Abstract
Background
We have reported that internal deletions in the nef, gag, and pol genes in HIV-1–infected patients are induced in those treated with Korean Red Ginseng (KRG). KRG delays the development of resistance mutations to antiretroviral drugs.
Methods
The vif-vpr genes over 26 years in 20 hemophiliacs infected with HIV-1 from a single source were sequenced to investigate whether vif-vpr genes were affected by KRG and KRG plus highly active antiretroviral therapy (ART) (hereafter called GCT) and compared the results with our previous data.
Results
A significantly higher number of in-frame small deletions were found in the vif-vpr genes of KRG-treated patients than at the baseline, in control patients, and in ART-alone patients (p < 0.001). These were significantly reduced in GCT patients (p < 0.05). In contrast, sequences harboring a premature stop codon (SC) were more significant in GCT patients (10.1%) than in KRG-alone patients, control (p < 0.01), and ART-alone patients (p = 0.078 for peripheral blood mononuclear cells). The proportion of SC in Vpr was similar to that in Vif, whereas the proportion of sequences revealing SC in the env-nef genes was significantly lower than that in the pol-vif-vpr genes (p < 0.01). The genetic distance was 1.8 times higher in the sequences harboring SC than in the sequences without SC (p < 0.001). Q135P in the vif gene is significantly associated with rapid progression to AIDS (p < 0.01).
Conclusion
Our data show that KRG might induce sΔ in the vif-vpr genes and that vif-vpr genes are similarly affected by lethal mutations.
Keywords: HIV-1, Korean Red Ginseng, Lethal mutations, Small deletion, vif-vpr genes
1. Introduction
It is presumed that the Korean subclade of human immunodeficiency virus type 1 (HIV-1) subtype B (KSB) was introduced to Korea by the founder effect in the mid-1980s [1,2]. During late 1989 to early 1990, 20 hemophiliacs (HPs) were diagnosed with KSB infection 1–2 years after exposure to domestic clotting factor IX manufactured using plasma from two cash-paid plasma donors in Korea [[2], [3], [4]]. They all were infected with a single source of HIV-1 [3,4]. Of the 20 HPs, about 70% did not progress to AIDS up to 10 years after HIV-1 infection [5]. They had been treated with Korean Red Ginseng (KRG) for a significant period before the introduction of highly active antiretroviral therapy (ART) in 2002, after which most of them were treated with KRG plus ART combination therapy (hereafter called GCT). Our previous studies have shown that KRG treatment induces nonspecific internal deletions (IDs) over the full genome of HIV-1 [[5], [6], [7], [8], [9], [10]] and clinically beneficial effects [5,11].
G-to-A hypermutation by APOBEC3 proteins (A3G) is represented as 5–12% of a collection of sequences [12,13] although HIV genetic variation is directed and restricted by DNA precursor availability. However, hypermutants are recovered from 1–2% of resting or activated peripheral blood mononuclear cells (PBMCs) in therapy-naive patients [12].
Recently, we reported that the proportion of premature stop codons (SCs) in the pol gene was 8.5% in the 20 HPs undergoing ART [5]. The median proportion of sequences harboring SC in the reverse transcriptase (RT) due to G-to-A hypermutation by A3G was 21% in the sample of patients on successful long-term ART [14]. A3G counteracts Vif proteins. It was recently discovered that Vpr also counteracts A3G [15]. A3G contributes 88.4% of the total mutation rate of HIV-1 in viral DNA sequences from PBMCs, whereas HIV-1 RT only contributes 2.0% [16]. Furthermore, the sublethal and lethal mutations caused by A3G have the potential to contribute significantly to HIV-1 evolution, pathogenesis, immune escape, and drug resistance [17]. HIV-1 has a significantly higher mutation rate and G-to-A hypermutation caused by A3G than HIV-2 [18], resulting in the asymptomatic periods of HIV-1 infection being shorter than those of HIV-2. Complete viral suppression by ART decreases the pool of replication-competent viruses and, consequently, thereby increases the detection of lethal mutations [14], promoting viral eradication. To date, there have been few studies on the effects of A3G on vif and vpr genes during ART, although there are a few studies on the effects of A3G on the pol gene during ART and on the vif gene in therapy-naive patients.
Thus, through analysis of lethal mutations due to A3G, we investigated whether variations in vif-vpr genes were equally affected and whether genetic defects were associated with KRG and GCT; we determined sequences of vif-vpr genes from baseline to GCT in 20 HPs infected with the same source of HIV-1 [[3], [4], [5]] and compared the results with the findings from our previous reports. Here, we first report on sequential changes in the vif-vpr genes over a period of 26 years before the commencement of ART and during ART.
These findings might provide us with significant implication of KRG and GCT in the treatment of AIDS as well as the importance of A3G in the pathogenesis of HIV-1 infection.
2. Materials and methods
2.1. Study population
The twenty patients with hemophilia (HP), identified in this study as HP 1–HP 20, were diagnosed with HIV-1 infection between 1990 and 1994 (Table S1) [[2], [3], [4], [5],19]. They had been treated primarily with imported clotting factors before the start of local domestic clotting factor production. The control patients (n = 80) for KRG-treated patients infected with subtype B had not been exposed to KRG or any antiretroviral therapies (e.g., zidovudine) at the time of sampling, and their PBMCs were available for gene amplification. As the control patients for GCT, 43 ART-alone patients were included. Informed written consent was obtained from the HPs. This study was approved by the Institutional Review Board of the Asan Medical Center (Code 2012-0390).
