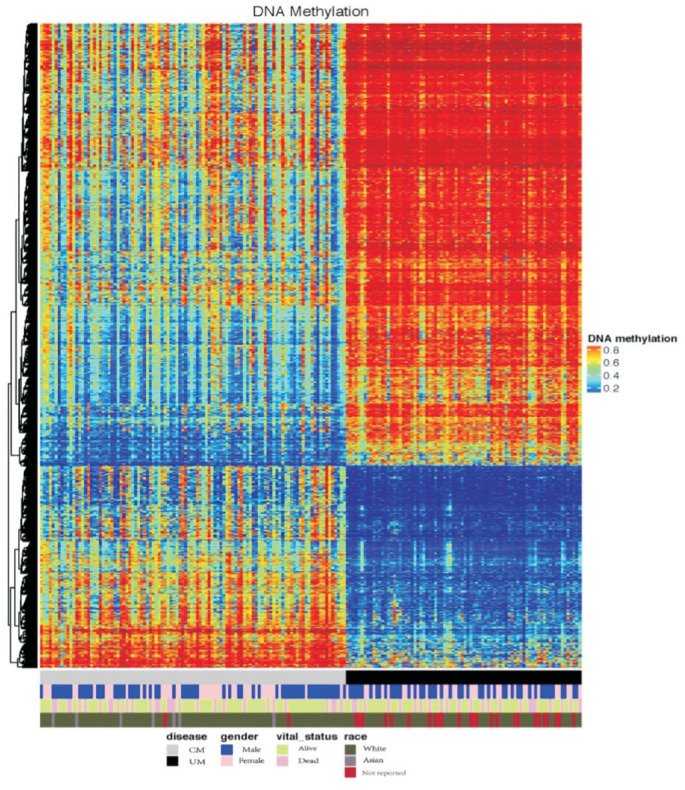
The heatmap shows the DNA methylation levels across CM and UM samples. Covariate tracks show the subtypes of diseases, genders, vital statuses and races.
DEGs and DNA Methylation
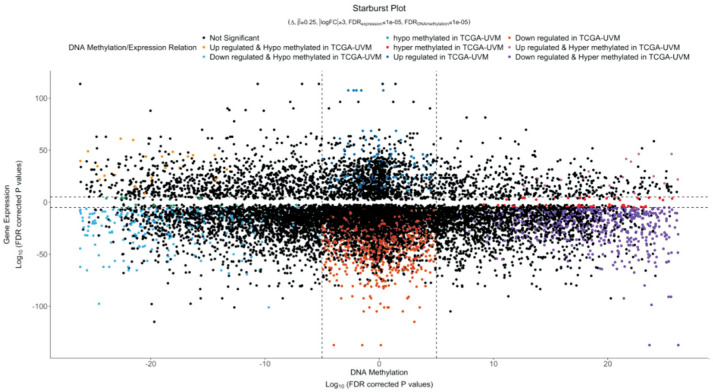
DNA methylation locating at promoters has been confirmed to have a silencing effect on the respective genes[10]. To investigate the methylation differences between CM and UM, a starburst plot was produced based on the differentially methylated CpG sites and nearby genes (Figure 5). DNA methylation [log10(FDR-corrected P-value)] and DEGs' expression [log10(FDR-corrected P-value)] were plotted on the x-axis and y-axis, respectively. Both of the horizontal and vertical black dashed lines presented an FDR-adjusted P value of 10−5. We noticed that most DEGs were downregulated in UM. Within the downregulated DEGs, hypo- and hypermethylation presented no obvious difference.
Figure 5. Starburst plot based on the differentially methylated CpG sites and nearby genes.
