Abstract
Squamous cell carcinoma in situ (SCCIS) is a prevalent precancerous lesion that can progress to cutaneous squamous cell carcinoma (cSCC). Although SCCIS is common, its pathogenesis remains poorly understood. To better understand SCCIS development, we performed laser-captured microdissection of human SCCIS and adjacent epidermis to isolate genomic DNA and RNA for next generation sequencing. Whole exome sequencing (WES) identified UV-signature mutations in multiple genes including Notch 1–3 in the epidermis and SCCIS and oncogenic TP53 mutations in SCCIS. Gene families, including SCHLAFEN genes, contained UV/oxidative-signature disruptive (UVD) epidermal mutations that manifested positive selection in SCCIS. The frequency and distribution of NOTCH and TP53 mutations indicate that NOTCH mutations may precede TP53 mutations.
RNA sequencing identified 1166 differentially expressed genes; the top 5 enriched GO biological processes included: 1) immune response, 2) epidermal development, 3) protein phosphorylation, 4) regulation of catalytic activity, 5) cytoskeletal regulation. The NEURL1 ubiquitin ligase, which targets Notch ligands for degradation, was upregulated in SCCIS. Neuralized 1 protein was found to be elevated in SCCIS suggesting that increased levels could represent a mechanism for downregulating Notch during UV-induced carcinogenesis. The data from DNA and RNA sequencing of epidermis and SCCIS provide insights regarding SCCIS formation.
Introduction
Cutaneous squamous cell carcinoma (cSCC) is the second most common human cancer with approximately 700,000 cases annually causing 3000 deaths (Rogers et al., 2010). Most cSCCs arise from precursors called actinic keratosis (AKs) and squamous cell carcinoma in situ (SCCIS) (Ratushny et al., 2012). The financial burden of treating these lesions is high given that approximately 60% of individuals over the age of 40 develop AKs/SCCIS (Czarnecki et al., 2002). Costs to treat UV-induced skin cancer approximate $8.1 billion/year; a sum likely to rise with an aging population and increased use of tanning salons (General, 2014; Marks, 1995; Wolff, 2008). Despite the prevalence of SCCIS and cSCC, significant questions remain regarding the pathogenesis of these lesions.
Targeted deep sequencing of 74 genes from blepharoplasty samples revealed multiple, potentially oncogenic, UV-signature mutations in NOTCH 1 and 2, TP53, FGFR3 and FAT1 in clinically unremarkable skin (Martincorena et al., 2015). This study correlates with data from earlier studies that showed UV-signature mutations in TP53 and NOTCH 1 and 2 genes in AKs and cSCC (Wang et al., 2011; Ziegler et al., 1994). Subsequent studies with exomic sequencing of UV-exposed skin, AKs and cSCCs showed mutations in TP53, NOTCH 1–2, FAT1 and MLL2 but not FGFR3 as likely drivers of cSCC formation (Chitsazzadeh et al., 2016). Although these studies implicate a common set of mutated genes in cSCC pathogenesis, sequential acquisition of UV-induced mutations leading to SCCIS and cSCC remains unclear.
To better understand the relationship between potential driver mutations in UV-exposed epidermis and SCCIS, we performed laser-capture micro-dissection (LCM) of SCCIS and adjacent epidermis, isolating genomic DNA and RNA for WES and RNA sequencing. Using this approach, we can better define the exomic and transcriptomic differences between UV-exposed epidermis and SCCIS.
Our whole exome sequencing (WES) data show a high frequency of UV-signature mutations in NOTCH 1–3 in both the epidermal and SCCIS samples with more mutations present in the former. Almost all UV-signature mutations in NOTCH genes were consistent with loss-of-function (LOF) mutations and were not shared between the epidermis and SCCIS. The minor allele frequency (MAF) of the UV-induced NOTCH LOF mutations were lower in the epidermis and higher in SCCIS indicating positive selection of a specific Notch-mutant keratinocyte to form SCCIS.
The data show prevalent UV-signature mutations in TP53 in SCCIS (6/10 samples) and none in the epidermis. This suggests acquisition of oncogenic TP53 mutation(s) may follow NOTCH 1–3 LOF mutation in promoting SCCIS. Oncogenic RAS mutations were not found in any of epidermal or SCCIS samples suggesting RAS mutations may not be involved in the early stages of UV-induced skin cancer. Additional gene families showing evidence of clonal selective pressure in SCCIS included the SCHLAFEN genes which regulate replicative stress.
RNA sequencing of the epidermal and SCCIS libraries identified 1166 genes that were differentially expressed and enriched for GO processes including: immune response, epidermal development, cell activation, protein phosphorylation and related gene categories. NEURL1, a ubiquitin ligase targeting Notch ligands, demonstrated increased expression in all SCCIS and was confirmed by immunostaining. Increased levels of Neuralized 1 could represent a mechanism for downregulating Notch signaling in keratinocytes. In summary, this WES and RNA-sequencing data provide insights into the development of SCCIS in UV-exposed skin.
Results
Comparative genomic and transcriptomic data for UV-exposed epidermis and SCCIS
Ten random biopsy specimens that contained SCCIS and adjacent epidermis were subjected to LCM to obtain distinct populations of epidermal keratinocytes and SCCIS cells (Figure 1a). Genomic DNA and total RNA were extracted from these samples and used to generate WES and RNA-seq libraries.
Figure 1.
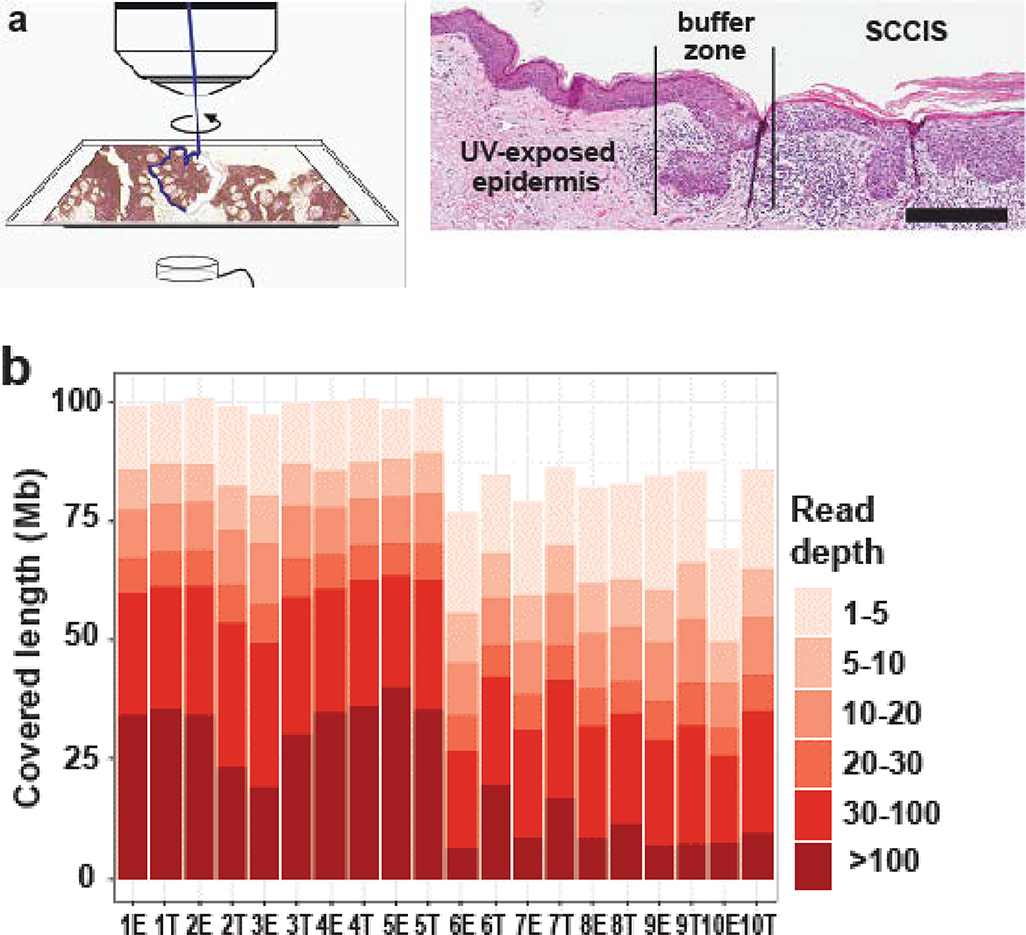
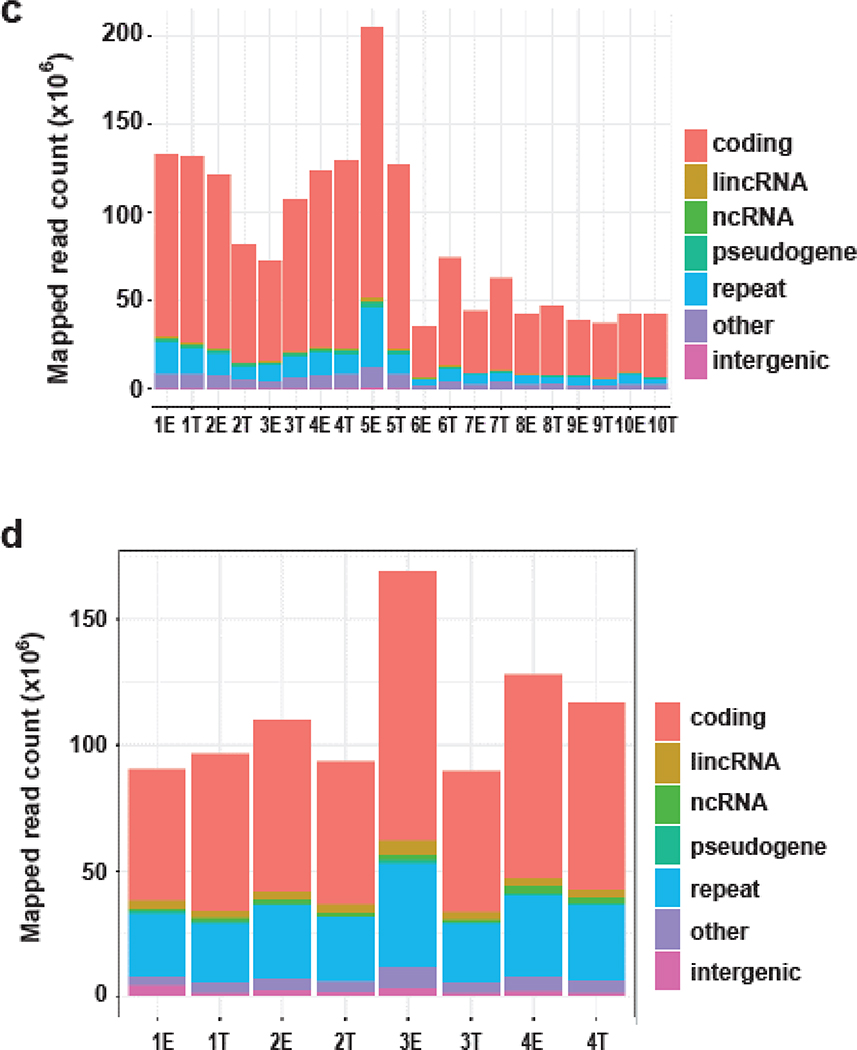
Approach and features of genomics and transcriptomics datasets. (a) Photo- micrograph of skin and detailing the laser capture microdissection approach to isolate epidermal and SCCIS samples. Scale bar = 0.25mm (b) Coverage summary of mapped reads for exome-seq samples; E-epidermal library, T-SCCIS library. (c, d): Summary of the genetic class for exome-seq (c) or RNA-seq (d) samples. ncRNA: non-coding RNAs = rRNA, tRNA, miRNA, snoRNA, etc. repeat: RMSK annotated transposons, tandem-repeats or RNAs. Intergenic: genomic regions without annotation.