2.2. DNA preparation and vif and vpr gene amplification
Viral DNA was isolated from PBMCs using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), and viral RNA was extracted from 300-μl serum samples using a QIAamp Ultra sense Viral RNA Kit (Qiagen, Hilden, Germany) as described in the study by Rawson et al [18]. The vif gene was amplified by nested polymerase chain reaction (PCR) using the TaKaRa R-Taq kit (Takara Bio Inc., Shiga, Japan). First and second PCR tests were performed in a 20-μl reaction mixture and 50-μl reaction mixture, respectively. The outer primer pairs were 545 (5′-GCAGTACAAATGGCAGTATTCATC-3′) and LA 106K (5′-TGRTAGAGRAACTTGATGRTYCTT-3′) or 545 and KMK2 (5′-ATGGGAATTGGTTCAAAGGA-3′) and the inner primer pairs were 547 (5′-GCTCCTCTGGAAAGGTGAAGG-3′) and LA100 (5′-AGTATCCCCGTAAGTTTCA-3': targeting at 758 bp) for the amplification of the vif gene [3] and 547K (5′-GCTTCTCTGGAAAGGTGAAGG-3′) and LA 102K (5′-TACAAGGAGTCTTGGGCTGACTTC-3`: targeting at 948 bp) and 547 and 566 (5′-GGCCCAAACATTATGTACCTCTGTA-3’: targeting at 1,240 bp) for the amplification of vif and vpr genes [20]. The first PCR cycling conditions were as follows: 95°C for 2 min, 35 cycles of 30 s at 95°C, 30 s at 52°C, 2 min and 30 s at 72°C, and a final extension at 72°C for 10 min. The second PCR was performed with 1 μL of the first PCR product; the cycling conditions were as follows: 95°C for 2 min, 35 cycles of 30 s at 95°C, 30 s at 57°C, 1 min and 30 s at 72°C, and a final extension at 72°C for 10 min. The subsequent sequences were directly sequenced using Applied Biosystems 3730XL (Applied Biosystems, Foster City, CA, USA).
2.3. ART and therapy with KRG
Outpatient-based KRG treatment in HIV-1–infected patients was initiated at the Korean National Institute of Health in late 1991. The daily dose of KRG for men was 5.4 g (six 300-mg capsules, three times per day) [5,9]. KRG has been supplied since November 1991, although the supply of KRG was not consistent before 2000 [5]. Most of our study patients had taken KRG for a variable period before the commencement of ART. The total amount of KRG used before the start of ART was 3,507 ± 5,468 g for 28 ± 36 months. The annual decrease of CD4+ T cells (AD) in the 20 HPs was 43 ± 27 per μL. There was a significant inverse correlation between the total amount of KRG and the AD (p < 0.01) [5]. The ART regimen has included integrase strand transfer inhibitors (INSTIs) in all living patients since 2014 in Korea. During ART, four HPs (1, 5, 9, and 13) did not take KRG (Fig. S1). The amount of KRG supplied to 16 HPs was 11,678 ± 9,593 g. The duration of ART was 171 ± 51 months in 18 HPs.
2.4. Statistical analysis
Data were expressed as mean ± standard deviation. Statistical significance was estimated by the Student two-tailed t test, the Chi-square test, Fisher's exact test, or correlation analysis and survival curve, using MedCalc software (Ostend, Belgium). Statistical significance was defined as p < 0.05.
2.5. Nucleotide sequences
GenBank accession numbers are AY581323-366, JF958013-081, JQ066823-7066, JQ327713-70, KX591061-219, MN364870-940, and MN792670-752.
3. Results
3.1. Patient demographics
The clinical characteristics of all patients were described in previous studies [[2], [3], [4], [5],19]. They all were infected with KSB from Plasma Donors O and P [[2], [3], [4], [5],19], and the earliest vif genes from Plasma Donors O and P were all wild-type (WT) ones [3]. In addition, we had obtained vif-vpr genes in the two plasma donors. In total, 28 sequences were obtained: 24 from seven sequential samples in donor O and four from two samples in donor P (Fig. S1). The 28 vif-vpr genes were all WT ones.
3.2. Distribution of defective genes at the patient, sample, and sequence level
At the patient level, irrespective of therapy, in-frame small deletion (SD), premature SCs, and ID including insertion or deletion of one or two nucleotides (hereafter called indel) in the vif gene were found in three (15%), 14 (70%), and seven patients (35%), respectively (Fig. 1A). The HPs revealed significantly higher proportions of SD, ID, and SC than the control group (p < 0.01) and the ART-alone group (p < 0.05) (Fig. 1A). Three patients (HPs 2, 3 and 9) did not reveal any defects in the vif gene (Fig. S1). There was no occurrence of SD or SC in two patients treated with the smallest amount of KRG (1,200 g in HP 9 and 1,800 g in HP 2) [5] although HP 3, treated with 3,980 g of KRG, revealed SC in the vpr gene only. However, they all revealed defects in the 5′ LTR/gag and nef genes (ID in HP 2 and HP 9) [7,9].
Fig. 1.
The proportion of the sequences harboring deletions and in-frame stop codon (SC) in the vif gene. (A) Proportion of patients with internal deletions (IDs), small deletions (SDs), and SCs. Despite the absence of ART in HP 1 and HP 5, all defects were significantly higher in HPs treated with KRG and/or GCT than in the control and ART-alone groups. The proportion of patients with SC was significantly higher in the ART patients than in the control patients. (B) At the sample level, the proportion of amplicon with SC in GCT patients was significantly higher than in the control and KRG patients, and the proportion of SD in KRG patients was significantly higher than in the control, GCT, and even ART-alone patients. (C) SD (≤15-bp) in the vif gene on KRG intake was significantly higher than at baseline and in the control patients (p < 0.001). Its detection was significantly decreased during GCT than on KRG treatment (p < 0.001). (D) The proportion of the sequences harboring SC on GCT was significantly higher than that on KRG treatment and in control patients, whereas it was similar to ART. The proportion was similar among control patients, at baseline, and in KRG patients. ART, antiretroviral therapy; GCT, KRG plus highly active antiretroviral therapy; HP, hemophiliac; KRG, Korean Red Ginseng; PBMC, peripheral blood mononuclear cell.
At the sample level, 38 (17.3%) of 220 samples revealed SC in HPs (Fig. S1). The proportion of PBMCs harboring SC was significantly higher in GCT patients (24.1%, 35/145) than in KRG (3.6%, 2/56) (p < 0.001), control (1.7%, 1/59) (p < 0.001), and ART-alone patients (12.7%, 7/55) (p = 0.066) (Fig. 1B).
With respect to sequences, we obtained 136 vif-vpr genes at baseline before KRG treatment in 20 HPs. Including 136 vif-vpr genes at baseline, we obtained 721 vif genes in HPs. Among the 721 vif genes, 51 genes (7.1%) were deleted: four deletions of a single nucleotide (in HPs 7, 12, 16, and 20), eight IDs (in HPs 1, 11, 12, 14, and 20), and 39 SDs (9 bp to 12 bp) [in HPs 5 (n = 9), 8 (n = 29), and 18 (n = 1)]. Of note, only two defective genes (an ID in HP 1 and a deletion of 1 bp in HP 12) were obtained at baseline (Fig. 1C, Fig. S1), whereas the remaining 49 deletions (8.5%: 49/577) were obtained during KRG treatment and GCT (p < 0.01). We also obtained 369 and 190 vif genes from the 80 control and 43 ART-alone patients, respectively.