DNA methylation [log10(FDR-corrected P-value)] and DEGs' expression [log10(FDR-corrected P-value)] were plotted on the x-axis and y-axis, respectively. Both of the horizontal and vertical black dashed lines presented an FDR-adjusted P value of 10−5.
miRNA
Totally 485 differentially expressed miRNAs were identified. The top 20 miRNAs were presented and ordered by the absolute value of logFC in Table 2. Despite miR-124-1, the other 19 miRNAs were all downregulated in UM. Recent investigations indicated that miR-205, miR-206, miR-203a, miR-124, miR-105-1, miR-200c and miR-516b was significantly downregulated in several types of malignancies[11]–[16].
Table 2. Top 20 differentially expressed miRNAs.
| miRNAs | logFC | logCPM | LR | P value | FDR | ABS (logFC) |
| miR-205 | -12.66967581 | 10.2741151 | 325.471682 | 9.31E-73 | 3.23E-70 | 12.6696758 |
| miR-1269b | -7.792267345 | 4.38439951 | 84.2362378 | 4.39E-20 | 5.21E-19 | 7.79226735 |
| miR-206 | -7.443771468 | 5.27880962 | 101.090716 | 8.79E-24 | 1.27E-22 | 7.44377147 |
| miR-4431 | -7.339842966 | 3.98045621 | 182.799675 | 1.19E-41 | 6.59E-40 | 7.33984297 |
| miR-615 | -7.238077 | 3.1900144 | 593.84844 | 3.65E-131 | 5.06E-128 | 7.238077 |
| miR-520c | -6.93553871 | 1.33684989 | 50.5823292 | 1.14E-12 | 8.35E-12 | 6.93553871 |
| miR-520b | -6.566167424 | 3.31422137 | 52.0846379 | 5.32E-13 | 4.01E-12 | 6.56616742 |
| miR-519a-1 | -6.529184223 | 5.01683563 | 71.5554615 | 2.70E-17 | 2.69E-16 | 6.52918422 |
| miR-203a | -6.507885127 | 13.6642122 | 212.183766 | 4.59E-48 | 3.54E-46 | 6.50788513 |
| miR-6510 | -6.46737802 | 1.68143693 | 61.7217392 | 3.96E-15 | 3.50E-14 | 6.46737802 |
| miR-767 | -6.317311885 | 7.78594041 | 150.26145 | 1.52E-34 | 4.49E-33 | 6.31731189 |
| miR-520f | -6.24965796 | 2.21361008 | 42.5022762 | 7.06E-11 | 4.23E-10 | 6.24965796 |
| miR-520g | -6.176848445 | 2.10505402 | 42.8267251 | 5.98E-11 | 3.63E-10 | 6.17684845 |
| miR-122 | -6.050454709 | 3.21884414 | 45.4044877 | 1.60E-11 | 1.03E-10 | 6.05045471 |
| miR-124-1 | 5.923243919 | 0.97794058 | 111.232904 | 5.26E-26 | 8.80E-25 | 5.92324392 |
| miR-519a-2 | -5.857701837 | 3.44673364 | 44.6804669 | 2.32E-11 | 1.46E-10 | 5.85770184 |
| miR-526b | -5.83801509 | 4.97960824 | 64.9939422 | 7.51E-16 | 7.06E-15 | 5.83801509 |
| miR-105-1 | -5.830972062 | 7.53994162 | 135.995571 | 2.00E-31 | 4.96E-30 | 5.83097206 |
| miR-200c | -5.820556883 | 9.58139161 | 293.602553 | 8.16E-66 | 1.89E-63 | 5.82055688 |
| miR-516b-1 | -5.782752272 | 2.0656276 | 34.4289993 | 4.42E-09 | 2.19E-08 | 5.78275227 |
FC: Fold change; CPM: Counts per million; LR: Likelihood ratio; FDR: False discovery rate; ABS: Absolute value.
Differentially Expressed Master Transcription Factors
In the hypomethylated (comparative hypomethylated in CM than in UM) direction, 39 enrichment motifs were identified while 66 were identified in the hypermethylated direction. Basing on the identified enriched motifs, we found 114 and 116 potential differentially expressed master TFs in hypomethylated and hypermethylated direction, respectively. Many of the identified master regulatory TFs were associated with the epithelial mesenchymal transition (EMT). Particularly, the acknowledged core EMT-TFs, i.e. SNAI1, SNAI2, TWIST1 and SIX1 were found to be relatively more hypomethylated in the CM than in the UM[17].