All exome libraries were constructed using the Agilent Haloplex platform. By performing up to five replicate runs of 100/125bp paired-end sequencing, all 20 WES libraries achieved sufficient depth for follow-up variant detection (Figure 1b and Supplementary Table S1). For RNA-seq, eight libraries were constructed from four paired SCCIS and epidermis samples, and subjected to NGS using the same platform.
NGS reads were mapped to the human reference genome (hg19 release) using AlignerBoost. At least a 10-fold read depth was achieved to identify variants in exome-seq experiments, for 50~75 Mbps of the Haloplex-Exome system targeted regions (Figure 1b) (Zheng and Grice, 2016a). Up to 40% of the exome had >100x coverage (Figure 1b, Supplementary Table S1), which permits identification of genetic mutations implicated in SCCIS development in a comprehensive, unbiased manner. A high majority of mapped exome-seq reads were associated with protein-coding genes, while a small number of reads mapped to non-coding elements (Figure 1c). These data indicate successful enrichment of exonic reads.
A similar read depth was achieved for the RNA-seq libraries with 30~60 million mapped reads (Supplementary Table S2). Beside protein-coding regions, a proportion of reads were mapped to non-coding elements as expected for total RNA (Figure 1d). All WES and RNA-seq datasets meet the quality criteria for identifying the genetic and transcriptional changes in SCCIS development. We achieved the minimum 10X or recommended 20X coverage for detecting SNPs using WES data (Meynert et al., 2014), and the recommended 10~25 M read requirement for detecting differentially expressed genes using RNA-seq data (Liu et al., 2014).
Frequent germline and somatic genetic variants detected in all skin samples
To identify genetic changes in SCCIS development, both germline and somatic variants were called from our exome-seq dataset using two distinct discovery models from the GATK toolkit (Van der Auwera et al., 2013). The joint model detects germline variants found in all samples compared to the reference genome using all samples as a cohort; while the de-novo model detects somatic variants by comparing each pair of epidermis vs. SCCIS sample. With this approach, we found over 1.3 million unique germline variants in the entire cohort of ten patients (Supplementary Table S3). These numbers are higher than most cancers reported previously (Frigola et al., 2017; Hao et al., 2016). However, this observation is consistent with prior data and is anticipated since our samples are from adults with long-term UV exposure (Frigola et al., 2017; Hao et al., 2016). We found fewer somatic variants using the paired discovery model, which yielded about 50 thousand total unique variants with 1–10 thousand per patient. Notably, the “germline” variants might not always occur in the germline since our “unremarkable” epidermal samples contain pre-existing mutations related to sun-exposure and age (Figure 1a). Therefore, we use the terms “joint” and “de-novo” variants instead of “germline” and “somatic” in this study.
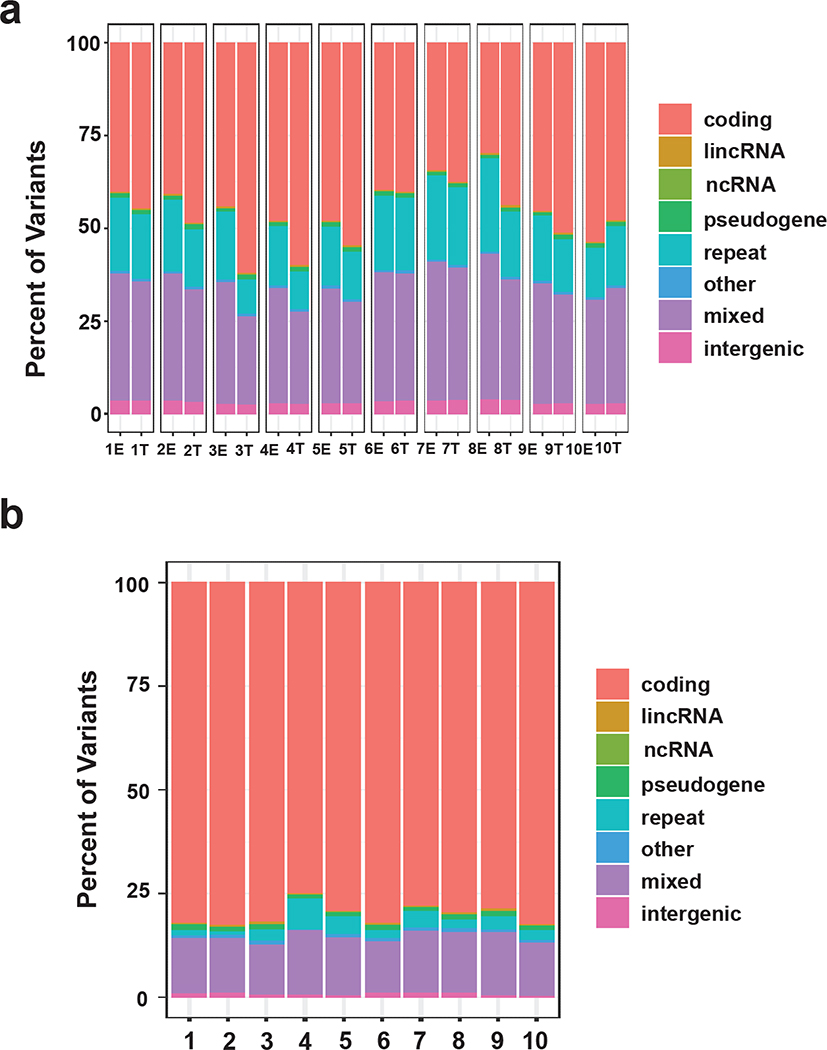
All joint and de-novo variants were classified into genetic types/classes according to their genomic locations (see Supplementary Methods). Most variants were protein-coding, but significant numbers were identified as ncRNAs and other non-coding elements (Figures 2a and b vs. 1c). For the joint variants, protein-coding variants are enriched in SCCIS compared to epidermis in almost all patients (Figure 2a), suggesting these mutations are not negatively selected against in SCCIS. In contrast, most of de-novo variants represent mutations in protein-coding genes (Figure 2b), and the SCCIS-specific variants may be important for lesion development.
Figure 2. Identification and characteristics of joint and de-novo variants.