Overall, there was no significant difference in defective genes between slow progressors (7/14) and progressors (1/6), limited to the period before GCT [5].
3.3. SDs are associated with KRG intake
We obtained 182 and 403 vif sequences from 20 HPs during the treatment with KRG and GCT, respectively. Thirty-nine SDs were found in 24 vif sequences (13.2%) from HP 5 and HP 8 during KRG treatment, whereas 15 vif sequences (3.7%) from HPs 8 and 18 were obtained during the GCT period (p < 0.001) (Fig. 1C). In detail, HP 1 and HP 5 had not taken ART. Detection of SDs was also significantly inhibited during GCT even when nine SDs of 28 sequences in the KRG intake in HP 1 and HP 5 were excluded and compared (9.7%) with GCT (3.8%) (p < 0.01). In addition, SDs were significantly higher during KRG than at baseline, in control patients, and in ART-alone (Fig. 1C) patients.
In contrast, compared with HIV-1 subtype B consensus, the same type of 12 and 21 SDs at AA185-187 was detected among 366 and 148 sequences in control patients and ART-alone patients, respectively (Fig. S2A). Two patients in control patients (HJiH and JSH) and two patients in ART-alone patients (HSHn and LHS) revealed it at least two samples per patient. All these 33 SDs were obtained from the first sample (Fig. S2A). Thus, all these 33 sequences revealing SDs could be considered as WT ones in view of personal baselines, suggesting that the two patients might be infected with the deleted viruses. This was quite different from the SD in KRG-treated patients. Thus, we can conclude that there was a significant difference in the frequency of SDs between patients being treated with KRG and ART-alone/control patients (Fig. 1A–C).
3.4. The proportion of SC depends on the duration of ART
Of the 721 sequences, 124 sequences obtained by RT-PCR were excluded in denominator for the proportion of sequence harboring SC. The proportion of SC was also significantly higher in GCT patients (10.1%, 40/397) than in control patients (p < 0.01) (Fig. 1D).
The earliest detection of SC in each patient was 69 ± 41 months (range; 10–137) since the introduction of ART. Regarding the time point of the first occurrence of SC during GCT, the proportion of sequences harboring SC significantly depended on the duration of ART. It was 2.3% (3/128) within three years and 13.8% (37/269) after three years in the vif gene (p < 0.001). The same result was obtained for the vpr gene (3.1% vs 13.6%, respectively) (p < 0.001). In other words, most SCs occurred three years after the introduction of ART. Three sequences harboring SC within three years were obtained at 10, 29, and 30 months in HP 15, HP 12, and HP 10, respectively (Fig. S1).
In addition, 1.9% of sequences obtained from control patients revealed SC [20], while 7.4% of sequences obtained from ART-alone patients revealed SC (p < 0.01, Fig. 1D). Thus, the proportion of sequences with SC in the GCT group was mildly higher in PBMCs or similar in the sequence level than in the ART-alone group (Fig. 1B and D). Taken together, our data suggest that KRG treatment might have had some effect on the occurrence of SC by additive antiviral effect.
3.5. ID and indel are not associated with KRG treatment
The proportion of sequences harboring ID (n = 8) or indel (n = 4) in the vif gene was 1.5% at baseline, 1.7% (3 IDs in HP 14) during KRG treatment, and 1.7% (4 IDs and 3 indels) during GCT (Fig. 1C). The size of eight IDs was Δ394 bp in HP 1 (before KRG treatment), Δ426 bp (off ART for 1 month) and Δ594 bp (compliance with 60–70%) in HP 11, Δ257 bp in HP 12 (five months since the introduction of ART), 3 Δ1,000 bp in HP 14 by 547/566 (positions of deletion not defined), and Δ210 bp in HP 20 (RNA copy = 14,700/ml by poor compliance) (Fig. S2A). Except for the 3 Δ1000 bp, all IDs were obtained by the inner primer 547K/LA102K. The ID in the Δ594 bp in HP 11 and Δ257 bp in HP 12 spans the vif-vpr genes (Fig. S2A and S2B). Seven of eight IDs and three of four indels were obtained during KRG treatment including the GCT period (1.8%; 10/566), whereas one ID and an indel were obtained in patients before KRG treatment or off-KRG treatment (1.2%; 2/161). There was no significant difference in the frequency of ID among the groups (Fig. 1C). However, the detection of ID was affected by the size of sequences. Actually, there was a significant difference in ID including data obtained in ART-alone patients (p < 0.001) (Fig. S3).
3.6. Q135P in the vif gene is associated with rapid progression to AIDS
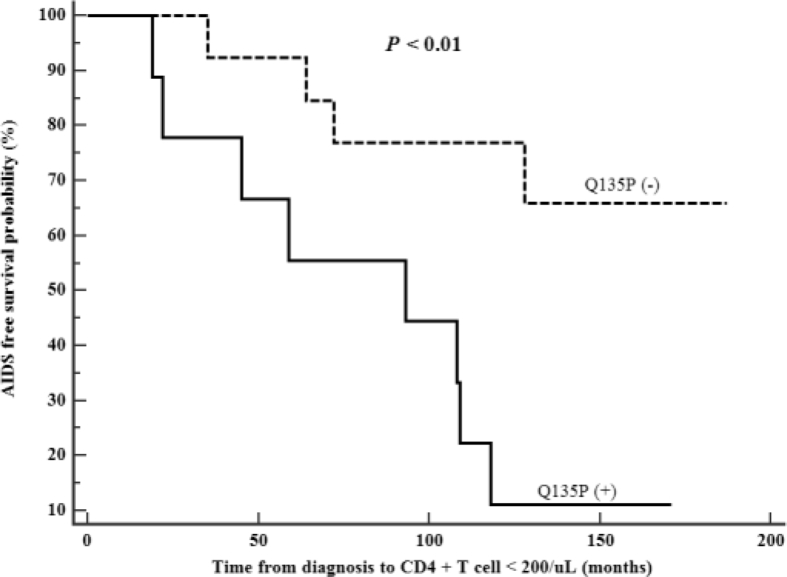
Twenty HPs revealed all lysine (K) at Position 22 in the earliest sample [3]. There was K22H in HP 9 (JQ066931-32), which is associated with low CD4+ T-cell counts and higher viral loads [21]. A report showed that mutation K22H was more frequent in patients failing to ART [22]. However, HP 9 consistently showed WT sequence in the pol gene [10]. Instead of the very rare K22H, we found an evolution from K22K to K22N in six HPs (3, 7, 8, 10, 11, and 19) (11.5%; 81/706 sequences) whose samples were not exposed to any antiretroviral drugs (except HP 19) (Fig. S2A). All first K22N was detected in AIDS or low CD4+ T-cell count, whereas it was also reported in long-term nonprogressors (LTNPs) [23]. Q135P [24] was detected in nine patients including Plasma Donor O (Fig. S1). Of note, in HP 17, Q135P developed during GCT. Interestingly, survival analysis revealed that Q135P was significantly associated with fast progression to AIDS, although K22N was not associated with it (Fig. 2). We did not find any specific changes in the nucleotide or amino acid sequences including insertion/deletion due to KRG intake.