DISCUSSION
Progress in elucidating the cellular and molecular distinctions between CM and UM has been made majorly in form of directly comparing results from isolated studies of the two tumors which might be restricted by the former studies' design and heterogeneity of the data. The publicly available data in the TCGA provide an optimal method to conduct a comprehensive study which can avoid the aforementioned problems. The findings of this multi-omics study enable us to elucidate the important problems concerning both malignancies like the role of EMT in CM and UM, the distinctive metastatic routes and organs' tropism.
The Role of Epithelial Mesenchymal Transition
The top 20 most frequently mutated genes supported the acknowledged key role of BRAF and GNAQ/11 in the etiopathogenesis of CM and UM, respectively[18]–[19]. While GNAQ/11 mutations were found almost uniquely in the UM, the majority of the other 18 mutated genes (including BRAF) were presented by the CM samples. Within the 18 mutated genes, MUC16, PCLO, DNAH5, DNAH7, DNAH9, ADGRV1, ANK3, FAT4 and XIRP2 encoded cytoskeleton- and extracellular matrix-related proteins (CECMPs) which were reported to provide the most common class of cancer mutants and estimated to play a role in the EMT[20]–[23].
On the transcriptomic level, it's noteworthy that eight of top 20-DEGs are keratin family members which were regarded as epithelial markers. And several previous studies implied that keratins may play a role in the progression of CM[24]. Besides, the DEGs-listed FN1, SERPINE 2, TNC, EEF2 and COL1A1 were also regarded as EMT-associated genes which could contribute to the proliferation, differentiation and migration of tumors[25]–[29].
Additionally, 13 of the top 20 differentially expressed miRNAs were previously reported to participate in the EMT. And they were all less expressed in CM samples than in UM samples. Ten of the 13 miRNAs, i.e. miR-205, 520c, 520b, 203a, 520f, 122, 124, 526b and 200c, were reported to negatively regulate the EMT through a variety of downstream genes like CDH1, ZEB1, ZEB2, ADAM9, TGFBR2, SNAI2, ERK, MYC, CCND1, JUN and SNAI1 etc[30]–[37]. Thus, we estimate that the relative low expression of these miRNAs in CM would promote the EMT. Nevertheless, the other three miRNAs, miR-520g, 105 and 1269a were reported to positively regulate the EMT through cooperating with SMAD7, TNF, TGFB and SOX4[38]–[40]. Therefore, more in-depth studies are needed to help elucidate the role of miRNAs in CM and UM.
In addition, the role of the EMT in differentiating CM and UM was also confirmed by the analysis of differentially expressed master regulatory TFs. In the 114 identified hypomethylated master regulatory TFs (TFs hypomethylated in CM compared to UM), the first three TFs were LEF1, E2F2 and TRIM62. They were all suggested to positively regulate the EMT[41]–[43]. After a brief MeSH (Medical Subject Headings) search on “Google Scholar” (https://scholar.google.com/) until Nov. 01, 2018, at least 65 TFs were previously reported to participate in the EMT, with 57 positively-regulating TFs (including three acknowledged key EMT-TFs, SNAI1, TWIST1 and SIX1) and 8 negatively-regulating TFs.
Among the 116 TFs, which were identified as relatively more hypermethylated in CM when compared with UM, we found at least 42 EMT-associated TFs after a brief MeSH search on “Google Scholar”. While 19 of them were reported to negatively regulate the EMT, 23 were found to promote the EMT. Theoretically, the hypermethylation of the 19 negatively-regulating TFs would contribute to the EMT which would support our hypothesis that CM undergoes a more obvious EMT than UM. On the contrary, the hypermethylation of the 23 positively-regulating TFs were supposed to suppress the EMT. Thus, more relevant studies are needed to present a more precise landscape of the TFs' regulating network in these two tumors.