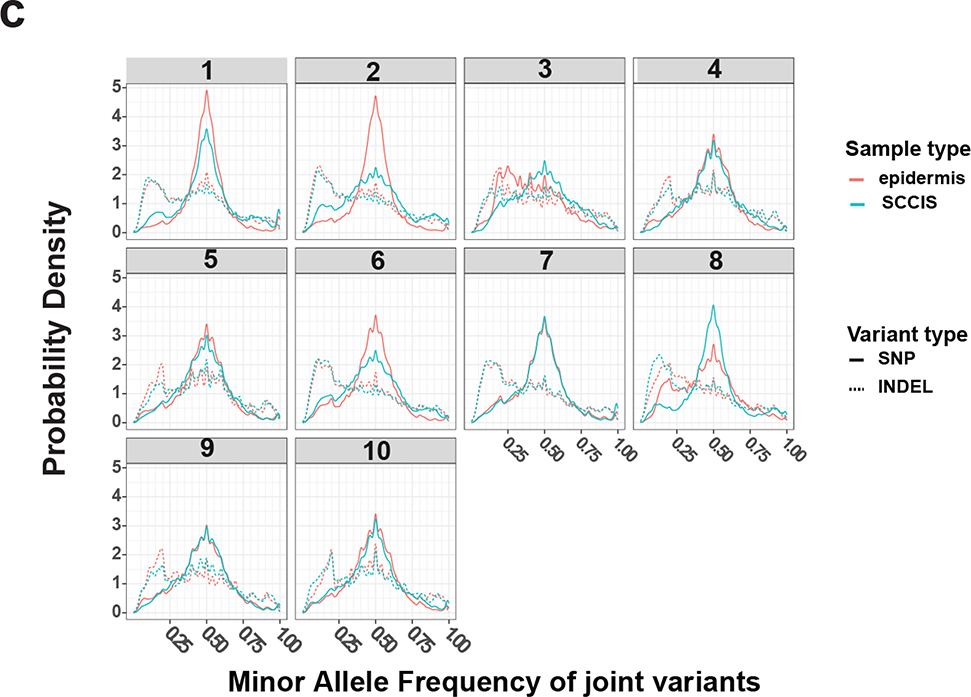
(a, b) Summary of the genetic class for joint (a) or de-novo (b) variants. Genetic classes are defined as for Fig. 1. Mixed: variants matching to multiple classes. Summaries are based on per-sample or per-patient for joint or de-novo variants, respectively. (c) Distribution densities of the minor allele frequency (MAF) of joint samples in each patient; only biallelic variants with coverage depth >= 10 are included.
Subclonal cell populations with mutations are common in UV-exposed epidermis
Given the large number of UV-induced joint variants in epidermal samples, we hypothesized that most joint variants were population polymorphisms (SNPs), technical errors (sequencing and/or mapping, although controlled) or spontaneous random mutations most of which should be selectively neutral for SCCIS development. However, by examining the MAF of those joint variants, we found that a group of variants with MAFs near 50% largely corresponds to population SNPs, and a number of joint variants with MAFs between 5% to 35% were enriched in all skin samples (Figure 2c). The enrichment of these low-to-medium level variants suggests cells with shared variants (subclonal variants/cells) are prevalent in these skin samples. Furthermore, we observed enrichment of subclonal variants in epidermal samples compared to SCCIS samples (Figures 1a and 2c). This was especially noted for the INDELs (insertion-deletion) variants (Figure 2c), presumably because the subclonal INDELs undergo negative selective pressure making them less common in SCCIS samples.
To distinguish subclonal variants from other joint variants, the MAF differences between the paired epidermis and SCCIS samples was determined based on the null assumption that most non-subclonal variants (SNPs, spontaneous mutations) are selectively neutral. Each joint variant with significant read coverage and non-reference minor allele was tested. Those with a significantly different MAF between sample types (epidermis vs. SCCIS) are defined as subclonal variants. This assumes most subclonal variants are either significantly expanded in SCCIS due to inherent growth advantages or eliminated by negative selective pressure from ‘fitter’ sub-clones of cells. We identified sub-clonal variants ranging from 1000~5000 per patient, likely representing potential drivers of SCCIS formation (Supplementary Table S3). With these subclonal variants, we estimated a lesion mutation burden (LMB) similar to the tumor mutation burden (TMB) found in comparable studies (Chalmers et al., 2017; Chan et al., 2019; Zehir et al., 2017). All ten patients were found to have quite high LMB (>= 10 mut/Mb), which only includes the sufficiently covered exome regions (Supplementary Table S4). The high mutation burden in these SCCIS samples supports previous findings where skin related cancers usually have the highest TMB among various cancer types (Alexandrov et al., 2013; Chalmers et al., 2017; Chan et al., 2019; Zehir et al., 2017).
Subclonal variants may play key roles in SCCIS development
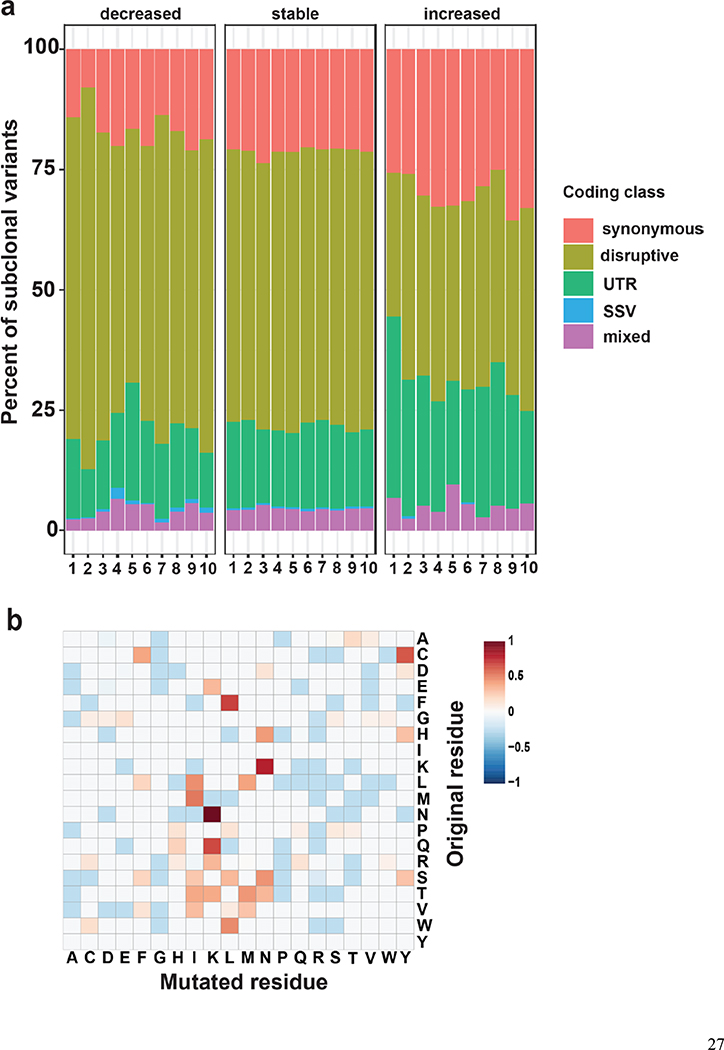
Subclonal variants are primarily classified as coding variants, but further analysis of the coding subclonal variants revealed that these variants, with either decreased or increased MAF in SCCIS, showed different sub-class distributions (Figure 3a). Compared to stable variants, subclonal variants with decreased MAF (decreased) are preferentially disruptive mutations; while subclonal variants with increased MAF (increased) are non-disruptive, synonymous or UTR types (Figure 3a), indicating positive selection of some non-disruptive variants in SCCIS.
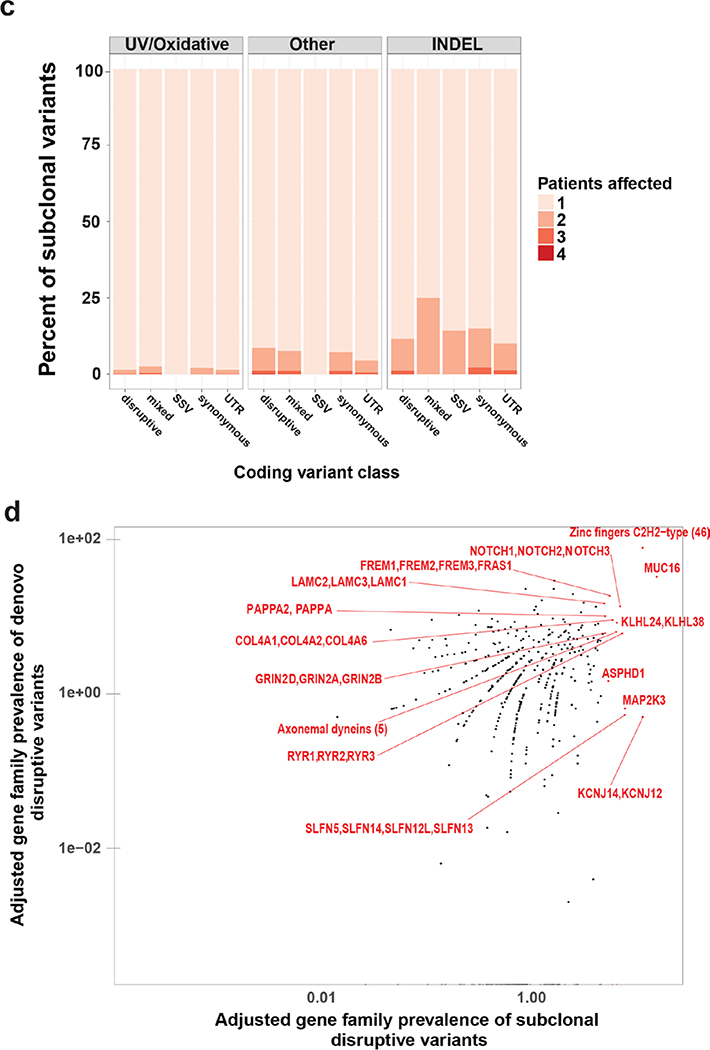
Figure 3.