Fig. 2.
Association of Vif Q135P with rapid progression. Q135P was found in Donor O and eight HPs (Fig. S1) and significantly associated with accelerated progression to AIDS. HP, hemophiliac.
3.7. Effect of ART on the vpr gene
We obtained 470 vpr genes. Except 75 sequences obtained by RT-PCR, 395 sequences were divided into pre-ART (n = 135) and ART (n = 260). Although there was the same deletion of 6 bp at the same position in six LTSPs and the insertion of amino acids “RAR” between AA90 and AA91 in a LTSP LSK [20] (Fig. S2B), 96 AAs of Vpr proteins were well conserved in all patients except HP 5, HP 8, and HP 18, who also revealed the same deletion of 12 bp and 9 bp in Vif proteins (AA12-15 and AA13-15) (Fig. S2B). The deletion is the same position as the 12-bp and 9-bp deletion in the vif gene previously mentioned. In addition, there was an indel in 92LCS3-6867 (JF957938). Except two sequences with only initial isoleucine instead of methionine in HP 7, the proportion of SC-containing sequences during ART (9.6%; 25/260) was significantly higher than the proportion of 2.3% found during the pre-ART period (p < 0.01) (Fig. 3). In our study of 470 Vpr proteins, there was no Q65R [25] and F72L as shown in LTNPs [26]. In addition, R77Q was reported in Western LTNPs [27,28]. However, it was found in 83 of 102 KSB-infected Korean patients (Fig. S2B). Interestingly, the vpr gene was also affected by A3G almost to the same extent as the vif gene (Fig. 3) as the two genes show the same extent of genetic diversity [29]. In HP 20, there was a single-nucleotide deletion in the vif-vpr gene in the same amplicon.
Fig. 3.
The proportion of the sequences harboring in-frame stop codons in the pol, vif, and vpr genes was significantly and similarly increased during ART, whereas there was no such increase in the env and nef genes [7]. ART, antiretroviral therapy.
Interestingly, 11 sequences of 40 amplicons with SC in the vif gene did not reveal SC at W residue in Vpr proteins, whereas two sequences of 25 amplicons with SC in the vpr gene did not reveal SC at W residue in Vif proteins (Table S1). Considering nine sequences with not determined (ND) in the vpr gene and a SC not at the W residue, there was a significant difference in the proportion of SC between two genes (11/33 versus 2/24, respectively; p < 0.05). Thus, to investigate whether there is a difference between Vif and Vpr proteins affected by A3G, we compared the proportion of SC of all tryptophan sites (W) on ART. All SCs occurred in W except one in Vpr in HP 18 (JQ067036). Vif and Vpr proteins had W at eight and three sites, respectively. The proportion of SC at W site in the same 260 sequences was similar in Vif proteins (3.9%; 82/2080) and in Vpr proteins (5.0%, 39/780) (Table S1).
Focusing on the sequences harboring SC during ART, Pol, Vif, and Vpr proteins revealed SC at 30.5% of position W (124/407W) with an additional SC at the site of lysine (AAA→TAA; HQ026608), 29.4% (87/296W), and 36% (38/105W) with an additional SC at arginine (AGA→TGA; JQ067036), respectively (Table S1). In contrast, there were only four SCs at the TGG (W) site in the 3 nef sequences. Thus, considering the nef gene containing 7–8 TGG sites, the proportion of SC at TGG was 0.23% (4/1768)–0.26% (4/1547) of all W sites of 221 nef genes. This is significantly lower than in the Vif proteins.
The frequency of the signature pattern nucleotide was also analyzed on the association of Plasma Donors O and P with the 20 HPs, compared with local controls (LCs). Regarding the epidemiological association, our previous report showed seven signature pattern nucleotides in the vif gene [3]. In the vpr gene, there were significant differences in the frequency at three nucleotide positions between Clusters O and P and LCs; synonymous change was at Position 5621 (GAG, all in Cluster O; GAA, all in Cluster P; and 78 GAG, 2 GAA, and 1 GGG in LCs), and two nonsynonymous changes were at 5633 (GAA→GAC/T) and 5741 (ATA→ACT). Thus, E25D and I61T showed a significantly high frequency in Cluster P, compared with that in the LCs and Cluster O (p < 0.0001) (Fig. S2B).