In summary, our multi-omics results indicate that many distinctions between CM and UM were associated with the EMT.
Metastatic Routes
While CM was estimated to spread through hematologic as well as lymphatic dissemination, UM was believed to only metastasize hematogenously[44]. The lymphatic endothelial cells were reported to secrete chemokines such as chemokine (C-C motif) ligand 21 (CCL21) and stroma cell derived factor-1 (SDF-1) which could stimulate the chemotaxis via the respective receptors C-C chemokine receptor 7 (CCR7) and leucine aminopeptidase 3 (LAP3) on tumor cells[45]–[46]. Besides, integrin α4β1 expressed on lymphatic vessels was also confirmed to play an essential role in the cancer-associated lymphangiogenesis through binding the corresponding ligands such as vascular cell adhesion molecule-1 (VCAM-1), fibronectin 1 (FN1)[47]–[48]. Because the lymphatic dissemination was not estimated to be involved in the metastasis of UM, we conject that the expression of CCR7, LAP3, VCAM-1 and FN1 might be lower expressed in UM when compared with CM. Supporting this, the DEGs' list showed that CCR7 (logFC=-2.378070086), LAP3 (logFC=-1.11), VCAM-1 (logFC=-1.63) and FN1 (logFC=-4.63) were all significantly lower expressed in UM. In addition, we also checked the expression of VEGFC and VEGFD in the DEGs' list because of the established involvement of VEGFC/D induced lymphangiogenesis in tumor metastasis[49]. While VEGFC and VEGFD were not presented in the DEGs, we observed that VEGFA was lower expressed in UM with a logFC of -1.69 (i.e. significantly higher expressed in CM). Basing on a study in mouse, Björndahl et al[50] and colleagues confirmed that the VEGFA induced lymphangiogenesis was independent from the VEGFC/D pathway. Thus, further studies are encouraged to examine the potential role of VEGFA in the lymph angiogenesis of CM.
Liver Tropism of the Metastatic UM
Though CM has no metastatic propensity for the liver, metastatic UM has an obvious unexplained liver tropism. Previous studies of UM have shown that the increased expression of Met gene in the primary tumors would promote the liver metastasis[51]. It was estimated that the liver propensity occurred because of the high HGF expression in the liver cell membrane surface. The activation of Met by binding hepatocyte growth factor (HGF) would lead to a series of downstream reactions which resulted in tumor cell proliferation and progression[52]. Our DEGs' results confirmed the relatively higher expression of Met in UM than in CM (logFC=2.04). Besides, Krüger et al[53] reported that the inhibition of matrix metalloproteinases (MMPs) would increase the expression level of HGF in the liver, thus promote the metastasis formation. In agreement with this, 13 MMP mRNAs in the DEGs were lower expressed in UM than in CM.
Moreover, basing on a study on UM, Laurent et al[54] have reported that the high expression of PTP4A3 could be seen as a predictor for liver tropism. Correspondingly, our study has proved that the PTP4A3 mRNA was significantly higher expressed in UM than in CM (logFC=1.91).
In summary, the comparison between CM and UM from multiple perspectives presented a comprehensive illustration which provides us a better understanding of these two tumors and new insights for finding better treatment strategies.
Acknowledgments
Foundations: Supported by the China Scholarship Council (CSC) Program (No.201708080023; No.201708080104); Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of University of Tübingen.
Conflicts of Interest: Zhang Q, None; Lin ZN, None; Chen J, None; Zheng WX, None.