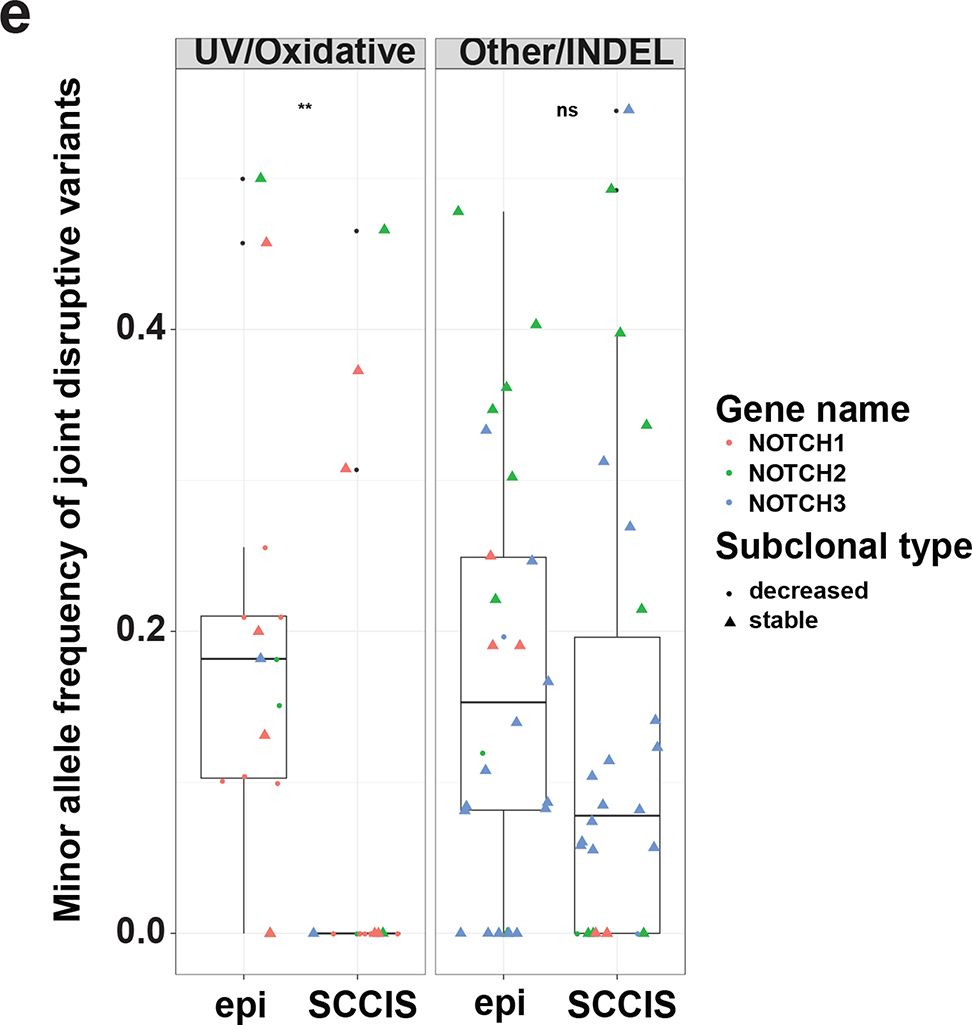
Characteristics of subclonal variants. (a) Summary of the coding class for subclonal and “stable” variants. disruptive: nonsynonymous, nonsense, readthrough or frame-shift types; SSV: splice-site variant; mixed: classified into more than one coding classes; stable: similar to subclonal variants but the MAF is not significantly different between epidermis and SCCIS; (b) Relative preference of amino acid substitution preference of nonsynonymous subclonal variants using nonsynonymous stable variants as background. Rows and columns represent the original and mutated amino acids, respectively. Values shown in cells are calculated LOD scores (in bits). LOD scores for non-observed substitutions were set to 0. (c) Commonality of subclonal variants reflected as number of shared patients regarding different coding class and variant types. UV: C->T(G->A) signature variant; oxidative: G->T(C->A) signature variant; INDEL: insertion/deletions; (d) Adjusted gene-family preference of subclonal (x-axis) and de-novo (y-axis) variants. Red highlighted dots: top 15 most preferred gene families for subclonal variants with all affected genes labeled. Large families with many affected genes are labeled with family names and number of affected genes in parentheses instead. (e) Distribution of the MAF of all subclonal and stable variants for different variant signatures (UV/oxidative, Other, INDEL) using the NOTCH gene family as an example. ** indicates statistically a significant difference with p-value < 0.01 based on Wilcoxon Rank-Sum Test, ns-indicates a non-significant difference with p-value > 0.05.
Focusing on nonsynonymous variants, specific amino acid (AA) substitutions were enriched in these disruptive subclonal variants. Using the stable nonsynonymous amino acid substitutions as a background, the most enriched AA substitutions were N->K, K->N, Q->K and F->L mutations (Figure 3b). These are substitutions of amino acids with similar properties because they are polar-to-polar substitutions or nonpolar F->L substitutions. The top enriched substitutions have non-negative scores in the BLOSUM62 AA substitution matrix for sequence homology (Johnson et al., 2008), suggesting conservation of amino acid biochemistry in many UV-driven mutation events.
Subclonal variants were found rarely in more than one patient; this was manifested by the UV/oxidative signature disruptive (UVD) mutations (Figure 3c). Instead, non-UV induced, non-disruptive variants appeared more frequently in multiple patients, suggesting they are likely germline variants or population polymorphisms. Together, these data indicate that subclonal UVD variants found in populations could provide a pool of mutated keratinocytes for UV-induced clonal selection.
`The heterogeneity of pre- or tumor lesion samples often results in dynamic clonal and subclonal features, regardless of whether true selection exists in the tumor microenvironment (Malikic et al., 2019). To test whether the clones in sun-exposed epidermis (SEE) or SCCIS are under true selection, we estimated the dN/dS ratios in each sample independently (see Supplemental Methods). We used only the subclonal UVD variants because they represent the UV/oxidative induced somatic mutations in the epidermal and SCCIS samples. Most of the epidermal samples (9 out of 10) exhibited an elevated dN/dS ratio greater than 1, implying significant positive selective pressure for the UVD subclonal variants in these UV-exposed epidermal samples (Supplementary Table S7). Interestingly, the estimated dN/dS ratios were lower in the paired SCCIS samples except for one patient, suggesting the positive effect of these variants were less profound once SCCIS formed (Supplementary Table S7). In fact, the dN/dS ratios in most SCCIS samples (8 out of 10) were less than 1, implying these mutations were under neutral or purifying selection. Therefore, we hypothesize that subclonal UVD variants in SEE are important for the formation but not maintenance of SCCIS.
Genes with subclonal UVD mutations regulate keratinocyte differentiation and cell growth
To identify genes with subclonal variants, the mutation preference of these variants was evaluated at the gene family level. The gene-family mutation preference was calculated for UVD variants, and normalized for coding region length and GC content (see methods and Figures 3d and Supplementary Figure S3). The top targets, usually with multiple variants in different family members, include NOTCH1–3, Axonemal dyneins (DNAH1/2/7/12/14), SLFN5/13/14, Ryanodine receptors (RYR1/2/3) and Laminins (LAMC1/2/3) (Figure 3d). Almost all (19/20) NOTCH1–3 mutations were located in the extracellular or transmembrane domains, consistent with a LOF mutation (Supplementary Figure S2). A total of 15 UVD NOTCH mutations were detected with nine and five in the epidermal and SCCIS libraries respectively (Supplementary Tables S6A and B). The number of NOTCH LOF mutations ranged from 0–4 per epidermal library and 0–2 per SCCIS library. The average MAF of NOTCH mutations in the epidermis was 14.2% and 42.8% in SCCIS, indicating positive selection for NOTCH LOF mutations in SCCIS. Sixty percent of SCCIS lesions contained oncogenic mutations in TP53 with none in the epidermal libraries (Supplementary Tables S6A and B). The bias for SCCIS TP53 mutations suggests this mutation may be key for lesion development. Oncogenic RAS mutations were not found in any library, indicating such mutations are not required for the early stages of keratinocytic neoplasia (Supplementary Tables S6 A and B).