3.8. The effect of SCs on virus evolution
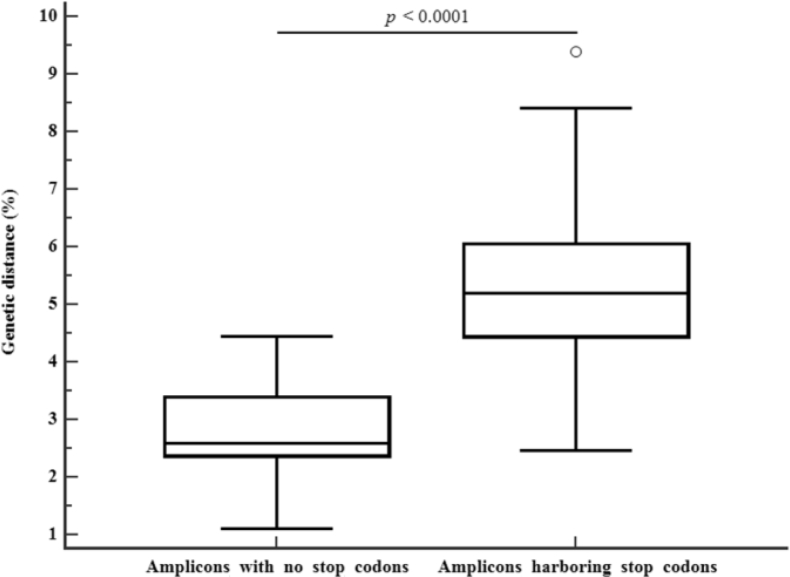
To analyze the effect of lethal mutations on virus evolution, we compared the sequences revealing SC and WT sequence alone with the earliest sequences of Donors O and P, respectively. The genetic distance was calculated based on January 1990, when Donors O and P were diagnosed as infected. Genetic distance in the sequences harboring SC or initial I was significantly higher (5.4 ± 1.7%) than that (2.9 ± 0.8%) in the WT sequences (p < 0.0001) (Fig. 4). The elapsed time from January 1990 to sampling time was 232 ± 33 months and 213 ± 21 months, respectively. When it was translated into annual mutation rate, they were in the order 0.28 ± 0.09% and 0.16 ± 0.04%, respectively. Thus, the genetic distance was 1.78 times higher in the sequences with SC than in the WT sequences. These factors lead to faster development of mutant strains that are resistant to therapeutic agents [22]. Intrapersonal genetic discrepancy in the earliest two sequences (October 1991 and February 1993) from Donors O and P was 0.37% and 0%, respectively. Sequence variation of WT sequences without SC or I showed a significant correlation with elapsed time (r = 0.45, p < 0.01 for the sequences obtained during ART) but that with SC showed no correlation. The correlation was more significant when the earliest sequences before ART were included (r = 0.70, p < 0.001).
Fig. 4.
Comparison of genetic distances in the sequences with no stop codons and in-frame stop codons. There were significant difference in the genetic distance between the sequences with no stop codons and in-frame stop codons (2.9 ± 0.8% versus 5.4 ± 1.7%, respectively) over 213 ± 21 and 232 ± 33 months compared with the corresponding earliest sequences from Donors O and P, respectively (p < 0.0001).
3.9. Effect of ART on pol genes
In the present study, the proportion of pol sequences harboring SC was 9.6% during GCT or ART, which was higher than that (1.2%) found during pre-ART (p < 0.01) (Fig. 3). However, the proportion of the sequences harboring SC in the env-nef genes increased a little during ART. These findings show that env-nef genes are significantly less affected by A3G than the pol-vif-vpr genes.
4. Discussion
Here, the frequency of the SC-containing vif gene in the KRG group was similar to the control group and baseline. However, it increased significantly to 9.0% during GCT. The proportion with SC depended significantly on the duration of GCT. Moreover, the genetic distance in the sequences harboring SC was significantly higher than that in the WT sequences (p < 0.001). The proportion of SDs was significantly higher during KRG treatment than in the control patients and at baseline, but the proportion was significantly inhibited during GCT. However, the level was still significantly higher than in the control group or at the baseline (Fig. 1A–C). Pol-vif-vpr genes were shown to be similarly affected by A3G, whereas env and nef genes were significantly less affected by 3G (Fig. 3). It is well known that hypermutation is not equally distributed along the HIV-1 genome [30], as shown in a report that the env gene is significantly less affected by A3G than the integrase-vif- vpr genes [31]. For this reason, unlike previous studies [[6], [7], [8], [9], [10]], ID was not affected by taking KRG, but SC may be affected to some extent, as shown in Fig. 1B. It might result from the synergistic effects of reductions on virus concentration by both KRG and ART. The second reason the proportion of SC is higher in GCT than in ART alone (Fig. 1B and D) might be a lower WT virus concentration as a result of KRG treatment, and thus, the possibility of amplifying the defective virus is increased. In the previous study, we did not include the ART-alone group, making it impossible to compare with the GCT group [20].
Consequently, despite increasing DNA concentration (2- to 4-fold), the success rate of PCR amplification significantly decreased 10 years after ART (data not shown) as shown in the pol gene [10] because we have not used the primer set designed to target at the hypermutated virus. In brief, it decreased from 63% before ART to 29% after 6 years of ART [10]. These findings support the view that that the longer the ART period, the lower the WT DNA concentration. Consequently, the ratio of defective DNA including SC is increased, and therefore, the PCR success rate is significantly decreased by primer mismatch. In addition, for this reason, it was very difficult to obtain PCR products in a few patients (HPs 6 and 14). Moreover, we could not obtain sequences even when we could rarely obtain PCR products in HP 14, suggesting that the reason might be the mismatch of primers due to G-to-A hypermutations. Thus, the proportion of sequences harboring SC might be underestimated. Irrespective of the kinds of the primer set, the success rate of PCR at pre-ART was 73% (101/138). However, it was significantly reduced to 41% (189/458) during the ART period (p < 0.0001). Furthermore, there was a significant inverse correlation between the duration of ART and the success rate of PCR (r = −0.20, p = 0.01).
Actually, the proportion of SC during GCT (10.1%) was similar to the proportion (7.4%) during ART alone. However, the proportion was significantly higher than that (1.4%) in the nef gene in the same patients (p < 0.001) (Fig. 3) [7]. In contrast, the proportion with ID in the vif gene on KRG treatment (1.7%) was significantly lower than that (20.6%; 62/301) in the nef gene (p < 0.0001) [7]. Probably, the first reason for this is significantly higher genetic stability in the vif gene than in the nef gene [32]. Another reason might be related to the size of the gene: the vif gene is 579 bp, whereas the nef gene is about 620 bp. Compared with the previous study with the same 1.2 kb of the integrase region (11.9% as of 84/704) [10], the proportion of IDs (4.9% as of 6/122) was significantly lower in the vif-vpr–containing 1,248 bp (p < 0.05) (Fig. S3).
Here, the two cysteines at Positions 113 and 132 were well conserved as shown in another report [31,33] except for two vif genes: each one in HP 8 and HP 15 revealed S132 and Y113, among 624 sequences, respectively. It is known that changes such as K22H in Vif protein are associated with the development of resistance to antiretroviral drugs [22,34]. In this study, however, there was no K22H in patients resistant to antiretroviral drugs [5], although two of six patients with K22N revealed resistant viruses (HPs 9 and 19). Further study is needed to determine whether this is associated with subtype difference (KSB versus subtype B) or KRG treatment.
In Korea, major resistance mutations (RMs) to INSTIs have been introduced since 2014. RMs to INSTIs [35] and the Q151M complex in 2014 [36] are frequent in KRG-naive patients (22%). In contrast, in the present study, despite further follow-up >four years than in the previous report in 2015 [5], most of these HPs had already developed RMs to previous monotherapy and two-drug combination therapy [5], and we could not find RMs to INSTIs (data not shown). These results are probably the result of the synergistic effects of taking KRG as shown in reversal to the WT sequence [37].