REFERENCES
- 1.van den Bosch T, Kilic E, Paridaens D, de Klein A. Genetics of uveal melanoma and cutaneous melanoma: two of a kind? Dermatol Res Pract. 2010;2010:360136. doi: 10.1155/2010/360136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belmar-Lopez C, Mancheno-Corvo P, Saornil MA, Baril P, Vassaux G, Quintanilla M, Martin-Duque P. Uveal vs. cutaneous melanoma. Origins and causes of the differences. Clin Transl Oncol. 2008;10(3):137–142. doi: 10.1007/s12094-008-0170-4. [DOI] [PubMed] [Google Scholar]
- 3.Pandiani C, Béranger GE, Leclerc J, Ballotti R, Bertolotto C. Focus on cutaneous and uveal melanoma specificities. Genes Dev. 2017;31(8):724–743. doi: 10.1101/gad.296962.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leiserson MDM, Vandin F, Wu HT, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu BF, McLellan M, Lawrence MS, Gonzalez-Perez A, Tamborero D, Cheng YW, Ryslik GA, Lopez-Bigas N, Getz G, Ding L, Raphael BJ. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2015;47(2):106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Cancer Genome Atlas Research Network. Hu H. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, Ceccarelli M, Bontempi G, Noushmehr H. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva TC, Colaprico A, Olsen C, Bontempi G, Ceccarelli M, Berman BP, Noushmehr H. TCGAbiolinksGUI: a graphical user interface to analyze cancer molecular and clinical data. F1000 Research. 2017;7:439. [Google Scholar]
- 8.Silva TC, Coetzee SG, Yao L, Hazelett DJ, Noushmehr H, Berman BP. Enhancer linking by methylation/expression relationships with the R package ELMER version 2. bioRxiv. 2017:148726. [Google Scholar]
- 9.Robertson AG, Shih J, Yau C, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2018;33(1):151. doi: 10.1016/j.ccell.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 10.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, Brenn T, Brown ER, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012;106(3):553–561. doi: 10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgantas RW, 3rd, Streicher K, Luo XB, Greenlees L, Zhu W, Liu Z, Brohawn P, Morehouse C, Higgs BW, Richman L, Jallal B, Yao YH, Ranade K. MicroRNA-206 induces G1 arrest in melanoma by inhibition of CDK4 and Cyclin D. Pigment Cell Melanoma Res. 2014;27(2):275–286. doi: 10.1111/pcmr.12200. [DOI] [PubMed] [Google Scholar]
- 13.Bu PY, Yang P. MicroRNA-203 inhibits malignant melanoma cell migration by targeting versican. Exp Ther Med. 2014;8(1):309–315. doi: 10.3892/etm.2014.1708. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Zhang W, Mao YQ, Wang H, Yin WJ, Zhu SX, Wang WC. MiR-124 suppresses cell motility and adhesion by targeting talin 1 in prostate cancer cells. Cancer Cell Int. 2015;15:49. doi: 10.1186/s12935-015-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu GX, Fu D, Jia CY, Chai L, Han Y, Liu J, Wu TM, Xie RT, Chang ZY, Yang HQ, Luo P, Lv Z, Yu F, Zhong XJ, Ma YS. Reduced miR-105-1 levels are associated with poor survival of patients with non-small cell lung cancer. Oncol Lett. 2017;14(6):7842–7848. doi: 10.3892/ol.2017.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao YF, Wang YQ, Xing GC. miR-516b functions as a tumor suppressor by directly modulating CCNG1 expression in esophageal squamous cell carcinoma. Biomedecine Pharmacother. 2018;106:1650–1660. doi: 10.1016/j.biopha.2018.07.074. [DOI] [PubMed] [Google Scholar]