To better understand how these genes acquired subclonal variants, we analyzed the MAF spectrum of all joint variants for gene families with a higher mutation frequency. Several gene families, including NOTCH, exhibited a statistically elevated MAF at low-to-medium levels (usually between 5% to 30%) in the epidermis compared to paired SCCIS samples (Figure 3e), (MAF in (Abbas et al., 2011) Wilcoxon Rank-Sum Test), a trend observed only for UVD variants but not for other non-UV/oxidative signature (such as C->G or A->T) SNPs or INDELs (Other/INDEL) (Figure 3e). Similar trends were observed for the Dyneins, Schlafen, Laminin and the Nucleoporin (NUP) genes, although not all reached statistical significance (Supplementary Figures S3–S6). Notably, more subclonal UVD variants in these gene families have a decreased MAF (Figure 3e, Supplementary Figures S3–S6). This indicates that SEE has a non-negligible pre-deposition of UV/oxidative induced mutations, and only a small proportion of these mutations were selected for “clonal” variants in the paired SCCIS. We hypothesize that although many subclonal variants in sun-exposed epidermis may be beneficial in that specific microenvironment, it appears that only a small fraction of cells with subclonal variants are selected to form an SCCIS; the SCCIS likely overgrows/displaces remaining non-selected, mutant keratinocytes eliminating other subclonal variants from the site of the lesion. In fact, none of the subclonal UVD variants in NOTCH genes were found to have an increased MAF in the SCCIS samples (Supplementary Table S6), suggesting that cells containing one specific subclonal variant in NOTCH genes likely grow to form an SCCIS, thus this variant is absent in the adjacent epidermis and does not show up on the subclonal variant list. Potentially, some NOTCH LOF variants in the epidermis may have not been called because of insufficient WES read coverage (10X minimum coverage for both epidermis and SCCIS samples, see Supplementary Methods). In contrast, the non-UV/oxidative SNPs or INDELs (Other/INDEL) likely represent population SNPs or stochastic germline mutations that are selectively neutral and did not exhibit a significant difference in their MAF spectrum between paired epidermis and SCCIS samples. Additionally, only 4.3%, 32%, 44% and 29% of those subclonal variants have decreased MAF for the Dyneins, Schlafen, Laminin and NUP gene families/super-families, respectively. This highlights the difficulty of identifying potential “causal” mutations even using LCM dissected, NGS-based assays like in this study. These results suggest that human SCCIS development selects keratinocytes harboring UVD mutations impacting gene families that regulate cancer development. The adjusted gene family prevalence for the de-novo variants correlated with the subclonal variants (Figure 3d), suggesting that genes containing subclonal variants gain additional de-novo mutations in SCCIS.
We performed gene ontology (GO) enrichment analysis for the protein-coding genes harboring subclonal UVD variants (Table S3). GO analysis showed alterations in cell morphogenesis, small GTPase regulation, cell development and microtubule based movement (Supplementary Figure S7), similar to other cancer related phenotypes (Baudot et al., 2010; Takebe et al., 2011).
RNA sequencing of epidermal and SCCIS libraries identifies 1166 differentially expressed genes
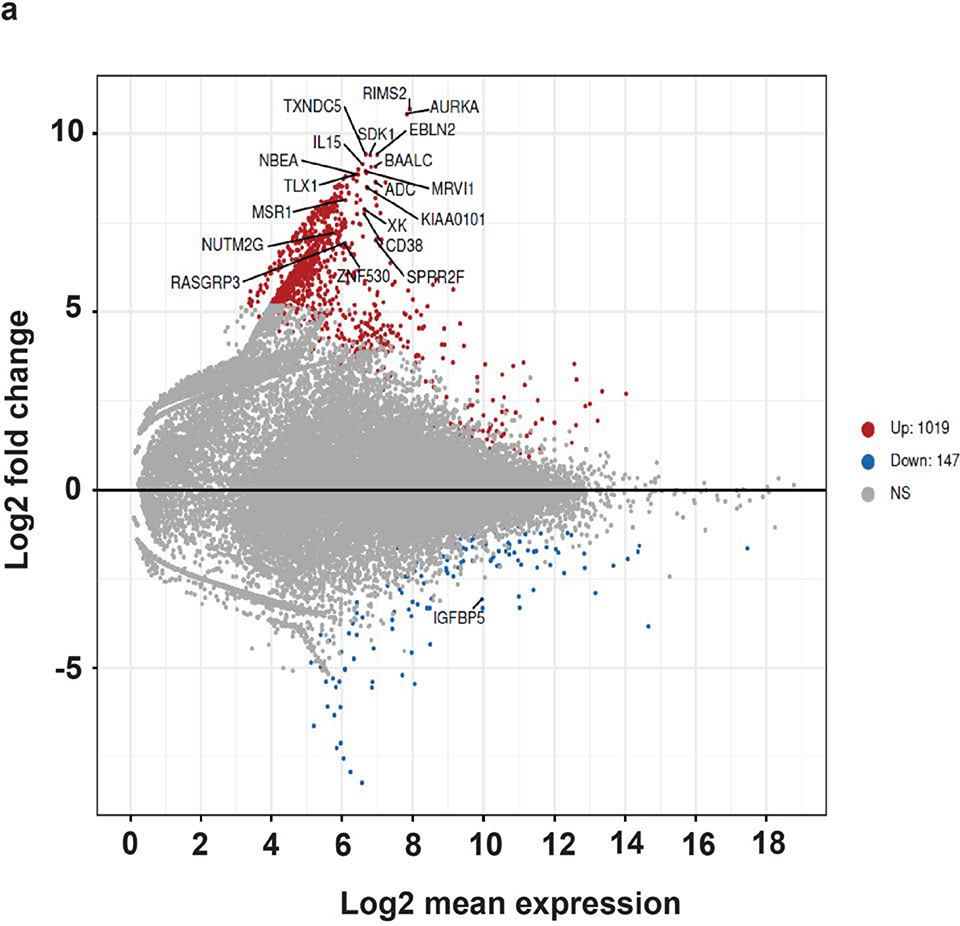
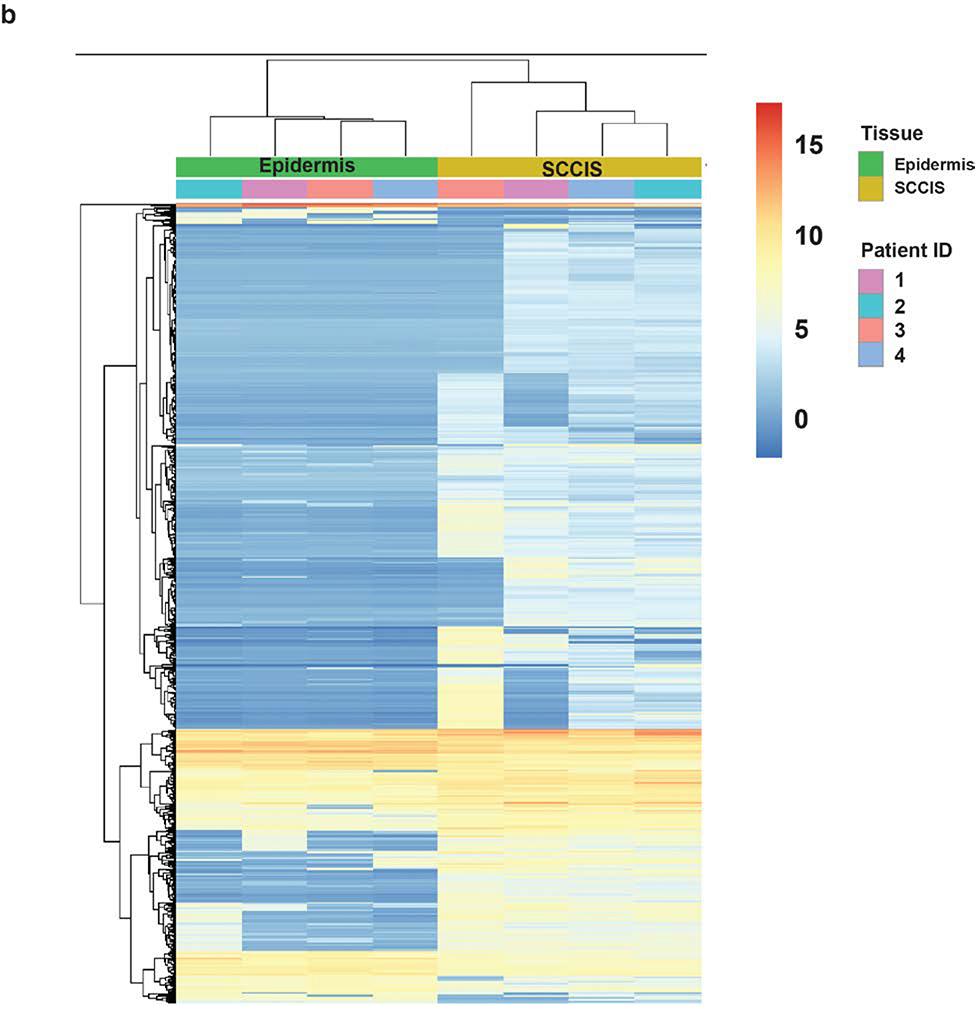
RNA sequencing of four epidermal and SCCIS libraries identified 1166 differentially expressed (DE) genes with an adjusted p-value (FDR) < 0.1 and 849 DE genes with an FDR < 0.05. A negative-binomial linear model (see Methods) revealed 1019 of 1166 genes (87.4%) were up-regulated in SCCIS while 147 (12.6%) were downregulated (Figure 4a). A heatmap of all 1166 DE-genes shows that gene-level clustering was driven by sample type (Figure 4b).
Figure 4. Transcriptomic analysis of the RNA-seq dataset.