Regarding potential mechanism for SD occurrence, it is difficult to point out which components of KRG are involved because we applied whole ginseng for patients. It contains many active components such as many kinds of ginsenosides and acid polysaccharides; some components of ginseng have inhibitory effects on HIV-1 RT [[38], [39], [40]]. It is possible that these inhibitory effects on RT might decrease its fidelity and result in a high frequency of genetic defects [41]. In addition, A3G disrupts the synthesis of cDNA [42] and is also targeted to the proteasomal degradation pathway by Vpr and Vif [15,43]. This finding might be the basis on which vif-vpr genes are similarly affected by lethal mutations.
The present study has the following limitations. There was a significant difference in the use of samples between the patients on GCT and ART alone. In other words, compared with the GCT group, a limited number of samples were used in most patients from the ART group.
Taken together, these data show that vif-vpr genes revealed similar proportions of sequences with SC due to G-to-A hypermutation on ART. Thus, the gene with SC showed about 1.8 times faster evolution than in WT sequences. This faster evolution can facilitate the emergence of some antiretroviral RMs. Further studies will be needed on the link between KRG treatment and SD.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by a grant from the Korean Society of Ginseng funded by the Korea Ginseng Corporation (2018).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2020.03.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Multimedia component 1Changes in the CD4+ T cell count, plasma viral load, and genetic defects according to Korean red ginseng (KRG) intake and highly active antiretroviral therapy (ART) or ginseng-based combination of ART (hereafter described as GCT) in 20 hemophiliacs (HP) and two plasma donors. The periods of KRG intake and ART are shown using a bar at the upper portion, respectively. We did not include 24 amplicons obtained from five samples in HP 9 and three samples in HP 13 in GCT because they did not take KRG during ART. The gross deletion in the nef gene is depicted at the bottom line. The upward black arrow (↑), Δ, Δ1, Δ+1 ▲, dotted line in the highest position (--), ins, dup, asterisk (∗) symbols and N/H at the base of the figure denote wild-type amplicon only, gross deletion, deletion of 1-bp, gross deletion plus deletion of 1-bp, in-frame small deletion (SD), poor compliance, insertion of a or two nucleotides, duplication, premature stop codon, and K22N and K22H in the vif gene, respectively. ZDV; zidovudine, ®;sequences were obtained by RT-PCR. DCF; domestic clotting factor.
Multimedia component 2Alignment of amino acids of the earliest Vif and Vpr proteins. The earliest viruses of plasma donor O (OSG) and P (KPS), which were the infectious agents, were both wild types. A. Alignment of two or three representative Vif protein sequences of 20 hemophiliacs (HPs) and two plasma donors. There were several deletions within amino acids (AA) positions 183-187. There was an inversion from AA94 in the Vif and Vpr in HP 4-93KGJ10-14599. KRG: Korean red ginseng, 7/7∗ denotes that seven of seven sequences contain in-frame small deletion. B. Alignment of Vpr protein sequences of 20 HPs, two plasma donors and 80 local controls infected with KSB (LCs). A significant high frequency of AA at two positions in cluster P (E25D and I61T) was observed compared to the earliest sequences in 80 LCs (p<0.001 for all). 92OSG1-6889 means the PCR amplicon number from the sample obtained from the patient OSG in January 1992. Two patients KJin and CJI with KRG 100% on the right column are a couple.
The proportion of amplicons with internal deletions (ID) according to target size of PCR amplicon. There was a significant difference in ID among internal primer sets (p< 0.001).
Multimedia component 4
References
- 1.Daniels R.S., Kang C., Patel D., Xiang Z., Douglas N.W., Zheng N.N., Cho H.W., Lee J.S. An HIV type 1 subtype B founder effect in Korea: gp160 signature patterns infer circulation of CTL-escape strains at the population level. AIDS Res Hum Retroviruses. 2003;19:631–641. doi: 10.1089/088922203322280847. [DOI] [PubMed] [Google Scholar]
- 2.Cho Y.K., Kim J.E., Foley B.T. Genetic analysis of the full-length gag gene from the earliest Korean subclade B of HIV-1: an outbreak among Korean hemophiliacs. Viruses. 2019;11 doi: 10.3390/v11060545. pii: E545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho Y.K., Jung Y.S., Lee J.S., Foley B.T. Molecular evidence of HIV-1 transmission in 20 Korean individuals with haemophilia: phylogenetic analysis of the vif gene. Haemophilia. 2012;18:291–299. doi: 10.1111/j.1365-2516.2011.02620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho Y.K., Foley B.T., Sung H.S., Kim Y.B., Kim J.H. Molecular epidemiologic study of a human immunodeficiency virus 1 outbreak in haemophiliacs B infected through clotting factor 9 after 1990. Vox Sang. 2007;92:113–120. doi: 10.1111/j.1423-0410.2006.00866.x. [DOI] [PubMed] [Google Scholar]
- 5.Kim B.R., Kim J.E., Sung H., Cho Y.K. Long-term follow up of HIV-1-infected Korean haemophiliacs, after infection from a common source of virus. Haemophilia. 2015;21 doi: 10.1111/hae.12527. http://www.ncbi.nlm.nih.gov/pubmed/?term=Long-term+follow+up+of+HIV-1-infected+Korean+haemophiliacs%2C+after+infection+from+a+common+source+of+virus comments e1-11. [DOI] [PubMed] [Google Scholar]
- 6.Cho Y.K., Lim J.Y., Jung Y.S., Oh S.K., Lee H.J., Sung H. High frequency of grossly deleted nef genes in HIV-1 infected long-term slow progressors treated with Korean red ginseng. Curr HIV Res. 2006;4:447–457. doi: 10.2174/157016206778560072. [DOI] [PubMed] [Google Scholar]
- 7.Cho Y.K., Jung Y.S., Sung H. Frequent gross deletion in the HIV type 1 nef gene in hemophiliacs treated with Korean red ginseng: inhibition of detection by highly active antiretroviral therapy. AIDS Res Hum Retroviruses. 2009;25:419–424. doi: 10.1089/aid.2008.0178. [DOI] [PubMed] [Google Scholar]