- 17.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 18.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363(23):2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spendlove HE, Damato BE, Humphreys J, Barker KT, Hiscott PS, Houlston RS. BRAF mutations are detectable in conjunctival but not uveal melanomas. Melanoma Res. 2004;14(6):449–452. doi: 10.1097/00008390-200412000-00003. [DOI] [PubMed] [Google Scholar]
- 20.Parry ML, Ramsamooj M, Blanck G. Big genes are big mutagen targets: a connection to cancerous, spherical cells? Cancer Lett. 2015;356(2 Pt B):479–482. doi: 10.1016/j.canlet.2014.09.044. [DOI] [PubMed] [Google Scholar]
- 21.Parry ML, Blanck G. Flat cells come full sphere: are mutant cytoskeletal-related proteins oncoprotein-monsters or useful immunogens? Hum Vaccin Immunother. 2016;12(1):120–123. doi: 10.1080/21645515.2015.1073428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fawcett TJ, Parry ML, Blanck G. A novel approach to evaluating cancer driver gene mutation densities: cytoskeleton-related gene candidates. Cancer Genomics Proteomics. 2015;12(6):283–290. [PubMed] [Google Scholar]
- 23.Kumar S, Das A, Sen S. Extracellular matrix density promotes EMT by weakening cell-cell adhesions. Mol Biosyst. 2014;10(4):838–850. doi: 10.1039/c3mb70431a. [DOI] [PubMed] [Google Scholar]
- 24.Koh SS, Wei JP, Li XM, Huang RR, Doan NB, Scolyer RA, Cochran AJ, Binder SW. Differential gene expression profiling of primary cutaneous melanoma and sentinel lymph node metastases. Mod Pathol. 2012;25(6):828–837. doi: 10.1038/modpathol.2012.32. [DOI] [PubMed] [Google Scholar]
- 25.Takano N, Kawakami T, Kawa Y, Asano M, Watabe H, Ito M, Soma Y, Kubota Y, Mizoguchi M. Fibronectin combined with stem cell factor plays an important role in melanocyte proliferation, differentiation and migration in cultured mouse neural crest cells. Pigment Cell Res. 2002;15(3):192–200. doi: 10.1034/j.1600-0749.2002.01079.x. [DOI] [PubMed] [Google Scholar]
- 26.Wu QW. Serpine2, a potential novel target for combating melanoma metastasis. Am J Transl Res. 2016;8(5):1985–1997. [PMC free article] [PubMed] [Google Scholar]
- 27.Song Y, Sun B, Hao LH, Hu J, Du S, Zhou X, Zhang LY, Liu L, Gong LL, Chi XM, Liu Q, Shao SJ. Elevated eukaryotic elongation factor 2 expression is involved in proliferation and invasion of lung squamous cell carcinoma. Oncotarget. 2016;7(36):58470–58482. doi: 10.18632/oncotarget.11298. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Katoh D, Nagaharu K, Shimojo N, Hanamura N, Yamashita M, Kozuka Y, Imanaka-Yoshida K, Yoshida T. Binding of αvβ1 and αvβ6 integrins to tenascin-C induces epithelial-mesenchymal transition-like change of breast cancer cells. Oncogenesis. 2013;2:e65. doi: 10.1038/oncsis.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosper NA, van den Berg PP, de Rond S, Popa ER, Wilmer MJ, Masereeuw R, Bank RA. Epithelial-to-mesenchymal transition in fibrosis: collagen type I expression is highly upregulated after EMT, but does not contribute to collagen deposition. Exp Cell Res. 2013;319(19):3000–3009. doi: 10.1016/j.yexcr.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 31.van Kampen JGM, van Hooij O, Jansen CF, Smit FP, van Noort PI, Schultz I, Schaapveld RQJ, Schalken JA, Verhaegh GW. miRNA-520f reverses epithelial-to-mesenchymal transition by targeting ADAM9 and TGFBR2. Cancer Res. 2017;77(8):2008–2017. doi: 10.1158/0008-5472.CAN-16-2609. [DOI] [PubMed] [Google Scholar]