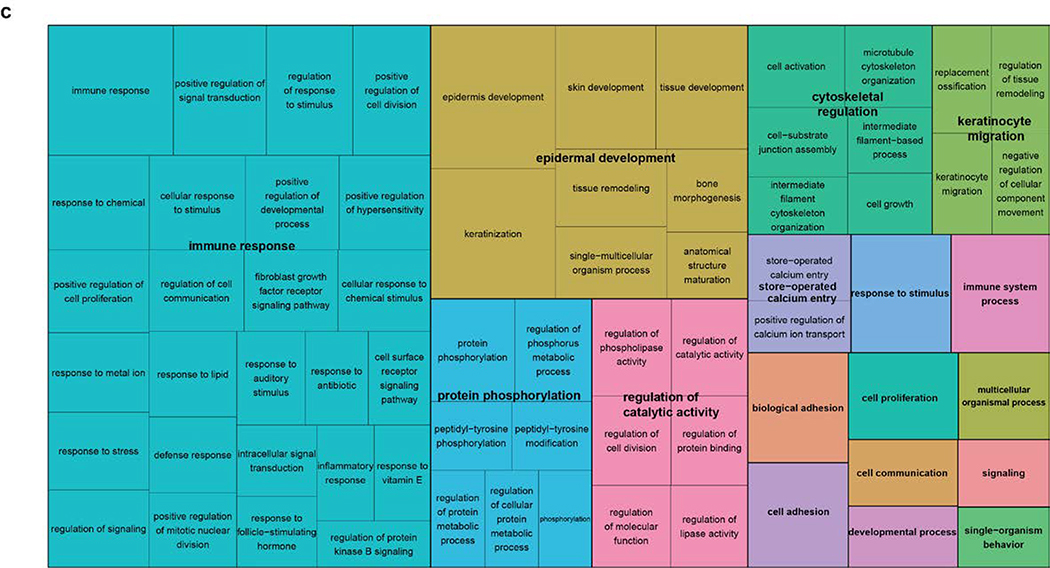
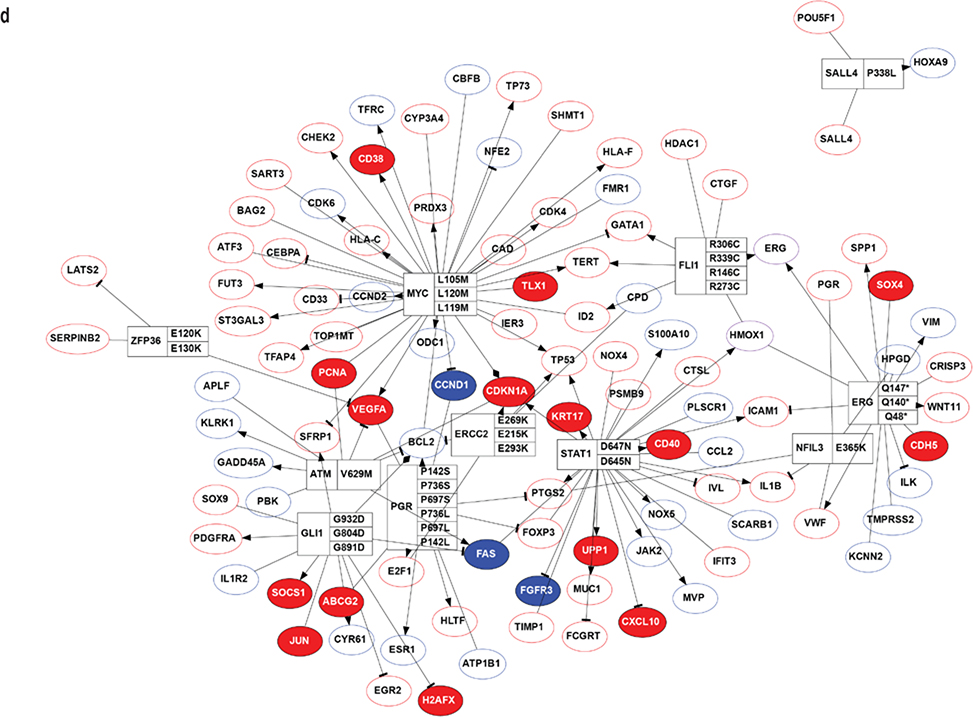
(a). MA-plot of all gene expression profiles revealed by RNA-seq data. X,Y-axes: A (average) and M (difference) of the gene expression between epidermis and SCCIS samples; Red and blue dots: up-regulated and down-regulated genes determined by DESeq2. Top 20 differentially expressed (DE) protein coding genes are labeled with their symbols. (b) Heatmap of top 1162 DE-genes, with their tissue type and patient ID information labeled by each column. Cell values are the normalized regularized log expression values calculated by DESeq2. Adj p value < 0.1 (c) Enriched Gene Ontology (GO) Biological Processes associated with DE-genes. Rectangle sizes reflect the significance of enrichment of each terms, with similar GO terms grouped into larger rectangles. (d) Core transcriptional regulator-target (TR-target) network for genes harboring subclonal UVD variants, only top genes with no less than 5 targets are included. Rectangles: transcriptional regulators with their names and associated subclonal UVD variants labeled; ellipses: known target genes from TRRUST database. Red or blue filled/bordered nodes represent genes with increased or decreased average expression in SCCIS compared to epidermis, respectively. Ovals with solid colors represent significantly differentially expressed genes (Adj p value < 0.1), ovals outlined color represent genes that were differentially expressed with an adjusted p value > 0.1. Arrows represent a positive regulatory relationship. T-bars represent a negative regulatory relationship.
Linking the genotypes of subclonal UVD variants with the gene expression changes in each sample indicated less bias in the number of gene pairs that demonstrated up/down regulation (6321 and 4391 up- and down-regulated pairs, respectively; see Methods), which is statistically significant (p < 9.2 × 10−80, χ2 test). Therefore, genes with subclonal UVD variants are not always associated with reduced transcription, supporting the hypothesis that transcriptomic changes occurred secondary to genomic changes. We speculate this is because short-term transcriptomic changes caused by prior genomic mutations may have been compensated for by cumulative cellular events that occurred in the years between the genomic mutations and the procurement of the patient samples for RNA-seq experiments. GO analysis revealed that the DE-genes were enriched in the following biological processes: 1) immune response, 2) epidermal development, 3) protein phosphorylation, 4) regulation of catalytic activity, 5) cytoskeleton organization, 6) keratinocyte migration (Fig. 4C). Genes that showed strong upregulation included Aurora A kinase (p≤ 2.5×10−10) which is critical for keratinocyte proliferation. The largest enriched GO category was immune response with upregulation of the following in SCCIS: 1) IL-15 (p≤ 9.8×10−7), 2) CXCL10 (p≤ 3.5× 10−5), 3) TSLP (p≤ 1.2×10−4), 4) CXCL9 (p≤ 0.008), 5) IFNAR2 (p≤ 0.05).
Two-hundred and twelve (212) out of 2844 (7.45%) genes harboring a subclonal UVD variant were transcriptional regulators. This represents a higher percentage than the known percentage of transcriptional regulators in the human genome (800 out of 20345 or 3.93%, p < 1.1e-17, χ2 test) (TRRUST) (Han et al., 2015), which suggests positive selective pressure for subclonal UVD variants in transcriptional regulators in SCCIS.
Transcription factors containing subclonal UVD variants are predicted to regulate more gene targets (6.34 targets) than the average of 3.53 targets for human transcription factors. Genes with subclonal UVD variants included proto-oncogenes and DNA damage repair genes, such as FLI1, MYC, ERCC2 and GLI1, which have significant numbers of known transcriptional targets (Figure 4d).