- 8.Cho Y.K., Jung Y.S. Dosage and duration effects of Korean red ginseng intake on frequency of gross deletions in the nef gene. J Ginseng Res. 2010;34:219–225. [Google Scholar]
- 9.Cho Y.K., Jung Y.S., Sung H., Joo C.H. Frequent genetic defects in the HIV-1 5’LTR/gag gene in hemophiliacs treated with Korean red ginseng: decreased detection of genetic defects by highly active antiretroviral therapy. J Ginseng Res. 2011;35:413–420. doi: 10.5142/jgr.2011.35.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho Y.K., Kim J.E., Woo J.H. Korean Red Ginseng increases defective pol gene in peripheral blood mononuclear cells of HIV-1-infected patients; inhibition of its detection during ginseng-based combination therapy. J Ginseng Res. 2019;43:684–691. doi: 10.1016/j.jgr.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho Y.K., Kim J.E. Effect of Korean Red Ginseng intake on the survival duration of human immunodeficiency virus type 1 patients. J Ginseng Res. 2017;41:222–226. doi: 10.1016/j.jgr.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vartanian J.P., Plikat U., Henry M., Mahieux R., Guillemot L., Meyerhans A., Wain-Hobson S. HIV genetic variation is directed and restricted by DNA precursor availability. J Mol Biol. 1997;270:139–151. doi: 10.1006/jmbi.1997.1104. [DOI] [PubMed] [Google Scholar]
- 13.Pace C., Keller J., Nolan D., James I., Gaudieri S., Moore C., Mallal S. Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. J Virol. 2006;80:9259–9269. doi: 10.1128/JVI.00888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fourati S., Lambert-Niclot S., Soulie C., Malet I., Valantin M.A., Descours B., Ait-Arkoub Z., Mory B., Carcelain G., Katlama C. HIV-1 genome is often defective in PBMCs and rectal tissues after long-term HAART as a result of APOBEC3 editing and correlates with the size of reservoirs. J Antimicrob Chemother. 2012;67:2323–2326. doi: 10.1093/jac/dks219. [DOI] [PubMed] [Google Scholar]
- 15.Norman J.M., Mashiba M., McNamara L.A., Onafuwa-Nuga A., Chiari-Fort E., Shen W., Collins K.L. The antiviral factor APOBEC3G enhances the recognition of HIV-infected primary T cells by natural killer cells. Nat Immunol. 2011;28:975–983. doi: 10.1038/ni.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuevas J.M., Geller R., Garijo R., López-Aldeguer J., Sanjuán R. Extremely high mutation rate of HIV-1 in vivo. PLoS Biol. 2015;13 doi: 10.1371/journal.pbio.1002251. e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadler H.A., Stenglein M.D., Harris R.S., Mansky L.M. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J Virol. 2010;84:7396–7404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawson J.M., Landman S.R., Reilly C.S., Mansky L.M. HIV-1 and HIV-2 exhibit similar mutation frequencies and spectra in the absence of G-to-A hypermutation. Retrovirology. 2015;12:60. doi: 10.1186/s12977-015-0180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho Y.K., Jung Y.S., Foley B.T. Phylogenetic analysis of full-length pol gene from Korean hemophiliacs and plasma donors infected with Korean subclade B of HIV-1. AIDS Res Hum Retroviruses. 2011;27:613–621. doi: 10.1089/AID.2010.0174. [DOI] [PubMed] [Google Scholar]
- 20.Cho Y.K., Kim B.R., Chang M.S., Kim J.E. Effects of Korean Red Ginseng and HAART on vif gene in 10 long-term slow progressors over 20 years: high frequency of deletions and G-to-A hypermutation. Evid Based Complement Alternat Med. 2013:871648. doi: 10.1155/2013/871648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villanova F., Barreiros M., Janini L.M., Diaz R.S., Leal É. Genetic diversity of HIV-1 gene vif among treatment-naive Brazilians. AIDS Res Hum Retroviruses. 2017;33:952–959. doi: 10.1089/AID.2016.0230. [DOI] [PubMed] [Google Scholar]
- 22.Fourati S., Malet I., Binka M., Boukobza S., Wirden M., Sayon S., Simon A., Katlama C., Simon V., Calvez V. Partially active HIV-1 Vif alleles facilitate viral escape from specific antiretrovirals. AIDS. 2010;24:2313–2321. doi: 10.1097/QAD.0b013e32833e515a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cruz N.V., Amorim R., Oliveira F.E., Speranza F.A., Costa L.J. Mutations in the nef and vif genes associated with progression to AIDS in elite controller and slow-progressor patients. J Med Virol. 2013;85:563–574. doi: 10.1002/jmv.23512. [DOI] [PubMed] [Google Scholar]
- 24.De Maio F.A., Rocco C.A., Aulicino P.C., Bologna R., Mangano A., Sen L. Effect of HIV-1 Vif variability on progression to pediatric AIDS and its association with APOBEC3G and CUL5 polymorphisms. Infect Genet Evol. 2011;11:1256–1262. doi: 10.1016/j.meegid.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 25.Jacquot G., Le Rouzic E., Maidou-Peindara P., Maizy M., Lefrère J.J., Daneluzzi V., Monteiro-Filho C.M., Hong D., Planelles V., Morand-Joubert L. Characterization of the molecular determinants of primary HIV-1 Vpr proteins: impact of the Q65R and R77Q substitutions on Vpr functions. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007514. e7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caly L., Saksena N.K., Piller S.C., Jans D.A. Impaired nuclear import and viral incorporation of Vpr derived from a HIV long-term non-progressor. Retrovirology. 2008;5:67. doi: 10.1186/1742-4690-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L., Huang Y., Yuan H., Tuttleton S., Ho D.D. Genetic characterization of vif, vpr, and vpu sequences from long-term survivors of human immunodeficiency virus type 1 infection. Virology. 1997;228:340–349. doi: 10.1006/viro.1996.8378. [DOI] [PubMed] [Google Scholar]
- 28.Yamada T., Iwamoto A. Comparison of proviral accessory genes between long-term nonprogressors and progressors of human immunodeficiency virus type 1 infection. Arch Virol. 2000;145:1021–1027. doi: 10.1007/s007050050692. [DOI] [PubMed] [Google Scholar]