- 32.Lu YC, Cheng AJ, Lee LY, You GR, Li YL, Chen HY, Chang JT. MiR-520b as a novel molecular target for suppressing stemness phenotype of head-neck cancer by inhibiting CD44. Sci Rep. 2017;7(1):2042. doi: 10.1038/s41598-017-02058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang CP, Zhou HJ, Qin J, Luo Y, Zhang T. MicroRNA-520c-3p negatively regulates EMT by targeting IL-8 to suppress the invasion and migration of breast cancer. Oncol Rep. 2017;38(5):3144–3152. doi: 10.3892/or.2017.5968. [DOI] [PubMed] [Google Scholar]
- 34.Liang YJ, Wang QY, Zhou CX, Yin QQ, He M, Yu XT, Cao DX, Chen GQ, He JR, Zhao Q. MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34(3):713–722. doi: 10.1093/carcin/bgs383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Yang L, Tu JF, Cai WW, Zhang MQ, Shou ZX, Yao YM, Xu QR. microRNA-526b servers as a prognostic factor and exhibits tumor suppressive property by targeting Sirtuin 7 in hepatocellular carcinoma. Oncotarget. 2017;8(50):87737–87749. doi: 10.18632/oncotarget.21209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin HF, Sha JP, Jiang CX, Gao XM, Qu LL, Yan HY, Xu TJ, Jiang QY, Gao HJ. miR-122 inhibits metastasis and epithelial-mesenchymal transition of non-small-cell lung cancer cells. Onco Targets Ther. 2015;8:3175–3184. doi: 10.2147/OTT.S91696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu DJ, Wu JL, Liu MZ, Yin H, He JT, Zhang B. Downregulation of miRNA-30c and miR-203a is associated with hepatitis C virus core protein-induced epithelial-mesenchymal transition in normal hepatocytes and hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2015;464(4):1215–1221. doi: 10.1016/j.bbrc.2015.07.107. [DOI] [PubMed] [Google Scholar]
- 38.Kan HP, Guo WB, Huang YQ, Liu DL. MicroRNA-520g induces epithelial-mesenchymal transition and promotes metastasis of hepatocellular carcinoma by targeting SMAD7. FEBS Lett. 2015;589(1):102–109. doi: 10.1016/j.febslet.2014.11.031. [DOI] [PubMed] [Google Scholar]
- 39.Shen ZT, Zhou R, Liu C, Wang YF, Zhan WQ, Shao ZY, Liu J, Zhang FF, Xu LJ, Zhou XY, Qi L, Bo F, Ding YQ, Zhao L. MicroRNA-105 is involved in TNF-α-related tumor microenvironment enhanced colorectal cancer progression. Cell Death Dis. 2017;8(12):3213. doi: 10.1038/s41419-017-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bu PC, Wang LH, Chen KY, Rakhilin N, Sun J, Closa A, Tung KL, King S, Kristine Varanko A, Xu YT, Huan Chen J, Zessin AS, Shealy J, Cummings B, Hsu D, Lipkin SM, Moreno V, Gümüş ZH, Shen XL. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-Β. Nat Commun. 2015;6:6879. doi: 10.1038/ncomms7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang JQ, Li YR, Daniels G, Sfanos K, de Marzo A, Wei JJ, Li X, Chen WQ, Wang JH, Zhong XL, Melamed J, Zhao J, Lee P. LEF1 targeting EMT in prostate cancer invasion is regulated by miR-34a. Mol Cancer Res. 2015;13(4):681–688. doi: 10.1158/1541-7786.MCR-14-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu TY, Chen J, Shang CL, Shen HW, Huang JM, Liang YC, Wang W, Zhao YH, Liu D, Shu M, Guo LY, Hu Z, Yao SZ. Tripartite motif containing 62 is a novel prognostic marker and suppresses tumor metastasis via c-Jun/Slug signaling-mediated epithelial-mesenchymal transition in cervical cancer. J Exp Clin Cancer Res. 2016;35(1):170. doi: 10.1186/s13046-016-0445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feliciano A, Garcia-Mayea Y, Jubierre L, Mir C, Hummel M, Castellvi J, Hernández-Losa J, Paciucci R, Sansano I, Sun YL, Ramón Y, Cajal S, Kondon H, Soriano A, Segura M, Lyakhovich A, LLeonart ME. miR-99a reveals two novel oncogenic proteins E2F2 and EMR2 and represses stemness in lung cancer. Cell Death Dis. 2017;8(10):e3141. doi: 10.1038/cddis.2017.