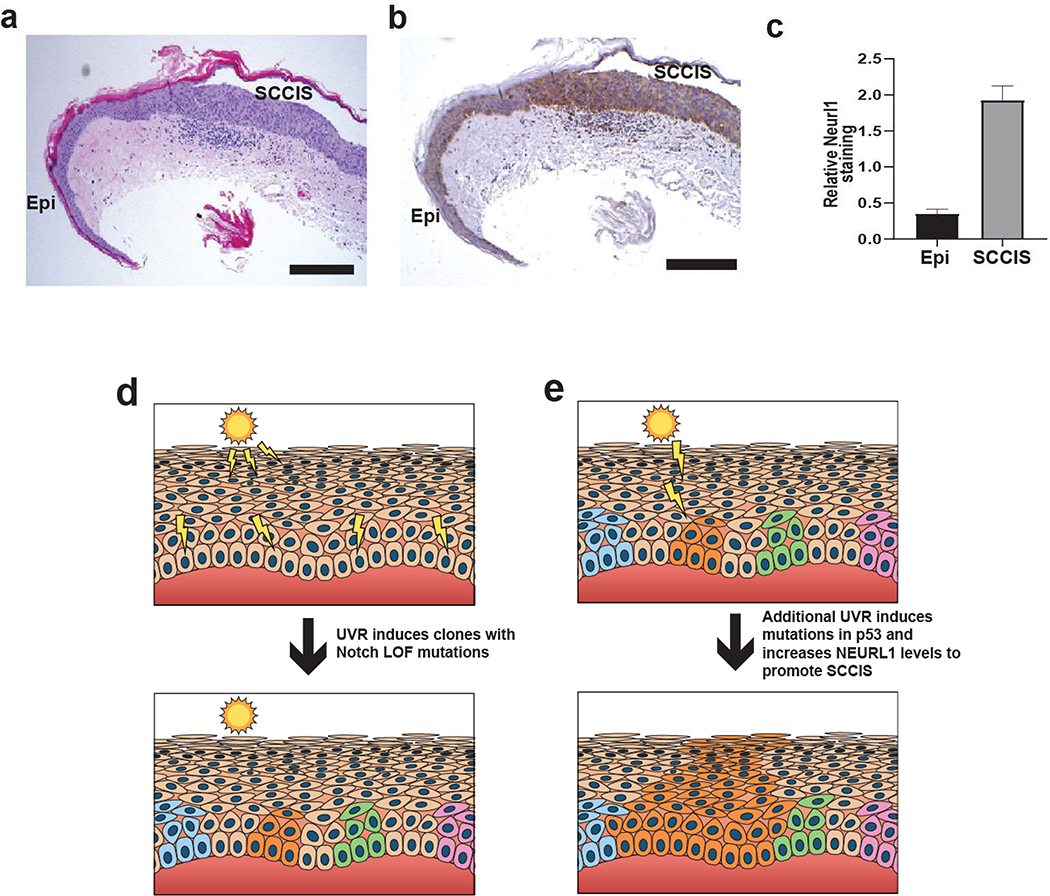
RNA seq data indicated upregulation of NEURL1 mRNA in all SCCIS libraries (p≤ 0.0015). NEURL1 is a ubiquitin ligase that targets Notch ligands resulting in lower expression of Notch target genes HES1 and HEY1. Increased NEURL1 expression may represent a mechanism for decreasing Notch signaling in SCCIS (Teider et al., 2010). Indeed, customized GSEA using Notch target genes found that they are enriched at gene-set level in the differentially expressed gene set (p<0.021, by GAGE package). CCND1, a known Notch target, was downregulated in all SCCIS samples on average 5.0-fold +/− 1.4 with an FDR = 0.0002. (See Supplementary Methods) (Katoh and Katoh, 2020). Immunohistochemical studies for Neurl1 in a prospective set of human biopsies with UV-exposed epidermis and SCCIS demonstrated increased Neurl1 staining in all SCCIS compared to epidermis, (p<0.0001) (Figures 5 a–c).
Figure 5. Immunohistochemical analysis of Neurl1 in epidermis and SCCIS with model of UV-induced SCCIS formation.
(a) H+E staining of representative specimen containing epidermis (Epi) and SCCIS. Scale bar = 0.25 mm. (b) Same representative immunohistochemical staining for Neurl1 showing higher levels of staining in SCCIS than in Epi. Scale bar = 0.25 mm. (c) Analysis of Nerul1b staining intensity. The relative staining intensity for Neurl1 is higher in SCCIS than adjacent epidermis. N = 10, p < 0.0001. (d) Scattered epidermal keratinocytes exposed to UVR acquire independent Notch LOF mutations and form small clones, clones highlighted in color. (e) A random clone acquires additional UV-induced mutations in p53 and/or increased levels of NEURL1 ubiquitin ligase that promotes progression to SCCIS manifested by full-thickness epidermal growth of lesional cells orange)
Discussion
SCCIS is a common precancerous lesion in humans, yet its pathogenesis remains unclear. To better understand SCCIS biology, LCM of UV-exposed epidermis and associated SCCIS was performed followed by WES and RNA seq.
The AlignerBoost algorithm permitted detection of not only traditional “somatic” variants (de-novo variants) using paired normal/tumor samples but also “subclonal” variants. The subclonal variants were present typically at an MAF less than 30% in the epidermal libraries and had a UVR/oxidative stress signature. Mutations in the same gene family in SCCIS had a higher MAF above 35% suggesting positive clonal selective pressure. For example, mutations in Notch genes (Supplementary Tables S6 a and b) generally showed that a specific NOTCH LOF UV signature mutation selected for in SCCIS was not present in the adjacent epidermis. The observation that UV signature LOF mutations in NOTCH were not typically shared between epidermis and SCCIS indicates that independent keratinocyte clones harboring such mutations may be randomly selected to form SCCIS. The UV signature mutations in NOTCH genes present in both epidermis and SCCIS were seen at MAF around 50% in the epidermis raising the possibility that these mutations could represent somatic mutations or population SNPs (Supplementary Table S6a).
Gene families with low frequency UV-signature mutations in the epidermis and positive selection of analogous mutations in SCCIS included NOTCH, Dyneins, SLFN (Schlafen genes), Laminins and NUP (Figure 3d). NOTCH LOF mutations have been found in UV-exposed epidermis from blepharoplasty specimens, AKs, and in a majority of invasive cSCCs (Chitsazzadeh et al., 2016; Martincorena et al., 2015; South et al., 2014; Wang et al., 2011). However, the relationship between UV-signature NOTCH LOF mutations in epidermis and SCCIS had not been clearly defined previously. By directly assessing the number and MAF of UV-signature Notch LOF mutations in epidermis and corresponding SCCIS, we show that specific NOTCH mutations are selected for in SCCIS that differ from those in adjacent epidermis. These data support the model that keratinocyte clones containing UV-signature NOTCH LOF mutations are positively selected to form SCCIS with continued UV exposure (Figures 5d and e).
The WES data indicate that acquisition of an oncogenic TP53 mutation promotes SCCIS formation as these mutations were present in 60% of SCCIS (Supplementary Tables S1 and 2). All TP53 mutations found in SCCIS have been identified in other tumors including esophageal SCCs (Cosmic database v89). Although no oncogenic TP53 mutations were detected in the epidermal libraries, we cannot exclude their rare presence in “clinically” unremarkable UV-exposed skin without targeted deep sequencing (Martincorena et al., 2015). Nevertheless, our data suggest such mutations are rare in epidermis verified to have no dysplasia. The distribution of NOTCH and TP53 mutations in our epidermal and SCCIS libraries indicates the transition from an epidermal Notch-deficient keratinocyte clone to SCCIS may be driven by a TP53 mutation (Figures 5d and e).
No oncogenic mutations in RAS genes were detected, which corroborates findings from other studies on UV-exposed skin and AKs (Chitsazzadeh et al., 2016; Martincorena et al., 2015). This suggests that activating RAS mutations are rare-to-absent in the early stages of human UV-induced skin cancer. This differs from the murine DMBA/TPA model of skin cancer where precancerous papillomas harbor activating HRAS mutations (Reeves et al., 2018). It may be the precancerous stages of the DMBA/TPA model are genetically different from human SCCIS.
The WES data indicated clonal selection of UV-signature mutations within the Schlafen gene family (Figure 3d). This is of interest as Schlafen 5 knockdown activates Akt and B-catenin signaling; pathways known to be activated in human SCCIS (Wan et al., 2019; Zhao et al., 2009). Alterations in Dynein function have been detected in lung squamous cell carcinoma primarily associated with a TRA2b-DNAH5 fusion protein which activates ERK1/2 and MMP-1 (Li et al., 2016). Also, genomic studies of nasopharyngeal carcinomas, a high-grade squamous cell carcinoma, implicated loss of dynein function in impaired primary cilium function and tumorigenesis (Chin et al., 2016).
The RNA-seq data revealed 1166 differentially expressed transcripts, and GO analysis highlighted immune regulatory genes as the most prominent category of differentially expressed genes. IL-15 was strongly upregulated in all SCCIS examined (p≤ 9.8×10−7). IL-15 appears to play a role in establishing functional T-reg cells in head and neck SCC, inhibiting immunosurveillance in the tumor microenvironment (Bergmann et al., 2008). Increased IL-15 in cutaneous SCCIS may be important for establishing immune tolerance for UV-exposed epidermal keratinocytes with potentially immunogenic mutations.
The RNA-seq data showed upregulation of TSLP at 6.4 fold (p≤ 1.2×10−4) and the NEURL1 ubiquitinase at 2.7-fold (p≤ 0.0015). It has been shown that TSLP expression increases in mice lacking epidermal Notch 1, 2 or 3 and such mice develop cSCCs (Demehri and Kopan, 2009). The NEURL1 ubiquitin protein ligase targets the Notch ligands Delta and Jagged for ubiquination, downregulating Notch signaling in cancerous cells (Teider et al., 2010). The increased NEURL1 expression in SCCIS could represent a mechanism for downregulating Notch signaling, and was detected in all SCCIS independent of NOTCH LOF mutations. Indeed, customized GSEA using Notch target genes found that they are enriched at gene-set level in the differentially expressed gene set (p<0.021, by GAGE package). CCND1, a known Notch target, was downregulated in all SCCIS samples on average 5.0-fold +/− 1.4 with an FDR = 0.0002. (See Supplementary Methods). However, given the complexity of Notch signaling, the biological significance of NEURL1 upregulation in SCCIS requires further mechanistic studies.
The SYNE1 (spectrin repeat containing nuclear envelope protein 1) gene was uniformly downregulated in SCCIS by 9.1-fold (p≤ 0.0078). SYNE1 is known to be commonly mutated in oral squamous cell carcinoma and loss of function correlates with aggressive behavior.