- 29.Saurya S., Lichtenstein Z., Karpas A. Characterization of pol, vif, vpr, and vpu genes of HIV type 1 in AIDS patients with high viral load and stable CD4+ T cell counts on combination therapy. AIDS Res Hum Retroviruses. 2002;18:1151–1155. doi: 10.1089/088922202320567905. [DOI] [PubMed] [Google Scholar]
- 30.Suspène R., Rusniok C., Vartanian J.P., Wain-Hobson S. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res. 2006;34:4677–4684. doi: 10.1093/nar/gkl555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geller R., Domingo-Calap P., Cuevas J.M., Rossolillo P., Negroni M., Sanjuán R. The external domains of the HIV-1 envelope are a mutational cold spot. Nat Commun. 2015;6:8571. doi: 10.1038/ncomms9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X.Y., Sova P., Chao W., Volsky D.J. Cysteine residues in the Vif protein of human immunodeficiency virus type 1 are essential for viral infectivity. J Virol. 1994;68:1714–1720. doi: 10.1128/jvi.68.3.1714-1720.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wieland U., Hartmann J., Suhr H., Salzberger B., Eggers H.J., Kühn J.E. In vivo genetic variability of the HIV-1 vif gene. Virology. 1994;203:43–51. doi: 10.1006/viro.1994.1453. [DOI] [PubMed] [Google Scholar]
- 34.Adekale M.A., Cane P.A., McCrae M.A. Changes in the Vif protein of HIV-1 associated with the development of resistance to inhibitors of viral protease. J Med Virol. 2005;75:195–201. doi: 10.1002/jmv.20256. [DOI] [PubMed] [Google Scholar]
- 35.Jeong W., Jung I.Y., Choi H., Kim J.H., Seong H., Ahn J.Y., Jeong S.J., Ku N.S., Kim J.M., Choi J.Y. Integrase strand transfer inhibitor resistance mutations in antiretroviral therapy-naive and treatment-experienced HIV patients in South Korea. AIDS Res Hum Retroviruses. 2019;35:213–216. doi: 10.1089/AID.2018.0213. [DOI] [PubMed] [Google Scholar]
- 36.Choi J.Y., Kwon O.K., Choi B.S., Kee M.K., Park M., Kim S.S. The prevalence of antiretroviral multidrug resistance in highly active antiretroviral therapy-treated patients with HIV/AIDS between 2004 and 2009 in South Korea. J Clin Virol. 2014;60:154–160. doi: 10.1016/j.jcv.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Cho Y.K., Kim B.R., Kim J.E., Woo J.H., Foley B.T. First report on a T69-ins insertion in CRF06_cpx HIV Type 1. AIDS Res Hum Retroviruses. 2013;29:1079–1084. doi: 10.1089/aid.2013.0013. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H., Lu Z., Tan G.T., Qiu S., Farnsworth N.R., Pezzuto J.M., Fong H.H.S. Polyacetyleneginsenoside-Ro, a novel triterpene saponin from Panax ginseng. Tetrahedron Lett. 2002;43:973e7. [Google Scholar]
- 39.Lam S.K., Ng T.B. Sanchi ginseng (Panax notoginseng) with inhibitory effects on human immunodeficiency virus-1 reverse transcriptase. Life Sci. 2002;70:3049e58. doi: 10.1016/s0024-3205(02)01557-6. [DOI] [PubMed] [Google Scholar]
- 40.Wang H.X., Ng T.B. Quinqueginsin, a novel protein with anti-human immunodeficiency virus, antifungal, ribonuclease and cell-free translation-inhibitory activities from American ginseng roots. Biochem Biophys Res Commun. 2000;269 doi: 10.1006/bbrc.2000.2114. 203e8. [DOI] [PubMed] [Google Scholar]
- 41.Cho Y.K., Kim J.E., Woo J.H. Genetic defects in the nef gene are associated with Korean Red Ginseng intake: monitoring of nef sequence polymorphisms over 20 years. J Ginseng Res. 2017;41:144–150. doi: 10.1016/j.jgr.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bishop K.N., Verma M., Kim E.Y., Wolinsky S.M., Malim M.H. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000231. e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou D., Wang Y., Tokunaga K., Huang F., Sun B., Yang R. The HIV-1 accessory protein Vpr induces the degradation of the anti-HIV-1 agent APOBEC3G through a VprBP-mediated proteasomal pathway. Virus Res. 2015;195:25–34. doi: 10.1016/j.virusres.2014.08.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1Changes in the CD4+ T cell count, plasma viral load, and genetic defects according to Korean red ginseng (KRG) intake and highly active antiretroviral therapy (ART) or ginseng-based combination of ART (hereafter described as GCT) in 20 hemophiliacs (HP) and two plasma donors. The periods of KRG intake and ART are shown using a bar at the upper portion, respectively. We did not include 24 amplicons obtained from five samples in HP 9 and three samples in HP 13 in GCT because they did not take KRG during ART. The gross deletion in the nef gene is depicted at the bottom line. The upward black arrow (↑), Δ, Δ1, Δ+1 ▲, dotted line in the highest position (--), ins, dup, asterisk (∗) symbols and N/H at the base of the figure denote wild-type amplicon only, gross deletion, deletion of 1-bp, gross deletion plus deletion of 1-bp, in-frame small deletion (SD), poor compliance, insertion of a or two nucleotides, duplication, premature stop codon, and K22N and K22H in the vif gene, respectively. ZDV; zidovudine, ®;sequences were obtained by RT-PCR. DCF; domestic clotting factor.
Multimedia component 2Alignment of amino acids of the earliest Vif and Vpr proteins. The earliest viruses of plasma donor O (OSG) and P (KPS), which were the infectious agents, were both wild types. A. Alignment of two or three representative Vif protein sequences of 20 hemophiliacs (HPs) and two plasma donors. There were several deletions within amino acids (AA) positions 183-187. There was an inversion from AA94 in the Vif and Vpr in HP 4-93KGJ10-14599. KRG: Korean red ginseng, 7/7∗ denotes that seven of seven sequences contain in-frame small deletion. B. Alignment of Vpr protein sequences of 20 HPs, two plasma donors and 80 local controls infected with KSB (LCs). A significant high frequency of AA at two positions in cluster P (E25D and I61T) was observed compared to the earliest sequences in 80 LCs (p<0.001 for all). 92OSG1-6889 means the PCR amplicon number from the sample obtained from the patient OSG in January 1992. Two patients KJin and CJI with KRG 100% on the right column are a couple.
The proportion of amplicons with internal deletions (ID) according to target size of PCR amplicon. There was a significant difference in ID among internal primer sets (p< 0.001).
Multimedia component 4