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh M, Durairaj P, Yeung J. Uveal melanoma: a review of the literature. Oncol Ther. 2018;6(1):87–104. doi: 10.1007/s40487-018-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emmett MS, Lanati S, Dunn DB, Stone OA, Bates DO. CCR7 mediates directed growth of melanomas towards lymphatics. Microcirculation. 2011;18(3):172–182. doi: 10.1111/j.1549-8719.2010.00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhuo W, Jia L, Song N, Lu XN, Ding YP, Wang XF, Song XM, Fu Y, Luo YZ. The CXCL12-CXCR4 chemokine pathway: a novel axis regulates lymphangiogenesis. Clin Cancer Res. 2012;18(19):5387–5398. doi: 10.1158/1078-0432.CCR-12-0708. [DOI] [PubMed] [Google Scholar]
- 47.Garmy-Susini B, Avraamides CJ, Desgrosellier JS, Schmid MC, Foubert P, Ellies LG, Lowy AM, Blair SL, Vandenberg SR, Datnow B, Wang HY, Cheresh DA, Varner J. PI3Kα activates integrin α4β1 to establish a metastatic niche in lymph nodes. Proc Natl Acad Sci U S A. 2013;110(22):9042–9047. doi: 10.1073/pnas.1219603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Alexander JS, Orr AW. Integrins and their extracellular matrix ligands in lymphangiogenesis and lymph node metastasis. Int J Cell Biol. 2012;2012:853703. doi: 10.1155/2012/853703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu Y, Qi XL, Guo SY. Lymphangiogenesis induced by VEGF-C and VEGF-D promotes metastasis and a poor outcome in breast carcinoma: a retrospective study of 61 cases. Clin Exp Metastasis. 2008;25(7):717–725. doi: 10.1007/s10585-008-9180-4. [DOI] [PubMed] [Google Scholar]
- 50.Björndahl MA, Cao RH, Burton JB, Brakenhielm E, Religa P, Galter D, Wu L, Cao YH. Vascular endothelial growth factor-a promotes peritumoral lymphangiogenesis and lymphatic metastasis. Cancer Res. 2005;65(20):9261–9268. doi: 10.1158/0008-5472.CAN-04-2345. [DOI] [PubMed] [Google Scholar]
- 51.Gardner FP, Serie DJ, Salomao DR, Wu KJ, Markovic SN, Pulido JS, Joseph RW. C-MET expression in primary and liver metastases in uveal melanoma. Melanoma Res. 2014;24(6):617–620. doi: 10.1097/CMR.0000000000000118. [DOI] [PubMed] [Google Scholar]
- 52.Bakalian S, Marshall JC, Logan P, Faingold D, Maloney S, di Cesare S, Martins C, Fernandes BF, Burnier MN., Jr Molecular pathways mediating liver metastasis in patients with uveal melanoma. Clin Cancer Res. 2008;14(4):951–956. doi: 10.1158/1078-0432.CCR-06-2630. [DOI] [PubMed] [Google Scholar]
- 53.Krüger A, Soeltl R, Sopov I, Kopitz C, Arlt M, Magdolen V, Harbeck N, Gänsbacher B, Schmitt M. Hydroxamate-type matrix metalloproteinase inhibitor batimastat promotes liver metastasis. Cancer Res. 2001;61(4):1272–1275. [PubMed] [Google Scholar]
- 54.Laurent C, Valet F, Planque N, Silveri L, Maacha S, Anezo O, Hupe P, Plancher C, Reyes C, Albaud B, Rapinat A, Gentien D, Couturier J, Sastre-Garau X, Desjardins L, Thiery JP, Roman-Roman S, Asselain B, Barillot E, Piperno-Neumann S, Saule S. High PTP4A3 phosphatase expression correlates with metastatic risk in uveal melanoma patients. Cancer Res. 2011;71(3):666–674. doi: 10.1158/0008-5472.CAN-10-0605. [DOI] [PubMed] [Google Scholar]