This LCM-directed WES and RNA-seq study of UV-exposed epidermis and SCCIS provides precise comparative genomic and transcriptomic data from human samples. The data are consistent with prior studies examining biopsies containing UV-exposed epidermis, AK and SCC not subjected to LCM. These studies showed a high prevalence of UV-signature mutations in NOTCH 1–2, TP53, FAT1 and MLL2 in cSCC (Chitsazzadeh et al., 2016). However, our data show a strong prevalence for oncogenic TP53 mutations in the SCCIS compared to adjacent UV-exposed epidermis, perhaps due to the precision of LCM. The data provide insights into gene families that are mutated in SCCIS and refine our understanding of what genes are selected for during SCCIS development. The data support a model where clinically unremarkable Notch-deficient clones acquire oncogenic p53 mutations and/or upregulate NEURL1 to form SCCIS (Figures 5d and e). Thus, dysregulation of the p53/Notch1,2,3/p21 pathway at multiple levels may be favorable for SCCIS formation. The data also provide support for the role of immunoregulatory genes in SCCIS development, as well as identification of additional genes as potential drivers and therapeutic targets.
Methods
Human materials, tissue isolation and total DNA/RNA extraction
Ten archived formalin fixed paraffin embedded biopsies containing both SCCIS and UV-exposed epidermis were retrieved from the Dermatopathology archives at the Hospital of the University of Pennsylvania under IRB protocol 808225. All specimens came from sun-exposed sites. Details regarding human materials, sectioning, LCM and nucleic acid isolation are provided in Supplementary Methods.
Next-Gen exome-seq and RNA-seq library construction
Testing of nucleic acid quality and library construction are detailed in Supplementary Methods.
NGS read pre-processing and mapping
All NGS runs were processed independently using AlignerBoost (Zheng and Grice, 2016b) with Bowtie2 (Langmead and Salzberg, 2012). See Supplementary Methods and Tables S3 and S4.
Variant identification
Variant identification followed the best practices approach in Supplementary Methods.
Functional classification and annotation of read and variants
All mapped reads and called variants were classified as described in Supplementary Methods.
Defining the subclonal variants
Identifying subclonal joint variants is outlined in Supplementary Methods.
Gene Ontology enrichment analysis and visualization
Gene Ontology (GO) enrichment analysis was performed as described in Supplementary Methods.
Gene family mutation preference analysis
RNA-seq data analysis
See Supplementary Methods and Table S4.
Linking the exome-seq and RNA-seq results
Immunostaining for Neuralized 1
Supplementary Material
Acknowledgments
This work was supported by NIH grants RO1-CA165836 (PI), RO1-ES02811 (PI) and P30-AR069589 (Core Director) to JTS, Abramson Cancer Center CCSG P30 CA016520 to WP and JTS. NIH grants P30-AR069589 (Cotsarelis PI) (Co-PI) to EG.
Abbreviations
- AA
amino acid
- AKs
actinic keratoses
- DE
differentially expressed
- DNA
deoxyribonucleic acid
- GO
gene ontology
- LCM
laser capture microdissection
- NGS
next generation sequencing
- RNA
ribonucleic acid
- RNA-seq
RNA sequencing
- cSCC
cutaneous squamous cell carcinoma
- SCCIS
squamous cell carcinoma in situ
- SEE
sun-exposed epidermis
- UVR
ultraviolet radiation
- UVD
UV/oxidative disruptive
- WES
whole exome sequencing
Footnotes
Conflict of Interest Statement: None of the authors have a conflict of interest regarding the research described in this manuscript.
Data Availability Statement: Datasets related to this article data have been submitted to NCBI dbGap under accession phs002019.v1.p1. The data will be available from dbGaP upon request.
References
- Abbas O, Richards JE, Yaar R, Mahalingam M (2011) Stem cell markers (cytokeratin 15, cytokeratin 19 and p63) in in situ and invasive cutaneous epithelial lesions. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 24:90–97. [DOI] [PubMed] [Google Scholar]
- Baudot A, de la Torre V, Valencia A (2010) Mutated genes, pathways and processes in tumours. EMBO Rep 11:805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C, Strauss L, Wang Y, Szczepanski MJ, Lang S, Johnson JT, et al. (2008) T regulatory type 1 cells in squamous cell carcinoma of the head and neck: mechanisms of suppression and expansion in advanced disease. Clin Cancer Res 14:3706–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin YM, Tan LP, Abdul Aziz N, Mushiroda T, Kubo M, Mohd Kornain NK, et al. (2016) Integrated pathway analysis of nasopharyngeal carcinoma implicates the axonemal dynein complex in the Malaysian cohort. Int J Cancer 139:1731–1739. [DOI] [PubMed] [Google Scholar]
- Chitsazzadeh V, Coarfa C, Drummond JA, Nguyen T, Joseph A, Chilukuri S, et al. (2016) Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat Commun 7:12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki D, Meehan CJ, Bruce F, Culjak G (2002) The majority of cutaneous squamous cell carcinomas arise in actinic keratoses. J Cutan Med Surg 6:207–209. [DOI] [PubMed] [Google Scholar]
- Demehri S, Kopan R (2009) Notch signaling in bulge stem cells is not required for selection of hair follicle fate. Development 136:891–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J, Sabarinathan R, Mularoni L, Muinos F, Gonzalez-Perez A, Lopez-Bigas N (2017) Reduced mutation rate in exons due to differential mismatch repair. Nat Genet 49:1684–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- General S (2014) The Surgeon General’s Call to Action to Prevent Skin Cancer. [PubMed] [Google Scholar]
- Han H, Shim H, Shin D, Shim JE, Ko Y, Shin J, et al. (2015) TRRUST: a reference database of human transcriptional regulatory interactions. Sci Rep 5:11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Wang L, Di LJ (2016) Distinct mutation accumulation rates among tissues determine the variation in cancer risk. Sci Rep 6:19458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Katoh M (2020) Precision medicine for human cancers with Notch signaling dysregulation (Review). Int J Mol Med 45:279–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Fang Z, Zhang J, Li C, Liu H, Xia J, et al. (2016) Identification of TRA2B-DNAH5 fusion as a novel oncogenic driver in human lung squamous cell carcinoma. Cell Res 26:1149–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou J, White KP (2014) RNA-seq differential expression studies: more sequence or more replication? Bioinformatics 30:301–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks R (1995) An overview of skin cancers. Incidence and causation. Cancer 75:607–612. [DOI] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. (2015) Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348:880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meynert AM, Ansari M, FitzPatrick DR, Taylor MS (2014) Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics 15:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratushny V, Gober MD, Hick R, Ridky TW, Seykora JT (2012) From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest 122:464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves MQ, Kandyba E, Harris S, Del Rosario R, Balmain A (2018) Multicolour lineage tracing reveals clonal dynamics of squamous carcinoma evolution from initiation to metastasis. Nat Cell Biol 20:699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HW, Weinstock MA, Harris AR, Hinckley MR, Feldman SR, Fleischer AB, et al. (2010) Incidence estimate of nonmelanoma skin cancer in the United States, 2006. Arch Dermatol 146:283–287. [DOI] [PubMed] [Google Scholar]
- South AP, Purdie KJ, Watt SA, Haldenby S, den Breems NY, Dimon M, et al. (2014) NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. J Invest Dermatol 134:2630–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe N, Harris PJ, Warren RQ, Ivy SP (2011) Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol 8:97–106. [DOI] [PubMed] [Google Scholar]
- Teider N, Scott DK, Neiss A, Weeraratne SD, Amani VM, Wang Y, et al. (2010) Neuralized1 causes apoptosis and downregulates Notch target genes in medulloblastoma. Neuro Oncol 12:1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. (2013) From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11 10 11–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Liu Y, Zhu J, Guo L, Li C, Yang Y, et al. (2019) SLFN5 suppresses cancer cell migration and invasion by inhibiting MT1-MMP expression via AKT/GSK-3beta/beta-catenin pathway. Cell Signal 59:1–12. [DOI] [PubMed] [Google Scholar]
- Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, et al. (2011) Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U S A 108:17761–17766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff KG, Katz L, Gilchrest S, Paller B, Leffell A, D. Dermatology in General Medicine, 7th edn. Mc Graw-Hill: New York, 2008, 1–1660, 1661–3002pp. [Google Scholar]
- Zhao L, Li W, Marshall C, Griffin T, Hanson M, Hick R, et al. (2009) Srcasm inhibits Fyn-induced cutaneous carcinogenesis with modulation of Notch1 and p53. Cancer Research 69:9439–9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Grice EA (2016a) AlignerBoost: A Generalized Software Toolkit for Boosting Next-Gen Sequencing Mapping Accuracy Using a Bayesian-Based Mapping Quality Framework. PLoS Comput Biol 12:e1005096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Grice EA (2016b) AlignerBoost: A Generalized Software Toolkit for Boosting Next-Gen Sequencing Mapping Accuracy Using a Bayesian-Based Mapping Quality Framework. PLoS Comput Biol 12:e1005096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Jonason AS, Leffell DJ, Simon JA, Sharma HW, Kimmelman J, et al. (1994) Sunburn and p53 in the onset of skin cancer. Nature 372:773–776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.